Abstract
Profiling post-translational modifications represents an alternative dimension to gene expression data in characterizing cellular processes. Many cellular responses to drugs are mediated by changes in cellular phosphosignaling. We sought to develop a common platform on which phosphosignaling responses could be profiled across thousands of samples, and created a targeted MS assay that profiles a reduced-representation set of phosphopeptides that we show to be strong indicators of responses to chemical perturbagens.
To develop the assay, we investigated the coordinate regulation of phosphosites in samples derived from three cell lines treated with 26 different bioactive small molecules. Phosphopeptide analytes were selected from these discovery studies by clustering and picking 1 to 2 proxy members from each cluster. A quantitative, targeted parallel reaction monitoring assay was developed to directly measure 96 reduced-representation probes. Sample processing for proteolytic digestion, protein quantification, peptide desalting, and phosphopeptide enrichment have been fully automated, making possible the simultaneous processing of 96 samples in only 3 days, with a plate phosphopeptide enrichment variance of 12%. This highly reproducible process allowed ∼95% of the reduced-representation phosphopeptide probes to be detected in ∼200 samples.
The performance of the assay was evaluated by measuring the probes in new samples generated under treatment conditions from discovery experiments, recapitulating the observations of deeper experiments using a fraction of the analytical effort. We measured these probes in new experiments varying the treatments, cell types, and timepoints to demonstrate generalizability. We demonstrated that the assay is sensitive to disruptions in common signaling pathways (e.g. MAPK, PI3K/mTOR, and CDK). The high-throughput, reduced-representation phosphoproteomics assay provides a platform for the comparison of perturbations across a range of biological conditions, suitable for profiling thousands of samples. We believe the assay will prove highly useful for classification of known and novel drug and genetic mechanisms through comparison of phosphoproteomic signatures.
Our understanding of disease mechanisms and therapeutic opportunities is rapidly expanding because of incredible advances in molecular profiling technologies. Within the last decade, the broad application of high-throughput transcriptional profiling has resulted in rich sets of gene expression data collected from biological samples subjected to drug and genetic perturbations (1, 2). As an example, the ambitious Connectivity Map (CMap)1 project (http://www.lincscloud.org/) collects transcriptional profiles from cells perturbed with biologically active compounds or genetic manipulations and enables cross-comparisons of these profiles to help develop insight into the biological mechanisms at play (3, 4).
High-throughput transcriptional profiling represents a novel approach to derive functional associations among drugs, genes, and diseases but only reflects one axis of cellular information (gene expression). The proteomic axis, and particularly the post-translational modifications to the proteome, may provide alternate and complementary information for discovering these connections. Initial and sustained signals to environmental changes (such as drug treatment and neomorphic disease states) are frequently mediated by changes of post-translational modifications on proteins. Protein phosphorylation in particular is known to be a strong mediator of cellular signaling (5, 6). Changes in the phosphoproteome can result in subsequent disruptions in protein-protein interactions (7, 8), alterations in protein stability, changes in cellular localization of proteins (9, 10), and potentiation of novel transcriptional programs. Importantly, dysregulation of phosphosignaling is also known to be involved in multiple diseases, including cancer (11–17). We propose that profiling phosphosignaling responses to drug treatments and other perturbations can generate cellular signatures that will expose novel functional connections complementary to gene expression profiles. Quantitative, mass spectrometry-based proteomics is one tool of choice for generating these profiles because it can provide direct observation of these post-translational events whereas nucleic acid sequence-based techniques cannot.
The majority of protein kinases are S/T-directed and the levels of phosphoserine (pS) and phosphothreonine (pT) are generally higher in abundance than phosphotyrosine (pY) sites. Although there are >70,000 known pS/pT sites in the human proteome (8, 18, 19), protein phosphorylation is typically present at sub-stoichiometric levels. Because of the level of phosphorylation and its role in many cell signaling processes, analytical techniques to enrich for protein phosphorylation have been developed. For example, antibody-based assays have been developed to study tyrosine phosphorylation (14, 20, 21), and metal affinity-based methods have been used to enrich pS, pT, and pY-containing peptides from proteolytic digests of cells and tissues (22, 23). In combination with highly sensitive mass spectrometry workflows, these enrichment techniques have facilitated global phosphoproteomic studies in many biological systems (24–27).
However, to facilitate modern “omics” analyses and leverage techniques pioneered in gene expression studies, it would be highly desirable to have reproducible observations of phosphopeptide analytes across large numbers of samples generated under different conditions. Yet, even with the fastest and most sensitive MS instruments currently available, it is not possible to reproducibly measure all of the same peptides or modified peptides across multiple experiments using data-dependent analysis methods. Comparisons across even small numbers of proteomic experiments are difficult because differences in sample processing protocols and mass spectrometry data acquisition methods can cause sampling variation (28–31). Variations in data production may result in the lack of phosphosite detection even when the modification is present. As a result, it is currently not possible to reproducibly and quantitatively monitor all known phosphosites in a large number of human phosphoproteome samples.
To overcome these challenges, we considered that phosphorylation is mediated by just a few hundred protein kinases and phosphatases that can modify hundreds of thousands of amino acid sites on various protein targets. We hypothesized that the implicit one-to-many relationship of kinases to substrates suggests that there is some redundancy in the cellular information conveyed by phosphorylation and that collapsing the number of monitored sites based on their coordinate activity could provide a core set of highly informative phosphopeptide probes. This idea is consistent with work published by Alcolea et al., where phosphosignaling events within acute myeloid leukemia cell lines with different sensitivities to kinase inhibitors were profiled to reveal several hundred correlated phosphorylation sites that were involved in parallel kinase pathways (32). In addition, a similar strategy was used to develop the “L1000” reduced-representation transcriptional profiling assay that is the basis for the transcriptional Connectivity Map project. The L1000 assay retains 80% of information content at <1% of the cost of microarray or RNA-Seq-based expression profiling (33).
Monitoring a reduced-representation set of phosphopeptide probes using a targeted MS approach could be a time- and cost-effective approach to monitor changes in phosphosignaling in response to multiple drug and genetic perturbations. Such an assay could identify connections between molecular perturbations and elucidate cell type-specific cell signal transduction using analysis methods similar to those utilized in the transcriptional profiling field. A similar approach was recently reported by Picotti and colleagues, who developed a targeted proteomic assay to probe biological processes in Saccharomyces cerevisiae in response to environmental perturbations by selecting sentinel proteins from existing data (34). Such targeted approaches could eliminate stochastic sampling effects and allow for accurate quantification across large sample sets without significant loss in information content relative to a full phosphoproteome.
The work described below explores this possibility and consists of three main sections: (1) a discovery arm where we identify a high-value set of phosphopeptide probes from traditional, data-dependent large scale SILAC-based phosphoproteomic data, (2) a configuration arm where we develop a targeted, internally standardized phosphopeptide assay that generates almost complete data (data that contains observations for all phosphopeptides), and (3) a proof-of-principle arm where we explore the sensitivity of the assay to diverse perturbations and biological systems and demonstrate its general utility. In our discovery arm we produced global phosphorylation data representing 156 samples that included 26 chemical perturbations in three different cell lines using conventional phosphopeptide enrichment and MS-based techniques. From these data we selected representative phosphosites (and their associated observable peptides) from clusters of sites that exhibited coordinated regulation across discovery experiments. We then configured a targeted parallel reaction monitoring (PRM) assay against these phosphopeptide probes using isotopically-labeled synthetic analogs, and in parallel, developed automated workflows that enabled proteolytic digestion, protein quantification, peptide desalting, and phosphopeptide enrichment from 96 samples simultaneously in anticipation of scaling the assay to generate a large corpus of data. Our initial evaluation of the configured assay was to regenerate samples under similar conditions to those used in our discovery experiments to see if we could recapitulate the previously observed relationships using only the reduced-representation phosphoproteome at a fraction of the effort required for deeper phosphoproteomic profiling. Finally, in our proof-of-principle arm we extended our experimental conditions to vary the treatments, cell types, and time points measured with the explicit goal of demonstrating that the assay could generate phosphoproteomic profiles outside of the parameters under which it was developed and to demonstrate that the assay is responsive to disruptions of signaling pathways of known biological importance.
We call this assay “P100” because it measures ∼100 phosphopeptide probes in a single 60 min assay. This assay is a phosphosignaling analog of our Global Chromatin Profiling assay (35, 36) that has already proven to be of great utility. We believe that the P100 assay will prove highly useful for classification and stratification of drug and genetic mechanisms as facilitated through comparison of phosphoproteomic signatures of known chemical and genetic perturbations to those of novel perturbations or those where mechanistic insight is currently lacking. Together, these proteomic signature generation assays can form the basis for proteomic Connectivity Maps to complement their transcriptional analogs.
EXPERIMENTAL PROCEDURES
Large-Scale, Discovery SILAC Cell Culture
MCF7 cells were cultured in DMEM GlutaMAX medium (Gibco, 10567–014, Waltham, MA) supplemented with 10% FBS (Sigma, F4135, St. Louis, MO), 1% penicillin/streptomycin (Gibco, 10378–016), and 0.8% glucose (Sigma, G8769). PC3 cells were cultured in RPMI medium (Gibco, 11875–093) supplemented with 10% FBS (Sigma, F4135), 1% penicillin/streptomycin (Gibco, 10378–016), 1% HEPES (Gibco, 15630), and 1% sodium pyruvate (Gibco, 11360). HL60 cells were cultured in RPMI medium (Gibco, 11875) supplemented with 10% FBS (Sigma, F4135), and 1% penicillin/streptomycin (Gibco, 10378–016). Either normal l-lysine (K0) and l-arginine (R0) or heavy-labeled 13C6-15N2 lysine (K8) and 13C6-15N4arginine (R10) were supplemented at concentrations of 40 mg/L and 120 mg/L, respectively, for SILAC cell culture.
Assay Validation Cell Culture
MCF7 cells were cultured in DMEM GlutaMAX medium (Gibco, 10567–014) supplemented with 10% FBS (Sigma, F4135), 1% penicillin/streptomycin (Gibco, 10378–016), and 0.8% glucose (Sigma, G8769). PC3 cells were cultured in RPMI medium (Gibco, 11875–093) supplemented with 10% FBS (Sigma, F4135), 1% penicillin/streptomycin (Gibco, 10378–016), 1% HEPES (Gibco, 15630), and 1% sodium pyruvate (Gibco, 11360). HL60 cells were cultured in RPMI medium (Gibco, 11875) supplemented with 10% FBS (Sigma, F4135), and 1% penicillin/streptomycin (Gibco, 10378–016). Lines were propagated according to standard tissue-culture practices.
Drug Treatment and Harvest
The following applies to discovery, configuration, and confirmation experiments. Compounds were obtained from Sigma (St. Louis, MO) or EMD Millipore (Darmstadt, Germany), with the exception of JQ-1 which was the generous gift of Dr. James Bradner (Dana-Farber Cancer Institute, Boston, MA). Once cells reached ∼95% confluence, they were treated with the compounds listed in Table I and supplemental Table S1 for 6 h at 37 °C. After 6 h, the cells were washed twice with cold PBS (Gibco, 10010–023) and harvested by scraping. Cells were pelleted at 1000 × g for 2 min. The supernatant was then removed, and the cell pellet was frozen in liquid nitrogen until cell lysis and phosphopeptide enrichment.
