Abstract
APOBEC3s (A3) are Zn2+ dependent cytidine deaminases with diverse biological functions and implications for cancer and immunity. Four of the seven human A3s restrict HIV by 'hypermutating' the reverse-transcribed viral genomic DNA. HIV Virion Infectivity Factor (Vif) counters this restriction by targeting A3s to proteasomal degradation. However, there is no apparent correlation between catalytic activity, Vif binding, and sequence similarity between A3 domains. Our comparative structural analysis reveals features required for binding Vif and features influencing polynucleotide deaminase activity in A3 proteins. All Vif-binding A3s share a negatively charged surface region that includes residues previously implicated in binding the highly-positively charged Vif. Additionally, catalytically active A3s share a positively charged groove near the Zn2+ coordinating active site, which may accommodate the negatively charged polynucleotide substrate. Our findings suggest surface electrostatics, as well as the spatial extent of substrate accommodating region, are critical determinants of substrate and Vif binding across A3 proteins with implications for anti-retroviral and anti-cancer therapeutic design.
Keywords: APOBEC3, Vif, HIV
INTRODUCTION
The APOBEC family of enzymes, present in all primate genomes(LaRue et al., 2009), has been under positive selective pressure over 33 million years of evolutionary history(Sawyer et al., 2004). This has given rise to multiple sub-family members through gene duplication events(LaRue et al., 2008). Located on chromosome 22 in humans, the APOBEC3 proteins exhibit diverse subcellular localization, tissue expression patterns and biological functions. The human APOBEC3 family of cytidine deaminase enzymes play diverse biological roles such as restricting endogenous retroelements(Lovsin and Peterlin, 2009),(Muckenfuss et al., 2006), mitigating HIV-1 infection(Albin and Harris, 2010; Sheehy et al., 2002), epigenetic regulation(Carpenter et al., 2012; Guidotti et al., 2013; Guo et al., 2011; Wijesinghe and Bhagwat, 2012), and repairing double-stranded breaks in cellular DNA(Nowarski et al., 2012) . These enzymes are also strongly associated with various cancers(Alexandrov et al., 2013; Burns et al., 2013a; Burns et al., 2013b; Roberts et al., 2013).
Differences in expression of the various APOBEC3 proteins across various tissues types are correlated with tissue function(Barbosa-Desongles et al., 2013; Guidotti et al.; Koning et al.; Refsland et al.), pathological condition(Burns et al., 2013a; Burns et al., 2013b; Liu et al., 2013; Taylor et al., 2013) or immunological role(Okeoma et al., 2010; Wang et al.). Cellular compartmentalization varies in a similar manner, with the various APOBEC3 proteins localizing to the cytoplasm(Land et al.; Stenglein et al.; Vetter and D'Aquila), forming complexes with RNA(McDougall and Smith; Zhang et al.; Zhen et al.) and cellular proteins(Aynaud et al.; Jager et al.; Jager et al.), or translocating to the nucleus(Lackey et al.; Lackey et al.; Mussil et al.). Substrate specificity, leading to further specialization of these cytidine deaminase enzymes, is determined by nucleotides flanking the target (deoxy/methyl) cytidine(Carpenter et al.; Iwatani et al.; Rathore et al.; Rausch et al.; Wijesinghe and Bhagwat) with implications for immunity and epigenetic regulation.
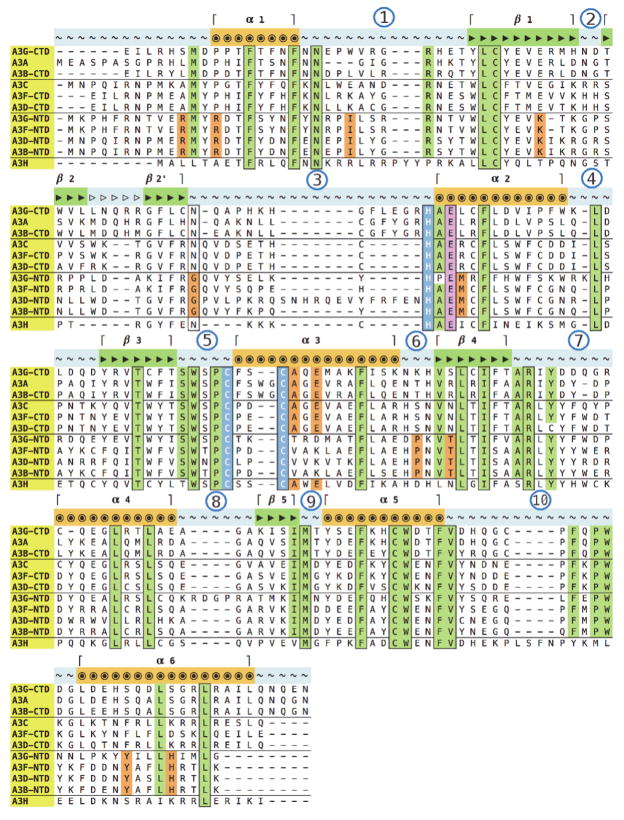
The structural basis underlying the physiological, cellular and biochemical characteristics mentioned above are not well understood. However, human APOBEC3 proteins are comprised of either one or two domains, subservient to the number of Zn2+ coordinating sites in the amino acid sequence(Conticello et al.; LaRue et al.; LaRue et al.). The single-domain proteins, APOBEC3A (A3A), APOBEC3C (A3C) and APOBEC3H (A3H), each have one Zn2+ coordinating site that is also the enzyme catalytic center. APOBEC3B (A3B), APOBEC3D (A3D), APOBEC3F (A3F) and APOBEC3G (A3G) each contain two Zn2+ coordinating sites located within nearly equal sized N- and C-terminal domains. The Zn2+ coordinating sites located within the C-terminal domains are known to be catalytically active in all double-domain proteins. However the Zn2+ coordinating sites within the N-terminal domains, which were never unequivocally demonstrated to be deamination capable. The Zn2+ coordinating sites in each APOBEC3 domain are classified based on signature sequence motifs into three domain classes, Z1, Z2 and Z3(LaRue et al.). A3A, A3B C-terminal domain (CTD) and A3G-CTD are Z1 domains. A3C, A3D-CTD, A3F-CTD and the N-terminal domains (NTD) of all double-domain APOBEC3 proteins are Z2 domains, whereas A3H is the sole member of the Z3 domain class (Fig. 1).
FIGURE 1.
