Abstract
G-protein-coupled receptors (GPCRs) are integral membrane proteins that initiate stimulus-dependent activation of cognate heterotrimeric G-proteins, triggering ensuing downstream cellular responses. Tight regulation of GPCR-evoked pathways is required because prolonged stimulation can be detrimental to an organism. Ste2, a GPCR in Saccharomyces cerevisiae that mediates response of MATa haploids to the peptide mating pheromone α-factor, is down-regulated by both constitutive and agonist-induced endocytosis. Efficient agonist-stimulated internalization of Ste2 requires its association with an adaptor protein, the α-arrestin Rod1/Art4, which recruits the HECT-domain ubiquitin ligase Rsp5, allowing for ubiquitinylation of the C-terminal tail of the receptor and its engagement by the clathrin-dependent endocytic machinery. We previously showed that dephosphorylation of Rod1 by calcineurin (phosphoprotein phosphatase 2B) is required for optimal Rod1 function in Ste2 down-regulation. We show here that negative regulation of Rod1 by phosphorylation is mediated by two distinct stress-activated protein kinases, Snf1/AMPK and Ypk1/SGK1, and demonstrate both in vitro and in vivo that this phospho-regulation impedes the ability of Rod1 to promote mating pathway desensitization. These studies also revealed that, in the absence of its phosphorylation, Rod1 can promote adaptation independently of Rsp5-mediated receptor ubiquitinylation, consistent with recent evidence that α-arrestins can contribute to cargo recognition by both clathrin-dependent and clathrin-independent mechanisms. However, in cells lacking a component (formin Bni1) required for clathrin-independent entry, Rod1 derivatives that are largely unphosphorylated and unable to associate with Rsp5 still promote efficient adaptation, indicating a third mechanism by which this α-arrestin promotes desensitization of the pheromone-response pathway.
Keywords: mating pheromone response, adaptation, desensitization, down-regulation, endocytosis
A cell must adapt rapidly to external stimuli and other changes in its environment. One mechanism to achieve an appropriate response is through remodeling of the repertoire of integral membrane proteins in the plasma membrane (PM), including receptors, channels, permeases, and other transporters. These transmembrane proteins are often shuttled between different cellular compartments in response to specific stimuli. This trafficking, especially endocytosis to remove these molecules from the PM, is controlled, in all cases examined, by regulated ubiquitinylation of the target protein (Horák 2003; Dupré et al. 2004; Nikko and Pelham 2009; Lauwers et al. 2010; Zhao et al. 2013; Crapeau et al. 2014; Ghadder et al. 2014).
In eukaryotes, G-protein-coupled receptors (GPCRs) are the most abundant class of cell-surface receptors (Granier and Kobilka 2012; Katritch et al. 2013). Internalization of a GPCR plays an important role in both rapid and long-term desensitization after exposure of a cell to the cognate agonist (Marchese and Trejo 2013; Irannejad et al. 2015). Aberrant GPCR signaling and dysregulation have been implicated in many pathophysiologies, including cancers, asthma, hypertension, neurological disorders, and autoimmune diseases (O’Hayre et al. 2014; West and Hanyaloglu 2015). For these reasons, GPCRs are the targets of the majority of clinically used pharmaceuticals (Shoichet and Kobilka 2012; Zhang and Xie 2012; Garland 2013). A model system that has served as a very informative experimental paradigm for investigating GPCR-initiated signaling and its regulation are the receptors in budding yeast (Saccharomyces cerevisiae) that mediate its response to peptide mating pheromones (Hao et al. 2007; Merlini et al. 2013).
It has been amply demonstrated that both basal and agonist-induced internalization of Ste2 (the GPCR on MATa cells that binds the mating pheromone α-factor) and Ste3 (the GPCR on MATα cells that binds the mating pheromone a-factor) requires ubiquitinylation on Lys residues in their cytosolic tails and that Rsp5 (mammalian ortholog is Nedd4L) is the ubiquitin ligase (E3) responsible for this modification (Dunn and Hicke 2001; Ballon et al. 2006; Rotin and Kumar 2009). Rsp5 catalyzes formation of K63-linked polyubiquitin chains on its substrates (Galan and Haguenauer-Tsapis 1997; Kim and Huibregtse 2009; Lauwers et al. 2009), leading to their recruitment into clathrin-coated pits and internalization (Weinberg and Drubin 2012; Myers and Payne 2013). Rsp5 associates via its WW domains with PPxY motifs (and variants thereof) in its targets. However, recruitment to many such targets is not direct, but mediated instead by intermediary “adaptor” proteins, and paramount among these molecular matchmakers are the α-arrestins (Lin et al. 2008; Léon and Haguenauer-Tsapis 2009; Nikko and Pelham 2009), a family of proteins found in all eukaryotes from yeast to humans (Alvarez 2008; Aubry and Klein 2013). In S. cerevisiae, these adaptors have been dubbed Art (for “Arrestin-Related Trafficking”) proteins (Lin et al. 2008), whereas in animal cells these are termed ARRDC (for “Arrestin-Domain-Containing”) proteins (Aubry and Klein 2013). In general, in these molecules, an arrestin fold (Aubry et al. 2009) situated near their N-terminal end mediates interaction with the target (Kang et al. 2015a,b), and PPxY motifs located in their C-terminal region associate with a WW domain-containing HECT-type E3 (Rotin and Kumar 2009).
The S. cerevisiae genome encodes 14 recognized α-arrestins, most of which have been implicated in endocytosis and trafficking of various nutrient permeases (Lin et al. 2008; Nikko and Pelham 2009; O’Donnell et al. 2010; Becuwe et al. 2012; Merhi and Andre 2012; O’Donnell et al. 2015). We demonstrated recently that specific α-arrestins also control internalization of both Ste2 (Alvaro et al. 2014) and Ste3 (Prosser et al. 2015). In both yeast and mammalian cells, the types of integral PM proteins greatly outnumber the α-arrestins present; hence, there is promiscuity in these interactions; that is, a given α-arrestin can have more than one target. However, in several respects, there is also considerable specificity: (i) most cargo are the target of several α-arrestins, but far from all (Lin et al. 2008; Nikko and Pelham 2009; Lauwers et al. 2010; Alvaro et al. 2014; Prosser et al. 2015); (ii) rapid internalization of a given cargo is triggered only in response to a specific stimulus and, as a result, often engages only one or just a few α-arrestins (Becuwe et al. 2012; O’Donnell et al. 2013; Zhao et al. 2013; Crapeau et al. 2014; Ghaddar et al. 2014; O’Donnell et al. 2015); and (iii) the function of an α-arrestin is often negatively regulated by phosphorylation (Shinoda and Kikuchi 2007; MacGurn et al. 2011; Becuwe et al. 2012; Jee et al. 2012; Merhi and Andre 2012; O’Donnell et al. 2013; Alvaro et al. 2014; Herrador et al. 2015). Arrestin phosphorylation raises important questions about what protein kinases are involved in these control circuits and under what conditions, and how such modifications affect the ability of an α-arrestin to promote internalization of its specific PM protein targets.
We have shown (Alvaro et al. 2014) that, in addition to all the other previously known mechanisms for down-regulating the mating pathway (Dohlman and Thorner 2001), three α-arrestins specifically contribute to desensitization of the pheromone response in MATa cells by mediating internalization of Ste2. Ldb19/Art1 participates mainly in basal Rsp5-dependent endocytosis of Ste2 (i.e., in the absence of pheromone), most likely through recognition of misfolded forms of the receptor, consistent with other evidence that this α-arrestin primarily serves a “quality control” function (Zhao et al. 2013). By contrast, Rod1/Art4 and its paralog Rog3/Art7, promote Rsp5-dependent endocytosis of pheromone-bound receptor; however, Rod1 function in Ste2 down-regulation obligatorily required its association with Rsp5, whereas forms of Rog3 unable to associate with Rsp5 were able to promote adaptation. Conversely, the ability of Rod1 to promote adaptation required its dephosphorylation by the Ca2+/calmodulin-stimulated phosphoprotein phosphatase calcineurin, whereas Rog3 did not. These findings focused our attention on the underlying mechanisms involved in phospho-regulation of Rod1. As described here, we identified two stress-responsive protein kinases that phosphorylate Rod1 in vivo and delineated the sites at which they exert their regulatory effect. Our studies also reveal that, in the absence of its phosphorylation, Rod1 can, like Rog3, also promote adaptation in an Rsp5-independent manner, suggesting that, in addition to negative regulation, phosphorylation may serve as a switch to control how Rod1 down-regulates mating pheromone response.
Materials and Methods
Strains and growth conditions
Yeast strains (Table 1) were grown at 30° in either rich (YPD) or synthetic complete (SC) medium containing 2% glucose (unless another carbon source is specified) and with appropriate nutrients to maintain selection for plasmids, if present (Sherman et al. 1986). Standard genetic methods were used for strain construction (Amberg et al. 2005).
Table 1. Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| sst2∆ GEV (JT5919) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 sst2∆::SpHIS5 leu2Δ0::GEV::NatMX | Alvaro et al. (2014) |
| snf1Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 snf1Δ::KanMX4 | This study |
| kin1Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 kin1Δ::KanMX4 | This study |
| kin2Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 kin2Δ::KanMX4 | This study |
| kin4Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 kin4Δ::KanMX4 | This study |
| kcc4Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 kcc4Δ::KanMX4 | This study |
| hsl1Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 hsl1Δ::KanMX4 | This study |
| frk1Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 frk1Δ::KanMX4 | This study |
| gin4Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 gin4Δ::KanMX4 | This study |
| cnb1Δ sst2∆ GEV (JT6694) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 cnb1Δ::KanMX4 | Alvaro et al. (2014) |
| cna1Δ cna2Δ sst2∆ GEV (JT6695) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 cna1Δ::KanMX4 cna1Δ::KanMX4 | Alvaro et al. (2014) |
| BJ5459 GEV (JT6743) | MATa ura3-52 trp1 lys2-801 leu2Δ1 his3∆200 pep4∆::HIS3 prb1∆1.6R can1 GAL leu2Δ1::GEV::NatMX | Alvaro et al. (2014) |
| rod1Δ rog3∆ ldb19∆ sst2∆ GEV (JT6716) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 rod1∆::KanMX4 rog3∆::KanMX4 ldb19∆::NatMX sst2∆::SpHIS5 | Alvaro et al. (2014) |
| STE2 sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 STE2::HPH | This study |
| STE27KR sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 STE27KR::HPH | This study |
| STE2∆296 sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 STE2∆296::HPH | This study |
| STE2-mCherry sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE2-mCherry::URA3 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 | This study |
| STE27KR-mCherry sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE27KR-mCherry::URA3 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 | This study |
| STE27KR-mCherry sst2∆ GEVb | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE27KR-mCherry::ura3 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 | This study |
| STE2-mCherry bar1∆ (JT6677) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE2-mCherry::URA3 bar1∆::CgLEU2 | Alvaro et al. (2014) |
| STE27KR-mCherry bar1∆ | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE27KR-mCherry::URA3 bar1∆::CgLEU2 | This study |
| STE2-mCherry rod1Δ rog3∆ bar1∆ (JT6679) | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE2-mCherry::URA3 rod1∆::KanMX4 rog3∆::KanMX4 bar1∆::CgLEU2 | Alvaro et al. (2014) |
| STE27KR-mCherry rod1Δ rog3∆ bar1∆ | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 STE27KR-mCherry::URA3 rod1∆::KanMX4 rog3∆::KanMX4 bar1∆::CgLEU2 | This study |
| bni1Δ sst2∆ GEVa | MATa leu2Δ0 ura3Δ0 his3Δ1 met15Δ0 leu2Δ0::GEV::NatMX sst2∆::SpHIS5 bni1Δ::KanMX4 | This study |
To generate a Gal4(1-93)-estrogen receptor (ER)-VP16 chimera (designated GEV)-expressing version of the indicated yeast strain, pACT1-GEV (Veatch et al. 2009; McIsaac et al. 2011) was digested with EcoRV and introduced into the cells of interest by DNA-mediated transformation (Amberg et al. 2005), and nourseothricin-resistant colonies were selected, in which GEV (expressed under control of an ACT1 promoter) is integrated at the leu2Δ0 locus.
The immediately preceding strain was streaked onto plates containing 5-FOA, and a resulting Ura- (ura3) derivative was selected.
Plasmids
Plasmids (Table 2) were constructed using standard procedures (Green and Sambrook 2012a,b). Briefly, DNA amplification by the polymerase chain reaction employed Phusion DNA polymerase (New England Biolabs, Ipswich, MA), and all constructs were verified by DNA sequencing. Site-directed mutagenesis was carried out using the same DNA polymerase and QuikChange methodology (New England Biolabs), according to the manufacturer’s instructions.
