Abstract
Members of the myosin superfamily are actin-based molecular motors that are indispensable for cellular homeostasis. The vast functional and structural diversity of myosins accounts for the variety and complexity of the underlying allosteric regulatory mechanisms that determine the activation or inhibition of myosin motor activity and enable precise timing and spatial aspects of myosin function at the cellular level.
This review focuses on the molecular basis of posttranslational regulation of eukaryotic myosins from different classes across species by allosteric intrinsic and extrinsic effectors. First, we highlight the impact of heavy and light chain phosphorylation. Second, we outline intramolecular regulatory mechanisms such as autoinhibition and subsequent activation. Third, we discuss diverse extramolecular allosteric mechanisms ranging from actin-linked regulatory mechanisms to myosin:cargo interactions. At last, we briefly outline the allosteric regulation of myosins with synthetic compounds.
Keywords: Actin, Cytoskeleton, Inhibitor, Posttranslational regulation
Introduction
Myosins constitute a large multigene family of actin-based molecular motors in eukaryotes. Family members are widely expressed and tightly integrated in cellular networks of interlinked biochemical pathways where they function as integrators between signaling and the dynamics of cytoskeletal mechanics [1–4]. As highly allosteric enzymes, the stringent regulation is a prerequisite for myosin’s function in active transport processes, cytoplasmic contractility, and mechanosensing in both physiological and pathological processes [2–5].
Each mammalian cell has a specific repertoire of more than a dozen members from different myosin classes which are associated with a unique set of functional capacities and limitations. The finely balanced regulation of a specific myosin is strictly required to meet the changing demands of cellular functions. This requires a plethora of regulatory targeting mechanisms to recruit a specific myosin to its site of action, to switch its enzymatic activity on and off and to determine the cargo that may be attached to it. This local myosin regulation can result in global changes of cellular function to produce a coherent cellular response. Defects in the allosteric myosin motor regulation are linked to diseases, including cardiomyopathies, cancer and diabetes and provides a means for the development of myosin-specific drugs.
This review describes the general principles of posttranslational allosteric mechanisms of myosin regulation across different classes and phyla. It focuses on selected, well-characterized myosin classes and is subdivided into sections to reveal reoccurring themes and fascinating variations of intrinsic and extrinsic regulatory features that determine myosin’s mechanoenzymology, oligomerization, interactome and cellular localization. As individual members of a myosin class are sometimes synergistically targeted by different regulatory mechanisms, a single myosin class may be discussed in multiple sections of this review. The detailed discussion of individual myosin classes is beyond the current work and can be found in exhaustive recent reviews [3, 4].
Myosin structure, mechanoenzymatic concepts and classification
The diversity of cellular functions in which myosins are involved is attributed to characteristic structural, enzymatic and regulatory properties of individual members of the myosin superfamily. At the level of primary sequence, a myosin heavy chain consists of a motor domain, a neck domain and a tail domain (Fig. 1). The myosin motor domain harbors both a prototypic ATP binding site as well as a binding region for filamentous actin and allosterically links a repeated cycle of ATPase activity to its translocation on actin (Fig. 2) [5]. While the motor domain is the most conserved in terms of amino acid sequence and structural homology, the maximal actin-activated ATPase activity of myosins varies by a factor of 3000 from ~0.13 s−1 to ~390 s−1. Large differences are also found in the rate of actin filament sliding in the in vitro motility assay that ranges from ~20 nm·s−1 to ~6000 nm·s−1 [6–8]. Moreover, some myosins have evolved into pseudoenzymes that do not have an actin-activated ATPase activity [9, 10].
Fig. 1.
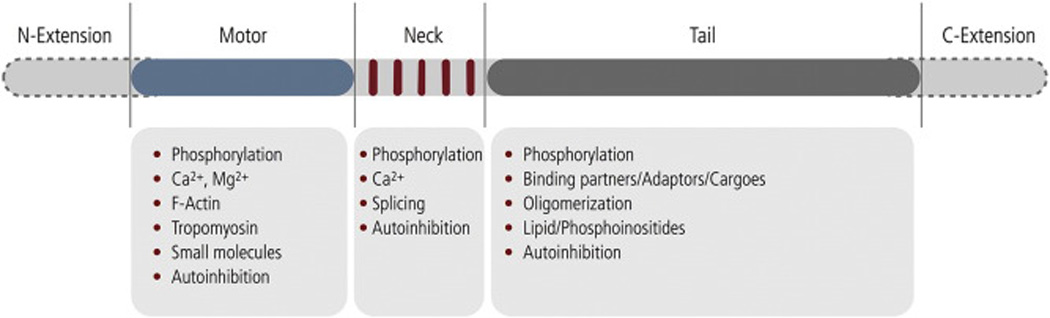
Basic building blocks of the myosin heavy chain: A myosin heavy chain consists of a prototypic motor domain (blue) which comprises the ATP binding site and an actin binding region, a light chain binding neck region containing multiple IQ-motifs (red), and a tail region (grey). N- and C-terminal extensions (grey dashed line) for the heavy chain are reported that may include N-terminal kinase domains, ATP-insensitive actin binding sites. Myosin light chains noncovalently associate with the myosin heavy chain to form the myosin holoenzyme. Moreover, myosin light chains stabilize and prime the neck region to competently function as a rigid lever that rotates relative to the myosin motor domain during the force generating power stroke that translocates myosin along actin [5]. Allosteric feedback mechanisms discussed in the present review target the motor domain, the neck domain and the tail region of the heavy chain and are listed below the respective domain. The N-terminal fraction of the molecule including the neck region and the associated light chains is referred to as S1. A longer fragment which additionally includes a portion of the tail domain is referred to as HMM. S1 fragments are monomeric whereas HMM fragments are dimeric. S1 is inherently active and serves as a powerful surrogate to study the transient-kinetic properties of the isolated myosin motor domain.
Fig. 2.
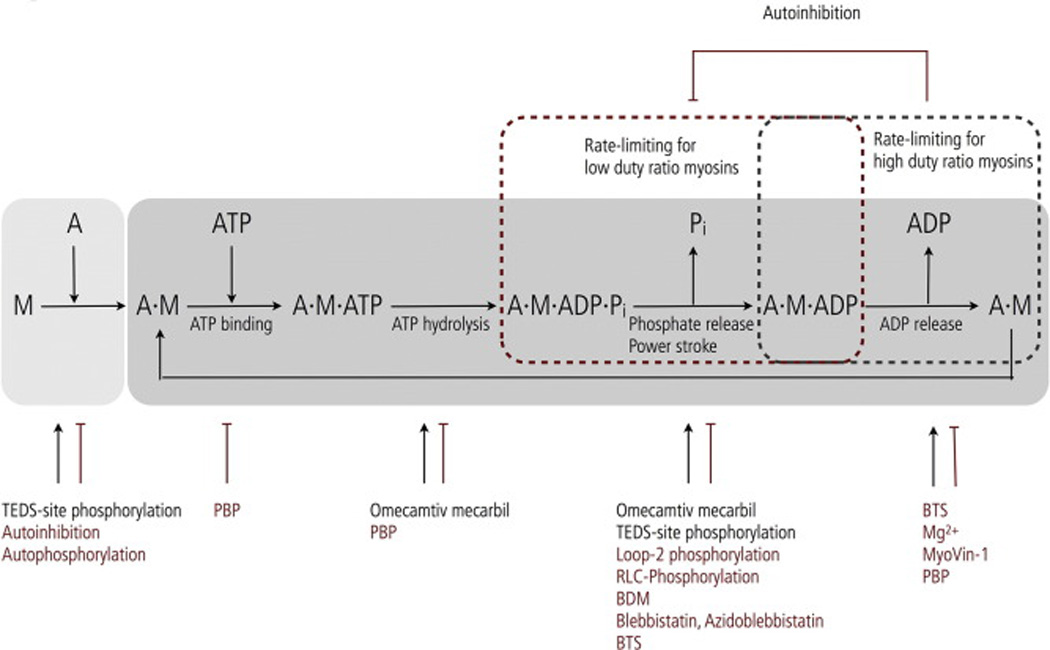
Simplified model of the basic actomyosin ATPase cycle and the kinetic steps altered by endogenous and exogenous regulatory mechanisms. Pi release (red dashed line) and ADP release (black dashed line) are entry and exit point to states during the ATP hydrolysis cycle in which the myosin motor domain is strongly bound to actin. The fraction of time of the ATP hydrolysis cycle myosin stays attached to actin is referred to as its duty ratio. Pi- and ADP release are commonly associated with the rate-limiting steps of low and high duty ratio myosins. The release of ADP also determines the speed myosins translocate on actin. Allosteric targeting mechanisms affecting key steps in the chemomechanical cycle are indicated. Black arrows and font color indicate activation; red blunt arrows and font color indicate inhibition. The abbreviations used are as follows: M = myosin, A = actin, Pi = inorganic phosphate.
Large structural and functional diversification is found in myosin non-motor domains: The neck domain typically contains one to six IQ motifs in vertebrate myosins which non-covalently associate with a specific set of calmodulin-like light chains [11]. Tail domains are most diverse and constitute a regulatory hotspot of the holoenzyme due to a vast domain complement including coiled-coils that mediate oligomerization, myosin tail homology; fourpoint-one, ezrin, radixin, moesin (MyTH/FERM) and pleckstrin homology (PH) domains that mediate the interaction with binding partners and phosphoinositides (PtdIns). Specifically, these domains confer unique physiological attributes including ligand interaction, thereby defining a requirement for individual regulatory features [5, 12, 13].
Sequence analysis of the conserved motor domain currently divides the eukaryotic myosin superfamily in ~35 phylogenetic classes, out of which 12 are expressed in humans [12]. The number of myosin classes is expected to increase steadily as more eukaryotic genomes are sequenced. Myosins can also be classified based on their biochemical and mechanical properties into four classes: fast movers, slow/efficient force holders, strain sensors or processive transporters [14]. Members of the myosin classes discussed here and their affiliation with these functional classes is shown in Fig. 3. For example, some dimeric mammalian myosins-5 and -6 are processive transporters, meaning that they can take multiple steps along actin without detaching. Besides structural prerequistites such as two motor domains and a long neck, processivity is facilitated by a high duty ratio, allowing myosin-5 and -6 to spend a long fraction of the kinetic cycle strongly bound to actin. Processivity is a prerequisite to transport cargoes such as melanosomes or endosomes along the actin cytoskeleton over long distances to its cellular destination [15].
Fig. 3.
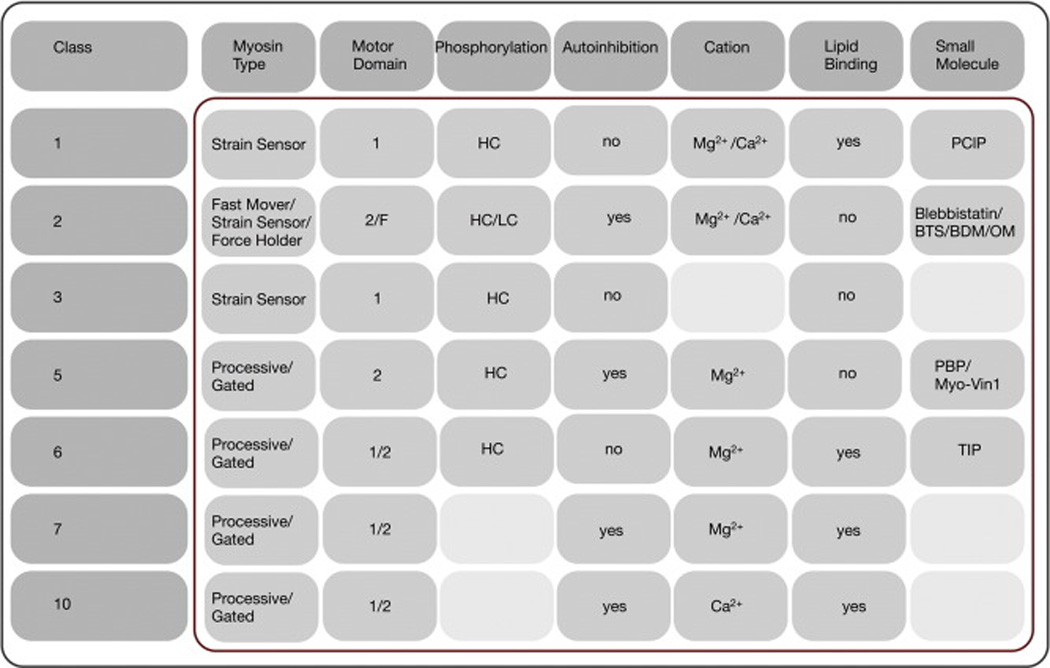
Selected myosin classes, their regulatory targeting mechanisms and cellular function. It is of note that not each of the regulatory mechanisms listed is applicable to each member in a given myosin class. The reader is redirected to comprehensive reviews for the detailed physiological function of individual members of a given myosin class [3, 4]. Motors indicate the number of myosin motor domains in the minimum functional unit of the protein. The notation 1/2 indicates that the myosin is a monomer but requires dimerization to become processive. The notation 2/F indicates that class-2 myosins are dimeric but assemble into filaments under physiological conditions inside cells. The column “small molecule” includes both, myosin activators and inhibitors. Light grey color indicated that regulatory events of phosphorylation, cations, and small molecule modulators have not be reported. Abbreviations are as follows: HC: heavy chain; LC: light chain.
