Abstract
Precisely knowing the stoichiometry of their components is critical for investigating structure, assembly, and function of macromolecular machines. This has remained a technical challenge in particular for large, hydrophobic membrane-spanning protein complexes. Here, we determined the stoichiometry of a type III secretion system of Salmonella enterica serovar Typhimurium using two complementary protocols of gentle complex purification combined with peptide concatenated standard and synthetic stable isotope-labeled peptide-based mass spectrometry. Bacterial type III secretion systems are cell envelope-spanning effector protein-delivery machines essential for colonization and survival of many Gram-negative pathogens and symbionts. The membrane-embedded core unit of these secretion systems, termed the needle complex, is composed of a base that anchors the machinery to the inner and outer membranes, a hollow filament formed by inner rod and needle subunits that serves as conduit for substrate proteins, and a membrane-embedded export apparatus facilitating substrate translocation. Structural analyses have revealed the stoichiometry of the components of the base, but the stoichiometry of the essential hydrophobic export apparatus components and of the inner rod protein remain unknown. Here, we provide evidence that the export apparatus of type III secretion systems contains five SpaP, one SpaQ, one SpaR, and one SpaS. We confirmed that the previously suggested stoichiometry of nine InvA is valid for assembled needle complexes and describe a loose association of InvA with other needle complex components that may reflect its function. Furthermore, we present evidence that not more than six PrgJ form the inner rod of the needle complex. Providing this structural information will facilitate efforts to obtain an atomic view of type III secretion systems and foster our understanding of the function of these and related flagellar machines. Given that other virulence-associated bacterial secretion systems are similar in their overall buildup and complexity, the presented approach may also enable their stoichiometry elucidation.
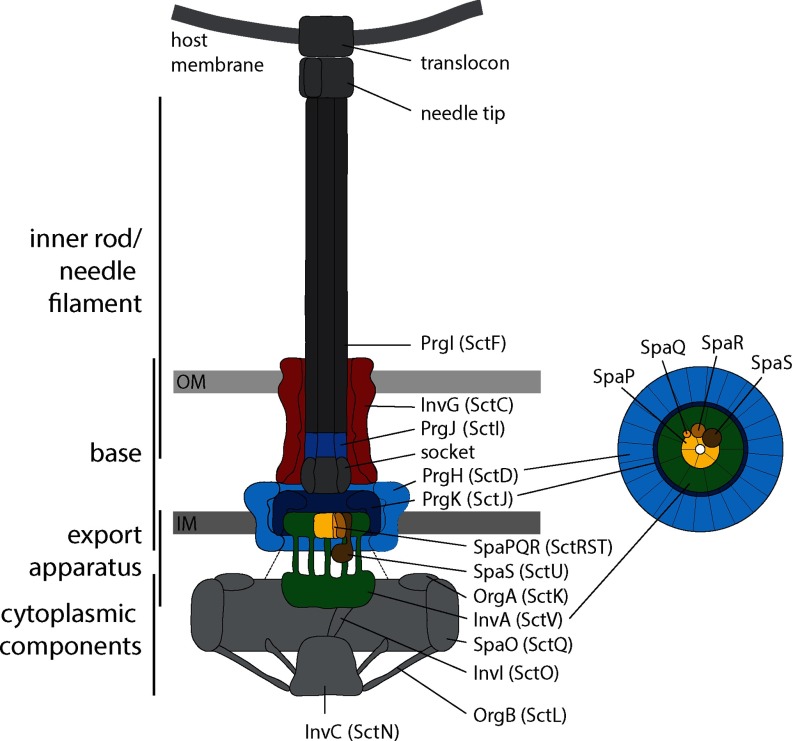
Type III secretion systems (T3SS), evolutionary and structurally related to bacterial flagella (1), are used by many pathogenic or symbiotic Gram-negative bacteria to inject effector proteins into eukaryotic host cells in order to promote bacterial survival and colonization (2). The core unit of T3SSs is a cell envelope-spanning macromolecular machine termed the needle complex. It consists of a base that anchors the complex in the bacterial inner and outer membranes (3), an inner membrane-embedded export apparatus facilitating substrate translocation located at the center of the base (4), and a filamentous inner rod and needle, which protrude from the bacterial surface and serve as conduit for substrates (2) (Fig. 1). The entire system, which also includes several cytoplasmic components involved in targeting and preparation of substrates (5–9), is composed of up to 20 different proteins with one to several hundred copies each.
Fig. 1.
Model of the type III secretion system needle complex. Base and export apparatus components whose stoichiometry was investigated are shown in color. Protein names of T3SS-1 of S. Typhimurium are indicated. Protein names according to the unified nomenclature by Hueck (50) are in brackets. On the right, a bottom view of the base and export apparatus is shown indicating stoichiometries obtained in this study. Abbreviations: IM: inner membrane; OM: outer membrane.
Studies using single particle cryo electron microscopy coupled to molecular docking of the atomic structures of domains of different components have begun to generate an atomic model of the type III secretion needle complex (10). However, many protein densities within the structure are still unaccounted for, leaving substantial knowledge gaps that have yet to be filled. A challenge for this effort has been the absence of the precise stoichiometry of all the components of the needle complex. The determination of an accurate stoichiometry for such a large and hydrophobic protein assembly is technically demanding but severely needed to facilitate approaches generating an atomic view of the machine. The stoichiometry that is known to date resulted from the analyses of the less hydrophobic components that have been amenable to high-resolution x-ray crystallography and NMR and low-resolution cryo electron microscopy. The low-resolution structural analysis of isolated needle complexes revealed a stoichiometry of 12–15 copies for the outer membrane secretin ring of T3SSs of different bacteria and of 12–24 copies for the two inner membrane ring proteins (11–14). The periodicity of the helical needle is 5.7 subunits per turn as assessed by solid-state NMR (15), but the stoichiometry of the inner rod has not been reported. So far, only the sizeable cytoplasmic domains of several homologs of the hydrophobic export apparatus components SpaS and InvA were amenable to high-resolution structural studies (16–24). While no stoichiometry could be deduced from structures of SpaSC, the C-terminal domain of the InvA homolog of Shigella (MxiA) crystallized as a nonameric ring (24). Besides SpaS, the stoichiometry of the minor export apparatus components SpaP, SpaQ, and SpaR is also unknown.
To evaluate the stoichiometry of the complete needle complex of the T3SS encoded within pathogenicity island 1 (SPI-1) of Salmonella enterica serovar Typhimurium (S. Typhimurium), including its hydrophobic export apparatus components (Fig. 1, see also for nomenclature), we have employed two complementing mass spectrometry (MS)-based strategies, using peptide concatenated standards (PCS) (25) and synthetic stable isotope-labeled peptides (26). Both strategies employ ratiometric comparison of isotope-labeled standard peptides of known quantity or stoichiometry with the quantities of the same peptides in assembled complexes (Fig. 2).
Fig. 2.
