Abstract
Bone and dentin are mineralized extracellular matrices produced by osteoblasts and odontoblasts, respectively, and their major organic portion is type I collagen. Dentinogenesis Imperfecta (DGI) is one of the most common clinically- and genetically-based disturbances of dentin formation, causing irreversible dentin defects. Among several types of DGI, patients with DGI type II exhibit opalescent dentin with partial or complete pulp obliteration. It has been previously reported that the non-sense mutation (c.133C>T) in Dentin Sialophosphoprotein (DSPP) was identified in DGI type II patients at glutamine residue 45, resulting in the premature stop codon (p.Q45X). DSPP is known to be synthesized as a single gene product and further processed at Gly462-Asp463, resulting in the production of Dentin Sialoprotein (DSP) and Dentin Phosphoprotein (DPP). We hypothesized that the shorter form (Q45X) of N-terminal Dentin Sialoprotein (N-DSP) may cause over-production of type I collagen protein as obliterated pulp is occupied by dentin. To test this hypothesis, we generated mouse recombinant Glutathione-S-Transferase (GST)-N-DSP fusion protein, and the effect of GST-N-DSP was investigated in calvarial bone explant culture and MC3T3-E1 osteoblastic culture systems. Here we show that a significant increase in calvarial bone formation is observed by GST-N-DSP. GST-N-DSP accelerates MC3T3-E1 osteoblast cell growth and proliferation and subsequent osteoblast differentiation by inducing the expression of certain osteogenic markers such as type I collagen, Runx2, Osterix and ATF4. Interestingly, GST-N-DSP significantly enhances dentinogenesis marker gene expression including Dspp and Dmp1 gene expression in non-odontogenic MC3T3-E1 cells. To rule out any artificial effect of GST-tag, we also used the synthetic peptide of N-DSP and confirmed the results of N-DSP peptide were essentially similar to those of GST-N-DSP. Taken together, our data suggest that N-DSP promotes bone formation by accelerating osteoblast cell proliferation and subsequent osteoblast differentiation accompanied by marked up-regulation of the dentin matrix markers, such as Dspp and Dmp1 genes.
Keywords: Dentinogenesis Imperfecta, Dentin Sialophosphoprotein (DSPP), Dentin Sialoprotein (DSP), MC3T3-E1 cells, Type I collagen
Highlights
-
•
Recombinant N-terminal DSP (N-DSP) protein was generated.
-
•
N-DSP mimics the non-sense mutation form of Dentinogenesis Imperfecta type II.
-
•
N-DSP enhances bone formation in clavarial ex vivo cultures.
-
•
N-DSP accelerates osteoblast proliferation.
-
•
N-DSP upregulates type I collagen and Dspp expression in non-odontogenic osteoblasts.
1. Introduction
Dentinogenesis Imperfecta (DGI, OMIM#125490) is one of major congenital dentin defects in humans [1]. According to the Shields's classification in 1973 [2], DGI is further divided into 3 types, and it is known that DGI type II is characterized by abnormal dentin mineralization affecting both primary and permanent dentition due to mutations in Dentin Sialophosphoprotein (DSPP) [3], [4], [5], [6]. The prevalence of DGI has been estimated at 1 in 6000 to 8000 children in the United States [7]. The classical clinical presentation is described as an opalescent discoloration of the teeth with enamel that cracks and fractures from the dentin. Exposed dentin undergoes rapid attrition leading to shortening the teeth being worn down to the gingival level. The pulp chamber is usually completely or partially obliterated by the continued deposition of type I collagen, the most abundant protein in dentin [8], [9], [10].
Dental pulp is a soft connective tissue that occupies a space in the central portion of the tooth where the pulp core is present and the root canal system surrounded by dentin is formed. The observation of completely obliterated pulp in DGI type II patients provides an interesting question; what induces and occupies pulp space. A non-sense mutation in the human DSPP gene was identified (glutamine at residue 45) in a large Chinese family, resulting in premature termination of the DSP peptide (c.133C>T, p.Q45X). Patients with this particular mutation had complete pulp chamber obliteration [3]. As DGI type II is an autosomal dominant disorder, the truncated protein is likely expressed, which may cause pulp obliteration. Since obliterated pulp is likely occupied by excessive dentin / type I collagen, in this current study, we sought to determine whether the N-terminal DSP fragment (referred to N-DSP, hereafter), the DSPP gene product with c.133C>T mutation, increases type I collagen production using calvarial bone explant and MC3T3-E1 cell culture models that are known to produce type I collagen.
Here we show proof-of-concept that N-DSP enhances calvarial bone formation ex vivo. Our results also demonstrate that N-DSP accelerates MC3T3-E1 osteoblast cell proliferation and subsequent differentiation with marked upregulation of type I collagen, Dentin sialophosphoprotein and Dmp1 gene expression.