In contrast, myosins such as certain class-1 myosins work as monomers and spend only a small fraction of time attached to actin. This feature does not allow them to transport cargoes but instead to function as strain sensors at the cytoskeleton-plasma membrane interface, a function essential to the regulation of membrane tension and cell migration. Other myosins with low duty ratios such as class-2 myosins assemble in bipolar filaments with sizes of 14–300 myosins. Smaller filaments are formed in the cytoplasm of nonmuscle and smooth muscle cells where myosins-2 power cytoplasmic contractility by sliding actin filaments past each other and function as force holders or strain sensors. Sarcomeric class-2 myosins assemble in large filaments made of hundreds of molecules and work together as fast movers to contract skeletal muscle.
Levels of myosin regulation
Myosin regulation occurs at three different levels: The first level includes transcriptional regulation, which dynamically affects the extent of myosin gene expression and alternate splicing in response to intra- and extracellular cues. The second and third levels of regulation includes substrate or effector binding and the regulation by posttranslational modifications, respectively. The latter mechanism is versatile and includes regulation at the myosin active site which is referred to as orthosteric and regulation at sites other than the active site which is referred to as allosteric. Orthosteric and allosteric events include covalent modifications such as phosphorylation and mutations to perturbation sites. Non-covalent targeting mechanisms include autoinhibition, ligand and effector binding [16]. Allosteric perturbations alter myosin’s mechanoenzymology and/or shift a preexisting myosin pool from an active to an inactive state or vice versa for example by the local accumulation of kinases and phosphatases or signatures of regulated actin [17].
Myosin regulation can follow either an “all-or-none” or a “modulatory” behavior. A classical example for the first is the phosphorylation-dependent autoinhibition relief of smooth myosin-2 in which the enzymatic activity is turned off in the absence of phosphorylation and is turned on by phosphorylation [18]. In contrast, Mg2+ has a modulatory effect on the enzymatic properties of some myosins, changing the activity by less than a factor of ten [19–23]. Moreover, no universal myosin regulation pattern exists: Myosins from one class, which are highly conserved at the levels of primary sequence and tertiary structure between protozoa and vertebrates exhibit multiple and entirely different regulatory aspects. This paradox underlines that evolution does not create new enzymes within a superfamily but rather new allosteric modes of protein regulation [24]. Fig. 3 summarizes modes of myosin regulation grouped by myosin class which are discussed in this review.
Regulation by phosphorylation
Phosphorylation is a reversible, covalent allosteric regulatory mechanism for myosins and occurs on the heavy chain and the associated light chain. Changes in phosphorylation patterns are rapid and local, induced by the finely balanced interplay of kinases and phosphatases at subcellular compartments and cytoskeletal structures.
Phosphorylation as regulatory targeting mechanism is best studied for class-2 myosins. These myosins uniquely polymerize from a monomeric form into bipolar or side polar filaments by means of a coil-coiled domain in their tail to generate contractile forces and movement by pulling on actin. Phosphorylation of the associated myosin-2 regulatory light chain (RLC) in response to upstream signaling pathways is the major regulatory mechanism of vertebrate smooth and nonmuscle myosins-2, but not the sarcomeric myosins-2 from skeletal and cardiac muscle [25]. Phosphorylation of Ser-19 of the smooth and nonmuscle myosin-2 RLC by kinases such as myosin light chain kinase (MLCK) or Rho-associated protein kinase is associated with increased (>1000 fold) actin-activated ATPase activity and allows myosin-2 to move actin in an in vitro motility assay [26–28]. Di-phosphorylation on Ser-19 and Thr-18 further increases the enzymatic activity but not the in vitro sliding velocity [29, 30]. RLC phosphorylation is further associated with the adaption of an assembly-competent conformation, contractility and stress-fiber formation, as outlined in greater detail below.
Protein kinase C (PKC) mediated phosphorylation of Ser-1, Ser-2 and Thr-9 on the RLC of smooth and nonmuscle myosin-2 that has already been phosphorylated at Ser-19 by MLCK is associated with decreased enzymatic activity due to decreased actin affinity in vitro but the rate of actin translocation in an in vitro motility assay is not affected [29, 31]. Phosphorylation at the PKC sites also allosterically renders Ser-19 of the RLC a poorer substrate for MLCK and may result in a decline in phosphorylation levels in cells [32]. Though the effect is evident in in vitro studies with purified proteins, PKC phosphorylation of the RLC has not been studied extensively in vivo. It is not regarded as major regulatory mechanism for nonmuscle myosin-2 function during cell division of HeLa cells and primary human keratinocytes as well as the assembly of nonmuscle myosin-2a in spreading and resting HeLa cells [33]. However, nonmuscle myosin-2a inhibition after phosphorylation of Ser-1/Ser-2 of the RLC at the leading edge is required for mesenchymal chemotaxis in fibroblasts [34].
In the social amoeba Dictyostelium, phosphorylation of the myosin-2 heavy chain is the major regulatory targeting mechanism of the actin-activated ATPase activity as discussed below. However, RLC phosphorylation at Ser-13 by MLCK increases the actin-activated ATPase activity of myosin-2 and the mechanical performance of the motor by a factor of ~5-fold. This modulatory regulation mechanism is dispensable for its in vivo function [35, 36].
RLC phosphorylation of mammalian sarcomeric myosins-2 has a minor impact on its enzymatic features but has modulatory effects on the mechanical performance of a muscle fiber. [37]. In cardiac muscle, RLC phosphorylation is associated with increased myocardial performance and enhances myosin motility under loaded conditions in vitro [38, 39]. Electron microscopic studies show that the motor domains of unphosphorylated skeletal and cardiac muscle myosin-2 filaments are held in close apposition to the filament backbone with a high degree of order. When the RLC is phosphorylated, the motor domains connected by the proximal coiled-coil regions lift off the surface of the filament and would be more likely to interact with actin [40, 41].
Interestingly, the paradigm that phosphorylation of striated muscle RLC is only modulatory does not extend to invertebrate muscle myosins-2. Myosins-2 from the striated muscle of Limulus and Tarantula are both regulated in an on/off manner as is seen with vertebrate smooth muscle myosin [42, 43].
Apart from myosin light chain phosphorylation, myosin-2 heavy chain phosphorylation in the tail coiled-coil and the non-helical tailpiece (NHT) is a regulatory targeting mechanism and implicated in the inhibition of filament formation and the interaction with binding partners [44–48].
Myosin-2 heavy chain phosphorylation is best studied in amoeba where multiple Ser and Thr residues in the tails have been identified as phosphorylation sites for myosin heavy chain kinases [49–52]. In Acanthamoeba, Ser phosphorylation of the NHT regulates filament assembly by inhibiting the dimerization of myosin monomers and the assembly pathway of dimers into filaments [44]. Recently, phosphorylation of a Ser in an actin-binding surface loop of the motor domain was shown to reduce the steady-state ATPase activity by the allosteric reduction of the rate-limiting Pi release ~ 3-fold (Fig. 2) [53, 54].
In Dictyostelium, myosin-2 heavy chain phosphorylation is associated with the disassembly of myosin filaments into monomers and/or small oligomers in vivo and in vitro [55, 56]. Conversely, the introduction of Ala as unphosphorylatable residues at the respective sites results in myosin overassembly in cells [55]. In contrast, Drosophila myosin-2 heavy chain phosphorylation in the NHT by PKC is not a major regulatory mechanism and dispensable for fly viability [57].
The contribution of heavy chain phosphorylation as a regulatory mechanism for mammalian myosin-2 filament formation and regulation is less well understood [48]. Several kinases including PKC and casein kinase 2 (CK2) were shown to phosphorylate isoform-specific Ser and Thr residues in the tail domain of the three mammalian nonmuscle myosin-2 paralogs [47, 58, 59]. Phosphorylation events are associated with the disassembly of existing myosin-2 filaments into monomers or the inhibition of myosin-2 monomers to form filaments in vitro, possibly by increasing the critical concentration for filament formation [46, 60]. The exclusive use of myosin-2 tail fragments, which are poor surrogates for full-length myosin-2 in in vitro studies, prevents a more detailed model of how heavy chain phosphorylation affects myosin-2 regulation at the molecular level. In cells, nonmuscle myosin-2 filament disassembly caused by phosphomimetic mutations in the tail domain is associated with increased migration rates and lamellipod extension in breast adenocarcinoma cells, suggesting that nonmuscle myosin-2 filaments act as a drag in biological processes such as cellular motility and chemotaxis [61]. In contrast, studies in osteosarcoma cells demonstrate that nonmuscle myosin-2a phosphorylated at a PKC site in the tail results in increased recruitment of this myosin to focal adhesions [62].
Class-1 myosins are single-headed and dynamically link the actin cytoskeleton to membranes. Phosphorylation of amoeboid myosin-1 molecules at a residue in the solvent exposed surface-loop in the myosin motor domain by STE20 family kinases is required for the actin-activation of their ATPase activity. The site was termed TEDS-site since virtually all myosins contain a Thr (T), Glu (E), Asp (D) or Ser (S) at this position [63].
Phosphorylation of TEDS-site residues in class-1 myosins is required for growth, actin organization and endocytosis in Dictyostelium [19, 64–68]. Similar, TEDS-site phosphorylation is indispensible for the localization of Myo1 to actin patches at the plasma membrane and endocytosis to occur in fission yeast. Those results imply increased actin affinity and motor activity of TEDS-site phosphorylated myosins-1 in this organism [69]. Concomitant, phosphomimetic TEDS-site mutants from Dictyostelium myosin-1d and -1e are characterized by increased (>20 fold) steady-state ATPase activity and actin affinity when compared to the unphosphorylated motor in vitro [19, 65]. For Dictyostelium myosin-1e, the speed with which it slides actin in the in vitro motility assay positively correlates with TEDS-site phosphorylation [65]. In contrast, the sole kinetic step increased by TEDS-site phosphorylation of Acanthamoeba myosin-1c is the rate-limiting phosphate Pi release which does not affect the speed of actin sliding [70].
Compared to amoeboid myosins-1, vertebrate class-1 myosins have a negatively charged amino acid at the TEDS-site which supersedes the phosphorylation requirement [63]. Further, allosteric and non-allosteric myosins can have similar evolutionary origins as seen for class-6 myosins which are phylogenetically closely related to class-1 myosins. Class-6 myosins have a Thr at the TEDS-site and are phosphorylated by PAK kinases in vitro [63, 71]. Strikingly, kinetic and functional studies of phosphomimetic mutants exclude TEDS-phosphorylation as a major regulatory mechanism in vitro, though cellular studies suggest phosphorylation-dependent myosin-6:actin interactions [72–74]. A possible explanation for discrepancy may be the use of phosphomimetic myosin mutants that do not always mimic bona fide phosphorylation [11, 75].
The importance of a negatively charged residue in the TEDS-site is also evident in in vitro studies, in which replacement of the endogenous TEDS-site Asp with in nonmuscle myosin-2a with a positive and neutrally charged residue reduces the enzymatic activity 8–10-fold, and the gliding speed of actin filaments in the in vitro motility assay 2–3-fold. Binding to actin is marginally affected by the mutations, suggesting that it does not contribute to the formation of a strongly-bound actomyosin complex [76]. This result is supported by recent studies showing that the TEDS-site is not part of the actomyosin interface, as previously speculated [77]. Instead, TEDS-site phosphorylation stabilizes and constrains the TEDS-loop which is crucial for the establishment of the actomyosin interface and allosterically accelerates the weak-to-strong transition of the myosin kinetic cycle (Fig. 2) [19, 77, 78].
