Abstract
Cse4, a histone H3-like centromeric protein, plays critical functions in chromosome segregation. Cse4 level is tightly regulated, but the underlying mechanism remains poorly understood. We employed a toxicity-based screen to look for the degradation components involved in Cse4 regulation. Here, we show that the F-box containing protein Rcy1 is required for efficient Cse4 turnover as Cse4 degradation is compromised in yeast cells lacking RCY1. Excessive Cse4 accumulation in rcy1Δ cells leads to growth retardation. Furthermore, the deletion of RCY1 is tied to enhanced chromosome instability and temperature-sensitive cell growth. Our results reveal the involvement of Rcy1 in chromosome regulation and another regulatory pathway controlling the Cse4 level and activity.
Keywords: centromere, proteasome, protein degradation, ubiquitin, ubiquitin ligase
Introduction
Chromosome segregation is key to equally divide the duplicated DNA to daughter cells. Following DNA replication, sister chromatids are joined at a highly condensed and constricted chromosome region called centromere (1, 2). A distinct feature of the specialized centromeric chromatin is the incorporation of an essential histone H3 variant CENP-A (called Cse4 in yeast Saccharomyces cerevisiae), instead of histone H3 in nucleosomes (1, 3, 4).
As altered localization and expression of CENP-A and its homologues have been linked to genome instability and cancers (1, 5, 6), CENP-A activity must be tightly regulated. Human CENP-A and yeast Cse4 have been found to be subject to proteasome-mediated degradation (1, 7, 8). Proteasome substrates are selected by a ubiquitin ligase E3, which collaborates with a ubiquitin-activating enzyme (E1) and a ubiquitin-conjugating enzyme (E2) to covalently attach ubiquitin molecules onto substrates (9, 10). Ubiquitin-marked substrates are then delivered to and degraded by the proteasome. The key to understanding the role of proteolysis in CENP-A regulation is to identify E3 enzymes involved.
Two groups previously isolated Psh1 as a yeast Cse4-binding protein via the affinity purification-mass spectrometry coupled approach and further demonstrated that Psh1 acts as an E3 ligase for Cse4 ubiquitylation and degradation (11, 12). The mechanism underlying Psh1-mediated Cse4 turnover remains to be elucidated. Because Cse4 degradation is partially impaired in psh1Δ cells, other E3s likely also promote Cse4 turnover (11–15).
We have previously developed a synthetic dosage lethality screen to uncover physiological targets of several proteolytic pathways (16–18). The synthetic lethality screen can be easily adapted to isolate relevant components involved in the degradation of a given substrate. Because Cse4 overexpression leads to toxicity in psh1Δ cells (11, 12), we looked for the degradation mutants that grow slower upon Cse4 overexpression. Yeast cells lacking RCY1 are found to be sensitive to Cse4 expression and exhibit compromised Cse4 degradation. RCY1 encodes a F-box containing protein, which is a substrate recognition component of the Skp1-Cdc53-F box (SCF)2 E3 complex (19). Rcy1 is one of the eight F-box proteins in yeast, but little is known about its degradation targets (19–21). Consistent with the involvement of Rcy1 in centromere regulation, rcy1Δ mutants display increased genome instability. Interestingly, Rcy1 and Psh1 appear to act in parallel pathways for Cse4 degradation. Our findings will help to unravel the intricate mechanisms that regulate Cse4/CENP-A level and activity.
Experimental Procedures
Yeast Strains and Plasmids
A collection of yeast S. cerevisiae strains lacking non-essential ubiquitylation or degradation components including various E2s and E3s in BY4741 background were obtained from Dr. Mark Hochstrasser (Yale University) (22). Haploid yeast mutant strains lacking RCY1, TOM1, PSH1, ATG1, ATG9, ATG14, YPT31, YPT32, and SNC1 in the BY4741 strain background were obtained from Open Biosystems (Huntsville, AL). GFP- or GST-tagged Cdc53 strain was obtained from Open Biosystems and ThermoFisher Scientific Inc., respectively. Strain YHR305 (psh1::LEU2 rcy1::KanMX4) was constructed by replacing PSH1 with LEU2 in the rcy1Δ strain. Sectoring strain YPH278 was obtained from Dr. P. Hieter and was used to delete the RCY1 gene by LEU2 to make strain YHR313. Endogenous RCY1 gene was tagged 9 Myc epitopes at its C-terminal using PCR-mediated homologous recombination to construct strain YHR297 in BY4741 background.
The plasmid pMB1458 bearing 3HA-tagged Cse4 and pYES2-HTH-HHT2 expressing HA-tagged histone H3 were previously described (12, 14, 23). The plasmid for V5-tagged Cse4 expression in Escherichia coli was kindly provided by Dr. Biggins. RCY1 was amplified by PCR to append the FLAG epitope at its C terminus and inserted into pRS425GAL1 to create the plasmid pRS425GAL1-Rcy1-Flag for FLAG-tagged Rcy1 expression in yeast. Yeast cells were grown in rich (YPD) or synthetic media containing standard ingredients and 2% glucose (SD medium), or 2% raffinose (SR medium), or 2% raffinose + 2% galactose (SRG medium).
