Abstract
In this study, we explored the existence of a transcriptional network co-regulated by E2F7 and HIF1α, as we show that expression of E2F7, like HIF1α, is induced in hypoxia, and because of the previously reported ability of E2F7 to interact with HIF1α. Our genome-wide analysis uncovers a transcriptional network that is directly controlled by HIF1α and E2F7, and demonstrates both stimulatory and repressive functions of the HIF1α -E2F7 complex. Among this network we reveal Neuropilin 1 (NRP1) as a HIF1α-E2F7 repressed gene. By performing in vitro and in vivo reporter assays we demonstrate that the HIF1α-E2F7 mediated NRP1 repression depends on a 41 base pairs ‘E2F-binding site hub’, providing a molecular mechanism for a previously unanticipated role for HIF1α in transcriptional repression. To explore the biological significance of this regulation we performed in situ hybridizations and observed enhanced nrp1a expression in spinal motorneurons (MN) of zebrafish embryos, upon morpholino-inhibition of e2f7/8 or hif1α. Consistent with the chemo-repellent role of nrp1a, morpholino-inhibition of e2f7/8 or hif1α caused MN truncations, which was rescued in TALEN-induced nrp1ahu10012 mutants, and phenocopied in e2f7/8 mutant zebrafish. Therefore, we conclude that repression of NRP1 by the HIF1α-E2F7 complex regulates MN axon guidance in vivo.
INTRODUCTION
Hypoxia (oxygen deprivation) is experienced by cells in fast growing tissues such as embryos or solid tumors that outgrow their vasculature or exhaust the local O2 pool. Transcriptional adaptation to hypoxia is regulated by the heterodimeric, hypoxia-inducible transcription factors HIF1 and HIF2, which consist of an oxygen-degradable HIFα, and an oxygen-independent HIFβ subunit. The importance of these factors for development is illustrated by the embryonic lethality of mice lacking Hif1α, Hif2α or Hif1β (1,2). In normoxia, HIFα isoforms are constitutively degraded by the O2-dependent PHD/VHL pathway (3,4). Below ∼5% O2, HIFα levels stabilize according to the level of hypoxia, resulting in functional HIF dimers that stimulate O2-homeostasis by inducing the expression of genes involved in glycolysis, erythropoiesis and angiogenesis (3,4). In mammals, the HIF-pathway is active over a wide physiological range as embryonic development occurs at 3–5% O2, and postnatal physiological oxygen levels range from 2–9% O2 (1,2). On the other hand, HIF factors are also regulated by O2-independent mechanisms leading to increased HIF activity under normoxia. For example, normoxic HIF1 activity can be enhanced by stimulation of HIF1α transcription as observed in macrophages or T cells (5), or by increased HIF1α translation, induced by the MAP kinase and mTOR pathways downstream of growth factor signaling in highly proliferative cells, or by augmented HIF1α protein stabilization induced by heatshock protein 90 and calcium signaling (3). These studies illustrate the versatile roles of HIF factors in regulating gene expression, being activated in response to a wide variety of stimuli.
The E2F family of transcription factors consists of eight members that are classified as activators (E2F1–3) or repressors E2Fs (E2F4–8). E2F7 and E2F8 (E2F7/8) are termed atypical as they harbor 2 instead of 1 DNA-binding domain and regulate transcription independent of DP proteins (6). E2F7/8 have overlapping functions as E2f7 or E2f8 deficient mice are viable and live to old age, while E2f7/8 double knockout mice die around embryonic day 10.5 (7). The RB/E2F pathway is well known for its role in cell cycle regulation, but recent studies demonstrate an emerging role in control of physiological processes, independent of its cell cycle functions, among which neuronal development. Interestingly, telencephalon-specific retinoblastoma 1 (Rb1) deletion in mice showed a cell-autonomous requirement for Rb1 in the migration of neuronal precursor cells (8). This appeared to be mediated by E2F3, independent of its ability to regulate proliferation, but through direct transcriptional control of the atypical E2F target neogenin (Neo1), a receptor for the netrin and repulsive guidance molecule (RGM) families of chemotropic ligands (9,10). In addition, the RB/E2F pathway also regulates neuronal differentiation. E2F3 controls interneuron differentiation in a cell cycle-independent manner upon retina-specific deletion of Rb1 (11), while E2F7 represses the expression of the early differentiation genes Dlx1 and Dlx2 upon telencephalon-specific deletion of Rb1 involved in neuronal differentiation in the brain (12). These data show that E2F factors are able to regulate physiological processes beyond their ability to control the cell cycle, but through the regulation of atypical E2F target genes.
HIF and E2F factors are essential for development by regulating distinct but also overlapping biological processes (e.g. apoptosis, autophagy, cell cycle progression and angiogenesis). Consistent with the notion that they have been reported to also regulate gene expression by cooperating with other transcription factors (6,13), we recently found that E2F7/8 regulate transcription of VEGFA by forming a complex with HIF1 (14). To investigate the existence of a transcriptional network that is regulated by this recently identified HIF1-E2F7/8 complex we performed ChIP-sequencing (ChIP-seq) and microarray analysis. Our data not only identify a previously unknown HIF1α and E2F7 co-regulated transcriptional network, but also reveal a unanticipated role for HIF1α in transcriptional repression, and discover an essential function for the HIF1α-E2F7 complex in regulating motor neuron guidance through direct transcriptional control of NRP1 in vivo.
MATERIALS AND METHODS
Cell culture and hypoxia
The cervical cancer (HeLa) cell line was cultured in DMEM (Invitrogen, 41966–052) supplemented with 10% FBS (Lonza, DE14–802F) and 1% Penicillin/Streptomycin (Lonza, DE17–602E). U2OS cells were cultured similarly. For hypoxia treatment, cells were incubated in the H35 Hypoxystation (Don Whitley Scientific) at 1% O2 for 16 h.
Microarray gene expression analysis
The following samples were analyzed by microarrays: RNA isolated from hypoxic HeLa cells transfected with either control (scr), E2F7, E2F7 and E2F8, HIF1α or E2F7 and HIF1α siRNAs (Figure 2A). Cells were harvested 48 h after transfection, and were grown the last 16 h in hypoxia. RNA isolated from scr transfected, normoxic HeLa cells was used common reference RNA. Within each group of two biological replicates, sample versus common reference hybridizations were performed in balanced dye-swap. Microarrays used were human whole genome gene expression microarrays V1 (Agilent, Belgium) representing 34127 H. sapiens 60-mer probes in a 4 × 44K layout. cDNA synthesis, cRNA amplification, labeling, quantification, quality control and fragmentation were performed with an automated system (Caliper Life Sciences NV/SA, Belgium), starting with 3 µg total RNA from each sample, all as previously described (15). Microarray hybridization and washing was with a HS4800PRO system with QuadChambers (Tecan, Benelux) using 1000 ng, 1–2% Cy5/Cy3 labeled cRNA per channel as described (15). Slides were scanned on an Agilent G2565BA scanner at 100% laser power, 30% PMT. After automated data extraction using Imagene 8.0 (BioDiscovery), Loess normalization was performed (16) on mean spot-intensities. Gene-specific dye bias was corrected by a within-set estimate (17). Data were further analyzed by MAANOVA, modeling sample, array and dye effects in a fixed effect analysis. P-values were determined by a permutation F2-test, in which residuals were shuffled 10 000 times globally. Gene probes with P < 0.05 after family wise error correction (FWER) were considered significantly changed. In cases of multiple probes per gene, the values from the most 3′ probe were used. Selection of targets was based on both a significant P-value (<0.05) and fold change cut-off of ≥2 or ≥1.5 (a cut-off m value of ±1.0 or 0.585 (2log fold change), respectively), as indicated in the text. Gene ontology analysis was performed using DAVID and PANTHER gene ontology tools (18,19). All microarray gene expression data have been deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66750).
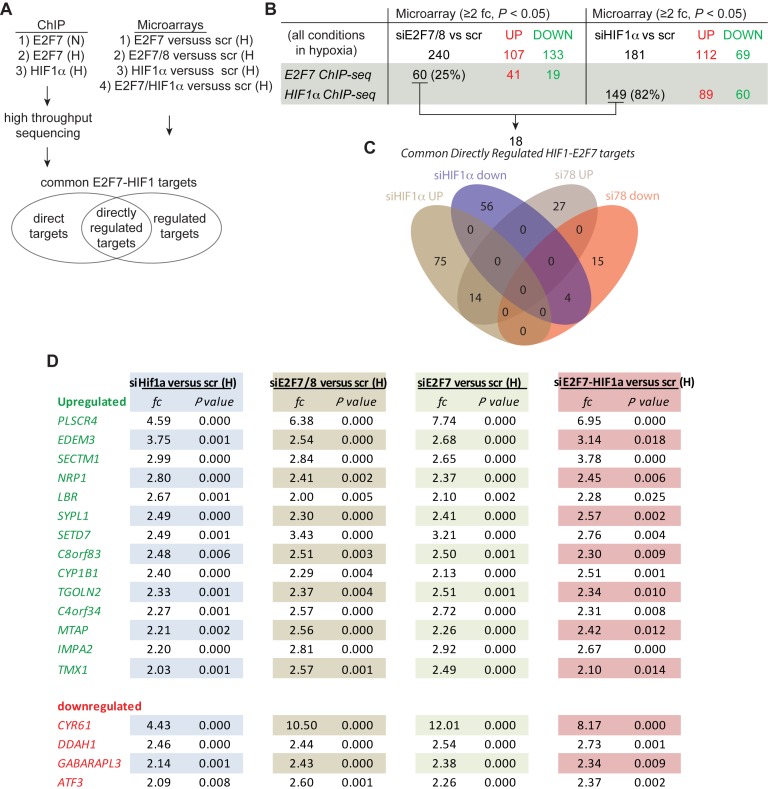
Figure 2.
Genome-wide analysis of HIF1α and E2F7 common targets by ChIP-seq and microarray analysis. (A) Flow chart showing the applied approach. The E2F7 ChIP-seq was performed both in normoxia (N) and hypoxia (H), all other experiments only in hypoxia. (B) Table that summarizes the overlap between the E2F7 and HIF1α targets identified in the microarray data (cut off: ≥ 2 fc; P < 0.05) and ChIP-seq. The E2F7 ChIP-seq experiment performed on hypoxic HeLa cells lysates was used for this analysis. Numbers between parentheses present the% overlap of the microarray and ChIP-seq data. Numbers of genes identified are separated in up- and down-regulated genes. (C) Venn diagram showing the overlap between HIF1α (149) and E2F7/8 (60) directly regulated targets, separated in up- and down-regulated genes. (D) Table showing 18 novel common direct and regulated (≥2 fc; P < 0.05) targets of HIF1α and E2F7/8. Numbers present fold up- or down-regulation of mRNA levels in the indicated conditions compared to controls, and associated P-values. HIF1α-E2F7 repressed genes are shown in green, activated genes in red.
siRNA transfection
HeLa cells were grown to confluence and re-seeded at 150 k/well (6 well plate, Greiner). Next day, cells were siRNA transfected as specified by the manufacturer using 5 μl/well RNAIMAX (Invitrogen, 13778–075) and a final siRNA concentration of 50 nM. Medium was replaced the next day and cells were grown overnight in normoxia or hypoxia and harvested 48 h after transfection. For harvesting, cells were washed twice with cold PBS on ice, scraped in cold PBS supplemented with protease inhibitors (Roche), and pelleted by centrifugation (2600×g, 2 min at 4°C). Protein samples were lyzed in 60 μl of lysisbuffer (0.05 M sodium phosphate pH7.3, 0.3M NaCl, 0.1% NP40, 10% Glycerol). Cell pellets for RNA isolation were frozen in liquid nitrogen and stored in −80°C. siRNAs used in this study: hHIF1α (L-004018–00–0005, Thermo Scientific), non-targeting siRNA #2 (D-001210–02, Thermo Scientific), hARNT siRNA (L-007207–00–0005), hE2F7 (HSS135118, HSS135119, HSS175354, Invitrogen), hE2F8 (HHS128758, HSS128759, HSS128760, Invitrogen), Negative control medium (Invitrogen, 12935–300). The negative control siRNAs from Thermo and Invitrogen were mixed and used at a 1:1 ratio. For double transfection, half of the amounts applied for the single siRNA transfections were used, ensuring a final concentration of 50 nM. For siRNA transfections in U2OS cells were seeded at a density of 100 k/well.
