Abstract
The ability of cells to detect and repair DNA double-strand breaks (DSBs) is dependent on reorganization of the surrounding chromatin structure by chromatin remodeling complexes. These complexes promote access to the site of DNA damage, facilitate processing of the damaged DNA and, importantly, are essential to repackage the repaired DNA. Here, we will review the chromatin remodeling steps which occur immediately after DSB production and which prepare the damaged chromatin template for processing by the DSB repair machinery. DSBs promote rapid accumulation of repressive complexes, including HP1, the NuRD complex, H2A.Z and histone methyltransferases at the DSB. This shift to a repressive chromatin organization may be important to inhibit local transcription and limit mobility of the break, and to maintain the DNA ends in close contact. Subsequently, the repressive chromatin is rapidly dismantled through a mechanism involving dynamic exchange of the histone variant H2A.Z. H2A.Z removal at DSBs alters the acidic patch on the nucleosome surface, promoting acetylation of the H4 tail (by the NuA4-Tip60 complex) and shifting the chromatin to a more open structure. Further, H2A.Z removal promotes chromatin ubiquitination and recruitment of additional DSB repair proteins to the break. Modulation of the nucleosome surface and nucleosome function during DSB repair therefore plays a vital role in processing of DNA breaks. Further, the nucleosome surface may function as a central hub during DSB repair, directing specific patterns of histone modification, recruiting DNA repair proteins and modulating chromatin packing during processing of the damaged DNA template.
Keywords: DSB repair, NuA4-Tip60, H2A.Z, chromatin remodeling, DNA damage
Graphical abstract
Introduction
DNA is constantly exposed to genotoxic agents which can modify the sugar and base residues, create DNA adducts, cross-link the DNA strands or even cleave the phosphate backbone to create single or double-strand breaks (DSBs). To counter these potentially mutagenic events, mammalian cells possess multiple DNA repair pathways which detect and remove modified bases or process and religate DNA strand breaks. DNA repair pathways, referred to collectively as the DNA damage response1, function as a highly regulated signal transduction pathway which recruit specific DNA repair complexes to damaged DNA. Further, the DNA damage response is intimately linked with the activation of checkpoints at several points in the cell cycle. Checkpoint activation can temporarily block DNA replication and prevent further damage or mutagenesis which may arise when attempting replication on a damaged DNA template. In addition, the complexity of the chromatin architecture at the site of DNA damage also plays a critical role in regulating DNA repair. Chromatin organization was originally proposed to present a “barrier” to repair, so that remodeling was required to gain access to the site, followed by repair and restoration of the original chromatin structure (the “access-repair-restore” model2; 3). However, it is now clear that chromatin organization, chromatin remodeling complexes and histone modifications are active players in DNA repair4; 5; 6, and that dynamic changes in nucleosome organization are critical for efficient repair of DNA damage. In this review, we will focus on how nucleosome dynamics, coupled with changes in histone acetylation, work together to create open, flexible chromatin domains which are required for the processing and repair of DNA double-strand breaks (DSBs).
DSB repair – signaling events
DSBs are lethal events which, if unrepaired, lead to chromosomal loss, translocations, genome instability and eventually cancer. DSBs can arise from many events, including collapse of replication forks or exposure to free radicals or ionizing radiation (IR). Many anti-cancer therapies, including radiation therapy and chemotherapy, specifically kill cancer cells by creating DSBs. Further, many tumors contain mutations in key DSB repair proteins, such as brca1 or p53. Consequently, there is significant clinical effort devoted to unraveling the mechanism of DSB repair in normal and tumor cell lines, and developing inhibitors of DNA repair which can target defective DNA repair in tumor cells.
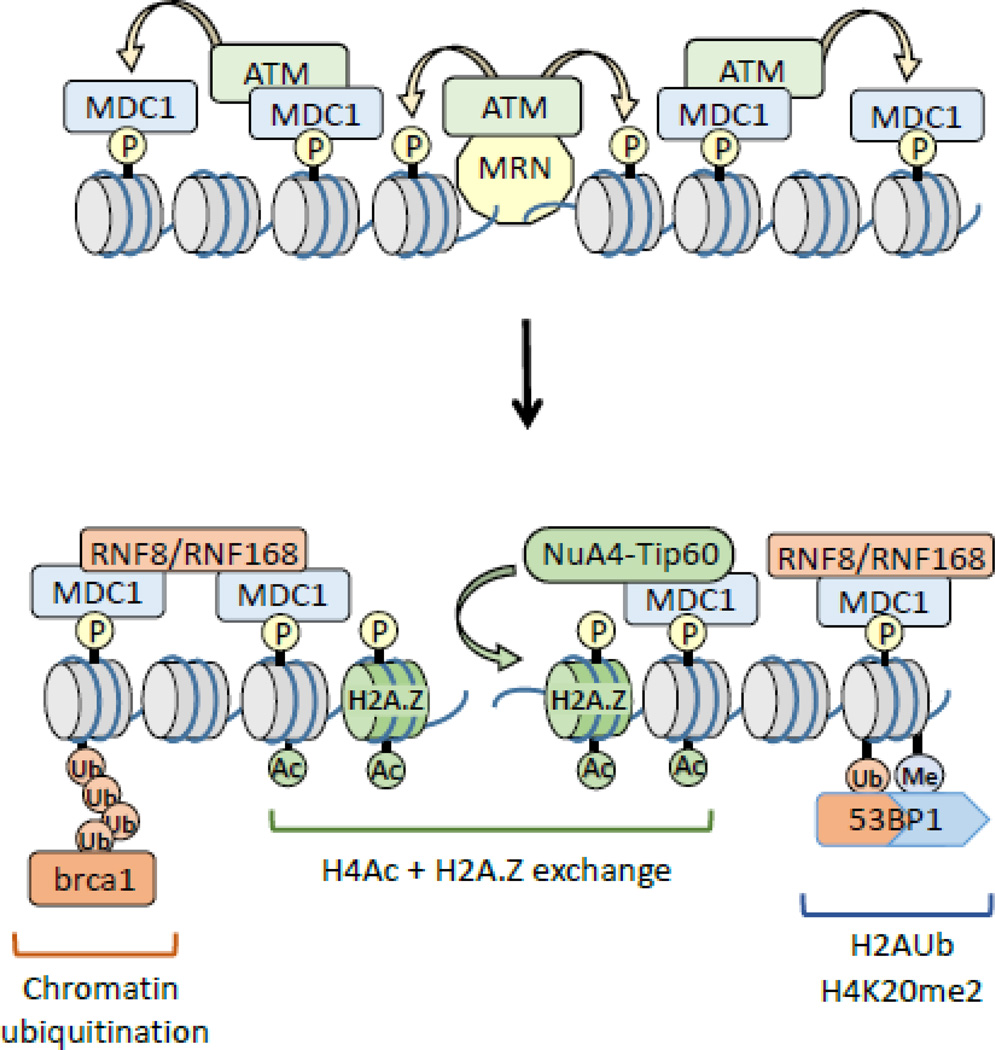
