ABSTRACT
After losing their protective peptidoglycan, bacterial spheroplasts can resynthesize a cell wall to recreate their normal shape. In Escherichia coli, this process requires the Rcs response. In its absence, spheroplasts do not revert to rod shapes but instead form enlarged spheroids and lyse. Here, we investigated the reason for this Rcs requirement. Rcs-deficient spheroids exhibited breaks and bulges in their periplasmic spaces and failed to synthesize a complete peptidoglycan cell wall, indicating that the bacterial envelope was defective. To determine the Rcs-dependent gene(s) required for shape recovery, we tested spheroplasts lacking selected RcsB-regulated genes and found that colanic acid (CA) biosynthesis appeared to be involved. Surprisingly, though, extracellular CA was not required for recovery. Instead, lysis was caused by mutations that interrupted CA biosynthesis downstream of the initial glycosyl transferase, WcaJ. Deleting wcaJ prevented lysis of spheroplasts lacking ensuing steps in the pathway, and providing WcaJ in trans to a mutant lacking the entire CA operon triggered spheroplast enlargement and lysis. Thus, CA is not required for spheroplast recovery. Instead, CA intermediates accumulate as dead-end products which inhibit recovery of wall-less cells. The results strongly imply that CA may not be required for the survival E. coli L-forms. More broadly, these findings mandate that previous conclusions about the role of colanic acid in biofilm formation or virulence must be reevaluated.
IMPORTANCE Wall-less bacteria can resynthesize their walls and recreate a normal shape, which in Escherichia coli requires the Rcs response. While attempting to identify the Rcs-dependent gene required for shape recovery, we found that colanic acid (CA) biosynthesis appeared to be involved. Surprisingly, though, cell death was caused by mutations that interrupted CA biosynthesis downstream of the initial step in the pathway, creating dead-end compounds that inhibited recovery of wall-less cells. When testing for the biological role of CA, most previous experiments used mutants that would accumulate these deadly intermediates, meaning that all prior conclusions must be reexamined to determine if the results were caused by these lethal side effects instead of accurately reflecting the biological purpose of CA itself.
INTRODUCTION
Bacterial cell shape is determined by coordinated and dynamic interactions among cytoskeletal elements, peptidoglycan synthesis, and cell division (as reviewed in references 1 and 2), and the wall and some of its morphological characteristics contribute to cell survival in suboptimal or hostile environments (3). For example, the host immune system elaborates several antibacterial factors, including lysozyme, cationic antimicrobial peptides, and protein complexes, which target the cell envelope and trigger bacterial lysis (4–6). In particular, lysozyme removes the peptidoglycan wall and leads to cell rupture. However, such cells may not lyse if they are immersed in an osmotically protective medium in which they may form wall-less cells of two broad types: those that can grow and divide independently (L-forms) and those that cannot (protoplasts or spheroplasts) (7). These entities can survive and regenerate a functional cell wall (8–13), which may enable them to avoid cell wall-specific host defenses until conditions allow for normal growth (14).
Recent work indicates that both Gram-positive and Gram-negative bacteria that have lost their cell walls can regenerate a defined morphology without the aid of a preexisting peptidoglycan template (12, 13). In particular, lysozyme-induced (LI) spheroplasts of Escherichia coli can regenerate their walls and morphology when grown in an osmotically protective medium (12, 15). However, this recovery process is impeded if the original rod-shaped cells lack one of several proteins, including penicillin-binding protein 1b (PBP 1b), LpoB, or Lpp, or if the cells cannot mount an active Rcs stress response (12). These mutants grow as normal rod-shaped cells when the wall is present, indicating that E. coli requires these additional proteins or processes to recreate a wild-type sacculus only when a preexisting peptidoglycan template is absent. Why these accessory processes are required is unknown.
L-forms are related to spheroplasts, in that they lack a peptidoglycan cell wall but retain functional membranes (7). The major difference between the two is that L-forms have gained the ability to grow and divide as cell wall-deficient forms, thereby perpetuating the strain, whereas spheroplasts may survive for a time but eventually die or revert to their original morphology (7). Though both spheroplasts and L-forms may sometimes regenerate a properly formed cell wall (13), L-forms must be able to divide and survive in the absence of a cell wall, and several genes and proteins are important for this transformation (15–17). One component reported to be required for L-form survival is the production of colanic acid (CA), a product of the Rcs stress response that regulates about 150 genes (15, 16, 18–20). CA is an extracellular capsular polysaccharide common to members of Enterobacteriaceae but absent from other bacteria (21), and its putative role in L-form survival is undefined.
Here, we attempted to identify the Rcs-dependent gene or genes required for spheroplast recovery. Of the known Rcs-regulated genes, 19 have RcsB-RcsB or RcsA-RcsB binding sites in or near their promoter regions (www.ecocyc.org), implying that these genes or operons are controlled directly by the Rcs response. We found that only one of these—the operon encoding the CA biosynthetic pathway—affected spheroplast recovery, suggesting that this extracellular capsular compound might be the pertinent Rcs-controlled component.
Surprisingly, we found that spheroplast recovery did not require extracellular colanic acid. Instead, spheroplasts lysed or died when cells carried mutations that interrupted CA biosynthesis downstream of WcaJ, the glycosyl transferase that initiates the pathway. Eliminating WcaJ allowed spheroplasts to survive even though no CA was produced, and spheroplasts enlarged and lysed if they contained WcaJ but no other CA synthetic enzymes. We conclude that CA intermediates are lethal to spheroplasts but that CA itself is not required for the survival of wall-less cells. Importantly, because of the way the CA pathway has been interrupted in previous experiments, it is now unclear whether CA plays a role in several other bacterial processes, including the survival of Gram-negative L-forms, the formation of biofilms, or virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids, DNA manipulation, and media.
Bacterial strains and plasmids are listed in Table S1 in the supplemental material. The primers and oligonucleotides used to create each plasmid and bacterial mutant are listed in Table S2 in the supplemental material. Routine cultures were grown in Luria-Bertani (LB) medium, and when appropriate, ampicillin (100 μg/ml) or kanamycin (50 μg/ml) was added. Standard DNA, PCR, and molecular biological techniques were utilized for cloning and plasmid construction (22), and E. coli DH5α was used as the intermediate cloning strain. All experiments were performed in the E. coli MG1655 background. The correctness of each plasmid was verified by DNA sequencing (UAMS DNA sequencing core).
Plasmid constructions.
Plasmids were constructed in the pDEV vector backbone (12). Using primers listed in Table S2 in the supplemental material, the following genes were amplified by PCR from the chromosome of E. coli MG1655 and cloned into pDEV to create different plasmids (in parentheses): ftsAZ (pFtsAZ), lolA (pLolA), wza (pWza), cpsG (pCpsG), ugd (pUgd), and wcaJ (pWcaJ). Restriction sites used to clone each gene are underlined in the primer sequences.
Strain constructions.
