Abstract
Peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC1α) is the primary regulator of mitochondrial biogenesis and was recently found to be highly expressed within the intestinal epithelium. PGC1α is decreased in the intestinal epithelium of patients with inflammatory bowel disease, but its role in pathogenesis is uncertain. We now hypothesize that PGC1α protects against the development of colitis and helps to maintain the integrity of the intestinal barrier. We selectively deleted PGC1α from the intestinal epithelium of mice by breeding a PGC1αloxP/loxP mouse with a villin-cre mouse. Their progeny (PGC1αΔIEC mice) were subjected to 2% dextran sodium sulfate (DSS) colitis for 7 days. The SIRT1 agonist SRT1720 was used to enhance PGC1α activation in wild-type mice during DSS exposure. Mice lacking PGC1α within the intestinal epithelium were more susceptible to DSS colitis than their wild-type littermates. Pharmacologic activation of PGC1α successfully ameliorated disease and restored mitochondrial integrity. These findings suggest that a depletion of PGC1α in the intestinal epithelium contributes to inflammatory changes through a failure of mitochondrial structure and function as well as a breakdown of the intestinal barrier, which leads to increased bacterial translocation. PGC1α induction helps to maintain mitochondrial integrity, enhance intestinal barrier function, and decrease inflammation.
Keywords: bacterial pathogenesis, inflammatory bowel disease (IBD), intestinal epithelium, mitochondria, oxidative stress
Introduction
It is estimated that over 1.4 million people are currently diagnosed with inflammatory bowel disease (IBD)3 in the United States (1–3). In 2008, the direct costs associated with the disease exceeded 6.8 billion dollars (4), and furthermore, the worldwide prevalence of IBD appears to be increasing (5). Although the pathophysiology of IBD is multifactorial, a growing body of literature has implicated oxidative stress within the intestinal epithelium and abnormal mitochondrial bioenergetics as important causative factors in the development of disease (6–10). Specifically, Santhanam et al. (11) and Sifroni et al. (12) demonstrated a decrease in mitochondrial electron transport chain complex activity in humans with IBD as well as in mice subjected to experimental colitis (13). This would suggest a bioenergetic failure of the intestinal mitochondria as a consequence of impaired oxidative phosphorylation. Interestingly, abnormalities in complex activity occur prior to the initiation of inflammatory changes, suggesting that a decrease in function is likely a primary defect within the intestinal epithelium as opposed to one occurring secondary to mucosal inflammation. Taken together, these data suggest that mitochondrial dysfunction may play a key role in the development of IBD and that a restoration of mitochondrial function may prevent and/or treat disease.
The modern view of the mitochondrion is that of a dynamic organelle that undergoes turnover in response to stimuli in a process known as mitochondrial biogenesis (14–16). The regulation of normal mitochondrial function requires the activity of peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC1α). Under conditions of stress, PGC1α is a potent stimulator of mitochondrial turnover and antioxidant activity. Recently, it was demonstrated that PGC1α is highly expressed within the intestinal epithelium in mice (17). A decrease in PGC1α within the intestinal epithelium of a patient with ulcerative colitis is thought to be a precursor of dysplastic changes (18).
Based on our following studies, we now hypothesize that PGC1α plays a critical role in the pathogenesis of experimental colitis in mice. Here, we show that PGC1α is decreased in humans with IBD as well as in mice subjected to experimental colitis. During murine colitis, PGC1α is acetylated to its inactive form and targeted for proteosomal degradation through ubiquitination. This decrease in PGC1α correlates with a down-regulation of the mitochondrial biogenesis pathway and a decreased expression of mtDNA and mitochondrial mass as well as abnormal mitochondrial structure. When PGC1α is deleted from the intestinal epithelium of mice, they develop dramatically worse experimental colitis. Furthermore, pharmacologic stimulation of PGC1α activity ameliorates experimental colitis. Additionally, loss of PGC1α was associated with a decrease in the tight junction protein occludin and an increase in bacterial translocation in the face of inflammation. Taken together, these results suggest that a failure of mitochondrial biogenesis and mitochondrial bioenergetics occurs during the pathogenesis of colitis and that this failure is dependent upon a pathologic decrease in PGC1α within the intestinal epithelium. Strategies aimed at enhancing PGC1α activity and mitochondrial health in humans may contribute to the treatment of human IBD.
Experimental Procedures
Materials and Reagents
Antibodies were obtained from the following: PGC1α, NRF-1, NRF-2, and MTCO2 (Abcam, Cambridge, MA); TFAM, TOM20, and ubiquitin (Santa Cruz Biotechnology, Dallas, TX); EUB338 (IDT Technologies, Coralville, IA); acetylated lysine (Cell Signaling Technology, Danvers, MA); and β-actin (Sigma). Other reagents used were SRT1720 (Selleck Biochem, Houston, TX), BrdU (Invitrogen), and dihydroethidium (DHE; Sigma).
Generation of PGC1αΔIEC Mice and Other Experimental Mice
C57BL/6, Rag1−/−, BALB/c, Vil-cre (B6.Cg-Tg(Vil-cre)997Gum/J), and PGC1αloxP/loxP mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and housed in accordance with the University of Pittsburgh animal care guidelines. To selectively remove PGC1α from the intestinal epithelium, the PGC1αloxP/loxP mice were bred with villin-cre transgenic mice (PGC1αΔIEC mice).
DSS-induced Colitis
A solution of 2 or 3% dextran sodium salt (DSS; 36,000-50,000 kDa, MP Biomedicals, Solon, OH) was administered in the drinking water for 7 days and subsequently replaced with regular water. Chronic colitis was induced by alternating between 2% DSS water for 7 days and regular water for 10 days, for a total of three cycles. Clinical signs of colitis were recorded daily, and the disease activity index (DAI), which includes weight loss (0–4), stool consistency (0–4), and blood in the stool (0–4), was determined (19). Weight loss was calculated as the change in percent change from the original weight before initiation of the experiment. 1 ml/100 g of BrdU was injected intraperitoneally 36 h prior to euthanasia.
TNBS-induced Colitis
Acute colitis was induced by the introduction of 5% 2,4,6-trinitrobenzene sulfonic acid (TNBS; Sigma) dissolved in 50% ethanol into the colons of BALB/c mice via a transrectal catheter advanced 4 cm proximal to the anus, as described previously (20). Vehicle control mice received 50% ethanol in the same manner. Clinical signs were recorded daily, and the DAI score was determined as described for the DSS model. Mice were euthanized on day 4, and weight loss was calculated as the change in grams as compared with the body weight before initiation of the experiment.
CD4+CD45+RBhigh T-cell Transfer Model of Chronic Colitis
Colitis was induced according to the protocol previously published by Ostanin et al. (21). In brief, splenic CD4+ T-cells were purified from splenocytes from donor C57BL/6 mice using CD4 (L3T4) MicroBeads (Miltenyi Biotec, San Diego) and autoMACS cell sorting. Cells with a CD4+ (L3T4; BD Pharmingen, San Diego) and CD45RBhigh (16A; BD Pharmingen) phenotype were further purified using a FACSAria cell sorter (BD Biosciences). The CD45RBhigh population is identified as the 40% of cells exhibiting the brightest CD45RBhigh staining. CD4+CD45RBhigh cells (5 × 105 cells in 200 ml of 1× Dulbecco's PBS (Life Technologies, Inc.)) were then injected intraperitoneally (i.p.) into recipient Rag1−/− mice. The DAI score for colitis was determined using a scoring system described above. Mice were euthanized 1 week after the onset of clinical colitis.
Quantitative Real Time-Polymerase Chain Reaction
Quantitative real time PCR (qRT-PCR) was performed as described previously using the CFX96 real time system (Bio-Rad) (22). The expression of the genes by qRT-PCR (Table 1) was measured relative to the housekeeping gene ribosomal protein L15 (Rplo). Total RNA was isolated from whole intestinal tissue or colonic mucosal scrapings from control and diseased mice and human intestine using the RNeasy kit (Qiagen, Valencia, CA) and reverse-transcribed (0.5 μg of RNA) using the QuantiTect reverse transcription kit (Qiagen).
TABLE 1.
qRT-PCR primer sequences
| Gene | Species | Forward sequence (5′–3′) | Reverse sequence (3′–5′) | Amplicon size |
|---|---|---|---|---|
| bp | ||||
| Il-1β | Mouse | AGTGTGGATCCCAAGCAATACCCA | TGTCCTGACCACTGTTGTTTCCCA | 175 |
| Il-6 | Mouse | GGCTAAGGACCAAGACCATCCAA | TCTGACCACAGTGAGGAATGTCCA | 138 |
| CxcL15 | Mouse | TGTTCACAGGTGACTGCTCC | AGCCCATAGTGGAGTGGGAT | 140 |
| inos | Mouse/rat | CTGCTGGTGGTGACAAGCACATTT | ATGTCATGAGCAAAGGCGCAGAAC | 167 |
| mtDNA | Mouse | CCTATCACCCTTGCCATCAT | GAGGCTGTTGCTTGTGTGAC | |
| Occludin | Mouse | ATGGCAAGCGATCATACCCAGAGT | AGGTTACCATTGCTGCTGTACCGA | 131 |
| Pecam | Mouse | ATGGAAAGCCTGCCATCATG | TCCTTGTTGTTCAGCATCAC | |
| PGC1α | Human | GATGCGCTGACAGATGGAGA | TAGAGACGGCTCTTCTGCCT | 102 |
| PGC1α | Mouse | TGGATGAAGACGGATTGCCC | TAGAGACGGCTCTTCTGCCT | 128 |
| PGC1α genotype | Mouse | TCCAGTAGGCAGAGATTTATGAC | TGTCTGGTTTGACAATCTGCTAGGTC | 360 (WT) |
| 400 (PGC1α ΔIEC) | ||||
| Rplo/RPLO | Mouse/human | GGCGACCTGGAAGTCCAACT | CCATCAGCACCACAGCCTTC | 143 |
| Tnfα | Mouse | CATCTTCTCAAAATTCGAGTGACAA | TGGGAGTAGACAAGGTACAACCC | 175 |
SDS-PAGE
Mouse tissue or whole cell lysates were prepared using 1× RIPA buffer (Boston Bio, Ashland, MA). Protein concentration was determined using a bicinchoninic acid assay (BCA) (Sigma). Protein lysates (30 μg/sample) were separated on 8–12% SDS gel by SDS-PAGE and transferred onto a PVDF membrane. Membranes were blocked with 5% nonfat dry milk in 1× TBST or 5% BSA in 1× TBST and probed with primary antibody at 4 °C overnight. An HRP-conjugated secondary antibody (Cell Signaling Technology) was used followed by incubation with a chemiluminescent HRP substrate (Thermo-Fisher Scientific, Waltham, MA).