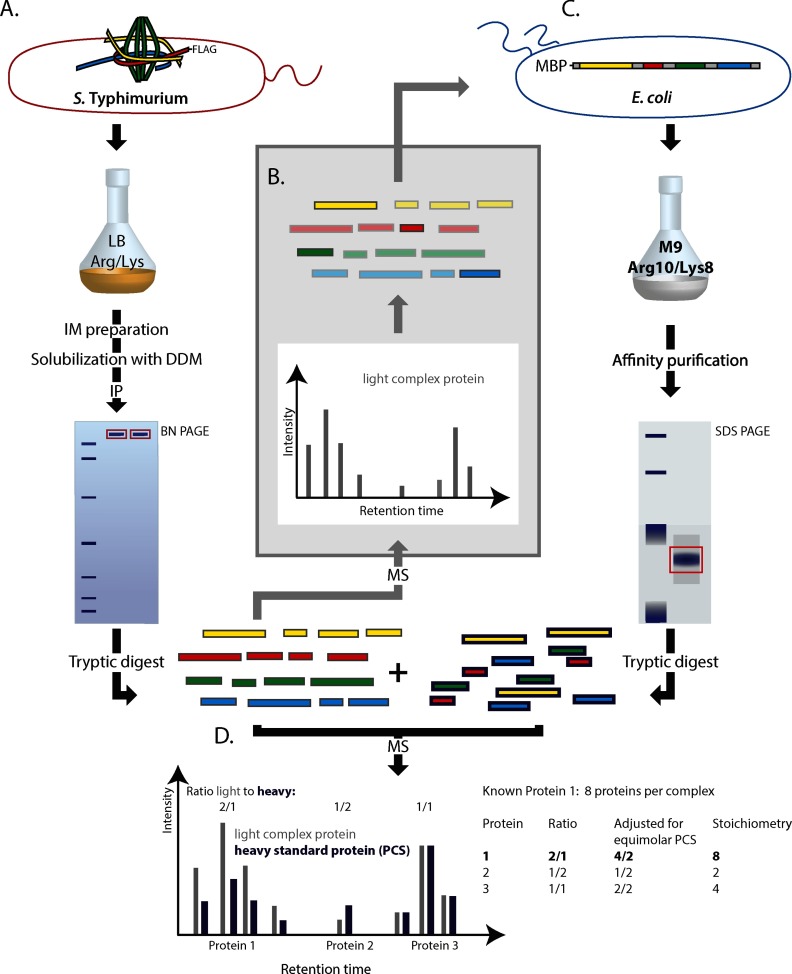
Experimental setup of the peptide concatenated standard strategy. (A) S. Typhimurium expressing needle complexes with FLAG-tagged bait protein (SpaSN258A or InvA) were grown in complex media. Inner membranes (IM) were purified, solubilized by DDM, and needle complexes were immunoprecipitated. Needle complexes were separated by blue native-PAGE, the corresponding bands were excised, and proteins were subsequently digested with trypsin. (B) Peptides were analyzed by MS and suitable peptides were selected for concatenation into the PCS. (C) The PCS was expressed as MBP-fusion in Arg and Lys auxotroph E. coli grown in defined medium containing heavy arginine and lysine. After purification via MBP, the PCS was run on a 10%/4% SDS-PAGE, mixed with purified needle complex and digested together with trypsin in gel. (D) Digested peptides from needle complex (A) and PCS (C) were mixed and analyzed by MS. The ratio of the evidence of light and heavy peptides was calculated for each protein. Ratios were transformed to absolute stoichiometries by normalization with the evidence-ratios of peptides of proteins of known stoichiometry. Abbreviations: IM: inner membrane.
These approaches have been used successfully to study the stoichiometry of protein assemblies of low complexity, hydrophobicity, and/or stoichiometric range (e.g. (25, 27–29), but the analysis of large, heterogeneous, and hydrophobic membrane protein complexes remains a challenge. Using two optimized complementary needle complex purification protocols and MS analysis, we have been able to reliably deduce the stoichiometry of the T3SS encoded by SPI-1 of S. Typhimurium. We reproduced the structure-based stoichiometry of 24:24:15 for PrgH, PrgK, and InvG, which validated our approach, and confirmed the proposed nonameric stoichiometry of InvA for the first time in situ. More importantly, we report evidence that the export apparatus of this T3SS contains five SpaP, one SpaQ, one SpaR, and one SpaS. Combined with the predicted transmembrane topologies of these proteins, this stoichiometry suggests that the inner membrane patch of the needle complex base houses 104 transmembrane domains in total, a dense assembly whose function in the secretion process can now be studied in greater detail. Furthermore, we present evidence that only one helical turn composed of six inner rod proteins PrgJ anchors the needle to the base, which may have implications for its suggested function in needle length control and substrate specificity switching (30).
The stoichiometry information provided in this study will facilitate further structural and functional studies of type III secretion and of the related flagellar systems and as such help to develop inhibitors of these machines central to the virulence of many pathogens. By combining gentle membrane protein complex purification and quantitative mass spectrometry, we show that MS-based stoichiometry determination can be extended to investigating highly hydrophobic complexes of a wide stoichiometric range such as multiple other bacterial protein secretion machines.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Detergent n-dodecyl-β-d-maltoside (DDM) was from Affimetrix-Anatrace (Maumee, OH). SERVA Blue G and SERVAGel™ TG PRiME™ 8–16% precast gels were from Serva (Heidelberg, Germany). NativePAGE Novex Bis-Tris 3–12% gels were from Life Technologies (Carlsbad, CA). Primers were synthetized by Eurofins (Ebersberg, Germany) and Integrated DNA Technologies (Coralville, IA). Arg10 (13C6, 15N4 - arginine) was from Cambridge Isotope Laboratories (Tewksbury, MA) and Lys8 (13C6, 15N2 - lysine) was from Silantes (Munich, Germany). Stable isotope-labeled peptides were from Thermo Fisher (Waltham, MA).
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table S1. All S. Typhimurium strains were derived from strain SL1344 (31). S. Typhimurium strains were grown at 37 °C in LB broth supplemented with 0.3 m NaCl with low aeration to enhance expression of genes of SPI-1. pMAL-c5x-PCS 1 and pMAL-c5x-PCS 2 were cloned by Gibson cloning according to published protocols (32) with primers listed in Table S2. For PCS purification, Escherichia coli strain AT713 (e.g. argA21, lysA22) was grown in defined M9 medium (Table S3) at 27 °C to 37 °C and 200 rpm, supplemented with 0.3 mm IPTG to induce expression of the maltose binding protein (MBP) fusion from plasmid pMAL-c5x. Cultures were supplemented as required with streptomycin (50 μg/ml), tetracycline (12.5 μg/ml), or ampicillin (100 μg/ml).
Secretion Assay
Analysis of type III-dependent secretion of proteins into the culture medium was carried out as described previously (33).
Immunoblotting
For protein detection, samples were subjected to SDS-PAGE using SERVAGel™ TG PRiME™ 8–16% precast gels (Serva), transferred onto a PVDF membrane (Bio-Rad, Hercules, CA), and probed with primary antibodies anti-SipB, anti-InvJ, anti-PrgH, antiSpaSN, or M2 anti-FLAG. Secondary antibodies were goat anti-mouse IgG DyLight 800 conjugate and goat anti-rabbit IgG DyLight 680 conjugate (Thermo Scientific Pierce, Rockford, IL). Detection was performed using the Odyssey imaging system (Li-Cor, Lincoln, NE).
Needle Complex Purification by Immunoprecipitation
Membrane fractionation, solubilization, and immunoprecipitation (IP) were carried out as previously described (4, 34).
Needle Complex Purification by CsCl Gradient Centrifugation
Purification of needle complexes of wild-type and SpaSN258AFLAG bacteria was carried out as published previously (3, 33), but n-dodecyl-N,N-dimethylamine-N-oxide was replaced by DDM (0.7% for lysis/extraction, 0.1% for maintenance) for lysis of cells and extraction of needle complexes throughout the protocol. Furthermore, an initial concentration of 35% (w/v) of CsCl was used to prepare the gradient.
Electron Microscopy
Purified needle complexes were negatively stained with 1% aqueous uranyl acetate on carbon-coated copper grids. Micrographs were recorded using a JEM-1400Plus (JEOL, Tokyo, Japan) microscope at 120 kV.
Blue Native-PAGE
Blue native-PAGE of purified needle complexes was carried out as previously described (4).
