Abstract
Despite representing an important source of genetic variation, tandem repeats (TRs) remain poorly studied due to technical difficulties. We hypothesized that TRs can operate as expression (eQTLs) and methylation (mQTLs) quantitative trait loci. To test this we analyzed the effect of variation at 4849 promoter-associated TRs, genotyped in 120 individuals, on neighboring gene expression and DNA methylation. Polymorphic promoter TRs were associated with increased variance in local gene expression and DNA methylation, suggesting functional consequences related to TR variation. We identified >100 TRs associated with expression/methylation levels of adjacent genes. These potential eQTL/mQTL TRs were enriched for overlaps with transcription factor binding and DNaseI hypersensitivity sites, providing a rationale for their effects. Moreover, we showed that most TR variants are poorly tagged by nearby single nucleotide polymorphisms (SNPs) markers, indicating that many functional TR variants are not effectively assayed by SNP-based approaches. Our study assigns biological significance to TR variations in the human genome, and suggests that a significant fraction of TR variations exert functional effects via alterations of local gene expression or epigenetics. We conclude that targeted studies that focus on genotyping TR variants are required to fully ascertain functional variation in the genome.
INTRODUCTION
Repetitive elements represent more than half of the human genome (1). These include tandem repeats (TRs), stretches of DNA comprised of two or more contiguous copies of a motif arranged in a head-to-tail pattern. The length of the repeated motif is variable and TRs can be classified based on their motif size: (i) TRs with repeat units of 1–6 bp are often referred to as short TRs or microsatellites; (ii) minisatellites have DNA motifs ranging in length from 10–100 bp; and (iii) larger repeats with unit sizes ≥100 bp are termed macrosatellites. Some macrosatellites can have unit sizes of several kb and may include entire genes (2), such that large macrosatellites spanning exons or entire genes are often referred as multi-copy genes. As a result of errors during replication or recombination, TRs can gain or lose copies of the repeated motif and, consequently, many TRs exhibit length polymorphism with multiple alleles observed at the population level. Such mutation events are several orders of magnitude more frequent than that seen for other forms of mutation, such as single nucleotide polymorphisms (SNPs) and copy number variants (3–5). Adding to their high polymorphism and mutation rate, TRs are abundant in the genome of most species. For example, there are over one million annotated TRs in the human genome, and as such TRs represent an abundant source of genetic variation.
Growing evidence supports the functional importance of TR variation. Analysis of genomes sequenced to date has revealed that TRs are often located within coding regions in many species, and that genes with specific biological functions are enriched for variable TRs (6). Targeted studies have revealed several examples of functional TRs in the human genome, length variations of which can alter disease susceptibility (7–10). Furthermore, variable TRs in coding and non-coding regions can modulate quantitative phenotypes in several other organisms including prokaryotes (6,11), yeast (12) and dogs (13,14). Additional evidence of the functional role of TRs comes from their association with disease. Several dozen human diseases are caused by large repeat expansions in either coding or non-coding regions (reviewed by (6)). Although the pathogenic effect of TRs has mostly been studied in humans, examples in other vertebrates (15,16) and plants (17) also exist.
Despite their biological relevance, TRs have been poorly studied, largely due to technical difficulties in their characterization resulting from their repetitive and multi-allelic nature. Even with the advent of high-throughput genotyping technologies all but the largest TRs cannot be effectively assayed by oligonucleotide probes, and are typically excluded from microarray designs. Similarly, short-read next-generation sequencing approaches usually fail to capture TR variations when standard mapping and variant calling pipelines are used, as their repetitive and highly polymorphic nature means that reads mapping to these regions of the genome are typically discarded. The problem of genotyping TR variations by short read technologies is compounded by the need for reads to completely span a repeat tract and have sufficient anchoring sequence at both flanks in order to be informative. Therefore, with currently used read lengths only smaller TR loci can be assayed with next-generation sequencing (18). As a result of these technical difficulties in their characterization, TRs are generally ignored in most studies of genetic variation, including GWAS. In the past few years new approaches for effectively genotyping repetitive elements using direct visualization, digital profiling and sequencing-based approaches have been developed (19–28), enabling their systematic characterization in the genome (29–32).
In spite of the challenges, studies of TRs are highly relevant given their abundance and functional associations. Previously, we characterized large TRs (repeat unit sizes >1 kb) including macrosatellites and multi-copy genes, characterizing their evolution and functional impact in humans and non-human primates (25). Here we complement our previous study by focusing on smaller TRs (unit sizes ranging from 1–45 bp) with the aim of assessing their functional impact in the human genome. Specifically, we aimed to identify functional TRs by searching for variations in repeat length that alter local gene expression and DNA methylation.
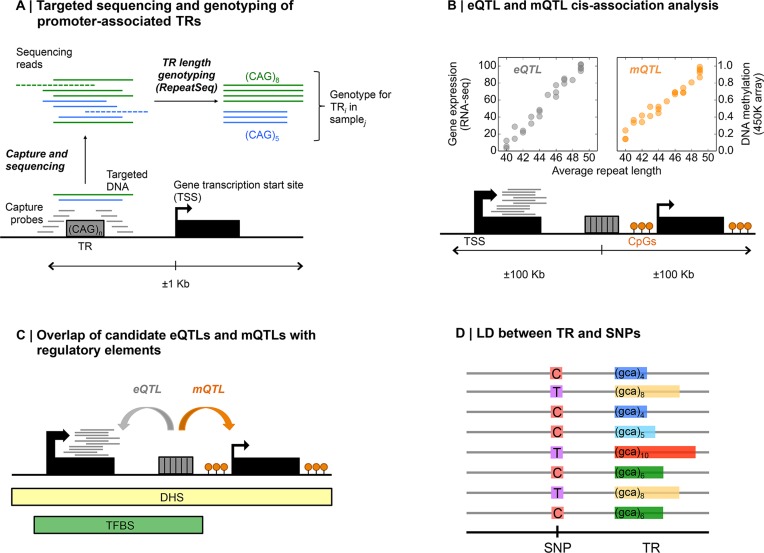
We first genotyped TRs located in gene promoters in the HapMap population using a targeted sequencing methodology (22) (Figure 1A), restricting our analysis to promoter-associated TRs with the assumption that these are more likely to exert effects on nearby gene activity. We then performed cis-association analysis to identify expression (eQTLs) and methylation (mQTLs) quantitative trait loci (Figure 1B) and analyzed their overlap with regulatory regions (Figure 1C). Finally, we investigated the extent to which SNP-based approaches can be used to tag potentially functional TR variations by estimating the degree of linkage disequilibrium (LD) between TRs and flanking SNPs (Figure 1D).
Figure 1.
Scheme of the TR genotyping strategy and downstream analyses. (A) Illustration of the TR targeted sequencing approach. Oligonucleotide probes are designed to hybridize and capture the genomic DNA of a promoter-associated repeat. After sequencing, only reads which span the entire repeat and have sufficient anchoring sequence at both flanks are informative and considered for genotyping, while other reads are discarded. TR length genotyping is performed using RepeatSeq (21). (B) For TR:gene and TR:CpG pairs separated by <100 kb we calculated Rho values between mean TR genotypes and (i) transcript expression levels derived from normalized RNA-sequencing (RNA-seq) data, (ii) CpG methylation levels derived from normalized microarray data. (C) To assess their functional impact, TRs were overlapped with functional elements such as transcription factor binding sites (TFBS) and DNaseI hypersensitivity sites (DHS). (D) In order to assess how effectively SNP arrays can tag TRs, we assessed levels of LD between TRs and flanking SNPs.