Immunoprecipitation and Immunoblot of PGC1α
Whole cell lysates of colonic tissue from control and DSS-subjected mice were prepared by homogenizing ∼50 mg of intestinal tissue in 250 μl of T-PER Tissue Protein Extraction Reagent (Thermo-Fisher Scientific). The protein solution was centrifuged for 15 min at 13,000 rpm at 4 °C. The supernatant was transferred to a new tube, and an aliquot of the protein lysates was subjected to a BCA to determine the protein concentration. 750 μg of protein lysate from each sample was individually incubated with 2.5 μg of PGC1α antibody overnight on a rotator at 4 °C. The antibody-protein complex was then incubated with protein G Dynabeads (Thermo-Fisher Scientific) and then immunoprecipitated and eluted according to the manufacturer's instructions. Immunoblot of the eluted proteins was performed as described above (see under “SDS-PAGE”).
Histological Analysis
Tissues from mice were fixed and stained with hematoxylin and eosin (H&E) for light microscopic examination. A blinded pathologist evaluated tissues for signs of disease as described previously (23).
Immunofluorescence
Tissues were fixed with 4% paraformaldehyde, dehydrated, and embedded in paraffin blocks. Tissue sections were cut at 5 μm, deparaffinized, and rehydrated through a gradient of xylene and ethanol baths. Citric acid was used as an antigen retrieval step, and sections were blocked in 1% BSA, 5% donkey serum for 1 h. Sections were incubated with primary antibody overnight at 4 °C and probed with a secondary antibody the following day. A coverslip was placed over the sections. The sections were allowed to dry and imaged using an Olympus fluorescent microscope (Olympus, Center Valley, PA). Bacterial translocation was determined by quantifying bacterial cell fluorescence (EUB338) using grayscale images of each JPEG file opened in ImageJ. The level of fluorescence in a given region was determined using the following equation: corrected total cell fluorescence = integrated density − (area of selected cell × mean fluorescence of background readings). The corrected total cell fluorescence values, which are arbitrary numbers, were graphed, and standard deviations were calculated.
Isolation of DNA from Murine Intestinal Crypts
The colons from 8-week-old WT C57BL/6 mice were isolated, cleared of all stool, and placed in cold PBS/antibiotic solution with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), 1% gentamycin (Life Technologies, Inc.), 1% penicillin/streptomycin (Life Technologies, Inc.), and 0.2% amphotericin B (Life Technologies, Inc.). The colons were then incised longitudinally, lavaged, cut into small (<5 mm) pieces, transferred into cell disruption media (DMEM (Life Technologies, Inc.), 10% FBS, 1% penicillin/streptomycin, 4 mm EDTA (Promega, Madison, WI), 1% gentamycin, and 0.2% amphotericin B), and placed on a rotator at 4 °C for 45 min. Following the cell disruption incubation, the tissue was centrifuged at 4 °C for 30 s at 200 × g. The supernatant was removed. The tissue pellet was resuspended in cold PBS/antibiotic solution and centrifuged at 4 °C for 5 min at 200 × g. This wash step was repeated three times. After the last wash, the crypts (the top “sandy” layer of the pellet) were collected and transferred to a new tube on ice. Total DNA was isolated from freshly harvested crypts utilizing the Qiagen DNeasy blood and tissue kit (Qiagen) following the manufacturer's instructions. The concentration and purity of each DNA sample were measured via spectrophotometry (ND-2000 spectrophotometer; Nanodrop Technologies, Inc., Wilmington, DE). The quality of each DNA sample was also assessed by gel electrophoresis.
Quantification of Mitochondrial DNA by Real Time PCR and Analysis
Total DNA isolated from murine intestinal crypts was used to determine whether DSS-treated mice had reduced amounts of mtDNA relative to nuclear DNA (nDNA) in intestinal epithelial cells. A primer set specific for mtDNA and a primer set specific for nDNA were previously published by Chen et al. (24) and were utilized in this study. Each DNA sample was amplified using a Bio-Rad CFX96 real time PCR detection system (Bio-Rad), with a final reaction volume of 10 μl containing 1 ng of total DNA, primers (mitochondrion-specific or nucleus-specific), and 1× iQ SYBR Green Supermix (Bio-Rad). The amplification conditions for the real time PCR reactions were as follows: 1 cycle of initial denaturation at 95 °C for 4 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 56 °C for 30 s, and elongation at 72 °C for 30 s. All real time reactions were carried out in triplicate. A validation experiment demonstrated that the PCR efficiencies of the mitochondrial and nuclear primers were similar, allowing the relative quantification of mtDNA to be calculated using the comparative Ct method (ΔΔCt method). The derived ΔΔCt values were converted into fold-difference values, and the range of the fold-difference (shown by bars on the graph) was derived by the incorporation of the standard deviation of the ΔΔCt value into the fold-difference calculation. Statistical analysis was performed using GraphPad Prism software (GraphPad, La Jolla, CA). An unpaired Student's t test was utilized to determine the significance between the control and DSS-treated groups. p values <0.05 were considered significant.
Mitochondrial Electron Transport Enzyme Activity
Whole tissue was used for the measurement of mitochondrial complexes I, II, and IV as follows: for complex I, the rotenone-sensitive rate of nicotinamide adenine dinucleotide oxidation was spectrophotometrically monitored at 340 nm in permeabilized tissue. Complex II activity was determined by measuring the reduction of dichloroindophenol at 600 nm, which was coupled to the oxidation of CoQ2 using succinate as a substrate. Thenoyltrifluoroacetone (complex II inhibitor) was used to determine specificity of the assay for complex II activity. Complex IV was measured by monitoring the oxidation of ferrocytochrome c at 550 nm. Potassium cyanide was used to determine specificity of oxidation by complex IV.
Electron Microscopy
Samples obtained from both control and experimental groups were fixed in 2.5% glutaraldehyde for analysis by electron microscopy. Transmission electron microscopy (TEM) was performed using a JEOL electron microscope (model number JSM6335F; JEOL, Peabody, MA) after tissues were placed into 8-mm3 blocks and post-fixed in 1% osmium tetroxide (OsO4) in 0.1 m PBS. Following alcohol dehydration, tissues were washed with hexamethyldisilizane and mounted on studs for sectioning. For transmission electron microscopy, tissues were placed on 1-mm3 blocks and post-fixed with 1% OsO4 containing 1% potassium ferricyanide. Following alcohol dehydration, tissues were further dehydrated with propylene oxide and then infiltrated with a 1:1 mix of propylene oxide and Epon overnight. Samples were then infiltrated with pure Epon overnight at 4 °C, embedded in pure Epon for 24 h at 37 °C, and cured for 48 h at 60 °C. Samples were 70 nm sectioned on an Ultra Microtome, placed on copper grids, and stained with urinal acetate and lead citrate.
DNA Extraction for 16S Analysis
All microbial DNA was extracted using the PowerSoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) in single tube extractions or PowerSoil-htp 96-well DNA isolation kit (MO BIO). For DNA extracted using the single tube format, fecal pellets were added directly into bead tubes and incubated at 65 °C for 10 min followed by 95 °C for 10 min. After addition of 60 μl of Solution C1, the bead tubes were then shaken horizontally on a laboratory mixer for 12–18 min at maximum speed using a MO BIO vortex adaptor. All remaining steps followed the manufacturer's protocol. For DNA extracted in a 96-well format, fecal pellets were added to individual wells of the bead plate and stored overnight at −80 °C. The following day, Bead Solution and Solution C1 were added, and the plates were incubated at 65 °C for 10 min. The plates were shaken on an Oscillating Mill MM400 (Retsch, Haan, Germany) with 96-well plate adaptors for 10 min at speed 20. The plates were rotated 180° and shaken again for 10 min at speed 20. All remaining steps followed the manufacturer's centrifugation protocol.
16S Amplicon PCR and Sequencing
PCR amplification of the small subunit ribosomal RNA gene (16S rRNA) was performed in triplicate 25-μl reactions. Amplicons were produced utilizing primers adapted for the Illumina MiSeq (Illumina, San Diego). Amplicons target the V4 region, and primers utilized either the Illumina adaptor, primer pad, and linker (forward primer) or the Illumina adaptor, Golay barcode, primer pad, and linker (reverse primer) followed by a sequence targeting a conserved region of the bacterial 16S rRNA gene as described by Caporaso et al. (25). The only deviation from the protocol was that PCR was run for 30 cycles. Individual PCR amplicons were purified, quantified, and pooled in equimolar ratios, and the library pool was gel-purified prior to submission for sequencing on the Illumina MiSeq at the Roy J. Carver Biotechnology Center High-Throughput Sequencing and Genotyping Unit, University of Illinois.
Analysis of 16S Sequences
To increase the size and improve the quality of 16S amplicons, forward and reverse reads were stitched together using PEAR. These were then demultiplexed using QIIME (version 1.9), and quality filtered using UPARSE (version 8.0). UPARSE was then used to dereplicate reads, cluster reads into operational taxonomic units at an identity threshold of 0.97, remove chimeric reads, and generate an operational taxonomic unit table. Taxonomic assignments to the predicted operational taxonomic units were made with QIIME using the UCLUST method. α diversity, β-diversity distances, and principal coordinate analyses (PcoA) were calculated using QIIME. The R vegan package was used to generate nMDS plots from β diversity unweighted unifrac distances and visualize within group heterogeneity of PGC1αloxP/loxP wild-type mice and PGC1αΔIEC mice. Statistical analysis of α diversity (observed species metric) was performed on samples rarefied to an even 1,600 reads, using a non-parametric test as implemented in QIIME. α diversity boxplots, PcoA plots, and taxon summary plots were generated using the ggplot2 package in R.
Human Tissue
Frozen specimens of human tissue were obtained from the Digestive Disease Tissue Resource of the University of Pittsburgh. These were surgical specimens from patients with ulcerative colitis or non-IBD pathology. All of the specimens received came from banked surgical specimens. Each sample was accompanied by minimal clinical data, including a pathology report that was de-identified by an honest broker.
Statistics
Results are expressed as the means ± S.E. of the mean (S.E.) or S.D. Statistical analysis was performed using SPSS 13.0 (IBM, Armonk, NY) software or Graphpad Prism software. Analysis of variance was used for comparisons in experiments involving more than two experimental groups. Two-tailed Student's t test was used for comparison in experiments consisting of two experimental groups. In all cases, statistical significance was accepted at p < 0.05 between groups.