The basic mechanism by which cells detect and repair DSBs is well-defined (figure 1). The mre11-rad50-nbs1 (MRN) complex binds directly to the DNA ends at DSBs. The MRN complex combines exo- and endo-nuclease activity (mre11), DNA binding functions (mre11) and ATPase activity (rad50) within a single complex7. A key function of MRN is to recruit the ATM kinase to the break site, activating ATM’s kinase activity and promoting the ATM-dependent phosphorylation of proteins required for DSB repair, checkpoint activation and apoptotic responses to DNA damage8. In particular, ATM can phosphorylate the c-terminal of histone H2AX (termed γH2AX;9), creating a binding site for the BRCT domain of the mdc1 platform protein10. Mdc1 recruits activated ATM, facilitating H2AX phosphorylation by ATM further from the break (figure 1). This, in turn, recruits additional mdc1 and active ATM, leading to spreading of phosphorylated H2AX for 100s of kb from the break4; 8; 9; 11. Mdc1 also serves to recruit additional DSB repair proteins to the site of damage. These include the ubiquitin ligases RNF8 and RNF168 (figure 1), which promote chromatin ubiquitination and ubiquitin dependent loading of the brca1 repair complex11. 53BP1, a key regulator of DSB repair, is also recruited to DSBs through a dual interaction with ubiquitinated H2AK13/14Ub (created by RNF168) and H4K20me212. Finally, the NuA4-Tip60 remodeling complex4; 13 exchanges H2A.Z onto nucleosomes at the break and promotes acetylation of histone H4 (figure 1). Further, modulation of nucleosome dynamics by NuA4-Tip60 is important for H2A ubiquitination, H4 acetylation and the loading of brca1 and 53BP1 at the DSB, and will be discussed in more detail in later sections.
Figure 1. DSB repair and chromatin organization.
The MRN (mre11-rad50-nbs1) complex is recruited to DSBs. MRN recruits the ATM kinase, which then phosphorylates the c-terminal region of H2AX (γH2AX). Subsequent loading of the mdc1 protein promotes spreading of γH2AX along the chromatin from the DSB. The RNF8 and RNF168 ubiquitin ligase complexes promote ubiquitination of chromatin proteins, including H2A. The NuA4-Tip60 complex promotes exchange of H2A.Z onto nucleosomes at the break as well as facilitating acetylation of histone H4. These modifications facilitate recruitment of brca1 (via chromatin ubiquitination) and 53BP1 (via dual interaction with H2AK13/15Ub and H4K20me2). P = phosphorylation; Ac = acetylation; Ub = ubiquitination.
DSB repair – HR vs NHEJ
Repair of DSBs can occur through 2 distinct mechanisms. In nonhomologous end-joining (NHEJ), the Ku70/80 DNA binding complex and several scaffold proteins are recruited to the break. Damaged bases may be removed and the DSB religated by DNA ligase IV14. Because processing during NHEJ can create short insertions/deletions, it is considered a low fidelity repair mechanism. In contrast, homologous recombination (HR) exploits the presence of sister chromatids in late S-phase and G2, which can provide a template for DSB repair. HR requires production of ssDNA at the break, a process referred to as end-resection. Resection is initiated by the CtIP-MRN complex, which then allows the exo1 and dna2 nucleases to extend this to create 3’-ssDNA overhangs15; 16. The ssDNA, bound by the rad51 protein, is then used to locate homologous sequences on the sister chromatid which can serve as a template for repair. HR is therefore considered to be a high fidelity DNA repair mechanism, although HR may still potentially create mutations17. Importantly, while NHEJ can occur throughout the cell cycle, HR is largely restricted to S-phase and G2-M, when adjacent sister chromatids are present to provide the template for the HR machinery.
DSB repair – the need for remodeling
Processing of the DNA for repair by e.g. HR requires significant remodeling of both the damaged chromatin to allow production of ssDNA, as well as remodeling involved in homology search on the sister chromatid. In contrast, NHEJ relies on direct end-ligation of the damaged DNA and is therefore likely to involve less extensive remodeling. Nevertheless, the detection and repair of DSBs within the complex organization of the chromatin is critically dependent on chromatin remodeling to drive efficient DSB repair. Here, we will discuss the earliest events that occur during the initial detection and processing of the damaged chromatin template. In particular, we will focus on how members of the Ino80 family promote progressive changes in nucleosome packing and structure, leading to increased histone acetylation and ubiquitination and promoting the formation of open chromatin structures which facilitate DSB repair.