2. Materials and methods
2.1. Molecular cloning of mouse DSP expression vectors
We used N-terminal DSP (N-DSP) constructs/protein (i.e. Q48X) in this study, which is derived from mouse DSP and equivalent to the human Q45X mutant form. The position of glutamine (Q) at residue 45 in human DSPP corresponds to residue 48 in mouse. Mouse DSP expression vector constructs in the pcDNA3.1-V5/His mammalian expression vector and the pGEX-4T-1 bacterial expression vector were generated by PCR methods as described [11]. Briefly, total RNA was extracted from mouse molar teeth (C57BL/6, 5-week-old, male), cDNA was then synthesized and used as PCR templates. The primers used were as follows; (pcDNA3.1-V5/His expression vector) forward primer: 5′-GCGAATTCATAATGAAAATGAAGATAATT-3′ and reverse primer: 5′-GCCTCGAGTGCTGCAGTTCCTGGATGTG-3′, (pGEX-4T-1 bacterial expression vector) forward primer: 5′-GCCGGATCCATTCCGGTTCCCCAGTTAG-3′ and reverse primer: 5′-GCCTCGAGCTATGCTGCAGTTCCTGGATG-3′. The PCR products were ligated into pcDNA3.1-V5/His and pGEX4T-1 vectors and sequenced. The plasmid harboring N-DSP cDNA including a signal peptide without stop codon followed by C-terminal V5/His-tag (pcDNA3.1-N-DSP-V5/His vector, thus encoding Met1–Ala47) was generated. The.
N-terminal Glutathione-S-Transferase (GST)-tag fused to N-DSP cDNA lacking a signal peptide but with stop codon (pGEX-4T-1-GST-N-DSP vector, thus encoding Ile18–Ala47 plus stop codon) was successfully generated.
2.2. Cell culture, transfection, immunoprecipitation and Western blot analysis
The human embryonic kidney (HEK) 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing a high concentration of glucose (4.5 mg/ml), supplemented with 10% fetal bovine serum (FBS, Life Technologies), 100 units/ml penicillin, 100 µg/ml streptomycin in a 5% CO2 atmosphere at 37 °C as previously described [11]. The HEK293 cells were plated onto 6 well culture plates at a density of 3×105 cells/well and transiently transfected with pcDNA3.1-N-DSP-V5/His or pcDNA3.1-V5/His A (a negative control) vector using X-tremeGENE 9 DNA transfection reagent (Roche Life Science). After 24 h of transfection, cells were washed with phosphate buffered saline (PBS) and incubated with serum-free media for 16 h. The conditioned media and cell lysates were collected, immunoprecipitated (IP) with anti-V5 antibody (Life Technologies) and subjected to Western blotting (WB) analysis using anti-V5 antibody. The same membrane was stripped by stripping buffer, and WB with anti-DSP antibody (sc-33587, Santa Cruz Biotechnology) was performed to verify the expression of N-DSP-V5 protein. Chemiluminescent detection of bound antibodies was determined using the ECL Western blotting detection reagents (Amersham ECL Prime, GE Healthcare Life Sciences).
2.3. Generation of recombinant GST-N-DSP fusion protein and N-DSP peptide
The pGEX-4T-1-GST-N-DSP or pGEX-4T-1-GST vector was transformed into the BL21-CodonPlus bacterial strain (Agilent Technologies), cultured, and GST-N-DSP fusion protein or GST protein alone was generated as previously reported [11]. Aliquots of each GST protein were mixed with SDS sample buffer, applied to 4–12% SDS-PAGE, and stained with Coomassie Brilliant Blue R-250 (CBB) (Bio-Rad). Western blot analysis with anti-DSP or anti-GST (GE Healthcare Life Sciences) antibody was also performed as described above and the immunoreactivity of the protein was confirmed. The synthetic mouse N-DSP peptide (IPVPQLVPLERDIVENSVAVPLLTHPGTAA) was obtained from Peptide 2.0, Inc.
2.4. Calvarial bone ex vivo culture
The use of animals and all animal procedures in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of Boston University Medical Campus. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Calvaria were collected from C57BL/6 neonates at postnatal day 7, cut into two halves along the sagittal suture and cultured with BGJb media containing 10% FBS, 50 µg/ml of ascorbic acid, and 100 units / ml of penicillin and 100 µg / ml of streptomycin. We initially treated calvarial explant cultures with various doses of GST or GST-N-DSP (with a range of 0.22–5.0 µg/ml) to determine the optimal concentration and found that 2 µg/ml was enough to detect the statistically significant increase of bone formation in the GST-N-DSP treatment group as compared to the GST group (data not shown). Calvarial bones were treated with 2 µg/ml of GST, GST-N-DSP, N-DSP peptide, or left untreated and five halves of calvaria for each protein were used (n=5) and cultured for up to 14 days. The media were refreshed twice a week with addition of exogenous proteins. At day 14, the cultured calvaria were fixed in 10% paraformaldehyde overnight at 4 °C, decalcified with 10% EDTA, embedded into paraffin, sectioned at 5 µm, and sections were subjected to hematoxylin and eosin (H&E) staining. Serial sections were also subjected to Masson Trichrome staining to visualize bone matrix.
2.5. Measurement of formed bone area by histomorphometric analysis
Sections were stained with H&E, and their images were taken under a light microscope. The whole area of each hemi-calvarium was imaged using Olympus DSU microscope within 4–5 image fields per section and these images were tiled. The margin of eosinophilic bone structures including frontal and parietal bones was then outlined using the free hand selection in the Image-J program, and the area was calculated and determined as the formed bone area. A total of 5 hemi-calvaria samples were used per treatment group, and 5 sections per sample were used for quantification. The values are shown as the mean (mm2)±S.D. (n=5) with statistical analysis.
2.6. Cell growth and proliferation measurement
MC3T3-E1 cells (Subclone 4) were maintained as described [12]. Cells were plated in triplicate onto 24-well culture plates at a density of 6×104 cells / well and on the following day, cells were treated with 2.0 µg/ml of GST, GST-N-DSP, N-DSP peptide, or left untreated and cultured for up to 14 days. At intervals (days 1, 3, 7, 10 and 14), the numbers of cells were counted using a hemocytometer, and cell proliferation was measured using CyQUANT Cell Proliferation Assay Kit (Life Technologies). Three independent experiments for each assay were performed. The values are shown as the mean±S.D. (n=3) with statistical analysis.