Table 2. Plasmids used in this study.
| Plasmid | Genotype | Source |
|---|---|---|
| pEGKG | GAL1prom-GST | Yeast Deletion Collection (Open Biosystems, Inc.) |
| 2µ, URA3 | ||
| pEGKG-Rod1 | GAL1prom-GST | Zhu et al. (2000) |
| 2µ, URA3 | ||
| pEGKG-Rod1315Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1447Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1641Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1706Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1720Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1781Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1447A 641Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod1447A 706Aa | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod13A (Rod1447A 641A 706A)a | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod14A (Rod1315A 447A 641A 706A)a | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod15A (Rod1315A 447A 641A 706A 720A)a | GAL1prom-GST | This study |
| 2µ, URA3 | ||
| pEGKG-Rod16SA (Rod1S315A S447A S641A S706A S720A S781A)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pGEX6P1-Rod16SE (Rod1S315E S447E S641E S706E S720E S781E)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pGEX6P1-Rod1ARR (Rod11-402)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pGEX6P1-Rod1TAIL (Rod1403-837)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pGEX6P1-Rod11SA ARR (Rod11-402 S315A)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pGEX6P1-Rod15SA TAIL (Rod1403-837 S447A S641A S706A S720A S781A)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pEGKG-Rod12SA (Rod1138A 807A)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pEGKG-Rod12SE (Rod1138E 807E)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pEGKG-Rod18SA (Rod1S315A S447A S641A S706A S720A S781A 138A 807A)a | GAL1prom-GST | This study |
| 2μ, URA3 | ||
| pEGKG-Rod1PANA (pJT4954) | GAL1prom-GST | Alvaro et al. (2014) |
| 2µ, URA3 | ||
| pEGKG-Rod1PASA (pJT4955) | GAL1prom-GST | Alvaro et al. (2014) |
| 2µ, URA3 | ||
| pEGKG-Rod1PPxY-less (pJT4956) | GAL1prom-GST | Alvaro et al. (2014) |
| 2µ, URA3 | ||
| pEGKG-Rod1V/PPxY-lessa | LDB19prom | This study |
| CEN, HIS3 | ||
| pEGKG-Rod12A, V/PPxY-lessa | LDB19prom | This study |
| CEN, HIS3 | ||
| pEGKG-Rod16A, V/PPxY-lessa | LDB19prom | This study |
| CEN, HIS3 | ||
| pEGKG-Rod18A, V/PPxY-lessa | LDB19prom | This study |
| CEN, HIS3 | ||
| pEGKG-Rog3 | GAL1prom-GST | Zhu et al. (2000) |
| 2µ, URA3 | ||
| pEGKG-Rog3∆400 | GAL1prom-GST | Alvaro et al. (2014) |
| 2µ, URA3 |
Generated by site-directed mutagenesis (Green and Sambrook 2012b) with synthetic oligonucleotides containing the desired codon alterations (using the wild-type sequences in pRS426 vectors as the template). DNA from the corresponding gene was amplified from genomic DNA by PCR (Green and Sambrook 2012a) and then cloned into pEGKG.
Pheromone-imposed growth arrest
Response to α-factor was assessed by an agar diffusion (halo) bioassay (Reneke et al. 1988). In brief, cells were plated in top agar on solid YPD or SC medium, as appropriate. On the resulting surface were laid sterile cellulose filter disks, onto which an aliquot (15 μl) of an aqueous solution (1 mg/ml) of synthetic α-factor (GeneScript, Piscataway, NJ) was aseptically spotted, and the plates were incubated at 30° for 4–5 days. In those experiments in which α-arrestin overexpression was induced, strains containing the tripartite S. cerevisiae Gal4-human estrogen receptor-herpes simplex virus transactivator VP16 fusion protein (Gal4-ER-VP16 or GEV) (Quintero et al. 2007) and a URA3-marked multi-copy (2 μm DNA) plasmid expressing from a GAL promoter the α-arrestin of interest [which was fused to the C terminus of glutathione S-transferase (GST)] were grown to midexponential phase, treated with β-estradiol (20 μM final concentration) for 3 hr, and then plated in top agar also containing β-estradiol (final concentration 200 nM). To confirm α-arrestin overexpression, samples of the same cultures were analyzed by immunoblotting.
Immunoblotting
Equal numbers of cells from midexponential phase cultures were collected by centrifugation and stored at −80°. The cell pellets were thawed on ice, and whole-cell protein extracts were prepared by alkaline lysis followed by collection of total protein by trichloroacetic acid precipitation (Volland et al. 1994). Protein precipitates were solubilized in SDS-urea gel sample buffer (5% SDS, fresh 8 M urea, 1% β-mercaptoethanol, 0.1 mM EDTA, 40 mM Tris–HCl, pH 6.8) with 0.1% bromophenol blue, heated at 37° for 15 min, resolved by SDS-PAGE, and analyzed by immunoblotting. To dephosphorylate phosphoproteins in extracts, protein precipitates were solubilized in sample buffer (80 mM Tris–HCl, pH 8.0, 8 mM EDTA, 120 mM DTT, 3.5% SDS, 0.29% glycerol, 0.08% Tris base, 0.01% Bromophenol blue), and then treated with 10 µl of calf intestinal phosphatase (CIP) (10,000 units/ml) for 1 hr at 37°. The resulting samples were then resolved by SDS-PAGE and analyzed by immunoblotting. Proteins in SDS-PAGE gels were transferred electrophoretically to nitrocellulose sheets using a semidry transfer apparatus (Transblot SD; Bio-Rad, Inc.). After blocking with carrier protein, the filters were incubated (generally overnight at 4°) with one of the following primary antibodies: rabbit polyclonal anti-GST (Sigma), rabbit polyclonal anti-Rsp5 (gift of Allyson F. O’Donnell, Duquesne University, Pittsburgh), or rabbit polyclonal anti-Pgk1 (this laboratory) as a loading control. The resulting immune complexes were then detected by incubation with infrared dye (IRDye 680/800)-labeled goat anti-rabbit IgG secondary antibody, followed by visualization using an infrared imager (Odyssey; Li-Cor).
Purification of GST fusion proteins from Escherichia coli
Freshly transformed BL21(DE3) cells carrying a plasmid expressing wild-type or mutant versions of GST-Rod1ARR (residues 1–403) or GST-Rod1TAIL (residues 402–837) were grown to A600 nm = 0.6, and protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (final concentration 0.5 mM). After aeration for 5 hr at 37°, cells were harvested and the GST fusion protein was purified by column chromatography on glutathione-agarose beads (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The beads were washed three times with 500 μl lysis buffer (150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM DTT, 50 mM Tris–HCl, pH 7.4). Bound protein was eluted from the beads in SDS-PAGE sample buffer, resolved by SDS-PAGE (7.5% acrylamide gel), and analyzed by immunoblotting.
In vitro kinase assay
Purified Snf1 (gift of Benjamin Turk, Yale University, New Haven, CT) or purified analog-sensitive Ypk1(L424A) (gift of Alexander Muir, this laboratory) was incubated at 30° in protein kinase assay buffer (20 mM Tris–HCl, pH 7.2, 125 mM potassium acetate, 12 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM DTT, 1% glycerol, 0.02% BSA, 25 mM β-glycerol phosphate, and 1 mM sodium orthovanadate) with 100 µM γ-[32P]ATP (∼5 × 105 cpm/nmol) and 0.5 µg of GST-fused substrate protein (prepared by expression in and purification from E. coli, as described above) with or without addition of Ypk1 inhibitor [1 μM 1-(tert-butyl)-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (3-MB-PP1)] (Burkard et al. 2007). After 30 min, reactions were terminated by addition of SDS-PAGE sample buffer containing 6% SDS followed by boiling for 5 min. Labeled proteins were resolved by SDS-PAGE and analyzed by autoradiography using a PhosphorImager (Molecular Dynamics Division, GE Healthcare).
Purification of GST fusion proteins from yeast
Rsp5 association with α-arrestins was assessed as described before (O’Donnell et al. 2013; Alvaro et al. 2014). Briefly, BJ5459 GEV cells carrying a plasmid vector (pEGKG) for expression of GST-Rod1 or GST-Rod1 derivatives containing a mutation(s) in its PPxY motifs (Rsp5-binding sites) were grown to midexponential phase and induced with β-estradiol (20 mM final concentration) for 3 hr. After harvesting by centrifugation, cells were washed and frozen in liquid N2. Cell pellets were resuspended in 600 μl co-immunoprecipitation (co-IP) buffer (100 mM NaCl, 0.2% Triton X-100, 15 nM EGTA, 50 mM Tris, pH 7.4) containing 5 mM N-ethylmaleimide and protease inhibitors [1 tablet of Complete protease inhibitor cocktail (Roche Applied Science) per 15 ml] and lysed at 4° by vigorous vortexing with ∼1 g glass beads (0.5 mm; BioSpec Products). After clarification, GST-tagged proteins were recovered from equal volumes of these extracts by incubation with GST-agarose beads for 4 hr at 4°. After washing two times with co-IP buffer containing 150 mM NaCl, liquid was removed by aspiration, and the beads were resuspended in SDS-PAGE sample buffer to elute the bound proteins, which were resolved by SDS-PAGE and analyzed by immunoblotting.
Fluorescence microscopy
Imaging of Ste2(7K-to-R)-mCherry was performed as described previously (Ballon et al. 2006). Cells were diluted in selective minimal medium, grown to midexponential phase, and treated with 20 μM β-estradiol for 3 hr to induce expression of the GST-arrestin variants of interest. After collection by brief centrifugation in a microfuge, the cell population was immediately examined using an Olympus BH-2 upright fluorescence microscope (Olympus, Tokyo) equipped with a 100× objective, illuminated with a SOLA light engine (Lumencore, Beaverton, OR), and images were recorded with a CoolSNAP MYO CCD camera (Photometrics, Tuscon, AZ). Images were analyzed using Micro-Manager software (Edelstein et al. 2010) and ImageJ (National Institutes of Health). All images grouped together in any given figure were always scaled identically and always adjusted identically for brightness using Photoshop (Adobe).
Data and reagent availability
We will freely send all plasmids, strains, antibodies, and other research materials and procedures generated from this research to investigators at any and all nonprofit institutions for research purposes upon request.
Results
Snf1 phosphorylates Rod1 and inhibits its function in mating pathway down-regulation
The preferred carbon source for S. cerevisiae is glucose under both fermentative and nonfermentative conditions (Fraenkel 2003); however, when the supply of glucose is exhausted and oxygen is present, the cells can utilize nonfermentable carbon sources, such as lactate (Schüller 2003). Entry of lactate is mediated by Jen1, a lactate-specific permease (Casal et al. 1999). It has been demonstrated by the prior work of others that Jen1 is endocytosed in a Rod1-dependent manner and that the role of Rod1 in promoting Jen1 internalization is blocked by phosphorylation of this α-arrestin by Snf1 (yeast AMPK) (Shinoda and Kikuchi 2007; Becuwe et al. 2012), a protein kinase strongly activated under glucose-limiting conditions (Rubenstein and Schmidt 2007; Hedbacker and Carlson 2008). In this way, Jen1 remains at the PM under conditions where uptake of lactate would be beneficial for continued growth of the cells. However, under other conditions that mimic glucose limitation and acutely activate Snf1 (addition of the nonmetabolizable analog 2-deoxyglucose), Rod1-dependent endocytosis of two low-affinity glucose transporters (Hxt1 and Hxt3) is stimulated (O’Donnell et al. 2015). Hence, it was not at all clear whether Snf1 phosphorylation of Rod1 has any effect, either positive or negative, on its ability to promote desensitization of mating pheromone response. Moreover, all of the sites in Rod1 phosphorylated by Snf1 have not been delineated previously.