Nucleosome dynamics and DSB repair
DSB repair and acetylated chromatin
Early studies demonstrated that DNA damage led to a decrease in chromatin compaction during DNA repair18. DSBs increase both the sensitivity of the chromatin to nuclease digestion19; 20 and the salt solubility of histones13. Other studies, using microscopy approaches, indicate a rapid expansion of chromatin at sites of DNA damage21 and, importantly, that depletion of histone H1 limits the DNA damage response whereas HDAC inhibitors, which promote more open structures, facilitate repair22. Further, the presence of more open chromatin at DSBs was associated with an increase in histone acetylation, particularly of histone H423; 24; 25; 26; 27. Thus DSBs promote the formation of open, accessible acetylated chromatin structures at the site of damage. These open chromatin structures would then allow the DNA repair machinery to gain access to the damaged DNA and carry out repair.
Compaction precedes chromatin relaxation
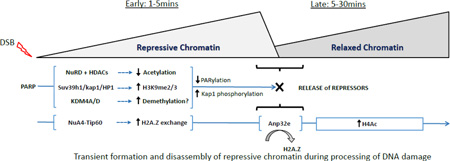
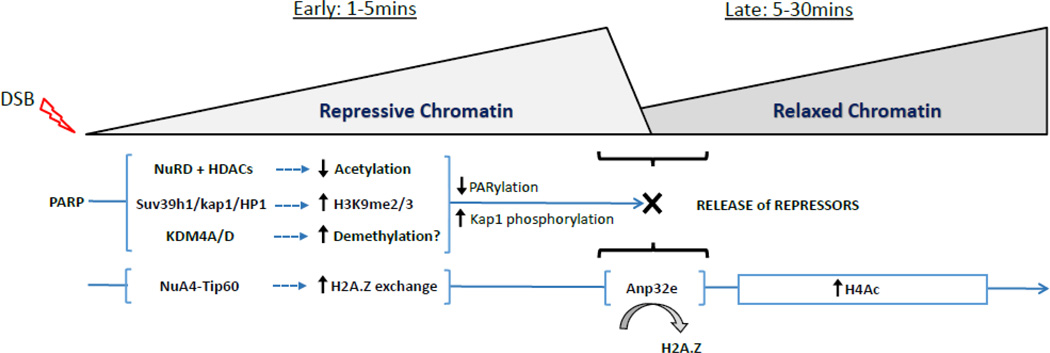
Recent work has suggested that the shift to an open, acetylated chromatin structure at DSBs may be preceded by a transient (seconds-minutes) compaction of the chromatin. Several repressive complexes are rapidly, but transiently recruited to DSBs through a mechanism regulated by PolyADP-ribose polymerases (PARP)28; 29; 30; 31. PARP family members create poly ADP-ribose (PAR) chains on the chromatin, which serve as binding sites for transcriptional regulators and DNA repair proteins32. Complexes containing HP1 and KAP-133; 34; 35, the H3K9 methyltransferases SUV39H131 and PRDM236 and several macroH2A variants36; 37 are all transiently loaded at DSBs in a PARP-dependent manner. This leads to rapid spreading of HP1 and H3K9me2/3 along the chromatin from the DSB31. Further, NuRD, a multi-subunit repressor complex containing both HDAC activity and the CHD3/CHD4 remodeling ATPase38, is recruited to PAR chains at DSBs28; 29; 39; 40, where it may function to deacetylate the chromatin. Loading of these repressor complexes will create local chromatin domains with low histone acetylation, increased H3K9me2/3 and increased density of HP1 and KAP-1 repressors adjacent to the DSBs. However, a critical regulatory element is that these repressive structures are transient, and are rapidly dismantled a few minutes after the initial DSB is detected28; 31; 35; 41. Removal of these repressive structures may involve chromatin dePARylation (or inactivation of PARP), phosphorylation of KAP-1 by the ATM kinase31 and demethylation of H3K9 by one or more of the histone demethylases which are recruited to DSBs42; 43.
The transient accumulation of repressive chromatin structures at DSBs may have several essential functions. They may be important for repressing local transcription to prevent passage of RNA Pol II through damaged regions44; 45; 46; 47. In addition, repressive structures may temporarily limit the mobility of the damaged chromatin template, reducing nucleosome mobility and forming structures which can keep the DNA ends at the break in close proximity during initial processing of the damage. Further, because many of these repressive proteins recruit essential activities, such as HDACs or remodeling ATPases28; 29; 38, this initial response may also serve to rewrite the local epigenetic code in preparation for repair.