Protein In-Gel Digestion
For identification and selection of suitable peptides for the design of peptide-concatenated standards (PCS), the stained protein bands corresponding to the needle complex were excised from blue native-PAGE gels and in-gel digested with trypsin (35). We performed a pilot experiment to evaluate whether two consecutive in-gel digests of the same gel piece would improve the yield of tryptic peptides qualitatively (additional unique peptides) and quantitatively (more evidence of high intensity of the same peptide) (data not shown). This procedure resulted in a quantitative improvement of up to 20% for all peptides identified and sequenced by tandem mass spectrometry; the yield of four critical peptides of needle complex components was even improved by 40% or more (SpaR GATHVLE, InvA AGIIDADAAR, PrgK LYSAIEQR, and SpaQ MDDLVFAGNK). Of the peptides identified in the second digest, up to 38% were not identified in the first digest. Because of these improvements, we decided to routinely use two consecutive digests and extractions. After each step, extracted peptides were desalted using C18 StageTips (36). Corresponding eluates were combined and subjected to LC-MS/MS analysis. For PCS analysis, blue native-gel bands containing the purified complex were mixed with the gel bands containing the “heavy” labeled purified PCS construct prior to in gel digestion.
For quantification using stable isotope-labeled peptides, combined eluates were divided into three equal parts. The synthetic stable isotope-labeled peptides QVIFLALAK (SpaQ) and VGVPVIVDIK (SpaS), labeled with Lys8 on the C terminus (Thermo Fisher Scientific), were added to the digested proteins in three different amounts: 0.375, 0.75, and 1.5 pmol. The peptide mixtures were subjected to LC-MS/MS analysis.
Mass Spectrometry
LC-MS/MS analyses were performed either on an EasyLC II nano-HPLC (Proxeon Biosystems) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific), or on an EasyLC 1000 nano-UHPLC (Proxeon Biosystems, Odense, Denmark) coupled to a Q Exactive HF mass spectrometer (Thermo Scientific), both as described previously (37, 38). Peptide mixtures were injected onto the column in HPLC solvent A (LTQ Orbitrap Elite: 0.5% acetic acid; Q Exactive HF: 0.1% formic acid) at a flow rate of 500 nl/min and subsequently eluted with either a 116 min (PCS analyses) of 5–33-50–90% of HPLC solvent B (80% acetonitrile in 0.5% acetic acid) or a 57 min gradient (quantification using stable isotope-labeled) of 10–33-50–90% of HPLC solvent B (80% acetonitrile in 0.1% formic acid). During peptide elution, the flow rate was kept constant at 200 nl/min. For Orbitrap Elite analysis, the 20 most intense precursor ions were sequentially fragmented in each scan cycle using collision-induced dissociation, and sequenced precursor masses were excluded from further selection for 60 s. In some cases, an inclusion list containing ions of light- and heavy-labeled PCS peptides was applied. For Q Exactive HF, the seven most intense precursor ions were sequentially fragmented in each scan cycle using HCD fragmentation. For quantification using stable isotope-labeled, full scan MS spectra were acquired in a mass range from m/z 480–705. An inclusion list containing ions of light- and heavy-labeled peptides QVIFLALAK and VGVPVIVDIK was applied, and no additional masses were allowed for fragmentation. Full scan resolution was set to 120,000. The target values for the MS scan and MS/MS fragmentation were 3 × 106 and 105 charges (Q Exactive HF) or 106 and 5 × 103 charges (Orbitrap Elite). The maximal injection time for MS/MS fragmentation was 110 ms and 25 ms, respectively.
Mass Spectrometry Data Processing
The MS data were processed with a setting of 1% for the false discovery rate using MaxQuant software (versions 1.2.2.9 and 1.5.2.8) as described previously (37, 39, 40) with slight modifications. A database search was performed using the Andromeda search engine (40), which is part of MaxQuant. MS/MS spectra were searched against a target database consisting of 10,152 protein entries from S. Typhimurium (uniprot 99287, March 1, 2009 and uniprot 216597, July 11, 2012), a database containing the PCS sequences and 248 commonly observed contaminants. In the database search, full tryptic specificity was required and up to two missed cleavages were allowed. Carbamidomethylation of cysteine was set as fixed modification, protein N-terminal acetylation and oxidation of methionine were set as variable modifications. Initial precursor mass tolerance was set to 6 ppm (versus 1.2.2.9) and 4.5 ppm (versus 1.5.2.8), respectively, and at the fragment ion level 0.5 Da was set for collision-induced dissociation fragmentation and 20 ppm for Higher-energy collisional dissociation (HCD) fragmentation. Quantitative MS data acquired using isotope-labeled peptides were processed either with or without restrictions at the false discovery rate. In the latter case, data were filtered manually by setting the posterior error probability (41) threshold to 0.01.
Design of Peptide-Concatenated Standards (PCS)
PCS candidates were chosen from a pool of peptides with a mascot score of >15, preferably without oxidized or miss cleaved forms (data not shown). Two PCSs were assembled containing a total of two to five tryptic peptides of each protein of interest. To retain native digestion behavior, one to four flanking amino acids were added before and behind each peptide. When the peptide was at the very N-terminal or C-terminal end of the protein of interest or when it was surrounded by arginine or lysine, alanins were added instead of the native sequence. In PCS 2, the native succession of peptides was retained when successive peptides or peptides in close proximity (separated by 10 amino acids or less) were found suitable for the PCS strategy. The synthetic PCS were fused to the C terminus of MBP using pMAL-c5x to enhance expression and solubility, and to facilitate purification.
PCS Purification
PCSs were expressed fused to MBP from a pMAL-c5x plasmid in AT713, grown in M9 medium containing Arg10 (13C6, 15N4-arginine) and Lys8 (13C6, 15N2-lysine). MBP-PCSs were purified using an amylose resin (New England Biolabs, Ipswich, MA). The eluted MBP-PCS was then run on a 10%/4% SDS-PAGE, which was subsequently stained with colloidal Coomassie. The 90 kDa band corresponding to PCS-MBP was cut from the 4% section of the gel and stored in 5% acetic acid until further use.
Selection of Stable Isotope-Labeled Peptides
Stable isotope-labeled peptides were chosen from the pool of selected PCS peptides. The criteria for selection were: a) no missed cleavage sites, b) no oxidized methionines, c) 6 to 15 amino acids in length, and d) a high intensity for reliable quantification. We selected peptides QVIFLALAK for SpaQ and VGVPVIVDIK for SpaS.
Ratiometric Quantification of Stoichiometry
For relative quantification of the components of the needle complex, we used the light/heavy ratios of the intensities of individual peptide sequencing events, comprising fragmentation, quantification, and identification, which are termed evidence hereafter. For needle complexes purified by IP or CsCl-gradient centrifugation and measured together with the PCSs, the following filter criteria and calculations were applied: a) Evidence measured in the last 10 min of each run were excluded. b) Evidence with a posterior error probability equal or higher than 0.01 were excluded. c) To compare the light/heavy intensity ratios of the evidence between the replicates, the values were transformed into a stoichiometry by normalizing with peptides from proteins of known stoichiometry: the InvG peptide SLLVGGYTR was used for normalization for data obtained with immunoprecipitated needle complexes, the PrgK peptide SDAQLQAPGTPVKR was used for normalization of data obtained with needle complexes purified by CsCl-gradient centrifugation. d) From these combined datasets for IP-PCS experiments and CsCl-PCS experiments, respectively, outliers were identified as two median absolute deviations from the median. For the analysis of the ratio of SpaQ and SpaS in purified needle complexes using stable isotope-labeled peptides, only the evidence with the highest intensity was used to calculate protein ratios, which typically accounted for 90–100% of the peptides total intensity. The resulting raw data identifiers used for calculating the mean stoichiometry of each protein can be found in Table S4.
