SUMMARY
Human genetic studies have identified the neuronal RNA binding protein, Rbfox1, as a candidate gene for autism spectrum disorders. While Rbfox1 functions as a splicing regulator in the nucleus, it is also alternatively spliced to produce cytoplasmic isoforms. To investigate the function of cytoplasmic Rbfox1, we knocked down Rbfox proteins in mouse neurons and rescued with cytoplasmic or nuclear Rbfox1. Transcriptome profiling showed that nuclear Rbfox1 rescued splicing changes, whereas cytoplasmic Rbfox1 rescued changes in mRNA levels. iCLIP-seq of subcellular fractions revealed that Rbfox1 bound predominantly to introns in nascent RNA, while cytoplasmic Rbox1 bound to 3′ UTRs. Cytoplasmic Rbfox1 binding increased target mRNA stability and translation, and Rbfox1 and miRNA binding sites overlapped significantly. Cytoplasmic Rbfox1 target mRNAs were enriched in genes involved in cortical development and autism. Our results uncover a new Rbfox1 regulatory network and highlight the importance of cytoplasmic RNA metabolism to cortical development and disease.
INTRODUCTION
Post-transcriptional regulation of RNA within neurons provides temporal and spatial control of gene expression during brain development and plasticity. RNA binding proteins (RBPs) play central roles in regulating each step of RNA processing. Mutations in RBPs have been found to cause and/or contribute to many human neurodevelopmental and neurologic disorders (Darnell and Richter, 2012; Lukong et al., 2008). Human genetic studies have focused attention on Rbfox1, a vertebrate homolog of the Caenorhabditis elegans Feminizing gene 1 on X (also known as Ataxin-2 Binding Protein 1, A2BP1) by associating chromosomal translocations and copy number variations in Rbfox1 with autism-spectrum disorders (ASDs) (Martin et al., 2007; Sebat et al., 2007). A gene co-expression network analysis of post mortem cerebral cortex from individuals with ASD identified Rbfox1 as a hub gene in a module of co-expressed transcripts involved in neuronal function and down-regulated in ASD (Voineagu et al., 2011). Other studies found Rbfox1 mutations associated with human epilepsy syndromes (Bhalla et al., 2004; Lal et al., 2013a; Lal et al., 2013b; Martin et al., 2007), which is often co-morbid with ASD.
Mammals express three Rbfox paralogs: Rbfox1 expressed in neurons, heart and muscle; Rbfox2 expressed in these three tissues as well as in stem cells, hematopoietic cells, and other cells; and Rbfox3 (also known as NeuN) expressed only in neurons (Kim et al., 2009b; Kuroyanagi, 2009). All Rbfox proteins contain a single RNA recognition motif (RRM)-type RNA binding domain that binds the hexanucleotide (U)GCAUG with great specificity (Auweter et al., 2006). When this sequence is present in regulated exons or flanking introns, Rbfox functions as a splicing regulator to promote or repress exon inclusion (Kuroyanagi, 2009). Studies in knockout (KO) mice revealed a role for Rbfox1 in regulating neuronal splicing networks involved in neurodevelopmental disorders and in controlling neuronal excitability (Gehman et al., 2011). A subsequent study identified several hundred additional Rbfox1 dependent splicing changes during neuronal differentiation of human fetal primary neural progenitor cells (Fogel et al., 2012). Crosslinking-immunoprecipitation and RNAseq (CLIP-seq) identified additional Rbfox1 splicing targets in mouse brain, including many ASD related genes (Lovci et al., 2013; Weyn-Vanhentenryck et al., 2014).
Rbfox1 is itself alternatively spliced to produce nuclear and cytoplasmic isoforms, referred to as Rbfox1_N and Rbfox1_C, respectively (Lee et al., 2009; Nakahata and Kawamoto, 2005). While the function of Rbfox1_N in splicing has been well studied, the function of Rbfox1_C is largely unexplored. The conservation of Rbfox binding sites in the 3′ UTRs of many neuronal transcripts suggests that Rbfox1 plays a role in regulating mRNAs in the cytoplasm (Ray et al., 2013; Xie et al., 2005). Ray et al (2013) correlated the expression of Rbfox1 in different tissues with the presence of Rbfox1 binding sites in 3′ UTRs and proposed that Rbfox1 stabilizes target mRNAs. Although they confirmed this by luciferase assay for one target using Rbfox1_N, it was not clear whether this stabilization arose from Rbfox1 actions in the nucleus or the cytoplasm. Other studies identifying Rbfox1 targets using CLIP-seq were done in whole tissue and thus could not differentiate between nuclear and cytoplasmic targets of Rbfox1 (Lovci et al., 2013; Weyn-Vanhentenryck et al., 2014).
To dissect the functions of Rbfox1 in the cytoplasm and nucleus, we profiled the transcriptome of neurons expressing either Rbfox1_C or Rbfox1_N. We complemented these experiments with Rbfox1 individual-nucleotide resolution CLIP (iCLIP) from subcellular fractions of neurons to identify the specific transcripts bound by Rbfox1 in the cytoplasm. Together, these findings identify a large number of transcripts regulated by cytoplasmically localized Rbfox1, independent of its effect on splicing. Our results indicate that the cytoplasmic portion of the Rbfox1 regulatory network controls many essential neuronal functions and further shows that these cytoplasmic targets overlap extensively with regulatory modules involved in cortical development and ASD.
RESULTS
Rbfox1_C is expressed at significant levels in mouse brain
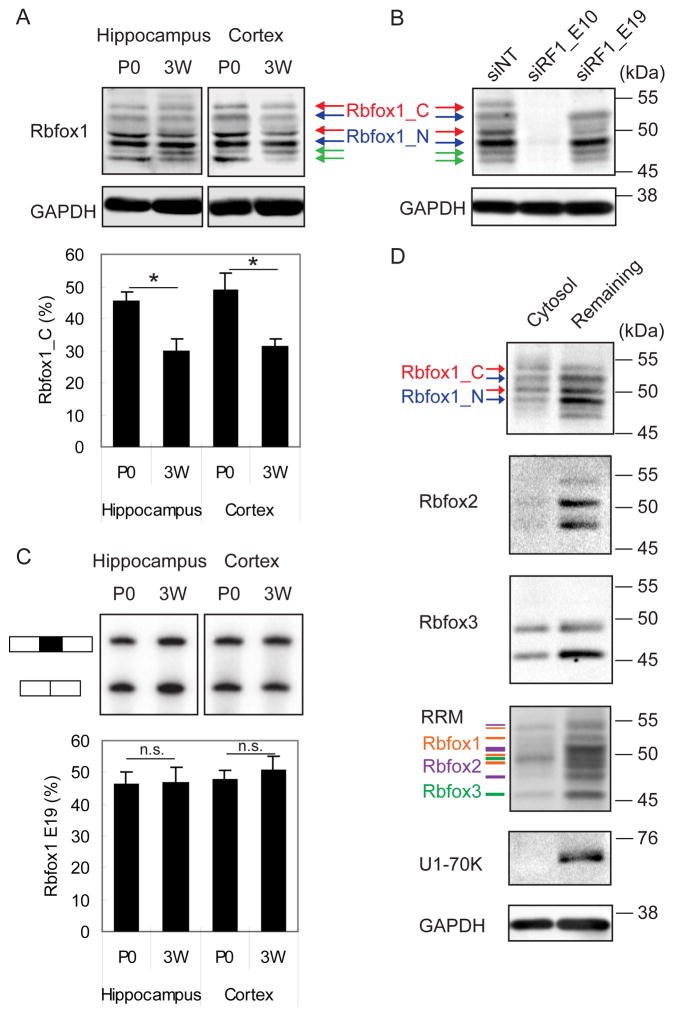
The Rbfox1 gene contains several promoters and alternatively spliced exons, which generate multiple protein isoforms (Damianov and Black, 2010). The skipping of mouse exon 19 generates an Rbfox1 mRNA encoding Rbfox1_N, a protein with a nuclear localization signal (NLS) in the C-terminus that is enriched in the nucleus (Hamada et al., 2013; Lee et al., 2009). In contrast, the mRNA including exon 19 encodes Rbfox1_C, a protein lacking the NLS that localizes to the cytoplasm. We examined Rbfox1 protein and mRNA in brain to compare the expression levels of Rbfox1_N and Rbfox1_C isoforms. Immunoblotting of lysates from hippocampus and cortex from P0 and P21 mice with an anti-Rbfox1 antibody revealed six bands ranging from 45 to 55 kDa (Figure 1A). All of the bands were reduced or abolished by an siRNA targeting a constitutive exon present in all Rbfox1 mRNAs (siRF1_E10, Figure 1B), indicating that they represent specific Rbfox1 products. Using an siRNA targeting alternative exon 19, two of the six protein bands were depleted, indicating that these represent Rbfox1_C (red arrows in Figure 1B). In P0 mouse hippocampus and cortex, Rbfox1_C constituted 46% and 49% of Rbfox1 protein (Figure 1A), respectively, and subsequently declined to the 30% from 3 weeks to 6 months of age in adult brain (data not shown). We also examined the splicing of exon 19 by semi-quantitative RT-PCR, and found that 47% and 48% of Rbfox1 mRNA included exon 19 in P0 hippocampus and cortex, respectively (Figure 1C). Rbfox1_C is thus expressed at significant concentrations in mouse brain.
Figure 1. Expression and localization of Rbfox1 isoforms in neurons.
(A) Immunoblot of Rbfox1 isoforms during development in mouse hippocampus and cortex at postnatal day 0 (P0), and 3 weeks (3W). Cytoplasmic Rbfox1 isoform (Rbfox1_C) and nuclear Rbfox1 isoform (Rbfox1_N) are indicated by red and blue arrows, respectively. The green arrow points to a lower molecular weight isoform that is reduced by Rbfox1 siRNA (panel B), but whose provenance is unknown. The percentage of Rbfox1_C in the two dominant bands in the middle is shown in the bar graph below. Error bars indicate SD. N = 3. Statistical significance was determined by student’s t-test. * indicates p value < 0.05.
