Abstract
RNA polymerase II (RNAPII) undergoes structural changes during the transitions from initiation, elongation, and termination, which are aided by a collection of proteins called elongation factors. NusG/Spt5 is the only elongation factor conserved in all domains of life. Although much information exists about the interactions between NusG/Spt5 and RNA polymerase in prokaryotes, little is known about how the binding of eukaryotic Spt4/5 affects the biochemical activities of RNAPII. We characterized the activities of Spt4/5 and interrogated the structural features of Spt5 required for it to interact with elongation complexes, bind nucleic acids, and promote transcription elongation. The eukaryotic specific regions of Spt5 containing the Kyrpides, Ouzounis, Woese domains are involved in stabilizing the association with the RNAPII elongation complex, which also requires the presence of the nascent transcript. Interestingly, we identify a region within the conserved NusG N-terminal (NGN) domain of Spt5 that contacts the non-template strand of DNA both upstream of RNAPII and in the transcription bubble. Mutating charged residues in this region of Spt5 did not prevent Spt4/5 binding to elongation complexes, but abrogated the cross-linking of Spt5 to DNA and the anti-arrest properties of Spt4/5, thus suggesting that contact between Spt5 (NGN) and DNA is required for Spt4/5 to promote elongation. We propose that the mechanism of how Spt5/NGN promotes elongation is fundamentally conserved; however, the eukaryotic specific regions of the protein evolved so that it can serve as a platform for other elongation factors and maintain its association with RNAPII as it navigates genomes packaged into chromatin.
Keywords: gene transcription, RNA polymerase II, transcription elongation factor, transcription factor, transcription regulation, DNA Protein Interactions, Spt4/5
Introduction
The conversion of DNA to RNA is a fundamental aspect of all life, and this process is carried out by RNA polymerases (RNAPs).2 These enzymatic powerhouses must maintain both high levels of fidelity and processivity over long distances to ensure that RNAs are accurately produced on a time scale amenable to life. Families of proteins called elongation factors have evolved to assist RNA polymerases during transcription elongation. The oldest and most conserved of these factors is the NusG/suppressor of Ty element (Spt) 5 family (1, 2). NusG is the eubacterial version of Spt5 and functions as a single polypeptide; however, archaea and eukaryotic Spt5 associate with an additional small protein, Spt4. In yeast, SPT5 is essential, but SPT4 is not. Deleting the gene encoding Spt4 impairs elongation, transcription-coupled repair, and mRNA processing (2–5). Some of the functions of Spt4 may be partially dependent on its ability to prevent degradation of Spt5 in cells (4).
The NusG/Spt5 family of proteins has been shown to enhance RNA polymerase transcription elongation in all domains of life (6–10). NusG regulates RNAP activity by stabilizing the post-translocated state thereby inhibiting backtracking and reducing long lifetime pauses (6, 11). The NusG homolog RfaH has also been implicated in regulating movement of the RNAP bridge helix suggesting that NusG and RfaH may function to alter RNAP conformational dynamics (6, 12). In fact, the movement of the trigger loop and bridge helix in the active site is a fundamental process in nucleotide incorporation and regulates arrest of active elongation complexes in both prokaryotes and eukaryotes (12–15). In prokaryotes, the movement of the trigger loop and bridge helix is linked to the formation of RNA hairpins, which regulate RNAP pausing (13, 16, 17). Although this method of pausing is not known to exist in eukaryotes, x-ray crystal structures of yeast RNAPII in different stages of elongation have generated a model in which the movement of the bridge helix and trigger loop can be coupled to translocation through the non-template strand of DNA (18). This information implies that the nucleic acid scaffold is critical in maintaining active RNAP during elongation.
Using a combination of crystal structures, cryo-EM, and model building of archaeal Spt4/5 bound to RNAP, it has been proposed that Spt4/5 closes the “crab claw”-like clamp of RNAP (8, 10) by binding across the jaws and interacting with a coiled-coil domain of RpoA (7). This binding may prevent the dissociation of the RNA polymerase from the template by encircling DNA and enhancing processivity (7, 8, 10). “Bridging” of the two lobes of RNAP occurs through the universally conserved NGN domain (NusG N-terminal region), and all known biomechanical properties of Spt5 are linked to this domain (1, 7, 19, 20).
Much of what we know about the biochemical activities of eukaryotic Spt4/5 arose from its identification as the DRB-sensitive inducing factor (DSIF) in HeLa cell extracts and was later found to be required to stably pause RNAPII in promoter proximal regions (9, 21). In this latter case, DSIF acts as a negative elongation factor working with the negative elongation factor (22). Conversion to a positive elongation factor requires the phosphorylation of Spt5 by positive transcription elongation factor b (P-TEFb) (23). Human Spt5 suppresses the arrest of RNAPII at poly(A) tracts (24), and the zebrafish version of Spt5 stimulated transcription elongation in extracts (25). In these two examples, eukaryotic Spt4/5 has been shown to function in a positive manner to support transcription elongation. Because the ability to induce promoter proximal pausing is unique to higher eukaryotes, much of the focus to understand DSIF function has been carried out in metazoans using depleted extracts or crude fractions. What is lacking is a highly defined biochemical reconstitution system to study eukaryotic Spt4/5 that can provide a deeper mechanistic understanding of how it affects RNAPII activity and promotes elongation. Studying yeast Spt4/5 provides an opportunity to understand the positive effect of Spt4/5 on transcription elongation, as yeast lacks promoter proximal pausing. Furthermore, genetic assays indicate that Spt4/5 functions in a purely positive function (3, 26, 27).
In prokaryotes, NusG of eubacteria and Spt4/5 from archaea bind to RNAP (7, 20). However, the association between fly and human DSIF and RNAPII is facilitated by the emergence of the nascent transcript (28, 29). Eukaryotic Spt5 contains an extended C terminus containing five Kyrpides, Ouzounis, Woese (KOW) domains that theoretically can reach anywhere on the surface of RNAPII. KOW domains are generally part of larger Tudor domains that are known to interact with nucleic acids (30, 31). In fact, a crystal structure of an isolated KOW1-linker (K1L1) fragment of Spt5 and biochemical analysis using isolated K1L1 provided evidence that it binds to nucleic acids (32). Furthermore, in vivo cross-linking data conducted on yeast Spt5 revealed that KOW domains 4 and 5 interact with surfaces on RNAPII, including the Rpb4/7 sub-module. Although more information is being gained on the functions of these KOW domains, how they affect RNAPII binding and activity is not clear. Eukaryotic Spt5 also contains an extended intrinsically disordered modifiable C-terminal region (CTR) that has been shown to be phosphorylated during transcription, which in turn recruits elongation and RNA-processing factors (4, 33–37). This raised the possibility that the elongation activity associated with Spt5 may have been delegated to other elongation factors during the course of evolution. The CTR of Spt5 does not contribute to the essential function of the protein because a CTR-less mutant of Spt5 is viable and displays minor phenotypes (36, 37); however, systematic deletion of KOW domains and the linker regions of Spt5 in yeast are lethal and result in reduced affinity for both RNAPI and RNAPII (38).
Spt4/5 likely associates with several regions of RNAPII and nucleic acids in the elongation complex. The consequences of these interactions with polymerase are not well understood. We show that the emerging RNA transcript is critical for the binding of yeast Spt4/5 to the elongation complex. Additionally, eukaryotic specific regions of Spt5 are required for stable binding to RNAPII elongation complexes. Furthermore, we provide direct evidence that interaction of the NGN domain of Spt5 with the non-template strand of DNA in the elongation complex is required for Spt4/5 to promote elongation. Together, these findings suggest a eukaryotic specific mode of binding to polymerase but a universally conserved mechanism of action to promote elongation. It also suggests an elongation promoting activity for Spt4/5 different from its ability to encircle the DNA and retain RNAP on DNA.
Experimental Procedures
Plasmid Construction
The coding sequences of SPT4 and SPT5 were amplified by PCR from genomic DNA with primers that introduced BamHI and BsrGI or BamHI and NgoMIV sites into the ends, respectively. The products were then ligated into the shuttle vectors pST50-N-term-Strep tag (39) and pST66-C-terminal His6 tag. The expression modules were then excised using unique restriction sites and ligated into the pST69 co-expression plasmid (a kind gift from Song Tan). Details are available upon request. The SPT5-HA3 sequence was amplified by PCR from pHQ1494 (a kind gift from Allen Hinnebusch) and then inserted into pRS313 plasmids using the In-fusion cloning method (Clontech). Mutations were constructed using the In-fusion mutagenesis procedure. Constructs were confirmed by sequencing.
Yeast Strains
Homologous recombination was used to fuse the FRB sequences in-frame with the endogenous SPT5 coding sequence in the anchor-away strain Y40343 (Euroscarf) as described previously (40). The strain, JR1734, genotype is MATα tor1-1 fpr1::NAT RPL13A-2xFKBP12::TRP1 SPT5xFRB::KANMX. Spotting of strains was performed by growing yeast cells transformed with pRS313 containing SPT5-HA3, and its mutant derivatives, to an OD of 1.0 and then spotting 10-fold serial dilutions onto synthetic media (−HIS) plates ±1 μg/ml rapamycin. For preparing extracts for Western blotting, cells were grown to an A600 of 0.7–0.8; rapamycin was added to 1 μg/ml, and after 1.5 h the cells were harvested, and whole cell extracts were prepared using the trichloroacetic acid method (41). Whole cell extract was then run on SDS-PAGE and subjected to Western blotting with anti-HA (Pierce) or anti-H2B (Active Motif) antibodies. Signals were detected using chemiluminescence detected on x-ray film.
Purification of Recombinant Spt4/5 and Yeast RNAPII
BL21 codon plus cells transformed with a plasmid co-expressing Strep-Spt4 and Spt5-His6 were grown to an OD of 0.4. Spt4/5 was induced by the addition of 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and 0.3 mm ZnSO4 overnight at 18 °C. Cells were then harvested, washed in lysis buffer (20 mm Tris-Cl, pH 8.0, 500 mm KCl, 0.010 mm ZnCl2, 5 mm imidazole, and 10% glycerol), lysed by adding lysozyme, and sonicated. The lysate was cleared by centrifugation at 14,000 rpm and incubated in Talon resin (Clontech) for 1 h at 4 °C. Resin was washed with lysis buffer and then eluted in lysis buffer containing 200 mm imidazole. The peak fractions were pooled and diluted to 200 mm salt in heparin buffer (20 mm Tris-Cl, pH 6.8, 100 mm NaCl, 8% glycerol). Protein was purified on heparin-agarose using step elution fractions of 0.5, 1.0, 1.5, and 2.0 m NaCl. Peak fractions were pooled and diluted to 100 mm NaCl and then passed over a Q-Sepharose column. Protein was eluted with step gradients from 200 to 800 mm KCl in 100 mm steps in Q buffer (20 mm HEPES, pH 7.5, 5 mm MgCl2, 1 mm β-mercaptoethanol, 0.01 mm ZnCl2, 10% glycerol). Peak fractions were pooled and dialyzed against 20 mm HEPES, pH 7.5, 200 mm KCl, 5 mm MgCl2, 1 mm β-mercaptoethanol, 0.01 mm ZnCl2, 10% glycerol. Protein was stored at −80 °C. Yeast RNAPII was purified as described previously (42).
Elongation Complex Reconstitution and Runoff Experiments
DNA templates for EC42 runoff experiments were prepared by annealing two chemically synthesized oligos corresponding to the non-template and template strands, producing a template with a 3′ overhang. The DNA template was gel-purified. The sequences are as follows: EC42 template strand, 5′-GCCACCGCGGTCTAGAGGATCCCCGGGAGTGGAATGAGAAATGAGTGTGAAGATAGAGGAGAGATCAAAAAAATTA-3′, and EC42 non-template strand, 5′-CTCCTCTATCTTCACACTCATTTCTCATTCCACTCCCGGGGATCCTCTAGACCGCGGTGGC-3′. EC70 templates were prepared using a PCR and tail ligation strategy (28, 43, 44)
Elongation complexes (ECs) were formed as described previously (28, 43, 44). Briefly, reactions were formed in 15 μl of transcription buffer (50 mm HEPES, pH 7.5, 100 mm KCl, 1 mm MnCl2, and 0.5 mm DTT, 10% glycerol, 0.5 mm UpG, 20 units of RNasin (Promega), 100 ng/μl BSA). Approximately 150 fmol of RNAPII was pre-incubated with DNA template, and then 5 μl of 0.1 mm ATP, 0.1 mm CTP, 0.005 mm UTP, and 8 μCi of [α-32P]UTP(6000 Ci/mmol) was added. For the anti-arrest assays, ECs were formed on EC42 templates in the presence of 0.1 mm ATP, 0.1 mm CTP, 0.005 mm UTP, and 8 μCi of [α-32P]UTP (6000 Ci/mmol) for 2.5 min, and then Spt4/5 was added to the reaction along with 100 μm UTP and 250 ng/μl salmon sperm DNA. ECs were then incubated for 5, 7.5, 10, and 20 min. After each time point, ECs were chased with 100 μm ATP, CTP, and GTP for 10 min. Reactions were terminated by transcription stop mix (20 mm EDTA, pH 8.0, 200 mm NaCl, 1% SDS, 0.5 μg/ml yeast total RNA). Samples were treated with 1 μg/ml proteinase K, and the RNA was purified by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and ethanol precipitation. RNA was resuspended in formamide dye solution (90% formamide, 1 mm EDTA, bromphenol blue, and xylene cyanol), heated, separated on 10% denaturing polyacrylamide gels, dried, and exposed to a phosphorimaging screen. Images were recorded using a Typhoon system (GE Healthcare) and analyzed using ImageJ. Percent (%) active complex was calculated by the following formula: % RNA runoff = (full-length RNA + EC RNA), plotted on the y axis versus time (x axis), and the data were then fit to an exponential decay curve for comparison using Excel.