2.7. Real time PCR analysis
MC3T3-E1 cells were plated onto 35 mm culture dishes at a density of 2×105 cells/dish, and when cells reached confluence, the media were supplemented with 50 μg/ml of ascorbic acid and 1 mM of β-glycerophosphate. Cells were treated with proteins in the same manner as described above and further maintained for up to 10 days. Total RNA was extracted at days 3 and 10, and cDNA was synthesized and used as PCR templates. Real time PCR was performed and the mean fold changes in the expression of each differentiation marker relative to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh, 4308313) were calculated using the values obtained from a control culture with GST treatment as a calibrator by means of 2−ΔΔCT method. Markers used were the following: Runt-related transcription factor 2 (Runx2, Mm00501578_m1); Osterix (Osx, Mm00504574_m1); Collagen type 1 alpha 2 chain (Col1a2, Mm00483888_m1); Activating transcription factor 4 (Atf4, Mm00515324_m1); Dspp (Mm00515667_g1); Dentin matrix protein1 (Dmp1, Mm01208363_m1). Three independent experiments were performed and the values are shown as mean±S.D. (n=3).
2.8. Statistical analysis
The difference between two groups was tested by Student t-test and a p value below 0.05 was considered as significant. The statistical significance was indicated by asterisks as follows; (*) for p<0.05 and (**) for p<0.001.
3. Results
3.1. N-DSP is a secretory protein
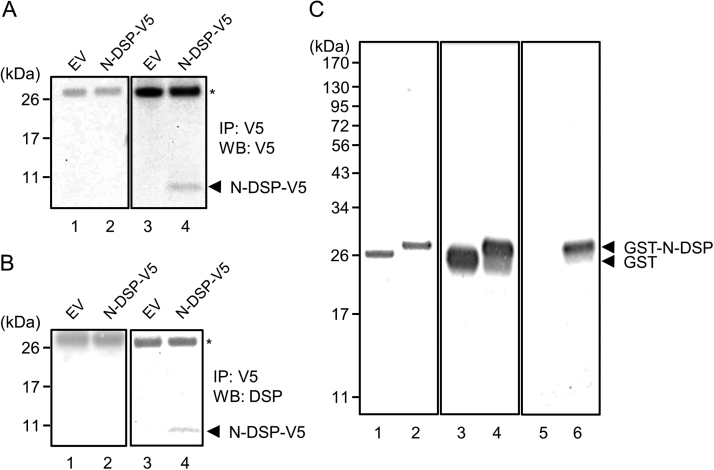
We first tested whether mouse N-DSP (i.e. Q48X) in mammalian cells was secreted. The mammalian expression vector encoding N-DSP (containing mouse N-terminal portion of DSP; 1–47 amino acids) fused to V5/His tag or an empty V5/His-tag vector (EV) was transiently transfected into HEK293 cells, the conditioned media or cell lysates were collected, immunoprecipitated (IP) with anti-V5 antibody and subjected to Western blot analysis (WB) with either anti-V5 (A) or anti-DSP (B) antibody. Our results showed that N-DSP was identified in the conditioned media (Fig. 1A, lane 4), but not in cell lysates (Fig. 1A, lane 2) by means of WB analysis with anti-V5 antibody at a molecular weight of approximately ~8 kDa (~3 kDa of DSP+5 kDa of V5/His tag). The immunoreactive band to anti-V5 antibody (indicated by an arrowhead as N-DSP-V5) was also positively recognized when anti-DSP antibody was used (Fig. 1B, lane 4). Thus, our results clearly demonstrate that N-DSP is a secretory protein.
Fig. 1.
Expression of mouse N-DSP in mammalian and bacterial expression systems. (A, B) Expression of N-DSP in HEK293 cells. The mammalian expression vector encoding N-DSP (containing mouse N-terminal portion of DSP; 1–47 amino acids) fused to V5/His tag or an empty V5/His-tag vector (EV) was transiently transfected into HEK293 cells, the conditioned media or cell lysates were collected, immunoprecipitated (IP) with anti-V5 antibody and subjected to Western blot analysis (WB) with either anti-V5 (A) or anti-DSP (B) antibody. *; IgG light chain. (C) Expression of GST-N-DSP in bacterial expression system. The equal amount (500 ng each) of GST (lane 1) and GST-N-DSP proteins (lane 2) were separated by 4–12% SDS-PAGE and stained with CBB. The presence of a single band in each lane confirms the purity of GST proteins. The same proteins were also analyzed by Western blot analysis (lanes 3–6). The immunoreactivities to anti-GST antibody were clearly detected in both GST proteins (lanes 3 and 4). The immunoreactivity to anti-DSP antibody was detected only in GST-N-DSP protein (lane 6), but not in GST alone (lane 5), demonstrating the successful generation of GST-N-DSP protein.