MATERIALS AND METHODS
Targeted sequencing of promoter-associated TRs
We used a previously published method using capture and high-throughput sequencing (22) to genotype 7851 TRs located in gene promoters in a total of 120 HapMap individuals of European (58 CEU individuals) and African (62 YRI individuals) ancestry (Figure 1A). Genomic DNA for these samples was extracted from lymphoblastoid cell lines (LCL). We defined promoter TRs as those TRs identified by the Tandem Repeats Finder algorithm (33) located within ±1 kb of the transcription start site (TSS) of a RefSeq gene annotated in the hg18 assembly of the human genome. Briefly, the method uses a custom Nimblegen EZ Capture system to enrich the genomic sequence flanking, and sometimes including, the target TRs to be genotyped prior to sequencing using an Illumina Hiseq2000 instrument. We multiplexed 24 individuals per sequencing lane and utilized 100 bp single-end reads.
Repeat length genotyping and quality assessment
RepeatSeq was used to determine genotypes for TR length (21). Reads were mapped to hg18 with Novoalign (http://www.novocraft.com/main/downloadpage.php) using parameters tolerant of indels, and only reads spanning the entire repeat and several flanking bases are used by RepeatSeq to predict the length of the two alleles at each TR that make an individual's genotype. In our set of 7851 TRs genotyped in 120 HapMap individuals, the number of flanking bases (the sum of both flanking ends, calculated as the read length minus the predicted repeat length) per genotyped TR allele had a mean of 62bp (5th percentile 28 bp, 95th percentile 74 bp). While our capture design targeted all annotated promoter-associated TRs irrespective of their total length or unit size, we were able to obtain genotype calls for 4849 of the targeted TRs in at least one individual (unit sizes ranging from 1–48 bp, median unit size 7 bp) (Supplementary File S1). To assess the quality of RepeatSeq genotypes generated by targeted sequencing, we compared them to genotypes generated by lobSTR as part of a catalog of TR variation in the 1000 Genomes Project based on low coverage whole-genome sequencing (29). We converted TR coordinates to hg19 using the liftOver tool and intersected loci with the lobSTR hg19 reference using BedTools intersectBed (34). Only loci annotated with the same motif and repeat tract length in both reference sets were included in the comparison, leaving 2603 distinct TRs with repeat motif lengths of 2–6 bp. Of these, 1381 loci were called in at least one overlapping sample in both datasets spanning 82 893 total genotype calls for comparison.
Gene expression and DNA methylation data
We used previously published RNA-seq gene expression data for 60 CEU (35) and 69 YRI (36) HapMap samples (GEO accession codes GSE25030 and GSE19480, respectively). RNA-seq for the CEU samples was expressed as reads per kilobase per million (RPKM). Short transcripts with low read counts can result in excessively elevated RPKM values. Therefore, we instead expressed RNA-seq data in reads per transcript per million (RPTM) using transcript lengths from the GENCODE Genes annotation, available for the hg19 version of the human genome assembly and downloaded from the UCSC Genome Browser (37); because our TR dataset was based on hg18 we previously lifted over gene transcript coordinates into hg18. To select transcripts showing variation in gene expression across samples, we log2-transformed RPTM values and selected those with median value across the 60 CEU samples greater than zero, leaving approximately one-third (50 716 transcripts) of the initial number of transcripts. On the other hand, YRI RNA-seq data was expressed as read counts per exon. To make data comparable between CEU and YRI datasets, we used the 50 716 gene transcript models retained in the CEU dataset to calculate RPTM in YRI samples: for each sample and transcript we aggregated read counts from exons comprised within the genomic coordinates of that transcript and normalized by millions of reads in that sample. RNA-seq data for YRI samples was generated in duplicate (referred to as ‘YRI Argonne’ and ‘YRI Yale’ sets). We calculated RPTM values in these two data sets separately and, as done for CEU transcripts, we selected transcripts with non-zero median expression across samples (44 591 and 44 513 transcripts in Argonne and Yale, respectively). In subsequent analysis we used 44 381 gene transcripts with non-zero median expression in either dataset.
We used PEER (38) to account for sources of variation in gene expression measures (e.g. batch effects, environmental variables) that can confound eQTL association analysis. Briefly, PEER first infers hidden covariates influencing gene expression measures as well as their weight. PEER then subtracts the component of the hidden covariates and produces a residual gene expression matrix that can be used for association analysis. We applied PEER to each of the three RNA-seq datasets independently (i.e. CEU, YRI Argonne and YRI Yale). RNA-seq data was log-transformed (RPTMtransformed = log2(2 + RPTM)) and we allowed PEER to fit 15 hidden covariates and account for the mean expression; all other inference parameters were used as default. We noted that 11 of the 69 YRI individuals had two technical replicates in both the YRI Argonne and YRI Yale datasets, and we only retained that replicate with the highest sequencing read depth. We observed a high concordance across samples in the expression residuals produced with PEER for YRI Argonne and YRI Yale (Supplementary Figure S1) and, therefore, we averaged expression residuals from the two datasets for the subsequent analyses. Of the 58 CEU and 62 YRI individuals genotyped in our study, RNA-seq data was available for all individuals (Supplementary Table S1).
We used published DNA methylation data (39) from 133 HapMap samples (60 CEU and 73 YRI) assayed with the Illumina Infinium HumanMethylation 450 BeadChip (GEO accession code GSE39672), which measures DNA methylation levels, expressed as β-values, at ∼482 000 individual CpGs. Data were first normalized and probes filtered (25), and samples with both DNA methylation data and TR genotypes were selected for subsequent association analysis (Supplementary Table S1).
Comparison of TR polymorphism and population variation in gene expression and DNA methylation
For gene expression, we calculated the per-transcript variance of the PEER-normalized gene expression values in samples of the same population (CEU or YRI). After excluding those genotyped TRs with >50% missing genotypes and those on chrX, we partitioned transcripts into those which had a TR within ±1 kb of their TSS and those that did not; the former were further classified into transcripts with an TR within ±1 kb of their TSS with non-modal allele frequency (NMAF, calculated as one minus the frequency of the major allele) greater than 0, 5, 10, 25 and 50%. For each NMAF distribution we tested whether its median was greater than the null expectation through permutation analysis: (i) from all N transcripts in which we calculated the population variance we randomly selected n transcripts, where n is the number of transcripts in a given NMAF category; (ii) we calculated the median population variance of the random sample and compared to the true median value of the distribution; (iii) the previous two steps are repeated a total of 1000 permutations an empirical P-value is calculated as the number of permutations in which the permuted median exceeds the true value. The sample analysis was performed for DNA methylation using instead the per-transcript variance of the normalized β-values in samples of the same population (CEU or YRI).