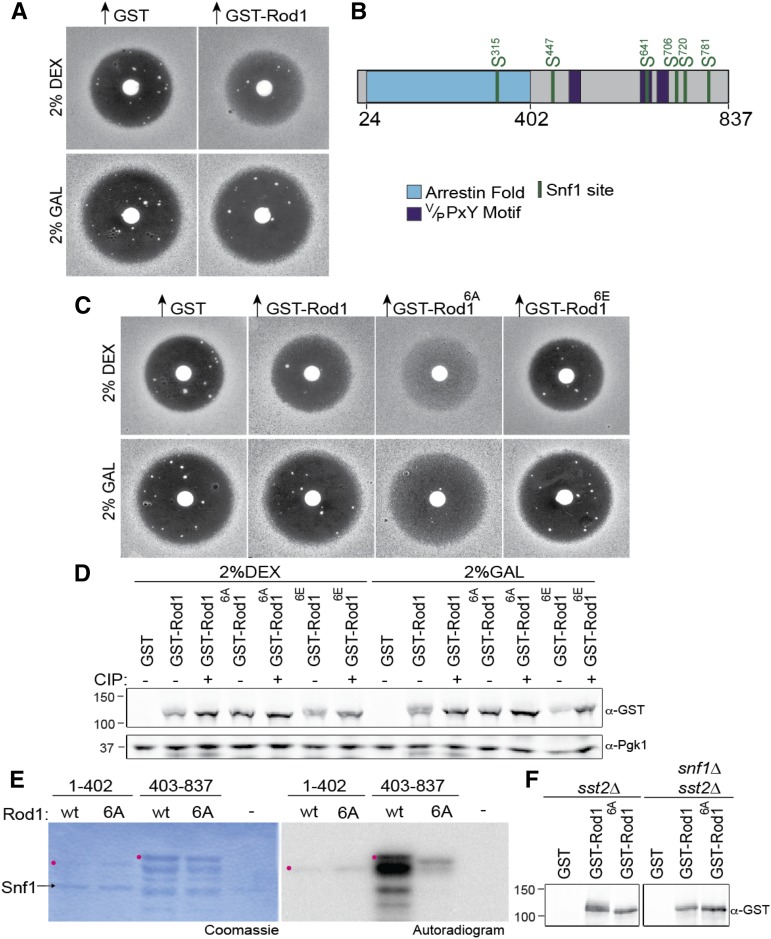
Snf1 is strongly activated when cells are shifted from glucose to a medium containing even another sugar, such as sucrose or galactose (Hedbacker and Carlson 2008). Hence, as a first means to examine the potential role of Snf1-mediated phosphorylation of Rod1 in desensitization of the mating pheromone response pathway, we compared the ability of Rod1 overexpression to promote adaptation on medium containing glucose vs. medium containing galactose. For this purpose, we used an agar diffusion bioassay that we have described before (Reneke et al. 1988; Alvaro et al. 2014). Specifically, in MATa cells lacking the RGS protein Sst2, upon exposure to pheromone, there is no way to prevent persistent receptor-initiated G-protein activation, and, hence, cells undergo a potent and sustained pheromone-induced G1 arrest (Chan and Otte 1982; Dohlman et al. 1996), manifest as a large clear zone in the lawn around a source of α-factor. Of course, if the receptor is efficiently removed by endocytosis, then there is no way to activate the G-protein, so cells have an opportunity to recover and resume growth, which is indicated by turbidity (“fill-in”) within the halo of initial growth inhibition. This fill-in is to be distinguished from the occasional large papillae that appear [which represent rare pheromone-resistant (ste) mutants that arise spontaneously at a significant frequency because a loss-of-function mutation in any gene product necessary for signal propagation, such as the MAPKKKK Ste20, the MAPKKK Ste11, the MAPKK Ste7, or the MAPK Fus3, for example, will confer a growth advantage when α-factor is present]. In any event, as we observed before (Alvaro et al. 2014), when GST-Rod1 overexpression was driven in a β-estradiol-induced manner in MATa sst2∆ cells grown on glucose, the halo displayed a faint, but readily detectable, turbidity compared to control cells expressing GST alone, as expected (Figure 1A, top). In striking contrast, when grown on galactose, but otherwise under the same conditions, the identical cells displayed much larger halos, and no fill-in was observed when GST-Rod1 was overexpressed (Figure 1A, bottom). These findings suggested that under conditions where Snf1 is expected to be highly active, Rod1 is ineffective in promoting desensitization.
Figure 1.
Snf1 phosphorylates Rod1 in vivo and in vitro. (A) MATa sst2∆ cells (JT6674) harboring the GEV chimera and a URA3-marked high-copy-number (2 µm DNA) plasmid expressing GST-Rod1 under GAL promoter control were grown in minimal medium (SC-Ura) with either 2% dextrose (top) or 2% galactose (bottom) as the carbon source, induced with β-estradiol as described in Materials and Methods, plated in top agar on the same medium, exposed to a filter disk containing 15 µg of α-factor, and incubated for 4 days at 30°. (B) Schematic diagram of Rod1. Arrestin fold (blue); Rsp5-binding motifs (purple); six Snf1 consensus motifs (green). (C) Same as in A, with inclusion of a nonphosphorylatable allele (Rod16A) and phospho-mimetic allele (Rod16E). (D) Samples of the cultures used in C were harvested and lysed, and the resulting extracts were divided and not treated (−) or treated (+), as indicated, with CIP, resolved by SDS-PAGE, and analyzed by immunoblotting with anti-GST or with anti-Pgk1 (loading control) antibodies. (E) GST fusions to the arrestin fold domain (residues 1–402) and the remaining C-terminal region (402–837) of either wild type (wt) or the 6A allele of Rod1 were purified from E. coli and incubated with [γ-32P]ATP and purified Snf1, and the resulting products were resolved by SDS-PAGE and analyzed by autoradiography. Position of the indicated full-length GST fragment (red dot). (F) GST alone, GST-Rod1, or GST-Rod16A, as indicated, were expressed in either SNF1+ sst2∆ cells (left) or snf1∆ sst2∆ cells (right) and then analyzed by SDS-PAGE.
As one approach to determine whether Snf1-mediated phosphorylation of Rod1 itself, and not some other target, is responsible for the observed inhibition of the ability of overexpressed Rod1 to promote adaptation on galactose medium, we sought to map and mutagenize all of the Snf1 sites in Rod1 and then test the ability of such variants to promote adaptation on both glucose and galactose. Based on phosphorylation of known physiological substrates, as well as synthetic peptides, both yeast Snf1 and mammalian AMPK phosphorylate at Ser exclusively (i.e., not Thr) within the context of a well-defined phospho-acceptor site consensus, ΦxR/KxxSxxxΦ (where Φ is a hydrophobic residue) (Hardie et al. 1998). This consensus phospho-acceptor site has been amply confirmed for yeast Snf1 using more advanced synthetic peptide library arrays (Mok et al. 2010). Hence, it was relatively straightforward to scan the Snf1 sequence and locate a total of six potential Snf1 sites (Ser315, Ser447, Ser641, Ser706, Ser720, and Ser781) (Figure 1B; Supplemental Material, Figure S1, A and B). The most N-terminal site is located within the arrestin fold (predicted using Phyre2.0; Kelley and Sternberg 2009), whereas the remaining five are found within or flanking the PPxY motifs in the C-terminal half of Rod1 (Figure 1B; Figure S1, A and B). Genome-wide proteomic analyses (Gnad et al. 2009; Soufi et al. 2009; Swaney et al. 2013) indicate that at least four of these sites (S447, S641, S706, and S720) are phosphorylated in vivo. Moreover, three (S447, S641, and S706) of these four sites are the most conserved in other sensu stricto Saccharomyces species (Figure S2A). Furthermore, one of these same sites (S447) was shown to be phosphorylated by Snf1 in vitro (Shinoda and Kikuchi 2007). In the same study, rod1 (“Resistance to o-Dinitrobenzene”) loss-of-function mutations caused yeast cells to exhibit increased sensitivity to the toxic effects of 1,2-dinitrobenzene, and a Rod1(S447A) mutant conferred a modest increase in resistance to this compound (Shinoda and Kikuchi 2007). These results are consistent with a function for Rod1 in down-regulating the (unidentified) transporter(s) that mediates entry of 1,2-dinitrobenzene and a role for Snf1-mediated phosphorylation in inhibiting Rod1 function.
Hence, we used site-directed mutagenesis to convert each of these six sites alone, and in various combinations, to either a nonphosphorylatable (Ala) residue or to a phospho-mimetic (Glu) residue. We found that, when overexpressed in our MATa sst2∆ tester cells, Rod1(S315A S447A S641A S706A S720A S781A), henceforth abbreviated Rod16A, was much more potent than wild-type Rod1 in promoting adaptation on glucose medium, as judged by the degree of turbidity of the halo fill-in and, very importantly, was able to support readily detectable halo fill-in even on galactose medium, unlike wild-type Rod1 (Figure 1C). In marked contrast, the Rod1(S315E S447E S641E S706E S720E S781E), henceforth abbreviated Rod16E, was unable to stimulate scarcely any adaptation on either carbon source (Figure 1C). These results are fully consistent with the conclusion that in vivo Snf1-mediated phosphorylation is responsible for inhibiting the ability of Rod1 to promote Ste2 down-regulation on galactose medium.
The observed differences in the adaptation-promoting phenotypes among wild-type Rod1, Rod16A, and Rod16E could not be attributed trivially to any dramatic differences in the expression levels of these proteins, as judged by immunoblotting of extracts of these same cells (Figure 1D). Moreover, and as expected, using purified Snf1 and bacterially expressed GST-Rod1, we found that the 6A mutations virtually abolished phosphorylation of this α-arrestin at its Snf1 sites in vitro (Figure 1E). Furthermore, in vivo, compared to glucose-grown SNF1+ cells, where the mobility of wild-type Rod1 is distinctly slower than that of Rod16A, in glucose-grown cells lacking Snf1, the mobility of wild-type Rod1 is increased and is very similar to that of Rod16A (Figure 1F). Thus, Snf1 is active at a physiologically relevant level even on glucose medium.
Under our standard conditions (glucose medium), three single-site mutants, Rod1(S447A), Rod1(S706A), and Rod1(S720A), displayed a slightly enhanced ability to promote adaptation, as compared to wild-type Rod1, whereas three others, Rod1(S315A), Rod1(S641A), and Rod1(S781A), did not (Figure S2B). Indeed, S447 seems to be largely responsible for the phosphorylation-dependent mobility shift of Rod1 (Figure S2C), in agreement with the findings of Shinoda and Kikuchi (2007). Combining together as few as two of the mutations that had a detectable effect led to at least an additive improvement in its adaptation-promoting ability; for example, Rod1(S447A S706A) was somewhat more effective in promoting adaptation than Rod1(S447A S641A) (Figure S2B). Most strikingly, however, as the number of sites mutated was increased from three, to four, to five, to all six, the adaptation-promoting potency of the corresponding mutant Rod1 was incrementally increased (Figure S2B). Although the differences between the 3A, 4A, 5A, and 6A mutants are not dramatic, we continued our analysis using the most extreme mutant (Rod16A) to eliminate the contribution from all putative sites. Again, these differences could not be attributed to differences in the level of expression of these proteins (Figure S2C). Together, these data demonstrate that phosphorylation at all six Snf1 sites occurs in vivo (albeit perhaps with different efficiencies at different sites) and, when phosphorylated at these sites, the ability of Rod1 to down-regulate Ste2 is markedly impeded.
The findings discussed above indicate that Snf1 is active at a physiologically relevant level even on glucose medium (although we cannot rule out that, in our halo bioassay, the glucose concentration may become depleted to a sufficiently low level to permit Snf1 activation during the rather protracted time required for growth of the lawn). In this regard, however, we noted that, even when grown in liquid culture on glucose medium, and especially on galactose medium, wild-type Rod1 runs as a very diffuse band, indicative of the presence of multiple phospho-isoforms (or other modifications) (Figure 1D). Treatment with phosphatase (CIP) collapsed these species to a single sharp band that comigrated with Rod16A (and the mobility of Rod16A was not significantly affected by CIP treatment) (Figure 1D). These data again indicate that wild-type Rod1 is phosphorylated at its Snf1 sites under normal growth conditions, even on glucose medium.
We also noted that, unlike the Rod16A mutant, the Rod16E mutant displayed a mobility shift that is collapsed by CIP treatment (Figure 1D). However, it is known that, in some yeast substrates (Lee et al. 2012), Snf1 phosphorylation installs a negative charge that can prime a nearby Ser for subsequent phosphorylation by casein kinase I (in S. cerevisiae, Yck1, Yck2, Yck3, and/or Hrr25), a protein kinase family that has a preference for phosphorylating at Ser where an Asp, Glu, or phosphorylated residue is located at position −3 (Vielhaber and Virshup 2001; Mok et al. 2010). We presume, therefore, that one or more of the six Glu residues present in Rod16E may create such a site(s). Moreover, at least one other yeast α-arrestin (Rim8) reportedly is a direct substrate for Yck1 and Yck2 (Herrador et al. 2015).
Snf1 is not solely responsible for negative regulatory phosphorylation of Rod1
Two observations indicated that, in cells growing on glucose, Snf1 is likely not the sole protein kinase responsible for negative regulatory phosphorylation of Rod1. First, if Snf1 was the major protein kinase controlling Rod1 activity on glucose, then, in a snf1∆ mutant, wild-type Rod1 would remain unphosphorylated and, when overexpressed, should be just as potent at promoting adaptation on glucose medium as Rod16A. However, that was clearly not the case (Figure S3).