3.2. Generation of recombinant mouse GST-N-DSP fusion protein
The position of glutamine at residue 45 in human DSPP is equivalent to glutamine at residue 48 in mouse DSPP. The mature portion of mouse DSP prior to the glutamine at residue 48, i.e. I18 to A47, followed by a stop codon was subcloned into the N-terminal Glutathione-S-Transferase (GST)-tag bacterial expression vector and GST-N-DSP fusion protein was generated. Equal amounts of GST and GST-N-DSP proteins were separated by SDS-PAGE and stained with CBB (Fig. 1C, lanes 1 and 2). A single band of GST-N-DSP protein was observed at the expected molecular weight size at ~29 kDa, i.e.~3 kDa DSP+26 kDa GST tag (Fig. 1C, lane 2). The same amounts of recombinant proteins were further analyzed by Western blot analyses using anti-GST (Fig. 1C, lanes 3 and 4) and anti-DSP (Fig. 1C, lanes 5 and 6) antibodies. Both recombinant proteins were immunoreactive to anti-GST antibody while only GST-N-DSP was detected by anti-DSP antibody. These results demonstrate that recombinant GST-N-DSP protein was successfully purified and that anti-DSP antibody recognizes GST-N-DSP protein.
3.3. Effect of N-DSP on calvarial bone formation
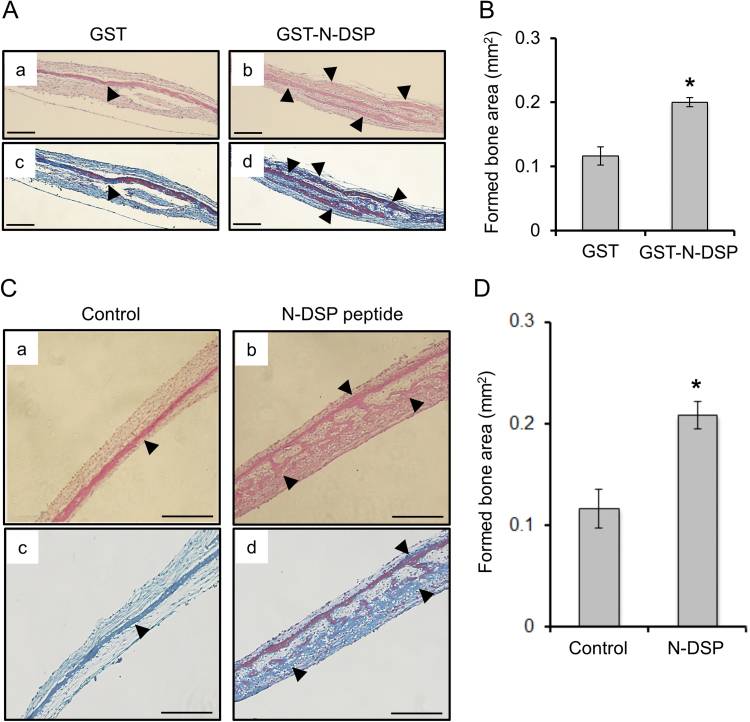
Mouse calvarial bone explant cultures were incubated with GST-N-DSP or GST for 14 days and processed for histological evaluation. Multiple eosinophilic layers (indicated by arrowheads) were observed in the calvarial culture with GST-N-DSP (Fig. 2A-b) or with N-DSP peptide (Fig. 2C-b) while there was a single eosinophilic layer with GST alone (Fig. 2A-a) or untreated (Control, Fig. 2C-a). By Masson Trichrome staining, these layers were visualized in blue (representing newly formed bone matrices) and red (representing existing bone matrices) (Fig. 2A-c and-d, C-c and-d), indicating that greater amount of bone matrices, likely composed of type I collagen were accumulated in N-DSP treated bone cultures (Fig. 2A-d and C-d) than controls (Fig. 2A-c and C-c). To quantitatively measure the areas of formed bone, histomorphometric analysis was performed. The result demonstrated that there was a significantly greater area of bone formation in GST-N-DSP group than in GST control, which was about 70% of the increase (Fig. 2B). The increase of bone formation was also confirmed when the N-DSP peptide was added to the culture, and the formed bone area in the N-DSP peptide group was about 80% more increased as compared to the untreated control group (Fig. 2D). There seemed no difference of formed bone areas between GST and Control groups (Fig. 2B and D), indicating the absence of any artificial effect of GST on bone formation.
Fig. 2.
Effect of N-DSP on bone formation in calvarial bone culture system. (A, C) Calvarial bones were treated with GST, GST-N-DSP, N-DSP peptide, or untreated (n=5 in each group) and paraffin-embedded sections were prepared. The representative H&E staining images of calvarial bone section incubated with GST (A-a), GST-N-DSP (A-b), untreated control (C-a), or N-DSP peptide (C-b) were shown. Serial sections were also stained with Masson's Trichrome and the representative images were shown (A-c; GST, A-d; GST-N-DSP, C-a; untreated control, C-b; N-DSP peptide treatment group). Greater numbers of eosinophilic layers were identified (indicated by arrowheads) when incubated with GST-N-DSP or N-DSP peptide, but not with GST or untreated control. These layers were Trichrome-positive, indicative of collagen. Scale bar; 0.2 mm. (B, D) The formed bone area in GST-N-DSP or N-DSP group was significantly increased as compared to GST or untreated control group by histomorphometric analysis. The mean value of formed bone areas was calculated, expressed as the mean±SD (mm2) and statistically analyzed (n=5). *; p<0.05.