Experimental Design and Statistical Rationale
Several measures were taken to minimize systematic errors. a) As IP baits, the two last-assembling export apparatus components were selected to ensure complete assembly of purified complexes. b) Two complementary strategies, IP and CsCl gradient centrifugation, were used for purification of T3SS needle complexes to account for potential purification biases. c) In-gel denaturing of proteins of native needle complexes by SDS and separation of MBP-PCS on a 4% SDS-PAGE were undertaken to match the state of the proteins for in-gel trypsinization and avoid biases because of the folding states of the proteins to be compared. d) To account for trypsinization biases between PCS and full-length analytes, four flanking residues were included at each side of a cleavage site for construction of the PCS. e) Analysis of selected proteins (SpaQ and SpaS) was performed using stable isotope-labeled peptides to validate results obtained using PCS.
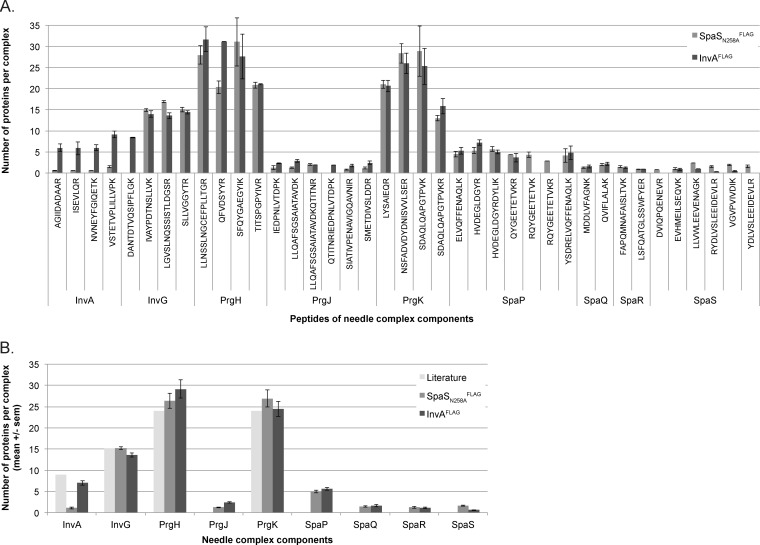
For quality control of analytical and biological reliability, two independent measures were taken: a) Three proteins of known stoichiometry were included in the analysis: PrgH, PrgK, and InvG. Since stoichiometry data obtained for these proteins were within 10% of the literature data, results obtained for proteins of hitherto unknown stoichiometry can be trusted. b) For PCS analysis, stoichiometries were determined from measurements of up to seven different peptides (Fig. 5). The very hydrophobic membrane proteins SpaQ and SpaR allowed for the inclusion of only 2 peptides, however.
Fig. 5.
Stoichiometry calculations based on normalized stable isotope labeling in cell culture ratios of evidence of peptides from immunoprecipitated needle complexes and peptide concatenated standards. (A) Normalized ratios of light and heavy evidence of the indicated peptides are shown. Normalization using InvG peptide SLLVGGYTR = 15. Error bars: standard error of the mean. (B) Literature values of stoichiometry and averages of normalized ratios of light and heavy evidence of the indicated proteins are shown. The literature value for InvA is based on the crystal structure of the C-terminal domain of the isolated Shigella homologue MxiA.
The experimental results presented were obtained with the following sample sizes: Sample sizes for or the central IP-PCS experiment were n = 5 for the IP-bait SpaSN258AFLAG and n = 5 for the IP-bait InvAFLAG, where n denotes biological replicates. Because of the accuracy of the protocol and instrumentation used, no technical replicates were performed. Sample sizes for the validating CsCl-gradient-PCS experiment were n = 1 for the IP-bait SpaSN258AFLAG and n = 1 for the IP-bait InvAFLAG. Sample sizes for the validating IP and CsCl-gradient experiments using stable isotope-labeled peptides were n = 3 for the immunoprecipitated analyte and n = 3 for the CsCl-gradient purified analyte, where n denotes technical replicates using three different concentrations of stable isotope-labeled peptides.
For the PCS strategy, statistical evaluation of the data was performed using Microsoft Excel (version 14.06112.5000) and RStudio (version 0.98.953). Evidence measured in the last 10 min of each run and evidence with posterior error probability ≥ 0.01 were removed from the dataset. Outliers were identified as two median absolute deviations from the median. The ratio of light to heavy intensities of each evidence pair was normalized to compare biological replicates. The mean of normalized evidence pairs of all peptides of a protein was used to calculate the stoichiometry for each experimental set.
The PCS approach was validated for the selected proteins SpaQ and SpaS by MS-based quantification using stable isotope-labeled peptides. For this quantification, light-to heavy-ratios of the evidence pairs were calculated from the most intense evidence of each peptide, representing 90–100% of the peptide's total intensity. Statistical analysis was performed using Microsoft Excel.
The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD003113.
RESULTS
Needle Complex Isolation and Identification of Suitable Standard Peptides
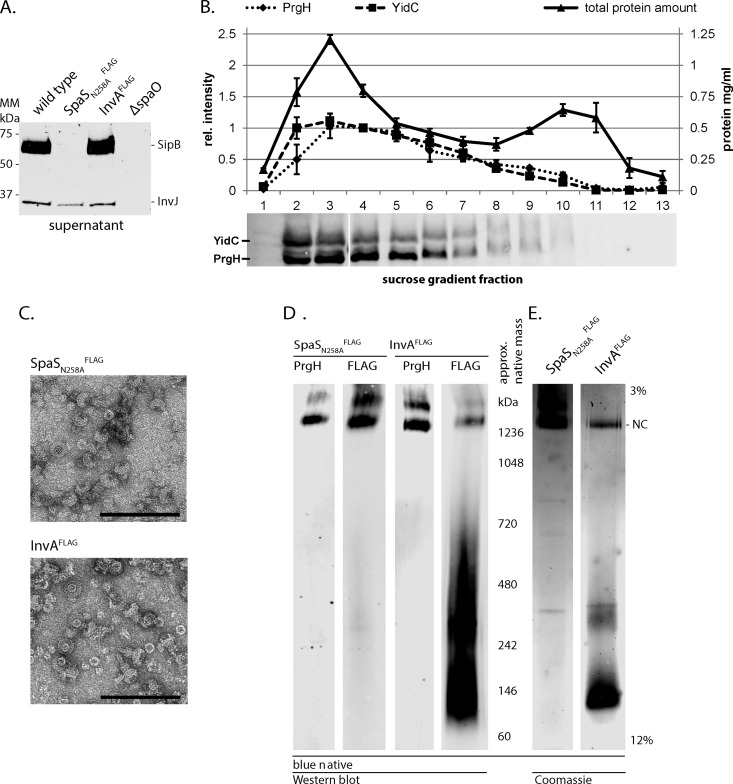
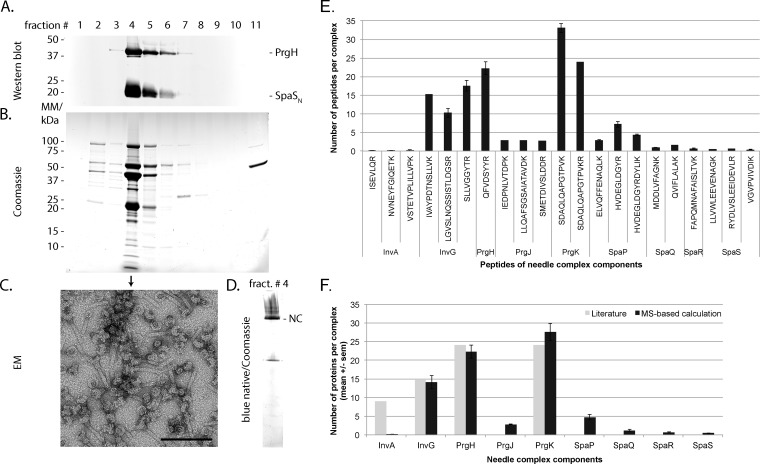
MS-based stoichiometry determination of complexes using PCS or synthetic stable isotope-labeled peptides requires the prior isolation of intact complexes and the identification of suitable standard peptides (Fig. 2). First, we employed a previously established immunoprecipitation (IP)-based protocol (4, 34) to isolate intact T3SS needle complexes from S. Typhimurium. As IP bait, we chose the two last-assembling export apparatus components, SpaS and InvA (brown and green in Fig. 1, respectively). These two proteins require the presence of the other three export apparatus components SpaP, SpaQ, and SpaR for assembly, thus ensuring completeness of the isolated complexes (4, 42). Inner and outer membranes of S. Typhimurium harboring C-terminally FLAG-tagged alleles of either autocleavage-deficient SpaS (Asn258Ala) or InvA, which are fully functional for type III-dependent secretion (Fig. 3A), were fractionated by sucrose density equilibrium centrifugation. Western blotting analysis of the 13 isolated fractions showed that needle complexes peak in fractions three to six as judged by the signal intensities of the inner membrane ring component PrgH (Fig. 3B). These fractions were pooled, membranes solubilized using the mild nonionic detergent