(A): Schematic representation of domain organization in human APOBEC3 proteins: Each oval represents a domain, with the N-terminal (NTD) and C-terminal (CTD) domains in double-domain APOBEC3 proteins indicated by a black linker. The domains are colored by Z-domain sequence signature (Z1: green, Z2: orange, Z3: blue) and indicated as catalytically active by the * symbol. Domains that bind to HIV-1 Vif are indicated by the yellow coloration. (B): Protein sequence homology based phylogram based on the multiple sequence alignment in (Fig. 2). Branch coloring represents Z-domain signature as in (A) and numbers indicate branch lengths. Domains with solved structures are highlighted within boxes (yellow). The dashed open bracket (red) indicates the N-terminal domain cluster. (C): Pairwise sequence identity (%) between the seven target APOBEC3 domains and corresponding template homologs. The target and template domains are colored according to the Z-domain signatures as in (A). Identity values are colored on a green-to-red scale, from the highest to lowest sequence identity.
Recent studies have advanced the structural understanding of this family of proteins by solving NMR and high-resolution crystal structures of A3G-CTD (Chen et al.; Furukawa et al.; Harjes et al.; Holden et al.; Li et al.; Shandilya et al.), A3C(Kitamura et al.), A3F-CTD(Bohn et al.; Siu et al., 2013) and A3A(Byeon et al.). These structures have elucidated details of the enzyme active-site region, adjacent putative substrate binding regions, and structural features that may influence multimer assembly or binding to other host/non-host proteins. However, no structural information regarding any of the N-terminal domains of double-domain enzymes is currently available. These domains have been characterized to be essential for activity of the full-length protein with well-defined residues that play numerous roles including binding to polynucleotides, such as single-stranded DNA (ssDNA) or RNA, and interaction with protein binding partners: The A3B-NTD carries residues that signal nuclear localization and import(Lackey et al.). The A3F-NTD & A3G-NTD contribute both to DNA binding as well as enzymatic processivity(Ara et al., 2014; Chelico et al., 2006) and contain sequences required not only for binding HIV-1 Vif (that leads to their proteasomal degradation)(Song et al.; Xu et al.), but also for packaging into HIV virions(Navarro et al.). Individual APOBEC3 domains can exhibit differences in subcellular localization compared to the full-length enzyme; for example either domain of A3D individually expressed, may localize to the nucleus, but the full-length protein is cytoplasmic(Lackey et al.). Thus much remains to be understood of the functional significance of the full-length double domain APOBEC3s.
The structural data on the APOBEC3 domain family enables detailed analyses of the biophysical characteristics of these enzymes never previously possible. Attempts at modeling the structures of some of these A3 domains, based on previously published structures, have helped expand our understanding of the structural basis underlying their functional roles(Aydin et al., 2014; Desimmie et al., 2014). Structural comparisons can identify putative determinants of substrate orientation in the active-site region and provide insights into residues influencing substrate binding, specificity and product release. Comparative analysis of surface electrostatic profiles may identify regions that influence charge-driven interactions with other proteins or control movement across membranes or charge gradients, with implications for cellular organelle localization, formation of large complexes and packaging into virions. Loop-lengths may be key in regulation of enzymatic activity by auto-inhibition, differences in substrate binding, domain assembly, oligomerization, or conformational plasticity accessible to an APOBEC3 protein domain.
In this study, we present a comprehensive structural analysis of all human APOBEC3 protein domains based on sequence comparisons and recently published structures. A sequence similarity driven and comparative approach using the most appropriate templates enabled modeling the human APOBEC3 with yet undetermined structures. The analysis of structural, biochemical and biophysical characteristics of human APOBEC3s provides a unique view of domain properties based on sequence analysis, surface electrostatic potential and active-site region landscape, supported with myriad experimental data from current literature. The comparative analyses provide insights into structural features that may influence substrate binding and catalytic activity, as well as the ability to bind HIV-1 Vif.
MATERIALS & METHODS
Sequence Analysis
Target sequences were obtained from the NCBI Reference Sequence database (RefSeq(Pruitt et al.)). Canonical sequences of the template domains from RefSeq were used for sequence alignment, instead of the actual crystallized protein's sequence, as in the case of A3G-CTD (4 mutations and a 4 residue deletion in 3V4K(Li et al.)) and A3F-CTD (11 mutations in 4IOU(Bohn et al.)). Multiple sequence alignment was calculated using MUSCLE(Edgar) with default parameters, followed by phylogenetic tree analysis using RapidNJ(Simonsen and Pedersen) with 10,000 bootstrapped trees. The resulting tree was visualized and re-rooted with A3H, the most distant sequence, as the root node using Dendroscope(Huson and Scornavacca). Needleman-Wunsch pairwise sequence alignment of all APOBEC3 domains were performed using the program 'needle' from the EMBOSS suite(Rice et al.), with parameters gapopen = 10.0 and gapextend = 0.5. The best templates for each target sequence were selected based on sequence identities, calculated from the pairwise sequence alignments. Target sequences with pairwise sequence identity greater than 50% to a template were modeled based solely on that template (A3B-CTD, A3D-CTD and A3B-NTD). Target sequences with identity less than 50% but greater than 40% were modeled based on two of the most identical templates (A3D-NTD, A3F-NTD and A3G-NTD). Only one target sequence (A3H), had no templates with greater than 40% identity and was modeled based on three most identical templates.
Homology Modeling
All template structures (A3A: 2M65(Byeon et al.); A3C: 3VOW(Kitamura et al.); A3F-CTD: 4IOU(Bohn et al.); A3G-CTD: 3V4K(Li et al.)) were processed with the 'Protein preparation Wizard' in Maestro(Maestro) (Schrödinger, LLC). Target sequences and corresponding template structures were loaded in the 'Multiple Sequence Viewer' module and used for Knowledge-based homology modeling using Prime(Jacobson et al.; Jacobson et al.). The resulting homology models were subjected to energy minimization using the Prime 'Protein Refinement' module with the OPLS2005(Banks et al.) force field and the 'automatic' approach that applies a conjugate gradient method with large gradients and a truncated Newton method with smaller gradients. Final quality of the models was estimated from z-DOPE score(Eramian et al.) and the Total PRIME energy(Banks et al.) (Table 1).
TABLE 1.