Synthetic Dosage Lethal Screen
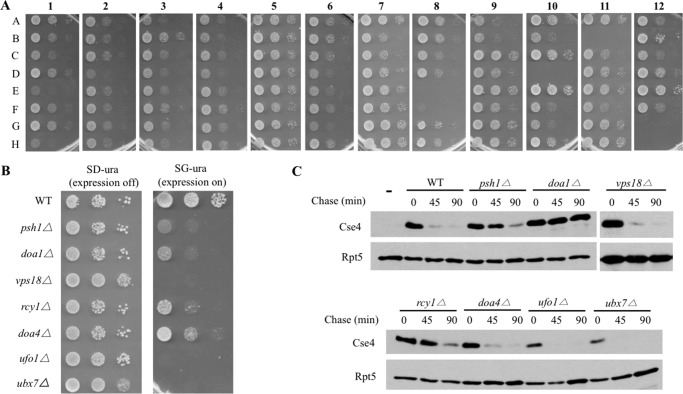
The plasmid pMB1458 was separately transformed into wild-type or 93 yeast mutants (see supplemental Table S1). Yeast cells bearing the plasmid were first grown in synthetic media containing glucose. Actively growing cells were spotted onto glucose-containing SD-ura and galactose-containing SG-ura media in serial 10-fold dilution. Cse4 expression is controlled by a GAL1 promoter and induced by galactose. The plates were incubated at 30 °C for 3 days. Thirteen mutants that exhibited reduced growth on SG-ura plates were deemed possible candidates from the initial screen and further analyzed (18).
Antibodies
Antibodies against HA and myc were obtained from BioLegend Inc. (San Diego, CA). Antibodies for V5 and FLAG were purchased from ThermoFisher Scientific and Sigma, respectively. Rpt5 antibody was obtained from Enzo Life Sciences (Farmingdale, NY).
Expression Shut-off Assay
As previously described (17), yeast cells expressing HA-tagged Cse4 were grown at 30 °C to an A600 of ∼1 in synthetic medium SR-uracil. The expression of HA-Cse4 was induced by galactose for 2 h (17). Protein translation was shut off by the addition of cycloheximide (150 μg/ml). Samples were collected at the indicated time points. Proteins were extracted by glass bead lysis of cells, processed for immunoblotting with anti-HA (BioLegend, MMS101P), followed by detection with goat anti-mouse HRP conjugate using ECL reagents (GE Healthcare). The stable protein Rpt5 was used as a loading control in the expression shutoff experiments.
Sectoring Assay
Yeast YPH278 cells contained the defective ade2–101 ochre mutation, which can be suppressed by the SUP11 gene, carried on a non-essential, 90-kb chromosome fragment. YPH278 cells that maintained this chromosomal fragment produced white colonies, whereas the loss of it resulted in a red color or sectors (17). The RCY1 gene was deleted in the YPH278 strain, and the resulting mutant YHR313 was compared with YPH278. As previously described (17), cells were first grown overnight in uracil-lacking medium that retained the SUP11-containing chromosome, followed by growing in non-selective rich YPD medium for 8 h and plating on YPD plates. The plates were incubated at 30 °C for 48 h and then kept at room temperature for ∼5 days for color development.
Results
The involvement of the proteasome in Cse4 turnover was previously demonstrated using proteasome mutant cim3-1 (7, 9), which has a number of defects that may potentially contribute to Cse4 stabilization indirectly. We assessed the effects of the proteasome inhibitor MG132 on Cse4 degradation. Cse4 turnover is impaired upon MG132 treatment (Fig. 1A), supporting the involvement of the proteasome in Cse4 regulation (7). Besides the proteasome, autophagy is the other major proteolytic system in eukaryotes (24). We evaluated whether autophagy is involved in Cse4 turnover. Whereas Cdc13 was degraded by autophagy as previously reported (Fig. 1C), Cse4 stability was not altered significantly in yeast cells lacking one of the core autophagy components (i.e. ATG1, ATG9, ATG14) (Fig. 1B), suggesting that autophagy is not involved in Cse4 degradation.
FIGURE 1.