RNA Cross-linking and Footprinting
RNA cross-linking was performed as described previously (28, 43), except that 3-O-MeGTP was omitted. Briefly, ECs were formed on DNA templates using ∼500 fmol of RNAPII in transcription buffer with 0.03 mm bromo-UTP, 0.03 mm ATP, 0.005 mm CTP, and 4 μCi of [α-32P]CTP (6000 Ci/mmol). ECs were formed for 20 min, and then Spt4/5 was added with 500 ng of competitor yeast total RNA. Samples were then UV-irradiated for 10 min at 308 nm, and 1 μg of RNase A and 10 units of DNase I (Roche Applied Science) were added and the samples incubated at 37 °C for 1 h. Samples were resolved on SDS-PAGE, dried, and exposed to a phosphorimaging screen.
For RNase protection, footprinting experiments were conducted on ECs formed on DNA templates immobilized on streptavidin M-280 Dynabeads. ECs were formed in transcription buffer using ∼100 fmol of RNAPII plus 0.1 mm ATP, 0.1 mm CTP, 0.1 mm for 20 min. Nucleotides and free protein were removed by magnetic collection and three washes in wash buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 1 mm DTT, 0.02% Nonidet P-40, 100 ng/μl BSA). Afterward, the beads were resuspended in reaction buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.01 mm ZnCl2, 100 ng/μl BSA, 10% glycerol), and [α-32P]GTP (6000 Ci/mmol) was added and incubated for 5 min to end label the RNA, and then cold GTP was added to 100 μm and incubated for an additional 5 min. The complexes were then collected and washed three times with wash buffer and resuspended in reaction buffer. Spt4/5 was then titrated into the binding buffer, and RNase I (New England Biolabs) was added to 1 unit/μl and incubated for 3 min. The reaction was terminated by the addition of transcription stop mixture, and then phenol/chloroform/isoamyl alcohol (25:24:1) was extracted and ethanol-precipitated. Samples were resuspended in formamide dye and run on 15% denaturing polyacrylamide gels.
DNA Gel Shift and ExoIII Footprinting
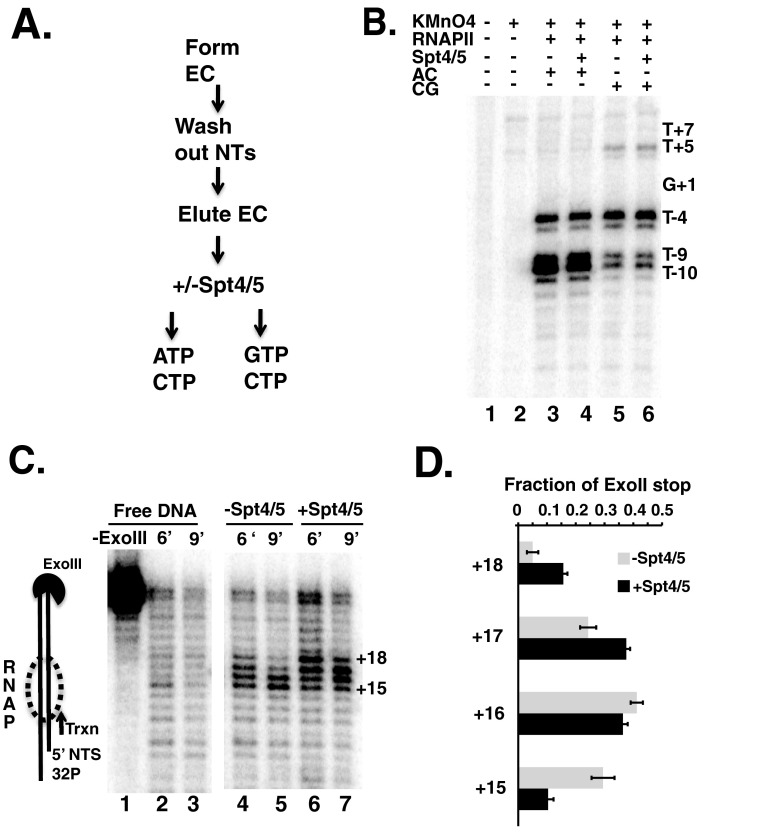
Transcription templates for EMSA and ExoIII nuclease footprinting were generating by annealing complementary gel-purified oligos. The top strand was end-labeled with [32P]ATP using polynucleotide kinase. EC formation was performed as described above, except no radiolabeled nucleotides were added, and 10 nm RNAPII for ExoIII footprinting the scale of the reaction was increased to 100 nm RNAPII. ECs (±Spt4/5) were loaded on a 4.5% native PAGE in 0.5× TBE and electrophoresed at room temperature for 2 h. For ExoIII footprinting, ExoIII was added to a final concentration of 10 units/μl and allowed to digest for varying amounts of time. Samples were quenched with 20 mm Tris-Cl, pH 7.5, 200 mm NaCl, 1% SDS, and 100 ng/μl single-stranded DNA. Samples were then treated with proteinase K; phenol/chloroform/isoamyl (25:24:1) was extracted, ethanol-precipitated, resuspended, and run on a 10% denaturing polyacrylamide gel. Quantification of the ExoIII footprinting was performed by taking the sum total of the bands comprising the RNAPII footprint at nucleotides +18, +17, +16, and +15. The signal in the free DNA digestion samples was considered background and was subtracted out. The sum of these four bands was then calculated, and each individual band was divided by the total (+18/Σ+18,+17,+16,+15), and this generated a fraction of the total population.
Immobilization of RNAPII and KMnO4 Footprinting
Protein A-Sepharose magnetic beads (GE Healthcare) were blocked with 20 mm Na-HEPES, pH 7.8, 150 mm NaCl, 10% glycerol, 250 ng/μl BSA and then bound to 8WG16 IgG for 1 h at room temperature. Beads were then washed three times with 10 bed volumes of bead wash buffer (50 mm Tris-HCl, pH 7.4, 250 mm KCl, and 100 ng/μl BSA) and then three times with IgG Wash buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl). RNAPII was then immobilized through the CTD of Rpb1 on protein A magnetic beads in transcription buffer for 1 h at 4 °C, washed three times with IgG wash buffer, and finally resuspended in transcription buffer. End-labeled DNA was added to form ECs for 20 min in the presence of 100 μm CTP, ATP, and UTP. Beads were washed two times with 20 mm HEPES, pH 7.5, 500 mm KCl, 5 mm MgCl2, 0.001 mm ZnCl2, 10% glycerol, and 100 ng/μl BSA and then two times with the same buffer except that the KCl was reduced to 100 mm. The immobilized ECs were resuspended in transcription reaction buffer supplemented with 2 mm CaCl2. Competitor salmon sperm DNA was added to a concentration of 100 ng/μl. For RNAPII walking experiments, complexes were eluted using a GST-CTD recombinant protein. Spt4/5 was either added or not, and either 100 μm CTP, GTP, or 100 μm CTP ATP was added back. 3-Fold molar excess of TFIIS was added back. Complexes were then treated with 5 mm KMnO4 for 2 min at room temperature and quenched with 20 mm EDTA, 250 mm β-mercaptoethanol, 300 ng/μl salmon sperm DNA, and 10% piperidine. Samples were then heated to 90 °C for 10 min and extracted once with water-saturated n-butanol, precipitated with ethanol, resuspended in 90% formamide buffer, and run on 10% denaturing polyacrylamide gels.
DNase I Footprinting
Elongation complexes were formed on immobilized RNAPII as described above. After washing, elongation complexes were eluted from the magnetic beads using 1 μg of recombinant GST-CTD in DNase I reaction buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.01 mm ZnCl2, 100 ng/μl BSA, 1 mm CaCl2, 10% glycerol). Spt4/5 or buffer was added for 5 min, and then 200 ng of salmon sperm DNA was added, and samples were treated with 0.0006 units of DNase I for 1 min at room temperature. Samples were then quenched with 10 mm EDTA and immediately loaded on 0.5× TBE, 4.5% native PAGE. After separation, the gel was exposed to x-ray film, and the bands corresponding to free DNA, ECs, and EC-Spt4/5 complexes were excised, and the DNA was recovered. The DNA was then purified and separated on 10% denaturing PAGE. Quantification of the DNase I footprint was performed using ImageQuant software (GE Healthcare). A trace of each individual lane was taken and adjusted for the total counts in each lane using Excel. Then the traces were generated in Excel and overlaid.
Generation of Photo-probes and 5-Iodo-CTP Photo-cross-linking
Photoreactive nucleotide was incorporated at unique positions by hybridizing an oligo to a long template strand immediately upstream of CA pairs. 10 μm 5-iodo-dCTP (Sigma) and 5 μCi of [α-32P]dATP (6000 μCi/mmol) were added along with Klenow exo− (New England Biolabs) and incubated for 10 min at 37 °C. Afterward, all dNTPs were added to a final concentration of 100 μm, and the reaction was allowed to extend for 30 min. Double-stranded modified probes were gel-purified on 10% native PAGE. ECs were formed as described above (EC42), and then Spt4/5 was added followed by 200 ng of carrier salmon sperm DNA. Samples were then irradiated using a 308 nm UV light source for 10 min. After irradiation, the samples were digested with DNase I (10 units) and RNase A (2.5 μg) at 37 °C for 60 min. SDS-loading buffer was then added, and samples were separated on SDS-polyacrylamide gels, dried, and exposed to phosphorimaging screens.
Results
Characterization of Elongation Complexes Formed from End-initiated Templates
We use a minimal system for assembling ECs from highly purified components (28, 43–46). RNAPII elongation was initiated from a 3′ single strand extension on a duplex template to assemble an EC with a defined transcript produced from a G-less cassette (Fig. 1A). ECs were formed with RNA transcripts between 35 and 70 nucleotides in length, where indicated. These lengths of RNA are sufficient to bind Spt4/5 (see below) (28, 29). To produce ECs with different length transcripts, the DNA sequence in the template between the initiation site and the first G was shortened. The sequences immediately upstream of the arrest site and the single strand-double strand junction were always identical. Previous studies have shown that initiating transcription from some 3′-extended templates produced aberrant ECs with extended RNA-DNA hybrids caused by the hybridization of the transcript to the template strand and a displacement of the non-template strand (NTS) from DNA (47–49). However, it was later found that the extent to which this occurs is dependent on a number of factors, including the source of RNAPII, the sequences of the 3′ tail, and the region encompassing the single strand-double strand junction on the template (47). Previously, it has been shown that templates using the same 3′ tail and duplex region as that described here produced transcripts that are resistant to RNase H (cuts RNA-DNA hybrids) and sensitive to RNase A, suggesting extended DNA-RNA hybrids are not produced (44). Nonetheless, because one of the goals of this study was to characterize interaction between Spt4/5 and the nucleic acid scaffold, we further characterized ECs built from this template to validate that a proper nucleic acid scaffold was forming.
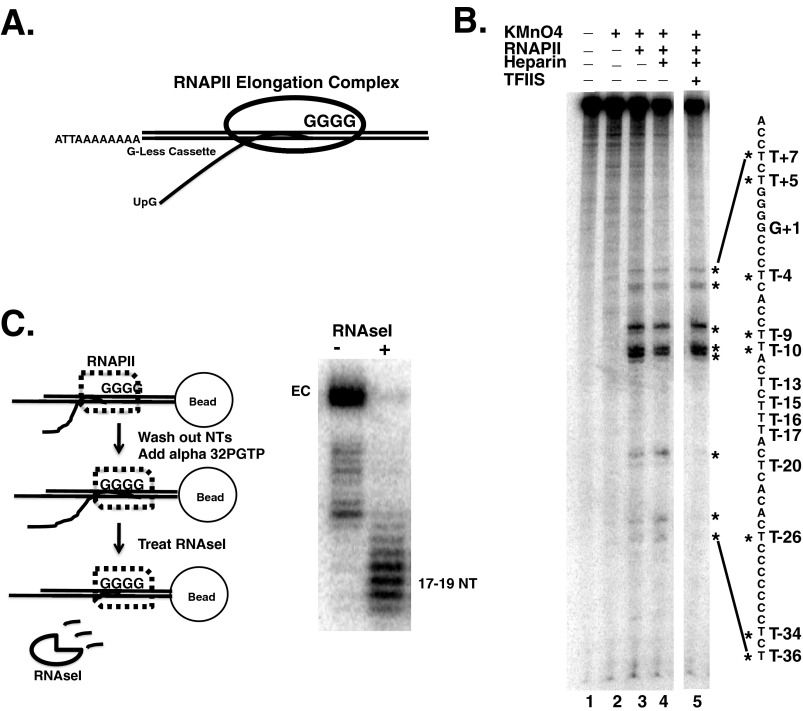
FIGURE 1.