The z-DOPE score(Eramian et al.) (as calculated by the Sali Lab ModEVAL server: http://modbase.compbio.ucsf.edu/modeval/) and Total PRIME energy(Banks et al.; Maestro) for the homology models used in this study (Fig. 4). All numbers rounded-off to one digit after decimal.
| A3 Domain | z-DOPE score | Total PRIME Energy (kCal/M) |
|---|---|---|
| A3B-NTD | −2 | −7.8 ×103 |
| A3B-CTD | −0.7 | −8.1 ×103 |
| A3D-NTD | −2.1 | −9.3 ×103 |
| A3D-CTD | −2.2 | −8.0 ×103 |
| A3F-NTD | −2 | −7.9 ×103 |
| A3G-NTD | −2.3 | −8.1 ×103 |
| A3H | −1.3 | −7.6 ×103 |
Structural Analysis
Molecular surfaces were calculated within Maestro (Schrödinger, LLC) and colored with electrostatic potential (ESP) in the range (−0.05 to +0.05 kcal/M) as red-white-blue, with red being negative ESP, blue positive and white neutral. SiteMap(Halgren) was used to identify and evaluate active-site/Zn2+-coordinating site volumes, using a 'Fine' grid to search around the Zn2+ atom in the template structures and homology models (pocket volumes reported in Table 3). Figures were generated using PyMOL(DeLano).
TABLE 3.
Pocket/groove volume around the active-site/Zn2+-coordinating-site in the 11 human APOBEC3 domains (solid green blobs in Figs. 7, 8) as calculated by SiteMap(Halgren; Maestro)(see Materials & Methods). In the context of double-domain APOBEC3 proteins, the catalytically active domain (indicated by *) consistently has a larger pocket volume than the inactive domain. All numbers rounded-off to one digit after decimal.
| A3 Domain | Pocket volume (A3) |
|---|---|
| A3A * | 4.1 ×102 |
| A3B-NTD | 3.0 ×102 |
| A3B-CTD * | 4.0 ×102 |
| A3C * | 2.1 ×102 |
| A3D-NTD | 0.7 ×102 |
| A3D-CTD * | 2.2 ×102 |
| A3F-NTD | 1.0 ×102 |
| A3F-CTD * | 1.9 ×102 |
| A3G-NTD | 0.6 ×102 |
| A3G-CTD * | 3.5 ×102 |
| A3H * | 1.6 ×102 |
RESULTS
Sequence and phylogenetic analysis reveal NTDs as a distinct subgroup
All APOBEC3 proteins/domains share regions of high sequence similarity as well as specific residues that are absolutely conserved, such as the catalytic center residues involved in coordinating Zinc as are revealed by multiple sequence alignment (Fig. 2). Certain regions are better conserved across specific subsets of these domains, indicating a stronger homology amongst a subset. These relationships are best illustrated by a phylogram (Fig. 1B) based on the multiple sequence alignment, that also reveals a grouping based on Z-domain signatures(LaRue et al.). The domains with Z2 signature sequences are further subdivided such that all the N-terminal domains form a distinct subgroup, suggestive of shared ancestry and structure/function characteristics, as suggested previously(LaRue et al., 2008).
FIGURE 2.
Multiple sequence alignment of all human APOBEC3 protein domain amino acid sequences. Residues conserved across all proteins are outlined in black with green highlight. Residues conserved across all, except one, sequence are highlighted in green and not outlined. Residues conserved only in either the C-terminal, or the N-terminal domain, are highlighted in orange. Zn2+-coordinating residues are outlined in black with blue highlights and the catalytic Glu residue is highlighted in pink. Secondary structure elements are indicated as per the A3G-CTD crystal structure (PDB: 3V4K), with α-helices indicated by circles, β-strands by filled right-handed arrows and loops by tilde symbols. The β-2/β-2' discontinuity (Fig. 3) is indicated by empty arrows, and loops are numbered by circled numerals.
Pairwise sequence alignments identified the best template structures for each of the seven target domain sequences (Fig. 1C). A3F-CTD and A3C, both Z2 domains, were determined as the best templates for the five Z2 targets and used individually or together for homology modeling (Materials & Methods). Similarly, A3A, a Z1 domain was identified as the best template for the only Z1 target, A3B-CTD. All template sequences were less than 35% identical to A3H, the only Z3 domain protein; therefore, one Z1 (A3G-CTD) and two Z2 (A3F-CTD, A3C) domain structures were used in combination as the best available templates. The sequence identity between all template and target sequence pairs is within range for building the main chain conformation with reasonable confidence(Chothia and Lesk), and is further supported by the absolute sequence conservation of the Zn2+ coordinating residues across all APOBEC3 domains (Fig. 2).
APOBEC3 domains share a common fold and Zinc-coordination architecture
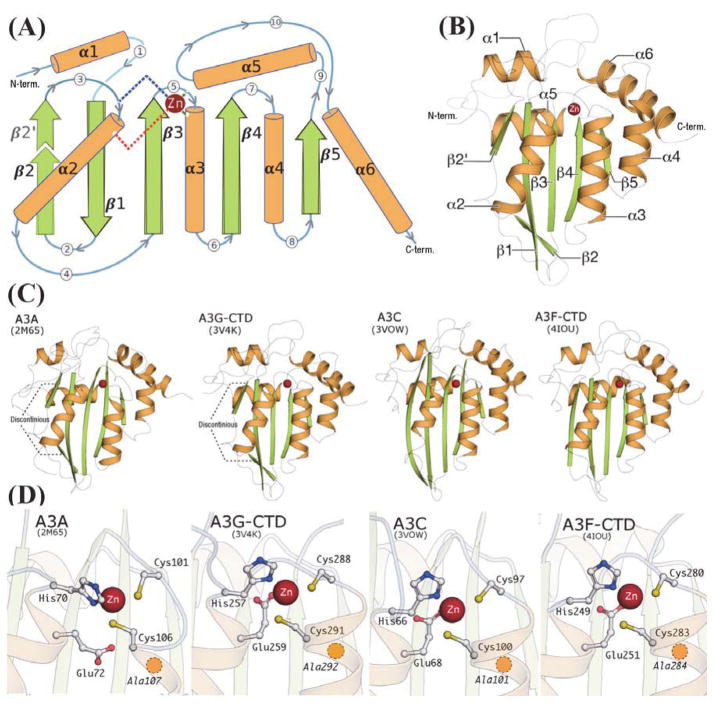
The template structures, irrespective of the Z-domain signature or sequence homology, share a common fold comprised of six α-helices and five β-strands (Fig. 3A, 3B). The relative arrangement of secondary structure elements, as demonstrated by the invariant α-helices, as well as the catalytic Zn2+-coordinating residues, one Histidine, two Cysteines and the catalytic Glutamate residue, follow a similar architecture across all template structures (Fig. 3D) as well as the homology models (Fig. 4).
FIGURE 3.