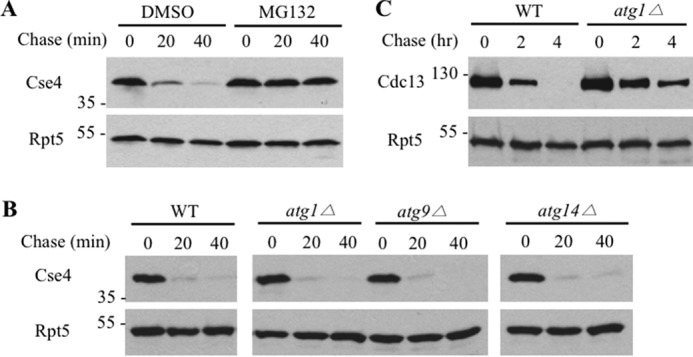
The involvement of the proteasome and autophagy in the degradation of Cse4. A, Cse4 turnover is compromised upon MG132 treatment. Wild-type yeast cells expressing HA-Cse4 were grown to an A600 of ∼1 and then added with the proteasome inhibitor MG132 (75 μm) or dimethyl sulfoxide (DMSO) control for 30 min. Samples were taken after expression shut-off by cycloheximide at the time points indicated and analyzed by Western blots using anti-HA antibody (BioLegend, MMS-101P). Equal amounts of samples were used and ascertained by blotting with Rpt5 antibody (Enzo Life Sciences, BML-PW8245) in all of the expression shutoff experiments (lower panels). Molecular mass standards (kDa) are indicated on the left. B, Cse4 is degraded efficiently in autophagy mutants. Cse4 turnover in wild-type and 3 mutant cells was determined as above. C, Cdc13 degradation is compromised in the atg1Δ mutant. GST-Cdc13 turnover was determined as previously described.
As Cse4 is still degraded in yeast cells lacking Psh1 E3 (11–15), other ubiquitin ligases likely are involved in Cse4 turnover. To this end, we employed an array of yeast mutant cells lacking known or putative ubiquitylation/degradation enzymes (22) (supplemental Table S1). Reasoning that Cse4 accumulation may lead to toxicity, we adopted a synthetic lethal screen to look for yeast mutants that show growth retardation upon Cse4 expression. The plasmid bearing the GAL1-promoter regulated CSE4 was introduced into wild-type and 93 yeast degradation mutants separately (Fig. 2A). Following the initial screen, seven mutants including psh1Δ were confirmed to show Cse4-induced slower growth by the serial spotting assay (Fig. 2B). It is worth noting that PSH1 and DOA1, both known to promote Cse4 destruction (11, 12, 14), were recovered in the screen, validating our screening approach. Doa1, also called Ufd3, is involved in ubiquitin homeostasis albeit the underlying mechanism remains enigmatic (14, 25–28).
FIGURE 2.
Identification of Rcy1 as a regulator of Cse4 turnover. A, isolation of yeast mutants sensitive to Cse4 overexpression. The plasmid bearing the GAL1-promoter-regulated Cse4 was transformed to wild-type and 93 mutants deficient in various ubiquitin pathways. Transformants were grown to the same density and spotted onto galactose-containing SG plates in 10-fold serial dilution. The labels (e.g. A–H, 1–12) indicate the corresponding positions of yeast mutants in the original 96-well plate provided by Dr. M. Hochstrasser. B, a spotting assay indicates Cse4 overexpression causes growth retardation in 7 mutants. The spotting assay was done as described above except yeast cells were spotted onto both glucose (SD, expression off) and galactose (SG, expression on) plates. The identities of these mutants are listed to the left of the panels. C, efficient Cse4 degradation requires Psh1, Rcy1, and Doa1. Cse4 turnover was determined as described in the legend to Fig. 1.
Next, we compared the degradation kinetics of Cse4 in wild-type and these mutants by an expression shut-off assay. Consistent with previous studies, Cse4 degradation is compromised in yeast cells missing PSH1 or DOA1 (11, 12, 14, 15). Four isolated mutants (i.e. vps18Δ, doa4Δ, ufo1Δ, ubx7Δ) did not exhibit Cse4 stabilization (Fig. 2C), indicating that they do not directly promote Cse4 turnover. The mechanisms underlying the Cse4-triggered toxicity in these mutants are unknown, but these genes may possibly be involved in a Cse4-related event indirectly.