Table I. Compounds and concentrations used for discovery experiments.
| Compound | Treatment concentration (μm) | Compound | Treatment concentration (μm) |
|---|---|---|---|
| Captopril | 17.2 | Mesalazine | 26.2 |
| Ambroxol | 9.6 | Anisomycin | 15 |
| Paclitaxel | 1 | Digitoxigenin | 10.6 |
| Carmustine | 100 | Digoxigenin | 10.2 |
| Fulvestrant | 1 | Digoxin | 5.2 |
| Clindamycin | 8.6 | Lanatoside C | 4 |
| Chlortetracycline | 7.8 | Irinotecan | 100 |
| Chlorzoxazone | 23.6 | Scriptaid | 10 |
| Doxorubicin | 6.8 | Trichostatin A | 1 |
| Daunorubicin | 7 | MS-275 | 10 |
| GW-8510 | 10 | Valproic acid | 1000 |
| Staurosporine | 1 | H-7 | 100 |
| Dexverapamil | 10 | Geldanamycin | 1 |
For discovery experiments, we used three-state SILAC labeling to determine quantification of phosphoproteomic changes. In these experiments, we held the “light” channel constant as DMSO and varied the drugs in the medium and heavy channels (as schematically depicted in Fig. 1C and listed in Table I). For each drug/cell type combination, two complete biological repeats (grown several weeks apart) were performed. Ratios were determined using MaxQuant (see below) of treatment versus DMSO for each drug. The entirety of the discovery data set, including a table that specifies SILAC labeling states for each of the 26 drug combinations, can be found in ftp://MSV000079524@massive.ucsd.edu. This data set has been deposited in MassIVE with accession ftp://MSV000079524@massive.ucsd.edu.
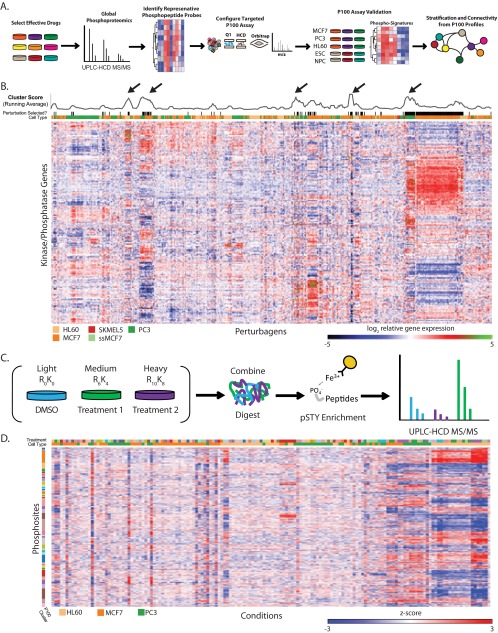
Fig. 1.
Large-scale transcriptional and phosphoproteomic profiling data for identification reduced-representation phosphoproteome probes. A, A schematic depicting the development of the P100 assay. Drug treatments that modulated phosphorylation in pleiotropic ways were selected, and large-scale global phosphorylation data were collected from multiple cell types treated with these drugs. Representative phosphopeptide probes were identified from these data and used to configure the P100 assay. Confirmation and proof-of-principle of the P100 assay's functionality were demonstrated via classification and stratification of samples from multiple biological contexts. Associations among samples were then identified using P100 data. B, Affymetrix gene expression levels corresponding to genes annotated as kinases or phosphatases were extracted from the CMap database and clustered (dendrograms omitted for clarity). Combinations of cell/compound treatments with similar kinase/phosphatase gene expression profiles are illustrated and groups with similar profiles are summarized by the “Running Average” graphic above. Arrows indicate strongly coordinated groups of samples. C, A workflow for the large-scale discovery experiments is depicted. Cells were grown in SILAC medium, lysed, digested, fractionated, and analyzed using high resolution UPLC-MS. D, A clustered heatmap representing the 1,200 commonly observed phosphosites that were present in >75% of all MS experiments is displayed. Groups of phosphosites with coordinate activity are clustered together along the vertical axis, whereas samples are clustered along the horizontal axis (dendrograms omitted for clarity). Data underlying the figure are available in Supplemental Data Set 1.
For all other experiments, cells were grown in typical growth medium without metabolic labeling of proteins. Instead, the synthetic versions of the P100 assay peptides (probes) are used to derive quantitative information.
Time-course Drug Treatment and Harvest
Compounds were obtained from Sigma or EMD Millipore (Darmstadt, Germany). Once cells reached ∼95% confluence, they were treated with the compounds in supplemental Table S5 for either 3, 6, or 24 h at 37 °C. After 3, 6, or 24 h, the cells were washed twice with cold PBS (Gibco, 10010–023), lysed in plate in urea buffer (8 m urea; 75 mm NaCl, 50 mm Tris HCl pH 8.0, 1 mm EDTA, 2 μg/ml aprotinin (Sigma, A6103), 10 μg/ml leupeptin (Roche, #11017101001, Basel, Switzerland), 1 mm PMSF (Sigma, 78830), 10 mm NaF, Phosphatase Inhibitor Mixture 2 (1:100, Sigma, P5726), Phosphatase Inhibitor Mixture 3 (1:100, Sigma, P0044), and harvested by scraping. Cells were lysed for 30 min at 4 °C and then frozen at −80 °C. Prior to phosphopeptide enrichment, cell lysates were clarified by centrifugation at 15,000 × g for 15 min.
Embryonic Stem Cell and Neural Progenitor Cell Culture and Treatment
Individual colonies of H9 human embryonic stem cells (ESCs) were cultured with mouse embryonic fibroblast-conditioned media (MEF-CM; provided by Sanford-Burhnam Research Institute, La Jolla, CA) in matrigel (BD, 356231)-coated plates. For neural progenitor cell (NPC) induction, ESC cell colonies of 60–80% confluence were incubated in MEF-CM containing each 5 μm of dorsomorphin (Sigma, P5499), A83–01 (Sigma, SML0788) and PNU 74654 (Sigma, P0052). Once NPC cells and reached ∼95% confluence, they were treated with the compounds listed in Supplemental Table S2 for 24 h at 37 °C. H9 cells or NPCs were harvested by washing twice with cold PBS (Gibco, 10010–023) and centrifugation at 1000 × g for 2 min. The supernatant was removed, and the cell pellets were frozen in liquid nitrogen until cell lysis and phosphopeptide enrichment.
Manual Sample Processing
Reduction, Alkylation, and Digestion
Frozen cell pellets were lysed for 30 min at 4 °C in urea buffer (8 m urea; 75 mm NaCl, 50 mm Tris HCl pH 8.0, 1 mm EDTA, 2 μg/ml aprotinin (Sigma, A6103), 10 μg/ml leupeptin (Roche, #11017101001), 1 mm PMSF (Sigma, 78830), 10 mm NaF, Phosphatase Inhibitor Mixture 2 (1:100, Sigma, P5726), Phosphatase Inhibitor Mixture 3 (1:100, Sigma, P0044). Lysates were centrifuged at 20,000 × g for 10 min, and protein concentrations of the clarified lysates were measured via 660 protein assay (Pierce, 22660). Five hundred micrograms of protein was reduced for 45 min with 5 mm dithiothreitol (DTT) and alkylated for 45 min with 10 mm iodoacetamide (IAA). Samples were diluted to 2 m urea with 50 mm Tris HCl, pH 8 (1:4 dilution). Lysates were digested overnight at room temperature with trypsin in a 1:50 enzyme-to-substrate ratio (Promega, V511X, Madison, WI) on a shaker. Digestion was stopped by bringing the samples to a final concentration of 0.5% TFA. Acidified samples were loaded onto a 100 mg capacity C18 SepPak (Waters, Milford, MA) for desalting. Samples were eluted using 50% ACN/0.1% formic acid and vacuum concentrated to dryness (37).
Discovery Phosphopeptide Enrichment
Phosphopeptide enrichment was completed using Phos-Select Iron Affinity gel (Sigma, P9740) that were prepared by washing four times with 40% acetonitrile/0.1% formic acid. Prior to enrichment, peptides were reconstituted in 40% acetonitrile/0.1% formic acid. Phosphorylated peptides were enriched for with 15 μl IMAC beads for each sample for 30 min. Phosphopeptide enrichment was completed as previously described (23). After enrichment, Phos-Select gel was loaded on Empore C18 silica-packed stage tips (3M, 2315, St. Paul, MN) and desalted (38). Briefly, StageTips were equilibrated with 2 × 100 μl washes of methanol, 2 × 50 μl washes of 50% acetonitrile/0.1% formic acid, and 2 × 100 μl washes of 1% formic acid. Samples were then loaded onto stage tips and washed twice with 50 μl of 80% acetonitrile/0.1% trifluoroacetic acid and 100 μl of 1% formic acid. Phosphorylated peptides were eluted from IMAC beads with 3 × 70 μl washes of 500 mm dibasic sodium phosphate, pH 7.0, and washed twice with 100 μl of 1% formic acid before being eluted from stage tips with 100 μl 50% acetonitrile/0.1% formic acid. All washes were performed on a tabletop centrifuge at a maximum speed of 3,500 × g.
Automated Sample Processing
Reduction, Alkylation, and Digestion
Frozen cell pellets were lysed for 30 min at 4 °C in lysis urea buffer (8 m urea; 75 mm NaCl, 50 mm Tris HCl pH 8.0, 1 mm EDTA, 2 μg/ml aprotinin (Sigma, A6103), 10 μg/ml leupeptin (Roche, #11017101001), 1 mm PMSF (Sigma, 78830), 10 mm NaF, Phosphatase Inhibitor Mixture 2 (1:100, Sigma, P5726), Phosphatase Inhibitor Mixture 3 (1:100, Sigma, P0044). Lysates were centrifuged at 20,000 × g for 10 min, and protein concentrations of the clarified lysates were measured via A660 protein assay (Pierce, 22660). 500 μg of protein was transferred to a Bravo robotic liquid handling platform (Agilent) for reduction in 100 mm DTT, alkylation in 200 mm IAA, dilution to 2 m urea in 50 mm Tris (pH 8.0), and digestion with 0.5 μg/μl (1:50) sequencing-grade modified trypsin (Promega) in 400μl volumes per sample at 37 °C overnight. Digestion was stopped by bringing the samples to a final concentration of 0.5% TFA (see Supplemental Protocol 1). Acidified samples were loaded onto a 25 mg capacity C18 SepPak in a 96-well plate format (Waters) for desalting. Samples were eluted using 50% ACN/0.1% formic acid and vacuum concentrated to dryness.
Phosphopeptide Enrichment
Desalted samples were reconstituted in 80% ACN/0.1% TFA that contained isotopically labeled standards for enrichment quality control and moved to an AssayMAP Bravo robotic system (Agilent, Santa Clara, CA). AssayMAP cartridges containing Ni-NTA-agarose packing material (Qiagen, 1018611) were washed with water, stripped with 100 mm EDTA, and loaded with 100 mm FeCl3. Fe-NTA cartridges were primed with 1:1:1 ACN/methanol/0.01% acetic acid, and samples were loaded at 5 μl/min. Flow-throughs were re-loaded onto cartridges at a flowrate of 2 μl/min. Cartridges were washed with 80% ACN/0.1% TFA, and peptides were eluted with 500 mm K2HPO4 (pH7) at 5 μl/min. Eluates were vacuum concentrated to dryness, and subsequently desalted using AssayMAP RP-S cartridges according to the manufacturer's instructions. A detailed protocol is shown as Supplemental Protocol 2.
