Abstract
The interindividual differences in response to Chlamydia trachomatis (CT) infections are for an important part based on the differences in our host genetic make-up. In the past, several genes and pathways have been identified and linked to protection against or risk for CT infection (i.e. susceptibility), and/or the severity of infection, with a major emphasis on the development of tubal pathology, one of the main causes of female infertility. In the current study, we analyzed in Dutch Caucasian women whether the carriage of HLA-A G>A SNP (rs1655900) was related to the susceptibility of CT infection in a STD cohort (n = 329) and to the severity of infection in a subfertility cohort (n = 482). We also investigated if this A-allele was linked to increase in severity of symptoms, from mild symptoms (lower genital infection) to lower abdominal pain (upper genital tract infection) to the most severe late complication of tubal pathology, including double-sided tubal pathology. We showed that the carriage of HLA-A SNP rs1655900 studied is not associated with the susceptibility to CT infection based on the data from the STD cohort, but might be protective to the development of late complications (p = 0.0349), especially tubal pathology could be relevant.
Keywords: Chlamydia trachomatis, SNP, HLA, host genetic factors, tubal pathology, susceptibility
The A-allele of the HLA-A SNP rs1655900 might protect against late complications of CT infections.
Graphical Abstract Figure.
The A-allele of the HLA-A SNP rs1655900 might protect against late complications of CT infections.
INTRODUCTION
Chlamydia trachomatis infection is the most prevalent sexually transmitted disease (STD), and the prevalence of the infection is on the rise globally, with roughly 100 million new infections occurring each year (Starnbach and Roan 2008; Vasilevsky et al. 2014). Untreated C. trachomatis can lead to pelvic inflammatory disease, ectopic pregnancy and infertility due to tubal pathology (Wizel et al. 2008). However, remarkable differences in the clinical course of infection with C. trachomatis are observed between different individuals (Morré, Karimi and Ouburg 2009). For these differences, environmental factors such as coinfections may play a role (Hillis et al. 1994), but the differences can also be attributed to immunogenetic characteristics of the host. Understanding the immune mechanisms that underlie the pathogenesis of C. trachomatis infection has major implications for diagnostic and therapeutic approaches.
During a C. trachomatis infection, adaptive immune responses are initiated, activating CD4+ and CD8+ T cells (Geisler 2010; Neefjes et al. 2011). The role of CD8+ T cells has received increased interest due to the intracellular nature of C. trachomatis (Starnbach et al. 2003; Wizel et al. 2008). Pathogen-derived factors of C. trachomatis that access the host's cytosol are explored by several studies, since intracellular proteins are presented on the cell surface by Human Leukocyte Antigen class I (HLA-I), triggering CD8+ T-cell response (Kim et al. 1999; Fling et al. 2001; Starnbach et al., 2003; Gervassi et al. 2004). A CD8+ T-cell response involves induction of apoptosis of the infected cell through perforin and granzyme, enabling the cytolytic potential of CD8+ T cells. However, it has been found that CD8+ T cells in the female genital tract have limited perforin expression, downgrading their cytolytic potential during an initial C. trachomatis infection (Ibana et al. 2012). Variance in CD8+ T-cell functionality was not found to have a significant influence on the clearance of a C. trachomatis infection, but it was found to influence the development of pathogenesis, including infertility (Igietseme et al. 2009; Murthy et al. 2011).
Adequate pathogen recognition is essential to initiate the immune response. There is evidence that host genetic variation affects the clinical course of infection with C. trachomatis (Den Hartog et al. 2006; Bailey et al. 2009; Jiang et al. 2012; Al-Kuhlani et al. 2014). At this point, these factors appear to be the most promising biological indicators of complication after a chlamydial infection (Ouburg et al. 2009; Malogajski et al. 2013; Brankovic et al. 2014). Associations with particular single nucleotide polymorphisms (SNPs) are typically confirmed for genes coding for a range of immunological factors, such as cytokines, chemokines and antigen presentation components (Morré et al. 2000).
