Abstract
The highly conserved polycomb group (PcG) proteins maintain heritable transcription repression of the genes essential for development from fly to mammals. However, sporadic reports imply a potential role of PcGs in positive regulation of gene transcription, although systematic investigation of such function and the underlying mechanism has rarely been reported. Here, we report a Pc-mediated, H3K27me3-dependent positive transcriptional regulation of Senseless (Sens), a key transcription factor required for development. Mechanistic studies show that Pc regulates Sens expression by promoting H4K20me1 at the Sens locus. Further bioinformatic analysis at genome-wide level indicates that the existence of H4K20me1 acts as a selective mark for positive transcriptional regulation by Pc/H3K27me3. Both the intensities and specific patterns of Pc and H3K27me3 are important for the fates of target gene transcription. Moreover, binding of transcription factor Broad (Br), which physically interacts with Pc and positively regulates the transcription of Sens, is observed in Pc+H3K27me3+H4K20me1+ genes, but not in Pc+H3K27me3+H4K20me1− genes. Taken together, our study reveals that, coupling with the transcription factor Br, Pc positively regulates transcription of Pc+H3K27me3+H4K20me1+ genes in developing Drosophila wing disc.
Keywords: polycomb group proteins, positive transcriptional regulation, H3K27me3, H4K20me1, Broad
Introduction
PcG proteins, initially shown to maintain the heritable repression of transcription of homeotic genes in Drosophila1,2,3, play an important role in controlling the expression of genes essential for development, differentiation and maintenance of cell fate4,5. The highly conserved PcG proteins form multi-protein complexes that occupy selected sites on the chromatin. Among PcG proteins, polycomb repressive complex (PRC) 1 and 2 have been the focus of investigation6,7. In Drosophila, PRC2 contains the Enhancer of zeste (E(z)), Suppressor of zeste 12 (Su(z)12), Extra sex combs (Esc) and Chromatin assembly factor 1 (Caf1). The E(z) catalytic subunit catalyzes H3K27me2/3 depending on Su(z)12 and Esc8. PRC1 has four core proteins: Pc, Polyhomeotic (Ph), Sex combs extra (Sce, also known as dRing) and Posterior sex combs (Psc) in Drosophila. Pc can specifically recognize H3K27me3 through its chromodomain, and recruit other components to selected chromatin sites9,10, although H3K27me3-independent recruitment of PRC1 has also been reported in mammals7,11. It is generally considered that PRC1 maintains the repression of their targets by either catalyzing H2A mono-ubiquitination12 or inducing local chromatin condensation13,14,15,16.
The majority of reports indicate that PcG proteins play repressive roles in transcriptional regulation17,18. However, several isolated studies in Drosophila and mammals suggest a positive regulatory role of PcGs in transcription19,20,21, but the underlying mechanism is largely unknown. Most of these studies have focused on the positive role of PcGs during carcinogenesis22,23,24,25,26, whereas such regulation during normal development has been poorly investigated27,28,29.
To explore the positive regulatory role of PcGs in gene expression and its underlying mechanism during development, we focused on Drosophila PRC1 and PRC2, which has a simpler complement of PcG proteins than mammals. We first searched for highly conserved genes involved in the developmental control of Drosophila that are positively regulated by Pc, and found several genes, including Sens, Rosy (Ry) and Serpin 100A (Spn100A) in our analysis. Pc positively regulates Sens in a PRC2/H3K27me3-dependent manner. Further investigation showed that Pc functions together with PR-Set7 (also known as SET8) to regulate Sens transcription by modulating H4K20me1. Bioinformatic analysis revealed that H4K20me1 acts as a selective mark for negative or positive transcriptional regulation by Pc/H3K27me3. In addition, the Br-binding motif was identified in Pc+H3K27me3+H4K20me1+ genes. We found Br forms a complex with Pc, prefers to bind to Pc+H3K27me3+H4K20me1+ genes than Pc+H3K27me3+H4K20me1− genes, and positively regulates the transcription of Sens, Ry and Spn100A. Our study reveals a multilevel engagement of transcriptional regulators and epigenetic marks that together underpin the Pc-mediated positive transcriptional regulation.
Results
Pc binding positively correlates with Sens transcription in developing Drosophila wing discs
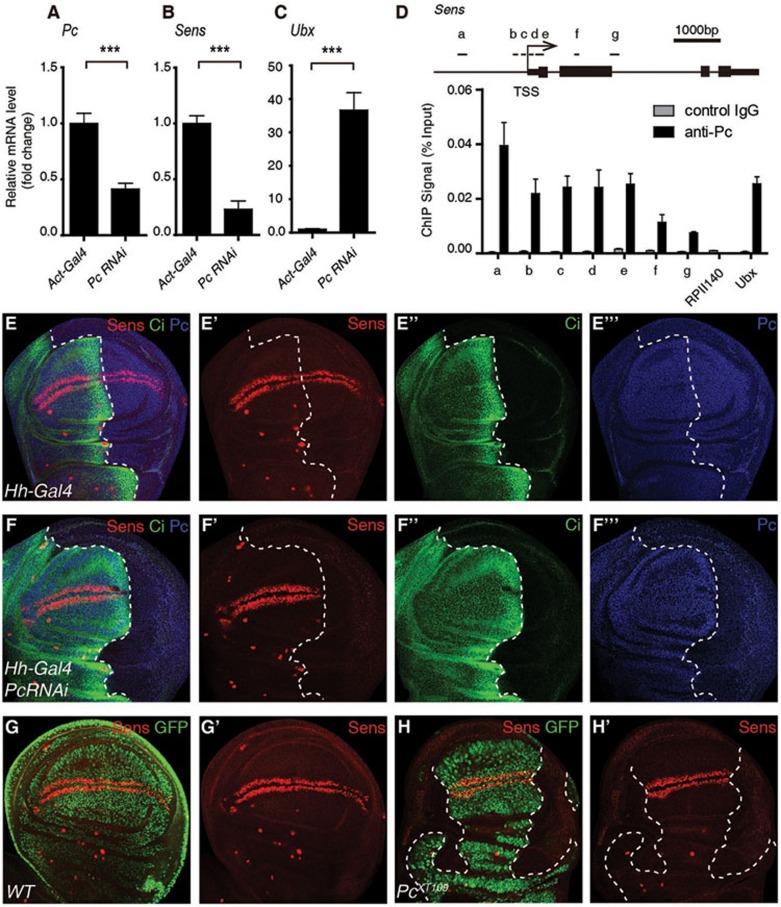
To explore novel targets positively regulated by Pc in Drosophila, uas-dsRNA of Pc driven by Act5c-Gal4 was employed to knock down expression of Pc throughout the fly. Wing discs from third instar larvae were dissected for mRNA extraction and sequencing (RNA-seq). The mRNA levels of Sens and some other highly conserved genes involved in developmental control, such as Ry and Spn100A, were significantly decreased when Pc expression was reduced, while mRNA of Ultrabithorax (Ubx), a classical target repressed by Pc in the wing discs, was dramatically increased as expected30,31 (Figures 1A-1C and 4D). Sens is a downstream target gene of the Wnt signaling pathway and encodes a C2H2-type zinc-finger transcription factor that is essential for development of sensory organ precursors in both the embryo and the imaginal disc32. We thus focused on the transcriptional regulation of Sens.
