Abstract
The humoral immune system is network of biological molecules designed to maintain a healthy homeostatic equilibrium. Because antibodies are an abundant and highly specific effector of immunological action, they are also an important reservoir of previous host exposures. Antibodies may play a major role in early detection of host challenge. Unfortunately, few practical methods exist for interpreting the information stored in antibody variable regions. Immunosignatures use a microarray of thousands of random sequence peptides to interrogate antibodies in a broad and unbiased fashion. The pattern of binding between antibody and peptide is reproducible. Once the system has been trained on a disease cohort, blinded samples can be reliably predicted. Although immunosignatures of both chronic and infectious disease have been extensively tested, less has been done to demonstrate how healthy immunosignatures change over time or between individuals. Here, we report the results of a study of immunosignatures of healthy persons over brief (12 h sampled once per hour), intermediate (32 days sampled once per day), and long (5 years sampled once every year) time spans. Using this information, we were also able to detect intentional and unintentional immunological perturbations in the form of a vaccine and an infection, respectively. Our findings suggest that, even with the variability inherent in healthy immunosignatures, a single person's immunosignature will remain constant over time. Over this healthy signature, vaccines and infections create subsignatures that are common across multiple people, even subsuming healthy fluctuations. These findings have implications for disease monitoring and early diagnosis.
The humoral immune system is a highly evolved network of biomolecules that captures information about environmental exposure. The goal is continuous testing and optimization of antibodies to eliminate a biological threat. Sometimes B-cells generate antihost antibodies, but generally, antibodies are benign information-containing markers of past exposure. The fact that the same molecule both captures information and exerts an effect has consequences on vaccine development and disease diagnosis (1). Since antibodies contain information about their target, we should be able to predict health status by monitoring for appearance of a new antibody species. However, without prior knowledge of which antibodies are important for a given disease, it becomes difficult to analyze the ∼1012 different antibody molecules en masse. An immunosignature provides a snapshot of many antibodies simultaneously. If immunosignature data are queried using statistical and machine learning methods, these seemingly random patterns of antibody–peptide interactions can diagnose disease, even many diseases simultaneously (2, 3). However, it has become clear that healthy controls play an important role in the ability to detect disease patterns. Is there a typical “healthy” immunosignature? Is a healthy immunosignature stable over time? How much variance exists between healthy individuals? How do infection and vaccine signatures differ? We created a number of experiments using human volunteers to address these questions.
How Do Immunosignatures Work?
An immunosignature is a pattern of binding between serum antibodies and a microarray of random-sequence peptides. Antibodies will bind to random peptides under permissive binding conditions (4). The binding is detected by a fluorescent antispecies secondary antibody. A high-resolution laser scanner provides an intensity value for each peptide. Currently, the immunosignature platform is either 10,420 unique random-sequence 20-mer peptides with an N-terminal Cys-Ser-Gly (CSG) linker (Gene Expression Omnibus or GEO accession# Gly-Ser-Gly (GSG)17600, aka CIM 10K) or 328,794 unique random-sequence peptides that average ∼14 amino acids in length with a C-terminal GSG linker (GEO accession# GPL17679, aka CIM 330K). Feature selection methods enable informative peptides to be selected, cross-validated, and then tested for diagnostic performance. Upon training with well-defined disease cohort(s) and relevant controls, prediction of blinded samples is often >90% accurate (5). Rather than measuring a single biomarker, immunosignatures measure hundreds of informative markers, yielding a reproducible and predictable disease-specific pattern with sufficient capacity to encompass variations in the normal population.
Disease Monitoring
One of the major impediments to any new diagnostic is the ability to cope with variations in large cohorts of nondisease patients (6). Generally, the trend for developing biomarkers is to reduce many promising candidates to a few or one biomolecule with the greatest promise (7, 8). In order to meet specificity requirements, a single biomarker must exhibit near-identical behavior for most patients, a prohibitive demand for rare or highly dilute molecules (9). In order to avoid dilution effects, one needs either an abundant biomolecule or a process of amplification. DNA and RNA can be very effective in this regard but suffer to some extent from degradation (10), dilution (9), environmental (11), racial (12), and personal variation (13). Antibodies may solve many of these issues. They are amplified during affinity maturation; they are stable, abundant, and can be detected using anticlass and antispecies antibodies. Immunosignatures measure many antibodies simultaneously, which enhances specificity. Sensitivity is enhanced due to the permissive binding conditions. By using data from many features, immunosignatures can accommodate variations in the nondisease population, which may include an endemic population having substantial subclinical pathogen exposures. Analysis can be very basic, with statistical methods used to select features and probabilistic classifiers for class prediction (14).
We examined several aspects of immunosignatures in this report. We designed the experiments to proceed from short to long term. First, a volunteer donated samples every hour for 12 h. We then collected blood from three healthy persons over 30 days with samples taken weekly then two different healthy persons over 32 days with samples taken daily. We switched to a higher-density immunosignature array to examine 73 different healthy donors. This provides sufficient numbers of unique signatures to detect any chance overlap. After establishing a baseline for variation in healthy controls, we next asked whether an immunosignature of a vaccine could be discerned from a population of healthy persons and whether a vaccine could be detected using time points from the same person. These investigations provided sensitivity measures. We deemed the sensitivity was sufficiently high to observe perturbations from unknown disease. Serendipitously, we obtained blood from a volunteer who was supplying samples over time. During the donation period, the volunteer self-reported symptoms of an upper respiratory disease midway through the collection. We attempted to triangulate on a signal that might correlate to the unknown disease.
EXPERIMENTAL PROCEDURES
Assay Conditions
We have published the general assay conditions and analytical methods for the immunosignature microarrays (4, 14–19) using serum. For the 10,000 arrays, we used Applied Microarrays (Tempe, AZ) to print 10,420 peptides onto aminosilane-coated glass slides in duplicate, creating a two-up single-use peptide microarray (4). A higher-density 330,000 array is made using lithography techniques, creating a 24-up single-use peptide microarray of 330,000 peptides per array (2). Secondary detection antibodies were purchased from Thermo-Fisher (Waltham, MA) who supplied the goat anti-human IgG (H+L) Alexa-Fluor 555 conjugate for IgG detection, and from Jackson ImmunoResearch (West Grove, PA) who supplied the rabbit anti-human IgM Fc5μ Alexa-Fluor 647 conjugate for IgM detection. Secondary antibodies were used at 5 nm final concentration for both the 10,000 and 330,000 arrays. Primary concentration of serum was 1:500 for the 10,000 arrays, 1:1500 for the 330,000 arrays and 1:50 for saliva for the 10,000 arrays.