Heterochromatin – The special one
The transient loading of repressive proteins at DSBs has significant parallels with the unique mechanism of DSB repair within heterochromatin. DSB repair in heterochromatin is slower than other regions of the chromatin20; 48, and spreading of γH2AX (figure 1) in heterochromatin is limited compared to the unrestricted spreading of γH2AX in more open regions (e.g.49; 50 reviewed in51). Sequencing of tumor cells indicates an excess of mutations in regions with elevated levels of H3K9me2/352. Further, repair in heterochromatin-like regions with elevated H3K9me2/3 occurs preferentially through error-prone NHEJ44. Because heterochromatin contains an excess of repetitive DNA elements, suppressing HR in heterochromatin may be important to limit unregulated HR between adjacent repeats. In fact, DSBs in heterochromatin may be relocated to the heterochromatin periphery for repair by HR53. Heterochromatin may therefore present a barrier to efficient DSB repair by the cell. Consequently, DSB repair in heterochromatin involves a unique mechanism in which phosphorylation of the KAP-1 repressor by the ATM kinase19; 20 releases the CHD3 remodeling ATPase from the damaged chromatin48; 54; 55. CHD3 may oppose the activity of an ISWI complex, comprised of the SNF2H catalytic sub-unit and the ACF1 protein54. Phosphorylation of KAP-1 and release of CHD3 at DSBs leads to significant relaxation of the chromatin19; 48; 54 and is required for efficient DSB repair in heterochromatin.
The rapid recruitment of repressor proteins to some DSBs indicates that, immediately after DNA damage, DSBs in heterochromatin and other more open regions share an overlapping set of repressive proteins. This includes increased H3K9me2/331; 36, repressive proteins such as HP1, KAP-1 and suv39h131; 33; 34; 35 and the CHD3 ATPase and HDACs (which are sub-units of the NuRD complex20; 28; 29; 48; 55). Further, phosphorylation of KAP-1 by ATM19; 20; 31; 54; 55 and release of the CHD3/NuRD complex28; 29; 48 are required in both heterochromatin and other chromatin domains to facilitate transition to the more open, flexible chromatin structures required for DSB repair. DSBs in more open chromatin domains may therefore be initially remodeled to create a chromatin structure resembling that found in more repressive domains. This may allow cells to rewrite the diverse epigenetic and structural organization in different chromatin domains to create a common template for the DSB repair machinery.
Transitioning from repressive to open chromatin
As discussed in the earlier section, DSB repair requires rapid removal of repressive proteins and a shift to a more open, acetylated structure4. This transition from repressive to open chromatin involves several remodeling complexes (discussed in several excellent reviews3; 5; 45; 56; 57). For example, RSC contributes to nucleosome sliding and ejection at DSBs58; 59, while FUN30 can regulate resection of the DSB60; 61. Here, we will focus on the key role played by the NuA4-Tip60 complex in facilitating the shift from repressive to open, acetylated chromatin at DSBs.
NuA4-Tip60
Mammalian NuA4 contains at least 16 sub-units, including 3 with catalytic activity – p400, a member of the Ino80 family of SWI/SNF ATPases; the ruvbl1 and ruvbl2 AAA+ ATPases; and the Tip60 (KAT5) acetyltransferase62. NuA4-Tip60 also contains multiple epigenetic readers containing chromodomains, PHD motifs and bromodomains, which may direct binding to specific epigenetic marks4; 63. Many of the sub-units of mammalian NuA4-Tip60, including Trrap26; 64, p40013; 65; 66, Tip6026; 27; 63; 67, DMAP168; 69 and ruvbl1/227; 70, are co-recruited to DSBs, implying that the intact NuA4-Tip60 is recruited to the DSB13; 26; 65; 71. Loss of functional NuA4-Tip60 leads to more compact chromatin at the DSB, defective DSB repair and increased sensitivity to DNA damaging agents26; 63; 66; 72. It is now clear that NuA4-Tip60 directs the shift from repressive to open chromatin through a mechanism dependent on 2 of its key sub-units – exchange of H2A.Z by the p400 SWI/SNF ATPase, coupled with rapid acetylation of histones H2A/H2AX and H4 by the Tip60 acetyltransferase after DNA damage13; 25; 26; 63; 73; 74.
H2A.Z exchange
p400 is a member of the Ino80 family of SWI/SNF remodelers, which includes Swr1 and Ino80 in yeast, and Ino80, p400 and SRCAP in mammalian cells56; 75. Ino80 family members are nucleosome sliders and histone H2A.Z exchangers75. H2A.Z has approximately 60% homology to H2A/H2AX, and has an extended acidic domain compared to H2A76; 77. This extended acidic domain creates an altered acidic patch on the surface of H2A.Z-nucleosomes, which has important consequences for DSB repair (discussed below). Ino80, swr1 and H2A.Z are important for the repair of persistent DSB breaks78, movement and tethering of damaged chromatin to the nuclear membrane75; 79; 80, translesion synthesis81 and overall repair of DSBs73; 79; 82; 83; 84. In yeast, Ino80 and swr1 facilitate the dynamic exchange of H2A.Z at DSBs78; 79; 82; 83, indicating that H2A.Z exchange is important for DSB repair. H2A.Z is also rapidly exchanged at DSBs in drosophila71 and mammalian cells41; 74; 85; 86; 87 by the p400 ATPase sub-unit of NuA4-Tip6041; 74. When H2A.Z exchange is blocked, either by targeting H2A.Z or inactivation of p400’s ATPase activity74, acetylation of histone H4 by Tip60 is blocked, and chromatin retains a more compact conformation and cells exhibit increased sensitivity to DNA damage78; 88; 89. Dynamic exchange of H2A.Z by members of the Ino80 family at DSBs is therefore required to promote histone H4 acetylation and to facilitate repair of DSBs.