(B) Immunoblot of Rbfox1 using an Rbfox1 pan siRNA (siRF1_E10) and an Rbfox1_C specific siRNA (siRF1_E19) in hippocampal neurons (DIV17).
(C) Semi-quantitative PCR analysis showing the splicing of Rbfox1 exon 19 in the same tissue and at the same time points as in (A). The percentage of exon 19 splicing is shown in histogram. Error bars indicate SD. N = 3. The percentage of Rbfox1 exon 19 inclusion was calculated; student’s t-test, p < 0.05. n.s. = not significant.
(D) Immunoblot showing that Rbfox1 and Rbfox3 but not Rbfox2 are present in a cytosolic fraction purified from mouse hippocampal neurons (DIV14, treated with AraC). In the RRM immunoblots, the orange lines correspond to Rbfox1 proteins, the purple lines correspond to Rbfox2 proteins, and the green lines correspond to Rbfox3 proteins.
To confirm that endogenous Rbfox1 localizes to the cytoplasm, we performed immunoblotting on subcellular fractions (Figure 1D). We prepared cytosolic fractions by permeabilizing the plasma membrane but not the nucleus of dissociated hippocampal cultures with digitonin (Mackall et al., 1979). After collecting the cytosolic fraction that leaked out of the cells, cultures were further solubilized with RIPA buffer. As shown in Figure 1D, the cytoplasmic protein GAPDH was present in the digitonin-solubilized cytosolic fraction and in the RIPA-solubilized remaining fraction, indicating that the lysis was partial. More importantly, the nuclear marker U1-70K was absent from the cytosolic fraction, indicating that digitonin permeabilization released cytosolic, but not nuclear proteins. Rbfox1 immunoblots of the cytosolic fraction detected both Rbfox1_C and Rbfox1_N isoforms, indicating that both Rbfox1 splice isoforms are present in the cytoplasm. However, the ratio of Rbfox1_C to Rbfox1_N was higher in the cytosolic fraction, with Rbfox1_C representing the dominant isoform in the cytoplasm. Using antibodies specific for Rbfox2, Rbfox3, and Rbfox RRM, we detected Rbfox3, but not Rbfox2, in the cytosolic fraction. Thus, Rbfox1 and Rbfox3 are the major Rbfox paralogs in the cytoplasm of hippocampal neurons.
Rbfox1 regulation of mRNA expression
Only a few changes in mRNA expression levels were detected in the brain of Rbfox1 KO mice (Gehman et al., 2011; Lovci et al., 2013), which we postulated was due to compensation by Rbfox3. To examine the cytoplasmic function of Rbfox1, we isolated dissociated mouse hippocampal neurons, which express Rbfox1_C at high levels. To obtain a neuron-enriched culture, we added AraC to eliminate glial cells and co-cultured with filter inserts containing glial cells (Figures 2A, S1A, and S1B). We performed double siRNA-mediated knockdown (KD) of Rbfox1 and Rbfox3 (Figure 2B). After acute KD, we used microarray analysis to detect changes in the transcriptome (n = 3 biological replicates). Analyzing for splicing changes, we identified 338 up-regulated and 208 down-regulated alternative cassette exons in the double KD neurons (Figure 2C). Our results recapitulated 11 of the 20 splicing changes identified in the whole brain of Rbfox1 KO mice (Gehman et al., 2011).
Figure 2. Rbfox1 regulates mRNA expression: knockdown and microarray analysis.
(A) Experimental flow chart.
(B) Immunoblot showing the knockdown (KD) of Rbfox1 and Rbfox3 proteins in neurons incubated with Accell siRNAs targeting Rbfox1 (siRF1) and Rbfox3 (siRF3). GAPDH is used as loading control.
(C) Histogram of up-regulated (Up) and down-regulated (Dn) exons in KD experiments.
(D) Histogram of up-regulated (Up) and down-regulated (Dn) gene in KD experiments.
(E) Histogram of percentage of genes that contain ≥1 UGCAUG motif conserved in the 3′ UTR in human, mouse, rat, dog, and chicken genomes (UGCAUG_5G), ≥1 UGCAUG motif conserved in human and mouse genomes (UGCAUG_2G), or ≥1 GCAUG motif conserved in human and mouse genomes (GCAUG_2G). *** p < 10−15 by hypergeometric test.
For additional data, see Figures S1 and S2.
In addition to splicing changes, we identified 746 genes exhibiting changes in abundance (expression) in the double KD (Figure 2D and Table S1), and validated a subset of genes by RT-qPCR analysis (20/23 = 87%, Figure S1C). The majority (676 genes, or 91%) were down-regulated in the double KD, indicating that their abundance is normally increased by Rbfox1 and Rbfox3. We also measured the protein products of several regulated genes by quantitative immunoblotting. As shown in Figure S2, mRNAs of calcium/calmodulin-dependent protein kinase (CaMK) family members, CaMK2A, CaMK2B, and CaMK4, were reduced in double KD cultures with a corresponding decrease in the concentration of CaMK2A and CaMK2B, but not CaMK4 protein. We also observed a slight increase in Rbfox2 mRNA and a 2-fold increase in Rbfox2 protein in the Rbfox1 and 3 knockdown cultures, as was reported in Rbfox1 KO mice (Gehman et al., 2011). These data suggest that Rbfox1 or Rbfox3 may repress Rbfox2 expression at the translational level to form a negative regulatory loop for Rbfox proteins in neurons.
To determine whether the changes in mRNA expression were caused directly or indirectly by the loss of Rbfox1 and Rbfox3, we searched for conserved (U)GCAUG sequence motifs in the 3′ UTRs of transcripts as an indicator of direct Rbfox1 regulation. The down-regulated gene set was significantly enriched for genes containing a 3′ UTR UGCAUG motif that was conserved across human, mouse, rat, dog, and chicken genomes (12%, p < 10−15, hypergeometric test, Figure 2E). Considering only conservation between human and mouse genomes, this enrichment increased to 41% (p < 10−15, hypergeometric test, Figure 2E). In contrast, we detected no enrichment of (U)GCAUG motifs in up-regulated genes. Together, these findings indicate that in addition to regulating alternative splicing, Rbfox1 and 3 regulate mRNA and protein abundance, and suggest that this regulation occurs by direct RNA binding to (U)GCAUG sites in the 3′ UTR.
Rbfox1_C but not Rbfox1_N rescues the expression changes induced by double KD of Rbfox1 and 3
To differentiate between the functions of Rbfox1_C and Rbfox1_N, we performed double KD of Rbfox1 and 3 and then rescued with either siRNA resistant Rbfox1_C (Flag-Rbfox1_C_siMt) or Rbfox1_N (Flag-Rbfox1_N_siMt). To facilitate these experiments, we performed them in cultures containing both glia and neurons, rather than in the filter co-culture system (after confirming that selected genes were regulated by double KD in both culture systems, Figure S3). To achieve cell-type specific rescue, we used AAV2/9 vectors with the human synapsin I promoter driving the expression of Flag-Rbfox1_C_siMt and Flag-Rbfox1_N_siMt in neurons (Figures S4A and S4B). The concentrations of virus were adjusted to match the expression of endogenous Rbfox1 and Rbfox3 (Figure S4C). Immunocytochemistry with Flag antibodies revealed that virally expressed Rbfox1_C localized predominantly to the cytoplasm and processes, with low but detectable expression in the nucleus (Figures S4D and S4F), while virally expressed Rbfox1_N localized predominantly in the nucleus, with low levels of detection in the somatic cytoplasm (Figures S4E and S4G). We performed RNAseq on: 1) control cells treated with non-targeting siRNAs and virus expressing EGFP; 2) cells treated with Rbfox1 and 3 siRNAs and virus expressing EGFP; 3) cells treated with Rbfox1 and 3 siRNAs and virus expressing Flag-Rbfox1_C_siMt; and 4) cells treated with Rbfox1 and 3 siRNAs and virus expressing Flag-Rbfox1_N_siMt (Figure S4C).
Strikingly, we found that changes in splicing were induced predominantly by Rbfox1_N while the large majority of changes in mRNA abundance (expression) were regulated by Rbfox1_C. Comparing Rbfox1_N rescue to double KD, we identified 146 up-regulated and 172 down-regulated alternative cassette exons (Figure 3A). Fewer exons were affected by the Rbfox1_C rescue of the double KD: 89 up-regulated and 61 down-regulated alternative cassette exons. Of the exons affected by either Rbox1 isoform, 81% changed in the same direction, and of this set 74% were more strongly affected by Rbfox1_N than by Rbfox1_C (more intense green and red signal in Figure 3B in the Rbfox1_N column than in the Rbfox1_C column). Exons affected by Rbfox1_C are potentially directly regulated by its low concentrations in the nucleus, or could be indirectly affected by changes in other proteins. An opposite pattern was observed in expression changes: the expression of 275 genes was altered by Rbfox1_C, whereas Rbfox1_N only affected the expression of 49 genes (Figure 3C). Of the genes whose expression was altered by either Rbfox1 isoform, 91% showed the same direction of change, and most of these genes (79%) showed a greater magnitude of change with Rbfox1_C rescue than with Rbfox1_N rescue (more intense green and red signal in Figure 3D in the Rbfox1_C column than in the Rbfox1_N column). These results indicate that Rbfox1_N predominately regulates pre-mRNA splicing, while Rbfox1_C predominately regulates mRNA abundance. We thus focused our subsequent analyses of splicing on the effects of Rbfox1_N and our analyses of overall mRNA abundance on Rbfox1_C.