Automated Sample Processing (Time-Course Experiment Only)
Reduction, Alkylation, and Digestion
Cells were lysed for 30 min at 4 °C in lysis urea buffer (8 m urea; 75 mm NaCl, 50 mm Tris HCl pH 8.0, 1 mm EDTA, 2 μg/ml aprotinin (Sigma, A6103), 10 μg/ml leupeptin (Roche, #11017101001), 1 mm PMSF (Sigma, 78830), 10 mm NaF, Phosphatase Inhibitor Mixture 2 (1:100, Sigma, P5726), Phosphatase Inhibitor Mixture 3 (1:100, Sigma, P0044). Lysates were centrifuged at 15,000 × g for 15 min, and protein concentrations of the clarified lysates were measured via 660 protein assay (Pierce, 22660). Five hundred micrograms of protein was transferred to a Bravo robotic liquid handling platform (Agilent) for reduction in 100 mm DTT, alkylation in 200 mm IAA, dilution to 2 m urea in 50 mm Tris (pH 8.0), and digestion with 0.5 μg/μl (1:50) sequencing-grade modified trypsin (Promega) in 400μl volumes per sample at 37 °C overnight. Digestion was stopped by bringing the samples to a final concentration of 0.5% TFA (see supplemental Protocol S1). Acidified samples were loaded onto a 25 mg capacity C18 SepPak in a 96-well plate format (Waters) for desalting. Samples were eluted using 50% ACN/0.1% trifluoroacetic acid and vacuum concentrated to dryness.
Phosphopeptide Enrichment
Desalted samples were reconstituted in 80% ACN/0.1% TFA that contained isotopically labeled standards for enrichment quality control and moved to an AssayMAP Bravo robotic system (Agilent). Agilent AssayMAP Fe-(III)-NTA cartridges (note that we changed the enrichment media from the previous experiment) were washed with water, stripped with 100 mm EDTA, and loaded with 100 mm FeCl3. Fe-(III)-NTA cartridges were primed with 1:1:1 ACN/methanol/0.01% acetic acid, samples were loaded at 20 μl/min and flow-throughs were re-loaded onto cartridges eight additional times. Cartridges were washed with 80% ACN/0.1% TFA, and peptides were eluted with 500 mm K2HPO4 (pH7) at 5 μl/min. Eluates were vacuum concentrated to dryness, and subsequently desalted using AssayMAP RP-S cartridges according to the manufacturer's instructions. A detailed protocol is shown in supplemental Protocol S3: Optimized Automated Phosphopeptide Enrichment SOP using Agilent NTA-polymeric resin.
Discovery Mass Spectrometry
Each sample was subjected to 3 consecutive LCMS analyses with the following rationale. First, data were acquired using a typical data-dependent top 12 method (parameters below). These data were searched using MaxQuant version 1.2.2.5 (see below). A second round of acquisition was performed using tailored exclusion lists (AMEx (30)) for each sample to minimize re-acquisition of the same precursors in consecutive analyses. Again, these data were searched in MaxQuant. When the first two rounds of acquisition and search were complete, we determined peptides (and their corresponding precursor m/z and retention times) that had been identified in a majority of the samples but lacked quantitative information in some samples. We rationalized that targeted MS/MS acquisition for these peptides would lead to a more complete data matrix for analysis. Therefore, we created individual precursors lists for each sample to “fill in the blanks” consistent with the principles of AIMS (39) data acquisition.
All three acquisition strategies employed the same LC separation conditions described below. Samples were chromatographically separated using a Proxeon Easy NanoLC 1000 (Thermo Scientific) fitted with a PicoFrit (New Objective, Woburn, MA) 75-μm inner diameter capillary with a 10 μm emitter tip was packed under pressure to ∼20 cm with C18 Reprosil beads (1.9 μm particle size, 200 Å pore size, Dr. Maisch GmBH) and heated at 50 °C during separation. Samples were loaded in 4 μl 3% ACN/1% formic acid and peptides were eluted with a linear gradient from 7–30% of Buffer B (0.1% FA and 90% ACN) over 82 min, 30–90% Buffer B over 6 min and then held at 90% Buffer B for 15 min at 200 nL/min (Buffer A, 0.1% FA and 3% ACN).
During data-dependent acquisition, eluted peptides were introduced into a Q-Exactive mass spectrometer (Thermo Scientific) via nanoelectrospray (2.15 kV). A full-scan MS was acquired at a resolution of 70,000 from 300 to 1800 m/z (AGC target 1e6, 5ms Max IT). Each full scan was followed by top 12 MS2 scans at a resolution 17,500 (Isolation width 2.5 m/z, ACG Target 5e4, 120 ms Max IT).
For AMEx (30) runs, individual exclusion lists were added and the match tolerance was set as the software default. Dynamic exclusion was set at 20 s.
For AIMS (37) runs, individual inclusion lists were added and the match tolerance was set as the software default. If fewer than 12 inclusion list targets were active, then the balance of MS/MS scans were selected from next most intense precursors subject to dynamic exclusion (20 s).
Targeted Mass Spectrometry
Targeted LCMS Data Acquisition
Samples were chromatographically separated using the same conditions as the Discovery Mass Spectrometry with the following changes. Samples were reconstituted in 10 μl 3% ACN/5% formic acid containing isotopically labeled versions of all phosphopeptide probes. Peptides were eluted using a shorter linear gradient from 3–40% of Buffer B over 45 min, 40–90% Buffer B over 5 min and then held at 90% Buffer B for 10 min at 200 nL/min.
Eluted peptides were introduced into a Q-Exactive mass spectrometer (Thermo Scientific) via nanoelectrospray (2.15 kV). A full-scan MS was acquired at a resolution of 35,000 from 300 to 1800 m/z (AGC target 3e6, 50 ms Max IT). Each full scan was followed by fully scheduled, targeted HCD MS/MS scans at resolution 17,500 (Isolation width 2 m/z, ACG Target 2e5, 50 ms Max IT). Each peptide species was subjected to targeting MS/MS for 3–5 min depending on the empirical chromatographic properties, centered on the average observed retention time of two scheduling runs containing synthetic versions of a subset of isotopically labeled phosphopeptide probes.
For the time-course experiments, eluted peptides were introduced into a Q-Exactive Plus mass spectrometer (Thermo Scientific) with the same parameters described above with the following exceptions: full-scan MS was acquired at 35,000 from 300 to 1200 m/z (AGC target 3e6, 20 ms Max IT) and HCD MS/MS scans at 17,500 (Isolation width 1.7 m/z with 0.3 m/z offset, AGC target 2e5, 50 ms Max IT).
The raw mass spectrometry data have been deposited in the public proteomics repository MassIVE and are accessible at ftp://MSV000079524@massive.ucsd.edu. Skyline files corresponding to the targeted analyses can be accessed at: https://panoramaweb.org/labkey/project/LINCS/AbelinSupplemental/begin.view?.
MaxQuant Data Analysis
Raw MS Data were searched with MaxQuant version 1.2.2.5 against the Uniprot Human Protein Database (Complete Isoforms, download date 20-MAR-2012) containing 81489 entries. Three-state SILAC was specified: Arg0:Lys0, Arg6:Lys4, and Arg10:Lys8, and the following modifications were allowed: Carbamidomethylation of C (fixed); Oxidation of M, Phosphorylation of S, T, and Y, Acetylation of N termini (variable). The mass tolerances allowed were 7 ppm for MS precursors and 20 ppm for MS/MS fragments. Peptide, protein, and site FDRs were 0.01 as calculated using by MaxQuant. H/L and M/L ratios were extracted for phosphosites, and log2-transformed. All MaxQuant tables, including parameters, can be found in ftp://MSV000079524@massive.ucsd.edu. Phosphosites that were observed in at least 75% of all experiments were retained. If a data point was missing for a particular phosphosite in a given condition, we imputed its value by random sampling of a normal distribution based on the mean and standard deviation of all other measurements of that phosphosite in other conditions (6.9% of values were imputed). Subsequently, variance was measured for each phosphosite, and sites with the lowest 15% of variance across all experiments were discarded as uninformative. The final yield of sites used for subsequent calculations and selections was ∼1000.
We performed hierarchical clustering of phosphosite ratios using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html), clustering both samples and sites using a distance metric of 1-Pearson's correlation. Principal Component Analysis of these data was performed using R. After reducing the data to 55 distinct clusters, representative phosphopeptides were selected from each cluster using an in-house Perl-script that combed through MaxQuant data tables, ranking each by heuristic rules designed to maximize success in automated proteomic workflows and data analysis (i.e. easy site localization, high re-observability, absence of missed tryptic cleavages, etc.). The peptides that were ultimately selected can be found in supplemental Table S3.
Skyline Data Analysis
Data Analysis
Files were imported into a Skyline-daily (40) with pre-selected charge states for all reduced-representation phosphopeptide probes (Supplemental Skyline Document 1) and for the quality control internal standard phosphopeptide probes (Supplemental Skyline Document 2). Transitions were chosen on the basis of selectivity for phosphosite localization and detectability. Each sample and phosphopeptide probe was manually validated using the criteria of retention-time agreement with other samples and the co-eluting presence of all transitions with corresponding isotopically labeled standards. Heavy/light ratios were extracted on the basis of transition area integration using Skyline defaults. Data for each modification were normalized by the median of all samples before clustering. Clustering was performed in GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html) using unsupervised hierarchical methods with the following methods: Pearson correlation, row and column clustering.
Computation and Visualization of Connectivity Maps
All possible intersample Pearson correlation coefficients were calculated using P100 profiles. A “sample group” was defined for each set of replicates for a given compound in a given cell type. An intergroup connectivity was assigned as the average correlation between all replicates in one group to another group. As an example, let us assign Group A as the three replicates of digoxin in MCF7 cells and Group B as the 3 replicates of lanatoside C in PC3 cells. It would follow that the intergroup connectivity between Group A and Group B is the average of the nine correlation coefficients possible between replicates of these groups.
Cytoscape (41) was used to visualize sample groups and the computed intergroup connectivity. Node arrangements were computed using preset spring-embedded and/or force-directed layouts options, using the intergroup correlations as the weights. Edge thickness is directly proportional to the intergroup connectivity.
RESULTS
We set out to develop a rapid and robust targeted proteomic assay that could generate molecular signatures based on the phosphorylation state of cellular proteins. An overview of this process is presented in Fig 1 A. We first identified small molecules that were predicted to modulate phosphorylation in pleiotropic (but non-uniform) ways. We then treated multiple cell types with these compounds and collected large-scale global phosphorylation data. We rationalized that, because a small number of kinases and phosphatases (<1000) seemingly modulate a large number of phosphosites (>100,000), many sites were likely to be coordinately regulated. Thus, traditional “deep” phosphoprofiling may convey redundant information when the levels of two or more sites are modulated similarly across a wide number of perturbations. We hypothesized that a smaller set of sites could serve as surrogate markers for a larger set. From our global phosphorylation data, we were able to identify groups of phosphosites that behaved similarly upon drug treatment, from which we selected highly-observable representative phosphopeptides. We then configured a reduced-representation targeted MS assay for these phosphopeptide probes (“P100”). We executed the P100 assay in new samples under similar treatment conditions used in our discovery experiments to confirm that the assay was functional. We also performed the P100 assay on samples from two new cell types and several additional compounds that were not used for assay configuration to further demonstrate the functionality of the assay under diverse conditions. Finally, we compared phosphosignaling profiles collected from cells treated with compounds of known mechanisms in a time-course experiment to demonstrate that the assay is sensitive to disruptions in multiple common signaling pathways, including MAPK, PI3K/mTOR, and CDK cell cycle. These experiments also allowed us to analyze the time-dependence of emergence of phosphosignatures. Overall, we established a data generation platform that enables the production of large-scale, phosphosignaling signatures that can be used to stratify drugs and other perturbations into classes and to compare known reference drugs to novel compounds, helping to further elucidate their mechanisms.
Discovery Experiments
To find small molecules with pleiotropic effects on phosphosignaling, we analyzed gene expression data in the Connectivity Map (CMap, https://www.broadinstitute.org/cmap/) for compound treatments that positively and/or negatively regulated the expression of groups of kinase and phosphatase genes. The hypothesis was that, if a set of kinases' expression is modulated, then the phosphosites that they regulate should be coordinately modulated as well. We avoided highly specific, single kinase inhibitors, as such compounds would not be likely to generate the desired pleiotropic effects on the phosphoproteome.