Analytical Methods
Data from the 10,000 and 330,000 arrays were obtained by first acquiring 16-bit TIFF images from an Agilent two-color 10 μm “C” scanner or an Innopsys two-color 0.5 μm 910 scanner, respectively. Raw data are extracted from the scanned images using GenePix Pro 6.0 (Molecular Devices, Santa Clara, CA), median-normalized to the 50th percentile of the foreground median signal, then log10 transformed. Linear models, statistical methods, and other calculations were performed in GeneSpring 7.3.1 (Agilent, Santa Clara, CA) or R (CRAN repository).
Samples
For saliva collection, we used the methods published in (20). All volunteers donated blood and saliva under IRB#0912004625 “Profiling Serum for Unique Antibody Signatures” by informed consent. IRB was renewed in 2015 by the Western Institutional Review Board, Olympia, WA. For Fig. 1, Volunteer #1 is a healthy male age 20–29 who provided both blood and serum. For Fig. 2, three different healthy volunteers donated blood over a 21-day time span. Volunteer #1 was used again, Volunteer #2 was a healthy female age 40–49, and Volunteer #3 was a healthy male age 40–49. For Fig. 3, Volunteer #2 and #3 were used again. Figure 4 used 73 different healthy donors, 18–41 years of age, male and female. Figure 5 used 49 healthy donors, 18–41 years of age, male and female, and Volunteer #5, a female age 30–39 who received the Vaccinia vaccine. Volunteer #6 received the hepatitis B vaccine and is a male age 50–59. Figure 6 used Volunteer #3 again. Figure 7 used Volunteer #2 and #3 again and added Volunteer #7, a female age 30–39; Volunteer #8, a male age 30–39; Volunteer #9, a female age 20–29; and Volunteer #10, a male age 50–59. For Fig. 8, Volunteer #11 is a female, age 40–49. All other volunteer information is protected by their right to privacy guaranteed by the collection protocol.
Fig. 1.
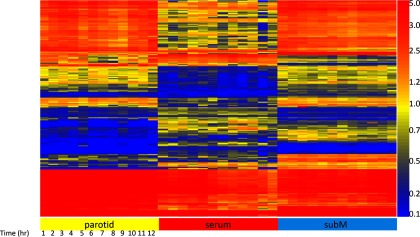
Analysis of serum and saliva taken from parotid and submandibular glands. This heatmap shows 3000 peptides out of 10,000 on the microarray that survived an ANOVA1 testing for differences between samples taken from Volunteer #1: saliva/submandibular, saliva/parotid, and serum, with p < 1.07 × 10−17. The Y-axis shows each of the 3000 peptides ordered by Euclidean distance. The X-axis shows each 1-h time point taken over 12 consecutive hours, ordered left to right. These 3000 peptides differentiate blood and saliva by the reactivity of antibodies that appear in blood versus those that appear in saliva. The average precision across technical replicates is r = 0.917 Pearson's correlation coefficient. Data shown are the average of two replicates.
Fig. 2.
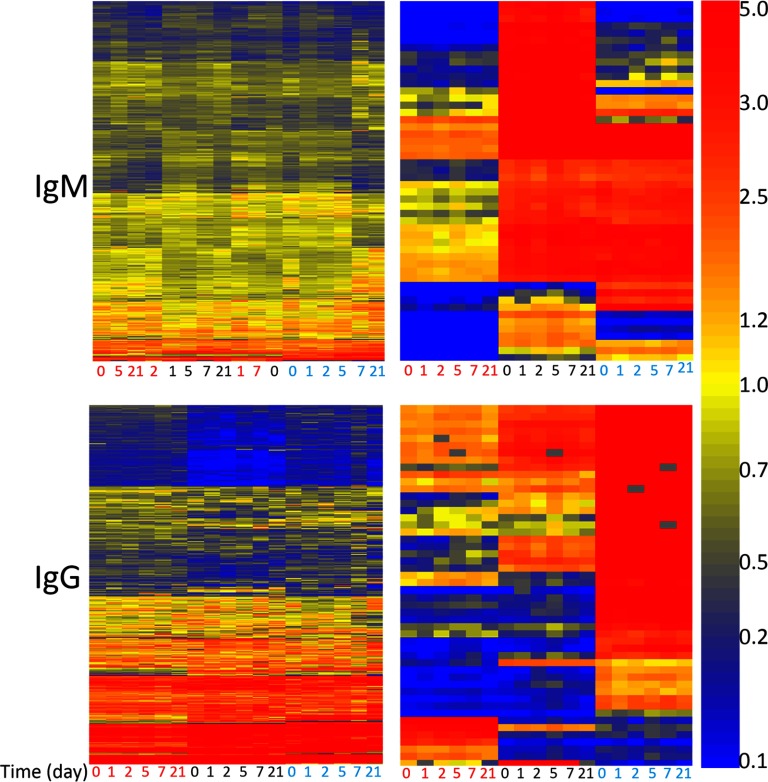
Heatmaps depicting IgG or IgM response in three different healthy persons (Volunteer #2, Volunteer #3, Volunteer #5) over the course of 21 days. IgM was detected with affinity-purified anti-human IgM antibody conjugated to red (647 nm) dye, IgG was detected with affinity-purified anti-human IgG antibody conjugated to green (555 nm) dye. X-axis is the day of donation of serum. Person 1 is labeled with red text, person 2 is labeled with black text, person 3 is labeled with cyan text. Clustering of the X-axis and Y-axis were done using Euclidean distance. The left heatmaps show all 10,000 peptides. Clustering of IgM signals shows little distinction per person or per day. Clustering of IgG signals shows more specificity across persons, and within each person signals order themselves by day of donation. Right heatmaps: 50 peptides were selected by ANOVA, p < 5.02 × 10−18 for IgM and p < 9.01 × 10−36 for IgG. At this cutoff, exactly 50 peptides pass the cutoff. Sequences are listed in Supplemental Table S1.