(A): Schematic representation of the secondary structure elements in APOBEC3 proteins: α-helices are depicted as cylinders (orange), β-strands as large arrows (green) and loops as curved lines (blue/grey) with encircled numerals indicating the loop-number as in Fig. 2. The β-2/β-2' discontinuity, observed only in the Z1-domain APOBEC3 proteins (A3A, A3G-CTD), is highlighted with a dotted outline and lighter coloration. The Zinc atom is represented as a sphere (dark red), with dashed lines representing the Zn2+-coordinating residues, Histidine (blue dashes), two Cysteines (dark green dashes) and the catalytic Glutamate (red dashes), drawn from their respective locations on the secondary structure. (B): Cartoon representation of A3G-CTD crystal structure (PDB: 3V4K), colored according to the secondary structure schematic in (A). (C): Cartoon representation of all four structures used as templates. The Z1-domain APOBEC3 proteins show the β-2/β-2' discontinuity. (D): Ball-and-stick representation of Zinc-coordinating residues at the active-site region of the template structures. The yellow circle with dashed outline, located just below C291 in A3G-CTD, indicates the position of A292.
FIGURE 4.
Cartoon representation of the homology models of human APOBEC3 domains depicting the overall domain architecture and secondary structure features (catalytic Zn2+ is represented by the dark red spheres).
However, differences in secondary structure features are correlated with Z-domain signature classes of the template structures (Fig. 3C). The Z1 domain templates, A3A and A3G-CTD have a distinct break or 'kink' in the β2-strand(Byeon et al.; Harjes et al.), whereas the Z2 domain structures, A3C and A3F-CTD have a continuous β2-strand (Fig. 3C). The model of A3B-CTD, a Z1 domain sequence, has a similar 'kink' much like the other Z1 domain structures A3G-CTD and A3A (Fig. 3, 4). This 'kink' is absent in all other models, with the exception of A3G-NTD and A3H that exhibit a somewhat shorter discontinuous segment in the β2-strand region compared to the Z1 domain proteins (Fig. 4). The homologous region in APOBEC2 is continuous, without any ‘kink’ or break and based on the arrangement of protein chains in the APOBEC2 crystal structure, this region was believed to influence dimerization(Prochnow et al., 2007), but there is no direct evidence of a similar role in the case of APOBEC3 proteins. The role for the varied structure β2-strand region remains elusive, but perhaps plays a role in differential recognition at a molecular level amongst the A3 proteins.
Loop lengths also differ across Z-domains, most significantly in the loop 3 region (Fig. 2, 3), with A3A and A3G-CTD having a longer loop 3 than A3C or A3F-CTD. The loop 3 of A3G-CTD is involved in coordinating an intermolecular Zn2+ ion in the A3G-CTD crystal structure and is a functionally relevant region based on cellular and biochemical evidence(Li et al.; Shandilya et al.). A3A loop 5 is extended by insertion of two residues 104WG105) between the two Zn2+-coordinating Cysteine residues (Fig. 2, 3). This feature, also observed in the A3B-CTD sequence, is a unique feature of this Z1- domain sub-branch.
Surface electrostatics of Vif-interacting regions differ in Vif-binders and non- binders
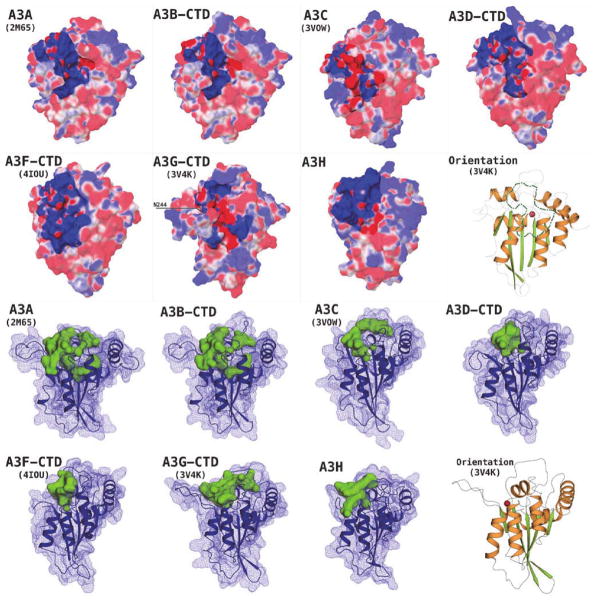
Biophysical characteristics, such as surface electrostatic potential, play an important role in protein-protein interactions. The isoelectric point (pI) of HIV-Vif, based on amino acid sequence, is estimated to be around 9.9, implying that Vif is likely highly positively charged at physiological pH. Similar calculations for the human APOBEC3 domains reveal a wide range of pI values (Table 2). Surprisingly, there is no apparent correlation between the pI of an APOBEC3 domain and ability to bind Vif (Fig. 1). Vif-binding APOBEC3 domains include A3C(Kitamura et al.; Smith and Pathak), A3D-CTD(Smith and Pathak), A3F-CTD(Albin and Harris; Smith and Pathak), A3G-NTD(Conticello et al.; Huthoff and Malim; Russell et al.) and A3H(Hultquist et al.; Zhen et al.). The HIV-1 Vif interaction generally results in proteasomal degradation of the corresponding APOBEC3(Hultquist et al., 2011; Wiegand et al., 2004; Yu et al., 2003). The pI values of these domains range from 5.9 (A3G-NTD) to 9.3 (A3D-CTD), indicating that interaction between Vif and these domains is driven by more nuanced factors than merely the overall charge. Similarly, the calculated pI values of Vif non-binding domains, including A3A(Jager et al.), A3B-NTD(Doehle et al.; Rose et al.), A3B-CTD(Doehle et al.; Rose et al.), A3D-NTD(Smith and Pathak; Zhang et al.), A3F-NTD(Russell et al.; Zhang et al.) and A3G-CTD(Conticello et al.; Schrofelbauer et al.), exhibit a range from 5.1 (A3B-NTD) to 9.4 (A3G-CTD), suggesting that overall charge alone is too simplistic and insufficient to explain the lack of binding. A closer inspection of the surface electrostatic potentials, when combined with experimental data from published literature, highlights the stark contrast in charge distribution across regions that include residues critical for binding Vif as compared to similar regions in domains that do not bind Vif (Fig. 5) and may help better comprehend the details of Vif-A3 binding.
TABLE 2.
Theoretical isoelectric point (pI) values for the human APOBEC3 domains, based on the primary amino acid sequence. (Calculated using the Compute pI/MW tool located at http://web.expasy.org/compute_pi/). Domains known to bind HIV-1 Vif (Fig. 1) are indicated by an asterisk (*). All numbers rounded-off to one digit after decimal.
| Protein | NTD (pI) | CTD (pI) | Full Length (pI) |
|---|---|---|---|
| A3A | 6.3 | ||
| A3B | 5.1 | 8.1 | 5.9 |
| A3C | * 7.5 | ||
| A3D | 7.2 | * 9.3 | 8.7 |
| A3F | 5.5 | * 8.9 | 6.9 |
| A3G | * 5.9 | 9.4 | 8.3 |
| A3H | * 9.2 |
FIGURE 5.