Characterization of ECs. A, schematic diagram of the formation of RNAPII ECs using 3′ end initiated templates. Radiolabeled UTP is incorporated into the transcript during the transcription of the G-less cassette. B, KMnO4 footprint of the non-template strand. ECs (EC70) were prepared on immobilized RNAPII (see under “Experimental Procedures”). DNA sequence is indicated on the right of the gel. The four G bases (+1 to +4) are equivalent to the G-tract shown in A. The reactive Ts are marked with an asterisk. Heparin and TFIIS was added to the samples shown in lanes 4 and 5, respectively. All samples were run on the same gel, but irrelevant lanes were removed between lanes 4 and 5. C, RNase I footprinting of the transcript in ECs. ECs were formed on a template that produced a 35-nucleotide (NT) transcript with radiolabeled GTP incorporated into the 3′ end of the transcript in the active site of RNAPII. Left, schematic representation of RNA protection experiment. Right, gel image of the labeled RNA produced with and without RNase I digestion.
We first tested whether the ECs produced here had a properly formed transcription bubble. Potassium permanganate (KMnO4) was used to interrogate the structure and location of the transcription bubble in RNAP (50, 51). The thymidine (T) bases in single-stranded DNA are prone to oxidation by KMnO4, whereas those located in double-stranded nucleic acids were resistant. If the NTS was being displaced and the transcription bubble failed to close, thymidines in the NTS behind RNAPII would be highly reactive to permanganate. This was not observed. ECs formed by this method resulted in strong permanganate reactivity immediately upstream of the G-tract (+1) at T−4, T−9, and T−10, which are expected to be in the transcription bubble when RNAPII arrests at the first G (Fig. 1B, compare lanes 2 and 3). Weaker activity is also observed at T+5 and T+7, which lie just on the downstream edge of the expected location of the transcription bubble. The weaker reactivity just ahead of the bubble of RNAPII ECs may be caused by bending of the template and enhanced base flipping in this region. Importantly, thymidines in the NTS immediately behind RNAPII are resistant to KnMO4 indicating bubble closure. Weak RNAPII-dependent reactivity of T−27, T−35, and T−36 was observed. However, these Ts are flanked by a C-tract found in the template. RNAPII stalls or arrests at polynucleotide tracts (52), and this reactivity may be caused by a fraction of RNAPII becoming arrested over this region due to backtracking. To test this hypothesis, the elongation factor TFIIS was added to the reaction. TFIIS would rescue the arrested complexes over the C-tract and lead to a loss of reactivity at these T bases. This was indeed the case; adding TFIIS eliminated the permanganate reactivity at these thymidines (Fig. 1B, compare lanes 3 and 5). Finally, we demonstrated the KMnO4 reactivity was resistant to heparin, suggesting that this was not due to a nonspecific interaction between RNAPII and the DNA (Fig. 1B, lanes 3 versus 4). Thus, KMnO4 reactivity indicates that a transcription bubble is produced and that the NTS is re-annealing with the template strand behind RNAPII.
Previous EC complex mapping revealed that RNAP protects 16–19 nucleotides of RNA from digestion by single-stranded RNases (53, 54). The structure and location of the emerging transcript in the ECs prepared by this method were examined. RNAPII ECs were formed on immobilized DNA templates by transcribing up to the G-tract using “cold” nucleotides lacking GTP; the complexes were washed to remove nucleotides, and then [α-32P]GTP was added to produce a 3′ end-labeled transcript. RNase I digested the transcript down to 17–19 nucleotides, a length expected if the RNA was threaded through the RNA exit channel (Fig. 1C). If it emerged aberrantly, such as through the main channel, the size of the protected transcript should be different. Additionally, the sensitivity of the transcript to single-stranded specific RNase indicates that extended RNA-DNA hybrids were not formed on the vast majority of the templates. All of the data described here, and elsewhere (28, 43–46), indicate that this template produces ECs with the expected nucleic acid scaffold.
Spt4/5 Reduces or Prevents RNAPII Arrest
After validating the structure of the ECs, we tested how Spt4/5 bound to ECs and affected the activity of RNAPII. Until now, it has been shown that human Spt4/5 (DSIF) stimulates elongation and mediates 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity of RNAPII in extracts (9, 24, 55, 56), but how Spt4/5 affects RNAPII activity is still unclear. We produced recombinant yeast Spt4/5 complex in Escherichia coli using a polycistronic co-expression system (39). Spt4/5 complexes containing a full-length Spt5 and another lacking the C-terminal region (ΔCTR) were isolated to high purity (Fig. 2A). Removing the disordered CTR increased expression of the complex and allowed for its separation from Rpb2 in gels in subsequent experiments (see below). Because the CTR is not required for viability in yeast or binding to RNAPII in vitro (36, 38), we suspected it might be dispensable for the elongation promoting activities of the complex, but this was not known. So the biochemical activities of these two forms were compared.
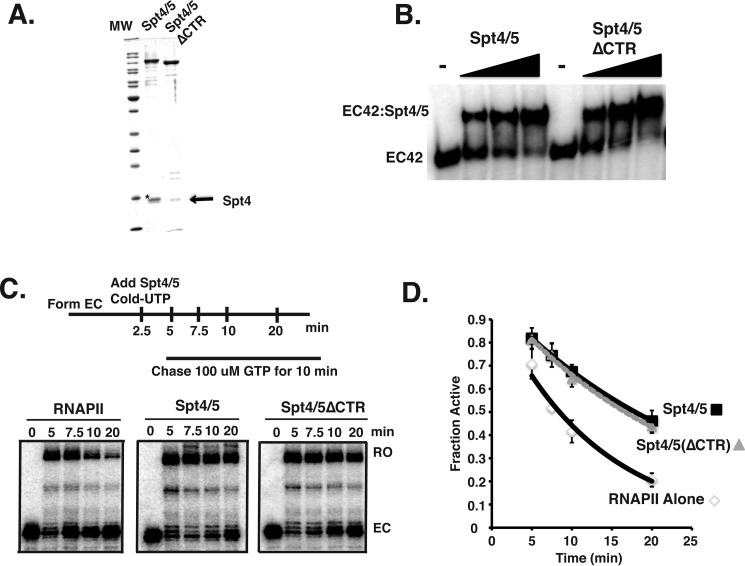
FIGURE 2.
Biochemical analysis of recombinant Spt4/5. A, Coomassie Blue-stained SDS-PAGE of recombinant of Spt4/5 and Spt4/5(ΔCTR). An arrow shows Spt4. The asterisk indicates a contaminating protein migrating just above Spt4 found in the preparation of full-length Spt4/5. B, EMSA of Spt4/5 and Spt4/Spt5 (ΔCTR) binding to RNAPII EC prepared on end-labeled DNA templates. The ratio of Spt4/5 to RNAPII was 0.75, 1.5, and 3.0. Free DNA migrated at the bottom of the gel and was cropped out of the image. C, RNAPII arrest assay comparing intact Spt4/5 and Spt4/5(ΔCTR). Saturating amounts of Spt4/5 were used in the assay, which was determined by shifting all of the EC in EMSA. D, RNAPII arrest assays were quantified as described under “Experimental Procedures” and are plotted as a fraction of active RNAPII as a function of time. The data were fit to an exponential decay curve. The data represent the averages and standard deviations of three independent assays.
We tested the ability of intact Spt4/5 and the CTR-less derivative to bind to ECs. Although it was known that the CTR-less version of Spt5 interacts with RNAPI and RNAPII (38), it was not known whether the CTR played a role in the binding of Spt4/5 to ECs. ECs (EC42) were assembled onto an end-labeled DNA template; Spt4/5 and Spt4/5(ΔCTR) were titrated into the binding assay, and the complexes were resolved on native gels. The results shown in Fig. 2B indicate that Spt4/5 complexes containing intact and CTR-less Spt5 bound to ECs to the same extent, indicating that the CTR region of Spt5 is not required for Spt4/5 to bind RNAPII ECs in vitro.
The bacterial homolog of Spt5, NusG, has been shown to reduce long lifetime pauses of RNAP and promote elongation (6, 57). Archaea Spt4/5 has been shown to enhance general processivity through interactions with the coiled-coil domain of RNAP (7). To determine whether yeast Spt4/5 promotes elongation, we established a transcription assay to measure the activity of Spt4/5 that relies on the propensity of RNAPII to become paused or arrested during prolonged periods of nucleotide depletion. In this assay, radiolabeled ECs are formed for 2.5 min in the absence of GTP, allowing RNAPII to transcribe up to the G-tract in the template (pulse). At that point, cold UTP and Spt4/5 or buffer was added; the excess cold UTP prevented the detection of newly formed ECs during the chase. The ECs were then incubated for increasing amounts of time before GTP was added to initiate the chase (Fig. 2C, upper panel). The fraction of full-length transcript (active RNAPII) was plotted as a function of time. As expected, the longer GTP was withheld from the reaction, the smaller the fraction of ECs converted into active complexes that produced runoff product (Fig. 2, C and D). When intact Spt4/5 (WT) or the CTR-less Spt4/5 (ΔCTR) was added, the amount of arrested complexes detected at the 5-, 7.5-, 10-, and 20-min time points was reduced compared when Spt4/5 was omitted. The percentage of runoff over time was fit to an exponential decay curve, and the slope of these lines was compared. This analysis indicated Spt4/5 reduced arrest by about half, and the CTR-less version showed the same level of activity (Fig. 2D). Complete runoff was not observed, however, even in the presence of Spt4/5. The fraction of ECs refractory to Spt4/5 activity (∼30%) likely represents RNAPII that reached the G-tract and arrested during the 2.5-min pulse before Spt4/5 was added, suggesting that Spt4/5 prevented arrest but cannot rescue pre-arrested complexes. In support of this hypothesis, if Spt4/5 was added after 20 min of GTP starvation, it could not rescue the pre-arrested RNAPII complexes.3 We conclude from these results that Spt4/5 reduces or prevents arrest of RNAPII, and the CTR of Spt5 is not required for its biochemical activities. Because we could generate higher quality complexes from the CTR-less Spt4/5, it was used for the remaining experiments unless noted otherwise. Following this section, the Spt4/5(ΔCTR) version will be referred to as Spt4/5 in the text for simplicity.
