Abstract
High-throughput genomic and proteomic studies have generated near-comprehensive catalogs of biological constituents within many model systems. Nevertheless, static catalogs are often insufficient to fully describe the dynamic processes that drive biology. Quantitative proteomic techniques address this need by providing insight into closely related biological states such as the stages of a therapeutic response or cellular differentiation. The maturation of quantitative proteomics in recent years has brought about a variety of technologies, each with their own strengths and weaknesses. It can be difficult for those unfamiliar with this evolving landscape to match the experiment at hand with the best tool for the job. Here, we outline quantitative methods for proteomic mass spectrometry and discuss their benefits and weaknesses from the perspective of the biologist aiming to generate meaningful data and address mechanistic questions.
Throughout the pioneering days of genomic and proteomic research, much effort was put into constructing comprehensive catalogs of biological data, exemplified by the sequencing of the human genome (1, 2) and proteome (3, 4). Although providing a necessary foundation, these catalogs remain insufficient for describing the complex biological mechanisms at work within cells. At its heart, biology is the study of dynamic processes in living organisms, and proteins are the operators that exert direct control over processes at the cellular level. Today, we seek to build upon these genomic and proteomic foundations to understand closely related biological states, including the stages of a therapeutic response, cellular differentiation, and cancer progression. Such studies promise to reveal the core of mechanistic cell biology by elucidating the relationships between proteins and their roles in cellular processes.
In the past half-century, key insights into many cellular processes have been revealed by demonstrating differential abundance of individual proteins across a small number of conditions. But as with a single photograph, information about a single protein in isolation provides only a narrow portal for viewing the dynamics of the cellular landscape. We now appreciate that the coordination of many proteins and the responses of multiprotein networks to the cellular environment define this landscape. To understand cellular mechanisms, experimental data showing the dynamic nature of the proteome under physiologic and experimentally manipulated conditions are required. In practice, this involves populating a multidimensional matrix of data “photographs” as either a function of time or of comparisons across many closely related states.
Over the last 50 years, our understanding of the levels, localization, interactions, and activation states for proteins has exploded alongside the emergence of increasingly complex biochemical, biophysical, and molecular tools. Biochemical assays using radioisotopes (5, 6) and spectrophotometric readouts (7) established a quantitative framework for understanding proteins and their functions, either individually or in small groups. With the emergence of monoclonal antibodies, molecular and cell biologists have gained the ability to probe any protein or any protein feature against which a specific reagent could be generated (8). A parallel explosion in molecular biology made it possible to overexpress or knock down genes, append affinity tags (9), and attach fluorescent reporters (10) to proteins in both cultured cells and in vivo. Additional techniques such as two-dimensional gel electrophoresis (11) allowed for the separation of complex protein mixtures. The most recent revolution in protein chemistry has come from the field of proteomic mass spectrometry (12), opening the door to the direct characterization of proteins and post-translational modifications (PTMs)1 with site-specific resolution.
Mass spectrometry (MS) provides access to the proteome through three main avenues: identifying the proteins present, assessing their post-translational modification states, and quantifying the relative abundance of each protein-modification state combination (13–17). Although characterization of protein-modification state combinations would ideally be done on intact proteins (18) to reveal the full repertoire of “proteoforms” (19), the majority of “proteomic” analyses are actually performed on peptides generated by proteolytic digestion of protein samples (12). With a focus on speed, sensitivity, and dynamic range in sequencing peptides from complex mixtures, improvements in mass spectrometry instrumentation have brought about dramatic improvements in cataloging the protein constituents of a sample; recent reports have shown the ability to identify the entire yeast proteome in an hour (20) and provided a draft of the human proteome (3, 4). Likewise, sophisticated metal- and immunoaffinity-based enrichment methods (21, 22) have proven useful in elucidating the repertoire of proteins carrying PTMs (23). Focused examples involving phosphorylation (17, 24), ubiquitination (25, 26), acetylation (27), and glycosylation (28, 29) profiling have been thoroughly reviewed elsewhere.
Obtaining abundance measurements for proteins and their various modification states across conditions produces a detailed picture of protein activity and is now possible via a myriad of technologies. In the simplest form of quantitative mass spectrometry, label-free analysis determines the signal intensities or peak areas associated with individual peptides (30). Binary and ternary comparisons provide additional accuracy over label-free techniques and have become routine using methods such as stable isotope-labeling of amino acids in cell culture (SILAC) (31) and reductive methylation (32), as have comparisons of 4–10 conditions using isobaric tags for relative and absolute quantification (iTRAQ) (33) and tandem mass tag (TMT) chemical-tagging reagents (34). In addition, isotopically labeled synthetic peptides and recombinant proteins have also proven valuable for more focused experiments. Further detailed information about these approaches can be found in a number of previous reviews (35–39). Finally, although a full description of such methods falls outside the scope of this review, significant advances have been made in data-independent analysis (40). These methods include both hypothesis-driven, high-throughput targeted analyses by selected and multiple reaction monitoring (37, 41–44) and further advances to the approach through parallel-reaction monitoring of multiple peptides at the same time (45, 46). Such methods can be leveraged to generate proteome maps through targeted post hoc data extraction (47), providing a complement to conventional data-dependent analyses. Proteomics technologies have reached the stage where studies comparing many samples are now feasible, facilitating time course experiments, multiple condition comparisons, and facile introduction of biological replicates.
Each quantitative MS technique has its own strengths and weaknesses, and the pairing of an experiment to the right quantitative model is essential to maximize the utility of the results. A biologist should consider the following questions. What are the key points in designing a successful proteomics analysis? How does one choose among the available proteomics technologies? How and when is multiplexing valuable? Here, we provide background on quantitative proteomic techniques useful for the parallel analysis of multiple samples, and we discuss their benefits and limitations in addressing biological questions.