Figure 1.
Pc binding positively correlates with Sens transcription in developing Drosophila wing disc. (A-C) RT-qPCR analysis of mRNA levels of Pc (A), Sens (B) and Ubx (C) in Act5c-Gal4 or Act5c-Gal4/uas-dsRNA Pc (labeled as Pc RNAi) wing discs. (D) ChIP-qPCR analysis using control IgG (grey) or anti-Pc antibody (black) with WT wing discs. Upper panel illustrates the positions of primers used for the qPCR assay at the Sens locus. In the lower panel, ChIP signal magnitudes are represented as percentages of input chromatin. RPII14037 and Ubx62 served as negative control and positive control, respectively. (E-F''') Wing discs with indicated genotypes were immunostained with anti-Sens antibody (red), anti-Ci antibody (green) and anti-Pc antibody (blue). The Ci-negative region defines the posterior (P) compartment, where Hh-Gal4 drives the expression of uas-dsRNA Pc. Dashed lines mark the anterior-posterior boundary. Sens is expressed along with the dorsal-ventral boundary and this expression is reduced dramatically in the P compartment where Pc is knocked down specifically. (G-H') Immunostaining of a WT wing disc (G-G') and a wing disc with chimeric PcXT109 null clones (H-H') using anti-Sens antibody (red). Sens staining is significantly reduced in the Pc-deficient region marked by dashed lines (GFP negative). Means ± SD; n = 3. ***P < 0.001. Unpaired, two-tailed Student's t-test. See also Supplementary information, Figure S1.
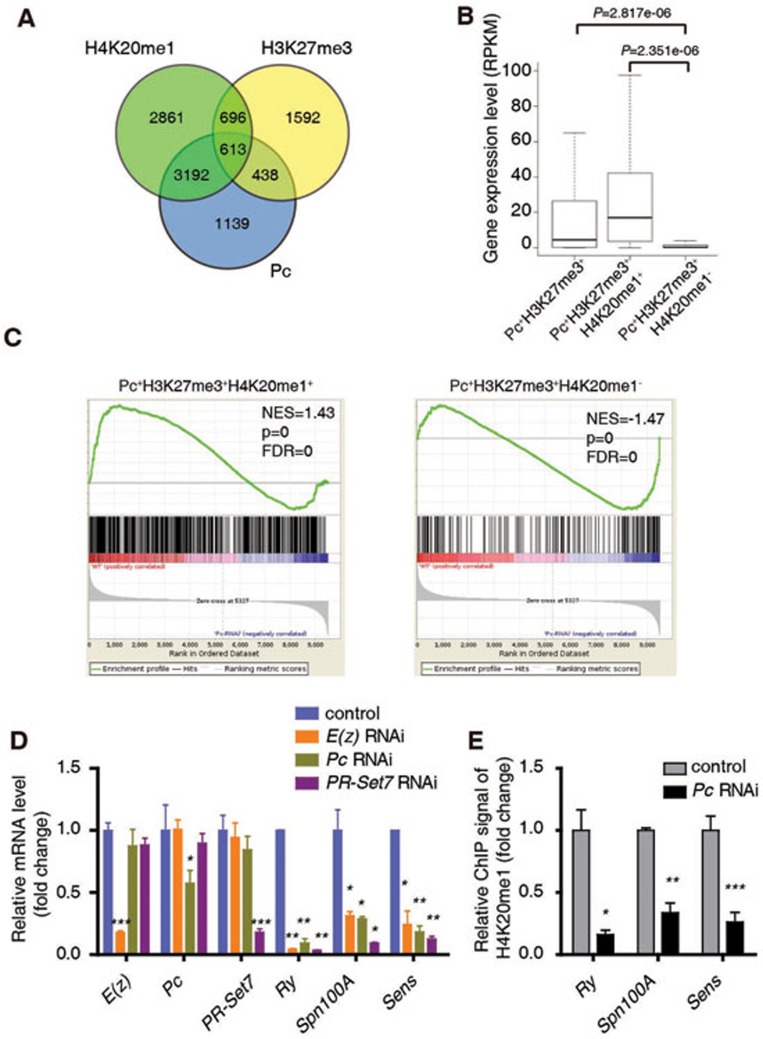
Figure 4.
H3K27me3, Pc and H4K20me1 function together to positively regulate common genes encoding developmental regulators. (A) A Venn diagram showing the overlap among target genes of Pc, H4K20me1 and H3K27me3 in WT 3rd instar larvae wing disc. (B) Expression analysis of all the Pc/H3K27me3 target genes (1 051 genes, Pc+H3K27me3+), and two subsets of these genes (438 genes without H4K20me1 named as Pc+H3K27me3+H4K20me1−, 613 genes with H4K20me1 named as Pc+H3K27me3+H4K20me1+). The RPKM values are based on RNA-seq data with 3rd instar larvae wing discs. (C) GSEA shows that Pc+H3K27me3+H4K20me1+ genes tend to be positively regulated by Pc while Pc+H3K27me3+H4K20me1− genes tend to be repressed by Pc in the wing disc. (D) RT-qPCR analysis of mRNA levels of E(z), Pc, PR-Set7, Ry, Spn100A and Sens in Act5c-Gal4 or Act5c-Gal4/uas-dsRNA E(z)/Pc/PR-Set7 wing disc. (E) A ChIP-qPCR analysis using anti-H4K20me1 antibody with control or Pc RNAi wing discs at Ry, Spn100A and Sens loci. ChIP signal levels are represented as percentages of input chromatin. The signal levels in control wing discs are defined as 1, and the fold change is shown. Means ± SD; n = 3. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired, two-tailed Student's t-test. See also Supplementary information, Figure S4.
To confirm this effect of Pc on the expression of Sens, we performed immunostaining with an anti-Sens antibody32. Knockdown of Pc led to an upregulation of Ubx (Supplementary information, Figure S1A-S1B''), but significantly repressed the expression of Sens (Figure 1E-1F'''). Consistent with these results, Ubx was activated while no obvious Sens protein was detected in PcXT109 null clones (Figure 1G-1H' and Supplementary information, Figure S1C-S1D'). To examine whether Pc was directly involved in the transcriptional regulation of Sens, we performed a chromatin immunoprecipitation (ChIP) assay with an anti-Pc antibody, followed by quantitative polymerase chain reaction (qPCR). We found that Pc bound to Sens locus in the wing disc (Figure 1D), suggesting that Pc binding positively correlates with Sens transcription in developing Drosophila wing disc.