Nascent RNA Transcript Is Required for Yeast Spt4/5 to Bind to ECs
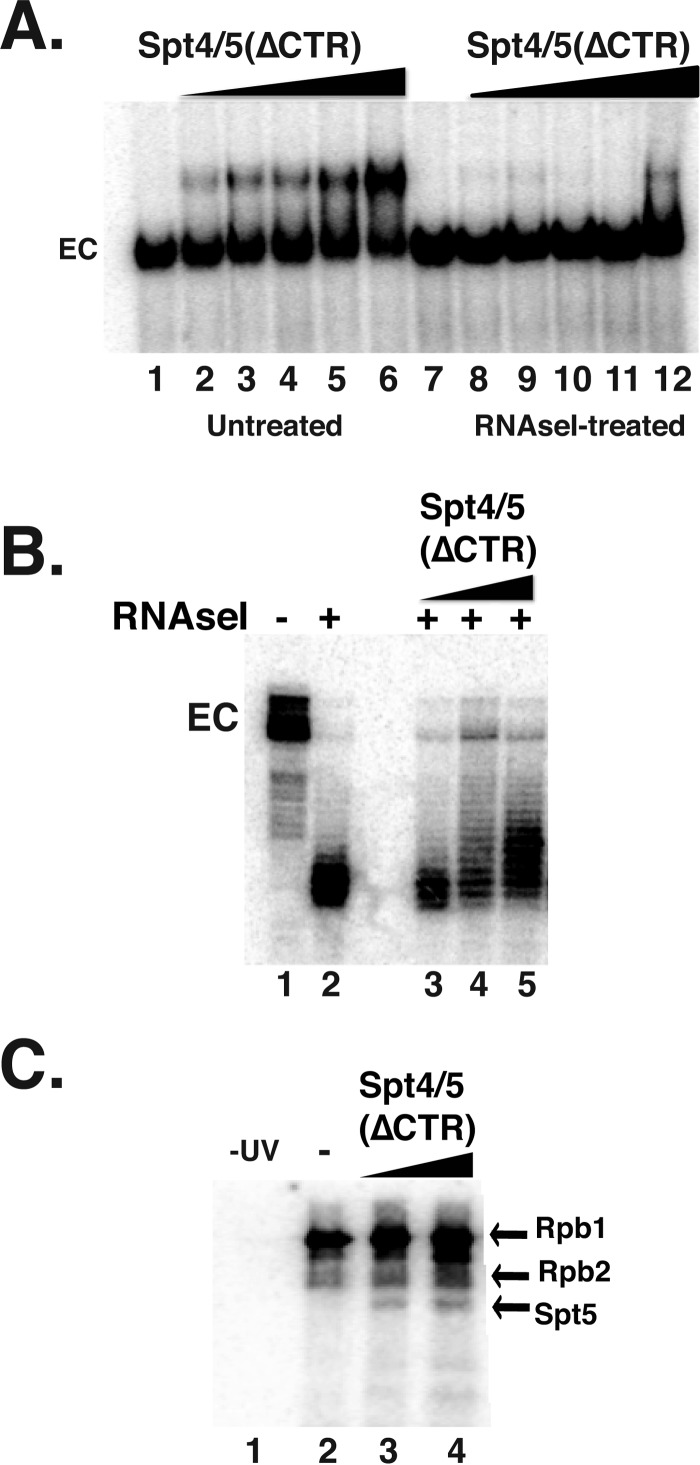
Previous studies indicated that the stable association of metazoan DSIF (Spt4/5) with ECs required RNA to emerge from RNAPII (28, 29). To test whether this was also the case for yeast Spt4/5, we measured the binding of Spt4/5 to ECs before and after digesting the transcript with RNase I. RNAPII ECs were formed on end-labeled DNA templates and then split into two pools; one pool was treated with RNase I to digest the emerging transcript, and the other was left untreated. Spt4/5 bound to the untreated ECs but not to those treated with RNase I (Fig. 3A). Even the small amount of binding observed in some lanes in this experiment could be attributed to a small fraction of ECs not digested with RNase.
FIGURE 3.
Spt4/5 contacts the emerging transcript. A, EMSA analysis of EC42 treated with (lanes 7–12) and without RNase I (lanes 1–6). Spt4/5 ΔCTR was titrated in and complexes were resolved on native polyacrylamide gels. B, RNase I footprint of the emerging transcript. Spt4/5 (ΔCTR) was titrated in prior to digestion. The experiment was conducted as described in the legend of Fig. 1C. C, ECs were prepared with body-labeled transcripts using [32P]CTP and bromo-UTP. After exposure to UV, where indicated, samples were digested with DNase I and RNase A and separated on SDS-PAGE. Labeled proteins were visualized by exposing dried gels to a phosphorimager screen. Two different amounts (1- and 3-fold molar excess) of Spt4/5 were added. The locations of the bands corresponding to Rpb1, Rpb2, and Spt5(ΔCTR) are indicated on the right.
It was estimated that PDB DSIF requires ∼4 nucleotides of RNA to associate with RNAPII ECs, suggesting that Spt4/5 contacts the transcript near the RNA exit channel (28). We conducted RNase I protection assay on ECs containing 3′ end-labeled transcripts in the presence and absence of Spt4/5 (Fig. 3B). As described above, RNAPII protected ∼17–19 nucleotides of the transcript from digestion (Fig. 3B); however, adding Spt4/5 to the assay extended the protection by ∼2–4 nucleotides (Fig. 3B). This result suggests that, like its Drosophila counterpart, yeast Spt4/5 interacts with RNA as it emerges from RNAPII.
UV cross-linking was performed to determine whether the RNase I protection is caused by direct contact between Spt5 and RNA. ECs were prepared in the presence of radiolabeled CTP and a photoreactive UTP analog, bromo-UTP, to prepare body-labeled transcripts. Complexes were then UV-irradiated and digested with nucleases, and the transfer of labeled RNA to proteins was detected by SDS-PAGE followed by autoradiography. The strongest cross-linking occurred between Rpb1 and RNA, with lower levels detected for Rpb2 (Fig. 3C). This is in agreement with other studies (28, 43, 58). Adding Spt4/5 resulted in the appearance of an additional radiolabeled band migrating at the expected position of Spt5 (Fig. 3C).
C-terminal KOW Domains of Spt5 Are Required for Spt4/5 Binding to RNAPII
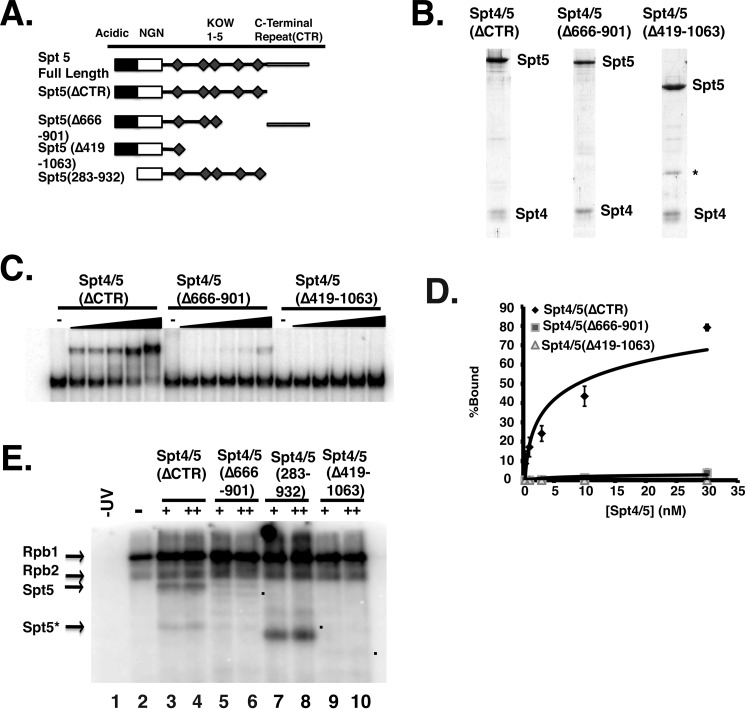
Prokaryotic NusG or Spt4/5 contains a minimal NGN-KOW1 region, and this is sufficient for it to associate with RNAP (7, 20). A previous study using glutathione S-transferase-Spt5 fusion proteins (GST-Spt5) determined that deleting several of the KOW domains resulted in a loss of Spt5 binding to RNAPI and RNAPII (38). Furthermore, in vivo cross-linking studies suggest that the region around KOW domains 4 and 5 cross-link to RNAPII (19). When these cross-link sites were modeled onto existing structures of RNAPII elongation complexes, KOW domains 4 and 5 fit between the Rpb4/7 subunits of RNAPII and the base of the Rpb1 clamp domain (19). Importantly, this model would position Spt5 to protect and physically interact with the emerging transcript. We next wanted to determine how removal of these eukaryotic specific domains would affect the association of Spt4/5 with ECs. Mutants lacking the eukaryotic specific regions of the C terminus of Spt5(Δ419–1063) and another that removed linkers 3 and 4 and KOW domains 4 and 5 (Δ666–901) (Fig. 4A) were produced. A gel showing the purified complexes is displayed as Fig. 4B. The ΔKOW4 and 5 mutant can only be expressed if the CTR region was included,3 so this construct contained the CTR. Because the effect of deleting these regions on the interaction between Spt4/5 and ECs is unknown, this was examined by EMSA. In these experiments, a range of Spt4/5 concentrations was used so that a Kd value for the association of Spt4/5 with RNAPII ECs can be estimated. Spt4/5 bound to RNAPII ECs with high affinity (Kd 8.9 ± 2.5 nm). However, deleting most of the C terminus of the Spt4/5(Δ419–1063) abolished the association of Spt4/5 with RNAPII ECs, and deleting KOW4 and 5 (Δ666–901) reduced the binding of the complex more than 10-fold (Fig. 4, C and D). When the RNA cross-linking was conducted on these binding mutants, no obvious bands arising from Spt5 were observed (Fig. 4E). These results confirm that the eukaryotic specific regions of Spt5 are required for Spt4/5 to bind to ECs. Additionally, to further confirm that the labeled protein detected in the RNA cross-linking experiments was indeed Spt5, we produced an Spt4/5 complex containing Spt5 lacking the N terminus (283–932). Cross-linking assays using this mutant resulted in a faster migrating band at the size expected of the Spt5 mutant lacking the N terminus, confirming that the labeled protein is Spt5 (Fig. 4E).
FIGURE 4.
C-terminal KOW domains of Spt5 are required for the binding to RNAPII ECs. A, schematic representation of Spt5 deletion mutants that were used in the figure. The area shaded in black is an acidic region in the N terminus; the white box is the NGN domain, and the diamonds are the KOW domains. B, Coomassie Blue-stained gel of the mutant complexes. C, EMSA comparing the binding of Spt4/5(ΔCTR), Spt4/5(Δ666–901), and Spt4/5(Δ419–1063) to ECs formed from 10 nm RNAPII. Increasing amounts of complex were titrated in and then resolved by native PAGE. D, quantification of EMSA experiments. Percent binding was plotted on the y axis as a function of Spt4/5 concentration. The data were then fit to a logarithmic binding curve, and a Kd value was estimated. Values represent the averages and standard deviations of three independent experiments (E). RNA cross-linking assay. The assay was conducted as described in Fig. 3C. A sample of RNAPII alone was untreated with UV (lane 1) (−UV) and all others were exposed to UV (lanes 2–10). Two amounts of each of the Spt4/5 complexes were titrated in. “+” indicates the amount of Spt4/5 required to shift ECs in EMSA assays and “++” is twice that amount (3- and 6-fold molar excess relative to RNAPII). Lane 1, −UV; lane 2, +UV; lanes 3 and 4, Spt4/5(ΔCTR); lanes 5 and 6, Spt4/5(Δ666–901); lanes 7 and 8, Spt4/5(284–931); and lanes 9 and 10, Spt4/5(Δ419–1063). The asterisk (Spt5*) marks a breakdown product in the Spt4/5 preparation. The dots mark the migration of the mutants in the gel.
Probing the Nucleic Acid Scaffold in Spt4/5-containing ECs
Next, we analyzed interactions between Spt4/5 and DNA in the EC. Current structural models generated from x-ray crystallographic data of archaeal Spt4/5 fused to the clamp domain of RNAP suggest that eukaryotic Spt4/5 may interact with upstream DNA exiting RNAPII. Additionally, two other structures, one of RNAPII including both strands of DNA (18) and another of the isolated KOW1-Linker1 region of Spt5(372–508, K1L1) (32), also suggest that Spt4/5 binds to upstream DNA. In addition, structural modeling of Spt4/5 in ECs suggests that Spt5 could contact the non-template strand of DNA. Although this function has not been documented for Spt5, it was recently reported that Bacillus subtilis NusG interacts with the non-template strand of DNA to promote sequence-specific pausing (59). Additionally, the recruitment of RfaH requires exposure of the operon polarity suppressor element within the non-template strand of DNA (60). Although it has been widely speculated that Spt4/5 binds DNA in the ECs, direct evidence for this is lacking.
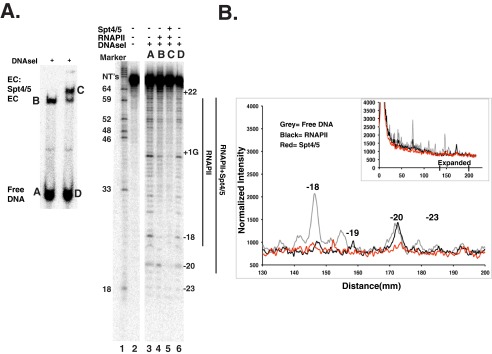
To identify contacts between Spt4/5 and DNA, DNase I footprinting of RNAPII ECs was conducted in the presence and absence of Spt4/5. Because only a fraction of the DNA templates will form ECs, ECs were formed on RNAPII immobilized on protein-A magnetic beads coated with 8WG16 antibody that binds the CTD of Rpb1. This allowed enrichment for RNAPII-engaged templates from the more abundant free templates. Complexes were then eluted from the beads with the recombinant glutathione S-transferase C-terminal domain (GST-CTD), treated with DNase I, and then resolved on preparative native gels to separate free DNA, ECs, and Spt4/5-containing ECs (Fig. 5A, left). The DNA contained in each band was purified and run on a denaturing gel (Fig. 5A, right). Consistent with published DNase I footprints of RNAPII, elongation complexes showed a region of protection of ∼35–40 bases (Fig. 5A, lanes 3 versus 4) (51, 61). Importantly, Spt4/5 extended the footprint upstream of RNAPII, and protection was extended to bases −19 to −21 or 22 (Fig. 5A, lanes 4 versus 5) when visualized on the gel. The footprinting result was presented graphically by scanning the gel, normalizing for the signal in the entire lane, and plotting it as a distance from the parental band (Fig. 5B). A close-up of the gel near the upstream side of polymerase reveals protection at −20 in the presence of Spt4/5 and weaker protection at −19 and upstream (Fig. 5B, compare the red versus black trace). The footprint is not strong but was observed in three experiments and on two different templates.3 This suggests Spt4/5 contacts DNA behind RNAPII, and additional evidence is presented below to support this claim.