Development of Quantitative Proteomic Mass Spectrometry
Many discovery proteomics experiments today utilize data-dependent tandem mass spectrometry. In data-dependent analysis, the instrument is programmed to first generate an MS1 spectrum that surveys the masses and signal intensities of intact peptide ions. As this information is frequently insufficient to conclusively match spectra to peptides, the instrument performs a series of secondary MS scans (termed MS/MS or MS2), where individual peptide ions are isolated and fragmented along their amide backbones. Differences in mass between these peptide fragment ions are used to decipher peptide sequence information (48).
Although the intensities of ions observed in an MS1 scan are generally proportional to peptide abundance in the sample, absolute signal intensities can vary depending on a number of factors. Run-to-run differences in sample complexity, chromatography, data-dependent sampling, and peptide ionization efficiency have historically limited the reliability of comparing peptide ion signals across runs. Even with biological replicates in hand, the prevalence of missing values in proteomic data has posed a challenge for downstream statistical analysis.
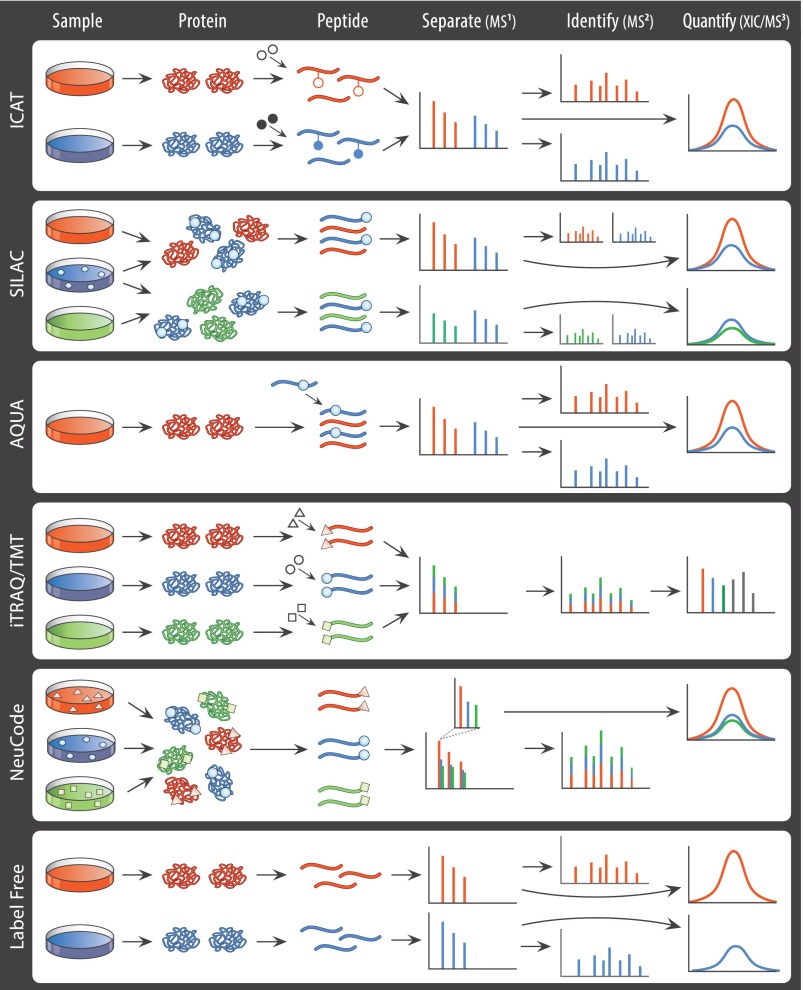
A breakthrough for quantitative proteomics came in the application of stable isotope dilution approaches to the quantitation of proteins and digested peptides. Stable isotopes such as 13C, 15N, 18O, and 2H (deuterium) can be introduced to proteomic samples in a variety of ways (Fig. 1). An early approach, isotope-coded affinity tags (ICAT), employs a biotin affinity tag coupled to a stable isotope-labeled (i.e. 8 deuterium; 2H8) linker and a thiol-reactive group (49). These tags allow for quantitation of cysteine-containing peptides via MS1 survey scans where intensities of peptide ions labeled with the “light” and “heavy” ICAT reagents are directly compared. This approach provides relative ratio measurements of peptides between samples as a proxy for overall proteoform abundance. Although effective, the reliance on cysteine-containing peptides means the majority of peptides lacking cysteine are unmeasured. Thus, alternative techniques such as dimethyl labeling, quantitative carbamylation, and the incorporation of other stable isotopes (18O and 15N) were subsequently developed (50–54).
Fig. 1.
Comparison of quantitative proteomic mass spectrometry techniques. The six techniques, more fully described in the text, differ in the methods of incorporation of stable isotopes (represented as filled shapes), the number of samples directly compared, and the method of quantification. In some techniques (SILAC and NeuCode), stable isotopes are metabolically introduced during sample growth, although in others the labels are introduced after proteins have been harvested and proteolytically digested into peptides, either through introduction of a synthetic labeled peptide (AQUA) or via covalent attachment of a molecule containing the label (ICAT, iTRAQ/TMT; ICAT also employs an unlabeled molecule indicated by an open circle). Samples are then combined as indicated and introduced to the mass spectrometer. After separation of different peptide ions by mass (MS1), all methods identify features from MS2 spectra. In most methods, the quantitative data are derived from an extracted ion chromatogram (XIC) formed by monitoring intact peptide ions from MS1 spectra over the time the feature elutes from liquid chromatography (in NeuCode, ultra-high resolution spectra are necessary to distinguish the near-isobaric masses). iTRAQ/TMT differs in this regard as relative abundances are measured directly from an MS3 spectrum.