Mammalian class-3 myosins are both, bona fide actin-based molecular motors and active STE20 Ser/Thr kinases and are best known for their function as cargo transporters in hair-cell stereocilia. Kinase activity is conferred by an N-terminal kinase domain prior to the myosin motor domain which is dependent on autophosphorylation. Phosphorylation of residues in an actin-binding surface loop of the myosin motor domain of human myosin-3a decreases the actin affinity under steady-state conditions > 100 fold along with the duty ratio of the motor [79, 80]. Deletion of the kinase domain increases the ATPase activity, actin affinity and duty ratio of human myosin-3a, implying that autophosphorylation reduces motor activity [81]. The kinetic data support a model for myosin-3a function in actin-based projections such as filopodia or stereocilia suggesting that the reduced motor activity of the phosphoprotein decreases the formation and number of filopodial protrusions and myosin accumulation to filopodial tips [81, 82].
Regulation by autoinhibition
Autoinhibition is a non-covalent allosteric mechanism and a reoccurring theme of myosin regulation at the single protein level for myosins from classes-2, -5, -7 and -10 which are subsequently discussed [83–86]. A mutual feature of autoinhibition is the adaption of a compact myosin conformer which is associated with reduced catalytic activity. Modes of autoinhibition at the structural level as well as mechanism of autoinhibition relief are myosin-specific.
In vitro, smooth and nonmuscle myosins-2 exist in a three-state equilibrium between a compact, autoinhibited conformation (10S, the sedimentation rate during ultracentrifugation assays), an extended conformation (6S), and filaments [83, 87]. In cells, the filamentous state likely represent the primary functional state of myosin [83, 88]. Electron micrographs of myosins-2 in the 10S conformation show that the two motor domains interact in an asymmetric manner and the tails are sharply bent at two locations to allow it to interact with and stabilize the motor-motor interaction [87, 89, 90]. The conformational restriction imposed by the motor:tail interaction allosterically shifts the myosin into a kinetically inert and assembly-incompetent state by trapping the motor in the weak actin-binding M.ADP·Pi state (Fig. 2) [91]. The motor-motor interaction required for the off state explains why S1 fragments of these myosins or asymmetric HMM molecules which contain only one motor domain are constitutively active in the absence of RLC phosphorylation [92, 93].
RLC phosphorylation at Ser-19 relieves autoinhibition, promotes filament formation and ATPase activity thereby triggering contractile forces on actin, as outlined above [87, 89]. Effects of RLC dephosphorylation are opposing. This widely accepted 10S-6S-filament three-state-equilibrium model is supported by cell biological studies demonstrating that smooth muscle myosin-2 exists in two functionally distinct cytoplasmic pools: A diffuse soluble 10S conformation and a filamentous one [94]. Interconversion can be exogenously induced and suggests a functional storage pool of monomeric 10S that could be readily mobilized to assemble via the transient 6S intermediate into filaments upon phosphorylation to meet changing demands of the cell [94].
The ability to adopt the asymmetric motor-motor autoinhibited conformation is conserved in myosin-2 molecules throughout evolution, as the motor domains of myosins-2 in relaxed Tarantula, Limulus and scallop thick filaments are in the same conformation as the motor domains of mammalian smooth and nonmuscle myosins-2 [95–97].
Recent studies suggest that the three-state equilibrium model for the regulation of nonmuscle myosin-2 has to be extended by a fourth state, as the three mammalian nonmuscle myosin-2 paralogs can form heterotypic filaments by co-polymerizing with each other. In addition, they form mixed heterotypic filaments with the enzymatically inactive pseudoenzyme myosin-18a in vitro and in vivo [98–100]. The physiological significance of heterotypic and mixed heterotypic filaments remains elusive but is hypothesized to regulate filament size and the collective mechanoenzymatic properties of the filament, thereby affecting force output. Other possible functional consequences of the formation of heterotypic and mixed heterotypic filaments are, besides altered cellular localizations, different interaction signatures with binding partners [99]. Moreover, this increased complexity and diversity in the myosin superfamily imposes different regulatory means which may contribute to the plethora of functions nonmuscle myosins-2 are involved in.
The dimeric cargo transporter myosin-5a is in a two-state equilibrium between an extended (14S) and an autoinhibited conformation (11S) [85, 101, 102]. Electron micrographs show a triangular structure of the autoinhibited myosin-5a holoenzyme with the two motor domains docked against the globular tail domains (GTD) [103, 104]. This intramolecular allosteric interaction reduces the kinetic competence of the motor domain which is characterized by a very low ATPase activity due to a reduced actin affinity [105]. A Ca2+-dependent autoinhibition relief is observed in vitro but is unlikely to be physiologically relevant as outlined in greater detail below [85]. A physiological cargo-dependent activation model suggests that interactions between the myosin GTD and binding partners such as melanophilin shift the equilibrium towards the catalytically active conformer while constituting a feedback mechanism that prevents futile ATP consumption under cargo-free conditions [106, 107].
Monomeric MyTH/FERM domain class-7 and -10 myosins adopt an autoinhibited conformation in which the GTD intramolecularly docks against the motor domain to form a tightly folded molecule with low actin-activated ATPase activity. Structural and biochemical studies indicate interactions between the second MyTH4 domain of the myosin-7 GTD, the motor and neck region [108, 109]. MyTH4 and PH domains of the myosin-10 tail also interact with the motor domain, respectively. Motor-tail crosstalk correlates with kinetically inertness in both myosins [84]. Ca2+ counteracts the tail-mediated inhibition of vertebrate class-7 but not class-10 myosins, where PtdIns(3,4,5)P3 relieves autoinhibition and renders the motor into a catalytically active dimer with cargo-transport function [84]. The presence of MyTH/FERM domains in the tail domain of class-15 myosins suggest autoinhibition as a possible regulatory mechanism.
Regulation by divalent cations
Divalent cations are second messengers, master regulators and integrators of myosin function: Transient Ca2+ gradients affect local cytoskeletal dynamics though the overall cytosolic cation concentration is tightly controlled and retains unchanged with a 1000-fold excess of Mg2+ over Ca2+ under resting conditions in mammalian cells [110, 111]. The effect of both, free Ca2+ and Mg2+ on the myosin holoenzyme is multimodal and either direct or indirect via interactions with the myosin heavy chain or the associated light chains [11]. Most identified myosin light chains have 4 EF-hand domains that may bind Ca2+ and/or Mg2+. For example MlcD, the native light chain of Dictyostelium myosin-1d, binds Mg2+ under physiological conditions. In contrast the versatile Ca2+ receptor calmodulin has four Ca2+-binding sites, two of which bind Ca2+ with high affinity even in the presence of excess Mg2+ [112]. Ca2+:calmodulin interactions trigger conformational rearrangements that may either result in the dissociation of a calmodulin subset from calcium-sensitive IQ-domains in the neck of some unconventional myosins from classes-1 and -5, or result in higher affinity for particular IQ motifs in some class-1 myosins [102, 113–116].
Ca2+-binding to the calmodulin occupying the first IQ motif of myosin-1c is associated with a conformational change that directly effects some transient kinetic parameters but not the steady-state ATPase activity [117]. Ca2+-bound myosin-1c has a lower duty ratio when compared to Ca2+-free myosin-1c which may be of physiological significance in the adaptation response of the inner ear [117]. A similar direct effect of Ca2+ has also been reported for mammalian myosin-1b [118].
In contrast to the direct effect on the enzymatic properties of some class-1 myosins, Ca2+-induced calmodulin dissociation and the associated rigidity changes alter the mechanochemical compliance of some myosin motors. For example, at least one calmodulin is released under physiological Ca2+ in vitro from myosins-1a and -1c [116, 119–123]. The in vitro sliding velocity decreased for both myosins, yet was restorable by the addition of exogenous calmodulin. The clutch-model suggests that calmodulin integrates force transduction from the motor to the tail domain in load-sensitive class-1 myosins [124]. Calmodulin is bound to the myosin neck in the absence of Ca2+ and provides mechanical stiffness: The clutch is engaged, the motor compliant. Ca2+-induced calmodulin dissociation from the first IQ-motif renders the neck floppy and disengages the clutch which results in idling of the myosin motor. Decreased neck stiffness prevents the motor domain to be restrained during the catalytic cycle, triggering increased catalytic activity. Noncompliance of the neck might be a regulatory mechanism to release myosin-1 from actin, a possible physiological scenario in Ca2+-dependent hair cell adaptation [124].
Motility defects, uncoupling of ATPase activity and lever arm mechanics induced by Ca2+ are also described for myosin-5a: Submicromolar Ca2+ concentrations allosterically relieves autoinhibition and potently activates the steady-state ATPase activity of full length myosin-5a in vitro [85]. This is likely due to the calmodulin-deficient myosin not being able to adopt the folded off state since Ca2+ does not affect the actin-activated ATPase activity of a two-headed fragment of myosin-5a which is missing the GTD [85]. Other means to prevent the formation of the myosin-5a off state include (i) the artificial increase or decrease in the neck length or tail, and (ii) single point mutations in the N-terminus of the myosin motor domain or the GTD [125, 126]. Caveats associated with the significance of Ca2+ activation as physiological regulatory mechanism are emerging: Ca2+ binding to the myosin-5a holoenzyme results in the dissociation of a calmodulin subset from the neck domain, as described for class-1 myosins. Calmodulin dissociation results in decreased mechanical stiffness of the neck region, which is paralleled by the impairment of a communication path that couples the conformation of the neck to the myosin motor domain [127]. Loss of mechanical compliance is reflected in motility defects in the presence of Ca2+ which can be restored in the presence of excess calmodulin in in vitro assays [127, 128]. That Ca2+ has no direct effect on the mechanoenzymatic properties of the myosin-5a other than through calmodulin dissociation is evident from experiments with calmodulin mutants that remain associated with the myosin heavy chain in the presence of Ca2+ and render the motor Ca2+-insensitive in the in vitro motility assay [127].
A Ca2+-dependent autoinhibition relief is likely to be a physiological regulatory mechanism for mammalian class-7 myosins which contain Ca2+-insensitive IQ domains. Ca2+:calmodulin interactions are proposed to induce conformational changes in the heavy chain bound calmodulin at the distal IQ motif which attenuate interactions with the tail, thus abolishing the tail-induced inhibition of myosin motor function [108].
Ca2+ also influences the enzymatic activity of conventional myosin-2s in a myosin-specific manner: In the absence of Ca2+, the two motor domains of molluscan muscle myosin-2 adopt a similar motor-motor autoinhibited state on the surface of the myosin filament as is seen in the monomeric state of unphosphorylated smooth and nonmuscle myosin-2. Ca2+ binding to the myosin essential light chain (ELC) activates the enzymatic activity, motility and tension generation of molluscan smooth muscle myosin-2. Structural studies indicated that Ca2+ mediates interactions between the ELC and the RLC. The Ca2+-bound ELC:RLC complex renders the neck into a rigid structure which does not allow the myosin motor domains to adopt an autoinhibited conformation with compromised enzymatic activity [129, 130].
Striated muscle myosins-2 in contrast are inherently unregulated but their activity is exogenously regulated by actin-linked Ca2+:troponin:tropomyosin interactions as reviewed in-depth elsewhere [131, 132]. The vertebrate skeletal muscle RLC can bind both Mg2+ or Ca2+, but preferentially binds Mg2+ under physiological conditions with high affinity. The Mg2+ dissociation rate is too slow to allow for transient binding of Ca2+ during the course of a muscle contraction [133].
The action of free Mg2+ on myosin is dual – cofactor-like in MgATP and allosteric. In vitro studies revealed that elevated levels of free Mg2+ reduce the actin-activated ADP release from Mg2+-sensitive class-1, -2, -5, -6 and -7 myosins in vitro (Fig. 2) [19–22, 65]. Effects of free Mg2+ differ depending on myosin’s mechanochemistry: High duty ratio motors such as myosin-5, -6 and -7a with a rate limiting actin-activated ADP release display reduced steady-state ATPase activity as the free Mg2+ concentration is increased [21–23]. Low duty ratio motors such as class-1 myosins and nonmuscle myosins-2 have a Mg2+-insensitive ATPase activities but display strong changes in their duty ratio paralleled with changes in free Mg2+ [19, 20, 65]. Universal characteristics of Mg2+-sensitive myosins are increased ADP affinity and reduced motility with increasing free Mg2+ concentration. Concomitantly, an Mg2+-dependent increase in run length of an artificially dimerized myosin-6 construct has been reported [134]. Allosteric regulatory targeting mechanisms can act simultaneously on a myosin as described for Dictyostelium myosin-1d where a high free Mg2+ concentration and TEDS-site dephosphorylation act synergistically and result in a > 40 fold reduction in motile activity [19].