Cis-association analysis of TR lengths with gene expression and methylation levels
For the CEU and YRI populations separately, we calculated Spearman Rho values between population values of TR length and per-transcript PEER-corrected RPTM gene expression values for TR:transcript pairs that were separated by <100 kb. We converted heterozygous TR genotypes to an average genotype of the length of the two alleles prior to calculating the correlation. To exclude TRs with low quality or low levels of polymorphism, we only used TR loci with genotypes available in ≥50% of individuals and with NMAF ≥10%. To avoid the potential confounder of X chromosome inactivation in females, we also removed TRs located on the X chromosome. These filters resulted in a final set of 1198 promoter-associated TRs that were used in the final association analyses. In each population we corrected nominal Rho P-values for multiple testing through permutations. For each TR:transcript pair, gene expression values were randomized and the correlation between these and the actual TR lengths was calculated; after repeating the randomization 1000 times, corrected P-values were calculated as the number of permutations that produced a correlation value greater, in absolute terms, than the actual Rho value. We calculated the Rho values between population values of TR length and methylation beta-values for autosomal TR:methylation probe pairs separated by <100 kb in the CEU and YRI populations separately. We treated heterozygous genotypes and filtered TRs as described above for the association analysis with gene expression data. We carried out multiple-testing correction through permutations as described above.
Distributions of TR:transcript and TR:CpG distances
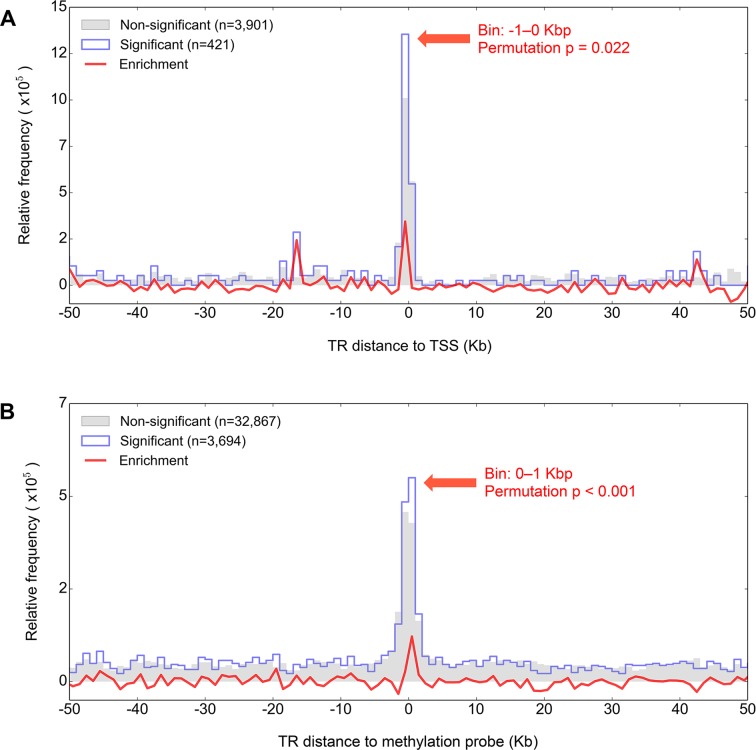
For the TR:transcript pairs included in the correlation analysis we calculated the distance between the midpoint of the TR (calculated using the start and end coordinates as annotated in hg18, that is, without using the actual range of allele lengths seen in our data) and the transcript TSS. We split TR:transcript distances into two mutually exclusive groups: (i) corresponding to a Rho values significant (nominal Rho P < 0.05) in any of the two populations (CEU or YRI) (i.e. nominally significant) and (ii) corresponding to a Rho values not significant in any of them (i.e. non-significant), and in each group we generated the distribution of TR:probe distances using bins of 1 kb. We proceeded in the same way to generate the distribution of TR:CpG distances, measured between the midpoint of the TR and the methylation probe tagging the corresponding CpG site. In the distributions of both TR:transcript and TR:CpG separations, we anticipated a higher relative frequency of small distances. This was because (i) our experimental design targeted TRs within 1 kb of the gene TSS, and (ii) the array used to generate the DNA methylation data has a higher coverage of CpG sites located within or nearby gene promoters. To account for this, we calculated the enrichment of significant correlations in each 1 kb bin as the difference between relative frequencies in the significant and non-significant distributions. In addition, we tested whether the calculated 1 kb enrichment values were greater than expected by chance as follows: in each type of data, we randomly split TR:TSS (or TR:CpG) distances into the two mutually exclusive significant and non-significant groups, keeping their sizes as in the true data, and calculated the permuted enrichment ratio; after repeating the randomization 1000 times, in each 1 kb bin we computed the permutation P-value for the true enrichment value as the number of permutations in which the permuted enrichment ratio was higher than the true value.
Overlap of promoter-associated TRs with TFBS and DHS
We downloaded predicted transcription factor binding sites (TFBS) in LCLs from http://centipede.uchicago.edu/ (posterior P > 0.99, with overlaps) (40). Because coordinates in this dataset correspond exclusively to the DNA motif that is recognized by the transcription factor, we extended these TFBS coordinates by ±10 bp the start and end of the TFBS genomic coordinates in order to be more inclusive. We downloaded from the UCSC Genome Browser the genomic coordinates of ENCODE DNaseI hypersensitivity sites (DHS) in the HapMap sample GM12878 (41). Since both TFBS and DHS were annotated in the hg19 human genome assembly we lifted the coordinates over to hg18. For each type of TR length correlation (i.e. with gene expression and with DNA methylation) and for each type of functional element (i.e. TFBS and DHS) we proceeded as follows. First, we filtered correlation values to only select those involving TRs with <5% missing genotypes in either population which were located within 5 kb of the gene TSS or methylation probe used in the calculation. We split the remaining correlation values into two mutually exclusive groups: (i) Rho values with nominal P < 0.05 in any of the two populations (CEU or YRI) (referred to as ‘significant’) and (ii) Rho values not significant in any of them (referred to as ‘non-significant’). After intersecting the genomic coordinates of the significant and non-significant TRs with those of the corresponding set of functional elements, we calculated the frequency of overlap in each category (Supplementary Table S2).
Linkage disequilibrium analysis
HapMap Phase II SNP genotypes for CEU and YRI individuals were downloaded (release 24, http://hapmap.ncbi.nlm.nih.gov), and SNPs with minor allele frequency <0.05, Hardy Weinberg Equilibrium (HWE) P < 0.05 or tri-allelic states were removed. After excluding TRs with NMAF < 0.05 (i.e. TRs in which the frequency of the major allele is >95%), we selected TR:SNP pairs separated by <250 kb, provided that the frequency of genotyped SNP and TR alleles in the pair was >75%. Variants were first phased using Beagle version 3.3.2 (42) with 1000 iterations, and after converting SNP genotypes to numerical format, for each TR:SNP pair we calculated the Pearson correlation between SNP and TR genotypes to derive the coefficient of determination (r2).