An inherent challenge with chemical labeling approaches is that they only account for differences in sample preparation that occur after the labeling step. Seeking to minimize the variability imparted through sample handling, metabolic labeling was developed for proteomics (55). A popular application of metabolic labeling is SILAC (31), where one or more naturally occurring amino acids are replaced with synthetic counterparts, enriched in the stable isotopes 13C and 15N. In common practice, fully labeled [13C615N2]lysine and [13C615N4]arginine are used in combination so that all peptides arising from trypsin digestion (except for those at the C terminus of the protein) can be systematically quantified. Incorporating 13C and 15N labels alleviates the chromatographic retention time shifts sometimes observed with 2H labeling. Amino acid reagents such as [13C6]lysine, [13C6]arginine, and [15N4]arginine (with lesser numbers of stable isotopes incorporated) are also useful and can be combined with fully labeled reagents to extend multiplexing beyond simple paired comparisons. In fact, proof-of-concept studies examining adipocyte differentiation and tyrosine phosphorylation dynamics have demonstrated 5-plex quantitation using these and additional forms of arginine (i.e. 13C615N42H7) (56) or combinations of labeled lysine, arginine, and tyrosine (57), respectively. Nevertheless, a limiting factor of multiplexing capacity in traditional SILAC has been the overlap in the mass dimension between different isotopically labeled forms, even in high resolution MS1 spectra. Moreover, splitting the signal across labeled forms of the same peptide increases the complexity of signals seen by the mass spectrometer, thereby impacting sensitivity and peptide identification rate.
One solution for multiplexing is to use standard metabolic labeling to “unroll” a large sample set into a series of binary comparisons. Instead of labeling multiple samples, this approach involves the preparation of an isotopically labeled reference standard that can be added to each experimental contrast. Comparing each sample against a single common reference mixture makes it possible to determine relative peptide abundance as a ratio of ratios. An advantage of this over label-free analysis is that the presence of reference features in each biological sample empowers longitudinal studies by permitting MS1-based quantification even when the corresponding features are absent (58–60). This approach is equally applicable in the context of both data-independent analysis and targeted experiments, where individual samples are compared with a common reference sample.
A variety of approaches for the construction of a common reference sample has been reported, ranging from focused studies using isotopically labeled synthetic peptides (i.e. AQUA) (61) or recombinant proteins (62) to more global studies employing labeled cell lysates (63) or mammalian tissues (64). In the case of cell lines, computational methods such as principal component analysis have been combined with initial screening experiments (either by proteome or microarray) to select an optimally comprehensive mix of cell lines (65, 66). By carefully combining cell lines that span the breadth and diversity of the proteome into a mixed protein reference standard, “Super-SILAC” (58) and similar approaches have been useful for comparative analysis of human tissues (67), primary neurons (68), cancer cells (69, 70), and model organisms such as nematodes (71), fly (72), zebrafish (73), and mice (74, 75).
Despite their utility, multiplexed approaches involving binary comparisons against a labeled reference sample have certain drawbacks. Considerable instrument acquisition time and bioinformatic analysis are necessary to analyze each biological sample and to select and optimize the common reference mixture. Undersampling of the proteome also remains a concern, as analysis time is spent examining redundant precursor ion species from the reference mix in each run (76). Furthermore, abundance determinations based upon the ratio-of-ratios approach are affected by the abundance of the feature in both the sample and the common reference. Nevertheless, the reliability and quantitative accuracy of these methods make them attractive methods for a variety of purposes.
Multiplexing Using Isobaric Reporter Approaches, iTRAQ and TMT
The metabolic and chemical labeling methods discussed thus far operate on the principle of incorporating stable isotopes into one or more samples to create peptide features that differ in their overall precursor ion mass. In contrast, isobaric tagging utilizes a carefully constructed molecular tag where each of the differentially labeled forms confers the same overall mass addition but yields a unique reporter ion upon fragmentation. Such isobaric tagging reagents consist of three parts as follows: a reactive group, a reporter region, and a balance region. The reactive group is used to couple the isobaric reagent onto the peptide, although the complete reagent remains attached through MS1 analysis. Upon fragmentation, the isobaric reagent is cleaved between the balance and reporter regions to liberate the low m/z reporter. By varying the distribution of stable isotopes between the reporter and balance regions, a series of reporter ions with varying masses can be generated. As reviewed by Rauniyar and Yates (77), iTRAQ (33) and TMT (34) have each made significant contributions in multiplexed proteomic analysis.
iTRAQ was first reported in 2004, where its use as a 4-plex reagent allowed for the simultaneous quantification of proteomic changes in yeast resulting from nonsense-mediated mRNA decay (33). iTRAQ reagents use an amine-reactive group to derivatize both the peptide N terminus and the ε-amine of lysines. Upon tandem MS analysis, iTRAQ reagents release singly charged reporter ions with masses between 114 and 117 m/z. Subsequent optimization extended iTRAQ to 8-plex, utilizing masses between 113 and 121 Da (avoiding potential interference from naturally occurring phenylalanine immonium ions at 120 Da) (78). The TMT reagent has a different structure than that of iTRAQ (79) but achieves largely the same goal, generating a 6-plex reagent with reporter ions of 126 to 131 m/z (34). TMT was later extended to 10-plex by exploiting the mass deficit between 15N and 13C (80, 81). Despite a mass difference of only 6.32 mDa, pairs of reporter ions ranging from 127 and 130 m/z are readily distinguishable from one another with standard high resolution instrumentation.
