Abstract
In Toxoplasma gondii, an intracellular parasite of humans and other animals, host mitochondrial association (HMA) is driven by a gene family that encodes multiple mitochondrial association factor 1 (MAF1) proteins. However, the importance of MAF1 gene duplication in the evolution of HMA is not understood, nor is the impact of HMA on parasite biology. Here we used within- and between-species comparative analysis to determine that the MAF1 locus is duplicated in T. gondii and its nearest extant relative Hammondia hammondi, but not another close relative, Neospora caninum. Using cross-species complementation, we determined that the MAF1 locus harbors multiple distinct paralogs that differ in their ability to mediate HMA, and that only T. gondii and H. hammondi harbor HMA+ paralogs. Additionally, we found that exogenous expression of an HMA+ paralog in T. gondii strains that do not normally exhibit HMA provides a competitive advantage over their wild-type counterparts during a mouse infection. These data indicate that HMA likely evolved by neofunctionalization of a duplicate MAF1 copy in the common ancestor of T. gondii and H. hammondi, and that the neofunctionalized gene duplicate is selectively advantageous.
Keywords: gene duplication, Toxoplasma gondii, Hammondia hammondi, Neospora caninum, neofunctionalization
GENE duplication is known to underlie the evolution of new gene functions and ultimately organismal phenotypes (Ohno 1970; Espinosa-Cantu et al. 2015). The expected outcome of most gene duplication events is that they will be lost by nonsense mutation and/or resolution of the locus (Ohno 1970; Lynch and Conery 2000; Lynch and Force 2000). However, those that confer a selective advantage through gene dosage, subfunctionalization, or neofunctionalization, can become fixed in the population (Ohno 1970; Lynch and Conery 2000; Lynch and Force 2000; Espinosa-Cantu et al. 2015). The phenotypic impact of locus expansions can be high in both natural and laboratory settings. When grown in noncompatible human cells, vaccinia virus was found to expand, diversify, and then contract the K3L locus, resulting in a highly adapted virus with a single K3L gene that could now disrupt the antiviral host protein Protein Kinase R (Elde et al. 2012). Laboratory studies with bacteria show that adaptation to selective conditions (stress or antibiotic exposure) via gene expansion and diversification occurs much more frequently than via point mutation (Kugelberg et al. 2006, 2010). Field studies with Drosophila spp. have identified Cyp6g1 duplication and diversification events as one source of resistance to insecticides such as dichlorodiphenyltrichloroethane (DDT) (Emerson et al. 2008; Cridland and Thornton 2010; Schmidt et al. 2010).
The examples above detail the importance of gene duplication in the evolution within species both in the laboratory and in the field. However, less is known about the impact of gene duplication and diversification events in defining species-specific traits (or even defining the species themselves, which was postulated by Ohno (1970)). It is certainly clear that there are specific gene duplication events that distinguish closely related species (such as humans and chimpanzees) (Bailey and Eichler 2006), but examples where species-specific gene expansions have been linked to species-specific traits are few.
Pathogens provide a unique setting in which to study the evolution and emergence of novel traits, given their large population size and the intense selective pressures placed upon them by the host. We use comparative approaches to understand the evolution of unique traits in members of Apicomplexa, a phylum of parasites of great importance in human and veterinary health. Our main focus is on Toxoplasma gondii and its near relatives. T. gondii is an important pathogen of humans, particularly in HIV/AIDS patients and the developing fetus. In addition, T. gondii is capable of infecting, causing disease in, and being transmitted by all warm-blooded animals studied to date (Dubey and Sreekumar 2003). In contrast, Hammondia hammondi and Neospora caninum have comparatively restricted host ranges and are not pathogenic in rodents or humans (Goodswen et al. 2013; Walzer et al. 2013). This is despite a high level of genetic similarity and genome-wide synteny across these three species (Reid et al. 2012; Walzer et al. 2013), and in the case of T. gondii and H. hammondi, extensive conservation of virulence effectors at both the sequence and functional levels (Walzer et al. 2013, 2014).
The unique phenotypic and life cycle features of T. gondii have most certainly contributed to its near global distribution and an incidence rate that ranges from 10 to 80% in humans. However, the genetic bases for these phenotypes are unknown, and to begin to address this question we have taken a comparative approach to identify genetic loci that are unique to T. gondii compared to H. hammondi and N. caninum. In doing so, we found that a small subset of T. gondii loci have undergone tandem duplication, expansion, and diversification only in the T. gondii lineage. Specifically, expanded loci are poorly conserved between T. gondii and its near relatives, having a higher propensity to be either missing, or not similarly expanded, in either N. caninum or H. hammondi (or both) (Adomako-Ankomah et al. 2014) than single-copy genes. On a gene-by-gene basis, expanded and diversified gene families are known to play important roles in parasite biology and within-species adaptation in T. gondii and Plasmodium spp. (reviewed in Reid 2015). For example, members of the var gene family are distributed throughout the P. falciparum genome and encode erythrocyte membrane antigens (PfEMPs) that are secreted into the host red blood cell during infection. PfEMPs are key determinants of parasite virulence and are under strong diversifying selection (Freitas-Junior et al. 2000; Deitsch et al. 2001; Pasternak and Dzikowski 2009). In both the field (Nair et al. 2008) and laboratory (Heinberg et al. 2013), copy number increases at the gch1 locus in P. falciparum confer resistance to pyrimethamine. In T. gondii, there are >50 members of the rhoptry protein 2 (ROP2) superfamily, and they are dispersed throughout the genome (Boothroyd and Dubremetz 2008). We recently showed that many members of the ROP2 superfamily are encoded by tandemly expanded gene clusters that have diversified significantly via positive selection (Reese et al. 2011; Adomako-Ankomah et al. 2014). One such example is the ROP5 locus, which is crucial for mouse virulence across the T. gondii phylogeny (Reese et al. 2011; Behnke et al. 2015). The ROP5 locus harbors multiple paralogs that are under strong diversifying selection both between and within strains (Reese et al. 2011). Importantly, individual ROP5 paralogs have synergistic, rather than additive, effects on mouse virulence, stressing the importance of paralog diversification in conferring the entire locus-driven phenotype (Reese et al. 2011). Importantly, duplicated and expanded loci represent a highly significant fraction of the genetic difference between T. gondii and its nearest relatives (Wasmuth et al. 2009; Adomako-Ankomah et al. 2014). Based on these data, our overall hypothesis is that selective locus expansion, and subsequent selection-driven diversification of individual paralogs, have played an important role in the evolution of traits that are unique to T. gondii.
One such locus is mitochondrial association factor 1 (MAF1) (Adomako-Ankomah et al. 2014; Pernas et al. 2014). The MAF1 locus is uniquely amplified in T. gondii relative to N. caninum and H. hammondi (annotated as Expanded Locus 4) (Adomako-Ankomah et al. 2014) and is required for host mitochondrial association (HMA) in T. gondii (Pernas et al. 2014). The MAF1 locus encodes a family of dense granule proteins that associate with the parasitophorous vacuolar membrane (PVM) that are necessary and sufficient for the HMA phenotype (Pernas et al. 2014), and MAF1 protein expression itself leads to significant changes in the host immune response to infection. The discovery of the MAF1 gene family as being responsible for HMA opens the door to solving a long-standing question in T. gondii biology regarding the importance of HMA in parasite infectivity and virulence.
In the present study, we use intra- and interspecies comparative analyses and molecular genetics to thoroughly trace the functional evolutionary history of the MAF1 gene in T. gondii. While the impact of MAF1 on HMA is clear, the evolutionary history of the MAF1 locus in terms of copy number and gene content is not, nor is the role of gene duplication itself in the evolution of the HMA phenotype. The MAF1 locus provides a unique opportunity to assess the relative impacts of gene duplication and subsequent diversification on a robust cellular phenotype. Moreover, the impact of MAF1 on parasite fitness in vivo has not been thoroughly investigated. To answer these questions, we have sequenced multiple MAF1 paralogs from three T. gondii clonotypes as well as the nearest extant relatives of T. gondii, H. hammondi, and N. caninum. We have determined that the MAF1 locus has undergone extensive sequence diversification and has been subjected to positive selection in T. gondii. We show that MAF1 copy number and gene content vary both between and within major T. gondii lineages and that not all copies of MAF1 mediate HMA. Through cross-species complementation experiments, we show that expression of an “HMA-competent” T. gondii MAF1 paralog in N. caninum is sufficient to confer the HMA phenotype in this species, indicating that the HMA competence of MAF1 emerged only recently in the parasites in question. Using additional genomic and cross-species complementation experiments, we also provide strong support for a model in which the MAF1 locus duplicated one time in a common ancestor of all three species, diversified one time in an ancestor to H. hammondi and T. gondii, and then amplified and diversified multiple times in the T. gondii lineage. Finally, we show that not only do different MAF1 paralogs differ in their ability to mediate HMA, but also in their ability to confer a selective advantage during infection in a mouse model. Taken together our data link a specific gene duplication and neofunctionalization event in the evolution of a novel trait (host mitochondrial association), and in doing so we have uncovered the selective advantage that likely fixed the MAF1 locus in most of the T. gondii population.
Materials and Methods
Sequence coverage and copy number analysis
Copy number analysis was performed as described previously (Adomako-Ankomah et al. 2014; Pernas et al. 2014). Briefly, raw sequence reads from multiple T. gondii strains and N. caninum (Liverpool strain) (Reid et al. 2012) were downloaded from the NCBI trace archive in fasta format (strains GT1, ME49, and VEG were derived from Sanger-based shotgun sequencing; MAS, P89, FOU, VAND, and RUB were generated using Roche 454 technology). T. gondii and N. caninum reads were aligned to the T. gondii ME49 genome (ToxoDB version 7.3; www.toxodb.org) using BLAT (parameters: -fastMap –minIdentity = 95 –minScore = 90) (Kent 2002), and coverage was calculated in each 500-bp window using coverageBed (from the Bedtools suite) (Quinlan and Hall 2010). H. hammondi reads (strain HH34) (Lorenzi et al. 2016) were aligned using Bowtie2 (using default parameters plus –end-to-end) (Langmead and Salzberg 2012), and sequence coverage calculations were made using the integrated genome browser (IGB) (Nicol et al. 2009). All coverage and annotation data were then plotted using custom scripts in R statistical software. To do this, start and end coordinates of regions of the MAF1 locus were noted and data were normalized to the average coverage of ∼20 Kb upstream of the locus (Lorenzi et al. 2016).