Successful presentation of chlamydial antigens enables the highly selective process of triggering a lymphocytic response. Determining the role of antigen presentation and elicitation of the immune responses is of crucial significance for the currently insufficient understanding of the immunopathology of genital infection with C. trachomatis (Geisler 2010). A number of alleles and suballeles in HLA genes have been found to be associated with susceptibility to chlamydial infection or associated pathologies (Morré, Karimi and Ouburg 2009). Since CD8+ T cells have been found to play an important role in complications after a C. trachomatis infection (Igietseme et al. 2009; Murthy et al. 2011; Ibana et al. 2012), an SNP in the gene region coding for HLA-I will be the focus of this study. A previous study presented by Kapil et al. (2013) stated that HLA-DQB1*05 had a protective effect for reinfection (p = 0.012, OR 2.6, 95% CI 1.2–5.6). In 2013, this work was presented orally at the STD & Aids World Congress and the HLA-A SNP rs1655900 was also presented as a possible candidate in their study in 199 African American subjects. We found the presented rs number to link to an SNP in the 3’UTR of HLA-A (ALFRED 2014). We studied this SNP (HLA-A rs1655900) in relation to (1) the susceptibility of genital infection with C. trachomatis, (2) occurrence of symptoms, and (3) for the severity of symptoms to the most severe form of double sided tubal pathology in infertile women.
METHODS
STD cohort
From a previous described cohort of 1150 Dutch Caucasian women, sufficient DNA was available from 329 samples to type the HLA-A SNP rs1655900 (Ouburg et al. 2005). In summary, between 2000 and 2004 data were collected from female patients who visited the STD outpatient clinic in Amsterdam, the Netherlands. Questionnaires responses were gathered about urogenital complaints, varying from increased discharge, having bloody discharge during and/or after coitus, recent lower abdominal pain (LAP)—not gastrointestinal or menstruation related—and/or dysuria. Chlamydia trachomatis status was assessed by Roche Amplicor PCR as previously described (Ouburg et al. 2005). Out of the 329 women, 128 (39%) were C. trachomatis positive (CT+) and 201 (61%) were C. trachomatis negative (CT−). Out of the CT+ women, 72 (56%) were asymptomatic, 56 (44%) were symptomatic and 16 (13%) had LAP.
Subfertility cohort
From 482 serum samples from Dutch Caucasian women, SNP data were available. These women were attending the fertility clinic of the University Medical Center Groningen, the Netherlands, and met the inclusion criteria for this study: C. trachomatis antibody test (CAT) result available, laparoscopic and/or hysterosalpingography (HSG) data available. Of the 482 serum samples, 471 had Chlamydia serology results available, and of these 413 (87.7%) were CT− and 58 (12.3%) were CT+ (pELISA, Medac Diagnostika mbH, Hamburg, Germany). Tubal pathology (TP) was defined as extensive periadnexal adhesions and/or distal occlusion of at least one tube. From 58 women who tested positive for C. trachomatis, 11 (19%) were diagnosed with TP. In this group, 55% were diagnosed with single-sided TP (TPss) and 45% double-sided TP (TPds). 378 (80%) women were diagnosed TP negative (TP−) through HSG. Of the CT+ women, 13 did not fulfill the criteria of TP and were not analyzed, of the CT− women 77 belonged to the HSG-0 group, which means that they were either not assessed, had mild HSG positivity but without reference to laparoscopy or other reasons, and were therefore not analyzed.
METC approval
The act ‘Medical Research Involving Human Subjects’ (WMO, Dutch Law) states that anonymous spare human materials and data may be used for research purposes after patients have been informed about this possible use and they have had the opportunity to object. All patients participating in the present study had not objected and therefore no ethical approval is required (MEC Letter reference: # 10.17.0046).
DNA isolation and SNP assessment
DNA was isolated with the High Pure PCR Template Preparation Kit (HPPTP Kit) according to the manufacturer's instructions (Roche Molecular, Mannheim, Germany). For the STD cohort 200 microliter cervical swab was resuspended in 1 ml 2SP, and for the subfertility cohort 200 microliter sera. The HLA-A G>A SNP rs1655900 was assessed using RT-PCR with detection on the LightCycler II (Roche Diagnostics, Basel, Switzerland) for the GG, AG and AA genotypes.