Statistical analysis
All statistical comparisons in this study were performed using a two tailed, independent t-test. Variances of two groups were compared with an F-test. Differences were considered significant with a P-value of <0.05.
Western blot analysis
Whole-cell lysates were prepared using a lysis buffer containing 150 mM NaCl, 1.0% NP40, 0.5% deoxycholate, 0.1% SDS and 50 mM Tris (pH 8.0), supplemented with Protease Inhibitor Cocktail (Roche). Protein lysates were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to a nitrocellulose membrane. Membranes were probed with the following antibodies: E2F7 (Santa Cruz, sc-66870), E2F8 (Abnova, H00079733-M01; Abcam AB109596), HIF1α (BD Biosciences, 610959), HDAC1 (sc-7872), E2F1 (sc-193), Mouse IgG HRP-linked whole Ab (GE Healthcare, NA931), Rabbit IgG HRP-linked whole Ab (GE Healthcare, NA934). As secondary antibodies, anti-rabbit-HRP (Amersham Biosciences, NA934; 1:5000) and anti-mouse-HRP (Amersham Biosciences, NA931; 1:5000) were used. All antibodies were diluted in 4% non-fat milk in TBST. Immuno-probed blots were subjected to standard ECL reagents as described by the manufacturer (GE Healthcare, RPN2106).
Chromatin immunoprecipitation (ChIP) and ChIP-sequencing
ChIP was performed according the EZ ChIP protocol (Upstate, 17–371) with the following specifications: HeLa cells were seeded at day 1 at a concentration of 7 × 106 cells per 145 mm plate. Next day, cells were cultured overnight in hypoxia, or continued to grow under normoxia, and harvested on day 3. For single and double ChIP, five 145 mm plates were used per condition. In vitro crosslinking was performed on a shaker at roomtemperature (RT) for 10 min, using 1% freshly made paraformaldehyde. Crosslinking was quenched for 5 min incubation at RT on a shaker. Next, cells were washed twice on ice with PBS (4°C, supplemented with protease inhibitors (11873580001, Roche)), and harvested, pelleted and resuspended in 2 ml lysis buffer (0.3% SDS, 10 mM EDTA, 50 mM Tris and protease inhibitors). Sonication (10 cycles of 10 s followed by 1 min incubation on ice) was performed in a FACS tubes using a Soniprep 150 sonicator (MSE). Ten microliters of sonicated sample was analyzed on gel to check for sonication efficiency. Sonicated lysates were centrifugated (10 min, 4°C) to remove insoluble components and large DNA fragments. Two hundred microliters lysate was used per ChIP sample. The DNA concentration in the sonicated lysate was measure using a Nanodrop to normalize the amount of input DNA between normoxic and hypoxic ChIP samples. Protein G agarose beads (16–266, Milipore) were coated overnight in 0.1% BSA (Sigma, A3294), and 60 μl was used for pre-clear (2 h, 4°C) and final precipitation of immune complexes (1 h, 4°C). For single ChIP, 5 μg and for double ChIP, 10 μg was used. Immunorecipitation were performed overnight at 4°C on a rotating platform. In case of double ChIP, eluates were diluted in dilution buffer and incubated with another 10 μg of antibody overnight. The following antibodies were used: ChIP grade HIF1α (ab2185, Abcam) E2F7 (sc-66870), E2F1 (sc-193), IgG (2729S; cell signaling). De-crosslinked DNA was purified over a column (Qiagen, 28106) and eluted in 65 μl H2O of which 2 μl was used for quantitative PCR. ChIP-sequencing was performed using the double ChIP protocol with approximately 100 million cells per sample. Lysates were sonicated for 7 min at maximum power on a Covaris S2 (Covaris). As ChIP-seq was used as a qualitative assay for target identification, the ChIP input material was not normalized. For validation of the targets in subsequent quantitative ChIP-qPCR experiments the input DNA was normalized as explained above. Library construction and sequencing were performed as described previously (20). All raw ChIP-seq data have been deposited in GEO with the accession number GSE66956 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66956).
Luciferase reporter assay
U2OS cells were seeded at 100 k/well (6-well plate, Greiner) and next day transfected (Superfect, Qiagen) following manufacturer's instructions. Per well (6 well plate) we used 2.5 μg reporter plasmid, 100 ng TK renilla, and 25–500 ng expression or control plasmid and 5 μl Superfect. For competition reporter assays, cells were transfected with 25 ng of E2F1 expression together with 475 ng empty vector, or 475 ng E2F7 or HIF1α expression plasmid. After 6 h, the medium was replaced and cells were cultured in hypoxia or normoxia. Next day reporter activity was measured using the Dual-Luciferase Reporter Assay System (Promega, E1910) on a microplate luminometer (Centro LB 960) 24 h after transfection. TK was used for normalization of the data.
Zebrafish
All zebrafish strains were maintained in the Hubrecht Institute (Utrecht Medical Center, The Netherlands) under standard husbandry conditions. Animal experiments were performed in accordance with the rules of the Animal Experimentation Committee of the Royal Netherlands Academy of Arts and Sciences (DEC). Transgenic line used: Tg(nrp1a:gfp)js12Tg, (21) expressing gfp form a ∼7.5 kb (−7.5 kb up to 1st ATG) nrp1a promoter fragment. Construction of TAL effector nucleases (TALENs): the TALEN-induced nrp1ahu10012 mutants, were generated and genotyped similarly and along with the previously published nrp1ahu9963 mutant, as previsouly described (22). TALEN sequences were identified in nrp1a using TAL effector Nucleotide Targeter 2.0 (https://tale-nt-cac-cornell.edu/) and chosen based on the location in the CUB domain and presence of an appropriate restriction enzyme site (PvuII) for genotyping purposes. Embryos were staged (23).
Morpholino (MO)
The following published (14,24) morpholino (MO) oligonucleotides (Genetools) were used: an E2f7 splice donor MO targeting exon 2–intron 2–3 (5′-ATAAAGTACGATTATCCAAATGCAC-3′); an E2f8 splice donor MO targeting exon 2-intron 2–3 (5′-CTCACAGGTATCCGAAAAAGTCATT-3′); a hif1ab ATGMO (5′-CAGGAATGGATACTGGAGTTGTCAC-3′); and a control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′). MO injections were performed using zebrafish embryos up to the 2-cell stage.
Imaging
Imaging was performed on live embryos mounted in 0.5–1% low melting point agarose (Invitrogen, 16520–050) dissolved in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4), supplemented with Tricaine mesylate (MS222)) on a cell view cell culture dish (627860, Greiner Bione). Imaging was performed with a Leica TCS SPE-II confocal microscope (Leica Microsystems) using a 10 or 20x objectives with 1.5 digital zoom, Laser 488 nM (7%), 600 Hz scan speed, frame average 4, format 1024 × 1024, 8-bit images. For imaging after 24 hpf embryos were raised in Phenylthiourea (PTU; Sigma). Imaging of gfp reporter expression in the eyes of Tg(nrp1a:gfp) fish was performed using identical settings (gain, offset) for high, medium and low classified embryos. Imaging and quantification of MN defects was performed on the ∼10–11 MN above the yolk sac extension.
In situ hybridization
In situ hybridization was performed as previously described (25). Briefly, the nrp1a ISH probe was PCR-amplified form zebrafish cDNA and cloned in the pGEM-T Easy vector (Promega). For probe generation, 5 μg of plasmid DNA was linearized overnight at RT using NcoI (Thermo Scientific). The linearized vector was excised from gel and purified using the QIAquick gel extraction kit as specified by the manufacturer. DIG-labeled (11277073910, Roche) RNA probes were generated by in vitro transcription using SP6 RNA polymerase (Promega) according to the manufacturer's instructions. Vector DNA was removed by DNAse treatment (Qiagen) at RT for 10 min after which RNA was column purified (74106, Qiagen) and eluted in 30 μl of RNAse free MilliQ, supplemented with 1 μl of RNAsin (N2511, Promega). The RNA concentration was measured on a NanoDrop 1000 (Thermo) and checked on agarose gel after denaturation at 80°C for 5 min.
RNA isolation, cDNA synthesis and quantitative PCR
Total RNA was extracted according to manufacturers’ instructions using the RNeasy Mini Kit (Qiagen, 74106). cDNA was synthesized with random hexamer primers according to manufacturers’ instructions (Fermentas, K1622). An additional on-column DNase treatment was performed using the RNAse-Free DNAse (79254; Qiagen). Quantitative PCR was performed on a MyiQ cycler (Biorad) using SYBRgreen chemistry (Biorad). Gene expression was calculated using the ΔΔCt method adapted for multiple-reference gene correction. In our in vitro studies two reference genes were used (ACTB, RPS18) and for zebrafish samples three reference genes were used (TBP, EF1α, β-Actin). MIQE standards were applied to our protocols (26). Oligonucleotides sequences use in this paper are listed in Supplementary Table S1.
RESULTS
Hypoxia induces E2F7 expression through HIF1
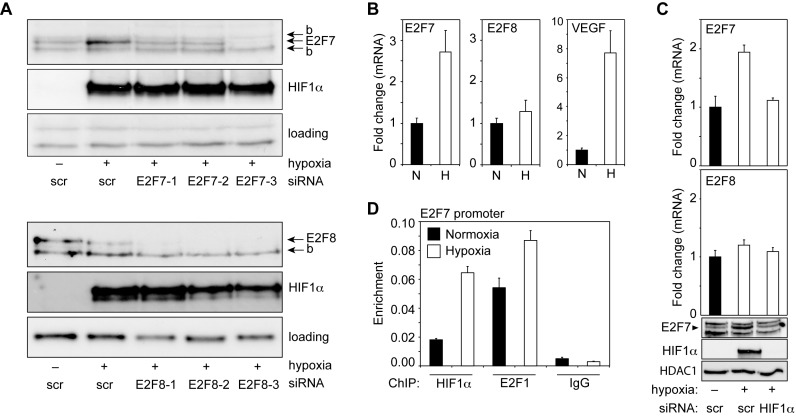
To explore a potential transcriptional cooperation between HIF and E2F factors we focused on HIF1 and E2F7/8, as E2F7/8 were suggested to regulate a set of ‘hypoxia genes’, as observed in E2f7/8 double knockout mice (7), and because of the recently reported interaction between E2F7/8 and HIF1 (14). Because HIF1α expression is regulated by hypoxia, we first analyzed if E2F7/8 expression was also regulated by hypoxia. E2F7/8 protein levels were examined by Western blot analysis using lysates from HeLa cells cultured under normoxia or hypoxia (16 h, 1% O2). This analysis showed that E2F7, but not E2F8 protein levels are induced in hypoxia (Figure 1A). The specificity of the E2F7 and E2F8 immuno-staining was confirmed using HeLa cell lysates transfected with three different siRNAs for E2F7 or E2F8 (Figure 1A). Throughout this study E2F7 siRNA #3 and E2F8 siRNA #1–3 were used because of their efficient depletion of E2F7 and E2F8, respectively. To investigate if hypoxia regulates E2F7 expression on the mRNA level, E2F7 mRNA levels were measured by quantitative PCR (qPCR), using total RNA isolated from normoxic and hypoxic HeLa cells. We found that E2F7, but not E2F8 mRNA levels are enhanced by hypoxia (Figure 1B). Treatment of HeLa cells with the hypoxia-mimetic agent desferrioxamine (DFO) also induced E2F7 protein and mRNA levels (Supplementary Figure S1A, B). The induction of E2F7 expression by hypoxia and DFO was also shown in U2OS cells (Supplementary Figure S1C–E), suggesting a general mechanism. Because HIF proteins are key regulators of the transcriptional response to hypoxia, we hypothesized a role for HIF in stimulating E2F7 transcription in hypoxia. Indeed, analysis of the E2F7 promoter (Supplementary Figure S1F) revealed the presence of multiple putative HIF-binding sites (HIF-BS), to which HIF1 binding was confirmed using ChIP assays (Figure 1C, Supplementary Figure S1G). Binding of E2F1 to the E2F7 promoter served as a positive control (14). To test if HIF1 also stimulates E2F7 transcription in hypoxia, E2F7 mRNA levels were determined in HIF1α depleted or control cells. Knockdown (KD) of HIF1α by siRNA abolished the hypoxic induction of E2F7 mRNA and protein in HeLa (Figure 1D) and U2OS cells (Supplementary Figure S1H). From these data we conclude that hypoxia stimulates E2F7 expression through direct promoter activation by HIF1.