Figure 3. Knockdown and rescue experiments identify distinct functions for Rbfox1_C and Rbfox1_N.
(A) Histogram of up-regulated (Up) and down-regulated (Dn) exons rescued by Rbfox1_N or Rbfox1_C compared to KD. p = 7.74e-15 by Pearson’s Chi-squared test with Yates’ continuity correction.
(B) Heatmap of the differences in percentage of splicing (Ψ) for the exons in (A).
(C) Histogram of up-regulated (Up) and down-regulated (Dn) genes rescued by Rbfox1_N or Rbfox1_C compared to KD. p = 3.125e-36 by Pearson’s Chi-squared test with Yates’ continuity correction.
(D) Heatmap of the Log2 fold change (Log2FC) in expression for the genes in (D).
(E) Histogram of the percentage of genes that contain conserved UGCAUG motifs in the 3′ UTR. Up = up-regulated, Dn = down-regulated, Non = not changed; In = mRNA concentration increased by Rbfox1, De = mRNA concentration decreased by Rbfox1. * p < 10−5, ** p < 10−10, and *** p < 10−15 by hypergeometric test.
(F) Weighted Venn diagram showing the overlap of genes regulated by Rbfox1 at the level of expression and splicing. P values are calculated by hypergeometric test.
For additional data, see Figures S3, S4, and S5.
To define a set of genes whose splicing is regulated by Rbfox1_N for downstream analyses, we selected exons showing opposite changes in splicing between 1) control and double KD and 2) double KD and Rbfox1_N rescue, and filtered for exons whose splicing change reaches statistical significance in either 1) or 2). Using these criteria, we defined 182 Rbfox1 activated exons and 184 Rbfox1 repressed exons in a total of 332 genes as Rbfox1_N splicing targets (Tables S1 and S2). Examining the flanking upstream and downstream introns of these exons, we found that GCAUG motif was the most enriched pentamer in these regions. Consistent with previous studies, the GCAUG motif was particularly enriched in the proximal region of introns downstream of the Rbfox1 activated exons (Underwood et al., 2005; Yeo et al., 2009; Zhang et al., 2008). Next, we examined the frequency of conserved (U)GCAUG motifs in the 3′ UTR sequences of transcripts whose expression was altered in double KD compared to control and in Rbfox1_C rescue compared to double KD. Consistent with the results in Figure 2E, genes whose expression was down-regulated by double KD of Rbfox1 and 3 were significantly enriched for conserved (U)GCAUG motifs in their 3′ UTRs. Correspondingly, genes whose expression was up-regulated by Rbfox1_C rescue were also enriched for conserved (U)GCAUG motifs in the 3′ UTR. This enrichment was not observed in genes whose expression was down-regulated by Rbfox1_C rescue (Figure 3E). We also compared the transcriptome data obtained by microarray and RNAseq methods. We found that the sets of genes down-regulated by double KD measured by these two methods overlapped significantly (odds-ratio (OR) = 2.97, p = 1.9e–9). There was also significant overlap between the set of down-regulated genes in the KD measured by microarray and the set of genes up-regulated by Rbfox1_C rescue measured by RNAseq (OR = 8.4, p = 1.1e–16). In contrast, we did not observe significant overlap between the up-regulated gene sets in the KD experiments measured by microarray and RNAseq (Figures 2E and 3E). Thus, gene expression changes that positively correlate with Rbfox1_C expression levels are reproducible between assays. These observations support a role for Rbfox1_C in increasing the stability, and thus the abundance of transcripts containing the (U)GCAUG motifs in their 3′ UTR.
To define a high confidence set of “expression” targets of Rbfox1_C (those whose mRNA abundance is regulated by Rbfox1_C), as opposed to the set of “splicing” targets of Rbfox1_N defined earlier, we used data from microarray and RNAseq experiments and selected genes that showed opposite changes in expression between 1) control and double KD and 2) double KD and Rbfox1_C rescue, filtering for genes whose change in expression reached statistical significance in either 1) or 2). This identified 613 genes whose mRNA abundance (“expression”) was down-regulated by Rbfox KD but up-regulated upon Rbfox1_C rescue and 403 genes whose abundance was up-regulated by Rbfox KD but down-regulated by Rbfox1_C rescue (Table S1). We termed the former “Rbfox1-increased genes” and the latter “Rbfox1-decreased genes” in the set of “expression” targets. A subset of each group was validated by RT-qPCR (Figure S3). We found that Rbfox1-increased transcripts, but not Rbfox1-decreased genes, were significantly enriched for conserved 3′UTR (U)GCAUG motifs (Figure 3E and Table S1, 40% of the Rbfox1-increased genes, p < 10−15 by hypergeometric test).
There was no significant overlap between the set of transcripts whose abundance was regulated by Rbfox1_C and the set of transcripts whose splicing was regulated by Rbfox1_N (Figures 3F and S5). The genetic programs controlled by these different Rbfox1 isoforms thus appear to be distinct.
Rbfox1 proteins bind to the 3′ UTR of target mRNAs in the cytoplasm
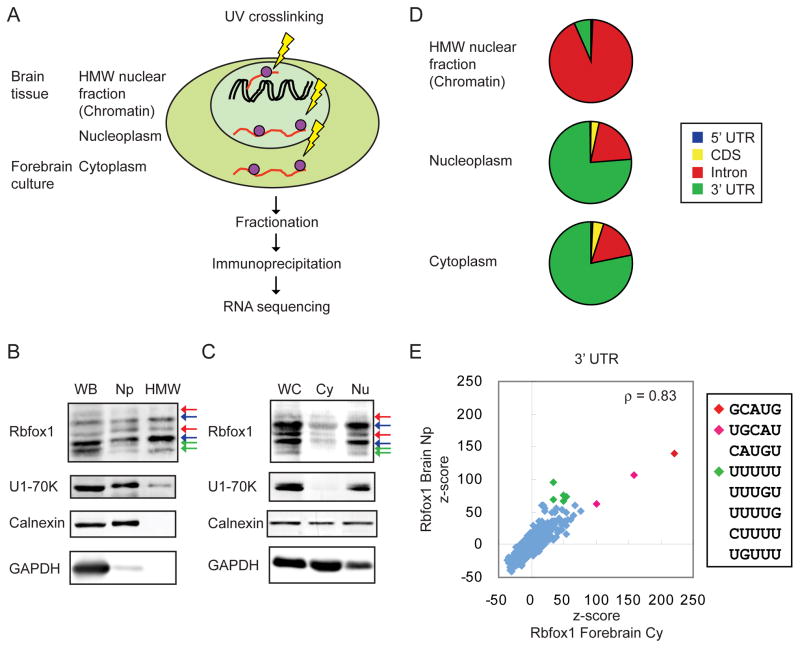
To identify mRNAs physically bound by Rbfox1, and to assess the location of Rbfox1 binding sites across the transcriptome, we performed iCLIP-seq analysis (Konig et al., 2010). Since our rescue experiments indicated that Rbfox1 has distinct nuclear and cytoplasmic functions (Figures 3A and 3C), we performed subcellular fractionation prior to immunoprecipitation to identify Rbfox1 targets (Figure 4A). We previously found that the majority of unspliced RNA fractionates with chromatin from isolated nuclei (Bhatt et al., 2012; Khodor et al., 2012; Pandya-Jones et al., 2013). To identify the binding of nuclear Rbfox proteins, we generated iCLIP data sets of Rbfox1, 2, and 3 in the high molecular weight (HMW) nuclear fraction containing chromatin, unspliced RNA, and nuclear speckle proteins, and the soluble nucleoplasm (Np) fraction from mouse brains (6 week old male mice). To profile the binding of cytoplasmic Rbfox1 proteins, we performed iCLIP on a cytoplasmic (Cy) fraction of cultured mouse neurons (DIV14). The analyses in this paper focus on the cytoplasmic fraction, and use the soluble Np and HMW fractions for comparison.
Figure 4. Characterization of Rbfox1 iCLIP tags in the 3′ UTR.
(A) Illustration of iCLIP experimental workflow. HMW: high molecular weight nuclear fraction that contains chromatin and unspliced RNA.
(B, C) Immunoblot analysis of purity of the fractions. WB: whole brain; Np: nucleoplasm; HMW: high molecular weight nuclear fraction containing chromatin; WC: whole culture; Cy: cytoplasm; Nu: nucleus.
(D) Pie charts of the percentage of Rbfox1 clustered tags mapped to 5′ UTR, codingsequence (CDS), intron, and 3′ UTR in different iCLIP experiments.
(E) Scatter plots of the z-scores of pentamers around the Rbfox1 crosslink sites in the 3′ UTR in the cytoplasm and nucleoplasm.
Immunoblotting showed that the soluble Np fraction from mouse brain was depleted of the cytoplasmic marker GAPDH and contained the nuclear marker U1-70K (Figure 4B). Both Rbfox1_C and Rbfox1_N isoforms were observed in the HMW and Np fractions. The ER marker protein calnexin was also detected in the soluble Np fraction, suggesting that ribosome-loaded mRNA transcripts associated with the ER copurified with the nuclei (Bhatt et al., 2012). To obtain sufficient material for the Cy fraction, we used forebrain neurons rather than hippocampal neurons. Immunoblot analysis showed that the Cy fraction was enriched for the cytoplasmic protein GAPDH and lacked the nuclear marker U1-70K (Figure 4C).