Parasite strains and host cell maintenance
All T. gondii and N. caninum strains used in this study were maintained by regular passage of tachyzoites from freshly lysed human foreskin fibroblast (HFF) onto new HFF monolayers and grown at 37° in 5% CO2. HFF and NRK-mitoRFP cells (a kind gift from Jennifer Lippincott-Schwartz, NIH, Bethesda, MD) (Mitra and Lippincott-Schwartz 2010) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM glutamine, and 50 µg/ml each of penicillin and streptomycin.
To produce H. hammondi oocysts, interferon-γ KO mice were fed 104 H. hammondi oocysts and killed ∼60 days postinfection (pi). Muscles from infected mice were then fed to 10- to 20-week-old cats, and feces were collected during days 5–11 postinfection. Unsporulated oocysts were isolated by sucrose flotation, and the resulting oocysts were allowed to sporulate at ambient temperature in 2% H2SO4 (Dubey and Sreekumar 2003). Oocyst preparations were stored at 4° for no longer than 6 months. Sporulated oocysts (40–80 million) were washed four times in Hank’s Buffered Saline Solution (HBSS) and treated with 10% bleach in PBS for 30 min. Pellets were resuspended in 4 ml HBSS and vortexed at maximum speed along with 1 g of sterile glass beads (710–1180 μM, Sigma) for 30 sec, allowed to cool for 30 sec, and then vortexed for 30 sec again. DNA was isolated directly from the pellet of cracked oocysts (containing sporocysts released from the oocysts and debris) using the DNAzol reagent (Invitrogen; Carlsbad, CA).
In other cases, we used the sporocyst preparation to generate in vitro cultures of H. hammondi. To do this, we exposed the cracked oocyst preparation to PBS containing trypsin (Sigma T4799; 12.5 mg/ml) and taurocholic acid (Sigma T4009; 50 mg/ml) at 37° for 30 min. The reaction was quenched by the addition of cDMEM (containing 10% FBS) and we removed debris from the preparation by filtration through 5-μm syringe filters (Millipore). The resulting sporozoites were used to infect confluent monolayers of HFFs seeded on 12-mm circle glass coverslips. Samples were fixed and processed for immunofluorescence (IF) as described below.
High molecular weight Southern blotting
Southern blotting was performed as previously described (Adomako-Ankomah et al. 2014). The six strains of T. gondii used were GT1 and RH (type I), ME49 and PRU (type II), and VEG and CTG (type III). Genomic DNA from each strain was digested with ScaI restriction enzyme in a 100-µl reaction volume for ∼12 hr and resolved by pulsed field gel electrophoresis (Bio-Rad CHEF-DR III system). Resolved fragments were probed with DIG-labeled (Roche) MAF1-specific probes followed by chromogenic detection as per manufacturer’s protocol.
Amplification and cloning of MAF1 paralogs from T. gondii, H. hammondi, and N. caninum, and construction of mutant constructs
Due to the fact that the MAF1 locus exhibits significant copy number variation across species and strains, we used long-extension PCR and cloning to identify MAF1 paralogs in T. gondii (strains RH, ME49, and CTG), H. hammondi (strain HhCatGer041), and N. caninum (strain NC-1). Primer sequences are listed in Supplemental Material, Table S1. Long extension PCR was used to minimize the potential for chimera formation between different MAF1 paralogs (as described in Pernas et al. 2014). For cloned sequences, all polymorphisms were validated by querying a local copy of the sequence read database for the presence of that polymorphism along with at least 40 bp of flanking sequence (RH was compared to GT1; ME49 was compared to ME49; CTG was compared to VEG) (Lorenzi et al. 2016). This served three purposes: validation of SNPs specific to a given clonal lineage, elimination of PCR-derived SNPs, and controlled for the possibility of generating interparalog chimeric sequences during PCR amplification when the polymophisms were ≤40 bp apart. Since our SNP curation method relied on comparing between members of the same clonal lineage (e.g., RH vs. GT1), it is possible that some isolate-specific SNPs were artificially eliminated during curation. We did not identify any evidence for chimerism in our sequences although this outcome cannot be completely ruled out.
Sequence analysis
Coding sequences of MAF1 paralogs from multiple T. gondii strains and other species were analyzed using algorithms implemented in MEGA6 (Tamura et al. 2013) as follows: Specifically, coding sequences were translated into protein and aligned using Muscle (default settings). Phylogenetic trees were constructed using maximum parsimony and the subtree-pruning-regrafting algorithm (Nei and Kumar 2000). Search level was 1 and the initial trees were obtained by the random addition of sequences (10 replicates were performed). Branch lengths were calculated using the average pathway method. All positions containing gaps and missing data were eliminated, and there were a total of 359 useful positions in the final dataset.
We calculated pairwise dN/dS ratios for all “b” paralogs (including the HMA-incompetent b0 paralogs) to determine if they had been under either positive or purifying selection. To do this, we used the modified Nei-Gojobori method with the assumed transition/transversion bias of 2 (Zhang et al. 1998), and as above, all positions containing gaps were eliminated. All analyses were conducted in MEGA6 (Tamura et al. 2013) and pairwise P-values for dN/dS ratios were deemed significant at P < 0.05.
Generation of expression constructs and transgenic parasites
Generation of pMAF1RHb1 (N-terminally hemagglutinin (HA)-tagged MAF1b) expression construct has been described previously (Pernas et al. 2014). For pMAF1RHa, the coding sequence for MAF1RHa was amplified from RH cDNA, cloned, and then used in a splicing by overlap extension (SOE) PCR reaction to fuse the N-terminal portion of MAF1RHb1 gene and the C-terminal portion of the MAF1RHa1 gene. The specific construct contained the MAF1RHb1 promoter, start codon, signal sequence, an HA tag (as in Pernas et al. 2014), and this was followed by the remainder of the C terminus encoding portion of the MAF1RHa1 gene. The TgMAF1RHb0, TgMAF1RHb1, HhMAF1a1, HhMAF1b1, and NcMAF1 constructs were made using SOE PCR to introduce an HA-tag following the predicted signal peptide for each isoform. Plasmid templates for the first round of PCR were generated from genomic DNA, which included 1116 bp upstream of the start site to include the putative promoter. Transgenic parasite lines were generated by transfecting TGME49Δhpt (MΔLuc) and NC-1Δhpt parental strains with 50 µg of HindIII-linearized plasmid. Stable expression lines were isolated by selection in mycophenolic acid (MPA)/xanthine followed by limiting dilution in 96-well plates.
TgMAF1RHa1 and TgMAF1RHb1 cloning, protein production, and purification
A construct encoding the predicted C-terminal domain of TgMAF1RHb1 (Thr159 to Asp435) was codon optimized for Escherichia coli and synthesized by GenScript. A construct of TgMAF1RHa1 containing the analogous C-terminal domain (TgMAF1RHa1; Ser173 to Ser443) was amplified from T. gondii cDNA. Each construct was subcloned into a modified pET28a vector encoding an N-terminal hexa-histidine tag separated from the sequence of interest by a tobacco etch virus (TEV) protease cleavage site. Constructs were produced recombinantly in E. coli BL21 cells. Following 4 hr of growth at 310 K and 12 hr at 303 K, the cells were harvested by centrifugation, resuspended, and lysed using a French press. TgMAF1 proteins were purified from the soluble fraction by Ni-affinity chromatography, the His tag was removed by TEV protease, and TgMAF1 proteins were further purified by size exclusion chromatography on a Superdex 75 16/60 HiLoad column in HBS (20 mM Hepes pH 7.5, 150–300 mM NaCl) with 1% glycerol and 1 mM dithiothreitol.
Generation of polyclonal antibodies
Female Balb/c mice were injected intraperitoneally (ip) with 100 µg of either TgMAF1RHa1 antigen or TgMAF1RHb1 antigen (purification described above) suspended in 100 µl PBS and mixed 1:1 with Sigma adjuvant (Sigma S6322) to a final volume of 200 µl. Additional injections of 50 µg of the appropriate antigen mixed 1:1 with Sigma adjuvant to a final volume of 200 µl were administered 14, 35, and 56 days after the initial injection. Sera were collected prior to initial injection, as well as on days 31, 81, and 88. All sera were tested for reactivity against both TgMAF1RHa1 and TgMAF1RHb1 by Western blot prior to use in Western blots or immunofluorescence assays.
Immunofluorescence assays and confocal microscopy
HFFs or NRK-mitoRFP cells were seeded on 12-mm coverslips in 24-well plates and grown to ∼80% confluency. NRK-mitoRFP cells were infected with N. caninum or T. gondii strains expressing GFP and incubated for 8 hr. For HFFs, MitoTracker staining was performed as follows: Growth medium on the HFF monolayer was replaced with DMEM containing MitoTracker (Red CMXRos, Invitrogen) at a 30-nM concentration and incubated for 30 min at 37°. Cells were then washed with PBS, infected with parasites in prewarmed DMEM, and incubated for 4 hr at 37°. After incubation, the infected cells were washed with PBS, fixed with 3% paraformaldehyde in PBS for 15 min, and blocked/permeabilized in PBS containing 5% BSA and 0.2% Triton X-100. Alternatively, NRK-mitoRFP infected cells were fixed with 3% PFA and either mounted directly or Hoechst stained prior to mounting followed by visualization. Fixed cells were then immunostained with rat monoclonal anti-HA (3F10 clone, Roche) at 1:1000, mouse anti-MAF1a/b polyclonal antibodies at 1:1000, or mouse monoclonal anti-MTCO2 (ab110258, Abcam) at 1:500.
Quantification of vacuole coverage
Percent vacuole coverage was determined using confocal microscopy and ImageJ. Populations transfected with HA-tagged TgMAF1RHb1 or HhMAF1b1 were fixed and stained with anti-HA and anti-MTCO2 primary antibodies. Confocal images were taken in three channels; 594 (anti-MTCO2), 488 (anti-HA), and DIC. All three images were converted to 8-bit images and merged using ImageJ. Vacuoles were traced while only the DIC and green channels were visible, and then pixel intensity along the vacuole was measured in the red channel. Pixel intensities >20 were considered to be mitochondria. Percent vacuole coverage was calculated by measuring the length of the vacuole trace with pixel intensity >20 and dividing it by total vacuole trace length. Twenty HA-positive vacuoles were measured for both the TgMAF1RHb1 and HhMAF1b1 populations. Ten HA-negative vacuoles were measured from each population (20 total) as a WT control.
Western blot analysis
Parasites were filtered away from host cell debris and lysed in 1× SDS lysis buffer. Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose membrane, and blocked for 1 hr in 5% (w/v) milk in TBS-Tween20 (TBS-T). Primary antibody incubation was performed in blocking buffer for 45–120 min followed by three washes in TBS-T. Anti-HA and anti-MAF1 (Pernas et al. 2014) antibodies were used at 1:1000 while anti-SAG1 was used at 1:2000 and rabbit anti-ROP5 (Behnke et al. 2011) was used at 1:40,000. Anti-TgMAF1RHa1 and anti-TgMAFRHb1 antibodies generated for this study were used at a 1:10,000 dilution. Secondary antibody incubation was performed with horseradish peroxidase-conjugated secondary antibodies to the respective primary antibodies in blocking buffer for 45 min. Bands were visualized with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). Densitometric analysis was performed using ImageJ.