Genes were deleted from E. coli MG1655 by moving each mutation from appropriate strains of the Keio collection (23), via P1 transduction. Other mutations were created by λ-red recombination (24), using individual mutagenic oligonucleotides (see Table S2 in the supplemental material). The completely null CA operon mutant (DR38) was constructed as follows. First, the kanamycin cassette from pKD13 was amplified by PCR, using the primers DP263 and DP273 (see Table S2 in the supplemental material). These primers contained 40-nucleotide extensions at their 5′ ends that were homologous to the region upstream of the promoter region of wza (DP263) and to the region downstream of wcaM (DP273), which are the first and last genes, respectively, of the 20-gene CA operon (see Fig. 2A). To delete the entire CA operon, this kanamycin cassette PCR product was introduced into electrocompetent MG1655 cells that carried the λ-red helper plasmid, which was then induced with l-arabinose (24). Primers from regions flanking the CA operon were used for diagnostic PCR to verify that the entire 22.7-kb segment was deleted, including all 20 genes from wza to wcaM. Subsequently, the kanamycin cassette from this null mutant was removed by site-specific excision by expressing the FLP recombinase from the helper plasmid pCP20 (24), thereby generating E. coli DR38 [E. coli MG1655 Δ(wza-wcaM)::frt]. This strain was unable to synthesize CA, as verified by deleting the yrfF gene via P1 transduction (E. coli strain BMKM111-3), since this mutation generates prodigiously mucoid colonies only when the CA operon is intact.
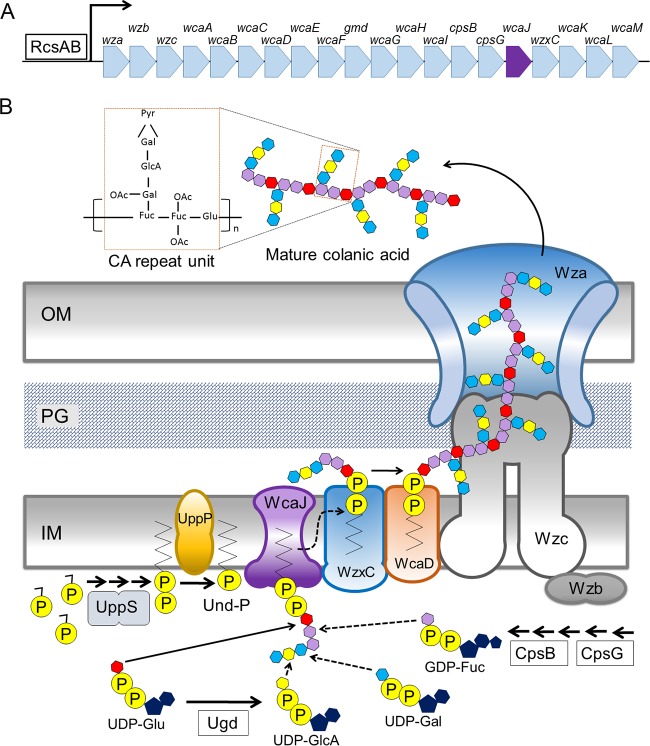
FIG 2.
Colanic acid genes and the CA biosynthetic pathway. (A) Genes of the CA operon. The promoter region contains a binding sequence that induces gene expression when occupied by the RcsAB heterodimer. (B) Selected substrates and enzymes in the CA biosynthesis and transport pathway (29, 39, 58). (Adapted from reference 34 with permission of the publisher.) Enzymes are devoted to precursor synthesis (CpsG, CpsG, Gmd, WcaG, and WcaH) or to CA assembly and transport (WcaJ, WcaA, WcaC, WcaE, WcaI, WcaL, WcaB, WcaF, WzxC, WcaD, Wzc, Wzb, and Wza). Undecaprenyl pyrophosphate synthase (UppS) catalyzes the condensation reaction of isopentenyl diphosphate and farnesyl diphosphate to generate undecaprenyl pyrophosphate (Und-PP). Undecaprenyl pyrophosphate phosphatase (UppP) converts Und-PP to undecaprenyl phosphate (Und-P). WcaJ, a glycosyl transferase, initiates the first dedicated step of CA assembly by adding glucose-1-phosphate from UDP-glucose (UDP-Glu) to Und-P, generating Und-PP-Glu. Additional sugar residues are added stepwise to Und-PP-Glu: GDP-fucose (GDP-Fuc), UDP-galactose (UDP-Gal), and UDP-glucaronic acid (UDP-GlcA). WzxC, a polysaccharide transporter, flips the lipid-linked CA repeat unit to the periplasmic face of the inner membrane (IM). WcaD polymerizes the repeat units to create CA polymers, and the Wzc-Wza complex transports mature CA through the outer membrane (OM) and out of the cell. OAc, O-acetyl; PG, peptidoglycan; Pyr, pyruvate.
Spheroplast recovery, peptidoglycan labeling, and microscopy.
The spheroplast recovery assay was performed as described previously (12). Briefly, cells from an exponentially growing LB broth culture were harvested at an optical density at 600 nm (OD600) of 0.2, washed with phosphate-buffered saline (PBS) (pH 8.0), and then plasmolyzed by resuspending them in 0.5 M sucrose plus lysozyme (20 μg/ml) for 10 min at 37°C. A mild osmotic shock was applied by diluting these cells with an equal volume of PBS (no sucrose) containing lysozyme (20 μg/ml) and incubating at 37°C for additional 10 min. To remove lysozyme, cells were centrifuged at 500 × g for 15 min and washed with sucrose recovery medium. The resulting cell pellet was transferred onto a sucrose recovery soft agar pad in a chambered slide. These slides were incubated and observed by using a Zeiss Axio imager Z1 microscope enclosed in a 37°C incubation chamber.
Peptidoglycan was labeled by feeding growing cells the fluorescent d-alanine derivative, hydroxy-coumarin-carbonyl-amino–d-alanine (HADA), a gift from Erkin Kuru and Michael S. VanNieuwenhze (25). Spheroplasts from rcsB mutant strains were grown for 1 h in spheroplast recovery broth containing HADA (500 μM, final concentration). These spheroplasts were fixed for 15 min in 2.8% formaldehyde and 0.04% glutaraldehyde, washed twice with PBS (pH 7.4), and prepared for microscopy. Spheroplasts and intact cells were visualized by using a wide-field epifluorescent Zeiss Axio imager Z1 microscope fitted with a 100× differential interference contrast objective (1.45 NA). Fluorescent images were captured by using 4′,6-diamidino-2-phenylindole (DAPI) filters (358-nm excitation, 461-nm emission) to image the HADA label. A green fluorescent protein (GFP) filter set (495-nm excitation, 519-nm emission) was used to image superfolder GFP. Images were acquired with a Zeiss Axiocam MRm camera and were processed with Axiovision software.
RESULTS
Rcs-deficient spheroplasts have defective envelopes.
If an LI spheroplast from an E. coli strain is to recover a rod-shaped morphology, the spheroplast must replace its missing cell wall in a way that regenerates the original cell shape. To do so, the spheroplast must have an intact and functional envelope, including an inner membrane (IM) and outer membrane (OM) with a periplasmic space, and must resynthesize an evenly distributed, rod-shaped layer of peptidoglycan. To determine the status of these components in recovering cells, we visualized the periplasmic space and peptidoglycan synthesis in wild-type and in Rcs-deficient spheroplasts (Fig. 1). (Note that, throughout the text, the term spheroplast refers to an LI spheroplast, that is, a spheroplast induced by lysozyme treatment, as described previously [12].)
FIG 1.