RESULTS
Genotyping of promoter-associated TRs by targeted sequencing
In order to characterize repeat length variation in promoter-associated TRs, we used a recently developed targeted sequencing methodology (22), which overcomes many of the technical difficulties in genotyping TRs. Briefly, the first step involves targeted enrichment of the genomic regions to be sequenced. Here we focused on TRs in gene promoters, reasoning that these are more likely to have a functional impact on gene activity. We defined promoter-associated TRs as those within ±1 kb of the TSS of RefSeq genes annotated in the human genome (Figure 1A). In total we targeted 7851 TRs with our assay, identifying 31% of RefSeq genes as having a promoter-associated TR. We then performed enrichment and sequencing of this set of promoter-associated TRs in 120 HapMap individuals with European (58 CEU samples) and African (62 YRI samples) ancestry (Supplementary Table S1). Using 100 bp Illumina reads, we multiplexed 24 individuals per sequencing lane, achieving a median coverage of 47× informative spanning reads per TR. Finally, we produced personal TR genotypes using RepeatSeq, a microsatellite variant caller for Illumina sequencing (21). After mapping of the reads using an aligner tolerant of indels, RepeatSeq uses aligned reads spanning the entire repeat length to determine genotypes for the two TR alleles in each sample. Supplementary File S1 contains the genotypes of the 7851 TRs for the 120 HapMap individuals. After filtering to retain those loci successfully genotyped in ≥50% of samples, we obtained genotype data for 3600 promoter-associated TRs (Supplementary Figure S2).
To measure the quality of our set of repeat length genotypes, we compared them to genotypes generated by lobSTR as part of a catalog of TR variation in the 1000 Genomes Project (29) (Supplementary Figure S3). We found that mean TR length was significantly correlated across datasets (r2 = 0.53, P-value smaller than the numerical precision of Python). Overall, 57% of TR calls showed concordant genotypes. Consistent with our previous analysis (22), the majority of discordant genotypes occurred at dinucleotide repeats that differed by a single repeat unit (Supplementary Table S3), with TRs with motif lengths >2 bp yielding 79% concordant genotypes. Furthermore, 43% of errors could be attributed to loss of one allele at heterozygous loci in the lobSTR calls, most likely due to the low sequencing coverage of the 1000 Genomes Project. Overall we conclude that TR genotypes generated by our targeted sequencing approach are generally robust.
Effects of TR polymorphism on local gene expression and DNA methylation
We hypothesized that polymorphic TRs can act as functional elements by altering gene expression and DNA methylation in the local vicinity. We took advantage of the fact that HapMap individuals included in this study had been previously characterized for gene expression through RNA-seq (35,36) and DNA methylation by microarray (39) experiments (Supplementary Table S1).
In order to gain global insight into functional effects of variation in promoter TRs, we first analyzed variance of gene expression and DNA methylation levels for genes lacking TRs within their promoters compared with those that have polymorphic promoter TRs. We found that genes with a promoter-associated TR showed significantly higher variation in both expression and DNA methylation levels, and that this effect was more pronounced for genes with highly polymorphic promoter TRs (Figure 2). Although this observation does not establish causation between repeat variation and changes in expression or methylation, it suggests that TR polymorphisms contribute to local variation in gene activity and epigenetics.
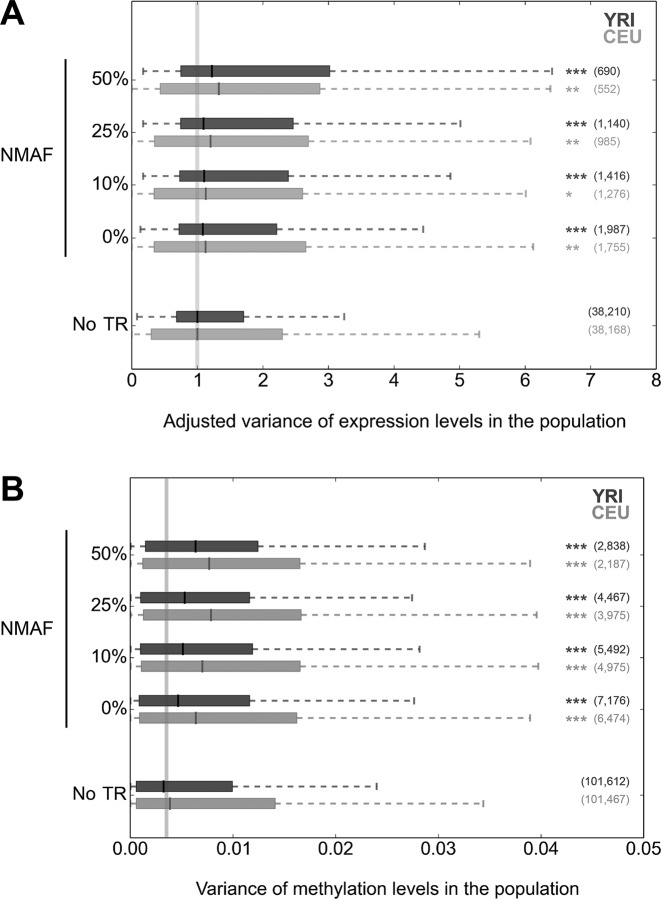
Figure 2.
Increased expression and methylation variation associated with polymorphic TRs. Distributions of variance in local (A) gene expression and (B) DNA methylation levels for CEU (light gray) and YRI (dark gray) samples based on increasing rates of TR polymorphism. NMAF refers to the non-modal allele frequency, the aggregated frequencies of all alleles other than the most common. To the right of each plot the number of genes in each category is indicated in parentheses, while asterisks indicate a significant difference of the median from the null distribution, as inferred through permutation analysis (*P < 0.05; **P < 0.01; ***P < 0.001). In the top panel (variance of gene expression), to allow for meaningful comparison in a single plot we normalized the ranges of values in CEU and YRI by dividing each by the median value of genes with no promoter-associated TR. In each figure the vertical gray line indicates the median value for the ‘No TR’ category. TR:gene and TR:CpG pairs were based on a separation of ≤1 kb.
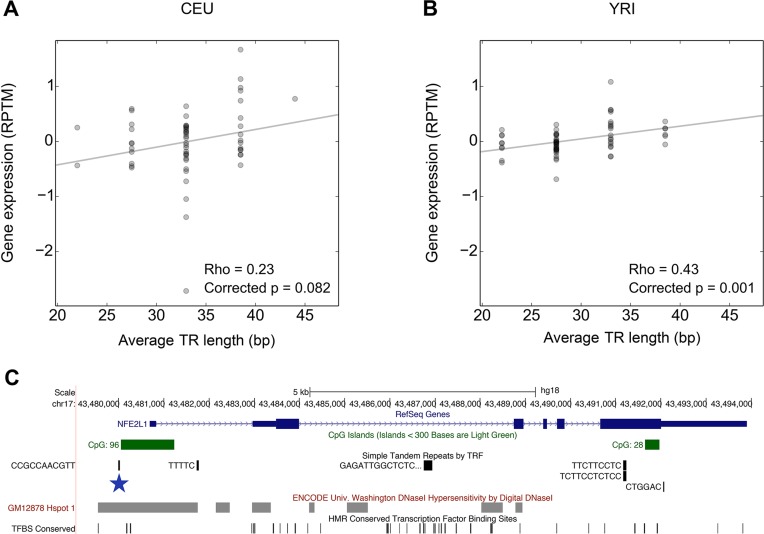
To address this question, we performed association studies to identify specific TR variations that correlate with changes in local gene expression and DNA methylation levels. To identify potential eQTLs and mQTLs we calculated pairwise Spearman correlations (Rho) between TR genotypes and expression levels for TR-transcript pairs in which the repeat was closer than ±100 kb of the TSS of the gene (Figure 1B), calculating correlations in CEU and YRI individuals separately. Of the 10 489 TR-transcript pairs (Supplementary File S2), 421 correlations corresponding to 183 unique TRs were significant in either CEU or YRI after correcting for multiple testing through permutation (Supplementary File S3). Eight of these putative 183 eQTL TRs (∼5%) were detected in both CEU and YRI populations and consistently showed the same effect on expression levels (Supplementary Table S4). For instance, we found an 11 bp repeat in which population values of TR length and gene expression were positively correlated in both CEU and YRI individuals, yet only in the former the correlation Rho value was significant (Figure 3A and B). This multi-allelic TR lies in the promoter region of the NFE2L1 gene [GeneBank Gene ID: 4779], in a region containing multiple regulatory elements (Figure 3C). In addition, we observed that methylation of NFE2L1 [GeneBank Gene ID: 4779] is also correlated with length variation of this TR:methylation levels at the CpG sites located in the promoter of this gene decrease with increasing number of copies of the 11 bp repeat (Supplementary Figure S4).