FIGURE 5.
Footprinting of Spt4/5 containing ECs. A, DNase I footprinting experiment. ECs were formed on RNAPII immobilized on 8WG16 antibody beads. Washed ECs were eluted with recombinant GST-CTD; Spt4/5 was added or not and then treated with DNase I. The samples were resolved native gels, and the DNA in the bands was gel-purified and analyzed by denaturing PAGE. Letters above the panel in the denaturing PAGE (right) correspond to lettered bands in the native gel (left). Numbers on the left of the denaturing polyacrylamide gel indicate the sizes of the DNA marker (GA) in base pairs. The numbers on the right mark the location of the bases in the template relative to the first G (+1). B, quantitative analysis of the DNase I footprinting gel plotted as intensity versus distance migrated on the gel. Analysis and normalization are described under “Experimental Procedures.” The scan of the entire gel is show in the inset, and the region on the upstream side of RNAPII is shown in the main panel. Free DNA trace is shown in gray; RNAPII alone is shown in black, and RNAPII+Spt4/5 is shown in red.
Next, we examined the effect of Spt4/5 on the size and structure of the transcription bubble by permanganate footprinting. Previous models of eukaryotic and archaeal Spt4/5 suggest it may play a role in maintaining bubble integrity by aiding in the closing of the bubble as DNA exits RNA polymerase (8, 10, 18, 32). The experiment was conducted under two conditions in the presence and absence of Spt4/5. In both cases, ECs were formed on immobilized RNAPII, washed of nucleotides, and then eluted from the protein A beads (Fig. 6A). At this point, ECs were separated into two pools. One pool was given ATP and CTP, which left RNAPII arrested at the G-tract, and allowed for the examination of the effect of Spt4/5 on the “static” EC (Fig. 6B, lanes 3 and 4). The other pool was given GTP and CTP, which allowed RNAPII to “walk” across the four G-nucleotides (Fig. 6B, lanes 5 and 6). This condition may reveal changes in the bubble brought about by Spt4/5 during/after movement of RNAPII.
FIGURE 6.
Footprinting elongation complexes with permanganate and exonuclease III. A, flow diagram explaining the experimental design. Details are given under “Experimental Procedures.” Briefly, immobilized RNAPII ECs were provided ATP and CTP (AC, lanes 3 and 4 in B) or provided CTP and GTP to allow transcription through the 4-base G-tract (CG, lanes 5 and 6 in B). Spt4/5(ΔCTD) was added where indicated. TFIIS was added after nucleotide addition to reduce the potential effects of backtracking on bubble progression. B, denaturing polyacrylamide gel showing the end-labeled non-template strand. Ts in the sequence are indicated on the right of the gel and labeled relative to the first G. C, ExoIII protection experiment to map the leading edge of RNAPII along the DNA. Denaturing PAGE comparing 6 and 9 min of cleavage by ExoIII. Free DNA (lanes 1–3) was compared with DNA + RNAPII-Spt4/5 (lanes 4 and 5) and DNA + RNAPII with Spt4/5(ΔCTD) (lanes 6 and 7). Positions of the bands are labeled to the right of the gel and represent the distance downstream from G+1. D, quantification of the ExoIII data from four independent experiments presented as the averages and S.E. The numbers on the left indicate the size of the band in nucleotides and correspond to those indicated in C.
As stated above, T−4, T−9, and T−10 lie within the transcription bubble when RNAPII arrests at the first G in the template. As observed in other experiments using RNAPII, T−4, T−9, and T−10 were the primary reactive species when only CTP and ATP were added, and RNAPII remained arrested at the first G (compare Figs. 1B and 6B, lanes 3 and 4). Adding Spt4/5 did not alter the footprinting pattern, suggesting that Spt4/5 did not change the transcription bubble under these conditions (Fig. 6B, compare lanes 3 and 4). Walking RNAPII forward by four bases across the G-tract by adding GTP would place T−9 and T−10 in the upstream edge of the duplex of the transcription bubble, reducing their reactivity to permanganate. Furthermore, T+5 would now be placed within the single-stranded region of the bubble and become more reactive. These changes were observed (Fig. 6B, compare lanes 3 versus 5), although it appears that not all of the RNAPII walked to the new location, because the increase in the reactivity of T+5 and reduction in reactivity of the −9 and −10 positions were not quantitative. Importantly, there were no differences in the reactivity of these locations in the presence of Spt4/5 (Fig. 6B, compare lanes 5 and 6). Although we cannot rule out that Spt4/5 may kinetically affect the opening or closing of the transcription bubble, we conclude that under equilibrium conditions Spt4/5 does not alter the size of the transcription bubble.
The results above suggest that Spt4/5 protects bases upstream of RNAPII. It is possible that Spt4/5 contacts DNA downstream (ahead of polymerase). Furthermore, nothing is known about how Spt4/5 affects the tracking of RNAPII along DNA. NusG causes the lateral movement of RNAP downstream by one nucleotide, which may represent the post-translocated state of RNAP (11). Either of these two changes brought about by Spt4/5 can be revealed by ExoIII footprinting of the leading edge of RNAPII. RNAPII ECs were formed on end-labeled DNA templates and incubated with or without Spt4/5, followed by digestion with ExoIII. Probing ECs with ExoIII generated an RNAPII-dependent footprint, as shown by the protection of the DNA compared with digestion of naked DNA (Fig. 6C, lanes 2 and 3 versus lanes 4 and 5). Adding Spt4/5 extended the footprint compared with RNAPII alone by approximately one or two nucleotides downstream (Fig. 6C, compare lanes 4 and 5 versus lanes 6 and 7). The shift in the distribution of ExoIII stops was quantified, and the data from multiple experiments are presented in Fig. 6B. This footprinting result argues against extensive protection of downstream DNA by Spt4/5, but it is strikingly similar to the lateral movement of RNAP caused by NusG (11). A conservative interpretation of this result is that Spt4/5 extends the leading edge of RNAPII downstream, which may be caused by a structural change in RNAPII.
Spt5 Contacts the Non-template Strand in the Transcription Bubble and DNA Upstream of RNAPII
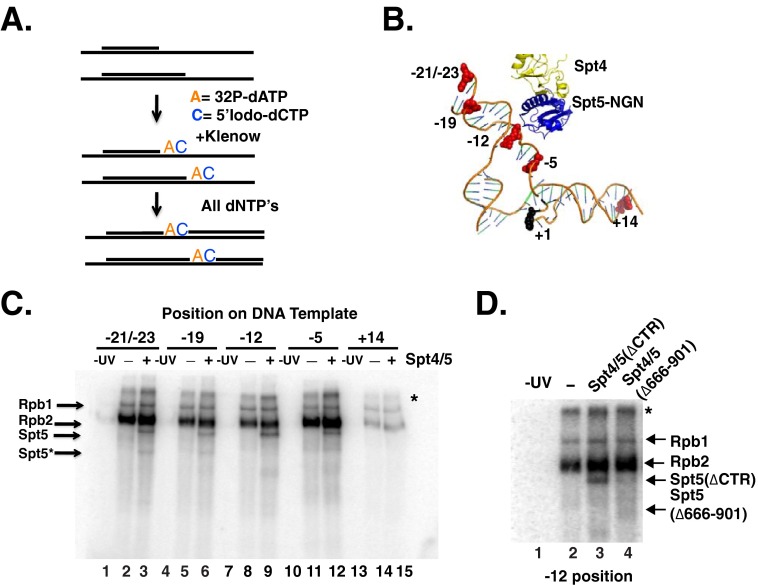
To provide further evidence that Spt4/5 contacts DNA within the nucleic acid scaffold, we turned to site-specific cross-linking. Using the findings from our DNase I footprinting experiments (Fig. 5) and the model of the Spt5(NGN)-Spt4 in the RNAPII EC (10), we selected locations to incorporate the photoreactive nucleotide analog 5-iodo-2′-dCTP adjacent to radiolabeled adenine nucleotide in the NTS (Fig. 7A). The photoreactive probes were placed into the NTS because structural models predicted that the NTS would be in close proximity to Spt4/5 and because incorporating the probes in the template strand could impede transcription of RNAPII up to the G-tract. Photoreactive nucleotides were incorporated at positions −21/23, −19, −12, −5, and +14 (+1 is the first G in the G-tract) (Fig. 7B). −21/23 and −19 are located upstream of RNAPII (14, 51); −12 is located near the expected site of strand re-annealing; −5 is located in the transcription bubble, and +14 is located downstream of RNAPII and is a control because we would expect lower levels of cross-linking of RNAPII at this location. The strategy to incorporate the radiolabeled and photoactive nucleotides produced templates with photoreactive nucleotides at both the −21 and −23 positions because of the DNA sequence. Rpb2 cross-linked to the DNA with the highest efficiency, with lower levels of Rpb1 cross-linking detected (Fig. 7C). The strong cross-linking of the NTS to Rpb2 is consistent with the recently published crystal structure of RNAPII with an intact transcription bubble, which revealed that the NTS passes along the surface of Rpb2 (18). A diffuse band above Rpb1 was also observed. It is unclear what this band is, but it may represent undigested DNA-protein products or UV-induced aggregates. As expected, low levels of Rpb1 and Rpb2 cross-linking were observed at the +14 site, which should be positioned just ahead of RNAPII. Spt5 cross-linked at −21/23, −19, −12, and −5. No cross-linking of Spt5 was detected at position +14 (Fig. 7C), suggesting that the cross-linking of Spt5 to DNA requires it to be properly positioned in the RNAPII EC. Weaker bands migrating faster than Spt5 were also observed in the lanes containing Spt4/5. These bands may be truncated forms of Spt5 contained in the preparations or breakdown of Spt5 during cross-linking. Interestingly, even though Spt4 is in close proximity to DNA in the model (Fig. 7B), we failed to conclusively identify a labeled band of the size of Spt4 in high percentage gels.3 However, this negative result does not definitely rule out the possibility that Spt4 interacts with another region of the nucleic acid scaffold. Furthermore, as a control, we performed cross-linking at the −12 position using a mutant form of Spt4/5(Δ666–901) that binds poorly to RNAPII ECs (see Fig. 4C), and we found that no cross-linking was observed (Fig. 7D). These data are the first evidence that eukaryotic Spt5 physically interacts with DNA upstream of RNAPII and contacts the non-template strand of the transcription bubble.
FIGURE 7.
Spt5 cross-links to the non-template strand in ECs. A, schematic outline of the strategy used to generate site-specific labeled DNA probes on the non-template strand of the EC42 DNA template. A short oligo complementary to the template strand was annealed and used to prime synthesis of the NTS. In the first step, [32P]dATP and 5-iodo-dCTP were incorporated using the Klenow (−exo) fragment of DNA polymerase. In a second step, cold dNTPs were added to complete the top strand (B). A model derived from overlaid crystal structures of RNAPII with the non-template strand, yeast Spt4/5, and archaeal RpoA fused to Spt5 (PDB codes 5C4X, 2EXU, and 3QQC) using PyMOL (version 1.7.4 Schrödinger, LLC). Highlighted in red are bases representing the position of photoreactive 5-iodo-dCTP nucleotide analog in the non-template strand of RNAPII. +1 is the site of NTP incorporation. C, SDS-PAGE analysis of cross-linked products. The location of the photoreactive nucleotide in each template is indicated above. The presence of Spt4/5(ΔCTD) (3-fold molar excess relative to RNAPII) is indicated in the panel. Arrows on the left of the gel indicate the migration of Rpb1, Rpb2, and Spt5(ΔCTD). Spt5* indicates the position of a breakdown product of Spt5 that cross-links to DNA. The band above Rpb1 marked with an asterisk may result from incomplete digestion of the nucleic acid template or products formed by protein-protein cross-links. D, SDS-PAGE comparing cross-linking of RNAPII only (lane 2), Spt4/5 (ΔCTR) (lane 3), and Spt4/5(Δ666–901) (lane 4) mutant to the −12 probe.