Isobaric tagging approaches mitigate several issues presented by MS1-based quantitation. With isobaric tags, the precursor ion signal for all samples accumulates in the same mass window at the MS1 level, aiding sensitivity. Multiplexed analysis within a single tandem mass spectrum allows for direct comparisons that minimize run-to-run variability, maximize analysis time, and help to eliminate missing values (82, 83). Early isobaric labeling experiments were carried out on time-of-flight (TOF)-based mass spectrometers, as the mechanics of ion-trap instrumentation and collision-induced dissociation rendered the low m/z reporter ions unmeasurable (84). The advent of higher energy collision dissociation within the LTQ-Orbitrap sidestepped this low mass cutoff and opened the door to the examination of isobaric tag reporter ions on Orbitrap instruments (85). At the same time, early studies showed TMT data to be affected by ratio compression, a phenomena in which observed ratios underestimate expected ratios for defined mixtures (86–90). This phenomenon stems from the presence of co-eluting peptides within the precursor ion isolation window used for MS/MS fragmentation. To rectify this issue, strategies involving gas-phase fragmentation (89) and MS3 analysis involving a second round of selection and fragmentation of one or more MS2 fragment ions (90, 91) have been introduced.
Technological advances promise to extend multiplexing beyond its current limits. For example, the combination of isobaric and metabolic labeling has been proposed to simultaneously measure up to 18 samples within a single instrument analysis. For an 18-sample comparison, Dephoure and Gygi (92) used a combination of triplex SILAC labeling along with 6-plex TMT tagging to study the effects of rapamycin in yeast. Pairing this approach with 10-plex TMT could conceivably empower 30-plex analysis, assuming that dedicated methods could be established to focus the mass spectrometer so that triplet features were consistently and equivalently sampled. Similar levels of multiplexing are proposed for the next-generation isobaric tagging reagents, including combinatorial isobaric mass tags (93).
Isobarically tagged protein-profiling methods have been employed successfully in a number of settings involving baseline proteomic profiling of ovarian cancer cell lines (94) and pancreatic organoid models (95), as well as in specialized applications such as validating protein-protein interaction networks (96) and characterizing site-specific acetylation (97). Grimsrud et al. (98) used TMT to characterize the in vivo mitochondrial phosphoproteome of over 50 mice across eight different conditions (strain, age, and obesity status) in a type 2 diabetes model. One particularly appealing application of multiplexed quantitation consists of matching small molecules with their cellular targets by thermal stability profiling. There, TMT-based quantitation has been used to generate pairs of melting curves for as many as 7000 proteins in the presence and absence of drug compounds (99).
Multiplexing Using Neutron-encoded Mass Signatures, NeuCode
Building on the concept of the neutron mass deficit between 15N and 13C (80, 81), Coon and co-workers (100) developed an MS1-centric multiplexing technique that takes advantage of isotopically labeled amino acids that differ in mass by at little as 6.32 mDa. The elegance of NeuCode stems from this compact representation of labeled samples within a narrow mass range. These isotopologues are spaced closely enough together that they appear as a single peak in MS1 scans at 30,000 resolution and are co-isolated for MS/MS fragmentation. Despite appearing isobaric in low resolution MS1 survey scans and ion trap MS2 scans, these isotopologues can be resolved in MS1 survey scans performed at ultra-high resolution (∼500,000 at 400 m/z) to reveal quantitative data. The reduction in apparent sample complexity that comes from co-isolation and co-fragmentation in the MS2 offers the potential to mitigate the sensitivity limitations of SILAC, which stems from the distribution of ion signals into multiple precursor ion populations (101). Although this approach combines the most desirable qualities from isobaric multiplexing with the quantitative readout in an MS1 survey scan, to successfully implement this technique, instruments capable of achieving ultra-high resolution on a chromatographic time scale are necessary. This need for high-resolving power also requires longer scan times than traditional SILAC, losing some of the time that was gained from simplifying the precursor ion population. Additional studies are also required to understand the phenomenon of peak coalescence, where closely spaced isotopologue signals from abundant or high m/z features can merge together. As with the ratio compression observed with isobaric tags, improvements to instrument performance and data analysis algorithms promise to mitigate these limitations as the technique matures.
Multiplexing without Isotopes, Label-free Analysis
Advances in instrument design continue to produce mass spectrometers that are faster, more accurate, and more sensitive. Just as these improvements have fueled the application of stable isotope-based quantitation methods, they have also improved the depth and reproducibility of peptide measurements without the use of isotopic labels. Through careful experimental design, sample replication, and improved data analysis procedures, many label-free experiments have proven capable of generating results on par with labeling methods (102). As label-free experiments require less complicated sample preparation and translate well to both in vivo models (103) and clinical specimens (104), they are often chosen for studying complex biological systems. The recent report from Humphrey et al. (103) describes the type of large, parallel study that is now possible, investigating in vivo phosphorylation dynamics upon insulin stimulation. In a brute-force approach, label-free analysis was carried out across at least eight biological replicates at five different time points, where each sample was interrogated using 2–4 h of instrument time. Similarly, global proteomic profiling of human colon and rectal tumors and spectral counting have revealed new tumor subtypes and candidate driver genes (104).