dodecyl-maltoside (DDM), and needle complexes were subsequently immunoprecipitated. Natively eluted needle complexes were evaluated by electron microscopy, SDS-PAGE, and Western blotting. The purified material contained mainly bases lacking needles (Fig 3C), possibly caused by the harsh mechanical disruption of bacterial cells by French pressing. For further purification and enabling of in-gel digestion, immunoprecipitated needle complexes were separated by blue native-PAGE. Needle complexes, identified by immunodetection of PrgH and SpaS or InvA, run at an apparent native mass of >1200 kDa (Fig. 3D) as reported previously (4, 34). Bands corresponding to InvA were also observed at apparent native masses of 150 and 250 kDa, respectively, suggesting that InvA is either expressed in excess or extracted easily from the needle complex (Fig. 3D). To identify suitable peptides for stoichiometry determination, the >1200 kDa bands corresponding to needle complexes were cut out (Fig. 3E); contained proteins were denatured by SDS, in-gel digested with trypsin and subsequently analyzed by MS.
Fig. 3.
Purification of needle complexes by immunoprecipitation and blue native-PAGE. (A) Type III secretion into culture supernatant of wild-type and indicated mutant strains was profiled by immunodetection of the early substrate InvJ and the intermediate substrate SipB. ΔspaO denotes a negative control strain defective in type III secretion. (B) Membrane fractions containing needle complexes were identified by immunodetection of the external inner membrane ring protein PrgH. The inner membrane protein YidC served as a marker for the inner membrane. Quantification of the protein bands and of total protein content of the fractions was graphed. The average of three independent experiments is shown. Error bars show the standard deviation. (C) Electron micrographs of immunoprecipitated needle complexes, scale bar = 100 nm. (D, E) Blue native-PAGE of immunoprecipitated needle complexes; (D) shows immunodetection of the indicated proteins; (E) shows a Coomassie-stained gel of immunoprecipitated material.
Construction, Expression, and Purification of the Peptide Concatenated Standard
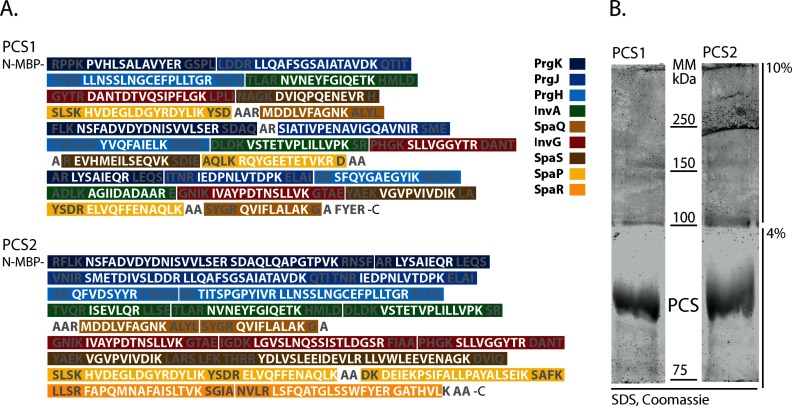
Suitable peptides (Table S5) were selected for concatenation into two different PCSs (Fig. 4A). Oxidation and acetylation-prone peptides were included in the PCSs if too few other suitable peptides were identified for the respective proteins and if the degree of the modification was stable across different experiments and comparable between sample and standard. Where appropriate, 1–4 flanking residues of the chosen peptides were included to accommodate possible effects of flanking residues on trypsinization efficiencies (Fig. 4A). To enhance the solubility of the synthetic protein, the PCSs were constructed as translational fusions with MBP. MBP-PCSs were expressed in the lysine and arginine auxotrophic E. coli strain AT713 in defined media supplemented with heavy arginine and lysine. Heavy-isotope-labeled PCSs facilitated MS-based discrimination of signature peptides of PCSs and needle complexes. MBP-PCSs were purified and subsequently run on a specially designed SDS-PAGE (Fig. 4B) composed of a 10% section for separation followed by a 4% section for extraction. This gel composition was used to achieve a similar acrylamide concentration for in-gel digestion of the SDS-PAGE-separated MBP-PCS and of the blue native-PAGE-separated needle complex. MBP-PCS bands were cut out, in-gel digested, and analyzed by MS. The amount of unlabeled PCS peptides was found to be negligible at below 2% of the total intensity of the respective peptides (Table S6).
Fig. 4.
Sequence and purification of peptide concatenated standards. (A) Sequence of PCS 1 and PCS 2. White: standard peptides. Gray: flanking residues added to provide natural context for trypsin cleavage. Color-coding for the individual needle complex components as indicated in the legend. (B) SDS-PAGE of MBP-PCS1 and MBP-PCS 2 with upper 10% acrylamide gel for separation and lower 4% acrylamide gel for extraction.
Stoichiometry Determination of Immunoprecipitated Needle Complexes by PCS Strategy
To determine the stoichiometry of needle complex components, blue native-gel bands containing needle complexes with light arginine and lysine and SDS-gel bands containing MBP-PCSs with heavy Arg10 and Lys8 were codigested and analyzed by MS. The light/heavy intensity ratios of evidence pairs from a total of 10 runs were used for calculation of stoichiometry. Because the amount of complex and PCS varied after each purification, light/heavy ratios were normalized using the robustly detected SLLVGGYTR peptide of InvG, for which a stoichiometry of 15 has been reported (12). Outliers were identified as two median absolute deviation from the median and disregarded for further analysis. We obtained a stoichiometry of the control proteins InvG:PrgH:PrgK of 15:26:27 and 14:28:24 for the SpaSN258AFLAG and InvAFLAG IPs, respectively (Fig. 5). These results are in good agreement with the reported ratio of 15:24:24 (12). The stoichiometry of the minor export apparatus components SpaP:SpaQ:SpaR:SpaS was 5:1:1:1 in both preparations. The major export apparatus protein InvA was largely absent in preparations immunoprecipitated with SpaSN258AFLAG as bait but showed a stoichiometry of seven proteins per needle complex when baiting with InvAFLAG. Finally, 2–3 proteins of the inner rod protein PrgJ were detected per needle complex.
