Abstract
The nuclear hormone receptor (NHR) superfamily is composed of a wide range of receptors involved in a myriad of important biological processes, including development, growth, metabolism, and maintenance. Regulation of such wide variety of functions requires a complex system of gene regulation that includes interaction with transcription factors, chromatin-modifying complex, and the proper recognition of ligands. NHRs are able to coordinate the expression of genes in numerous pathways simultaneously. This review focuses on the role of nuclear receptors in the central nervous system and, in particular, their role in regulating the proper development and function of the brain and the eye. In addition, the review highlights the impact of mutations in NHRs on a spectrum of human diseases from autism to retinal degeneration.
Keywords: central nervous system, nuclear hormone receptors, regulation, brain, eye, human disease
Introduction
The nuclear hormone receptor (NHR) superfamily is a group of transcriptional regulators that play a key role in numerous pathways, including development, growth, metabolism, and maintenance. These receptors belong to a large family of genes that regulate gene expression. NHRs are activated by ligands, such as hormones and vitamins, and a subset of these NHRs fall under the category of orphan receptors with unidentified ligands. NHRs share a common structure characterized by dimeric zinc fingers that can bind to a lipid-soluble hormone and form a complex with other proteins, such as cofactors and histones, to regulate gene expression. NHRs modulate numerous processes, such as central nervous system (CNS) development, homeostasis, reproduction, differentiation, metabolism, circadian functions, steroidogenesis, cell differentiation, and lipid metabolism.1,2 The goal of this review is to discuss the role of NHRs in the development of the CNS, with an emphasis on their function in the brain and the eye and their impact on human disease.
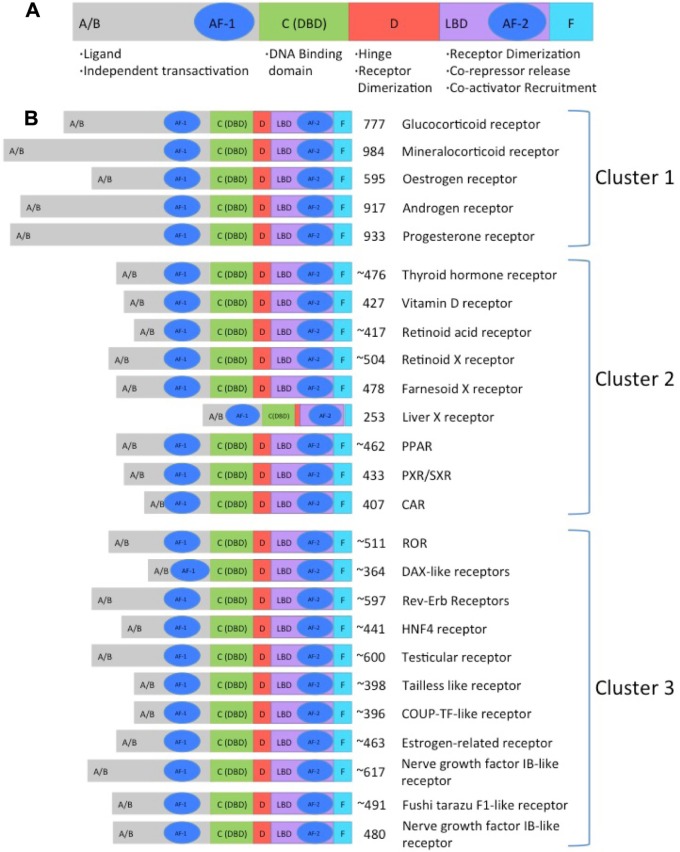
While being involved in a wide range of functions, NHRs have very conserved structure of functional domains (Fig. 1A). There are two activation function (AF) domains that bind to DNA. The variable NH2 terminal (A/B) contains an activation function AF-1 domain, and the AF-2 domain is found in the E domain also known as the ligand-binding domain (LBD). Modifying the A/B domain produces different isoforms of the receptors. A conserved DNA-binding domain (C) binds to specific DNA sequences known as the hormone response element (HREs). In addition, the DNA-binding domain is composed of two zinc fingers and is the most conserved region of the NHR. A P-box region is found on the first zinc finger and functions to recognize the half-site of a response element. The second zinc finger contains a D-box, which mediates receptor dimerization. The most variable and unique sequence of NHRs is the linker region (D), which aids in the conformational changes of the binding ligands. Finally, there is a conserved region with LBD (E) that contains the activation function AF-2. Some receptors also include a COOH-terminal region (F) that aids in the folding of the receptor into a transcriptionally active form.3–5
Figure 1.
Schematic representation of the functional domains of NHRs in each classification cluster. The A/B domain is the most variable region, and it contains the activation function-1 (AF-1) site. The activation functions AF-1 and -2 are important in the regulation of receptor transcriptional activity. The C domain is one of the most highly conserved regions across the superfamily on nuclear receptors and consists of the DNA-binding region. The D domain, also known as the hinge region, is involved in the conformational changes that occur after ligand binding allowing the coactivators or corepressors to bind. LBD is the second most conserved region and is in charge of the recognition and binding of the ligands. The F region is a carboxyl terminal that is not present in all receptors.2,4,73
Regulation
NHRs are involved in a wide spectrum of functions and are key components of a great number of signaling pathways. Nuclear receptors have the ability to enhance or repress the expression of a gene in response to changes in the environment or to endocrine signals. When a ligand binds to its receptor, it alters the receptor activity and thus the expression of entire gene networks. However, ligands are not the only mechanism through which receptors can regulate genes. A set of proteins known as coregulators also interact with the receptors to regulate gene expression. Coregulators facilitate or truncate the receptors’ ability to bind to the transcriptional complex as well as the promoter.6 Receptors use several mechanisms such as interaction with the HREs, coregulators, interaction with the transcription factor complex, and histone modification to finely orchestrate an appropriate gene expression.6 A collection of HREs and coregulators and corepressors for each nuclear receptor is given in Table 1.
Table 1.
Hormone response elements’ (HREs) sequences and their coactivators and cosuppressors.
| NUCLEAR HORMONE RECEPTOR | HRE SEQUENCE | RESPONSE ELEMENT | CO-ACTIVATORS | CO-SUPPRESORS |
|---|---|---|---|---|
| Thyroid hormone receptor alpha (NR1A1) | AGGTCA | NCOA1237 | NCOR1238 | |
| NCOA2239 | NCOR2240 | |||
| NCOA3241 | ||||
| MED1242 | ||||
| Thyroid hormone receptor beta (NR1A2) | AGGTCA | NCOA1237 | NCOR1240 | |
| NCOA2239 | ||||
| NCOA3241 | ||||
| Vitamin D receptor | Not known | Not known | ||
| Retinoid acid receptor alpha (NR1B1) | 5′-PuG(G/T)TCA-3′ | Heterodimer | CREBBP243 | NCOR122,238,240,244 |
| EP300243,245 | NCOR2246 | |||
| MED1247 | ||||
| NCOA1248,249 | ||||
| NCOA2250 | ||||
| NCOA3241 | ||||
| Retinoid acid receptor beta (NR1B2) | 5′-PuG(G/T)TCA | Heterodimer | MED122,251–253 | |
| NCOA1248,254,255 | ||||
| NCOA2250 | ||||
| NCOA3241,254,256 | ||||
| Retinoid acid receptor gamma (NR1B3) | 5′-PuG(G/T)TCA-3′ | Heterodimer | NCOA122,248,255 | NCOR122,240,246,254 |
| NCOA222,250,257 | NCOR2238 | |||
| NCOA3241,254,256 | ||||
| Retinoid X alpha (NR2B1) | 5′-AGGTCA | Homodimer, Heterodimer | CREBBP258,259 | |
| EP300258 | ||||
| MED122,253 | ||||
| NCOA122,248 | ||||
| NCOA2250,257 | ||||
| NCOA322,241 | ||||
| PPARGC1A260 | ||||
| TAF11261 | ||||
| TAF422,262,263 | ||||
| TBP22,262,263 | ||||
| Retinoid X beta (NR2B2) | 5′-AGGTCA | Homodimer, Heterodimer | NCOA122,248,255 | |
| NCOA2250,255,257 | ||||
| NCOA322 | ||||
| PNR (NR2E3) | AAGTCA n AAGTCA | Homodimer | CRX171 | |
| Retinoid X gamma (NR3B3) | 5′-AGGTCA | Homodimer, Heterodimer | NCOA122,248,255 | |
| NCOA2250,255,257 | ||||
| NCOA322 | ||||
| Farnesoid X receptor (NR1H4) | AGTTCAnTGAACT | CARM1264 | ||
| MED1265 | ||||
| NCOA1266,267 | ||||
| PPARGC1A268–270 | ||||
| PRMT1271 | ||||
| TRRAP272 | ||||
| Farnesoid X receptor beta (NR1H5) | AGTTCA N TGAACT | Heterodimer | NCOA1273 | |
| Liver X receptor alpha (NR1H3) | AGGTCANNNNAGGTCA | EP300274 | NCOR1275 | |
| NCOA1274 | NCOR2275 | |||
| NCOA2276 | ||||
| PPARGC1A277 | ||||
| PPARGC1B278 | ||||
| TRRAP272 | ||||
| Liver X receptor beta (NR1H2) | AGGTCANNNNAGGTCA | EP300274 | ||
| NCOA1279 | ||||
| NCOR1275 | ||||
| PPAR alpha (NR1C1) | 5′-AACTAGGNCA A AGGTCA-3′ | Heterodimer | CITED2280 | NCOR1281–283 |
| CREBBP284,285 | NRIP1286–288 | |||
| HADHA289 | ||||
| MED1290 | ||||
| NCOA1291 | ||||
| NCOA3292 | ||||
| NCOA6293,294 | ||||
| PPARGC1A295 | ||||
| PPARGC1B295 | ||||
| SMARCA2296 | ||||
| PPAR beta (NR1C2) | NCOA1297 | NCOR1298–300 | ||
| NCOA3292 | NCOR2298,301 | |||
| NCOA6293 | ||||
| PPARGC1A302 | ||||
| PPAR gamma (NR1C3) | 5′-AACTAGGNCA A AGGTCA-3′ | Heterodimer | CITED2280 | WWTR1303 |
| CREBBP285,304 | SAFB305 | |||
| EP300306 | NRIP1307 | |||
| MED1308–310 | NCOR2298,301 | |||
| NCOA1285,297,310 | NCOR1281,300,301 | |||
| NCOA2311 | ||||
| NCOA3392 | ||||
| NCOA4312 | ||||
| NCOA6294,313 | ||||
| NCOA7314 | ||||
| PPARGC1A315–319 | ||||
| PPARGC1B317–321 | ||||
| PRMT2322 | ||||
| SCAND1323 | ||||
| SMARCA1324,325 | ||||
| TGS1310 | ||||
| PXR/SXR receptor (NR1I2) | AGGTCA | Heterodimer | FOXO1326 | NCOR2327,328 |
| GRIP1329 | NR0B2330 | |||
| NCOA1331 | ||||
| NRIP1332 | ||||
| PPARGC1A333 | ||||
| CAR (NR1I3) | AGGTCA | Heterodimer | MED1334 | |
| NCOA1335 | ||||
| PPARGC1A336 | ||||
| ROR alpha (NR1F1) | T/A A/T T/A C A/T A/GGGTCA | Monomer, Homodimer | EP300337 | HR338 |
| NCOA2339 | NCOR1340 | |||
| MED1339 | NCOR2340 | |||
| ROR beta (NR1F2) | T/A A/T T/A C A/T A/GGGTCA | NCOR1341 | NCOR1342 | |
| NRIP2343 | ||||
| ROR gamma (NR1F3) | AGGTCA nnnnn AGGTCA | NCOA1344 | HR342 | |
| DAS-like receptors | Not known | Not known | ||
| Rev-Erb alpha (NR1D1) | A/T A A/T N T PuGGTCA | NCOA5345 | C1D176 | |
| HDAC3346 | ||||
| NCOA5345 | ||||
| NCOR1347 | ||||
| Rev-Erb beta (NR1D2) | A/T A A/T N T PuGGTCA | NCOA5345 | NCOA5345 | |
| NCOR1347,348 | ||||
| HNF4 alpha (NR2A1) | CREBBP349–351 | NCOR2352 | ||
| GRIP1349,350 | ||||
| MED1353,354 | ||||
| PPARGC1A355,356 | ||||
| PPARGC1B356,357 | ||||
| HNF4 gamma (NR2A2) | AGGTCA n AGGTCA | Not known | Not known | |
| Testicular receptor 2 (NR2C1) | AGGTCA n AGGTCA | Monomer, Homodimer | HDAC3358 | |
| HDAC4358 | ||||
| NRIP1359 | ||||
| Testicular receptor 4 (NR C2) 2 | Monomer, Homodimer, Heterodimer | JAZF 1360 | ||
| NR2C2AP361 | ||||
| Tailless like receptor (NR2E1 and NR2E3) | AAGTCA n AAGTCA | Not known | Not known | |
| COUP-TF1 (NR2F1) | AGGTCA n AGGTCA | CREBBP362 | BCL11A363 | |
| NCOA1364 | BCL11B363 | |||
| NCOR1365 | ||||
| NCOR2365 | ||||
| COUP-TF2 (NR2F2) | A/GGGTCA n AGGGTCA | BCL11B363 | BCL11A363 | |
| SQSTM1366 | NCOR1365 | |||
| NCOR2365 | ||||
| ZFPM2367 | ||||
| V-erbA-related gene (NR2F6) | AGGTCA n AGGTCA | NCOA1368 | ||
| Estrogen-related receptor alpha (NR3B1) | TNA AGGTCA | NCOA1369 | ||
| NCOA2369 | ||||
| NCOA3369 | ||||
| PNRC2370 | ||||
| PPARGC1A315,316,371,372 | ||||
| Estrogen-related receptor beta (NR3B2) | TNA AGGTCA | NCOA1373 | ||
| NCOA2373 | ||||
| NCOA3373 | ||||
| PNRC1374 | ||||
| Estrogen-related receptor gamma (NR3B3) | TNA AGGTCA | NCOA1375 | ||
| PNRC2376 | ||||
| PPARGC1A377 | ||||
| PPARGC1B377 | ||||
| TLE1376 | ||||
| Nerve growth factor IB-like receptor (NR4A1) | AAAGGTCA | Homodimer, Heterodimer | EP300378 | |
| KAT2B378 | ||||
| NCOA1378 | ||||
| NCOA2378 | ||||
| NCOA3378 | ||||
| MED1378 | ||||
| Nuclear receptor related 1 (NR4A2) | AAAGGTCA | Not known | Not known | |
| Neuron-derived orphan receptor 1 (NR4A3) | AAAGGTCA | SIX3379,380 | ||
| MED1381 | ||||
| EP300378 | ||||
| NCOA2378 | ||||
| KAT2B378 | ||||
| Steroidogenic factor 1 (NR5A1) | YCA AGG YCR | Monomer | CREBBP382 | NCOR2383 |
| EDF1384 | ||||
| NCOA1385 | ||||
| NCOA2383,386 | ||||
| PNRC2370 | ||||
| Liver receptor homolog-1 (NR5A2) | YCA AGG YCR | Monomer | NCOA1387 | PROX1388,389 |
| NCOA3390 | ||||
| EP300391 | ||||
| EID1392 | ||||
| Germ cell nuclear factor (NR6A1) | TCA AGGTCA | Homodimer | NCOR1393,394 | |
| NCOR2393,394 | ||||
| SHP (NR0B2) | SIN3A395,396 | |||
| HDAC3397 | ||||
| HDAC1395,397,398 | ||||
| EID1399 | ||||
| NCOR1395 | ||||
| NCOR2395,400 | ||||
| Estrogen receptor-α (NR3A1) | GGTCAnnnTGACC | Homodimer | NCOA1401 | NCOR1401 |
| NCOA2401 | NRIP1401 | |||
| NCOA3401 | ||||
| CREBBP401 | ||||
| EP300401 | ||||
| MED1401 | ||||
| DDX5401 | ||||
| SRA1401 | ||||
| Estrogen receptor-β (NR3A2) | GGTCAnnnTGACC | Homodimer | NCOA1401 | NCOR1401 |
| NCOA2401 | NRIP1401 | |||
| NCOA3401 | ||||
| EP300401 | ||||
| CREBBP401 | ||||
| MED1401 | ||||
| DDX5401 | ||||
| SRA1401 | ||||
| Androgen receptor (NR3C4) | GGTACANNNTGTTCT | Homodimer | CARM1402 | NCOR1403,404 |
| CCND1405 | NCOR2403,404 | |||
| EP300406 | ||||
| FHL2407,408 | ||||
| KAT2B409 | ||||
| KAT5410 | ||||
| MED1411 | ||||
| NCOA122,412 | ||||
| NCOA2413–415 | ||||
| NCOA322 | ||||
| NCOA4413–415 | ||||
| RAN415,416 | ||||
| RNF14417–419 | ||||
| SMARCD1420 | ||||
| TGFB1I1421–423 | ||||
| UBE3A424 | ||||
| Glucocorticoid receptor (NR3C1) | GGTACANNNTGTTCT | Monomer | CREBBP259 | BAG1425 |
| MED1426 | NCOR1427,428 | |||
| NCOA2250,429,430 | ||||
| NCOA6431 | ||||
| PTMS432 | ||||
| SMARCA1433 | ||||
| SMARCD1434 | ||||
| TADA2A435 | ||||
| Mineralocorticoid receptor (NR3C2) | ACAAGANNNTGTTCT | Homodimer, Heterodimer | CASP8AP2436 | DAXX436 |
| ELL437 | NCOR1438 | |||
| EP300439 | NCOR2438 | |||
| FAF1436 | NFYC440 | |||
| MTL5441 | PIAS1442 | |||
| NCOA1248,438,443,444 | ||||
| NCOA2438,439,445 | ||||
| NRIP1444 | ||||
| PPARGC1A439 | ||||
| PPARGC1A437 | ||||
| PPARGC1A443,446 | ||||
| TRIM24444 | ||||
| UBE2I447 | ||||
| Progesterone receptor (NR3C3) | GGTACANNNTGTTCT | Homodimer | NCOA3130,448 | |
| SRA1449,450 | ||||
| NCOA1358,451–454 | ||||
| NCOR2372,455–457 | ||||
| CREBBP241,458 | ||||
| JDP2459 |
Regulation by HREs
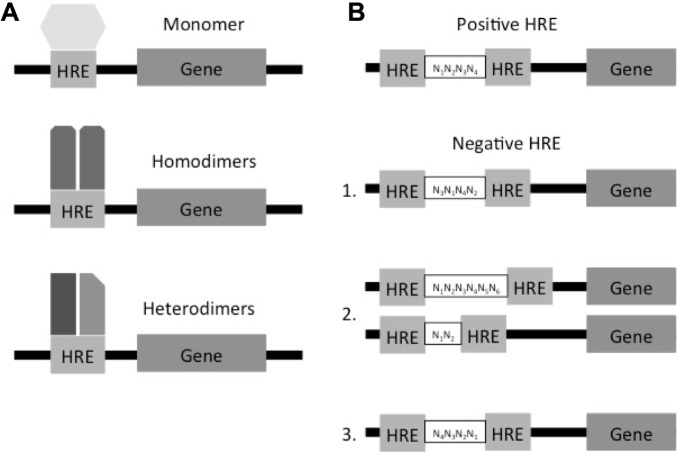
HREs are small sequences of DNA commonly located near the promoter region of a gene, yet they may also be found up to several hundred kilobases upstream of the promoter and even within the first intron of a gene.7 Each receptor subfamily has a preferred HRE motif it binds, although significant variation has been observed in the ability of NHRs to bind to particular HRE sequences (Table 1). For example, steroids predominantly bind to the motif AGAACA, while thyroid hormones bind to AGG/TTCA.8 The motif AGGTCATGACCT is recognized by both the thyroid hormone and the retinoic acid (RA) receptor (RAR). In addition to the motif, the number of nucleotides between motifs and the direction of the motif contribute to binding capacity. For example, the arrangement of direct repeats (DR) AGGTCAnxAGGTCA, inverted repeats AGGTCAnxT GACCT, and reverted repeats (ER) TGACCTnxAGGTCA has anything from 0 to 8 nucleotides as spacers.9 The different combination of spacing and orientation ensures specificity. Nuclear receptors can bind to more than one half-site configuration, allowing coregulation and cross talk between nuclear receptors.10 The optimal binding HRE site for the orphan hormone receptor Nr2e3, identified by evaluating a series of synthetic duplex DNA fragments acquired from a library of random segments, is a hexamer core binding site of AG(G/T)TCA.11 Receptors can bind to the HREs as monomers as in the case of many orphan receptors, as homodimers as in the case of the majority of steroid receptors, or as heterodimers as in the case of several nonsteroid receptors, which usually bind to the promiscuous retinoid X receptor (RXR) (Fig. 2A).3,12,13 There are nonpermisive heterodimer complexes that are activated only by the partner receptor and not by RXR, and permissive heterodimer complexes that can be activated by either of the receptors.14,15
Figure 2.
HREs’ potential patterns associated with NHRs. Schematic representation of (A) NHRs binding to HREs and (B) positive HRE and nHRE patterns.
HREs are classified by their ability to repress (negative HREs [nHREs]) or activate (positive HREs) gene expression. HRE sequences have been identified near the 5′ or downstream of the TATA box and even been within the 3′-untranslated region.6,7 nHREs have been identified in genes that are expressed in the pituitary gland highlighting the importance of the feedback mechanism in hormone regulation.16 nHREs differ from pHREs in (1) the nucleotide sequence of the half-site, (2) in the orientation of the half-site, and (3) in the number of nucleotides that separates them (Fig. 2B).17 It is interesting to note that unoccupied nHREs increase the transcription of a gene, while binding to the ligand has the opposite effect of repressing gene expression. Some sequences of the nHREs overlap with the binding site of different activation factors, therefore, creating a physical competition between the receptors and the transcription factors in the regulation gene expression.18 HREs are one of the several mechanisms by which receptors regulate genes. Another important mechanism is the regulation through coregulators.