3.4. Effect of N-DSP on cell proliferation
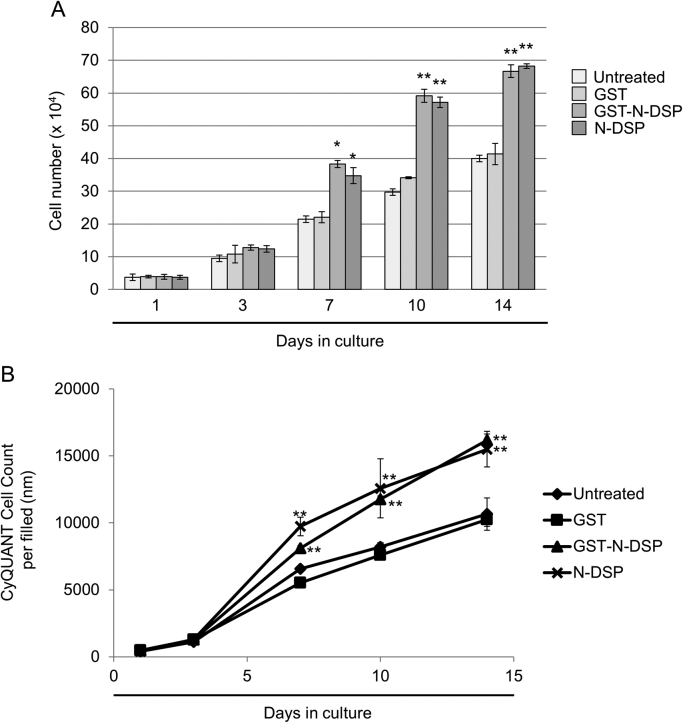
To further gain the biological insights into how N-DSP increases bone formation at a cellular level, we used an immortalized osteoblastic cell line, MC3T3-E1 cells isolated from calvaria [13]. The effect of N-DSP on MC3T3-E1 cell growth was analyzed by cell counting. As the DNA content is an indicator of cell numbers, we also used the cyanine dye CyQUANT, which binds to DNA to analyze cell proliferation. The results of cell growth by cell counting demonstrated that both GST-N-DSP and N-DSP peptide treatment accelerated osteoblast cell growth at days 7, 10, and 14 as compared to GST and untreated controls (Fig. 3A). The accelerated proliferation of osteoblasts by GST-N-DSP and N-DSP peptide treatment was also verified by means of CyQUANT Cell Proliferation Assay (Fig. 3B). The data showed a significant increase by N-DSP treatment at days 7, 10 and 14 as compared to controls, further demonstrating the proliferative effect of N-DSP on osteoblasts.
Fig. 3.
Effect of N-DSP on MC3T3-E1 osteoblastic cell growth and proliferation. MC3T3-E1 cells were left untreated or treated with GST, GST-N-DSP, or N-DSP peptide, and further cultured for up to 14 days. At Day 1, 3, 7, 10 and 14, cell numbers were counted (A) or cell proliferation was measured by the CyQUANT Cell Proliferation Assay (B). Data are expressed as the mean±SD (n=3) with statistical analysis. *; p<0.05,**; p<0.001.
3.5. Effect of N-DSP on cell differentiation
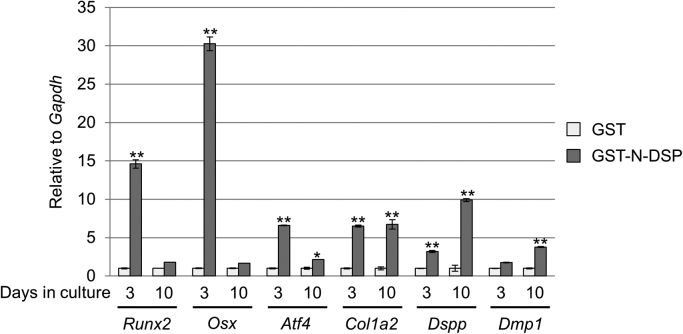
We next investigated the effect of N-DSP on osteoblast differentiation by measuring the levels of osteogenic and dentinogenesis markers (Fig. 4). The data showed that GST-N-DSP treatment significantly upregulated osteogenic marker expressions including Runx2 (~14.6 fold increase), Osx (~30.3 fold) and Atf4 (~6.6 fold) at day 3 as compared to GST, while at day 10, the extent of upregulation in these transcription factor gene expression in GST-N-DSP group was moderate (Runx2;~1.8 fold increase, Osx;~1.7 fold, and Atf4;~2.2 fold). The expression of Col1a2 and Dspp was significantly upregulated at day 3 (~6.5 fold and ~3.2 fold, respectively) and the expression of Dmp1, another dentinogenesis marker was moderately upregulated (~1.8 fold). The level of higher Col1a2 gene expression was sustained (~6.7 fold) in GST-N-DSP group at day 10. Intriguingly, the expression of Dspp and Dmp1 was further increased (~9.9 fold and ~3.8 fold, respectively) at day 10, indicating that GST-N-DSP positively regulates the dentinogenesis marker expression in non-odontogenic cells.
Fig. 4.
Effect of GST-N-DSP on MC3T3-E1 osteoblast differentiation. MC3T3-E1 cells were treated with GST or GST-N-DSP, and further cultured for up to 10 days. The expression of osteogenic/dentinogenesis markers was analyzed by real-time PCR. The normalized values are shown as the mean±S.D. and statistically analyzed (n=3). *; p<0.05, **; p<0.001. Runx2; Runt-related transcription factor 2, Osx; Osterix, Atf4; Activating transcription factor 4; Col1a2; Collagen type 1 alpha 2 chain, Dspp; Dentin sialophosphoprotein, Dmp1; Dentin matrix protein 1, Gapdh; Glyceraldehyde-3-phosphate dehydrogenase.