Figure 1.
E2F7 expression is induced in response to hypoxia by HIF1. (A) Westernblot analysis of E2F7, E2F8 and HIF1α expression in lysates from HeLa cells transfected with control (scrambled, scr) siRNA, or one of three different E2F7 or E2F8-specific siRNAs (numbered 1, 2 and 3), as indicated. Cells were maintained in normoxia (−) or hypoxia (+) as indicated. Non-specific and background (‘b’) bands serve as loading controls. (B) Graphs showing E2F7, E2F8 or VEGFA mRNA levels (depicted as fold change compared to normoxic mRNA levels) isolated from HeLa cells grown in normoxia or hypoxia, and determined by qPCR. In this and all subsequent figures black bars present normoxic (N), and white bars hypoxic (H) conditions. (C) ChIP assay, using normoxic or hypoxic HeLa cells, showing enrichment of HIF1α, E2F1 (positive control) or IgG (negative control) on the E2F7 promoter (element 2). (D) Graphs showing E2F7 or E2F8 mRNA levels as determined by qPCR. RNA was isolated from HeLa cells grown in normoxia or hypoxia, transfected with control (scr) or HIF1α siRNA as indicated. Lower panels are Western blots showing E2F7, HIF1α and HDAC1 (loading) protein levels. All quantified data present the average ± S.D. compared to the indicated controls in at least three independent experiments.
Identification of HIF1α-E2F7 target genes by genome-wide analysis
To investigate if the hypoxia-induced factors HIF1 and E2F7 co-regulate a transcriptional network, ChIP-seq and microarray experiments were conducted. As outlined in Figure 2A, ChIP-seq was performed for E2F7 using normoxic or hypoxic HeLa cells, while HIF1α ChIP-seq experiments were only performed using HeLa cells cultured in hypoxia. ChIP-seq for E2F7 revealed 3761 and 2682 target genes in normoxia and hypoxia, respectively, of which the majority (2381) are bound by E2F7 under both conditions (Supplementary Figure S2A, Supplementary data set S1). The observed slight reduction of the number of E2F7 targets in hypoxia likely results from lower amounts of input DNA as hypoxia reduces the proliferation of the cells, demonstrated by an average cell number reduction of 24.3% in six independent experiments. The HIF1α ChIP-seq revealed 11666 HIF1α targets of which 2258 overlap with the identified E2F7 targets (Supplementary Figure S2A). Approximately 60–65% of the E2F7, and 30% of the HIF1α peaks are located within 1000 base pairs (bp) from an annotated transcriptional start sites, and both E2F and HIF DNA binding motifs were significantly enriched in E2F7 and HIF1α peaks, respectively (Supplementary Figure S2B, Supplementary data set S2). Notably, gene-ontology analysis (Supplementary data set S3) revealed many significantly HIF1α and E2F7 co-regulated biological processes (among which cell cycle, DNA replication and nucleoside, nucleotide and nucleic acid metabolism). Examples of overlapping peaks of common HIF1α and E2F7 bound targets are shown in Supplementary Figure S3A. ChIP-seq analysis also confirmed E2F7 as a HIF1α target (not shown).
Microarray analysis was next performed (in duplicate, using biological replicates) to analyze which of the bound targets are also regulated by HIF1α and E2F7. As we previously noted that depletion of E2F7 in hypoxia derepresses E2F8 expression (14), through which E2F8 may compensate for the loss of E2F7 (7), we not only analyzed gene expression in E2F7 depleted cells, but also in E2F7/8 depleted cells. To analyze a possible synergistic cooperation between E2F7 and HIF1α we also simultaneously knocked-down these proteins. In addition to these conditions, RNA was also isolated from hypoxic HeLa cells transfected with a control or HIF1α siRNA (Figure 2A). For the identification of E2F7 and HIF1α common targets described below we used the microarray data set from the E2F7/8 depleted cells, although the E2F7 microarray data set in general yielded the same results (as will be discussed below).
Using a stringent threshold of ≥2-fold change and a P-value of <0.05, microarray analysis identified 240 E2F7/8, and 181 HIF1α regulated genes (Figure 2B). Sixty out of these 240 E2F7/8 regulated genes were also identified by E2F7 ChIP-seq, whereas 149 out of 181 HIF1α regulated genes were also identified by HIF1α ChIP-seq (Figure 2B). The overlap between the 60 E2F7, and 149 HIF1α direct and regulated genes revealed 18 common HIF1α-E2F7 targets of which fourteen are repressed and four are stimulated (Figure 2C,D). Similar analysis, now using a microarray threshold of ≥1.5-fold change (and a P-value of <0.05), revealed 56 direct and regulated HIF1α-E2F7 targets, of which 37 are repressed and 17 are activated (Supplementary Figure S2D–F). The fact that almost all of these common targets are not differentially regulated by E2F7/8 and HIF1α, irrespective of their opposite roles in gene regulation, strongly suggests that they are regulated by a single HIF1α-E2F7 complex and are not independent acts of two transcription factors on the same set of promoters. This is also underscored by an almost complete lack of differentially regulated HIF1α and E2F7/8 targets in the microarray data (Supplementary Figure S2C–D). The microarray analysis also demonstrates a predominant role for E2F7 in gene regulation in hypoxia, as the 18 novel HIF1α-E2F7 targets were comparably regulated between E2F7 and E2F7/8 depleted cells (Figure 2D), and because siRNA-knockdown of E2F7 or E2F7/8 not only resulted in a comparable number of deregulated genes (980 versus 951, respectively (cut-off ≥ 1.5; P < 0.05), Supplementary Figure S2E), but also in a significant overlap (76% of the genes identified in the siE2F7/8 microarrays are also identified in the siE2F7 microarrays, S4 Data set). In addition we show that inactivation of one component of the HIF1α-E2F7 complex is sufficient to disable its function, and that no synergistic gene regulatory effects are observed in E2F7 and HIF1α co-depleted cells (Figure 2D). Together these data demonstrate the existence of a HIF1α and E2F7 regulated transcriptional network in which the HIF1α-E2F7 complex can function as a repressor or activator.
The transcriptional repressive activity of the HIF1α-E2F7 complex is enhanced in hypoxia
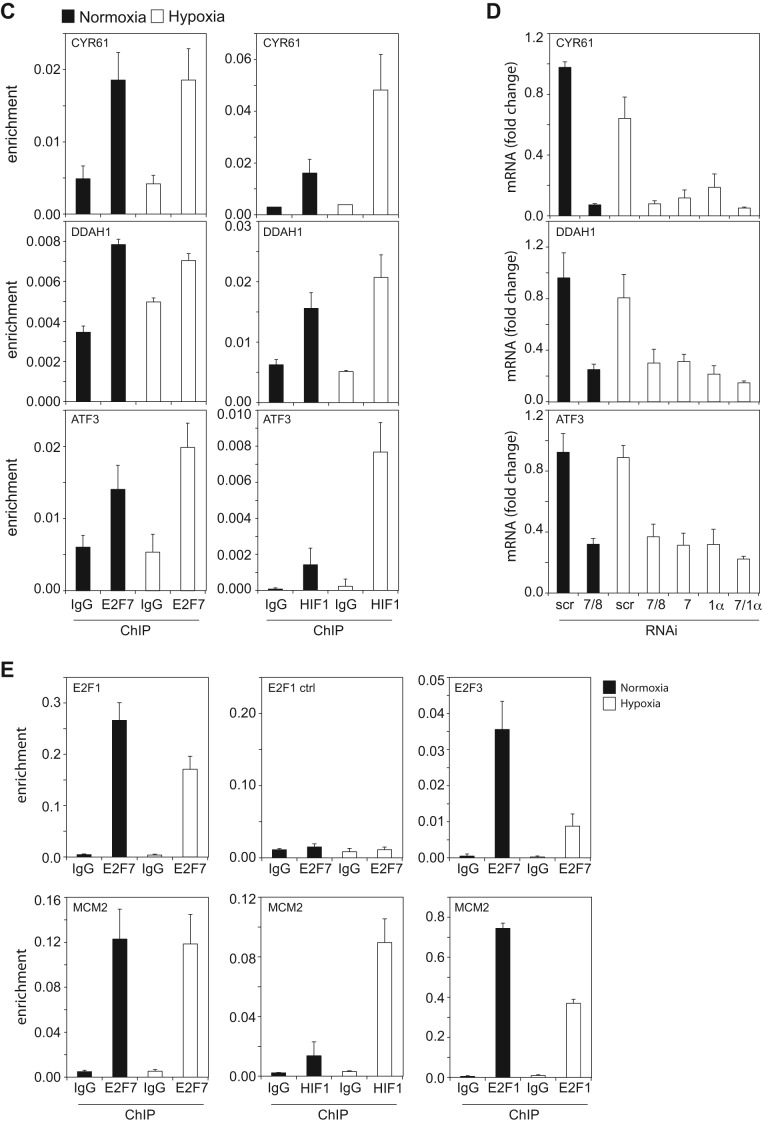
To investigate the binding of the HIF1α-E2F7 complex to the 18 common targets (Figure 2D) in normoxia and hypoxia, we performed ChIP-qPCR assays. To correct for the reduced proliferation in hypoxia, ChIP lysates were normalized for the amount of input DNA. Multiple independent E2F7 ChIP-qPCR assays confirmed E2F7 binding to the common targets and demonstrated that E2F7 binding is comparable between normoxia and hypoxia, although binding to NRP1 (Figure 4B), MTAP, LBR (Figure 3A) and ATF3 (Figure 3C) was slightly enhanced in hypoxia. E2F7 binding to a previous reported E2F site in the E2F1 promoter served as a positive control (14), whereas no E2F7 binding was detected to a non-specific region 700 bp upstream of this site, showing a ChIP-resolution of <700 bp (Figure 3E). HIF1α binding was also confirmed to all common target promoters, showing enhanced enrichment in hypoxia (Figures 3A, C and 4B).