Snf1 is the founding member of a subfamily of protein kinases, present in both yeast and mammalian cells (Alessi et al. 2006; Rubenstein and Schmidt 2007), that includes closely related enzymes called AMPK-like protein kinases (AMPKLs). In S. cerevisiae, the AMPKLs are the paralogous sets Kin1 and Kin2, Frk1 and Kin4, and Hsl1, Gin4, and Kcc4. We reasoned that, if any one AMPKL was primarily responsible for phosphorylation of Rod1 at its Snf1 sites when cells are grown on glucose medium that, in a loss-of-function mutant of that kinase, overexpressed wild-type Rod1 would be as potent at stimulating desensitization as Rod16A. However, in every case, Rod16A was significantly more efficacious at promoting adaptation than wild-type Rod1 in kin1∆, kin2∆, frk1∆, kin4∆, hsl1∆, gin4∆, and kcc4∆ cells (Figure S3). Of course, one or more of the AMPKLs may act redundantly with each other, or with Snf1, with regard to Rod1 phosphorylation on glucose medium.
Three upstream kinases (Elm1, Tos3, and Sak1) all contribute to activation loop phosphorylation of Snf1 (Sutherland et al. 2003; Elbing et al. 2006) and the AMPKLs (Asano et al. 2006; Szkotnicki et al. 2008). Hence, as an alternative to constructing strains carrying a snf1∆ mutation and all possible combinations of AMPKL loss-of-function mutations, we examined an elm1∆ tos3∆ sak1∆ triple mutant. Again, we found that Rod16A is more efficacious at promoting adaptation than wild-type Rod1 in the elm1∆ tos3∆ sak1∆ sst2∆ strain (Figure S3), although the elm1∆ tos3∆ sak1∆ sst2∆ mutant cells are rather slow-growing, making the distinctions a bit harder to discern unambiguously. Nonetheless, these findings suggested that yet another class of protein kinase might be involved in controlling Rod1 function in cells growing on glucose.
Indeed, a second observation supported the conclusion that an additional protein kinase must negatively regulate Rod1 function on glucose medium. Specifically, despite the fact that Rod16A already lacks phosphorylation at all of its Snf1 sites, its potency in promoting adaptation is lost almost completely in calcineurin (CN)-deficient cells (see later section Calcineurin dephosphorylates the Ypk1 sites in Rod1), indicating that phosphorylation(s) at another position(s) also needs to be removed to allow Rod1 to function. In this regard, we noted that Rod1 (and several other α-arrestins) were recovered in a global screen that we conducted for potential substrates of the target-of-rapamycin (TOR) complex-2 (TORC2)-activated protein kinase Ypk1 (Muir et al. 2014).
Ypk1 phosphorylates Rod1 and inhibits its function in mating pathway down-regulation
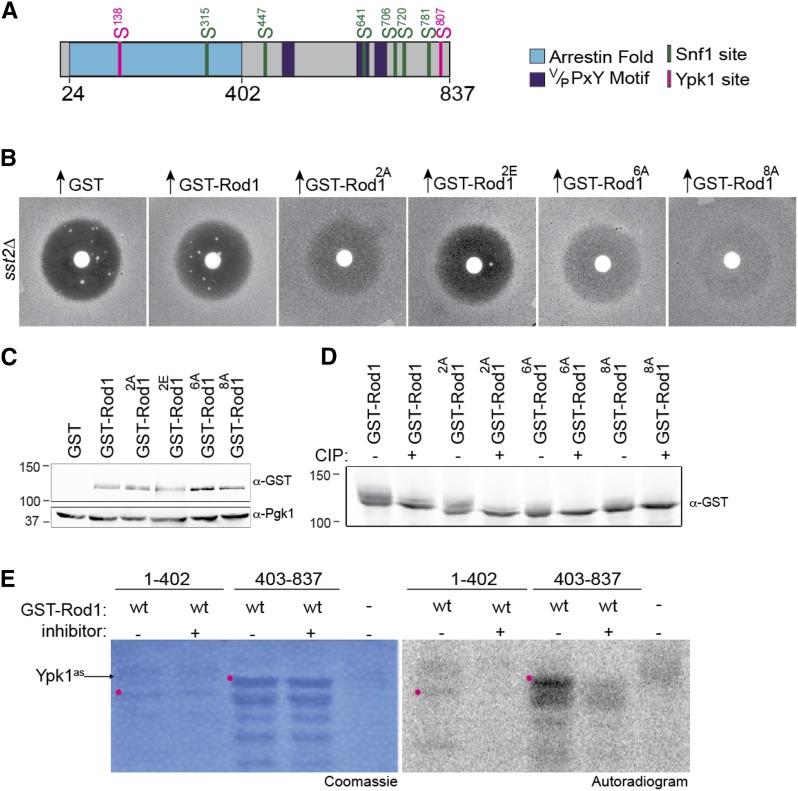
It has been well established that the TORC2-Ypk1 signaling axis regulates the sphingolipid content and other aspects of the lipid composition of the PM (Olson et al. 2016). Hence, it was an intriguing possibility that, through effects on the function of α-arrestins, that TORC2-Ypk1 signaling may also regulate the protein composition of the PM. Like Snf1, Ypk1 has a well-defined phospho-acceptor site motif, RxRxxS(Φ) (Casamayor et al. 1999; Mok et al. 2010; Muir et al. 2014), and Rod1 contains two matches to this consensus: Ser138 within the arrestin fold and Ser807 near its C terminus (Figure 2A; Figure S1, A and B). Genome-wide proteomic analyses (Gnad et al. 2009; Swaney et al. 2013) indicate that both sites are phosphorylated in vivo and both sites are conserved in other sensu stricto Saccharomyces species (Figure S4).
Figure 2.
Ypk1 phosphorylates Rod1 in vivo and in vitro. (A) Schematic diagram of Rod1. Arrestin fold (blue); Rsp5-binding motifs (purple); six Snf1 consensus motifs (green); two Ypk1 consensus motifs (pink). (B) The adaptation-promoting capacity of the indicated alleles of Rod1 was assessed as in Figure 1A. 2A, Rod1(S138A S807A); 2E, Rod1(S138E S807E); 6A, Rod1(S315A S447A S641A S706A S720A S781A); 8A, Rod1(S138A S315A S447A S641A S706A S720A S781A S807A). (C) Expression of the Rod1 variants shown in B was assessed by harvesting the indicated cultures just prior to plating, preparing whole-cell extracts, resolving samples of those lysates by SDS-PAGE (7.5% gel), and analyzing immunoblots of the resulting gels with anti-GST or anti-Pgk1 (loading control) antibodies. (D) Phosphorylation status of the Rod1 variants shown in B was assessed as described in Figure 1D, except that the SDS-PAGE separation was performed on a 5% gel to exaggerate band shifts. (E) In vitro phosphorylation assay, conducted as in Figure 1E, except that purified Ypk1-as was the protein kinase added, not Snf1, in the absence (−) and presence (+) of the Ypk1-as-specific inhibitor 3-MB-PP1. Position of the indicated full-length GST fragment (red dot).
As we did with the predicted Snf1 sites, we used site-directed mutagenesis to generate Rod1(S138A S807A), hereafter Rod12A, and Rod1(S138E S807E), hereafter Rod12E, and tested their ability to promote recovery from pheromone-induced growth arrest, compared to wild-type Rod1 and the Snf1-site mutant Rod16A, using the halo bioassay. Strikingly, Rod12A was significantly more potent than wild-type Rod1 and just as potent, if not more so, than Rod16A, in stimulating adaptation on glucose medium (Figure 2B). Conversely, Rod12E promoted scarcely any desensitization, nearly comparable to the large clear halo observed for the control (GST alone) cells (Figure 2B). The dramatic difference in the phenotypes between Rod12A and Rod12E could not be attributed to any difference in their level of expression (Figure 2C). Therefore, phosphorylation of Rod1 at its Ypk1 sites clearly has a role in negatively regulating the function of this α-arrestin in post-pheromone response adaptation.
Unlike removal of the six Snf1 phosphorylation sites, which largely eliminated the smear of phospho-isoforms exhibited by wild-type Rod1 when examined by SDS-PAGE (Figure 1D and Figure 2D), removal of both Ypk1 phosphorylation sites did not change the migration pattern markedly, and treatment with CIP collapsed the species present to a single more prominent band. Thus, these data suggest that phosphorylation occurs independently at both the Ypk1 and Snf1 sites in vivo.
In the global screen that identified Rod1 as a candidate Ypk1 substrate, a fragment of Rod1 containing the C-terminal Ypk1 site purified from bacteria was phosphorylated in a Ypk1-dependent manner in an in vitro protein kinase assay that utilized purified Ypk1(L424A) (Ypk1-as), a derivative that is sensitive to inhibition by the adenine analog 1-MB-PP1 (Muir et al. 2014). Using the same approach, we reproduced this result (Figure 2E). We also found that a fragment of Rod1 containing its N-terminal Ypk1 site was phosphorylated much less efficiently and only very weakly above the inhibiter-containing sample (Figure 2E). However, the in vitro assay may be misleading if the N-terminal fragment is a poor substrate simply because it lacks a high-affinity docking site for Ypk1. Hence, in intact Rod1, both its N-terminal and C-terminal Ypk1 sites may be phosphorylated in a Ypk1-dependent manner in vivo.
If both Snf1- (and/or AMPKL-) and Ypk1-dependent phosphorylation contributes to negative regulation of the desensitization-promoting function of Rod1, the combination of the Rod16A and Rod12A alleles should generate a molecule the potency of which in stimulating adaptation is further enhanced. Indeed, overexpression of the resulting octuple mutant, hereafter Rod18A, exhibited an ability to stimulate recovery after pheromone-induced growth arrest that was reproducibly more robust than either Rod12A or Rod16A (Figure 2B and Figure 3A). These data corroborate genetically that phosphorylation by both Ypk1 and Snf1 (and/or a AMPKL) inhibits Rod1 function at different sets of Ser residues. Furthermore, various global phospho-proteomics analyses (Gnad et al. 2009; Soufi et al. 2009; Swaney et al. 2013) indicate that other sites in Rod1 are phosphorylated in vivo. Consistent with this, even the Rod18A derivative displays a small, but detectable, trail of slower mobility isoforms that are removed upon CIP treatment (Figure 2D); nonetheless, in the Rod18A mutant, the majority of the phosphorylations responsible for the mobility shifts displayed by wild-type Rod1 have been largely eliminated.
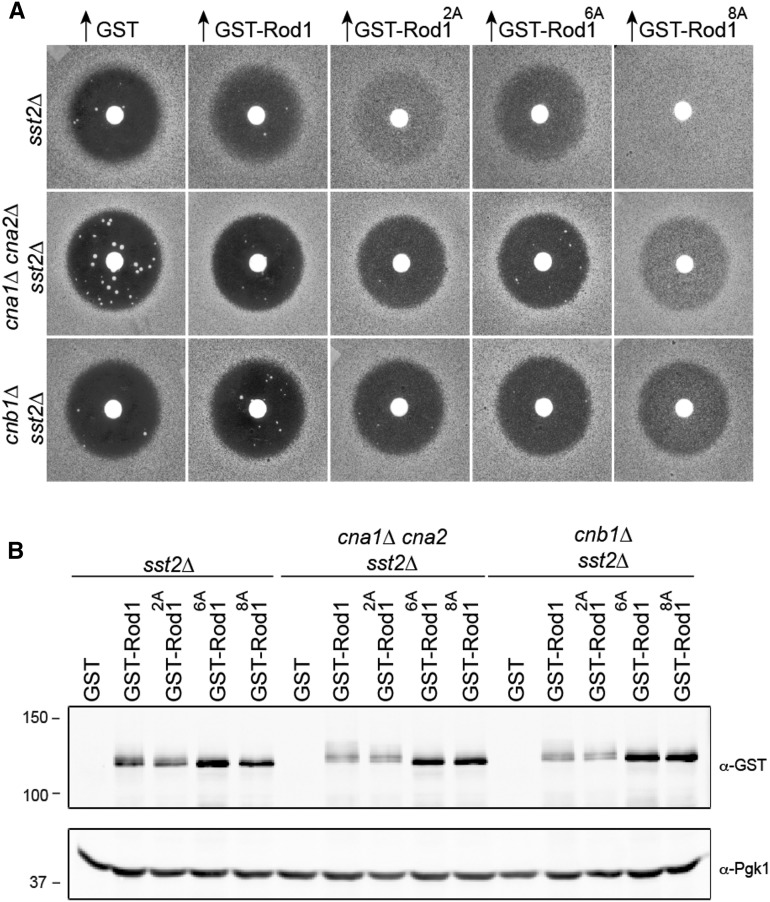
Figure 3.
The requirement for calcineurin-dependent dephosphorylation of Rod1 to promote adaptation is bypassed by nonphosphorylatable Rod1 alleles. (A) The adaptation-promoting capacity of the indicated alleles of Rod1 was assessed, as in Figure 1A, in otherwise isogenic sst2∆ tester cells that were wild type or lacked the paralogous catalytic subunits (cna1∆ cna2∆) or the small regulatory subunit (cnb1∆) of phosphoprotein phosphatase 2B/calcineurin. (B) Expression of the Rod1 variants shown in A was confirmed as in Figure 2C.