Label-free approaches provide an appealing alternative to metabolic labeling approaches while still retaining the definition of the MS1 peptide ion signal as an area-under-curve measurement. However, in label-free methods, samples are compared across different instrument analyses without employing a stable isotope-based reference. Although comparisons across multiple analyses can introduce variability at the individual observation level, repeated measurements across biological replicates and data aggregation at the peptide or protein level can empower modeling approaches using established statistical methods. A variety of methods has been proposed to normalize quantitative data from area-based label-free experiments, including empirical normalization (105, 106), regression-based approaches (107), correlation-based approaches combined with regression (108), and statistical inference (60), among others (30, 109). In 2012, Clough et al. (110) reported a generalized statistical model for label-free analysis while characterizing hypoxia-induced proteomic changes in breast cancer cell lines. This approach was released to the community as the MSstats package for the R statistical programming language (111). We and others have applied similar statistical approaches to the study of cysteine oxidation (112), phosphorylation dynamics (59), inducible ubiquitination (113–116), and cell surface proteins (117). When sufficient data can be generated to overcome the inherent variability of individual peptide measurements, label-free analyses provide an attractive option for quantification while avoiding laborious or technically challenging metabolic labeling techniques.
Choosing the Right Quantitative Approach
From a biologist's perspective, choosing between the available methods for quantitative proteomic analysis can be a daunting task, as one seeks to balance optimal experimental design against the practicalities of time, cost, and available technology. Throughout this process, the importance of an active dialogue between the experimental biologist, the analytical mass spectrometrist, and the computational proteomics specialist is paramount. An early, important consideration is whether the proposed experiments aim to stand alone in testing a biological hypothesis or instead represent a screening approach to generate lead candidates for orthogonal validation. Screening-type experiments provide more freedom in experimental design and have proven to be enormously valuable in revealing the key players in cellular signaling, but these results come at the expense of variability and incomplete information. Common screening approaches involve the profiling of post-translationally modified peptides, whereby differentially abundant phospho-, acetyl-, or ubiquitinated peptides can quickly reveal when and where PTMs are being added to or removed from proteins of interest. It is important to determine early on whether abundance changes measured at the aggregate protein level are sufficient or whether information at the peptide or site level is necessary for follow-up study. As compared with global proteome analyses, where many peptide observations are aggregated to estimate protein abundance, PTM site-level quantitation is often supported by a limited number of measurements for a single feature per sample. Moreover, in the absence of data showing the baseline protein levels, numerous mechanisms involving altered gene expression, protein synthesis, degradation, or alternative PTMs can confound apparent abundance changes in the PTM of interest. Our experience in studying ubiquitination suggests that label-free analyses are powerful at revealing protein level effects even in small sample sizes but that stable isotope-based approaches or large sample sizes are more required for quantification of site-specific features.
In practice, decisions about which quantitative proteomics technology to use are often bounded by the nature of the model system being studied. Issues encountered with primary or non-proliferating cell culture models can limit the amount of protein available or the feasibility of metabolic labeling, forcing consideration of alternative approaches. Despite the potential of these technologies, it is not possible to visualize a dynamic protein signal before it has emerged or after it has been resolved. Careful experimental protocols are required to ensure synchronization of the biological response between individuals and the timely collection of samples to maintain transient proteoforms in vivo. Given the importance of the time dimension, a pre-emptive definition of the temporal dynamics of known markers or phenotypes within the model system can dramatically improve the outcome of MS proteomics investigations. Similar considerations also apply to the realm of peptide abundance and instrument sensitivity. Discovery experiments must be performed on a scale large enough to ensure that low level analytes can be detected after enrichment and/or fractionation procedures. Scaling-up can insidiously affect signaling dynamics within cells and tissues. It can also result in unforeseen variability in the effectiveness of sample preparation protocols; the additional time and handling steps required to harvest experimental protein samples en masse can have consequences on protein, PTM, and inhibitor stability in solution (118), in addition to downstream steps such as digestion, enrichment, and desalting.
Beyond sample-processing considerations, a constant challenge in experimental design is determining how much instrument time should be devoted to an analysis. As undersampling in proteomics remains a key limitation, extending the instrument time devoted to an experiment can produce increased data density and yield improvements in signal-to-noise, especially when coupled with fractionation. At the same time, practical considerations such as the cost of MS instrumentation require that resources be allocated thoughtfully. By collecting quantitative information for each “isotopically bar-coded” feature within individual scan events, multiplexing methods such as isobaric tagging maximize the amount of useful data generated per unit of time. However, it has been shown that the quality of quantitative data from isobarically tagged samples scales with ion-injection time and the purity of the ion population (89, 90). Although recent reports are encouraging (119), additional careful studies are required to fully benchmark these technologies against the more traditional approaches for baseline protein profiling. Additionally, reagent cost, larger input sample sizes, and the nature of enrichment protocols can make isobaric labeling less attractive for multiplexed PTM analysis. For example, immunoaffinity enrichment of K-GG peptides in ubiquitin-substrate profiling requires the free N terminus of the PTM remnant that is blocked by isobaric labeling. The cost of the isotopically labeled amino acid reagent is a consideration in metabolic labeling approaches as well. It is important to consider these costs not only for pilot experiments but also for full-scale studies involving replicates and controls.
Irrespective of the method chosen, multiplexed quantitative proteomics provides a powerful tool that complements genetic approaches to allow for direct insight in the molecular mechanisms underlying biological processes. In recent years, a steady stream of studies illuminating specific areas of biology have marked the successful transition of these approaches from proof-of-concept applications to essential components of a molecular biologist's toolkit. Although the approaches will undoubtedly improve in sensitivity, precision, and ease of use, the current landscape of techniques allows one to gain rich knowledge about the molecular basis of biology and disease.
Footnotes
Author contributions: C. E. B. and D. S. K. wrote the paper.
The authors declare that they have no conflicts of interest with the contents of this article.
1 The abbreviations used are:
- PTM
- post-translational modification
- iTRAQ
- isobaric tags for relative and absolute quantification
- TMT
- tandem mass tag
- SILAC
- stable isotope-labeling of amino acids in cell culture.