4. Discussion
There have been so far ~40 cases of reported DGI patients affected by DSPP mutations, which include mis-sense, non-sense, insertion and deletion mutations [14]. It has been reported that many inherited diseases resulting from non-sense mutations activate nonsense-mediated mRNA decay (NMD), leading to the degradation of such premature termination codon (PTC)-containing mRNAs [15]. In some cases such as β-thalassemia, mutations in β-GLOBIN do not elicit NMD pathway, resulting in the presence of truncated β-GLOBIN protein which functions as a dominant-negative form [16]. In order for aberrant mRNA to escape from NMD activation, it is considered that the PTC has to be located less than 50–55 nucleotides upstream of a splicing from exon-exon junction [17]. Glutamine at residue 45, where nonsense DSP-Q45X mutation is located, is the last amino acid in exon 3. This points out the potential pathological mechanism that PTC located in this position unlikely elicits NMD pathway, resulting in the N-DSP mRNA translation and protein synthesis, which at least in part leads to the DGI type II pulp phenotype. It has been previously shown that the DSP “IPV” mutant protein, where the DGI mutations occur within the first three amino acids of the mature DSPP portion, was intracellularly accumulated in the secretory pathway likely at rough endoplasmic reticulum (rER), which presented from protein secretion [18]. In contrast to this, our results demonstrated that N-DSP-V5/His protein was identified in the conditioned media, but not in the cell lysates (Fig. 1), indicating that these DSP mutants may likely have different mechanisms to lead to the DGI phenotypes.
Odontoblasts secrete extracellular matrix proteins, including type I collagen and non-collagenous proteins (NCPs) during dentinogenesis. Among several NCPs, DSPP appears to be the major dentin specific protein despite its temporal and less abundant expression in other tissues such as bone, cementum, and non-skeletal tissues [19], [20], [21], [22]. It is known that DSPP is proteolytically processed and 3 major cleaved products are identified, namely DSP, dentin glycoprotein (DGP) and dentin phosphoprotein (DPP) [23], [24], [25]. While it is generally accepted, based on its chemical composition, that DPP likely plays a crucial role in controlling dentin mineralization [26] with association with type I collagen [27], [28], the roles of DSP and DGP are not fully understood. It has been previously reported that recombinant human DSP protein produced in bacteria (referred as rhDSP hereafter) induces growth and differentiation on human primary [29] and immortalized dental pulp cells [30], and rhDSP protein used in these studies (residues from I376 to G462) encodes a C-terminal portion of DSP (I376–S382) and full length of DGP (S383–G462), suggesting the biological effect of the rhDSP on dental pulp cells may be due to the function of DGP. In their studies, rhDSP treatment induces type I collagen [29] and DSPP [30] upregulation in human dental pulp cells. More recently, it has been reported that the recombinant mouse C-terminal DSP fragment (rC-DSP, residues from 183 to 457) produced in bacteria enhances cell proliferation in human periodontal ligament (PDL) cells and PDL stem cells [31]. Consistent with our gene expression data (Fig. 4), a few studies have reported that overexpression of DSPP or an exogenous rC-DSP treatment increased the expression of osteogenic markers including Runx2 and Osx [31], [32]. Thus, it is possible to suggest that the DSP portion within DSPP protein may play a modulatory role in cellular functions, i.e. cell proliferation and differentiation. Our results support this potential DSP function in cell proliferation and differentiation, although the portion of DSP in our study, e.g. N-DSP, differs from those previous reports (rhDSP and rC-DSP). It has been previously shown that deletion of the entire Dspp coding exons in mice by replacing with LacZ-neo cassette leads to the widened predentin zone, defective dentin mineralization and altered levels of proteoglycans [33]. Although Dspp deficiency does not change the level of type I collagen gene expression in odontoblasts of Dspp null mice as compared to those in wild type mice [33], overexpression of Dsp (residues from I18–S370) in the Dspp null background induces dentin volume, but not dentin mineral density [34]. As the Dsp transgene includes a portion of N-DSP, it is possible to speculate, at least in part, that DSP plays an important role in type I collagen gene regulation in vivo. It is likely that the molecular mechanisms by which rhDSP, rC-DSP and N-DSP affect cell functions may differ. Our results together with previous findings support the biological importance of DSP portion in type I collagen synthesis possibly through enhancing cell proliferation.