Fig. 3.
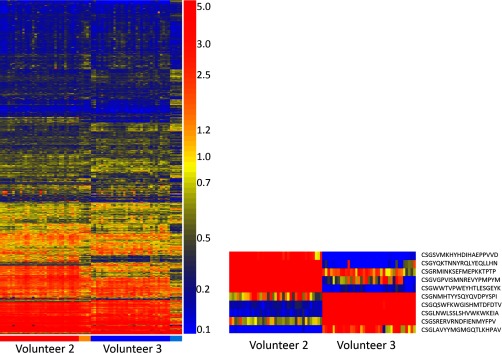
Heatmaps showing immunosignatures for volunteers who donated blood every day for 32 days. Left: heatmap of all 10,000 peptides for Volunteer #2 (red bar) and Volunteer #3 (blue bar). Daily immunosignatures are shown in order from left to right, from Nov. 9, 2013 to Dec. 10, 2013. Orange and cyan bars correspond to blood donated on 1 day in 2008, 2009, 2010, 2011, and 2012 for Volunteers #2 and #3, respectively. Volunteer #3 received the seasonal influenza vaccine on Nov. 17, 2013.
Fig. 4.
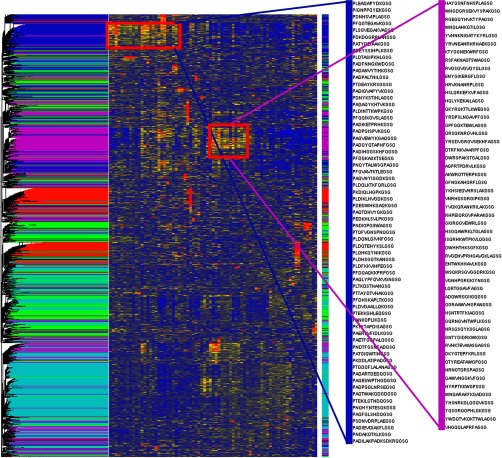
Heatmap of 73 healthy donors. 73 serum samples from healthy donors were processed on the 330,000 immunosignature array. No statistical tests were performed. Rather, a random sample of 100,000 peptides were hierarchically clustered using Euclidean distance as the separation metric on both the Y-axis (peptides) and X-axis (human donors). A five-group k-means clustering was also performed on the peptides. Results of the k-means cluster were used to color the dendrogram arms. Two k-means groups of peptides are highlighted. The blue group shown immediately left of the heatmap represents a group of peptides common across at least 26 healthy volunteers. The three amino acid sequence PAD/PLD/PDN appears commonly in the blue group. At least 10 donors shared peptides from the magenta group, but motifs in these peptides are more cryptic than the blue group. In the absence of disease-specific peptides, healthy donors can group by common antibody reactivity.
Fig. 5.
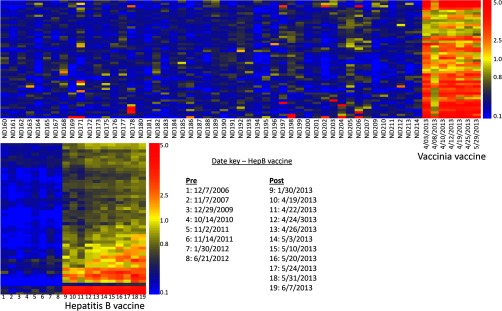
Immunosignature of vaccines. Two different comparisons are shown. Top heatmap depicts 50 different healthy volunteers who donated blood once are compared with a single laboratory worker, Volunteer #5, who received the Vaccinia vaccine on Mar. 23, 2013 as part of his or her research program, and 50 peptides selected by comparing postvaccine samples to healthy donors using a t test (p < 3.03 × 10−38). Seven postvaccine time points were taken, with dates shown on the X-axis. Bottom heatmap represents Volunteer #6 who received a three-dose hepatitis B vaccine on Jan. 25, 2013 with a boost on Apr. 23, 2013 and July 15, 2013. Prevaccine time points were collected from previous years and listed to the right of the heatmap. A t test between pre- and postvaccine samples yielded 50 peptides with p < 9.41 × 10−28 that define the hepatitis B vaccine signature. No samples drawn after the third boost.
Fig. 6.
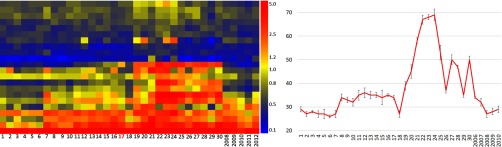
Heatmap from vaccine trial. In 2009, six volunteers received the seasonal trivalent influenza vaccine (A/Brisbane/59/2007, A/Brisbane/10/2007, B/Brisbane/60/2008) donated blood before and 21 days following receipt of the vaccine. 28 peptides were selected using a paired t test to find peptides different on day 21 versus prevaccine (p < 4.60 × 10−16). These 28 peptides were filtered to find 21 that were higher on day 21 for every volunteer (see Supplemental Table S5). For patient 73, we collected only two time points, and for patients 112 and 113, we were unable to draw blood on day 2. Each patient has slightly different peptides that were most reactive against the influenza vaccine components. Graph to the right is the sum of all fluorescence intensity. Error bars represent the upper and lower 95% CI.
Fig. 7.
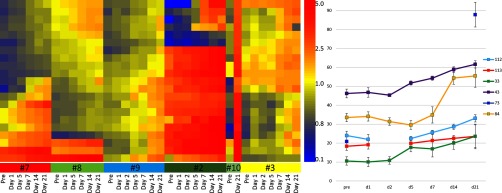
Heatmap of a time course for Volunteer #3 across 32 days in 2013 (Nov. 9, 2013 to Dec. 10, 2013) and across the years 2008 to 2012 (the last 2 days were excluded). Left: Volunteer #3 received the 2013 seasonal influenza vaccine (A/California/7/2009, A/Victoria/361/2011, and B/Massachusetts/2/2012) on Nov. 17, 2013. Without using information about the date of immunization, 22 peptides were selected that changed intensity for at least 2 days consecutively, either up or down (Y-axis). The X-axis is an ordered list of immunosignatures from Volunteer #3 for 30 consecutive days (left part of heatmap) and also once per year for 5 years (right part of heatmap). Right: sum of the intensities for all 22 peptides displayed in a line graph. Error bars represent the upper and lower 95% CI.