Coregulators
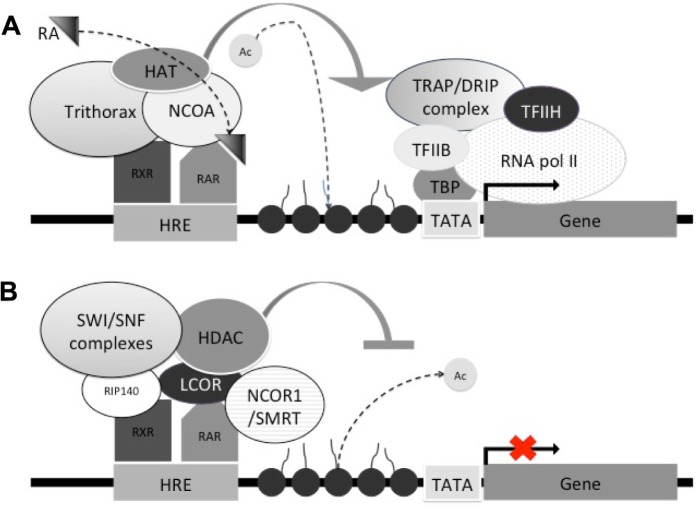
Accessory proteins, known as coregulators, can repress (corepressors) or activate (coactivators) the expression of a given gene (Fig. 3 and Table 1). Coactivators have several mechanisms of action. For example, they can directly interact with the nuclear receptor activation domain in the absence of an antagonist and do not directly bind to the promoter, therefore, preventing them from initiating transcription on their own.19,20 Another way in which coactivators operate is by interacting with components of the transcription machinery and other transcription factors or directly with the promoter changing chromatin conformation via histone acetyltransferase (HAT) proteins (Fig. 3A).20 Coactivators bind predominantly to the AF-2 and AF-1 domains and interact with transcription factors, such as transcription factor II B and TATA box-binding protein. A coactivator can contain more than one motif allowing the molecule to interact with receptors found in dimeric form. One of the most common known motifs is the Leucine LXXLL motif; however; not all coactivators have this motif. For example, the coactivator SUG-1, an ATPases of the 26s proteasome, lacks the LXXLL motif and, therefore, does not bind to the AF-2 region.6,21,22 Several coactivators have been identified, including the steroid hormone receptor coactivator group (SRC-1, -2, and -3), the E3 ubiquitin protein ligase E6-AP, and the RFP-1 coactivator. It is important to highlight that coactivators can also be found in complexes, such as the TRAPs/DRIPs and TRAP220/PBP that also interact with the transcription factors.23 Expression of genes can be repressed in the presence of an antagonist or in the absence of the ligand.
Figure 3.
Transcription repression or activation mediated by RAR–RXR nuclear receptors. Coregulator binding can alter chromatin structure, aiding gene transcription or blockage via histone acetylation or deacetylation. (A) Transcriptional activation by coactivators NCOA and trithorax and histone acetylation by HAT proteins, in the presence of RA. (B) Transcriptional repression by corepressors NCOR, SMRT, LCOR, and RIP140 and histone deacetylation by HDAC proteins, in the absence of RA.
Corepressors are known for their ability to bind to a receptor as well as to elements of the transcription machinery, thus reducing the promoter activity (Fig. 3B and Table 1).22 Examples of corepressors includes the nuclear receptor corepressor (NCoR), also known as the silencing mediator for the retinoid and thyroid hormones (SMRT) (Fig. 3B). Steroid hormones, such as glucocorticoid receptor (GR), mineralocorticoid receptor (MR, Nr3c2), and estrogen receptor (ER), do not interact with NCoR; they instead bind to the corepressors in the presence of an antagonist molecules silencing the expression of a gene.24 The mechanism of action of the coactivators has been defined largely by crystallographic studies that illustrate the molecular conformation changes of NHRs upon ligand binding, thereby releasing the corepressor complex and leaving the site open for the recruitment of a coactivator complex.25
Classification
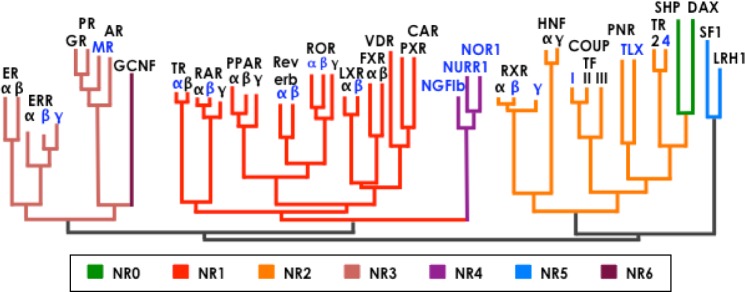
The classification of NHRs has proven to be as complex as the myriad of interactions that they regulate. Another way of grouping the more than 50 members of this group is by focusing on the dimerization and DNA-binding properties of each receptor, grouping the NHRs into four classes. Receptors that belong to Class I bind as homodimers to a palindromic HRE sequence of two half-sites separated by a variable DNA sequence. Class II receptors commonly bind as heterodimers to RXR direct repeats. Class III receptors bind as homodimers to direct repeats. Finally, Class IV receptors bind as monomers to a single half-site.26,27 This form of grouping does not reflect the great number of functions in which the receptors are involved. Taking into considerations the function they perform, the nuclear receptors are divided into three major clusters. The first cluster contains the families of the glucocorticoid receptors, estrogen receptors, mineralocorticoid receptors, androgen receptors, and progesterone receptors. The second cluster includes the families of the thyroid receptors (THRs), vitamin D receptors (VDRs), retinoic acid receptors (RARs), peroxisome proliferator-activated receptors (PPARs), farnesoid X receptor (FXRs), liver x receptor (LXR), pregnane X receptor/steroid and zenobiotic receptor (PXR/SXR), RXR, and constitutive androstate receptor (CAR). The third cluster is composed of the receptors whose ligands have not yet been discovered and are commonly known as the family of the orphan nuclear receptors (ONRs).28–30 Figure 1 illustrates the functional domains of NHRs in each classification cluster. Interestingly, the phylogeny of NHRs depicted in Figure 4 shows the cluster of receptors independent of their expression profile or dimerization and DNA-binding properties, highlighting the importance to put in place another classification system. This classification leads to a different nomenclature where a number from 0 to 6 identifies each clade. In this review, we focus on the major families of NHRs that play important roles in the CNS development. Table 2 describes NHRs expressed in adult mice and in the human brain. It is important to highlight that the receptors involved in the CNS (Fig. 4, blue colored) are common and can be found throughout all the clades.
Figure 4.
Phylogenetic depiction of the nuclear receptor families. Receptors expressed in the CNS are highlighted in blue.122
Table 2.
Nuclear receptors expressed in the different areas of the human or mouse brain.
| RECEPTOR | LOCATION |
|---|---|
| Pparα | BC, C, BS |
| Pparβ/δ | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Pparγ | OA, BC, CO, C |
| Rxrα | OAM, CP, H, HY, CO, C, BS |
| Rxrβ | OA, BC, H, T, HY, AN, CO, C, BS |
| Rxrγ | OA, BC, CP, H, HY, AN, BS |
| Rorα | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Rorβ | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Rorγ | BC, H, HY, CO, C, BS |
| Tr2 | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Tr4 | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Tlx | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Nurr1 | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Nor1 | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Rarα | OA, BC, CP, H, HY, CO, C, BS |
| Rarβ | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Rarγ | OA, BC, H, HY, CO, C, BS |
| Trα | OA, BC, CO, H, T, HY, CO, C, BS |
| Trβ | OA, BC, CP, H, HY, AN, CO, C, BS |
| Mr | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Ngf1B | OA, BC, CP, H, T, HY, CO, C, BS |
| Nr1d1 | OA, BC, CP, H, T, HY, CO, C, BS |
| Nr1d2 | BC, CP, H, T, HY, AN, CO, C, BS |
| Nr2f1 | OA, BC, T, HY, C, BS |
| Nr2f2 | BC, H, T, HY, CO, C, BS |
| Lxrα | OA, BC, H, HY, AN, CO, C, BS |
| Lxrβ | BC, CP, H, T, HY, AN, CO, C, BS |
| Lxrγ | OA |
| Vdr | OA, BS |
| Gcnf | BC, H, AN, CO, C, BS |
| Ear2 | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Errα | OA, BC, H, HY, CO, C, BS |
| Errβ | BC, H, HY, CO, C, BS |
| Errγ | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Gr | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Pr | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Car | OA, BC, HY, C, BS |
| Erα | OA, BC, H, HY, AN, CO, C, BS |
| Erβ | OA, BC, HY, AN, CO, BS |
| Ar | OA, BC, H, HY, CO, C, BS |
| Shp | OA, BC, H, T, HY, AN, CO, C, BS |
| Fxrα | OA, BC, CP, H, T, HY, AN, CO, C, BS |
| Trb | T |
| Dax1 | HY, AN |
| Sf-1 | HY |
| Lrh-1 | AN, BS |
Abbreviations: BC, brain cortex; C, cerebellum; BS, brain stem; OA, olfactory areas; CP, caudate putamen; H, hyppocamps; T, thalamus; HY, hypothalamus; AN, arcuate nucleus; CO, colliculi.
Nuclear Retinoid Receptors Family and the CNS
There are two families of nuclear retinoid receptors: RARs and RXRs. Both families of receptors can bind to target genes as either heterodimers or homodimers allowing them the capacity to target genes in different pathways.6 Humans have the following three types of RAR genes: RARα, RARβ, and RARγ; there are the following three types of RXR: RXRα, RXRβ, and RXRγ. Thirteen different proteins can be produced from the six different genes identified thus far as a consequence of alternative splicing and different promoters.31 An important characteristic that differentiates the two classes of retinoid receptors is the unique amino acid sequence found in LBD conferring each receptor with specific individual characteristics, thereby allowing them to modulate different biological pathways.32
The unique LBD sequence establishes varying affinity toward a particular ligand. For example, RARs bind with high affinity to 9-cis-RA as well as all-trans-RA, while RXR only binds to the 9-cis-RA.33 Along with these differences in the ligand binding, there is also variation in tissue expression. RARα and RARβ are found in the majority of tissues, while RARγ is predominantly expressed in the skin.32 Similarly, RXRα and RXRβ are widely distributed, while RXRγ is expressed in the muscles and brain and in the differentiation of the cartilage and epithelial cells.34,35 RXRγ is not only expressed in the CNS, it is also involved in lipid sensing and modulates gene expression accordingly.36 Retinoic receptors can form heterodimers with both the thyroid hormone nuclear receptors (THRs) and vitamin D receptors (VDRs).
When RXR binds to a THRs or VDR it increases the affinity of DNA biding.37 Similarly, the interaction of THRs with RARs also increases the affinity of the DNA binding. A study performed in monkey kidneys showed that the interaction of both receptors had a positive effect in the transcriptional rate of the gene whose promoter contains a response element for the thyroid hormone, while it also represses those genes that did not have it. This result suggests that the formation of the heterodimers allows for a better control of the transcription rate, and it also hints to the idea that both RAR and THR control the expression of genes in overlapping networks.32,38 Retinoid receptors are involved in a wide range of biological processes that include embryonic development, growth, differentiation, apoptosis, and spermatogenesis. Their functions are not only limited to development, but they also play important roles in adults to maintain proper function of the lungs, liver, and neuronal and immune systems.39 In brain, RARs and RXRs are expressed in multiple stages (Table 2) and play a major role in the development and function of the CNS.
During brain cortex development, RA is synthesized in a tissue-specific manner by the aldehyde dehydrogenase (ALDH) family of enzymes40–42 that is composed of the following three members: ALDH1A1, ALDH1A2, and ALDH1A3. ALDHs have distinct and specific expression patterns that correlate with RA activity as detected by reporter transgenes.43 In brain, ALDH1A3 is primarily expressed in the lateral ganglionic eminence, and ALDH1A1 is expressed in the dopamine-containing mesostriatal system and meninges, while ALDH1A2 is present only in the meninges. Consistent with the presence of ALDH1A3 and ALDH1A1 in the lateral ganglionic eminence and its derivative striatum, where high levels of RA and other retinoids are detected in the developing striatum.40–42 RA is derived from the liposoluble vitamin A and is critical for vision, since its derivative (retinaldehyde) is the light-sensitive molecule, which triggers the phototransduction process in photoreceptor cells in the retina.44 Vitamin A and RA are essential for eye development and maintenance and serve a similar role in brain cortex development. Young animals raised on a vitamin A-deficient diet grow poorly, become blind, and have a low survival rate.45,46 The teratogenic effect of RARs was demonstrated in a study using the antagonist of RARs (AGN193109). An- or microphthalmia was observed in 75% of treated mice.47 Additionally, a recent study revealed mutations in human ALDH1A3 is associated with microphthalmia and anophthalmia.48–53
RA biological action is to bind to RARs, switching them from transcription repressive elements to transcription activator elements, while they are bound to RAR elements (RAREs).54,55 In contrast, RARβ has been involved in brain structure development and maintenance; especially in ventral and dorsal telencephalon development, hippocampal plasticity, and brain barrier development.56–59 In addition, RARβ is expressed with other RARs and RXRs in adult mice and human brains (Table 2). Other studies revealed that Rarβ expression is inducible by adding external RA at stage E15 to rat brain cortex cultures. Therefore, increases in Rarβ expression by RA in brain cortex may be due to a partial depressive mechanism as RARβ is autoregulated by RA through a RARE-dependent mechanism.60–62 Moreover, in the E15 mouse, Rarβ enhances protein tyrosine phosphatase, nonreceptor type 5 (PTPN5) expression, a brain-specific tyrosine phosphatase that has been shown to dephosphorylate several key neuronal signaling proteins in brain cortex.56,63 Interestingly, Rarβ seems to enhance or repress gene transcription, depending not only on the presence (enhance) or lack (repress) of RA but also on the isoform tissue-specific transcription. Rarβ 1 expression in rats has been shown to upregulate Ptpn5 gene expression. In contrast, Rarβ 4, an isoform composed of only four amino acids at the N terminus, resulted in a lower transcription of Ptpn5.56 The development and maintenance of the brain demonstrate the complexity of the networks that the hormone receptors regulate, which requires the interaction of the receptors with enzymes. Lack of Rarβ, as observed in the Rarbtm1Vgi knockout mouse, presents with memory and spatial learning problems. Rarβ deficiency in these mice virtually eliminates hippocampal CA1 long-term potentiation and long-term depression.64
RARβ and RA signaling pathway also play an important role in mediating eye development. RA signal depends on the transcription factor Pax-6 to activate the α crystalline β gene, which codes for the formation of the lens in the eye. Inhibition of RA synthesis as a consequence of antagonist results in the inhibition or abnormal morphogenesis of the lens in murine models.65 RA signaling is detected in the lens placode and lens pit and is also expressed during the differentiation of the primary lens fibers. RA signaling is also active in the surface ectoderm and the corneal epithelium. Finally, during the development of the retina, RA signaling is detected in the optic vesicle and the retinal pigment ephitelium (RPE).66 Interestingly, Rarβ transcripts were discovered at the onset (E12) of retinal pigment epithelium formation, in the vitreous body, and in the condensed mesenchyme around the eye cup, which forms the choroid layer in mice.67 Rarβ−/− (Table 3) mice have abnormal retrolenticular mass of pigmented tissue in the vitreous body, which presented a large base adherent to the lens, containing a persistent hyaloid artery and vein.68 This abnormal structure is found bilaterally in most of the homozygous mice but rarely seen in heterozygosis.68 By E13.5, mouse fibroblasts of the primary vitreous body expand and thin as the secondary vitreous forms and is found at the optic disk by E15.5, forming Bergmeister’s papilla.68 In E14.5, Rarβ−/− mice, the number of cell nuclei in the secondary vitreous is four to six times higher than the number of nuclei in wild-type animals.69 These observations indicate that the loss of Rarβ results in overproliferation of the fibroblastic neural crest cells of the primary vitreous body in mice.69 Additional eye defects observed in Rarβ−/−-null mice include congenital folds of the retina and cataracts, which are likely to be secondary to mechanical and/or metabolic stresses resulting from the presence of the persistent hyperplastic primary vitreous.68
Table 3.
Knockout mice models excising that effect the CNS function.
| MODEL NAME | BRAIN/EYE DEFECTS |
|---|---|
| Thrbtm2Df/tm2Df | – Lack of M-opsin expressing cones.106 |
| – S cone gradient in the retina disturbed.106 | |
| TRβ mut | – Impairment in balance and coordination.95 |
| – Reduce cerebellum mass.95 | |
| – Decrease in Purkinje cell layer.95 | |
| Rorasg | – Deficiency of granule cells and Purkinje cells.460 |
| – Cerebellar cortex underdeveloped.460 | |
| Nr4a2tm1Tpe | – Failure to develop dopaminergic neurons.218 |
| Nr2e3rd7/J | – White spots evenly distributed in the retina.461 |
| – Whorls and rossetts can be detected in the retina –Progressive loss of cones and rods.461 | |
| Rorβ−/− | – Loss of rods.167 |
| – Overproduction of primitive S cones.167 | |
| Nr1d1tm1Ven | – Delay cerebellum development.462 |
| – Abnormal Purkinje ad granule cells.462 | |
| RAR β2 γ2−/− | – Marked atrophy and dysplasia of the retina.463 |
| – In the central retina only 2 or 3 rows of nuclei are found.463 | |
| RXRα−/− and RXR β−/− | – Fetus diet at E12.5–16.5.464 |
| – Fetus have conotruncal and ocular defects.464 | |
| – Failure to close the neural tube.464 | |
| TLX KO | – Thickness of the retina is reduces.465 |
| – Increased number of apoptotic cells in the inner nuclear layer of the retina.465 | |
| – Decreased proportion of mitotic cells.465 | |
| Rarbtm2Ipc|Rargtm3Ipc |Tg (Wnt1-cre)11Rth | – Severe ocular abnormalities.213 |
| Rxratm4Ipc|Tg (Tyrp1-cre)1Ipc | – Decreased number of photoreceptors.466 |
| – Abnormal photoreceptor outer segment morphology.466 | |
| – Reduce light response.466 | |
| Nr1h2tm1.1Gstr|Nr1h3tm1.1Gstr | – Abnormal astrocyte morphology.467 |
| – Abnormal brain vasculature morphology.467 |
Thyroid Hormone Receptor Family and the CNS
The thyroid hormone receptor (THR) family plays a major role in the development of brain, regulation of metabolic rate, and the development of photoreceptor cells in the vertebrate retina. THRs are involved in a great number of biological activities that are both hormone dependent and independent. Some of the activities in which this receptor is involved include regulation of the Rous sarcosoma virus and the human immunodeficiency virus type 1,27,70 initiation of the metamorphosis in amphibians by controlling the formation of tissues,71 regulation of body temperature,72 differentiation of muscle cells by regulating genes of the Ca2+ ATPase,73 and migration of cells in the hippocampus, cerebellum, and cortex.74 An important characteristic of this family is their ability to also regulate expression in the absence of a hormone.70 However, when they bind to a hormone, it is either with triiodothyronine (T3) or with thyroxin (T4). The nonligand-dependent THR form can regulate transcription of a gene by recruiting corepressors or coactivators to either repress or activate expression by forming protein–protein interactions with members of the transcription initiation machinery, such as the transcription factor IIB.71,72
TRs are members of Class II, meaning that they predominantly bind to RXR. However, THRs do not bind exclusively to RXR, and they can also form heterodimers with VDR and PPAR.73,74 The receptors of this family are coded by two genes, such as thyroid hormone receptor α (THRα) and thyroid hormone receptor β (THRβ); as a result of splicing differentiation, two isoforms are formed, THRα1 and THRα2, THRβ2 and as a result of different promoter usage, two more isoforms are formed, THRβ1 and THRβ2.70 THRβ1 and THRβ2 have the exact same LBD, yet the amino-terminal domain is different, therefore making two proteins that have different biological function and are expressed in different tissues. The difference in the amino acid terminal influences the affinity in which the ligand interacts with the initiation complex or, in the case of THRβ2, it has another activation domain.75 The level of expression of each receptor varies greatly in different tissues according to the respective stage of development. THRβ2 is predominantly found in the hypothalamus, anterior pituitary gland, and cochlea formation. TRα1 is involved in the development of the immune system as well as signaling of thyroid hormone receptor during postnatal development.76,77
Thyroid hormone receptors regulate several developmental events in the CNS, including neuronal differentiation and migration, synaptogenesis, and myelination.78–80 T3, T4, and TRs are present in the developing cortex, prior to the onset of fetal thyroid gland activity, before week 12 in humans. This suggests that maternal thyroid hormone (TH) concentration is important during brain development.81,82 Thyroid hormone influence in the CNS is well documented through studies of TH in the cerebellum.83 Two genes encode at least three high-affinity THRs in mammals, THRβ1 (Thrb), THRβ2 (Thrb), and THRα1 (Thra).84 Different isoforms of TRs are expressed across the brain, cerebellum, and retina,85,86 and they continue to be expressed in adult mice and human brains in other brain areas (Table 2). Studies in rats show that THRα1 binds to nearly 80% of T3 and is the main isoform expressed both prenatally and postnatally across the brain, including the developing cerebellum.87,88 THRβ expression, is not as common, and is confined to a few postnatal neuronal populations in the CNS.89 In rat cerebellum, both Thrα and Thrβ are expressed; Thrα is mostly expressed early in the cerebellar neuroephithelium and granular cell precursors and later in the transient external granular layer, whereas Thrβ is predominantly expressed during the later stages.85,90 Knockout studies of Thrb and Thra mice (Table 3) depict milder neurological malformations compared to that of hypothyroid animals, demonstrating that T3 has a greater function in the developing CNS than the THR isoforms.91–94 In contrast, ∆337T Thrb mice, harboring a deletion in LBD that prevents T3 to bind the receptor, presented cerebral deformities similar to the hyt/hyt mouse model affected with congenital hypothyroidism.95 These mice displayed impairments in balance and coordination and in the Purkinje cell layers and decreases in the number and branching of Purkinje cells, which may account for the decreased cerebellar size observed in these mutant animals.91,96–98 The role of THRα1 in brain development has been described recently in humans, since descriptions of patients with mutations in THRα gene present cognitive impairment, similar to those observed in congenital hypothyroidism.99,100 These data suggest that both THRα and THRβ function synergistically to mediate cerebellar development via THs.101
Several isoforms of the thyroid nuclear receptors are expressed in the eye. THR form complex with activators, such as SMART and histone deacetylases (HDACs) to regulate the transcription activity of the target genes.102 The receptor THRα is found in the progenitor layer, while THRβ1 is found in the inner nuclear layer, and THRβ2 is expressed in the photoreceptor layer in particular in the developing cones.85,102,103 TRs play an important role in the development of the retina by participating in the signaling system by which the progenitor cells are differentiated. It has been determined that the abundance of T3 in the environment can induce the proliferation of cone photoreceptor cells.104 In general, thyroid hormone receptors act as coactivators in the presence of T3, and in the absence, they repress the expression of their target gene.105 It has been demonstrated that in Thrβ2 knockout animals, the S-opsin gene is expressed earlier leading to a failure of the correct expression of the M-opsin gene leading to a failure in the development of the M-cones.106 The Thrβ2−/− mice have a cell fate switch of their cone photoreceptor cells such that all cones are blue or S cones.