Electrostatic surface potential (ESP) of the Vif-binding APOBEC3 domains, delineating putative Vif-binding regions in green/white outline. ESP range (−0.05 to +0.05 kcal/M) as red-white-blue, with red being negative ESP, blue positive and white neutral. All domains oriented as indicated by the cartoon representation of A3F-CTD crystal structure.
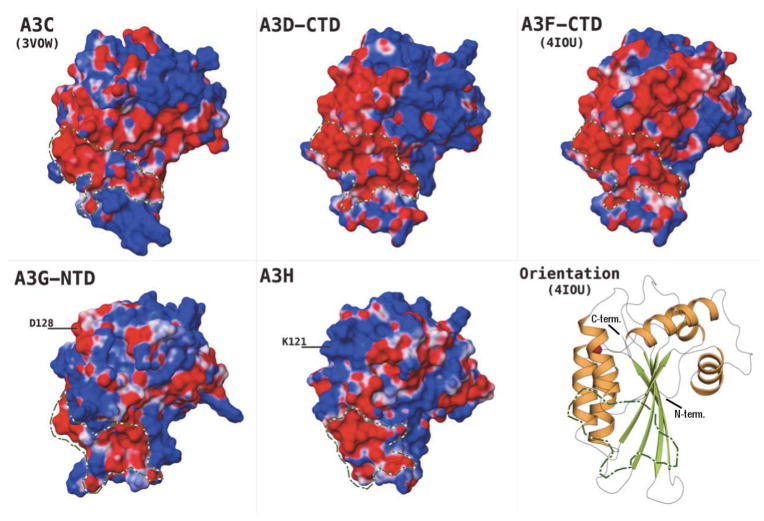
Based on surface electrostatic potential, a contiguous, negatively charged region is identifiable in the Vif-binding domains (Fig. 5). The majority of this region can be traced to include parts of the α–3 and α–4 helices, loops 6 and 8, and surface accessible regions of the β–3, β–4 and β–5 sheets, in nearly all these domains. These regions have been identified in previous studies by single point mutations as well as a protein-wide MAPPIT study(Lavens et al., 2010), to include residues required for interaction with Vif in A3C(Kitamura et al.; Smith and Pathak), A3D-CTD(Smith and Pathak) and A3F-CTD(Albin and Harris; Smith and Pathak). The primary residue implicated for Vif-binding in the case of A3G-NTD is D128(Huthoff and Malim; Santa-Marta et al.; Xu et al.), located on loop 7, is diametrically opposite the region being discussed (Fig. 5). This apparent discrepancy between the single-domain A3C and double-domain A3G/F suggests Vif-binding may be influenced by inter-domain packaging and oligomerization in the double-domain A3G/F. Based on the role played by oligomerization in double-domain A3 function, for example from Atomic Force Microscopy (AFM)(Shlyakhtenko et al., 2014; Shlyakhtenko et al., 2011; Shlyakhtenko et al., 2012, 2013) and Fluorescence Fluctuation Spectroscopy (FFS) (Li et al., 2014), homodimer assembly and/or interactions between NTD and CTD may help create the surface shape and charge characteristics that are eventually required to bind Vif. Thus, the role of single, binding-critical residues, such as D128 in A3G, may be exerted not only by the currently accepted, direct interaction of Vif-A3G, but also by indirectly influencing the interaction interface. Similarly, residue E121 in A3H, a critical determinant for Vif binding(Zhen et al.), is located on the α–4 helix and close to loop 7 in the A3H homology model (labeled as K121 in Fig. 5 as the target A3H sequence used for his study is the isoform 1 (NP_001159475.1) sequence). A3H isoforms that contain K105 and E121 are known to show stronger anti-viral activity against HIV-1(Zhen et al.). Even though residues in remote locations are documented to be critical for binding to Vif, the contiguous, negatively charged region (Fig. 5) identified here, appears to be a feature shared amongst Vif-binding A3 domains, and is absent or interrupted by positively charged patches in the non Vif-binding domains (Fig. 6). This observation provides additional clues to the role that surface charge may play in Vif-APOBEC3 interactions, but further biochemical studies and structural data from Vif-A3 complexes are required for their verification.
FIGURE 6.
Electrostatic surface potential (ESP) of the Vif non-binding APOBEC3 domains, delineating in green/white outline the regions contrasting with Vif-binding domains in Fig. 5. ESP range (−0.05 to +0.05 kcal/M) as red-white-blue, with red being negative ESP, blue positive and white neutral. All domains oriented identical to the cartoon representation of A3F-CTD crystal structure in Fig. 5.
Surface charge & substrate accommodating pocket volume may influence binding affinity & catalytic ability
APOBEC3 proteins are known to bind polynucleotide sequences, primarily ssDNA, the catalytic substrate(Chelico et al.; Conticello et al.; Jarmuz et al.; Suspene et al.), as well as specific types of RNA(Belanger et al.; Huthoff et al.; Khan et al.), such as the 7SL RNA(Bach et al.). Binding is a prerequisite for catalysis of target cytidine(s) on the substrate ssDNA, which may be influenced by multiple biophysical factors, including surface electrostatic potential and the presence or absence of favorable inter-atomic contacts in the substrate binding regions. To investigate likely determinants of substrate binding and subsequent catalysis, the human APOBEC3 domain structures and homology models were compared with respect to surface electrostatic potential and substrate accommodating residues that may contribute to favorable interactions with polynucleotides.