Animal experiments
All mouse experiments were performed with 4- to 8-wk-old BALB/C mice. All animal procedures in this study meet the standards of the American Veterinary Association and were approved locally under Institutional Animal Care and Use Committee protocol no 12010130.
In vitro and in vivo competition assays
In vitro competition assays were performed as follows: An ME49 strain engineered to express an N-terminal HA-tagged type I (RH) MAF1 (ME49:TgMAF1RHb1) was mixed with ME49:WT in ratios 4:1 and 1:4. These two mixed populations were used to infect HFFs at an MOI of 3. Flasks were passed via syringe lysis every 3 days. At the 0-, 4-, and 8-wk time marks, HFFs grown on 12-mm glass coverslips were infected at an MOI of 3, and the proportion of HA+ and HA− parasites was calculated by immunofluorescence using rat α-HA (as above) and serum from a mouse chronically infected with T. gondii at 1:1000 dilution. The ratio of HA+ to HA− was determined by counting at least 200 vacuoles. The entire experiment was repeated two times, each time with a genetically distinct clone set (WT and complemented).
In vivo competition assays were performed as follows: Using the same genetically engineered ME49 clone sets, we again created mixed populations at ratios of 1:4, 1:1, 4:1, 100% ME49:TgMAF1RHb1, and 100% ME49:WT. We injected 105 tachyzoites intraperitoneally of these five populations into Balb/c mice in 200 µl of PBS (three to five mice per population). In a separate experiment, we transfected ME49 with the same pTgMAF1RHa1, grew the population under MPA/xanthine selection for 2 weeks, and then injected 105 tachyzoites of this mixed population into Balb/c mice as above. On the day of injection, we used the same parasite preparation to infect HFFs seeded on glass coverslips to quantify the exact input proportions using IF imaging as described above. Parasite burden and location were assessed daily for the next 5 days using in vivo bioluminescence imaging (Walzer et al. 2013) since the parental ME49 strain expressed click beetle luciferase off of a dihydrofolate reductase promoter (Walzer et al. 2013). On day 5 pi, all mice were killed and an intraperitoneal lavage was performed to harvest peritoneal cells and associated parasites. Samples were spun down and resuspended in cDMEM and used to infect HFFs. After one passage, parasites were used to infect HFFs seeded onto glass coverslips at an MOI of 3 to quantify proportions using IF imaging as above.
Data availability
All strains and plasmids available upon request. All MAF1 paralog and ortholog sequences obtained for this study have been deposited in GenBank (accession numbers KU761333-KU761342).
Results
MAF1 is uniquely expanded in T. gondii and exhibits inter- and intralineage copy number variation
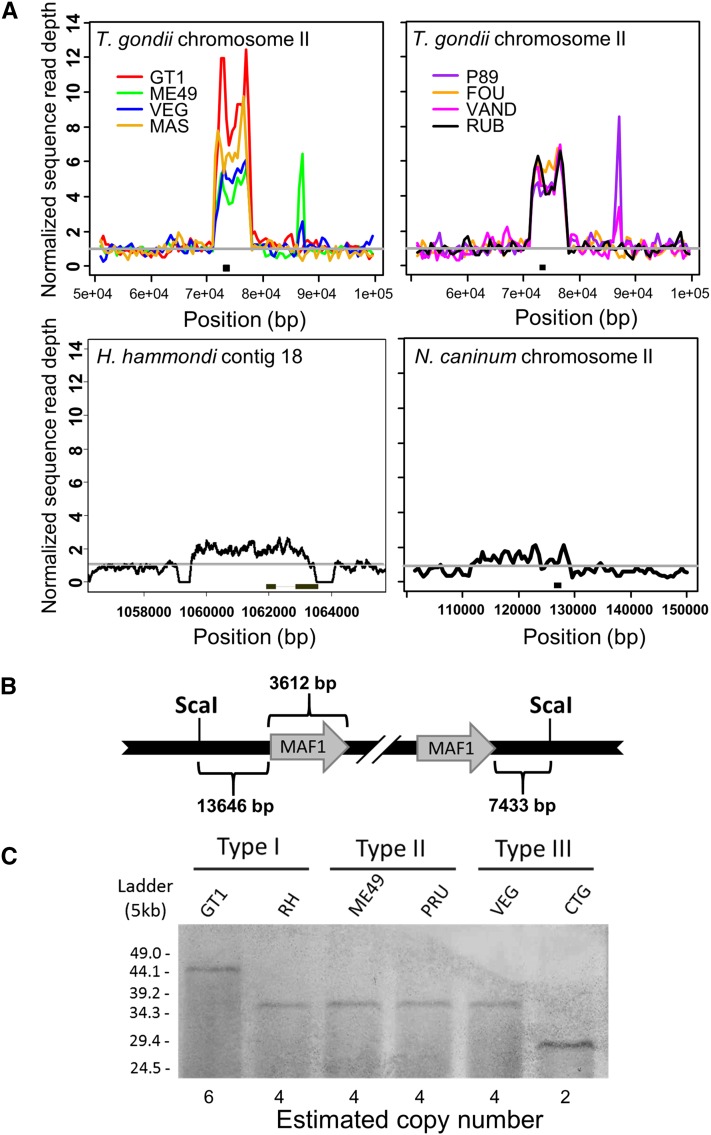
We previously reported that MAF1 is a multicopy locus in T. gondii, and based on sequence read coverage, exhibits strain-specific copy number variation between representatives of the canonical T. gondii lineages (types I, II, and III: GT1, ME49, and VEG) (Adomako-Ankomah et al. 2014; Pernas et al. 2014). We have extended these copy number analyses to five additional T. gondii clonotypes outside of the three major lineages and also find that MAF1 is similarly expanded in these strains (Figure 1A). Similar to types I, II, and III, there is significant copy number variation between strains at this locus, ranging from an estimated 8–10 copies for MAS to 4–6 copies for P89, FOU, VAND, and RUB (Figure 1A). While these data provide only an estimate of copy number differences between strains, they do confirm that the multicopy state of the MAF1 locus is conserved across highly diverse T. gondii isolates.
Figure 1.
The MAF1 locus exhibits copy number variation across strains of T. gondii and has comparatively low copy number in H. hammondi and N. caninum. (A) Coverage depth analysis for the MAF1 locus in eight T. gondii strain types and for the syntenic locus in H. hammondi and N. caninum. T. gondii sequences are from ToxoDB v7.3. Portions of the upper left panel of this figure were similarly represented in Pernas et al. (2014). Raw reads were plotted as described in Materials and Methods and normalized to the coverage 20 Kb upstream of the repetitive locus. Arrowheads indicate the location of predicted gene sequences based on ToxoDB (v7.3 for T. gondii; v26 for all other species). Asterisks indicate smaller repetitive sequence unrelated to MAF1 (see Materials and Methods for further explanation). (B) Schematic representation of the MAF1 locus showing ScaI restrictions sites outside of the locus, the size of the regions flanking the MAF1 locus, and the size of the repeat unit used to estimate copy number based on Southern blotting. The most relevant T. gondii ME49 gene name is indicated (from ToxoDB v7.3), although it does not fully match the sequenced paralogs. (C) ScaI-digested gDNA from each of six T. gondii strains was resolved by PFGE and probed with a MAF1-specific probe. The blot shows copy number variation consistent with predictions from sequence coverage analysis for strain types GT1, ME49, and VEG. Copy number for each strain was determined based on the schematic presented in B.
To further confirm differential expansion of the MAF1 locus in T. gondii, and to identify differences in MAF1 copy number between them, we performed high molecular weight Southern blot analysis of the MAF1 locus in six Toxoplasma strains. These strains comprised two each from the type I (GT1, RH), type II (ME49, PRU), and type III (VEG, CTG) lineages. Genomic DNA from each strain was digested with ScaI, which cuts on either side of the entire locus but not within, allowing for locus size (and therefore copy number) to be estimated (Figure 1B) (Reese et al. 2011; Adomako-Ankomah et al. 2014). Sequence coverage analysis shows higher copy number for GT1 compared to ME49 and VEG (Figure 1A), and the Southern blot was consistent with this observation: GT1 has the largest MAF1 locus (∼44.9 Kb), while the MAF1 loci in ME49 and VEG were smaller (28.4 Kb; Figure 1C). No other bands were visible on the blot (which resolved fragments ranging in size from 4.9 Kb to 53.9 Kb), indicating that the entire locus was intact for all strains. Moreover, the ScaI sites flanking the expanded locus were conserved in all six strains tested (Figure S1, A and B), indicating these differences are not due to mutations within the flanking sequences.
Based on the size of the MAF1 repeat unit (3612 bp) and the known size of the regions between the locus and the ScaI sites (see Figure 1B), we estimate that there are six copies of MAF1 in GT1; four in RH, ME49, PRU, and VEG; and two in CTG. Similar to what we have observed previously at other expanded loci in T. gondii (Adomako-Ankomah et al. 2014), MAF1 exhibits copy number variation within members of the same clonal lineage (i.e., GT1 vs. RH and VEG vs. CTG), suggesting that expanded loci change more rapidly in size and copy number compared to the single nucleotide polymorphism rate at single-copy.
We also performed copy number analysis of the MAF1 loci in both H. hammondi and N. caninum, two relatives of T. gondii with distinct virulence and host range phenotypes (Dubey et al. 2002; Dubey and Sreekumar 2003). For H. hammondi, copy number analysis suggested the presence of two copies of MAF1, which is consistent with the presence of two predicted MAF1 paralogs in the H. hammondi genome (HHA_220950 and HHA_279100). For N. caninum, we predicted the existence of one to two copies of MAF1 (Liverpool strain; Figure 1A; www.toxodb.org). In version 10.0 of the N. caninum genome there is only a single predicted MAF1 ortholog (NCLIV_004730).