PPARs Family and the CNS
The PPAR family was first discovered for its role in increasing the quantities of peroxisomes in different animal species and increasing the transcription of genes that encode for fatty acids.107 Since this discovery, several additional roles have been discovered for these receptors, such as lipid metabolism, inflammation, glucose homeostasis, and cell differentiation. It has become clear that PPARs have an important role in body’s response toward exogenous ligands.108 Three different isoforms have been identified for PPAR: α, β/δ, and γ. Each isoform has a unique pattern of expression and has affinity to different ligands. PPARα is an important factor regulating lipid metabolism and is mostly found in the heart, kidney, intestine, and fat tissue.12 In contrast, PPARβ/δ is found predominantly in different areas of adult brain (Table 2), kidney, and small intestine, is involved in cell survival, and is often present in the formation of tumors in the colon. For this reason, several studies have focused on understanding how PPAR might be an important therapeutic target. It was discovered that the nonsteroidal anti-inflammatory drugs compete with PPARβ/δ for its responsive elements inhibiting the action of PPARβ/δ and eventually reducing tumorigenesis.109 PPARγ is mostly found in the spleen, intestine, and adipose tissue.12 PPARs have the ability to bind to different types of fatty acids as well as synthesized compounds. Their ability to bind to different types of molecules is most likely due to the spacious ligand-binding pocket that is observed in the crystallographic structure.110 While PPARs can be found in different tissues, they are predominantly expressed in tissues that regulate energy homeostasis. The fact that the majority of ligands are fatty acids and other metabolic molecules is evidence that PPARs have an important role in the regulation of metabolism. For example, the signal molecule eicosanoids is a product of fatty acid metabolism and is also known for being an inflammatory agent. This led to the discovery of a second function of PPARs, which is inflammatory signaling regulated in particular by PPARα and PPARγ.111,112 Since Alzheimer’s disease has an inflammatory component to its pathology as a consequence of the neurodegeneration and the extracellular deposition of β-amyloid peptides, recent studies have focused on the protective effects of nonsteroidal anti-inflammatory drugs in delaying or reducing the risk of Alzheimer’s disease.113 PPARs as many other receptors form heterodimers with RXR. Therefore, there is evidence of competition between PPARs and receptors, such as TRs and vitamin D, which also partner with RXR. As an evidence of this competition, it was observed that the transcription of the genes that are regulated by PPARs was repressed in the presence of T3.114 The fact that different receptors compete for their heterodimer partners is evidence of the complexity of gene regulation and the dual roles that the nuclear receptors play. The PPARs are an excellent example of previous orphans whose ligands were finally discovered; for all other receptors whose ligands still remain to be discern, they conform the ONR family.115
ONRs and the CNS
Orphan receptors are a unique class of NHRs in that they do not have a ligand identified and often lack all of the conserved domains typical of NHRs. Furthermore, a high degree of diversification has been observed in this group of NHRs, as many of them are not evolutionary or functionally related. This is evidenced in a phylogenetic analysis when aligning the DBD and LBD sequences. The resulting evolutionary tree has six major branches with the orphan receptors distributed across all branches.116,117 The degree of diversification of orphan NHRs is further illustrated by studies revealing that some orphan receptors in invertebrate species such as Drosophila and nematodes lack DBD yet have LBD, and other receptors, such as Nr2e3, retain DBD.118,119
Another striking difference of orphan NHRs is the size of the A/B region that can range from a minimal eight amino acids in the retinoid orphan receptor (ROR) β to 280 amino acids in the NGFI-B receptor.30 Some orphan receptors lack the activation function AF-1, while others lack an AF-2 domain. Many orphan receptors form heterodimers with RXR, while the remaining orphan receptors bind to DNA as homodimers.3 The classification system divides the ONRs into seven different groups starting with group 0. Group I is comprised of seven different families, Group II is comprised of 5 families while group III, IV, V and VI contain one family.2,12,120
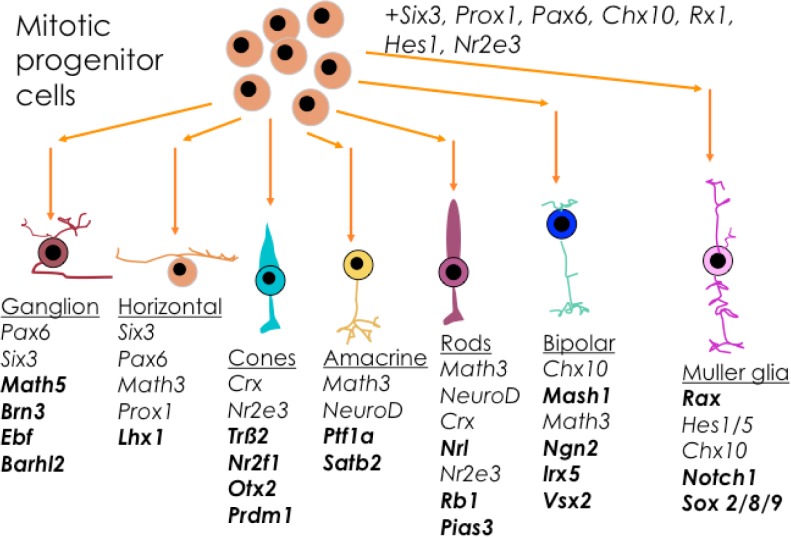
The NHR Nr2e3 was first identified as photoreceptor-specific nuclear receptor. Unlike most NHRs that exhibit ubiquitous expression, expression of NR2e3 is restricted solely to the outer nuclear layer of the neurosensory retina (cones and rods; Fig. 5).121–123 During human eye development, Nr2e3 transcription is observed at week 11.7 in the putative immature human rods on the foveal edge.124 Many studies performed using the Nr2e3rd7/rd7 mouse, which lacks a functional Nr2e3 gene, indicate that Nr2e3 has a dual regulation role regulating rod and cone development and function.60,121,125 Lack of Nr2e3 leads to abnormal proliferation of blue cones (the least populous of the photoreceptors) as a consequence of the loss of Nr2e3 in the mitotic progenitor cell program.126 In rods, Nr2e3 does not affect rhodopsin expression per se, although it seems that rhodopsin expression needs both Nr1d1 and Nr2e3 potentiated by the orthodenticle-like cone–rod homeobox transcription factor (CRX)- and the neural retina basic motif-leucine zipper transcription factor (NRL)-driven activities.127 NR2E3 works in conjunction with multiple transcription factors, including CRX, which is detected initially at day E12.5 in retinal progenitor cells and newly postmitotic cells,128–130 and NRL, which is detected at day E14.5 and directs the generation of rod photoreceptor cells.131,132 Although previous studies have identified several genes that are differentially expressed between normal and knockout mice, the transcriptional network regulated by Nr2e3 is not fully understood. Some potential genes that are regulated by Nr2e3 include the following cone-specific phototransduction genes: blue opsin (Opn-SW), Gnαt2 and Gnβ3 cone transducing subunits, the rod-specific gene guanine nucleotide-binding protein 1 (Gnb1), and the NHR Nr1d1.121 It is interesting to highlight that, even though in the Nr2e3 regulation network the majority of genes were upregulated, a significant number were also downregulated supporting the hypothesize that Nr2e3 functions as both a suppressor and an enhancer of transcription.133
Figure 5.
List of NHRs involved in the differentiation of the mitotic progenitor cells and the retinal cell types. Genes marked in bold correspond to NHRs unique for each cell type.
The RORs and the Rev-Erb are two of the receptor families that belong to Group I. The ROR family is made up of the following three members: RORα, RORβ, and RORγ. RORα is ubiquitously expressed and is involved in the development of Purkinje cells (Table 2), bone metabolism, immune response, development of cone photoreceptors of the retina, bone morphogenesis, angiogenesis, and lipid metabolism.134 RORβ is mostly found in the retina,135 brain (Table 2),135–138 and spleen and functions in maintenance of the circadian rhythm.139–141 RORγ is found in the liver, kidney, thymus, and lungs, and its functions consist of lymph node organogenesis and thymopoiesis.1,141–145 Each member of the family also has different isoforms that are formed by alternative splicing and alternative promoters.146 The isoforms differ in the A/B domain: four ROR isoforms have been identified in humans and two have been identified in mice. RORα1 and 4 are localized in the cerebellum.146 RORβ has two known isoforms; RORβ2 is restricted to the pineal gland and the retina (Fig. 5).135
Both RORα and RORβ have an important role in the CNS, as observed by knockout mouse models that have severe cerebral abnormalities.147,148 Cerebellum development is regulated by RORs and takes a large period during fetal development. The main types of cells in the cerebellum develop at different stages and positions. The Purkinje cells arise from the ventricular zone of the metencephalic alar plates at the fifth or sixth week of development.149 Granule cells are added from the rhombic lip at the end of the embryonic period, and their migration is facilitated by Bergmann glia cells to their deeper definitive site.150 During this process, RORα expression is restricted to Purkinje cells and not to the granule cell layer, as observed in mice at age E12.5.109–116
The staggerer mouse, which lacks functional RORα (RORαsg/sg or RORα−/−), presents with fewer Purkinje cells and a loss of cerebellar granule cells.147,151–157 RORα heterozygous mice (RORα+/sg) with only one functional allele exhibit a milder phenotype and present with an accelerated Purkinje and granule cells loss in aged animals.154,158–160 RORα regulates genes related to Purkinje cells maturity, in particular its dendritic differentiation. RORα−/−-deficient mice show downregulation of several genes involved in signal-dependent calcium release, including the calmodulin inhibitors Pcp4 and Pcp2, the IP3 receptor (ITPr1) and its coreceptor Cals1, the calcium-transporting ATPase (Atp2a2) and major intracellular calcium buffer 1 (Calb1), Shh, and solute carrier family 1 (high-affinity aspartate/glutamate transporter), member 6 (Slc1a6).151,161,162 Moreover, most of the downregulated genes in RORαsg/sg or RORα−/− mice have retinoic orphan receptor elements, a type of HRE recognized by the ROR family of NHRs. The retinoic orphan receptor element pattern sequence consists of the sequence RGGTCA core motif followed by a 6 bp A/T-rich sequence in the regulatory region of RORs’ target genes, which explains their downregulation by RORα absence.135,163,164 As observed by the use of the RORαsg/sg, it is clear that animal models are essential for understanding the functions of NHRs and the genes that they regulate.
RORβ plays an important role in retina cell differentiation as studies in murine models have described. Differentiation of cone photoreceptor cells in mice is regulated by the nuclear receptors Rorβ and Rxrγ (Fig. 5), which control the generation of short(S)-opsin-expressing cones (also referred to as blue cones).165,166 RORβ also stimulates rod photoreceptor differentiation by activating the rod-selective transcription factor neural retina leucine zipper (Nrl).167 The function and survival of rod photoreceptor are also regulated by the estrogen-related receptor β (Errb).168 Rorβ is expressed in the formative ganglion cell layer, and Rorβ−/− has disorganized cone cell topography and lacks the inner and outer segments, suggesting that RORB exerts an important role in the proper development of cone photoreceptor cells.165
NHRs regulate gene expression in a common pool of multipotent retinal progenitor cells found in the neuroephithelium of the optic cup to form all retinal cell types (Fig. 5).169 Retinal proliferation is a highly conserved dynamic process in vertebrates that occurs in the following two overlapping phases: (1) the first phase begins with the proliferation of ganglion cells followed by horizontal cells, cone photoreceptors, and amacrine cells and (2) the second phase overlaps with the generation of rod photoreceptors, bipolar cells, and Müller glial cells. The differentiation of retinal cells also has overlapping phases.170 The formation and maintenance of the different types of cells in the retina requires specific gene regulation. The transcription and regulation of NHRs, including Nr2e3, RARβ, RORβ, Nr1d1, RXRγ, and Trβ (Fig. 5), in synergy with several transcription factors, including CRX, NRL, and Chx10, help determine retinal cell fate and pluripotent capacity of retinal progenitors.171,172
The Rev-Erb members are characterized for having unique features, such as their transcription from the opposite strand of the T3 receptor gene.173 They also lack the AF-2 domain and mostly function as repressors of gene expression by associating with the corepressors NCoR.24,174–176 Rev-Erb has been associated with several functions, such as being a mediator of adiponogenesis,177 the regulator of myogenesis,178 involvement with cell proliferation,34 the regulator of the circadian clock,179 and effects on lipid metabolism.180
The testicular nuclear receptors (TRs) and tailless (TLX) receptor are two of the five members of Group II of the orphan receptors. There are two members of the testicular nuclear receptors (TR2 and TR4). TR2 is highly expressed in the testes as well as in several other tissues, including brain, retina, kidney, and intestine, with the highest peak of expression during embryonic time points.181 TR2 can bind as a homodimer or as a heterodimer, and it has the ability to act as a repressor of gene expression.182 TR4 is expressed in several tissues from the CNS, adrenal gland, spleen, and prostate gland with the highest expression observed in the skeleton and testes, as indicated by the name of the family.181 TR4 is expressed in a temporal and spatial manner during brain development,183 and TR4 knockout mice show growth defects, and females exhibited poor maternal behavior skills.184
These behavioral traits in TR4 knockout rodent animals seem to be the consequence of a poor cerebellum development. During this process, TR4 expression patterns correlate directly with cerebellum neurogenesis, suggesting a role in this stage.183,185 During postnatal cerebellum development, several changes occur in the cerebellum laminar construction with engrossing of the external granule layer followed by granule cells migration toward the internal granule layer. Precursors of these cells migrate as well to the internal granule layer guided by Bergmann fibers, which expresses the astrocyte-specific glutamate transporter, GLAST.150,186,187 This migration process is affected fundamentally by the order of the Bergmann fibers. If these fibers are disorganized or atrophied, migration will not occur. Additionally, migration impairment can also be the consequence of defects in granule cells.188,189 Mouse models lacking TR4 exhibit abnormal cerebellar development in lobules VI–VII, which correlate with poor granule cell migration and Bergmann glia maturation.190–195 These alterations are related to a lack of GLAST, causing a long-lasting negative impact on neural connectivity pathways during brain development.196 As a consequence of TR4 knockout, aberrant neuronal connectivity has been observed due to the delay in granule cell migration and the disarrangement of the cerebellar Purkinje neurons. The result is an abnormal response to environmental changes and also problems with social contact, startle activity, and deficits in prepulse startle inhibition and cerebellar motor learning when no motor coordination seems to be present.196
The TLX receptor is the human homolog of the tailless gene in Drosophila that is uniquely expressed in the neuroephithelium of Drosophila embryonic brain. A unique characteristic of TLX is a highly conserved substitution of a lysine residue in the P-box of DBD.197,198 TLX is highly conserved in vertebrates and plays a major role in the development of the forebrain and brain function, retinal development, and aggressive behavior3,199,200 and is expressed in human and mice adult brain areas (Table 2). TLX is a key component necessary to create the superficial cortical layers of the brain in embryos, and it also regulates the timing of neurogenesis in the cortex. TLX knockout mice have reduced cerebral hemispheres and reduced ability to learn. An important function of TLX is the maintenance and self-renewal of neural stem cells in the brain.201,202 The development of the retina is also influenced by TLX receptors that are expressed in the neuroblastic layer (Fig. 5). Mice that lack the expression of TLX do not respond to light stimuli, suggesting a key role for TLX in the formation of several retinal cell types, such as photoreceptors and bipolar and amacrine cells.203
Group IV is composed of one receptor member with three different subtypes, α, β, and γ. This family, also known as Nr4a, is expressed in adult mice and human brain (Table 2). Nr4a receptors play an important role in the retina and cerebellum development and N-methyl-d-aspartate (NMDA) receptor-mediated neuroprotection. Barneda-Zahonero et al observed that Nur77 protects hippocampal neurons in culture from staurosporine and growth factor removal-induced toxicity in vitro, and it also protects hippocampal neurons from kainate-induced toxicity in vivo.204,205 The role of Nr4a subfamily in the survival of cerebral granule cells (CGCs) was studied in cell culture assays and revealed that Nurr1 is involved in the development and differentiation of the midbrain dopaminergic neurons and regulates several of the tyrosine hydroxylase and dopamine transporter gene networks.66–68 Nurr1 has been shown to regulate genes, such as the vasoactive intestinal peptide (VIP),206 α-synuclein,207 and brain-derived neurotrophic factor (BDNF).208 It was also observed that after silencing Nurr1 expression by shRNA, BDNF transcription levels decreased in CGC cultures treated with the glutamatergic agonist, and NMDA activation promoted Nurr1 binding to promoter IV of BDNF in a specific manner. These studies demonstrated that Nurr1 is involved in the prosurvival of CGC neurons via NMDA receptor activation by enhancing BDNF transcription. Furthermore, CREB shRNA studies revealed a reduction in Nurr1 expression and protein concentration due to a blockage on the NMDA receptor-CREB cascade, leading to lower BDNF transcription levels.204 These studies provide valuable insight into the biological processes and genes regulated by the NH receptors.
NHRs Associated with Diseases in the Eye and Brain
Animal models provide valuable insight into the biological pathways and roles of the receptors in normal conditions (Table 3). Nuclear receptors have been associated with a wide range of diseases, such as diabetes, obesity, metabolic syndromes, autism, Parkinson’s disease, ocular disease, and cancer. In this review, we focus on diseases caused by disrupting the NHR function in the CNS, particularly eye or brain development (Table 4). RARs play an important role in the development of the CNS, and mutations in the RAR networks are associated with complex human diseases. RA is derived from vitamin A (retinoid), which is supplied by mothers to embryos transplacentally.209 Two classes of enzymes, the cytosolic alcohol dehydrogenases and the microsomal short-chain dehydrogenases/reductases, oxidize retinol into retinaldehyde.210 Retinaldehyde is then oxidized to RA by the ALDHs (ALDH1A1, ALDH1A2, and ALDH1A3). ALDHs have distinct and specific expression patterns that correlate with RA activity as detected by reporter transgenes.43 In the CNS, ALDH1A3 is primarily expressed in the lateral ganglionic eminence, while ALDH1A1 is expressed in the dopamine-containing mesostriatal system and meninges, consistent with the presence of ALDH1A3 and ALDH1A1 in the lateral ganglionic eminence and its derivative striatum, where high levels of RA and other retinoid are detected in the developing striatum, eye, and olfactory system.40–42 ALDH1A3−/− mice present an aberrant development in nasal and ocular systems and die at birth due to respiratory distress.211 CNS human diseases have been linked to mutations in ALDH1A3, which have been directly associated with microphthalmia and anophthalmia. A recent report has also linked variants in ALDH1A3 with risk for autism.212 ALDH1A1−/− mice, in contrast, are viable but present minor defects in the dorsal retina.213,214 As RA regulates genes via RARs, depletion of this metabolite causes a poor gene regulation of RARs, which is the main cause of the diseases exposed, though there are other diseases caused directly by alterations in RARs family.
Table 4.
Nuclear hormone receptor and associated diseases in humans.
| GENE | HUMAN DISEASE | OMIM ID |
|---|---|---|
| Nr1b2 (RAR-β) | Syndromic microphthalmia468 | 615524 |
| Schizophrenia469 | 181500 | |
| Nr1c1 (PPAR-α) | Alzheimer’s disease470 | 104300 |
| Nr2c2 (Tr4) | Deficit in motor coordination471 | |
| Nr2e3 (Pnr) | Enhanced S cone syndrome472 | 268100 |
| Retinitis pigmentosa232 | 611131 | |
| Nr2f1 (Coup-TFI) | Bosch-Boonstra optic atrophy syndrome473 | 615722 |
| Nr3a1 (ER α) | Migraine474,475 | 157300 |
| Nr3a2 (ER-β) | Alzheimer’s disease476 | 104300 |
| Nr3c1 (GR) | Depression477 | 608520 |
| Nr3c2 | Stress478 | |
| Nr3c3 (PR) | Vertigo479 | 193007 |
| Nr4a2 | Parkinson disease480 | 300557 |
In the brain, RARs are expressed with different location patterns: RARα is found ubiquitously, and RARβ and RARγ are found in the striated regions that have dopaminergic neurons.215 Dopamine receptor 2 knockout mouse phenotype resembles the human Parkinson’s disease.216 Parkinson’s disease is characterized by the degeneration of the dopaminergic pathway. Both trans-RA and 9-cis-RA induce the expression of dopamine receptor 2. RAR forms a heterodimer with RXR to bind directly to the dopamine receptor gene linking this gene and the nuclear receptors to Parkinson’s disease.217 RXRs also form a heterodimer with the orphan receptor Nurr1 that is expressed in the mesenphalic flexure of the brain, the region that develops the dopamine-producing neurons.218,219
The most common disease associated with RARα mutations is acute promyelocytic leukemia (MIM: 612376), caused by gene rearrangements. Often, a gene fusion of promyelocytic leukemia (PML) and RARα occurs due to the translocation t(15,17)(q21;q22).220 However, though most of the rearrangements involving RARα are associated with different types of leukemia, one individual diagnosed with Smith–Magenis syndrome presented a region translocation of chromosome 17 q11.2–q21.3 that was inserted into the short arm of the same chromosome at region p11.2.221 Patients with Smith–Magenis syndrome have multiple congenital anomalies, including mental retardation, which are associated with a deletion involving band p11.2 of chromosome 17.222 Though the report given by Park et al221 did not expose RARα translocation as causative of Smith–Magenis syndrome, it shows how chromosome 17 q11.2–q21.3 region is a breakage and rearrangement hotspot, same as chromosome 17 p11.2 region, which may cause different related syndromes221 not directly associated with the CNS.
Unlike RARα, RARβ is important for the proper development of the brain cortex, and variants in this gene have been associated with locomotor disease and ocular disorders. RXRB and RARβ expression have been observed in the striatum.223 Several polymorphisms in RARβ are associated with microphthalmia in humans (MIM: 615524). Double knockout mutants for RARβ and RXRγ have a 30%–40% reduction in D1 and D2 dopamine receptor expression, though they are the most abundant type of receptors in the striatum,223 and exhibit reduction in forward locomotion. In the case of D2, its lower expression is possibly explained by the presence of RAREs in its promoter region.224 This fact permits hypothesize that RARβ and RXRγ are essential in developing the proper locomotion, and it is involved in controlling the dopaminergic system during development and adult stages in CNS.
RORα gene networks appear to play an important role in the neurodegenerative disease spinocerebellar ataxia-1. A microarray analysis from mutant and normal mouse brain showed that genes regulated by RORα were downregulated in the first stages of development of the Purkinje cells. Loss of RORα leads to an increase expression of the ataxia gene ATXN1. A further analysis by coimmunoprecipitation revealed that ATXN1, RORα, and the RORα coactivator Tip60 form a complex to modulate cerebellar development. The RORα1 knockout model lacks the LDB region and is characterized as having abnormal Purkinje cells and abnormal morphology and ataxic.225,226 The staggerer mouse, which has a deletion in the RORα gene that blocks the translation of LBD, models cerebellar ataxia affecting the development of the Purkinje cells. RORα−/− also shows atrophy and loss of the dendritic cells and synaptic rearrangement of the Purkinje cells.227 The phenotype of the staggerer mouse suggests that RORα is involved in the thyroid hormone signaling pathway in charge of the maturation of the Purkinje cells.227 Different studies have demonstrated that Rorα is also involved in the protection of neurons, suppresses inflammation, and is important in regulating circadian rhythm.228
NR2E3 has been associated with several retinal diseases, including enhanced S-cone syndrome (ESCS),229 Goldmann-Favre syndrome (GFS), and retinitis pigmentosa.230–232 ESCS and GFS are autosomal recessive diseases characterized by an increase in the sensitivity of the S (blue wavelength)-cone photoreceptor cells accompanied with a decreased in the rods and L/M (red/green) cones.11,233,234 GFS is a milder form of ESCS. Patients with autosomal dominant retinitis pigmentosa suffer from early onset loss of night vision followed by retinal spots in the peripheral field, and eventually loss of central vision. By analyzing the phenotypes of patients with different diseases caused by mutations in the Nr2e3, it is possible to conclude that the location of the mutation has a major impact on disease outcome. The diseases are viewed as a collection of Nr2e3-associated retinal diseases.235 NHRs are involved in a myriad of functions that cover more than just the CNS normal functioning. Yet, it is still important to understand how NHRs participate in diseases, as NHRs play a key role in normal CNS development.