Study Approval
All animal studies were approved by the Institutional Review Board at the University of Pittsburgh and conducted in accordance with the guidelines set forth by the Animal Research and Care Committee at the University of Pittsburgh and the Children's Hospital of Pittsburgh of UPMC. Human tissue samples were obtained after approval by the Institutional Review Board of the University of Pittsburgh.
Results
Peroxisome Proliferator-activated Receptor-γ Coactivator 1-α Is Decreased in Human Colitis and Murine Experimental Colitis
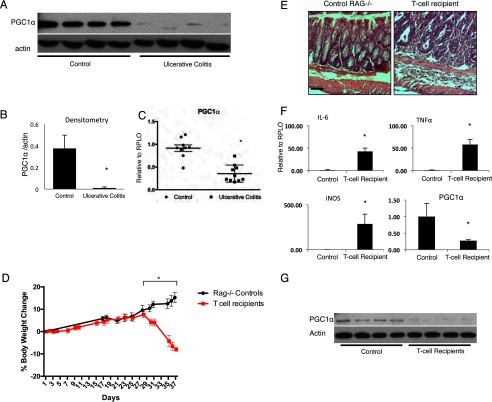
To characterize PGC1α expression in human IBD, we obtained surgical samples from patients with severe surgical ulcerative colitis. We found a dramatic decrease in protein levels of PGC1α in samples from patients with ulcerative colitis as compared with control patients (Fig. 1, A and B). A decrease in the expression of PGC1α in ulcerative colitis samples was demonstrated using qRT-PCR (Fig. 1C). We therefore hypothesized that PGC1α would be similarly decreased in the intestines of mice subjected to a physiologically relevant model of experimental colitis. To demonstrate this, CD4+CD45RBhigh cells were obtained from the spleens of C57BL/6 mice and transferred into Rag1−/− mice via i.p. injection. Over 6 weeks, these mice developed a robust colitis (Fig. 1, D and E). The expression of inflammatory cytokines was dramatically up-regulated in the intestines of these mice, although PGC1α expression declined (Fig. 1F). Protein levels of PGC1α, as determined by Western blot, were similarly decreased in T-cell recipient mice as compared with control mice (Fig. 1G). Similar to findings in human disease, mice subjected to T-cell transfer colitis demonstrated a decrease in intestinal PGC1α. However, this occurred early on during the pathogenesis of disease. We next sought to define the expression pattern of PGC1α in the pathogenesis of experimental colitis in mice.
FIGURE 1.
Human tissues from patients with ulcerative colitis and control patients were analyzed for PGC1α protein levels via Western blot with densitometry (A and B, n = 9–10/group), and PGC1α expression was analyzed using qRT-PCR (C). Rag1−/− mice received CD4+CD45RBhigh cells obtained from the spleens of C57BL/6 mice via intraperitoneal injection (n = 4/group). Over 6 weeks, these mice lost weight and demonstrated inflammatory changes on H&E (D and E, scale bars, 100 μm). They demonstrated a dramatic increase in the expression of inflammatory tissue cytokines. As seen in humans with ulcerative colitis, there was an overall decrease in RNA expression of PGC1α as well as PGC1α protein levels (F and G). *, p ≤ 0.05.
PGC1α Demonstrates a Bimodal Pattern of Expression during the Pathogenesis of Colitis
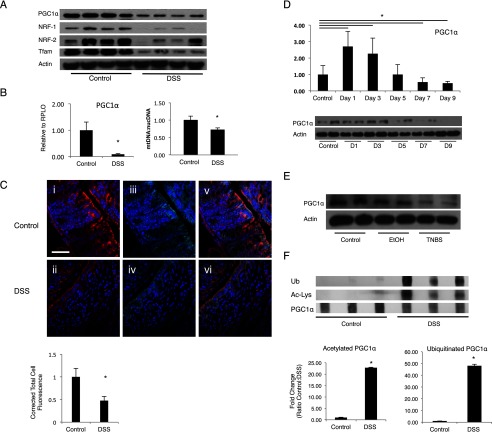
We evaluated PGC1α protein levels and RNA expression in a model of 3% DSS colitis. Similar to what was seen in human IBD and murine T-cell transfer colitis, PGC1α is down-regulated at day 7 of DSS exposure as demonstrated by Western blot (Fig. 2A). PGC1α is the key regular of mitochondrial biogenesis, a process whereby new mitochondria are made and damaged mitochondria are repaired. This process is initiated by PGC1α activation and driven largely by the transcription factors, nuclear respiratory factor (NRF)-1 and 2. Ultimately, this leads to activation of mitochondrial transcription factor A (TFAM), a nucleus-encoded transcription factor that moves to the mitochondria, promoting transcription of the mitochondrial genome. Until recently, no evidence of abnormal mitochondrial biogenesis in the intestine had been reported (17). To assess how PGC1α down-regulation affects mitochondrial biogenesis in the intestinal epithelium during experimental colitis, we evaluated its downstream mediators in the biogenesis pathway, which were similarly down-regulated (Fig. 2A). PGC1α expression as determined by qRT-PCR demonstrated a similar pattern of RNA expression over the same time period, although the down-regulation of the mitochondrial biogenesis pathway resulted in a decrease in mtDNA copy number (Fig. 2B). Immunofluorescence for PGC1α and an epithelial protein (E-cad) confirms that the decrease in PGC1α occurs primarily within the intestinal epithelium (Fig. 2C). To demonstrate the pattern of PGC1α expression during the development of murine colitis, mice were subjected to 3% DSS colitis for 7 days after which mice were then transitioned to regular drinking water for 2 more days. Mice were euthanized at 2-day intervals over the course of the protocol, and their respective tissues were analyzed. We observed an early up-regulation of PGC1α expression at day 1 of DSS exposure (Fig. 2D). However, PGC1α expression falls progressively as animals begin to demonstrate clinical signs of colitis between days 3 and 5. By day 7, levels are decreased as compared with control mice. Protein levels of PGC1α followed a similar but delayed pattern with levels peaking at 3 days before decreasing by day 5. We hypothesize that this may represent an early adaptive response to oxidative stress that ultimately fails in the presence of ongoing insult. To determine whether the observed changes in PGC1α are seen in other chemically induced models of colitis, we performed TNBS colitis on BALB/c mice. We similarly see a profound decrease in PGC1α by day 4 in TNBS-treated animals (Fig. 2E). These findings reveal that a decrease in PGC1α within the intestinal epithelium is associated with a down-regulation of the machinery of mitochondrial biogenesis and progressive colitis.
FIGURE 2.
C57BL/6 mice were subjected to 3% DSS colitis for 7 days (n = 8/group). There was an overall decrease in PGC1α, Nrf-1, Nrf-2, and Tfam protein in intestinal tissue after DSS exposure (A). PGC1α expression was decreased in mice subjected to DSS and assessed using qRT-PCR. DSS-treated mice demonstrated a 30% reduction in mtDNA content relative to nDNA content as compared with control mice. Relative quantification of mtDNA/nDNA was determined by real time PCR (B). Immunofluorescence for PGC1α (C, panels i and ii; Cy3, red), E-cad (C, panels iii and iv; Cy5, cyan), and nuclear stain (DAPI, blue) illustrates a decrease in PGC1α within the intestinal epithelium after 7 days of 3% DSS exposure (C, panels v and vi, merge; scale bars, 100 μm). Quantification of immunofluorescence reveals a statistically significant decrease in PGC1α staining. Mice were then subjected to 3% DSS over 7 days followed by a transition to normal drinking water for 2 days and sacrificed at 2-day intervals (n = 4/group, 6 groups). mRNA expression of PGC1α demonstrates an early increase, followed by a decrease over time with levels decreasing significantly as compared with control mice at day 7 (D). Protein levels demonstrate a similar but delayed trend with levels peaking at day 3 before declining. BALB/c mice were treated with TNBS or ethanol (EtOH) vehicle and euthanized on day 4 after treatment (n = 4/group). Mice treated with EtOH demonstrated a non-significant decrease in PGC1α protein in intestinal tissue, whereas mice treated with TNBS demonstrated a significant decrease (E). PGC1α protein was immunoprecipitated from the intestinal tissue of C57BL/6 mice subjected to DSS colitis (n = 8). Lysine residues on PGC1α protein in mice treated with DSS were highly acetylated as compared with PGC1α from control mice (F). PGC1α protein in mice treated with DSS was highly ubiquitinated as compared with that from control mice. Densitometry confirms statistical significance. *, p ≤ 0.05.
It is not currently known what causes this decrease in PGC1α during colitis. PGC1α is known to be activated by deacetylases, such as sirtuin 1 (SIRT1). By immunoprecipitating PGC1α, we show here that PGC1α is highly acetylated within the intestinal epithelium after a murine model of DSS colitis (Fig. 2F). Additionally, there is an increase in ubiquitinated PGC1α in mice after DSS exposure. Thus, we demonstrate that although PGC1α expression is dramatically decreased in the intestinal epithelium after experimental colitis, the PGC1α protein still present after DSS exposure exhibits decreased activity and has been targeted for proteosomal degradation through ubiquitination.
Evidence of Mitochondrial Dysfunction during Experimental Colitis
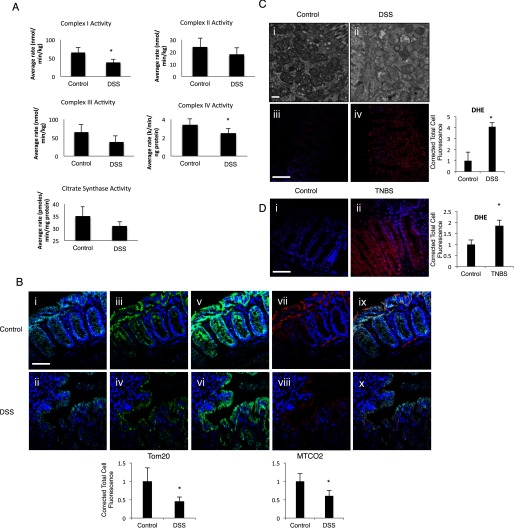
It has been reported previously that mitochondrial electron transport chain complex activity is altered in the setting of human colitis as well as murine DSS colitis (11, 12). To assess evidence of mitochondrial dysfunction in our model, we subjected C57BL/6 wild-type mice to 3% DSS for 7 days. Similar to other published reports, we found a decrease in the electron transport chain complex activity after 7 days of DSS exposure (Fig. 3A). As seen in Fig. 2, we have also noted a decrease in mitochondrial biogenesis within the intestinal epithelium of mice subjected to DSS colitis. To further determine the effect of DSS colitis and a down-regulation in mitochondrial biogenesis on mitochondrial mass and structure, we analyzed colonic tissue from the same mice via immunofluorescence for Mtco2 and Tom20, two mitochondrial outer membrane proteins along with an epithelial protein (E-cad). There was a decrease in mitochondrial staining within the intestinal epithelium of DSS-treated mice, suggesting a decrease in overall mitochondrial mass after DSS colitis (Fig. 3B, panels i–x). TEM of the same samples demonstrates a severe disruption in mitochondrial structure with membrane disruption, obliteration of the cristae, and an accumulation of intra-mitochondrial vesicles upon DSS exposure (Fig. 3C, panels i and ii). These changes closely resemble those seen in humans with Crohn's disease (26). This suggests the propensity of mitochondria to leak more ROS per unit of ATP generated, similar to what has been previously described (27, 28). In support of this hypothesis, we found a significant increase in the superoxide content within the intestinal epithelium of DSS-treated mice as measured by DHE oxidation (Fig. 3C, panels iii and iv). Finally, tissues from BALB/c mice subjected to TNBS colitis were similarly evaluated for DHE oxidation (Fig. 3D, panels i and ii). These mice were also found to have a dramatic increase in superoxide content upon completion of the model. These data taken in aggregate suggest that PGC1α depletion and a subsequent disruption of mitochondrial biogenesis in the setting of experimental colitis contributes to a concomitant derangement of mitochondrial structure and function as well as an overall decrease in mitochondrial mass.