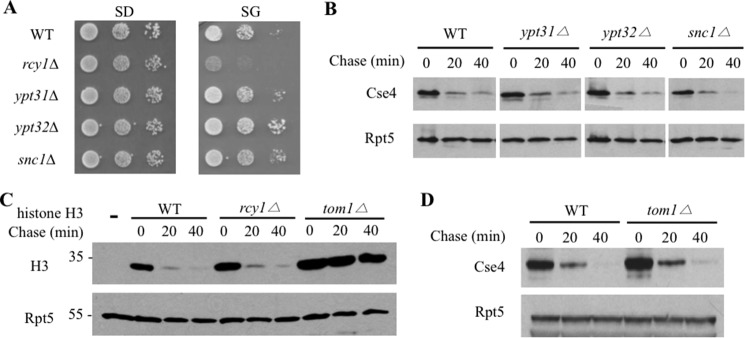
Interestingly, Cse4 degradation is impaired in the rcy1Δ mutant (Fig. 2C). Rcy1 contains a F-box domain and can be assembled into a Skp1-Cdc53-Rcy1 complex with demonstrated ubiquitin ligase activity (19). RCY1 was originally identified in a screen for genes involved in a endocytic process and was shown to regulate protein trafficking from endosomes to the Golgi network (20, 21, 29, 30). Although yeast Rcy1 was shown to regulate Snc1 ubiquitylation (21) and the Aspergilus nidulans Rcy1 homologue promotes kinesin KipA degradation (31), the physiologic function of Rcy1 in the ubiquitin-proteasome pathway remains elusive. It is possible that the Rcy1-mediated protein trafficking may indirectly affect Cse4 turnover. To determine the specificity of Rcy1 in Cse4 regulation, we evaluated Cse4 degradation in yeast cells lacking Ypt31 and Ypt32, two GTPases that work with Rcy1 in the endosome-Golgi transport (21, 30, 32). Interestingly, Cse4 did not elicit toxicity and was not stabilized in ypt31Δ or ypt32Δ cells (Fig. 3, A and B). Moreover, Cse4 was unaffected in cells missing the SNARE Snc1 (Fig. 3, A and B), which interacts with Rcy1 and regulates protein trafficking (21). Combined, the data suggest that the role of Rcy1 in Cse4 regulation is likely separate from its involvement in protein trafficking outside of nucleus. Histone H3 was previously shown to be ubiquitylated and degraded by the ubiquitin ligase Tom1 pathway (23). We found that histone H3 degradation was compromised in tom1Δ (Fig. 3C) as expected (23), but was little affected in rcy1Δ cells (Fig. 3C), suggesting the specific involvement of Rcy1 in Cse4 turnover. Although Tom1 regulates histone H3, Tom1 was not involved in Cse4 turnover (Fig. 3D).
FIGURE 3.
Specific involvement of Rcy1 in Cse4 regulation. A, Cse4 expression is not toxic in other protein trafficking mutants. The spotting assay was done as described in the legend to Fig. 2B. B, Cse4 is efficiently degraded in yeast cells deficient in protein transport. Cse4 stability in ypt31Δ, ypt32Δ, and snc1Δ was determined as described in the legend to Fig. 2C. C, histone H3 turnover is not affected in rcy1Δ cells. The empty vector (lane 1) or the plasmid expressing HA-tagged histone H3 was transformed into the indicated yeast cells. As the ubiquitin ligase Tom1 is known to be involved in histone H3 turnover, tom1Δ cell serves as a positive control. D, Tom1 is not involved in Cse4 degradation. Cse4 turnover in wild-type or tom1Δ was determined as described in the legend to Fig. 2C.
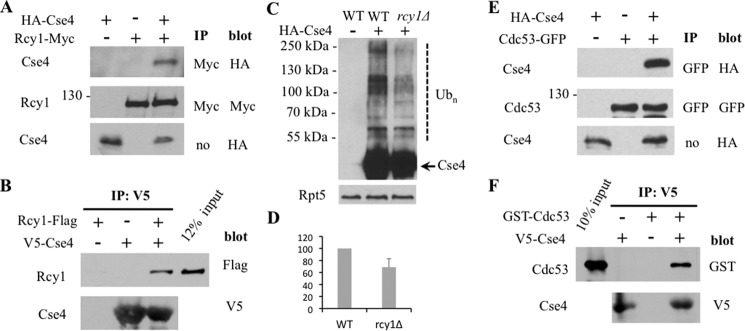
As the F-box protein is the substrate-recognition component of SCF E3s (9), we examined whether Cse4 associates with Rcy1 in yeast by the co-immunoprecipitation experiment (Fig. 4A). We found that endogenous Rcy1 immunoprecipitates Cse4 (Fig. 4A), supporting a close link between Rcy1 and Cse4 in vivo. Furthermore, we also carried out the in vitro binding experiment. Cse4 purified from E. coli can interact with Rcy1 (Fig. 4B), suggesting that Rcy1 recognizes Cse4. Consistent with the role of Rcy1 in Cse4 ubiquitylation, Cse4 ubiquitylation was reduced in rcy1Δ cells (Fig. 4, C and D). We also evaluated whether other SCF components are involved. We found that Cse4 interacts with Cdc53, a core component of SCF E3, in vivo and in vitro (Fig. 4, E and F).
FIGURE 4.