Fig. 8.
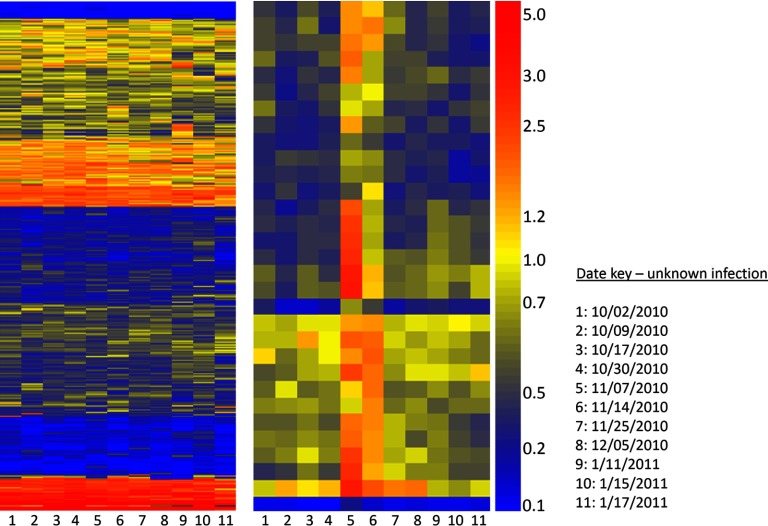
Time course of volunteer-submitted blood-samples over 3 months. Left: Heatmap represents Volunteer #11 who collected blood spots over a 3-month period between Oct. 2, 2010 and Jan. 17, 2011 (X-axis, time points are in order left to right, see Date Key) and self-reported health status. Y-axis shows all 10,000 peptides on the microarray, ordered by Euclidean distance. Right: Heatmap shows 31 peptides (Y-axis) selected by filtering for a consecutive change in signal existed across at least two subsequent time points. Peptides sequences are listed in Supplemental Information Table S6. Volunteer reported symptoms on or about Nov. 24, 2010.
RESULTS
Short-Term Analysis of Serum and Saliva—1 Day
In the interest of thoroughness, we tested the possibility that changes in antibody profiles could occur during the course of a single day. We did not assume a priori that antibody abundance would change hour to hour. In order to maximize sensitivity, we assayed each sample in duplicate and averaged the two technical replicates. We had not thoroughly tested whether saliva contained the same antibody profile as serum in previous experiments, so we combined these questions into a single time-course experiment. We asked Volunteer #1 to draw venous blood (ser) and saliva from both the parotid (par) and submandibular (subM) glands every hour for 12 h. We processed the samples on the 10,000 immunosignature peptide microarrays using two technical replicates for each of the samples tested. Figure 1 displays the average of the replicates. If the Pearson's correlation coefficient across technical replicates was <0.90, the sample was rerun on a new microarray. Technical replicate correlations for the 12 serum samples averaged 0.942, the subM samples averaged 0.904, and the par samples averaged 0.911. The average R across all replicates was 0.917. Figure 1 is a heatmap for the top 3000 most significant peptides for each of the three sample types based on a two-way ANOVA with p < 1.07 × 10−17 across the three sample types (Bonferonni cutoff is 4.76 × 10−6). According to these results, there are specific antibody populations missing from saliva that are present in serum, as well as antibodies in saliva that are missing from serum. In some cases, there are antibodies that appeared only in samples from the submandibular gland and not the parotid. Results suggest that saliva can be source of IgG for disease detection in order to avoid finger sticks or venous blood draws. However, without altering assay conditions, the precision of replicates when using saliva is significantly lower (at p < 0.05) than the precision across replicates of serum.
Short-Term Analysis of Serum—21 Days
We collected blood from three healthy volunteers (Volunteers #2, #3, #5) who donated 5 ml of blood on days 0, 1, 2, 5, 7, and 21 in August and September of 2013. We tested both IgG and IgM using the appropriate αhu-IgG or αhu-IgM secondary antibody (see Methods). Figure 2, left shows heatmaps for all 10,000 peptides for IgM (top) and IgG (bottom). On the right, we show heatmaps for 50 peptides that were selected by a one-way ANOVA (p < 5.02 × 10−18 for IgM and p < 9.01 × 10−36 for IgG) using the three volunteers as the groups. These peptides differ most across the three volunteers and are listed in Supplemental Table S1. In the absence of disease, antibodies that appear in healthy individuals but are constant across time may reflect natural or autoantibodies (21) or a long-lasting vaccine or pathogen exposure.
Short-Term Analysis of Serum—30 Days
Figure 3 illustrates how unsupervised analysis of all array peptides appears. Two people donated blood over 32 days. Volunteer #2 (red bar, shown on the left) and Volunteer #3 (blue bar, shown on the right) provided sequential daily blood samples. The peptides are grouped using Euclidean distance hierarchical clustering on the peptides (Y-axis). The X-axis is ordered by each person's signature Nov. 9, 2013 to Dec. 10, 2013. The orange bar (left) and cyan bar (right) represent a single time point from 2008, 2009, 2010, 2011, and 2012 for the corresponding volunteers. Volunteer #3 (blue) received the yearly influenza vaccine on day 17, although this is not obvious in the unsupervised analysis (see detail in Fig. 6). The small heatmap to the right is each of 10 peptides that differ between Volunteer 2 and Volunteer 3 by t test at p < 5.18 × 10−47. Together, these data represent a more granular time division than Fig. 2 but confirm that there are a number of antibodies highly unique to these two individuals in the midst of a large number of similar reactivities.