At the time of our analyses, the CMap data set consisted of Affymetrix array data for >700 samples across cell lines representing 4 lineages: breast cancer (with both regular and serum stripped growth conditions), skin cancer, prostate cancer, and leukemia, where each sample is the collapsed gene expression profile (derived from >6000 individual profiles, or 8–9 profiles/sample) of a compound treatment in a particular cell type. We began by extracting all Affymetrix probes corresponding to genes annotated as kinases or phosphatases (a total of 317 probes). We then clustered these data in the sample dimension, such that cell/compound treatment combinations with similar kinase/phosphatase gene expression profiles segregated together (Fig. 1B). We scored each cluster for the number of genes positively or negatively regulated. Examples of high scoring clusters are indicated by arrows along the top in Fig. 1B. Representative compounds were selected from high-scoring clusters, with an emphasis on diversity of the compounds (indicated by tick marks in the track below the running average in Fig 1B). In some cases we selected several structural analogs of a compound family to compare their effects on cells. Ultimately, we selected 26 compounds (including 1 broad spectrum kinase inhibitor and 1 broad spectrum phosphatase inhibitor; Table I) that we deemed to be strong candidates for general modulation of kinase and phosphatase genes across a range of cell types.
Next, we measured the cellular phosphoproteome after perturbation by these 26 different compound treatments (compared with DMSO control) across three cell lines (MCF7, PC3, and HL60) in biological duplicate, for a total of 156 experiments (scheme in Fig. 1C). The amalgamated data set quantified more than 10,000 unique phosphosites. Importantly, we found over 1200 sites that were quantified in >75% of all experiments. Using only these 1200 sites, we imputed missing values as described above, and low variance phosphosites were discarded as uninformative (bottom 15%). To better visualize the data set, we performed hierarchical clustering of both the conditions (x axis) and the detected phosphosites (y axis) (Fig. 1D).
We analyzed the discovery data set with an eye toward finding reproducible responses (signatures) of drug treatments coordinately exhibited by multiple phosphosites for single or related conditions (ftp://MSV000079524@massive.ucsd.edu). If such signatures and coordinated groups could be found, we could then select representative or “proxy” phosphopeptide marker(s) and thereby formulate a condensed phosphoproteomic assay. Selected regions of the heatmap from Fig. 1D are shown in Fig. 2 to illustrate these concepts. We show a unified response to paclitaxel treatment across all three cell lines studied in Fig. 2A, with up-regulation of several phosphosites across a diverse set of proteins. Note that all 6 biological replicates of paclitaxel treatment cluster together in the heatmap regardless of the cell line being studied (MCF7, PC3, or HL60). We call this a lineage-independent signature, and it is notable as these cell lines are of diverse origin (including a hematological line that grows in suspension, HL60). Although many other phosphosites also contribute to these treatments clustering together, these particular sites (shown in Fig. 2A) are a major driver of the clustering.
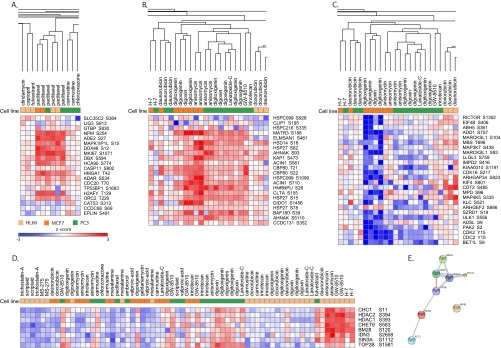
Fig. 2.
Diverse compound treatments produce unique phosphosignatures in discovery data. Selected regions of the large-scale discovery heatmap (Fig. 1D) are shown. A, Phosphosignatures of paclitaxel-treated cells cluster together regardless of cell line (MCF7, PC3, or HL60), and an up-regulation of several phosphosites across a diverse set of proteins is shown. Both lineage-independent (B) and lineage-specific signatures (C) across a set of cardiac glycoside structural analogs (digoxin, digoxigenin, digitoxigenin, and lanatoside (C) are shown. D, A cluster of phosphosites that appear to be coordinately regulated across a diverse set of conditions. The parent genes of these sites have a strong bias toward chromatin function. E, A tight network of protein-protein interactions in which 7 of the 8 parent genes in the chromatin binding cluster participate.
We demonstrate both lineage-independent and lineage-specific signatures across a set of structural analogs in Fig. 2B and 2C. Digoxin, commonly known as Digitalis, is a cardiac glycoside widely used in the treatment of various cardiovascular conditions (42). Its structural analogs digoxigenin, digitoxigenin, and lanatoside C are reported to have similar bioactivity. Nearly all biological replicates of these compounds cluster together across the MCF7 and PC3 cell lines and elicit a common up-regulation of a series of phosphosites (Fig. 2B). Interestingly, the antibiotic protein synthesis inhibitor anisomycin also clusters together with the cardiac glycosides, although this observation is of unknown significance. In parallel, we can also detect a lineage-specific down-regulation of a series of sites that only seems to occur in MCF7 cells (Fig. 2C). These examples show that phosphoproteomic signatures are potentially useful in recognizing common mechanisms of actions of related compounds yet retain enough information to differentiate subtleties in cellular response.
Although we believe that these phosphoproteomic signatures are generalizable and are useful on their own, the derivation of specific biological information from these data is also desirable. We noticed an interesting cluster of phosphosites in the data set that seemed to be coordinately regulated across a diverse number of conditions (Fig. 2D). By “coordinately regulated,” we mean that the group of sites went up or down as a group in response to different treatment contexts and across multiple cell types. We immediately recognized that the parent genes of the sites shown in Fig. 2D seemed to have a strong bias toward chromatin function, and this was borne out by performing a functional enrichment test against the Gene Ontology categories which showed that the parent genes were enriched for Chromatin Binding (GO:0003682) with an FDR-adjusted p value of 7 × 10−3 when compared with all parent genes in the data set. We further learned that seven of the eight parent genes in the cluster are known to participate in protein-protein interactions in a tight network (Fig. 2E) in the STRING database (43). This observation raises the intriguing possibility of direct regulation of the chromatin machinery by phosphosignaling in a coordinated manner. Although there are likely numerous other biological insights to be gained from deeper analysis of the discovery data set, this was beyond the scope the present study, the aim of which was to develop an efficient phosphosignaling panel for wider deployment.
Reducing Representation and Configuring the P100 Assay
We employed a reductionist approach to collapse the large universe of phosphosites that could be monitored into a relatively small number of detectable surrogate targets, referred to as the reduced representation set. This innovation will allow retention of important signaling information at a fraction of the effort normally associated with deep phosphoproteomic profiling. This idea draws inspiration from the L1000 assay developed for the Library of Integrated Network-based Cellular Signatures (LINCS) at Broad Institute that reduces the number of transcripts needed for monitoring global expression profiling. LINCS efforts have resulted in a 1000-plex Luminex-based gene expression assay that retains 80% of information content at <1% of the cost of microarray or RNA-Seq-based expression profiling. A similar proteomic approach has recently been illustrated by Soste et al., in the context of signaling in yeast (34).
We began by performing hierarchical clustering on our high quality phosphoproteomic data as depicted in Fig. 1D and Fig. 2. Using principal component analysis, we estimated that it would take at least ∼40–50 components to explain 80% of the total variance in the data set. We therefore made simple linear models and measured the sum-squared-error of these models versus a partial hold-out of the data (cross-validation) for increasing numbers of components above 40. We found a local minimum in the error at 55 components and therefore went on to set thresholds on our hierarchical clustering data that divided the phosphosites into 55 different components. We note that this achieves approximately the same degree of compression (1200 sites to 55 sites, or ∼20 fold) of the L1000 gene expression assay (20,000 genes to 1000 genes). Lastly, we selected 2 phosphopeptide probes from each of the 55 clusters for a total of 110 phosphopeptides (the “P100 probes”). These probes were chosen using heuristic rules designed to maximize mass spectrometric ease of observation (i.e. minimize missed tryptic cleavages, bias toward minimal phosphosite localization possibilities, etc.).
Building on the work above, we developed a high resolution, accurate mass targeted assay to identify and quantify ∼100 highly informative and representative phosphopeptides across multiple cell types treated with various drugs. Several of our initial 110 probes peptide probes could not be successfully configured into the targeted assay for various reasons (i.e. failed peptide synthesis, poor peptide stability, unacceptable MS performance, etc.), resulting in a final panel of 96 phosphopeptide probes in supplemental Table S3. An analysis of the peptides that we selected (using PhosphoMotif Finder) suggests quite a diversity of possible kinase substrates, with 27 unique known motifs spanning a range of kinases (including CDKs, GSK3, PKA, and PKC) among our 96 probes (44) (supplemental Table S4).
Anticipating deploying this assay at a large scale, we automated the sample processing protocol as depicted in Fig. 3A. After a manual cell lysis step, the automated sample processing protocol encompassed protein quantification (using a colorimetric assay), protein digestion, IMAC phosphopeptide enrichment, and sample desalting using Agilent Bravo liquid handling and AssayMAP platforms. Automation facilitated the simultaneous processing of 96 samples in only 3 days, with a plate phosphopeptide enrichment variance of 12% as determined using stable-isotope-labeled internal standards (supplemental Fig. S1). This highly reproducible process allowed ∼95% detection of the reduced-representation phosphopeptides in close to 200 samples. Detailed protocols for the automated portions of the method are provided in supplemental Protocols S1, S2, and S3. We were able to generate phosphosignaling profiles in under 2 weeks from the time of cell harvest to processed results using this assay by directly measuring the reduced-representation set of phosphopeptides across multiple cell lineages and perturbations.
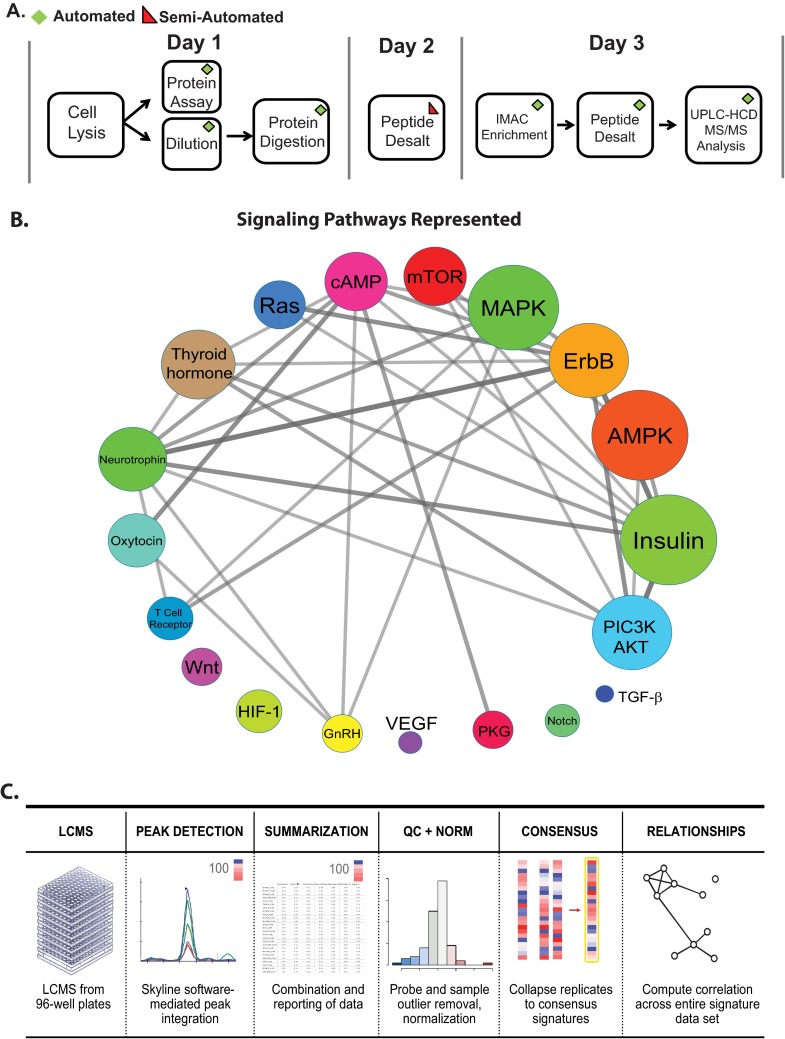
Fig. 3.