MAF1 paralogs are uniquely divergent and under diversifying selection in T. gondii
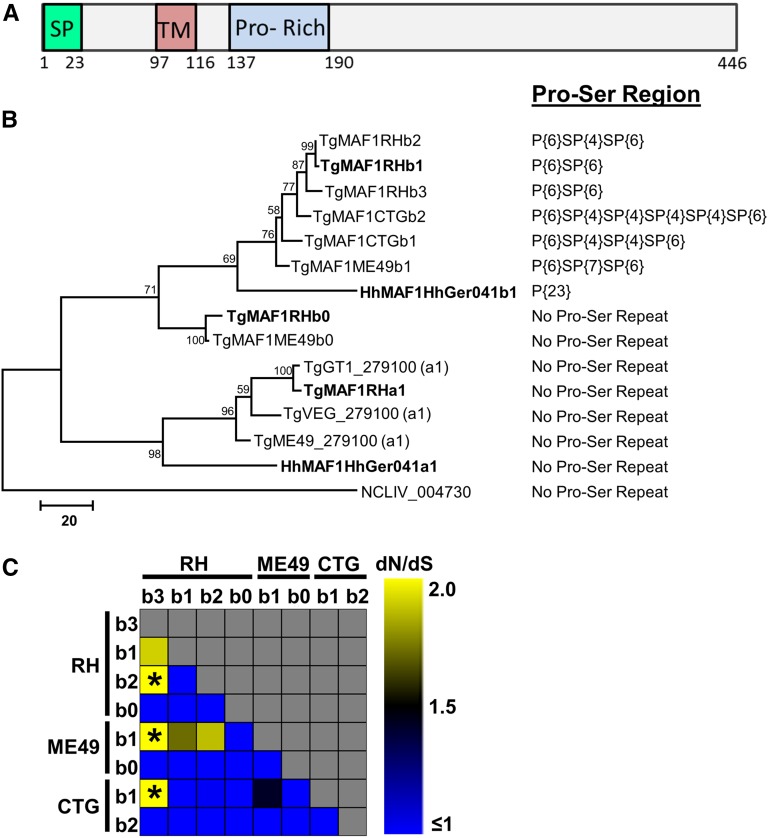
To further characterize the MAF1 locus across strains and species, we sequenced six PCR-derived MAF1 clones from each of three representative strains from the type I (RH), II (ME49), and III (CTG) T. gondii lineages, 13 clones from H. hammondi HhCatGer041, and 10 clones from N. caninum strain NC-1 (Rettigner et al. 2004). We found that in T. gondii, the MAF1 locus harbors multiple diverse paralogs of the MAF1 gene, indicating that the locus has both amplified and diversified. Importantly, all cloned T. gondii MAF1 paralogs were distinct from those found in existing annotation datasets for the T. gondii genome, including the putative MAF1 paralogs TGME49_020950 and TGME49_220950 (Figure 2B). Therefore, we have resorted to a new nomenclature that can be found in Figure 2B. In addition to a putative signal peptide, each paralog is predicted to encode a single transmembrane domain located in the N-terminal region (Figure 2A). The most significant distinguishing feature across the sequenced MAF1 paralogs is the presence or absence of a repetitive stretch of four to seven prolines followed by a serine (P{4:7}S), as well as the amino acids surrounding the proline motif (∼20 N terminal to the motif and ∼10 C terminal to the motif). Importantly, the MAF1 paralog that was shown previously to complement the host mitochondrial association phenotype in type II T. gondii (TgMAF1RHb1) also harbors this P{4:7}S motif (Pernas et al. 2014). This motif is either completely missing or repeated up to six times depending on the paralog (Figure 2B, Figure S2).
Figure 2.
The T. gondii and H. hammondi MAF1 loci harbor two distinct isoforms while only one isoform is present in N. caninum. (A) Schematic representation of the predicted MAF1 protein. The signal peptide (SP) was predicted using SignalP v4.0 and the putative transmembrane domain (TM) was predicted by TMHMM v2.0. The proline-rich region (Pro-Rich) stretches from AA152 to 164 of TgMAF1RHb1 and is not found within all MAF1 paralogs (e.g., TgMAF1RHa1, a2). (B) Phylogram of either cloned MAF1 amino acid sequences from T. gondii, H. hammondi, and N. caninum, or those downloaded directly from ToxoDB (with TG Gene nos.). Cloned sequences of all of the “b” paralogs from T. gondii did not match any predicted gene models in ToxoDB in terms of predicted coding region length and were left out of the analysis. Paralog family is indicated at the end of each name (e.g., a1, b1, b2, etc.) (C) dN/dS ratio calculations for all T. gondii “b” MAF1 paralogs, including b0. * indicates significant evidence for diversifying selection for that particular paralog comparison (P < 0.05).
For RH, five of the six sequenced clones contain the P{4:7}S motif, while all six CTG clones, which represent only two unique coding sequences, have some form of the repeat motif. Interestingly, of the six clones sequenced from ME49, three are pseudogenes with premature stop codons, and all three of these clones are predicted to encode MAF1 paralogs with the P{4:7}S motif. Of the remaining three clones, two harbored the P{4:7}S motif while the other did not. Based on amino acid identity of 15 nonpseudogenized genes from RH, ME49, and CTG, we identified a total of eight unique coding sequences (four for RH, two for ME49, and two for CTG). However, there is also significant variation across these sequences outside of the P{4:7}S motif. We calculated pairwise dN/dS ratios for all unique T. gondii paralogs, and find significantly higher dN/dS ratios in TgMAF1RHb3 when compared to other RH paralogs as well as those from ME49 and CTG (P < 0.05; Figure 2C). While not significant (P > 0.05), TgMAF1ME49b1 also shows a higher dN/dS ratio when compared to TgMAF1RHb1 and b2 (Figure 2C).
When we sequenced 13 distinct clones for the H. hammondi MAF1 locus, we identified only two distinct sequences. One contained a stretch of 23 prolines in the same region as the P{4:7}S motif in T. gondii MAF1 paralogs (Figure 2B, Figure S2; as found in HHA_220950), and the other lacked this proline-rich region (Figure 2, B and C; similar to HHA_279100). This suggests that, consistent with the sequence coverage analysis and genome annotations, the MAF1 locus harbors only two paralogs in H. hammondi, but that these paralogs also differ in the presence or absence of a proline-rich motif. Finally, based on the current genome assembly and direct sequencing of 10 clones, the sole N. caninum MAF1 paralog (NCLIV_004730) is more similar to the T. gondii and H. hammondi MAF1 paralogs that do not have a proline-rich region (Figure 2B). A maximum likelihood tree of amino acid sequences for all unique MAF1 paralogs from T. gondii, H. hammondi, and N. caninum is shown in Figure 2B and illustrates these relationships.
Given this diversity of sequences both within and between species, we have named the identified MAF1 paralogs and deposited them in GenBank. As shown in Figure 2B, based on sequence similarity MAF1 has two major groups, and we have dubbed these “a” and “b,” and all of the “a” paralogs lack the P{4:7}S motif. For the “b” paralogs, we identified two sequences without the P{4:7}S motif and have named these “b0” and then named all other MAF1 paralogs with the P{4:7}S motif as b1, b2, etc. (Figure 2B). We feel this nomenclature accurately reflects the relationships between the various sequences in terms of broad groupings as well as the presence or absence of the P{4:7}S motif. From this point onward, when we use “MAF1” without further indication of paralog, it is because the exact paralog is unknown or the statement applies to all known paralogs. Paralogs belonging to the “a” subfamily are most closely related to TGME49_279100, and the “b” subfamily is related to TGME49_020950 (www.toxodb.org). All MAF1 paralogs are located in tandem on chromosome II as shown in Figure 1B, and the locus was previously identified as Expanded Locus 4 (EL4) (Adomako-Ankomah et al. 2014). Individual paralog numbers vary by strain; identified paralogs are named in Figure 2B. We do not assert that this represents the full complement of MAF1 paralogs from all queried strains. Indeed, further analysis of sequences and genomic sequence reads from the strains of interest will be necessary to determine this.
MAF1a and MAF1b gene families have distinct interstrain transcriptional profiles
We reported previously that MAF1 transcript levels were of lower abundance in a type II T. gondii strain (ME49) compared to types I and III (RH and CTG, respectively) (Boyle et al. 2008; Pernas et al. 2014). The previously reported data were derived from spotted cDNA microarray experiments, which would not distinguish transcripts for MAF1a and b paralogs. Therefore we used the sequence alignments shown in Figure 2 to determine if probes for the MAF1a and b paralog families could be found on the T. gondii Affymetrix array (Bahl et al. 2010) and therefore could be used to assess paralog-specific transcript levels. MAF1a was most similar to TGME49_279100 and the MAF1b family was most similar to TGME49_220950, respectively (Figure 2B; www.toxodb.org). Similar to what was reported previously (Pernas et al. 2014 and Figure S3A), we found that MAF1b transcript levels were lower in type II strains (ME49 and PRU; www.toxodb.org and Figure S3, A and B) compared to type I strains (GT1 and RH) and type III strains (CTG and VEG; Figure S3, A and B). In contrast, we found that transcript levels for TGME49_279100 (MAF1a) were of comparatively high abundance (>90th percentile; data not shown) across all six queried T. gondii strain types (Figure S3B). These data indicate that MAF1a and MAF1b have significantly diverged in terms of their transcript abundance in the type II T. gondii lineage.