Figure 3.
Identification of a TR eQTL associated with expression of NFE2L1. In (A) CEU and (B) YRI populations, scatter plots of TR length genotypes against gene expression values inferred from RNA-seq for the ENST00000361665 transcript of the NFE2L1 gene [GeneBank Gene ID: 4779]; RPTM correspond to reads per transcript per million of reads after PEER normalization (see ‘Materials and Methods’ section). Shown are the Rho values with P-values corrected for multiple testing through permutations and the best linear fit of the data (trend line). (C) The CCGCCAACGTT repeat (chr17:43,479,993–43,480,025, hg19) (blue star) is located upstream of the NFE2L1 gene [GeneBank Gene ID: 4779] and overlaps a predicted DHS.
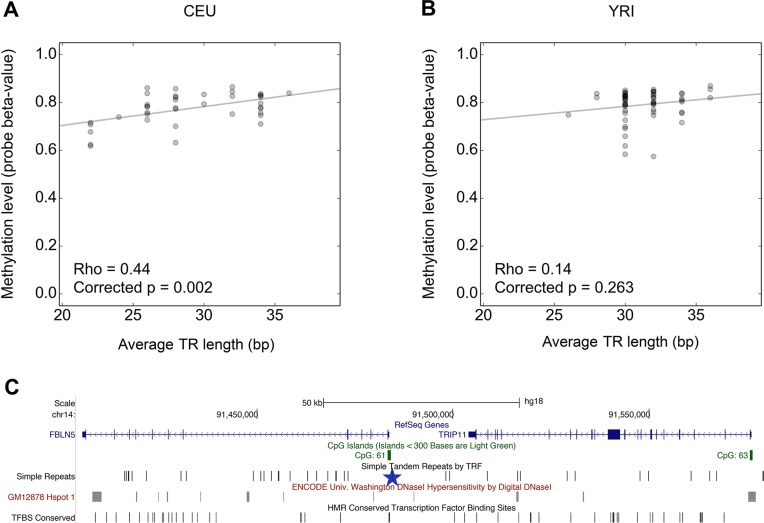
In the mQTL association analysis we calculated correlation Rho values between repeat lengths and methylation β-values for all TR:CpG pairs separated by ≤10 0kb (Figure 1B). Of the 88 492 TR:CpG pairs (Supplementary File S4), 3694 correlations corresponding to 463 unique TRs were significant in at least one of the two populations after correcting for multiple testing (Supplementary File S5). However, only 39 of these 462 loci (∼8%) were detected independently in both the CEU and YRI populations and with consistent directionality in the two groups (Supplementary Table S4). We found a TR located downstream of the TRIP11 gene [GeneBank Gene ID: 9321] in which the number of tandemly repeated copies of a TGTT motif correlates with methylation levels at the promoter of this gene. Although this correlation only remained significant after multiple-testing correction in CEU individuals, the same trend was also observed in YRI samples (Figure 4). We also observed a weak correlation of TRIP11 [GeneBank Gene ID: 9321] expression levels with this same TR in CEU samples (Rho = −0.26, nominal P = 0.05). TRIP11 [GeneBank Gene ID: 9321] encodes for the thyroid hormone receptor interactor 11 protein, which acts as a co-activator of transcription activity. Intersection of TRs scored as significant mQTLs and probes on the Infinium 450 k array used to interrogate methylation levels of each CpG showed that only 13 of the 463 mQTL TRs (2.8%) directly overlap with the methylation probe, indicating that probe:TR overlaps do not significantly influence our results.
Figure 4.
Identification of an TR mQTL affecting methylation at TRIP11. Scatter plots of population values of TR length against methylation values of probe cg14294158 in (A) CEU and (B) YRI individuals. Shown are correlation Rho values with P-values corrected for multiple testing through permutation, and the best linear fit of the data. (C) Location of the TGTT-repeat (blue star) at chr14:91,484,391–91,484,491, downstream of the TRIP11 gene [GeneBank Gene ID: 9321].
Overall, we observed a strong concordance for TRs that were significant eQTLs to also be mQTLs. Between 90 and 96%, depending on the population compared, of TRs scored as eQTLs were also significantly associated with changes in local DNA methylation levels (Supplementary Figure S5). We performed gene ontology analysis to search for functional annotation terms enriched among genes associated with eQTLs and mQTLs using GOrilla (http://cbl-gorilla.cs.technion.ac.il/), but did not detect any significant enrichment.
We investigated the characteristics of functional TRs, comparing the frequency of motif length and sequence for TRs scored as significant QTLs (either eQTLs, mQTLs or both) to TRs that were non-significant QTLs. Grouping TRs by motif length (Supplementary Table S5) indicated a strong positive relationship of the size of a TR unit on propensity to be a QTL, with TRs with motif length ≥4 bp being overall 3.4-fold more likely to act as QTLs compared to TRs with shorter dinucleotide motifs (P = 4.3 × 10−23, Fisher's Exact Test). Considering motif lengths with at least 30 occurrences in the set of 1198 TRs that were used in our association analyses, we observed suggestive evidence for a linear relationship between motif length and odds ratio for TRs to act as a QTL (Supplementary Figure S6), although this did not reach statistical significance (r2 = 0.54, P = 0.16). We then searched for specific TR motifs that were associated with probability of being a significant QTL. We first grouped motifs that were the same when inverted or reverse complemented (e.g. AC, CA, GT and TG were considered as a single motif) and then calculated relative enrichment for each motif that was represented ≥10 times in the dataset (Supplementary Table S6). Results indicate that TRs comprised of AAC, AAAC and AAAT motifs were significantly over-represented among significant QTLs, while AC dinucleotide TRs were significantly under-represented. However, it is possible that the enrichments for these specific TR motifs may simply reflect the underlying relationship between motif length and probability of being a QTL.
Previous work indicates that the majority of SNPs that act as eQTLs are preferentially located close to the TSS of the gene they regulate (43). Similarly, SNPs that act as mQTLs tend to be located close to the CpG sites that they are associated with (44). We therefore analyzed the distribution of separation between significant TR:gene and TR:CpG pairs relative to the background. We first generated a null distribution of TR:TSS and TR:CpG pairwise distances from those TRs which had no significant association with gene expression and DNA methylation, respectively (Rho nominal P > 0.05 in both CEU and YRI), and compared this to TRs with nominal evidence of eQTL or mQTL activity (P < 0.05 in at least one of the two populations). We observed a significant enrichment for TR eQTLs and mQTLs within ±1 kb of their target compared to the null (permutation P < 0.05) (Figure 5). This preferential co-localization of functional TRs with their associated TSSs/CpGs is consistent with the hypothesis that some TR variants influence local levels of DNA methylation and gene expression.