H2A.Z exchange regulates H4 acetylation by altering the acidic patch on the nucleosome surface. The acidic patch is a charged groove on the nucleosome surface formed by a conserved acidic domain in the c-terminal of H2A along with residues provided by H2B76. The acidic patch plays a critical role in regulating nucleosome function, and provides a binding site for regulatory proteins such as HMGN290, the KSHV LANA protein91, and the PRC1 ubiquitin ligase complex92. Binding of the n-terminal tail of histone H4 to the acidic patch on adjacent nucleosomes can promote the formation of packed nucleosomal arrays93; 94. Acetylation of H4 on lysine 16 blocks this interaction4; 76; 93; 94; 95; 96, preventing 30nm fiber formation and shifting nucleosomes to a more open, flexible structure. Tip60 can acetylate H4 on chromatin domains which spread up to 50Kb from the DSB, decreasing nucleosome stability and favoring the formation of open, flexible chromatin domains4; 13; 23; 26; 65; 71. This suggests a simple model in which NuA4-Tip60 drives acetylation of H4 at DSBs, blocking interaction of H4 with the acidic patch, and thereby creating open, flexible chromatin domains which promote DSB repair. However, it now appears that H2A.Z exchange itself provides the key regulatory step which controls H4 acetylation by Tip60 at DSBs4; 13; 74.
Dynamic H2A.Z removal by ANP32E
Recent work revealed that H2A.Z is only transiently retained at DSBs41; 85. H2A.Z is rapidly removed from the DSB through the combined action of ANP32E, an H2A.Z-specific histone chaperone41; 97; 98 and the Ino80 remodeling complex85. ANP32E is a member of the acidic, leucine rich phosphoprotein ANP32 family implicated in apoptosis, phosphatase inhibition, intracellular transport and many cancers99. ANP32E binds to a unique sequence in the c-terminal docking domain of H2A.Z, catalyzing the removal of the entire H2A.Z-H2B dimer97; 98. Interestingly, the unique ANP32E binding site on H2A.Z is located directly adjacent to the acidic domain of H2A.Z76; 100. When the ANP32E binding domain of H2A.Z was swapped with the equivalent domain from H2A (which does not bind ANP32E), the chimeric H2A.Z protein was exchanged onto the nucleosomes at DSBs, but was not removed, and H2A.Z was retained at the site of DNA damage for an extended time41. Further, under this condition, H4 acetylation was blocked. This implies that H2A.Z exchange actually suppresses H4Ac, and that removal of H2A.Z by ANP32E is required to increase H4 acetylation at DSBs.
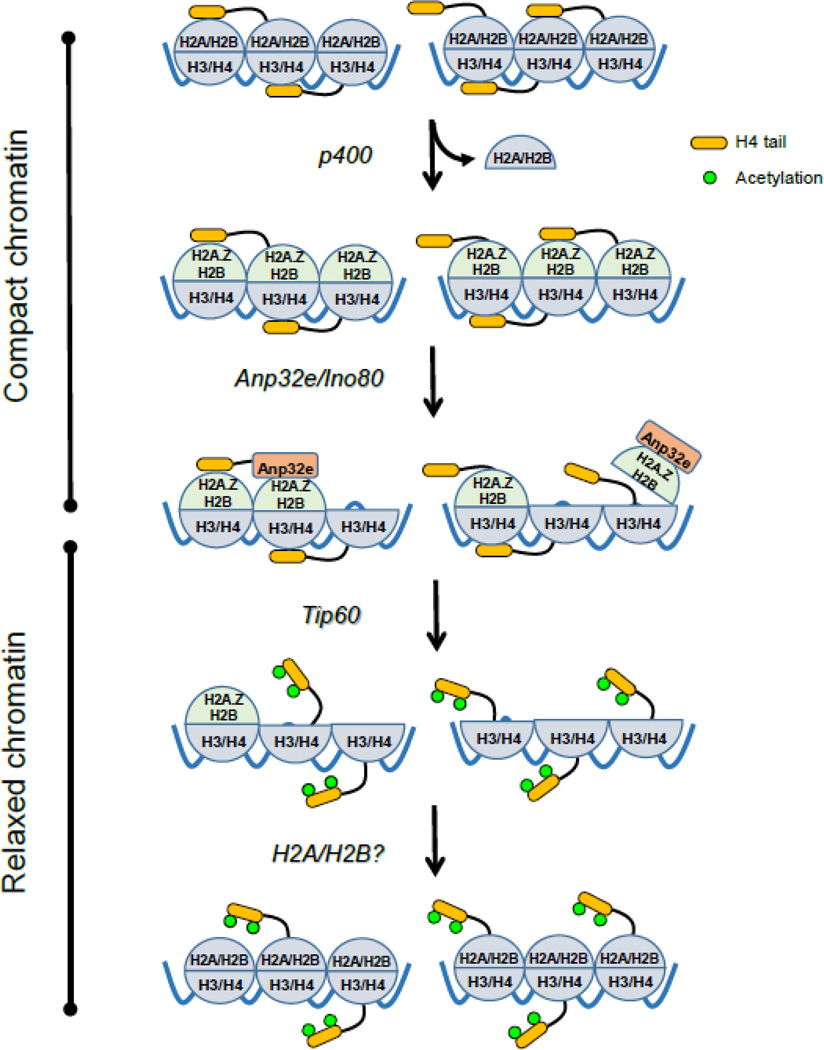
A potential explanation for this is that H2A.Z exchange at DSBs will increase the charged surface of the acidic patch, stabilizing binding of the H4 tail and promoting interaction between adjacent nucleosomes (figure 2). This is consistent with the observation that H2A.Z stabilizes nucleosomes101; 102 and favors binding of the H4 tail and packing of nucleosome fibers102; 103. DSBs also promote the rapid, but temporary recruitment of repressive complexes such as NuRD to DSBs with the same time course as the accumulation of H2A.Z28; 29; 35; 41; 85. The NuRD complex, which contains HDAC activity38, may potentially deacetylate H4, contributing to increased binding of the H4 tail to the acidic patch. The combination of transient loading of repressive factors coupled with transient exchange of H2A.Z may then favor H4 tail binding to the acidic patch (figure 2). This will tend to increase nucleosome packing, suppress local transcription and maintain the broken DNA ends in close proximity. H2A.Z exchange by NuA4-Tip60 therefore contributes to the formation of more compact chromatin by reinforcing binding of the H4 tail to the acidic patch immediately following DNA damage.