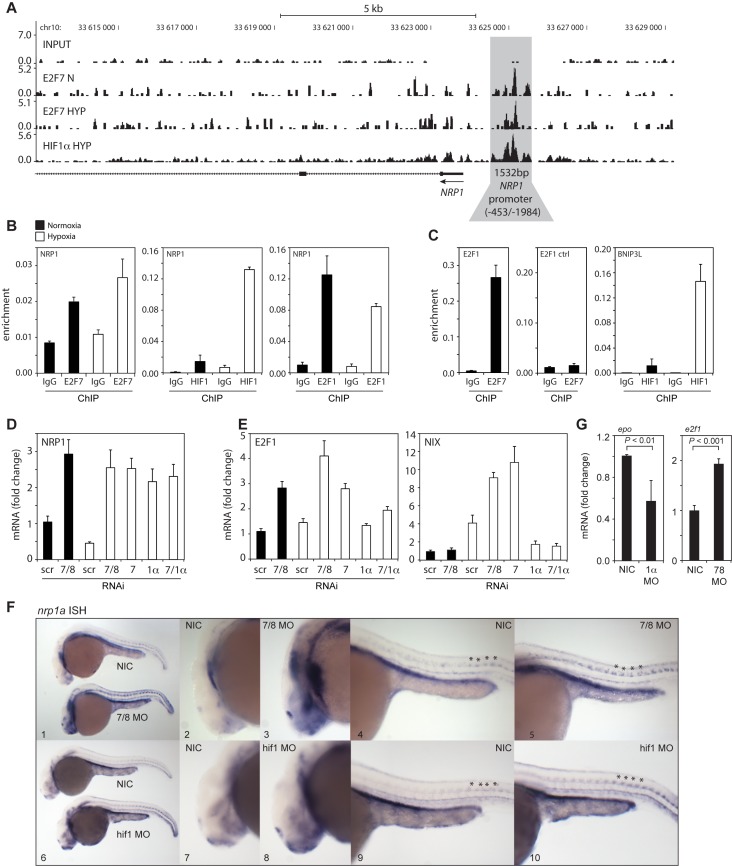
Figure 4.
In vitro and in vivo validation of NRP1 regulation by HIF1α and E2F7. (A) ChIP-seq signal (y-axis: peak height) shown for E2F7 (N and HYP) and HIF1α (HYP) on the NRP1 promoter (indicated in grey). Input DNA was sequenced as a control. Lines underneath the graphs indicate annotated genes, boxes present exons and lines with arrows indicate introns. Arrow indicates direction of transcription. (B) Validation of HIF1α, E2F7 and E2F1 enrichment on the NRP1 promoter as analyzed by ChIP-qPCR in normoxic or hypoxic HeLa cells. Isotype matched IgG served as a negative control. (C) Positive controls for ChIP: binding of E2F7 to the E2F1 promoter, and HIF1α binding to the BNIP3L promoter. A non-specific region 700 bp upstream of the E2F binding site in the E2F1 promoter served as a negative control. (D) Graph shows NRP1 mRNA levels as determined by qPCR in lysates isolated from HeLa cells transfected with control (scr), E2F7 and E2F8 (7/8), E2F7 (7), HIF1α (1α) or E2F7 and HIF1α (7/1α). (E) Same as (D) but now for E2F1 and NIX mRNA levels, positive controls for E2F7 and HIF1α, respectively. (F) In situ hybridizations (ISH) for nrp1a using non-injected control (NIC), e2f7/8 MO (5 + 5 ng, n = 46), or hif1ab MO (5 ng, n = 70) injected zebrafish embryos, obtained from three independent experiments. e2f7/8 MO and NIC littermates were analyzed at 26 hpf, hif1ab MO injected and NIC littermate embryos at 28 hpf. 100% of the e2f7/8 MO injected embryos, and 69% of the hif1ab MO injected embryos showed enhanced nrp1a expression in MN. All panels show lateral views. Panels 2, 3, 7 and 8 show magnifications of the head region. Panels 4, 5, 9 and 10 show magnifications of the trunk region. Asterisks show examples of spinal motorneurons. (G) Graphs show fold change of epo or e2f1 mRNA expression in hif1ab MO (1α) or e2f7/8 (7/8) injected zebrafish embyros, respectively, compared to NIC. mRNA levels were determined in >30 embryos from three independent experiments. All quantified data present the average ± S.D. compared to the indicated controls in at least three independent experiments.
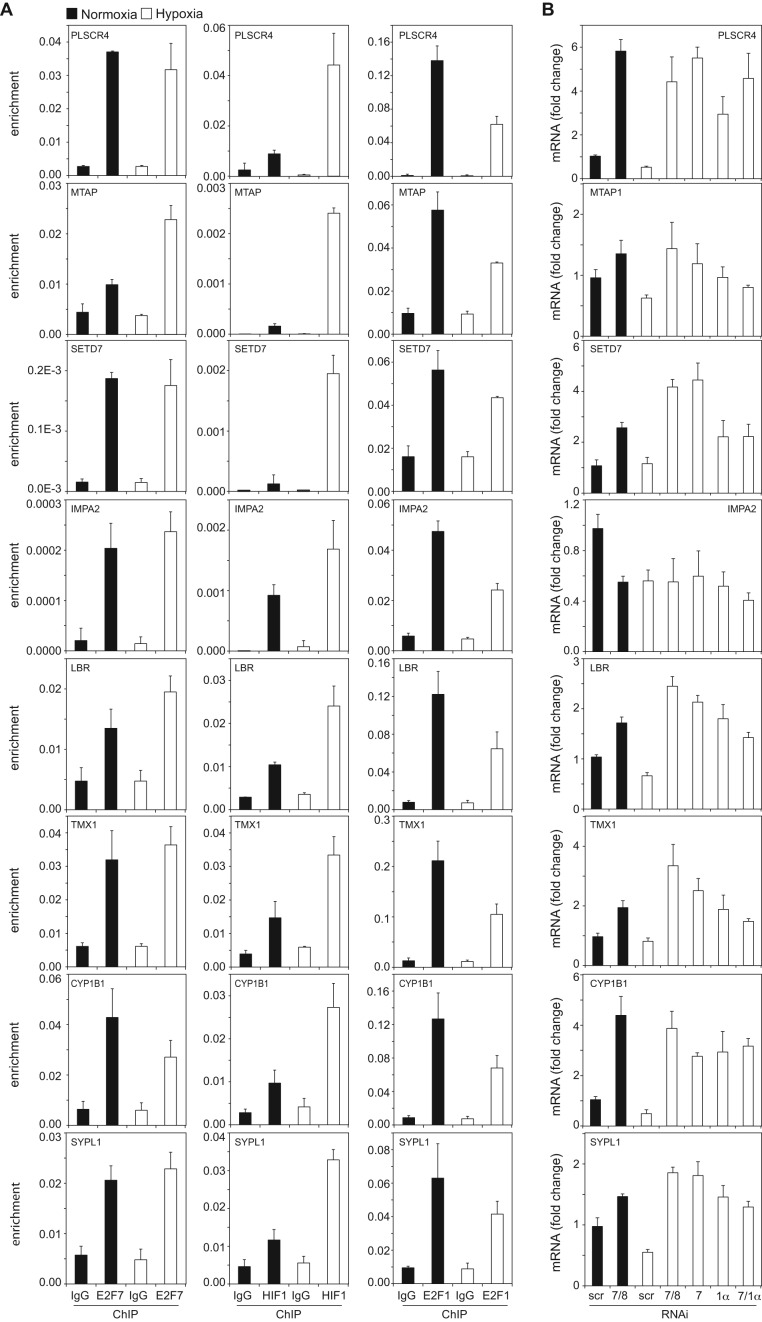
Figure 3.
Binding and regulation of the common targets by the HIF1α and E2F7. (A) Graphs show ChIP-qPCR analysis of E2F7 (left panels), HIF1α (middle panels) and E2F1 (right panels) enrichment on the common repressed target promoters. Non-specific, isotype matched IgG serve as a negative control. (B) Graphs showing mRNA levels as determined by qPCR and presented as fold change comparing to scr normoxia, of the common repressed targets. Messenger RNA levels were analyzed in HeLa cells transfected with control (scr), E2F7 and E2F8 (7/8), E2F7 (7), HIF1α (1α) or E2F7 and HIF1α (7/1α) siRNAs and grown in normoxia or hypoxia, as indicated. (C) Similar as in (A) but now for the HIF1α-E2F7 induced targets. (D) Similar as in (B) but now for the HIF1α-E2F7 induced targets. (E). Upper graphs present E2F7 binding to the E2F1 promoter, to a control region (in the E2F1 promoter) and the E2F3 promoter. Lower graphs show E2F7, HIF1α and E2F1 enrichment to the MCM2 promoter. All quantified data present the average ± S.D. compared to the indicated controls in at least three independent experiments.
As E2F7 inhibits transcription upon binding to E2F-binding sites (BS) (20) and HIF activates transcription when acting through HIF-BS (27,28) we hypothesized that the HIF1α-E2F7 complex regulates the repressed targets via E2F-BS, through which E2F7 may engage HIF1α in transcriptional repression. This hypothesis predicts that other E2F family members may also bind the promoters of HIF1α-E2F7 repressed targets. We tested this hypothesis for E2F1, by performing ChIP-qPCR. These experiments demonstrate that E2F1 binds the NRP1 (Figure 4B) and all other common repressed target promoters both in normoxia and hypoxia, although binding was significantly diminished in hypoxia (Figure 3A). As hypoxia increased HIF1α, but decreased E2F1 binding to the common targets, these data suggest that HIF1 might compete with E2F1 for promoter binding in hypoxia.
To explore the expression of the common targets in normoxia and hypoxia, we performed siRNA-qPCR experiments. Compared to control transfected cells, siRNA ablation of either E2F7/8, E2F7, HIF1α or HIF1α and E2F7 verified the repressive (Figures 3B and 4D, Supplementary Figure S4A) or stimulatory (Figure 3D) regulation of the targets by HIF1α-E2F7. The predominant role of E2F7 over E2F8 in regulating gene expression in hypoxia was also confirmed, as knockdown of E2F7 alone resulted in a comparable response compared to E2F7/8 depleted cells (Figures 3B, D and 4D, E). For IMPA2 and GABARAPL3 no qPCR protocols could be designed meeting MIQE standards (26). The E2F7 repressed target E2F1 (7,29), and the HIF1 stimulated target NIX served as positive controls, confirming functional E2F7 and HIF1α KD, respectively (Figure 4E). By comparing the normoxic and hypoxic expression of the common targets we noted that 9 out of 13 of the HIF1α-E2F7 repressed targets are downregulated in hypoxia (Figure 3B), consistent with the hypoxic induction of E2F7 and HIF1α. In addition, 12 out of 13 repressed HIF1α-E2F7 targets were more significantly repressed by the complex in hypoxia compared to normoxia (Figures 3B and 4D, Supplementary Figure S4A). For example, compared to their condition specific controls, KD of E2F7/8 in hypoxia resulted in a higher NRP1 derepression (∼5-fold) than E2F7/8 KD in normoxia (∼3-fold, Figure 4D). Together these data confirm the binding and regulation of the common targets by HIF1α and E2F7, and show that the increased expression of E2F7 (and HIF1α) in hypoxia results in enhanced repression of the common repressed targets under these conditions.
Because HIF1α is primarily known as part of the HIF1 transcription complex, in which HIF1α cooperates with HIF1β/ARNT (3,4), and because we recently demonstrated that E2F7 binds to the N-terminal 80 amino acids of HIF1α, a region to which ARNT also binds (14), we hypothesized that ARNT could be part of the HIF1α-E2F7 complex. To test this we knocked down ARNT in normoxic and hypoxic HeLa cells using siRNA, after which HIF1α-E2F7 common target gene expression was analyzed by qPCR. Efficient siRNA knockdown of ARNT (Supplementary Figure S4B) resulted in the expected reduced hypoxic induction of the known HIF1 targets PGK1 and NIX (Supplementary Figure S4C). Interestingly, ARNT differentially affected expression of the common target genes. Knockdown of ARNT (and HIF1α) derepressed NRP1, and reduced CYR61 mRNA expression (Supplementary Figure S4C), similar to the HIF1α and E2F7 regulation of these genes (Figures 3D and 4D), while the HIF1α-E2F7 repressed targets CYP1B1 and PLSCR4 (Figure 3B) were not affected (Supplementary Figure S4C). These data suggest a differential requirement for ARNT in HIF1α-E2F7 target gene regulation.