Cse4 interacts with Rcy1 and Cdc53. A, Cse4 binds endogenous Rcy1 in vivo. The plasmid bearing HA-Cse4 or a control vector was transformed into yeast cells expressing Rcy1-tagged with the Myc epitope at its chromosomal locus. Proteins were extracted from the cells indicated and incubated with the protein G beads coated with Myc antibody (BioLegend, MMS150P). Samples were resolved by SDS-PAGE and visualized by Western blotting using Myc or HA antibody as indicated. Cse4 levels in the extract were also determined. The identity of the bands is shown on the left of the panels. The antibodies for immunoprecipitation (IP) and Western blotting are indicated to the right. B, Cse4 interacts with Rcy1 in vitro. The V5 antibody (ThermoFisher Scientific, R960-25)-coated beads were mixed with V5-tagged Cse4 expressed in E. coli and yeast extracts with or without FLAG-tagged Rcy1 at 4 °C. The bound proteins were resolved by SDS-PAGE and immunoblotted with antibody to FLAG (upper panel; Sigma, F1804) or V5 (lower panel). Lane 4 contains ∼12% input extract expressing FLAG-Rcy1. C, reduced Cse4 ubiquitylation in rcy1Δ cells. The experiment was carried out in a similar previously described procedure (12, 37). Briefly, wild-type or rcy1Δ cells bearing a vector plasmid or a plasmid expressing Cse4 tagged with both HA and His6 epitopes were grown in galactose media. Cse4 was enriched by incubating extracts with nickel-nitrilotriacetic acid-agarose. Cse4 and its ubiquitin conjugates were detected by immunoblotting with anti-HA. Ubiquitylated and unmodified Cse4 are indicated by a dashed line and an arrow, respectively. Rpt5 was employed as a loading control. D, quantitation of the data in C. Values are derived from three independent experiments and SD is shown. p value is 0.062. E, Cse4 associates with Cdc53 in vivo. The experiment was carried out as described in the legend to Fig. 4A except the yeast strain bearing Cdc53 tagged with GFP at its chromosomal locus was used. F, Cse4 binds Cdc53 in vitro. Similar to the experiment described in Fig. 4B, Cse4 purified from E. coli was mixed with yeast extracts containing GST-tagged Cdc53. Lane 1 contains 10% input extract expressing GST-Cdc53.
Because both Psh1 and Rcy1 promote Cse4 destruction, we wondered whether they work the same or different pathways. To determine the functional relationship between PSH1 and RCY1, we assessed the degradation of Cse4 in cells lacking either or both RCY1 and PSH1. Consistent with earlier studies, deletion of PSH1 slightly stabilized Cse4 (11, 12, 14, 15). RCY1 deletion appeared to cause stronger Cse4 stabilization (Fig. 5, A and B). Interestingly, the double mutant psh1Δ rcy1Δ showed more compromised Cse4 degradation than single mutants, suggesting that Psh1 and Rcy1 likely work in parallel pathways in Cse4 regulation.
FIGURE 5.
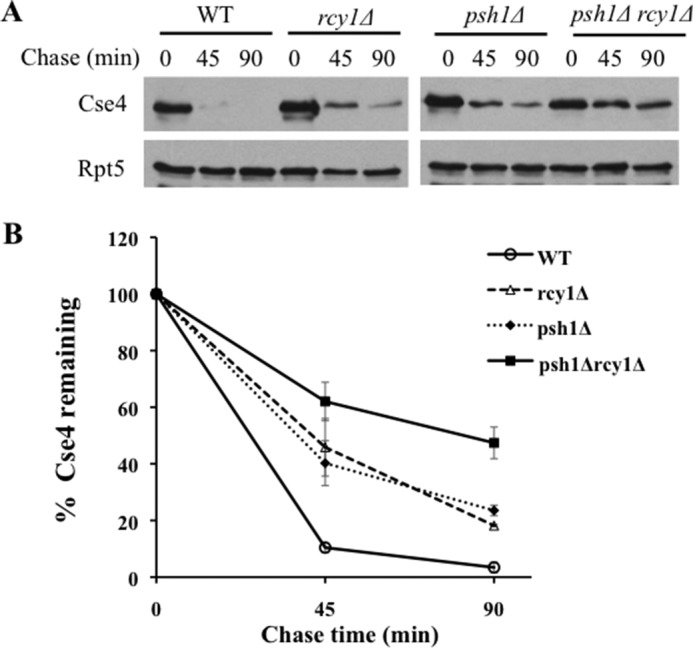
Stronger stabilization of Cse4 in rcy1Δ psh1Δ double mutant cells. A, Cse4 degradation in wild-type, rcy1Δ, psh1Δ, rcy1Δ psh1Δ cells was determined as described above. B, quantitation of the data in A. The experiments were done more than 3 times, and the average values with S.D. are shown.