Figure 5.
TRs that are significant eQTLs and mQTLs preferentially co-localize with their associated target. After first dividing eQTL and mQTLs into those that were either nominally significant (P < 0.05 in either CEU or YRI, blue outline) or non-significant (P > 0.05 in both CEU and YRI, gray shading), we plotted the separation of (A) TR:TSS and (B) TR:CpG pairs for the two groups in 1 kb bins. The red line shows the frequency difference between the non-significant and significant distributions, with the significance of the enrichment determined through permutations (see ‘Materials and Methods’ section). For both eQTLs and mQTLs, significant associations are enriched for separations of <1 kb. These results mirror those from previous studies using SNPs, which have shown a strong enrichment for eQTLs occurring in close proximity to the TSS of the associated gene (43), and for mQTLs to colocalize with their associated CpG (44).
Overlap of TRs with regulatory elements
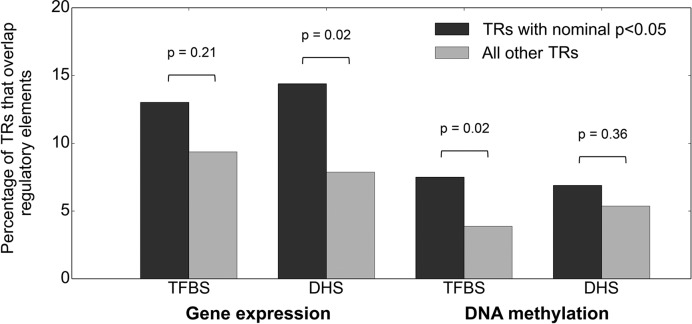
Previous analyses have identified thousands of SNPs that affect nearby chromatin accessibility, a genomic mark of regulatory potential, and that such SNPs often also act as eQTLs (45). Reasoning that functional TR variants that alter the expression and methylation patterns of adjacent genes would preferentially overlap regulatory elements, we investigated the co-localization of our TR eQTLs and mQTLs with TFBS and DHS. Consistent with this hypothesis, in each category tested we observed that TRs scored as significant eQTLs/mQTLs overlapped TFBS and DHS regulatory elements more frequently than other TRs, although due to limited sample size this enrichment achieved statistical significance in only two of the four cases (2.3-fold enrichment for eQTL TRs overlapping DHS, P = 0.02 and 1.9-fold enrichment for mQTL TRs overlapping TFBS, P = 0.02) (Figure 6). Among those TFBS overlapping with eQTL and mQTL, none showed a frequency higher than 10% nor was enriched compared to level of overlapping seen for non-eQTL and non-mQTL loci (data not shown).
Figure 6.
Significant eQTL and mQTL TRs preferentially overlap regulatory elements. We divided eQTL and mQTLs into those that were either nominally significant (P < 0.05 in either CEU or YRI, black) or non-significant (P > 0.05 in both CEU and YRI, gray), and performed overlaps with high-confidence TFBS and DHS assayed in LCLs. Significance between pairs of frequencies were calculated with the two-sided Fisher test.
Linkage disequilibrium between TRs and SNPs
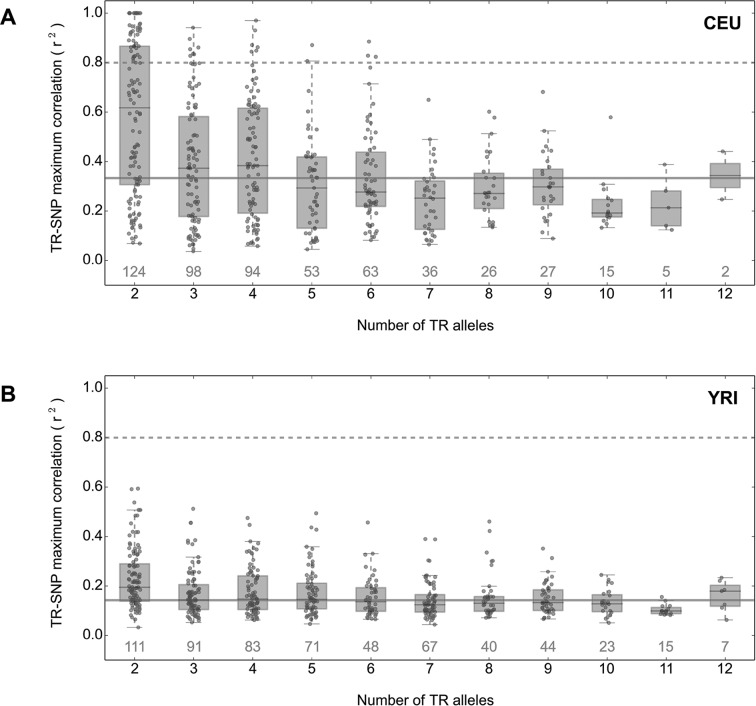
Given that GWAS utilize SNP markers in an effort to identify genetic variants that modify disease susceptibility, we evaluated the extent to which TR variants are ‘tagged’ by local SNP markers by estimating patterns of LD between TRs and nearby SNP variants. We first phased TR and SNP genotypes using Beagle (42), and then calculated pairwise correlations between TR and SNP alleles (r2) in both the CEU and YRI populations for all TRs and SNPs separated by ≤250 kb. We considered the maximum observed r2 for each TR as a measure of how well its variation could be captured by local SNP markers (Figure 7). Overall we observed low levels of LD between SNP and TR alleles and a sharp decay in LD with TR length diversity. In the CEU population (Figure 7A), even for TRs with only two alleles there was broad variation in the best TR:SNP correlation: the median r2 for bi-allelic TRs was 0.65, but some TR loci were poorly tagged by nearby SNPs. For TRs with more than four alleles, the best tag SNPs for most of these multi-allelic variants showed r2 < 0.4. The low LD between TRs and SNPs was even more pronounced in the YRI individuals (Figure 7B). No TR achieved an r2 value >0.60, even in bi-allelic TRs, and for multi-allelic TRs the median r2 value was <0.2. Overall, these results indicate that most TR variants, particularly those with multiple alleles, are poorly tagged by SNP markers.
Figure 7.
Decay of LD between TRs and SNPs with TR length diversity. We calculated the LD (r2) between TRs and SNPs separated by <250kb, and after binning TRs based on the number of observed alleles (x-axis), plotted the maximum r2 observed for each TR in the (A) CEU and (B) YRI populations. The number of TRs represented in each boxplot is indicated in gray above the x-axis. Horizontal lines correspond to r2 = 0.8 (dashed) and the population median for all TRs analyzed (solid gray).