Figure 2. Dynamic H2A.Z exchange at DSBs by NuA4-Tip60.
The p400 ATPase sub-unit of NuA4-Tip60 exchanges H2A-H2B dimers for H2A.Z-H2B. This increases the charge density of the acidic patch on the nucleosome surface and favors binding of the H4 tail. Subsequent binding of ANP32E/Ino80 to H2A.Z leads to removal of the entire H2A.Z-H2B dimer. H2A.Z removal disrupts the acidic patch, releasing the H4 tail and promoting its acetylation by Tip60. Intact nucleosomes are then reformed by addition of H2A-H2B, although the remodeling complex involved in this is not known. Whether p400 creates homotypic (H2A.Z-H2B dimers) or heterotypic (H2A.Z-H2B /H2A-H2B) nucleosomes has not been established.
Promoting H4 acetylation through H2A.Z removal
H2A.Z exchange also creates a binding domain for the H2A.Z-specific histone chaperone ANP32E. Because the acidic domain and ANP32E-binding regions on H2A.Z are contiguous, ANP32E binding may simply displace the H4 tail and thereby facilitate acetylation by Tip60. However, ANP32E binding to H2A.Z actually removes the entire H2A.Z-H2B dimer from nucleosomes at the DSB41; 97; 98 (figure 2). Further, removal of H2A.Z-H2B at DSBs by ANP32E also requires the Ino80 ATPase85, which, like NuA4-Tip60, is recruited to DSBs and is important for DSB repair104; 105. Although purified ANP32E can remove H2A.Z-H2B dimers from reconstituted nucleosomes97, Ino80 may provide the energy required for ANP32E to extract H2A.Z-H2B from intact nucleosomes in cells. ANP32E/Ino80 removal of the entire H2A.Z-H2B dimer will effectively eliminate the acidic patch, thereby releasing the H4 tail for acetylation by Tip60 (figure 2).
How ANP32E is loaded onto the chromatin at DSBs and co-operates with Ino80 is unclear. Hamiche et al identified ANP32E as a sub-unit of NuA497, suggesting that it is co-recruited to DSBs with NuA4-Tip60. Previous purification of mammalian NuA4 did not reveal the presence of ANP32E106; 107, suggesting that interaction with NuA4 may be weak or require the presence of chromatin. Alternatively, ANP32E may be loosely associated with the Ino80 complex, or ANP32E interactions with either NuA4-Tip60 or Ino80 may be regulated by DNA damage. Further, whereas H2A.Z is only transiently retained at DSBs41; 85, ANP32E remains at the break site for an extended period, even in the absence of H2A.Z41. This suggests that ANP32E may have additional functions during DSB repair. Of relevance to this is the observation that, of the 2 genes for H2A.Z, H2A.Z-1 and H2A.Z-2108, only H2A.Z-2 was specifically exchanged onto the chromatin at DSBs86 and plays a key role in regulating sensitivity to chemotherapy109. Although these 2 H2A.Z variants differ by only 3 amino-acids, it raises the possibility that H2A.Z-2 is specific for DSB repair, while H2A.Z-1 participates in other H2A.Z driven events, such as transcription. Identifying specific remodeling factors which discriminate between H2A.Z variants, and understanding how small sequence differences between H2A.Z-1 and H2A.Z-2 impact their function during DSB repair are needed.
Nucleosome dynamics at the DSB
Dynamic H2A.Z exchange and removal by ANP32E therefore creates hyperacetylated chromatin domains at the break. Interestingly, nucleosome organization at DSBs has similarity to that seen at Transcriptional Starts Sites (TSSs). The TSS of many poised genes contain well positioned H2A.Z-nucleosomes at the +1 positon, adjacent to the nucleosome free region77; 110. H2A.Z may reduce the stability of the +1 nucleosome, promoting ejection of H2A.Z-H2B and allowing progression of RNA polymerase II. Similar to TSSs, DSBs are a nucleosome depleted region111; 112; 113; 114. H2A.Z exchange will position H2A.Z-nucleosomes either side of the break, providing positional stability to the damaged chromatin. The presence of H2A.Z-nucleosomes may then facilitate removal of H2A.Z-H2B dimers by ANP32E (figure 2). However, removal of H2A.Z-H2B dimers would create extended chromatin domains containing only tetrasomes (comprised of H3–H4 dimers (figure 2)). In both yeast and mammalian cells, histones are significantly depleted around DSBs111; 112; 113; 114. However, these nucleosome depleted regions are generally limited to 1000–2000bp from the DSB in mammalian cells74; 111, whereas H2A.Z exchange can spread up to 20kb from the break74. This implies that removal of H2A.Z by ANP32E/Ino80 must be coupled to an additional exchange reaction in which H2A.Z-H2B dimers are actively replaced with e.g. H2A-H2B dimers (figure 2). This may be carried out by Ino8085 or other remodeling complexes, which may function to restore intact nucleosomes at the DSB. Acetylation of the H4 tail would then prevent H4 from re-binding to the acidic patch on these newly formed nucleosomes. H2A.Z exchange by NuA4-Tip60 is therefore tightly coupled to H2A.Z removal, and it is H2A.Z removal that promotes both acetylation of H4 (by Tip60) and the shift to more open chromatin structures at DSBs (figure 2).