Hierarchically recruited PcGs are essential for the transcription of Sens
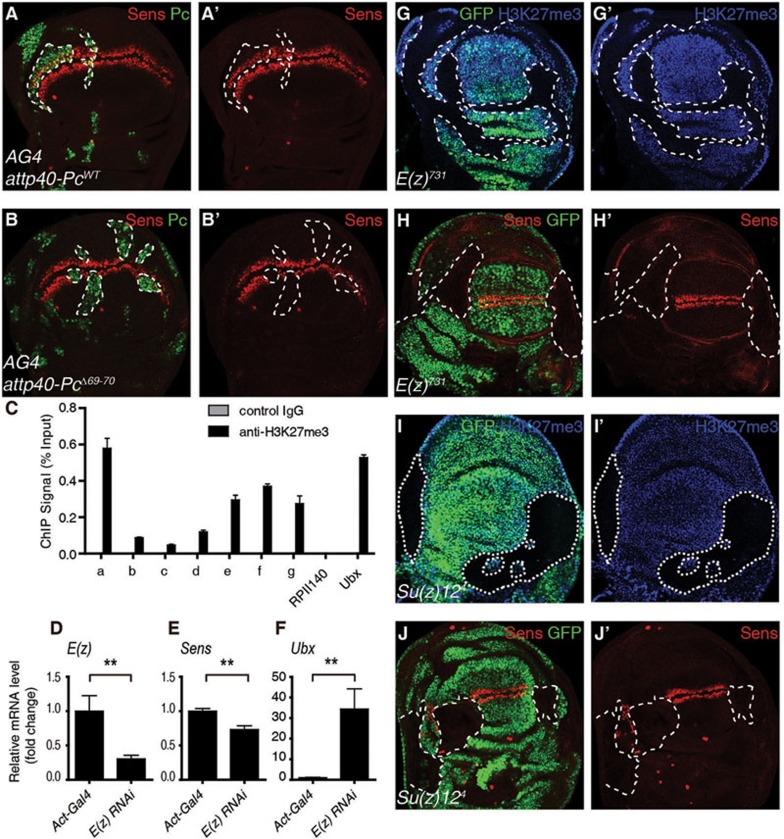
Previous studies have established a hierarchical recruitment model of PcG-mediated gene repression6,33. H3K27me3 is catalyzed by PRC234,35,36,37, and Pc can specifically recognize H3K27me3 through its chromodomain38 and recruit other components of PRC1 to selected chromatin sites9 to repress the expression of its target genes. Ile69 and Asp70 in Pc chromodomain are essential for the function of Pc both in vitro and in vivo as revealed by the phenotype of their deletion39,40. We asked whether the positive transcriptional regulatory role of Pc on Sens also requires its chromodomain. Over-expression of wild-type (WT) Pc did not significantly affect Ubx expression, whereas PcΔ69-70 induced an ectopic expression of Ubx (Supplementary information, Figure S2A-S2B'), indicating that PcΔ69-70 has a dominant negative effect. Expression of Sens was not altered by the over-expression of PcWT but was dramatically repressed by the over-expression of PcΔ69-70 (Figure 2A-2B'), indicating that the chromodomain is essential for Sens expression during the development of the wing disc. This prompted us to investigate whether Pc positively regulates Sens transcription in an H3K27me3-dependent manner using a ChIP-qPCR assay with an anti-H3K27me3 antibody. We detected a relatively strong H3K27me3 signal in the promoter and gene body of Sens, but a relatively weak signal around its transcription start site (TSS) (Figure 2C). This suggests that H3K27me3 is directly involved in the transcriptional regulation of Sens.
Figure 2.
Hierarchically recruited PcGs are essential for the transcription of Sens. (A-B) Immunostaining of wing discs expressing attp40-PcWT (A, A') or attp40-PcΔ69-70 (B, B') driven by AG4 using anti-Sens antibody (red) and anti-Pc antibody (green). Overexpression of PcWT has no obvious effect on the level of Sens while PcΔ69-70 downregulates the Sens level in the clones marked by dashed lines (Pc positive). (C) ChIP-qPCR analysis using control IgG (grey) or anti-H3K27me3 antibody (black) with WT wing discs (Act5c-Gal4). ChIP signal levels are represented as percentages of input chromatin. RPII140 and Ubx serve as negative and positive controls, respectively. (D-F) RT-qPCR analysis of mRNA levels of E(z) (D), Sens (E) and Ubx (F) in Act5c-Gal4 or Act5c-Gal4/uas-dsRNA E(z) wing discs. (G-J') Wing discs carrying E(z)731 or Su(z)124 mutant clones were immunostained with anti-Sens antibody (red) or anti-H3K27me3 antibody (blue); GFP negative clones are marked by dashed lines. Means ± SD; n = 3. **P < 0.01. Unpaired, two-tailed Student's t-test. See also Supplementary information, Figure S2.
To further investigate the role of the H3K27me3 modification in the transcription of Sens, we knocked down E(z), the histone methyltransferase in PRC2 that catalyzes H3K27me3, and examined Sens expression. Depletion of E(z) decreased H3K27me3 levels and Sens expression in vivo (Supplementary information, Figure S2C-S2D'''). Consistent with this result, RT-qPCR showed that mRNA levels of E(z) and Sens were decreased whereas that of Ubx was increased upon suppression of E(z) (Figure 2D-2F). These observations were further confirmed in E(z)731 mutant clones, where H3K27me3 levels and the expression of Sens were reduced, whereas the expression of Ubx was increased (Figure 2G-2H' and Supplementary information, Figure S2E-S2E'). Furthermore, dramatic reduction in both the H3K27me3 levels and Sens expression was observed in clones of Su(z)124 (Figure 2I-2J' and Supplementary information, Figure S2F-S2F'), a gene essential for the histone methyltransferase activity of E(z) in catalyzing H3K27me341,42,43. These results together suggest that Pc positively regulates transcription of Sens by recognizing H3K27me3 modification catalyzed by the intact PRC2.