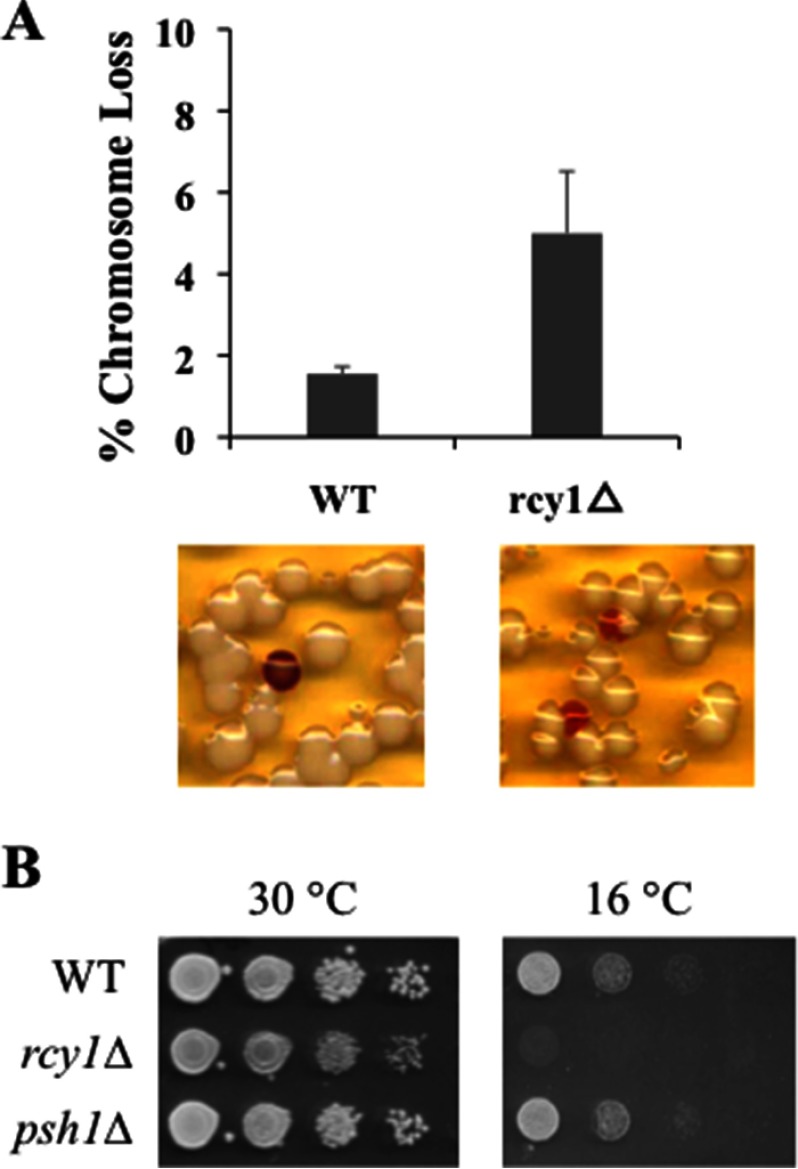
As Cse4 plays a critical role in chromosome segregation, altered Cse4 regulation may affect genome stability (5). To determine whether Rcy1 may regulate genome maintenance, we employed a colony sectoring assay to compare the rate of chromosome loss in wild-type and rcy1Δ cells (Fig. 6A) (17). These cells contain a non-essential chromosome fragment bearing the SUP11 gene, which can suppress the ade2–101 mutation-induced red pigment accumulation. As expected, wild-type cells can efficiently maintain the non-essential SUP11-bearing chromosome, leading to few cells (less than 2%) with red sectors. In contrast, RCY1 deletion leads to a significant increase of red sectors in rcy1Δ cells (∼5%) (Fig. 6A), suggesting enhanced chromosome instability in rcy1Δ cells, a phenotype that is consistent with the RCY1-CSE4 link.
FIGURE 6.
Phenotypes associated with rcy1Δ mutant. A, increased chromosome loss in rcy1Δ mutant cells. Sectoring assay was done as described (17). Actively growing wild-type or rcy1Δ mutant cells were plated onto YPD plates and incubated for 48 h at 30 °C. Quantification of three independent experiments is shown in the upper panel (p value 0.023), and representative images from the sectoring assay are shown in lower panels. B, rcy1Δ mutant is cold-sensitive. Mid-log phase yeast cultures were spotted onto the plates in serial 10-fold dilution and incubated at either normal temperature, 30 °C, or 16 °C as indicated. The genotypes of the strains are shown on the left of the panels.
We noticed that rcy1Δ cells grow slower than wild-type cells at room temperature. Indeed, upon closer examination, we found that rcy1Δ cells are sensitive to cold temperature (Fig. 6B). In line with temperature sensitivity, a drastic increase of chromosome loss was observed in rcy1Δ cells (∼40%) that were grown at room temperature (data not shown).
Discussion
Here we identify another degradation component, Rcy1, involved in Cse4 regulation. Rcy1 partnered with Skp1 and Cdc53 exhibits the ubiquitin E3 ligase activity (19). Rcy1 was previously known to promote intracellular trafficking from the endosome to the Golgi (20, 21, 29, 30), an endocytic recycling pathway important for the localization of several proteins including a GTPase-activating protein Bem3, a Golgi resident Kex2 protease, and a SNARE protein Snc1 (21, 30, 33). Without RCY1, Snc1 ubiquitylation was impaired, leading to its entrapment on endosomes (21, 30). Nevertheless, the physiological functions of RCY1 remain far from clear. We found that Rcy1 is specifically involved in the degradation of Cse4, but not histone H3. Our study expands the function of Rcy1 in proteolysis, distinct from its involvement in protein trafficking, and reveals new phenotypes associated with the RCY1 deletion (Fig. 3).