MAF1a and MAF1b differ in protein expression between T. gondii strains
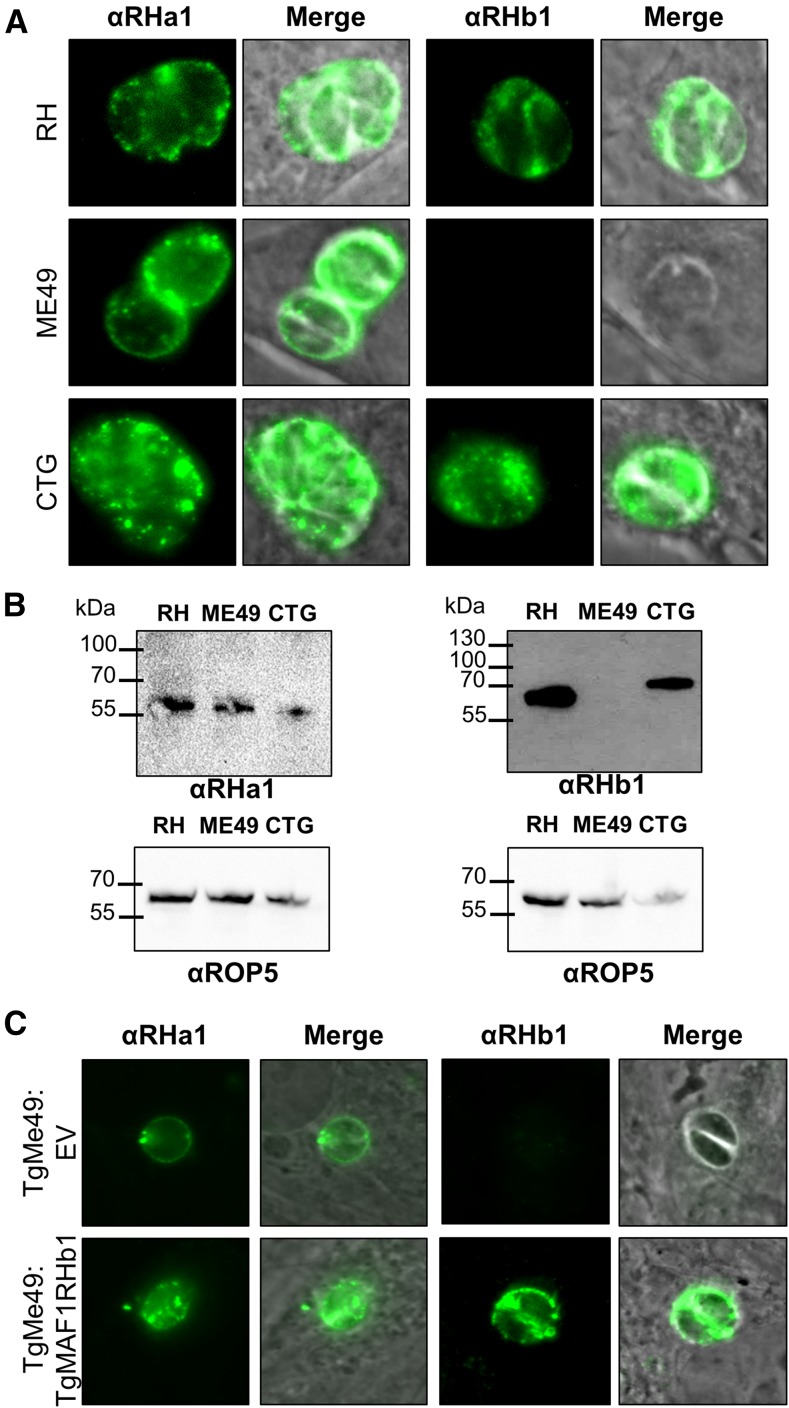
We previously demonstrated a lack of MAF1 protein expression in TgME49 compared to TgRH and TgCTG using a polyclonal anti-MAF1 antibody raised against the C terminus of TgMAF1RHb1 (Pernas et al. 2014). Using this same antibody, we compared MAF1 protein expression in the six T. gondii strains examined in the Southern blot analysis and observed MAF1b expression in the type I and III strains, but did not detect any MAF1b protein in either of the type II strains (Figure S3, C and D). Additionally, we observed that MAF1b protein from both type III strains had a slightly higher apparent molecular weight compared to those in the type I strains (Figure 3B, Figure S3C). This is consistent with the observation that the two clones of MAF1b sequenced from the CTG strain encode either four or six P{4:7}S repeat motifs, while the highest number of P{4:7}S repeat motifs in RH was three (Figure 2B, Figure S2). To determine if types I, II, and III all express a MAF1a isoform, we generated new polyclonal antibodies against the C terminus of TgMAF1RHa1 (Ser173 to Ser443) or TgMAF1RHb1 (Thr159 to Asp435) (indicated in Figure S2). We exposed two mice to the TgMAF1RHa1 and three mice to TgMAF1RHb1. Polyclonal serum from four of the five mice was specific for the input antigen, while one mouse exposed to TgMAF1RHb1 harbored antibodies that bound to both MAF1 paralogs (Figure S3E and data not shown). Given the amount of similarity between the two antigens, it is likely that the epitopes recognized by sera from most of the mice were derived from the dissimilar regions. Additionally, it is likely that each polyclonal serum is capable of recognizing multiple “a” or “b” paralogs. In Western blots against the input antigen, a higher molecular weight band is detected by all of these antisera in addition to the major species at the expected molecular weight (Figure S3E). The antisera recognize the higher MW band with similar specificity as the purified protein. This higher MW band may be a dimer of the purified protein, as it is approximately twice the size of the major species, can be seen upon Coomassie staining, and its quantity is reduced after longer boiling times of the purified protein (Figure S4). Using antibodies from mouse 5 for immunofluorescence, we detected MAF1a protein in all three strains, while once again we did not detect MAF1b in type II when using antibodies from mouse 1 (Figure 3A). We also saw a similar pattern of expression by Western blot (Figure 3B). The specificity of the MAF1b antiserum to MAF1b and not MAF1a was further confirmed by the fact that the MAF1b antiserum bound to type II T. gondii when expressing an ectopic copy of TgMAF1RHb1 (Figure 3C). These data indicate significant strain-specific variation between major clonotypes in both MAF1 protein level and in the qualitative nature of the paralogs that are expressed.
Figure 3.
T. gondii MAF1 paralog expression differs between lineages. (A and B) Polyclonal antibodies were generated specifically against the C termini of TgMAF1RHa1 (Ser173 to Ser443) or TgMAF1RHb1 (Thr159 to Asp435). Protein expression was compared by immunofluorescence across three strains representing clonotypes I, II, and III. Based on immunofluorescence and Western blotting, antibodies against TgMAF1RHa1 detected protein in all three strains, while antibodies against TgMAF1RHb1 detected protein only in RH and CTG (and not ME49). (C) Antibodies against TgMAF1RHb1 are able to detect TgMAF1RHb1 expression in transgenic type II parasites expressing the TgMAF1RHb1 protein.
T. gondii MAF1 paralogs differ in their ability to mediate host mitochondrial association in T. gondii and N. caninum
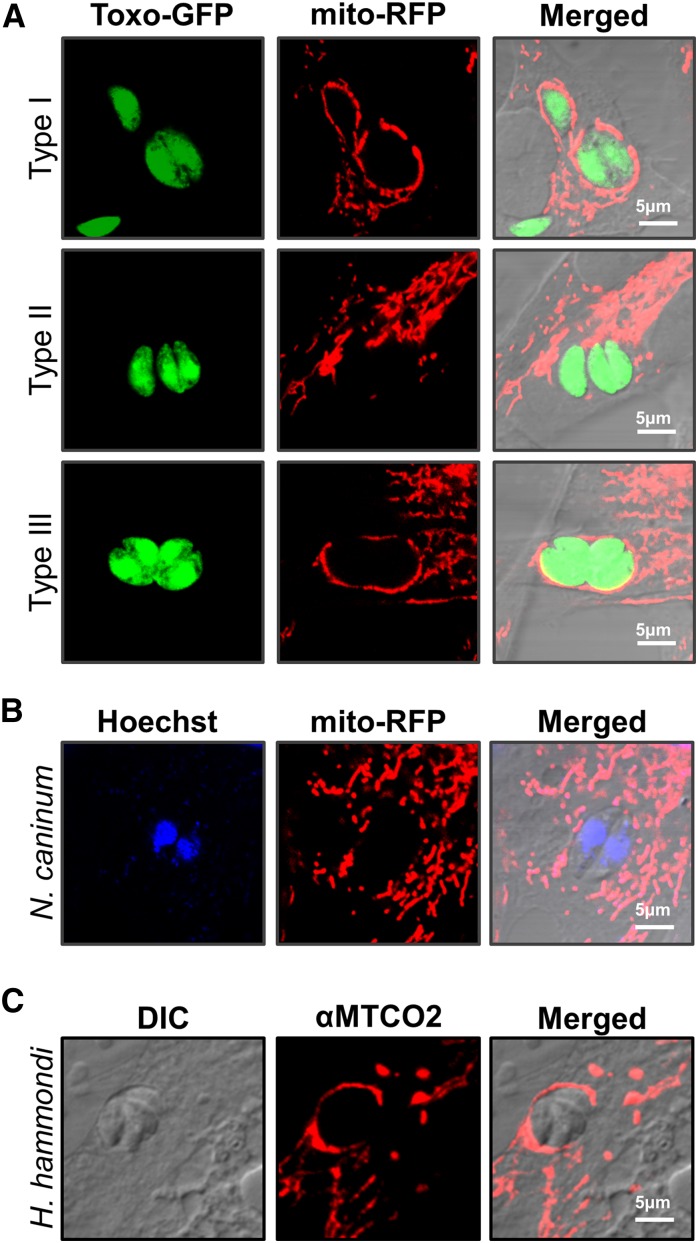
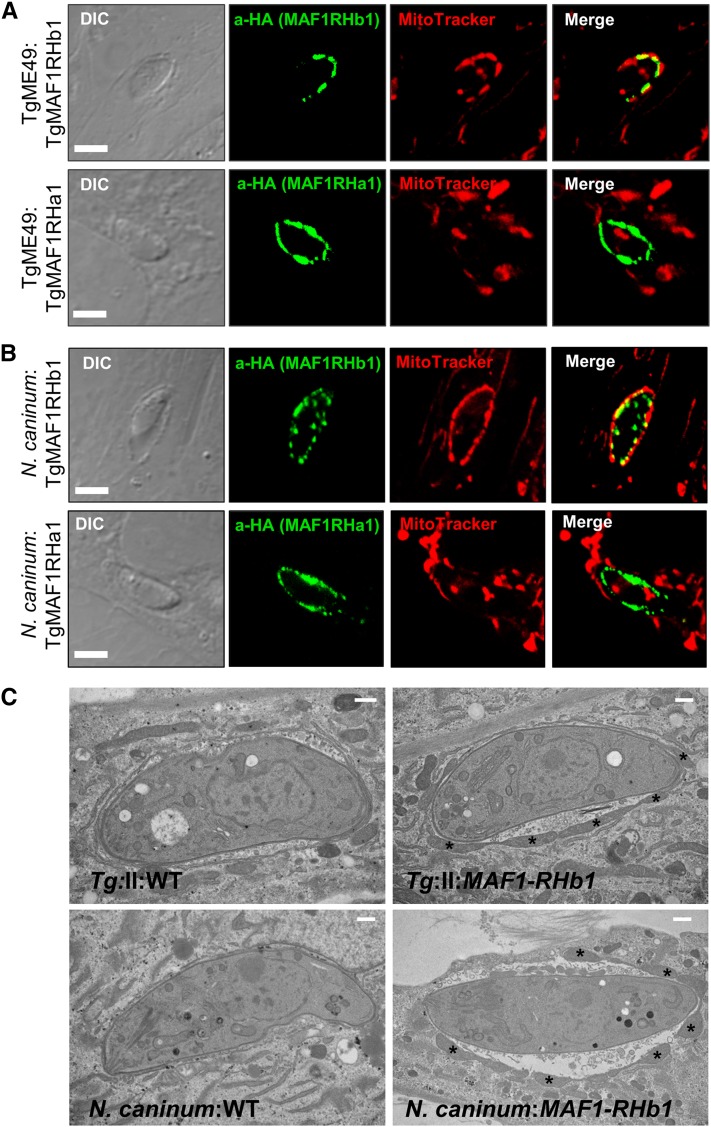
HMA is a strain-specific trait in T. gondii (lacking in type II stains; Figure 4A), and this trait is consistent with reduced MAF1b transcript and protein levels in members of the type II lineage (Figure 3, Figure S3, A–D). In contrast the MAF1a gene is highly expressed at the transcript and protein level equally well across multiple T. gondii strains. To determine if the MAF1a and b genes differed in their ability to confer HMA in HMA- parasites, we generated N-terminally HA-tagged clones of the two paralogs that differed in the absence or presence of the P{4:7}S motif (TgMAF1RHa1 and TgMAF1RHb1, respectively). To do this, we cloned TgMAF1RHa1 in place of TgMAF1RHb1, while retaining the TgMAF1RHb1 promoter in the construct to ensure equal expression between paralogs. We expressed these genes in both a type II strain (TgME49) and in N. caninum (NC-1) (Rettigner et al. 2004) and used confocal microscopy and mitochondrial staining to determine the impact on HMA. TgMAF1RHb1 expression was sufficient to mediate HMA in T. gondii strain ME49 (Figure 5A) and also in N. caninum (Figure 5B) 18 hr postinfection. In contrast, TgMAF1RHa1 was unable to mediate HMA in either TgME49 or N. caninum although its protein localization profile was similar to that of MAF1RHb1 (Figure 5, A and B, bottom). We also generated clones of TgME49:MAF1RHb1 and NC-1:MAF1RHb1 for electron microscopy. Both wild-type TgME49 and NC-1 have little, if any, HMA (Figure 5C, left). However, when these strains express MAF1RHb1 they become HMA+ and there is an increase in host mitochondria directly adjacent to the PVM (Figure 5C, right).