DISCUSSION
TR variations are often excluded from genomic studies, and as a result their potential functional impacts are largely unexplored. Previously, we have demonstrated that copy number variation of macrosatellites is often associated with functional impacts on the local genomic environment (25). In the present study, we hypothesized that repeat length variation of smaller TRs might show similar functional effects. To test this hypothesis we profiled repeat length variation at thousands of promoter-associated TRs, and performed cis-association analyses with nearby gene expression and DNA methylation. We found that genes containing polymorphic TRs in their promoters exhibit higher variance of gene expression and DNA methylation. This mirrors previous observations made among primates in which genes with polymorphic repeats have increased expression divergence (31), suggesting a mechanistic link between TR variation and local genome function. We then identified ∼500 specific promoter-associated TRs linked to changes in the expression or methylation levels of nearby genes and thus putatively acting as cis eQTLs/mQTLs. TRs scored as eQTLs and mQTLs tend to be located close (<1 kb) to the genes or CpGs that they influence, in agreement with previous SNP-based studies (43,44). Moreover, our results suggest that many of these putative cis QTL TRs influence both gene expression and methylation, with almost all TRs scored as eQTLs also being identified as putative mQTLs. Finally, we observed that putatively functional TRs are enriched for overlaps with TFBS and DHS, which mirrors similar observations made for SNPs that are associated with altered chromatin accessibility and gene expression (45). Furthermore, by analyzing LD patterns of TRs with flanking SNPs we show that the majority of TRs, particularly those that are highly polymorphic, are not effectively tagged by nearby SNP markers.
We hypothesize several possible mechanisms whereby insertion/deletion of TR units could exert functional effects on the local genomic environment: (i) the direct addition or removal functional DNA motifs, such as TFBS, present within each repeat unit. In support of this notion, we observed that significant eQTL/mQTL TRs are enriched for overlaps with TFBS and DHS compared to the null. (ii) Modification of local DNA/RNA secondary structure. Pertinent to this, many TRs, particularly those composed of AT- or GC-rich motifs, are capable of forming stable structures such as hairpins and G-quadruplexes via intra-molecular base pairing. In some cases the modulation of these secondary structures has been shown to have functional consequences (46). (iii) Epigenetic modification of the local region, for example through changes in CpG content. Such mechanisms are known to operate in the case of some triplet repeat diseases such as Fragile X, where extreme expansions of a CGG repeat become highly methylated, resulting in local transcriptional suppression (47). (iv) Altered spacing of cooperative elements that function optimally at certain separation. This mechanism has been shown to operate in bacterial promoters, where expansion and contraction of promoter-associated TRs can be associated with altered mRNA levels (48). Similarly in mRNA splicing, the intronic branch site has an optimal separation (typically 15–55 bp) from the end of intron for efficient for lariat formation (49). (v) Alterations of local nucleosome positioning. Nucleosome occupancy is influenced by the underlying DNA sequence, and the presence of certain polynucleotides that are commonly observed in TRs, such as poly(dA/dT) motifs, can modify local nucleosome occupancy (50). (vi) Modifying the relative spatial orientation of cooperative motifs on the helical turn of the DNA strand, where one turn is made every ∼10 bp (51). For example, paired protein binding sites may work optimally when arranged in a specific relative orientation to each other on the DNA helix (52). These mechanisms are not mutually exclusive, and we speculate that diverse functional mechanisms likely operate at different TR loci depending on their specific sequence and genomic context.
The approach used here to assess the effect of TR variation in the human genome has several limitations. First, despite the use of specialized approaches, the accuracy of TR genotyping by targeted sequencing is imperfect (22), and such errors would create additional noise in our association testing, resulting in reduced power. Second, our use of read lengths of 100 bp creates a limitation that TRs with a span exceeding ∼80–90 bp cannot be effectively assayed (22). We note however, that our previous analysis indicates that 100 bp reads are sufficiently long to capture the majority of TR alleles in gene promoters, and that the use of longer reads provides only marginal increases in genotype yields (22). Third, despite that fact that our study generated genotypes for individual TR alleles, our association testing utilized averaged TR lengths and linear regression. This method is conservative, and assumes that any quantitative effects of TR variation are directly proportional to repeat length. As a result, more complex non-linear effects of TR variation on gene expression and epigenetics would not be accurately modeled, thus potentially reducing the power of our analysis. In particular, previous in vitro manipulations of TR lengths in yeast have indicated non-linear relationships between the length of promoter TRs and gene expression levels (53). However, in a discovery setting attempts to model more complex relationships have a strong likelihood of over-fitting, which would create large numbers or false positive associations. Moreover, there may be other genetic variation between individuals that confound the link between repeat variation and gene expression or methylation. For instance, the phenotypic effect of polyglutamine expansions is modified by secondary mutations in Arabidopsis thaliana (54). Finally, our association analyses utilized a relatively small sample size, with only ∼60 individuals in both the CEU and YRI populations. As a result, our study has relatively low power, which likely explains why only a small proportion of the eQTLs/mQTLs identified were shared across both CEU and YRI populations (Supplementary Table S4). Given these caveats, and the fact that our study only analyzed a small fraction of all TRs in the genome, future studies that assay larger populations with increased power will be needed to uncover the full spectrum of functional TR variation in the genome.
If a significant number of TR variants have functional effects on the genome, what are the chances that these can be identified in conventional disease-mapping studies? GWAS typically use dense SNP panels typed using microarrays, and rely on the notion that putative functional variants will either be directly interrogated or tagged by genotyped SNPs that are in LD with the functional variant. However, as a result, GWAS provide little information on variants that are not well tagged by the available SNP markers. Our results provide evidence that TRs have generally low LD with nearby SNP markers, and that LD is generally lower both for TR loci with many different alleles and in African populations (Figure 7). Considering that the rule-of-thumb is that variation at one locus can be effectively tagged by a second marker if these show an r2 > 0.8, our results imply that many functional TR alleles are not effectively tagged by SNP-based GWAS approaches. The limited ability of GWAS to effectively assay genomic variants which are not well tagged by SNPs has been proposed as a possible explanation for the so-called ‘missing heritability’, that is, the gap between estimates of heritability and phenotypic variation explained by loci identified in GWAS studies (55). We propose that variations of TRs contribute to this phenomenon.
To conclude, here we performed targeted sequencing and genotyping of thousands of promoter-associated TRs. Our results suggest that there are potentially thousands of TR variants in the human genome that exert functional effects via alterations of local gene expression or epigenetics. Our results also suggest that conventional SNP-based mapping approaches have limited ability to assay many of these functional TR variants, as repeat length variation is poorly tagged by nearby SNPs. We conclude that specific studies that focus on genotyping TR variants are required to fully ascertain functional variation in the genome.
Supplementary Material
Footnotes
Present addresses:
Javier Quilez, Centre for Genomic Regulation (CRG), Dr Aiguader 88, 08003 Barcelona, Spain.