Following UV crosslinking and cellular fractionation, we immunoprecipitated Rbfox1 with the Rbfox1-specific monoclonal antibody 1D10 and used anti-Flag antibodies as a negative control. We sequenced the RNA fragments crosslinked to immunoprecipitated Rbfox1 and obtained 2.2 and 3.2 million unique Rbfox1 iCLIP tags from the Np fraction and the Cy fraction, respectively (Table S3). These iCLIP tags were mapped to the longest transcript of each gene in the UCSC Known Gene Table (Hsu et al., 2006). In iCLIP, the UV crosslink site is located one nucleotide (nt) upstream of the 5′ end of the aligned iCLIP tag. To define reproducible and clustered crosslink sites, the probability of each site was calculated based on the number of tags mapping to that particular crosslink site compared to random sites. Crosslinked sites with an FDR < 0.01 were selected, and those located within 20 nts of one another were clustered. Clusters with width > 1 nt were used for downstream analyses. With these criteria, significant iCLIP clusters were identified in the Rbfox1 Np and Cy fractions, comprised of 136,483 and 162,916 tags respectively (Table S3).
For the Rbfox1 Cy fraction, 78% of the clustered tags mapped to the 3′ UTR and 17% mapped to introns (Figure 4D). The distribution of mapped tags from the Np fraction was similar to that in the Cy fraction. In contrast, 93% of the Rbfox1 clustered tags mapped to introns in the HMW nuclear fraction. By comparison, analysis of Rbfox targets in whole cells in mouse brain reported that about 70% of clustered tags mapped to introns and that 20% to 27% mapped to 3′UTR (Lovci et al., 2013; Weyn-Vanhentenryck et al., 2014).
To evaluate the specificity of the iCLIP data, we examined the enrichment of pentamer motifs in the sequences surrounding Rbfox1 crosslinking sites in 3′ UTRs. In the Cy fraction, GCAUG was the most enriched pentamer within the sequence extending 40 nucleotides on either side of the crosslink site. The 50 most-enriched pentamers also included 6 that differed by one nucleotide from GCAUG, suggesting that Rbfox1 can bind to sub-optimal GCAUG motifs in vivo, as has been seen by others (Lambert et al., 2014).
As in the Cy fraction, GCAUG was also the most enriched pentamer for Rbfox1 in the Np fraction, and the rankings of pentamers were highly correlated between the Cy and the Np fractions (Figure 4E; ρ= 0.83, p < 2.2×10−16, Spearman correlation). Interestingly, several U-rich motifs were more enriched in the Np than in the Cy fraction. This could be caused by differences in Rbfox1 interacting proteins in different cellular compartments. The Rbfox1 iCLIP pentamers were also highly correlated with Rbfox2 and Rbfox3 iCLIP pentamers in the Np fraction, as expected from their similar RNA binding properties (data not shown). The extensive overlap of 3′ UTR iCLIP clusters from the soluble Np and Cy fractions indicates that Rbfox likely binds fully processed mRNAs in the nucleus and accompanies them to the cytoplasm.
Cytoplasmic Rbfox1 increases the expression levels of iCLIP target genes
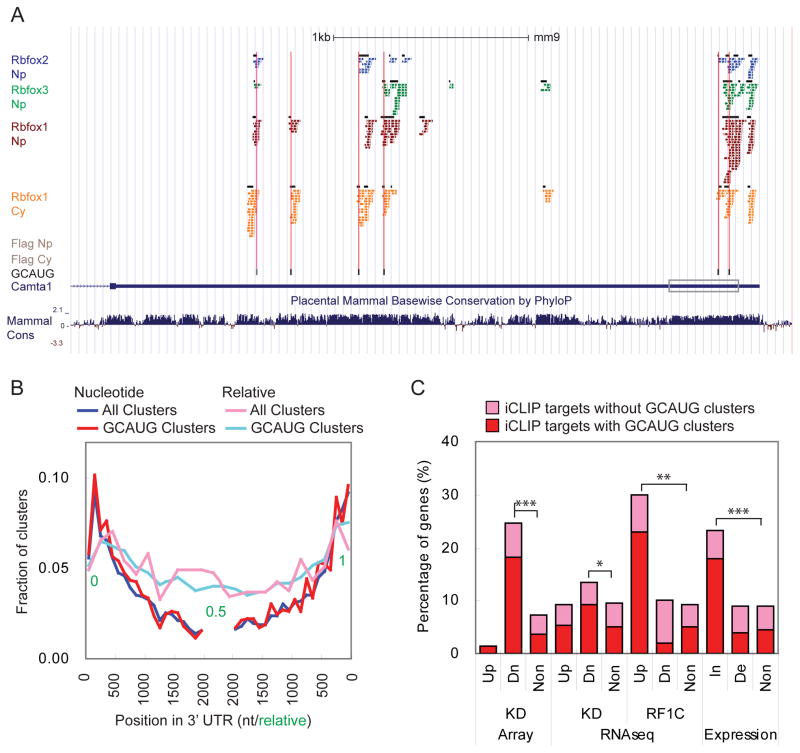
We next characterized the properties of the iCLIP clusters. We defined clusters (“GCAUG clusters”) as high confidence Rbfox binding sites in vivo if they contained at least one GCAUG motif within 10 nts upstream or downstream of the cluster. Binding of Rbfox1 within a GCAUG cluster is illustrated by the mapped clusters in the Camta1 3′ UTR (Figure 5A). Camta1 was identified as an Rbfox1-increased gene in the KD and rescue experiments. Ten cytoplasmic Rbfox1 iCLIP clusters were identified in its 3′ UTR and 5 of them overlapped with conserved GCAUG motifs, indicating direct binding of Rbfox1 to these sites (Figure 5A). The first Rbfox1 cluster in the Camta 3′UTR did not overlap with a GCAUG motif, but had a sub-optimal GCAcG motif, suggesting that this cluster also reflected a direct Rbfox1 binding site. Other clusters that did not directly overlap with a GCAUG motif were located within 50 nts of GCAUG motifs, suggesting that crosslinking at these sites might result from an Rbfox1 interaction with a GCAUG motif.
Figure 5. Identification of Rbfox1 iCLIP 3′ UTR target genes.
(A) Screenshot of UCSC genome browser showing iCLIP tags and clusters (black box) in Camta1 3′ UTR. GCAUG motifs are underlined in black and a UGCAcG motif is underlined in gray. The boxed region indicates the part of 3′ UTR subcloned to the luciferase reporters in Figure 6A.
(B) The distribution of the clusters within binned 3′ UTR locations relative to the 5′ and 3′ ends shown as nucleotide (nt) and relative positions.
(C) Histogram of the percentage of genes that contain iCLIP clusters in the 3′ UTR in different experiments. Up: up-regulated in the experiment; Dn: up-regulated in the experiment; In: mRNA increased by Rbfox1; De: mRNA decreased by Rbfox1. * p < 10−5, ** p < 10−10, and *** p value < 10−15 by hypergeometric test.
For additional data, see Figure S6.
Examining the distribution of GCAUG clusters in cytoplasmic Rbfox1 target transcripts, we found that GCAUG clusters were enriched at the 5′ and 3′ ends of the 3′ UTRs (Figure 5B). Similar 5′ and 3′ enrichment has been observed for proteins and miRNAs controlling mRNA stability and translation (Bartel, 2009; Boudreau et al., 2014; Chi et al., 2009). The binding of Rbfox1 to these regions is consistent with our observations of cytoplasmic Rbfox1 affecting mRNA abundance. We defined a set of 3′ UTR target genes as containing at least one significant iCLIP cluster and a total of 11 tags in their 3′ UTRs. Of these 1490 genes identified in the Cy fraction, 788 (53%) contained a cluster with a GCAUG motif (Table S2). In the Np fraction, 915 genes were identified as Rbfox1 3′ UTR target genes in brain tissue. Of these genes, 400 (44%) contained GCAUG clusters in 3′ UTR (Table S4 and Figures S6A and S6B).
We next asked whether Rbfox1-mediated changes in mRNA abundance correlated with Rbfox1 binding to the 3′ UTR. We found that Rbfox1-increased genes were significantly enriched for genes containing 3′ UTR clusters compared to non-regulated genes (23%, p < 10−15, hypergeometric test, Figure 5C). In contrast, Rbfox1-decreased genes were not enriched for Rbfox1 iCLIP targets. Subdividing iCLIP target genes into those with or without a GCAUG cluster in the 3′ UTR, we found those with GCAUG clusters were enriched in Rbfox1-increased genes. These results indicate that Rbfox1 binding to 3′ UTR GCAUG motifs increases the level of the target mRNAs in the cytoplasm. While binding of Rbfox1 to mRNAs lacking 3′ UTR GCAUG motifs were identified by iCLIP, these interactions were not correlated with any changes in mRNA abundance. We thus focused on iCLIP targets with GCAUG clusters and used these as high confidence Rbfox1 binding targets for downstream analyses.