Perspectives and Conclusions
This review discusses the important role of NHRs in the development, function, and maintenance of the CNS. There is great variation in the types of isoforms, as well as the abundance and time of expression of NHRs throughout the CNS. Furthermore, it is not uncommon to see members of a particular family with multiple functions as illustrated by the ROR and TR families of NHRs. All members of the THR family, for example, are involved in the development and/or function of the CNS. In contrast, RARβ simply has one member involved in a specific tissue of the CNS, and even more restrictive is the orphan receptor NR2E3 that functions in only one type of retinal cell, the photoreceptor cell. Thus, while most NHRs are ubiquitously expressed, others have family members or isoforms with expression patterns limited to particular in organs or cell types. There is a clear overlap in NHRs signaling of the developing eye and brain, suggesting a common template for CNS development. The majority of NHRs have a dual role as both transcription activators and suppressors. Their function depends on their temporal and spatial expression, cellular context, as well as the interaction with other factors, ligands, and DNA sequences of HREs. Furthermore, NHRs can modulate gene expression, via HRE recognition, by several mechanisms, including ligand binding and cofactor recruitment. A comprehensive reconstruction of the complex, multimodal interacting gene networks and identifying the target genes they regulate will provide greater insight into overall neuronal signaling of the CNS.
It is well established that NHRs regulate numerous biological pathways and processes. NHRs interact with several factors, such as transcription factors, chromatin-modifying complexes, and hormones. NHRs often serve as master regulators simultaneously modulating several signaling pathways in a temporal/spatial/cell type-specific manner. The highly conserved nature of NHRs and their numerous isoforms throughout the CNS suggests functional redundancy and a possible mechanism to avoid disease. This is evidenced in data demonstrating that NHRs can serve as genetic modifiers that alter disease onset, progression, or severity.236 This new role for NHRs as genetic modifiers of disease is important in understanding normal CNS development and has great therapeutic implications. NHRs that function as master regulators of key biological pathways may have a profound impact on disease treatment as they have the potential to impact multiple mutations and multiple pathways.
Footnotes
ACADEMIC EDITOR: Lora Watts, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,060 words, excluding any confidential comments to the academic editor.
FUNDING: This review was funded by RO1 EY017653, the Edward N. and Della L. Thome Memorial Foundation, the Bank of America N.A., Trustee, the CHASE Fund, Hope for Vision, RPB, the Webster Foundation, and the American Macular Degeneration Foundation. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: AMO, OAM-R, and NBH. Analyzed the data: AMO, OAM-R. Contributed to the writing of the article: AMO, OAM-R, and NBH. Agreed the manuscript results and conclusions: AMO, OAM-R, and NBH. Jointly developed the structure and arguments for the article: AMO and OAM-R. Made critical revisions and approved the final version: NBH. All authors reviewed and approved the final article.
REFERENCES
- 1.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 2.Committee NR. A unified nomenclature system for the nuclear receptor superfamily. [Accessed March 10, 2015];Cell. 1999 97(2):161–163. doi: 10.1016/s0092-8674(00)80726-6. http://www.ncbi.nlm.nih.gov/pubmed/10219237. [DOI] [PubMed] [Google Scholar]
- 3.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. [Accessed July 31, 2015];Steroids. 1999 64(5):310–319. doi: 10.1016/s0039-128x(99)00014-8. http://www.ncbi.nlm.nih.gov/pubmed/10406480. [DOI] [PubMed] [Google Scholar]
- 4.Pawlak M, Lefebvre P, Staels B. General molecular biology and architecture of nuclear receptors. [Accessed August 5, 2015];Curr Top Med Chem. 2012 12(6):486–504. doi: 10.2174/156802612799436641. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3637177&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selmi-Ruby S, Casanova J, Malhotra S, Roussett B, Raaka BM, Samuels HH. Role of the conserved C-terminal region of thyroid hormone receptor-alpha in ligand-dependent transcriptional activation. [Accessed October 6, 2015];Mol Cell Endocrinol. 1998 138(1–2):105–114. doi: 10.1016/s0303-7207(98)00016-1. http://www.ncbi.nlm.nih.gov/pubmed/9685219. [DOI] [PubMed] [Google Scholar]
- 6.Smirnov AN. Nuclear receptors: nomenclature, ligands, mechanisms of their effects on gene expression. [Accessed July 31, 2015];Biochem Biokhimiia. 2002 67(9):957–977. doi: 10.1023/a:1020545200302. http://www.ncbi.nlm.nih.gov/pubmed/12387709. [DOI] [PubMed] [Google Scholar]
- 7.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. [Accessed June 17, 2015];Physiol Rev. 2001 81(3):1269–1304. doi: 10.1152/physrev.2001.81.3.1269. http://www.ncbi.nlm.nih.gov/pubmed/11427696. [DOI] [PubMed] [Google Scholar]
- 8.Stunnenberg HG. Mechanisms of transactivation by retinoic acid receptors. Bioessays. 1993;15(5):309–315. doi: 10.1002/bies.950150504. [DOI] [PubMed] [Google Scholar]
- 9.Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19(3):595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- 10.Perlmann T, Umesono K, Rangarajan PN, Forman BM, Evans RM. Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol. 1996;10(8):958–966. doi: 10.1210/mend.10.8.8843412. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25(1):118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giguère V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20(5):689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XK, Hoffmann B, Tran PB, Graupner G, Pfahl M. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature. 1992;355(6359):441–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]
- 14.Kurokawa R, DiRenzo J, Boehm M, et al. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371(6497):528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 15.Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355(6359):446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr FE, Wong NC. Characteristics of a negative thyroid hormone response element. [Accessed July 31, 2015];J Biol Chem. 1994 269(6):4175–4179. http://www.ncbi.nlm.nih.gov/pubmed/8307979. [PubMed] [Google Scholar]
- 17.Ou XM, Storring JM, Kushwaha N, Albert PR. Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J Biol Chem. 2001;276(17):14299–14307. doi: 10.1074/jbc.M005363200. [DOI] [PubMed] [Google Scholar]
- 18.Malkoski SP, Dorin RI. Composite Glucocorticoid Regulation at a Functionally Defined Negative Glucocorticoid Response Element of the Human Corticotropin-Releasing Hormone Gene. Mol Endocrinol. 1999;13(10):1629–1644. doi: 10.1210/mend.13.10.0351. [DOI] [PubMed] [Google Scholar]
- 19.Kalkhoven E, Valentine JE, Heery DM, Parker MG. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17(1):232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robyr D, Wolffe AP, Wahli W. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol. 2000;14(3):329–347. doi: 10.1210/mend.14.3.0411. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Ryan F, Swaffield JC, Johnston SA, Moore DD. Interaction of thyroidhormone receptor with a conserved transcriptionalmediator. Nature. 1995;374(6517):91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 22.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20(3):321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 23.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. [Accessed October 5, 2015];Trends Endocrinol Metab. 2001 12(3):127–134. doi: 10.1016/s1043-2760(00)00355-6. http://www.ncbi.nlm.nih.gov/pubmed/11306338. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Jeyakumar M, Petukhov S, Bagchi MK. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol Endocrinol. 1998;12(4):513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]
- 25.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. [Accessed July 31, 2015];Mol Cell. 2000 5(2):289–298. doi: 10.1016/s1097-2765(00)80424-4. http://www.ncbi.nlm.nih.gov/pubmed/10882070. [DOI] [PubMed] [Google Scholar]
- 26.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. [Accessed July 31, 2015];Cell. 1995 83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. http://www.ncbi.nlm.nih.gov/pubmed/8521508. [DOI] [PubMed] [Google Scholar]
- 27.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. [Accessed April 7, 2015];Cell. 1995 83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. http://www.ncbi.nlm.nih.gov/pubmed/8521507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans RM. The steroid and thyroid hormone receptor superfamily. [Accessed October 5, 2015];Science. 1988 240(4854):889–895. doi: 10.1126/science.3283939. http://www.ncbi.nlm.nih.gov/pubmed/3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore DD, Kato S, Xie W, et al. International Union of Pharmacology. LXII. The NR1H and NR1I Receptors: Constitutive Androstane Receptor, Pregnene X Receptor, Farnesoid X Receptor, Farnesoid X Receptor beta, Liver X Receptor, Liver X Receptor beta, and Vitamin D Receptor. Pharmacol Rev. 2006;58(4):742–759. doi: 10.1124/pr.58.4.6. [DOI] [PubMed] [Google Scholar]
- 30.Benoit G, Cooney A, Giguere V, et al. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev. 2006;58(4):798–836. doi: 10.1124/pr.58.4.10. [DOI] [PubMed] [Google Scholar]
- 31.Leid M, Kastner P, Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. [Accessed August 31, 2015];Trends Biochem Sci. 1992 17(10):427–433. doi: 10.1016/0968-0004(92)90014-z. http://www.ncbi.nlm.nih.gov/pubmed/1333659. [DOI] [PubMed] [Google Scholar]
- 32.Rowe A, Brickell PM. The nuclear retinoid receptors. [Accessed August 31, 2015];Int J Exp Pathol. 1993 74(2):117–126. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2002115&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 33.Heyman RA, Mangelsdorf DJ, Dyck JA, et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. [Accessed October 5, 2015];Cell. 1992 68(2):397–406. doi: 10.1016/0092-8674(92)90479-v. http://www.ncbi.nlm.nih.gov/pubmed/1310260. [DOI] [PubMed] [Google Scholar]
- 34.Dollé P, Ruberte E, Kastner P, et al. Differential expression of genes encoding alpha, beta and gamma retinoic acid receptors and CRABP in the developing limbs of the mouse. Nature. 1989;342(6250):702–705. doi: 10.1038/342702a0. [DOI] [PubMed] [Google Scholar]
- 35.Mangelsdorf DJ, Borgmeyer U, Heyman RA, et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. [Accessed August 31, 2015];Genes Dev. 1992 6(3):329–344. doi: 10.1101/gad.6.3.329. http://www.ncbi.nlm.nih.gov/pubmed/1312497. [DOI] [PubMed] [Google Scholar]
- 36.Chawla A. Nuclear Receptors and Lipid Physiology: Opening the X-Files. Science (80−. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 37.Yu VC, Delsert C, Andersen B, et al. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. [Accessed September 26, 2015];Cell. 1991 67(6):1251–1266. doi: 10.1016/0092-8674(91)90301-e. http://www.ncbi.nlm.nih.gov/pubmed/1662118. [DOI] [PubMed] [Google Scholar]
- 38.Glass CK, Lipkin SM, Devary OV, Rosenfeld MG. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. [Accessed October 5, 2015];Cell. 1989 59(4):697–708. doi: 10.1016/0092-8674(89)90016-0. http://www.ncbi.nlm.nih.gov/pubmed/2555064. [DOI] [PubMed] [Google Scholar]
- 39.Allenby G, Bocquel MT, Saunders M, et al. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. [Accessed July 31, 2015];Proc Natl Acad Sci U S A. 1993 90(1):30–34. doi: 10.1073/pnas.90.1.30. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=45593&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCaffery P, Dräger UC. High levels of a retinoic acid-generating dehydrogenase in the meso-telencephalic dopamine system. [Accessed July 31, 2015];Proc Natl Acad Sci U S A. 1994 91(16):7772–7776. doi: 10.1073/pnas.91.16.7772. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=44484&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zetterström RH, Lindqvist E, Mata de Urquiza A, et al. Role of retinoids in the CNS: differential expression of retinoid binding proteins and receptors and evidence for presence of retinoic acid. [Accessed July 31, 2015];Eur J Neurosci. 1999 11(2):407–416. doi: 10.1046/j.1460-9568.1999.00444.x. http://www.ncbi.nlm.nih.gov/pubmed/10051741. [DOI] [PubMed] [Google Scholar]
- 42.Toresson H, Mata de Urquiza A, Fagerström C, Perlmann T, Campbell K. Retinoids are produced by glia in the lateral ganglionic eminence and regulate striatal neuron differentiation. [Accessed July 31, 2015];Development. 1999 126(6):1317–1326. doi: 10.1242/dev.126.6.1317. http://www.ncbi.nlm.nih.gov/pubmed/10021349. [DOI] [PubMed] [Google Scholar]
- 43.Rhinn M, Dollé P. Retinoic acid signalling during development. Development. 2012;139(5):843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 44.Parker RO, Crouch RK. Retinol dehydrogenases (RDHs) in the visual cycle. Exp Eye Res. 2010;91(6):788–792. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Underwood BA. Vitamin A and cancer prevention conference—an introduction. [Accessed August 1, 2015];J Natl Cancer Inst. 1984 73(6):1371–1372. http://www.ncbi.nlm.nih.gov/pubmed/6595446. [PubMed] [Google Scholar]
- 46.Wolf G. A history of vitamin A and retinoids. [Accessed August 1, 2015];FASEB J. 1996 10(9):1102–1107. doi: 10.1096/fasebj.10.9.8801174. http://www.ncbi.nlm.nih.gov/pubmed/8801174. [DOI] [PubMed] [Google Scholar]
- 47.Kochhar DM, Jiang H, Penner JD, Johnson AT, Chandraratna RA. The use of a retinoid receptor antagonist in a new model to study vitamin A-dependent developmental events. [Accessed August 1, 2015];Int J Dev Biol. 1998 42(4):601–608. http://www.ncbi.nlm.nih.gov/pubmed/9694631. [PubMed] [Google Scholar]
- 48.Fares-Taie L, Gerber S, Chassaing N, et al. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet. 2013;92(2):265–270. doi: 10.1016/j.ajhg.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mory A, Ruiz FX, Dagan E, et al. A missense mutation in ALDH1A3 causes isolated microphthalmia/anophthalmia in nine individuals from an inbred Muslim kindred. Eur J Hum Genet. 2014;22(3):419–422. doi: 10.1038/ejhg.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semerci CN, Kalay E, Yıldırım C, et al. Novel splice-site and missense mutations in the ALDH1A3 gene underlying autosomal recessive anophthalmia/microphthalmia. Br J Ophthalmol. 2014;98(6):832–840. doi: 10.1136/bjophthalmol-2013-304058. [DOI] [PubMed] [Google Scholar]
- 51.Aldahmesh MA, Khan AO, Hijazi H, Alkuraya FS. Mutations in ALDH1A3 cause microphthalmia. Clin Genet. 2013;84(2):128–131. doi: 10.1111/cge.12184. [DOI] [PubMed] [Google Scholar]
- 52.Roos L, Fang M, Dali C, et al. A homozygous mutation in a consanguineous family consolidates the role of ALDH1A3 in autosomal recessive microphthalmia. Clin Genet. 2013 Sep; doi: 10.1111/cge.12277. [DOI] [PubMed] [Google Scholar]
- 53.Yahyavi M, Abouzeid H, Gawdat G, et al. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet. 2013;22(16):3250–3258. doi: 10.1093/hmg/ddt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9(7):541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 55.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134(6):921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao W-L, Wang H-F, Tsai H-C, et al. Retinoid signaling competence and RARbeta-mediated gene regulation in the developing mammalian telencephalon. Dev Dyn. 2005;232(4):887–900. doi: 10.1002/dvdy.20281. [DOI] [PubMed] [Google Scholar]
- 57.Liao W-L, Liu FC. RARbeta isoform-specific regulation of DARPP-32 gene expression: an ectopic expression study in the developing rat telencephalon. Eur J Neurosci. 2005;21(12):3262–3268. doi: 10.1111/j.1460-9568.2005.04178.x. [DOI] [PubMed] [Google Scholar]
- 58.Bonhomme D, Pallet V, Dominguez G, et al. Retinoic acid modulates intrahippocampal levels of corticosterone in middle-aged mice: consequences on hippocampal plasticity and contextual memory. Front Aging Neurosci. 2014;6:6. doi: 10.3389/fnagi.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizee MR, Wooldrik D, Lakeman KAM, et al. Retinoic acid induces blood-brain barrier development. J Neurosci. 2013;33(4):1660–1671. doi: 10.1523/JNEUROSCI.1338-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sucov HM, Murakami KK, Evans RM. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. [Accessed July 31, 2015];Proc Natl Acad Sci U S A. 1990 87(14):5392–5396. doi: 10.1073/pnas.87.14.5392. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=54330&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Thé H, Tiollais P, Dejean A. The retinoic acid receptors. [Accessed July 31, 2015];Nouv Rev Fr Hematol. 1990 32(1):30–32. http://www.ncbi.nlm.nih.gov/pubmed/2161511. [PubMed] [Google Scholar]
- 62.Wu TC, Wang L, Wan YJ. Retinoic acid regulates gene expression of retinoic acid receptors alpha, beta and gamma in F9 mouse teratocarcinoma cells. [Accessed July 31, 2015];Differentiation. 1992 51(3):219–224. doi: 10.1111/j.1432-0436.1992.tb00699.x. http://www.ncbi.nlm.nih.gov/pubmed/1334013. [DOI] [PubMed] [Google Scholar]
- 63.Reinhart VL, Nguyen T, Gerwien R, Kuhn M, Yates PD, Lanz TA. Downstream effects of striatal-enriched protein tyrosine phosphatase reduction on RNA expression in vivo and in vitro. Neuroscience. 2014;278:62–69. doi: 10.1016/j.neuroscience.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Chiang MY, Misner D, Kempermann G, et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. [Accessed November 7, 2015];Neuron. 1998 21(6):1353–1361. doi: 10.1016/s0896-6273(00)80654-6. http://www.ncbi.nlm.nih.gov/pubmed/9883728. [DOI] [PubMed] [Google Scholar]
- 65.Tini M, Fraser RA, Giguere V. Functional Interactions between Retinoic Acid Receptor-related Orphan Nuclear Receptor (ROR) and the Retinoic Acid Receptors in the Regulation of the F-Crystallin Promoter. J Biol Chem. 1995;270(34):20156–20161. doi: 10.1074/jbc.270.34.20156. [DOI] [PubMed] [Google Scholar]
- 66.Cvekl A, Wang WL. Retinoic acid signaling in mammalian eye development. Exp Eye Res. 2009;89(3):280–291. doi: 10.1016/j.exer.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dollé P, Ruberte E, Leroy P, Morriss-Kay G, Chambon P. Retinoic acid receptors and cellular retinoid binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. [Accessed August 1, 2015];Development. 1990 110(4):1133–1151. doi: 10.1242/dev.110.4.1133. http://www.ncbi.nlm.nih.gov/pubmed/1966045. [DOI] [PubMed] [Google Scholar]
- 68.Ghyselinck NB, Dupé V, Dierich A, et al. Role of the retinoic acid receptor beta (RARbeta) during mouse development. [Accessed October 2, 2015];Int J Dev Biol. 1997 41(3):425–447. http://www.ncbi.nlm.nih.gov/pubmed/9240560. [PubMed] [Google Scholar]
- 69.Johnston MC, Noden DM, Hazelton RD, Coulombre JL, Coulombre AJ. Origins of avian ocular and periocular tissues. Exp Eye Res. 1979;29(1):27–43. doi: 10.1016/0014-4835.(79)90164.7. [DOI] [PubMed] [Google Scholar]
- 70.Muñoz A, Bernal J. Biological activities of thyroid hormone receptors. [Accessed August 31, 2015];Eur J Endocrinol. 1997 137(5):433–445. doi: 10.1530/eje.0.1370433. http://www.ncbi.nlm.nih.gov/pubmed/9405018. [DOI] [PubMed] [Google Scholar]
- 71.Saatcioglu F, Deng T, Karin M. A novel cis element mediating ligand-independent activation by c-ErbA: implications for hormonal regulation. [Accessed August 31, 2015];Cell. 1993 75(6):1095–1105. doi: 10.1016/0092-8674(93)90319-l. http://www.ncbi.nlm.nih.gov/pubmed/7903219. [DOI] [PubMed] [Google Scholar]
- 72.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O’Malley BW. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. [Accessed October 5, 2015];Proc Natl Acad Sci U S A. 1993 90(19):8832–8836. doi: 10.1073/pnas.90.19.8832. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=47454&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schräder M, Müller KM, Nayeri S, Kahlen JP, Carlberg C. Vitamin D3-thyroid hormone receptor heterodimer polarity directs ligand sensitivity of transactivation. Nature. 1994;370(6488):382–386. doi: 10.1038/370382a0. [DOI] [PubMed] [Google Scholar]
- 74.Bogazzi F, Hudson LD, Nikodem VM. A novel heterodimerization partner for thyroid hormone receptor. Peroxisome proliferator-activated receptor. [Accessed October 5, 2015];J Biol Chem. 1994 269(16):11683–11686. http://www.ncbi.nlm.nih.gov/pubmed/8163464. [PubMed] [Google Scholar]
- 75.Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. [Accessed October 8, 2015];J Biol Chem. 1996 271(45):28516–28520. doi: 10.1074/jbc.271.45.28516. http://www.ncbi.nlm.nih.gov/pubmed/8910480. [DOI] [PubMed] [Google Scholar]
- 76.Oberste-Berghaus C, Zanger K, Hashimoto K, Cohen RN, Hollenberg AN, Wondisford FE. Thyroid Hormone-independent Interaction between the Thyroid Hormone Receptor 2 Amino Terminus and Coactivators. J Biol Chem. 2000;275(3):1787–1792. doi: 10.1074/jbc.275.3.1787. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 78.Anderson GW, Schoonover CM, Jones SA. Control of thyroid hormone action in the developing rat brain. Thyroid. 2003;13(11):1039–1056. doi: 10.1089/105072503770867219. [DOI] [PubMed] [Google Scholar]
- 79.de Escobar GM, Obregón MJ, del Rey FE. Maternal thyroid hormones early in pregnancy and fetal brain development. Best Pract Res Clin Endocrinol Metab. 2004;18(2):225–248. doi: 10.1016/j.beem.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 80.Bernal J. Thyroid hormones and brain development. Vitam Horm. 2005;71:95–122. doi: 10.1016/S0083-6729(05)71004-9. [DOI] [PubMed] [Google Scholar]
- 81.Piosik PA, van Groenigen M, van Doorn J, Baas F, de Vijlder JJ. Effects of maternal thyroid status on thyroid hormones and growth in congenitally hypothyroid goat fetuses during the second half of gestation. Endocrinology. 1997;138(1):5–11. doi: 10.1210/endo.138.1.4843. [DOI] [PubMed] [Google Scholar]
- 82.Obregon MJ, Mallol J, Pastor R, Morreale de Escobar G, Escobar del Rey F. L-thyroxine and 3,5,3′-triiodo-L-thyronine in rat embryos before onset of fetal thyroid function. Endocrinology. 1984;114(1):305–307. doi: 10.1210/endo-114-1-305. [DOI] [PubMed] [Google Scholar]
- 83.Faustino LC, Ortiga-Carvalho TM. Thyroid hormone role on cerebellar development and maintenance: a perspective based on transgenic mouse models. Front Endocrinol (Lausanne) 2014;5:75. doi: 10.3389/fendo.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tata JR. The road to nuclear receptors of thyroid hormone. Biochim Biophys Acta. 2013;1830(7):3860–3866. doi: 10.1016/j.bbagen.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 85.Bradley DJ, Towle HC, Young WS. Spatial and temporal expression of alpha- and beta-thyroid hormone receptor mRNAs, including the beta 2-subtype, in the developing mammalian nervous system. [Accessed August 31, 2015];J Neurosci. 1992 12(6):2288–2302. doi: 10.1523/JNEUROSCI.12-06-02288.1992. http://www.ncbi.nlm.nih.gov/pubmed/1607941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chan S, Kachilele S, McCabe CJ, et al. Early expression of thyroid hormone deiodinases and receptors in human fetal cerebral cortex. [Accessed August 31, 2015];Brain Res Dev Brain Res. 2002 138(2):109–116. doi: 10.1016/s0165-3806(02)00459-5. http://www.ncbi.nlm.nih.gov/pubmed/12354639. [DOI] [PubMed] [Google Scholar]