Audrey Guilmatre, Institut Pasteur, Human Genetics and Cognitive Functions Unit, Paris, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [DA033660, HG006696, HD073731, MH097018 to A.J.S.]; March of Dimes Research Grant [6-FY13-92 to A.J.S.]; Scientific Interface from the Burroughs Wellcome Fund Career Award (to Y.E.); Gift from Andria and Paul Heafy; NIJ [2014-DN-BX-K089]; Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai (in part). Funding for open access charge: NIH [DA033660, HG006696, HD073731, MH097018 to A.J.S.]; March of Dimes Research Grant [6-FY13-92 to A.J.S.]; Scientific Interface from the Burroughs Wellcome Fund Career Award (to Y.E.); Gift from Andria and Paul Heafy; NIJ [2014-DN-BX-K089].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Warburton P.E., Hasson D., Guillem F., Lescale C., Jin X., Abrusan G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 2008;9:533. doi: 10.1186/1471-2164-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell C.D., Chong J.X., Malig M., Ko A., Dumont B.L., Han L., Vives L., O'Roak B.J., Sudmant P.H., Shendure J., et al. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 2012;44:1277–1281. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondrashov A.S. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum. Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 5.Sun J.X., Helgason A., Masson G., Ebenesersdóttir S.S., Li H., Mallick S., Gnerre S., Patterson N., Kong A., Reich D., et al. A direct characterization of human mutation based on microsatellites. Nat. Genet. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemayel R., Vinces M.D., Legendre M., Verstrepen K.J. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 7.Warpeha K.M., Xu W., Liu L., Charles I.G., Patterson C.C., Ah-Fat F., Harding S., Hart P.M., Chakravarthy U., Hughes A.E. Genotyping and functional analysis of a polymorphic (CCTTT)(n) repeat of NOS2A in diabetic retinopathy. FASEB J. 1999;13:1825–1832. doi: 10.1096/fasebj.13.13.1825. [DOI] [PubMed] [Google Scholar]
- 8.Contente A., Dittmer A., Koch M.C., Roth J., Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 9.Hirakawa S., Lange E.M., Colicigno C.J., Freedman B.I., Rich S.S., Bowden D.W. Evaluation of genetic variation and association in the matrix metalloproteinase 9 (MMP9) gene in ESRD patients. Am. J. Kidney Dis. 2003;42:133–142. doi: 10.1016/s0272-6386(03)00416-5. [DOI] [PubMed] [Google Scholar]
- 10.Amador M.L., Oppenheimer D., Perea S., Maitra A., Cusatis G., Cusati G., Iacobuzio-Donahue C., Baker S.D., Ashfaq R., Takimoto C., et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139–9143. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- 11.Yi H., Song H., Hwang J., Kim K., Nierman W.C., Kim H.S. The tandem repeats enabling reversible switching between the two phases of β-lactamase substrate spectrum. PLoS Genet. 2014;10:e1004640. doi: 10.1371/journal.pgen.1004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verstrepen K.J., Jansen A., Lewitter F., Fink G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fondon J.W., Garner H.R. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. U.S.A. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baranowska Körberg I., Sundström E., Meadows J.R.S., Rosengren Pielberg G., Gustafson U., Hedhammar A., Karlsson E.K., Seddon J., Söderberg A., Vilà C., et al. A simple repeat polymorphism in the MITF-M promoter is a key regulator of white spotting in dogs. PLoS One. 2014;9:e104363. doi: 10.1371/journal.pone.0104363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drögemüller C., Karlsson E.K., Hytönen M.K., Perloski M., Dolf G., Sainio K., Lohi H., Lindblad-Toh K., Leeb T. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- 16.Lohi H., Young E.J., Fitzmaurice S.N., Rusbridge C., Chan E.M., Vervoort M., Turnbull J., Zhao X.-C., Ianzano L., Paterson A.D., et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- 17.Sureshkumar S., Todesco M., Schneeberger K., Harilal R., Balasubramanian S., Weigel D. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323:1060–1063. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 18.Fondon J.W., Martin A., Richards S., Gibbs R.A., Mittelman D. Analysis of microsatellite variation in Drosophila melanogaster with population-scale genome sequencing. PLoS One. 2012;7:e33036. doi: 10.1371/journal.pone.0033036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkan C., Kidd J.M., Marques-Bonet T., Aksay G., Antonacci F., Hormozdiari F., Kitzman J.O., Baker C., Malig M., Mutlu O., et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gymrek M., Golan D., Rosset S., Erlich Y. lobSTR: A short tandem repeat profiler for personal genomes. Genome Res. 2012;22:1154–1162. doi: 10.1101/gr.135780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Highnam G., Franck C., Martin A., Stephens C., Puthige A., Mittelman D. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 2013;41:e32. doi: 10.1093/nar/gks981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilmatre A., Highnam G., Borel C., Mittelman D., Sharp A.J. Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum. Mutat. 2013;34:1304–1311. doi: 10.1002/humu.22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tessereau C., Buisson M., Monnet N., Imbert M., Barjhoux L., Schluth-Bolard C., Sanlaville D., Conseiller E., Ceppi M., Sinilnikova O.M., et al. Direct visualization of the highly polymorphic RNU2 locus in proximity to the BRCA1 gene. PLoS One. 2013;8:e76054. doi: 10.1371/journal.pone.0076054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi K., Monjo T., Hoang P.H., Yoshimura J., Yurino H., Mitsui J., Ishiura H., Takahashi Y., Ichikawa Y., Goto J., et al. Rapid detection of expanded short tandem repeats in personal genomics using hybrid sequencing. Bioinformatics. 2013;30:815–822. doi: 10.1093/bioinformatics/btt647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brahmachary M., Guilmatre A., Quilez J., Hasson D., Borel C., Warburton P., Sharp A.J. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet. 2014;10:e1004418. doi: 10.1371/journal.pgen.1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ummat A., Bashir A. Resolving complex tandem repeats with long reads. Bioinformatics. 2014;30:3491–3498. doi: 10.1093/bioinformatics/btu437. [DOI] [PubMed] [Google Scholar]
- 27.Carlson K.D., Sudmant P.H., Press M.O., Eichler E.E., Shendure J., Queitsch C. MIPSTR: a method for multiplex genotyping of germline and somatic STR variation across many individuals. Genome Res. 2015;25:750–761. doi: 10.1101/gr.182212.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handsaker R.E., Van Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., McCarroll S.A. Large multiallelic copy number variations in humans. Nat. Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willems T.F., Gymrek M., Highnam G., Mittelman D., Erlich Y. The landscape of human STR variation. Genome Res. 2014;24:1894–1904. doi: 10.1101/gr.177774.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duitama J., Zablotskaya A., Gemayel R., Jansen A., Belet S., Vermeesch J.R., Verstrepen K.J., Froyen G. Large-scale analysis of tandem repeat variability in the human genome. Nucleic Acids Res. 2014;42:5728–5741. doi: 10.1093/nar/gku212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilgin Sonay T., Carvalho T., Robinson M., Greminger M., Krutzen M., Comas D., Highnam G., Mittelman D.A., Sharp A.J., Marques-Bonet T. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 2015;25:1591–1599. doi: 10.1101/gr.190868.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stegle O., Parts L., Durbin R., Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moen E.L., Zhang X., Mu W., Delaney S.M., Wing C., McQuade J., Myers J., Godley L.A., Dolan M.E., Zhang W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–996. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutierrez-Arcelus M., Lappalainen T., Montgomery S.B., Buil A., Ongen H., Yurovsky A., Bryois J., Giger T., Romano L., Planchon A., et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.-B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E., et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 48.Egbert R.G., Klavins E. Fine-tuning gene networks using simple sequence repeats. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16817–16822. doi: 10.1073/pnas.1205693109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corvelo A., Hallegger M., Smith C.W.J., Eyras E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 2010;6:e1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J.C. Helical repeat of DNA in solution. Proc. Natl. Acad. Sci. U.S.A. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- 53.Vinces M.D., Legendre M., Caldara M., Hagihara M., Verstrepen K.J. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Undurraga S.F., Press M.O., Legendre M., Bujdoso N., Bale J., Wang H., Davis S.J., Verstrepen K.J., Queitsch C. Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19363–19367. doi: 10.1073/pnas.1211021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.