As a cofoactor for ATP, magnesium binds together with the nucleotide as MgATP to the myosin active site. A structural model of Mg2+-sensitivity implies that interactions between active site residues with the bound nucleotide and Mg2+ need to be broken before the hydrolysis products can be sequentially released: Pi exits the nucleotide binding pocket first, followed by Mg2+ and ADP (Fig. 2) [23]. Changes in the free Mg2+ concentration allosterically shift the equilibrium between Mg2+-free and Mg2+-bound states by mass action. Accordingly, elevated free Mg2+ favors and stabilizes the tension and load-bearing A·M·ADP state and hinders the motor’s ability to produce rapid movement [19, 65]. Structural and functional conservation within the myosin motor domain suggests Mg2+-sensitivity to be a general regulatory mechanism. However, Dictyostelium myosins-1b, myosins-2 from Dictyostelium and Drosophila, as well as rabbit skeletal muscle myosin-2 are Mg2+-insensitive in the physiological concentration range in vitro, indicating different allosteric targeting mechanisms and intramolecular communication pathways [135, 136].
The effect of Mg2+ on the kinetic and functional activity of several myosins has been demonstrated in vitro, however, the physiological significance of Mg2+ as regulatory targeting mechanism has not been elucidated in detail in cells.
Actin-linked regulation
Six actins, more than forty tropomyosins and numerous other actin-binding proteins diversify and dynamically compartmentalize the mammalian actin cytoskeleton. Specifically, actin-binding proteins determine the speed of actin polymerization, depolymerization, and the length of actin filaments. Moreover, some actin-binding proteins induce the formation of higher-order actin structures such as bundles in response to a plethora of signal transduction cascades. This causes spatiotemporal patterning and segregation of actin-populations with different signatures which directly influence myosin’s track-selectivity and mechanochemisty to meet changing requirements of cellular functions [17]. Signatures of actin such as age and higher-order actin structures such as bundles are believed to guide myosin to its correct cellular destination, an important feature for example in active transport where misdelivery of a cargo or an interruption of a supply chain can result in pathologies.
Human nonmuscle myosin-2 isoforms and myosin-7a distinctly interact with isoactins [137]. In vitro, the enzymatic activity of nonmuscle myosin-2a and -2b is 4-fold greater with cytoplasmic β- and γ-actin than with skeletal muscle α-actin, as is the mechanical competence [137]. The paralog nonmuscle myosin-2c shows the greatest stimulation of its in vitro ATPase activity in the presence of β-actin, as does the myosin-7a:γ-actin pairing [137]. In a physiological context, intracellular isoactin:myosin cognate pairs are predicted to locally fine-tune the mechanochemical properties and target the motor to areas with a specialized actin composition such as stress fibers or filopodia. It was further shown that not only the isoactin but also its age, determined by its nucleotide state, is involved in myosin sorting: The transporter myosin-5a has a longer run length on “young” actin which is the ADP·Pi state, concomitant with the site of actin polymerization at the plus-end at the cell periphery. Myosin-6, a minus-end directed motor takes longer runs on “old” actin in the ADP state, consistent with its movement to the center of the cell [138]. Sorting mechanism of the actin cytoskeleton result in regions that are enriched for actin signatures that either promote or exclude the interaction with a specific myosin class are cell type specific and dependent on cellular needs [139]. Regional selective motility of unconventional myosins has been observed in detergent-extracted, permeabilized cells, underlining that the actin cytoskeleton fine-tunes myosin motor function and localization [139, 140].
Actin cross-linking proteins increase the complexity of cytoskeletal architecture by organizing actin into higher-order structures such as bundles with different spacing, register and polarity. Though the current data are controversial, myosins such as the cargo transporter myosin-10 may recognize these structural differences, adapt their motile behavior and move selectively on a specific population in the actin cytoskeleton. Myosin-10 was initially proposed to move processively only on actin bundles [141], but other studies demonstrate that it also moves processively along single actin filaments [142–144]. High resolution measurements of the myosin-10 stepping mechanism along fascin-bundled actin filaments shows that while it most commonly steps from one actin monomer to another in the same filament, it also take side steps to an adjacent actin filament [142]. Speculatively, bundle-selectivity might also be an actin-linked regulatory mechanism for members of the cargo transporting class-3, -6, -7 and -15 myosins which are commonly found in actin protrusions and sensory hair cell stereocilia [145].
Mammalian skeletal muscle myosin-2 is inherently unregulated and the cyclical interaction with actin is instead regulated by the Ca2+-sensitive tropomyosin-troponin regulatory complex as reviewed elsewhere [146, 147]. By contrast, the interaction between nonsarcomeric myosins-2 and regulated actin filaments is strikingly different due to the lack of troponin. The actin binding protein caldesmon which binds to actin filaments in the absence, but not the presence of Ca2+-calmodulin, might play a similar role as troponin in nonmuscle and smooth muscle cells but its regulatory effect on the actomyosin interaction has not been elucidated in detail [148].
Diversification of the decoration status of actin with tropomyosins is a major regulatory mechanism of the mechanochemistry and recruitment of myosin motors in nonmuscle cells. Tropomyosin is a coiled-coil forming protein that binds along the entire length of the actin filament. Conventional smooth and nonmuscle myosins-2 show a tropomyosin-dependent actin-activated ATPase activity and actin gliding in vitro as well as tropomyosin-dependent cellular localization to stress fibers, which result in decreased cell migration [29, 31, 149, 150]. In vitro, tropomyosin isoforms have differential effects on the actin-activated ATPase activity and motile properties of nonmuscle myosins-2a, -2b and -2c [149].
Tropomyosin also promotes the motor activity and increases the actin affinity of class-5 myosins from fission yeast and switches myosin-5 from budding yeast, Myo2p, from a nonprocessive to a processive motion by increasing the duty ratio [73, 151]. Strikingly, Myo2p exhibits processive movement with slower speeds but increased run length on tropomyosin decorated actin bundles when compared to single actin-tropomyosin filaments [151]. Processivity, run length and frequency are tropomyosin-isoform-dependent and isoactin-independent [151]. In contrast, in vitro reconstitution of the yeast Myo4p messenger ribonucleoprotein complex (mRNP), an essential molecular transport complex important for mRNA localization, shows a significant increased run length and run frequencies on parallel fascin-actin-tropomyosin bundles over single actin-tropomyosin filaments without speed changes [152]. Processive movement of Myo4p is tropomyosin-independent [152]. Strikingly, mammalian myosin-5a displays no track-selectivity for single actin filaments or bundles [142].
Tropomyosin is a negative effector of the mechanoenzymatic properties of the monomeric strain sensor myosin-1b, which is excluded from tropomyosin-decorated actin and regulated actin structures such as stress-fibers and bundles [153, 154]. In agreement, tropomyosin-decorated actin does not support in vitro motility of brush border myosin-1a and inhibits its steady-state ATPase activity [116]. The actin-crosslinking protein fimbrin relieves the tropomyosin-mediated inhibition of Myo1p in fission yeast by displacement of tropomyosin from the actin filament [155]. A model for the tropomyosin-linked myosin regulation suggests that by virtue of actin binding characteristics and terminal amino acid composition, different tropomyosin isoforms control the accessibility of myosin to the respective binding site on actin and directly regulate myosin mechanochemistry and localization.
Actin-linked regulatory mechanisms challenge the notion that the tail solely determines myosin’s spatial distribution since the motor domain discriminates the actin tracks that lead the myosin to its correct cellular destination. In line with this interpretation, a structural model for the tropomyosin-based regulation of myosin binding to regulated actin suggest direct myosin:tropomyosin interactions [77]. The surface loop-4 in the myosin motor domain is a central element of the actomyosin interface and contributes to non-covalent interactions with actin and tropomyosin [77]. Loop-4 sterically interferes with myosin binding to regulated actin in tropomyosin-sensitive class-1 myosins [78]. Concomitantly, mutational analysis reveals that truncation of loop-4 or the exchange of the loop with a corresponding loop from a tropomyosin-insensitive donor myosin promotes the interaction with regulated actin in vitro but is not sufficient to target class-1 myosins to different actin-tropomyosin structures in the cell [156].
Regulation by PtdIns
PtdIns recruit and anchor some myosins at cytoskeleton-membrane interfaces. On-site, the bilateral interactions impact myosin’s mechanochemistry which vice versa regulates membrane tension in biological processes including endocytosis, exocytosis and cell migration that require membrane deformability [157]. Myosin:PtdIns interactions are mediated by PH domains and clusters of polybasic residues in the myosin tail [158–160]. The balancing act to function as a divalent crosslinker between the actin cytoskeleton and the membrane impacts the mechoenzymatic properties of some class-1 myosins. For example, myosin-1c powers actin gliding on fluid lipid bilayers in vitro. The mechanical activity decreases parallel to the content of PtdIns(4,5)P2 in the bilayer while the membrane affinity increases [161]. Moreover, myosin-1c, but not myosins-1a and -1b, drives asymmetric, counterclockwise motility on lipid bilayers, suggesting a nonaxial component to the powerstrokes of some class-1 myosins. The physiological significance remains elusive but suggest myosin-1 isoforms to be involved in symmetry breaking events in vivo [161].
The myosin-6 heavy chain contains a lever arm extension that adopts a stable and monomeric three-helix bundle in solution [162, 163]. Exposure to lipid membranes triggers a reversible conformational change of this compact three-helix bundle to a rod-shaped structure. A proposed kiss-and-run model suggest the synergistic action of adaptor-regulated dimerization and cargo-vesicle induced unfolding of the three-helix bundle to render monomeric myosin-6 into a dimeric, processive transporter with large and variable step size [162, 164]. In line with this finding, structural changes in the myosin-6 tail are also induced by Ca2+-dependent PtdIns(4,5)P2-binding [165]. Crosslinking experiments suggest a lipid-mediated dimerization of two myosin tail fragments. Mutational studies indicate that both, PtdIns(4,5)2 and the binding partner disabled-2 (Dab2) are required for cellular positioning and the functional role of myosin-6 in clathrin-coated structures in the early endocytic pathway [165].
The monomeric transporter myosin-10 directly binds PtdIns(3,4,5)P3 through its tail PH domains which is required for its localization, the frequency of filopodia formation and function in macrophage phagocytosis [84, 166, 167]. PtdIns(3,4,5)P3 binding to myosin-10 activates the steady-state ATPase activity by relieving the tail-induced autoinhibition and promotes the motile activity of myosin-10 in the in vitro actin gliding assay [84]. Further, PtdIns(3,4,5)P3-mediates myosin-10 dimerization which is crucial for processive motion in vitro and in vivo and its function as intracellular transporter that shuttles components required for the filopodial growth, maintenance and bona fide function to the filopodial tip [142, 144, 168, 169].
Regulation by binding partners and cargos
Many non-filamentous unconventional myosins from classes-3, -5, -6 and -7 and -10 are essential intracellular transporters that actively shuttle their cargo between different cellular compartments. Myosin cargoes are structurally and functionally diverse and include proteins, organelles, mRNA and membrane compartments [152, 170–173]. Cargo-tethering to the myosin tail domain is mediated and specified by adaptors which provide a functional link between the molecular motor and its cargo. Myosin:cargo interactions determine the mechanochemistry, oligomerization and localization of the motor complex. Pertinent questions including the processes of cargo recognition, selection, loading and unloading are still elusive for most myosin:cargo complexes.
The well-studied class-5 myosins are best known for the transport of membranous structures such as melanosomes, recycling endosomes and the endoplasmic reticulum along the actin cytoskeleton [172–175]. For example, the myosin-5a GTD binds the adaptor melanophilin which couples the motor protein to the Rab27a-melanosome. The myosin-5a:melanophilin complex exhibits slower movement, enhanced run length and uniform processive stepping when compared to cargo-free myosin-5a in single molecule assays in vitro [173, 176, 177]. Melanophilin is an active component of the transport complex and dually functions as myosin-5a adaptor protein and tether that links the motor:cargo-complex to the actin track via its actin binding site [27]. Although the receptor that links the melanosome to Xenopus myosin-5 is unknown, phosphorylation of the myosin GTD by CK2 triggers cargo unloading in a cell cycle-dependent manner [178].