Comparing catalytically active and inactive APOBEC3 domains revealed differential structural patterns surrounding the potential substrate binding regions. The catalytically active APOBEC domains, A3A(Bulliard et al.), A3B-CTD(Bogerd et al.), A3C(Bourara et al.; Harris et al.), A3D-CTD(Dang et al.), A3F-CTD(Hache et al.), A3G-CTD(Newman et al.; Shindo et al.) and A3H(OhAinle et al.), display a deep, well-formed groove carrying a primarily positive charge on the surface (Fig. 7). The larger part of this groove, formed by residues from loops 1, 3, 5 and 7, lies across and spans the active-site pocket, containing the Zinc and the catalytic Glutamate residue (Fig. 2, pink highlight; Fig. 3), suggesting a role in accommodating the negatively charged phosphodiester backbone of the substrate ssDNA during substrate binding and subsequent deamination of the target cytidine. The groove further extends along the outer surfaces of α–2 and α–3 helices (Figs. 3, 7), which although comparatively less positively charged exhibit a similar shape profile (Fig. 7). Loop 7, which has been implied in DNA motif recognition, in A3G and A3F (5’–CCC–3’ vs 5’–TC–3’ nucleotide preference), in a number of previous studies (Carpenter et al., 2010; Holden et al., 2008; Kohli et al., 2009; Siu et al., 2013), forms one of the boundaries of this groove. The N-terminal domains are catalytically inactive (with the catalytic capability of A3B-NTD debated(Bogerd et al.; Hakata and Landau; Pak et al.)). These inactive domains reveal comparatively shallower grooves, with A3F-NTD and A3G-NTD being the most explicit examples (Fig. 8). The groove profile can be characterized by SiteMap analysis of the active and inactive domains, which highlights the extent of the pocket that exists around the Zn2+-coordinating site. Although the absolute volume (Table 3) does not directly distinguish the pockets, the shapes of the pockets are quite distinct (Figs. 7, 8). With the exception of A3B-NTD, the pocket volume of a catalytically inactive domain is less than any of the active domains (Table 3). An explanation for A3B-NTD being the outlier may lie with the seven-residue insertion in the loop 3 region, that borders the active-site pocket region and in the absence of template residues in the A3C structure (Fig. 1), is modeled using de-novo approach by the homology modeling algorithm, driven primarily by avoidance of energetically unfavorable clashes, resulting in a larger pocket than is observed in other NTDs.
FIGURE 7.
Upper panel Electrostatic surface potential (ESP) of catalytically active human APOBEC3 domains, highlighting the putative substrate-binding region around the Zn2+-coordinating (active) site. ESP range (−0.05 to +0.05 kcal/M) as red-white-blue, with red being negative ESP, blue positive and white neutral. Lower panel: The substrate-binding groove/pocket at the active-site region is larger and extends deep into the protein (solid green blobs; Table 3), in catalytically active APOBEC3 proteins; the protein surface is shown as a dark-blue mesh and secondary structure in cartoon representation. The domain orientation is indicated by the cartoon representation of A3G-CTD crystal structure with the catalytic Zn2+ atom shown as red sphere, in both panels.
FIGURE 8.
Left panel Catalytically inactive human APOBEC3 domains, contrasting the Zn2+-coordinating region's electrostatic surface potential (ESP) to the catalytically active domains in Fig. 7. ESP range (−0.05 to +0.05 kcal/M) as red-white-blue, with red being negative ESP, blue positive and white neutral. Right panel: The pocket/groove around the Zn2+-coordinating region is smaller and does not extend deep into the protein (solid green blobs; Table 3), in the catalytically inactive APOBEC3 proteins; the protein surface is shown as a dark-blue mesh and secondary structure in cartoon representation. The domain orientation is indicated by the cartoon representation of A3G-CTD crystal structure, as in the corresponding panels of Fig. 7.
The sequence of loop 3, containing N244 in A3G-CTD and located near the active-site region, also distinguishes active from inactive domains. Multiple sequence alignment reveal that Asn is conserved at this position in all active domains, and Gly in all inactive domains (Fig. 2, outlined, start of loop 3), confirming a role in substrate recognition and processing, consistent with previous reports(Bransteitter et al.; Holden et al.). Another conserved change is linked with A292 in the A3G-CTD (Fig. 2, residue after the last blue box) that lies near the “floor” of the active-site region (Fig. 3), where a bulkier residue is located within inactive domains. These bulkier side chains appear to result in a shallower cytidine non-accommodating pocket (Fig. 8). Thus, structural features combined with sequence comparisons suggest residues 244 and 292 (A3G-CTD numbering) are likely contributory determinants of catalytic activity in APOBEC3 domains.
DISCUSSION
This comprehensive computational analysis of all human APOBEC3 proteins provides insights into structural similarities and differences across this family of domains are likely relevant to substrate binding, catalysis and binding to the HIV-1 protein Vif. Recently determined structures of some APOBEC3 protein domains enabled generating reliable models of the remaining APOBEC3 domains using the most homologous domains as templates. Although there have been previous efforts modeling APOBEC3 protein structures(Albin et al.; Albin et al.; Aydin et al., 2014; Belanger et al.; Bulliard et al.; Bulliard et al.; Chelico et al.; Chen et al.; Desimmie et al., 2014; Harjes et al.; Lackey et al.; Lavens et al.; Shirakawa et al.; Stauch et al.; Stenglein et al.; Zhang et al.; Zhen et al.; Zhen et al.), a comprehensive and comparative analysis encompassing all human A3 domains has not been published. Our analysis focuses on comparing the key characteristics relevant to the primary biological functions of the APOBEC3s.
The surface electrostatic characteristics common to putative Vif-binding regions suggest charge complementarity may play a critical role in Vif interactions of APOBEC3 domains. Vif is an intrinsically disordered protein with a very high isoelectric point, and is likely highly positive charged under physiological conditions(Auclair et al.). The contiguous negatively charged region identified in Vif-binding APOBEC3 proteins is thus the most probable determinant driving the interaction with Vif, and correlates well with residues implicated in Vif binding in A3C(Kitamura et al.) and A3F-CTD(Bohn et al.; Kitamura et al.), the two Vif-binding APOBEC3 domains of known structure. More recently, negative surface charge at the region of α–3 and α–4 helices in A3F, was shown to mediate sensitivity to Vif(Land et al., 2014). Complementing this observation, the published structures of APOBEC3 domains that do not bind Vif, A3A(Byeon et al.) and A3G-CTD(Li et al.; Shandilya et al.), have a largely positive charge distribution across the same regions. The outstanding exception to this pattern appears in the A3H homology model, which is overwhelmingly positively charged as per the calculated electrostatic surface, including near known Vif-binding residues (K121) in the homologs.
Surface electrostatic potential is also a likely critical factor in binding the negatively charged ssDNA substrate, as can be observed in nearly all APOBEC3 domains. Experimental mutagenesis of residues in and around the active-site region supports the role of positively charged residues in binding the negatively charged substrate, as demonstrated specifically for A3G-CTD, where this region was also referred to as the Arginine rich “brim-domain”(Chen et al.; Furukawa et al., 2009; Shindo et al., 2012). Other studies have shown that introducing negative charge in this region can abrogate binding to ssDNA, for example, by phosphorylation of residue T218(Demorest et al.). Since the primary negative charge is carried by the phosphodiester backbone in ssDNA, the presence of substrate (dC) or product (dU) bases in the polynucleotide sequence is less likely to impact this primarily charge-driven interaction when compared to other biochemical factors like pH. Surface charge is likely to be strongly influenced by pH in the local environment, with implications for ssDNA binding, as reported recently for A3A (Pham et al., 2013), and thus play a critical role in enzymatic activity.