- 87.Ercan-Fang S, Schwartz HL, Oppenheimer JH. Isoform-specific 3,5,3′- triiodothyronine receptor binding capacity and messenger ribonucleic acid content in rat adenohypophysis: effect of thyroidal state and comparison with extrapituitary tissues. Endocrinology. 1996;137(8):3228–3233. doi: 10.1210/endo.137.8.8754744. [DOI] [PubMed] [Google Scholar]
- 88.Berbel P, Ausó E, García-Velasco JV, Molina ML, Camacho M. Role of thyroid hormones in the maturation and organisation of rat barrel cortex. [Accessed August 31, 2015];Neuroscience. 2001 107(3):383–394. doi: 10.1016/s0306-4522(01)00368-2. http://www.ncbi.nlm.nih.gov/pubmed/11718994. [DOI] [PubMed] [Google Scholar]
- 89.Bradley DJ, Young WS, Weinberger C. Differential expression of alpha and beta thyroid hormone receptor genes in rat brain and pituitary. [Accessed August 31, 2015];Proc Natl Acad Sci U S A. 1989 86(18):7250–7254. doi: 10.1073/pnas.86.18.7250. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=298035&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strait KA, Schwartz HL, Seybold VS, Ling NC, Oppenheimer JH. Immunofluorescence localization of thyroid hormone receptor protein beta 1 and variant alpha 2 in selected tissues: cerebellar Purkinje cells as a model for beta 1 receptor-mediated developmental effects of thyroid hormone in brain. [Accessed August 31, 2015];Proc Natl Acad Sci U S A. 1991 88(9):3887–3891. doi: 10.1073/pnas.88.9.3887. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=51558&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morte B, Manzano J, Scanlan T, Vennström B, Bernal J. Deletion of the thyroid hormone receptor alpha 1 prevents the structural alterations of the cerebellum induced by hypothyroidism. Proc Natl Acad Sci U S A. 2002;99(6):3985–3989. doi: 10.1073/pnas.062413299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flamant F, Samarut J. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. [Accessed August 31, 2015];Trends Endocrinol Metab. 2003 14(2):85–90. doi: 10.1016/s1043-2760(02)00043-7. http://www.ncbi.nlm.nih.gov/pubmed/12591179. [DOI] [PubMed] [Google Scholar]
- 93.Göthe S, Wang Z, Ng L, et al. Mice devoid of all known thyroid hormone receptors are viable but exhibit disorders of the pituitary-thyroid axis, growth, and bone maturation. [Accessed August 31, 2015];Genes Dev. 1999 13(10):1329–1341. doi: 10.1101/gad.13.10.1329. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=316730&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mellström B, Naranjo JR, Santos A, Gonzalez AM, Bernal J. Independent expression of the alpha and beta c-erbA genes in developing rat brain. Mol Endocrinol. 1991;5(9):1339–1350. doi: 10.1210/mend-5-9-1339. [DOI] [PubMed] [Google Scholar]
- 95.Hashimoto K, Curty FH, Borges PP, et al. An unliganded thyroid hormone receptor causes severe neurological dysfunction. Proc Natl Acad Sci U S A. 2001;98(7):3998–4003. doi: 10.1073/pnas.051454698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heuer H, Mason CA. Thyroid Hormone Induces Cerebellar Purkinje Cell Dendritic Development via the Thyroid Hormone Receptor {alpha}1. [Accessed August 31, 2015];J Neurosci. 2003 23(33):10604–10612. doi: 10.1523/JNEUROSCI.23-33-10604.2003. http://www.jneurosci.org/content/23/33/10604.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez R, Gomes FCA. Proliferation of cerebellar neurons induced by astrocytes treated with thyroid hormone is mediated by a cooperation between cell contact and soluble factors and involves the epidermal growth factor-protein kinase a pathway. J Neurosci Res. 2005;80(3):341–349. doi: 10.1002/jnr.20463. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Zhong J, Xu H, et al. Perinatal iodine deficiency and hypothyroidism increase cell apoptosis and alter doublecortin and reelin protein expressions in rat cerebellum. Arch Med Res. 2012;43(4):255–264. doi: 10.1016/j.arcmed.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 99.Bochukova E, Schoenmakers N, Agostini M, et al. A mutation in the thyroid hormone receptor alpha gene. N Engl J Med. 2012;366(3):243–249. doi: 10.1056/NEJMoa1110296. [DOI] [PubMed] [Google Scholar]
- 100.van Mullem A, van Heerebeek R, Chrysis D, et al. Clinical phenotype and mutant TRα1. N Engl J Med. 2012;366(15):1451–1453. doi: 10.1056/NEJMc1113940. [DOI] [PubMed] [Google Scholar]
- 101.Quignodon L, Grijota-Martinez C, Compe E, et al. A combined approach identifies a limited number of new thyroid hormone target genes in post-natal mouse cerebellum. J Mol Endocrinol. 2007;39(1):17–28. doi: 10.1677/JME-06-0054. [DOI] [PubMed] [Google Scholar]
- 102.Trimarchi JM, Harpavat S, Billings NA, Cepko CL. Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC Dev Biol. 2008;8(1):101. doi: 10.1186/1471-213X-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forrest D, Sjöberg M, Vennström B. Contrasting developmental and tissuespecific expression of alpha and beta thyroid hormone receptor genes. [Accessed January 4, 2016];EMBO J. 1990 9(5):1519–1528. doi: 10.1002/j.1460-2075.1990.tb08270.x. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=551844&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kelley MW, Turner JK, Reh TA. Ligands of steroid/thyroid receptors induce cone photoreceptors in vertebrate retina. [Accessed November 19, 2015];Development. 1995 121(11):3777–3785. doi: 10.1242/dev.121.11.3777. http://www.ncbi.nlm.nih.gov/pubmed/8582287. [DOI] [PubMed] [Google Scholar]
- 105.Havis E, Le Mevel S, Morvan Dubois G, et al. Unliganded thyroid hormone receptor is essential for Xenopus laevis eye development. EMBO J. 2006;25(20):4943–4951. doi: 10.1038/sj.emboj.7601356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ng L, Hurley JB, Dierks B, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27(1):94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 107.Reddy JK, Goel SK, Nemali MR, et al. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. [Accessed October 5, 2015];Proc Natl Acad Sci U S A. 1986 83(6):1747–1751. doi: 10.1073/pnas.83.6.1747. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=323161&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr. 1994;14:343–370. doi: 10.1146/annurev.nu.14.070194.002015. [DOI] [PubMed] [Google Scholar]
- 109.He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. [Accessed September 25, 2015];Cell. 1999 99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3779681&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanderson LM, Kersten S. In: PPARs: important rgulators in metabolism and inflammation in: Nuclear Receptors. Bunce CM, Campbell MJ, editors. Dordrecht: Springer Netherlands; 2010. pp. 259–285. [DOI] [Google Scholar]
- 111.Varga T, Czimmerer Z, Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta. 2011;1812(8):1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clark RB. The role of PPARs in inflammation and immunity. [Accessed August 31, 2015];J Leukoc Biol. 2002 71(3):388–400. http://www.ncbi.nlm.nih.gov/pubmed/11867676. [PubMed] [Google Scholar]
- 113.Heneka MT, Landreth GE. PPARs in the brain. Biochim Biophys Acta. 2007;1771(8):1031–1045. doi: 10.1016/j.bbalip.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 114.Chu R, Madison LD, Lin Y, et al. Thyroid hormone (T3) inhibits ciprofibrate-induced transcription of genes encoding beta-oxidation enzymes: cross talk between peroxisome proliferator and T3 signaling pathways. [Accessed October 5, 2015];Proc Natl Acad Sci U S A. 1995 92(25):11593–11597. doi: 10.1073/pnas.92.25.11593. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=40448&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Göttlicher M, Widmark E, Li Q, Gustafsson JA. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. [Accessed August 31, 2015];Proc Natl Acad Sci U S A. 1992 89(10):4653–4657. doi: 10.1073/pnas.89.10.4653. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=49141&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Amero SA, Kretsinger RH, Moncrief ND, Yamamoto KR, Pearson WR. The origin of nuclear receptor proteins: a single precursor distinct from other transcription factors. Mol Endocrinol. 1992;6(1):3–7. doi: 10.1210/mend.6.1.1738368. [DOI] [PubMed] [Google Scholar]
- 117.Escriva H, Delaunay F, Laudet V. Ligand binding and nuclear receptor evolution. Bioessays. 2000;22(8):717–727. doi: 10.1002/1521-1878(200008)22:8. <717: AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 118.Burris TP, Guo W, McCabe ER. The gene responsible for adrenal hypoplasia congenita, DAX-1, encodes a nuclear hormone receptor that defines a new class within the superfamily. [Accessed October 5, 2015];Recent Prog Horm Res. 1996 51:241–259. discussion 259–260. http://www.ncbi.nlm.nih.gov/pubmed/8701082. [PubMed] [Google Scholar]
- 119.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. [Accessed October 5, 2015];Science. 1996 272(5266):1336–1339. doi: 10.1126/science.272.5266.1336. http://www.ncbi.nlm.nih.gov/pubmed/8650544. [DOI] [PubMed] [Google Scholar]
- 120.Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. [Accessed October 2, 2015];J Mol Endocrinol. 1997 19(3):207–226. doi: 10.1677/jme.0.0190207. http://www.ncbi.nlm.nih.gov/pubmed/9460643. [DOI] [PubMed] [Google Scholar]
- 121.Cheng H, Khanna H, Oh ECT, Hicks D, Mitton KP, Swaroop A. Photoreceptorspecific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13(15):1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 122.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haider NB, Jacobson SG, Cideciyan AV, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24(2):127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 124.Bumsted O’Brien KM, Cheng H, Jiang Y, Schulte D, Swaroop A, Hendrickson AE. Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Invest Ophthalmol Vis Sci. 2004;45(8):2807–2812. doi: 10.1167/iovs.03-1317. [DOI] [PubMed] [Google Scholar]
- 125.Nystuen AM, Sachs AJ, Yuan Y, Heuermann L, Haider NB. A novel mutation in Prph2, a gene regulated by Nr2e3, causes retinal degeneration and outersegment defects similar to Nr2e3 rd7/rd7 retinas. Mamm Genome. 2008;19(9):623–633. doi: 10.1007/s00335-008-9138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haider NB, Demarco P, Nystuen AM, et al. The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis Neurosci. 2006;23(6):917–929. doi: 10.1017/S095252380623027X. [DOI] [PubMed] [Google Scholar]
- 127.Mollema NJ, Yuan Y, Jelcick AS, et al. Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS One. 2011;6(3):e17494. doi: 10.1371/journal.pone.0017494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. [Accessed September 3, 2015];Cell. 1997 91(4):531–541. doi: 10.1016/s0092-8674(00)80439-0. http://www.ncbi.nlm.nih.gov/pubmed/9390562. [DOI] [PubMed] [Google Scholar]
- 129.Freund CL, Gregory-Evans CY, Furukawa T, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. [Accessed September 3, 2015];Cell. 1997 91(4):543–553. doi: 10.1016/s0092-8674(00)80440-7. http://www.ncbi.nlm.nih.gov/pubmed/9390563. [DOI] [PubMed] [Google Scholar]
- 130.Chen S, Wang QL, Nie Z, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. [Accessed September 3, 2015];Neuron. 1997 19(5):1017–1030. doi: 10.1016/s0896-6273(00)80394-3. http://www.ncbi.nlm.nih.gov/pubmed/9390516. [DOI] [PubMed] [Google Scholar]
- 131.Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. [Accessed September 3, 2015];Oncogene. 1996 12(1):207–211. http://www.ncbi.nlm.nih.gov/pubmed/8552394. [PubMed] [Google Scholar]
- 132.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29(4):447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 133.Haider NB, Mollema N, Gaule M, et al. Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res. 2009;89(3):365–372. doi: 10.1016/j.exer.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silveira AC, Morrison MA, Ji F, et al. Convergence of linkage, gene expression and association data demonstrates the influence of the RAR-related orphan receptor alpha (RORA) gene on neovascular AMD: A systems biology based approach. Vision Res. 2011;50(7):698–715. doi: 10.1016/j.visres.2009.09.016.Convergence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.André E, Gawlas K, Becker-André M. A novel isoform of the orphan nuclear receptor RORbeta is specifically expressed in pineal gland and retina. [Accessed August 1, 2015];Gene. 1998 216(2):277–283. doi: 10.1016/s0378-1119(98)00348-5. http://www.ncbi.nlm.nih.gov/pubmed/9729429. [DOI] [PubMed] [Google Scholar]
- 136.André E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-André M. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17(14):3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Azadi S, Zhang Y, Caffé AR, Holmqvist B, van Veen T. Thyroid-beta2 and the retinoid RAR-alpha, RXR-gamma and ROR-beta2 receptor mRNAs; expression profiles in mouse retina, retinal explants and neocortex. [Accessed December 10, 2015];Neuroreport. 2002 13(6):745–750. doi: 10.1097/00001756-200205070-00003. http://www.ncbi.nlm.nih.gov/pubmed/11997680. [DOI] [PubMed] [Google Scholar]
- 138.Nakagawa Y, O’Leary DDM. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25(2–4):234–244. doi: 10.1159/000072271. doi:72271. [DOI] [PubMed] [Google Scholar]
- 139.Schaeren-Wiemers N, André E, Kapfhammer JP, Becker-André M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. [Accessed August 1, 2015];Eur J Neurosci. 1997 9(12):2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. http://www.ncbi.nlm.nih.gov/pubmed/9517474. [DOI] [PubMed] [Google Scholar]
- 140.Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. Rhythmic expression of ROR beta mRNA in the mice suprachiasmatic nucleus. [Accessed December 10, 2015];Neurosci Lett. 2002 320(1–2):13–16. doi: 10.1016/s0304-3940(02)00011-3. http://www.ncbi.nlm.nih.gov/pubmed/11849752. [DOI] [PubMed] [Google Scholar]
- 141.Ueda HR, Chen W, Adachi A, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 142.Yang XO, Pappu BP, Nurieva R, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28(1):29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 144.Jetten AM, Joo JH. Retinoid-related Orphan Receptors (RORs): Roles in Cellular Differentiation and Development. Adv Dev Biol. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ueda HR, Hayashi S, Chen W, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 146.Matysiak-Scholze U, Nehls M. The structural integrity of ROR alpha isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43(1):78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- 147.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguère V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. [Accessed July 31, 2015];Mech Dev. 1998 70(1–2):147–153. doi: 10.1016/s0925-4773(97)00187-1. http://www.ncbi.nlm.nih.gov/pubmed/9510031. [DOI] [PubMed] [Google Scholar]
- 148.Jetten AM, Ueda E. Retinoid-related orphan receptors (RORs): roles in cell survival, differentiation and disease. Cell Death Differ. 2002;9(11):1167–1171. doi: 10.1038/sj.cdd.4401085. [DOI] [PubMed] [Google Scholar]
- 149.Bayer SA, Wills KV, Wei J, et al. Phenotypic effects of the weaver gene are evident in the embryonic cerebellum but not in the ventral midbrain. [Accessed August 1, 2015];Brain Res Dev Brain Res. 1996 96(1–2):130–137. doi: 10.1016/0165-3806(96)00107-1. http://www.ncbi.nlm.nih.gov/pubmed/8922675. [DOI] [PubMed] [Google Scholar]
- 150.Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. [Accessed August 1, 2015];Curr Opin Neurobiol. 1997 7(1):40–47. doi: 10.1016/s0959-4388(97)80118-3. http://www.ncbi.nlm.nih.gov/pubmed/9039803. [DOI] [PubMed] [Google Scholar]
- 151.Gold DA, Baek SH, Schork NJ, et al. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. [Accessed August 1, 2015];Neuron. 2003 40(6):1119–1131. doi: 10.1016/s0896-6273(03)00769-4. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2717708&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. [Accessed June 11, 2015];Trends Neurosci. 1998 21(9):375–382. doi: 10.1016/s0166-2236(98)01313-7. http://www.ncbi.nlm.nih.gov/pubmed/9735945. [DOI] [PubMed] [Google Scholar]
- 153.Doulazmi M, Frédéric F, Capone F, Becker-André M, Delhaye-Bouchaud N, Mariani J. A comparative study of Purkinje cells in two RORalpha gene mutant mice: staggerer and RORalpha(−/−) [Accessed August 1, 2015];Brain Res Dev Brain Res. 2001 127(2):165–174. doi: 10.1016/s0165-3806(01)00131-6. http://www.ncbi.nlm.nih.gov/pubmed/11335003. [DOI] [PubMed] [Google Scholar]
- 154.Doulazmi M, Frédéric F, Lemaigre-Dubreuil Y, Hadj-Sahraoui N, Delhaye-Bouchaud N, Mariani J. Cerebellar Purkinje cell loss during life span of the heterozygous staggerer mouse (Rora(+)/Rora(sg)) is gender-related. [Accessed August 1, 2015];J Comp Neurol. 1999 411(2):267–273. http://www.ncbi.nlm.nih.gov/pubmed/10404252. [PubMed] [Google Scholar]
- 155.Herrup K, Mullen RJ. Intercellular transfer of beta-glucuronidase in chimeric mice. [Accessed August 1, 2015];J Cell Sci. 1979 40:21–31. doi: 10.1242/jcs.40.1.21. http://www.ncbi.nlm.nih.gov/pubmed/536387. [DOI] [PubMed] [Google Scholar]
- 156.Landis DM, Reese TS. Structure of the Purkinje cell membrane in staggerer and weaver mutant mice. J Comp Neurol. 1977;171(2):247–260. doi: 10.1002/cne.901710208. [DOI] [PubMed] [Google Scholar]
- 157.Landis DM, Sidman RL. Electron microscopic analysis of postnatal histogenesis in the cerebellar cortex of staggerer mutant mice. J Comp Neurol. 1978;179(4):831–863. doi: 10.1002/cne.901790408. [DOI] [PubMed] [Google Scholar]
- 158.Hadj-Sahraoui N, Frederic F, Zanjani H, Delhaye-Bouchaud N, Herrup K, Mariani J. Progressive atrophy of cerebellar Purkinje cell dendrites during aging of the heterozygous staggerer mouse (Rora(+/sg)) [Accessed August 1, 2015];Brain Res Dev Brain Res. 2001 126(2):201–209. doi: 10.1016/s0165-3806(01)00095-5. http://www.ncbi.nlm.nih.gov/pubmed/11248354. [DOI] [PubMed] [Google Scholar]
- 159.Hadj-Sahraoui N, Frederic F, Zanjani H, Herrup K, Delhaye-Bouchaud N, Mariani J. Purkinje cell loss in heterozygous staggerer mutant mice during aging. [Accessed August 1, 2015];Brain Res Dev Brain Res. 1997 98(1):1–8. doi: 10.1016/s0165-3806(96)00153-8. http://www.ncbi.nlm.nih.gov/pubmed/9027398. [DOI] [PubMed] [Google Scholar]
- 160.Zanjani HS, Mariani J, Delhaye-Bouchaud N, Herrup K. Neuronal cell loss in heterozygous staggerer mutant mice: a model for genetic contributions to the aging process. [Accessed August 1, 2015];Brain Res Dev Brain Res. 1992 67(2):153–160. doi: 10.1016/0165-3806(92)90216-j. http://www.ncbi.nlm.nih.gov/pubmed/1511513. [DOI] [PubMed] [Google Scholar]
- 161.Messer A, Plummer-Siegard J, Eisenberg B. Staggerer mutant mouse Purkinje cells do not contain detectable calmodulin mRNA. [Accessed August 1, 2015];J Neurochem. 1990 55(1):293–302. doi: 10.1111/j.1471-4159.1990.tb08851.x. http://www.ncbi.nlm.nih.gov/pubmed/2355223. [DOI] [PubMed] [Google Scholar]
- 162.Serinagaoglu Y, Zhang R, Zhang Y, et al. A promoter element with enhancer properties, and the orphan nuclear receptor RORalpha, are required for Purkinje cell-specific expression of a Gi/o modulator. Mol Cell Neurosci. 2007;34(3):324–342. doi: 10.1016/j.mcn.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 163.Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-André M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8(6):757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 164.Giguère V, Beatty B, Squire J, Copeland NG, Jenkins NA. The Orphan Nuclear Receptor RORα (RORA) Maps to a Conserved Region of Homology on Human Chromosome 15q21–q22 and Mouse Chromosome 9. Genomics. 1995;28(3):596–598. doi: 10.1006/geno.1995.1197. [DOI] [PubMed] [Google Scholar]
- 165.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Mol Endocrinol. 2006;20(8):1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 166.Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46(8):2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- 167.Jia L, Oh ECT, Ng L, et al. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci U S A. 2009;106(41):17534–17539. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Onishi A, Peng G-H, Poth EM, et al. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc Natl Acad Sci U S A. 2010;107(25):11579–11584. doi: 10.1073/pnas.1000102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. [Accessed August 1, 2015];Trends Neurosci. 2002 25(1):32–38. doi: 10.1016/s0166-2236(00)02028-2. http://www.ncbi.nlm.nih.gov/pubmed/11801336. [DOI] [PubMed] [Google Scholar]
- 170.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 171.Peng G-H, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14(6):747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 172.Marquardt T. Transcriptional control of neuronal diversification in the retina. [Accessed October 6, 2015];Prog Retin Eye Res. 2003 22(5):567–577. doi: 10.1016/s1350-9462(03)00036-3. http://www.ncbi.nlm.nih.gov/pubmed/12892642. [DOI] [PubMed] [Google Scholar]
- 173.Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. [Accessed October 2, 2015];Mol Cell Biol. 1989 9(3):1128–1136. doi: 10.1128/mcb.9.3.1128. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=362703&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Forman BM, Chen J, Blumberg B, et al. Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8(9):1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 175.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. [Accessed October 2, 2015];Mol Cell Biol. 1995 15(9):4791–4802. doi: 10.1128/mcb.15.9.4791. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=230723&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zamir I, Dawson J, Lavinsky RM, Glass CK, Rosenfeld MG, Lazar MA. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. [Accessed October 2, 2015];Proc Natl Acad Sci U S A. 1997 94(26):14400–14405. doi: 10.1073/pnas.94.26.14400. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=24996&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. [Accessed October 2, 2015];J Biol Chem. 1993 268(22):16265–16269. http://www.ncbi.nlm.nih.gov/pubmed/8344913. [PubMed] [Google Scholar]
- 178.Downes M, Carozzi AJ, Muscat GE. Constitutive expression of the orphan receptor, Rev-erbA alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Mol Endocrinol. 1995;9(12):1666–1678. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- 179.Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. 2011;3(5):623–638. doi: 10.4155/fmc.11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gervois P, Chopin-Delannoy S, Fadel A, et al. Fibrates Increase Human REV-ERBα Expression in Liver via a Novel Peroxisome Proliferator-Activated Receptor Response Element. Mol Endocrinol. 1999;13(3):400–409. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- 181.Lee Y-F, Lee H-J, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. [Accessed July 31, 2015];J Steroid Biochem Mol Biol. 2002 81(4–5):291–308. doi: 10.1016/s0960-0760(02)00118-8. http://www.ncbi.nlm.nih.gov/pubmed/12361719. [DOI] [PubMed] [Google Scholar]
- 182.Chinpaisal C, Lee CH, Wei LN. Mechanisms of the mouse orphan nuclear receptor TR2-11-mediated gene suppression. [Accessed October 5, 2015];J Biol Chem. 1998 273(29):18077–18085. doi: 10.1074/jbc.273.29.18077. http://www.ncbi.nlm.nih.gov/pubmed/9660764. [DOI] [PubMed] [Google Scholar]
- 183.van Schaick HS, Rosmalen JG, Lopes da Silva S, Chang C, Burbach JP. Expression of the orphan receptor TR4 during brain development of the rat. [Accessed November 23, 2015];Brain Res Mol Brain Res. 2000 77(1):104–110. doi: 10.1016/s0169-328x(00)00046-2. http://www.ncbi.nlm.nih.gov/pubmed/10814836. [DOI] [PubMed] [Google Scholar]