Transitioning from repressive to relaxed chromatin - a model
We propose a model in which the earliest events after DSB production involve rapid PARylation of the chromatin by PARP family members (figure 3). PARylation functions to recruit multiple repressive complexes, including NuRD and HDACs, the suv39h1/KAP-1/HP1 methyltransferase complex and several lysine demethylases onto the chromatin at the break. This leads to rapid histone deacetylation and increased H3K9me2/3. Further, NuA4-Tip60 rapidly exchanges H2A.Z onto nucleosomes at the DSB, further limiting nucleosome mobility by promoting interaction of the H4 tail with the acidic patch. This creates temporary repressive chromatin structures which can stabilize the break and limit local transcription. However, as repair proceeds, these repressive complexes must be rapidly disassembled. This is particularly important since repressive chromatin tends to inhibit DSB repair20; 51; 56. First, because PARylation is transient after DNA damage32; 115, rapid dePARylation leads to loss of NuRD and other complexes from the DSB. Further, activation of the ATM kinase (driven by H3K9me2 mediated activation of Tip6063), leads to phosphorylation of KAP-1 and release of the suv39h1/KAP-1/HP1 complex from the chromatin31. Finally, recruitment of ANP32E promotes removal of H2A.Z, potentially increasing nucleosome mobility as well as making the H4 tail available for acetylation by Tip60. This combination of events serves to promote the shift from repressive to more open, acetylated chromatin at the DSB. This rapid transition is critical for efficient DSB repair, since failure to remove H2A.Z or complexes such as the suv39h1/KAP-1/HP1 from the DSB significantly impairs DSB repair28; 31; 41.
Figure 3. A model for chromatin dynamics during DSB repair.
Loading of repressive complexes and H2A.Z exchange by NuA4-Tip60 leads to the rapid spread of repressive chromatin away from the DSB. DePARylation and H2A.Z removal (by ANP32E) results in removal of repressive chromatin, releasing the H4 tail and allowing rapid H4 acetylation by the Tip60 sub-unit of NuA4-Tip60.
Several key points remain to be addressed in this model. NuA4-Tip60 recruitment does not require PARP4; 13. However, recent work suggests that NuA4-Tip60 can promote PARylation116, suggesting potential cross-regulation between NuA4-Tip60 and other repressor complexes during DSB repair. This may indicate that certain functions of NuA4-Tip60, such as H2A.Z exchange, may be influenced by chromatin PARylation immediately after DNA damage. In addition, how lysine demethylases such as KDM4A and KDM4D42; 117, which are recruited to DSBs, contribute to chromatin organization during DSB repair is unclear. It is possible that they are required to remove H3K9me2/3 generated by suv39h131 (figure 3), or to remove other methylation sites which may block DNA repair.
Finally, it is important to note that the model in figure 3 represents a generalized view of how DSB repair promotes chromatin reorganization. Many of the repressive complexes in figure 3 are already associated with heterochromatin, implying that DSB repair in heterochromatin only requires the removal (relaxation) step to transition to open chromatin48; 55; 118. Thus DSBs in transcribed genes may require transient repression to block transcription, while silent compact regions require shifting to more open structures. Further, recent work has shown that, while DNA damage promotes an initial chromatin decompaction, this is followed by loading of macroH2A1 and subsequent chromatin condensation at later times36; 119. This indicates that chromatin reorganization during repair is dynamic, with rapid transition between open and more compact structures occurring as repair progresses. DSBs in different chromatin structures are therefore unlikely to be processed by a common, undirectional chromatin remodeling pathway. Instead, chromatin reorganization during DSB repair will be dependent on the pre-existing chromatin architecture, the nature and extent of the damage and the choice of DSB repair pathway (NHEJ vs HR). For example, monitoring changes in chromatin structure after damage in live cells is complex, relying on expansion of micro-irradiated regions21; 36 or the mobility of tagged proteins at enzymatic breaks53; 120. Such approaches monitor changes occurring over large, megabase domains of chromatin, as opposed to approaches based on enzymatically generated DSBs and ChIP, which tend to focus on changes occurring directly at, or within a few 10s of kilobases, of the DSB13; 74. It would not difficult to envision that DNA damage can promote chromatin decompaction, extending over megabase/whole chromosome domains, while simultaneously loading repressive factors at the DSB, which compact nucleosomes for a few kilobases either side of the break. This may allow cells to compact and stabilize the local nucleosome organization directly at the DSB, while simultaneously relaxing the surrounding higher order chromatin structure to allow for large scale chromatin mobility and access to the DSB itself. Thus chromatin dynamics during DSB repair are likely to be highly fluid119, transitioning between open and condensed conformations both at the break and in the surrounding chromatin neighborhood. Given the recent advances in our understanding of higher chromatin structure, chromatin looping and the proteins which maintain this fluid structure, it should be possible to unravel how DNA breaks impact chromatin organization at all levels.
H2A.Z turnover and DSB repair
In addition to regulating nucleosome dynamics at DSBs, it is now clear that H2A.Z and acetylation of histone H4 are also important for promoting additional histone modifications and for the recruitment of several bromodomain proteins to the H4 tail. Here, we will examine how H2A.Z and NuA4-Tip60: (i) regulate the processing of the damaged DNA ends; (ii) direct further histone modification, including ubiquitination; and (iii) promote recruitment of proteins to the acetylated H4 tail at DSBs.