Binding of Rbfox to 3′ UTR GCAUG motifs in the cytoplasm increases target mRNA concentration and translation
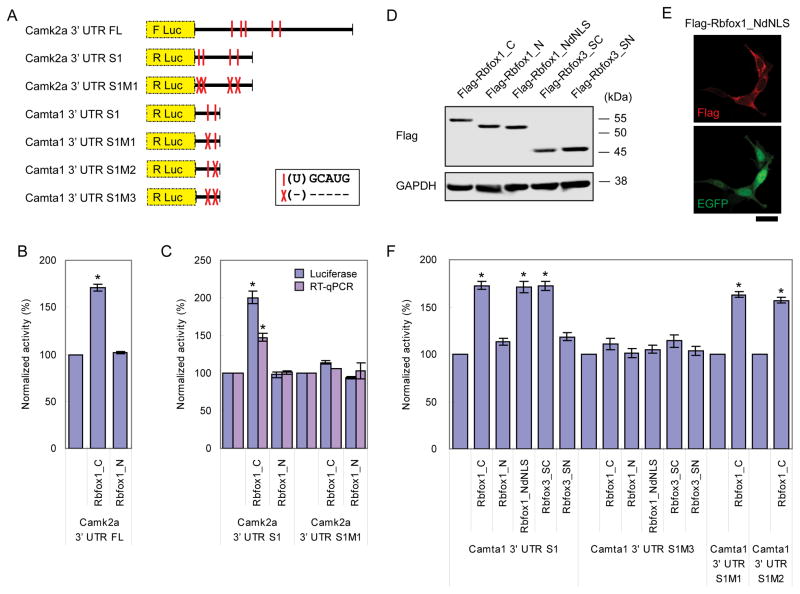
We next tested whether binding of Rbfox1 to the 3′ UTR was sufficient to alter target mRNA concentration and translation. We focused on Camk2a because of its roles in memory and in synaptic plasticity (Lisman et al., 2002). Both Camk2a mRNA and protein were decreased by approximately 40% upon Rbfox1 and 3 KD (Figure S2). The Camk2a 3′ UTR is 3372 nt long and contains five GCAUG motifs. Two of the five motifs overlapped with Rbfox1 iCLIP clusters (Figure S6C). We coexpressed a luciferase reporter gene containing the full length Camk2a 3′ UTR (Figure 6A) with Rbfox1 in HEKT cells and found that Rbfox1_C but not Rbfox1_N induced a 70% increase in luciferase activity (Figure 6B). To test the role of the Rbfox binding sites, we generated a reporter containing a shorter 1.1 kb fragment of the Camk2a 3′ UTR and deleted the 4 (U)GCAUG motifs within this fragment (Figure 6A). When the wildtype Camk2a reporter was coexpressed with Rbfox1_C or Rbfox1_N in HEKT cells, a twofold increase of luciferase activity was observed with Rbfox1_C but not with Rbfox1_N (Figure 6C). RT-qPCR analysis of the luciferase mRNA revealed a somewhat smaller increase in mRNA, indicating that the change in luciferase expression is at least partially due to changes in mRNA stability. Deletion of the (U)GCAUG motifs abolished the Rbfox1_C-induced increase in luciferase activity and in reporter mRNA levels.
Figure 6. Rbfox1 and Rbfox3 proteins increase gene expression by binding to the 3′ UTR.
(A) Diagram depicting the luciferase reporters containing part of Camk2a 3′ UTR or Camta 3′ UTR as labeled in Figures S6 and 5A, respectively. (U)GCAUG motifs are denoted by vertical red lines, with red crosses marking deletion of these motifs.
(B) Histogram of normalized luciferase activity of the reporter containing full length (FL) Camk2a 3′ UTR when coexpressed with Rbfox1_C or Rbfox1_N in HEKT cells. Error bars indicate SD. * p < 0.001 by student’s t-test. N = 3.
(C) Histogram of normalized luciferase activity and mRNA concentration (RT-qPCR) of Camk2a 3′ UTR S1 reporters when coexpressed with Rbfox1_C or Rbfox1_N in HEKT cells. Error bars indicate SD. * p < 0.001 by student’s t-test. N = 3.
(D) Immunoblot showing of Rbfox1 and Rbfox3 splice isoforms and a mutant expressed in HEKT cells. dNLS = delete nuclear localization signal, SC = short cytoplasmic isoform, and SN = short nuclear isoform.
(E) Immunocytochemistry of Flag-Rbfox1_NdNLS detected with anti-Flag antibodies (red) and the whole cell labeled with EGFP (green) in HEKT cells. Scale bar = 20 μm.
(F) Histogram of normalized luciferase activity of Camta1 3′ UTR S1 reporters when coexpressed with Rbfox1 or Rbfox3 isoform or mutant in HEKT cells. Error bars indicate SD. * p < 0.001 by student’s t-test. N = 3.
Cell fractionation and overexpression data indicated that a portion of Rbfox1_N is cytoplasmically localized (Figures 1D and S4G). This motivated us to test the activity of Rbfox1_N in the cytoplasm by deleting the NLS peptide sequence FAPY (Rbfox1_NdNLS, Figure 6E). Coexpression of Rbfox1_NdNLS with a luciferase reporter gene containing part of the Camta1 3′ UTR (containing iCLIP GCAUG clusters) revealed that the mutant, cytoplasmically localized Rbfox1_NdNLS significantly increased luciferase activity of the reporter (Figure 6F). Alternative splicing of exon 15 of Rbfox3 also generates a cytoplasmically localized Rbfox3_SC isoform (short cytoplasmic Rbfox3) (Kim et al., 2009b). We found that Rbfox3_SC also increased luciferase activity when expressed with the Camta1 3′UTR luciferase reporter (Figure 6F). Together, these results indicate that multiple isoforms of Rbfox1 or 3 can increase mRNA concentration and promote translation as long as they localize to the cytoplasm.
Rbfox1_C increases the expression of genes affecting synaptic activity and autism
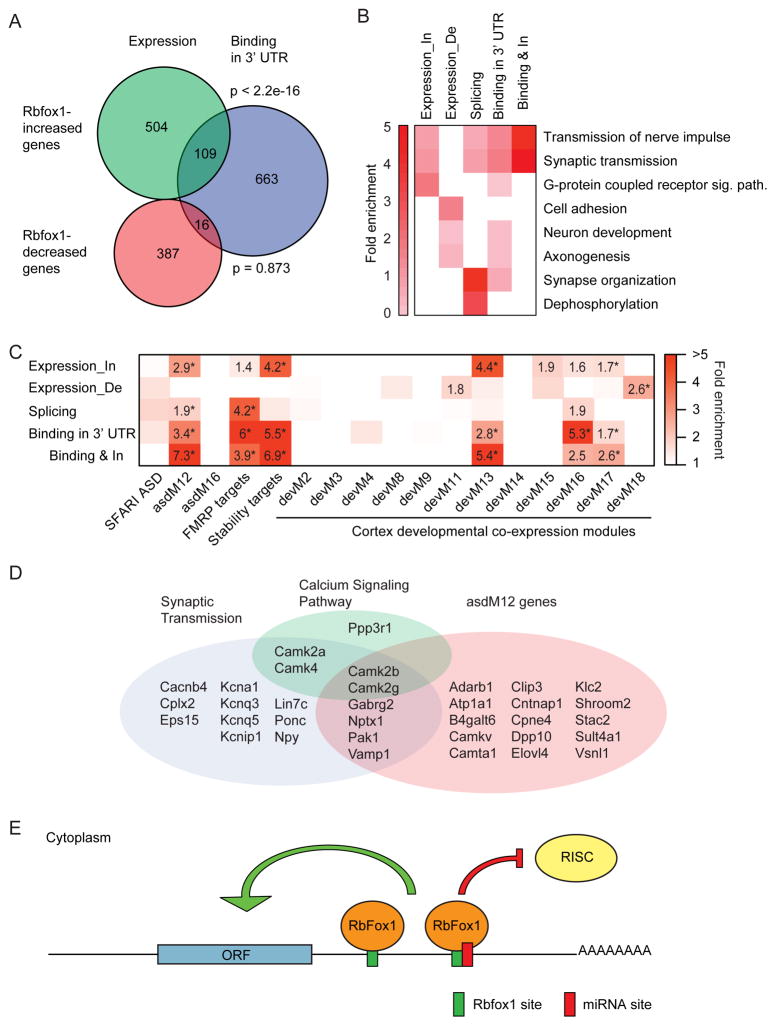
To define a high confidence set of genes whose expression is directly regulated by Rbfox1 in the cytoplasm, we combined our iCLIP data with the transcriptome data to identify genes that had opposite expression changes in the KD and Rbfox1_C rescue experiments and that had cytoplasmic Rbfox1 iCLIP GCAUG clusters in their 3′ UTRs. The overlap between Rbfox1-increased genes and iCLIP target genes was highly significant, whereas the overlap between Rbfox1-decreased genes and iCLIP targets was not significant (Figure 7A). These analyses identified a set of 109 directly bound Rbfox1-increased transcripts for downstream functional analyses (Table S5).
Figure 7. Rbfox1 increases the expression level of autism genes in the cytoplasm.
(A) Weighted Venn diagram showing the overlap of genes whose expression is increased or decreased by Rbfox1 and iCLIP target genes with Rbfox1 bound in 3′ UTR. P values calculated by hypergeometric test.
(B) Heatmap of the fold enrichment of GO terms in different experiments. In = increased. De = decreased. Sig. path. = signaling pathway.
(C) Heatmap of the enrichment of Rbfox1 regulated genes in gene sets of candidate autism genes, SFARI ASD (Basu et al., 2009), ASD-associated co-expression modules from human cortex ,asdMs (Voineagu et al., 2011), FMRP targets (Darnell et al., 2011), Rbox1 3′ UTR stability targets (Ray et al., 2013), and co-expression modules from fetal cortex, devMs (Parikshak et al., 2013). All enrichment values for overrepresenting sets with odds ratio > 1.5 are shown. * FDR adjusted two-sided Fisher’s exact test p value < 0.05.
(D) Venn diagram showing the overlap of direct Rbfox1_C -increased genes in three functional categories.
(E) A model for the cytoplasmic function of Rbfox1 in neurons. Rbfox1 binds to the 3′ UTR of target mRNAs and increases their concentration in the cytoplasm of neurons. Rbfox1 binding is predicted to antagonize miRNA binding to a miRNA binding site overlapping or neighboring an Rbfox1 binding site in the 3′ UTR.
For additional data, see Figure S7.
Gene ontology (GO) analysis of the Rbfox1 regulated genes shown in Figure 3F and 7A revealed that the Rbfox1-increased genes and iCLIP 3′ UTR targets were enriched for terms of transmission of nerve impulse and synaptic transmission (Figure 7B and Table S6). These enrichments were even greater in the high confidence set of 109 direct Rbfox1_C-increased genes. Analyzing the 109 genes using the Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed a significant enrichment for calcium signaling pathways, including 4 CaM kinases (Camk2a, Camk2b, Camk2g, and Camk4) and one calcineurin B (Ppp3r1) (Table S7). Enrichment analysis using Mammalian Phenotype Ontology (Smith and Eppig, 2012) revealed that the 109 direct Rbfox1_C-increased genes were enriched for phenotypes related to seizure (Table S8). Together, these results indicate that Rbfox1_C target mRNAs play an important role in controlling synaptic activity, in particular via the calcium signaling pathway.