Figure 4.
Host mitochondrial association is a feature of T. gondii and H. hammondi infections, but not N. caninum. (A) NRK-mitoRFP cells were infected with GFP-expressing type I, II, and III (RH, PRU, and CTG) parasites. Type II parasites are HMA−, while types I and III are HMA+. (B) NRK-mitoRFP cells were infected with N. caninum strain NC-1. Cells were fixed and counterstained with Hoechst stain. Wild-type N. caninum are HMA−. (C) HFFs were infected with H. hammondi sporozoites for 8 days before fixation. Host mitochondria were visualized using an antibody to human MTCO2. H. hammondi is HMA+.
Figure 5.
MAF1RHa1 and MAF1RHb1 differ in their ability to complement HMA in T. gondii and N. caninum. (A) HFFs were labeled with MitoTracker and infected with parasites transiently transfected with either HA-MAF1RHa1 or HA-MAF1RHb1. MAF1RHb1 but not MAF1RHa1 is able to confer the HMA phenotype in TgME49. (B) Identical results were obtained for N. caninum. Bar, 5.0 μm. (C) HA-MAF1RHb1 was transfected into either TgME49 (top) or N. caninum (bottom), and HA-positive clones were isolated by limiting dilution. Wild type (WT, left) and TgMAF1RHb1 complemented (right) were grown for 18 hr in HFFs and processed for electron microscopy. Asterisks indicate host mitochondria. Bar, 500 nm.
TgMAF1b1 from T. gondii and H. hammondi can confer the HMA phenotype in T. gondii type II, while TgMAF1b0 and HhMAF1a1 cannot
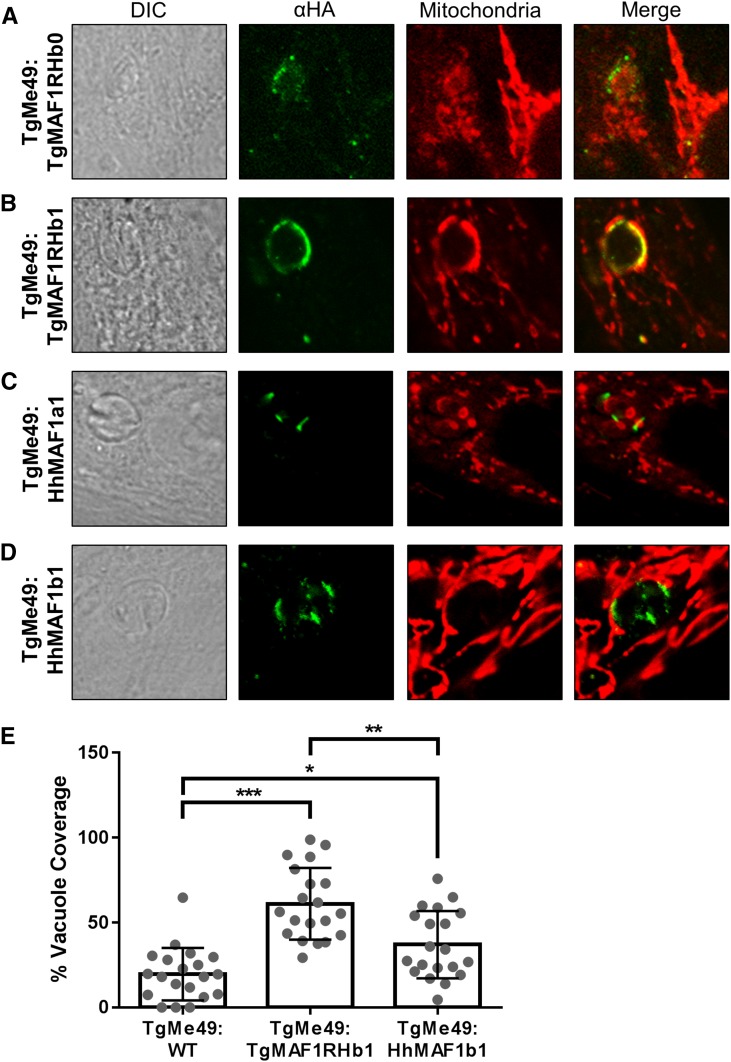
HMA is greatly reduced in the closely related N. caninum (Figure 4B) (Pernas and Boothroyd 2010), but the HMA phenotype of the nearest extant relative of T. gondii, H. hammondi, is unknown. To test this we assessed HMA in sporozoite-derived tachyzoites of H. hammondi (strain HhCatEth1) (Dubey et al. 2013), and found clear evidence for HMA in this species (Figure 4C). Therefore we hypothesized that T. gondii and H. hammondi would harbor MAF1 paralogs that could complement the HMA defect in type II T. gondii, while N. caninum would not. To test this hypothesis, we cloned N-terminally tagged MAF1 paralogs from T. gondii, H. hammondi, and N. caninum. For T. gondii, the coding sequences for TgMAF1RHb0 and TgMAF1RHb1 with the endogenous promoters were cloned directly from RH strain genomic DNA. Similar constructs were made for H. hammondi and N. caninum. Each construct was transfected into the HMA− TgME49 strain and the ability of each isoform to mediate HMA was assessed by immunofluorescence. Similar to our results with TgMAF1RHa1, TgMAF1RHb0 was unable to mediate HMA (Figure 6A), indicating that not all “b” paralogs are capable of mediating this phenotype. Importantly, the same was true for HhMAF1a1: When transfected into type II T. gondii, this protein did not confer the HMA phenotype, although it did have an expression profile that was distinct from other MAF1 paralogs (Figure 6C). In contrast, both TgMAF1RHb1 (as shown previously) and HhMAF1b1 could confer the HMA phenotype when ectopically expressed in type II T. gondii (Figure 6, B and D). We quantified percent vacuole coverage for 20 vacuoles for each MAF1 paralog using confocal microscopy and found that parasites expressing TgMAF1RHb1 or HhMAF1b1 had significantly more vacuole membrane associated with host mitochondria than wild-type type II parasites (Figure 6E). The localization of TgMAF1RHb0, TgMAF1RHb1, and HhMAF1b1 are all similar. We also performed the same experiment with NcMAF1 (based on NCLIV_ 004730), but we were unable to detect any protein following multiple (more than three) transfections. Whether this is due to upstream regulatory sequences or some other species-specific factor is unknown.
Figure 6.
N-terminally HA-tagged MAF1 isoforms were expressed in TgME49 parasites and HMA was assessed using MitoTracker or immunofluorescence assay using antibodies against the mitochondrial marker MTCO2. TgMAF1RHb0 and HhMAF1a1 (A and C) did not mediate HMA, while TgMAF1RHb1 and HhMAF1b1 (B and D) are both able to mediate HMA. (E) Quantification of percent vacuole coverage, determined by confocal microscopy. Twenty vacuoles were quantified for each of the MAF1 paralogs indicated, as well as wild-type TgME49. χ2 P-values: *0.0144; **0.0005; ***<0.0001.
Expression of MAF1RHb1, but not MAF1RHa1, in type II T. gondii increases competitive advantage during infection in vivo
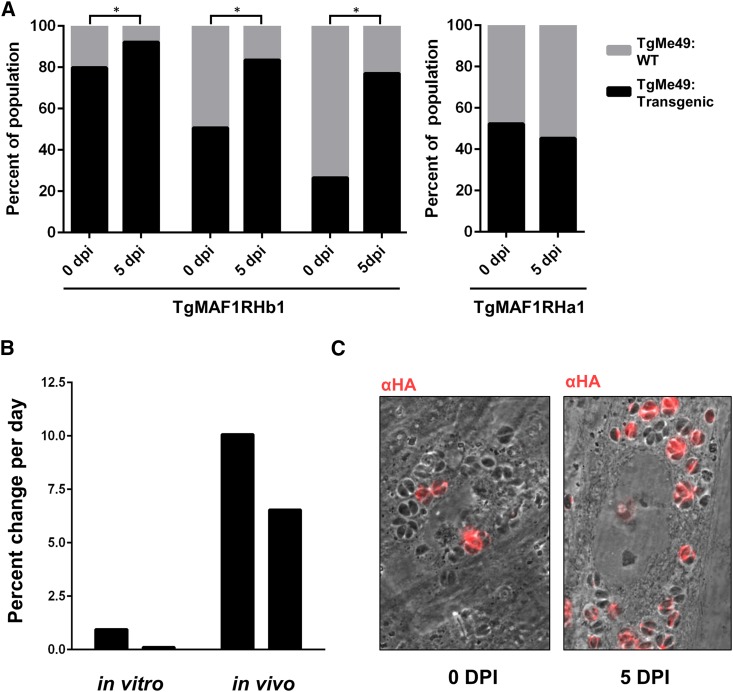
To directly examine the impact of MAF1 in an in vivo infection system, we infected Balb/c mice with TgME49 wild-type or a TgMAF1RHb1-complemented line and measured their rates of proliferation and dissemination in vivo using bioluminescence imaging (Walzer et al. 2013). We used a sublethal dose for a type II strain (100 tachyzoites) (Saeij et al. 2005) to allow the mice to survive the full course of the infection and enable us to detect any subtle differences in parasite dissemination. We observed marginally higher, but statistically insignificant, parasite burdens in infection with TgME49:TgMAF1RHb1 (Figure S5). Given the marginally higher parasite burden observed in mice infected with TgME49:TgMAF1RHb1 compared to wild-type TgME49, we hypothesized that this increase in parasite growth, although marginal, could provide a competitive advantage during an infection with a mixed population. To test this hypothesis, we created mixed populations of TgME49 and TgME49:TgMAF1RHb1 and infected female Balb/c mice with these populations of known proportions. Mice were infected with 105 tachyzoites of 1:4 or 4:1 proportions of TgME49 to TgME49:TgMAF1RHb1 parasites. Initial population proportions were quantified by immunofluorescence assay (IFA). Parasite burden was monitored by bioluminescence imaging for 5 days, after which mice were euthanized and parasites were collected by peritoneal lavage and the population proportions were determined by IFA. After 5 days the proportion of TgMAF1RHb1-expressing parasites increased significantly, regardless of initial proportion (Figure 7A). This competitive advantage was not observed when mice were infected with a mixed population of TgME49 and TgME49:TgMAF1RHa1 parasites (Figure 7A). While we did not notice any differences in growth rate in vitro between these strains, we also constructed mixed populations of TgME49 and TgME49:TgMAF1RHb1 and maintained them in HFFs by serial syringe lysis and passage in vitro for 8 weeks. Population proportions were determined by IFA at days 0, 28, and 54–59. In three of four populations, TgMAF1RHb1-expressing parasites significantly increased in proportion compared to their wild-type counterparts (Figure S6). However the competitive advantage of TgMAF1RHb1 expression appears much greater in vivo than in vitro, as evidenced by the 15-fold greater percent change per day of TgMAF1RHb1 expressing parasites within the population during in vivo compared to in vitro (Figure 7B).