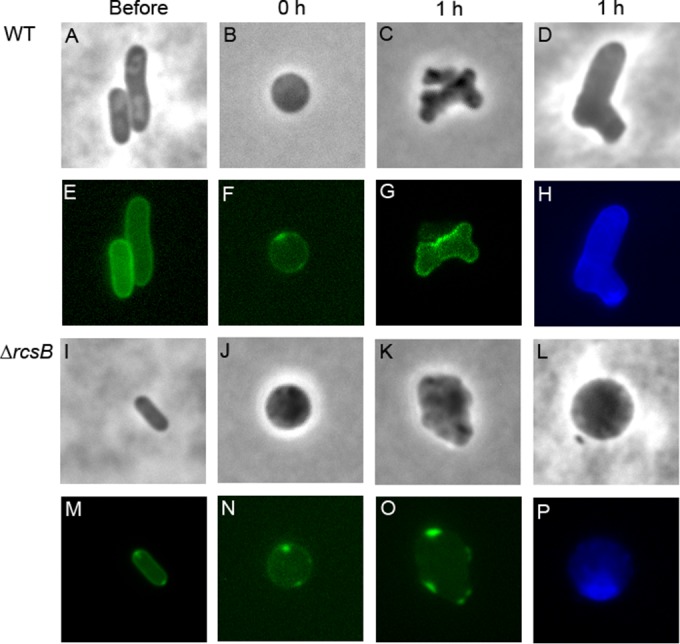
Organization of the periplasm and peptidoglycan in recovering LI spheroplasts. Wild-type E. coli strain MG1655 (A to H) and the isogenic ΔrcsB mutant DR5 (I to P) were visualized immediately before being transformed into LI spheroplasts (Before; n = 8 to 10), immediately after being transformed into LI spheroplasts (0 h; n = 5 to 6), and 1 h into the recovery period (1 h; n = 5 to 6). Cells expressing the periplasmic fluorescent protein DsbA-ss-sfGFP were visualized by phase (A to C, I to K) and fluorescence (E to G, M to O) microscopy. Newly synthesized peptidoglycan in recovering spheroplasts was labeled with the fluorescent d-alanine derivative HADA (D, H, L, P). The images are representative.
The periplasmic space of each cell was visualized by expressing the fluorescent protein DsbA-ss-sfGFP (the DsbA signal sequence fused to the amino terminus of superfolder GFP), which is transported to and accumulates within the periplasm (26). Wild-type spheroplasts grew, divided, and eventually returned to their normal wild-type rod shapes (Fig. 1B to D). In contrast, spheroplasts lacking rcsB never divided but, instead, enlarged (Fig. 1J to L) until they lysed (not shown), consistent with previous observations (12).
The periplasmic spaces in the two types of spheroplast were quite different. In untreated, wild-type, rod-shaped cells, the periplasm appeared as a continuous, evenly distributed fluorescent line that followed the circumference of each cell (Fig. 1E). The same circumferential distribution of periplasm was observed in each wild-type spheroplast immediately after the wall had been removed and in each recovering cell, no matter how aberrantly shaped these intermediate cells appeared (Fig. 1F and G). Similarly, cells lacking rcsB exhibited the same even distribution of periplasm around the cell periphery in untreated rod-shaped cells (Fig. 1M) and in newly formed spheroplasts (Fig. 1N). However, the periplasmic space was clearly aberrant in Rcs-deficient spheroplasts as they expanded, when the periplasm became discontinuous and formed a broken or spotty line around the periphery. In all cases, the periplasmic space was distended or bulged so that the fluorescent marker accumulated in distinctive and brightly lit sacs (Fig. 1M). Thus, the periplasmic space in Rcs-deficient spheroplasts was abnormal, implying that the organization of the cell envelope was defective and that, in the absence of the Rcs response, the envelope could not be reconstructed properly.
Because the periplasm was abnormal in Rcs-deficient spheroplasts, it seemed likely that peptidoglycan synthesis might also be irregular. We therefore visualized newly synthesized peptidoglycan in recovering spheroplasts by labeling cells with the fluorescent d-alanine derivative HADA (12, 25). Spheroplasts were incubated for 40 min in recovery broth, HADA was added for an additional 20 min to label newly synthesized peptidoglycan, and the labeled spheroplasts were visualized by fluorescence microscopy. As reported previously (12), wild-type spheroplasts recovered by forming a series of aberrantly shaped cells (e.g., Fig. 1C and D). HADA staining was distributed evenly in these cells, with a barely discernible thin line around the circumference indicating the presence of an intact and continuous layer of newly synthesized peptidoglycan (Fig. 1H). In contrast, spheroplasts lacking rcsB enlarged, but the peptidoglycan in the resulting spheroids was not distributed evenly, appeared to be fibrous, and was concentrated in only a fraction of each cell (e.g., Fig. 1P). Thus, Rcs-deficient spheroplasts failed to synthesize a continuous and normal protective layer of peptidoglycan, consistent with the view that such spheroplasts do not recreate the wild-type cell envelope.
Screening Rcs-regulated genes to identify those required for spheroplast recovery.
The Rcs stress response system affects the expression of over 150 genes (18, 19), at least one of which is required for spheroplast recovery (12). As a first step toward identifying the relevant element(s), we scanned databases for genes whose promoter regions contained RcsB binding sites, on the assumption that such genes would be regulated directly by the Rcs response. According to the EcoCyc database (www.ecocyc.org), 13 genes or operons have promoters with RcsB-RcsB binding sites (Table 1), and 5 have promoters with RcsA-RcsB binding sites (Table 2). We first mutated each of the 11 nonessential genes preceded by RcsB-RcsB sites (osmB, bdm, osmC, gadB, safA, ydeP, rprA, hdeA, hdeD, gadY, and gadA). Spheroplasts lacking any one of these genes grew normally and recovered their wild-type rod shapes (Table 1), indicating that none of these protein products were required for Rcs-dependent recovery. The remaining two genes preceded by RcsB-RcsB sites were essential (ftsAZ and lolA), and it was possible that the Rcs response enhances expression of these genes under spheroplast conditions. To test this, we enhanced gene expression by introducing cloned ftsAZ or lolA genes into an rcsF mutant. Expressing either ftsAZ or lolA did not rescue these Rcs-deficient spheroplasts (Table 1). Thus, none of the 13 genes preceded by RcsB-RcsB binding sites was required for spheroplast recovery.
TABLE 1.
Spheroplast recovery phenotypes of mutants lacking RcsB-RcsB-regulated genes or operonsa
| Operon | Associated function or characteristic | Gene(s) tested | Strain | Spheroplast recoveryb |
|---|---|---|---|---|
| ftsA ftsZ | Cell division | ΔrcsF Plac-ftsAZ | DR5-PF | − |
| lolA rarA | Transport lipoprotein to OM | ΔrcsF Plac-lolA | DR5-PL | − |
| osmB | Lipoprotein induced by osmotic stress | ΔosmB | DR20 | + |
| bdm sra | Biofilm modulation | Δbdm::kan | DR21 | + |
| osmC | Induced by osmotic stress | ΔosmC::kan | DR22 | + |
| gadB gadC | Glutamate decarboxylase | ΔgadB::kan | DR23 | + |
| safA yedO | Membrane connector for EvgS/EvgA | ΔsafA::kan | DR24 | + |
| ydeP | Oxidoreductase | ΔydeP::kan | DR25 | + |
| rprA | Regulatory RNA | ΔrprA::kan | DR26 | + |
| hdeA hdeB yhiD | Periplasmic acid resistance | ΔhdeA::kan | DR27 | + |
| hdeD | IM acid resistance | ΔhdeD::kan | DR28 | + |
| gadY | Regulator of gadX and gadW | ΔgadY::kan | DR29 | + |
| gadA gadX | Glutamate decarboxylase | ΔgadA::kan | DR30 | + |
kan, kanamycin resistance gene; OM, outer membrane; IM, inner membrane.