We next compared Rbfox1-regulated genes to modules within a gene co-expression network derived from human cortical gene expression data from fetal brain to 3 years of postnatal development. Three of these coexpression modules, devM13, devM16, and devM17, were enriched for the GO term of synaptic transmission, as well as ASD susceptibility genes (Parikshak et al., 2013). As shown in Figure 7C, we found that the 109 direct Rbfox1_C-increased genes were highly enriched in the same three synaptic modules. To directly examine the correlation with ASD, we compared the direct Rbfox1_C-increased genes to several sets of ASD candidate genes and found that the Rbfox1_C-increased genes were enriched in an ASD coexpression module (asdM12) that is down regulated in post mortem cerebral cortex from patients with ASD (Figure 7C) (Voineagu et al., 2011). Notably, Rbfox1 was characterized as a hub gene in asdM12 and while its putative splicing targets were only modestly enriched, it was subsequently hypothesized to increase the mRNA stability of these ASD genes (Ray et al., 2013). Our finding that genes whose expression was directly increased by Rbfox1_C were significantly enriched for these ASD related genes supports this hypothesis. Rbfox1_C-increased genes also showed high enrichment of Fragile X mental retardation protein (FMRP) targets (Darnell et al., 2011), further connecting post-transcriptional regulation with ASD biology. The substantially stronger correlation of the ASD module with cytoplasmic Rbfox1 regulation than with its nuclear splicing targets underscores the need to understand this portion of the Rbfox1 program.
Rbfox1_C may compete with microRNAs to regulate mRNA stability and translation
Several RBPs have been shown to regulate microRNA (miRNA) activity by binding to the 3′ UTRs of target mRNAs (Ciafre and Galardi, 2013; Srikantan et al., 2012; Xue et al., 2013). Since Rbfox1 showed an opposite activity to that of miRNAs, but exhibited similar enriched binding at the 5′ and 3′ ends of 3′ UTR (Boudreau et al., 2014; Chi et al., 2009), we hypothesized that the binding of Rbfox1 may interfere with the binding of miRNAs and thereby antagonize miRNA activity. To test this hypothesis, we compared the Rbfox1 iCLIP data to an Ago CLIP dataset generated from P13 mouse neocortex (Chi et al., 2009). We searched for miRNA binding sites located within 50 nucleotides of the Rbfox1-bound GCAUG motifs (Tables S9). Since one Rbfox1 binding site can be close to multiple miRNA sites and vice versa, the number of single Rbfox1 site and miRNA site pairs with the same 50 nt interval was counted. By this criterion, 196 pairs of Rbfox1 and miRNA sites were identified. This number was significantly higher than the number of pairs identified after randomization of miRNA sites within the same 3′ UTR where the miRNA sites were identified, indicating that the proximity of Rbfox1 binding sites to miRNA sites within the 3′ UTR was not by chance (Figure S7A, z score = 5.38). These 196 pairs included 173 miRNA sites and 109 GCAUG motifs in 87 genes (Table S10). We found that Rbfox1-bound GCAUG motifs were more conserved than GCAUG motifs present in the same 3′ UTR but not bound by Rbfox1 (Figure S7B). The conservation scores of GCAUG motifs and their flanking sequences were greater for Rbfox1-bound GCAUG motifs that were adjacent to or overlapping a miRNA site, indicating that the colocalization is under high selection pressure, consistent with it playing an important role in the regulation of gene expression. The Ago CLIP data used here identified the binding sites of the 20 most abundant miRNAs (Chi et al., 2009). We found that 14 of these 20 miRNA sites overlapped with a GCAUG motif in at least one 3′ UTR (Figure S7C). For example, the Camk2a 3′ UTR was found to contain 11 miRNA binding sites, three of which, miR-26, miR-124, and miR-30 binding sites, were located within 50 nucleotides of the most upstream GCAUG motif bound by Rbfox1 (Figures S6C and S7C), with the miR-124 site overlapping with the GCAUG motif. In the context of our data showing that Rbfox1 binding stabilizes mRNAs and promotes translation (Figure 6), this finding suggests that Rbfox1 blocks Ago binding to these three miRNA sites. Within the 50 nts surrounding the GCAUG motif, the number of miRNA seed sites was greatest in regions overlapping the GCAUG motif and gradually decreased with distance from GCAUG motif (Figure S7D), indicating that the miRNA sites were concentrated in regions in which Rbfox1 would be expected to interfere with miRNA binding.
DISCUSSION
A cytoplasmic function for Rbfox proteins
The goal of this study was to delineate the function of cytoplasmic Rbfox. Using KD and rescue approaches together with Rbfox1 iCLIP of subcellular neuronal fractions, we identified 109 genes whose abundance was directly regulated by Rbfox1_C. Our data indicate that cytoplasmically localized Rbfox1 promotes the stability and/or translation of target transcripts by binding to their 3′ UTRs (Figure 6). We also show that cytoplasmic Rbfox1 targets are enriched in human cortical development modules affecting synaptic function and ASD (Figure 7C). Our findings highlight the importance of considering the cytoplasmic arm of Rbfox1 regulation in linking Rbfox1 to synaptic function and to neurodevelopmental disorders such as ASD.
We focused on one nuclear and one cytoplasmic Rbfox1 isoform, but there are others. Using a monoclonal antibody 1D10 that targets the N-terminal sequence of Rbfox1 and a polyclonal antibody against Rbfox RRM (Figure 1D), we detected six Rbfox1 bands ranging from 45 and 55 kDa that were all reduced by a pan-Rbfox1 siRNA (Figure 1B), and eliminated in the Rbfox1 KO mouse (data not shown). Two of these bands were eliminated by an siRNA targeting the exon of the cytoplasmic isoforms (Figure 1B). We previously found that the two of the middle bands likely represent N-terminally cleaved Rbfox1 isoforms (Lee et al., 2009). Additional bands of Rbfox1 may represent differentially phosphorylated proteins. The diverse species of Rbfox1 imply many levels of regulation and/or function. It will be interesting to examine whether and how neuronal activity regulates Rbfox1 splicing, proteolysis and/or post-translational modifications, as such modifications could alter Rbfox1 regulation of gene expression.
Over 90% of the changes in mRNA level detected by microarray analysis represented decreases in expression induced by KD of Rbfox1 and 3, reflecting a role for Rbfox in stabilizing mRNAs (Figure 2D and Table S1). In contrast, the RNAseq experiments identified more similar numbers of up and down-regulated genes (Figure 3C), although significantly more down-regulated genes contained (U)GCAUG sequences in their 3′UTRs. One difference that could lead to differences in the gene expression was that the microarray analyses were performed on pure neurons cultured with glial cells in inserts, while the RNAseq analyses were performed on mixed neuronal-glial cultures. Thus, the cell environment and culture conditions may be reflected in the transcriptome analyses. We addressed this variation by considering all of our datasets together – including the KD in both culture conditions, the rescue experiments, and the iCLIP target sets (Table S1) – and focused on genes that showed consistent changes in all of the experiments. Most importantly, we focused on transcripts that underwent opposite directions of regulation in the KD and Rbfox1_C rescue experiments and that were bound by Rbfox1 in iCLIP experiments. The results of these analyses identify a large class of transcripts whose expression was positively regulated by Rbfox1_C (Table S5). There may yet be some transcripts that are negatively regulated by Rbfox1, but these will require additional experiments to identify.
Our data add to a growing literature revealing the multifunctionality of RBPs (Bielli et al., 2011; Heraud-Farlow and Kiebler, 2014; Turner and Hodson, 2012; Vanharanta et al., 2014). The differences in activity between Rbfox1 isoforms are reflected in the iCLIP data. While the majority of the Rbfox1 binding was detected in introns in the HMW fraction, the binding of Rbfox1 shifted to 3′ UTR when assayed in the soluble nucleoplasm and the cytoplasm. Although not always specifically localized to the nucleus or cytoplasm, other RNA binding proteins have been found to bind introns to affect splicing, as well as 5′ or 3′ UTRs to affect translation (Ince-Dunn et al., 2012; Licatalosi et al., 2008; Xue et al., 2009). In many cases, it is not clear how these functions are segregated and whether different isoforms are involved or differently modified. In the case of Rbfox1 and Rbfox3, the nuclear and cytoplasmic functions arise at least in part from differentially spliced isoforms. We note, however, that some Rbfox1_N is constitutively present in the cytoplasm (Figure 1D), and that a low but detectable amount of Rbfox1_C is present in the nucleus. The finding that the NLS deleted Rbfox1_N mutant can regulate stability and translation of target mRNAs (Figure 6F) indicates that the primary determinant of target mRNA regulation in the cytoplasm is simply cytoplasmic localization of Rbfox1 rather than any other feature of the Rbfox1_C or Rbfox1_N.
Several RBPs are known to regulate mRNA stability using different mechanisms. For example, HuR proteins and Ataxin-2 proteins can bind to AU-rich elements (AREs) in the 3′ UTR and stabilize target mRNAs (Lebedeva et al., 2011; Mukherjee et al., 2011; Yokoshi et al., 2014), while PTB and hnRNP L compete with miRNA binding in the 3′ UTR and stabilize target mRNAs (Rossbach et al., 2014; Xue et al., 2013). HuR can stimulate or inhibit miRNA binding and thus regulate mRNA decay (Kim et al., 2009a; Young et al., 2012). Our results show that Rbfox1 proteins can increase mRNA concentration in the cytoplasm, and suggest that one mechanism for this is to stabilize mRNAs by competing with miRNA binding. However, this mechanism is likely only active on a subset of Rbfox1 target transcripts. For example, we find that Rbfox1 increased expression of the luciferase Camta1 3′ UTR reporter (Figure 6F), which lacks identified miRNA binding sites. In Xenopus Oocytes, Rbfox2 (XRbm9) is exclusively expressed in the cytoplasm and directly interacts with XGld2 in the cytoplasmic polyadenylation complex to promote translation (Papin et al., 2008). Identification of the cytoplasmic interacting partners of Rbfox1 may provide insights into the molecular mechanisms of cytoplasmic Rbfox1 regulation.