Stoichiometry Determination of CsCl Gradient Centrifugation-Purified Needle Complexes by PCS Strategy
The SpaS/InvA IP-based purification of needle complexes was chosen to obtain completely assembled needle complexes in respect to export apparatus components. Unfortunately, this protocol also yielded complexes with broken needles, probably due to bacterial lysis by French pressing. Shearing of needles might also result in a loss of inner rod protein and thus be responsible for the low number of detected PrgJ in the samples. To accommodate for this problem, we chose to also analyze the stoichiometry of needle complexes extracted from bacteria without mechanical disruption but enzymatic (lysozyme) digestion and detergent solubilization, followed by CsCl density equilibrium centrifugation-based needle complex purification. This classical protocol (3) typically results in a severe loss of some export apparatus components due to the use of the stringent detergent n-dodecyl-N,N-dimethylamine-N-oxide (4). To retain the export apparatus components and at the same time keep needles intact, we replaced n-dodecyl-N,N-dimethylamine-N-oxide by the milder detergent DDM. Using this modified protocol, CsCl gradient fractions 4 and 5 showed an enrichment of intact needle complexes and bases as judged by electron microscopy, SDS-PAGE, and Western blotting (Figs. 6A-6C). Purified needle complexes were separated by blue native-PAGE (Fig. 6D) and their stoichiometry analyzed by MS as described above, except that the PrgK peptide SDAQLQAPGTPVKR was used for normalization.
Fig. 6.
Stoichiometry determination of CsCl gradient-purified needle complexes using peptide concatenated standards. (A) CsCl gradient fractions containing needle complexes were identified by immunodetection of the external inner membrane ring protein PrgH and of the switch protein SpaS. (B) Coomassie-stained polyacrylamide gels of CsCl fractions. (C) Electron micrographs showing bases and needle complexes in fraction 4 of the CsCl gradient, scale bar = 100 nm. (D) Coomassie-stained blue native-polyacrylamide gel of purified needle complexes. Abbreviations: EM: electron microscopy. (E) Normalized ratios of light and heavy evidence of the indicated peptides are shown. Normalization using PrgK peptide SDAQLQAPGTPVKR = 24. Error bars: standard error of the mean. (F) Literature values of stoichiometry and averages of normalized ratios of light and heavy evidence of the indicated proteins are shown. The literature value for InvA is based on the crystal structure of the C-terminal domain of the isolated Shigella homologue MxiA.
Corroborating the stoichiometry calculations based on immunoprecipitated needle complexes, we obtained average stoichiometries of the control proteins InvG:PrgH:PrgK of 14:22:27 and stoichiometries of the minor export apparatus proteins SpaP:SpaQ:SpaR:SpaS of 5:1:1:1 (Figs. 6E and 6F). The major export apparatus protein InvA was entirely absent from these preparations, and hence, its stoichiometry could not be calculated. The stoichiometry of PrgJ was three per complex as obtained by analysis of immunoprecipitated needle complexes.
Stoichiometry Determination of SpaQ and SpaS Using Stable Isotope-Labeled Peptides
To validate the obtained results for the export apparatus components SpaQ and SpaS, we chose to quantify the ratio of the two proteins using synthetic stable isotope-labeled peptides. Tryptic digests of needle complexes purified by both methods, IP and CsCl gradients, were spiked with three different concentrations of the standard peptides QVIFLALAK (for SpaQ) and VGVPVIVDIK (for SpaS) at equimolar ratios, and the mixtures were subsequently analyzed by MS. Heavy-to-light ratios were calculated from the most intense evidence pair of each peptide, representing 90–100% of the peptide's total intensity. Based on these results, we obtained average ratios of SpaQ to SpaS of 0.99 and 0.76, respectively, for analysis of needle complexes isolated by the two different methods (Fig. 7). These results corroborate the calculated stoichiometry of 1:1 deduced from PCS-based experiments.
Fig. 7.
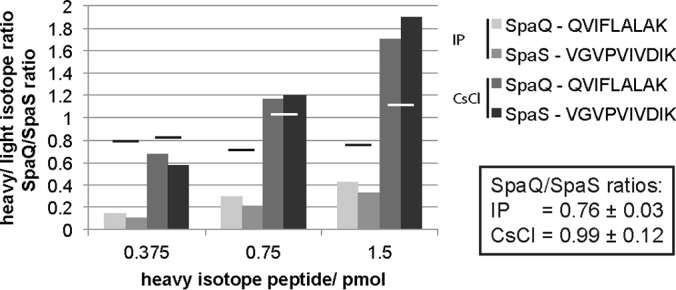
Determination of the ratio of SpaQ and SpaS using synthetic stable isotope-labeled peptides. Trypsin digests of needle complexes purified by IP or CsCl-gradient, respectively, were spiked with stable isotope-labeled peptides at indicated concentrations and analyzed by MS. Columns show the ratio of heavy (stable isotope-labeled peptides) to light (needle complex) evidence for each measurement. Bars show the ratio of SpaQ to SpaS for each pair in a measurement. Average values ± S.D. of SpaQ/SpaS ratios are presented in the box.
DISCUSSION
Needle complexes of T3SSs are complex macromolecular machines consisting of up to 20 different proteins with one to several hundred copies each. While the stoichiometry of some components had been structurally elucidated, the stoichiometry of the membrane-embedded export apparatus components was largely unknown, primarily because of technical challenges.
We adapted two MS-based relative quantification strategies to evaluate the stoichiometry of complete T3SS needle complexes. The preparation of intact and homogeneous complexes is key to the successful implementation of these approaches (43) but particularly difficult for large membrane protein complexes. We solved this critical problem using a combination of mild detergent extraction, immunoprecipitation or CsCl-gradient-based purification, and blue native-PAGE. The subsequent analysis of these hydrophobic complexes by quantitative mass spectrometry was challenged by two major obstacles: a) the detection of only a limited number of peptides that were suitable for quantification of the highly hydrophobic membrane proteins and b) the bias in the accessibility of peptides from intact complexes and standards. Membrane proteins contain only a few charged residues, hence also only a few cleavage sites for trypsin, cutting after arginine or lysine. In addition, the presence of tightly bound lipids or detergents may preclude the efficient cleavage of membrane proteins by site-specific proteases, further reducing the yield of suitable peptides for mass spectrometrical analysis. We were able to increase the yield of four critical peptides of the needle complex by implementing two consecutive tryptic digests and extractions of each gel piece. However, despite this effort, for some proteins, only few peptides were found, and therefore, modification-prone peptides were considered for quantification. We identified four suitable peptides containing oxidation-prone methionines and two peptides prone to N-terminal acetylation. Since the degree of oxidation and acetylation was stable throughout different experiments, we could include these peptides for quantification of the stoichiometry.
Even though suitable tryptic peptides were identified for the proteins of interest, several factors needed consideration that could potentially influence the yield of these peptides and could bias quantification: Trypsinization efficiency depends on the sequence context of the respective peptides (25), bound lipids or detergents (44), and the folding state of proteins (45); and the acrylamide concentration of gels may affect the peptide extraction efficiency. The use of PCSs allows minimization of differences in peptide yield because the peptide context and the preparation of PCS and sample are largely congruent. To align the digestion efficiencies of PCS and sample, we retained the sequence context around the tryptic digestion sites of the peptides and denatured blue native-PAGE-separated complex components by SDS treatment prior to in-gel digestion. Furthermore, we aligned the polyacrylamide concentrations of the PCS-containing gel and of the complex-containing blue native-gel to achieve similar efficiencies of peptide extraction. In contrast, synthetic stable isotope-labeled peptides are not subjected to the same preparative procedures as the complex components of interest. While concentrations of the synthetic peptides are exactly known and invariable, a benefit for absolute quantification, variations in the efficiencies of sample protein digestion and peptide extraction, which are not matched by the synthetic peptides, are likely to skew stoichiometry determination. Therefore, we implemented only two suitable synthetic stable isotope-labeled peptides for the validation of the stoichiometry of the export apparatus components SpaQ and SpaS but refrained from using this approach for the global analysis of the stoichiometry of the complete needle complex.