The HIF1α-E2F7 complex represses NRP1 during zebrafish development
To examine if the HIF1α-E2F7 complex also regulates its targets in vivo, we selected Neuropilin 1 (NRP1) for further analysis, as the emerging role of E2Fs in neuronal development (see introduction) could involve transcriptional control of NRP1. NRP1 is a non-tyrosine kinase transmembrane receptor that was originally identified in the Xenopus nervous system (30). Gene knock-out studies in mice showed an essential role for this receptor in neuronal (and vascular) development (31–33). In neuronal development, NRP1 functions in a holoreceptor complex with plexins, serving as the main receptor for class III semaphorin 3A (SEMA3A) (33,34). Binding of SEMA3A to the NRP1 receptor complex results in growth cone collapse and repulsive axon guidance signals (35,36), as displayed by target overshooting of cranial and spinal nerves in Nrp1 or Sema3a knockout mice (31,37), which can ultimately also lead to neuronal cell death (38,39).
Because nrp1a mRNA expression can efficiently be detected by in situ hybridization (ISH) during zebrafish development (40), and established zebrafish MO directed against e2f7/8 (14) or hif1α mRNA (24) are available, we used this model system to verify the regulation of NRP1 by HIF1α-E2F7/8 in vivo. As it was recently shown that MO-induced phenotypes are not necessarily recapitulated in corresponding genetic mutants (22), it is important to note that the specificity of the e2f7/8 MO and hif1α MO was extensively tested. The e2f7/8 splice site MO were demonstrated to interfere with e2f7/8 mRNA splicing, resulting in the deregulation of classic e2f7/8 target genes in vivo (14). In addition, angiogenic defects induced by these e2f7/8 MO were rescued upon co-injection of wild-type mRNA, and were phenocopied in corresponding e2f7/8 mutant zebrafish (14). Comparably, the hif1α ATG-MO effectively blocked hif1α translation, and inhibited hif1 target genes expression in vivo, while its effects on neural crest development could be rescued upon co-injection of wild-type hif1a mRNA (24). Whether hif1α morphants phenocopy hif1α mutants is unknown as these mutants are currently unavailable. Together, these experiments validate the use of these MO to characterize the regulation of NRP1 by E2F7/8 and HIF1α during zebrafish development.
To investigate this, zebrafish embryos were injected with e2f7/8 MO or hif1α MO, after which nrp1a mRNA levels were analyzed at day 1 post fertilization (dpf) by ISH. MO inhibition of endogenous e2f7/8 resulted in increased nrp1a mRNA levels compared to non-injected control embryos (NIC), displayed by enhanced expression in the head region (Figure 4F, panel 1–3) and in the trunk motorneurons (MN) (Figure 4F, panel 1,4,5). Similarly, MO inhibition of endogenous hif1 also enhanced nrp1a expression in the head and trunk MN (Figure 4F). Consistent, with the previous studies (14,24), MO depletion of e2f7/8 derepressed expression of its classical targets e2f1 expression, while inhibition of hif1α reduced expression of its classical target epo, indicating the functional MO inhibition of e2f7/8 and hif1 (Figure 4G). These data show that e2f7/8 and hif1α repress nrp1a during zebrafish development.
We also tested if hypoxia repressed nrp1a and nrp1b expression in zebrafish. Similar to hypoxic NRP1 mRNA repression in HeLa and U2OS cells, hypoxia significantly repressed nrp1a and nrp1b mRNA expression in zebrafish embryos (Supplementary Figure S5A). The hif1 target gene phd3 (41) served as a hypoxia marker. Interestingly, MO-inhibition of hif1a significantly derepressed hypoxic nrp1a and nrp1b expression (Supplementary Figure S5B), while MO-inhibition of e2f7/8 also derepressed hypoxic nrp1a and nrp1b expression, although to a non-significant level. These data suggest an evolutionary conserved hypoxic regulation of NRP1 by the HIF1α-E2F7 complex.
The HIF1α-E2F7 complex represses NRP1 expression through an E2F-hub
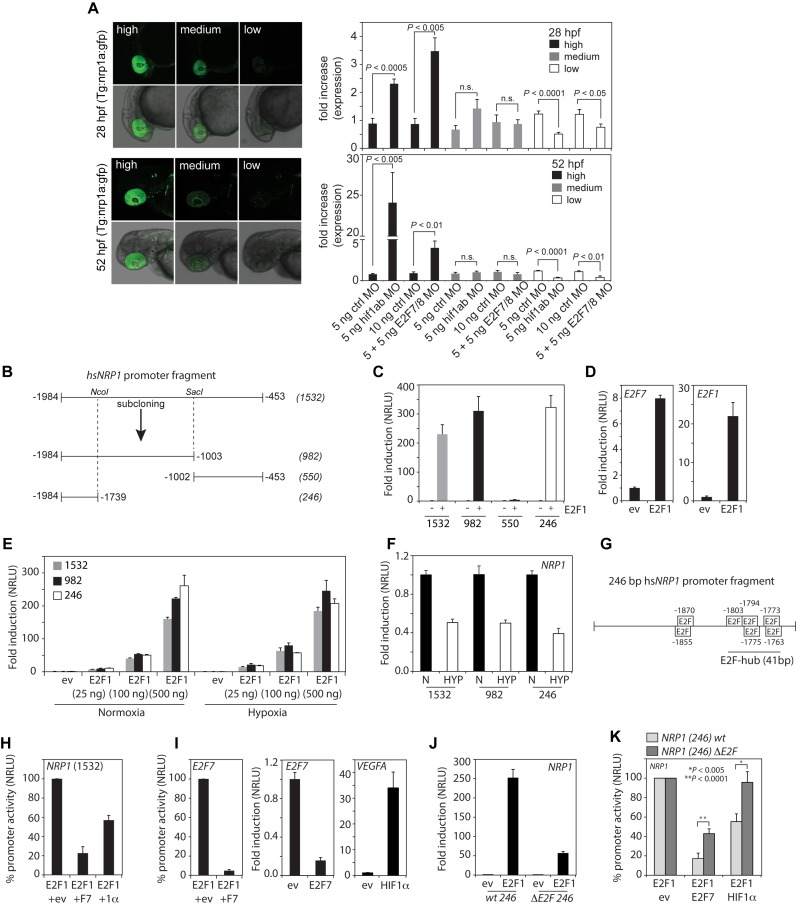
To investigate mechanistically how the HIF1α-E2F7 represses NRP1 expression, we first performed in vivo reporter assays using Tg(nrp1a:gfp)js12 reporter fish (21), in which gfp is expressed from a 7.5 kb nrp1a promoter fragment displaying gfp expression in multiple tissues, including the retina. A Tg(nrp1a:gfp)js12 line was used exhibiting low transgene copy numbers, resulting in embryos having variable retinal gfp expression, classified as high, medium or low (Figure 5A). Compared to injection of control MO, injection of e2f7/8 MO or hif1α MO significantly enhanced retinal gfp expression compared to control MO injected fish both at 1 and 2 dpf (Figure 5A). Consistently, the number of fish with low retinal gfp expression were decreased upon MO-depletion of e2f7/8 or hif1α (Figure 5A). These data demonstrate that e2f7/8 and hif1α repress the nrp1a promoter in vivo.
Figure 5.
The HIF1α-E2F7 complex regulates NRP1 through an E2F-hub. (A) Left panels show confocal images of the head region of Tg(nrp1a:gfp)js12 zebrafish at 28 and 52 hpf. Fish are divided in three groups (high, medium, low) based on the level of transgene expression in the eye (showing 2 examples per group). Graphs present quantification of the three groups at the indicated times after injection of control MO, e2f7/8 MO (5 + 5 ng) or hif1α MO (5 ng) in zebrafish embryos (n ≥ 154 embryos in ≥4 independent experiments for each specific condition). (B) Schematic figure of the cloned 1532 bp human NRP1 promoter, and promoter regions subcloned from it, using NcoI and SacI restriction sites. (C) Luciferase reporter assays showing the fold induction of normalized relative luciferase units (NRLU) of different NRP1 promoter constructs by E2F1 compared to controls. (D) Similar as in (C) but now for the E2F1 control promoters E2F7 and E2F1. (E) Reporter assay showing the dose-dependent induction of the NRP1 promoter activity by different amounts of E2F1. (F) Reporter assay comparing the activity of the indicated NRP1 promoter constructs in hypoxia compared normoxia. (G) Representation of the 246 bp NRP1 promoter and putative E2F binding sites as identified by MatInspector. HIF binding sites were not identified in the 246 bp fragment. (H) Reporter assay showing activity of the 1532 bp NRP1 promoter in the presence of E2F1 alone (set at 100%) or together with E2F7 or HIF1α (1α). (I) Control reporter assays showing regulation of the E2F7 promoter (left panel) promoter in the presence of E2F1 alone (set at 100%), or together with E2F7. Middle and right panels show positive controls to check for functional E2F7 or HIF1 activity using E2F1 or VEGFA promoter constructs respectively. (J) Reporter assay showing induction of the wild-type and the ΔE2F-hub NRP1 promoter by E2F1 compared to controls. (K) Similar as in (H) but now for the wildtype or 41 bp E2F-hub deleted (ΔE2F) 246 bp NRP1 promoter. All quantified data present the average ± S.D. (except for (A) in which the average ± S.E.M. as shown) compared to the indicated controls in at least three independent experiments.
To examine the promoter regulation in more detail, a 1532-human NRP1 (-1984/-453) promoter fragment was cloned encompassing the E2F7 and HIF1 binding peaks (Figure 4A, grey box). Based on available restriction sites, three other fragments were subcloned from this fragment (Figure 5B). These promoter fragments were used to further explore the regulation of the NRP1 promoter using in vitro reporter assays. As we found that E2F1 binds the NRP1 promoter, and we hypothesized that the E2F7-HIF1α complex may particularly inhibit E2F1-induced NRP1 promoter activation (judging from the differential E2F1 and HIF1α binding to the common repressed targets, Figures 3A and 4B), we first analyzed the ability of E2F1 to induce the NRP1 promoter fragments. Overexpression of E2F1 equally induced the 1532, 982 and 246 NRP1 promoter fragments, but not the 550-bp fragment (Figure 5C), demonstrating the E2F-responsiveness of the NRP1 promoter is contained in the 246-fragment. Notably, the published E2F1-responsive promoters E2F1 and E2F7 were induced 20-fold and 8-fold, respectively (Figure 5D), while the NRP1 246 fragment was induced 300-fold, showing the highly E2F-responsiveness of this promoter. Ectopic E2F1 induced the NRP1 promoter in a dose dependent manner both in normoxia and hypoxia (Figure 5E). Consistent with the down-regulation of NRP1 mRNA levels in hypoxia (Figure 3D), hypoxia also down-regulated the 246 bp NRP1 promoter (Figure 5F). Notably, E2F7 and HIF1α repressed E2F1-induced NRP1 promoter activation (set at 100%) by 80% or 55%, respectively (Figure 5H). As positive controls, E2F7 repressed E2F1-induced E2F7 promoter activity, whereas expression of E2F7 or HIF1 alone, repressed the E2F7, or activated the VEGFA promoter, respectively (Figure 5I).