+, recovered; −, did not recover.
TABLE 2.
Spheroplast recovery phenotypes of mutants lacking RcsA-RcsB-regulated genes or operons
| Operon | Associated function or characteristic | Gene(s) testedb | Strain | Spheroplast recoverya |
|---|---|---|---|---|
| csgD csgE csgF csgG | Curli assembly and secretion | ΔcsgD::kan ΔrcsF | DR31 | − |
| flhD flhC | Regulator of flagellum biogenesis | ΔflhD::kan ΔrcsF | DR33 | − |
| rcsA | Auxiliary activator of colanic acid synthesis | ΔrcsA::kan | DR6 | + |
| yjbE yjbG yjbH | Biofilm formation | ΔyjbE::kan | DR34 | + |
| wza wzb wzc wcaA wcaB | Colanic acid biosynthesis | Δwza::kan | DR35 | − |
+, recovered; −, did not recover.
kan, kanamycin resistance gene.
Of the 5 genes containing RcsA-RcsB binding sites, the RcsA-RcsB heterodimer negatively regulates csgD and flhD (27, 28). Thus, an Rcs-deficient mutant may produce higher levels of CsgD or FlhD than the wild type, which might in turn prevent spheroplast recovery. If so, then deleting csgD or flhD should restore the ability to recover to Rcs-deficient spheroplasts. Instead, ΔrcsF spheroplasts lacking csgD or flhD did not recover (Table 2), indicating that these gene products do not inhibit the process. The remaining 3 genes (rcsA, yjbE, and wza) are positively regulated by RcsA-RcsB (18, 29–31), meaning that deleting one of these from an Rcs-proficient spheroplast might prevent recovery. Spheroplasts lacking either rcsA or yjbE recovered normally (Table 2), indicating that these two genes did not affect recovery. However, spheroplasts lacking wza did not recover their normal rod shapes (Table 2), implying that this gene might encode the pertinent Rcs-dependent factor required for spheroplast recovery.
Spheroplast recovery does not depend on extracellular colanic acid.
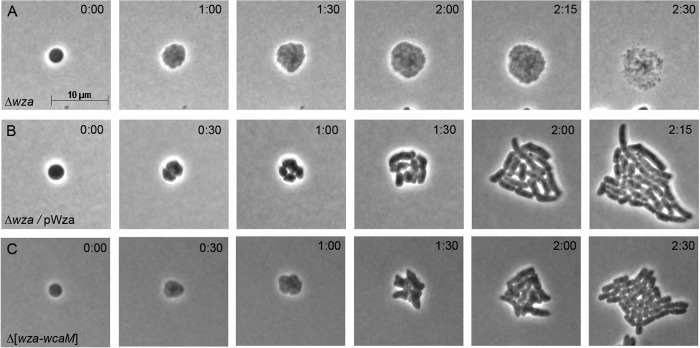
wza is the first gene of the cps operon (Fig. 2A), which encodes proteins that synthesize and export CA (Fig. 2B). Triggering the Rcs system induces expression of this operon, leading to the production of copious amounts of this extracellular capsular carbohydrate (21, 32). Because CA was reported to be essential for creating L-forms (15, 16, 20), it seemed possible that Rcs-deficient spheroplasts would not recover, because they could not synthesize this compound. Consistent with this expectation, spheroplasts lacking wza did not revert to a normal rod shape but instead expanded to become giant spheroids that lysed (Fig. 3A). (Note, though, that there was no detrimental effect on the growth or morphology of the wza mutant as long as the strain did not pass through a spheroplast stage.) Expressing wza in trans rescued the mutant so that spheroplasts divided and recovered (Fig. 3B), indicating that genes downstream of wza were expressed normally. Wza is the outer membrane component of the CA export channel, and the absence of Wza blocks extracellular CA secretion (33, 34). Thus, the inability of Wza-deficient spheroplasts to revert to a rod shape suggested either that extracellular CA was required for spheroplast recovery or that the accumulation of CA intermediates was deleterious to such cells.
FIG 3.
Extracellular colanic acid is not required for spheroplast recovery. Spheroplasts lacking Wza were incubated on recovery medium, and the recovery process was monitored by time-lapse phase-contrast microscopy. Time after plating (hour:minute) is displayed in each panel. The images are representative. (A) E. coli DR36 Δwza (n = 16); (B) E. coli DR37 Δwza, complemented with the pWza plasmid (n = 9); (C) E. coli DR38 Δ(wza to wcaM) (n = 16).
To determine if spheroplasts required CA to recover a normal morphology, we deleted 22.7 kb of chromosomal DNA to remove all 20 genes of the cps operon, from wza to wcaM (Fig. 2A), thereby creating a strain lacking the entire CA biosynthetic pathway (Fig. 2B). We confirmed the absence of these genes by diagnostic PCR and by the inability of the strain to produce CA (see Materials and Methods). To our surprise, spheroplasts derived from this strain divided and recovered normally (Fig. 3C), despite the fact that no CA could be synthesized, polymerized, or transported. Thus, the production of extracellular CA was not essential for the recovery process, nor did the absence of CA lead to spheroplast enlargement and lysis.
Colanic acid intermediates inhibit spheroplast recovery.
Though CA was not required for spheroplast recovery, a wza mutant prevented recovery. Thus, it seemed likely that, in the absence of the CA transporter Wza, intermediate periplasmic or cytoplasmic compounds might accumulate and inhibit recovery. To determine which intermediates were responsible, we interrupted CA assembly by working backward through the steps of CA synthesis, transport, and polymerization, which are illustrated schematically in Fig. 2B. In brief, assembly of the CA repeat unit is initiated by WcaJ, a glycosyl transferase that adds glucose (from UDP-Glu) to undecaprenyl phosphate (Und-P) at the cytoplasmic face of the inner membrane. Subsequently, fucose (from GDP-Fuc), galactose (from GDP-Gal), and glucosamine (from GDP-GlcA) are added to create the basic glycosyl repeat unit. This repeat unit is believed to be flipped across the inner membrane into the periplasm by the action of WzxC (29, 35). WcaD polymerizes these units, after which the CA polymer is transported across the outer membrane via the Wza-Wzc complex (29, 35).