- 184.Collins LL, Lee Y-F, Heinlein CA, et al. Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc Natl Acad Sci. 2004;101(42):15058–15063. doi: 10.1073/pnas.0405700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen Y-T, Collins LL, Chang S-S, Chang C. The roles of testicular orphan nuclear receptor 4 (TR4) in cerebellar development. Cerebellum. 2008;7(1):9–17. doi: 10.1007/s12311-008-0006-3. [DOI] [PubMed] [Google Scholar]
- 186.Rakic P, Sidman RL. Weaver mutant mouse cerebellum: defective neuronal migration secondary to abnormality of Bergmann glia. [Accessed November 23, 2015];Proc Natl Acad Sci U S A. 1973 70(1):240–244. doi: 10.1073/pnas.70.1.240. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=433223&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shibata T, Yamada K, Watanabe M, et al. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. [Accessed November 23, 2015];J Neurosci. 1997 17(23):9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. http://www.ncbi.nlm.nih.gov/pubmed/9364068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Goldowitz D. The weaver granuloprival phenotype is due to intrinsic action of the mutant locus in granule cells: evidence from homozygous weaver chimeras. [Accessed November 23, 2015];Neuron. 1989 2(6):1565–1575. doi: 10.1016/0896-6273(89)90045-7. http://www.ncbi.nlm.nih.gov/pubmed/2627379. [DOI] [PubMed] [Google Scholar]
- 189.Hamre KM, Goldowitz D. meander tail acts intrinsic to granule cell precursors to disrupt cerebellar development: analysis of meander tail chimeric mice. [Accessed November 23, 2015];Development. 1997 124(21):4201–4212. doi: 10.1242/dev.124.21.4201. http://www.ncbi.nlm.nih.gov/pubmed/9334269. [DOI] [PubMed] [Google Scholar]
- 190.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. [Accessed November 23, 2015];Development. 1999 126(14):3089–3100. doi: 10.1242/dev.126.14.3089. http://www.ncbi.nlm.nih.gov/pubmed/10375501. [DOI] [PubMed] [Google Scholar]
- 191.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004;270(2):393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 192.Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. [Accessed November 23, 2015];J Neurosci. 2002 22(1):218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. http://www.ncbi.nlm.nih.gov/pubmed/11756505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Eiraku M, Tohgo A, Ono K, et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8(7):873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- 194.Tohgo A, Eiraku M, Miyazaki T, et al. Impaired cerebellar functions in mutant mice lacking DNER. Mol Cell Neurosci. 2006;31(2):326–333. doi: 10.1016/j.mcn.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 195.Sadakata T, Kakegawa W, Mizoguchi A, et al. Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J Neurosci. 2007;27(10):2472–2482. doi: 10.1523/JNEUROSCI.2279-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim Y-S, Harry GJ, Kang HS, et al. Altered cerebellar development in nuclear receptor TAK1/TR4 null mice is associated with deficits in GLAST(+) glia, alterations in social behavior, motor learning, startle reactivity, and microglia. Cerebellum. 2010;9(3):310–323. doi: 10.1007/s12311-010-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jackson A, Panayiotidis P, Foroni L. The Human Homologue of theDrosophila taillessGene (TLX): Characterization and Mapping to a Region of Common Deletion in Human Lymphoid Leukemia on Chromosome 6q21. Genomics. 1998;50(1):34–43. doi: 10.1006/geno.1998.5270. [DOI] [PubMed] [Google Scholar]
- 198.Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370(6488):375–379. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- 199.Kobayashi M, Yu RT, Yasuda K, Umesono K. Cell-type-specific regulation of the retinoic acid receptor mediated by the orphan nuclear receptor TLX. [Accessed August 31, 2015];Mol Cell Biol. 2000 20(23):8731–8739. doi: 10.1128/mcb.20.23.8731-8739.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=86495&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Abrahams BS, Kwok MCH, Trinh E, Budaghzadeh S, Hossain SM, Simpson EM. Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J Neurosci. 2005;25(27):6263–6270. doi: 10.1523/JNEUROSCI.4757-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Land PW, Monaghan AP. Expression of the transcription factor, tailless, is required for formation of superficial cortical layers. [Accessed August 31, 2015];Cereb Cortex. 2003 13(9):921–931. doi: 10.1093/cercor/13.9.921. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2724011&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shi Y, Chichung Lie D, Taupin P, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427(6969):78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 203.Yu RT, Chiang MY, Tanabe T, et al. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci U S A. 2000;97(6):2621–2625. doi: 10.1073/pnas.050566897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Barneda-Zahonero B, Servitja J-M, Badiola N, et al. Nurr1 protein is required for N-methyl-D-aspartic acid (NMDA) receptor-mediated neuronal survival. J Biol Chem. 2012;287(14):11351–11362. doi: 10.1074/jbc.M111.272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang S-J, Steijaert MN, Lau D, et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53(4):549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 206.Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, Federoff HJ. VIP is a transcriptional target of Nurr1 in dopaminergic cells. Exp Neurol. 2007;203(1):221–232. doi: 10.1016/j.expneurol.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 207.Yang YX, Latchman DS. Nurr1 transcriptionally regulates the expression of alphasynuclein. Neuroreport. 2008;19(8):867–871. doi: 10.1097/WNR.0b013e3282ffda48. [DOI] [PubMed] [Google Scholar]
- 208.Volpicelli F, Caiazzo M, Greco D, et al. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem. 2007;102(2):441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- 209.Simões-Costa MS, Azambuja AP, Xavier-Neto J. The search for non-chordate retinoic acid signaling: lessons from chordates. J Exp Zool B Mol Dev Evol. 2008;310(1):54–72. doi: 10.1002/jez.b.21139. [DOI] [PubMed] [Google Scholar]
- 210.Parés X, Farrés J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families. Cell Mol Life Sci. 2008;65(24):3936–3949. doi: 10.1007/s00018-008-8591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dupé V, Matt N, Garnier J-M, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100(24):14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Moreno-Ramos OA, Olivares AM, Haider NB, de Autismo LC, Lattig MC. Whole-Exome Sequencing in a South American Cohort Links ALDH1A3, FOXN1 and Retinoic Acid Regulation Pathways to Autism Spectrum Disorders. PLoS One. 2015;10(9):e0135927. doi: 10.1371/journal.pone.0135927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Matt N, Dupé V, Garnier J-M, et al. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132(21):4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- 214.Molotkov A, Molotkova N, Duester G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development. 2006;133(10):1901–1910. doi: 10.1242/dev.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ruberte E, Friederich V, Chambon P, Morriss-Kay G. Retinoic acid receptors and cellular retinoid binding proteins. III. Their differential transcript distribution during mouse nervous system development. [Accessed September 2, 2015];Development. 1993 118(1):267–282. doi: 10.1242/dev.118.1.267. http://www.ncbi.nlm.nih.gov/pubmed/8397079. [DOI] [PubMed] [Google Scholar]
- 216.Baik JH, Picetti R, Saiardi A, et al. Parkinsonian-like locomotor impairment in mice lacking dopamine D2 receptors. Nature. 1995;377(6548):424–428. doi: 10.1038/377424a0. [DOI] [PubMed] [Google Scholar]
- 217.Valdenaire O, Vernier P, Maus M, Dumas Milne, Edwards JB, Mallet J. Transcription of the rat dopamine-D2-receptor gene from two promoters. [Accessed September 2, 2015];Eur J Biochem. 1994 220(2):577–584. doi: 10.1111/j.1432-1033.1994.tb18658.x. http://www.ncbi.nlm.nih.gov/pubmed/8125117. [DOI] [PubMed] [Google Scholar]
- 218.Zetterström RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. [Accessed September 2, 2015];Science. 1997 276(5310):248–250. doi: 10.1126/science.276.5310.248. http://www.ncbi.nlm.nih.gov/pubmed/9092472. [DOI] [PubMed] [Google Scholar]
- 219.Le W, Conneely OM, He Y, Jankovic J, Appel SH. Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. [Accessed September 2, 2015];J Neurochem. 1999 73(5):2218–2221. http://www.ncbi.nlm.nih.gov/pubmed/10537083. [PubMed] [Google Scholar]
- 220.Vitoux D, Nasr R, de The H. Acute promyelocytic leukemia: new issues on pathogenesis and treatment response. Int J Biochem Cell Biol. 2007;39(6):1063–1070. doi: 10.1016/j.biocel.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 221.Park JP, Moeschler JB, Davies WS, Patel PI, Mohandas TK. Smith-Magenis syndrome resulting from a de novo direct insertion of proximal 17q into 17p11.2. [Accessed September 2, 2015];Am J Med Genet. 1998 77(1):23–27. http://www.ncbi.nlm.nih.gov/pubmed/9557889. [PubMed] [Google Scholar]
- 222.Smith AC, McGavran L, Robinson J, et al. Interstitial deletion of (17) (p11.2p11.2) in nine patients. Am J Med Genet. 1986;24(3):393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- 223.Krezel W, Ghyselinck N, Samad TA, et al. Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. [Accessed September 2, 2015];Science. 1998 279(5352):863–867. doi: 10.1126/science.279.5352.863. http://www.ncbi.nlm.nih.gov/pubmed/9452386. [DOI] [PubMed] [Google Scholar]
- 224.Samad TA, Krezel W, Chambon P, Borrelli E. Regulation of dopaminergic pathways by retinoids: activation of the D2 receptor promoter by members of the retinoic acid receptor-retinoid X receptor family. [Accessed September 2, 2015];Proc Natl Acad Sci U S A. 1997 94(26):14349–14354. doi: 10.1073/pnas.94.26.14349. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=24972&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Giguère V, McBroom LD, Flock G. Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. [Accessed August 20, 2015];Mol Cell Biol. 1995 15(5):2517–2526. doi: 10.1128/mcb.15.5.2517. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=230482&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Serra HG, Duvick L, Zu T, et al. RORα-Mediated Purkinje Cell Development Determines Disease Severity in Adult SCA1 Mice. Cell. 2006;127(4):697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 227.Hamilton BA, Frankel WN, Kerrebrock AW, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379(6567):736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 228.Sarachana T, Hu VW. Genome-wide identification of transcriptional targets of RORA reveals direct regulation of multiple genes associated with autism spectrum disorder. Mol Autism. 2013;4(1):14. doi: 10.1186/2040-2392-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jacobson SG. Nuclear receptor NR2E3 gene mutations distort human retinal laminar architecture and cause an unusual degeneration. Hum Mol Genet. 2004;13(17):1893–1902. doi: 10.1093/hmg/ddh198. [DOI] [PubMed] [Google Scholar]
- 230.Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol (Chicago, Ill 1960) 2003;121(9):1316–1323. doi: 10.1001/archopht.121.9.1316. [DOI] [PubMed] [Google Scholar]
- 231.Boughman JA, Conneally PM, Nance WE. Population genetic studies of retinitis pigmentosa. [Accessed October 8, 2015];Am J Hum Genet. 1980 32(2):223–235. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1686021&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 232.Coppieters F, Leroy BP, Beysen D, et al. Recurrent mutation in the first zinc finger of the orphan nuclear receptor NR2E3 causes autosomal dominant retinitis pigmentosa. Am J Hum Genet. 2007;81(1):147–157. doi: 10.1086/518426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Haider NB, Zhang W, Hurd R, et al. Mapping of genetic modifiers of Nr2e3 rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm Genome. 2008;19(3):145–154. doi: 10.1007/s00335-008-9092-2. [DOI] [PubMed] [Google Scholar]
- 234.Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. [Accessed May 6, 2014];Hum Mol Genet. 2001 10(16):1619–1626. doi: 10.1093/hmg/10.16.1619. http://www.ncbi.nlm.nih.gov/pubmed/11487564. [DOI] [PubMed] [Google Scholar]
- 235.Schorderet DF, Escher P. NR2E3 mutations in enhanced S-cone sensitivity syndrome (ESCS), Goldmann-Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP) Hum Mutat. 2009;30(11):1475–1485. doi: 10.1002/humu.21096. [DOI] [PubMed] [Google Scholar]
- 236.Cruz NM, Yuan Y, Leehy BD, et al. Modifier genes as therapeutics: the nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS One. 2014;9(1):e87942. doi: 10.1371/journal.pone.0087942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nevado J, Tenbaum SP, Aranda A. hSrb7, an essential human Mediator component, acts as a coactivator for the thyroid hormone receptor. Mol Cell Endocrinol. 2004;222(1–2):41–51. doi: 10.1016/j.mce.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 238.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 239.Ma D, Tian H, Sun H, Kozikowshi AP, Pshenichkin S, Wroblewski JT. Synthesis and biological activity of cyclic analogues of MPPG and MCPG as metabotropic glutamate receptor antagonists. Bioorg Med Chem Lett. 1997;7(9):1195–1198. doi: 10.1016/S0960-894X(97)00177-7. [DOI] [Google Scholar]
- 240.Hörlein AJ, Näär AM, Heinzel T, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377(6548):397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 241.Chen H, Lin RJ, Schiltz RL, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. [Accessed December 24, 2015];Cell. 1997 90(3):569–580. doi: 10.1016/s0092-8674(00)80516-4. http://www.ncbi.nlm.nih.gov/pubmed/9267036. [DOI] [PubMed] [Google Scholar]
- 242.Ito M, Yuan C-X, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 Component of the TRAP/SMCC Coactivator Complex in Embryonic Development and Thyroid Hormone Action. Mol Cell. 2000;5(4):683–693. doi: 10.1016/S1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 243.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. [Accessed August 9, 2015];Genes Dev. 2000 14(13):1553–1577. http://www.ncbi.nlm.nih.gov/pubmed/10887150. [PubMed] [Google Scholar]
- 244.Sande S, Privalsky ML. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10(7):813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 245.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) [Accessed November 18, 2015];Blood. 1991 77(5):1080–1086. http://www.ncbi.nlm.nih.gov/pubmed/1995093. [PubMed] [Google Scholar]
- 246.Wong CW, Privalsky ML. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. [Accessed December 24, 2015];Mol Cell Biol. 1998 18(10):5724–5733. doi: 10.1128/mcb.18.10.5724. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=109158&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mark M, Chambon P. Functions of RARs and RXRs in vivo: Genetic dissection of the retinoid signaling pathway. Pure Appl Chem. 2003;75:11–12. doi: 10.1351/pac200375111709. [DOI] [Google Scholar]
- 248.Oñate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. [Accessed December 3, 2015];Science. 1995 270(5240):1354–1357. doi: 10.1126/science.270.5240.1354. http://www.ncbi.nlm.nih.gov/pubmed/7481822. [DOI] [PubMed] [Google Scholar]
- 249.Westin S, Rosenfeld MG, Glass CK. Nuclear receptor coactivators. [Accessed December 24, 2015];Adv Pharmacol. 2000 47:89–112. doi: 10.1016/s1054-3589(08)60110-6. http://www.ncbi.nlm.nih.gov/pubmed/10582085. [DOI] [PubMed] [Google Scholar]
- 250.Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17(2):507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lemon BD, Freedman LP. Nuclear receptor cofactors as chromatin remodelers. [Accessed December 24, 2015];Curr Opin Genet Dev. 1999 9(5):499–504. doi: 10.1016/s0959-437x(99)00010-6. http://www.ncbi.nlm.nih.gov/pubmed/10508693. [DOI] [PubMed] [Google Scholar]
- 252.Pogenberg V, Guichou J-F, Vivat-Hannah V, et al. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J Biol Chem. 2005;280(2):1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 253.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. [Accessed December 24, 2015];Proc Natl Acad Sci U S A. 1998 95(14):7939–7944. doi: 10.1073/pnas.95.14.7939. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=20908&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Farboud B, Hauksdottir H, Wu Y, Privalsky ML. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. [Accessed November 18, 2015];Mol Cell Biol. 2003 23(8):2844–2858. doi: 10.1128/MCB.23.8.2844-2858.2003. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=152560&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Laudet V, Gronemeyer H. The Nuclear Receptor FactsBook. London: Elsevier; 2002. [DOI] [Google Scholar]
- 256.Li H, Tierney C, Wen L, Wu JY, Rao Y. A single morphogenetic field gives rise to two retina primordia under the influence of the prechordal plate. [Accessed August 1, 2015];Development. 1997 124(3):603–615. doi: 10.1242/dev.124.3.603. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2041934&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. [Accessed December 29, 2015];EMBO J. 1996 15(14):3667–3675. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=452006&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 258.Chakravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383(6595):99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 259.Kamei Y, Xu L, Heinzel T, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. [Accessed December 29, 2015];Cell. 1996 85(3):403–414. doi: 10.1016/s0092-8674(00)81118-6. http://www.ncbi.nlm.nih.gov/pubmed/8616895. [DOI] [PubMed] [Google Scholar]
- 260.Delerive P, Wu Y, Burris TP, Chin WW, Suen CS. PGC-1 functions as a transcriptional coactivator for the retinoid X receptors. J Biol Chem. 2002;277(6):3913–3917. doi: 10.1074/jbc.M109409200. [DOI] [PubMed] [Google Scholar]
- 261.May M, Mengus G, Lavigne AC, Chambon P, Davidson I. Human TAF(II28) promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. [Accessed December 29, 2015];EMBO J. 1996 15(12):3093–3104. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=450252&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 262.Lala DS, Mukherjee R, Schulman IG, et al. Activation of specific RXR heterodimers by an antagonist of RXR homodimers. Nature. 1996;383(6599):450–453. doi: 10.1038/383450a0. [DOI] [PubMed] [Google Scholar]
- 263.Schulman IG, Chakravarti D, Juguilon H, Romo A, Evans RM. Interactions between the retinoid X receptor and a conserved region of the TATA-binding protein mediate hormone-dependent transactivation. [Accessed December 29, 2015];Proc Natl Acad Sci U S A. 1995 92(18):8288–8292. doi: 10.1073/pnas.92.18.8288. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=41142&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem. 2004;279(52):54348–54357. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
- 265.Pineda Torra I, Freedman LP, Garabedian MJ. Identification of DRIP205 as a coactivator for the Farnesoid X receptor. J Biol Chem. 2004;279(35):36184–36191. doi: 10.1074/jbc.M405126200. [DOI] [PubMed] [Google Scholar]
- 266.Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. [Accessed December 3, 2015];Science. 1999 284(5418):1362–1365. doi: 10.1126/science.284.5418.1362. http://www.ncbi.nlm.nih.gov/pubmed/10334992. [DOI] [PubMed] [Google Scholar]
- 267.Parks DJ. Bile Acids: Natural Ligands for an Orphan Nuclear Receptor. Science (80−) 1999;284(5418):1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 268.Kanaya E, Shiraki T, Jingami H. The nuclear bile acid receptor FXR is activated by PGC-1alpha in a ligand-dependent manner. Biochem J. 2004;382(Pt 3):913–921. doi: 10.1042/BJ20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Savkur RS, Thomas JS, Bramlett KS, Gao Y, Michael LF, Burris TP. Ligand-dependent coactivation of the human bile acid receptor FXR by the peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Pharmacol Exp Ther. 2005;312(1):170–178. doi: 10.1124/jpet.104.072124. [DOI] [PubMed] [Google Scholar]
- 270.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18(2):157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol. 2005;68(2):551–558. doi: 10.1124/mol.105.012104. [DOI] [PubMed] [Google Scholar]
- 272.Unno A, Takada I, Takezawa S, et al. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem Biophys Res Commun. 2005;327(3):933–938. doi: 10.1016/j.bbrc.2004.12.095. [DOI] [PubMed] [Google Scholar]
- 273.Otte K, Kranz H, Kober I, et al. Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. [Accessed December 24, 2015];Mol Cell Biol. 2003 23(3):864–872. doi: 10.1128/MCB.23.3.864-872.2003. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=140718&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Huuskonen J, Fielding PE, Fielding CJ. Role of p160 coactivator complex in the activation of liver X receptor. Arterioscler Thromb Vasc Biol. 2004;24(4):703–708. doi: 10.1161/01.ATV.0000121202.72593.da. [DOI] [PubMed] [Google Scholar]
- 275.Hu X, Li S, Wu J, Xia C, Lala DS. Liver X receptors interact with corepressors to regulate gene expression. Mol Endocrinol. 2003;17(6):1019–1026. doi: 10.1210/me.2002-0399. [DOI] [PubMed] [Google Scholar]
- 276.Song C, Kokontis JM, Hiipakka RA, Liao S. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. [Accessed December 24, 2015];Proc Natl Acad Sci U S A. 1994 91(23):10809–10813. doi: 10.1073/pnas.91.23.10809. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=45115&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Oberkofler H, Schraml E, Krempler F, Patsch W. Potentiation of liver X receptor transcriptional activity by peroxisome-proliferator-activated receptor gamma co-activator 1 alpha. Biochem J. 2003;371(Pt 1):89–96. doi: 10.1042/BJ20021665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lin J, Yang R, Tarr PT, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120(2):261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 279.Menke JG, Macnaul KL, Hayes NS, et al. A novel liver X receptor agonist establishes species differences in the regulation of cholesterol 7alpha-hydroxylase (CYP7a) Endocrinology. 2002;143(7):2548–2558. doi: 10.1210/endo.143.7.8907. [DOI] [PubMed] [Google Scholar]
- 280.Tien ES, Davis JW, Vanden Heuvel JP. Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator-activated receptor alpha coregulator. J Biol Chem. 2004;279(23):24053–24063. doi: 10.1074/jbc.M401489200. [DOI] [PubMed] [Google Scholar]
- 281.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 282.DiRenzo J, Söderstrom M, Kurokawa R, et al. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. [Accessed December 24, 2015];Mol Cell Biol. 1997 17(4):2166–2176. doi: 10.1128/mcb.17.4.2166. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=232065&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor alpha interacting protein. [Accessed December 24, 2015];J Biol Chem. 1999 274(22):15901–15907. doi: 10.1074/jbc.274.22.15901. http://www.ncbi.nlm.nih.gov/pubmed/10336495. [DOI] [PubMed] [Google Scholar]
- 284.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. p300 functions as a coactivator for the peroxisome proliferator-activated receptor alpha. [Accessed December 24, 2015];J Biol Chem. 1997 272(52):33435–33443. doi: 10.1074/jbc.272.52.33435. http://www.ncbi.nlm.nih.gov/pubmed/9407140. [DOI] [PubMed] [Google Scholar]