Cse4/CENP-A is crucial to establish a specialized chromatin structure at the centromere (1, 2), the site of kinetochore assembly, which is key to chromosome segregation that allows proper distribution of duplicated genetic information to daughter cells. Excessive or nonrestraint Cse4 can lead to its occupancy at the euchromatic region, which may cause aneuploidy and promote tumorigenesis (1, 5, 6, 11, 12). The discovery of enhanced chromosome instability in rcy1Δ cells suggest that altered regulation of the Rcy1-Cse4 pathway may be involved in cancer.
Doa1, but not Rcy1, was isolated in a similar synthetic dosage lethality screen employed to identify potential regulators of Cse4 (5). Whereas the early screen scanned the entire set of yeast deletion mutants available (>4000) (5), we probed a selected group of mutants linked to ubiquitin-mediated proteolysis. Our targeted approach allowed a more careful examination of mutant phenotypes. As Doa1 regulates intracellular levels of ubiquitin in general, Doa1 affects ubiquitin-mediated processes much more broadly than Rcy1 (14, 25–27). Thus, it is not surprising that Cse4 turnover is more severely compromised in doa1Δ than in rcy1Δ.
Our results suggest that Psh1 and Rcy1 likely work in parallel pathways to tune down the Cse4 level (Fig. 5, A and B). Because Cse4 remains partially degraded in psh1Δ rcy1Δ double mutant cells, other E3s involved in Cse4 degradation exist. It is not unusual for a protein targeted for degradation by multiple E3s. For example, histone H3 degradation has been shown to involve five E3s including Tom1, Snt2, Pep5, Hel1, and Hel2 (34). A more “extreme” case is the p53 tumor suppressor, for which more than 15 E3 enzymes have been implicated in its turnover under various conditions (35). The employment of multiple E3s in substrate degradation allows more regulation. Identification of additional E3s involved in the degradation of Cse4, especially its human homolog CENP-A, remains a future challenge.
Besides ubiquitin-mediated proteolysis, a variety of approaches are adopted to restrain Cse4 activity with the participations of casein kinase 2, the proline isomerase Fpr3, the H3 chaperones HIR and CAF-1, and several chromatin modulators (e.g. SWI/SNF remodeling enzymes, the FACT complex) (15, 36–39). Unraveling the functional cross-talks among these factors/pathways will be key to revealing the molecular basis underlying Cse4 regulation. Given the intimate tie between CENP-A and cancer, the insights gained on Cse4 may shed light into elusive principles governing human CENP-A regulation and potential strategies against CENP-A-triggered diseases.
Author Contributions
H. C. designed and carried out most of the experiments described. X. B. performed the experiments in Fig. 4. H. R. designed the experiments. All three authors were involved in data analysis and manuscript preparation.
Supplementary Material
Acknowledgments
We are grateful to Drs. M. Hochstrasser, J. Gerton, S. Biggins, M. A. Basrai, A. Gunjan, C. Wu, and S. Lee for strains and plasmids. We thank Dr. M. Strand for support and members of the Rao laboratory for discussions.
This work was supported by Welch Foundation Grant AQ 1747, the William & Ella Owens Medical Research Foundation, the Helen F. Kerr Foundation, Department of Defense Grant W911NF-11-10466, and National Institutes of Health Grant GM 078085 (to H. R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that there is no conflict of interest for this study.

This article contains supplemental Table S1.
- SCF
- Skp1-Cdc53-F box.