FIGURE 3.
C57BL/6 mice were subjected to 3% DSS colitis for 7 days. Activity of the electron transport chain complexes I and IV was significantly decreased in intestinal tissue, although complex II, III, and citrate synthase activities showed non-significant decreases in activity (A, n = 4/group). Immunofluorescence was performed in mice subjected to DSS to evaluate the mitochondrial outer membrane proteins Mtco2 and Tom20 (B, panels i–x, n = 8/group, scale bars, 100 μm). E-cad (Cy5, cyan) stained intestinal epithelium in both control and DSS tissue (B, panels i and ii). Tom20 (FITC, green) is decreased in mice subjected to DSS colitis as compared with WT mice (B, panels ii–iv). E-cad demonstrates colocalization with Tom20 (B, panels v and vi). Similarly, Mtco2 (Cy3, red) is significantly decreased in mice subjected to DSS colitis as compared with WT mice (B, panels vii and viii). E-cad demonstrates colocalization with Mtco2 (B, panels ix and x). All images contain nuclear stain (DAPI, blue). Mitochondrial structure was deranged in the same mice after DSS treatment as seen on TEM (C, panel i and ii, scale bars, 1 μm). DHE staining demonstrated a significant increase in superoxide ions in the intestinal tissue of mice subjected to DSS, which was confirmed by quantification (C, panels iii and iv, blue to red indicates increase in superoxide ions). Intestinal tissue from BALB/c mice subjected to TNBS colitis was similarly analyzed for DHE staining, and an increase in superoxide ions is seen in mice with colitis (D, panels i and ii). *, p ≤ 0.05.
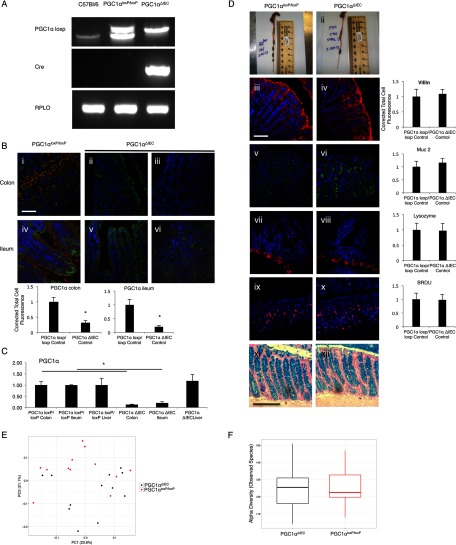
Mice Lacking PGC1α in the Intestinal Epithelium Develop Dramatically Worse Colitis than Their Wild-type Littermates
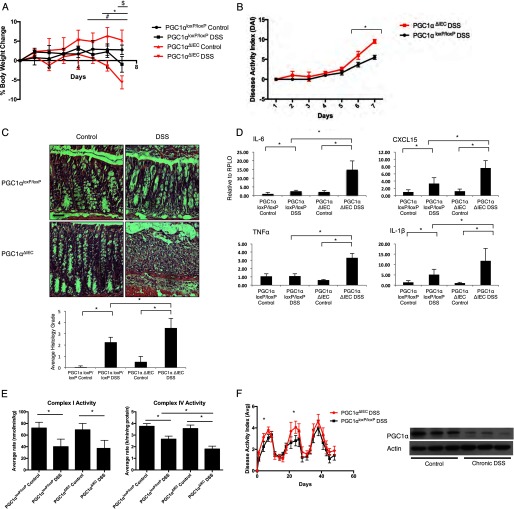
To evaluate whether the loss of PGC1α can be a cause as opposed to a consequence of colitis, we developed the intestinal epithelium-specific PGC1α knock-out mouse (PGC1αΔIEC) by mating a PGC1αloxP/loxP mouse with a villin-cre mouse (Fig. 4A). As predicted, immunofluorescence demonstrated that PGC1α was found to be predominantly located at apical enterocytes and colonocytes with less expression within the crypts of WT mice (Fig. 4B, panels i–vi). PGC1αΔIEC mice demonstrated a near-total loss of PGC1α within the intestinal epithelium with some islands of expression remaining, which is consistent with a mosaic pattern of villin expression in the colon. Minimal expression of PGC1α in the intestinal epithelium of PGC1αΔIEC mice was confirmed by qRT-PCR (Fig. 4C). PGC1α expression was preserved in other tissues, such as the liver. To demonstrate that cre-recombination or the absence of PGC1α within the intestinal epithelium has not altered the overall structure or cellular makeup of the intestinal epithelium, we performed immunofluorescence for villin and Muc2 in the colon, as well as Alcian blue staining. Our staining would suggest a similar content of epithelial cells and goblet cells between strains (Fig. 4D, panels iii–iv and xi–xii). We also stained the ileum for lysozyme and saw no differences in Paneth cell mass (Fig. 4D, panels vii and viii). A similar proliferative index is noted in both the PGC1αΔIEC mice and the PGC1αloxP/loxP mice as demonstrated by BrdU staining (Fig. 4D, panels ix and x). Similar results were seen with Ki-67 and proliferating cell nuclear antigen staining (data not shown). We do not see significant apoptotic indices in either strain at baseline (data not shown). Finally, we used 16S rRNA gene sequencing to determine whether any differences in a response to experimental colitis could be attributable to strain-specific differences in the gut microbiota. Fecal samples were obtained for analysis after mice were weaned. Neither α diversity nor β diversity (community composition) of fecal microbial communities differed between the PGC1αΔIEC mice and their PGC1αloxP/loxP littermates (Fig. 4, E and F). These studies suggest that PGC1α deletion in the PGC1αΔIEC mice is near complete in the colon and small intestine. Furthermore, the deletion is indeed specific to the intestinal epithelium. These mice do not appear to differ from wild-type mice with regard to colonic anatomy, cellular makeup, proliferative index, or their gut microbiome. Individual PGC1αΔIEC mice have survived for more than 1 year and have proven to be healthy, without demonstrating any specific phenotype in the absence of an insult (data not shown). To determine the extent of colitis that PGC1αΔIEC mice would develop, we subjected them to 2% DSS. A lower concentration of DSS is often chosen in susceptible strains to prevent mortality, preserve intestinal architecture, and maintain discernible differences between strains. PGC1αΔIEC mice developed a more severe clinical colitis than their PGC1αloxP/loxP littermates, as evidenced by a greater weight loss and higher DAI (Fig. 5, A and B). Histology demonstrated dramatically worse inflammatory changes in the PGC1αΔIEC mice (Fig. 5C). When intestinal tissues were evaluated by qRT-PCR, we found that PGC1αΔIEC mice expressed much higher levels of intestinal inflammatory cytokines that are known to be elevated in experimental colitis and human IBD, including Il-6, Cxcl15, Tnfα, and Il-1β (Fig. 5D). Based on the role of PGC1α in promoting mitochondrial biogenesis, we evaluated activity of the electron transport chain in mice subjected to DSS. PGC1αΔIEC mice demonstrated a non-significant decrease in complex I activity and a significant decrease in complex IV activity as compared with PGC1αloxP/loxP mice (Fig. 5E). As seen in Fig. 3A, these are the key complexes that were found to be compromised in our model of DSS colitis. To further characterize the inflammatory response of PGC1αΔIEC mice subjected to experimental colitis, we performed a chronic relapsing model of DSS exposure consisting of three 7-day cycles of 2% DSS exposure followed by a transition to regular drinking water for 10 days. As expected, PGC1αΔIEC mice developed earlier clinical manifestations of the disease and suffered a more significant clinical colitis during each of the first two cycles of DSS compared with PGC1αloxP/loxP littermates (Fig. 5F). Interestingly, during the third cycle of exposure, only a slight non-significant difference in DAI was noted between the two strains. This may suggest that compensatory mechanisms develop over the course of a chronic relapsing disease that may help to mitigate, in part, the contribution of PGC1α depletion to inflammation. Alternatively, we noted a decrease in PGC1α protein levels within the intestinal epithelium of WT mice at the end of the model similar to that seen after a 7-day DSS exposure, indicating that the down-regulation of PGC1α remains stable over time. Thus, by the third cycle of DSS colitis, the PGC1αloxP/loxP mice may come to resemble the PGC1αΔIEC mice in terms of PGC1α protein levels in the intestine and may in turn respond to DSS exposure similarly. These data suggest that PGC1α plays a key role in preventing or ameliorating experimental colitis and that its down-regulation during disease contributes to the initiation and propagation of inflammation.
FIGURE 4.