The monomeric class-5 myosin Myo4p is involved in posttranslational events of gene expression by transporting non-membranous mRNP into the emerging bud tip in budding yeast [170]. To accomplish active transport function with a single motor domain, Myo4p forms a heterodimer with the adaptor protein She3p and couples to a cargo mRNA via the tetrameric mRNA-binding protein She2p. In vitro reconstitution of the mRNP demonstrates cargo-linked myosin regulation since cargo mRNA is required for the formation of a functional, processive transport complex whereas cargo-free Myo4p is immotile [152]. Artificially increasing the number of localizing elements on the mRNA and multiple motors recruitment to the mRNP increases the frequency and supports longer processive runs [152]. In yeast, cargo unloading from Myo2 has is triggered through adaptor protein degradation and subsequent proteolytic degradation or GTP-hydrolysis within the myosin:cargo complex [179, 180].
Different from the high duty ratio motor myosin-5a which dimerizes via an extended coiled-coil region in the tail and exhibits processive movement along actin, both monomeric high duty ratio motors myosin-6 and -7a require external dimerization factors to function as cargo transporters [21, 22, 72]. The endocytic clathrin cargo adaptor Dab2 for example induces the regulated dimerization of the cargo-binding domain (CBD) of two myosins-6 in anterograde trafficking pathways [164, 165]. Adaptor-induced dimerization converts the non-processive myosin monomer into a processive dimer and is required for the localization of myosin-6 to clathrin-coated vesicles [164, 181]. A mutation within the myosin-6 motor domain that disrupts the necessary communication between the motor domains of the dimer is correlated with a strongly perturbed mechanoenzymatic regulation which renders the myosin to a nonfunctional transporter. Physiological consequences are the disrupted transport of nascent uncoated vesicles to the early endosome and impaired myosin-6 motor function in stereocilia resulting in deafness in mice [182].
As outlined in greater detail above, myosin-6 binds lipids via a three-helix bundle immediately preceding the CBD [162]. A proposed kiss-and-run model suggests that lipid binding of the three-helix bundle to cargo vesicles increases the local pool of monomeric myosin-6 at the membrane, thereby facilitating the cargo-induced dimerization [164]. Further, optical trapping experiments with artificially dimerized myosin-6 show that under low load, the myosin-6 functions as an efficient transporter while increasing loads render the motor to a dynamic actin tether - a prerequisite for its function as stereocilia cargo transporter and anchor of the apical cell membrane to the stereocilia base [183].
Artificially dimerized myosin-7a from Drosophila moves processively in single molecule in vitro assays, a requirement for its function as intracellular cargo transporter [86, 184]. In agreement, cell biological studies indicate that solely dimerized and not monomeric myosin-7a transports the MyRIP/Rab27 complex to the filopodial tip of retinal cells [184]. The model for the myosin-7a–dependent cargo transport suggests that the motor associates with the adaptor protein MyRIP which binds Rab27 thereby promoting the association with membrane vesicles such as melanosomes. Lateral movements on the membrane results in clustering of ternary myosin-7a/MyRIP/Rab27 complexes and crowding. The increase in the local concentration of monomeric myosin-7a on membrane vesicles might be sufficient to transport the cargo to filopodial tips. Alternatively, crowding-induced self-dimerization of myosin-7a could promote processive movement along the actin cytoskeleton [184]. Regulated cargo- or clustering-induced dimerization of two myosins is a conceptually similar regulatory mechanism for class-10 myosins [84, 144, 167].
Nonmuscle myosin-2 isoforms operate as teams in small bipolar ensembles with characteristic filament and functional signatures [185]. S1004A, a small Ca2+ binding protein associates with the myosin tail domain and promotes myosin filament disassembly and preventing filament assembly [186]. The nonmuscle myosin-2a:S100A4 interaction has physiological significance in metastasis-associated cellular motility [187]. Structurally, shifting of the monomer-filament equilibrium is triggered by partial unwinding of the myosin coiled-coil upon S100A4 binding and steric constraints [188]. The S100A4 interaction is inhibited by myosin tail phosphorylation by CK2 [46]. In contrast, the actin binding protein fesselin stabilizes smooth muscle myosin-2 filaments and increases filament length and thickness. Fesselin also decreases the rate of actomyosin dissociation in the presence of ATP and inhibits the actin-activation of the myosin ATPase activity in a concentration-dependent and tropomyosinindependent manner with an IC50 of 0.6 µM [189, 190].
Mammalian class-3 myosins challenge the existing dogma that monomeric myosins can’t exhibit effective transport functions. Different from the monomeric high duty ratio motors myosin-6 and -7a, myosin-3 exhibits dimerization-independent transport functions. An additional ATP-insensitive actin binding site in the myosin-3a tail which minimizes diffusion of the motor off the track is proposed to allow the motor:cargo complete inchworm to the tip of actin protrusions. Association with the cargo such as the actin-binding protein espin-1 actively enhances the length of actin rich protrusions such as filopodia and stereocilia [171, 191]. Myosin-3b lacks the tail actin binding site but utilizes a actin binding site in its cargo protein espin-1 to target the protrusion tip. Utilization of the biochemical properties of the cargo is indispensable for myosin-3b tip-localization to actin protrusions such as stereocilia and filopodia [191]. The cargo-assisted motility of the myosin-3b:espin-1 complex is another example of cooperative myosin regulation.
Regulation by small molecules
Myosins are druggable and promising targets for therapeutic intervention due to their implications in normal and pathological physiology [5]. Small-molecule druggability of the myosin motor domain gives valuable insight in myosin’s chemomechanical cycle (Fig. 2) at the molecular level and pharmacological block and release approaches have fundamentally advanced our understanding of myosin function in cells.
Allosteric extrinsic myosin regulation with covalent and non-covalent myosin inhibitors and activators has been described. The Pi-analog vanadate (Vi) is a nonspecific, competitive myosin inhibitor in the presence of ADP. Incorporation of ADP.Vi in the nucleotide binding pocket of the myosin motor domain leads to the formation of the kinetically inert ternary M.ADP.Vi state, which has been widely used in x-ray crystallography to trap myosin in a state that resembles the ATP bound conformation [192].
Mutual features of all other small molecule effectors are a non-competitive mode of action on the myosin motor domain. 2,3-butanedione monoxime (BDM), N-benzyl-p-toluenesulphonamide (BTS) and blebbistatin are inhibitors of class-2 myosins [193–198]. The inhibitory effect of the low-affinity compound BDM is skeletal-muscle myosin-2 specific (IC50 ~5 mM) and associated with the allosteric inhibition of Pi release [197]. BDM has many reported off targets and it is not recommended for use in cell biological experiments [197]. The inhibitory effect of BTS is specific for fast-twitch skeletal muscle myosin-2 (IC50 ~ 5 µM) caused by the reduction in the release rates of the ATP hydrolysis products [195].
Primary myosin targets of the small molecule compound blebbistatin are nonmuscle and skeletal muscle myosins-2 with isoform-dependent IC50 between ~ 0.5 and 5 µM [199]. Blebbistatin does not inhibit Drosophila nonmuscle myosin-2 and human unconventional myosins-1b, -5a and -10 [136, 198, 199]. Mechanistically, blebbistatin binds with high affinity to the M·ADP·Pi state and inhibits the Pi release (Fig. 2). The M·ADP·blebbistatin complex has a very low actin affinity and adopts a structural state that resembles the starting point of the powerstroke [193, 196, 200]. Structural studies show the blebbistatin binding site in the apex of the myosin cleft [201]. Blebbistatin is widely used in in vitro and cell biological studies to dissect processes which depend of the contractility of the actin cytoskeleton in nonmuscle cells such as cytokinesis and cell migration. The major disadvantages, which limit the use of blebbistatin in live cell experiments to study dynamic aspects of nonmuscle myosin-2 motor function, are its blue-light sensitivity and cytotoxic side effects which precludes its use with GFP-tagged myosins [198, 202, 203]. To circumvent this problem, blebbistatin-derivatives have been synthesized. Among those, para-nitroblebbistatin is the most latest described non-cytotoxic and photostable blebbistain derivative that displays the same inhibitory potency to myosins-2 as blebbistatin and allows studies of GFP-nonmuscle myosin-2 inhibition in live cells [204]. Another blebbistatin derivative, photoreactive azidoblebbistatin, covalently crosslinks to the myosin-2 motor domain after UV-irradiation [205]. This photoreactivity improves the in vitro and in vivo effectiveness when compared to blebbistatin but its binding to the myosin-2 motor domain is irreverible [205].
Halogenated pseudilins bind to an allosteric site near the tip of the 50 kDa subdomain and reversibly inhibit the enzymatic activity of unconventional myosins. Pentabromopseudilin (PBP) potently reduced the steady-state ATPase activity (>50-fold) and motile activity (>50-fold) of eukaryotic class-5 myosins (IC50 ~ 0.4 µM) and exhibits less potency for class-1 and -2 myosin [206]. Mechanistically, PBP interferes with ATP binding, hydrolysis and ADP release (Fig. 2) and alters the myosin-5a dependent distribution of mitochondria between mother and daughter cells during cytokinesis in yeast [206]. The pyrazolopyrimidine compound MyoVin-1 also inhibits the actin-activated ADP release of myosin-5 but with a weak IC50 ~ 6 µM in vitro [207]. Its inhibitory potency to inhibit myosin-5a function in cells has not been firmly established yet.
Another pseudilin derivative, pentachloropseudilin (PClP), is a class-1 myosin specific inhibitor (IC50 ~ 1–5 µM) with drastically reduced or no affinity for class-2, -5, -6 and -7 myosins (IC50 ≥ 100 µM) [208]. PClP allosterically lowers the coupling efficiency (> 30-fold) between the nucleotide binding site and the actin binding region and the motile behavior of class-1 myosins [208]. PClP-treatment of HeLa cells results in changes in lysosome morphology and distribution – an effect similar to that observed after siRNA-mediated myosin-1c depletion [208].
The motor activity of the closely related myosin-6 is inhibited ~ 3-fold by the small molecule 2,4,6-triiodophenol (TIP) [22]. In the cell, TIP-mediated myosin-6 inhibition is associated with defects of vesicle fusion at the plasma membrane in the final stages of the secretory pathways [22].
Solely one myosin activator is described: Omecamtiv mecarbil (OM) specifically targets cardiac myosin-2 (EC50 ~ 1.6 µM) and enhances the efficiency of myocyte contractility independent from Ca2+ in vitro and in vivo [209, 210]. The drug binds in a cleft between the 25 kDa and the lower 50 kDa subdomain of the myosin motor domain and allosterically increases the actin-activated steady-state ATPase activity of cardiac myosin-2 in a dose-dependent manner by increasing the rates of both, the ATP hydrolysis the rate-limiting Pi release in vitro (Fig. 2) [209] [211]. An activating effect of omecamtiv mecarbil is not reported for other muscular myosins-2 [209].
The small molecule EMD 57033 is predicted to bind to the same region as omecamtiv mercarbil in cardiac myosin-2. EMD 57033 is not a direct activator of cardiac myosin-2 and rather acts as pharmacological chaperone. By binding to and refolding of stress-induced misfolded protein, EMD57033 can restore the enzymatic activity of the motor, a function that may improve therapeutic approaches of protein-misfolding diseases including myosinopathies [212].
Conclusion and Perspectives
One myosin - numerous regulation signatures. Posttranslational allosteric myosin regulation provides a mechanism for increased protein diversity and reveals fascinating insights in basic mechanisms that influence the mechanoenzymatic properties of a motor in vitro and in vivo. The awareness of multifaceted modes of myosin regulation drastically expands the knowledge of how myosin motors are functioning within a cell and how they are integrated in complex signaling networks. At present, no comprehensive information is available on how much the levels of distinct myosin motors are spatially and temporally distributed to cellular subcompartments and tissues, their regulatory state and how they fluctuate over time in living cells. Is there a correlation between myosin dysregulation and pathogenic events? Exploring these emerging challenges with the innovative integration of experimental and computational approaches will relate myosin’s mechanoenzymology and interactome to its cellular function and address the effect of myosin regulation on the systems-level. Future research will focus on posttranslational regulation signatures of myosins to address the ‘who’, ‘what’, ‘when’, ‘where’, and ‘how’ does it work.