P100 automated sample processing, pathway coverage, and data analysis pipeline. A, A schematic of automated P100 sample processing. On the first day, cells are lysed, subjected to protein quantification and diluted to a uniform concentration. Proteins are then reduced, alkylated, and digested. On the second day, samples are desalted using a 96-well device and dried overnight. On the third day, phosphopeptide enrichment by IMAC and desalting occurs. Finally, samples are analyzed using high resolution UPLC-HCD MS/MS. B, The top signaling pathways represented by the larger set of phosphopeptides used to identify the reduced-representation phosphopeptide probe set are shown. Each signaling pathway is depicted as a circle that is sized to indicate the number of source proteins involved in a specific pathway. A larger size indicates that a signaling pathway is represented by a larger number of phosphopeptides. The color of each pathway is only meant to show the diversity of the signaling pathways represented. C, The data analysis pipeline for the P100 assay is shown. Data are collected in a 96-well plate format, analyzed within Skyline, and exported for summarization. Phosphopeptide probes and sample outliers are removed and the light/heavy peptide ratios are normalized. Quality controlled data are hierarchically clustered and molecular signatures for different perturbations are revealed.
The top signaling pathways from the KEGG database (45) represented by the 1200 phosphopeptides that were used to generate the reduced-representation phosphopeptide probe set are shown in (Fig. 3B). The abbreviations for each signaling pathway are shown on a circle that is sized to indicate the number of source proteins involved in a specific pathway that are present in the set of 1200 phosphopeptides. A larger size indicates that a signaling pathway is represented by a larger number of phosphopeptides. The edges of the signaling network in Fig. 3B represent source proteins that are shared among different signaling pathways, and thicker lines indicate a higher number of source proteins are shared between two pathways. We believe that these pathways should be covered in the targeted assay by proxy through the reduced-representation set, and indeed demonstrate this for several of them (see results and discussion of Fig. 7 below). All targeted P100 phosphoproteomics data are analyzed using Skyline (40), and subsequent data reduction, QC, and visualizations are achieved through a series of R-scripts to facilitate reproducible research. These R-scripts are automatically executed upon uploading of P100 Skyline data documents into our Panorama server (https://panoramaweb.org/labkey/LINCS.url). A schematic illustrating the final P100 data analysis pipeline is presented in Fig. 3C.
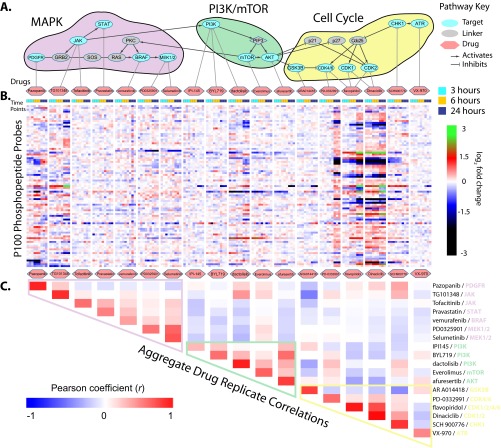
Fig. 7.
Time-resolved signatures from P100 reveal modularity of biologically important signaling pathways. A, Pathway reconstruction showing the targets of a set of drugs known to inhibit important nodes in key signaling pathways. These drugs were administered for 3, 6, and 24 h to MCF7 cells. B, P100 molecular signatures from samples treated with the drugs. No clustering of samples has been performed, but P100 probes (rows) have been clustered (dendrogram omitted). In each major column corresponding to one drug, individual profiles from each sample are ordered according to time point from left to right. C, Correlation matrix of profiles from comparing profiles of samples at the same time point. The boundaries shown are meant to draw attention to the off-diagonal adjacent enrichment of correlation. These boundaries happen to correspond to the modules in the reconstructed pathway, and are colored by pathway module as in Fig. 7A.
P100 Assay Confirmation and Proof-of-Principle
To confirm that the P100 assay could produce molecular signatures of comparable utility to those from larger scale phosphoproteomic data, we replicated a subset of the perturbational conditions from our discovery experiments (supplemental Table S1 and supplemental Data Set S2). We illustrate the results of these experiments in Fig. 4. Fig. 4A presents an overview of the entire data set, where all of the samples are clustered by their P100 signatures (columns) and all of the P100 probes are also clustered (rows). The value in each cell is the ratio of the endogenous (light) version of the peptide to the internal standard (heavy) version, after row-based normalization and Z-score transformation. Therefore, the value is equivalent to the number of standard deviations away from the mean a given phosphopeptide is in a sample relative to all other samples. Importantly, very few missing values are present in this data set (they appear as gray cells when present). After data QC and filtering, we were able to derive acceptable measurements from 141 of 144 possible samples while retaining sufficient data quality from 92 of the 96 P100 probes. Samples were rejected if less than 80% of the probes were observed, and probes were rejected if they were present in less than 90% of samples. It should be noted that a missing value does not indicate that we failed to detect the peptide via its internal standard, but rather that the endogenous level was too low to detect. For the future, we could either assign these values as zeros or use another technique to substitute a suitable value.
Fig. 4.
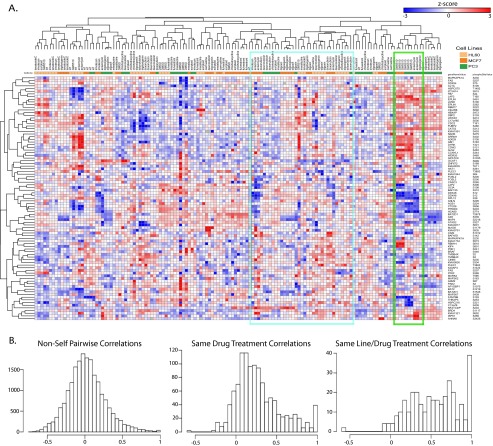
Reduced-representation P100 data recapitulates large-scale discovery data. P100 reduced representation validation data set. A, A heatmap of P100 phosphoprofiles produced from samples generated under the same conditions as the initial large-scale phosphoproteomic experiments. Many of the same sample associations observed in large-scale data can be seen using the reduced-representation P100 assay. Cyan box: Biological replicates and structurally related compounds clustered closely together, as shown for digoxigenin, digitoxigenin, digoxin, and lanatoside C treatments. Green box: Phosphoprofiles of GW8510 treatments cluster across cell lines, while retaining some cell line-specific responses. B, A summary of the pairwise correlations between non-self (a sample of one cell type treated with one compound compared with all other cell line and treatment combinations), the same drug treatment across cell lines (all biological replicates of the same drug compared in two different cell lines), and the same drug treatment within each cell line are shown (all biological replicates of the same drug compared in a single cell line).
We isolated regions of this heatmap from Fig. 4A, shown as cyan and green boxes, to draw attention to how samples treated with similar compounds clustered together. In general, we observed that biological replicates and structurally related compounds clustered closely together in the assay. For example, phosphosignaling profiles that arose from digoxigenin, digitoxigenin, digoxin, and lanatoside C treatments all clustered together in the assay (cyan box). We also observed that phosphoprofiles of the same treatments clustered across cell lineages, while retaining some lineage-specific determinants, as shown in the green box for GW8510 treatments. We present an alternative visualization of these results in https://panoramaweb.org/labkey/project/LINCS/AbelinSupplemental/begin.view?, with accompanying text in supplemental Methods and Results 1. We think that this alternative “connectivity map” view allows intuitive insight into biological results.
A summary of the pairwise correlation comparisons among non-self (a sample of one cell type treated with one compound compared with other cell line and treatment combinations), the same drug treatments across cell lines, and the same drug treatment within each cell line are presented in Fig. 4B. In the left panel, the distribution of non-self pairwise Pearson correlations among phosphoprofiles is plotted. If a sufficient number of samples are profiled, and the phosphosignaling signatures obtained are not systematically biased in some way, one would expect that comparison of any two random samples (through their signatures) would not yield a strong correlation. Thus, the distribution of pairwise correlations of all samples should be approximately normal and centered on zero. Indeed, we observe such a distribution of non-self pairwise correlations as expected, which demonstrates that the assay is not systematically biased. Intriguingly, the long tail at the right of the distribution shows some samples do demonstrate strong correlations despite being treated with different compounds. Based on the observations from Fig. 4A, these strong correlations likely occur among class members or structural analogs. This suggests that the P100 profiles may be useful in classifying and stratifying novel compounds and perturbations.
The middle panel of Fig. 4B shows the correlations among the phosphoprofiles produced from the same drug treatment across differing cell types, characterizing the lineage-independent response in the assay. If a drug treatment impacts cell signaling pathways similarly across cell types, the distribution of pairwise correlations should shift toward a positive correlation. The distribution of pairwise correlations between the same drug treatments across different cell lines does shift toward 1 indicating that the same drug has similar effects on phosphosignaling across cell types. However, the majority of the pairwise correlations reside between 0 and 0.5, demonstrating that, although detectable, lineage-independent responses to the same drug probably only represent a portion of the signatures obtained through P100.
In the right panel of Fig. 4B, the pairwise correlations among the phosphoprofiles of the same drug treatment within each cell line (biological replicates) are displayed. The distribution of pairwise correlations now shifts even more toward a positive correlation demonstrating the reproducibility of the entire P100 assay process across different drug treatments. The strong rightward shift toward 1 demonstrates excellent “replicate recall,” meaning that biological replicates are likely to be strongly correlated. Taken together, the middle and bottom panels suggest that there is a strong lineage-specific component to signaling responses in cells although some commonality persists at a lineage-independent level.
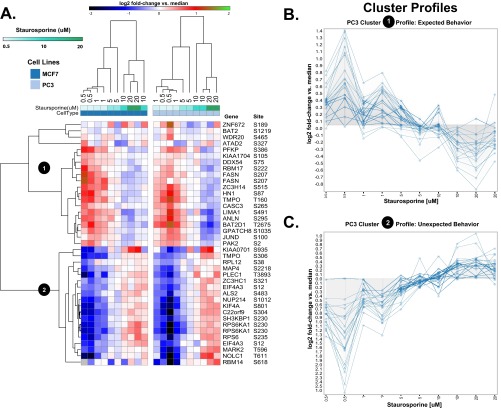
To further explore the performance characteristics of the assay, we performed a dose response experiment with staurosporine. A heatmap illustrating the molecular signatures from a subset of P100 probes is shown in Fig. 5A. Although staurosporine is generally annotated as a broad-spectrum kinase inhibitor, we observed examples of phosphopeptide probes whose levels decrease and others that unexpectedly increase in response with increasing staurosporine dose (Fig. 5A–5C). The fact that we observe a diversity in the responses of probes, where some follow the expected behavior whereas others demonstrate unique behaviors, suggests that we have selected analytes that have informative, dynamic responses under a wide variety of cellular conditions, making them excellent proxies for the signaling state of the cell in general. Furthermore, unsupervised hierarchical clustering almost perfectly organized the doses in order of increasing concentration in both MCF7 and PC3 cells with both the decreasing cluster (Fig. 5B) and the increasing cluster (Fig. 5C) making contributions. This suggests that the strength of P100 signatures (overall deviation from unchanged) may be proportional to dose and will be useful for determining dose responses, IC50 values, and other concentration-dependent phenotypic phenomena that project into phosphosignaling.
Fig. 5.