Rbfox1_C regulates genes involved in synaptic function, calcium signaling, and autism
Recent studies have focused attention on Rbfox1 as a critical regulator of gene expression in cortical development (Parikshak et al., 2013; Weyn-Vanhentenryck et al., 2014), and as a candidate ASD susceptibility gene (Fogel et al., 2012; Martin et al., 2007; Sebat et al., 2007; Voineagu et al., 2011; Weyn-Vanhentenryck et al., 2014). While these studies focused on Rbfox1’s role as a splicing factor, we show that the mRNAs that are regulated by Rbfox1_C are significantly enriched for genes involved in cortical development and autism (Figure 7C) (Parikshak et al., 2013; Voineagu et al., 2011). In a coexpression analysis of the brain transcriptome from patients with autism, RBFOX1, CNTNAP1, CHRM1, APBA2 were identified as 4 hub genes, genes that are defined as being highly connected in the ASD-associated co-expression module, asdM12. These four genes, along with SCAMP5 and KLC2, were ranked highest in this module (Voineagu et al., 2011). Here, we show that Rbfox1 bind the 3′UTRs of CNTNAP1, CHRM1, SCAMP5 and KLC2 transcripts, and that CNTNAP1 and KLC2 mRNA levels are both increased by Rbfox1_C (Table S1). These results suggest that Rbfox1 is upstream of the two hub genes, CNTNAP1 and CHRM1, in a molecular pathway that is altered in autism. Mutations in FMRP are the most common single gene cause of autism (Talkowski et al., 2014), and we find that Rbfox1 3′ UTR target genes overlap significantly with FMRP target genes (Darnell et al., 2011). Together, these results link gene expression changes associated with sporadic autism with a single gene cause of autism. Our findings thus add to the emerging recognition that post-transcriptional RNA metabolism plays a critical role in cortical development and neurodevelopmental disorders (Darnell and Richter, 2012).
We find that Rbfox1_N and Rbfox1_C regulate two different sets of genes (Figure 3F). Both sets are enriched for the same GO terms of transmission of nerve impulse and synaptic transmission, but their targets can be quite different. For example, in the CaM kinase family, the splicing of Camk2d is regulated by Rbfox1_N. However, the expression of Camk2a, Camk2b, and Camk4 is regulated by Rbfox1_C, and Camk2g is regulated both at the level of splicing by Rbfox1_N and at the level of expression by Rbfox1_C (Figure S5). Taken together, our results identify a coherent and intricate gene network regulated by two distinct Rbfox1 splice isoforms and exemplify the functional consequences of alternative splicing for this RNA binding protein. The existence of multiple additional Rbfox1, Rbfox2 and Rbfox3 isoforms and variants indicates that our analysis likely uncovers only a fraction of the total complexity of Rbfox-mediated RNA regulation. Understanding the mechanisms by which RBP mutations give rise to neural circuit abnormalities and disease will require consideration of their multiple functions in RNA metabolism, in the nucleus and in the cytoplasm.
EXPERIMENTAL PROCEDURES
Tissues for Immunoblotting and RT-PCR
All experiments with animals were performed using approaches approved by the UCLA Institutional Animal Care and Use Committee. Hippocampi and cortices at postnatal day 0 (P0) and 3 weeks were dissected from C57BL/6J mice. Protein was purified from half of the tissues for immunoblotting. RNAs were purified from the other half of the tissues for RT-PCR analysis.
Subcellular Fractionation of Neuronal Cultures
Hippocampal cultures used in Figure 1 were prepared from postnatal day 0 C57BL/6J mice (Jackson Laboratory) as previously described (Ho et al., 2014) and incubated with 2 μM Cytosine beta-D-arabinofuranoside (AraC) (Sigma Aldrich, c1768) from postnatal day 3 to day 6. On day 13, the cultures were incubated with 15 μM of digitonin (Sigma Aldrich, D141-100MG) and complete protease inhibitor (Roche #05892791001) in 1X PBS buffer for 3 minutes at room temperature to permeabilize the cells and the eluate was collected as the cytosol fraction. An equal volume of RIPA buffer was then used to completely lyse the cells.
RNAi Knockdown in Neuronal Cultures
Primary mouse hippocampal and cortical cultures were prepared from postnatal day 0 C57BL/6J mice. 160,000 hippocampal cells were plated in one 12-well well and 120,000 cortical cells were plated in a cell culture insert with pore size of 3 μm (Corning Life Sciences, 353181). On day 3, hippocampal cultures were incubated with 2 μM AraC for 3 days and then co-cultured with the cortical culture from day 6. Two Rbfox1 and two Rbfox3 Accell siRNAs (Dharmacon, sequences in Supplemental Information) were added to the co-culture on day 10 at a total concentration of 1.2 mM and incubated for 4 days prior to RNA and protein extraction.
Microarray
RNA from three biological replicates for each condition was probed for gene expression and alternative splicing changes using Affymetrix MJAY microarrays (Affymetrix). Array analysis was performed using the OmniViewer method (http://metarray.ucsc.edu/omniviewer/) (Sugnet et al., 2006).
Adeno-Associated Virus (AAV) 2/9 Transduction, RNAi, cDNA Library Preparation, and RNA Sequencing
The siRNA target site in the Rbfox1 coding sequence was mutated to generate a silent mutation and the coding sequence of the mutated Flag-tagged Rbfox1_C or Rbfox1_N was then cloned into pAAV-hSyn-eNpHR 3.0-EYFP plasmid (Addgene plasmid # 26972), downstream of the hSynI promoter between AgeI and EcoRI restriction sites to replace eNpHR 3.0-EYFP coding sequence. AAV2/9 vectors containing hSyn.Flag-Rbfox1_C_siMt and hSyn.Flag-Rbfox1_N_siMt were generated at the University of Pennsylvania Vector Core Facility. An AAV2/9 vector expressing hSyn.EGFP was used as a control for transduction (Penn Vector Core, #AV-9-PV1696). Hippocampal neurons (9 DIV) were transduced with AAV2/9 vectors expressing EGFP, Flag-Rbfox1_C_siMt, or Flag-Rbfox1_N_siMt at a concentration of 1.5×103 genomic copies (GC)/cell for 12 h and then removed. The neurons (10 DIV) were then incubated with non-targeting (Dharmacon #D001910-01 and #D001910-02) or Rbfox1 and Rbfox3 Accell siRNA for 4 days at concentration of 1.2 mM. Total RNA was extracted using RNeasy Micro Kit (Qiagen). Ribosomal RNA was removed using Ribo-Zero™ rRNA Removal Kits (Epicentre), and the cDNA libraries were prepared using TruSeq RNA Sample Preparation Kit (Illumina) and sequenced in HiSeq 2000 (Illumina) (pair end, 50 nt) by the Southern California Genotyping Consortium (SCGC) Gene Expression Core Facility (Los Angeles, California). Alternative splicing changes were analyzed by SpliceTrap (Wu et al., 2011). Splicing changes where percent spliced in (psi) > 10, and reads of exon 1 > 50, exon 2 > 20, exon 3 > 50, exon 1-exon 2 junction > 5, exon 2-exon 3 junction > 5, and exon 1-exon 3 junctions > 5 in both samples in the comparison were considered significant. Gene expression changes were analyzed by Cufflinks-2.0.2. (Trapnell et al., 2010) and q < 0.3 was set to detect significant expression changes. Gene expression changes were validated by RT-qPCR on RNA samples from two independent biological replicates from those used for RNA sequencing. The expression levels of each gene were normalized to Tuj1 (Tubb3) expression for comparison. The means of normalized expression were calculated and the statistical significance was determined using a paired, one-tailed Student’s t test with significance set to p < 0.05. N = 2 biological replicates.
Criteria for Defining Expression and Splicing Gene Sets
Genes selected for inclusion in the splicing set were required to have exons that showed significant splicing changes in Rbfox1 and 3 double knockdown (KD) or Rbfox1_N rescue experiment as measured by RNA sequencing, and that showed opposite direction of splicing changes in the KD and Rbfox1_N rescue experiments. This identified 366 Rbfox1 regulated alternative exons in 332 genes as shown in Tables S1 and S2.