REFERENCES
- 1. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 2. Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., Gocayne J. D., Amanatides P., Ballew R. M., Huson D. H., Wortman J. R., et al. (2001) The sequence of the human genome. Science 291, 1304–1351 [DOI] [PubMed] [Google Scholar]
- 3. Kim M.-S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., et al. (2014) A draft map of the human proteome. Nature 509, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 5. Yalow R. S., and Berson S. A. (1960) Immunoassay of endogenous plasma insulin in man. J. Clin. Invest. 39, 1157–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walsh D. A., Perkins J. P., and Krebs E. G. (1968) An adenosine 3′,5′-monophosphate-dependent protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243, 3763–3765 [PubMed] [Google Scholar]
- 7. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 8. Köhler G., and Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 9. Munro S., and Pelham H. R. (1984) Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. EMBO J. 3, 3087–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsien R. Y. (1998) The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 11. O'Farrell P. H. (1975) High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 12. Hunt D. F., Yates J. R. 3rd., Shabanowitz J., Winston S., and Hauer C. R. (1986) Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 83, 6233–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aebersold R., and Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 14. Ong S.-E., and Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 15. Domon B., and Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., Fonslow B. R., Shan B., Baek M.-C., and Yates J. R. 3rd (2013) Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 113, 2343–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimsrud P. A., Swaney D. L., Wenger C. D., Beauchene N. A., and Coon J. J. (2010) Phosphoproteomics for the masses. ACS Chem. Biol. 5, 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tran J. C., Zamdborg L., Ahlf D. R., Lee J. E., Catherman A. D., Durbin K. R., Tipton J. D., Vellaichamy A., Kellie J. F., Li M., Wu C., Sweet S. M., Early B. P., Siuti N., LeDuc R. D., Compton P. D., Thomas P. M., and Kelleher N. L. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith L. M., Kelleher N. L., and Consortium for Top Down Proteomics (2013) Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hebert A. S., Richards A. L., Bailey D. J., Ulbrich A., Coughlin E. E., Westphall M. S., and Coon J. J. (2014) The one hour yeast proteome. Mol. Cell. Proteomics 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., and White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 22. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X.-M., Polakiewicz R. D., and Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 23. Olsen J. V., and Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macek B., Mann M., and Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 25. Bustos D., Bakalarski C. E., Yang Y., Peng J., and Kirkpatrick D. S. (2012) Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11, 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ordureau A., Münch C., and Harper J. W. (2015) Quantifying ubiquitin signaling. Mol. Cell 58, 660–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 28. Thaysen-Andersen M., and Packer N. H. (2014) Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 1844, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 29. Levery S. B., Steentoft C., Halim A., Narimatsu Y., Clausen H., and Vakhrushev S. Y. (2015) Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 [DOI] [PubMed] [Google Scholar]
- 30. Neilson K. A., Ali N. A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S. C., and Haynes P. A. (2011) Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 [DOI] [PubMed] [Google Scholar]
- 31. Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 32. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 33. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 34. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., and Sanchez J.-C. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 35. Bantscheff M., Schirle M., Sweetman G., Rick J., and Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 36. Bantscheff M., Lemeer S., Savitski M. M., and Kuster B. (2012) Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–965 [DOI] [PubMed] [Google Scholar]
- 37. Liebler D. C., and Zimmerman L. J. (2013) Targeted quantitation of proteins by mass spectrometry. Biochemistry 52, 3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Law K. P., and Lim Y. P. (2013) Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev. Proteomics 10, 551–566 [DOI] [PubMed] [Google Scholar]
- 39. Chapman J. D., Goodlett D. R., and Masselon C. D. (2014) Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 33, 452–470 [DOI] [PubMed] [Google Scholar]
- 40. Venable J. D., Dong M.-Q., Wohlschlegel J., Dillin A., and Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 41. Anderson L., and Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 42. Keshishian H., Addona T., Burgess M., Kuhn E., and Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lange V., Picotti P., Domon B., and Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whiteaker J. R., Zhao L., Yan P., Ivey R. G., Voytovich U. J., Moore H. D., Lin C., and Paulovich A. G. (2015) Peptide immunoaffinity enrichment and targeted mass spectrometry enables multiplex, quantitative pharmacodynamic studies of phospho-signaling. Mol. Cell. Proteomics 14, 2261–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson A. C., Russell J. D., Bailey D. J., Westphall M. S., and Coon J. J. (2012) Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11, 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gallien S., Duriez E., Crone C., Kellmann M., Moehring T., and Domon B. (2012) Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteomics 11, 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steen H., and Mann M. (2004) The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711 [DOI] [PubMed] [Google Scholar]
- 49. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., and Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 50. Hsu J.-L., Huang S.-Y., Chow N.-H., and Chen S.-H. (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 51. Boersema P. J., Aye T. T., van Veen T. A., Heck A. J., and Mohammed S. (2008) Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 8, 4624–4632 [DOI] [PubMed] [Google Scholar]