To explore through which motifs NRP1 is regulated by HIF1α-E2F7, the 246-bp fragment was analyzed for the presence of HIF- and E2F-BS using MatInspector software. This revealed the presence of multiple putative E2F, but no putative HIF-BS (Figure 5G). Deletion of a 41 bp region containing 5 closely located, putative E2F-BS (hereafter referred to as ‘E2F-binding site hub’ (E2F-hub)) reduced the ability of E2F1 to stimulate the NRP1 promoter by 80% (Figure 5J). Notably, the capacity of E2F7 and HIF1α to repress the E2F1-induction of the NRP1 promoter was significantly impaired in the ΔE2F-hub mutant promoter (Figure 5K). In conclusion, these data show that the HIF1α-E2F7 complex inhibits NRP1 promoter activation through an E2F-hub.
The HIF1α-E2F7 complex regulates axon guidance of spinal motorneurons through NRP1
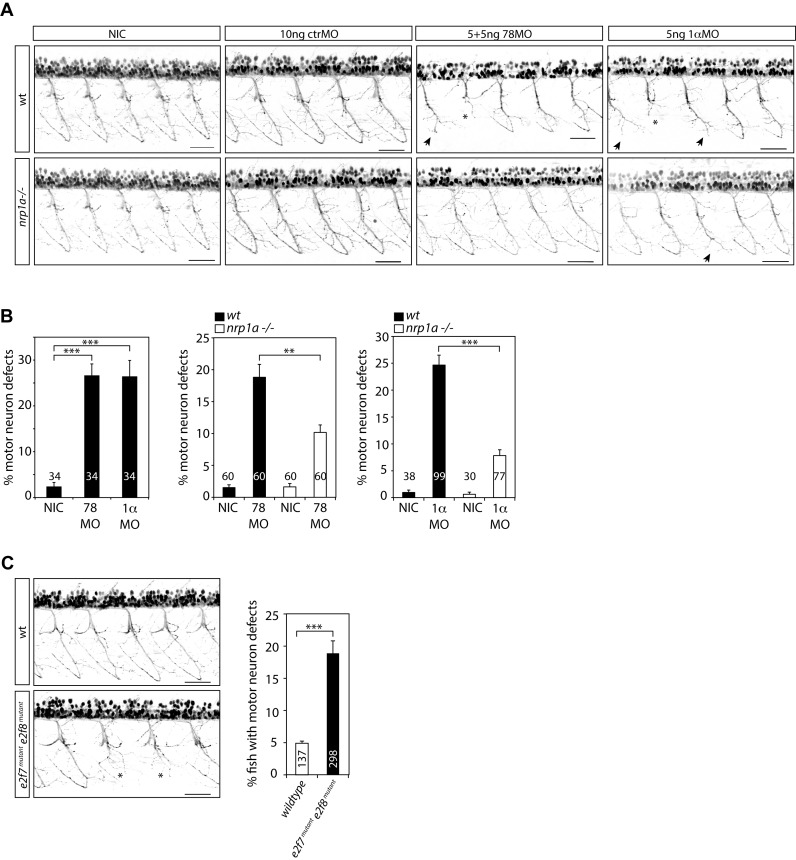
Because NRP1 functions in a holoreceptor complex with plexins, serving as the main receptor for SEMA3A in neuronal development (33,34), transducing repulsive axon guidance signals (35,36), we next explored if the HIFα-E2F7 complex may regulate MN axon guidance through regulation of NRP1. To this end, we injected Tg(nrp1a:gfp)js12 zebrafish with e2f7/8 MO, hif1α MO or control MO after which ventral MN projections were analyzed in the trunk at 2 dpf. Interestingly, while injection of a control MO (10 ng) did not affect the formation of caudal primary MN, injection of e2f7/8 MO (5 ng each) or hif1α MO (5 ng) significantly affected MN guidance, resulting in MN truncation in approximately 25% of all MN analyzed (Figure 6A, B).
Figure 6.
The HIF1α-E2F7 complex regulates MN development in an NRP1-dependent manner. (A) Confocal images of MN in the trunk regions above the yolk sac extension of Tg(nrp1a:gfp)js12 zebrafish at 48 hpf. Zebrafish embryos were non-injected (NIC: non-injected control), or injected with e2f7/8 (5 + 5 ng), hif1ab (5 ng), or control (10 ng) MO. Stunted MN are indicated with an asterisks, truncations resulting in the absence of the hinge are indicated with an arrow. Black bar presents 50 μM. (B) Quantification of MN defects in all MN analyzed, as described under (A). Left graph shows quantification of MN defects in e2f7/8 MO or hif1ab MO injected or non-injected wild-type Tg (nrp1a:gfp)js12 zebrafish. The two right graphs show quantification of MN defects in e2f7/8 MO or hif1ab MO injected, and non-injected wild-type (black bars) or nrp1ahu10012 mutant (white bars) Tg(nrp1a:gfp)js12 zebrafish. The numbers in the graphs present the number of analyzed zebrafish (obtained from at least three independent experiments). Per fish, all MN above the yolk sac extension (10–11 MN) were analyzed. (C) Analysis of MN defects in wild-type Tg(nrp1a:gfp)js12 zebrafish embryos, or in embryos obtained from crossing e2f7A207/A207; e2f8A196/A196; Tg(nrp1a:gfp)js12 zebrafish with e2f7A207/A207; e2f8WT/A196; Tg(nrp1a:gfp)js12 zebrafish. MN defects were analyzed in the trunk regions above the yolk sac extension at 48 hpf. Left panels show representative confocal images of analyzed MN for both groups. Graph presents quantification of the number of fish with MN defects (presented as%), analyzed in 137 wild-type, or 220 e2f7/8 mutant zebrafish embryos. All quantified data present the average ± S.E.M. compared to the indicated controls in at least three independent experiments. *P < 0.01, **P < 0.001, ***P < 0.0001.
To investigate if E2F7/8 and HIF1α regulate MN guidance through NRP1, we performed similar experiments in a TALEN-induced nrp1ahu10012 mutants, which were generated similarly and along with the previously published nrp1ahu9963 mutant, and result in an identically truncated protein (Supplementary Figure S6A) (22). Similar to the previously published nrp1ahu9963 mutant, nrp1ahu10012 mutant zebrafish are viable and do not display vascular defects (22). Analysis of ventral MN in nrp1ahu10012 mutant zebrafish did not reveal MN defects around 2 dpf (Figure 6A, left panels), showing that loss of nrp1a itself is not essential for MN axon guidance. These data are in contrast with the essential function of Nrp1 in neuronal development in mice (31,32), and suggest that nrp1b may compensate for the loss of nrp1a in zebrafish. Besides this, in our experiments, MO-depletion of hif1 or e2f/8 results instead in significantly enhanced nrp1a expression, suggesting that the chemo-repulsive action of nrp1a must not exceed a certain threshold in order for MN development to proceed normally. Apparently, the HIF1α-E2F7 complex is required to limit NRP1 expression in this process. In line with our observations, ectopic sema3 levels also causes truncated (or missing) ventral motorneurons during zebrafish development (21,42).
Interestingly, MN defects induced by injection of e2f7/8 MO or hif1α MO, were reduced by 50% in nrp1ahu10012 mutant zebrafish (Figure 6A and B), demonstrating that ablation of hif1α or e2f7/8 causes MN truncation, at least partially in a nrp1a dependent manner, and that the chemo-repulsive action of nrp1a must not exceed a certain threshold in order for MN development to proceed normally, which is ensured by the transcriptional repression by HIF1α-E2F7.
Although VEGFA regulates neuronal development (34), and is regulated by HIF1 (3) and E2F7/8 (14), were are confident that regulation of VEGFA by HIF1 or E2F7/8 does not affect the observed MN phenotype for several reasons. First, we used a e2F7/8 MO concentration (5 ng each) that did not significantly affect vegfAa expression and angiogenesis as previsouly shown (14). Second, others reported that MO depletion of vegfa in zebrafish abolished angiogenesis, but did not disturb ventral MN guidance (43), and that reduced neuronal Vegfa expression in mice also did not disturb MN guidance during embryogenesis, although it did cause late-onset progressive degeneration of lower motor neurons after 5 months of age (44). Therefore, we conclude that the regulation of MN guidance by HIF1α-E2F7 does not implicate VEGFA, but is at least partially dependent on NRP1, as the MN defects are partly rescued in nrp1ahu10012 mutant fish (Figure 6A and B). In this light it is interesting to mention that MO-inhibition of hif1α both derepressed nrp1a and nrp1b expression in hypoxic zebrafish embryos (Supplementary Figure S5), suggesting that the HIF1α-E2F7 complex may control MN development also through nrp1b.
Because e2f7/8 double mutant fish (e2f7A207/A207; e2f8A196/A196) phenocopy angiogenic defects induced by e2f7/8 MO (14), we next analyzed if these e2f7/8 mutant zebrafish also phenocopy the MN defects. Because deletion of either E2F7/8 or HIF1α alone deregulates NRP1 expression in vitro (Figures 2D and 4D) and in vivo (Figures 4F and 5A), it can be expected that mutation of e2f7/8 in zebrafish is sufficient to cause a MN defect. At 48 hpf, approximately 5% of 137 analyzed wildtype Tg(nrp1a:gfp)js12 zebrafish displayed a MN defect. Notably, similar MN analysis of 220 embryos obtained by crossing e2f7−/−; e2f8−/− with e2f7−/−; e2f8+/− Tg(nrp1a:gfp)js12 zebrafish, revealed that these embryos displayed significantly more MN defects (∼20%) compared to wild-type embryos (Figure 6C). Sequencing of the e2f7/8 mutant embryos revealed that the MN defect occurred both in e2f7/8 double knockout (DKO) embryos (e2f7−/−; e2f8−/−) and in e2f7−/−; e2f8+/− zebrafish (Supplementary Figure S6B), although the phenotype was significantly enhanced in DKO compared to e2f7−/−; e2f8+/− embryos (58.6% versus 41.4%, P < 0.05). This is consistent with the enhanced occurrence of angiogenic defects in e2f7/8 DKO, compared to e2f7−/−; e2f8+/− embryos (14). Sequencing of embryos derived from e2f7−/−; e2f8+/− intercrosses did not reveal a significant defect in e2f7−/−; e2f8+/+ embryos, showing that two wild-type alleles of e2f8 can compensate for the loss of e2f7 (data not shown). In conclusion, our data show that e2f7/8 mutant zebrafish phenocopy the e2f7/8 MO-induced MN phenotype, and demonstrate that the HIF1α-E2F7/8 complex regulates guiding of ventral MN during zebrafish development at least partially in an NRP1-dependent manner.
DISCUSSION
By performing ChIP-seq we have identified the existence of a HIF1α-E2F7 co-regulated transcriptional network consisting of 2258 target genes, although the majority of these targets are not or only marginally regulated in HeLa cells, or during normal proliferation. By combining our ChIP-seq data with microarray analysis we have identified 18 direct transcriptionally controlled HIF1α-E2F7 targets (Figure 2D), using a cut-off of ≥ 2 fold change regulation and a P-value of <0.05 in the microarrays (or 56 targets using a cut-off of ≥1.5-fold change and a P-value of <0.05; S2F Figure). Notably, these combined data, as well as the microarray data alone (Supplementary Figure S2C,D) demonstrate the almost complete lack of differential regulation of common targets (knowing that HIF1 is defined as an activator, and E2F7 as a repressor), excluding that HIF1 and E2F7 act independent on a common set of promoters, at the same time providing clear evidence for the existence of the HIF1α-E2F7 transcription complex. Interestingly, gene-ontology analysis identified many HIF1α-E2F7 regulated biological processes (Supplementary data set S3), suggesting versatile yet unexplored functions of the complex.