Signature strength in the P100 assay is dose responsive. A, A heatmap illustrating a condensed signature derived from a subset of P100 probes as a result of treating cells (MCF7, PC3) with varying doses of the kinase inhibitor staurosporine (0.5–20 mm) is shown. Examples of phosphopeptide probes that decrease in response to increased staurosporine dose (cluster 1 and profile (B) and probes that increase in response to increasing dose (cluster 2 and profile (C) are observed.
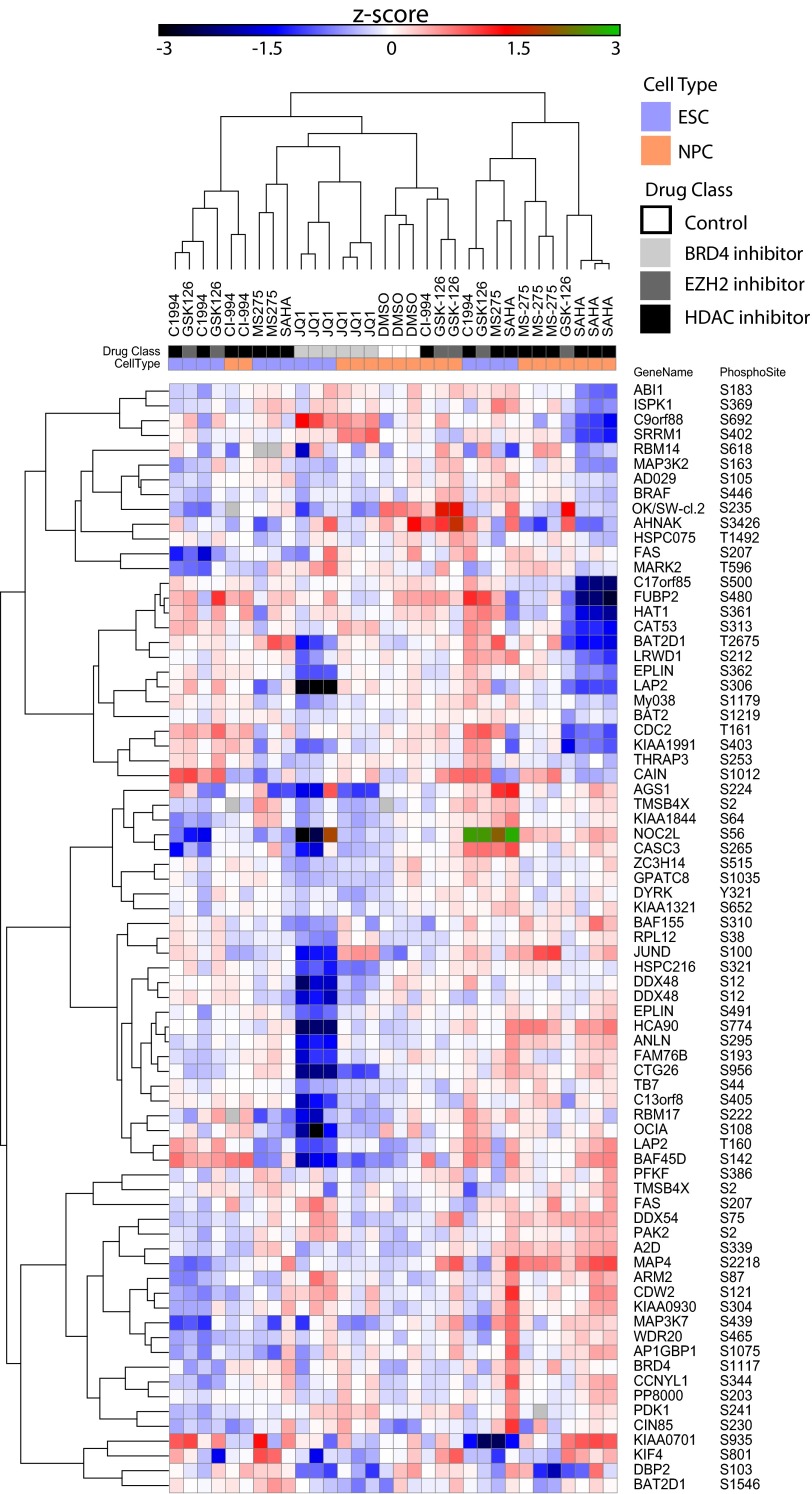
To confirm that the P100 assay could retain utility in biological systems beyond those used for development, we conducted a study using cell lines and perturbations that were not used during assay configuration. We selected human embryonic stem cells (ESC) and derived neuronal precursor cells (NPC) from them as these represent extremely active areas of research in developmental biology. We chose to treat the ESCs and NPCs with compounds known to affect levels of chromatin modifications because epigenetic regulation has been shown to be important for both maintenance of and exit from pluripotency (46). Moreover, mutations in some epigenetic modifier genes have been identified as genetic risk factors associated with autism spectrum disorders and other neurodevelopmental etiologies (47). The drug classes represented in this study include a BRD4 inhibitor, an EZH2 inhibitor, and HDAC inhibitors. The P100 phosphosignaling profiles generated from epigenetically active drug-treated ESCs and NPCs are shown in Fig. 6 (see also https://panoramaweb.org/labkey/project/LINCS/AbelinSupplemental/begin.view?). As we observed in our first P100 confirmation experiment, biological replicates of compound treatments generally cluster together. With the BRD4-inhibitor JQ1, we observe that sample profiles cluster together across both cell types while retaining lineage-specific determinants. We also observed general co-clustering of HDAC inhibitors, although lineage-specific responses seem to indicate that the signaling consequences of administering these compounds may vary widely in differing cell types. Notably, the structurally similar SAHA and MS-275 HDAC inhibitors developed visibly distinct signatures in the space of phosphosignaling despite having ostensibly the same target (in contrast, we have shown that their chromatin modification signatures are largely the same using our chromatin profiling assay (35); data not shown). Remarkably, we show that these epigenetically active compounds develop distinct lineage-specific signatures in the space of phosphosignaling, lending further evidence to the notion that there may be direct linkages between the chromatin and signaling machinery in cells, as earlier suggested by the observations in Fig. 2D–2E. Moreover, these observations were garnered using the reduced-representation assay in a biological context that was not considered during its design. Even so, we were able stratify samples and demonstrate good replicate recall, indicating that the P100 assay can be applied to diverse perturbations in multiple cell types, further validating its general utility.
Fig. 6.
P100 generates molecular signatures in embryonic stem cells (ESC) and neuronal precursor cells (NPC) in response to multiple compound treatments. The heatmap illustrates phosphosignaling signatures in ESC and NPC when treated with different classes of epigenetically active drugs including a Brd4 inhibitor, an EZH2 inhibitor, and HDAC inhibitors. As observed in the P100 validation experiments, replicates of the same drug treatment cluster together within each cell type. Additionally, molecular signatures of the same treatment cluster across NPC and ES, as seen with the Brd4 inhibitor JQ1. Neither cell type nor any drug treatment (with the exception of MS-275) depicted above was used in the development of the P100 assay.
As a final proof-of-principle, we designed an experiment to demonstrate that the P100 assay is responsive to perturbations of known biological pathways. We selected a panel of small molecules that were known to inhibit members of MAPK, PI3K/mTOR, and Cell Cycle signaling pathways. The mapping of the compounds onto the enzymes that they inhibit, along with a partial pathway reconstruction (as assembled from KEGG (45, 48)), is shown in Fig. 7A. Additionally, this experiment was designed to measure the time-dependence of P100 signature evolution, and we collected samples after 3, 6, and 24 h of compound treatment. We carried out these experiments in the breast cancer line MCF7, which has the notable characteristics being Her2−, ER+, and PR+/−; and also bears helical region PIK3CA mutations, increased phosphorylation of AKT, and overall hyperploidy up to 4n (49, 50).
The P100 molecular profiles derived from these experiments are shown in Fig. 7B (data are available in https://panoramaweb.org/labkey/project/LINCS/AbelinSupplemental/begin.view?). The profiles are ordered according to the linear pathway reconstruction, and within each major column, all of the replicates at each time point are shown (increasing in time from left to right). It is visually apparent from these profiles that the different compound treatments generated a diverse set of molecular responses in the P100 assay. We also saw that most of the drug perturbations have only modest effects with increasing time of treatment, with the greatest effects generally observed at the 24 h time point but clearly observable at the 3 h time point. This is reminiscent of the effect observed with increasing doses of staurosporine (Fig. 5), and thus P100 signatures are both dose- and time-responsive. There a few exceptions, however. The upstream inhibitors Pazopanib (PDGFR) and TG101348 (JAK2) seem to have dramatic alterations to their profiles at 24 h. As well, the CDK4/6 inhibitor PD-0332991 only manifests a strong signature after 24 h of administration, at which time its signature appears largely like the other CDK inhibitors tested. Interestingly, PD-0332991 (now called Palbociclib) was developed as a specific treatment for Her2−, ER+ breast cancer, matching the genetic status of MCF7 (50). We also note that, despite the cytotoxicity of many of these compounds after 24 h, unique signatures persist for the different drugs. In other words, the profiles do not converge to a universal “signature of death” at the 24 h time point and diverse signaling mechanisms still seem to be at play.
We noticed that, serendipitously, signatures derived from compound treatments that were immediately proximal with respect to their projections onto the reconstructed pathway seemed to be visually similar at most time points. To further investigate this observation, we calculated all of the pairwise correlations of samples at each time point (i.e. all 3 h samples compared with other 3 h samples, 6 h with 6 h, etc.), and then calculated the mean correlation for all compound pairs. The resulting correlation matrix is shown in Fig. 7C. Several interesting features emerged from this analysis. First, the strongest average correlations are on the diagonal, which again demonstrates the good replicate recall of the assay. Second, however, we noticed that there were blocks of off-diagonal correlation that appeared to be immediately adjacent to the diagonal. In fact, when we attempted to draw boundaries around these blocks of off-diagonal correlation (lavender, green, and yellow polygons), we noticed that they almost perfectly corresponded to the pathway modularity that we had assembled from the KEGG database. This is not to say that all of the perturbations within the same pathway module generate the same signature in the P100 assay. However, the correlation matrix suggests that near-neighbors in the pathways share components of the same phosphosignaling response, even with our reduced-representation of the phosphoproteome, thus allowing us to reassemble some notion of pathway modularity from the P100 data alone.
DISCUSSION
Our goal was to develop a framework for a high-throughput, standardized proteomic assay that could be used to interrogate the effects of molecular perturbations on cell signaling across large sample sets generated under disparate conditions. In general, most proteomic investigations of cellular responses to drugs involve shotgun data acquisition(26, 28–30, 39). Shotgun proteomics enables the identification of thousands of proteins in complex samples, but large-scale comparisons across disparate conditions are difficult and yield incomplete data sets. In addition, discovery experiments that implement shotgun phosphoproteomic strategies are typically slow and labor intensive.
In order to generate almost “complete” data with few missing values, we configured a targeted proteomic assay for a set phosphopeptide probes that provides a reduced but highly informative representation of the phosphoproteome. We identified these probes from large-scale, discovery cell perturbation studies and optimized the assay for robustness under several biologically relevant paradigms (i.e. multiple cell types, dose response, time-series). We hope that targeted methods for phosphoproteomic investigations, such as our P100 assay, will enable more rapid data generation and will increase the throughput of MS-based phosphosignaling studies at a fraction of the effort of traditional discovery experiments.