- 285.Zhou G, Cummings R, Li Y, et al. Nuclear receptors have distinct affinities for coactivators: characterization by fluorescence resonance energy transfer. Mol Endocrinol. 1998;12(10):1594–1604. doi: 10.1210/mend.12.10.0176. [DOI] [PubMed] [Google Scholar]
- 286.Joyeux A, Cavaillès V, Balaguer P, Nicolas JC. RIP 140 enhances nuclear receptor-dependent transcription in vivo in yeast. Mol Endocrinol. 1997;11(2):193–202. doi: 10.1210/mend.11.2.9884. [DOI] [PubMed] [Google Scholar]
- 287.Miyata KS, McCaw SE, Meertens LM, Patel HV, Rachubinski RA, Capone JP. Receptor-interacting protein 140 interacts with and inhibits transactivation by, peroxisome proliferator-activated receptor alpha and liver-X-receptor alpha. [Accessed December 24, 2015];Mol Cell Endocrinol. 1998 146(1–2):69–76. doi: 10.1016/s0303-7207(98)00196-8. http://www.ncbi.nlm.nih.gov/pubmed/10022764. [DOI] [PubMed] [Google Scholar]
- 288.Yan ZH, Karam WG, Staudinger JL, Medvedev A, Ghanayem BI, Jetten AM. Regulation of peroxisome proliferator-activated receptor alpha-induced transactivation by the nuclear orphan receptor TAK1/TR4. [Accessed December 24, 2015];J Biol Chem. 1998 273(18):10948–10957. doi: 10.1074/jbc.273.18.10948. http://www.ncbi.nlm.nih.gov/pubmed/9556573. [DOI] [PubMed] [Google Scholar]
- 289.Juge-Aubry CE, Kuenzli S, Sanchez JC, Hochstrasser D, Meier CA. Peroxisomal bifunctional enzyme binds and activates the activation function-1 region of the peroxisome proliferator-activated receptor alpha. [Accessed December 24, 2015];Biochem J. 2001 353(Pt 2):253–258. doi: 10.1042/0264-6021:3530253. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1221566&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. [Accessed December 24, 2015];J Biol Chem. 1997 272(41):25500–25506. doi: 10.1074/jbc.272.41.25500. http://www.ncbi.nlm.nih.gov/pubmed/9325263. [DOI] [PubMed] [Google Scholar]
- 291.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor gamma. [Accessed December 24, 2015];Gene Expr. 1996 6(3):185–195. http://www.ncbi.nlm.nih.gov/pubmed/9041124. [PMC free article] [PubMed] [Google Scholar]
- 292.Molnár F, Matilainen M, Carlberg C. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. J Biol Chem. 2005;280(28):26543–26556. doi: 10.1074/jbc.M502463200. [DOI] [PubMed] [Google Scholar]
- 293.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson JA. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. [Accessed December 24, 2015];J Biol Chem. 2000 275(8):5308–5317. doi: 10.1074/jbc.275.8.5308. http://www.ncbi.nlm.nih.gov/pubmed/10681503. [DOI] [PubMed] [Google Scholar]
- 294.Zhu Y. Isolation and Characterization of Peroxisome Proliferator-activated Receptor (PPAR) Interacting Protein (PRIP) as a Coactivator for PPAR. J Biol Chem. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 295.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. [Accessed November 30, 2015];Mol Cell Biol. 2000 20(5):1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=85369&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U S A. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. [Accessed December 24, 2015];Cell Biochem Biophys. 2000 Spring;32:187–204. doi: 10.1385/cbb:32:1-3:187. http://www.ncbi.nlm.nih.gov/pubmed/11330046. [DOI] [PubMed] [Google Scholar]
- 298.Jackson TA, Richer JK, Bain DL, Takimoto GS, Tung L, Horwitz KB. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol Endocrinol. 1997;11(6):693–705. doi: 10.1210/mend.11.6.0004. [DOI] [PubMed] [Google Scholar]
- 299.Krogsdam A-M, Nielsen CAF, Neve S, et al. Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor delta-mediated transactivation. The Biochemical journal. [Accessed December 24, 2015]. https://www.google.com.co/?gfe_rd=cr&ei=7Xh7VtDWGNKuzQLDuJzABg&gws_rd=ssl#q=Nuclear+receptor+corepressor-dependent+repression+of+peroxisome-proliferator-activated+receptor+delta-mediated+transactivation. Published April 1, 2002. [DOI] [PMC free article] [PubMed]
- 300.Zamir I, Harding HP, Atkins GB, et al. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. [Accessed October 2, 2015];Mol Cell Biol. 1996 16(10):5458–5465. doi: 10.1128/mcb.16.10.5458. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=231546&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280(14):13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 302.Wang Y-X, Lee C-H, Tiep S, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. [Accessed December 24, 2015];Cell. 2003 113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. http://www.ncbi.nlm.nih.gov/pubmed/12705865. [DOI] [PubMed] [Google Scholar]
- 303.Hong J-H, Hwang ES, McManus MT, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 304.Mizukami J, Taniguchi T. The antidiabetic agent thiazolidinedione stimulates the interaction between PPAR gamma and CBP. Biochem Biophys Res Commun. 1997;240(1):61–64. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- 305.Debril M-B, Dubuquoy L, Feige J-N, et al. Scaffold attachment factor B1 directly interacts with nuclear receptors in living cells and represses transcriptional activity. J Mol Endocrinol. 2005;35(3):503–517. doi: 10.1677/jme.1.01856. [DOI] [PubMed] [Google Scholar]
- 306.Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and—dependent manner, respectively. [Accessed December 29, 2015];J Biol Chem. 1999 274(12):7681–7688. doi: 10.1074/jbc.274.12.7681. http://www.ncbi.nlm.nih.gov/pubmed/10075656. [DOI] [PubMed] [Google Scholar]
- 307.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson JA. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12(6):864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 308.Kodera Y, Takeyama K, Murayama A, Suzawa M, Masuhiro Y, Kato S. Ligand type-specific interactions of peroxisome proliferator-activated receptor gamma with transcriptional coactivators. J Biol Chem. 2000;275(43):33201–33204. doi: 10.1074/jbc.C000517200. [DOI] [PubMed] [Google Scholar]
- 309.Yang W, Rachez C, Freedman LP. Discrete roles for peroxisome proliferator-activated receptor gamma and retinoid X receptor in recruiting nuclear receptor coactivators. [Accessed December 29, 2015];Mol Cell Biol. 2000 20(21):8008–8017. doi: 10.1128/mcb.20.21.8008-8017.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=86411&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zhu Y, Qi C, Cao WQ, Yeldandi AV, Rao MS, Reddy JK. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc Natl Acad Sci U S A. 2001;98(18):10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Leers J, Treuter E, Gustafsson JA. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. [Accessed December 29, 2015];Mol Cell Biol. 1998 18(10):6001–6013. doi: 10.1128/mcb.18.10.6001. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=109186&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Heinlein CA, Ting HJ, Yeh S, Chang C. Identification of ARA70 as a ligand-enhanced coactivator for the peroxisome proliferator-activated receptor gamma. [Accessed December 29, 2015];J Biol Chem. 1999 274(23):16147–16152. doi: 10.1074/jbc.274.23.16147. http://www.ncbi.nlm.nih.gov/pubmed/10347167. [DOI] [PubMed] [Google Scholar]
- 313.Qi C, Surapureddi S, Zhu Y-J, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem. 2003;278(28):25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- 314.Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. [Accessed December 29, 2015];Mol Cell Biol. 2002 22(10):3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=133794&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277(43):40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 316.Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24(20):9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Knutti D, Kralli A. PGC-1, a versatile coactivator. [Accessed December 29, 2015];Trends Endocrinol Metab. 2001 12(8):360–365. doi: 10.1016/s1043-2760(01)00457-x. http://www.ncbi.nlm.nih.gov/pubmed/11551810. [DOI] [PubMed] [Google Scholar]
- 318.Oberkofler H, Esterbauer H, Linnemayr V, Strosberg AD, Krempler F, Patsch W. Peroxisome proliferator-activated receptor (PPAR) gamma coactivator-1 recruitment regulates PPAR subtype specificity. J Biol Chem. 2002;277(19):16750–16757. doi: 10.1074/jbc.M200475200. [DOI] [PubMed] [Google Scholar]
- 319.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. [Accessed October 21, 2015];Cell. 1998 92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. http://www.ncbi.nlm.nih.gov/pubmed/9529258. [DOI] [PubMed] [Google Scholar]
- 320.Guan H-P, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19(4):453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kamei Y, Ohizumi H, Fujitani Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A. 2003;100(21):12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277(32):28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 323.Castillo G, Brun RP, Rosenfield JK, et al. An adipogenic cofactor bound by the differentiation domain of PPARgamma. EMBO J. 1999;18(13):3676–3687. doi: 10.1093/emboj/18.13.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Debril M-B, Gelman L, Fayard E, Annicotte J-S, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279(16):16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 325.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 326.Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol. 2004;24(18):7931–7940. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7(5):584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 328.Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J Biol Chem. 2002;277(36):32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- 329.Sugatani J, Nishitani S, Yamakawa K, et al. Transcriptional regulation of human UGT1A1 gene expression: activated glucocorticoid receptor enhances constitutive androstane receptor/pregnane X receptor-mediated UDP-glucuronosyltransferase 1A1 regulation with glucocorticoid receptor-interacting protei. Mol Pharmacol. 2005;67(3):845–855. doi: 10.1124/mol.104.007161. [DOI] [PubMed] [Google Scholar]
- 330.Ourlin JC, Lasserre F, Pineau T, et al. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17(9):1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- 331.Kliewer SA, Moore JT, Wade L, et al. An Orphan Nuclear Receptor Activated by Pregnanes Defines a Novel Steroid Signaling Pathway. Cell. 1998;92(1):73–82. doi: 10.1016/S0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 332.Cavaillès V, Dauvois S, L’Horset F, et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. [Accessed December 24, 2015];EMBO J. 1995 14(15):3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=394449&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279(43):45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- 334.Jia Y, Guo GL, Surapureddi S, et al. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc Natl Acad Sci U S A. 2005;102(35):12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Forman BM, Tzameli I, Choi HS, et al. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395(6702):612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- 336.Shiraki T, Sakai N, Kanaya E, Jingami H. Activation of orphan nuclear constitutive androstane receptor requires subnuclear targeting by peroxisome proliferator-activated receptor gamma coactivator-1 alpha. A possible link between xenobiotic response and nutritional state. J Biol Chem. 2003;278(13):11344–11350. doi: 10.1074/jbc.M212859200. [DOI] [PubMed] [Google Scholar]
- 337.Medvedev A, Yan ZH, Hirose T, Giguère V, Jetten AM. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. [Accessed December 24, 2015];Gene. 1996 181(1–2):199–206. doi: 10.1016/s0378-1119(96)00504-5. http://www.ncbi.nlm.nih.gov/pubmed/8973331. [DOI] [PubMed] [Google Scholar]
- 338.Harding HP, Atkins GB, Jaffe AB, Seo WJ, Lazar MA. Transcriptional activation and repression by RORalpha, an orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11(11):1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- 339.Bitsch F, Aichholz R, Kallen J, Geisse S, Fournier B, Schlaeppi JM. Identification of natural ligands of retinoic acid receptor-related orphan receptor alpha ligand-binding domain expressed in Sf9 cells—a mass spectrometry approach. [Accessed December 24, 2015];Anal Biochem. 2003 323(1):139–149. doi: 10.1016/j.ab.2003.08.029. http://www.ncbi.nlm.nih.gov/pubmed/14622968. [DOI] [PubMed] [Google Scholar]
- 340.Atkins GB, Hu X, Guenther MG, Rachez C, Freedman LP, Lazar MA. Coactivators for the orphan nuclear receptor RORalpha. Mol Endocrinol. 1999;13(9):1550–1557. doi: 10.1210/mend.13.9.0343. [DOI] [PubMed] [Google Scholar]
- 341.Stehlin C, Wurtz JM, Steinmetz A, et al. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J. 2001;20(21):5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Moraitis AN, Giguère V, Thompson CC. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. [Accessed December 24, 2015];Mol Cell Biol. 2002 22(19):6831–6841. doi: 10.1128/MCB.22.19.6831-6841.2002. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=134023&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Greiner EF, Kirfel J, Greschik H, et al. Differential ligand-dependent protein-protein interactions between nuclear receptors and a neuronal-specific cofactor. [Accessed December 24, 2015];Proc Natl Acad Sci U S A. 2000 97(13):7160–7165. doi: 10.1073/pnas.97.13.7160. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=16516&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Kurebayashi S, Nakajima T, Kim S-C, et al. Selective LXXLL peptides antagonize transcriptional activation by the retinoid-related orphan receptor RORgamma. Biochem Biophys Res Commun. 2004;315(4):919–927. doi: 10.1016/j.bbrc.2004.01.131. [DOI] [PubMed] [Google Scholar]
- 345.Sauvé F, McBroom LD, Gallant J, Moraitis AN, Labrie F, Giguère V. CIA, a novel estrogen receptor coactivator with a bifunctional nuclear receptor interacting determinant. Mol Cell Biol. 2001;21(1):343–353. doi: 10.1128/MCB.21.1.343-353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 347.Downes M, Burke LJ, Bailey PJ, Muscat GE. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA alpha and RVR: physical association is dependent on the E region of the orphan receptors. [Accessed November 29, 2015];Nucleic Acids Res. 1996 24(22):4379–4386. doi: 10.1093/nar/24.22.4379. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=146280&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Burke LJ, Downes M, Laudet V, Muscat GE. Identification and characterization of a novel corepressor interaction region in RVR and Rev-erbA alpha. Mol Endocrinol. 1998;12(2):248–262. doi: 10.1210/mend.12.2.0061. [DOI] [PubMed] [Google Scholar]
- 349.Sladek FM, Ruse MD, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. [Accessed December 24, 2015];Mol Cell Biol. 1999 19(10):6509–6522. doi: 10.1128/mcb.19.10.6509. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=84621&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Wang JC, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. [Accessed December 24, 2015];J Biol Chem. 1998 273(47):30847–30850. doi: 10.1074/jbc.273.47.30847. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3968904&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Yoshida E, Aratani S, Itou H, et al. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Commun. 1997;241(3):664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 352.Ruse MD, Privalsky ML, Sladek FM. Competitive cofactor recruitment by orphan receptor hepatocyte nuclear factor 4alpha1: modulation by the F domain. [Accessed December 24, 2015];Mol Cell Biol. 2002 22(6):1626–1638. doi: 10.1128/MCB.22.6.1626-1638.2002. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=135595&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Maeda Y, Rachez C, Hawel L, Byus CV, Freedman LP, Sladek FM. Polyamines modulate the interaction between nuclear receptors and vitamin D receptorinteracting protein 205. Mol Endocrinol. 2002;16(7):1502–1510. doi: 10.1210/mend.16.7.0883. [DOI] [PubMed] [Google Scholar]
- 354.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. [Accessed December 24, 2015];Mol Cell Biol. 2002 22(15):5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=133960&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Lehto M, Bitzén PO, Isomaa B, et al. Mutation in the HNF-4alpha gene affects insulin secretion and triglyceride metabolism. [Accessed December 24, 2015];Diabetes. 1999 48(2):423–425. doi: 10.2337/diabetes.48.2.423. http://www.ncbi.nlm.nih.gov/pubmed/10334325. [DOI] [PubMed] [Google Scholar]
- 356.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 357.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC- 1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277(3):1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 358.Li G, Franco PJ, Wei LN. Identification of histone deacetylase-3 domains that interact with the orphan nuclear receptor TR2. [Accessed December 24, 2015];Biochem Biophys Res Commun. 2003 310(2):384–390. doi: 10.1016/j.bbrc.2003.08.145. http://www.ncbi.nlm.nih.gov/pubmed/14521922. [DOI] [PubMed] [Google Scholar]
- 359.Lee CH, Chinpaisal C, Wei LN. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. [Accessed December 24, 2015];Mol Cell Biol. 1998 18(11):6745–6755. doi: 10.1128/mcb.18.11.6745. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=109258&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Nakajima T, Fujino S, Nakanishi G, Kim Y-S, Jetten AM. TIP27: a novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Res. 2004;32(14):4194–4204. doi: 10.1093/nar/gkh741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Yang Y, Wang X, Dong T, Kim E, Lin W-J, Chang C. Identification of a novel testicular orphan receptor-4 (TR4)-associated protein as repressor for the selective suppression of TR4-mediated transactivation. J Biol Chem. 2003;278(9):7709–7717. doi: 10.1074/jbc.M207116200. [DOI] [PubMed] [Google Scholar]
- 362.Pipaón C, Tsai SY, Tsai MJ. COUP-TF upregulates NGFI-A gene expression through an Sp1 binding site. [Accessed December 24, 2015];Mol Cell Biol. 1999 19(4):2734–2745. doi: 10.1128/mcb.19.4.2734. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=84066&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Avram D, Fields A, Pretty On, Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. [Accessed November 24, 2015];J Biol Chem. 2000 275(14):10315–10322. doi: 10.1074/jbc.275.14.10315. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2819356&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Sugiyama T, Wang JC, Scott DK, Granner DK. Transcription activation by the orphan nuclear receptor, chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI). Definition of the domain involved in the glucocorticoid response of the phosphoenolpyruvate carboxykinase gene. [Accessed December 24, 2015];J Biol Chem. 2000 275(5):3446–3454. doi: 10.1074/jbc.275.5.3446. http://www.ncbi.nlm.nih.gov/pubmed/10652338. [DOI] [PubMed] [Google Scholar]
- 365.Shibata H, Nawaz Z, Tsai SY, O’Malley BW, Tsai MJ. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11(6):714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 366.Marcus SL, Winrow CJ, Capone JP, Rachubinski RA. A p56(lck) ligand serves as a coactivator of an orphan nuclear hormone receptor. [Accessed December 24, 2015];J Biol Chem. 1996 271(44):27197–27200. doi: 10.1074/jbc.271.44.27197. http://www.ncbi.nlm.nih.gov/pubmed/8910285. [DOI] [PubMed] [Google Scholar]
- 367.Huggins GS, Bacani CJ, Boltax J, Aikawa R, Leiden JM. Friend of GATA 2 physically interacts with chicken ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and represses COUP-TF2-dependent activation of the atrial natriuretic factor promoter. J Biol Chem. 2001;276(30):28029–28036. doi: 10.1074/jbc.M103577200. [DOI] [PubMed] [Google Scholar]
- 368.Zhu XG, Park KS, Kaneshige M, et al. The orphan nuclear receptor Ear-2 is a negative coregulator for thyroid hormone nuclear receptor function. [Accessed December 24, 2015];Mol Cell Biol. 2000 20(7):2604–2618. doi: 10.1128/mcb.20.7.2604-2618.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=85476&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Xie G, Drachenberg C, Yamada M, Wess J, Raufman JP. Cholinergic agonist-induced pepsinogen secretion from murine gastric chief cells is mediated by M1 and M3 muscarinic receptors. Am J Physiol Gastrointest Liver Physiol. 2005;289(3):G521–G529. doi: 10.1152/ajpgi.00105.2004. [DOI] [PubMed] [Google Scholar]
- 370.Zhou D, Chen S. PNRC2 is a 16 kDa coactivator that interacts with nuclear receptors through an SH3-binding motif. [Accessed December 24, 2015];Nucleic Acids Res. 2001 29(19):3939–3948. doi: 10.1093/nar/29.19.3939. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=60244&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Ichida M, Nemoto S, Finkel T. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha) J Biol Chem. 2002;277(52):50991–50995. doi: 10.1074/jbc.M210262200. [DOI] [PubMed] [Google Scholar]
- 372.Nagy L, Kao HY, Chakravarti D, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. [Accessed December 29, 2015];Cell. 1997 89(3):373–380. doi: 10.1016/s0092-8674(00)80218-4. http://www.ncbi.nlm.nih.gov/pubmed/9150137. [DOI] [PubMed] [Google Scholar]
- 373.Xie W, Hong H, Yang NN, et al. Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Mol Endocrinol. 1999;13(12):2151–2162. doi: 10.1210/mend.13.12.0381. [DOI] [PubMed] [Google Scholar]
- 374.Suetsugi M, Su L, Karlsberg K, Yuan Y-C, Chen S. Flavone and isoflavone phytoestrogens are agonists of estrogen-related receptors. [Accessed December 29, 2015];Mol Cancer Res. 2003 1(13):981–991. http://www.ncbi.nlm.nih.gov/pubmed/14638870. [PubMed] [Google Scholar]
- 375.Greschik H, Wurtz J-M, Sanglier S, et al. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. [Accessed December 29, 2015];Mol Cell. 2002 9(2):303–313. doi: 10.1016/s1097-2765(02)00444-6. http://www.ncbi.nlm.nih.gov/pubmed/11864604. [DOI] [PubMed] [Google Scholar]
- 376.Hentschke M, Borgmeyer U. Identification of PNRC2 and TLE1 as activation function-1 cofactors of the orphan nuclear receptor ERRgamma. [Accessed December 29, 2015];Biochem Biophys Res Commun. 2003 312(4):975–982. doi: 10.1016/j.bbrc.2003.11.025. http://www.ncbi.nlm.nih.gov/pubmed/14651967. [DOI] [PubMed] [Google Scholar]
- 377.Hentschke M, Süsens U, Borgmeyer U. PGC-1 and PERC, coactivators of the estrogen receptor-related receptor gamma. [Accessed December 29, 2015];Biochem Biophys Res Commun. 2002 299(5):872–879. doi: 10.1016/s0006-291x(02)02753-5. http://www.ncbi.nlm.nih.gov/pubmed/12470660. [DOI] [PubMed] [Google Scholar]
- 378.Wansa KDSA, Harris JM, Muscat GEO. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem. 2002;277(36):33001–33011. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- 379.Laflamme C, Filion C, Bridge JA, Ladanyi M, Goldring MB, Labelle Y. The homeotic protein Six3 is a coactivator of the nuclear receptor NOR-1 and a corepressor of the fusion protein EWS/NOR-1 in human extraskeletal myxoid chondrosarcomas. [Accessed December 29, 2015];Cancer Res. 2003 63(2):449–454. http://www.ncbi.nlm.nih.gov/pubmed/12543801. [PubMed] [Google Scholar]
- 380.Ohkura N, Ohkubo T, Maruyama K, Tsukada T, Yamaguchi K. The orphan nuclear receptor NOR-1 interacts with the homeobox containing protein Six3. Dev Neurosci. 2001;23(1):17–24. doi: 10.1159/000048692. doi:48692. [DOI] [PubMed] [Google Scholar]
- 381.Wansa KDSA, Harris JM, Yan G, Ordentlich P, Muscat GEO. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. J Biol Chem. 2003;278(27):24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- 382.Monté D, DeWitte F, Hum DW. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. [Accessed December 24, 2015];J Biol Chem. 1998 273(8):4585–4591. doi: 10.1074/jbc.273.8.4585. http://www.ncbi.nlm.nih.gov/pubmed/9468515. [DOI] [PubMed] [Google Scholar]