Single Time-Point Analysis of Multiple Healthy Donors
Figure 4 is a heatmap of samples taken from 73 different healthy donors at a single time point from both males and females of mixed ages. In this experiment, the 330,000 peptide microarray was used. This microarray provides 33 times more peptides than the 10,000 array and more coverage of random sequence space. The array contains 328,794 random-sequence peptides in a smaller form-factor than the 10,000 arrays [1]. After processing the 73 330,000 arrays, we randomly selected 100,000 out of the 330,000 peptides to cluster. As there are no diseases being tested, one would expect little to no similarities across individuals. k-means clustering with k = 5 was used to identify clusters of peptides that behave similarly. The results of the k-means clustering overlapped to some degree with the results from the vertical hierarchy of volunteers. The five clusters were assigned five colors and were used to color the dendrogram bars. In this list of 100,000 peptides, some individuals share common reactivity to an unknown epitope. We chose two clusters, blue and magenta, that had obvious similarity across multiple people. The blue and magenta clusters are shown to the right of the heatmap. Peptides from the blue and magenta clusters are listed in Supplemental Table S2. A simple alignment in CLUSTALW of the blue peptides shows a dominant but not complex N-term biased 3-mer motif of PAD/PLD/PAE in nearly all of the peptides. The magenta peptides have a more cryptic and complex relationship to each other. A simple CLUSTALW alignment shows at least seven different complex motifs that can be found throughout the peptide, rather than near the C-term or N-term. Motifs include WNQ, HRVK/YRVN/YRVK, VNRH, HSL/HGL/HSG, RVG/RVH, and others with varying sized gaps. This suggests that the blue peptides may represent a simple linear antigen, common to many people (or perhaps to residents of Arizona), and the magenta peptides may represent a mimotope or mimotopes to a nonlinear or nonprotein antigen.
Detecting Vaccine Immunosignatures—Training
Figure 5 represents two immunosignature sensitivity tests. The first test asked whether the variance in multiple different healthy people exceeds the detection of a single person's vaccine. We obtained blood from ND183 (aka Volunteer #5) several months prior to the Vaccinia immunization on Mar. 23, 2013. After immunization, seven different time points were collected. We compared these time points to 49 other healthy individuals and ND183 prior to vaccination. There were 50 peptides that survived a t test between these groups, with p < 3.03 × 10−38. These peptides were plotted in a heatmap format and are listed in Supplemental Table S3. The vaccination samples appear on the same dendrogram when using hierarchical clustering with Euclidean distance as the separation metric, and are perfectly classified using SVM with leave-one-out cross-validation.
The second test asked whether the variance in a single person over time exceeds the detection of a vaccine event. Volunteer#6 received a commercial hepatitis B vaccine and subsequent boosts in 2013. Blood draw dates are shown in Fig. 5. In the years prior to receiving the vaccine, Volunteer#6 had donated blood from 2006 to 2012. A t test was used to identify 50 peptides that differed between prevaccine samples and postvaccine samples, with p < 9.41 × 10−28. These 50 peptides are plotted in a heatmap format with time on the X-axis and peptides on the Y-axis. Colors indicate the relative fluorescence intensity after normalization; the key is drawn to the right of each heatmap.
Results suggest that vaccines disturb the immune profiles of recipients, such that a vaccine signature can be distinguished from immunosignatures of many different healthy people, even with the variation seen in Fig. 4. Results also suggest that immunosignatures from the same person, even with the variation seen in Fig. 3, can detect a vaccine signature. We then asked whether a combination of time and person could overwhelm the ability to detect and distinguish a vaccine signature.
We collect time-course samples of volunteers who receive the seasonal influenza vaccine every year and have since 2006. In 2009, the influenza vaccine was composed of A/Brisbane/59/2007, A/Brisbane/10/2007, and B/Brisbane/60/2008. Six different people donated blood in 2009 before and after immunization. We asked for a difference between pre- and postvaccine signatures as in Fig. 5 but with the restriction that the signature is common to every person. We performed a paired t test between prevaccine and postvaccine day 21. There were 28 peptides with p < 4.60 × 10−15. 21 peptides were higher postvaccine than prevaccine and are listed in Supplemental Table S4 and shown as a heatmap in Fig. 6. The peptides that increased between pre- and postvaccine differed somewhat from person to person, but the vaccine signature was identified even given the personal variance over time and the variance across people. Figure 6 right panel shows that the cumulative signal for each person has a linear increasing trend by 5 days postvaccination, even with the variation in peptide specificity.
Detecting Vaccine Immunosignatures—Test/Validation
Given the vaccine signals seen in Figs. 5 and 6, we asked whether a vaccine could be detected if the analysis team was blinded to the date of vaccination. A t test can identify peptides that react to a vaccination if the date of vaccination is known, but we wished to test whether an immune disturbance could be identified without knowledge of the vaccination date. To do this, we used data from Fig. 3. Volunteer #3 received the 2013 seasonal trivalent influenza vaccine on Nov. 17, 2013. We queried each of the 10,000 peptides across 30 days of signatures, asking for signals that changed more than 10% over the previous signal but only changed 0–10% up or 0–5% down from the subsequent day. Any signal that changed up or down more than these values was considered spurious. By restricting signals using this rule, the filtered peptides tended to follow a smoothed trend. This rule left 22 peptides that rose postvaccine (see Fig. 7, left). The intensity of the median-normalized values for each of these 22 peptides was summed and plotted in a line graph (see Fig. 7, right panel). There was an obvious peak in total signal 3 days postvaccine then a gradual drop in the weeks following immunization. The volunteer had received prior influenza vaccines, possibly leading to a memory response or pre-existing signal from the 2013 reactive peptides, but even so, there was a biologically relevant trend in the 22 peptides shown (see Supplemental Table S5).
Detecting an Unknown Disease
A volunteer in California who was collecting sequential blood samples over a period of 3 months using Whatman ProteinSaver® 903 cards (20) self-reported flu-like symptoms on or about Nov. 25, 2010 and lasting 3–4 days. 39 peptides were selected using the algorithm from Fig. 7. 31 peptides were kept that possessed a trend that increased over time. The heatmap in Fig. 8 shows the full 10,000 peptides (left) and 31 peptides that changed in intensity over two or more consecutive time points (see Supplemental Table S6). These 31 peptides spike upward after Nov. 7, 2010 in a consistent fashion. The patient reported being ill “just before the Thanksgiving weekend”; detected signature waned just prior to symptoms. No other peptide list was found using these criteria. No significance test was done with these samples.