There are several reasons why we took advantage of using MC3T3-E1 cells and calvaria instead of odontogenic cell types. (1) There is almost no information regarding which cells are responsible for making excessive amounts of dentin in DGI type II, resulting in the pulp obliteration, i.e. pulp cells or odontoblasts. (2) Odontoblasts usually secrete type I collagen/ECM molecules towards their apical direction, not basal, thus it is presumed that pulp cells are likely responsible for DGI type II pulp phenotype. (3) Even if the cell type responsible for pulp obliteration is likely pulp cells, they are heterozygous population in nature. So it is difficult to determine which cell sub-population is responsible for DGI type II pulp phenotype. (4) There are almost no “best-characterized” cell lines of odontoblasts or pulp cells which have been reported to produce Dspp as a minimum requirement of dentin marker. (5) If cells are immortalized and forced to proliferate, they may not enter into differentiation stage of odontoblasts, as cell differentiation is expected when cells exit the cell cycle. Instead of dealing with such open questions/technical concerns described, we decided to use the spontaneously immortalized MC3T3-E1 osteoblast cell line which is a very established cell line and well-known to produce type I collagen. Although this is not an odontoblast-cell line, our results showed a proof-of-concept and, to the best of our knowledge, our current study was the first report to show a remarkable increase of endogenous Dspp expression by an exogenous N-DSP treatment in non-odontogenic MC3T3-E1 cells, a lineage-committed osteoblast cell line (Fig. 4). A similar finding, but in PDL cells, has also been reported by Lee et al. [30] that DSPP expression was modestly induced by the exogenous addition of rhDSP determined by a conventional RT-PCR method. Although there have been only limited numbers of studies reported, it appears that non-odontogenic cells expressing DSPP are considered as differentiated odontoblast-like cells [32], [35]. It is thus reasonable to speculate that a high level of DSPP expression is probably the only prerequisite to define odontoblasts, despite a lack of a clear standard threshold. To support this notion, no other gene mutations than DSPP are known to cause non-syndromic dentin phenotype in humans. It is well known that mutations in other gene markers of odontoblast (such as DMP1, type I collagen, etc.) cause syndromic dental and bone phenotypes in humans. Further studies are clearly warranted to investigate the usefulness of DSP-Q45X fragment for odontoblast lineage induction as well as de novo dentin formation, which may further address the pathogenesis of DGI type II phenotype.
In summary, to the best of our knowledge, our study was the first to show that N-DSP fragment induces bone formation in ex vivo bone culture, at least in part by stimulating osteoblast cell growth and differentiation levels, thereby providing a mechanism for DSP induced anabolic effects on bone.
Acknowledgements
The authors thank Dr. Philip Trackman (Boston University) for his valuable comments. This study was supported in part by grants from NIH/DE019527 (YM) and the American Association of Endodontists Foundation (HJ).
Footnotes
Transparency Document associated with this article can be found in the online version at 10.1016/j.bbrep.2016.04.004.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Witkop C.J., Jr. Hereditary defects of dentin. Dent. Clin. N. Am. 1975;19:25–45. [PubMed] [Google Scholar]
- 2.Shields E.D., Bixler D., el-Kafrawy A.M. A proposed classification for heritable human dentine defects with a description of a new entity. Arch. Oral Biol. 1973;18:543–553. doi: 10.1016/0003-9969(73)90075-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Zhao J., Li C., Gao S., Qiu C., Liu P., Wu G., Qiang B., Lo W.H., Shen Y. DSPP mutation in dentinogenesis imperfecta shields type II. Nat. Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 4.Xiao S., Yu C., Chou X., Yuan W., Wang Y., Bu L., Fu G., Qian M., Yang J., Shi Y., Hu L., Han B., Wang Z., Huang W., Liu J., Chen Z., Zhao G., Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 5.Malmgren B., Lindskog S., Elgadi A., Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum. Genet. 2004;114:491–498. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- 6.Kim J.W., Nam S.H., Jang K.T., Lee S.H., Kim C.C., Hahn S.H., Hu J.C., Simmer J.P. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum. Genet. 2004;115:248–254. doi: 10.1007/s00439-004-1143-5. [DOI] [PubMed] [Google Scholar]
- 7.Witkop C.J., Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J. Oral Pathol. 1988;17:547–553. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 8.Takagi Y., Koshiba H., Kimura O., Kuboki Y., Sasaki S. Dentinogenesis imperfecta: evidence of qualitative alteration in the organic dentin matrix. J. Oral Pathol. 1980;9:201–209. doi: 10.1111/j.1600-0714.1980.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 9.Kida M., Tsutsumi T., Shindoh M., Ikeda H., Ariga T. De novo mutation in the DSPP gene associated with dentinogenesis imperfecta type II in a Japanese family. Eur. J. Oral Sci. 2009;117:691–694. doi: 10.1111/j.1600-0722.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- 10.Wright J.T., Gantt D.G. The ultrastructure of the dental tissues in dentinogenesis imperfecta in man. Arch. Oral Biol. 1985;30:201–206. doi: 10.1016/0003-9969(85)90116-5. [DOI] [PubMed] [Google Scholar]
- 11.Mochida Y., Parisuthiman D., Yamauchi M. Biglycan is a positive modulator of BMP-2 induced osteoblast differentiation. Adv. Exp. Med. Biol. 2006;585:101–113. doi: 10.1007/978-0-387-34133-0_7. [DOI] [PubMed] [Google Scholar]