KOW1 Linker of Spt5 Is Dispensable for the Biochemical Activities of Spt4/5
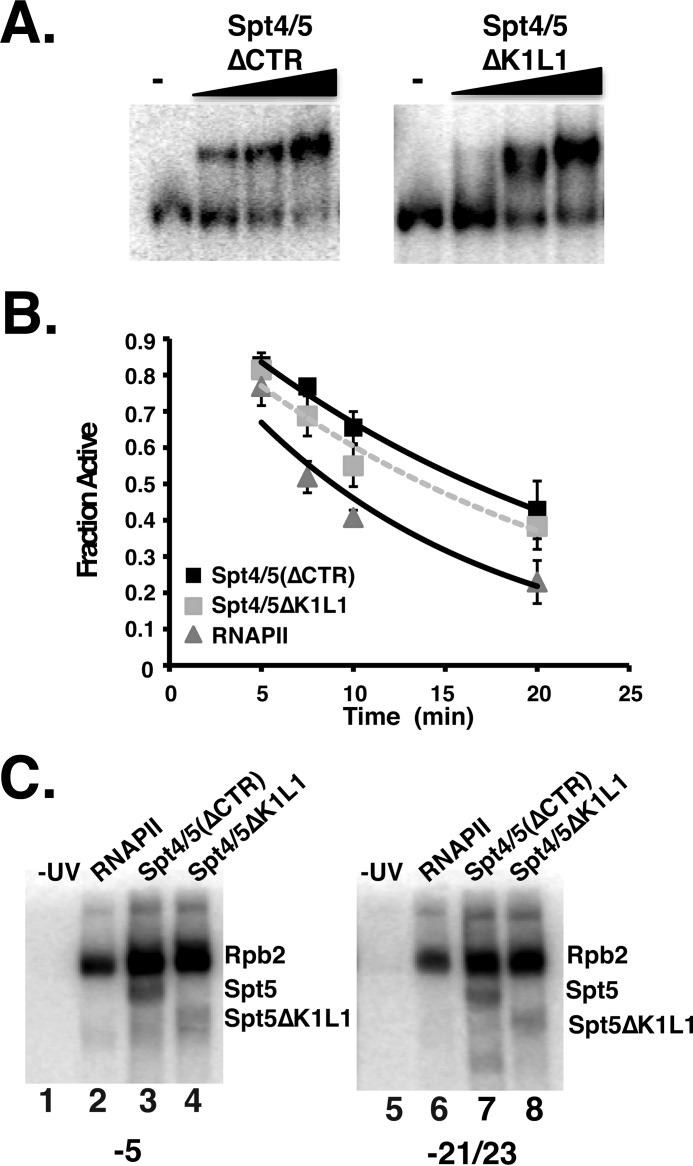
A high resolution x-ray crystal structure of the KOW1-linker (K1L1) region of Spt5 was solved, and this fragment of the protein bound single- and double-stranded nucleic acids (32). However, the importance of this region, which is believed to be the counterpart of the KOW domain in prokaryotic Spt5 homologs, for the biochemical activities of the complex is not known. We examined the association of an Spt4/5 complex containing Spt5 lacking the K1L1 region (ΔK1L1) with ECs. Fig. 8A shows that the mutant complex bound to ECs, and a titration of different amounts of the complex suggests that it has a slightly reduced affinity compared with the intact complex (about 2-fold). Thus, the K1L1 region plays a minor role in the binding of Spt4/5 to RNAPII elongation complexes in vitro.
FIGURE 8.
Conserved KOW1-linker region is dispensable for the biochemical activities of Spt4/5. A, EMSA comparing the binding of Spt4/5(ΔCTR) and Spt4/5ΔK1L1 to ECs. Assays were conducted as described in the legend of Fig. 2B. The Spt4/5ΔK1L1 mutants also have the CTR removed, but it is labeled more simply in the figure. B, RNAPII arrest assay comparing the activity of RNAPII alone or RNAPII plus Spt4/5(ΔCTR) or Spt4/5ΔK1L1. Data are plotted as the fraction of active ECs as a function of time. The data were fit to an exponential decay curve. Error bars represent the standard deviation of three independent experiments. C, DNA photo-cross-linking of Spt4/5 to the −5 (lanes 1–4) and −21/−23 (lanes 5–8) positions.
The anti-arrest activity of the K1L1 mutant was examined using the EC arrest assay described above. The results show that the Spt5 K1L1 mutant can prevent arrest but may not be as effective as the wild type complex (Fig. 8B). The slight reduction in activity, which is reproducible, may be related to the small decrease in binding observed in the EMSA experiments. Nonetheless, although the K1L1 region may be required for optimal interaction with RNAPII ECs, it is not essential for the biochemical activities of Spt4/5 examined here.
One potential function of the K1L1 region in Spt5 is binding to DNA in the transcription scaffold. DNA cross-linking assays were conducted using the Spt4/5(ΔK1L1) mutant under saturated binding conditions. We examined cross-linking within the transcription bubble (−5) and upstream DNA (−21/−23). It was proposed that the K1L1 of Spt5, together with Spt4, “cradles” upstream DNA (32). However, the results show that the K1L1 mutant cross-linked at the −5 and −21/−23 positions (Fig. 8C). This does not rule out the possibility that K1L1 binds to DNA, but deleting it does not affect Spt5 cross-linking in the regions tested here.
Highly Conserved Basic Surface on the NGN Domain of Spt5 Contacts the Non-template Strand and Is Required for Spt4/5 to Prevent RNAPII Arrest
The recent publication of the structure of an RNAPII EC with the complete transcription bubble provided an opportunity to more precisely position Spt4/5 within the RNAPII EC (18). Based on this model, we identified two regions within the NGN domain of Spt5 that contained stretches of positively charged amino acids that could form a DNA-binding surface. One of these surfaces (302–308) is highly conserved in all Spt5 homologs and is predicted to face the transcription bubble (Fig. 9A, green residues), whereas the other (316–321) is not universally conserved and projects away from the NTS (Fig. 9A, orange). Spt4/5 complexes containing Spt5 with alanine substitutions in the basic residues on these surfaces, 302–308A and 316–321A, were purified and analyzed.
FIGURE 9.
Charged surface on the NGN domain is required for the anti-arrest activity of Spt4/5. A, model derived as in Fig. 7B of Spt4/5(NGN) with the transcription bubble. Spt4 is colored in yellow and Spt5-NGN in blue. The region of Spt5-NGN encompassing the conserved basic patch (302–308) is colored in green, and the region Spt5-NGN containing the non-conserved patch (316–321) is colored in orange. B, SDS-PAGE of DNA cross-linking products using a template with a probe at positions −5 (lanes 1–8) and −12 (lanes 9–16). Note, each of the point mutations contained the CTR removed like the “wild type” Spt4/5 but is labeled only to indicate the amino acid changes in the NGN. C, EMSA testing binding of Spt4/5(ΔCTR) and Spt4/5(302–308A) to RNAPII ECs. D, RNAPII arrest assay comparing RNAPII alone and RNAPII plus the Spt4/5 derivatives indicated in the panel. The assay was conducted with saturating amounts of Spt4/5, estimated by the results of the EMSA. Data were plotted as the fraction of active RNAPII as a function of time and fit to an exponential decay curve for comparison. Error bars represent the standard deviation of four independent experiments.
The mutants were subjected to site-specific DNA cross-linking on ECs with 5-iodo-2′-dC at the −5 and −12 positions in the NTS. Consistent with our modeling results, mutating the highly conserved patch of basic residues (302–308) prevented the cross-linking of Spt5 to the NTS, whereas the mutant with substitutions in the non-conserved patch facing away from the transcription bubble (316–321) cross-linked to both positions (Fig. 9B). These results indicate that the NTS binds to this basic surface of the NGN domain of Spt5 and provides stronger evidence that eukaryotic Spt5 binds to DNA in the EC. The 302–308A Spt5 mutant was able to form a complex with Spt4,3 suggesting the mutations did not cause gross structural changes to the NGN domain. However, a trivial explanation for why the Spt4/5(302–308A) derivative failed to cross-link to the NTS is that the mutations prevented the binding of the mutant complex to ECs. However, this was not the case as the mutant complex bound as well as the wild type version (Fig. 9C).
We next wanted to understand the significance of the binding of the NTS to the NGN domain. Based on work on NGN homologs from prokaryotes (60), interaction between the NGN domain and DNA may be necessary for Spt4/5 to prevent arrest of RNAPII. As described above, Spt4/5 prevented the pausing or arrest of RNAPII over the G-tract during prolonged GTP omission (Fig. 9D). However, the Spt4/5(302–308A) mutant lost this activity. Because this mutant associated with RNAPII ECs (Fig. 9C), we conclude that contact between this conserved surface of Spt5 and the NTS is required for Spt4/5 to prevent arrest of RNAPII. The significance of this interaction will be addressed under the “Discussion.”
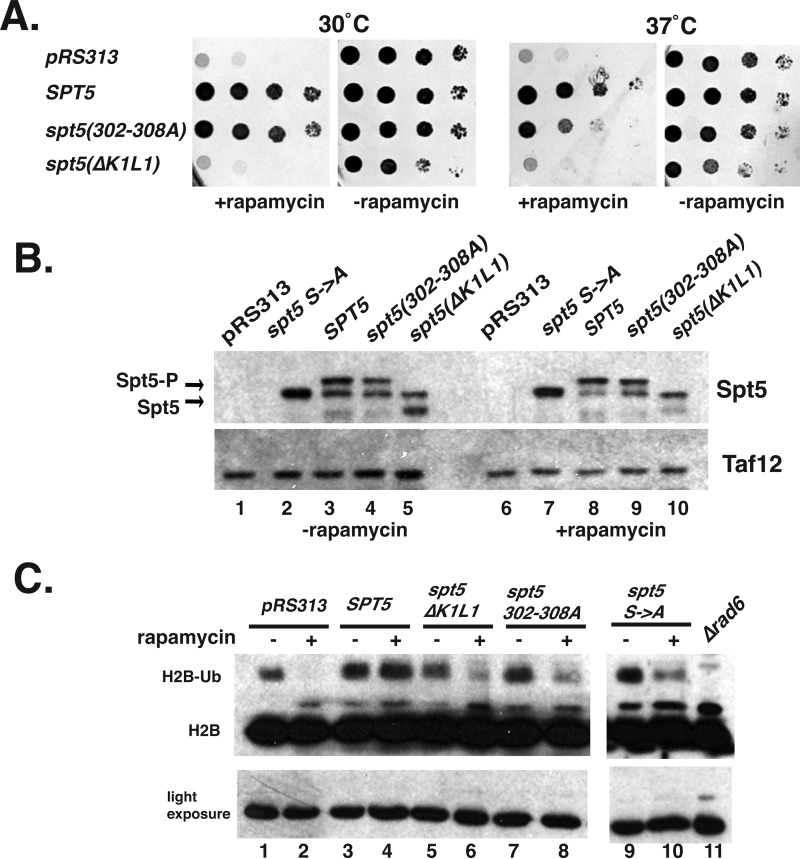
Phenotypic Effects of 302–308A and K1L1 Mutations
SPT5 is an essential gene (27). The ability of the K1L1 deletion and the NGN 302–308A mutant to support viability was examined. To do so, we utilized the “anchor-away” approach (62) to conditionally deplete Spt5 from the nucleus. A strain whose only copy of SPT5 contains an in-frame fusion with the FRB fragment (SPT5-FRB) and expressing an FKBP12 fusion of the ribosomal protein Rpl13 was constructed. Rapamycin induced dimerization of Spt5-FRB, and FKBP12-Rpl13 conditionally sequesters Spt5 in the cytoplasm, and cells transformed with the empty vector (pRS313) were not viable on rapamycin plates (Fig. 10A). However, a plasmid containing a wild type copy SPT5 fully complemented the growth defect (Fig. 10A). The spt5 K1L1Δ mutant did not support viability (Fig. 10A). Interestingly, expression of the K1L1 mutant caused slightly slowed growth of the strain even in the absence of rapamycin. This can be explained by competition between the mutant and the Spt5-FRB protein that supported the functions. Thus, even though this mutant displayed only mild biochemical defects, it could not complement the loss of Spt5 from the nucleus. This suggests it has additional functions in the cell. In contrast, the spt5(302–308A) mutant was viable but displayed temperature-sensitive growth. The viability of this mutant suggests that the essential functions of Spt5 are not impacted by mutation of these residues (see under “Discussion”).
FIGURE 10.
Analysis of the growth properties of Spt5 mutants. A, anchor-away yeast strains (SPT5-FRB) containing either pRS313, pRS313-SPT5-HA3 (WT), pRS313-spt5(302–308A)-HA3, or pRS313-spt5(ΔK1L1)-HA3 were spotted onto synthetic media-His ± rapamycin (1 μg/ml) and incubated at either 30 or 37 °C for 3 days. B, Western blot analysis of yeast extracts from cells grown in the absence or presence of 1 μg/ml rapamycin. Cells were grown to an A600 of 0.7, and then rapamycin was added for 1.5 h. Yeast bearing a plasmid containing an SPT5 mutant with the phosphorylated serine residues changed to alanines (spt5-CTR Ser → Ala) was used to discriminate the phosphorylated from the un-phosphorylated forms of Spt5 in the blot. C, analysis of H2B ubiquitylation. Western blot of cell extracts using an antibody to yeast H2B. A light exposure panel (below) serves as a control for total histone levels. Extracts from a rad6Δ mutant was run on the gel as a control to identify the ubiquitylated form of H2B.