DISCUSSION
Immunosignaturing is well-suited for detection and classification of diseases and vaccines (4, 5, 22–26). In each of these demonstrations, there were controls representing an “unaffected” state, but the clinical definition of unaffected varied by researcher. Unaffected may mean healthy from a nonendemic region, healthy from an endemic region, or simply “unaffected by disease X,” which has other health implications. Immunosignatures work best when most of the expected variance in the projected test population is measured during training. Poorly selected controls used to train the system can diminish the test performance substantially. Sequential samples taken over time from one person may increase the ability to detect disease in that person since only one baseline is used, but this limits retrospective studies. We designed a set of experiments that tested whether variance across time and variance across people affected the ability to detect an immune perturbation.
We first asked whether variation in immunosignatures occurs over brief periods of time. We asked a volunteer to donate blood and saliva every hour for 12 h to see whether variation in short-term immunosignatures exceeded the technical variability of the platform (Fig. 1). If saliva could replace serum for short time-series collections, less pain would be involved and might yield greater volunteer rates. The variation in serum immunosignatures from hour to hour averaged ∼8% coefficient of variation for serum and ∼15% for saliva from both parotid and submandibular glands. The technical variation across technical replicates averaged ∼10%. The high variation in saliva immunosignatures suggests that our assay conditions were not optimized for saliva or the collection method introduced random noise. These conditions may be improved for future studies.
We next examined IgG and IgM in healthy individuals. IgM is widely considered a “first responder” antibody. It is quickly up-regulated following infection by a pathogen. However, it is not widely known how circulating IgM behaves before an infection or after convalescence. We first showed that the peptides that make up the signature from patient for IgG and IgM are quite different (Fig. 2). This is supported by immunology, as IgG and IgM are often raised against nonidentical epitopes from the same pathogen (27). We then demonstrated that there are patterns in IgG from healthy volunteers that are highly specific to that individual. Although IgM shows personal differences as well, it is known that IgG circulates in the blood longer than IgM and may exist years or decades after infection or vaccination.
We then tested a longer, more granular time course. We looked at two individuals over 32 days plus a comparison of serum taken from these volunteers 1, 2, 3, 4, and 5 years prior (Fig. 3 large heatmap). We showed many similarities in signatures when examined at a gross level, both day to day and person to person, but there were also peptides that are highly unique (Fig. 3 small heatmap). The implications are that some exposures might be unique to one person, but it is also possible that two people exposed to the same antigen might produce dissimilar immunosignature profiles. Different people can raise antibodies to different parts of a pathogen, which typically will not affect diagnostics like whole-pathogen ELISAs but would affect a partitioned diagnostic like immunosignatures. This may not be an insurmountable problem. Many diseases have been tested using immunosignatures. There are numerous instances where antibody reactivity to a pathogen is highly conserved person to person (25) but other examples where even published disease epitopes are not wholly conserved. In (28), a known epitope for dengue could be detected in less than 6% of the cases. However, there were some epitopes present in almost every dengue patient tested, suggesting that some antigens produce antibodies useful for ELISA-like assays, while others may work best for immunosignatures.
These time-course experiments prompted a test comparing multiple healthy normals. Would many randomly selected people possess a similar background of vaccines and pathogen exposures, or would each person have a unique immunosignature? Figure 4 shows 73 different donors, mostly college-age students split nearly evenly between male and female. We tested these samples on a higher-density array, the CIM 330,000 array, which has shorter peptides (median length of 14.1 amino acids including the three amino acid GSG linker) than the 20-mers found on the CIM 10,000 arrays (all peptides are 20-mers, including the three amino acid CSG linker). In Fig. 4, we sampled 100,000 of the 330,000 peptides to see whether any common signatures appeared in the healthy population. By clustering the samples using both distance (hierarchical cluster) and k-means (k = 5 clusters), we can observe relationships that suggest some groups of individuals possess similar reactivity to certain peptides. The k-means cluster was used to color the dendrogram bars (X-axis), and two obvious clusters were picked out by eye. The first group that was selected (blue bar, Fig. 4 and Supplemental Table S2) appeared to have common peptide intensities across at least 26 different healthy persons. None of the clinical information provided by the volunteers (race, gender, age) correlated to this block. Using CLUSTALW alignment of the peptides in the blue group, we see that much of the N terminus of each peptide had similar motifs, suggesting that an antibody or antibodies in these 26 people had some shared reactivity to a PAD/PLD/PDN motif. Conversely, the magenta group that was shared by at least 10 different volunteers had almost no common linear motifs as measured by CLUSTALW. These peptides may correspond to mimotopes that react with structural or nonprotein epitope(s), or perhaps this cluster results from a pool of antibodies that are binding nonspecifically. This simple demonstration suggests that one can test immunological effects from geography, prior vaccine history, or perhaps for something as esoteric as predicting vaccine efficacy, pathogen resistance, or autoantibody prevalence. Mining the random-peptide sequences is arduous but can lead to unique insight. Figure 3 suggests that much of our immunosignature remains constant for years. If so, this would enhance efforts to diagnose disease using time-course data and routine monitoring.
Strong patterns of reactivity appeared in a random sampling of only 73 people. This might damp out an immune perturbation if it appeared in a population. Figure 5 top shows a heatmap with 50 different healthy volunteers (a subset of those from Fig. 4) versus a single volunteer, ND183, who donated blood prior to receiving a Vaccinia immunization on Mar. 12, 2013. A t test between postvaccine samples and all of the healthy controls yielded 50 peptides with p < 3.03 × 10−38 (see Supplemental Table S3). This suggests that the normal variation seen in healthy people might still allow a vaccine to be detectable over the biological noise from different people.