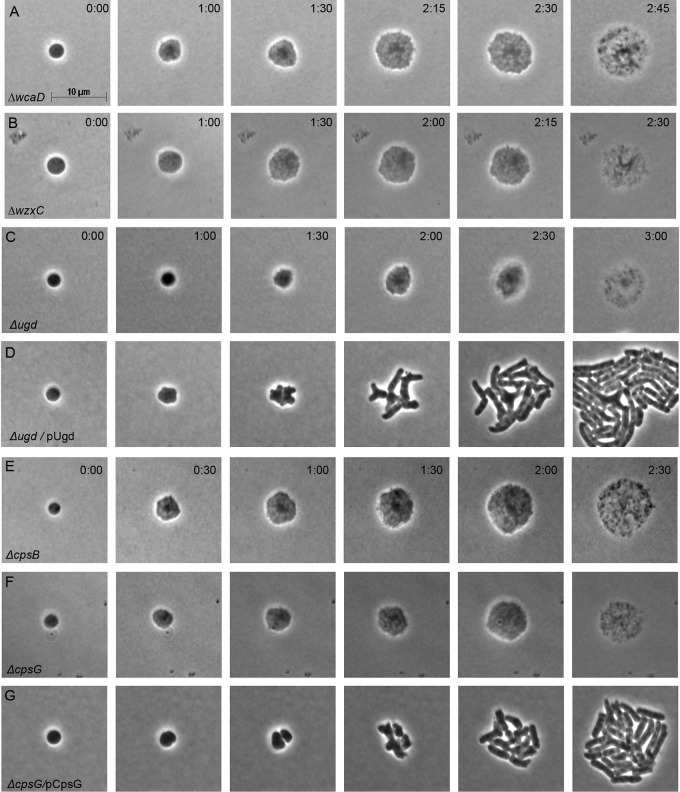
Spheroplasts lacking wcaD enlarged and lysed (Fig. 4A), as did spheroplasts lacking wxzC (Fig. 4B). Thus, the putative inhibitory intermediate(s) does not have to be periplasmic, because, in the absence of WxzC, such an intermediate would remain in the cytoplasm. We then attempted to determine which parts of the repeat unit were needed to create an inhibitory cytoplasmic intermediate. UDP-glucose (UDP-Glu) dehydrogenase (encoded by ugd) produces UDP-glucaronic acid (UDP-GlcA), which supplies glucaronic acid as the next-to-last carbohydrate in the CA repeat unit (Fig. 2B) (29). The Rcs system regulates the expression of ugd in Salmonella enterica, and an identical promoter sequence precedes ugd in E. coli (36), suggesting that the gene in E. coli is also controlled by the Rcs system, even though it was absent from the list of such genes in the EcoCyc database. E. coli spheroplasts lacking ugd did not recover but enlarged and lysed (Fig. 4C). The ugd phenotype was complemented by providing the gene in trans (Fig. 4D), indicating that the mutation had no polar effects on genes in its operon. The second carbohydrate added to the CA repeat unit is fucose (Fig. 2B), which is synthesized by gene products encoded by cpsB, cpsG, gmd, wcaG, and wcaH (Fig. 2A and B) (29, 37).
FIG 4.
Blocking colanic acid transport inhibits spheroplast recovery. Spheroplasts unable to assemble or transport CA were grown on osmotically protected sucrose recovery medium, and the recovery process was monitored by time-lapse phase-contrast microscopy. Time after plating (hour:minute) is displayed in each panel. The images are representative. (A) E. coli DR39 ΔwcaD (n = 9); (B) E. coli DR40 ΔwzxC (n = 22); (C) E. coli DR44 Δugd (n = 5); (D) E. coli DR45, in which ugd is complemented with the pUgd plasmid (n = 4); (E) E. coli DR41 ΔcpsB (n = 7); (F) E. coli DR42 ΔcpsG (n = 4); (G) E. coli DR43, in which ΔcpsG is complemented with the pCpsG plasmid (n = 8).
Spheroplasts lacking either cpsB or cpsG failed to recover and instead enlarged and lysed (Fig. 4E and F). The cpsG phenotype was complemented by supplying cpsG in trans (Fig. 4G), again indicating the absence of polar expression effects. These results indicated that the minimally relevant inhibitory compound did not contain any carbohydrate residues beyond the initial glucose residue.
A WcaJ-dependent intermediate prevents spheroplast recovery and triggers lysis.
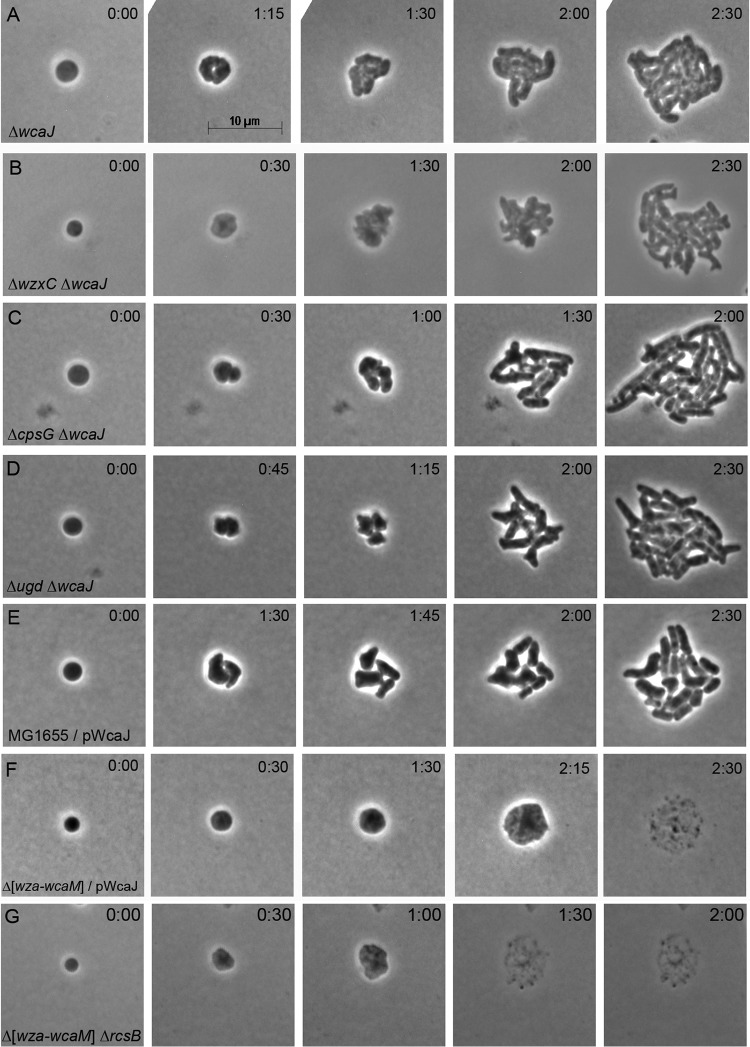
The foregoing experiments strongly suggested that the initial step in making the CA repeat unit was responsible for creating an inhibitory compound. WcaJ catalyzes this reaction by moving glucose-1-phosphate from UDP-glucose (UDP-Glu) onto the lipid carrier undecaprenyl phosphate (Und-P), thereby generating Und-PP-Glu (Fig. 2B) (38, 39). In fact, spheroplasts lacking wcaJ did divide and recover just as well as wild-type cells (Fig. 5A), indicating that the WcaJ-bound Und-P-P-Glu subunit was the minimum-sized inhibitor.
FIG 5.
Deleting wcaJ restores recovery to spheroplasts lacking components of the colanic acid biosynthesis pathway but not to spheroplasts lacking rcsB. Spheroplasts from the indicated mutants were grown on osmotically protected sucrose recovery medium, and the recovery process was monitored by time-lapse phase-contrast microscopy. Time after plating (hour:minute) is displayed in each panel. The images are representative. (A) E. coli DR46 ΔwcaJ (n = 13); (B) E. coli DR47 ΔwzxC ΔwcaJ (n = 15); (C) E. coli DR48 ΔcpsG ΔwcaJ (n = 14); (D) E. coli DR49 Δugd ΔwcaJ (n = 18); (E) E. coli MG1655 carrying the pWcaJ plasmid (n = 9); (F) E. coli DR51 Δ(wza to wcaM) carrying the pWcaJ plasmid (n = 15); (G) E. coli DR57 Δ(wza to wcaM) ΔrcsB (n = 12).