Pc positively regulates Sens expression by regulating H4K20me1 catalyzed by PR-Set7
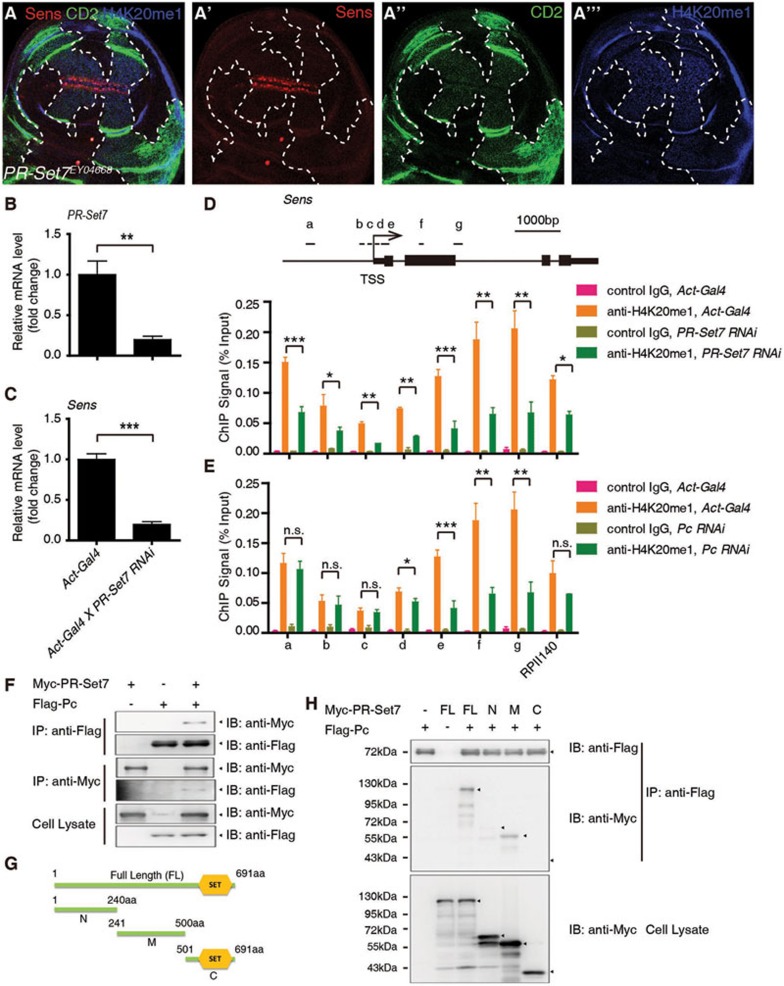
To probe the underlying mechanism by which Pc positively regulates Sens transcription, we performed an RNAi screen for epigenetic regulator(s) of Sens, and found PR-Set7, the only known enzyme catalyzing H4K20me1 in Drosophila44,45,46, as a strong candidate. Immunostaining showed that H4K20me1 was decreased dramatically upon PR-Set7 depletion, and Sens expression was also significantly reduced (Supplementary information, Figure S3). Furthermore, a mutant allele of PR-Set7 resulted in a dramatic reduction in the H4K20me1 levels and Sens expression (Figure 3A-3A'''). In addition, RT-qPCR showed that the mRNA level of Sens was decreased upon PR-Set7 knockdown (Figure 3B-3C). These results indicate that PR-Set7 positively regulates Sens transcription in Drosophila wing disc. Consistently, PR-Set7 has been reported to activate Wnt signaling in zebrafish and mammalian cell lines47.
Figure 3.
Pc positively regulates Sens expression by regulating H4K20me1 catalyzed by PR-Set7. (A-A''') A wing disc carrying PR-Set7EY04668 mutant clones and immunostained with anti-Sens antibody (red), anti-CD2 (green) and anti-H4K20me1 antibody (blue). The clones are CD2 negative, and their boundary is demarcated with dashed lines. (B-C) RT-qPCR analysis of mRNA levels of PR-Set7 (B) and Sens (C) in Act5c-Gal4 or Act5c-Gal4/uas-dsRNA PR-Set7 wing discs. (D) ChIP-qPCR analysis using control IgG or anti-H4K20me1 antibody at the Sens locus with Act5c-Gal4 wing discs or Act5c-Gal4/uas-dsRNA PR-Set7 wing discs. Upper panel illustrates the positions of the primers used for the qPCR assay at the Sens locus. In the lower panel, ChIP signal levels are represented as percentages of input chromatin. (E) ChIP-qPCR analysis using control IgG or anti-H4K20me1 antibody with Act5c-Gal4 wing discs or Act5c-Gal4/uas-dsRNA Pc wing discs at the Sens locus. ChIP signal levels are represented as percentages of input chromatin. (F) S2 cells were transfected with combinations of DNA constructs as indicated. After 48 h of transfection, lysates from transfected S2 cells were immunoprecipitated with anti-Flag antibody or anti-Myc antibody. Western blots were performed to analyze the presence of Flag- or Myc-tagged proteins. (G-H) Mapping of individual domains responsible for the interaction of PR-Set7 with Pc. Schematic drawings of PR-Set7 full length (FL) protein and different fragments (G). The SET domain that catalyzes H4K20me1 is located in the C-terminal (C) part of PR-Set7, while no significant domain is in the N-terminal (N) and middle (M) regions. A co-IP-WB shows that PR-Set7 interacts with Pc mainly through its M region (H). Means ± SD; n = 3. n.s., no significant difference. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired, two-tailed Student's t-test. See also Supplementary information, Figure S3.
Since H4K20me1 is reduced in PR-Set7 mutant clones, and H4K20me1 is important for transcriptional regulation47,48,49,50, we asked whether PR-Set7 regulates transcription of Sens through H4K20me1 modification at the Sens locus. ChIP-qPCR of the wing disc with anti-H4K20me1 antibody showed a strong H4K20me1 signal in the gene body of Sens, and this modification was decreased upon depletion of PR-Set7 (Figure 3D). These results suggest that PR-Set7 regulates Sens expression by catalyzing H4K20me1 at the Sens locus.
Next, we asked if there is any functional interaction between Pc and PR-Set7 in modulating the transcription of Sens. First we tested whether H4K20me1 catalyzed by PR-Set7 at the Sens locus is affected by Pc. ChIP-qPCR results showed that H4K20me1 was dramatically reduced in the gene body of Sens upon Pc knockdown (Figure 3E), suggesting that both Pc and PR-Set7 are required for H4K20me1 at the Sens locus, and that Pc is functionally associated with PR-Set7 in regulating Sens transcription. We then asked whether Pc physically interacts with PR-Set7, and indeed, this was confirmed using co-immunoprecipitation (co-IP) (Figure 3F). Further domain mapping suggest that PR-Set7 interacts with Pc mainly through its middle part adjacent to the catalytic SET domain44,45 (Figure 3G-3H). In addition, Pc and PR-Set7 in cell nuclear extract could be co-eluted in a gel filtration assay (Figure 5E), further supporting the physical interaction between these two proteins. Taken together, these results indicate that Pc regulates transcription of Sens by promoting PR-Set7-mediated H4K20me1 at the Sens locus.
Figure 5.