We then examined time points from one person using samples collected in previous years versus an immunization with the commercial hepatitis B vaccine. Figure 5 bottom is a heatmap displaying the pre- and postvaccine time points. 50 peptides with p < 9.41 × 10−28 are displayed (see Supplemental Table S3). The results suggest that the normal variation seen in one healthy person across time does not obscure a vaccine response. For 12 of these 50 peptides (lower part of heatmap), there is a continuing increase in fluorescence over time, suggesting that some antibodies to the hepatitis B vaccine were still increasing their affinity, their abundance, or both. Even the peptides that do not show an increase over time still have a large ratio between prevaccine and postvaccine. We created another test that combined these two factors—variance over time and variance across people. Figure 6 displays the results when we asked for a common signature across time and across people for the same vaccine, the three-part seasonal influenza vaccine. This test combines the requirements for consistency over time and adds a requirement for a common signature across different people. The results show that different people respond slightly differently to the influenza vaccine rather than expressing common reactivity. This may create a problem if one was monitoring a population for a common signature, when in fact a group of people may show different patterns of reactivity.
We tested the system in two ways: First, we asked if we could find a vaccine signature using an unsupervised method of feature selection with a single person (Volunteer#3) who received an influenza vaccine in 2013. Using a simple moving window, we identified 22 peptides that correlated with the immunization schedule. Even summing the peptides to provide a single value, like an ELISA, still provided some resolution in identifying the vaccine date. Figure 7 is a heatmap and line graph illustrating these statements, and Supplemental Table S4 lists the peptides that were identified. We attempted to identify the influenza sequences from the random peptide sequences, but as we reported in (16), the 10,000 microarray may not provide enough sequences to accurately map a linear epitope.
With all of the information gathered about variation of healthy normal controls and detection of immune perturbations over time, we were fortuitously presented with a real-world challenge. We examined samples from a volunteer who donated blood over a period of several months. This person self-reported an illness in November 2010. Using the same simple moving window strategy used in Fig. 6, we looked for peptides that moved in concert over a window of at least three time points rather than two. We identified the 31 peptides shown in Fig. 8 and listed in Supplemental Table S5 that showed an increase in signal prior to the reported symptoms then diminished during and slightly after symptoms waned. Unsupervised clustering of all peptides on the 10,000 array revealed no signal that corresponded to the symptoms (Fig. 8, left), but the moving window averaging revealed peptides that seemed to correlate slightly ahead of reported symptoms. It may be possible that production of IgG preceded symptoms, or we were detecting immature IgG that matured and no longer bound to the same peptides. Had we measured IgM at the same time, we may have had a traditional IgM/IgG onset of antibodies, even though the peptides for each isotype would be different.
A general hypothesis created by the preceding work is that early detection of disease may be possible by monitoring persons over time. It is also possible that a large pool of healthy controls can be used for training disease diagnostics since there is a great deal of personal variation in the immunosignature. It is uncertain whether a time-course study using many different people would yield the same sensitivity in detecting a disease as using a single person, but this may vary by disease, and the influenza vaccine study presented here used too few people to make a general statement. Regardless, the emphasis of this study suggests that immunosignatures can be sensitive and well-powered using sequential samples. Routine monitoring may be a way to detect disease before it becomes difficult to treat.
Another important issue is correct selection of controls. The strong overlapping signatures we see in Fig. 4 shows how easily a group of controls can form a common pattern. These patterns can cause statistical issues if the control arm is underpowered, especially when examining diseases where an endemic healthy population expresses a signature to local pathogens. We have previously shown that the Valley Fever immunosignature exists to a greater or lesser extent in most healthy controls who have lived in the Phoenix area for more than 6 y (25). Over time, exposure to local pathogens, even in the absence of infection, can create a disease-like immunosignature that could lead to false positives if not properly trained.
We propose a number of applications for immunosignatures that hinge on the ability to correctly identify an infection using either time-series data or large control populations. Healthcare workers entering a region of infection might wish to provide samples prior to travel. During deployment, saliva or blood could be taken routinely and checked against previous samples to monitor for any sign of change in the signature, even using the simple moving-window measurement. No training need be done for a disease; the peptides that change after exposure have the potential to point to possible epitopes, eliminating the need for a specific diagnostic.
If enough individuals in a large population center were monitoring their own immunosignatures, it may be possible to detect an emerging pathogen or a biothreat, using normal controls from other regions for comparison. Since immunosignatures can reveal common reactivity, this method might be sensitive enough to detect a new infectious agent before it becomes an epidemic or pandemic. These opportunities are available to us because of the behavior of immunosignatures of healthy people and the signal to noise capability of the immunosignature system.
Footnotes
Author contributions: P.S. and S.J. designed the research; P.S. and D.W. performed the research; P.S. and D.W. analyzed data; and P.S. and D.W. wrote the paper.
 This article contains supplemental material Supplemental Tables S1-S6.
This article contains supplemental material Supplemental Tables S1-S6.
Unpublished papers cited: None.
1 The abbreviations used are:
- ANOVA
- Analysis of Variance
- CRAN
- Comprehensive R Archive Network
- IRB
- Institution Review Board
- SVM
- support vector machines
- TIFF
- tagged image file format.