Calcineurin dephosphorylates the Ypk1 sites in Rod1
We demonstrated before (Alvaro et al. 2014) that CN-mediated dephosphorylation of Rod1 is required for its function in desensitization of mating pheromone response. Specifically, overexpression of Rod1 in wild-type cells promotes adaptation, whereas Rod1 overexpression in cells lacking either the paralogous CN catalytic subunits (cna1∆ cna2∆) or their shared Ca2+-binding regulatory subunit (cnb1∆) fails to display any detectable recovery (Figure 3A) and, based on electrophoretic mobility smearing, Rod1 clearly remains more heavily phosphorylated in cells lacking CN than in wild-type cells (Figure 3B), as we showed before (Alvaro et al. 2014). Remarkably, the Rod12A mutant was able to promote faint, but detectable, halo fill-in in cells lacking CN, whereas Rod16A was barely effective at promoting adaptation in CN-deficient cells (Figure 3A), even though Rod12A remained more heavily phosphorylated overall than Rod16A in cells lacking CN (Figure 3B). More striking still, the Rod18A mutant was substantially more potent at promoting adaptation in CN-deficient cells than either Rod12A or Rod16A (Figure 3A). These findings suggest that CN is responsible for dephosphorylation of both the Ypk1 and Snf1 sites in Rod1, but that CN action at the former is somewhat more important to alleviate Rod1 inhibition than dephosphorylation at the latter.
As assessed by electrophoretic mobility, the sites removed from Rod18A bypass the need for CN-mediated dephosphorylation (Figure 3B). However, as efficacious as Rod18A is in promoting recovery in CN-deficient cells, Rod18A overexpression is even more potent in promoting adaptation in wild-type cells, where other cellular phosphatases can act in conjunction with CN (Figure 3A). This finding indicates that, even though the Ypk1 and Snf1 sites are clearly major points of control, Rod18A is subject to additional (albeit more minor) negative regulatory phosphorylation, consistent with the fact that, in wild-type cells, Rod18A displays a small but detectable trail of slower mobility isoforms that are removed upon CIP treatment (Figure 2D).
In any event, we have clearly pinpointed at least eight sites that are controlled by specific dephosphorylation by CN. In this regard, it has been demonstrated that all bona fide CN substrates possess a conserved motif (PxIxIT and variants thereof), usually accompanied by another conserved motif (ΦLxVP and variants thereof) that can be situated up to 200 or more residues away, which serve, respectively, as primary and secondary docking sites for the binding of CN to its target protein (Grigoriu et al. 2013). In this regard, Rod1 possesses readily discernible matches to both sequences: 545-PQIKIE-550 and 688-LLPLP-692. We demonstrated before that a corresponding Rod1AQAKAA mutant in the apparent PxIxIT site is no longer able to bind CN and displays a defect in promoting adaptation (Alvaro et al. 2014).
Unphosphorylated Rod1 can act in an Rsp5-independent manner
The HECT domain E3 Rsp5 and its orthologs bind via their multiple WW folds to PPxY motifs (or variants thereof) in α-arrestins (Qi et al. 2014a). Rsp5 possesses three WW domains (Watanabe et al. 2015) and Rod1 possesses two PPxY sites and one variant in its C-terminal half (residues indicated): PPNY (487–490), VPSY (639–642), and PPAY (656–659) (Figure 1B). We previously showed, in otherwise wild-type MATa cells growing in glucose medium, that mutants lacking either the first, the third, or both sites (Rod1PANA, Rod1PAAA, and Rod1PPxY-less) were, unlike wild-type Rod1, incapable of promoting adaptation (Alvaro et al. 2014). Moreover, compared to wild-type Rod1, GST-Rod1PPxY-less exhibited markedly reduced binding to Rsp5 in vivo, as judged by pull-down assays from cell extracts, and displayed drastically reduced in vitro modification by purified Rsp5 in ubiquitinylation assays (Alvaro et al. 2014). Therefore, we concluded that, to mediate desensitization to pheromone, Rod1 must associate with Rsp5 and deliver this E3 to its target, which other evidence indicated was the α-factor receptor Ste2.
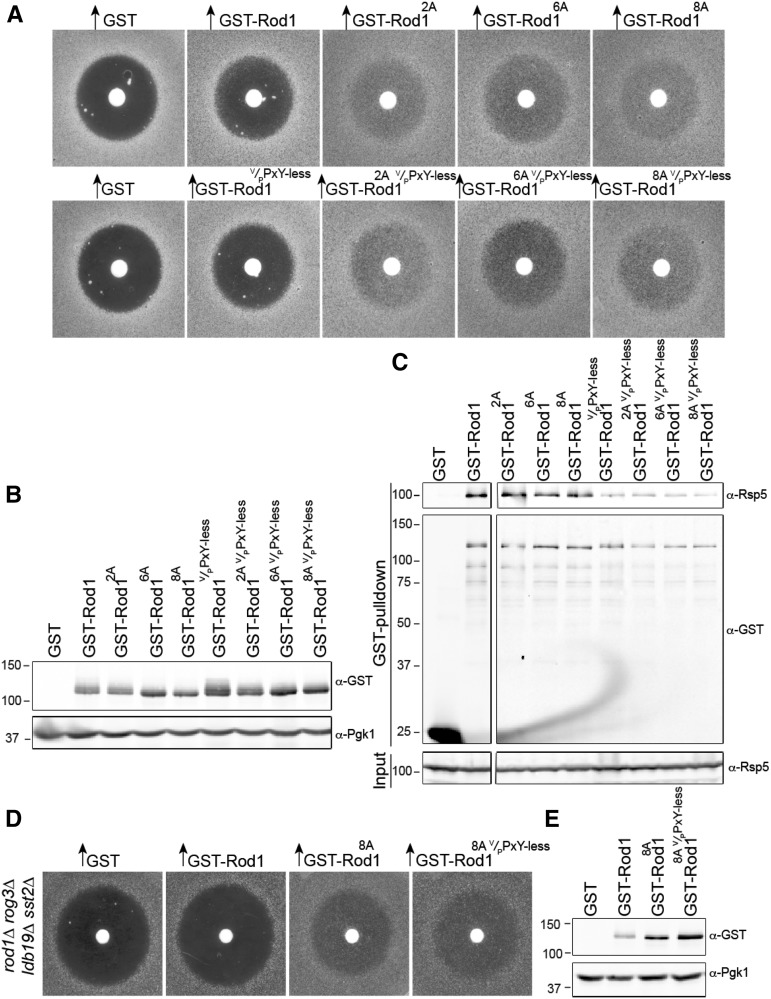
As we demonstrated here, Rod12A, Rod16A, and Rod18A are considerably more potent in promoting recovery from pheromone-induced G1 arrest than wild-type Rod1. One possible explanation for this enhancement of function is that the lack of phosphorylation allows for higher-affinity binding of Rsp5. As one means to address this issue, we tested whether the function of Rod12A, Rod16A, or Rod18A requires intact V/PPxY motifs. Quite unexpectedly, we found that derivatives of Rod12A, Rod16A, and Rod18A in which all three motifs were mutated (PPNY→PANA, VPSY→VASA, and PPAY→PAAA), hereafter Rod1V/PPxY-less, retained their ability to promote adaptation more robustly than wild-type Rod1 (Figure 4A). These properties were not due to any differences in the level of expression of these proteins (Figure 4B). Remarkably, however, the adaptation-promoting ability of Rod12A, Rod16A, or Rod18A clearly does not require intact V/PPxY motifs in these proteins, and, thus, the ability to interact with Rsp5 is not necessary for their potent desensitization of pheromone response (Figure 4A). This finding suggests that, when unphosphorylated, Rod1 acts more like its paralog Rog3, in that it becomes able to promote adaptation in an Rsp5-independent manner, as we demonstrated for Rog3 previously (Alvaro et al. 2014). Indeed, we confirmed that the V/PPxY-less versions of Rod12A, Rod16A, and Rod18A all lost high-affinity binding to Rsp5 (Figure 4C).
Figure 4.
Hypophosphorylated Rod1 does not require Rsp5 binding to squelch mating pheromone-evoked growth arrest. (A) The adaptation-promoting capacity of the indicated 2A, 6A, and 8A alleles of Rod1 was assessed, as in Figure 1A, with or without mutation of all three Rsp5-binding motifs (V/PPxY-less). (B) Expression of the Rod1 variants shown in A was confirmed as in Figure 2C. (C) Cultures of a GEV-carrying derivative of the protease-deficient strain BJ5459 expressing the indicated Rod1 mutant were grown to midexponential phase. Protein expression was induced with β-estradiol and, after 3 hr, the cells were harvested by centrifugation and ruptured by vigorous vortex mixed with glass beads, and the GST-fusion proteins in the resulting extracts were captured by binding to glutathione-agarose beads. After washing, bead-bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. (D) The adaptation-promoting capacity of the indicated Rod1 alleles was assessed, as in Figure 1A, in mutant cells lacking endogenous Rod1, Rog3 and Ldb19. (E) Expression of the Rod1 variants shown in A was confirmed as in Figure 2C.
We used the Rod1V/PPxY-less, instead of the Rod1PPxY-less (Alvaro et al. 2014) because we found that when the Rod18A allele was combined with the PANA PAAA double mutation (i.e., Rod1PPxY-less) it retained its recovery-promoting ability (data not shown). One possibility to explain this result was that the remaining VPSY site might be sufficient to recruit Rsp5, a similar concern we had for its paralog Rog3 (Alvaro et al. 2014). Indeed, using GST pull-downs, it was clear that the VPSY site contributes to Rsp5 binding to Rod1 in vivo (Figure S5A). To eliminate the contribution of the VPSY site, therefore, we additionally mutated it, creating Rod18A V/PPxY-less, and found that it retained its ability to robustly promote adaptation (Figure 4A). Thus, a nonphosphorylatable version of Rod1 bypasses the need for Rsp5 binding.
Interestingly, when we compared wild-type Rod1, Rod1PPxY-less, and Rod1V/PPxY-less (with none of the eight serines mutated), we found that Rod1V/PPxY-less causes a degree of adaptation similar to that of wild-type Rod1, unlike Rod1PPxY-less (Figure S5B). However, we attribute this difference to the fact that Rod1V/PPxY-less was expressed at a higher level than either wild-type Rod1 or Rod1PPxY-less (Figure S5C).
Another possibility to explain the fact that the V/PPxY-less versions of Rod12A, Rod16A, and Rod18A retain their potency in promoting adaptation is that these α-arrestin mutants are still able to recruit Rsp5 by forming homo-oligomers with endogenous Rod1, or hetero-oligomers with its paralog Rog3/Art7 or with the more distantly related α-arrestin Ldb19/Art1, both of which we previously showed contribute to Ste2 down-regulation (Alvaro et al. 2014). If so, then the partner α-arrestin could still bind Rsp5 and thereby deliver this E3 in trans to its target. However, even in triple-mutant cells (rod1∆ rog3∆ ldb19∆) lacking all three of these other potential partners, Rod18A and Rod18A V/PPxY-less were equally efficacious in promoting recovery from pheromone-induced growth arrest (Figure 4D) and were expressed at an equivalent level (Figure 4E). Thus, the Rod18A V/PPxY-less mutant is able to act alone to promote adaptation without recruitment of Rsp5. Thus, Rod1 has both Rsp5-dependent and Rsp5-independent mechanisms for down-regulation of mating pathway signaling, and these different adaptation-promoting functions are clearly modulated by the state of phosphorylation of this α-arrestin.