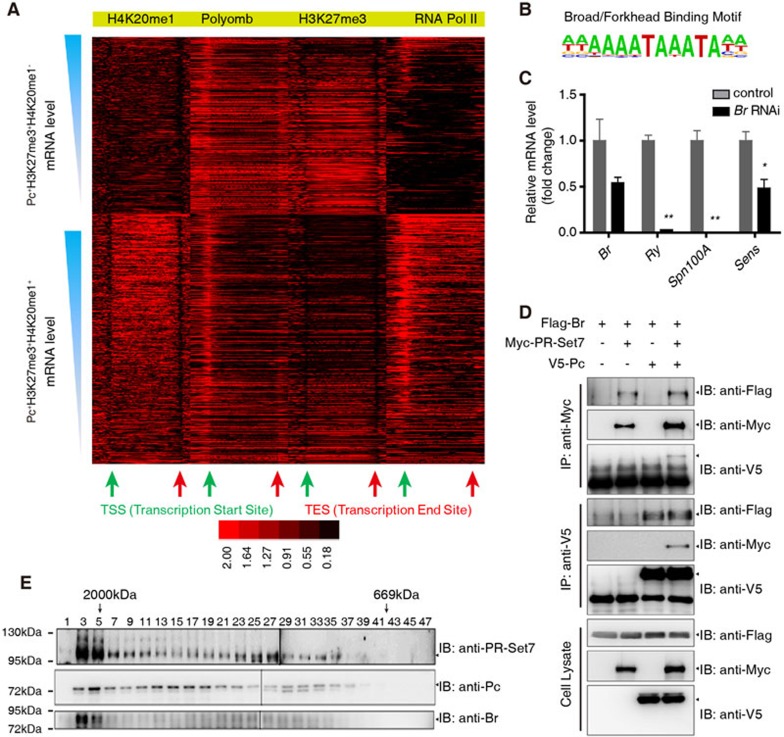
Selective markers for the functional switch of Pc from a transcription repressor to an activator. (A) Heat maps of ChIP signals at Pc+H3K27me3+H4K20me1− and Pc+H3K27me3+H4K20me1+ genes. (B) The Broad/Forkhead motif identified in Pc+H3K27me3+H4K20me1+ genes. (C) RT-qPCR analysis of mRNA levels of Br, Ry, Spn100A and Sens in Act5c-Gal4 or Act5c-Gal4/uas-dsRNA Br wing discs. (D) S2 cells were transfected with combinations of DNA constructs as indicated. After 48 h of transfection, lysates from transfected S2 cells were immunoprecipitated with anti-V5 antibody or anti-Myc antibody. Western blots were performed to analyze the presence of Flag-, V5- or Myc-tagged proteins. (E) Gel filtration chromatography on a Superose 6 column and western blot of Cl-8 nuclear extract. Molecular weight standards are shown on top (in kDa). Means ± SD; n = 3. *P < 0.05, **P < 0.01. Unpaired, two-tailed Student's t-test. See also Supplementary information, Figure S5.
H3K27me3, Pc and H4K20me1 function together to positively regulate common genes encoding developmental regulators
We next explored if what we show above represents a general mechanism regulating other genes in addition to Sens. First we combined the ChIP-seq data for H4K20me1 generated in this study with the ChIP-chip data for Pc and ChIP-seq data for H3K27me3 reported previously21,51. A total of 1 051 genes were chosen for the subsequent analysis since they seem to be the targets of both PRC2 and PRC1, i.e., regulated by the hierarchically recruited PcG proteins (Figure 4A, Pc+H3K27me3+ genes). We divided these genes into two sets based on the status of H4K20me1: 613 genes in the first set were Pc+H3K27me3+H4K20me1+, and 438 genes in the second set were Pc+H3K27me3+H4K20me1− (Figure 4A).
We also classified every gene in the genome as either a silent gene or an expressed gene according to the abundance of its mRNA in the wing disc. The expression level of each expressed gene was further categorized as low, median or high. We found that about half of the Pc+H3K27me3+ genes were expressed, while 26.98% of the Pc+H3K27me3+H4K20me1− genes and 80.16% of the Pc+H3K27me3+H4K20me1+ genes were expressed (Supplementary information, Figure S4A). Moreover, the average expression levels of Pc+H3K27me3+ and Pc+H3K27me3+H4K20me1+ genes were significantly higher than that of Pc+H3K27me3+H4K20me1− genes (Figure 4B). It thus appears that H4K20me1 acts as a selective mark to determine the transcription state of Pc+H3K27me3+ genes. This notion is further supported by a gene set enrichment analysis (GSEA) showing that Pc+H3K27me3+H4K20me1+ genes tend to be positively regulated by Pc while Pc+H3K27me3+H4K20me1− genes tend to be repressed by Pc in the wing disc (Figure 4C). Collectively, these data indicate that Pc, H3K27me3 and H4K20me1 are three key factors that function together to activate gene transcription. This effect of Pc in promoting the transcription of a subset of target genes (Pc+H3K27me3+H4K20me1+) is likely to be direct, because, if it were an indirect effect, one would expect similar numbers of genes are upregulated or downregulated in both gene sets after Pc knockdown.
Thus, a key role of H4K20me1 is to convert the negative role of Pc in transcriptional regulation to a positive role. Analysis of the gene functions indicate that the most enriched ontologies of Pc+H3K27me3+H4K20me1+ genes are DNA binding, development or morphogenesis, while those of Pc+H3K27me3+H4K20me1− genes are homeobox and DNA binding (Supplementary information, Figure S4B), suggesting that Pc and H3K27me3 are involved in different biological processes in the wing disc by differentially controlling the expression of distinct target genes. Of note, the canonical PcG target genes, such as Ubx, Abdominal A (Abd-A) and Abdominal B (Abd-B), belong to the Pc+H3K27me3+H4K20me1− group, while genes positively regulated by PcG, such as Sens, Ry and Spn100A, belong to the Pc+H3K27me3+H4K20me1+ group. It seems that the Pc+H3K27me3+H4K20me1− state defines the canonical PcG target genes directly repressed by Pc, while the Pc+H3K27me3+H4K20me1+state defines a novel subset of PcG targets, whose transcription also requires the direct binding of Pc.
Considering that Pc+H3K27me3+H4K20me1+ genes are mainly involved in development, we selected several highly conserved regulators of development to strengthen our observation. RT-qPCR analysis confirmed that these genes were positively regulated by E(z), Pc and PR-Set7 (Figure 4D). ChIP-qPCR showed strong signals for H4K20me1 at these gene loci (Supplementary information, Figure S4C), and Pc knockdown dramatically decreased H4K20me1 levels (Figure 4E). Taken together, these results indicate that Pc positively regulates a subset of targets via H4K20me1. Taking into account that Spn100A is actively and ubiquitously transcribed in entire wing pouch of WT 3rd instar larvae (Supplementary information, Figure S4D), and Pc knockdown reduced Spn100A mRNA level (Figure 4D and Supplementary information, Figure S4E), we conclude that Pc can bind to actively transcribed genes and positively regulate their transcription by modulating H4K20me1. Of note, we have observed similar regulatory mechanism for Sens in salivary gland where Sens is actively expressed in every cell, and for a subset of Pc target genes in S2 cell line (Supplementary information, Figure S4F-S4M).
The existence of H4K20me1, specific patterns of Pc and H3K27me3, and binding of the transcription factor Br, together serve as selective marks for the functional switch of Pc from a transcriptional repressor to an activator
The binding of Pc and H3K27me3 modification are known to mediate transcriptional repression52, why do Pc+H3K27me3+H4K20me1+ genes tend to be positively regulated by Pc while Pc+H3K27me3+H4K20me1− genes tend to be repressed by Pc in the wing disc? We sought to determine the mechanism by which Pc, H3K27me3 and H4K20me1 positively regulate transcription. To this end, we generated binding heat maps of H4K20me1, Pc, H3K27me3 and RNA Pol II at the Pc+H3K27me3+H4K20me1+and Pc+H3K27me3+H4K20me1− genes. Genes were sorted according to their mRNA levels, and graphed against the enrichments of H4K20me1, Pc, H3K27me3 and RNA Pol II at their loci (Figure 5A). As expected, the RNA Pol II signal mainly appeared around the TSSs, and exhibited positive correlations with gene expression levels. The RNA Pol II signal was markedly enriched in the Pc+H3K27me3+H4K20me1+ gene set, consistent with higher expression levels of these genes compared with the Pc+H3K27me3+H4K20me1− genes (Figure 4B and Supplementary information, Figure S4A). Meanwhile, the H4K20me1 modification was mostly enriched across the gene body but not at the promoter and was at a very low level around the TSS. More importantly, the H4K20me1 signal at the gene body positively correlated with the transcription level of the target genes (Figure 5A).