The plasmid copy of the SPT5 derivatives contained an epitope tag (HA) at the C terminus to allow for the measurement of protein levels by Western blotting. When Spt4/5 is recruited to genes, the CTR of Spt5 is phosphorylated by Kin28 and Bur1, thereby changing its migration in gels (36). Analyzing the phosphorylation state of the Spt5 derivatives can provide information on its incorporation into elongation complexes in vivo. Spt5 migrated as two species, a lower un-phosphorylated form and a slower migrating phosphorylated form (Fig. 10B). Previous studies confirmed that the upper species is phosphorylated Spt5 (36, 37, 63). As a control, extracts from cells expressing a version of Spt5 with all the phosphorylation sites in the CTR (CTR Ser → Ala) mutated were included to help identify the un-phosphorylated form of Spt5 in gels (37). A few interesting things were noted. First, when rapamycin was added the proportion of the phosphorylated species increased. This makes sense because as the Spt5-FRB leaves the nucleus, it no longer competes with the plasmid copy of Spt5. This increases the proportion of Spt5-HA recruited to genes where it can become phosphorylated. Second, the mutants were expressed to similar levels as the wild type cells. Finally, both mutants were phosphorylated. This suggests that these mutants are incorporated into ECs in vivo. This is consistent with the data showing that both mutants bound to RNAPII ECs in vitro (Figs. 8 and 9).
The phosphorylation of the CTR of Spt5 recruits Paf1c (RNA polymerase-associated factor 1), which in turn leads to the co-transcriptional ubiquitylation of histone H2B by Rad6 (35, 63–65). H2B ubiquitylation is a mark of active transcription elongation and is used as an indicator of ongoing transcription elongation on a global scale. Ubiquitylated H2B appears as a slower migrating species in Western blots that is dependent on the RAD6 gene. The Spt5 anchor-away strain containing plasmid copies of wild type or Spt5 mutants were grown to log phase and then rapamycin was added to half of the culture. Cell extracts were prepared after 90 min and analyzed by Western blotting using an antibody to H2B (Fig. 10C). Treating cells transformed with an empty vector (pRS313) with rapamycin caused a dramatic reduction of H2B ubiquitylation (Fig. 10C, lanes 1 versus 2), but introducing a plasmid containing a wild type copy of SPT5 fully restored H2B ubiquitylation levels. However, the 302–308A and the ΔK1L1 mutants only partially complemented the loss of H2B ubiquitylation (Fig. 10C, lanes 5 versus 6 and lanes 7 versus 8). The reduced H2B ubiquitylation suggests that transcription elongation is impaired in these SPT5 mutants and that the basic patch in the NGN of Spt5(302–308) contributes to elongation in vivo. As controls, a strain expressing an Spt5 derivative where all of the serines in the CTR were changed to alanines (spt5 Ser → Ala) was analyzed. As expected, this mutant caused reduced H2B ubiquitylation (35, 63).
Discussion
Changes occur to RNAPII as it transitions from the pre-initiation complex into a productive elongation complex. One such change is its association with elongation factors, including the primordial elongation factor Spt4/5 (66). The recruitment of DSIF (metazoan Spt4/5 complex) requires a minimal length of mRNA emerging from RNAPII, and this event is thought to be among the earliest changes in the elongation complex during the transition from initiation to elongation into the body of genes. Although it has been shown that Spt5 can bind across the jaw and the clamp of archaeal RNAP, which may close the clamp to encircle DNA to enhance processivity (67), little was known about how the binding of Spt4/5 to the nucleic acid scaffold in ECs affects elongation. The interaction between Spt5 and upstream DNA was predicted from structural modeling studies based on x-ray crystal structures and high resolution cryo-EM of Spt5 homologs (8, 10); however, prior to our study no direct evidence for the binding of Spt5 to the DNA has been presented nor has the significance of these interactions been tested. Here, we used a defined, highly purified system to demonstrate that a surface on the NGN domain of Spt5 contacts the non-template strand in the transcription bubble and that this interaction is important for Spt4/5 to promote elongation by reducing arrest or pausing of RNAPII.
Importance of Spt5-DNA Interactions
It has been shown that RfaH, a NusG homolog, and NusG from eubacteria interact with the NTS of DNA in a sequence-specific manner, where it controls pausing of RNAP (59, 60). Whether or not the interaction between NGN domains of Spt5 and the non-template strand is important for function was unknown, especially because sequence-specific pausing is not known to occur in eukaryotes. We used site-specific photoreactive DNA cross-linking to show for the first time that a highly conserved basic patch on the NGN domain of Spt5 contacts the NTS in the transcription bubble. Contact between the NGN domain of eukaryotic Spt5 and the NTS strongly suggests that this function is conserved throughout evolution, and we propose that it may be a fundamental requirement for Spt5 homologs to regulate RNAP elongation. Recently, Spt4/5 from the archaea Methanocaldococcus jannaschii was shown to bind to double-stranded DNA. They described a region of the NGN domain as containing an alkaline surface that is responsible for binding to free DNA that overlaps with residues 302–308 of yeast Spt5 (68). Additionally, mutations of basic amino acids in this region affect the activity of RfaH (69). Thus, the DNA binding properties of the NGN domain and that of its paralog appear to be conserved.
It has been speculated that Spt4/5 plays a role in stabilizing the upstream duplex, assisting in bubble closure (8, 10, 18, 32). Until recently, DNA upstream of the RNAPII elongation complex was a virtual unknown, which lacked structural definition. This led many to view it as a semi-stable region of duplexed DNA, which could be stabilized by Spt4/5. Recent crystallization of the RNAPII complete transcription bubble seems to indicate all of the structural requirements to promote strand re-annealing is located within Rpb1 and Rpb2. In particular, the wedge/hairpin loop within Rpb2 was shown to contact the upstream duplex and likely works with “the arch,” composed of the rudder and FL1 residues, to close the bubble. Mutations in the tip of the loop reduce transcription elongation rates in vitro, and the mutants display a number of phenotypes consistent with impaired elongation in cells (18). Clearly, RNAPII can close the bubble on its own without Spt4/5, and our permanganate footprinting experiment failed to detect changes in the size and/or closure of the transcription bubble in the presence of Spt4/5 (Fig. 6B). However, as stated under the “Results,” such experiments can only provide a static picture of the bubble. Whether or not Spt4/5 stabilizes upstream of DNA during the dynamic act of transcription remains to be tested.
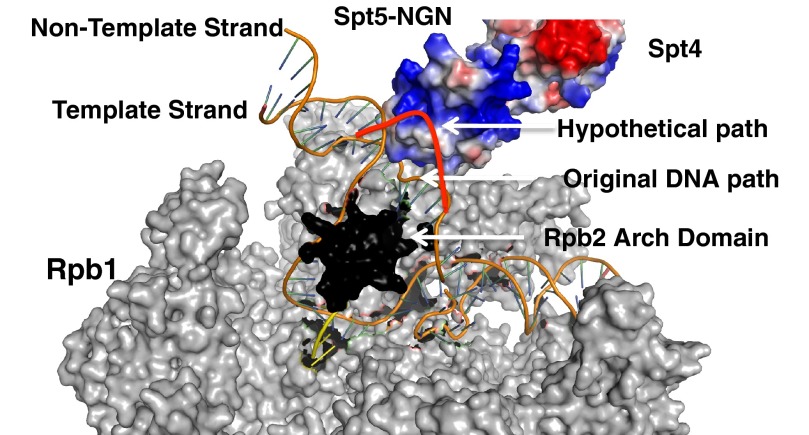
So what is the role of the interaction between the basic patch on the NGN domain of Spt5 and the non-template strand? We propose a modification of the bubble closure models proposed by others, called the bubble chaperone mechanism. In RNAPII, the NTS passes over the arch (rudder and FL1) and rejoins the template strand behind it (18). The basic patch on the NGN provides an additional surface to guide the NTS around the arch, in essence “taming” the NTS to ensure it proceeds along the most productive path to rejoin the NTS (Fig. 11). Additionally, the binding of the NGN domain to the NTS could also prevent the association of the NTS with surfaces on the wall or arch that stabilize a paused/arrested state, thus reducing the pausing/arrest of RNAPII. This could explain why mutating the residues on the basic patch (302–308) of the NGN abolished the anti-arrest properties of Spt4/5. This model also suggests a role for Spt4/5 beyond encircling the DNA and closing the clamp and suggests that Spt4/5 plays a more extensive role in promoting elongation.
FIGURE 11.
Bubble chaperone model of Spt4/5 action on ECs. A model illustrating the bubble chaperone mechanism. Model was generated by overlaying crystal structures of the archaeal clamp fused to archaeal Spt5 (data not shown, PDB code 3QQC), RNAPII with template and non-template strand (PDB code 5C4X) (18), and yeast Spt4/5 (PDB code 2EXU) (74). Spt4 and Spt5 are shown in electrostatic space-filling mode. Gray indicates all subunits of RNAPII except for Rpb2. Rpb2 was omitted from this structure except the Arch domain, which is shown in black. The template and non-template strand are shown as orange, and the RNA is in yellow. Hypothetical DNA path in the presence of Spt4/5 was drawn manually in red over the existing structure.
Modeling, footprinting, and cross-linking data suggest Spt4/5 contacts DNA as it exits RNAPII, so binding to the NTS strand in the transcription bubble is one part of the equation. Contact between Spt5 and upstream DNA may stabilize the association of RNAPII with DNA. A recent crystal structure of the KOW1-Linker1 (K1L1) region of yeast Spt5 identified a nucleic acid interaction surface, called the positively charged patch (32). The authors proposed, based on genetic evidence that showed mutations in the positively charged patch of K1L1 are synthetically lethal when Spt4 was absent, that the K1L1 wraps up along the DNA on the opposite side of the helix where Spt4 is positioned to act like pinchers in a claw to stabilize the interaction of Spt4/5 in the EC (32). We found that deleting the entire K1L1 region had a modest ∼2-fold effect on the binding of the mutant Spt4/5 complex to ECs (Fig. 8A), which generally supports a role for this domain in stabilizing the interaction of Spt4/5 with elongation complexes. Surprisingly, this mutant displayed very little reduction in anti-pausing/arrest activity and still interacted with the NTS in the transcription bubble. Thus, its appears that the K1L1 is dispensable for most known biochemical activities of the Spt4/5 complex. Interestingly, the ΔK1L1 mutant is expressed and phosphorylated in vivo but is unable to support viability (Fig. 10A). These results suggest that the K1L1 has functions in cells beyond preventing RNAPII arrest. Further characterization of the ΔK1L1 mutant in vivo would be interesting but is beyond the scope of our study. Compare that with the NGN(302–308A) mutant, which could support viability. The ability of the SPT5(302–308A) mutant to support viability seems at odds with its inability to prevent arrest in vitro. However, most bona fide elongation factors are non-essential in yeast; SPT5 is the exception because it participates in a number of essential functions such as splicing, capping, and ribosomal RNA production (5, 34, 38, 70, 71). The elongation promoting activity of Spt4/5 may not be an essential function for cell survival under idealized growth conditions, but it imparts a competitive advantage to the organism or is more important during stress. The latter explanation would be consistent with the temperature-sensitive phenotype of the SPT5(302–308A) mutant and the H2B ubiquitylation defects observed in both Spt4/5 mutants.
Extended Regions of Eukaryotic Spt5 May Coordinate Changes in RNAPII Structure
We provide evidence that contact between the NGN and the NTS is important for the activity of Spt4/5. However, this is not likely the only way Spt4/5 promotes elongation. NusG and archaeal Spt5 completely bridge the two lobes of RNAP, interacting with the clamp coil of Rpb1/β′ and the β gate loop of β and RpoB. Mutation of these surfaces compromises the activity of NusG/Spt5 (7, 20). From this study and other data, a model has been proposed that NusG/Spt5 acts to reduce “fluttering” of the two subunits, which in turn controls the movement of the RNAP trigger loop (12, 13, 17). NusG acts to stabilizes RNAP in a post-translocated state (11). Likewise, the binding of Spt4/5 to the clamp could affect structures such as the trigger loop or increase processivity by binding across the jaws of RNAPII.