To summarize how multifaceted the regulation of a given myosin can be, we will revisit the global regulation of nonmuscle myosin-2b. The primary regulation of this myosin is via phosphorylation of its RLC. In the absence of phosphorylation of nonmuscle myosin-2b is inactive and adopts a monomeric conformation where the two heads make an asymmetric interaction and where the tail folds in hairpins at two locations to give rise to a compact conformation lacking actin-activated ATPase activity and the ability to form filaments. Phosphorylation of Ser-19 on the RLC disrupts this conformation, allowing the myosin to assemble into bipolar filaments where the myosin motor domains can interact productively with actin that result in stimulation of the ATPase activity and translocation of actin filaments. Several kinases which are targets of different signal transduction pathways can affect this phosphorylation including Rho-associated protein kinase and MLCK, which is Ca2+-calmodulin dependent. The RLC can also be phosphorylated at Thr-18 which is further stimulatory and at Ser-1/2 and Thr-9 which are inhibitory to a certain extent. The activity of individual myosin-2b filaments can be modulated by co-polymerization with nonmuscle myosins-2a and -2c or with the pseudoenzyme myosin-18a. The monomer-filament equilibrium can further be shifted for example by heavy chain phosphorylation and binding partners. The nonmuscle myosin-2b heavy chain can be alternatively spliced at two locations in the motor domain which either slightly increase or significantly decreases motor activity depending on the location. The activity may also be modulated by the isoform of actin, the actin age, by the tropomyosin isoform bound to the actin or higher-order actin structures. All those regulatory mechanisms follow spatiotemporal patterns, suggesting that each nonmuscle myosin-2 filament is constantly exposed to different regulatory mechanism that fine-tune it mechanoenzymatic activity which in the context of a cell results in a coherent response.
Highlights.
Numerous intrinsic and extrinsic allosteric mechanisms regulate myosin activity
Regulatory properties are myosin-specific
Multiple regulatory mechanism can synergistically act on one myosin
Acknowledgments
We apologize in advance to all authors whose work was not cited due to length considerations of the manuscript. This work was funded by the Intramural Program of the National Heart, Lung, and Blood Institute.
Abbreviations
- CBD
Cargo-binding Domain
- CK2
Casein Kinase 2
- EC50
half maximal effective concentration
- ELC
Essential Light Chain
- GTD
Globular Tail Domain
- HMM
heavy meromyosin
- IC50
half maximal inhibitory concentration
- mRNP
messenger ribonucleoprotein complex
- NHT
non-helical tailpiece
- PKC
protein kinase C
- PtdIns
Phosphatidylinsositol
- RLC
Regulatory Light Chain
- S
Svedberg
- S1
Myosin subfragment-1
- TEDS
Thr-Glu-Asp-Ser-site
Glossary
- Allostery
The communication of distinct sites in a protein that results in a functional change of its activity.
- Duty ratio
The fraction of time during the ATPase cycle a myosin spends strongly bound to actin.
- Processivity
The ability of a myosin to undergo subsequent steps on actin without detaching.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 2.Myosins. Springer Netherlands: [Google Scholar]
- 3.Hartman MA, Spudich JA. The myosin superfamily at a glance. J Cell Sci. 2012;125:1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heissler SM, Sellers JR. Myosins. In: Bradshaw RA, Stahl PD, editors. Encyclopedia of Cell Biology. Academic Press; 2016. [Google Scholar]
- 5.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 6.Sellers JR. Myosins. 2. Oxford University Press; 1999. [Google Scholar]
- 7.Ito K, Ikebe M, Kashiyama T, Mogami T, Kon T, Yamamoto K. Kinetic mechanism of the fastest motor protein, Chara myosin. J Biol Chem. 2007;282:19534–19545. doi: 10.1074/jbc.M611802200. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, White HD, Li XD. Drosophila myosin-XX functions as an actin-binding protein to facilitate the interaction between Zyx102 and actin. Biochemistry. 2014;53:350–360. doi: 10.1021/bi401236c. [DOI] [PubMed] [Google Scholar]
- 10.Guzik-Lendrum S, Heissler SM, Billington N, Takagi Y, Yang Y, Knight PJ, et al. Mammalian myosin-18A, a highly divergent myosin. J Biol Chem. 2013;288:9532–9548. doi: 10.1074/jbc.M112.441238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heissler SM, Sellers JR. Myosin Light Chains: Teaching Old Dogs New Tricks. Bioarchitecture. 2014;4:169–188. doi: 10.1080/19490992.2015.1054092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q, Li J, Zhang M. Cargo recognition and cargo-mediated regulation of unconventional myosins. Acc Chem Res. 2014;47:3061–3070. doi: 10.1021/ar500216z. [DOI] [PubMed] [Google Scholar]
- 14.Bloemink MJ, Geeves MA. Shaking the myosin family tree: Biochemical kinetics defines four types of myosin motor. Semin Cell Dev Biol. 2011;22:961–967. doi: 10.1016/j.semcdb.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 16.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 17.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Sellers JR. Mechanism of the phosphorylation-dependent regulation of smooth muscle heavy meromyosin. J Biol Chem. 1985;260:15815–15819. [PubMed] [Google Scholar]
- 19.Fujita-Becker S, Durrwang U, Erent M, Clark RJ, Geeves MA, Manstein DJ. Changes in Mg2+ ion concentration and heavy chain phosphorylation regulate the motor activity of a class I myosin. J Biol Chem. 2005;280:6064–6071. doi: 10.1074/jbc.M412473200. [DOI] [PubMed] [Google Scholar]
- 20.Heissler SM, Manstein DJ. Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J Biol Chem. 2011;286:21191–21202. doi: 10.1074/jbc.M110.212290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heissler SM, Manstein DJ. Functional characterization of the human myosin-7a motor domain. Cell Mol Life Sci. 2012;69:299–311. doi: 10.1007/s00018-011-0749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heissler SM, Selvadurai J, Bond LM, Fedorov R, Kendrick-Jones J, Buss F, et al. Kinetic properties and small-molecule inhibition of human myosin-6. FEBS Lett. 2012;586:3208–3214. doi: 10.1016/j.febslet.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenfeld SS, Houdusse A, Sweeney HL. Magnesium regulates ADP dissociation from myosin V. J Biol Chem. 2005;280:6072–6079. doi: 10.1074/jbc.M412717200. [DOI] [PubMed] [Google Scholar]
- 24.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 25.Heissler SM, Manstein DJ. Nonmuscle myosin-2: mix and match. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholey JM, Taylor KA, Kendrick-Jones J. Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature. 1980;287:233–235. doi: 10.1038/287233a0. [DOI] [PubMed] [Google Scholar]
- 27.Sellers JR, Pato MD, Adelstein RS. Reversible phosphorylation of smooth muscle myosin, heavy meromyosin, and platelet myosin. J Biol Chem. 1981;256:13137–13142. [PubMed] [Google Scholar]
- 28.Trybus KM. Filamentous smooth muscle myosin is regulated by phosphorylation. The Journal of cell biology. 1989;109:2887–2894. doi: 10.1083/jcb.109.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umemoto S, Bengur AR, Sellers JR. Effect of multiple phosphorylations of smooth muscle and cytoplasmic myosins on movement in an in vitro motility assay. J Biol Chem. 1989;264:1431–1436. [PubMed] [Google Scholar]
- 30.Ikebe M, Hartshorne DJ, Elzinga M. Identification, phosphorylation, and dephosphorylation of a second site for myosin light chain kinase on the 20,000-dalton light chain of smooth muscle myosin. J Biol Chem. 1986;261:36–39. [PubMed] [Google Scholar]
- 31.Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishikawa M, Sellers JR, Adelstein RS, Hidaka H. Protein kinase C modulates in vitro phosphorylation of the smooth muscle heavy meromyosin by myosin light chain kinase. J Biol Chem. 1984;259:8808–8814. [PubMed] [Google Scholar]
- 33.Beach JR, Licate LS, Crish JF, Egelhoff TT. Analysis of the role of Ser1/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 2011;12:52. doi: 10.1186/1471-2121-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, et al. Mesenchymal chemotaxis requires selective inactivation of myosin II at the leading edge via a noncanonical PLCgamma/PKCalpha pathway. Dev Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrow BD, Chen P, Chisholm RL. Expression of a myosin regulatory light chain phosphorylation site mutant complements the cytokinesis and developmental defects of Dictyostelium RMLC null cells. The Journal of cell biology. 1994;127:1945–1955. doi: 10.1083/jcb.127.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffith LM, Downs SM, Spudich JA. Myosin light chain kinase and myosin light chain phosphatase from Dictyostelium: effects of reversible phosphorylation on myosin structure and function. The Journal of cell biology. 1987;104:1309–1323. doi: 10.1083/jcb.104.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamm KE, Stull JT. Signaling to myosin regulatory light chain in sarcomeres. J Biol Chem. 2011;286:9941–9947. doi: 10.1074/jbc.R110.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toepfer C, Caorsi V, Kampourakis T, Sikkel MB, West TG, Leung MC, et al. Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. J Biol Chem. 2013;288:13446–13454. doi: 10.1074/jbc.M113.455444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karabina A, Kazmierczak K, Szczesna-Cordary D, Moore JR. Myosin regulatory light chain phosphorylation enhances cardiac beta-myosin in vitro motility under load. Arch Biochem Biophys. 2015;580:14–21. doi: 10.1016/j.abb.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine RJ, Kensler RW, Yang Z, Stull JT, Sweeney HL. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine RJ, Kensler RW, Yang Z, Sweeney HL. Myosin regulatory light chain phosphorylation and the production of functionally significant changes in myosin head arrangement on striated muscle thick filaments. Biophys J. 1995;68:224S. [PMC free article] [PubMed] [Google Scholar]
- 42.Craig R, Padron R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellers JR. Phosphorylation-dependent regulation of Limulus myosin. J Biol Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- 44.Liu X, Hong MS, Shu S, Yu S, Korn ED. Regulation of the filament structure and assembly of Acanthamoeba myosin II by phosphorylation of serines in the heavy-chain nonhelical tailpiece. Proc Natl Acad Sci U S A. 2013;110:E33–E40. doi: 10.1073/pnas.1219727110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang W, Warrick HM, Spudich JA. A structural model for phosphorylation control of Dictyostelium myosin II thick filament assembly. The Journal of cell biology. 1999;147:1039–1048. doi: 10.1083/jcb.147.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 47.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17:2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dulyaninova NG, Bresnick AR. The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins JH, Cote GP, Korn ED. Localization of the three phosphorylation sites on each heavy chain of Acanthamoeba myosin II to a segment at the end of the tail. J Biol Chem. 1982;257:4529–4534. [PubMed] [Google Scholar]
- 50.Collins JH, Kuznicki J, Bowers B, Korn ED. Comparison of the actin binding and filament formation properties of phosphorylated and dephosphorylated Acanthamoeba myosin II. Biochemistry. 1982;21:6910–6915. doi: 10.1021/bi00269a045. [DOI] [PubMed] [Google Scholar]
- 51.Kuczmarski ER, Spudich JA. Regulation of myosin self-assembly: phosphorylation of Dictyostelium heavy chain inhibits formation of thick filaments. Proc Natl Acad Sci U S A. 1980;77:7292–7296. doi: 10.1073/pnas.77.12.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahmsdorf HJ, Malchow D, Gerisch G. Cyclic AMP-induced phosphorylation in Dictyostelium of a polypeptide comigrating with myosin heavy chains. FEBS Lett. 1978;88:322–326. doi: 10.1016/0014-5793(78)80203-8. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Lee DY, Cai S, Yu S, Shu S, Levine RL, et al. Regulation of the actin-activated MgATPase activity of Acanthamoeba myosin II by phosphorylation of serine 639 in motor domain loop 2. Proc Natl Acad Sci U S A. 2013;110:E23–E32. doi: 10.1073/pnas.1219713110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heissler SM, Liu X, Korn ED, Sellers JR. Kinetic Characterization of the ATPase and Actin-activated ATPase Activities of Acanthamoeba castellanii Myosin-2. J Biol Chem. 2013;288:26709–26720. doi: 10.1074/jbc.M113.485946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 56.Pasternak C, Flicker PF, Ravid S, Spudich JA. Intermolecular versus intramolecular interactions of Dictyostelium myosin: possible regulation by heavy chain phosphorylation. J Cell Biol. 1989;109:203–210. doi: 10.1083/jcb.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su Z, Kiehart DP. Protein kinase C phosphorylates nonmuscle myosin-II heavy chain from Drosophila but regulation of myosin function by this enzyme is not required for viability in flies. Biochemistry. 2001;40:3606–3614. doi: 10.1021/bi010082j. [DOI] [PubMed] [Google Scholar]
- 58.Conti MA, Sellers JR, Adelstein RS, Elzinga M. Identification of the serine residue phosphorylated by protein kinase C in vertebrate nonmuscle myosin heavy chains. Biochemistry. 1991;30:966–970. doi: 10.1021/bi00218a012. [DOI] [PubMed] [Google Scholar]