Intestinal epithelium-specific PGC1α knock-out mouse (PGC1αΔIEC) was created by mating a PGC1αloxP/loxP mouse with a villin-cre mouse. Mice were genotyped using RT-PCR (A). Immunofluorescence for PGC1α (Cy3, red), E-cad (FITC, green), and nuclear stain (DAPI, blue) demonstrates a significant decrease of PGC1α protein within the intestinal epithelium of PGC1αΔIEC mice (B, n = 8/group, scale bars, 100 μm). Staining in both the colon and ileum was quantified. Representative images are provided from the colon of PGC1αloxP/loxP mice (B, panel i) and PGC1αΔIEC mice (B, panels ii and iii) as well as the ileum of PGC1αloxP/loxP mice (B, panel iv) and PGC1αΔIEC mice (B, panels v and vi). qRT-PCR of intestinal tissue demonstrates PGC1α expression in the colon, ileum, and liver of PGC1αΔIEC and PGC1αloxP/loxP mice. Expression is decreased in the intestines of PGC1αΔIEC mice as compared with PGC1αloxP/loxP mice but not in the liver (C). No differences in intestinal morphology were noted between strains (D, panels i and ii). Immunofluorescence was performed for colonic villin (D, panels iii and iv, scale bars, 100 μm), colonic Muc2 (D, panels v and vi), ileal lysozyme (D, panels vii and viii), and colonic BrdU (D, panels ix and x). When quantification was performed, no differences were noted between strains. Alcian blue staining for goblet cells was similar between strains (D, xi and xii). β diversity comparisons of microbial communities within fecal samples from PGC1αΔIEC mice and their PGC1αloxP/loxP littermates are not significant. Shown are the observed species indices for each sample group (E). Displayed are principal coordinate analyses of unweighted Unifrac distances between all samples analyzed. Axis labels indicate the proportion of variance explained by each principal coordinate axis. Fecal samples of the two strains do not cluster separately within the principal coordinate space. Diversity within each group is as great as the diversity between sample types. α diversity comparisons of microbial communities within fecal samples from PGC1αΔIEC mice and their PGC1αloxP/loxP littermates are not significant. Shown are the observed species indices for each sample group (F). *, p ≤ 0.05.
FIGURE 5.
Intestinal epithelium-specific PGC1α knock-out mouse (PGC1αΔIEC) was subjected to 2% DSS for 7 days along with PGC1αloxP/loxP littermates (n = 8/group). When the % body weight was evaluated, significant differences were noted between PGC1αΔIEC control versus PGC1αΔIEC DSS-treated mice (#, p = ≤0.05), PGC1αΔIEC DSS versus PGC1αloxP/loxP DSS-treated mice (*, p ≤ 0.05), and PGC1αloxP/loxP control versus PGC1αloxP/loxP DSS-treated mice ($, p ≤ 0.05) (A). DAI score for PGC1αΔIEC mice was significantly higher than that for PGC1αloxP/loxP mice subjected to DSS colitis (B). Histologically, PGC1αΔIEC mice developed a more dramatic colitis, as quantified by average histology grade (C, scale bars, 100 μm). PGC1αΔIEC mice also demonstrated a significant increase in pro-inflammatory tissue cytokine release as compared with PGC1αloxP/loxP mice (D). Activity of the electron transport chain complexes I and IV were significantly decreased in both strains of mice after DSS exposure. However, complex IV activity was significantly decreased in PGC1αΔIEC mice subjected to DSS as compared with PGC1αloxP/loxP mice similarly treated (E). PGC1αΔIEC mice and PGC1αloxP/loxP mice were subjected to a chronic model of 2% DSS exposure (n = 16/group). Significant differences in DAI were noted in the first two cycles of DSS treatment but not the third cycle (F). As seen after acute DSS colitis, PGC1α is decreased in the intestines of PGC1αloxP/loxP mice after the completion of the chronic DSS model. *, p ≤ 0.05.
SRT1720 Stimulates an Increase in Intestinal PGC1α and Ameliorates Experimental Colitis
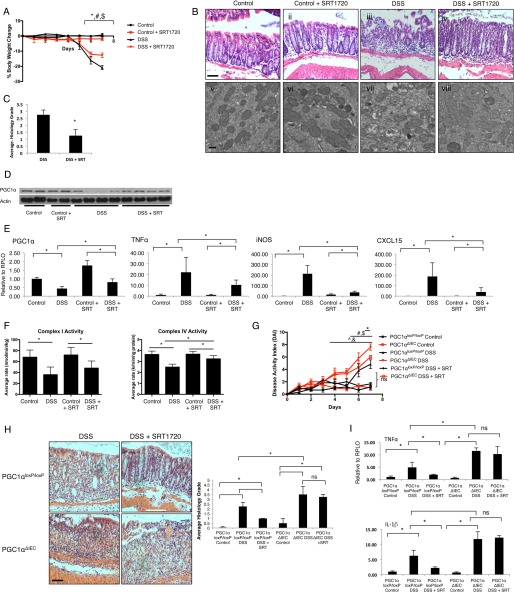
Having demonstrated that the deletion of PGC1α from the intestinal epithelium increases the severity of experimental colitis in mice, we next sought to further confirm the protective role of PGC1α in the intestine using pharmacologic means. Thus, we treated C57BL/6 wild-type mice undergoing DSS colitis with 100 mg/kg/day of the Sirt1 activator SRT1720. SIRT1 is a powerful deacetylase that is known to activate PGC1α and drive mitochondrial biogenesis (29). SRT1720 has been shown to enhance mitochondrial biogenesis in a SIRT1-dependent process (30–35). SRT1720-treated mice were found to be protected from experimental colitis as predicted (Fig. 6, A and B). H&E sections of colonic tissue demonstrated a preservation of epithelial architecture in mice receiving SRT1720, whereas TEM demonstrated a relative preservation of mitochondrial structure (Fig. 6, B and C). As shown in Fig. 6D, we observed an increase in PGC1α protein in the intestines of mice receiving SRT1720 while subjected to DSS colitis. This was an unexpected finding, although PGC1α is thought to be able to drive its own expression through a positive feedback loop (36). A modest increase in RNA expression of PGC1α is also seen in these mice (Fig. 6E). Additionally, expression of Tnfα, inos, and Cxcl15 was significantly decreased within the intestinal tissue of DSS-treated mice receiving SRT1720. To evaluate the effects of PGC1α activation on mitochondrial function, we once again performed an analysis of electron transport chain activity. SRT1720 was shown to have minimal impact on complex I activity, although it significantly increased complex IV activity (Fig. 6F). When we treated PGC1αΔIEC mice with SRT1720 while undergoing DSS colitis, there was a minimal effect on DAI and no significant changes to histology or inflammatory cytokine expression (Fig. 6, G–I), suggesting that SRT1720 protects against DSS colitis, at least in part, through an up-regulation and activation of PGC1α in the intestinal epithelium and not in other cell types.
FIGURE 6.
C57BL/6 mice were subjected to 3% DSS colitis and SRT1720 100 mg/kg/day via oral gavage versus vehicle (water). Significant differences in % body weight change were seen in control versus DSS (*, p ≤ 0.05), control + SRT1720 versus DSS + SRT1720 (#, p ≤ 0.05), and DSS versus DSS + SRT1720 ($, p ≤ 0.05) (A, n = 8/group). Histopathology demonstrated significant manifestations of disease in DSS-treated animals with a preservation of architecture in SRT1720-treated animals (B, panels i–iv, scale bar, 100 μm). TEM demonstrated a predicted derangement of mitochondrial structure in mice subjected to DSS colitis with a partial preservation of structure in SRT1720-treated mice (B, panels v–viii, scale bar, 1 μm). The average histology grade for DSS + SRT-treated mice is significantly lower that that for DSS-treated mice (C, *, p ≤ 0.05). No changes were seen in control mice (data not shown). Interestingly, PGC1α was increased in SRT-treated DSS animals as compared with DSS-treated mice, as demonstrated by Western blot and qRT-PCR (D and E). Similarly, tissue cytokine release was significantly decreased in DSS + SRT-treated mice as compared with DSS-treated mice (E). Activity of the electron transport chain complexes I and IV were significantly decreased in both DSS and DSS + SRT-treated mice. However, complex IV activity was significantly decreased in DSS as compared with DSS + SRT-treated mice (F). PGC1αΔIEC mice treated with SRT1720 along with DSS exposure demonstrated minimal protection from DSS colitis (G and I). Significant differences in DAI were noted between PGC1αΔIEC control versus PGC1αΔIEC DSS-treated mice (^p = ≤ 0.05), PGC1αΔIEC DSS versus PGC1αloxP/loxP DSS-treated mice (&, p ≤ 0.05), PGC1αloxP/loxP control versus PGC1αloxP/loxP DSS-treated mice (#, p ≤ 0.05), and PGC1αloxP/loxP control versus PGC1αloxP/loxP DSS + SRT-treated mice ($, p ≤ 0.05) (G). A small but significant difference in DAI score was noted between PGC1αΔIEC DSS versus PGC1αΔIEC DSS + SRT-treated mice only at day 7 (*, p ≤ 0.05). Average histology grades were not significantly different between PGC1αΔIEC DSS-treated mice and PGC1αΔIEC DSS + SRT-treated mice (H, scale bar, 100 μm; *, p ≤ 0.05; ns, not significant). Similarly, tissue cytokine levels were not significant between PGC1αΔIEC DSS-treated mice and PGC1αΔIEC DSS + SRT-treated mice (I, *, p ≤ 0.05; ns, not significant).
Loss of PGC1α Leads to a Decrease in Intestinal Barrier Function, Leading to Increased Bacterial Translocation
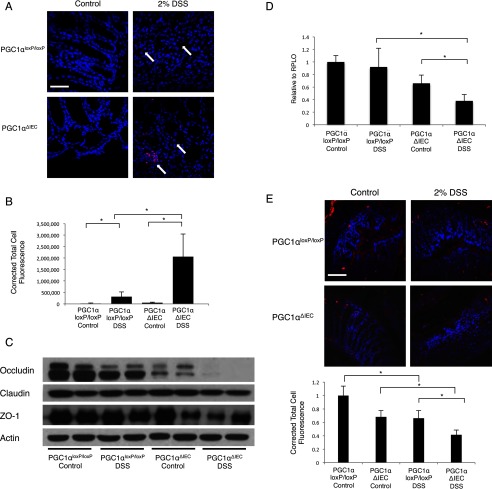
Defects in intestinal barrier function are known to be characteristic features of IBD (37). Furthermore, it has been hypothesized that the maintenance of the intestinal barrier requires healthy mitochondria and robust ATP availability (38). Thus, we hypothesized that changes in PGC1α expression with a resultant disruption in mitochondrial biogenesis would lead to abnormal barrier function, resulting in bacterial translocation. In situ staining of bacterial 16S rRNA using the EUB338 probe demonstrated mild bacterial translocation in PGC1αloxP/loxP mice subjected to DSS colitis (Fig. 7, A and B). PGC1αΔIEC mice demonstrated significantly greater translocation after DSS exposure. This suggests that PGC1α contributes to the maintenance of barrier function during intestinal inflammation. To investigate how a decrease in PGC1α may lead to increased bacterial translocation, we evaluated tight junction protein levels in intestinal tissue. At baseline, we noted a slight decrease in occludin levels in PGC1αΔIEC control mice as compared with PGC1αloxP/loxP mice. Following exposure to 2% DSS, occludin protein levels were mildly decreased in PGC1αloxP/loxP mice but were nearly undetectable in PGC1αΔIEC mice (Fig. 7C). Expression levels of occludin RNA were not significantly decreased in PGC1αloxP/loxP mice subjected to DSS colitis. However, expression levels were significantly decreased in PGC1αΔIEC mice (Fig. 7D). Immunofluorescence for occludin confirms that occludin protein is decreased in PGC1αloxP/loxP mice subjected to experimental colitis but that occludin levels are significantly lower in PGC1αΔIEC mice (Fig. 7E). A similar decrease in Zo-1 protein was noted, but not in Claudin-1. Differences in Zo-1 expression and immunofluorescence, however, were not seen (data not shown). Taken together, these data suggest that PGC1α plays a key role in maintaining the intestinal barrier, likely through its effects on mitochondrial biogenesis and cellular energetics. A decrease in PGC1α with the initiation of inflammation leads to a decrease in expression of the tight junction protein occludin, contributing to enhanced bacterial translocation.