- 52. Angel P. M., and Orlando R. (2007) Quantitative carbamylation as a stable isotopic labeling method for comparative proteomics. Rapid Commun. Mass Spectrom. 21, 1623–1634 [DOI] [PubMed] [Google Scholar]
- 53. Murphy R. C., and Clay K. L. (1979) Synthesis and back exchange of 18O labeled amino acids for use as internal standards with mass spectrometry. Biomed. Mass Spectrom. 6, 309–314 [DOI] [PubMed] [Google Scholar]
- 54. Conrads T. P., Alving K., Veenstra T. D., Belov M. E., Anderson G. A., Anderson D. J., Lipton M. S., Pasa-Tolić L., Udseth H. R., Chrisler W. B., Thrall B. D., and Smith R. D. (2001) Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal. Chem. 73, 2132–2139 [DOI] [PubMed] [Google Scholar]
- 55. Oda Y., Huang K., Cross F. R., Cowburn D., and Chait B. T. (1999) Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 96, 6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Molina H., Yang Y., Ruch T., Kim J.-W., Mortensen P., Otto T., Nalli A., Tang Q.-Q., Lane M. D., Chaerkady R., and Pandey A. (2009) Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J. Proteome Res. 8, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tzouros M., Golling S., Avila D., Lamerz J., Berrera M., Ebeling M., Langen H., and Augustin A. (2013) Development of a 5-plex SILAC method tuned for the quantitation of tyrosine phosphorylation dynamics. Mol. Cell. Proteomics 12, 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., and Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 59. Kirkpatrick D. S., Bustos D. J., Dogan T., Chan J., Phu L., Young A., Friedman L. S., Belvin M., Song Q., Bakalarski C. E., and Hoeflich K. P. (2013) Phosphoproteomic characterization of DNA damage response in melanoma cells following MEK/PI3K dual inhibition. Proc. Natl. Acad. Sci. U.S.A. 110, 19426–19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gerber S. A., Rush J., Stemman O., Kirschner M. W., and Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hanke S., Besir H., Oesterhelt D., and Mann M. (2008) Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J. Proteome Res. 7, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 63. Ishihama Y., Sato T., Tabata T., Miyamoto N., Sagane K., Nagasu T., and Oda Y. (2005) Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat. Biotechnol. 23, 617–621 [DOI] [PubMed] [Google Scholar]
- 64. Wu C. C., MacCoss M. J., Howell K. E., Matthews D. E., and Yates J. R. 3rd (2004) Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76, 4951–4959 [DOI] [PubMed] [Google Scholar]
- 65. Gilmore J. M., Milloy J. A., and Gerber S. A. (2013) SILAC surrogates: rescue of quantitative information for orphan analytes in spike-in SILAC experiments. Anal. Chem. 85, 10812–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pozniak Y., and Geiger T. (2014) Design and application of super-SILAC for proteome quantification. Methods Mol. Biol. 1188, 281–291 [DOI] [PubMed] [Google Scholar]
- 67. Lund R. R., Terp M. G., Laenkholm A.-V., Jensen O. N., Leth-Larsen R., and Ditzel H. J. (2012) Quantitative proteomics of primary tumors with varying metastatic capabilities using stable isotope-labeled proteins of multiple histogenic origins. Proteomics 12, 2139–2148 [DOI] [PubMed] [Google Scholar]
- 68. Liao L., Sando R. C., Farnum J. B., Vanderklish P. W., Maximov A., and Yates J. R. (2012) 15N-labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J. Proteome Res. 11, 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang W., Wei Y., Ignatchenko V., Li L., Sakashita S., Pham N.-A., Taylor P., Tsao M. S., Kislinger T., and Moran M. F. (2014) Proteomic profiles of human lung adeno and squamous cell carcinoma using super-SILAC and label-free quantification approaches. Proteomics 14, 795–803 [DOI] [PubMed] [Google Scholar]
- 70. Schweppe D. K., Rigas J. R., and Gerber S. A. (2013) Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteomics 91, 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Larance M., Bailly A. P., Pourkarimi E., Hay R. T., Buchanan G., Coulthurst S., Xirodimas D. P., Gartner A., and Lamond A. I. (2011) Stable-isotope labeling with amino acids in nematodes. Nat. Methods 8, 849–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sury M. D., Chen J.-X., and Selbach M. (2010) The SILAC fly allows for accurate protein quantification in vivo. Mol. Cell. Proteomics 9, 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Westman-Brinkmalm A., Abramsson A., Pannee J., Gang C., Gustavsson M. K., von Otter M., Blennow K., Brinkmalm G., Heumann H., and Zetterberg H. (2011) SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J. Proteomics 75, 425–434 [DOI] [PubMed] [Google Scholar]
- 74. Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., and Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 75. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., et al. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mertins P., Udeshi N. D., Clauser K. R., Mani D. R., Patel J., Ong S., Jaffe J. D., and Carr S. A. (2012) iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 11, M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rauniyar N., and Yates J. R. 3rd (2014) Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 13, 5293–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Choe L., D'Ascenzo M., Relkin N. R., Pappin D., Ross P., Williamson B., Guertin S., Pribil P., and Lee K. H. (2007) 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics 7, 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., and Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 80. Werner T., Becher I., Sweetman G., Doce C., Savitski M. M., and Bantscheff M. (2012) High-resolution enabled TMT 8-plexing. Anal. Chem. 84, 7188–7194 [DOI] [PubMed] [Google Scholar]
- 81. McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., and Gygi S. P. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zieske L. R. (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 83. Li Z., Adams R. M., Chourey K., Hurst G. B., Hettich R. L., and Pan C. (2012) Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J. Proteome Res. 11, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 84. Köcher T., Pichler P., Schutzbier M., Stingl C., Kaul A., Teucher N., Hasenfuss G., Penninger J. M., and Mechtler K. (2009) High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J. Proteome Res. 8, 4743–4752 [DOI] [PubMed] [Google Scholar]