In our ChIP-seq experiments we identify more E2F7 targets (2381) compared to our previous study (737) (20). Although the methodology was similar, here we used non-synchronized, rather than S-phase synchronized cells, and used 4 times more ChIP input material, allowing the discovery of a wider array of targets. The large amount of HIF1α ChIP-seq targets identified in this study is comparable to a previous study where 7704 HIF1α target genes were identified in T cells (45), although another study identified 356 high-stringency HIF1-binding sites in MCF-7 breast cancer cells (27). These differences may be explained by alternative technical approach.
The large number of HIF1 binding targets identified in this and another study (45), suggest that HIF1α may serve a general role in regulating gene expression in response to hypoxia. With respect to this it is interesting to mention that the HIF binding motif 5′-RCGTG-3′overlaps with the E-box motif 5′-CACGTG-3′ to which MYC binds, a factor which has been reported to accumulate on all active promoters, amplifying the output of the existing gene expression (46). Because HIF1 is capable of replacing MYC from promoters (47), it is tempting to speculate that HIF1 could counteract MYC in hypoxia, inhibiting the overall gene expression. Consistently, HIF1 is indeed required for the reported hypoxia-induced cell cycle arrest in response to O2-deprivation (48). Furthermore, the HIF1α-E2F7 complex could play a particular role in the hypoxia-induced cell cycle arrest as E2F7 may recruit HIF1α to classic E2F targets involved in cell cycle progression.
Although HIF factors generally function as activators (27,28) and E2F7/8 as repressors (20), here we provide evidence that the HIF1α -E2F7 complex can have both transcriptional properties (Figure 7). These properties are at least in part determined by the presence of E2F or HIF binding motifs present in the common target promoters, because we show that E2F7 engages HIFα in transcriptional repression of NRP1 by acting directly on a 41-bp promoter region that contains 5 E2F, but no HIF motifs. HIF on the other hand can engage E2F7 in transcriptional activation, as we previously showed for VEGFA, acting independent of consensus E2F motifs, but dependent on a HIF motif (14). In line with these results, ChIP-seq experiments for E2F1, 4 and 6 showed that the majority of their target regions do not contain the E2F motifs, as previously mentioned (20), suggesting that the recruitment of E2Fs to E2F motif-less regions can indeed be performed by other transcription factors, such as nuclear factor–κβ (NF–κβ), MYC and CAAT enhancer/binding protein–α (CEBPα) (6). We identified HIF1α as such a factor, an also show that HIF1α instead can be recruited to the HIF motif-less NRP1 promoter by E2F7.
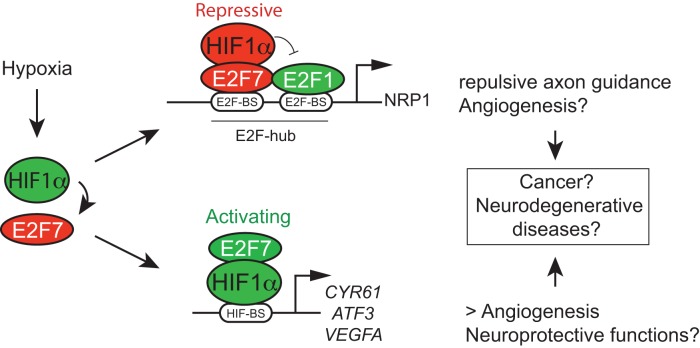
Figure 7.
Dualistic functions of the HIF1α-E2F7 complex in gene regulation, and biological implications. Hypoxia induces E2F7 expression through transcriptional activation by HIF1. The almost complete absence of differential gene regulation of common targets by the classified transcriptional activator HIF1α and the repressor E2F7, as observed in our microarray data (Supplementary Figure S2C and D), as well as in the combined ChIP-seq and microarray data (Figure 2C and D; Supplementary Figure S2F), unequivocally demonstrated the existence of the transcriptional network regulated by the HIF1α-E2F7 complex, in which the complex can either function as a repressor or activator. We reveal a direct role for HIF1α in transcriptional repression by acting independent of HIF-binding sites, but instead through an E2F-hub, as we show for NRP1. We expect that the HIF1α-E2F7 complex stimulates gene expression through HIF-binding sites, as we recently showed for VEGFA (14). Although not shown, NRP1 is also repressed, and CYR61 stimulated by HIF1β/ARNT (Supplementary Figure S4B,C). The HIF1α-E2F7 complex may counterbalance the expression of common repressed targets by replacing E2F1 from these promoters in hypoxia, when expression of HIF1α and E2F7 is induced. This mechanism regulates MN axon guidance during normal development, but could also serve neuroprotective functions, as growth cone collapse may eventually result in neuronal death. Whether HIF1-E2F7 induction of VEGFA expression also serves neuroprotective functions remains to be shown, which is also true for the potential role of the HIF1α-E2F7/NRP1 pathway in regulating (tumor) angiogenesis.
In this study we demonstrate a hypoxia-specific role for E2F7, by showing that specifically E2F7 expression is induced by HIF1 in hypoxia (Figure 1, Supplementary Figure S1). In addition, our input-corrected ChIP-qPCR experiments demonstrate a comparable or sometimes increased E2F7 binding to the common targets in hypoxia (Figures 3A, C and 4B), whereas E2F7 binding to the classic E2F targets E2F1 and E2F3 was reduced (Figure 3E). Furthermore, by analyzing mRNA expression of the HIF1α-E2F7 repressed targets we reveal that these genes are more significantly repressed by E2F7 in hypoxia (Figures 3B and 4D, Supplementary Figure S4A). Therefore, we conclude that the increased E2F7 expression in hypoxia results in enhanced downregulation of the HIF1α-E2F7 repressed targets.
Mechanistically, the HIF1α-E2F7 complex may repress the hypoxia downregulated targets by replacing the activator E2F1 from their promoters, as we observed a reversed correlation between the binding of HIF1α and E2F1 to these promoters (Figures 3A and 4B), and a repression of E2F1-induced NRP1 reporter activity by E2F7 and HIF1α (Figure 5H, K). Interestingly, others reported that E2F1 and E2F7 can form a heterodimeric complex on the DNA through binding to adjacent E2F motifs, in which E2F7 binding eventually leads to the dissociation of E2F1 from the promoter, switching the promoter from active to inactive (49). Thus the initial E2F1 binding to the E2F-hub in the NRP1 promoter, may recruit the HIF1α-E2F7 complex to adjacent E2F motifs, leading to the dissociation of E2F1 from the promoter (Figure 7), a mechanism that could be relevant for all common repressed targets. However, the regulation of NRP1 may be more complex, involving other E2F family members, as analysis of existing ChIP-seq data also showed binding of E2F4 and 6 to the same NRP1 promoter region (Supplementary Figure S7).
Interestingly, several studies have observed a downregulation of Neuropilin gene expression in response to hypoxia. Hypoxic repression of NRP1 has been reported in human astrocytoma cells (50), and in bone marrow derived macrophages (51), while hypoxic NRP2 repression has been observed in glioblastoma and melanoma cells (52). Although ectopic HIF1 levels could reduce NRP2 promoter activity in glioblastoma cells (52), and hypoxic Nrp1 mRNA expression in macrophages was suggested to be mediated through activation of the NF-κB pathway downstream of HIF2α (51), the exact molecular mechanism of how Neuropilin genes are repressed by hypoxia has so far remained elusive. Our data now demonstrate that E2F7 engages HIF1α in direct transcriptional repression of NRP1 in hypoxia, acting independent of HIF-binding sites, but instead through an ‘E2F-binding site hub’ located in the proximal NRP1 promoter region. Thus, providing evidence for a molecular mechanism of how expression of NRP genes is repressed by hypoxia.
Here, we report that regulation of NRP1 by the HIF1α-E2F7 complex is essential for axon guidance of spinal MN during zebrafish development, also providing evidence for a novel role for E2Fs in the regulation of neuronal development through the direct transcriptional control of atypical E2Fs targets (see introduction). Notably, the relatively minor MN phenotype observed in e2f7/8 mutant zebrafish may be explained by the fact that other E2F proteins may compensate for the loss of E2F7/8. For example, the E2F repressors E2F4 and E2F6 were also observed to bind the NRP1 promoter (Supplementary Figure S7). The identification of NRP1 as a target of the E2F and HIF pathways provides a novel mechanism of how these pathways may mediate vascular and neuronal development, whereas their deregulation may contribute to neurodegenerative diseases and cancer (Figure 7). With respect to neuronal development it has recently been put forward that transcriptional regulation of neuropilins presents a significant molecular mechanism in control of axon guidance (35). Our data identify the HIF and E2F pathways as novel and essential players in this process. In neurodegenerative diseases the HIF1α-E2F7 complex may have neuroprotective functions by inhibiting E2F-induced (NRP1-dependent) neuronal death, e.g. in response to cerebral ischemia (38). In this respect it is interesting to mention that inactivation of E2F7/8 in zebrafish and mice results in widespread apoptosis, including neuronal tissues (7,14). In mice, this cell death phenotype of E2f7/8 deficient mice could be rescued upon additional deletion of E2f1 (7), providing further support that the balance of transcriptional activity between the activator E2F1, and the repressor HIF1α-E2F7 complex may be critical for the survival and development of neuronal cells. Interestingly, the HIF1α-E2F7 complex may also be involved in amyotrophic lateral sclerosis (ALS), a neurodegenerative MN disease characterized by progressive distal axonopathy that precedes actual motor neuron death. Notably, deletion of the HIF-responsive element in the VEGFA promoter in mice results in late-onset (>5 months) progressive degeneration of lower motor neurons, indicative of ALS (44). Adaptation to hypoxia is indeed crucial for MN as they are extremely susceptible to ischemia (53). Deregulation of axon guidance proteins has been proposed as a major cause of ALS (54), as was recently also suggested for SEMA3A/NRP1 signaling in a mouse model for ALS (55). Therefore, deregulation of VEGFA (14) and NRP1 (this study) by the cooperative action of HIF1α and E2F7 may be implicated in ALS.
Deregulation of HIF1α-E2F7/NRP1 pathway may also be involved in cancer. For example, as part of a VEGR signaling complex through which NRPs stimulate tumor angiogenesis (33). Increased activator E2F activity, as detected in a variety of human cancers (6), may thus promote tumor angiogenesis through stimulation of NRP1 expression. In addition, HIF1α and E2F7 mediated NRP1 expression may also affect tumor vascularization and immunity by regulating the NRP1-dependent tumor infiltration capacity of macrophages and regulatory T cells (51,56). Interestingly, hypoxia-induced downregulation of NRP1 expression in tumor infiltrating macrophages, caused their retention in hypoxic tumor areas (51), suggesting that the HIF1α-E2F7/NRP1 pathway may be responsible for the homing of tumor associated macrophages towards hypoxic tumor regions. For these reasons it will be interesting to investigate the role of the novel HIF1α-E2F7/NRP1 pathway not only in neurodegenerative diseases, but also in cancer.
ACCESSION NUMBERS
All microarray gene expression data have been deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66750). All raw ChIP-seq data have been deposited in GEO with the accession number GSE66956 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66956).
Supplementary Material
Acknowledgments
We thank Wataru Shoji (Tohoku University, Japan) for generously providing us with Tg(nrp1a:gfp)js12Tg zebrafish, Jeroen den Hertog (Hubrecht Institute) for generously providing us the ability to conduct zebrafish experiments in his lab, and Jeroen Pasterkamp (University Medical Center Utrecht) for critically reviewing the manuscript. The study was stimulated by the HypoxiaNet (COST Action: TD0901) - http://www.hypoxianet.com.