A wcaJ mutant not only abolishes the production of extracellular CA (39) but also eliminates the generation of subsequent CA intermediates (Fig. 2B). Thus, removing WcaJ would be expected to eliminate the deleterious effects observed in mutants lacking downstream components of the CA pathway. Consistent with this expectation, spheroplasts lacking both wcaJ and wzxC divided and recovered normally (Fig. 5B), thus reversing the lethal effects observed in spheroplasts lacking only wzxC (Fig. 4B). Similarly, when wcaJ was deleted from cells lacking either cpsG or ugd, the resulting spheroplasts divided and recovered normally (Fig. 5C and D), once again reversing the lethal effects observed in spheroplasts lacking only one of the latter genes (Fig. 4C and F). These results substantially strengthened the idea that any of several CA intermediates were inhibitory.
Finally, we determined whether the presence of WcaJ inhibited recovery in the absence of all other enzymes in the CA biosynthetic pathway. To that end, we overexpressed wcaJ in E. coli DR38, which lacks the entire CA pathway. Overexpressing wcaJ in spheroplasts derived from wild-type E. coli did not inhibit recovery (Fig. 5E), and, as described above, spheroplasts derived from the CA-negative E. coli strain DR38 recovered normally in the absence of additional WcaJ (Fig. 3C). However, when WcaJ was overproduced in CA-negative cells, the resulting spheroplasts did not recover but instead enlarged and lysed (Fig. 5F). Thus, active WcaJ prevented spheroplast recovery without the participation of downstream components of the pathway.
In short, the presence of WcaJ prevented spheroplast recovery in strains unable to complete and export a mature CA or in strains that could not synthesize a complete CA repeat unit. The results also further confirmed that the production of extracellular CA was not required for spheroplast recovery.
WcaJ does not prevent spheroplast recovery by sequestering Und-P.
The preceding results suggested the existence of deleterious intermediate products in the CA biosynthetic pathway, products that would not normally accumulate because they would be utilized and removed by later steps. The simplest explanation was that, in the presence of WcaJ and in the absence of a downstream CA pathway component, undecaprenyl-phosphate (Und-P)-linked compounds were accumulating as dead-end products. Spheroplast recovery might be inhibited if these intermediates reduced the pool of Und-P available for other, essential pathways (most notably, that portion of the pool required to synthesize peptidoglycan). Just such a competition for Und-P contributes to the creation of aberrantly shaped cells in mutants that accumulate dead-end Und-P-linked intermediates in the pathway that synthesizes enterobacterial common antigen (40) and in Bacillus subtilis mutants that accumulate teichoic acid intermediates (41).
If the above-described scenario was correct, then increasing the cellular pool of Und-P should suppress the effects of WcaJ, as it does in the enterobacterial common antigen (ECA) and teichoic acid cases (40, 41). To test this possibility, we increased the Und-P pool by overexpressing genes encoding enzymes that synthesize Und-P. However, overexpressing uppS (ispU), the undecaprenyl diphosphate synthase, did not restore the ability to recover for spheroplasts lacking wza or ugd (see Fig. S1A and B in the supplemental material). Similarly, overexpressing uppP (bacA), an undecaprenyl pyrophosphate phosphatase involved in recycling Und-P, did not restore recovery to cells lacking wza or ugd (see Fig. S1C and D in the supplemental material). As a complementary approach, we deleted uppP (bacA) to see if reducing the Und-P pool by this means might prevent spheroplast recovery, since UppP (BacA) accounts for ∼75% of the undecaprenyl pyrophosphate phosphatase activity in a cell (42). However, the resulting ΔuppP spheroplasts divided and recovered normally (see Fig. S1E in the supplemental material), suggesting either that the amount of Und-P had no effect on spheroplast recovery or that the uppP mutation did not reduce the pool of Und-P to a sufficient degree. In short, WcaJ-derived CA intermediates appeared to inhibit spheroplast recovery by a mechanism other than by sequestering the pool of Und-P.
The Rcs requirement for spheroplast recovery is independent of colanic acid and its intermediates.
The above results proved that extracellular CA was not required for spheroplast recovery (e.g., Fig. 3C); instead, CA intermediates were toxic to these cells. However, an active Rcs response is definitely required for spheroplast recovery. Therefore, it was formally possible that an intact Rcs response prevented the accumulation of toxic CA intermediates (e.g., if CA biosynthesis was interrupted for some reason). To investigate this possibility, we tested the recovery of spheroplasts lacking both rcsB and the entire CA operon [Δ(wza- wcaM)]. If the death of rcsB spheroplasts was mediated by CA or its intermediates, then the complete absence of these compounds should allow this mutant to recover normally. Instead, these doubly mutated spheroplasts failed to divide but grew into large spheroids and lysed (Fig. 5G) in exactly the same manner as rcsB mutants containing a wild-type CA gene cluster (Fig. 1) (12). This result indicates that the Rcs response is required for spheroplast recovery because it controls one or more processes unrelated to CA biosynthesis. At the moment, this Rcs-dependent mechanism remains unknown.
DISCUSSION
The role of colanic acid in wall-less E. coli and L-forms.
CA has been deemed to be essential for the continued growth and long-term survival of wall-less E. coli cells (variously described as L-forms or L-form-like cells) (15, 16, 20). However, we find that, in the complete absence of the entire CA biosynthetic pathway, LI spheroplasts survive, grow, and revert to a wild-type rod shape. In contrast, spheroplasts enlarge and lyse if they carry mutations that interrupt CA synthesis at any point after the first step catalyzed by WcaJ. Because all L-forms of E. coli, by definition, must survive the loss of the cell wall, previous experiments arguing that CA is essential for this process must be reevaluated in light of the present results.
Joseleau-Petit et al. first reported that CA was absolutely required for propagating E. coli without its cell wall (20). However, in these experiments, CA production was blocked by inactivating the Rcs stress response or by inserting a cpsE::Tn10 mutation (20). Here, though, we show that the Rcs response seems to be required for reasons that are unrelated to the production of CA, meaning that Rcs-negative mutants should not be used to prove that CA is required for a phenotype. As for the cpsE::Tn10 mutation, this allele was originally unmapped (43) but actually inactivates the wza gene (44; D. K. Ranjit and K. D. Young, unpublished data). Since a wza (≡cpsE) mutation impedes CA biosynthesis downstream of WcaJ and prevents spheroplast recovery, the cpsE::Tn10 allele cannot be used to infer that CA is necessary for L-form growth.
Glover et al. also concluded that CA was “essential for L-form colony formation” (16). These authors created unstable L-forms by growing E. coli in the presence of penicillin G and found that the Rcs stress response was induced 5- to 53-fold and that genes involved in CA synthesis were induced from 10- to 34-fold (16). Evidence supporting the importance of CA was that, of 42 mutants unable to grow as unstable L-forms, 3 inactivated the Rcs response and 10 inactivated CA production (including mutations in cpsB, wcaA, wzxC, and related genes). However, as argued above, Rcs-negative mutations have effects beyond the failure to synthesize CA. More to the point, though, is that all CA mutants that were isolated inhibited synthesis after the first WcaJ-initiated step in the pathway. In fact, Glover et al. reported that a wcaJ mutant did survive and formed unstable L-forms (16), a result that should not occur if CA was itself required for L-form survival. Instead, the failure of CA mutants to produce unstable L-forms, and the ability of a wcaJ mutant to do so, can now be explained more easily by the presence or absence of deleterious Und-P-linked CA intermediates.