In the case of prokaryotes, the NGN domain is sufficient for association with the elongation complex and controlling elongation activity (7, 72). Our data, and that of others (38), suggest that this is not the case in eukaryotes and that additional domains in Spt5 are required for its association with RNAPII. Elegant in vivo site-specific cross-linking studies found that a region of Spt5 containing KOW domains 4 and 5 cross-linked to Rpb4/7, the stalk, of RNAPII (19, 58, 73). Our biochemical data support the important role that KOW domains 4 and 5, and other regions in the C terminus, play in the binding of Spt4/5 to RNAPII (Fig. 4). It is interesting that KOW domains 4 and 5 contact Rpb4/7, because Spt5 cross-links to RNA near the exit channel (this study), and the binding of the emerging transcript to Rpb7 has been observed (58, 73). Interactions between Spt4/5 and ECs may be stabilized through contact with Rpb4/7 and the emerging transcript. Because these regions are physically linked to the NGN domain, this could allow “communication” between the clamp, the stalk of RNAPII, and the emerging transcript.
Spt5 Links Different Features of the EC Complex to the Elongation Process
Our data show that Spt4/5 makes contact with several different structural features of the RNAPII elongation complex required for it to bind to ECs and prevent arrest. It could simply be that Spt4/5 requires additional binding surfaces contributed by the eukaryotic specific regions of Spt5 to stabilize its interaction with elongating RNAPII as it navigates the chromatin barrier or as different co-transcriptional RNA and chromatin-modifying factors are exchanged during elongation. A more interesting idea is that Spt5 serves as a sensor of multiple steps in the transcription process. Spt5 contacts upstream DNA, the transcription bubble, the clamp, the Rpb4/7 module, and the emerging transcript. Spt4/5 may coordinate the translocation of RNAPII with the growth of the emerging transcript, the movement of the Rpb4/7 and clamp, and the tracking of the DNA in the transcription bubble. Conceptually, the ability of the Spt4/5 complex to interact with all of the “moving parts” of the ECs (nucleic acids and RNAPII) provides a mechanical rationale for Spt4/5 to stabilize the RNAPII ground state after each translocation event during elongation. This could prevent arrest by preventing non-productive, uncoordinated movements among the different structures that would cause arrest.
Author Contributions
J. B. C. and J. C. R. designed the studies and wrote the manuscript. J. B. C. performed the experiments and analyzed the data. J. F. provided RNAPII, edited the manuscript, and shared unpublished data.
Acknowledgments
Alan Hinnebusch, Steve Hahn, and Song Tan are recognized for providing plasmids used in this study. Dave Gilmour is acknowledged for input during the course of the work and reading the manuscript. The members of the Center for Eukaryotic Gene Regulation are recognized for their comments and feedback during the completion of this work.
This work was supported by National Institutes of Health Grants GM058672 (to J. C. R.) and GM100997 (to J. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.
J. B. Crickard and J. C. Reese, unpublished results.
- RNAP
- RNA polymerase
- RNAPII
- RNA polymerase II
- RNAPI
- RNA polymerase I
- CTD
- C-terminal region of Rpb1
- TFIIS
- transcription factor II S
- EC
- elongation complex
- CTR
- C-terminal region of Spt5
- KOW
- Kyrpides, Ouzounis, Woese
- NGN
- NusG N terminus
- DSIF
- 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor
- NTS
- non-template strand
- FRB
- FKB12-rapamycin-binding domain of human mTor
- NusG
- N utilization substance G
- H2B
- histone 2B
- ExoIII
- exonuclease III
- oligo
- oligonucleotide
- PDB
- Protein Data Bank.
References
- 1. Werner F. (2012) A nexus for gene expression—molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 417, 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartzog G. A., and Fu J. (2013) The Spt4–Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta 1829, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rondón A. G., García-Rubio M., González-Barrera S., and Aguilera A. (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ding B., LeJeune D., and Li S. (2010) The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J. Biol. Chem. 285, 5317–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lindstrom D. L., Squazzo S. L., Muster N., Burckin T. A., Wachter K. C., Emigh C. A., McCleery J. A., Yates J. R. 3rd, and Hartzog G. A. (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23, 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herbert K. M., Zhou J., Mooney R. A., Porta A. L., Landick R., and Block S. M. (2010) E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 399, 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirtreiter A., Damsma G. E., Cheung A. C., Klose D., Grohmann D., Vojnic E., Martin A. C., Cramer P., and Werner F. (2010) Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 38, 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klein B. J., Bose D., Baker K. J., Yusoff Z. M., Zhang X., and Murakami K. S. (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc. Natl. Acad. Sci. U.S.A. 108, 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., and Handa H. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez-Rucobo F. W., Sainsbury S., Cheung A. C., and Cramer P. (2011) Architecture of the RNA polymerase–Spt4/5 complex and basis of universal transcription processivity. EMBO J. 30, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bar-Nahum G., Epshtein V., Ruckenstein A. E., Rafikov R., Mustaev A., and Nudler E. (2005) A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 [DOI] [PubMed] [Google Scholar]
- 12. Nayak D., Voss M., Windgassen T., Mooney R. A., and Landick R. (2013) Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol. Cell 50, 882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toulokhonov I., Zhang J., Palangat M., and Landick R. (2007) A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419 [DOI] [PubMed] [Google Scholar]
- 14. Cheung A. C., and Cramer P. (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 [DOI] [PubMed] [Google Scholar]
- 15. Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., and Kornberg R. D. (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hein P. P., Kolb K. E., Windgassen T., Bellecourt M. J., Darst S. A., Mooney R. A., and Landick R. (2014) RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol. 21, 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekine S., Murayama Y., Svetlov V., Nudler E., and Yokoyama S. (2015) The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol. Cell 57, 408–421 [DOI] [PubMed] [Google Scholar]
- 18. Barnes C. O., Calero M., Malik I., Graham B. W., Spahr H., Lin G., Cohen A. E., Brown I. S., Zhang Q., Pullara F., Trakselis M. A., Kaplan C. D., and Calero G. (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol. Cell 59, 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li W., Giles C., and Li S. (2014) Insights into how Spt5 functions in transcription elongation and repressing transcription coupled DNA repair. Nucleic Acids Res. 42, 7069–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sevostyanova A., Belogurov G. A., Mooney R. A., Landick R., and Artsimovitch I. (2011) The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell 43, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamaguchi Y., Wada T., Watanabe D., Takagi T., Hasegawa J., and Handa H. (1999) Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274, 8085–8092 [DOI] [PubMed] [Google Scholar]
- 22. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., and Handa H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 [DOI] [PubMed] [Google Scholar]
- 23. Wada T., Takagi T., Yamaguchi Y., Watanabe D., and Handa H. (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17, 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bourgeois C. F., Kim Y. K., Churcher M. J., West M. J., and Karn J. (2002) Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22, 1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., and Rosenthal A. (2000) A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408, 366–369 [DOI] [PubMed] [Google Scholar]
- 26. Mason P. B., and Struhl K. (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17, 831–840 [DOI] [PubMed] [Google Scholar]
- 27. Swanson M. S., Malone E. A., and Winston F. (1991) SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11, 3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Missra A., and Gilmour D. S. (2010) Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl. Acad. Sci. U.S.A. 107, 11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng B., and Price D. H. (2008) Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 36, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lasko P. (2010) Tudor domain. Curr. Biol. 20, R666–R667 [DOI] [PubMed] [Google Scholar]
- 31. Charier G., Couprie J., Alpha-Bazin B., Meyer V., Quéméneur E., Guérois R., Callebaut I., Gilquin B., and Zinn-Justin S. (2004) The tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure 12, 1551–1562 [DOI] [PubMed] [Google Scholar]
- 32. Meyer P. A., Li S., Zhang M., Yamada K., Takagi Y., Hartzog G. A., and Fu J. (2015) Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol. Cell. Biol. 35, 3354–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wier A. D., Mayekar M. K., Héroux A., Arndt K. M., and VanDemark A. P. (2013) Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc. Natl. Acad. Sci. U.S.A. 110, 17290–17295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayer A., Schreieck A., Lidschreiber M., Leike K., Martin D. E., and Cramer P. (2012) The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol. Cell. Biol. 32, 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mayekar M. K., Gardner R. G., and Arndt K. M. (2013) The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol. Cell. Biol. 33, 3259–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., and Hahn S. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qiu H., Hu C., Gaur N. A., and Hinnebusch A. G. (2012) Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 31, 3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viktorovskaya O. V., Appling F. D., and Schneider D. A. (2011) Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J. Biol. Chem. 286, 18825–18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan S., Kern R. C., and Selleck W. (2005) The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 [DOI] [PubMed] [Google Scholar]
- 40. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 41. Cox J. S., Chapman R. E., and Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suh M.-H., Ye P., Zhang M., Hausmann S., Shuman S., Gnatt A. L., and Fu J. (2005) Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc. Natl. Acad. Sci. U.S.A. 102, 17314–17319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kruk J. A., Dutta A., Fu J., Gilmour D. S., and Reese J. C. (2011) The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes Dev. 25, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Z., Fu J., and Gilmour D. S. (2005) CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 19, 1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dutta A., Babbarwal V., Fu J., Brunke-Reese D., Libert D. M., Willis J., and Reese J. C. (2015) Ccr4-Not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol. Cell. Biol. 35, 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babbarwal V., Fu J., and Reese J. C. (2014) The Rpb4/7 of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-Not) complex to promote elongation. J. Biol. Chem. 289, 33125–33130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dedrick R. L., and Chamberlin M. J. (1985) Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry 24, 2245–2253 [DOI] [PubMed] [Google Scholar]
- 48. Kadesch T. R., and Chamberlin M. J. (1982) Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem. 257, 5286–5295 [PubMed] [Google Scholar]
- 49. Sluder A. E., Price D. H., and Greenleaf A. L. (1988) Elongation by Drosophila RNA polymerase II. Transcription of 3′-extended DNA templates. J. Biol. Chem. 263, 9917–9925 [PubMed] [Google Scholar]
- 50. Kireeva M. L., Komissarova N., Waugh D. S., and Kashlev M. (2000) The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275, 6530–6536 [DOI] [PubMed] [Google Scholar]
- 51. Saeki H., and Svejstrup J. Q. (2009) Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell 35, 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reines D., Chamberlin M. J., and Kane C. M. (1989) Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 264, 10799–10809 [PubMed] [Google Scholar]
- 53. Komissarova N., and Kashlev M. (1998) Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 95, 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gu W., Wind M., and Reines D. (1996) Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl. Acad. Sci. U.S.A. 93, 6935–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu W., Wada T., Okabe S., Taneda T., Yamaguchi Y., and Handa H. (2007) DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 35, 4064–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim D.-K., Inukai N., Yamada T., Furuya A., Sato H., Yamaguchi Y., Wada T., and Handa H. (2003) Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells 8, 371–378 [DOI] [PubMed] [Google Scholar]
- 57. Burova E., Hung S. C., Sagitov V., Stitt B. L., and Gottesman M. E. (1995) Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J. Bacteriol. 177, 1388–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ujvári A., and Luse D. S. (2006) RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 13, 49–54 [DOI] [PubMed] [Google Scholar]
- 59. Yakhnin A. V., Murakami K. S., and Babitzke P. (2016) NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J. Biol. Chem. 291, 5299–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Artsimovitch I., and Landick R. (2002) The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203 [DOI] [PubMed] [Google Scholar]
- 61. Buratowski S., Hahn S., Guarente L., and Sharp P. A. (1989) Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561 [DOI] [PubMed] [Google Scholar]
- 62. Haruki H., Nishikawa J., and Laemmli U. K. (2008) The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31, 925–932 [DOI] [PubMed] [Google Scholar]
- 63. Zhou K., Kuo W. H., Fillingham J., and Greenblatt J. F. (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 106, 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiao T., Kao C.-F., Krogan N. J., Sun Z.-W., Greenblatt J. F., Osley M. A., and Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., and Greenblatt J. F. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., and Cramer P. (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 67. Hartzog G. A., and Kaplan C. D. (2011) Competing for the clamp: promoting RNA polymerase processivity and managing the transition from initiation to elongation. Mol. Cell 43, 161–163 [DOI] [PubMed] [Google Scholar]
- 68. Guo G., Gao Y., Zhu Z., Zhao D., Liu Z., Zhou H., Niu L., and Teng M. (2015) Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII. J. Struct. Biol. 192, 418–425 [DOI] [PubMed] [Google Scholar]
- 69. Belogurov G. A., Sevostyanova A., Svetlov V., and Artsimovitch I. (2010) Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol. 76, 286–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pei Y., and Shuman S. (2002) Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277, 19639–19648 [DOI] [PubMed] [Google Scholar]
- 71. Anderson S. J., Sikes M. L., Zhang Y., French S. L., Salgia S., Beyer A. L., Nomura M., and Schneider D. A. (2011) The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J. Biol. Chem. 286, 18816–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sevostyanova A., and Artsimovitch I. (2010) Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 38, 7432–7445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Andrecka J., Lewis R., Brückner F., Lehmann E., Cramer P., and Michaelis J. (2008) Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 105, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guo M., Xu F., Yamada J., Egelhofer T., Gao Y., Hartzog G. A., Teng M., and Niu L. (2008) Core structure of the yeast Spt4-Spt5 complex: a conserved module for regulation of transcription elongation. Structure 16, 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]