In contrast, the intensity of H3K27me3 modification negatively correlated with the transcription levels of target genes (Figure 5A), which is consistent with previous studies52,53,54,55. Furthermore, we noticed that the H3K27me3 signal patterns differed in these two gene sets, which is an interesting but unexpected phenomenon. The average enrichment of H3K27me3 at the Pc+H3K27me3+H4K20me1− gene loci was higher, and the modification was distributed almost evenly throughout the promoter and gene body. In contrast, the levels of H3K27me3 modification were generally lower in the Pc+H3K27me3+H4K20me1+ genes, and such modification mainly occurred at the promoter regions but not the TSSs, especially for those genes with high and median expression levels (Figure 5A). Another study in mammalian cells also found a similar phenomenon56.
We found that Pc signals tended to peak around TSSs in the Pc+H3K27me3+H4K20me1+ genes, whereas at the gene bodies the Pc signals were significantly reduced; but, interestingly, this reduction was not significant in the Pc+H3K27me3+H4K20me1− genes (Figure 5A). In addition, the ratio of the Pc signal around TSS to that at the gene body (Pcpromoter/Pcgene body) positively correlated with the gene's expression level (Figure 5A). Taken together, these observations indicate that not only the intensities but also the specific patterns of Pc binding and of H3K27me3 and H4K20me1 modifications are important for the control of the target gene expression.
To further explore the mechanism of Pc-mediated positive regulation of gene transcription, we performed motif analysis. We found a significant enrichment of the binding motif for transcription factors Br and Forkhead (FkH)57,58 in the Pc+H3K27me3+H4K20me1+ genes (Figure 5B). Based on a reported Br ChIP-chip data59, about half of the genes harboring this binding motif are actually targets of Br. Consistent with these results, analysis of the average enrichment of the Br signal revealed that Br mainly bound to Pc+H3K27me3+H4K20me1+ genes but not Pc+H3K27me3+H4K20me1− genes (Supplementary information, Figure S5A), supporting a hypothesis that the binding of Br is important for transcriptional regulation of Pc, H3K27me3 and H4K20me1 co-targets.
Since FkH is not expressed in the wing disc (data not shown), we next tested whether Br could regulate expression of Sens. Knockdown of Br indeed decreased Sens protein level (Supplementary information, Figure S5B-S5C'), and the mRNA levels of Sens, Ry and Spn100A were decreased upon Br suppression (Figure 5C). We next revealed that Br physically associated with Pc and PR-Set7 in a co-IP assay (Figure 5D), and this observation was confirmed by a gel filtration assay showing that Br, Pc and PR-Set7 could be co-eluted (Figure 5E).
These results suggest that Br binding may determine the regulatory outcome of Pc on transcription. Br binding is positively correlated with H4K20me1 levels, and also the transcription levels of Pc+H3K27me3+H4K20me1+ target genes in the wing disc, revealing Br binding as another layer of regulation in Pc-mediated transcriptional control. Thus Pc functions together with specific transcription factors such as Br, and epigenetic enzymes such as PR-Set7, to positively regulate gene transcription.
Discussion
Recently, Dr Reinberg and colleagues reported that PRC1-AUTS2 activates the cell-based luciferase reporter directly, and regulates the transcription of several neuronal genes in mouse central nervous system60, supporting our notion that Pc can directly and positively regulate the transcription of a subset of its direct target genes, which, in our case, are Pc+H3K27me3+H4K20me1+ genes involved in the developing Drosophila wing disc.
However, how PRC1 attains the target specificity for activation or repression remains largely unknown. Our study reveals a hierarchical and combinatorial recruitment of transcriptional regulators and epigenetic marks that ultimately determines the outcome of Pc-mediated transcriptional regulation. We base our conclusion on the following experimental observations. First, the expression levels of Pc+H3K27me3+H4K20me1+ genes are higher than those of Pc+H3K27me3+H4K20me1− genes in WT flies, suggesting H4K20me1 serves as a selective mark that influences Pc's effect on transcriptional regulation. Second, GSEA data show that Pc+H3K27me3+H4K20me1+ genes tend to be positively regulated by Pc while Pc+H3K27me3+H4K20me1− genes tend to be negatively regulated by Pc. Third, GO analysis indicates the Pc+H3K27me3+H4K20me1− genes are the canonical PcG target genes repressed by Pc while the Pc+H3K27me3+H4K20me1+ genes are a different subset of PcG targets, whose transcription requires the direct regulation of Pc. Fourth, the intensities and specific patterns of Pc and H3K27me3 in these two sets of genes suggest that they act as another selective mark to affect Pc function. Fifth, the transcript factor Br specifically binds to Pc+H3K27me3+H4K20me1+ genes but not Pc+H3K27me3+H4K20me1− genes, and our functional study indicates that Br binding is instrumental in determining the fates of Pc-mediated transcriptional regulation. These results together unveil a mechanism by which Pc is directly involved in positive transcriptional regulation of Pc+H3K27me3+H4K20me1+ genes.
Materials and Methods
RT-qPCR and RNA-seq
Wing discs with indicated genotypes were lysed in TRIzole (Invitrogen) for total RNA isolation following standard protocol. For RT-qPCR, 0.5 μg RNA was used for reverse transcription with ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, FSQ-301). Real-time PCR was performed on CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with SYBR® Green Realtime PCR Master Mix (TOYOBO, QPK-201). The 2−ΔΔCt method was used for quantification. The primer pairs used are listed in Supplementary information, Table S1. RNA-seq experiments were performed using total polyA mRNA isolated from both WT and knockdown samples as described previously61 with some modification. Briefly, 10 ng mRNA was extracted from Act5c-Gal4 or Pc knockdown sample of Drosophila wing disc and enriched using polyT beads, and was converted into cDNA, which was fragmented and sequenced from single end on Illumina HiSeq 2000.