Statistical analyses
In the STD cohort, CT+ and CT− women were compared to each other, and within the CT+ group women with and without symptoms were compared. In the subfertility cohort the CT−TP− and the CT+TP+ were compared to each other, as well as women with and without tubal pathology within the CT+ group. To study increased severity, the following groups were compared for a trend in the occurrence of the HLA-A SNP studied: CT+ women with symptoms, to CT+ women with LAP, to women with TPss, and to women with TPds.
Data were compared between groups using Chi-square test and Fisher's exact test when appropriate. Risk factors were described as odds ratio (OR) with 95% confidence interval (CI). P-values < 0.05 were considered statistically significant. Analyses were performed using IBM SPSS Statistics. The regression coefficient (r2) for the line between CT+ women with symptoms, LAP, TPss and TPds was calculated in Microsoft Office Excel. In general, the higher the r2, the better the model fits the data. 80% explained variation is considered good, >90% is a very good fitting line and an association can be assumed.
RESULTS
The HLA-A SNP G>A (rs1655900) are shown in Table 1. The STD and Subfertility cohorts were in Hardy Weinberg Equilibrium.
Table 1.
Genotype distribution of rs1655900 in susceptibility and severity cohorts.
| RS1655900 G>A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| GG | % | GA | % | AA | % | Tot. | |||
| Susceptibility | CT− | 139 | 69.2% | 54 | 26.9% | 8 | 4.0% | 201 | |
| CT+ | 91 | 71.1% | 33 | 25.8% | 4 | 3.1% | 128 | ||
| AS | 51 | 70.8% | 17 | 23.6% | 4 | 5.6% | 72 | ||
| S | 28 | 70.0% | 12 | 30.0% | 0 | 0.0% | 40 | ||
| LAP | 12 | 75.0% | 4 | 25.0% | 0 | 0.0% | 16 | ||
| Severity | CT− | 278 | 67.3% | 113 | 27.3% | 22 | 5.3% | 413 | |
| CT− HSG− | 251 | 66.0% | 105 | 28.0% | 22 | 5.8% | 378 | ||
| CT+ | 42 | 72.4% | 14 | 24.1% | 2 | 3.4% | 58 | ||
| CT+ TP+ s.s. | 5 | 83.3% | 0 | 0.0% | 1 | 16.7% | 6 | ||
| CT+ TP+ d.s. | 5 | 100.0% | 0 | 0.0% | 0 | 0.0% | 5 | ||
| TP- | 21 | 61.8% | 13 | 38.2% | 0 | 0.0% | 34 | ||
CT: C. trachomatis; AS: asymptomatic; S: symptomatic; LAP: lower abdominal pain; TP: tubal pathology; s.s.: single-sided occlusion; d.s.: double-sided occlusion; +: positive; –: negative.
STD cohort: susceptibility to infections
There were no significant associations with the HLA-A SNP studied, between the CT+ and CT− women enrolled in this cohort or between those with and without symptoms (Table 1). The distribution of the genotypes (GG, GA, AA) is comparable in the two groups.
Subfertility cohort: severity of infection
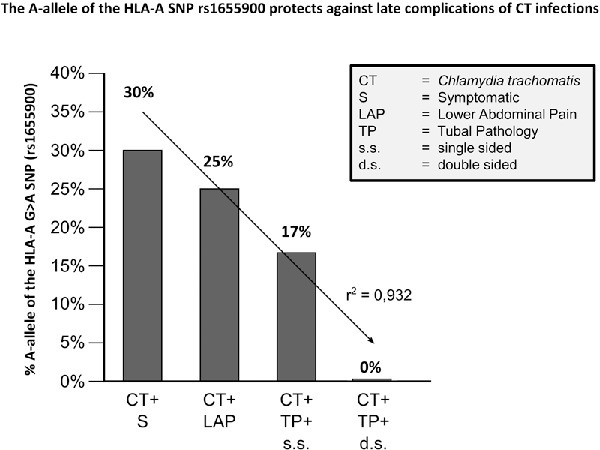
Women who tested negative for C. trachomatis and who had a negative result on the HSG were compared to women with C. trachomatis and tubal pathology for the occurrence of carriage of HLA-A rs1655900 * A. In this comparison, a protective trend is seen for women carrying the A-allele versus women with the G-allele (Table 1), since there are relatively more women with the A-allele in the CT− group. In the CT− group, there are 34% (127 GA&AACT-HSG-/378 GG&GA&AACT-HSG-) versus 9% (1 GA&AACT+TP+/11 GG&GA&AACT+TP+). The same protective trend was observed when solely looking at CT+ women with and without tubal pathology 38% (13 GA&AATP-/34 GG&GA*AATP-) versus 9% (1 GA&AACT+TP+/11 GG&GA&AACT+TP+). In all women with TP (n = 11), all women with TPds (n = 5) had the wild-type genotype (Table 1 and Fig. 1).