- 12.Mochida Y., Parisuthiman D., Pornprasertsuk-Damrongsri S., Atsawasuwan P., Sricholpech M., Boskey A.L., Yamauchi M. Decorin modulates collagen matrix assembly and mineralization. Matrix Biol. 2009;28:44–52. doi: 10.1016/j.matbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D., Christensen K., Chawla K., Xiao G., Krebsbach P.H., Franceschi R.T. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J. Bone Miner. Res. 1999;14:893–903. doi: 10.1359/jbmr.1999.14.6.893. [DOI] [PubMed] [Google Scholar]
- 14.de La Dure-Molla M., Philippe Fournier B., Berdal A. Isolated dentinogenesis imperfecta and dentin dysplasia: revision of the classification. Eur. J. Hum. Genet. 2015;23:445–451. doi: 10.1038/ejhg.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maquat L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 16.Neu-Yilik G., Amthor B., Gehring N.H., Bahri S., Paidassi H., Hentze M.W., Kulozik A.E. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA. 2011;17:843–854. doi: 10.1261/rna.2401811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 18.von Marschall Z., Mok S., Phillips M.D., McKnight D.A., Fisher L.W. Rough endoplasmic reticulum trafficking errors by different classes of mutant dentin sialophosphoprotein (DSPP) cause dominant negative effects in both dentinogenesis imperfecta and dentin dysplasia by entrapping normal DSPP. J. Bone Miner. Res. 2012;27:1309–1321. doi: 10.1002/jbmr.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin C., Brunn J.C., Cadena E., Ridall A., Tsujigiwa H., Nagatsuka H., Nagai N., Butler W.T. The expression of dentin sialophosphoprotein gene in bone. J. Dent. Res. 2002;81:392–394. doi: 10.1177/154405910208100607. [DOI] [PubMed] [Google Scholar]
- 20.Baba O., Qin C., Brunn J.C., Jones J.E., Wygant J.N., McIntyre B.W., Butler W.T. Detection of dentin sialoprotein in rat periodontium. Eur. J. Oral Sci. 2004;112:163–170. doi: 10.1111/j.0909-8836.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 21.Alvares K., Kanwar Y.S., Veis A. Expression and potential role of dentin phosphophoryn (DPP) in mouse embryonic tissues involved in epithelial-mesenchymal interactions and branching morphogenesis. Dev. Dyn. 2006;235:2980–2990. doi: 10.1002/dvdy.20935. [DOI] [PubMed] [Google Scholar]
- 22.Ogbureke K.U., Fisher L.W. SIBLING expression patterns in duct epithelia reflect the degree of metabolic activity. J. Histochem. Cytochem. 2007;55:403–409. doi: 10.1369/jhc.6A7075.2007. [DOI] [PubMed] [Google Scholar]
- 23.MacDougall M., Simmons D., Luan X., Nydegger J., Feng J., Gu T.T. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J. Biol. Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 24.Feng J.Q., Luan X., Wallace J., Jing D., Ohshima T., Kulkarni A.B., D’Souza R.N., Kozak C.A., MacDougall M. Genomic organization, chromosomal mapping, and promoter analysis of the mouse dentin sialophosphoprotein (Dspp) gene, which codes for both dentin sialoprotein and dentin phosphoprotein. J. Biol. Chem. 1998;273:9457–9464. doi: 10.1074/jbc.273.16.9457. [DOI] [PubMed] [Google Scholar]
- 25.Yamakoshi Y., Hu J.C., Fukae M., Zhang H., Simmer J.P. Dentin glycoprotein: the protein in the middle of the dentin sialophosphoprotein chimera. J. Biol. Chem. 2005;280:17472–17479. doi: 10.1074/jbc.M413220200. [DOI] [PubMed] [Google Scholar]
- 26.Prasad M., Butler W.T., Qin C. Dentin sialophosphoprotein in biomineralization. Connect. Tissue Res. 2010;51:404–417. doi: 10.3109/03008200903329789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stetler-Stevenson W.G., Veis A. Type I collagen shows a specific binding affinity for bovine dentin phosphophoryn. Calcif. Tissue Int. 1986;38:135–141. doi: 10.1007/BF02556873. [DOI] [PubMed] [Google Scholar]
- 28.Traub W., Jodaikin A., Arad T., Veis A., Sabsay B. Dentin phosphophoryn binding to collagen fibrils. Matrix. 1992;12:197–201. doi: 10.1016/s0934-8832(11)80062-4. [DOI] [PubMed] [Google Scholar]
- 29.Yun Y.R., Kim H.W., Kang W., Jeon E., Lee S., Lee H.Y., Kim C.H., Jang J.H. Expression and purification recombinant human dentin sialoprotein in Escherichia coli and its effects on human dental pulp cells. Protein Expr. Purif. 2012;83:47–51. doi: 10.1016/j.pep.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.Y., Kim S.Y., Park S.H., Kim J.J., Jang J.H., Kim E.C. Effects of recombinant dentin sialoprotein in dental pulp cells. J. Dent. Res. 2012;91:407–412. doi: 10.1177/0022034511436113. [DOI] [PubMed] [Google Scholar]
- 31.Ozer A., Yuan G., Yang G., Wang F., Li W., Yang Y., Guo F., Gao Q., Shoff L., Chen Z., Gay I.C., Donly K.J., MacDougall M., Chen S. Domain of dentine sialoprotein mediates proliferation and differentiation of human periodontal ligament stem cells. PLoS One. 2013;8:e81655. doi: 10.1371/journal.pone.0081655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L., Zhu F., Wu Y., Lin Y., Nie X., Jing W., Qiao J., Liu L., Tang W., Zheng X., Tian W. Dentin sialophosphoprotein-promoted mineralization and expression of odontogenic genes in adipose-derived stromal cells. Cells Tissues Organs. 2008;187:103–112. doi: 10.1159/000110079. [DOI] [PubMed] [Google Scholar]
- 33.Sreenath T., Thyagarajan T., Hall B., Longenecker G., D’Souza R., Hong S., Wright J.T., MacDougall M., Sauk J., Kulkarni A.B. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J. Biol. Chem. 2003;278:24874–24880. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki S., Sreenath T., Haruyama N., Honeycutt C., Terse A., Cho A., Kohler T., Muller R., Goldberg M., Kulkarni A.B. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol. 2009;28:221–229. doi: 10.1016/j.matbio.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayanan K., Srinivas R., Ramachandran A., Hao J., Quinn B., George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc. Natl. Acad. Sci. USA. 2001;98:4516–4521. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material