References
- 1. Earnshaw W. C. (2015) Discovering centromere proteins: from cold white hands to the A, B, C of CENPs. Nat. Rev. Mol. Cell Biol. 16, 443–449 [DOI] [PubMed] [Google Scholar]
- 2. Westhorpe F. G., and Straight A. F. (2015) The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7, a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Earnshaw W. C., and Rothfield N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 [DOI] [PubMed] [Google Scholar]
- 4. Meluh P. B., Yang P., Glowczewski L., Koshland D., and Smith M. M. (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613 [DOI] [PubMed] [Google Scholar]
- 5. Au W. C., Crisp M. J., DeLuca S. Z., Rando O. J., and Basrai M. A. (2008) Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amato A., Schillaci T., Lentini L., and Di Leonardo A. (2009) CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 8, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collins K. A., Furuyama S., and Biggins S. (2004) Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14, 1968–1972 [DOI] [PubMed] [Google Scholar]
- 8. Lomonte P., Sullivan K. F., and Everett R. D. (2001) Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276, 5829–5835 [DOI] [PubMed] [Google Scholar]
- 9. Finley D., Ulrich H. D., Sommer T., and Kaiser P. (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 11. Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., Florens L., Workman J. L., and Gerton J. L. (2010) Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40, 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., and Biggins S. (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hewawasam G. S., and Gerton J. L. (2011) Cse4 gets a kiss-of-death from Psh1. Cell Cycle 10, 566–567 [DOI] [PubMed] [Google Scholar]
- 14. Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., and Basrai M. A. (2013) A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hewawasam G. S., Mattingly M., Venkatesh S., Zhang Y., Florens L., Workman J. L., and Gerton J. L. (2014) Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J. Biol. Chem. 289, 29297–29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu C., Choe V., and Rao H. (2010) Genome-wide approaches to systematically identify substrates of the ubiquitin-proteasome pathway. Trends Biotechnol. 28, 461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C., van Dyk D., Choe V., Yan J., Majumder S., Costanzo M., Bao X., Boone C., Huo K., Winey M., Fisk H., Andrews B., and Rao H. (2011) Ubiquitin ligase Ufd2 is required for efficient degradation of Mps1 kinase. J. Biol. Chem. 286, 43660–43667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C., van Dyk D., Li Y., Andrews B., and Rao H. (2009) A genome-wide synthetic dosage lethality screen reveals multiple pathways that require the functioning of ubiquitin-binding proteins Rad23 and Dsk2. BMC Biol. 7, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kus B. M., Caldon C. E., Andorn-Broza R., and Edwards A. M. (2004) Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54, 455–467 [DOI] [PubMed] [Google Scholar]
- 20. Wiederkehr A., Avaro S., Prescianotto-Baschong C., Haguenauer-Tsapis R., and Riezman H. (2000) The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149, 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S. H., Shah A. H., and Segev N. (2011) Ypt31/32 GTPases and their F-Box effector Rcy1 regulate ubiquitination of recycling proteins. Cell Logist 1, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ravid T., and Hochstrasser M. (2007) Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat. Cell. Biol. 9, 422–427 [DOI] [PubMed] [Google Scholar]
- 23. Singh R. K., Kabbaj M. H., Paik J., and Gunjan A. (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell. Biol. 11, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Athané A., Buisson A., Challier M., Beaumatin F., Manon S., Bhatia-Kiššova I., and Camougrand N. (2015) Insights into the relationship between the proteasome and autophagy in human and yeast cells. Int. J. Biochem. Cell Biol. 64, 167–173 [DOI] [PubMed] [Google Scholar]
- 25. Ghislain M., Dohmen R. J., Levy F., and Varshavsky A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15, 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 26. Mullally J. E., Chernova T., and Wilkinson K. D. (2006) Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain. Mol. Cell. Biol. 26, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russell N. S., and Wilkinson K. D. (2004) Identification of a novel 29-linked polyubiquitin binding protein, Ufd3, using polyubiquitin chain analogues. Biochemistry 43, 4844–4854 [DOI] [PubMed] [Google Scholar]
- 28. Böhm S., Lamberti G., Fernández-Sáiz V., Stapf C., and Buchberger A. (2011) Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol. Cell. Biol. 31, 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lafourcade C., Galan J. M., and Peter M. (2003) Opposite roles of the F-box protein Rcy1p and the GTPase-activating protein Gyp2p during recycling of internalized proteins in yeast. Genetics 164, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen S. H., Chen S., Tokarev A. A., Liu F., Jedd G., and Segev N. (2005) Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol. Biol. Cell 16, 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herrero S., Takeshita N., and Fischer R. (2014) F-box protein RcyA controls turnover of the kinesin-7 motor KipA in Aspergillus nidulans. Eukaryot. Cell 13, 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., and Tanaka K. (2007) Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 18, 295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mukherjee D., Sen A., Boettner D. R., Fairn G. D., Schlam D., Bonilla Valentin F. J., Michael McCaffery J., Hazbun T., Staiger C. J., Grinstein S., Lemmon S. K., and Claudio Aguilar R. (2013) Bem3, a Cdc42 GTPase-activating protein, traffics to an intracellular compartment and recruits the secretory Rab GTPase Sec4 to endomembranes. J. Cell Sci. 126, 4560–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh R. K., Gonzalez M., Kabbaj M. H., and Gunjan A. (2012) Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS ONE 7, e36295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love I. M., and Grossman S. R. (2012) It takes 15 to tango: making sense of the many ubiquitin ligases of p53. Genes Cancer 3, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gkikopoulos T., Singh V., Tsui K., Awad S., Renshaw M. J., Scholfield P., Barton G. J., Nislow C., Tanaka T. U., and Owen-Hughes T. (2011) The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 30, 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deyter G. M., and Biggins S. (2014) The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 28, 1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopes da Rosa J., Holik J., Green E. M., Rando O. J., and Kaufman P. D. (2011) Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohkuni K., Abdulle R., and Kitagawa K. (2014) Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl cis-trans isomerase. Genetics 196, 1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.