Cambré et al. recently created wall-less E. coli by treating cells with cefsulodin, an antibiotic that inhibits penicillin-binding proteins 1a and 1b (PBPs 1a and 1b) (15). The resulting cells are technically not L-forms but are instead β-lactam-induced spheroplasts, similar to the LI spheroplasts that we discuss here. However, because antibiotic pressure can be maintained, these β-lactam-induced L-form-like cells grow and produce colonies. By using a transposon mutagenesis screen, Cambré et al. isolated 14 mutants that could not grow as L-forms, and each mutation perturbed either the Rcs stress response or CA synthesis; 5 mutations were in one of two genes of the Rcs pathway (rcsC, rcsD), while the remaining 9 were in genes encoding steps in CA biosynthesis (wzc, wcaD, wcaE, wcaK, wcaL, galE, or ugd) (15). These results led to the conclusion that CA was “crucial … for L-form growth” and that “all … requirements for L-form growth and multiplication can be attributed to the formation of the capsular polysaccharide colanic acid” (15). Indeed, this is exactly what we believed at first. The results do confirm and reinforce our previous observation that the Rcs system must be active if this type of spheroplast is to survive (12). However, again, Rcs involvement does not mean that CA itself is required. Also, every nongrowing CA mutant isolated by Cambré et al. interrupted CA synthesis downstream of WcaJ (15). The simplest explanation, then, is that these mutants failed to survive the loss of their cell wall because lethal Und-P-linked intermediates accumulated, which is the same reason that LI spheroplast CA mutants lysed in the present work. The fact that Cambré et al. recovered no mutations in wcaJ is consistent with this interpretation, because a wcaJ mutant would produce L-form-like colonies and would therefore escape notice in their genetic screen (15).
Therefore, reinterpreting the available evidence strongly suggests that CA itself is not required for the survival of wall-less E. coli. On the other hand, although an active Rcs stress response is essential for such survival, the identity of this Rcs-dependent factor remains unknown.
Role of colanic acid in virulence.
Beyond its supposed role in sustaining Gram-negative L-forms, CA has been implicated as important for several other bacterial traits and activities, including those related to virulence. For example, uropathogenic E. coli (UPEC) strains with mutations in either the wcaM or wcaL gene of the CA pathway are only 11% as effective in colonizing mouse spleens, suggesting that CA makes a significant contribution to UPEC virulence (45). Similarly, a wcaE mutant of an avian pathogenic E. coli strain survives very poorly in an in vivo chicken septicemia model (46). The mutant was especially deficient in colonizing the heart tissue, the lung, and the spleen, leading the authors to infer that this was “direct evidence for an association of colanic acid with virulence and fitness” (46). CA also reportedly imparts serum resistance to extraintestinal pathogenic E. coli (ExPEC) (45, 47–49). For example, only 3% of an ExPEC ΔwcaDE mutant and <20% of an rcsB mutant survived exposure to human serum (48), and a strain lacking any of three CA genes (wcaF, wcaH, or wcaI) was more sensitive to serum killing (47). Finally, wcaD and wcaE mutants of enterohemorrhagic E. coli (EHEC) strain O157:H7 grow less well under acidic conditions, at high temperatures, or in simulated stomach acid, suggesting that CA protects EHEC against these conditions (50, 51).
Unfortunately (in retrospect), all of the above-described experiments tested mutants that impaired CA biosynthesis at stages downstream of WcaJ (Fig. 2). Therefore, the observed phenotypes may have arisen either because CA was absent or because the mutants accumulated metabolic intermediates that impeded growth so that the cells were less able to survive the test conditions. To our knowledge, only one virulence experiment has used a mutant lacking the entire CA operon, in Salmonella enterica serovar Typhimurium (52). In this case, the loss of CA caused no difference at all in virulence (52), supporting the idea that CA itself may play no role in these situations. In any event, CA cannot be said to contribute to virulence unless appropriate mutants are created and retested.
The role of colanic acid in biofilms and other phenotypes.
One of the earliest functions proposed for CA was that it contributed to the formation of E. coli biofilms, in both E. coli K-12 and E. coli O157:H7 (49, 53, 54). CA-deficient E. coli strains adhere to surfaces just as well as wild-type cells, but they fail to mature into the normal three-dimensional biofilm architecture (53), and CA-deficient E. coli O157:H7 adheres less well to alfalfa sprouts (54). Similarly, CA is reported to be required for biofilm formation in Salmonella species (55). Once again, these conclusions were based on the behavior of strains carrying either wcaF or wcaD mutations (53, 54) or on the behavior of strains carrying mutations in wcaM, wcaA, or wza (55), all of which interrupt CA biosynthesis downstream of WcaJ (Fig. 2). Because biofilm formation requires cell growth, the accumulation of toxic CA intermediates may have simply impeded the survival of E. coli or Salmonella in these multicellular environments, making CA only appear to be important for biofilm formation. To our knowledge, only one E. coli biofilm experiment has tested a strain lacking the entire CA operon, and this CA mutation reduced biofilm formation by only ∼40% (56). In short, mature CA may contribute only moderately to biofilm formation.
Finally, CA-negative cpsB mutants of E. coli, Acinetobacter calcoaceticus, and Erwinia stewartii are much less resistant to desiccation, as is the cpsE mutant (57), leading to the speculation that CA increases survival in the wake of desiccation or during osmotic or acid stress (50, 57). Because the cpsB and cpsE (≡wza) mutations interrupt CA synthesis downstream of WcaJ, these conclusions should be revisited to be certain that CA is relevant.
Possible mechanisms.
In theory, CA intermediates could inhibit spheroplast recovery because these dead-end metabolic compounds compete for and sequester part of the pool of undecaprenyl-phosphate that is also required for peptidoglycan synthesis. We recently showed that just such a sequestration accounts for the inhibitory effects of similar intermediates in the pathway that synthesizes enterobacterial common antigen (40). However, increasing the pool of Und-P by overproducing UppS (IspU) did not restore the ability of spheroplasts to recover a normal shape, suggesting that this simple form of the sequestration model was not the explanation. The remaining alternative is that CA intermediates are toxic in and of themselves by a mechanism that remains unknown.
What is clear is that some property or process controlled by the Rcs stress response is required for the survival and morphological conversion of E. coli LI spheroplasts (12). Although we tried to identify the relevant Rcs-driven activity by mutating genes known to be controlled directly by RcsB, this approach has been unsuccessful so far. One would have thought that such a gene might have been represented in the 5,760 mutants described by Cambré et al., who screened for undirected mutants unable to grow as wall-less cells (15). The fact that no such mutation was isolated suggests either that more than one Rcs-dependent gene product is required for spheroplast survival or that too few mutants have been screened by us and by others.
Summary.