The P100 assay enables investigations into how perturbations affect cellular phosphosignaling phenotypes through generation of standardized profiles that can be compared across many conditions. Perturbations may take several forms, such as small molecule treatment, genetic manipulation, and induction of disease or differentiation models. In the most basic sense, the assay can help to stratify compounds into classes based on mechanism of action while denoting cell-type specific responses. For example, we identified strong connectivity among cardiac glycosides across multiple cell types using P100 data but are able to recognize cell-type of origin of each of the samples. Importantly, we can compare profiles obtained from any mode of perturbation to one another, and perhaps generate hypotheses about specific genetic mechanisms through which small molecules are acting (by similarity of profiles), or potential therapeutic options could counter a disease state (by anti-correlation of profiles). Thus, the assay espouses the “Connectivity Map” principle of using molecular profiles as an abstract means of associating cellular phenotypes (3, 4).
In alignment with our view of the P100 assay as a secondary screening tool, the robustness and throughput of the assay were enhanced by automating sample processing steps (Fig. 3A). We developed automated protocols for protein reduction, alkylation, digestion, phosphopeptide enrichment, and sample desalting using Agilent Bravo liquid handling and AssayMAP platforms. We also introduced important process monitoring controls, such as enrichment standards, to monitor for peptide losses and systematic errors that are crucial to a production effort. We even demonstrated that we could simultaneous process 96 samples in only 3 days with a plate enrichment CV of 12% and detect ∼95% of the reduced-representation phosphopeptides in close to 200 samples using our isotopically labeled quality control standards (supplemental Fig. S1). The assay itself is a single injection LCMS experiment with a 60 min nominal run time, enabling significant annual throughput to generate large data sets.
We qualified the targeted P100 assay and the use of a reduced-representation phosphoproteome by comparing measurements of our limited probe set to deeper data acquired under the conditions used for our large-scale discovery experiments (Fig. 4A). Using these data, we were able to replicate sample correlations observed in our discovery experiments demonstrating that a smaller set of phosphopeptide probes can be used as surrogates for a larger phosphopeptide set. These connections are intuitively visualized using a network graph architecture (supplemental Fig. S2) that efficiently summarizes sample correlations and helps to evaluate the biological relevance of results. Above and beyond these re-observations, we systematically evaluated the performance of the assay statistically. We calculated compared pairwise correlations among treatments and cell lines (Fig. 4B) and demonstrated that the null background is in line with expectations while replicate treatment can be detected both within a single cell type and even across cell types.
We went on to show that the strengths of the profiles in the assay have dose- and time-dependent correlations (Figs. 5, 7). Demonstration of dose-responsiveness is important for judging relative efficacies of similar compounds and downstream efforts in lead optimization for compounds in development. Time-responsiveness also demonstrates that there is sufficient dynamic range in the assay to study processes as they evolve, and that cellular responses to drug treatments are sustained over time. Although we could distinguish treatment time (via profile strength) via the P100 profiles, we saw very few instances of radical dynamic shifts in the profiles among the 3, 6, and 24 h time points that we measured. These observations argue against wholesale changes in protein levels over the assay time course. We acknowledge the shortcoming of not measuring the related “nonphospho” form of the P100 probes, but this turned out to be extremely difficult because of the complexity of the “nonphospho” proteome given that we wanted to keep the assay to a single LCMS experiment. In any case, the overall amount of phosphorylation of a given site is still fit-for-purpose as a component of the signature as we define it, despite it reflecting both the site-specific changes and the overall protein abundance. We do not claim that the assay represents site stoichiometry.
Further analysis of the time course experiments shows that signatures are largely established by 3 h, and that the strongest correlations occur between the 3 and 6 h time points. We therefore believe that this “assay window” is the ideal time to measure phosphosignaling responses, before processes like global changes in gene- or protein-expression have a chance to exert strong effects on the signatures in the assay. We extended the assay window to 24 h to see if secondary effects would dominate the signatures. Surprisingly, most signatures simply increased in strength rather than changed in major ways (with exceptions, discussed in results), despite cytotoxicity of some compounds. When we measured signatures of epigenetically active compounds in ESCs and NPCs (further discussed below), we chose to do so at 24 h with the idea of generating the strongest possible signatures in cell types that we had not yet tested. At the same time, we acknowledge that some of these data may not be ideal because some molecular perturbations, such as treatment with the kinase inhibitor staurosporine, can result in the death of some cell types after 24 h. Therefore, we only presented those data as an illustration of our ability to collect P100 phosphosignaling profiles in diverse cell types that include NPC and ESC.
We believe that the P100 assay will be widely applicable in many areas of biology. We demonstrated that the assay was responsive in diverse biological backgrounds by collecting data from new cell types treated with compounds not used during the assay configuration phase. We were able to measure P100 phosphosignaling profiles in both embryonic stem cells (ESC) and neuronal precursor cells (NPC) in response to different classes of epigenetically active drugs, including a BRD4 inhibitor, an EZH2 inhibitor, and HDAC inhibitors (Fig. 6). As with our cancer cell line validation experiments, biological replicates of drug treatments clustered together by their P100 molecular signatures across NPCs and ESCs. Our ability to generate molecular signatures using P100 data for drug treated ESCs and NPCs illustrates the potential for the P100 assay to be applied in a broad range of cells and perturbation conditions. We also demonstrated that compounds with epigenetic mechanisms of action can elicit cell signaling phenotypes, suggesting that monitoring phosphosites is important for many biological contexts. Furthermore, the phosphosignaling profiles we generated from ESCs and NPCs may provide insight into neuronal lineage development and neuropsychiatric disorders.
Our use of reduced-representation phosphopeptide probes is similar to the use of “sentinel proteins” reported by Soste et al. of the Picotti laboratory. Picotti and colleagues developed a targeted proteomic assay to probe biological responses to environmental perturbations in a less complex biological system, Saccharomyces cerevisiae, by selecting sentinel proteins from existing data (34). Like Soste et al., we selected probes that were highly detectable in our discovery data set. However, we selected probes to specifically monitor signaling through the lens of protein phosphorylation, whereas Soste et al. selected a mix of probes for general protein abundance and phosphoproteins, thus requiring two separate mass spectrometry experiments and biochemical workflows to execute the assay.
Although we selected a specific set of phosphopeptide probes to monitor with the P100 assay, the concept of reduced-representation phosphoprofiling can be extended to different sets of probes as long as they can be reproducibly detected and have varying responses to molecular perturbations. Extending reduced-representation profiling to alternate probes or adding more peptides enables researchers to tailor their assays to investigate specific biological questions, while retaining the ability to reproducibly measure targets across large sample sets. However, tailoring the panel may reduce the general utility of the assay. It is important to note that the selection of the P100 reduced-representation set was completely data driven, without preconceived notions of cellular pathways or biological function. This may seem counterintuitive; as high value is typically placed on readouts of “known pathways.” Here, we challenge that notion by demonstrating that many phosphosites with poorly understood or unknown “functions” provide a great deal of information, in the entropic sense, about the state of the cellular response to perturbations that helps to classify responses to drug perturbations. In that way, the P100 is complementary to readouts of “known pathways” and may help to define novel networks of cellular signaling.
Nevertheless, the discovery data used to configure the P100 assay contained a wide selection of phosphoproteins that belong to known pathways. Thus, it can be argued that the P100 assay “projects” into these pathways by the proxy, because the selected probes have correlations to the underlying discovery data. The top 20 pathways described by our coordinately regulated P100 groups are shown in Fig. 3B. Many of these cell signaling pathways have been implicated in disease etiology and are of interest to both biologist and clinicians. For example, dysregulation within the MAPK signaling pathway has been implicated in Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, and multiple types of cancer (51, 52). The AMPK pathway controls metabolic functions within cells, and has been linked to obesity, cardiovascular disease, neurogenesis and stroke (53–55). In addition, inefficient ErbB signaling is associated with neurodegenerative diseases including multiple sclerosis and Alzheimer's disease (56, 57).
Perhaps most dramatically, we have demonstrated that the P100 assay is sensitive to disruptions of (at least) the MAPK, PI3K/mTOR, and cell cycle cascades despite not being explicitly designed to monitor these pathways (Fig. 7). In these experiments, we intentionally disrupted important kinases in these signaling pathways and observed unique P100 signatures that simultaneously allowed us to distinguish the treatments while reconstructing the modularity of the linear pathways. For example, phosphoprofiles produced from JAK and PI3K inhibition positively correlated with one another because inhibition of JAK will prevent phosphorylation of insulin receptor substrate and p85, which leads to inhibition of the PI3K pathway (58). These data illustrate how the P100 assay can stratify samples and reinforce our understanding of network architecture according to direct and indirect interactions among the pathways perturbed by drug treatments.
These results validate the idea that the assay is a good proxy for monitoring diverse cellular processes. Furthermore, our ability to monitor alterations in these pathways is in good agreement with the pathways onto which we believe the P100 assay projects (Fig. 3B) based on the underlying configuration data. Of course, there is certainly room for more focused, “known” target-driven phosphosignaling assay panels in the future for specific applications. Another possible improvement might be a hybrid P100-DIA assay that allows focused detection and quantification of our P100 analytes while retaining the ability to mine primary data for novel phosphopeptide analytes of interest in defined pathways.
The P100 assay is a robust screening tool on its own, but is also intended to be used in conjunction with other molecular profiling techniques (such as gene expression profiling) to develop comprehensive pictures of cellular responses to perturbation. For example, it would be interesting to generate chromatin signature data on epigenetically active compounds in our global chromatin profiling assay (35, 59) along with P100, thus providing measures of processes that are directly targeted by the molecules together with measures of secondary process that might also be of interest. Our demonstration of signaling responses for these compound classes (Fig. 6) hints at this possibility. We plan to test this hypothesis by collaborating with others under the auspices of the Library of Integrated Network-based Cellular Signatures (LINCS) program to systematically collect molecular profiling data from a diverse set of chemical, genetic, and environmental manipulations across a range of cell types representing important disease models (www.lincsproject.org). A preview of these data has been made available even in advance of publication (https://panoramaweb.org/labkey/LINCS.url). We believe that these types of large-scale data integration efforts will further our understanding of how genomics, epigenetics, and phosphosignaling contribute to disease progression.
In conclusion, we developed and qualified a high-throughput assay that can be used as a tool to interrogate the effects of molecular perturbations on cell signaling in various cell types. Using P100 assay data, we demonstrated that reduced-representation phosphoprofiles are informative by reproducing sample classifications observed in our high-throughput discovery data. We recapitulated prior expected cell signaling correlations in a reduced assay space and demonstrated that signature strength is time- and dose-responsive. We were also able to identify correlations among perturbations that are known to target the same pathways, as well as correlations among perturbations that have overlapping downstream signaling events. Therefore, we believe the P100 assay can produce a large number of foundational data sets that can be queried to identify correlations among phosphosignaling profiles produced by disparate drug treatments even though only ∼100 phosphopeptide probes are monitored. Overall, we demonstrated the tractability of large-scale phosphoproteomic studies by presenting a generalizable, high-throughput MS assay that can be used as a robust tool that offers the ability to make longitudinal comparisons among thousands of samples. These comparisons, especially when married with other molecular profiling data, should be an important contribution from the phosphoproteomic sphere to our understanding of the molecular underpinning of cellular machinery and its response to biologically-relevant perturbations.
Supplementary Material
Acknowledgments
We thank Bang Wong, Broad Institute for allowing us to adapt his data workflow figure concept (Fig 3C). We also thank Jaime Conway for her excellent technical assistance.
Footnotes
Author contributions: J.G.A., J.P., X.L., R.S., E.Y.S., S.A.C., and J.D.J. designed research; J.G.A., J.P., X.L., C.M.F., L.F., A.L.C., D.L., D.D., A.O., J.L., and J.D.J. performed research; J.G.A., R.H., A.S., R.S., E.Y.S., and J.D.J. contributed new reagents or analytic tools; J.G.A., J.P., C.M.F., A.L.C., R.H., A.S., and J.D.J. analyzed data; J.G.A., J.P., C.M.F., S.A.C., and J.D.J. wrote the paper; C.M.F., L.F., L.P., J.W.Q., E.K., and S.A. automation development.