For the expression set, two sets of genes were selected and combined. The first set of genes was required to have significant expression changes in the KD experiment measured by microarray and showed opposite expression changes in the Rbfox1_C rescue experiment measured by RNA sequencing. The second set of genes was required to have significant expression changes in the knockdown or Rbfox1_C rescue experiment measured by RNA sequencing and showed opposite direction of expression changes in the KD and Rbfox1_C rescue experiments. This identified 613 Rbfox1-increased genes and 403 Rbfox1-decreased genes as shown in Table S1.
iCLIP Data Analyses
A high molecular weight (HMW) nuclear fraction and soluble nucleoplasm fraction were purified from the brains of 6 weeks old male C57BL/6J mice. Briefly, nuclei were purified as described (Grabowski, 2005) and lysed. Soluble and HMW fractions were separated by centrifugation. A cytoplasmic fraction was purified from mouse forebrain cultures at DIV14. iCLIP was performed according to the original protocol (Konig et al., 2010), with some modifications described in Supplemental Experimental Procedures. iCLIP sequencing results were analyzed as described in Konig et al. (Konig et al., 2010) with a few modifications. In brief, the iCLIP tags generated by PCR duplication were discarded based on the comparisons of the random barcodes in the tags. The unique iCLIP tags were then mapped to mouse genome (mm9/NCBI37) using Bowtie allowing 2 nucleotide mismatches (Langmead et al., 2009). Mapped tags were further mapped to the longest transcripts in Known Gene table (Hsu et al., 2006) and divided into four regions of 5′ UTR, CDS, intron, and 3′ UTR for downstream analyses. The first nucleotide in the genome upstream of the iCLIP tags was defined as UV-crosslink site and the significance of crosslinking at each crosslink site was evaluated by the false discovery rate (FDR) calculated as described in Konig et al. (Konig et al., 2010). Crosslink sites with FDR ≤ 0.01 were used for clustering. Any two crosslink sites located within 20 nucleotides on the mouse genome were clustered together. The width of a cluster was defined as the distance between the first and the last crosslinked sites and clusters with width > 1 nt were selected for downstream analyses. A cluster was defined as a GCAUG cluster if it overlapped with a GCAUG motif with at least one nucleotide within -10 to 10 nt genomic sequences of the cluster. The iCLIP tags in these clusters were called clustered tags.
Motif Analyses
Crosslink sites with FDR ≤ 0.01 were used for motif analyses. The genomic sequences of the crosslink site plus 40 nucleotides upstream and 40 nucleotides downstream were used for the motif enrichment analyses. The z-scores of each pentamer were calculated by comparing the occurrence of the pentamers around the crosslink sites to the occurrence around randomized sites in the same genomic region (i.e., within the same intron or 3′ UTR) to control for the differences in expression of different genes.
Identification of Rbfox 3′ UTR Target Genes
To identify 3′ UTR target genes for Rbfox1, 2, and 3 in different experiments, the genes were first ranked by the number of clustered tags in 3′ UTR. Next, to account for the differences in the number of clustered tags generated in different experiments, genes contained the top 95% of clustered tags were selected as 3′ UTR target genes. By this criterion, the top 55% of genes that contained clustered tags in 3′ UTR were selected. This set the cutoff as 11 tags/3′ UTR for the identification of Rbfox1, 2, and 3 3′ UTR target genes in the forebrain cultures and the brain tissues. The cutoffs for Rbfox1 in the forebrain and hindbrain tissues were set higher at 21 tags/3′ UTR and 16 tags/3′ UTR, respectively, due to the greater numbers of clustered tags generated in these experiments.
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed with candidate gene lists and co-expression networks using a two-tailed Fisher’s exact test followed by Benjamini-Hochberg FDR adjustment (Benjamini and Hochberg, 1995). All mouse gene set overlaps were performed using mouse Ensembl IDs, all human lists were converted to their homologous mouse Ensembl IDs using Ensembl 73 (Gencode v18), using only one-to-one orthologs. FDR adjustment took into consideration all gene set overlaps performed. The following gene lists and co-expression modules were used: candidate genes from SFARI with a gene score of S or 1–4 (Basu et al., 2009), two ASD associated co-expression modules from post-mortem human cortex (asdM12 and asdM16; (Voineagu et al., 2011)), FMRP binding targets in mouse brain (Darnell et al., 2011), predicted Rbfox1 3′ UTR stability targets (Ray et al., 2013), and 12 co-expression modules reflecting cortical developmental processes (Parikshak et al., 2013) and enriched for protein interactions and GO terms, including cellular proliferation (devM8, and devM11), transcriptional/chromatin regulation (devM2, and devM3), and synaptic development (devM13, devM16, and devM17).
Supplementary Material
Highlights.
Nuclear and cytoplasmic Rbfox1 isoforms regulate distinct neuronal mRNAs.
Cytoplasmic Rbfox1 regulates the stability and translation of its target mRNAs.
Rbfox1 and miRNA binding sites overlap significantly in target mRNA 3′UTRs.
Cytoplasmic Rbfox1 targets are enriched in cortical development and autism genes.
Acknowledgments
We thank J. König and J. Ule (UCL Institute of Neurology) for the iCLIP protocol; L. Zipursky (UCLA) and members of the Martin and Black labs for helpful discussions; and V. Ho, L. Zipursky and K. Otis (UCLA) for comments on the manuscript. This work was supported by R21 MH101684 to K.C.M., P50-HD-055784 pilot grant to K.C.M., and a NARSAD Young Investigator Award to J.A.L. Work in the Black lab was supported by HHMI, and by NIH grant R01 GM084317. E.S.A. was supported by the UCLA Medical Scientist Training Program, and by NIH Graduate Fellowship 4F30AG033993. D.L.B. is an Investigator of HHMI. N.N.P was supported by NRSA Fellowship F30MH099886. M.F was supported by Fundação para a Ciência e a Tecnologia through the Graduate Program in Areas of Basic and Applied Biology.
Footnotes
AUTHOR CONTRIBUTIONS
J.A.L. and K.C.M. designed the experiments with input from D.L.B.. J.A.L. performed co-culture, RNAi, and rescue experiments for microarray and RNA sequencing experiments, and all biochemical experiments. E.S.A. prepared cDNA for microarray experiments. A.D. and J.A.L. performed iCLIP experiments. J.A.L. and M.F. performed luciferase assays. C.H.L. and J.A.L performed bioinformatic analyses. N.N.P performed gene set enrichment analyses. Figures were prepared by J.A.L., C.H.L., and N.N.P. The manuscript was written by J.A.L and K.C.M with input from the other authors.
ACCESSION NUMBERS
The microarray, RNAseq, and iCLIP data from cytoplasmic fraction were deposited to the NCBI Gene Expression Omnibus under accession number GSE71917.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. The EMBO journal. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic acids research. 2009;37:D832–836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. Journal of human genetics. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli P, Busa R, Paronetto MP, Sette C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocrine-related cancer. 2011;18:R91–R102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Jiang P, Gilmore BL, Spengler RM, Tirabassi R, Nelson JA, Ross CA, Xing Y, Davidson BL. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA biology. 2013;10:935–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. Rna. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harbor perspectives in biology. 2012;4:a012344. doi: 10.1101/cshperspect.a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Human molecular genetics. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ. Splicing-active nuclear extracts from rat brain. Methods. 2005;37:323–330. doi: 10.1016/j.ymeth.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Hamada N, Ito H, Iwamoto I, Mizuno M, Morishita R, Inaguma Y, Kawamoto S, Tabata H, Nagata K. Biochemical and morphological characterization of A2BP1 in neuronal tissue. Journal of neuroscience research. 2013;91:1303–1311. doi: 10.1002/jnr.23266. [DOI] [PubMed] [Google Scholar]
- Heraud-Farlow JE, Kiebler MA. The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends in neurosciences. 2014;37:470–479. doi: 10.1016/j.tins.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho VM, Dallalzadeh LO, Karathanasis N, Keles MF, Vangala S, Grogan T, Poirazi P, Martin KC. GluA2 mRNA distribution and regulation by miR-124 in hippocampal neurons. Molecular and cellular neurosciences. 2014;61:1–12. doi: 10.1016/j.mcn.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. Rna. 2012;18:2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes & development. 2009a;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. The Journal of biological chemistry. 2009b;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cellular and molecular life sciences : CMLS. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Reinthaler EM, Altmuller J, Toliat MR, Thiele H, Nurnberg P, Lerche H, Hahn A, Moller RS, Muhle H, et al. RBFOX1 and RBFOX3 mutations in rolandic epilepsy. PloS one. 2013a;8:e73323. doi: 10.1371/journal.pone.0073323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Trucks H, Moller RS, Hjalgrim H, Koeleman BP, de Kovel CG, Visscher F, Weber YG, Lerche H, Becker F, et al. Rare exonic deletions of the RBFOX1 gene increase risk of idiopathic generalized epilepsy. Epilepsia. 2013b;54:265–271. doi: 10.1111/epi.12084. [DOI] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Molecular cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Molecular cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes & development. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nature reviews Neuroscience. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends in genetics : TIG. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Mackall J, Meredith M, Lane MD. A mild procedure for the rapid release of cytoplasmic enzymes from cultured animal cells. Analytical biochemistry. 1979;95:270–274. doi: 10.1016/0003-2697(79)90216-1. [DOI] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Molecular cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic acids research. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Bhatt DM, Lin CH, Tong AJ, Smale ST, Black DL. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. Rna. 2013;19:811–827. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin C, Rouget C, Mandart E. Xenopus Rbm9 is a novel interactor of XGld2 in the cytoplasmic polyadenylation complex. The FEBS journal. 2008;275:490–503. doi: 10.1111/j.1742-4658.2007.06216.x. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Khrameeva E, Schreiner S, Konig J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA biology. 2014;11:146–155. doi: 10.4161/rna.27991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Current protein & peptide science. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS computational biology. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Minikel EV, Gusella JF. Autism spectrum disorder genetics: diverse genes with diverse clinical outcomes. Harvard review of psychiatry. 2014;22:65–75. doi: 10.1097/HRP.0000000000000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Hodson DJ. An emerging role of RNA-binding proteins as multifunctional regulators of lymphocyte development and function. Advances in immunology. 2012;115:161–185. doi: 10.1016/B978-0-12-394299-9.00006-0. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Molecular and cellular biology. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massague J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife. 2014;3 doi: 10.7554/eLife.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Akerman M, Sun S, McCombie WR, Krainer AR, Zhang MQ. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011;27:3010–3016. doi: 10.1093/bioinformatics/btr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Molecular cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoshi M, Li Q, Yamamoto M, Okada H, Suzuki Y, Kawahara Y. Direct binding of Ataxin-2 to distinct elements in 3′ UTRs promotes mRNA stability and protein expression. Molecular cell. 2014;55:186–198. doi: 10.1016/j.molcel.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Molecular cancer research : MCR. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes & development. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.