- 59.Murakami N, Chauhan VP, Elzinga M. Two nonmuscle myosin II heavy chain isoforms expressed in rabbit brains: filament forming properties, the effects of phosphorylation by protein kinase C and casein kinase II, and location of the phosphorylation sites. Biochemistry. 1998;37:1989–2003. doi: 10.1021/bi971959a. [DOI] [PubMed] [Google Scholar]
- 60.Murakami N, Kotula L, Hwang YW. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry. 2000;39:11441–11451. doi: 10.1021/bi000347e. [DOI] [PubMed] [Google Scholar]
- 61.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasapera AM, Plotnikov SV, Fischer RS, Case LB, Egelhoff TT, Waterman CM. Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr Biol. 2015;25:175–186. doi: 10.1016/j.cub.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bement WM, Mooseker MS. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- 64.Novak KD, Titus MA. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol Biol Cell. 1998;9:75–88. doi: 10.1091/mbc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durrwang U, Fujita-Becker S, Erent M, Kull FJ, Tsiavaliaris G, Geeves MA, et al. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci. 2006;119:550–558. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- 66.Brzeska H, Knaus UG, Wang ZY, Bokoch GM, Korn ED. p21-activated kinase has substrate specificity similar to Acanthamoeba myosin I heavy chain kinase and activates Acanthamoeba myosin I. Proc Natl Acad Sci U S A. 1997;94:1092–1095. doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brzeska H, Lynch TJ, Martin B, Korn ED. The localization and sequence of the phosphorylation sites of Acanthamoeba myosins I. An improved method for locating the phosphorylated amino acid. J Biol Chem. 1989;264:19340–19348. [PubMed] [Google Scholar]
- 68.Albanesi JP, Hammer JA, 3rd, Korn ED. The interaction of F-actin with phosphorylated and unphosphorylated myosins IA and IB from Acanthamoeba castellanii. J Biol Chem. 1983;258:10176–10181. [PubMed] [Google Scholar]
- 69.Attanapola SL, Alexander CJ, Mulvihill DP. Ste20-kinase-dependent TEDS-site phosphorylation modulates the dynamic localisation and endocytic function of the fission yeast class I myosin, Myo1. J Cell Sci. 2009;122:3856–3861. doi: 10.1242/jcs.053959. [DOI] [PubMed] [Google Scholar]
- 70.Ostap EM, Lin T, Rosenfeld SS, Tang N. Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation. Biochemistry. 2002;41:12450–12456. doi: 10.1021/bi0262193. [DOI] [PubMed] [Google Scholar]
- 71.Yoshimura M, Homma K, Saito J, Inoue A, Ikebe R, Ikebe M. Dual regulation of mammalian myosin VI motor function. J Biol Chem. 2001;276:39600–39607. doi: 10.1074/jbc.M105080200. [DOI] [PubMed] [Google Scholar]
- 72.De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 73.Buss F, Kendrick-Jones J, Lionne C, Knight AE, Cote GP, Paul Luzio J. The localization of myosin VI at the golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naccache SN, Hasson T. Myosin VI altered at threonine 406 stabilizes actin filaments in vivo. Cell Motil Cytoskeleton. 2006;63:633–645. doi: 10.1002/cm.20150. [DOI] [PubMed] [Google Scholar]
- 75.Tarrant MK, Cole PA. The chemical biology of protein phosphorylation. Annu Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang F, Harvey EV, Conti MA, Wei D, Sellers JR. A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- 77.Behrmann E, Muller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kollmar M, Durrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. Embo J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kempler K, Toth J, Yamashita R, Mapel G, Robinson K, Cardasis H, et al. Loop 2 of limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry. 2007;46:4280–4293. doi: 10.1021/bi062112u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dalal JS, Stevens SM, Jr, Alvarez S, Munoz N, Kempler KE, Dose AC, et al. Mouse class III myosins: kinase activity and phosphorylation sites. J Neurochem. 2011;119:772–784. doi: 10.1111/j.1471-4159.2011.07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dose AC, Ananthanarayanan S, Moore JE, Corsa AC, Burnside B, Yengo CM. The kinase domain alters the kinetic properties of the myosin IIIA motor. Biochemistry. 2008;47:2485–2496. doi: 10.1021/bi7021574. [DOI] [PubMed] [Google Scholar]
- 82.Quintero OA, Moore JE, Unrath WC, Manor U, Salles FT, Grati M, et al. Intermolecular autophosphorylation regulates myosin IIIa activity and localization in parallel actin bundles. J Biol Chem. 2010;285:35770–35782. doi: 10.1074/jbc.M110.144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trybus KM, Huiatt TW, Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci U S A. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Umeki N, Jung HS, Sakai T, Sato O, Ikebe R, Ikebe M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18:783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 85.Wang F, Thirumurugan K, Stafford WF, Hammer JA, 3rd, Knight PJ, Sellers JR. Regulated conformation of myosin V. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 86.Yang Y, Kovacs M, Sakamoto T, Zhang F, Kiehart DP, Sellers JR. Dimerized Drosophila myosin VIIa: a processive motor. Proc Natl Acad Sci U S A. 2006;103:5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 88.Beach JR, Hammer JA., 3rd Myosin II isoform co-assembly and differential regulation in mammalian systems. Exp Cell Res. 2015;334:2–9. doi: 10.1016/j.yexcr.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung HS, Billington N, Thirumurugan K, Salzameda B, Cremo CR, Chalovich JM, et al. Role of the tail in the regulated state of myosin 2. J Mol Biol. 2011;408:863–878. doi: 10.1016/j.jmb.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. The Journal of cell biology. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cross RA, Jackson AP, Citi S, Kendrick-Jones J, Bagshaw CR. Active site trapping of nucleotide by smooth and non-muscle myosins. J Mol Biol. 1988;203:173–181. doi: 10.1016/0022-2836(88)90100-3. [DOI] [PubMed] [Google Scholar]
- 92.Cremo CR, Sellers JR, Facemyer KC. Two heads are required for phosphorylation-dependent regulation of smooth muscle myosin. J Biol Chem. 1995;270:2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- 93.Sweeney HL, Chen LQ, Trybus KM. Regulation of asymmetric smooth muscle myosin II molecules. J Biol Chem. 2000;275:41273–41277. doi: 10.1074/jbc.M008310200. [DOI] [PubMed] [Google Scholar]
- 94.Milton DL, Schneck AN, Ziech DA, Ba M, Facemyer KC, Halayko AJ, et al. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc Natl Acad Sci U S A. 2011;108:1421–1426. doi: 10.1073/pnas.1011784108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woodhead JL, Zhao FQ, Craig R. Structural basis of the relaxed state of a Ca2+-regulated myosin filament and its evolutionary implications. Proc Natl Acad Sci U S A. 2013;110:8561–8566. doi: 10.1073/pnas.1218462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao FQ, Craig R, Woodhead JL. Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J Mol Biol. 2009;385:423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, et al. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA., 3rd Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Billington N, Beach JR, Heissler SM, Remmert K, Guzik-Lendrum S, Nagy A, et al. Myosin 18A coassembles with nonmuscle myosin 2 to form mixed bipolar filaments. Curr Biol. 2015;25:942–948. doi: 10.1016/j.cub.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shutova MS, Spessott WA, Giraudo CG, Svitkina T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 102.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. The Journal of cell biology. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thirumurugan K, Sakamoto T, Hammer JA, 3rd, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 105.Sato O, Li XD, Ikebe M. Myosin Va becomes a low duty ratio motor in the inhibited form. J Biol Chem. 2007;282:13228–13239. doi: 10.1074/jbc.M610766200. [DOI] [PubMed] [Google Scholar]
- 106.Sckolnick M, Krementsova EB, Warshaw DM, Trybus KM. More Than Just a Cargo Adapter, Melanophilin Prolongs and Slows Processive Runs of Myosin Va. J Biol Chem. 2013;288:29313–29322. doi: 10.1074/jbc.M113.476929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li XD, Ikebe R, Ikebe M. Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J Biol Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 108.Umeki N, Jung HS, Watanabe S, Sakai T, Li XD, Ikebe R, et al. The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc Natl Acad Sci U S A. 2009;106:8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang Y, Baboolal TG, Siththanandan V, Chen M, Walker ML, Knight PJ, et al. A FERM domain autoregulates Drosophila myosin 7a activity. Proc Natl Acad Sci U S A. 2009;106:4189–4194. doi: 10.1073/pnas.0808682106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 111.Grabarek Z. Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. Biochim Biophys Acta. 2011;1813:913–921. doi: 10.1016/j.bbamcr.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De La Roche MA, Lee SF, Cote GP. The Dictyostelium class I myosin, MyoD, contains a novel light chain that lacks high-affinity calcium-binding sites. Biochem J. 2003;374:697–705. doi: 10.1042/BJ20030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bahler M, Kroschewski R, Stoffler HE, Behrmann T. Rat myr 4 defines a novel subclass of myosin I: identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity. J Cell Biol. 1994;126:375–389. doi: 10.1083/jcb.126.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Williams R, Coluccio LM. Novel 130-kDa rat liver myosin-1 will translocate actin filaments. Cell Motil Cytoskeleton. 1994;27:41–48. doi: 10.1002/cm.970270105. [DOI] [PubMed] [Google Scholar]
- 115.Houdusse A, Silver M, Cohen C. A model of Ca(2+)-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure. 1996;4:1475–1490. doi: 10.1016/s0969-2126(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 116.Collins K, Sellers JR, Matsudaira P. Calmodulin dissociation regulates brush border myosin I (110-kD-calmodulin) mechanochemical activity in vitro. The Journal of cell biology. 1990;110:1137–1147. doi: 10.1083/jcb.110.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adamek N, Coluccio LM, Geeves MA. Calcium sensitivity of the cross-bridge cycle of Myo1c, the adaptation motor in the inner ear. Proc Natl Acad Sci U S A. 2008;105:5710–5715. doi: 10.1073/pnas.0710520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Coluccio LM, Geeves MA. Transient kinetic analysis of the 130-kDa myosin I (MYR-1 gene product) from rat liver. A myosin I designed for maintenance of tension? J Biol Chem. 1999;274:21575–21580. doi: 10.1074/jbc.274.31.21575. [DOI] [PubMed] [Google Scholar]
- 119.Manceva S, Lin T, Pham H, Lewis JH, Goldman YE, Ostap EM. Calcium regulation of calmodulin binding to and dissociation from the myo1c regulatory domain. Biochemistry. 2007;46:11718–11726. doi: 10.1021/bi700894h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wolenski JS, Hayden SM, Forscher P, Mooseker MS. Calcium-calmodulin and regulation of brush border myosin-I MgATPase and mechanochemistry. The Journal of cell biology. 1993;122:613–621. doi: 10.1083/jcb.122.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu T, Beckingham K, Ikebe M. High affinity Ca2+ binding sites of calmodulin are critical for the regulation of myosin Ibeta motor function. J Biol Chem. 1998;273:20481–20486. doi: 10.1074/jbc.273.32.20481. [DOI] [PubMed] [Google Scholar]
- 122.Lin T, Tang N, Ostap EM. Biochemical and motile properties of Myo1b splice isoforms. J Biol Chem. 2005;280:41562–41567. doi: 10.1074/jbc.M508653200. [DOI] [PubMed] [Google Scholar]
- 123.Wolenski JS, Hayden SM, Forscher P, Mooseker MS. Calcium-calmodulin and regulation of brush border myosin-I MgATPase and mechanochemistry. J Cell Biol. 1993;122:613–621. doi: 10.1083/jcb.122.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends in cell biology. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li XD, Jung HS, Wang Q, Ikebe R, Craig R, Ikebe M. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc Natl Acad Sci U S A. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 127.Trybus KM, Gushchin MI, Lui H, Hazelwood L, Krementsova EB, Volkmann N, et al. Effect of calcium on calmodulin bound to the IQ motifs of myosin V. J Biol Chem. 2007;282:23316–23325. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- 128.Nguyen H, Higuchi H. Motility of myosin V regulated by the dissociation of single calmodulin. Nat Struct Mol Biol. 2005;12:127–132. doi: 10.1038/nsmb894. [DOI] [PubMed] [Google Scholar]
- 129.Xie X, Harrison DH, Schlichting I, Sweet RM, Kalabokis VN, Szent-Gyorgyi AG, et al. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature. 1994;368:306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]
- 130.Houdusse A, Cohen C. Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation. Structure. 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- 131.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 132.Geeves MA. Thin Filament Regulation. In: Goldman YE, Ostap EM, editors. Comprehensive Biophysics. Amsterdam: Elsevier; 2012. pp. 251–268. [Google Scholar]
- 133.Bagshaw CR, Reed GH. The significance of the slow dissociation of divalent metal ions from myosin 'regulatory' light chains. FEBS Lett. 1977;81:386–390. doi: 10.1016/0014-5793(77)80560-7. [DOI] [PubMed] [Google Scholar]