Figure 1.

Carriership of rs1655900 * A in C. trachomatis negative and positive women shows a decreasing trend in C. trachomatis induced complications. CT: C. trachomatis; HSG: hysterosalpingography; AS: asymptomatic; S: symptomatic; LAP: lower Abdominal Pain; TP: tubal pathology; s.s.: single-sided occlusion of the tubae; d.s.: double-sided occlusion of the tubae. Black line: trend based on the severity of Chlamydia infections (dark gray bars).
Increasing severity of infection among CT+ women
The occurrence of the carriage of HLA-A rs1655900 * A in patient groups with increasing severity—from CT+ women with symptoms to CT+ women with TPds—showed a statistically significant regression coefficient (r2) of 0.932 (P = 0.0349) (Fig. 1). This suggests that there is a correlation between the mutant allele and a less severe disease progression.
DISCUSSION
When analyzing the carriage of HLA-A rs1655900 * A in patient groups with respectively an increasing degree of general and mild symptoms, LAP, TPss and TPds, we observed a decreasing trend of carriage of the A-allele in our cohorts. Our findings point to a protective role of allele A over G, especially for symptoms and late complications in women who tested positive for C. trachomatis (r2: 0.932; P = 0.0349).
The SNP lies in the 3′-UTR intergenic region of HLA-A gene, which encodes for the heavy chain of the heterodimeric HLA-I molecule. The 3′-UTR region is not translated into the protein sequence, hence the SNP is unable to lead to a change of amino acids. It may, however, work by changing the level of post-transcriptional expression. A seemingly protective effect observed in our study would correspond to a reduction of expression of HLA-A transcript, resulting in less assembled HLA-I dimers and insufficient antigen presentation. A long-term consequence would be lower inflammation levels and less damage incurred by immune reactions to the local tissue. CD8+ T cells contribute to immune defense against C. trachomatis (Wizel et al. 2008); however, excessive cytolytic activity of these cells and the accompanying inflammation may be a major factor to contribute to the tubal damage and scarring (Cohen and Brunham 1999; Vasilevsky et al. 2014). Nevertheless, what Reddy et al. (2004) reported is a significant increase in the number of CD8+ T cells in Chlamydia-positive infertile women versus Chlamydia-negative women, which does point to their role in Chlamydia-related complications. Therefore, alleles of the HLA genes whose protein products present chlamydial antigens more successfully are expected to lead to a more pronounced inflammation and vice versa.
Our results indicate that the number of patients carrying A-allele increases based on the presence of symptoms, LAP and the degree of tubal damage. These findings would be congruent with a carriership of such allele that impairs antigen presentation to CD8+ T cells. The CD8+ T cells are specifically important in the immune response leading to infertility, and less perforin and tumor necrosis factor α production could protect the host from tissue damage (Murthy et al. 2011; Ibana et al. 2012). The implications of this type of study are, however, insufficient to make firm conclusions about biological mechanisms at hand and the findings warrant further studies. In earlier studies, other immunogenetic factors such as SNPs have been showed to be associated with the course of infection of C. trachomatis (Den Hartog et al. 2006; Morré, Karimi and Ouburg 2009; Ouburg et al. 2009). The current study contributes to the promising implications of SNPs as biological indicators in the diagnosis of complicated chlamydial infection (Malogajski et al. 2013; Brankovic et al. 2014).