ChIP-qPCR and ChIP-seq
ChIP assays were performed as previously described62,63 with modification. Briefly, 3rd instar larvae with indicated phenotypes were dissected in PBS and, after removing the guts and fat body, the carcasses with discs still attached were cross-linked for 20 min at room temperature in 1 ml 1% formaldehyde in PBS buffer. The cross-linking solution was changed three times during fixation and, after final change, cross-linking was stopped by washing for 10 min in 1 ml PBS/0.01% Triton X-100/0.125 M glycine. Then, fixed carcasses were washed for 10 min in 1 ml PBS/0.01% Triton X-100 for three times. Wing discs were hand-dissected in PBS and pooled. Sonication of discs was performed in 200 μl sonication buffer (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and proteinase cocktail) with a Bioruptor sonicator. The sonication yielded genomic DNA fragments with an average size of about 250 bp. After centrifugation, lysates were incubated with 2 μg antibody for 4 h (or overnight); 20 μl protein A/G PLUS agarose (Santa Cruz) was then added and incubated for another 4 h (or overnight) on a rotator at 4 °C. Beads were washed three times with the ChIP wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 150 mM NaCl, and 20 mM Tris-Cl, pH 8.0), and then washed with ChIP final wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 500 mM NaCl, and 20 mM Tris-Cl, pH 8.0). Genomic DNA was eluted with elution buffer (1% SDS and 100 mM NaHCO3) at 65 °C for 30 min. 5 M NaCl was added to a final concentration of 200 mM for further incubation at 65 °C for 4 h or overnight. Then, 0.5 M EDTA and 20 mg/ml proteinase K were added until their final concentrations were 5 mM and 0.25 mg/ml, respectively. The resulting mixture was incubated at 55 °C for 2 h to digest the protein. Genomic DNA was purified with a DNA purification kit (QIAGEN) and sent for real-time PCR or sequencing. Antibodies used were rabbit anti-Pc antibody (Santa Cruz), rabbit anti-H3K27me3 antibody (Millipore), rabbit anti-H4K20me1 antibody (Abcam) and control rabbit IgG (Santa Cruz). Primer pairs used in this study are listed in Supplementary information, Table S2. ChIP-seq experiments were conducted as previously described with some modifications64. Briefly, 10 ng ChIP-enriched genomic DNA with anti-H4K20me1 antibody was used for library preparation and sequenced from single-end on Illumina HiSeq 2000.
ChIP-seq and RNA-seq data processing
Both ChIP-seq and RNA-seq reads were mapped to dm3 using bowtie65 and Tophat266 with default parameters except specifically indicated. For H4K20me1 ChIP-seq, only uniquely mapped and at most three duplicated reads were kept for further analysis. Peaks were identified using SICER67 with a window size of 200 bp, and gap size of 400 bp and E value of 0.01. Peaks were annotated to dm3 refSeq genes using an in-house program with promoters extended to 2 kb upstream from TSSs. Co-targeted genes were defined as genes that have H4K20me1, Pc and H3K27me3 peaks in their gene bodies or promoter regions. ChIP-seq reads were extended to 200 bp and then converted into the bedGraph format and normalized by the total number of reads to generate genome-wide binding profiles, and then used to calculate average modification levels for different gene expression groups and to generate heat maps. Motifs were searched for using an online tool, RSAT68, in different sets of genes. For RNA-seq datasets, reads were first mapped to the refSeq transcriptome and the left parts were then mapped to the genome. Reads that mapped to more than 5 locations were excluded. Gene expression levels were calculated using cufflinks66 and GSEA was performed as reported previously69.
DNA constructs
To construct pUAST-attb-PcWT/PcΔ69-70, full-length Pc cDNA was cloned from Drosophila Melanogaster embryonic RNA using RT-PCR and inserted into the pUAST-attb vector. PCR-based site-directed mutagenesis was used to delete the 69th and 70th amino acids of Pc. Full-length Pc was inserted into pUAST-Flag or pUAST-V5 vectors to generate pUAST-Flag-Pc or pUAST-V5-Pc, respectively. Full-length PR-Set7 cDNA was obtained as was Pc. The full-length or indicated fragments of PR-Set7 were inserted into the pUAST-Myc vector to generate pUAST-Myc-PR-Set7 FL/N/M/C (FL indicates residues 1-691; N, M and C indicate residues 1-240, 241-500, and 501-691, respectively). Full-length Br cDNA was obtained as was Pc and inserted into the pUAST-Flag vector to generate pUAST-Flag-Br.
Cell culture, transfection, immunoprecipitation, gel filtration and western blot analysis
Drosophila wing disc-derived Cl-8 cells and Drosophila embryo-derived S2 cells were cultured following standard protocols. Specifically, S2 cells were cultured at 25 °C in Schneider's Drosophila Medium (Invitrogen) with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Transfection, immunoprecipitation (IP) and western blot (WB) analysis were carried out using standard protocols. Specifically, we used the Calcium Phosphate Transfection kit (Specialty Media) following manufacturer's instructions. IP was performed at 4 °C. Cells were lysed in standard NP-40 buffer; lysates were first incubated with 2 μg of antibody for 2 h and then with 20 μL protein A/G PLUS agarose (Santa Cruz) for 1 h. Beads were washed three times and boiled in SDS loading buffer. Cl-8 cell nuclear extract was subjected to gel filtration (Superose 6 HR column, GE Healthcare) as reported previously70 after WB. Anti-Pc and anti-Myc antibodies used for co-IP-WB were purchased from Santa Cruz and Sigma, respectively. An anti-PR-Set7 antibody (which was generated in this study by using the 345-691 aa of PR-Set7 as an antigen to elicit the immune response in the rabbit, ABclonal technology) was generated in this study. Anti-Br antibody was purchased from DSHB.
Drosophila strains
Fly stocks used in this study were maintained under standard culture conditions. AP4, AP4GFP, Hh-Gal4 and AG4 have been described (flybase). PcXT109, E(z)731 and Su(z)124 are gifts from Dr Rongwen Xi at NIBS71. PR-Set7EY04668 (B15761), uas-dsRNA Pc (32443R-4), uas-dsRNA E(z) (6502R-4), uas-dsRNA PR-Set7 (3307R-1), uas-dsRNA Br (V38526) and Act5c-Gal4 (B4414) were obtained from Bloomington, NIG or VDRC. The attp40-PcWT and attp40-PcΔ69-70 transgenes were generated with a P element-mediated insertion at the 25C6 site (attp40) in this study. Mutant clones were generated with Flp-mediated mitotic recombination. In order to generate large clones, the Minute technique was applied72. With this method, suitable PcXT109, E(z)731, Su(z)124 and PR-Set7EY04668 clones were generated to analyze the expression levels of the indicated genes.
Immunostaining of the wing disc
3rd instar larvae wing discs were dissected for immunostaining using the standard protocol as reported previously73,74. Larvae with indicated genotypes were fixed in 4% formaldehyde in PBS buffer, washed and incubated with a specific combination of primary and secondary antibodies in PBS/0.1% Triton X-100, and mounted in 40% glycerol. Pictures were taken with the confocal microscope (LAS SP8; Leica) using a 40×/1.25 NA oil objective (Leica). Antibodies used in this study were rat anti-Ci antibody (DSHB), rabbit anti-Pc antibody (which was generated in this study by using the peptide RKAEVLKESGKIG), mouse anti-Ubx antibody (DSHB), guinea pig anti-Sens antibody (a kind gift from Dr Hugo J Bellen), rabbit anti-H3K27me3 antibody (Millipore), mouse anti-CD2 antibody (AbD Serotec), rabbit anti-H4K20me1 antibody (Abcam), and secondary antibodies were bought from Millipore.