- 85. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., and Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 86. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., and Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly.” J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 87. Savitski M. M., Sweetman G., Askenazi M., Marto J. A., Lang M., Zinn N., and Bantscheff M. (2011) Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal. Chem. 83, 8959–8967 [DOI] [PubMed] [Google Scholar]
- 88. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., and Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wenger C. D., Lee M. V., Hebert A. S., McAlister G. C., Phanstiel D. H., Westphall M. S., and Coon J. J. (2011) Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 8, 933–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ting L., Rad R., Gygi S. P., and Haas W. (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., and Gygi S. P. (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dephoure N., and Gygi S. P. (2012) Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci. Signal. 5, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Braun C. R., Bird G. H., Wühr M., Erickson B. K., Rad R., Walensky L. D., Gygi S. P., and Haas W. (2015) A new isobaric labeling architecture supporting reporter ion fragmentation enables improved multiplexing capacity. Anal. Chem. 87, 9855–9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fan G., Wrzeszczynski K. O., Fu C., Su G., Pappin D. J., Lucito R., and Tonks N. K. (2015) A quantitative proteomics-based signature of platinum sensitivity in ovarian cancer cell lines. Biochem. J. 465, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Boj S. F., Hwang C.-I., Baker L. A., Chio I. I., Engle D. D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M. S., Gracanin A., Oni T., Yu K. H., van Boxtel R., Huch M., et al. (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Roe J.-S., Mercan F., Rivera K., Pappin D. J., and Vakoc C. R. (2015) BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol. Cell 58, 1028–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grimsrud P. A., Carson J. J., Hebert A. S., Hubler S. L., Niemi N. M., Bailey D. J., Jochem A., Stapleton D. S., Keller M. P., Westphall M. S., Yandell B. S., Attie A. D., Coon J. J., and Pagliarini D. J. (2012) A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab. 16, 672–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Savitski M. M., Reinhard F. B., Franken H., Werner T., Savitski M. F., Eberhard D., Martinez Molina D., Jafari R., Dovega R. B., Klaeger S., Kuster B., Nordlund P., Bantscheff M., and Drewes G. (2014) Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784. [DOI] [PubMed] [Google Scholar]
- 100. Hebert A. S., Merrill A. E., Bailey D. J., Still A. J., Westphall M. S., Strieter E. R., Pagliarini D. J., and Coon J. J. (2013) Neutron-encoded mass signatures for multiplexed proteome quantification. Nat. Methods 10, 332–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Merrill A. E., Hebert A. S., MacGilvray M. E., Rose C. M., Bailey D. J., Bradley J. C., Wood W. W., El Masri M., Westphall M. S., Gasch A. P., and Coon J. J. (2014) NeuCode labels for relative protein quantification. Mol. Cell. Proteomics 13, 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tebbe A., Klammer M., Sighart S., Schaab C., and Daub H. (2015) Systematic evaluation of label-free and super-SILAC quantification for proteome expression analysis. Rapid Commun. Mass Spectrom. 29, 795–801 [DOI] [PubMed] [Google Scholar]
- 103. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 104. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., et al. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bakalarski C. E., Elias J. E., Villén J., Haas W., Gerber S. A., Everley P. A., and Gygi S. P. (2008) The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Griffin N. M., Yu J., Long F., Oh P., Shore S., Li Y., Koziol J. A., and Schnitzer J. E. (2010) Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W. J., Webb-Robertson B.-J., Smith R. D., and Lipton M. S. (2006) Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Park S. K., Venable J. D., Xu T., and Yates J. R. 3rd (2008) A quantitative analysis software tool for mass spectrometry–based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wong J. W., Sullivan M. J., and Cagney G. (2008) Computational methods for the comparative quantification of proteins in label-free LCn-MS experiments. Brief. Bioinform. 9, 156–165 [DOI] [PubMed] [Google Scholar]
- 110. Clough T., Thaminy S., Ragg S., Aebersold R., and Vitek O. (2012) Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics 13, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 [DOI] [PubMed] [Google Scholar]
- 112. Zaccarin M., Falda M., Roveri A., Bosello-Travain V., Bordin L., Maiorino M., Ursini F., and Toppo S. (2014) Quantitative label-free redox proteomics of reversible cysteine oxidation in red blood cell membranes. Free Radic. Biol. Med. 71, 90–98 [DOI] [PubMed] [Google Scholar]
- 113. Bingol B., Tea J. S., Phu L., Reichelt M., Bakalarski C. E., Song Q., Foreman O., Kirkpatrick D. S., and Sheng M. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 [DOI] [PubMed] [Google Scholar]
- 114. Cunningham C. N., Baughman J. M., Phu L., Tea J. S., Yu C., Coons M., Kirkpatrick D. S., Bingol B., and Corn J. E. (2015) USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 17, 160–169 [DOI] [PubMed] [Google Scholar]
- 115. Varfolomeev E., Izrael-Tomasevic A., Yu K., Bustos D., Goncharov T., Belmont L. D., Masselot A., Bakalarski C. E., Kirkpatrick D. S., and Vucic D. (2015) Ubiquitination profiling identifies sensitivity factors for IAP antagonist treatment. Biochem. J. 466, 45–54 [DOI] [PubMed] [Google Scholar]
- 116. Yu K., Phu L., Varfolomeev E., Bustos D., Vucic D., and Kirkpatrick D. S. (2015) Immunoaffinity enrichment coupled to quantitative mass spectrometry reveals ubiquitin-mediated signaling events. J. Mol. Biol. 427, 2121–2134 [DOI] [PubMed] [Google Scholar]
- 117. Shimogawa M. M., Saada E. A., Vashisht A. A., Barshop W. D., Wohlschlegel J. A., and Hill K. L. (2015) Cell surface proteomics provides insight into stage-specific remodeling of the host-parasite interface in Trypanosoma brucei. Mol. Cell. Proteomics 14, 1977–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies, et al. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics 13, 1690–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Murphy J. P., Stepanova E., Everley R. A., Paulo J. A., and Gygi S. P. (2015) Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol. Cell. Proteomics 14, 2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]