Using the above-discussed strategy, we confirmed the reported stoichiometry of the base components PrgH, PrgK, and InvG and of the major export apparatus protein InvA. We further report a stoichiometry of 5:1:1:1 for the minor export apparatus components SpaP, SpaQ, and SpaR and for the switch protein SpaS. Our data also suggest a loose association of InvA with the other needle complex components, and we present evidence for a low stoichiometry of the inner rod protein PrgJ in the needle complex.
Cryo electron microscopy structural analysis of needle complexes from the SPI-1 T3SS of S. Typhimurium revealed a stoichiometry of PrgH and PrgK of 24 and of InvG of 15 (12). In this study, we yielded MS-based stoichiometries of PrgH, PrgK, and InvG that deviate in average only by 8.4% from the literature values, which imparts confidence for the obtained stoichiometries of the other needle complex components.
Based on a crystal structure, it has been proposed that the isolated Shigella InvA homologue MxiA forms a nonamer, but confirmation based on direct measurements of InvA as part of the assembled needle complex was missing. Here, we determined a stoichiometry of seven InvA per assembled needle complex, which is close to the proposed MxiA stoichiometry and to the reported stoichiometry of the flagellar homolog FlhA (24, 46). Interestingly, it was only possible to detect InvA in needle complexes coimmunoprecipitated when baiting with InvAFLAG itself. InvA was almost entirely lost after IP of needle complexes by SpaSN258AFLAG or needle complex isolation using mild extraction and CsCl gradient centrifugation, and needle complexes coimmunoprecipitated at a lower efficiency with InvAFLAG as bait than with SpaSN258AFLAG. The easy extraction of InvA from needle complexes, even by mild detergents, indicates that InvA is only loosely associated with the base and the other export apparatus components, and it might also mean that the herein reported stoichiometry of seven per needle complex is still an underestimation.
Early studies of the flagellar system proposed a stoichiometry of 4–5 of the SpaP homolog FliP and 1–3 of the SpaR homolog FliR based on quantitative Western-blotting and densitometry of autoradiograms (47). The data presented in this study suggest a similar stoichiometry of five SpaP and one SpaR for Salmonella's virulence-associated T3SS on SPI-1. We furthermore deduced a stoichiometry of one SpaQ and one SpaS, for which no previous reports exist. Clostridium encodes a natural fusion of the flagellar homologs of SpaR and SpaS, FliR and FlhB, respectively. An artificial fusion of these proteins is also functional in the flagellar system of Salmonella (48). These observations suggest an equimolar stoichiometry of FliR and FlhB, which, together with our data, support a stoichiometry of one SpaQ, one SpaR, and one SpaS. We could show previously that SpaP and SpaR form the cup substructure at the center of the base of the needle complex (4). Given a stoichiometry of five SpaP, it is conceivable that a circular assembly of SpaP, possibly in conjunction with SpaR, constitutes the substrate translocation pore of the system in the inner membrane.
PrgJ is the inner rod protein anchoring the needle to the base (49). As the term “inner rod” implies, it was originally thought to constitute the substrate conduit inside the base while the needle extends this conduit on the outside. However, the number of PrgJ subunits forming the inner rod is unclear and so is its length. It is thought that PrgJ assembles into a helix analogous to the needle structure, and crosslinks of up to six consecutive PrgJ were reported (30), suggesting at least one complete turn of PrgJ. Here, we report the detection of three PrgJ proteins per needle complex, independent of the method of needle complex isolation. Different to the other investigated needle complex components, PrgJ itself is a substrate of the T3SS and can only be secreted and assembled into the needle complex after completion of the principal secretion competent machine. It is possible that not all measured complexes were competent for secretion, and hence, we might underestimate the number of PrgJ in needle complexes. We chose two complementing approaches of needle complex isolation to minimize this problem. As mentioned before, IP of needle complexes with InvA as bait ensured that only complete base-export apparatus assemblies were isolated; however, nothing can be said about the state of assembly and the functionality of the associated cytoplasmic components. Since needles were sheared off during the course of this isolation protocol, the presence of needles cannot be taken as an indicator for functional secretion. The latter can, however, be used to estimate the number of fully functional needle complexes in preparations from CsCl-gradients. More than half of all needle complexes purified by CsCl-gradient fractionation contained needles (Fig. 6C). If also half of the measured complexes contain inner rods, a stoichiometry of six PrgJ results for these needle complexes, which supports the view that PrgJ forms only one turn of the inner rod helix that anchors the needle to the base. This low stoichiometry of the inner rod has conceptual implications for the timer model of needle length control, which presumes that the duration of PrgJ assembly determines the duration of needle assembly and hence needle length (2, 30, 49).
In summary, we have presented evidence that the export apparatus of bacterial T3SSs is composed of nine InvA, five SpaP, one SpaQ, one SpaR, and one SpaS, which suggests a total of 104 transmembrane domains within the membrane patch of the base of these systems. We furthermore show that InvA is only loosely associated with the other needle complex components and that not more than six PrgJ form the inner rod of the needle complex. This information will facilitate further structural and functional analyses of these and of the related flagellar systems and as such help to develop antiinfective strategies targeting these machines central to the virulence of many pathogens. In addition, the herein presented extension of MS-based stoichiometry determination to investigating highly hydrophobic complexes of very heterogeneous composition and wide stoichiometric range may foster the analysis of other membrane-spanning protein complexes, in particular of different virulence-associated bacterial secretion systems.
Supplementary Material
Acknowledgments
We thank Silke Wahl for technical assistance in MS sample preparation. Thomas Marlovits is acknowledged for providing antisera against SpaSN. Jan-Willem de Gier is acknowledged for providing antisera against YidC. Lea Krampen, Julia Monjarás Feria, and Mehari Tesfazgi Mebrhatu are thanked for critically reading the manuscript.
Footnotes
Author contributions: S.Z., M.F., J.E.G., B.M., and S.W. designed the research; S.Z., M.F., Y.S., and S.W. performed the research; S.Z., M.F., Y.S., B.M., and S.W. analyzed data; and S.Z., M.F., J.E.G., B.M., and S.W. wrote the paper.
* This work was supported by a postdoctoral fellowship of the Human Frontiers Science Program (to S.W.) and by the Alexander von Humboldt Foundation in the framework of the Sofja Kovalevskaja Award endowed by the Federal Ministry of Education and Research (to S.W.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- DDM
- n-dodecyl-β-D-maltoside
- HCD
- Higher-energy collisional dissociation
- IM
- inner membrane
- IP
- immunoprecipitation
- IPTG
- isopropyl-β-D-thiogalactopyranoside
- MBP
- maltose-binding protein
- OM
- outer membrane
- PCS
- peptide concatenated standard
- SPI-1m
- Salmonella pathogenicity island 1
- T3SS
- type III secretion system.