Accession numbers
The RNA-seq and H4K20me1 ChIP-seq datasets were deposited in the Gene Expression Omnibus (GEO) database with accession number GSE59924. Pc and Pol II ChIP-chip datasets are available from GSE42106. H3K27me3 data are available from GSE24115.
Author Contributions
XL conceived and designed the project, performed most of the experiments, analyzed the data, and drafted the manuscript; ZH performed the bioinformatics analysis, and interpreted the data; HC, BY, YX, CP, LF, and SZ contributed reagents and materials, and performed experiments; XY and HH contributed to the experimental design and data analysis; MW, ZZ, LZ, LL, and GW contributed to data analysis and manuscript preparation, and YZ conceived and designed the project, analyzed data, and prepared the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We are grateful to Drs Ruth Steward, Gary H Karpen, Jeffrey A Simon, Jürg Müller, Rongwen Xi, Degui Chen, Guoliang Xu, Miguel Vidal, Sharon E Bickel, DSHB, VDRC, NIG and the Bloomington Stock Center for fly stocks and reagents. We also thank Drs Jinqiu Zhou, Hai Jiang, Dangsheng Li, Yingzi Yang, Chi-chung Hui, Guoliang Xu and Gaoxiang Ge for discussions and comments on the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31171414, 31371492, and 31271354), 'Strategic Priority Research Program' of the Chinese Academy of Sciences (XDA01010405), and the Chinese Academy of Sciences (2012OHTP09).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Loss-of Pc leads to the derepression of Ubx in Drosophila wing disc. Related to Figure 1
The hierarchically recruited PcGs repress expression of Ubx but stimulate that of Sens. Related to Figure 2
Knockdown of PR-Set7 decreases H4K20me1 and Sens levels. Related to Figure 3
Transcriptional regulation and functional analysis of the Pc+H3K27me3+H4K20me1− genes and Pc+H3K27me3+H4K20me1+ genes. Related to Figure 4
Function of Br in positive transcriptional regulation. Related to Figure 5
Primers for RT-qPCR
Primers for ChIP-qPCR
References
- Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the polycomb perspective. Dev Cell 2010; 19:651–661. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The DrosophilaFab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 1998; 93:505–518. [DOI] [PubMed] [Google Scholar]
- Struhl G, Brower D. Early role of the Esc+ gene-product in the determination of segments in Drosophila. Cell 1982; 31:285–292. [DOI] [PubMed] [Google Scholar]
- Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 2014; 14:735–751. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 2009; 19:150–158. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Morey L. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 2010; 35:323–332. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet 2011; 27:454–464. [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 2011; 12:123–135. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, et al. RYBP-PRC1 Complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012; 148:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004; 431:873–878. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science 2004; 306:1574–1577. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010; 38:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Endo TA, Ku M, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 2013; 26:565–577. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011; 144:214–226. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet 2013; 14:853–864. [DOI] [PubMed] [Google Scholar]
- Eun SH, Shi Z, Cui K, Zhao K, Chen X. A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker. Science 2014; 343:1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob E, Hod-Dvorai R, Schif-Zuck S, Avni O. Unconventional association of the polycomb group proteins with cytokine genes in differentiated T helper cells. J Biol Chem 2008; 283:13471–13481. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 2007; 27:3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Gause M, et al. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet 2013; 9:e1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Moore HM, Li X, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA 2014; 111:3098–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HY, Jun S, Lee M, et al. PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol Cell 2013; 52:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Li Z, Wu Z, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43:798–810. [DOI] [PubMed] [Google Scholar]
- Shi B, Liang J, Yang X, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol 2007; 27:5105–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KX, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science 2012; 338:1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shao Z, Li D, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 2015; 57:304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Isono K, Kondo K, et al. Polycomb potentiates meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev Cell 2014; 28:94–101. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell 2012; 45:255–262. [DOI] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of DrosophilaHox genes. Development 2001; 128:993–1004. [DOI] [PubMed] [Google Scholar]
- Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J 1985; 4:3259–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 2000; 102:349–362. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 2004; 14:637–646. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002; 111:197–208. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111:185–196. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 2002; 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 2003; 17:1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev 1992; 6:1241–1254. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 2003; 17:1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23:4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 2004; 15:57–67. [DOI] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol 2005; 25:6857–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 2002; 9:1201–1213. [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 2002; 12:1086–1099. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA 2011; 108:3116–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 2005; 19:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 2006; 26:9185–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S, Blanco E, Carbonell A, et al. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res 2011; 39:4628–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 2011; 471:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA 2006; 103:15782–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349–353. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006; 125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 2011; 39:7415–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J 1994; 13:5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kalm L, Crossgrove K, Von Seggern D, Guild GM, Beckendorf SK. The Broad-Complex directly controls a tissue-specific response to the steroid hormone ecdysone at the onset of Drosophila metamorphosis. EMBO J 1994; 13:3505–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, et al. A cis-regulatory map of the Drosophila genome. Nature 2011; 471:527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 2014; 516:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang QS, Zhao KJ. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res 2009; 37:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 2006; 20:2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 1997; 11:205–214. [DOI] [PubMed] [Google Scholar]
- Wei G, Hu G, Cui K, Zhao K. Genome-wide mapping of nucleosome occupancy, histone modifications, and gene expression using next-generation sequencing technology. Methods Enzymol 2012; 513:297–313. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang CZ, Schones DE, Zeng C, Cui KR, Zhao KJ, Peng WQ. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res 2012; 40:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du Y, Ward James M, et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 2014; 14:575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strübbe G, Popp C, Schmidt A, et al. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci USA 2011; 108:5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Han Y, Xi R. Polycomb group genes Psc and Su(z)2 restrict follicle stem cell self-renewal and extrusion by controlling canonical and noncanonical Wnt signaling. Genes Dev 2010; 24:933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. Parameters of cell competition in the compartments of the wing disc of Drosophila. Dev Biol 1979; 69:182–193. [DOI] [PubMed] [Google Scholar]
- Shi D, Lv X, Zhang Z, et al. Smoothened oligomerization/higher order clustering in lipid rafts is essential for high Hedgehog activity transduction. J Biol Chem 2013; 288:12605–12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lv X, Yin WC, et al. Ter94 ATPase complex targets k11-linked ubiquitinated Ci to proteasomes for partial degradation. Dev Cell 2013; 25:636–644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss-of Pc leads to the derepression of Ubx in Drosophila wing disc. Related to Figure 1
The hierarchically recruited PcGs repress expression of Ubx but stimulate that of Sens. Related to Figure 2
Knockdown of PR-Set7 decreases H4K20me1 and Sens levels. Related to Figure 3
Transcriptional regulation and functional analysis of the Pc+H3K27me3+H4K20me1− genes and Pc+H3K27me3+H4K20me1+ genes. Related to Figure 4
Function of Br in positive transcriptional regulation. Related to Figure 5
Primers for RT-qPCR
Primers for ChIP-qPCR