Figure 7.
Expression of TgMAF1RHb1, but not TgMAF1RHa1, in type II T. gondii increases competitive advantage. (A) Mice were infected with mixed populations of TgME49:EV and TgME49:MAF1 with the indicated isoforms and ratios. Infection was allowed to progress for 5 days and population proportions before and after infection were quantified by IFA. Both HMA+ and HMA− MAF1 isoforms were assessed. TgME49:TgMAF1RHb1 significantly increases in proportion to TgME49:EV. *χ2 P-value <0.05. The proportion of TgMAF1RHa1-expressing parasites did not increase during the infection. (B) Percent change per day was calculated for the populations that started with 4:1 TgME49:EV to TgME49:TgMAF1RHb1 both in vitro and in vivo by dividing the total percent increase of TgMAF1RHb expressing parasites within the population by the number of days of infection. The first bar of both the in vitro and in vivo infections represent one clone set, while the second bar for each represents a second clone set. (C) Representative images of a mixed population from A before and after a 5-day in vivo infection. HA staining indicates TgMAF1RHb1-positive vacuoles.
Discussion
One of the many ways in which T. gondii interacts differently with host cells, compared to its relative N. caninum, is the close association formed between the PVM of T. gondii and the host cell mitochondria during the first hours after invasion (Pernas and Boothroyd 2010). HMA (Jones and Hirsch 1972; de Melo et al. 1992; Sinai et al. 1997) is not unique to T. gondii and has been described in other intracellular pathogens including Legionella pneumophila and Chlamydia psittaci (Horwitz 1983; Matsumoto et al. 1991; Scanlon et al. 2004). However, the biological relevance of HMA in these pathogens remains unclear partly due to the lack of understanding of the molecular mechanisms involved. The T. gondii gene responsible for this phenomenon is MAF1, and the MAF1 protein appears to mediate interactions not only with the host mitochondria but also the host immune response (Pernas et al. 2014). The discovery of MAF1 has removed initial barriers to understanding the biological relevance of HMA, yet the impact of MAF1-mediated HMA and host immune alterations on parasite biology has remained unclear. Here we demonstrate a clear competitive advantage of expressing an HMA+ paralog of MAF1, providing the first evidence of selective pressure maintaining HMA within parasite populations. During preliminary acute mouse infections with HMA+ and HMA− parasites, we did not observe any significant MAF1-dependent alterations in parasite virulence as assayed by parasite burden and mouse morbidity. This is consistent with published work showing that deletion of the MAF1 locus in a type I strain (which abolishes HMA) also had no dramatic impact on acute parasite virulence (Pernas et al. 2014). However, when we competed strains head to head, we did observe highly significant growth advantages in TgMAF1RHb1-expressing type II T. gondii parasites compared with wild-type controls. During a 5-day mouse infection, TgMAF1RHb1+ parasites outcompeted wild-type parasites by 7–10% per day (Figure 7). In contrast, when we performed a similar in vivo experiment with TgMAF1RHa1-expressing parasites, we observed no competitive advantage in the complemented strain vs. wild type. These data provide strong evidence for a selective advantage of HMA itself, since only TgMAF1RHb1-complemented parasites, and not TgMAF1RHa1-complemented parasites, had a growth advantage in vivo.
In addition to the selective pressure maintaining HMA within T. gondii populations, we have traced the evolutionary history of the MAF1 locus with respect to HMA. We propose a model in which the MAF1 gene (the “a” paralog) duplicated initially in a common ancestor to H. hammondi and T. gondii, and that this duplication event was followed by diversification and eventual neofunctionalization of one of the copies (into the “b” paralog) that had the capacity to mediate HMA. These data are consistent with the observation that T. gondii and H. hammondi (Figure 4C) are HMA+, while N. caninum is HMA− (Pernas and Boothroyd 2010), and that MAF1b paralogs from T. gondii and H. hammondi can confer the HMA phenotype to type II T. gondii, while their MAF1a counterparts cannot. It is also possible that N. caninum is HMA− due to secondary loss of a functional MAF1b paralog. However, electron micrographs of intracellular N. hughesi (a close relative of N. caninum) demonstrate a clear lack of HMA (Dubey et al. 2001), particularly in comparison to the very tight association between host mitochondria and the PVM that we see in TgMAF1RHb1-expressing T. gondii and N. caninum (Figure 5C, right). This suggests that N. hughesi is in fact HMA−, providing further evidence for the neofunctionalization of MAF1b in T. gondii and H. hammondi after they split from the Neospora lineage. In addition, we currently do not know the role that MAF1a protein plays in parasite biology. However, we do know that the MAF1a gene is maintained within parasite populations and is expressed at high levels (Figure 3, Figure S3, A–D), despite evidence that it is not essential for parasite replication in vitro (Pernas et al. 2014), and that it does not mediate HMA (Figure 5).
It was reported (Nolan et al. 2015) that an N. caninum strain different from that used in this study (N. caninum Liverpool) associated with host mitochondria after 24 hr of infection. However compared to T. gondii strain RH (an HMA+ strain) (Pernas et al. 2014), mitochondria were fewer in number, significantly further away from the PV, and did not accumulate until at least 24 hr postinfection. It may be that MAF1b paralogs (which are missing from N. caninum Liverpool) (Reid et al. 2012) mediate a very rapid and early association with host mitochondria (during the first 24 hr postinfection) but that mitochondria accumulate on N. caninum vacuoles due to other as-yet-unidentified factors. Strain differences between Liverpool and NC-1 also cannot yet be ruled out. Regardless, our confocal (Figure 5, A and B) and electron microscopy (Figure 5C) data show that in the first 18 hr postinfection there is a dramatic difference in HMA in T. gondii and N. caninum, and that we can convert N. caninum from being HMA− to HMA+ by complementation with a single gene product (TgRHMAF1b1).
While we have performed an extensive survey of the MAF1 paralogs found across multiple T. gondii strains and in two other species, our PCR-based methods could still be subject to creating chimeric artifacts during amplification. We did test for this by comparing sequences with existing Sanger-based complete genome sequence reads and found no evidence of chimeric sequences. To determine this with 100% certainty, however, we would need to clone MAF1 paralogs directly from T. gondii genomic DNA (as we did for T. gondii ROP5) (Reese et al. 2011).
In our isolated sequences there are significant sequence differences between the MAF1a and MAF1b paralogs, the most striking of which is the existence of a proline-rich repeat that is interspersed with serines in T. gondii, while H. hammondi MAF1 encodes a similar proline-rich region but without the interspersed serines. Some MAF1b paralogs from both species are functional with respect to HMA, while all MAF1 paralogs tested (whether derived from the “b” or “a” lineage) that lack this proline-rich region are not HMA competent. Additionally, there is a great diversity of MAF1 paralogs in T. gondii that can eventually be exploited to identify key residues that functionally distinguish HMA-competent and incompetent MAF1 paralogs. Structural biology comparisons between paralogs may also be particularly useful to identify polymorphic residues that are exposed and to determine the impact of specific mutations on the overall structure.
What remains to be determined is the relative importance of MAF1 locus amplification, which occurred only in members of the T. gondii lineage and not H. hammondi or N. caninum. Since H. hammondi is HMA+ and harbors a MAF1 paralog that is capable of mediating HMA in HMA- parasites (type II; Figure 6) locus expansion (to more than two copies) is not a prerequisite to drive HMA during infection, so the question remains as to the utility of having up to six copies of MAF1 (as shown for GT1; Figure 1C). Moreover T. gondii strain CTG is also HMA+ (Figure 4A) and is predicted to have only two MAF1 copies. The duplication of genes encoding secreted proteins in T. gondii, and importantly their subsequent diversification and optimization, may be a means of rapid and flexible adaptation to different hosts or even niches within a given host. In poxviruses, this process has been observed in real time under selective pressure at the K3L locus. Under strong host-induced pressure, K3L copy number increases and individual mutations accumulate in the duplicate copies. Once a mutation in a single copy of K3L emerges that is highly selective for virus survival, the locus resolves back to a single copy due to the negative impact of increased genome size (Elde et al. 2012). Based on our comparisons of closely related members of the same clonal lineage using both Southern blotting and sequence coverage analysis, expanded loci like MAF1 appear to be in comparatively rapid flux, undergoing multiple rounds of expansion and contraction on relatively short evolutionary time scales. It remains to be seen, however, if the changes in MAF1 locus size (or other T. gondii-specific expanded loci encoding secreted effectors) (Adomako-Ankomah et al. 2014) are of functional consequence as for the K3L locus in poxviruses. The observed differences in locus size (and therefore gene content) could be driven by selection-driven expansion/diversification/contraction as for K3L or simply be a result of stochastic changes during DNA replication and/or crossing over that are more common in repetitive DNA regions. A more detailed characterization of individual MAF1 copies, as well as their impact on HMA and parasite biology, will be necessary to determine the full impact of the observed strain- and species-specific features of the MAF1 locus.