- 134.Elting MW, Bryant Z, Liao JC, Spudich JA. Detailed tuning of structure and intramolecular communication are dispensable for processive motion of myosin VI. Biophys J. 2011;100:430–439. doi: 10.1016/j.bpj.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsiavaliaris G, Fujita-Becker S, Durrwang U, Diensthuber RP, Geeves MA, Manstein DJ. Mechanism, regulation, and functional properties of Dictyostelium myosin-1B. J Biol Chem. 2008;283:4520–4527. doi: 10.1074/jbc.M708113200. [DOI] [PubMed] [Google Scholar]
- 136.Heissler SM, Chinthalapudi K, Sellers JR. Kinetic characterization of the sole nonmuscle myosin-2 from the model organism Drosophila melanogaster. FASEB J. 2015;29:1456–1466. doi: 10.1096/fj.14-266742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muller M, Diensthuber RP, Chizhov I, Claus P, Heissler SM, Preller M, et al. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS One. 2013;8:e70636. doi: 10.1371/journal.pone.0070636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zimmermann D, Santos A, Kovar DR, Rock RS. Actin age orchestrates myosin-5 and myosin-6 run lengths. Curr Biol. 2015;25:2057–2062. doi: 10.1016/j.cub.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc Natl Acad Sci U S A. 2009;106:9685–9690. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hariadi RF, Sommese RF, Sivaramakrishnan S. Tuning myosin-driven sorting on cellular actin networks. Elife. 2015;4 doi: 10.7554/eLife.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, et al. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci U S A. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bao J, Huck D, Gunther LK, Sellers JR, Sakamoto T. Actin Structure-Dependent Stepping of Myosin 5a and 10 during Processive Movement. PLoS One. 2013;8:e74936. doi: 10.1371/journal.pone.0074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sun Y, Sato O, Ruhnow F, Arsenault ME, Ikebe M, Goldman YE. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shin JB, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, et al. Molecular architecture of the chick vestibular hair bundle. Nat Neurosci. 2013;16:365–374. doi: 10.1038/nn.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 147.Gordon AM, Regnier M, Homsher E. Skeletal and cardiac muscle contractile activation: tropomyosin"rocks and rolls". News Physiol Sci. 2001;16:49–55. [PubMed] [Google Scholar]
- 148.Sobue K, Sellers JR. Caldesmon, a novel regulatory protein in smooth muscle and nonmuscle actomyosin systems. J Biol Chem. 1991;266:12115–12118. [PubMed] [Google Scholar]
- 149.Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochemistry. 2014;53:4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chacko S, Eisenberg E. Cooperativity of actin-activated ATPase of gizzard heavy meromyosin in the presence of gizzard tropomyosin. J Biol Chem. 1990;265:2105–2110. [PubMed] [Google Scholar]
- 151.Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE, Trybus KM. Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Current biology : CB. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol. 2013;20:952–957. doi: 10.1038/nsmb.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Current biology : CB. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 154.Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- 155.Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Current biology : CB. 2010;20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 156.Lieto-Trivedi A, Dash S, Coluccio LM. Myosin surface loop 4 modulates inhibition of actomyosin 1b ATPase activity by tropomyosin. Biochemistry. 2007;46:2779–2786. doi: 10.1021/bi602439f. [DOI] [PubMed] [Google Scholar]
- 157.Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proc Natl Acad Sci U S A. 2009;106:11972–11977. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brzeska H, Hwang KJ, Korn ED. Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J Biol Chem. 2008;283:32014–32023. doi: 10.1074/jbc.M804828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci U S A. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pyrpassopoulos S, Feeser EA, Mazerik JN, Tyska MJ, Ostap EM. Membrane-bound myo1c powers asymmetric motility of actin filaments. Current biology : CB. 2012;22:1688–1692. doi: 10.1016/j.cub.2012.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu C, Lou J, Wu J, Pan L, Feng W, Zhang M. Membrane-induced lever arm expansion allows myosin VI to walk with large and variable step sizes. J Biol Chem. 2012;287:35021–35035. doi: 10.1074/jbc.M111.328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mukherjea M, Llinas P, Kim H, Travaglia M, Safer D, Menetrey J, et al. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yu C, Feng W, Wei Z, Miyanoiri Y, Wen W, Zhao Y, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 165.Spudich G, Chibalina MV, Au JS, Arden SD, Buss F, Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, et al. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 167.Plantard L, Arjonen A, Lock JG, Nurani G, Ivaska J, Stromblad S. PtdIns(3,4,5)P(3) is a regulator of myosin-X localization and filopodia formation. J Cell Sci. 2010;123:3525–3534. doi: 10.1242/jcs.069609. [DOI] [PubMed] [Google Scholar]
- 168.Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, et al. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Current biology : CB. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 170.Chang W, Zaarour RF, Reck-Peterson S, Rinn J, Singer RH, Snyder M, et al. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid-protein complex that contains mRNAs and subunits of the RNA-processing body. Rna. 2008;14:491–502. doi: 10.1261/rna.665008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Salles FT, Merritt RC, Jr, Manor U, Dougherty GW, Sousa AD, Moore JE, et al. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat Cell Biol. 2009;11:443–450. doi: 10.1038/ncb1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wagner W, Brenowitz SD, Hammer JA., 3rd Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol. 2011;13:40–48. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu X, Rao K, Bowers MB, Copeland NG, Jenkins NA, Hammer JA., 3rd Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J Cell Sci. 2001;114:1091–1100. doi: 10.1242/jcs.114.6.1091. [DOI] [PubMed] [Google Scholar]
- 174.Lindsay AJ, Jollivet F, Horgan CP, Khan AR, Raposo G, McCaffrey MW, et al. Identification and characterization of multiple novel Rab-myosin Va interactions. Mol Biol Cell. 2013;24:3420–3434. doi: 10.1091/mbc.E13-05-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang Z, Edwards JG, Riley N, Provance DW, Jr, Karcher R, Li XD, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fukuda M, Kuroda TS. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 177.Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 178.Karcher RL, Roland JT, Zappacosta F, Huddleston MJ, Annan RS, Carr SA, et al. Cell cycle regulation of myosin-V by calcium/calmodulin-dependent protein kinase II. Science. 2001;293:1317–1320. doi: 10.1126/science.1061086. [DOI] [PubMed] [Google Scholar]
- 179.Donovan KW, Bretscher A. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Dev Cell. 2012;23:769–781. doi: 10.1016/j.devcel.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yau RG, Peng Y, Valiathan RR, Birkeland SR, Wilson TE, Weisman LS. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev Cell. 2014;28:520–533. doi: 10.1016/j.devcel.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Phichith D, Travaglia M, Yang Z, Liu X, Zong AB, Safer D, et al. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci U S A. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hertzano R, Shalit E, Rzadzinska AK, Dror AA, Song L, Ron U, et al. A Myo6 mutation destroys coordination between the myosin heads, revealing new functions of myosin VI in the stereocilia of mammalian inner ear hair cells. PLoS Genet. 2008;4:e1000207. doi: 10.1371/journal.pgen.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chuan P, Spudich JA, Dunn AR. Robust mechanosensing and tension generation by myosin VI. J Mol Biol. 2011;405:105–112. doi: 10.1016/j.jmb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sakai T, Umeki N, Ikebe R, Ikebe M. Cargo binding activates myosin VIIA motor function in cells. Proc Natl Acad Sci U S A. 2011;108:7028–7033. doi: 10.1073/pnas.1009188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013 doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ford HL, Silver DL, Kachar B, Sellers JR, Zain SB. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–16327. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- 187.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 188.Kiss B, Duelli A, Radnai L, Kekesi KA, Katona G, Nyitray L. Crystal structure of the S100A4-nonmuscle myosin IIA tail fragment complex reveals an asymmetric target binding mechanism. Proc Natl Acad Sci U S A. 2012;109:6048–6053. doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kingsbury NL, Renegar RH, Chalovich JM. Avian Synaptopodin 2 (Fesselin) Stabilizes Myosin Filaments and Actomyosin in the Presence of ATP. Biochemistry. 2013 doi: 10.1021/bi401013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schroeter MM, Chalovich JM. Fesselin binds to actin and myosin and inhibits actin-activated ATPase activity. J Muscle Res Cell Motil. 2005;26:183–189. doi: 10.1007/s10974-005-9009-6. [DOI] [PubMed] [Google Scholar]
- 191.Merritt RC, Manor U, Salles FT, Grati M, Dose AC, Unrath WC, et al. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Current biology : CB. 2012;22:320–325. doi: 10.1016/j.cub.2011.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Goodno CC. Inhibition of myosin ATPase by vanadate ion. Proc Natl Acad Sci U S A. 1979;76:2620–2624. doi: 10.1073/pnas.76.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Takacs B, Billington N, Gyimesi M, Kintses B, Malnasi-Csizmadia A, Knight PJ, et al. Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke. Proc Natl Acad Sci U S A. 2010;107:6799–6804. doi: 10.1073/pnas.0907585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Higuchi H, Takemori S. Butanedione monoxime suppresses contraction and ATPase activity of rabbit skeletal muscle. J Biochem. 1989;105:638–643. doi: 10.1093/oxfordjournals.jbchem.a122717. [DOI] [PubMed] [Google Scholar]
- 195.Cheung A, Dantzig JA, Hollingworth S, Baylor SM, Goldman YE, Mitchison TJ, et al. A small-molecule inhibitor of skeletal muscle myosin II. Nat Cell Biol. 2002;4:83–88. doi: 10.1038/ncb734. [DOI] [PubMed] [Google Scholar]
- 196.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 197.Ostap EM. 2,3-Butanedione monoxime (BDM) as a myosin inhibitor. J Muscle Res Cell Motil. 2002;23:305–308. doi: 10.1023/a:1022047102064. [DOI] [PubMed] [Google Scholar]
- 198.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 199.Limouze J, Straight AF, Mitchison T, Sellers JR. Specificity of blebbistatin, an inhibitor of myosin II. J Muscle Res Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 200.Ramamurthy B, Yengo CM, Straight AF, Mitchison TJ, Sweeney HL. Kinetic mechanism of blebbistatin inhibition of nonmuscle myosin IIb. Biochemistry. 2004;43:14832–14839. doi: 10.1021/bi0490284. [DOI] [PubMed] [Google Scholar]
- 201.Allingham JS, Smith R, Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat Struct Mol Biol. 2005;12:378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- 202.Sakamoto T, Limouze J, Combs CA, Straight AF, Sellers JR. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- 203.Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 204.Kepiro M, Varkuti BH, Vegner L, Voros G, Hegyi G, Varga M, et al. para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor. Angew Chem Int Ed Engl. 2014;53:8211–8215. doi: 10.1002/anie.201403540. [DOI] [PubMed] [Google Scholar]
- 205.Kepiro M, Varkuti BH, Bodor A, Hegyi G, Drahos L, Kovacs M, et al. Azidoblebbistatin, a photoreactive myosin inhibitor. Proc Natl Acad Sci U S A. 2012;109:9402–9407. doi: 10.1073/pnas.1202786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fedorov R, Bohl M, Tsiavaliaris G, Hartmann FK, Taft MH, Baruch P, et al. The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat Struct Mol Biol. 2009;16:80–88. doi: 10.1038/nsmb.1542. [DOI] [PubMed] [Google Scholar]
- 207.Islam K, Chin HF, Olivares AO, Saunders LP, De La Cruz EM, Kapoor TM. A myosin V inhibitor based on privileged chemical scaffolds. Angew Chem Int Ed Engl. 2010;49:8484–8488. doi: 10.1002/anie.201004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chinthalapudi K, Taft MH, Martin R, Heissler SM, Preller M, Hartmann FK, et al. Mechanism and specificity of pentachloropseudilin-mediated inhibition of myosin motor activity. J Biol Chem. 2011;286:29700–29708. doi: 10.1074/jbc.M111.239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Teerlink JR. A novel approach to improve cardiac performance: cardiac myosin activators. Heart Fail Rev. 2009;14:289–298. doi: 10.1007/s10741-009-9135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Winkelmann DA, Forgacs E, Miller MT, Stock AM. Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity. Nat Commun. 2015;6:7974. doi: 10.1038/ncomms8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Radke MB, Taft MH, Stapel B, Hilfiker-Kleiner D, Preller M, Manstein DJ. Small molecule-mediated refolding and activation of myosin motor function. Elife. 2014;3:e01603. doi: 10.7554/eLife.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]