Our study has several limitations. Most importantly, the precise function of rs1655900 is unknown based on the available literature. The reported findings could well be the outcome of the base change in the region involved in the regulation of translation; however, we might instead be observing the effect of another polymorphism that is in strong linkage to the examined SNP. Since HLA-A is not simply an HLA class 1a locus, it is an HLA class Ia locus embedded within HLA class Ib genes and its tightest linkage to non-self is with HLA class 1b, meaning a simple explanation on neither its function nor its potential linkage is easy to be given or analyzed. Furthermore, despite the considerable size of our total cohort in comparison to previous studies, the size of our severity groups does not offer sufficient statistical power.
It would therefore be valuable to attempt a reproduction of this study in a cohort with a higher number of patients with increasing symptomatology and especially tubal pathology. Also, functional analysis of the consequence of the base substitute on HLA-A expression levels would be useful to potentially corroborate these findings. In conclusion, the HLA-A G>A SNP studied is not associated with the susceptibility to infection but appears protective to the development of late complications, especially tubal pathology, after a C. trachomatis infection.
FUNDING
This work was supported by the following grants: Dutch NGI Life Sciences Pre-Seed Grant [Registration number 93611006]; USA NIH R21 Grant Prime [award number 1R21AI.098660-01; subaward number 0025996 (120407-1)] the European EuroTransBio Grant [Reference number 110012 ETB], and the Eurostars grant [ESTAR15106].
Acknowledgments
We want to thank the Public Health Laboratory, Cluster Infectious Diseases, Public Health Service Amsterdam (Amsterdam, the Netherlands) for the STD cohort included in this study. We want to thank Prof. Dr J.A. Land for the tuba factor infertility cohort enrolled in this study. We want to thank R. Heijmans, BASc for advice on HLA.
CONFLICT OF INTEREST
Prof. Dr S.A. Morré (S.A.M.), fulltime employee of the VU University Medical Center Amsterdam, is involved in the technical development of the SNP based assay to predict tubal pathology. This is done via TubaScan Ltd. (S.A.M. is Founder and Director), a spin-in company of the VU University Medical Center, Amsterdam, the Netherlands.
None of the other authors report a potential conflict of interest.
REFERENCES
- Al-Kuhlani M, Rothchild J, Pal S, et al. TRAIL-R1 is a negative regulator of pro-inflammatory responses and modulates long-term sequelae resulting from Chlamydia trachomatis infections in humans. PLoS One. 2014;9:e93939. doi: 10.1371/journal.pone.0093939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALFRED (The Allele Frequency Database) Polymorphism Information. Yale University; New Haven, Connecticut, USA: 2014. http://alfred.med.yale.edu/alfred/recordinfo.asp?condition=sites.site_uid=% (30 September 2015, date last accessed) [Google Scholar]
- Bailey RL, Natividad-Sancho A, Fowler A, et al. Host genetic contribution to the cellular immune response to Chlamydia trachomatis: heritability estimate from a Gambian twin study. Drugs Today. 2009;45:45–50. [PubMed] [Google Scholar]
- Brankovic I, Malogajski J, Morré SA. Biobanking and translation of human genetics and genomics for infectious diseases. Appl Transl Genomics. 2014;3:30–5. doi: 10.1016/j.atg.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CR, Brunham RC. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex Transm Infect. 1999;75:21–4. doi: 10.1136/sti.75.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hartog JE, Ouburg S, Land JA, et al. Do host genetic traits in the bacterial sensing system play a role in the development of Chlamydia trachomatis-associated tubal pathology in subfertile women? BMC Infect Dis. 2006;6:122. doi: 10.1186/1471-2334-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling SP, Sutherland RA, Steele LN, et al. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. P Natl Acad Sci USA. 2001;98:1160–5. doi: 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler WM. Duration of untreated, uncomplicated Chlamydia trachomatis genital infection and factors associated with chlamydia resolution: a review of human studies. J Infect Dis. 2010;201:S104–13. doi: 10.1086/652402. [DOI] [PubMed] [Google Scholar]