REFERENCES
- 1. Abby S. S., and Rocha E. P. C. (2012) The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8, e1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galán J. E., Lara-Tejero M., Marlovits T. C., and Wagner S. (2014) Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kubori T., Matsushima Y., Nakamura D., Uralil J., Lara-Tejero M., Sukhan A., Galán J. E., and Aizawa S. I. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 [DOI] [PubMed] [Google Scholar]
- 4. Wagner S., Königsmaier L., Lara-Tejero M., Lefebre M., Marlovits T. C., and Galán J. E. (2010) Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U.S.A. 107, 17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akeda Y., and Galán J. E. (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915 [DOI] [PubMed] [Google Scholar]
- 6. Lara-Tejero M., Kato J., Wagner S., Liu X., and Galán J. E. (2011) A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Notti R. Q., Bhattacharya S., Lilic M., and Stebbins C. E. (2015) A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat. Commun. 6, 7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diepold A., Kudryashev M., Delalez N. J., Berry R. M., and Armitage J. P. (2015) Composition, formation, and regulation of the cytosolic C-ring, a dynamic component of the type III secretion injectisome. PLoS Biol. 13, e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu B., Morado D. R., Margolin W., Rohde J. R., Arizmendi O., Picking W. L., Picking W. D., and Liu J. (2015) Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl. Acad. Sci. U.S.A. 112, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burkinshaw B. J., and Strynadka N. C. J. (2014) Assembly and structure of the T3SS. Biochim. Biophys. Acta 1843, 1649–1663 [DOI] [PubMed] [Google Scholar]
- 11. Hodgkinson J. L., Horsley A., Stabat D., Simon M., Johnson S., da Fonseca P. C. A., Morris E. P., Wall J. S., Lea S. M., and Blocker A. J. (2009) Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schraidt O., and Marlovits T. C. (2011) Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331, 1192–1195 [DOI] [PubMed] [Google Scholar]
- 13. Kowal J., Chami M., Ringler P., Müller S. A., Kudryashev M., Castaño-Díez D., Amstutz M., Cornelis G. R., Stahlberg H., and Engel A. (2013) Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure 21, 2151–2161 [DOI] [PubMed] [Google Scholar]
- 14. Kudryashev M., Stenta M., Schmelz S., Amstutz M., Wiesand U., Castaño-Díez D., Degiacomi M. T., Münnich S., Bleck C. K., Kowal J., Diepold A., Heinz D. W., Dal Peraro M., Cornelis G. R., and Stahlberg H. (2013) In situ structural analysis of the Yersinia enterocolitica injectisome. Elife 2, e00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loquet A., Sgourakis N. G., Gupta R., Giller K., Riedel D., Goosmann C., Griesinger C., Kolbe M., Baker D., Becker S., and Lange A. (2012) Atomic model of the type III secretion system needle. Nature 486, 276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zarivach R., Deng W., Vuckovic M., Felise H. B., Nguyen H. V., Miller S. I., Finlay B. B., and Strynadka N. C. J. (2008) Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453, 124–127 [DOI] [PubMed] [Google Scholar]
- 17. Deane J. E., Graham S. C., Mitchell E. P., Flot D., Johnson S., and Lea S. M. (2008) Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol. Microbiol. 69, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiesand U., Sorg I., Amstutz M., Wagner S., van den Heuvel J., Lührs T., Cornelis G. R., and Heinz D. W. (2009) Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J. Mol. Biol. 385, 854–866 [DOI] [PubMed] [Google Scholar]
- 19. Lountos G. T., Austin B. P., Nallamsetty S., and Waugh D. S. (2009) Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci 18, 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saijo-Hamano Y., Imada K., Minamino T., Kihara M., Shimada M., Kitao A., and Namba K. (2010) Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol. 76, 260–268 [DOI] [PubMed] [Google Scholar]
- 21. Bange G., Kümmerer N., Engel C., Bozkurt G., Wild K., and Sinning I. (2010) FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 107, 11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore S. A., and Jia Y. (2010) Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J. Biol. Chem. 285, 21060–21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Worrall L. J., Vuckovic M., and Strynadka N. C. J. (2010) Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 19, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abrusci P., Vergara-Irigaray M., Johnson S., Beeby M. D., Hendrixson D. R., Roversi P., Friede M. E., Deane J. E., Jensen G. J., Tang C. M., and Lea S. M. (2013) Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kito K., Ota K., Fujita T., and Ito T. (2007) A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J. Proteome Res. 6, 792–800 [DOI] [PubMed] [Google Scholar]
- 26. Gerber S. A., Rush J., Stemman O., Kirschner M. W., and Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nanavati D., Gucek M., Milne J. L., Subramaniam S., and Markey S. P. (2008) Stoichiometry and absolute quantification of proteins with mass spectrometry using fluorescent and isotope-labeled concatenated peptide standards. Mol. Cell. Proteomics 7, 442–447 [DOI] [PubMed] [Google Scholar]
- 28. Olinares P. D., Kim J., Davis J. I., and van Wijk K. J. (2011) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23, 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. von Appen A., Kosinski J., Sparks L., Ori A., DiGuilio A. L., Vollmer B., Mackmull M.-T., Banterle N., Parca L., Kastritis P., Buczak K., Mosalaganti S., Hagen W., Andres-Pons A., Lemke E. A., Bork P., Antonin W., Glavy J. S., Bui K. H., and Beck M. (2015) In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lefebre M. D., and Galán J. E. (2014) The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 111, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoiseth S. K., and Stocker B. A. (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 [DOI] [PubMed] [Google Scholar]
- 32. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 33. Monjarás Feria J. V., Lefebre M. D., Stierhof Y.-D., Galán J. E., and Wagner S. (2015) Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. MBio 6, e01459–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fischer M., Zilkenat S., Gerlach R. G., Wagner S., and Renard B. Y. (2014) Pre- and post-processing workflow for affinity purification mass spectrometry data. J. Proteome Res. 13, 2239–2249 [DOI] [PubMed] [Google Scholar]
- 35. Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., Sommer R. J., and Macek B. (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 37. Carpy A., Krug K., Graf S., Koch A., Popic S., Hauf S., and Macek B. (2014) Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (fission yeast). Mol. Cell. Proteomics 13, 1925–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelstrup C. D., Jersie-Christensen R. R., Batth T. S., Arrey T. N., Kuehn A., Kellmann M., and Olsen J. V. (2014) Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J. Proteome Res. 13, 6187–6195 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 41. Käll L., Storey J. D., MacCoss M. J., and Noble W. S. (2008) Posterior error probabilities and false discovery rates: Two sides of the same coin. J. Proteome Res. 7, 40–44 [DOI] [PubMed] [Google Scholar]
- 42. Diepold A., and Wagner S. (2014) Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 38, 802–822 [DOI] [PubMed] [Google Scholar]
- 43. Ori A., Andrés-Pons A., and Beck M. (2014) The use of targeted proteomics to determine the stoichiometry of large macromolecular assemblies. Methods Cell Biol. 122, 117–146 [DOI] [PubMed] [Google Scholar]
- 44. Mirza S. P., Halligan B. D., Greene A. S., and Olivier M. (2007) Improved method for the analysis of membrane proteins by mass spectrometry. Physiol. Genomics 30, 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hubbard S. J. (1998) The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382, 191–206 [DOI] [PubMed] [Google Scholar]
- 46. Morimoto Y. V., Ito M., Hiraoka K. D., Che Y.-S., Bai F., Kami-Ike N., Namba K., and Minamino T. (2014) Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol. Microbiol. 91, 1214–1226 [DOI] [PubMed] [Google Scholar]
- 47. Jones C. J., Macnab R. M., Okino H., and Aizawa S. (1990) Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212, 377–387 [DOI] [PubMed] [Google Scholar]
- 48. Van Arnam J. S., McMurry J. L., Kihara M., and Macnab R. M. (2004) Analysis of an engineered Salmonella flagellar fusion protein, FliR-FlhB. J. Bacteriol. 186, 2495–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., and Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 [DOI] [PubMed] [Google Scholar]
- 50. Hueck C. J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.