Rod1 and Rog3 action do not require the C-terminal tail of Ste2
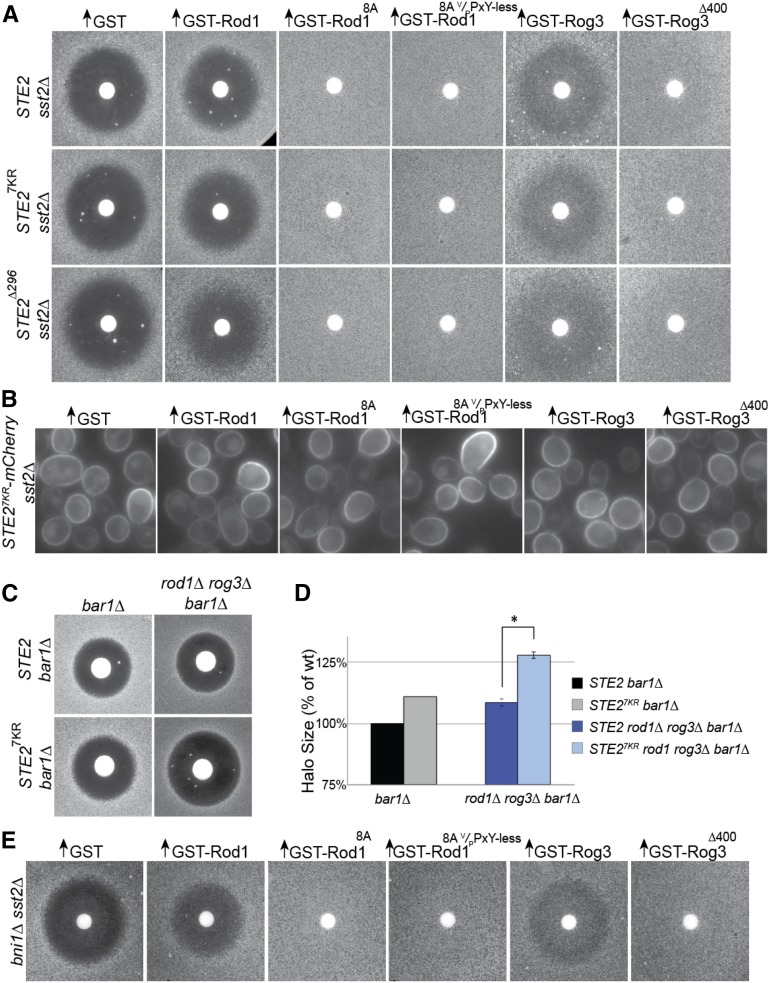
We demonstrated before that, in cells lacking Rod1, Rog3, and Ldb19, internalization of Ste2 from the PM is greatly impeded and that, normally, the actions of these α-arrestins contribute to Rsp5-mediated ubiquitinylation-dependent endocytosis of this GPCR (Alvaro et al. 2014). Indeed, prior work had demonstrated that seven Lys residues in the C-terminal cytosolic tail of Ste2 are sites of ubiquitinylation (Hicke et al. 1998; Terrell et al. 1998; Toshima et al. 2009) and are required for its clathrin-mediated endocytosis (Ballon et al. 2006; Dores et al. 2010). Likewise, truncations of Ste2 that remove its entire 134-residue C-terminal cytosolic tail just after a stop-transfer sequence installed after its seventh transmembrane helix, such as Ste2(∆296-431), also prevent endocytosis of Ste2 (Reneke et al. 1988; Ballon et al. 2006). Furthermore, we obtained some evidence that interactions with the C-terminal cytosolic tail of Ste2 contribute to association of Ldb19, Rod1, and Rog3 with this receptor (Alvaro et al. 2014). However, the abilities of Rod12A V/PPxY-less, Rod16A V/PPxY-less, and Rod8A V/PPxY-less to promote adaptation quite potently (Figure 4A) suggested that, in the absence of phosphorylation, a desensitization mechanism distinct from decoration of the tail of the receptor with ubiquitin and its recognition by the endocytosis machinery was occurring.
As one means to address this issue, we asked whether the Rod18A V/PPxY-less mutant was still able to potently promote recovery from pheromone-induced G1 arrest in cells where either Ste2(7K-to-R) or Ste2(∆296-431) was the sole source of this receptor. We have shown previously that these receptor variants are poorly internalized and localize predominantly to the PM (Ballon et al. 2006). Indeed, we found that Rod18A V/PPxY-less was able to stimulate recovery as efficiently in cells expressing Ste2(7K-to-R) or Ste2(∆296-431) as in cells expressing wild-type Ste2 and to do so much more effectively than wild-type Rod1 (Figure 5A). Similar to what we observed before in cells expressing wild-type Ste2 (Alvaro et al. 2014), both Rog3 and a Rog3 truncation mutant (∆400) that removes all three of its V/PPxY motifs also effectively promoted recovery in cells expressing Ste2(7K-to-R) or Ste2(∆296-431) as the sole source of this receptor (Figure 5A). Although there were some differences in the level of expression of these proteins that may contribute to their observed phenotypes (Figure S6B), these differences are clearly not sufficient to explain their relative efficacy in promoting adaptation. Specifically, despite the level of Rod18A V/PPxY-less being much lower than that of Rog3∆400 (Figure S6B), they both promote robust adaptation to the point where the halo of initial growth has become obscured nearly completely.
Figure 5.
Hypophosphorylated Rod1 and Rog3 can stimulate adaptation independently of Ste2 ubiquitinylation. (A) The adaptation-promoting capacity of the indicated alleles of Rod1 and Rog3 was assessed, as in Figure 1A, in otherwise isogenic cells expressing either wild-type Ste2, Ste2(7K-toR) (Ste27KR), or Ste2(∆296-431) (Ste2∆296) as the sole source of this receptor. (B) Expression of the indicated GST-α-arrestins from the GAL promoter on URA3-marked 2-μm DNA vectors was induced with 20 µM β-estradiol for 3 hr in a ura3 derivative of a strain expressing Ste2(7K-to-R)-mCherry from the chromosomal STE2 locus and then examined by fluorescence microscopy. (C) Pheromone sensitivity of MATa bar1∆ cells, either containing or lacking endogenous Rod1 and Rog3 and expressing mCherry-tagged versions of either wild-type Ste2 or Ste2(7K-to-R), as indicated, was assessed on SC-Ura medium with sterile filter disks containing 600 ng α-factor and photographed after incubation for 2 days at 30°. (D) Quantification and statistical analysis of the change in halo diameter for independent trials (n = 6) of the comparative halo assays shown in C. Average halo diameter for control cells was set at 100%, and the other halo sizes of each mutant were normalized to the control. Error bars, ±SEM; *P < 0.001. (E) The adaptation-promoting capacity of the indicated alleles of Rod1 and Rog3 was assessed, as in Figure 1A, in MATa sst2∆ tester cells lacking the formin Bni1.
Furthermore, overexpression of these four α-arrestin variants had no effect on the PM localization of Ste2(7K-to-R)-mCherry (Figure 5B), indicating that the adaptation-promoting potency of these α-arrestin variants was not due to greater efficacy in driving receptor internalization. Moreover, as judged by the halo bioassay, these α-arrestin variants promoted the same degree of adaptation when the sole source of the receptor was Ste2(7K-to-R)-mCherry (Figure S6A) as when it was either wild-type Ste2 or Ste2(7K-to-R) (Figure 5A), confirming that the mCherry tag had no interfering effect. Collectively, these data indicate that both nonphosphorylatable Rod1 and Rog3 are able to promote desensitization of the mating pheromone response pathway via a mechanism independent of Rsp5-dependent ubiquitin-mediated receptor internalization.
A prediction of the conclusion that both Rod1 and Rog3 act to promote adaptation via both Rsp5-dependent and Rsp5-independent mechanisms is that loss of Rod1 and Rog3 function in cells expressing Ste2(7K-to-R) as the sole source of this receptor should display an increase in sensitivity to α-factor-induced growth arrest, compared to either rod1∆ rog3∆ cells or Ste2(7K-to-R) cells. Indeed, as judged by the halo bioassay, we observed an additive effect of combining a rod1∆ rog3∆ double mutation with the Ste2(7K-to-R) mutation (Figure 5C) that was both reproducible and statistically significant (Figure 5D).
The fact that, in the absence of its phosphorylation, Rod1 can still promote adaptation independently of Rsp5-mediated receptor ubiquitinylation is consistent with recent evidence that α-arrestins can contribute to cargo recognition by both clathrin-dependent and clathrin-independent mechanisms (Prosser et al. 2015). However, in cells lacking a component (the formin Bni1) required for the clathrin-independent route (Prosser et al. 2011, 2015), derivatives of Rod1 that are largely unphosphorylated and unable to associate with Rsp5, as well as Rog3 and a derivative that is unable to associate with Rsp5, still promote efficient adaptation (Figure 5E), indicating a third means by which this α-arrestin is able to promote desensitization of the pheromone-response pathway.
Discussion
Because endocytosis of many integral PM proteins in yeast is regulated by one or more of its 14 identified α-arrestins (Lin et al. 2008; Nikko et al. 2008; Becuwe et al. 2012; O’Donnell et al. 2010, 2015), including the GPCRs Ste2 (Alvaro et al. 2014) and Ste3 (Prosser et al. 2015), a current question in the field is how, when, and where any given α-arrestin is recruited to a particular target. Recent studies demonstrate that phosphorylation of an α-arrestin either inhibits its ability to stimulate internalization of its target (Shinoda and Kikuchi 2007; Lin et al. 2008; Nikko et al. 2008; MacGurn et al. 2011; Becuwe et al. 2012; Merhi and Andre 2012; O’Donnell et al. 2013) or causes the α-arrestin to function in a different way (Crapeau et al. 2014; O’Donnell et al. 2015).
As we demonstrate here, phosphorylation of Rod1 has a profound effect in blocking the ability of this α-arrestin to promote adaptation in the mating pheromone response pathway, where its apparent target is the α-factor receptor Ste2 (Alvaro et al. 2014). Phosphoproteomic analysis by others (Gnad et al. 2009; Soufi et al. 2009; Swaney et al. 2013) and the mutational approach described here show that under normal growth conditions Rod1 is inhibited by phosphorylation at its predicted Snf1 and Ypk1 sites because preventing phosphorylation at each of the six Snf1 sites and its two Ypk1 sites (by mutating the corresponding Ser residues to Ala) caused Rod1 to be more and more potent in promoting adaptation in an additive manner. Conversely, conversion of the same sites to Glu, mimicking its permanently phosphorylated state, ablated the ability of Rod1 to stimulate adaptation. In this same regard, using N- and C-terminal fragments of Rod1, we found that Snf1-mediated phosphorylation of Rod1 in vitro occurs primarily on its C-terminal sites, and not on the one site (S315) in its arrestin fold domain. This finding suggested that, when Rod1 phosphorylation occurs in vivo, modification of the Snf1 sites might block Rod1 function in the main by impeding its recruitment of Rsp5 (rather than by preventing its association with Ste2). However, in pull-down experiments, Rod12A, Rod16A, and Rod18A did not bind more Rsp5 than wild-type Rod1, indicating that phosphorylation of wild-type Rod1 does not impede its association with Rsp5 per se. Moreover, here again the in vitro assay may be misleading if the N-terminal fragment is an inefficient substrate simply because it lacks a high-affinity docking site for Snf1 and/or has one-fifth the number of sites as the C-terminal fragment.
Unexpectedly, and revealingly, we found that, when phosphorylation of Rod1 is prevented on its Ypk1 sites, its Snf1 sites, or both, the corresponding Rod1 derivatives were able to promote adaptation potently, even when Rod1 was unable to associate with the E3 Rsp5 due to mutation of all three of its V/PPxY motifs. These observations revealed that Rod1 is able to promote adaptation in an Rsp5-independent manner, similarly to what we have previously shown for its paralog Rog3 (Alvaro et al. 2014). Our findings thus suggest that the phosphorylation state of Rod1 dictates the mechanism by which it regulates the mating pathway.
Although phosphorylation of Rod1 by the AMPK Snf1 was shown previously to inhibit internalization of the lactate permease Jen1 (Becuwe et al. 2012) and stimulate internalization of the low-affinity glucose transporters Hxt1 and Hxt3 (O’Donnell et al. 2015), the specific phosphorylation sites in Rod1 that mediate these effects where not identified in those studies. Here, we identified six Snf1 consensus sites that are phosphorylated both in vivo and in vitro, all of which contribute to blocking the adaptation-promoting function of Rod1. When cells are grown in galactose, a condition that markedly activates Snf1 (Hardie et al. 1998; Hedbacker and Carlson 2008), Rod1 cannot promote adaptation; however, a Rod16A mutant that is immune to Snf1-mediated phosphorylation was able to promote adaptation on galactose medium. This finding indicates that Snf1 action inhibits the ability of Rod1 to down-regulate the mating pathway. This phosphorylation-based mechanism makes physiological sense because it helps ensure that haploid cells will have the highest level of receptor and, hence, the greatest responsiveness to pheromone, on carbon sources other than glucose, where the capacity to mate and form diploid cells (which can sporulate when carbon is limiting) will have the greatest survival value for this organism.
We also observed that Rod16A, in which all the sites for Snf1 were converted to Ala, promoted adaptation more robustly than wild-type Rod1 even when cells are grown in glucose, a condition where Snf1 activity is quite low. This result suggested that, on glucose (i) basal Snf1 activity is nonetheless sufficient to inhibit Rod1 and/or (ii) a related protein kinase of the AMPKL family is responsible for phosphorylation of these sites. Although Snf1 displays detectable basal activity under high-glucose conditions (McCartney et al. 2014; O’Donnell et al. 2015), Rod16A still exhibited much more potent adaptation than wild-type Rod1 in cells lacking Snf1. This result favors the latter possibility; however, deletion of no one AMPKL caused any dramatic enhancement in the adaptation-promoting ability of wild-type Rod1. Hence, it is possible that there is some degree of redundancy among the AMPKLs to phosphorylate Rod1 at its Snf1 sites. To address this possibility, we examined cells that lack the three upstream protein kinases (Elm1, Sak1, and Tos3) that are known activators of Snf1 and the other AMPKLs, which again did not cause any significant enhancement in the adaptation-promoting ability of wild-type Rod1. However, several of the AMPKLs are known to possess significant activity even in the absence of their T-loop phosphorylation (Asano et al. 2006; Szkotnicki et al. 2008; B. Gullbrand and J. Thorner, unpublished data); hence, it is still possible that certain AMPKLs redundantly phosphorylate Rod1 at its Snf1 sites when cells are grown in glucose.