FIGURE 7.
After 7 days of 2% DSS exposure, in situ staining for bacterial 16S rRNA using EUB338 (red) and DAPI nuclear stain (blue) was performed (A, n = 8/group, scale bar, 100 μm). A significant increase in bacterial translocation is seen in PGC1αΔIEC mice when compared with PGC1αloxP/loxP mice. Quantification of immunofluorescence demonstrates statistical significance (B). The tight junction protein occludin is mildly decreased in PGC1αloxP/loxP mice subjected to DSS colitis. Occludin levels in PGC1αΔIEC mice are decreased at baseline and are nearly absent after DSS colitis. Zo-1 levels are also decreased in PGC1αΔIEC mice subjected to DSS colitis as compared with control mice (C). qRT-PCR analysis reveals stable expression of occludin in PGC1αloxP/loxP mice subjected to DSS as compared with control mice, whereas a significant decrease is seen in PGC1αΔIEC mice subjected to colitis (D). Immunofluorescence for occludin demonstrates a diffuse decrease in staining after DSS induction in PGC1αloxP/loxP mice with a greater decrease in PGC1αΔIEC mice (E, scale bar, 100 μm). *, p ≤ 0.05.
Discussion
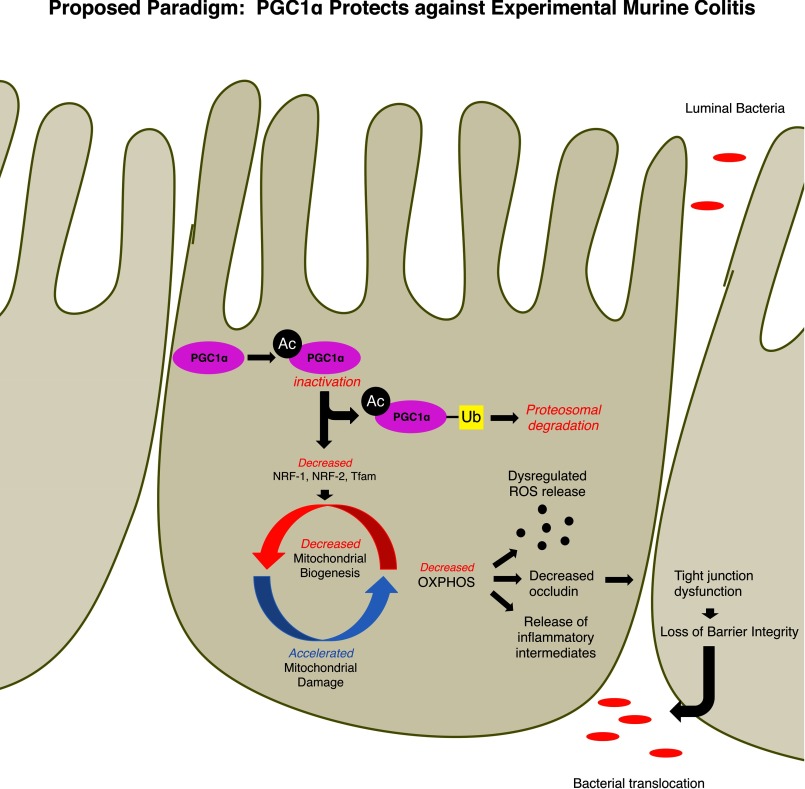
We show here that PGC1α protects against colonic inflammation and promotes mitochondrial homeostasis within the intestinal epithelium. Moreover, an observed decrease in PGC1α in the intestine contributes to the pathogenesis of colitis through a derangement of mitochondrial function and a breakdown of the intestinal barrier (Fig. 8). In support of this paradigm, we have demonstrated for the first time that mitochondrial biogenesis is significantly decreased within the intestine during the early stages of experimental colitis. Furthermore, we show that during experimental colitis there is a significant increase in the acetylation status and the ubiquitination state of PGC1α protein in the intestinal epithelium. We propose that the increase in acetylated PGC1α is indicative of inactive protein, which eventually becomes ubiquitinated for proteosomal degradation in an attempt to prevent a large cellular buildup of inactive protein. PGC1α is known to contain at least 13 acetylation sites, and acetylation status of these residues is strongly associated with repression of PGC1α activity (29). This is consistent with our results demonstrating a down-regulation of gene and protein expression mediated by PGC1α. In contrast, NAD+-dependent deacetylases, such as SIRT1, catalyze the deacetylation of PGC1α, converting it to its active form and initiating the mitochondrial biogenesis pathway, regulating oxygen consumption in cells and increasing the transcriptional activity of PGC1α (29, 39–42). The PGC1α-SIRT1 interaction has been shown to boost mitochondrial activity in neurons, although decreased deacetylation of PGC1α by SIRT1 leads to altered metabolism in cardiomyocytes (43, 44). Furthermore, activation of the SIRT1-PGC1α axis has proven beneficial in trauma-hemorrhage, oxidant-induced renal injury, and neurodegenerative diseases among others (33, 45, 46). Our key finding is that a loss of PGC1α in the intestinal epithelium, as seen in human IBD, predisposes mice to mitochondrial stress and inflammation during experimental murine colitis. Stimulation of PGC1α transcription and activation protects against inflammation in our model of colitis. Key intermediates of mitochondrial biogenesis are similarly down-regulated with PGC1α depletion during colitis, including NRF-1 and -2 (47, 48). Tfam, a nuclearly encoded mitochondrial transcription factor that provides the critical link between the nucleus and the mitochondrion during mitochondrial biogenesis, is similarly decreased in mice subjected to experimental colitis. This results in a decrease in both mtDNA copy number and the number of healthy mitochondria in the intestinal epithelium. This down-regulation of the mitochondrial biogenesis machinery is accompanied by a decrease in mitochondrial function, a derangement of mitochondrial structure, and an increase in oxidative stress. Although decreased PGC1α contributed to a morphologic and bioenergetic failure of mitochondria, it may also contribute to oxidative stress, which is a hallmark of colitis. PGC1α-dependent mitochondrial failure is associated with a potentiation of bacterial translocation in the gut, likely through a decrease in the tight junction protein occludin. Taken together, these data highlight how colitis develops, in part, through a disruption of mitochondrial integrity.
FIGURE 8.
We propose that PGC1α is inactivated through acetylation and targeted for degradation by ubiquitination within the intestinal epithelium during intestinal inflammation. A decrease in PGC1α protein down-regulates the mitochondrial biogenesis pathway and results in an overall decrease in healthy mitochondria. This potentiates inflammation through a dysregulated release in ROS, decreased oxidative phosphorylation, and an accumulation of unhealthy mitochondria. Consequently, there is a decrease in the tight junction protein occludin, and the intestinal barrier is compromised leading to translocation of luminal bacteria.
The current findings extend our knowledge regarding the role of mitochondrial failure in the development of intestinal inflammation. ROS and reactive nitrogen species (RNS) have long been known to serve as key intermediates in the pathogenesis of IBD; however, the cause of dysregulated ROS/RNS release during intestinal inflammation remains unknown. Here, we provide evidence outlining a potential mechanism for increased oxidative stress during colitis. PGC1α is known to play a key role in the control of ROS homeostasis. It is required for the induction of some antioxidants and is known to help coordinate skeletal muscle tissue adaptation during exercise (49–51). Under conditions of oxidative stress, PGC1α transcriptional activity can increase as a means of buffering ROS production (52). This may explain why PGC1α is up-regulated early on during acute DSS colitis. This early adaptive response to increasing oxidative stress ultimately fails with ongoing insult. As the key regulator of mitochondrial biogenesis, PGC1α loss contributes to the degradation of mitochondrial function, resulting in the propensity to leak ROS (27, 28). Through acetylation and ubiquitination, PGC1α is deactivated and removed from the intestinal epithelium, contributing to inflammation largely through ROS release. Previous studies have suggested that mitochondrial function is compromised in the setting of intestinal inflammation. ROS/RNS have long been implicated in the pathogenesis of colitis (6). Multiple studies have demonstrated increased ROS/RNS in the colonic mucosa of animals subjected to models of colitis, and levels seem to correlate with severity of disease (7). Similarly, there is a depletion of endogenous antioxidants and increased accumulation of biomarkers of oxidative damage (8, 9). Endogenous and exogenous antioxidant compounds have been demonstrated to ameliorate experimental colitis using both chemical and genetic methods (10, 53). Trials of some antioxidant compounds in humans with IBD have demonstrated promising yet limited results. A decrease in both expression and function of the components of the mitochondrial electron transport chain has also been demonstrated in humans with IBD and mice subjected to experimental colitis (11–13). With changes appearing in inflamed and non-inflamed segments of bowel, these data may suggest underlying changes in mitochondrial function, which may contribute to not only the pathogenesis of colitis, but also to the susceptibility to disease. Finally, some mtDNA polymorphisms, which increase ATP levels in mice, have been shown to protect mice from experimental colitis (54). This adds relevance to previous studies demonstrating decreased ATP levels within the intestinal epithelium of humans with IBD (55, 56). Our studies demonstrate a PGC1α-dependent decrease in mitochondrial mass and a severe derangement of mitochondrial structure during colitis. Based on these results, we now speculate that mitochondrial dynamics, in particular mitochondrial biogenesis, are disrupted during colitis, which contributes to the pathogenesis of disease (Fig. 8).