REFERENCES
- 1. Notkins A. L. (2007) New predictors of disease. Scientific American. 2007, 72–80 [PubMed] [Google Scholar]
- 2. Legutki J. B., Zhao Z.-G., Greving M., Woodbury N., Johnston S. A., and Stafford P. (2014) Scalable high-density peptide arrays for comprehensive health monitoring. Nat Commun. 5, 4785 doi: 10.1038/ncomms5785 [DOI] [PubMed] [Google Scholar]
- 3. Navalkar K. A., Johnston S. A., and Stafford P.(2015) Peptide based diagnostics: Are random-sequence peptides more useful than tiling proteome sequences? J. Immunolog. Meth. 417, 10–21. doi: http://dx.doi.org/10.1016/j.jim.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 4. Stafford P., Halperin R., Legutki J. B., Magee D. M., Galgiani J., and Johnston S. A. (2012) Physical characterization of the ‘immunosignaturing effect.’ Mol. Cell. Proteomics doi: 10.1074/mcp.M111.011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stafford P., Cichacz Z., Woodbury N. W., and Johnston S. A. (2014) Immunosignature system for diagnosis of cancer. Proc. Natl. Acad. Sci. U.S.A. 111, E3072-E80 doi: 10.1073/pnas.1409432111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bossuyt P. M., Reitsma J. B., Bruns D. E., Gatsonis C. A., Glasziou P. P., Irwig L. M, Moher D., Rennie D., de Vet H. C., and Lijmer J. G. (2003) The STARD statement for reporting studies of diagnostic accuracy: Explanation and elaboration. Clin. Chem. 49, 7–18 [DOI] [PubMed] [Google Scholar]
- 7. Weigelt B., Pusztai L., Ashworth A., and Reis-Filho J. S. (2012) Challenges translating breast cancer gene signatures into the clinic. Nat. Rev. Clin. Oncol. 9, 58–64 [DOI] [PubMed] [Google Scholar]
- 8. Bidard F.-C., Pierga J.-Y., Soria J.-C., and Thiery J. P. (2013) Translating metastasis-related biomarkers to the clinic - progress and pitfalls. Nat. Rev. Clin. Oncol. 10, 169–79 [DOI] [PubMed] [Google Scholar]
- 9. Hori S. S., and Gambhir S. S. (2011) Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci. Translational Med. 3, 109ra16 doi: 10.1126/scitranslmed.3003110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ngoka L. (2008) Sample prep for proteomics of breast cancer: Proteomics and gene ontology reveal dramatic differences in protein solubilization preferences of radioimmunoprecipitation assay and urea lysis buffers. Proteome Sci. 6, 30 doi: 10.1186/1477-5956-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibson G. (2008) The environmental contribution to gene expression profiles. Nat. Rev. Genet. 9, 575–581 [DOI] [PubMed] [Google Scholar]
- 12. Sharma S., Murphy A., Howrylak J., Himes B., Cho M. H., Chu J.-H., Hunninghake G. M., Fuhlbrigge A., Klanderman B., Ziniti J., Senter-Sylvia J., Liu A., Szefler S. J., Strunk R., Castro M., Hansel N. N., Diette G. B., Vonakis B. M., Adkinson N. F. Jr., Carey V. J. and Raby B. A. (2011) The impact of self-identified race on epidemiologic studies of gene expression. Genetic Epidemiol. 35, 93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams R. B. H., Chan E. K. F., Cowley M. J., and Little P. F. R. (2007) The influence of genetic variation on gene expression. Genome Res. 17, 1707–1716. doi: 10.1101/gr.6981507 [DOI] [PubMed] [Google Scholar]
- 14. Kukreja M., Johnston S. A., and Stafford P. (2012) Comparative study of classification algorithms for immunosignaturing data. BMC Bioinformatics 13 doi: doi: 10.1186/1471-2105-13-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown J., Stafford P., Johnston S., and Dinu V. (2011) Statistical methods for analyzing immunosignatures. BMC Bioinformatics 12, 349 doi: 10.1186/1471-2105-12-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halperin R. F., Stafford P., Legutki J. B., and Johnston S. A. (2010) Exploring antibody recognition of sequence space through random-sequence peptide microarrays. Mol. Cell. Proteomics 28, e101230.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kukreja M., Johnston S. A., and Stafford P. (2012) Immunosignaturing microarrays distinguish antibody profiles of related pancreatic diseases. Proteomics Bioinformatics S6 doi: doi: 10.4172/jpb.S6-001 [DOI] [Google Scholar]
- 18. Stafford P., editor. Biological Interpretation of Microarray Normalization Selection. 1st ed Boca Raton: CRC Press, 2008, pp. 151–173 [Google Scholar]
- 19. Stafford P. Data normalization selection. In: Hardiman G, editor, Microarray Innovations: Technology and Experimentation. Drug Discovery. 11. Boca Raton: CRC Press, 2009, pp. 97–119 [Google Scholar]
- 20. Chase B. A., Johnston S. A., and Legutki J. B. (2012) Evaluation of biological sample preparation for immunosignature-based diagnostics. Clin. Vaccine Immunol. 19, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacroix-Desmazes S., Kaveri S. V., Mouthon L., Ayouba A., Malanchère E., Coutinho A, and Kazatchkine M. D. (1998) Self-reactive antibodies (natural autoantibodies) in healthy individuals. J. Immunol. Meth. 216, 117–137. doi: http://dx.doi.org/10.1016/S0022-1759(98)00074-X. [DOI] [PubMed] [Google Scholar]
- 22. Legutki J. B., Magee D. M., Stafford P., and Johnston S. A. (2010) A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine. 28, 4529–4537 [DOI] [PubMed] [Google Scholar]
- 23. Restrepo L., Stafford P., Magee D. M., and Johnston S. A. (2011) Application of immunosignatures to the assessment of Alzheimer's disease. Ann. Neurol. 70, 286–295 doi: doi: 10.1002/ana.22405 [DOI] [PubMed] [Google Scholar]
- 24. Hughes A. K., Cichacz Z., Scheck A. C., Coons S. W., Johnston S. A., and Stafford P. (2012) Immunosignaturing can detect products from molecular markers in brain cancer. PLoS ONE 7, e40201 doi: doi: 10.1371/journal.pone.004020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Navalkar K. A., Johnston S. A., Woodbury N., Galgiani J. N., Magee D. M., Cichacz Z., and Stafford P. (2014) Application of immunosignatures to diagnosis of Valley Fever. Clin. Vaccine Immunol. 21, 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Williams S., Stafford P., and Hoffman S. A. (2014) Diagnosis and early detection of CNS-SLE in MRL/lpr mice using peptide microarrays. BMC Immunol. 15, 15–23 doi: 10.1186/1471-2172-15-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dillner J. (1990) Mapping of linear epitopes of human papillomavirus type 16: The E1, E2, E4, E5, E6 and E7 open reading frames. Int. J. Cancer 46, 703–711 doi: 10.1002/ijc.2910460426 [DOI] [PubMed] [Google Scholar]
- 28. Richer J., Johnston S. A., and Stafford P. (2015) Epitope identification from fixed-complexity random-sequence peptide microarrays. Mol. Cell. Proteomics 14, 136–147 doi: 10.1074/mcp.M114.043513 [DOI] [PMC free article] [PubMed] [Google Scholar]