Author contributions: W.J.B. carried out all in vitro and in vivo (zebrafish) experiments, assisted by P.W.A.C. (generation of ChIP-seq samples, ChIP-qPCR), E.I. (ChIP-qPCR), E.N. (ISH, cloning), K.H. (DFO experiments), K.T.S. (primer design and validation, ChIP-qPCR), A.V. (primer design and validation, siRNA-qPCR). ChIP-seq was performed by M.M. and E.C. M.J.G.K. and F.C.H. performed microarray analysis. B.C.K. generated the nrp1aHU10012 mutant. W.J.B. and A.d.B. designed experiments, S.S.M. co-designed zebrafish experiments. W.J.B. and A.d.B. wrote the manuscript, which was edited and reviewed by B.C.K., M.M., M.J.G.K., F.C.H. and S.S.M.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dutch Cancer Society [UU2009–4353 to W.J.B.]; Association of International Cancer Research [09–0718 to W.J.B.]; Netherlands Organisation for Scientific Research [NWO-ALW 11–28 to A.d.B.]. Funding for open access charge: University Utrecht.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dunwoodie S.L. The role of hypoxia in development of the mammalian embryo. Dev. Cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Simon M.C., Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat. Rev. Mol. Cell Biol. 2008;9:285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza G.L. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 2009;24:97–106. doi: 10.1152/physiol.00045.2008. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin W.G., Jr, Ratcliffe P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Palazon A., Goldrath A.W., Nizet V., Johnson R.S. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H.Z., Tsai S.Y., Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Ran C., Li E., Gordon F., Comstock G., Siddiqui H., Cleghorn W., Chen H.Z., Kornacker K., Liu C.G., et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson K.L., McClellan K.A., Vanderluit J.L., McIntosh W.C., Schuurmans C., Polleux F., Slack R.S. A cell-autonomous requirement for the cell cycle regulatory protein, rb, in neuronal migration. EMBO J. 2005;24:4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClellan K.A., Ruzhynsky V.A., Douda D.N., Vanderluit J.L., Ferguson K.L., Chen D., Bremner R., Park D.S., Leone G., Slack R.S. Unique requirement for Rb/E2F3 in neuronal migration: Evidence for cell cycle-independent functions. Mol. Cell. Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrusiak M.G., McClellan K.A., Dugal-Tessier D., Julian L.M., Rodrigues S.P., Park D.S., Kennedy T.E., Slack R.S. Rb/E2F regulates expression of neogenin during neuronal migration. Mol. Cell. Biol. 2011;31:238–247. doi: 10.1128/MCB.00378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D., Opavsky R., Pacal M., Tanimoto N., Wenzel P., Seeliger M.W., Leone G., Bremner R. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghanem N., Andrusiak M.G., Svoboda D., Al Lafi S.M., Julian L.M., McClellan K.A., De Repentigny Y., Kothary R., Ekker M., Blais A., et al. The Rb/E2F pathway modulates neurogenesis through direct regulation of the Dlx1/Dlx2 bigene cluster. J. Neurosci. 2012;32:8219–8230. doi: 10.1523/JNEUROSCI.1344-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lendahl U., Lee K.L., Yang H., Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 2009;10:821–832. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- 14.Weijts B.G., Bakker W.J., Cornelissen P.W., Liang K.H., Schaftenaar F.H., Westendorp B., de Wolf C.A., Paciejewska M., Scheele C.L., Kent L., et al. E2F7 and E2F8 promote angiogenesis through transcriptional activation of VEGFA in cooperation with HIF1. EMBO J. 2012;31:3871–3884. doi: 10.1038/emboj.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wageningen S., Kemmeren P., Lijnzaad P., Margaritis T., Benschop J.J., de Castro I.J., van Leenen D., Groot Koerkamp M.J., Ko C.W., Miles A.J., et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143:991–1004. doi: 10.1016/j.cell.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margaritis T., Lijnzaad P., van Leenen D., Bouwmeester D., Kemmeren P., van Hooff S.R., Holstege F.C. Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol. Syst. Biol. 2009;5:266. doi: 10.1038/msb.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westendorp B., Mokry M., Groot Koerkamp M.J., Holstege F.C., Cuppen E., de Bruin A. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 2012;40:3511–3523. doi: 10.1093/nar/gkr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato-Maeda M., Tawarayama H., Obinata M., Kuwada J.Y., Shoji W. Sema3a1 guides spinal motor axons in a cell- and stage-specific manner in zebrafish. Development. 2006;133:937–947. doi: 10.1242/dev.02268. [DOI] [PubMed] [Google Scholar]
- 22.Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S., van Impel A., Kirchmaier B.C., Peterson-Maduro J., Kourkoulis G., Male I., et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 24.Barriga E.H., Maxwell P.H., Reyes A.E., Mayor R. The hypoxia factor hif-1alpha controls neural crest chemotaxis and epithelial to mesenchymal transition. J. Cell Biol. 2013;201:759–776. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte-Merker S. Looking at embryos. In: Nusslein-Volhard C, editor. Zebrafish: A Practical Approach. NY: Oxford University Press; 2002. pp. 39–58. [Google Scholar]
- 26.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 27.Schodel J., Oikonomopoulos S., Ragoussis J., Pugh C.W., Ratcliffe P.J., Mole D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mole D.R., Blancher C., Copley R.R., Pollard P.J., Gleadle J.M., Ragoussis J., Ratcliffe P.J. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalmas L.P., Zhao X., Graham A.L., Fisher R., Reilly C., Coutts A.S., La Thangue N.B. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008;9:252–259. doi: 10.1038/sj.embor.7401158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takagi S., Hirata T., Agata K., Mochii M., Eguchi G., Fujisawa H. The A5 antigen, a candidate for the neuronal recognition molecule, has homologies to complement components and coagulation factors. Neuron. 1991;7:295–307. doi: 10.1016/0896-6273(91)90268-5. [DOI] [PubMed] [Google Scholar]
- 31.Kitsukawa T., Shimizu M., Sanbo M., Hirata T., Taniguchi M., Bekku Y., Yagi T., Fujisawa H. Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron. 1997;19:995–1005. doi: 10.1016/s0896-6273(00)80392-x. [DOI] [PubMed] [Google Scholar]
- 32.Gu C., Rodriguez E.R., Reimert D.V., Shu T., Fritzsch B., Richards L.J., Kolodkin A.L., Ginty D.D. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev. Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Djordjevic S., Driscoll P.C. Targeting VEGF signalling via the neuropilin co-receptor. Drug Discov. Today. 2013;18:447–455. doi: 10.1016/j.drudis.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Raimondi C., Ruhrberg C. Neuropilin signalling in vessels, neurons and tumours. Semin. Cell Dev. Biol. 2013;24:172–178. doi: 10.1016/j.semcdb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Pasterkamp R.J. Getting neural circuits into shape with semaphorins. Nat. Rev. Neurosci. 2012;13:605–618. doi: 10.1038/nrn3302. [DOI] [PubMed] [Google Scholar]
- 36.Bagri A., Tessier-Lavigne M. Neuropilins as semaphorin receptors: in vivo functions in neuronal cell migration and axon guidance. Adv. Exp. Med. Biol. 2002;515:13–31. [PubMed] [Google Scholar]
- 37.Taniguchi M., Yuasa S., Fujisawa H., Naruse I., Saga S., Mishina M., Yagi T. Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron. 1997;19:519–530. doi: 10.1016/s0896-6273(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 38.Jiang S.X., Sheldrick M., Desbois A., Slinn J., Hou S.T. Neuropilin-1 is a direct target of the transcription factor E2F1 during cerebral ischemia-induced neuronal death in vivo. Mol. Cell. Biol. 2007;27:1696–1705. doi: 10.1128/MCB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirvan A., Ziv I., Fleminger G., Shina R., He Z., Brudo I., Melamed E., Barzilai A. Semaphorins as mediators of neuronal apoptosis. J. Neurochem. 1999;73:961–971. doi: 10.1046/j.1471-4159.1999.0730961.x. [DOI] [PubMed] [Google Scholar]
- 40.Martyn U., Schulte-Merker S. Zebrafish neuropilins are differentially expressed and interact with vascular endothelial growth factor during embryonic vascular development. Dev. Dyn. 2004;231:33–42. doi: 10.1002/dvdy.20048. [DOI] [PubMed] [Google Scholar]
- 41.Santhakumar K., Judson E.C., Elks P.M., McKee S., Elworthy S., van Rooijen E., Walmsley S.S., Renshaw S.A., Cross S.S., van Eeden F.J. A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 2012;72:4017–4027. doi: 10.1158/0008-5472.CAN-11-3148. [DOI] [PubMed] [Google Scholar]
- 42.Roos M., Schachner M., Bernhardt R.R. Zebrafish semaphorin Z1b inhibits growing motor axons in vivo. Mech. Dev. 1999;87:103–117. doi: 10.1016/s0925-4773(99)00153-7. [DOI] [PubMed] [Google Scholar]
- 43.Feldner J., Becker T., Goishi K., Schweitzer J., Lee P., Schachner M., Klagsbrun M., Becker C.G. Neuropilin-1a is involved in trunk motor axon outgrowth in embryonic zebrafish. Dev. Dyn. 2005;234:535–549. doi: 10.1002/dvdy.20520. [DOI] [PubMed] [Google Scholar]
- 44.Oosthuyse B., Moons L., Storkebaum E., Beck H., Nuyens D., Brusselmans K., Van Dorpe J., Hellings P., Gorselink M., Heymans S., et al. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat. Genet. 2001;28:131–138. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- 45.Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkurst C.N., Muratet M., et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang C.V., Kim J.W., Gao P., Yustein J. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 48.Goda N., Ryan H.E., Khadivi B., McNulty W., Rickert R.C., Johnson R.S. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B., Shats I., Angus S.P., Gatza M.L., Nevins J.R. Interaction of E2F7 transcription factor with E2F1 and C-terminal-binding protein (CtBP) provides a mechanism for E2F7-dependent transcription repression. J. Biol. Chem. 2013;288:24581–24589. doi: 10.1074/jbc.M113.467506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding H., Wu X., Roncari L., Lau N., Shannon P., Nagy A., Guha A. Expression and regulation of neuropilin-1 in human astrocytomas. Int. J. Cancer. 2000;88:584–592. doi: 10.1002/1097-0215(20001115)88:4<584::aid-ijc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 51.Casazza A., Laoui D., Wenes M., Rizzolio S., Bassani N., Mambretti M., Deschoemaeker S., Van Ginderachter J.A., Tamagnone L., Mazzone M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Coma S., Shimizu A., Klagsbrun M. Hypoxia induces tumor and endothelial cell migration in a semaphorin 3F- and VEGF-dependent manner via transcriptional repression of their common receptor neuropilin 2. Cell. Adh Migr. 2011;5:266–275. doi: 10.4161/cam.5.3.16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lang-Lazdunski L., Matsushita K., Hirt L., Waeber C., Vonsattel J.P., Moskowitz M.A., Dietrich W.D. Spinal cord ischemia. development of a model in the mouse. Stroke. 2000;31:208–213. doi: 10.1161/01.str.31.1.208. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt E.R., Pasterkamp R.J., van den Berg L.H. Axon guidance proteins: novel therapeutic targets for ALS. Prog. Neurobiol. 2009;88:286–301. doi: 10.1016/j.pneurobio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Venkova K., Christov A., Kamaluddin Z., Kobalka P., Siddiqui S., Hensley K. Semaphorin 3A signaling through neuropilin-1 is an early trigger for distal axonopathy in the SOD1G93A mouse model of amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2014;73:702–713. doi: 10.1097/NEN.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen W., Hutzler M., Abel S., Alter C., Stockmann C., Kliche S., Albert J., Sparwasser T., Sakaguchi S., Westendorf A.M., et al. Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. J. Exp. Med. 2012;209:2001–2016. doi: 10.1084/jem.20111497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.