Mitochondrial dysfunction alone is most likely not enough to cause significant inflammatory changes in the intestine. Specifically, increased oxidative stress is not thought to be a primary activator of disease (6). Similarly, low ATP levels within areas of active inflammation are known to be a hallmark of disease; however, the molecular causes of a low energy state have until now been poorly understood (54, 55). Recent evidence has suggested a critical link between cellular bioenergetics and the maintenance of the mucosal barrier (57–61). It is postulated that the perijunctional cytoskeleton that associates with and supports tight junction proteins demonstrates ATP-dependent contractility (62). Furthermore, tight junction stability is thought to be dependent on adequate energy stores (63). Thus, any perturbation in mitochondrial dynamics within the intestinal epithelium could lead to changes in the intestinal barrier integrity. We investigated intestinal barrier function through an analysis of bacterial infiltration and translocation through the mucosal barrier as well as an analysis of tight junction proteins. As expected, the PGC1αΔIEC mice demonstrated a dramatic increase of bacterial infiltration into the intestinal tissue. Increased translocation is associated with a decrease in the tight junction protein occludin. A down-regulation of occludin has been noted in both ulcerative colitis and Crohn's disease, and evidence would suggest that occludin is a key regulator of tight junction function (64–66). Thus, we now hypothesize that changes in PGC1α expression, with a resultant disruption in mitochondrial biogenesis, lead to abnormal barrier function, in part through a loss of the tight junction protein occludin. These findings lend increased significance to the role of PGC1α in the development of IBD.
It is possible that PGC1α may be exerting a protective role in the intestinal epithelium through other mechanisms as well. Although these data would suggest that the dominant contribution of PGC1α comes from its effects on mitochondrial dynamics and function, PGC1α can also play a role in regulating cellular metabolism. For instance, through its interaction with peroxisome proliferator-activated receptor γ and estrogen-related receptor α, PGC1α is known to stimulate the uptake and oxidation of fatty acids and cholesterol, driving gluconeogenesis (67, 68). In skeletal muscle, PGC1α is required for the expression of glucose transporter type 4 (GLUT4), which can drive glucose uptake in the presence of insulin (68). Furthermore, PGC1α is known to have direct effects on oxidative phosphorylation, enhancing efficient energy production from mitochondria (69).
The mitochondria themselves may play key roles in innate immune signaling in the intestinal epithelium. Damaged or dysregulated mitochondria as well as mitochondrially derived ROS have been demonstrated to activate the NLRP3 inflammasome (70–72). Mitochondrial disruption during the pathogenesis of IBD may similarly activate the mitochondrial unfolded protein response. Additionally, damaged mitochondria may release mitochondrial damage-associated molecular patterns, which would potentiate further damage within the epithelium (73).
In summary, we now show that the documented decrease in PGC1α within the intestinal epithelium is not only a marker of pre-neoplastic ulcerative colitis but also a significant contributor to intestinal inflammation. Furthermore, the activation of PGC1α and mitochondrial biogenesis appears to contribute to gut homeostasis and barrier function at baseline. Such findings not only offer insights into the molecular mechanisms that lead to the development of colitis, but also potentially identify novel therapeutic targets for this highly morbid disease.
Author Contributions
K. E. C. and G. V. conducted experiments and acquired data. C. P. S. and E. A. N. analyzed and interpreted data and provided statistical analysis. C. E. E. provided technical support and analyzed and interpreted data. D. B. S. acquired and analyzed data. B. S. Z. acquired data and provided critical revision of the manuscript. G. K. G. and D. J. H. supervised the study and provided critical revision of the manuscript. K. P. M. supervised, designed, and interpreted experiments and wrote the manuscript.
This work was supported by National Institutes of Health Grants DK101753 (to K. P. M.), GM078238 (to D. J. H.), and DK083752 (to D. J. H.) and the Association for Academic Surgery (to K. P. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IBD
- inflammatory bowel disease
- DHE
- dihydroethidium
- DSS
- dextran sodium sulfate
- MTCO2
- cytochrome c oxidase subunit II
- NRF
- nuclear respiratory factor
- PGC1α
- peroxisome proliferator-activated receptor-γ coactivator 1-α
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- TNBS
- 2,4,6-trinitrobenzenesulfonic acid
- TEM
- transmission electron microscopy
- Tfam
- mitochondrial transcription factor A
- qRT
- quantitative RT
- DAI
- disease activity index
- E-cad
- E-cadherin.
References
- 1. Shikhare G., and Kugathasan S. (2010) Inflammatory bowel disease in children: current trends. J. Gastroenterol. 45, 673–682 [DOI] [PubMed] [Google Scholar]
- 2. Long M. D., Hutfless S., Kappelman M. D., Khalili H., Kaplan G. G., Bernstein C. N., Colombel J. F., Gower-Rousseau C., Herrinton L., Velayos F., Loftus E. V. Jr., Nguyen G. C., Ananthakrishnan A. N., Sonnenberg A., Chan A., et al. (2014) Challenges in designing a national surveillance program for inflammatory bowel disease in the United States. Inflamm. Bowel Dis. 20, 398–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kappelman M. D., Rifas-Shiman S. L., Kleinman K., Ollendorf D., Bousvaros A., Grand R. J., and Finkelstein J. A. (2007) The prevalence and geographic distribution of Crohn's disease and ulcerative colitis in the United States. Clin. Gastroenterol. Hepatol. 5, 1424–1429 [DOI] [PubMed] [Google Scholar]
- 4. Kappelman M. D., Rifas-Shiman S. L., Porter C. Q., Ollendorf D. A., Sandler R. S., Galanko J. A., and Finkelstein J. A. (2008) Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology 135, 1907–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molodecky N. A., Soon I. S., Rabi D. M., Ghali W. A., Ferris M., Chernoff G., Benchimol E. I., Panaccione R., Ghosh S., Barkema H. W., and Kaplan G. G. (2012) Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54 [DOI] [PubMed] [Google Scholar]
- 6. Zhu H., and Li Y. R. (2012) Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidence. Exp. Biol. Med. 237, 474–480 [DOI] [PubMed] [Google Scholar]
- 7. Pavlick K. P., Laroux F. S., Fuseler J., Wolf R. E., Gray L., Hoffman J., and Grisham M. B. (2002) Role of reactive metabolites of oxygen and nitrogen in inflammatory bowel disease. Free Radic. Biol. Med. 33, 311–322 [DOI] [PubMed] [Google Scholar]
- 8. Işman C. A., Yeğen B. C., and Alican I. (2003) Methimazole-induced hypothyroidism in rats ameliorates oxidative injury in experimental colitis. J. Endocrinol. 177, 471–476 [DOI] [PubMed] [Google Scholar]
- 9. Damiani C. R., Benetton C. A., Stoffel C., Bardini K. C., Cardoso V. H., Di Giunta G., Pinho R. A., Dal-Pizzol F., and Streck E. L. (2007) Oxidative stress and metabolism in animal model of colitis induced by dextran sulfate sodium. J. Gastroenterol. Hepatol. 22, 1846–1851 [DOI] [PubMed] [Google Scholar]
- 10. Seguí J., Gironella M., Sans M., Granell S., Gil F., Gimeno M., Coronel P., Piqué J. M., and Panés J. (2004) Superoxide dismutase ameliorates TNBS-induced colitis by reducing oxidative stress, adhesion molecule expression, and leukocyte recruitment into the inflamed intestine. J. Leukocyte Biol. 76, 537–544 [DOI] [PubMed] [Google Scholar]
- 11. Santhanam S., Rajamanickam S., Motamarry A., Ramakrishna B. S., Amirtharaj J. G., Ramachandran A., Pulimood A., and Venkatraman A. (2012) Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis. 18, 2158–2168 [DOI] [PubMed] [Google Scholar]
- 12. Sifroni K. G., Damiani C. R., Stoffel C., Cardoso M. R., Ferreira G. K., Jeremias I. C., Rezin G. T., Scaini G., Schuck P. F., Dal-Pizzol F., and Streck E. L. (2010) Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis. Mol. Cell. Biochem. 342, 111–115 [DOI] [PubMed] [Google Scholar]
- 13. Taylor C. T., and Moncada S. (2010) Nitric oxide, cytochrome c oxidase, and the cellular response to hypoxia. Arterioscler. Thromb. Vasc. Biol. 30, 643–647 [DOI] [PubMed] [Google Scholar]
- 14. Kuznetsov A. V., and Margreiter R. (2009) Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int. J. Mol. Sci. 10, 1911–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liesa M., Palacín M., and Zorzano A. (2009) Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89, 799–845 [DOI] [PubMed] [Google Scholar]
- 16. Lehman J. J., Barger P. M., Kovacs A., Saffitz J. E., Medeiros D. M., and Kelly D. P. (2000) Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D'Errico I., Salvatore L., Murzilli S., Lo Sasso G., Latorre D., Martelli N., Egorova A. V., Polishuck R., Madeyski-Bengtson K., Lelliott C., Vidal-Puig A. J., Seibel P., Villani G., and Moschetta A. (2011) Peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC1α) is a metabolic regulator of intestinal epithelial cell fate. Proc. Natl. Acad. Sci. U.S.A. 108, 6603–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ussakli C. H., Ebaee A., Binkley J., Brentnall T. A., Emond M. J., Rabinovitch P. S., and Risques R. A. (2013) Mitochondria and tumor progression in ulcerative colitis. J. Natl. Cancer Inst. 105, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siegmund B., Lehr H. A., Fantuzzi G., and Dinarello C. A. (2001) IL-1β-converting enzyme (caspase-1) in intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 98, 13249–13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wirtz S., Neufert C., Weigmann B., and Neurath M. F. (2007) Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546 [DOI] [PubMed] [Google Scholar]
- 21. Ostanin D. V., Bao J., Koboziev I., Gray L., Robinson-Jackson S. A., Kosloski-Davidson M., Price V. H., and Grisham M. B. (2009) T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G135–G146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leaphart C. L., Cavallo J., Gribar S. C., Cetin S., Li J., Branca M. F., Dubowski T. D., Sodhi C. P., and Hackam D. J. (2007) A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J. Immunol. 179, 4808–4820 [DOI] [PubMed] [Google Scholar]