We have not yet explored the impact of MAF1 expression in type II strains (or MAF1 locus deletion in HMA-competent strains like RH and CTG) on the virulence or persistence of other life cycle stages, including bradyzoites and sporozoites. These experiments will be important to fully assess the impact and importance of HMA in T. gondii biology in addition to the clear selective advantage of MAF1b expression (and presumably HMA) demonstrated here. It is conceivable that T. gondii contains built-in redundancies, which allow the parasite to assemble manipulative strategies that are targeted to specific hosts. For example, the IRG pathway targeted for neutralization by ROP5 is missing in humans and other systems where T. gondii is equally capable of surviving (Niedelman et al. 2012). This seems less likely for MAF1 since HMA occurs in multiple cell types from multiple species (Pernas and Boothroyd 2010; Pernas et al. 2014).
Finally it is important to note that in the type II strains examined MAF1b protein is undetectable and this correlates with the HMA− phenotype of strains from this lineage. This demonstrates that while MAF1b expression (and presumably HMA) is selectively advantageous, it is certainly not essential, and in the case of type II strains, is dispensable. This unique phenotype with respect to MAF1b expression and HMA is consistent with other strain-specific phenotypes in type II parasites, including a unique lack of ROP16-driven manipulation of the STAT3/6 pathway and a unique ability to modulate NFκB activation via the secreted effector GRA15 (Saeij et al. 2007; Rosowski et al. 2011). However, in eukaryotic parasites like T. gondii, it is the collection of virulence alleles at multiple locations in the genome that determine overall pathogenicity, and it may be that the silencing of MAF1b expression in the type II lineage may have been selected for as this strain evolved in a distinct niche. It will be interesting to determine the mechanism of MAF1b silencing in type II strains of T. gondii as a first step toward answering these questions.
Acknowledgments
The authors thank Abby Primack for critical reading of the manuscript. This work was funded by a Pew Scholarship in the Biomedical Sciences and National Institutes of Health (NIH) grant AI114655 to J.P.B. NIH grant AI73756 (awarded to John C. Boothroyd, Stanford University) supported L.F.P.
Footnotes
Communicating editor: J. Heitman
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.186270/-/DC1.
Literature Cited
- Adomako-Ankomah Y., Wier G. M., Borges A. L., Wand H. E., Boyle J. P., 2014. Differential locus expansion distinguishes Toxoplasmatinae species and closely related strains of Toxoplasma gondii. MBio 5: e01003–e01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl A., Davis P. H., Behnke M., Dzierszinski F., Jagalur M., et al. , 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. A., Eichler E. E., 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 7: 552–564. [DOI] [PubMed] [Google Scholar]
- Behnke M. S., Khan A., Wootton J. C., Dubey J. P., Tang K., et al. , 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl. Acad. Sci. USA 108: 9631–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke M. S., Khan A., Lauron E. J., Jimah J. R., Wang Q., et al. , 2015. Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent South American strains of Toxoplasma gondii. PLoS Genet. 11: e1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd J. C., Dubremetz J. F., 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6: 79–88. [DOI] [PubMed] [Google Scholar]
- Boyle J. P., Saeij J. P., Harada S. Y., Ajioka J. W., Boothroyd J. C., 2008. Expression quantitative trait locus mapping of Toxoplasma genes reveals multiple mechanisms for strain-specific differences in gene expression. Eukaryot. Cell 7: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Thornton K. R., 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol. Evol. 2: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo E. J., de Carvalho T. U., de Souza W., 1992. Penetration of Toxoplasma gondii into host cells induces changes in the distribution of the mitochondria and the endoplasmic reticulum. Cell Struct. Funct. 17: 311–317. [DOI] [PubMed] [Google Scholar]
- Deitsch K. W., Calderwood M. S., Wellems T. E., 2001. Malaria. Cooperative silencing elements in var genes. Nature 412: 875–876. [DOI] [PubMed] [Google Scholar]
- Dubey J. P., Sreekumar C., 2003. Redescription of Hammondia hammondi and its differentiation from Toxoplasma gondii. Int. J. Parasitol. 33: 1437–1453. [DOI] [PubMed] [Google Scholar]
- Dubey J. P., Liddell S., Mattson D., Speert C. A., Howe D. K., et al. , 2001. Characterization of the Oregon isolate of Neospora hughesi from a horse. J. Parasitol. 87: 345–353. [DOI] [PubMed] [Google Scholar]
- Dubey J. P., Barr B. C., Barta J. R., Bjerkas I., Bjorkman C., et al. , 2002. Redescription of Neospora caninum and its differentiation from related coccidia. Int. J. Parasitol. 32: 929–946. [DOI] [PubMed] [Google Scholar]
- Dubey J. P., Tilahun G., Boyle J. P., Schares G., Verma S. K., et al. , 2013. Molecular and biological characterization of first isolates of Hammondia hammondi from cats from Ethiopia. J. Parasitol. 99: 614–618. [DOI] [PubMed] [Google Scholar]
- Elde N. C., Child S. J., Eickbush M. T., Kitzman J. O., Rogers K. S., et al. , 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150: 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Cardoso-Moreira M., Borevitz J. O., Long M., 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631. [DOI] [PubMed] [Google Scholar]
- Espinosa-Cantu A., Ascencio D., Barona-Gomez F., DeLuna A., 2015. Gene duplication and the evolution of moonlighting proteins. Front. Genet. 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior L. H., Bottius E., Pirrit L. A., Deitsch K. W., Scheidig C., et al. , 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018–1022. [DOI] [PubMed] [Google Scholar]
- Goodswen S. J., Kennedy P. J., Ellis J. T., 2013. A review of the infection, genetics, and evolution of Neospora caninum: from the past to the present. Infect. Genet. Evol. 13: 133–150. [DOI] [PubMed] [Google Scholar]
- Heinberg A., Siu E., Stern C., Lawrence E. A., Ferdig M. T., et al. , 2013. Direct evidence for the adaptive role of copy number variation on antifolate susceptibility in Plasmodium falciparum. Mol. Microbiol. 88: 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. A., 1983. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. C., Hirsch J. G., 1972. The interaction between Toxoplasma gondii and mammalian cells. II. The absence of lysosomal fusion with phagocytic vacuoles containing living parasites. J. Exp. Med. 136: 1173–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Kofoid E., Reams A. B., Andersson D. I., Roth J. R., 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 103: 17319–17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Kofoid E., Andersson D. I., Lu Y., Mellor J., et al. , 2010. The tandem inversion duplication in Salmonella enterica: selection drives unstable precursors to final mutation types. Genetics 185: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi H., Khan A., Behnke M. S., Namasivayam S., Swapna L. S., et al. , 2016. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun. 7: 10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Bessho H., Uehira K., Suda T., 1991. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J. Electron Microsc. (Tokyo) 40: 356–363. [PubMed] [Google Scholar]
- Mitra, K., and J. Lippincott-Schwartz, 2010 Analysis of mitochondrial dynamics and functions using imaging approaches. Curr. Protoc. Cell Biol. Chapter 4: Unit 4.25.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Miller B., Barends M., Jaidee A., Patel J., et al. , 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet. 4: e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S., 2000. Molecular Evolution and Phylogenetics, Oxford University Press, Oxford. [Google Scholar]
- Nicol J. W., Helt G. A., Blanchard S. G., Jr, Raja A., Loraine A. E., 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedelman W., Gold D. A., Rosowski E. E., Sprokholt J. K., Lim D., et al. , 2012. The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon-gamma response. PLoS Pathog. 8: e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan S. J., Romano J. D., Luechtefeld T., Coppens I., 2015. Neospora caninum recruits host cell structures to its parasitophorous vacuole and salvages lipids from organelles. Eukaryot. Cell 14: 454–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication, Springer-Verlag, Berlin. [Google Scholar]
- Pasternak N. D., Dzikowski R., 2009. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int. J. Biochem. Cell Biol. 41: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Pernas L., Boothroyd J. C., 2010. Association of host mitochondria with the parasitophorous vacuole during Toxoplasma infection is not dependent on rhoptry proteins ROP2/8. Int. J. Parasitol. 40: 1367–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L., Adomako-Ankomah Y., Shastri A. J., Ewald S. E., Treeck M., et al. , 2014. Toxoplasma effector MAF1 mediates recruitment of host mitochondria and impacts the host response. PLoS Biol. 12: e1001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M. L., Zeiner G. M., Saeij J. P., Boothroyd J. C., Boyle J. P., 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl. Acad. Sci. USA 108: 9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. J., 2015. Large, rapidly evolving gene families are at the forefront of host-parasite interactions in Apicomplexa. Parasitology 142(Suppl 1): S57–S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. J., Vermont S. J., Cotton J. A., Harris D., Hill-Cawthorne G. A., et al. , 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog. 8: e1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettigner C., Leclipteux T., De Meerschman F., Focant C., Losson B., 2004. Survival, immune responses and tissue cyst production in outbred (Swiss white) and inbred (CBA/Ca) strains of mice experimentally infected with Neospora caninum tachyzoites. Vet. Res. 35: 225–232. [DOI] [PubMed] [Google Scholar]
- Rosowski E. E., Lu D., Julien L., Rodda L., Gaiser R. A., et al. , 2011. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208: 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij J. P., Boyle J. P., Boothroyd J. C., 2005. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 21: 476–481. [DOI] [PubMed] [Google Scholar]
- Saeij J. P., Coller S., Boyle J. P., Jerome M. E., White M. W., et al. , 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445: 324–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon M., Leitch G. J., Visvesvara G. S., Shaw A. P., 2004. Relationship between the host cell mitochondria and the parasitophorous vacuole in cells infected with Encephalitozoon microsporidia. J. Eukaryot. Microbiol. 51: 81–87. [DOI] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai A., Webster P., Joiner K., 1997. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 110: 2117–2128. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer K. A., Adomako-Ankomah Y., Dam R. A., Herrmann D. C., Schares G., et al. , 2013. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc. Natl. Acad. Sci. USA 110: 7446–7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer K. A., Wier G. M., Dam R. A., Srinivasan A. R., Borges A. L., et al. , 2014. Hammondia hammondi harbors functional orthologs of the host-modulating effectors GRA15 and ROP16 but is distinguished from Toxoplasma gondii by a unique transcriptional profile. Eukaryot. Cell 13: 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J., Daub J., Peregrin-Alvarez J. M., Finney C. A., Parkinson J., 2009. The origins of apicomplexan sequence innovation. Genome Res. 19: 1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Rosenberg H. F., Nei M., 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and plasmids available upon request. All MAF1 paralog and ortholog sequences obtained for this study have been deposited in GenBank (accession numbers KU761333-KU761342).