In agreement with a global screen that identified Rod1 (as well as two other α-arrestins, Rog3 and Aly2) as potential substrates for protein kinase Ypk1 (Muir et al. 2014), we also pinpointed two sites in Rod1 that are indeed phosphorylated by Ypk1 both in vivo and in vitro and showed that phosphorylation at these sites is also strikingly inhibitory to the adaptation-promoting function of Rod1. Optimal activity of Ypk1 requires its phosphorylation by TORC2 (Roelants et al. 2010, 2011), and TORC2 and Ypk1 activity are upregulated under certain stressful conditions (e.g., elevated temperature) (Sun et al. 2012) where again enhancing the mating proficiency of haploid cells to form diploid cells with the capacity to form heat-resistant spores would offer survival value.
Although our evidence indicates that Ypk1 and Snf1 (and/or one or more AMPKLs) are protein kinases that make major contributions to the phospho-regulation of Rod1, we also found that even a Rod18A mutant lacking both its Ypk1 and Snf1 sites exhibited minor amounts of additional isoforms that were eliminated by CIP treatment, indicating that Rod1 function may also be controlled to at least some degree via phosphorylation by yet other protein kinases. Consistent with this possibility, in at least one global phosphoproteomic study (Swaney et al. 2013), phosphate was detected on Ser and/or Thr residues other than the Ypk1 and Snf1 sites that we mutated. For example, four such sites fit the SP/TP consensus that could make them potential CDK or MAPK targets. In this regard, it would be interesting to determine whether Rod1 function also is controlled in a cell cycle-dependent manner and/or subject to feedback phoshorylation by Fus3, the MAPK specifically activated by the mating pheromone response pathway (Hao et al. 2007; Merlini et al. 2013). If Rod1 is a target for Fus3, and phosphorylation by Fus3 is also inhibitory to Rod1-mediated stimulation of Ste2 internalization, such a circuit would provide a self-reinforcing mechanism for maintaining Ste2 at the PM and thereby more sustained pheromone signaling at least in the early phase of mating pathway activation. However, at the latter stage of pheromone response, there is a marked influx of Ca2+ (Ohsumi and Anraku 1985; Nakajima-Shimada et al. 2000; Martin et al. 2011) sufficient to stimulate activation of CN (Withee et al. 1997), which we showed previously is necessary to activate the adaptation-promoting function of Rod1 (Alvaro et al. 2014). As we documented here, CN activates Rod1 function by removing the phosphorylations at both the Ypk1 and Snf1 sites. An open question is whether this Ca2+ influx also activates any calcium-activated protein kinase that may also influence Rod1 function or other aspects of the mating process at this stage.
Perhaps the most striking aspect of our current findings is that, in the absence phosphorylation of Rod1, even at as few as its two Ypk1 sites, its adaptation-promoting ability is markedly enhanced and, most surprisingly, no longer requires Rod1 association with the E3 Rsp5. In our prior work, we found that Rod1PPxY-less, which lacks two of its Rsp5-binding sites, is unable to stimulate recovery from pheromone-induced growth arrest (Alvaro et al. 2014). Here we found that, although mutating the third Rsp5-binding motif (VPSY) further reduced Rsp5 binding, Rod1V/PPxY-less displayed a slight increase in its ability to promote adaptation, suggesting that, like the absence of phosphorylation, elimination of Rsp5 binding further promotes the Rsp5-independent mechanism by which Rod1 promotes desensitization.
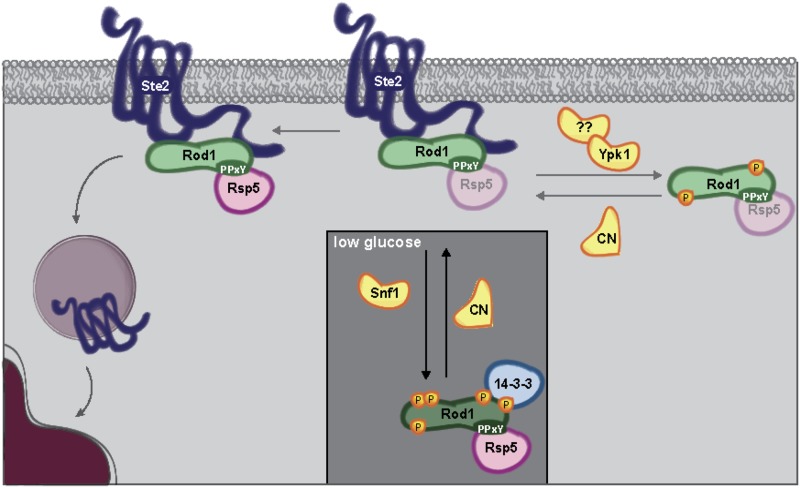
Collectively, our results support a model (Figure 6) in which Rod1 has at least two distinct mechanisms for blocking the function of Ste2 and thus preventing the mating pheromone response. First, it is incontrovertible that, in otherwise normal cells, a primary mechanism for down-regulation is that Rod1 delivers the ubiquitin ligase Rsp5 to the receptor, permitting its ubiquitinylation and engagement of the clathrin-dependent endocytosis machinery, followed by internalization and destruction of Ste2 in the vacuole (Alvaro et al. 2014). However, our mutational studies revealed that, when hypophosphorylated, Rod1 can potently dampen pheromone-initiated signaling in a manner that does not require its association with Rsp5. We propose the following explanation for this second adaptation-promoting mechanism.
Figure 6.
Phospho-regulation of Rod1 function in mating pathway desensitization. Under normal growth conditions, Rod1 is phosphorylated at multiple sites that do not prevent its interaction with Rsp5, but do prevent its productive association with Ste2. Conditions that activate the phosphoprotein phosphatase calcineurin, or that diminish the activities of the protein kinases Snf1 and Ypk1, or both, permit Rod1-receptor association, promoting the Rsp5-dependent ubiquitinylation and clathrin-mediated endocytosis of Ste2. When phosphorylation of Rod1 at its Snf1 and Ypk1 sites is blocked, the only way it can be removed from the receptor is via its own Rsp5- and ubiquitin-dependent and proteasome-mediated destruction. When Rod1 cannot be phosphorylated at its Snf1 and Ypk1 sites and its V/PPxY are mutated (preventing Rsp5 recruitment), Rod1 remains bound to Ste2, blocking the ability of the receptor to stimulate its cognate G-protein and thereby potently squelching mating pheromone-evoked growth arrest.
In the absence of the steric and electrostatic interference imposed by both phosphorylation and Rsp5 binding, we speculate that the N-terminal arrestin fold in Rod1 is freed structurally to adopt more facilely a conformation similar to that of the N-terminal arrestin fold found both in β-arrestin (Shukla et al. 2014) and in visual arrestin (Kang et al. 2015b) when bound to their target receptors. In these molecules, which lack a PPxY-containing C-terminal extension that is the hallmark of the α-arrestins, the N- and C-lobes of their arrestin folds undergo a dramatic rotation with respect to one another to engage their target receptors (rhodopsin and β2-adrenergic receptor, respectively) (Kang et al. 2015a). Thereby, visual arrestin and β-arrestin hold their cognate receptors in an intimate embrace, where most of the contacts do not include interactions with the C-terminal cytosolic tails of these receptors. Importantly, this binding prevents any further signaling because it is mutually exclusive with occupancy of these receptors by their cognate G-proteins (Attramadal et al. 1992; Lohse et al. 1992; Craft et al. 1994). Indeed, consistent with this same kind of β-arrestin-like role for unphosphorylated Rod1, we found that Rod18A V/PPxY-less could robustly promote adaptation even in cells that express a Ste2 mutant lacking its entire C-terminal tail as the sole source of this receptor.
Because it has been shown recently that, in yeast, some α-arrestins can promote a Rho1- and formin-requiring, but clathrin-independent, mechanism for internalization of certain integral PM proteins (Prosser et al. 2011, 2015), we considered the possibility that absence of phosphorylation and Rsp5 binding allows Rod18A V/PPxY-less to engage this clathrin-independent route for Ste2 internalization more efficiently. However, this does not appear to be the case because Rod18A V/PPxY-less-promoted adaptation was not at all reduced in cells lacking a component (the formin Bni1) required for the clathrin-independent internalization route.
What, then, is the normal role of α-arrestin phosphorylation? Given the fact that Rod1 action is involved in the endocytosis of quite a number of other integral PM proteins (at least Jen1, Hxt1, Hxt3, and Hxt6), and when unimpeded by phosphorylation or association with Rsp5, the arrestin fold of Rod1 appears to bind very tightly to Ste2, it is possible that a primary and physiologically relevant function for phosphorylation of Rod1 is to prevent this potential sequestration by promoting dissociation of Rod1 from Ste2 (and from its other targets). Viewed in this way, control by phosphorylation enhances the dynamic recycling of Rod1 as a means to maintain an adequate cytosolic pool so that at least some Rod1 is always available for action on each of its targets in response to the correct stimulus. In the case of Rod1 in pheromone response, Rod1 action provides a mechanism to ensure clearance of Ste2 from the surface of mating cells only in response to its CN-mediated dephosphorylation triggered by the influx of Ca2+ that occurs at a late stage in pheromone response.
Of course, more complicated models for how phosphorylation might control Rod1 function in the processes that promote desensitization to mating pheromone are possible. In this regard, it has been reported that phosphorylation of the α-arrestins Bul1 and Bul2 alters the way in which these adaptors bind to and regulate internalization of the general amino acid permease Gap1 (Crapeau et al. 2014). Thus, in the same way, it is possible that differential phosphorylation, or the lack thereof, allows Rod1 to interact with components in the mating pheromone response pathway other than Ste2 in ways that may also help to squelch signaling and promote pathway down-regulation.
GPCRs are initiators of vital signal transduction pathways in all eukaryotes, and their association with arrestins (both α- and β-arrestins in animal cells) is important to understand the control of both signal propagation and signal dampening at the molecular level. Several of the six currently recognized α-arrestins in mammalian cells have been implicated in GPCR internalization (Nabhan et al. 2010; Puca et al. 2013; Qi et al. 2014b). Our work sheds new light on the roles of phospho-regulation of α-arrestins in GPCR down-regulation. Thus, S. cerevisiae continues to serve as a useful model to explore α-arrestin function and related mechanistic aspects of GPCR biology.
Acknowledgments
We thank Benjamin Turk (Yale University) for the gift of purified Snf1; Alexander Muir (former member of this laboratory) for the gift of purified Ypk1as; current members of the Thorner Lab, especially Gregory C. Finnigan and Françoise M. Roelants, for useful advice about strain construction and other help; and Allyson F. O’Donnell (Duquesne University) for her encouragement, stimulating discussions, and other support. This work was supported by an National Institutes of Health (NIH) National Research Service Award Predoctoral Traineeship GM07232 (to C.G.A.) and by NIH R01 Research grant GM21841 (to J.T.).
Footnotes
Communicating editor: D. J. Lew
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.186122/-/DC1.