Topographical surface charge complementarity may contribute a significant component of the A3 domains’ affinity for positively charged Vif and negatively charged substrate ssDNA backbone, but is not sufficient to explain catalytic ability. The deamination reaction likely requires flipping the substrate cytidine base into the Zn2+-coordinating (“active-site”) region, as is observed in the case of other polynucleotide deaminases; for example the S. aureus t-RNA adenosine deaminase (TadA), where the substrate base is flipped out of the stacking arrangement and inserted into the enzyme active site(Losey et al.). The catalytically inactive domains may bind to the negatively charged ssDNA backbone based on general charge complementarity, but may fail to position the targeted cytidine’s (-NH2) group at the enzymatic center to trigger the catalytic cascade(Snider et al., 2002). The positioning of the target base for catalysis would also require sufficient spatial volume to correctly orient the base at the reaction center and also accommodate adjacent portions of the polynucleotide chain. The Zn2+-coordinating region, in the catalytically inactive APOBEC3 domains, appears to have smaller pocket volumes for substrate binding than the catalytically active domains, as evidenced by the shallower and narrower grooves in the corresponding region. A deeper exploration of the role played by pocket depth and substrate accommodating region by future experiments will help in understanding the mechanistic basis of this observation and potential biological significance.
The surface charge profiles, as well as the substrate accommodating volume around the Zn2+-coordinating region appear capable of influencing APOBEC3 binding to Vif as well substrate ssDNA. However, the same features may also be important in other roles, such as oligomerization, for instance in the case of A3G which forms High Molecular Mass complexes(Chiu et al.) in the cytoplasm, binding to non-substrate polynucleotides, such as RNA, and possibly play a role in packaging of these potent anti-HIV proteins into the virions(Li et al., 2014; Wang et al.; Zhang et al.). Further details of the functional importance of these features will emerge as experimental data on various APOBEC3 domains become available from future investigations. Regardless, the structural features identified here open the possibility of the design and development of specific agents targeting the Vif-APOBEC3 interaction for anti-retroviral effect as well as strategies targeting the disruption of polynucleotide binding interaction to develop anti-cancer agents.
The models generated for this study have been deposited at the “Model Archive”, part of the “Protein Model Portal”(Haas et al., 2013), and are available for download as PDB format files.
Research highlights (for review).
We generated homology models and analyzed published structures of A3 domains
Vif-binding A3 domains have a contiguous, negatively-charged surface patch
Catalytically-active domains have positively-charged regions around active-site
Catalytically-active A3 domains have larger active-site pockets vs inactive domains
Acknowledgments
The authors thank Dr. Mohan Somasundaran, Dr. Nese Kurt-Yilmaz and Ms. Tania Silvas for helpful discussions during the preparation of this manuscript. This work was supported by National Institutes of Health Institute of General Medical Sciences Grant P01 GM091743.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albin JS, Anderson JS, Johnson JR, Harjes E, Matsuo H, Krogan NJ, Harris RS. Dispersed sites of HIV Vif-dependent polyubiquitination in the DNA deaminase APOBEC3F. Journal of molecular biology. 2013 doi: 10.1016/j.jmb.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert reviews in molecular medicine. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, LaRue RS, Weaver Ja, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. The Journal of biological chemistry. 2010;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara A, Love RP, Chelico L. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS pathogens. 2014;10:e1004024. doi: 10.1371/journal.ppat.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair JR, Green KM, Shandilya S, Evans JE, Somasundaran M, Schiffer CA. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–284. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H, Taylor MW, Lee JE. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Aynaud MM, Suspene R, Vidalain PO, Mussil B, Guetard D, Tangy F, Wain-Hobson S, Vartanian JP. Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A. The Journal of biological chemistry. 2012;287:39182–39192. doi: 10.1074/jbc.M112.372722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM. Integrated Modeling Program, Applied Chemical Theory (IMPACT) Journal of computational chemistry. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Desongles A, Hernandez C, Simo R, Selva DM. Testosterone induces cell proliferation and cell cycle gene overexpression in human visceral preadipocytes. American journal of physiology. Cell physiology. 2013;305:C355–359. doi: 10.1152/ajpcell.00019.2013. [DOI] [PubMed] [Google Scholar]
- Belanger K, Savoie M, Rosales Gerpe MC, Couture JF, Langlois MA. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic acids research. 2013 doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourara K, Liegler TJ, Grant RM. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS pathogens. 2007;3:1477–1485. doi: 10.1371/journal.ppat.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cellular and molecular life sciences : CMLS. 2009;66:3137–3147. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, Michielin O, Weitzman MD, Trono D. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J Virol. 2009;83:12611–12621. doi: 10.1128/JVI.01491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013a;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nature genetics. 2013b;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nature communications. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL, Harris RS. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. The Journal of biological chemistry. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA repair. 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3'--> 5' on single-stranded DNA. Nature structural & molecular biology. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chelico L, Prochnow C, Erie Da, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. The Journal of biological chemistry. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Letters. 2007;581:4761–4766. doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. The EMBO journal. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Current biology : CB. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Molecular Biology and Evolution. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WLea. The PyMOL Molecular Graphics System. Schrodinger, LLC; 2014. [Google Scholar]
- Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. The Journal of biological chemistry. 2011;286:26568–26575. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. Journal of molecular biology. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramian D, Eswar N, Shen MY, Sali A. How well can the accuracy of comparative protein structure models be predicted? Protein science : a publication of the Protein Society. 2008;17:1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO Journal. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR. DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcoholism, clinical and experimental research. 2013;37:417–424. doi: 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas J, Roth S, Arnold K, Kiefer F, Schmidt T, Bordoli L, Schwede T. The Protein Model Portal--a comprehensive resource for protein structure and model information. Database : the journal of biological databases and curation. 2013;2013:bat031. doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Liddament MT, Harris RS. The Retroviral Hypermutation Specificity of APOBEC3F and APOBEC3G is Governed by the C-terminal DNA Cytosine Deaminase Domain. Journal of Biological Chemistry. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. The Journal of biological chemistry. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- Halgren TA. Identifying and characterizing binding sites and assessing druggability. Journal of chemical information and modeling. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- Harjes E, Gross PJ, Chen KMM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. Journal of molecular biology. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Molecular cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic biology. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS pathogens. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical Activities of Highly Purified, Catalytically Active Human APOBEC3G: Correlation with Antiviral Effect. Journal of Virology. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. Journal of molecular biology. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Global landscape of HIV-human protein complexes. Nature. 2012a;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012b;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nature structural & molecular biology. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. The Journal of biological chemistry. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L, Demorest ZL, Land AM, Hultquist JF, Brown WL, Harris RS. APOBEC3B and AID have similar nuclear import mechanisms. Journal of molecular biology. 2012;419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, Harris RS. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. The Journal of biological chemistry. 2013;288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AM, Shaban NM, Evans L, Hultquist JF, Albin JS, Harris RS. APOBEC3F Determinants of HIV-1 Vif Sensitivity. J Virol. 2014 doi: 10.1128/JVI.02362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. Journal of Virology. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Jonsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hotzel I, Andresdottir V, Smith TP, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavens D, Peelman F, Van der Heyden J, Uyttendaele I, Catteeuw D, Verhee A, Van Schoubroeck B, Kurth J, Hallenberger S, Clayton R, Tavernier J. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic acids research. 2010;38:1902–1912. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. Journal of molecular biology. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS chemical biology. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Calhoun VD, Chen J, Claus ED, Hutchison KE. Effect of homozygous deletions at 22q13.1 on alcohol dependence severity and cue-elicited BOLD response in the precuneus. Addiction biology. 2013;18:548–558. doi: 10.1111/j.1369-1600.2011.00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nature structural & molecular biology. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- Lovsin N, Peterlin BM. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Annals of the New York Academy of Sciences. 2009;1178:268–275. doi: 10.1111/j.1749-6632.2009.05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro. Maestro. Schro dinger, LLC; New York, NY: 2011. [Google Scholar]
- McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochemical and biophysical research communications. 2011;412:612–617. doi: 10.1016/j.bbrc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. Journal of Biological Chemistry. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Mussil B, Suspene R, Aynaud MM, Gauvrit A, Vartanian JP, Wain-Hobson S. Human APOBEC3A Isoforms Translocate to the Nucleus and Induce DNA Double Strand Breaks Leading to Cell Stress and Death. PloS one. 2013;8:e73641. doi: 10.1371/journal.pone.0073641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Current biology : CB. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nowarski R, Wilner OI, Cheshin O, Shahar OD, Kenig E, Baraz L, Britan-Rosich E, Nagler A, Harris RS, Goldberg M, Willner I, Kotler M. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–375. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive Evolution and Antiviral Activity of the Conserved Mammalian Cytidine Deaminase APOBEC3H. Journal of Virology. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Huegel AL, Lingappa J, Feldman MD, Ross SR. APOBEC3 proteins expressed in mammary epithelial cells are packaged into retroviruses and can restrict transmission of milk-borne virions. Cell host & microbe. 2010;8:534–543. doi: 10.1016/j.chom.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V, Heidecker G, Pathak VK, Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J Virol. 2011;85:8538–8547. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Landolph A, Mendez C, Li N, Goodman MF. A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer. The Journal of biological chemistry. 2013;288:29294–29304. doi: 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore A, Carpenter MA, Demir O, Ikeda T, Li M, Shaban NM, Law EK, Anokhin D, Brown WL, Amaro RE, Harris RS. The Local Dinucleotide Preference of APOBEC3G Can Be Altered from 5'-CC to 5'-TC by a Single Amino Acid Substitution. Journal of molecular biology. 2013 doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. The Journal of biological chemistry. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic acids research. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in genetics : TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature genetics. 2013 doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS research and human retroviruses. 2005;21:611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. Journal of Virology. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Marta M, da Silva FA, Fonseca AM, Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. The Journal of biological chemistry. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient Adaptive Evolution of the Primate Antiviral DNA-Editing Enzyme APOBEC3G. PLoS Biology. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya SMD, Nalam MNL, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Shindo K, Li M, Gross PJ, Brown WL, Harjes E, Lu Y, Matsuo H, Harris RS. A Comparison of Two Single-Stranded DNA Binding Models by Mutational Analysis of APOBEC3G. Biology. 2012;1:260–276. doi: 10.3390/biology1020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. The Journal of biological chemistry. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Shirakawa K, Takaori-Kondo A, Yokoyama M, Izumi T, Matsui M, Io K, Sato T, Sato H, Uchiyama T. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nature structural & molecular biology. 2008;15:1184–1191. doi: 10.1038/nsmb.1497. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AJ, Li M, Harris RS, Lyubchenko YL. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PloS one. 2014;9:e99354. doi: 10.1371/journal.pone.0099354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. The Journal of biological chemistry. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Nanoscale structure and dynamics of ABOBEC3G complexes with single-stranded DNA. Biochemistry. 2012;51:6432–6440. doi: 10.1021/bi300733d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Atomic force microscopy studies of APOBEC3G oligomerization and dynamics. Journal of structural biology. 2013;184:217–225. doi: 10.1016/j.jsb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen M, Pedersen CNS. Rapid Computation of Distance Estimators from Nucleotide and Amino Acid Alignments. Proceedings of 26th Annual ACM Symposium on Applied Computing (SAC 2011); ACM Press; 2011. [Google Scholar]
- Siu KK, Sultana A, Azimi FC, Lee JE. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nature communications. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Pathak VK. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol. 2010;84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider MJ, Reinhardt L, Wolfenden R, Cleland WW. 15N kinetic isotope effects on uncatalyzed and enzymatic deamination of cytidine. Biochemistry. 2002;41:415–421. doi: 10.1021/bi011410i. [DOI] [PubMed] [Google Scholar]
- Song C, Sutton L, Johnson ME, D'Aquila RT, Donahue JP. Signals in APOBEC3F N-terminal and C-terminal Deaminase Domains Each Contribute to Encapsidation in HIV-1 Virions and Are Both Required for HIV-1 Restriction. The Journal of biological chemistry. 2012;287:16965–16974. doi: 10.1074/jbc.M111.310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauch B, Hofmann H, Perkovic M, Weisel M, Kopietz F, Cichutek K, Münk C, Schneider G. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12079–12084. doi: 10.1073/pnas.0900979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. Journal of Virology. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic acids research. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter ML, D'Aquila RT. Cytoplasmic APOBEC3G Restricts Incoming Vif-positive HIV-1 and Increases 2-LTR Circle Formation In Activated T Helper Subtype Cells. Journal of Virology. 2009 doi: 10.1128/JVI.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang X, Zhang H, Zhou L, Liu S, Kolson DL, Song L, Ye L, Ho WZ. Expression and regulation of antiviral protein APOBEC3G in human neuronal cells. Journal of Neuroimmunology. 2009;206:14–21. doi: 10.1016/j.jneuroim.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. The EMBO journal. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic acids research. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen G, Niewiadomska AM, Xu R, Yu XF. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PloS one. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du J, Yu K, Wang T, Yong X, Yu XF. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV-1 virions. J Virol. 2010;84:12903–12913. doi: 10.1128/JVI.01632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant antiviral activities are associated with altered RNA binding activities. PloS one. 2012;7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. Journal of virology. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]