- 383.Hammer GD, Krylova I, Zhang Y, et al. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. [Accessed December 24, 2015];Mol Cell. 1999 3(4):521–526. doi: 10.1016/s1097-2765(00)80480-3. http://www.ncbi.nlm.nih.gov/pubmed/10230405. [DOI] [PubMed] [Google Scholar]
- 384.Kabe Y, Goto M, Shima D, et al. The Role of Human MBF1 as a Transcriptional Coactivator. J Biol Chem. 1999;274(48):34196–34202. doi: 10.1074/jbc.274.48.34196. [DOI] [PubMed] [Google Scholar]
- 385.Crawford PA, Polish JA, Ganpule G, Sadovsky Y. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol Endocrinol. 1997;11(11):1626–1635. doi: 10.1210/mend.11.11.9970. [DOI] [PubMed] [Google Scholar]
- 386.Li Y, Choi M, Cavey G, et al. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17(4):491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 387.Xu Q, Wang Y, Dabdoub A, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. [Accessed December 24, 2015];Cell. 2004 116(6):883–895. doi: 10.1016/s0092-8674(04)00216-8. http://www.ncbi.nlm.nih.gov/pubmed/15035989. [DOI] [PubMed] [Google Scholar]
- 388.Qin J, Gao D-M, Jiang Q-F, et al. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol Endocrinol. 2004;18(10):2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- 389.Steffensen KR, Holter E, Båvner A, et al. Functional conservation of interactions between a homeodomain cofactor and a mammalian FTZ-F1 homologue. EMBO Rep. 2004;5(6):613–619. doi: 10.1038/sj.embor.7400147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ortlund EA, Lee Y, Solomon IH, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12(4):357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 391.Weck J, Mayo KE. Switching of NR5A proteins associated with the inhibin alpha-subunit gene promoter after activation of the gene in granulosa cells. Mol Endocrinol. 2006;20(5):1090–1103. doi: 10.1210/me.2005-0199. [DOI] [PubMed] [Google Scholar]
- 392.Brendel C, Gelman L, Auwerx J. Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol Endocrinol. 2002;16(6):1367–1377. doi: 10.1210/mend.16.6.0843. [DOI] [PubMed] [Google Scholar]
- 393.Fuhrmann G, Chung AC, Jackson KJ, et al. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. [Accessed December 24, 2015];Dev Cell. 2001 1(3):377–387. doi: 10.1016/s1534-5807(01)00038-7. http://www.ncbi.nlm.nih.gov/pubmed/11702949. [DOI] [PubMed] [Google Scholar]
- 394.Yan Z, Jetten AM. Characterization of the repressor function of the nuclear orphan receptor retinoid receptor-related testis-associated receptor/germ cell nuclear factor. J Biol Chem. 2000;275(45):35077–35085. doi: 10.1074/jbc.M005566200. [DOI] [PubMed] [Google Scholar]
- 395.Bae Y, Kemper JK, Kemper B. Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP) DNA Cell Biol. 2004;23(2):81–91. doi: 10.1089/104454904322759894. [DOI] [PubMed] [Google Scholar]
- 396.Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24(17):7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Gobinet J, Carascossa S, Cavaillès V, Vignon F, Nicolas J-C, Jalaguier S. SHP represses transcriptional activity via recruitment of histone deacetylases. Biochemistry. 2005;44(16):6312–6320. doi: 10.1021/bi047308d. [DOI] [PubMed] [Google Scholar]
- 398.Boulias K, Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32(20):6096–6103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Båvner A, Johansson L, Toresson G, Gustafsson J-A, Treuter E. A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep. 2002;3(5):478–484. doi: 10.1093/embo-reports/kvf087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Seol W, Chung M, Moore DD. Novel receptor interaction and repression domains in the orphan receptor SHP. [Accessed December 29, 2015];Mol Cell Biol. 1997 17(12):7126–7131. doi: 10.1128/mcb.17.12.7126. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=232569&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5(6):343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 402.Majumder S, Liu Y, Ford OH, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66(12):1292–1301. doi: 10.1002/pros.20438. [DOI] [PubMed] [Google Scholar]
- 403.Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol Endocrinol. 2002;16(7):1492–1501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- 404.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. [Accessed December 24, 2015];Mol Cell. 2002 9(3):601–610. doi: 10.1016/s1097-2765(02)00471-9. http://www.ncbi.nlm.nih.gov/pubmed/11931767. [DOI] [PubMed] [Google Scholar]
- 405.Burd CJ, Petre CE, Morey LM, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103(7):2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Fu M, Wang C, Reutens AT, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 407.Chan KK, Tsui SK, Ngai SM, et al. Protein-protein interaction of FHL2, a LIM domain protein preferentially expressed in human heart, with hCDC47. [Accessed December 24, 2015];J Cell Biochem. 2000 76(3):499–508. http://www.ncbi.nlm.nih.gov/pubmed/10649446. [PubMed] [Google Scholar]
- 408.Müller JM, Isele U, Metzger E, et al. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 2000;19(3):359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Reutens AT, Fu M, Wang C, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15(5):797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 410.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277(29):25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 411.Coulthard VH, Matsuda S, Heery DM. An extended LXXLL motif sequence determines the nuclear receptor binding specificity of TRAP220. J Biol Chem. 2003;278(13):10942–10951. doi: 10.1074/jbc.M212950200. [DOI] [PubMed] [Google Scholar]
- 412.Powell SM, Christiaens V, Voulgaraki D, Waxman J, Claessens F, Bevan CL. Mechanisms of androgen receptor signalling via steroid receptor coactivator-1 in prostate. [Accessed December 29, 2015];Endocr Relat Cancer. 2004 11(1):117–130. doi: 10.1677/erc.0.0110117. http://www.ncbi.nlm.nih.gov/pubmed/15027889. [DOI] [PubMed] [Google Scholar]
- 413.Geserick C, Meyer H-A, Barbulescu K, Haendler B. Differential modulation of androgen receptor action by deoxyribonucleic acid response elements. Mol Endocrinol. 2003;17(9):1738–1750. doi: 10.1210/me.2002-0379. [DOI] [PubMed] [Google Scholar]
- 414.Hu Y-C, Yeh S, Yeh S-D, et al. Functional domain and motif analyses of androgen receptor coregulator ARA70 and its differential expression in prostate cancer. J Biol Chem. 2004;279(32):33438–33446. doi: 10.1074/jbc.M401781200. [DOI] [PubMed] [Google Scholar]
- 415.Yeh S, Sampson ER, Lee DK, et al. Functional analysis of androgen receptor N-terminal and ligand binding domain interacting coregulators in prostate cancer. [Accessed December 29, 2015];J Formos Med Assoc. 2000 99(12):885–894. http://www.ncbi.nlm.nih.gov/pubmed/11155740. [PubMed] [Google Scholar]
- 416.Hsiao PW, Lin DL, Nakao R, Chang C. The linkage of Kennedy’s neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. [Accessed December 5, 2015];J Biol Chem. 1999 274(29):20229–20234. doi: 10.1074/jbc.274.29.20229. http://www.ncbi.nlm.nih.gov/pubmed/10400640. [DOI] [PubMed] [Google Scholar]
- 417.Ito K, Adachi S, Iwakami R, et al. N-Terminally extended human ubiquitinconjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. [Accessed December 29, 2015];Eur J Biochem. 2001 268(9):2725–2732. doi: 10.1046/j.1432-1327.2001.02169.x. http://www.ncbi.nlm.nih.gov/pubmed/11322894. [DOI] [PubMed] [Google Scholar]
- 418.Kang HY, Yeh S, Fujimoto N, Chang C. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. [Accessed December 29, 2015];J Biol Chem. 1999 274(13):8570–8576. doi: 10.1074/jbc.274.13.8570. http://www.ncbi.nlm.nih.gov/pubmed/10085091. [DOI] [PubMed] [Google Scholar]
- 419.Miyamoto H, Rahman M, Takatera H, et al. A dominant-negative mutant of androgen receptor coregulator ARA54 inhibits androgen receptor-mediated prostate cancer growth. J Biol Chem. 2002;277(7):4609–4617. doi: 10.1074/jbc.M108312200. [DOI] [PubMed] [Google Scholar]
- 420.van de Wijngaart DJ, Dubbink HJ, Molier M, de Vos C, Trapman J, Jenster G. Functional screening of FxxLF-like peptide motifs identifies SMARCD1/BAF60a as an androgen receptor cofactor that modulates TMPRSS2 expression. Mol Endocrinol. 2009;23(11):1776–1786. doi: 10.1210/me.2008-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Fujimoto N, Yeh S, Kang HY, et al. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. [Accessed December 29, 2015];J Biol Chem. 1999 274(12):8316–8321. doi: 10.1074/jbc.274.12.8316. http://www.ncbi.nlm.nih.gov/pubmed/10075738. [DOI] [PubMed] [Google Scholar]
- 422.Mestayer C, Blanchère M, Jaubert F, Dufour B, Mowszowicz I. Expression of androgen receptor coactivators in normal and cancer prostate tissues and cultured cell lines. Prostate. 2003;56(3):192–200. doi: 10.1002/pros.10229. [DOI] [PubMed] [Google Scholar]
- 423.Miyoshi Y, Ishiguro H, Uemura H, et al. Expression of AR associated protein 55 (ARA55) and androgen receptor in prostate cancer. Prostate. 2003;56(4):280–286. doi: 10.1002/pros.10262. [DOI] [PubMed] [Google Scholar]
- 424.Khan OY, Fu G, Ismail A, et al. Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol Endocrinol. 2006;20(3):544–559. doi: 10.1210/me.2005-0110. [DOI] [PubMed] [Google Scholar]
- 425.Kullmann M, Schneikert J, Moll J, et al. RAP46 Is a Negative Regulator of Glucocorticoid Receptor Action and Hormone-induced Apoptosis. J Biol Chem. 1998;273(23):14620–14625. doi: 10.1074/jbc.273.23.14620. [DOI] [PubMed] [Google Scholar]
- 426.Rachez C, Lemon BD, Suldan Z, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398(6730):824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 427.Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol Cell Biol. 2013;33(9):1711–1722. doi: 10.1128/MCB.01151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Surjit M, Ganti KP, Mukherji A, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145(2):224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 429.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. [Accessed December 29, 2015];Proc Natl Acad Sci U S A. 1996 93(10):4948–4952. doi: 10.1073/pnas.93.10.4948. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=39385&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Torchia J, Rose DW, Inostroza J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387(6634):677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 431.Lee S-K, Anzick SL, Choi J-E, et al. A Nuclear Factor, ASC-2, as a Cancer-amplified Transcriptional Coactivator Essential for Ligand-dependent Transac-tivation by Nuclear Receptors in Vivo. J Biol Chem. 1999;274(48):34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 432.Okamoto K, Isohashi F. Macromolecular translocation inhibitor II (Zn(2+)-binding protein, parathymosin) interacts with the glucocorticoid receptor and enhances transcription in vivo. J Biol Chem. 2005;280(44):36986–36993. doi: 10.1074/jbc.M506056200. [DOI] [PubMed] [Google Scholar]
- 433.Chan SW, Hong W. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem. 2001;276(30):28402–28412. doi: 10.1074/jbc.M100313200. [DOI] [PubMed] [Google Scholar]
- 434.Hsiao P-W, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. [Accessed December 17, 2015];Mol Cell Biol. 2003 23(17):6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=180928&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Henriksson A, Almlöf T, Ford J, McEwan IJ, Gustafsson JA, Wright AP. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. [Accessed December 29, 2015];Mol Cell Biol. 1997 17(6):3065–3073. doi: 10.1128/mcb.17.6.3065. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=232159&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Obradović D, Tirard M, Némethy Z, Hirsch O, Gronemeyer H, Almeida OFX. DAXX FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells—toward a basis for the opposite actions elicited by two nuclear receptors? Mol Pharmacol. 2004;65(3):761–769. doi: 10.1124/mol.65.3.761. [DOI] [PubMed] [Google Scholar]
- 437.Pascual-Le Tallec L, Simone F, Viengchareun S, Meduri G, Thirman MJ, Lombès M. The elongation factor ELL (eleven-nineteen lysine-rich leukemia) is a selective coregulator for steroid receptor functions. Mol Endocrinol. 2005;19(5):1158–1169. doi: 10.1210/me.2004-0331. [DOI] [PubMed] [Google Scholar]
- 438.Wang Q, Anzick S, Richter WF, Meltzer P, Simons SS. Modulation of transcriptional sensitivity of mineralocorticoid and estrogen receptors. J Steroid Biochem Mol Biol. 2004;91(4–5):197–210. doi: 10.1016/j.jsbmb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 439.Fuse H, Kitagawa H, Kato S. Characterization of transactivational property and coactivator mediation of rat mineralocorticoid receptor activation function-1 (AF-1) Mol Endocrinol. 2000;14(6):889–899. doi: 10.1210/mend.14.6.0467. [DOI] [PubMed] [Google Scholar]
- 440.Murai-Takeda A, Shibata H, Kurihara I, et al. NF-YC functions as a corepressor of agonist-bound mineralocorticoid receptor. J Biol Chem. 2010;285(11):8084–8093. doi: 10.1074/jbc.M109.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Rogerson FM, Yao Y-Z, Young MJ, Fuller PJ. Identification and characterization of a ligand-selective mineralocorticoid receptor coactivator. FASEB J. 2014;28(10):4200–4210. doi: 10.1096/fj.13-242479. [DOI] [PubMed] [Google Scholar]
- 442.Tallec LP-L, Kirsh O, Lecomte M-C, et al. Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: implication of small ubiquitin-related modifier 1 modification. Mol Endocrinol. 2003;17(12):2529–2542. doi: 10.1210/me.2003-0299. [DOI] [PubMed] [Google Scholar]
- 443.Hultman ML, Krasnoperova NV, Li S, et al. The ligand-dependent interaction of mineralocorticoid receptor with coactivator and corepressor peptides suggests multiple activation mechanisms. Mol Endocrinol. 2005;19(6):1460–1473. doi: 10.1210/me.2004-0537. [DOI] [PubMed] [Google Scholar]
- 444.Zennaro MC, Souque A, Viengchareun S, Poisson E, Lombès M. A new human MR splice variant is a ligand-independent transactivator modulating corticosteroid action. Mol Endocrinol. 2001;15(9):1586–1598. doi: 10.1210/mend.15.9.0689. [DOI] [PubMed] [Google Scholar]
- 445.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. [Accessed December 29, 2015];Mol Cell Biol. 1997 17(5):2735–2744. doi: 10.1128/mcb.17.5.2735. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=232124&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. [Accessed December 29, 2015];Mol Cell Biol. 2000 20(7):2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=85422&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Yokota K, Shibata H, Kurihara I, et al. Coactivation of the N-terminal transactivation of mineralocorticoid receptor by Ubc9. J Biol Chem. 2007;282(3):1998–2010. doi: 10.1074/jbc.M607741200. [DOI] [PubMed] [Google Scholar]
- 448.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. [Accessed December 29, 2015];Science. 1997 277(5328):965–968. doi: 10.1126/science.277.5328.965. http://www.ncbi.nlm.nih.gov/pubmed/9252329. [DOI] [PubMed] [Google Scholar]
- 449.Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. [Accessed December 29, 2015];Cell. 1999 97(1):17–27. doi: 10.1016/s0092-8674(00)80711-4. http://www.ncbi.nlm.nih.gov/pubmed/10199399. [DOI] [PubMed] [Google Scholar]
- 450.Shi Y, Downes M, Xie W, et al. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15(9):1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Liu Z, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor coactivator-1 (SRC-1) enhances ligand-dependent and receptor-dependent cell-free transcription of chromatin. [Accessed December 29, 2015];Proc Natl Acad Sci U S A. 1999 96(17):9485–9490. doi: 10.1073/pnas.96.17.9485. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=22235&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Sheppard HM, Harries JC, Hussain S, Bevan C, Heery DM. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol. 2001;21(1):39–50. doi: 10.1128/MCB.21.1.39-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Shiozawa T, Shih H-C, Miyamoto T, et al. Cyclic changes in the expression of steroid receptor coactivators and corepressors in the normal human endometrium. J Clin Endocrinol Metab. 2003;88(2):871–878. doi: 10.1210/jc.2002-020946. [DOI] [PubMed] [Google Scholar]
- 454.Spencer TE, Jenster G, Burcin MM, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389(6647):194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 455.Giangrande PH, Kimbrel EA, Edwards DP, McDonnell DP. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. [Accessed December 29, 2015];Mol Cell Biol. 2000 20(9):3102–3115. doi: 10.1128/mcb.20.9.3102-3115.2000. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=85605&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Liu Z, Auboeuf D, Wong J, et al. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc Natl Acad Sci U S A. 2002;99(12):7940–7944. doi: 10.1073/pnas.122225699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8–bromocyclic AMP-dependent transcriptional activity of the human progesterone receptor. [Accessed December 29, 2015];Mol Cell Biol. 1998 18(3):1369–1378. doi: 10.1128/mcb.18.3.1369. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=108850&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Smith CL, Oñate SA, Tsai MJ, O’Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. [Accessed December 29, 2015];Proc Natl Acad Sci U S A. 1996 93(17):8884–8888. doi: 10.1073/pnas.93.17.8884. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=38563&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. [Accessed December 29, 2015];Mol Cell Biol. 2002 22(15):5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=133955&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Mamontova A, Séguret-Macé S, Esposito B, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. [Accessed December 30, 2015];Circulation. 1998 98(24):2738–2743. doi: 10.1161/01.cir.98.24.2738. http://www.ncbi.nlm.nih.gov/pubmed/9851961. [DOI] [PubMed] [Google Scholar]
- 461.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. [Accessed December 30, 2015];Vision Res. 2002 42(4):517–525. doi: 10.1016/s0042-6989(01)00146-8. http://www.ncbi.nlm.nih.gov/pubmed/11853768. [DOI] [PubMed] [Google Scholar]
- 462.Chomez P, Neveu I, Mansén A, et al. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor. [Accessed December 30, 2015];Development. 2000 127(7):1489–1498. doi: 10.1242/dev.127.7.1489. http://www.ncbi.nlm.nih.gov/pubmed/10704394. [DOI] [PubMed] [Google Scholar]
- 463.Grondona JM, Kastner P, Gansmuller A, Décimo D, Chambon P, Mark M. Retinal dysplasia and degeneration in RARbeta2/RARgamma2 compound mutant mice. [Accessed December 30, 2015];Development. 1996 122(7):2173–2188. doi: 10.1242/dev.122.7.2173. http://www.ncbi.nlm.nih.gov/pubmed/8681798. [DOI] [PubMed] [Google Scholar]
- 464.Wendling O, Chambon P, Mark M. Retinoid X receptors are essential for early mouse development and placentogenesis. [Accessed December 30, 2015];Proc Natl Acad Sci U S A. 1999 96(2):547–551. doi: 10.1073/pnas.96.2.547. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=15173&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Miyawaki T, Uemura A, Dezawa M, et al. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci. 2004;24(37):8124–8134. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Mori M, Metzger D, Picaud S, et al. Retinal dystrophy resulting from ablation of RXR alpha in the mouse retinal pigment epithelium. [Accessed December 30, 2015];Am J Pathol. 2004 164(2):701–710. doi: 10.1016/s0002-9440(10)63157-4. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1602254&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99(21):13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Chitayat D, Sroka H, Keating S, et al. The PDAC syndrome (pulmonary hypoplasia/agenesis, diaphragmatic hernia/eventration, anophthalmia/microphthalmia, and cardiac defect) (Spear syndrome, Matthew-Wood syndrome): report of eight cases including a living child and further evidence for autosomal. Am J Med Genet A. 2007;143A(12):1268–1281. doi: 10.1002/ajmg.a.31788. [DOI] [PubMed] [Google Scholar]
- 469.Goodman AB. Three independent lines of evidence suggest retinoids as causal to schizophrenia. [Accessed October 8, 2015];Proc Natl Acad Sci U S A. 1998 95(13):7240–7244. doi: 10.1073/pnas.95.13.7240. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=33865&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Sjölander A, Minthon L, Bogdanovic N, Wallin A, Zetterberg H, Blennow K. The PPAR-α gene in Alzheimer’s disease: Lack of replication of earlier association. Neurobiol Aging. 2009;30(4):666–668. doi: 10.1016/j.neurobiolaging.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 471.Chen Y-T, Collins LL, Uno H, Chang C. Deficits in motor coordination with aberrant cerebellar development in mice lacking testicular orphan nuclear receptor 4. Mol Cell Biol. 2005;25(7):2722–2732. doi: 10.1128/MCB.25.7.2722-2732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Jacobson SG, Marmor MF, Kemp CM, Knighton RW. SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. [Accessed October 8, 2015];Invest Ophthalmol Vis Sci. 1990 31(5):827–838. http://www.ncbi.nlm.nih.gov/pubmed/2335450. [PubMed] [Google Scholar]
- 473.Bosch DGM, Boonstra FN, Gonzaga-Jauregui C, et al. NR2F1 mutations cause optic atrophy with intellectual disability. Am J Hum Genet. 2014;94(2):303–309. doi: 10.1016/j.ajhg.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Featherstone HJ. Migraine and muscle contraction headaches: a continuum. [Accessed October 8, 2015];Headache. 1985 25(4):194–198. doi: 10.1111/j.1526-4610.1985.hed2504194.x. http://www.ncbi.nlm.nih.gov/pubmed/4019180. [DOI] [PubMed] [Google Scholar]
- 475.Carlsson A, Forsgren L, Nylander P-O, et al. Identification of a susceptibility locus for migraine with and without aura on 6p12.2-p21.1. [Accessed October 8, 2015];Neurology. 2002 59(11):1804–1807. doi: 10.1212/01.wnl.0000036617.04943.96. http://www.ncbi.nlm.nih.gov/pubmed/12473779. [DOI] [PubMed] [Google Scholar]
- 476.Kelly JF, Bienias JL, Shah A, et al. Levels of estrogen receptors alpha and beta in frontal cortex of patients with Alzheimer’s disease: relationship to Mini-Mental State Examination scores. [Accessed October 8, 2015];Curr Alzheimer Res. 2008 5(1):45–51. doi: 10.2174/156720508783884611. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3268687&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Young EA, Haskett RF, Murphy-Weinberg V, Watson SJ, Akil H. Loss of glucocorticoid fast feedback in depression. [Accessed October 8, 2015];Arch Gen Psychiatry. 1991 48(8):693–699. doi: 10.1001/archpsyc.1991.01810320017003. http://www.ncbi.nlm.nih.gov/pubmed/1652926. [DOI] [PubMed] [Google Scholar]
- 478.de Quervain DJ, Roozendaal B, McGaugh JL. Stress and glucocorticoids impair retrieval of long-term spatial memory. Nature. 1998;394(6695):787–790. doi: 10.1038/29542. [DOI] [PubMed] [Google Scholar]
- 479.Lee H, Sininger L, Jen JC, Cha Y-H, Baloh RW, Nelson SF. Association of progesterone receptor with migraine-associated vertigo. Neurogenetics. 2007;8(3):195–200. doi: 10.1007/s10048-007-0091-3. [DOI] [PubMed] [Google Scholar]
- 480.Decressac M, Volakakis N, Björklund A, Perlmann T. NURR1 in Parkinson disease—from pathogenesis to therapeutic potential. Nat Rev Neurol. 2013;9(11):629–636. doi: 10.1038/nrneurol.2013.209. [DOI] [PubMed] [Google Scholar]