Literature Cited
- Alessi D. R., Sakamoto K., Bayascas J. R., 2006. LKB1-dependent signaling pathways. Annu. Rev. Biochem. 75: 137–163. [DOI] [PubMed] [Google Scholar]
- Alvarez, C. E., 2008 On the origins of arrestin and rhodopsin. BMC Evol. Biol. 8: 222.1–222.13. [DOI] [PMC free article] [PubMed]
- Alvaro C. G., O’Donnell A. F., Prosser D. C., Augustine A. A., Goldman A., et al. , 2014. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell. Biol. 34: 2660–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N., 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Asano S., Park J. E., Yu L. R., Zhou M., Sakchaisri K., et al. , 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 281: 27090–27098. [DOI] [PubMed] [Google Scholar]
- Attramadal H., Arriza J. L., Aoki C., Dawson T. M., Codina J., et al. , 1992. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J. Biol. Chem. 267: 17882–17890. [PubMed] [Google Scholar]
- Aubry L., Klein G., 2013. True arrestins and arrestin-fold proteins: a structure-based appraisal. Prog. Mol. Biol. Transl. Sci. 118: 21–56. [DOI] [PubMed] [Google Scholar]
- Aubry L., Guetta D., Klein G., 2009. The arrestin fold: variations on a theme. Curr. Genomics 10: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon D. R., Flanary P. L., Gladue D. P., Konopka J. B., Dohlman H. G., et al. , 2006. DEP-domain-mediated regulation of GPCR signaling responses. Cell 126: 1079–1093. [DOI] [PubMed] [Google Scholar]
- Becuwe M., Vieira N., Lara D., Gomes-Rezende J., Soares-Cunha C., et al. , 2012. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell Biol. 196: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard M. E., Randall C. L., Larochelle S., Zhang C., Shokat K. M., et al. , 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 104: 4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal M., Paiva S., Andrade R. P., Gancedo C., Leão C., 1999. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 181: 2620–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R., 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9: 186–197. [DOI] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A., 1982. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a-factor and α-factor pheromones. Mol. Cell. Biol. 2: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft C. M., Whitmore D. H., Wiechmann A. F., 1994. Cone arrestin identified by targeting expression of a functional family. J. Biol. Chem. 269: 4613–4619. [PubMed] [Google Scholar]
- Crapeau M., Merhi A., Andre B., 2014. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J. Biol. Chem. 289: 22103–22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J. W., 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70: 703–754. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Song J., Ma D., Courchesne W. E., Thorner J., 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit). Mol. Cell. Biol. 16: 5194–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores M. R., Schnell J. D., Maldonado-Baez L., Wendland B., Hicke L., 2010. The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic 11: 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R., Hicke L., 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12: 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., 2004. Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1695: 89–111. [DOI] [PubMed] [Google Scholar]
- Edelstein, A., N. Amodaj, K. Hoover, R. Vale, and N. Stuurman, 2010 Computer control of microscopes using micromanager. Curr. Protoc. Mol. Biol.: Chap. 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbing K., McCartney R. R., Schmidt M. C., 2006. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 393: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel D. G., 2003. The top genes: on the distance from transcript to function in yeast glycolysis. Curr. Opin. Microbiol. 6: 198–201. [DOI] [PubMed] [Google Scholar]
- Galan J. M., Haguenauer-Tsapis R., 1997. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16: 5847–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland S. L., 2013. Are GPCRs still a source of new targets? J. Biomol. Screen. 18: 947–966. [DOI] [PubMed] [Google Scholar]
- Ghaddar K., Merhi A., Saliba E., Krammer E. M., Prevost M., et al. , 2014. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol. Cell. Biol. 34: 4447–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F., de Godoy L. M. F., Cox J. R., Neuhauser N., Ren S., et al. , 2009. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9: 4642–4652. [DOI] [PubMed] [Google Scholar]
- Granier S., Kobilka B., 2012. A new era of GPCR structural and chemical biology. Nat. Chem. Biol. 8: 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Sambrook J., 2012a Molecular Cloning: A Laboratory Manual, Vol. 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Green M. R., Sambrook J., 2012b Molecular Cloning: A Laboratory Manual, Vol. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Grigoriu, S., R. Bond, P. Cossio, J. A. Chen, N. Ly et al., 2013 The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. PLoS Biol. 11: e1001492.1–e1001492.13. [DOI] [PMC free article] [PubMed]
- Hao N., Behar M., Elston T. C., Dohlman H. G., 2007. Systems biology analysis of G protein and MAP kinase signaling in yeast. Oncogene 26: 3254–3266. [DOI] [PubMed] [Google Scholar]
- Hardie D. G., Carling D., Carlson M., 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67: 821–855. [DOI] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M., 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A., Livas D., Soletto L., Becuwe M., Léon S., et al. , 2015. Casein kinase 1 controls the activation threshold of an α-arrestin by multisite phosphorylation of the interdomain hinge. Mol. Biol. Cell 26: 2128–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Zanolari B., Riezman H., 1998. Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J., 2003. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim. Biophys. Acta 1614(2): 139–155. [DOI] [PubMed] [Google Scholar]
- Irannejad R., Tsvetanova N. G., Lobingier B. T., von Zastrow M., 2015. Effects of endocytosis on receptor-mediated signaling. Curr. Opin. Cell Biol. 35: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee C., Choi T.-W., Kalichamy K., Yee J. Z., Song H.-O., et al. , 2012. CNP-1 (ARRD-17), a novel substrate of calcineurin, is critical for modulation of egg-laying and locomotion in response to food and lysine sensation in Caenorhabditis elegans. J. Mol. Biol. 417: 165–178. [DOI] [PubMed] [Google Scholar]
- Kang Y., Gao X., Zhou X. E., He Y., Melcher K., et al. , 2015a A structural snapshot of the rhodopsin-arrestin complex. FEBS J. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Zhou X. E., Gao X., He Y., Liu W., et al. , 2015b Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V., Cherezov V., Stevens R. C., 2013. Structure-function of the G protein–coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53: 531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J. E., 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kim H. C., Huibregtse J. M., 2009. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29: 3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., Andre B., 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B., 2010. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20: 196–204. [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Jeschke G. R., Roelants F. M., Thorner J., Turk B. E., 2012. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 32: 4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S., Haguenauer-Tsapis R., 2009. Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315: 1574–1583. [DOI] [PubMed] [Google Scholar]
- Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D., 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725. [DOI] [PubMed] [Google Scholar]
- Lohse M. J., Andexinger S., Pitcher J., Trukawinski S., Codina J., et al. , 1992. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J. Biol. Chem. 267: 8558–8564. [PubMed] [Google Scholar]
- MacGurn J. A., Hsu P.-C., Smolka M. B., Emr S. D., 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147: 1104–1117. [DOI] [PubMed] [Google Scholar]
- Marchese A., Trejo J., 2013. Ubiquitin-dependent regulation of G protein-coupled receptor trafficking and signaling. Cell Signal. 25: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. C., Kim H., Mackin N. A., Maldonado-Baez L., Evangelista C. C., et al. , 2011. New regulators of a high affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J. Biol. Chem. 286: 10744–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney R. R., Chandrashekarappa D. G., Zhang B. B., Schmidt M. C., 2014. Genetic analysis of resistance and sensitivity to 2-deoxyglucose in Saccharomyces cerevisiae. Genetics 198: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R. S., Silverman S. J., McClean M. N., Gibney P. A., Macinskas J., et al. , 2011. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22: 4447–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhi A., Andre B., 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14–3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32: 4510–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini, L., O. Dudin, and S. G. Martin, 2013 Mate and fuse: how yeast cells do it. Open Biol. 3: 130008.1–130008.13. [DOI] [PMC free article] [PubMed]
- Mok, J., P. M. Kim, H. Y. Lam, S. Piccirillo, X. Zhou et al., 2010 Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3: ra12.1–ra12.13. [DOI] [PMC free article] [PubMed]
- Muir, A., S. Ramachandran, F. M. Roelants, G. Timmons, and J. Thorner, 2014 TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife 3: e03779.1–e03779.34. [DOI] [PMC free article] [PubMed]
- Myers M. D., Payne G. S., 2013. Clathrin, adaptors and disease: insights from the yeast Saccharomyces cerevisiae. Front. Biosci. 18: 862–891. [DOI] [PubMed] [Google Scholar]
- Nabhan J. F., Pan H., Lu Q., 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 11: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima-Shimada J., Sakaguchi S., Tsuji F. I., Anraku Y., Iida H., 2000. Ca2+ signal is generated only once in the mating pheromone response pathway in Saccharomyces cerevisiae. Cell Struct. Funct. 25: 125–131. [DOI] [PubMed] [Google Scholar]
- Nikko E., Pelham H. R. B., 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10: 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Sullivan J. A., Pelham H. R. B., 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Apffel A., Gardner R. G., Cyert M. S., 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21: 3552–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Huang L., Thorner J., Cyert M. S., 2013. A calcineurin-dependent switch controls the trafficking function of alpha-arrestin Aly1/Art6. J. Biol. Chem. 288(33): 24063–24080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., McCartney R. R., Chandrashekarappa D. G., Zhang B. B., Thorner J., et al. , 2015. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and α-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 35: 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hayre M., Degese M. S., Gutkind J. S., 2014. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr. Opin. Cell Biol. 27: 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y., Anraku Y., 1985. Specific induction of Ca2+ transport activity in MATa cells of Saccharomyces cerevisiae by a mating pheromone, alpha-factor. J. Biol. Chem. 260: 10482–10486. [PubMed] [Google Scholar]
- Olson D. K., Fröhlich F., Farese R. V., Walther T., 2016. Taming the sphinx: mechanisms of cellular sphingolipid homeostasis. Biochim. Biophys. Acta (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser D. C., Drivas T. G., Maldonado-Baez L., Wendland B., 2011. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J. Cell Biol. 195: 657–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser D. C., Pannunzio A. E., Brodsky J. L., Thorner J., Wendland B., et al. , 2015. Alpha-arrestins participate in cargo selection for both clathrin-independent and clathrin-mediated endocytosis. J. Cell Sci. 128: 4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca L., Chastagner P., Meas-Yedid V., Israel A., Brou C., 2013. Alpha-arrestin 1 (ARRDC1) and beta-arrestins cooperate to mediate Notch degradation in mammals. J. Cell Sci. 126: 4457–4468. [DOI] [PubMed] [Google Scholar]
- Qi S., O’Hayre M., Gutkind J. S., Hurley J. H., 2014a Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3). J. Biol. Chem. 289: 4743–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., O’Hayre M., Gutkind J. S., Hurley J. H., 2014b Insights into β2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3. Protein Sci. 23: 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, M. J., D. Maya, M. Arévalo-Rodríguez, Á. Cebolla, and S. Chávez, 2007 An improved system for estradiol-dependent regulation of gene expression in yeast. Microb. Cell Fact. 6: 10.1–10.9. [DOI] [PMC free article] [PubMed]
- Reneke J. E., Blumer K. J., Courchesne W. E., Thorner J., 1988. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell 55: 221–234. [DOI] [PubMed] [Google Scholar]
- Roelants F. M., Baltz A. G., Trott A. E., Fereres S., Thorner J., 2010. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. USA 107: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J., 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 19222–19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., Kumar S., 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10: 398–409. [DOI] [PubMed] [Google Scholar]
- Rubenstein E. M., Schmidt M. C., 2007. Mechanisms regulating the protein kinases of Saccharomyces cerevisiae. Eukaryot. Cell 6: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H. J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43: 139–160. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B., (Editors), 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shinoda J., Kikuchi Y., 2007. Rod1, an arrestin-related protein, is phosphorylated by Snf1-kinase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 364: 258–263. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K., Kobilka B. K., 2012. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 33: 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A. K., Westfield G. H., Xiao K., Reis R. I., Huang L. Y., et al. , 2014. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512: 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B., Kelstrup C. D., Stoehr G., Fröhlich F., Walther T. C., et al. , 2009. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5: 1337–1346. [DOI] [PubMed] [Google Scholar]
- Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K. M., et al. , 2012. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55–PP2A pathways. Mol. Biol. Cell 23: 2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C. M., Hawley S. A., McCartney R. R., Leech A., Stark M. J., et al. , 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13: 1299–1305. [DOI] [PubMed] [Google Scholar]
- Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., et al. , 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkotnicki L., Crutchley J. M., Zyla T. R., Bardes E. S., Lew D. J., 2008. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell 19: 4675–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J., Shih S., Dunn R., Hicke L., 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell 1: 193–202. [DOI] [PubMed] [Google Scholar]
- Toshima J. Y., Nakanishi J.-I., Mizuno K., Toshima J., Drubin D. G., 2009. Requirements for recruitment of a G protein-coupled receptor to clathrin-coated Pits in budding yeast. Mol. Biol. Cell 20: 5039–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch J. R., McMurray M. A., Nelson Z. W., Gottschling D. E., 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137: 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber E., Virshup D. M., 2001. Casein kinase I: from obscurity to center stage. IUBMB Life 51: 73–78. [DOI] [PubMed] [Google Scholar]
- Volland C., Urban-Grimal D., Géraud G., Haguenauer-Tsapis R., 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269: 9833–9841. [PubMed] [Google Scholar]
- Watanabe D., Murai H., Tanahashi R., Nakamura K., Sasaki T., et al. , 2015. Cooperative and selective roles of the WW domains of the yeast Nedd4-like ubiquitin ligase Rsp5 in the recognition of the arrestin-like adaptors Bul1 and Bul2. Biochem. Biophys. Res. Commun. 463: 76–81. [DOI] [PubMed] [Google Scholar]
- Weinberg J., Drubin D. G., 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C., Hanyaloglu A. C., 2015. Spatial programming of G protein-coupled receptor activity: decoding signaling in health and disease. Mol. Endocrinol. 29: 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J. L., Mulholland J., Jeng R., Cyert M. S., 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Xie X., 2012. Tools for GPCR drug discovery. Acta Pharmacol. Sin. 33: 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., J. A. MacGurn, M. Liu, and S. Emr, 2013 The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife 2(0): e00459.1–e00459.18. [DOI] [PMC free article] [PubMed]
- Zhu H., Klemic J. F., Chang S., Bertone P., Casamayor A., et al. , 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26: 283–289. [DOI] [PubMed] [Google Scholar]