- Gervassi AL, Grabstein KH, Probst P, et al. Human CD8+ T cells recognize the 60-kDa cysteine-rich outer membrane protein from Chlamydia trachomatis. J Immunol. 2004;173:6905–13. doi: 10.4049/jimmunol.173.11.6905. [DOI] [PubMed] [Google Scholar]
- Hillis SD, Nakashima A, Marchbanks PA, et al. Risk factors for recurrent Chlamydia trachomatis infections in women. Am J Obstet Gynecol. 1994;170:801–6. doi: 10.1016/s0002-9378(94)70286-1. [DOI] [PubMed] [Google Scholar]
- Ibana JA, Myers L, Porretta C, et al. The major CD8 T cell effector memory subset in the normal and Chlamydia trachomatis-infected human endocervix is low in perforin. BMC Immunol. 2012;13:66. doi: 10.1186/1471-2172-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme JU, He Q, Joseph K, et al. Role of T lymphocytes in the pathogenesis of Chlamydia disease. J Infect Dis. 2009;200:926–34. doi: 10.1086/605411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Karimi O, Ouburg S, et al. Interruption of CXCL13-CXCR5 axis increases upper genital tract pathology and activation of NKT cells following chlamydial genital infection. PLoS One. 2012;7:e47487. doi: 10.1371/journal.pone.0047487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapil R, Tang J, Press C, et al. O01. 1 Association of genetic variants with Chlamydia trachomatis reinfection. Sex Transm Infect. 2013;89:A26. [Google Scholar]
- Kim S-K, Angevine M, Demick K, et al. Induction of HLA class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J Immunol. 1999;162:6855–66. [PubMed] [Google Scholar]
- Malogajski J, Brankovic I, Verweij SP, et al. Translational potential into health care of basic genomic and genetic findings for human immunodeficiency virus, Chlamydia trachomatis, and human papilloma virus. BioMed Res Int. 2013;2013 doi: 10.1155/2013/892106. http://dx.doi.org/10.1155/2013/892106 (18 December 2015, date last accessed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré S, Rozendaal L, Van Valkengoed I, et al. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J Clin Microbiol. 2000;38:2292–6. doi: 10.1128/jcm.38.6.2292-2296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré SA, Karimi O, Ouburg S. Chlamydia trachomatis: identification of susceptibility markers for ocular and sexually transmitted infection by immunogenetics. FEMS Immunol Med Micr. 2009;55:140–53. doi: 10.1111/j.1574-695X.2009.00536.x. [DOI] [PubMed] [Google Scholar]
- Murthy AK, Li W, Chaganty BK, et al. Tumor necrosis factor alpha production from CD8+ T cells mediates oviduct pathological sequelae following primary genital Chlamydia muridarum infection. Infect Immun. 2011;79:2928–35. doi: 10.1128/IAI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J, Jongsma ML, Paul P, et al. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–36. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- Ouburg S, Lyons J, Land J, et al. TLR9 KO mice, haplotypes and CPG indices in Chlamydia trachomatis infection. Drugs Today. 2009;45:83. [PubMed] [Google Scholar]
- Ouburg S, Spaargaren J, den Hartog JE, et al. The CD14 functional gene polymorphism -260 C>T is not involved in either the susceptibility to Chlamydia trachomatis infection or the development of tubal pathology. BMC Infect Dis. 2005;5:114. doi: 10.1186/1471-2334-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BS, Rastogi S, Das B, et al. Cytokine expression pattern in the genital tract of Chlamydia trachomatis positive infertile women–implication for T‐cell responses. Clin Exp Immunol. 2004;137:552–8. doi: 10.1111/j.1365-2249.2004.02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnbach MN, Loomis WP, Ovendale P, et al. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–9. doi: 10.4049/jimmunol.171.9.4742. [DOI] [PubMed] [Google Scholar]
- Starnbach MN, Roan NR. Conquering sexually transmitted diseases. Nat Rev Immunol. 2008;8:313–7. doi: 10.1038/nri2272. [DOI] [PubMed] [Google Scholar]
- Vasilevsky S, Greub G, Nardelli-Haefliger D, et al. Genital Chlamydia trachomatis: understanding the roles of innate and adaptive immunity in vaccine research. Clin Microbiol Rev. 2014;27:346–70. doi: 10.1128/CMR.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizel B, Nyström-Asklin J, Cortes C, et al. Role of CD8+ T cells in the host response to Chlamydia. Microbes Infect. 2008;10:1420–30. doi: 10.1016/j.micinf.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