- 23. Berg D. J., Davidson N., Kühn R., Müller W., Menon S., Holland G., Thompson-Snipes L., Leach M. W., and Rennick D. (1996) Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J. Clin. Invest. 98, 1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H., Vermulst M., Wang Y. E., Chomyn A., Prolla T. A., McCaffery J. M., and Chan D. C. (2010) Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J. A., Smith G., and Knight R. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nazli A., Yang P. C., Jury J., Howe K., Watson J. L., Söderholm J. D., Sherman P. M., Perdue M. H., and McKay D. M. (2004) Epithelia under metabolic stress perceive commensal bacteria as a threat. Am. J. Pathol. 164, 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davies K. J., Quintanilha A. T., Brooks G. A., and Packer L. (1982) Free radicals and tissue damage produced by exercise. Biochem. Biophys. Res. Commun. 107, 1198–1205 [DOI] [PubMed] [Google Scholar]
- 28. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., and Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., and Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 30. Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., et al. (2010) SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamazaki Y., Usui I., Kanatani Y., Matsuya Y., Tsuneyama K., Fujisaka S., Bukhari A., Suzuki H., Senda S., Imanishi S., Hirata K., Ishiki M., Hayashi R., Urakaze M., Nemoto H., et al. (2009) Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am. J. Physiol. Endocrinol. Metab. 297, E1179–E1186 [DOI] [PubMed] [Google Scholar]
- 32. Yao H., Chung S., Hwang J. W., Rajendrasozhan S., Sundar I. K., Dean D. A., McBurney M. W., Guarente L., Gu W., Rönty M., Kinnula V. L., and Rahman I. (2012) SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J. Clin. Invest. 122, 2032–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Funk J. A., Odejinmi S., and Schnellmann R. G. (2010) SRT1720 induces mitochondrial biogenesis and rescues mitochondrial function after oxidant injury in renal proximal tubule cells. J. Pharmacol. Exp. Ther. 333, 593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., et al. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minor R. K., Baur J. A., Gomes A. P., Ward T. M., Csiszar A., Mercken E. M., Abdelmohsen K., Shin Y. K., Canto C., Scheibye-Knudsen M., Krawczyk M., Irusta P. M., Martín-Montalvo A., Hubbard B. P., Zhang Y., et al. (2011) SRT1720 improves survival and healthspan of obese mice. Sci. Rep. 1, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Handschin C., Rhee J., Lin J., Tarr P. T., and Spiegelman B. M. (2003) An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. U.S.A. 100, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merga Y., Campbell B. J., and Rhodes J. M. (2014) Mucosal barrier, bacteria and inflammatory bowel disease: possibilities for therapy. Dig. Dis. 32, 475–483 [DOI] [PubMed] [Google Scholar]
- 38. Wang A., Keita A. V., Phan V., McKay C. M., Schoultz I., Lee J., Murphy M. P., Fernando M., Ronaghan N., Balce D., Yates R., Dicay M., Beck P. L., MacNaughton W. K., Soderholm J. D., and McKay D. M. (2014) Targeting mitochondria-derived reactive oxygen species to reduce epithelial barrier dysfunction and colitis. Am. J. Pathol. 184, 2516–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., and Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., and Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nemoto S., Fergusson M. M., and Finkel T. (2005) SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 42. Rodgers J. T., Lerin C., Gerhart-Hines Z., and Puigserver P. (2008) Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wareski P., Vaarmann A., Choubey V., Safiulina D., Liiv J., Kuum M., and Kaasik A. (2009) PGC-1α and PGC-1β regulate mitochondrial density in neurons. J. Biol. Chem. 284, 21379–21385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garcia M. M., Guéant-Rodriguez R. M., Pooya S., Brachet P., Alberto J. M., Jeannesson E., Maskali F., Gueguen N., Marie P. Y., Lacolley P., Herrmann M., Juillière Y., Malthiery Y., and Guéant J. L. (2011) Methyl donor deficiency induces cardiomyopathy through altered methylation/acetylation of PGC-1α by PRMT1 and SIRT1. J. Pathol. 225, 324–335 [DOI] [PubMed] [Google Scholar]
- 45. Jian B., Yang S., Chaudry I. H., and Raju R. (2012) Resveratrol improves cardiac contractility following trauma-hemorrhage by modulating Sirt1. Mol. Med. 18, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pallàs M., Casadesús G., Smith M. A., Coto-Montes A., Pelegri C., Vilaplana J., and Camins A. (2009) Resveratrol and neurodegenerative diseases: activation of SIRT1 as the potential pathway towards neuroprotection. Curr. Neurovasc. Res. 6, 70–81 [DOI] [PubMed] [Google Scholar]
- 47. Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., and Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 48. Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., and Clayton D. A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18, 231–236 [DOI] [PubMed] [Google Scholar]
- 49. St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., and Spiegelman B. M. (2006) Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 50. Aquilano K., Baldelli S., Pagliei B., and Ciriolo M. R. (2013) Extranuclear localization of SIRT1 and PGC-1α: an insight into possible roles in diseases associated with mitochondrial dysfunction. Curr. Mol. Med. 13, 140–154 [PubMed] [Google Scholar]
- 51. Akimoto T., Pohnert S. C., Li P., Zhang M., Gumbs C., Rosenberg P. B., Williams R. S., and Yan Z. (2005) Exercise stimulates Pgc-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 280, 19587–19593 [DOI] [PubMed] [Google Scholar]
- 52. Baldelli S., Aquilano K., and Ciriolo M. R. (2014) PGC-1α buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 5, e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oku T., Iyama S., Sato T., Sato Y., Tanaka M., Sagawa T., Kuribayashi K., Sumiyosi T., Murase K., Machida T., Okamoto T., Matsunaga T., Takayama T., Takahashi M., Kato J., et al. (2006) Amelioration of murine dextran sulfate sodium-induced colitis by ex vivo extracellular superoxide dismutase gene transfer. Inflamm. Bowel Dis. 12, 630–640 [DOI] [PubMed] [Google Scholar]
- 54. Bär F., Bochmann W., Widok A., von Medem K., Pagel R., Hirose M., Yu X., Kalies K., König P., Böhm R., Herdegen T., Reinicke A. T., Büning J., Lehnert H., Fellermann K., Ibrahim S., and Sina C. (2013) Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterology 145, 1055–1063 e1053 [DOI] [PubMed] [Google Scholar]
- 55. Roediger W. E. (1980) The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet 2, 712–715 [DOI] [PubMed] [Google Scholar]
- 56. Kameyama J., Narui H., Inui M., and Sato T. (1984) Energy level in large intestinal mucosa in patients with ulcerative colitis. Tohoku J. Exp. Med. 143, 253–254 [DOI] [PubMed] [Google Scholar]
- 57. Glover L. E., Bowers B. E., Saeedi B., Ehrentraut S. F., Campbell E. L., Bayless A. J., Dobrinskikh E., Kendrick A. A., Kelly C. J., Burgess A., Miller L., Kominsky D. J., Jedlicka P., and Colgan S. P. (2013) Control of creatine metabolism by HIF is an endogenous mechanism of barrier regulation in colitis. Proc. Natl. Acad. Sci. U.S.A. 110, 19820–19825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rodenburg W., Keijer J., Kramer E., Vink C., van der Meer R., and Bovee-Oudenhoven I. M. (2008) Impaired barrier function by dietary fructo-oligosaccharides (FOS) in rats is accompanied by increased colonic mitochondrial gene expression. BMC Genomics 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma C., Wickham M. E., Guttman J. A., Deng W., Walker J., Madsen K. L., Jacobson K., Vogl W. A., Finlay B. B., and Vallance B. A. (2006) Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: role of mitochondrial associated protein (Map). Cell Microbiol. 8, 1669–1686 [DOI] [PubMed] [Google Scholar]
- 60. Lewis K., and McKay D. M. (2009) Metabolic stress evokes decreases in epithelial barrier function. Ann. N.Y. Acad. Sci. 1165, 327–337 [DOI] [PubMed] [Google Scholar]
- 61. Schürmann G., Brüwer M., Klotz A., Schmid K. W., Senninger N., and Zimmer K. P. (1999) Transepithelial transport processes at the intestinal mucosa in inflammatory bowel disease. Int. J. Colorectal Dis. 14, 41–46 [DOI] [PubMed] [Google Scholar]
- 62. Novak E. A., and Mollen K. P. (2015) Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell Dev. Biol. 3, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zheng B., and Cantley L. C. (2007) Regulation of epithelial tight junction assembly and disassembly by AMP-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 104, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heller F., Florian P., Bojarski C., Richter J., Christ M., Hillenbrand B., Mankertz J., Gitter A. H., Bürgel N., Fromm M., Zeitz M., Fuss I., Strober W., and Schulzke J. D. (2005) Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 129, 550–564 [DOI] [PubMed] [Google Scholar]
- 65. Zeissig S., Bürgel N., Günzel D., Richter J., Mankertz J., Wahnschaffe U., Kroesen A. J., Zeitz M., Fromm M., and Schulzke J. D. (2007) Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut 56, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen Y., Zhang H. S., Fong G. H., Xi Q. L., Wu G. H., Bai C. G., Ling Z. Q., Fan L., Xu Y. M., Qin Y. Q., Yuan T. L., Sun H., and Fang J. (2015) PHD3 stabilizes the tight junction protein occludin and protects intestinal epithelial barrier function. J. Biol. Chem. 290, 20580–20589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mehlem A., Palombo I., Wang X., Hagberg C. E., Eriksson U., and Falkevall A. (2016) PGC-1α coordinates mitochondrial respiratory capacity and muscular fatty acid uptake via regulation of VEGF-B. Diabetes 2016, db151231. [DOI] [PubMed] [Google Scholar]
- 68. Leick L., Fentz J., Biensø R. S., Knudsen J. G., Jeppesen J., Kiens B., Wojtaszewski J. F., and Pilegaard H. (2010) PGC-1α is required for AICAR-induced expression of GLUT4 and mitochondrial proteins in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 299, E456–E465 [DOI] [PubMed] [Google Scholar]
- 69. LeBleu V. S., O'Connell J. T., Gonzalez Herrera K. N., Wikman H., Pantel K., Haigis M. C., de Carvalho F. M., Damascena A., Domingos Chinen L. T., Rocha R. M., Asara J. M., and Kalluri R. (2014) PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang X., Jiang W., Yan Y., Gong T., Han J., Tian Z., and Zhou R. (2014) RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 15, 1126–1133 [DOI] [PubMed] [Google Scholar]
- 71. Heid M. E., Keyel P. A., Kamga C., Shiva S., Watkins S. C., and Salter R. D. (2013) Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J. Immunol. 191, 5230–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu D., Xu M., Ding L. H., Lv L. L., Liu H., Ma K. L., Zhang A. H., Crowley S. D., and Liu B. C. (2014) Activation of the Nlrp3 inflammasome by mitochondrial reactive oxygen species: A novel mechanism of albumin-induced tubulointerstitial inflammation. Int. J. Biochem. Cell Biol. 57, 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., and Hauser C. J. (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]