* This work was supported in part through NIH Grants U01CA164186 and U54HG008097 to J.D.J and by a Thought Leader Award and gift to S.A.C. from the Agilent Technologies Foundation and Agilent Technologies, Inc. E.K and S.A.C also acknowledge financial support from NCI Clinical Proteomics Tumor Analysis Consortium initiative grant U24CA160034 to S.A.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental materials.
This article contains supplemental materials.
Supplemental Mass Spectrometry Data: Raw data and MaxQuant results are available at ftp://MSV000079524@massive.ucsd.edu. Skyline data documents are available at https://panoramaweb.org/labkey/project/LINCS/AbelinSupplemental/begin.view?.
1 The abbreviations used are:
- CMap
- Connectivity Map
- AMEx
- Accurate Mass Exclusion-Based Data-Dependent Acquisition
- AIMS
- Accurate Inclusion Mass Screening
- DTT
- dithiothreitol
- IAA
- iodoacetamide
- RGE
- Regularized Gaussian Estimation
- DIA
- Data Independent Acquisition
- LINCS
- Library of Integrated Network-based Cellular Signatures
- PRM
- parallel reaction monitoring.
REFERENCES
- 1. Edgar R., Domrachev M., and Lash A. E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. (2002) Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Muertter R. N., Holko M., Ayanbule O., Yefanov A., and Soboleva A. (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., and Golub T. R. (2006) The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 4. Lamb J. (2007) The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer 7, 54–60 [DOI] [PubMed] [Google Scholar]
- 5. Freeman M. (2000) Feedback control of intercellular signalling in development. Nature 408, 313–319 [DOI] [PubMed] [Google Scholar]
- 6. Lalli E., and Sassone-Corsi P. (1994) Signal transduction and gene regulation: the nuclear response to cAMP. J. Biol. Chem. 269, 17359–17362 [PubMed] [Google Scholar]
- 7. Nishi H., Hashimoto K., and Panchenko A. R. (2011) Phosphorylation in protein-protein binding: effect on stability and function. Structure 19, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ubersax J. A., and Ferrell J. E. Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 9. Ozer R. S., and Halpain S. (2000) Phosphorylation-dependent Localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell 11, 3573–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suter B., and Steward R. (1991) Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell 67, 917–926 [DOI] [PubMed] [Google Scholar]
- 11. Jimeno A., Rubio-Viqueira B., Rajeshkumar N. V., Chan A., Solomon A., and Hidalgo M. (2010) A fine-needle aspirate–based vulnerability assay identifies polo-like kinase 1 as a mediator of gemcitabine resistance in pancreatic cancer. Mol. Cancer Ther. 9, 311–318 [DOI] [PubMed] [Google Scholar]
- 12. Firestein R., Bass A. J., Kim S. Y., Dunn I. F., Silver S. J., Guney I., Freed E., Ligon A. H., Vena N., Ogino S., Chheda M. G., Tamayo P., Finn S., Shrestha Y., Boehm J. S., Jain S., Bojarski E., Mermel C., Barretina J., Chan J. A., Baselga J., Tabernero J., Root D. E., Fuchs C. S., Loda M., Shivdasani R. A., Meyerson M., and Hahn W. C. (2008) CDK8 is a colorectal cancer oncogene that regulates [bgr]-catenin activity. Nature 455, 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma S. V., Bell D. W., Settleman J., and Haber D. A. (2007) Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 [DOI] [PubMed] [Google Scholar]
- 14. Amit I. W., and Ron Yarden Y. (2007) Evolvable signaling networks of receptor tyrosine kinases: relevance of robustness to malignancy and to cancer therapy. Mol. Syst. Biol. 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zoncu R., Efeyan A., and Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholl C., Fröhling S., Dunn I. F., Schinzel A. C., Barbie D. A., Kim S. Y., Silver S. J., Tamayo P., Wadlow R. C., Ramaswamy S., Döhner K., Bullinger L., Sandy P., Boehm J. S., Root D. E., Jacks T., Hahn W. C., and Gilliland D. G. (2009) Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 [DOI] [PubMed] [Google Scholar]
- 17. Barbie D. A., Tamayo P., Boehm J. S., Kim S. Y., Moody S. E., Dunn I. F., Schinzel A. C., Sandy P., Meylan E., Scholl C., Fröhling S., Chan E. M., Sos M. L., Michel K., Mermel C., Silver S. J., Weir B. A., Reiling J. H., Sheng Q., Gupta P. B., Wadlow R. C., Le H., Hoersch S., Wittner B. S., Ramaswamy S., Livingston D. M., Sabatini D. M., Meyerson M., Thomas R. K., Lander E. S., Mesirov J. P., Root D. E., Gilliland D. G., Jacks T., and Hahn W. C. (2009)Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E. and Zhang B.. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 (2004) [DOI] [PubMed] [Google Scholar]
- 19. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002)The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 20. Tinti M., Nardozza A. P., Ferrari E., Sacco F., Corallino S., Castagnoli L., and Cesareni G. (2012)The 4G10, pY20 and p-TYR-100 antibody specificity: profiling by peptide microarrays. Affin. Proteomics 29, 571–577 [DOI] [PubMed] [Google Scholar]
- 21. Bergström Lind S., Molin M., Savitski M. M., Emilsson L., Aström J., Hedberg L., Adams C., Nielsen M. L., Engström A., Elfineh L., Andersson E., Zubarev R. A., and Pettersson U. (2008)Immunoaffinity enrichments followed by mass spectrometric detection for studying global protein tyrosine phosphorylation. J. Proteome Res. 7, 2897–2910 [DOI] [PubMed] [Google Scholar]
- 22. Ficarro S. B., Adelmant G., Tomar M. N., Zhang Y., Cheng V. J., and Marto J. A. (2009)Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal. Chem. 81, 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villen J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies S. R., Ruggles K. V., Fenyo D., Kitchens R. T., Li S., Olvera N., Dao F., Rodriguez H., Chan D. W., Liebler D., White F., Rodland K. D., Mills G. B., Smith R. D., Paulovich A. G., Ellis M., and Carr S. A. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics 13, 1690–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choudhary A., Hu H. K., Mertins P., Udeshi N. D., Dančík V., Fomina-Yadlin D., Kubicek S., Clemons P. A., Schreiber S. L., Carr S. A., and Wagner B. K. (2014) Quantitative-Proteomic comparison of alpha and beta cells to uncover novel targets for lineage reprogramming. PLoS ONE 9, e95194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macek B., Mann M., and Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 27. Olsen J. V. V. Santamaria M., Kumar A., Miller C., Jensen M. L., Gnad L. J., Cox F., Jensen J., Nigg T. S., Brunak E. A., and Mann S. M. (2010)Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3–ra3 [DOI] [PubMed] [Google Scholar]
- 28. Jedrychowski M. P., Huttlin E. L., Haas W., Sowa M. E., Rad R., and Gygi S. P. (2011) Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol. Cell. Proteomics 10, M111.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bodenmiller B., Mueller L. N., Mueller M., Domon B., and Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Meth. 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 30. Rudomin E. L., Carr S. A., and Jaffe J. D. (2009) Directed sample interrogation utilizing an accurate mass exclusion-based data-dependent acquisition strategy (AMEx) J. Proteome Res. 8, 3154–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu H., Sadygov R. G., and Yates J. R. (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 32. Alcolea M. P., Casado P., Rodríguez-Prados J.-C., Vanhaesebroeck B. and Cutillas P. R. (2012) Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol. Cell. Proteomics 11, 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duan Q., Flynn C., Niepel M., Hafner M., Muhlich J. L., Fernandez N. F., Rouillard A. D., Tan C. M., Chen E. Y., Golub T. R., Sorger P. K., Subramanian A. and Ma'ayan A. (2014) LINCS canvas browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 42, W449–W460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soste M., Hrabakova R., Wanka S., Melnik A., Boersema P., Maiolica A., Wernas T., Tognetti M., von Mering C., and Picotti P. (2014) A sentinel protein assay for simultaneously quantifying cellular processes. Nat. Meth. 11, 1045–1048 [DOI] [PubMed] [Google Scholar]
- 35. Creech A. L., Taylor J. E., Maier V. K., Wu X., Feeney C. M., Udeshi N. D., Peach S. E., Boehm J. S., Lee J. T., Carr S. A., and Jaffe J. D. (2015) Building the connectivity map of epigenetics: chromatin profiling by quantitative targeted mass spectrometry. Methods 72, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaffe J. D., Wang Y., Chan H. M., Zhang J., Huether R., Kryukov G. V., Bhang H. E., Taylor J. E., Hu M., Englund N. P., Yan F., Wang Z., Robert McDonald E. 3rd, Wei L., Ma J., Easton J., Yu Z., deBeaumount R., Gibaja V., Venkatesan K., Schlegel R., Sellers W. R., Keen N., Liu J., Caponigro G., Barretina J., Cooke V. G., Mullighan C., Carr S. A., Downing J. R., Garraway L. A., and Stegmeier F. (2013) Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 45, 1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., and Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Meth. 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 39. Jaffe J. D., Keshishian H., Chang B., Addona T. A., Gillette M. A., and Carr S. A. (2008) Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteomics 7, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gheorghiade M., Adams K. F., and Colucci W. S. (2004) Digoxin in the management of cardiovascular disorders. Circulation 109, 2959–2964 [DOI] [PubMed] [Google Scholar]
- 43. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., and Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotech. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 45. Kanehisa M., and Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. StemBook [Internet]. Cambridge (MA): Harvard Stem Cell Institute; 2008-. Available from: http://www.ncbi.nlm.nih.gov/books/NBK27044/ [PubMed] [Google Scholar]
- 47. Krumm N., O'Roak B. J., Shendure J., and Eichler E. E. (2014) A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 37, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu G., Xing M., Mambo E., Huang X., Liu J., Guo Z., Chatterjee A., Goldenberg D., Gollin S. M., Sukumar S., Trink B., and Sidransky D. (2005) Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 7, R609–R616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holliday D. L., and Speirs V. (2011) Choosing the right cell line for breast cancer research. Breast Cancer Res. BCR 13, 215–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim E. K., and Choi E. J. (2010) Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 1802, 396–405 [DOI] [PubMed] [Google Scholar]
- 52. Lawrence M. C., Jivan A., Shao C., Duan L., Goad D., Zaganjor E., Osborne J., McGlynn K., Stippec S., Earnest S., Chen W., and Cobb M. H. (2008) The roles of MAPKs in disease. Cell Res 18, 436–442 [DOI] [PubMed] [Google Scholar]
- 53. Mihaylova M. M., and Shaw R. J. (2011) The AMP-activated protein kinase (AMPK) signaling pathway coordinates cell growth, autophagy, and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Steinberg G. R., and Kemp B. E. (2009) AMPK in health and disease. Physiol. Rev. 89, 1025–1078 [DOI] [PubMed] [Google Scholar]
- 55. Salminen A., and Kaarniranta K. (2012) AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 11, 230–241 [DOI] [PubMed] [Google Scholar]
- 56. Bublil E. M., and Yarden Y. (2007) The EGF receptor family: spearheading a merger of signaling and therapeutics. Cell Regul. 19, 124–134 [DOI] [PubMed] [Google Scholar]
- 57. Wieduwilt M. J., and Moasser M. M. (2008) The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell. Mol. Life Sci. 65, 1566–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foster F. M., Traer C. J., Abraham S. M., and Fry M. J. (2003) The phosphoinositide (PI) 3-kinase family. J. Cell Sci. 116, 3037–3040 [DOI] [PubMed] [Google Scholar]
- 59. Tang H., Fang H., Yin E., Brasier A. R., Sowers L. C., and Zhang K. (2014) Multiplexed parallel reaction monitoring targeting histone modifications on the QExactive mass spectrometer. Anal. Chem. 86, 5526–5534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.