Colanic acid is not required for the survival or morphological reversion of wall-less E. coli cells, and it is almost certainly not required for the survival of the Gram-negative L-forms tested so far. In fact, all conclusions regarding the biological roles for CA must now be considered uncertain until the underlying experiments eliminate CA by interrupting its biosynthesis at the first committed step performed by WcaJ. Unfortunately, virtually all previous experiments pertaining to the function of CA examined the behavior of mutants in which CA biosynthesis was blocked downstream of WcaJ, which we now posit leads to the accumulation of deleterious UndP-linked intermediates. These intermediates may seriously obstruct bacterial growth under particular experimental conditions, a result that could then be falsely ascribed to a lack of CA. CA might be important for some of the phenotypes listed above, but the relevant experiments must be repeated with appropriate mutants before drawing such conclusions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kerry Evans for providing E. coli strain KE27 and for the ΔyrfF P1 lysate from E. coli strain BMKM111-3. We thank Erkin Kuru and Michael S. VanNieuwenhze for providing hydroxyl-coumarin-carbonyl-amino–d-alanine (HADA). We also thank Ry Young, Chris Whitfield, Suresh Kannan, Daniel Vega, Matthew Jorgenson, and William MacCain for helpful discussions.
Research reported in this publication was supported by award number R01-GM061019 from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01034-15.
REFERENCES
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Shaevitz JW. 2013. The mechanics of shape in prokaryotes. Front Biosci (Schol Ed) 5:564–574. [DOI] [PubMed] [Google Scholar]
- 3.Young KD. 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clin Microbiol Rev 19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 6.Sedov SA, Belogurova NG, Shipovskov S, Levashov AV, Levashov PA. 2011. Lysis of Escherichia coli cells by lysozyme: discrimination between adsorption and enzyme action. Colloids Surf B Biointerfaces 88:131–133. doi: 10.1016/j.colsurfb.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 7.Allan EJ, Hoischen C, Gumpert J. 2009. Bacterial L-forms. Adv Appl Microbiol 68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- 8.Lederberg J, St Clair J. 1958. Protoplasts and L-type growth of Escherichia coli. J Bacteriol 75:143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zinder ND, Arndt WF. 1956. Production of protoplasts of Escherichia coli by lysozyme treatment. Proc Natl Acad Sci U S A 42:586–590. doi: 10.1073/pnas.42.9.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makemson JC, Darwish RZ. 1972. Calcium requirement and magnesium stimulation of Escherichia coli L-form induction. Infect Immun 6:880–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruthe HJ, Adler J. 1985. Fusion of bacterial spheroplasts by electric fields. Biochim Biophys Acta 819:105–113. doi: 10.1016/0005-2736(85)90200-7. [DOI] [PubMed] [Google Scholar]
- 12.Ranjit DK, Young KD. 2013. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J Bacteriol 195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai Y, Mercier R, Errington J. 2014. Bacterial cell morphogenesis does not require a preexisting template structure. Curr Biol 24:863–867. doi: 10.1016/j.cub.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorbara MT, Philpott DJ. 2011. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol Rev 243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 15.Cambré A, Zimmermann M, Sauer U, Vivijs B, Cenens W, Michiels CW, Aertsen A, Loessner MJ, Noben JP, Ayala JA, Lavigne R, Briers Y. 2015. Metabolite profiling and peptidoglycan analysis of transient cell wall-deficient bacteria in a new Escherichia coli model system. Environ Microbiol 17:1586–1599. doi: 10.1111/1462-2920.12594. [DOI] [PubMed] [Google Scholar]
- 16.Glover WA, Yang Y, Zhang Y. 2009. Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS One 4:e7316. doi: 10.1371/journal.pone.0007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercier R, Kawai Y, Errington J. 2014. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 3:e04629. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrières L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol 50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 19.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. 2007. Unstable Escherichia coli L-forms revisited: growth requires peptidoglycan synthesis. J Bacteriol 189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol 185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vianney A, Jubelin G, Renault S, Dorel C, Lejeune P, Lazzaroni JC. 2005. Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151:2487–2497. doi: 10.1099/mic.0.27913-0. [DOI] [PubMed] [Google Scholar]
- 28.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol 178:4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stout V. 1996. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol 178:4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wehland M, Bernhard F. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 275:7013–7020. [DOI] [PubMed] [Google Scholar]
- 32.Brill JA, Quinlan-Walshe C, Gottesman S. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol 170:2599–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickerson NN, Mainprize IL, Hampton L, Jones ML, Naismith JH, Whitfield C. 2014. Trapped translocation intermediates establish the route for export of capsular polysaccharides across Escherichia coli outer membranes. Proc Natl Acad Sci U S A 111:8203–8208. doi: 10.1073/pnas.1400341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. 2006. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen IT, Beness AM, Saier MH Jr. 1997. Computer-based analyses of the protein constituents of transport systems catalyzing export of complex carbohydrates in bacteria. Microbiology 143(Part 8):2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 36.Mouslim C, Groisman EA. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol 47:335–344. doi: 10.1046/j.1365-2958.2003.03318.x. [DOI] [PubMed] [Google Scholar]
- 37.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 38.Furlong SE, Ford A, Albarnez-Rodriguez L, Valvano MA. 2015. Topological analysis of the Escherichia coli WcaJ protein reveals a new conserved configuration for the polyisoprenyl-phosphate hexose-1-phosphate transferase family. Sci Rep 5:9178. doi: 10.1038/srep09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, Bernards MA, Valvano MA. 2012. Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J Bacteriol 194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 22 December 2015. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol doi: 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Elia MA, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, Moreno-Hagelsieb G, Brown ED. 2009. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol 16:548–556. doi: 10.1016/j.chembiol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 42.El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. 2004. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J Biol Chem 279:30106–30113. doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- 43.Trisler P, Gottesman S. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol 160:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May T, Okabe S. 2008. Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and curli. J Bacteriol 190:7479–7490. doi: 10.1128/JB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li G, Laturnus C, Ewers C, Wieler LH. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun 73:2818–2827. doi: 10.1128/IAI.73.5.2818-2827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Gomes Moriel D, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug-resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miajlovic H, Cooke NM, Moran GP, Rogers TR, Smith SG. 2014. Response of extraintestinal pathogenic Escherichia coli to human serum reveals a protective role for Rcs-regulated exopolysaccharide colanic acid. Infect Immun 82:298–305. doi: 10.1128/IAI.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol 5:1173–1184. doi: 10.2217/fmb.10.83. [DOI] [PubMed] [Google Scholar]
- 50.Mao Y, Doyle MP, Chen J. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 183:3811–3815. doi: 10.1128/JB.183.12.3811-3815.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao Y, Doyle MP, Chen J. 2006. Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett Appl Microbiol 42:642–647. [DOI] [PubMed] [Google Scholar]
- 52.Wang S, Shi H, Li Y, Shi Z, Zhang X, Baek CH, Mothershead T, Curtiss R, 3rd 2013. A colanic acid operon deletion mutation enhances induction of early antibody responses by live attenuated Salmonella vaccine strains. Infect Immun 81:3148–3162. doi: 10.1128/IAI.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthysse AG, Deora R, Mishra M, Torres AG. 2008. Polysaccharides cellulose, poly-beta-1,6-n-acetyl-d-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ledeboer NA, Jones BD. 2005. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J Bacteriol 187:3214–3226. doi: 10.1128/JB.187.9.3214-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung BH, Lee CH, Yu BJ, Lee JH, Lee JY, Kim MS, Blattner FR, Kim SC. 2006. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl Environ Microbiol 72:3336–3342. doi: 10.1128/AEM.72.5.3336-3342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ophir T, Gutnick DL. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol 60:740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatar LD, Marolda CL, Polischuk AN, van Leeuwen D, Valvano MA. 2007. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology 153:2518–2529. doi: 10.1099/mic.0.2007/006312-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.