Nucleosome dynamics, H2A.Z and end resection
Nucleosomes can be barriers to end processing of DNA breaks and require remodeling to promote repair16. DSB repair can occur through 2 pathways. In NHEJ, the DNA ends are minimally processed and then religated16. HR requires the production of ssDNA intermediates, which become coated with the ssDNA binding complex RPA, through a mechanism regulated by MRN-CtIP and the EXO1/SGS1 nucleases16. In mammalian cells, failure to remove H2A.Z13; 41; 65; 74 leads to defects in end processing of the break, consistent with a direct role for NuA4-Tip60 in DSB repair by HR. In particular, depletion of ANP32E, which leads to H2A.Z retention on the chromatin41, tends to increase ssDNA at the break41; 74; 85. This, in turn prevents loading of the Ku70/80 complex (which requires dsDNA ends to bind), thereby reducing NHEJ activity4; 87. The increase in end resection when H2A.Z is retained can be blocked by depletion of CtIP13; 74, which acts to initiate end resection. Further, EXO1-dependent ssDNA production is increased on H2A.Z-nucleosomes or tetrasomes lacking H2A-H2B dimers121. Together, this suggests that the presence of H2A.Z on nucleosomes facilitates ssDNA production by EXO1 and related nucleases. Rapid exchange and removal of H2A.Z-H2B dimers may therefore be tightly coupled with processivity of the MRN-CtIP and EXO1 nucleases along the chromatin.
However, H2A.Z may also indirectly regulate end resection. For example, depletion of Ino80 leads to a decrease in ssDNA at DSBs85; 104; 105, despite the retention of H2A.Z at the DSB in these cells85. This may reflect an active role for Ino80 in end resection or the influence of Ino80 on nucleosome dynamics at DSBs. However, it is also possible that the presence of H2A.Z on the nucleosome does not directly promote end-resection. H2A.Z can promote H4 acetylation, chromatin ubiquitination and loading of e.g. 53BP113; 41; 74. H2A.Z may therefore indirectly influence end-resection by directing specific patterns of histone and chromatin modification which favor end-resection. How the positioning of nucleosomes and H2A.Z at the DSB impacts ssDNA production therefore remains unclear. Further, it is unclear if nucleosomes/tetrasomes must be displaced to allow DNA end-resection and ssDNA production, nor is it clear if the resulting ssDNA, which is coated with ssDNA binding protein RPA, can associate with the nucleosome. How H2A.Z impacts these processes also requires clarification. An additional consideration is that regions of the chromatin with high density of H2A.Z may be less dependent on NuA4-Tip60 to increase H2A.Z at the DSB. Instead, ANP32E and Ino80 may be important to actively remove H2A.Z in these regions. Clearly, a better understanding of how nucleosome dynamics and histone variants such as H2A.Z are linked to the process of end resection is needed. Further, how pre-existing levels of H2A.Z, and indeed other chromatin binding factors, influence end resection also require further investigation.
Nucleosome dynamics and ubiquitination
Chromatin ubiquitination in response to DSBs is complex (reviewed in11), and is required to load DSB repair proteins such as brca1 and 53BP1 (figure 1) onto the chromatin. 53BP1 recruitment requires dual binding of its tudor domain to H4K20me2 and Ubiquitin Interaction Motif (UIM) to H2A ubiquitinated on K13/15Ub12. Inducible ubiquitination of H2A at DSBs is carried out by the combined action of the RNF8 and RNF168 ligases12; 122. Further, RNF168 requires the acidic patch on the nucleosome surface to ubiquitinate H2A123; 124. RNF168 does not bind directly to the acidic patch; instead, it provides specificity to allow RNF168 to ubiquitinate K13/K15 of H2A123. When NuA4-Tip60 or H2A.Z exchange are inhibited, the H4 tail remains bound to the acidic patch on the nucleosome surface41 and there is a loss of both chromatin ubiquitination and 53BP1 loading13; 23; 26; 65. H2A.Z exchange and H4 acetylation may therefore also function to expose the acidic patch41 and facilitate H2A ubiquitination by RNF168 and recruitment of 53BP1123; 124. Further, because 53BP1 inhibits ssDNA production125, the increase in ssDNA seen when H2A.Z removal is blocked may reflect failure to increase H2AK13/15Ub and load 53BP141; 85. However, it is not known if the ability of RNF168 to ubiquitinate H2A122; 123; 124 is influenced by the unique acidic patch present on the surface of H2A.Z-nucleosomes. H2A.Z lacks one of the lysine residues ubiquitinated by RNF168 on H2A100, suggesting that H2A.Z itself may not be a substrate for RNF168 during DSB repair. Finally, exchange of H2A or H2A.Z may provide a mechanism for controlling overall H2AUb levels (by removing H2AUb), or H2AUb may itself regulate the exchange and removal of H2A.Z. Addressing these issues will help to elucidate the link between nucleosome dynamics, histone modifications and DSB repair.
The acidic patch and the nucleosome surface may therefore be an important hub for regulating DNA damage mediated histone modifications. In fact, recent work indicates that H2A.Z exchange can recruit fumarase to the nucleosome surface, leading to localized increases in fumarate, which in turn inhibits the KDM2B demethylase87. The acidic patch on the nucleosome surface may therefore provide a critical landing pad for many of the chromatin modifiers involved in DSB repair. We note that RNF168 ubiquitinates other proteins in addition to H2A during DSB repair, indicating that only a sub-set of RNF168 targets may be dependent on H2A.Z removal. Further, there are several other ubiquitin ligase complexes which ubiquitinate chromatin during DSB repair and which are not directly regulated by the acidic patch11.
Nucleosome dynamics and the H4 tail
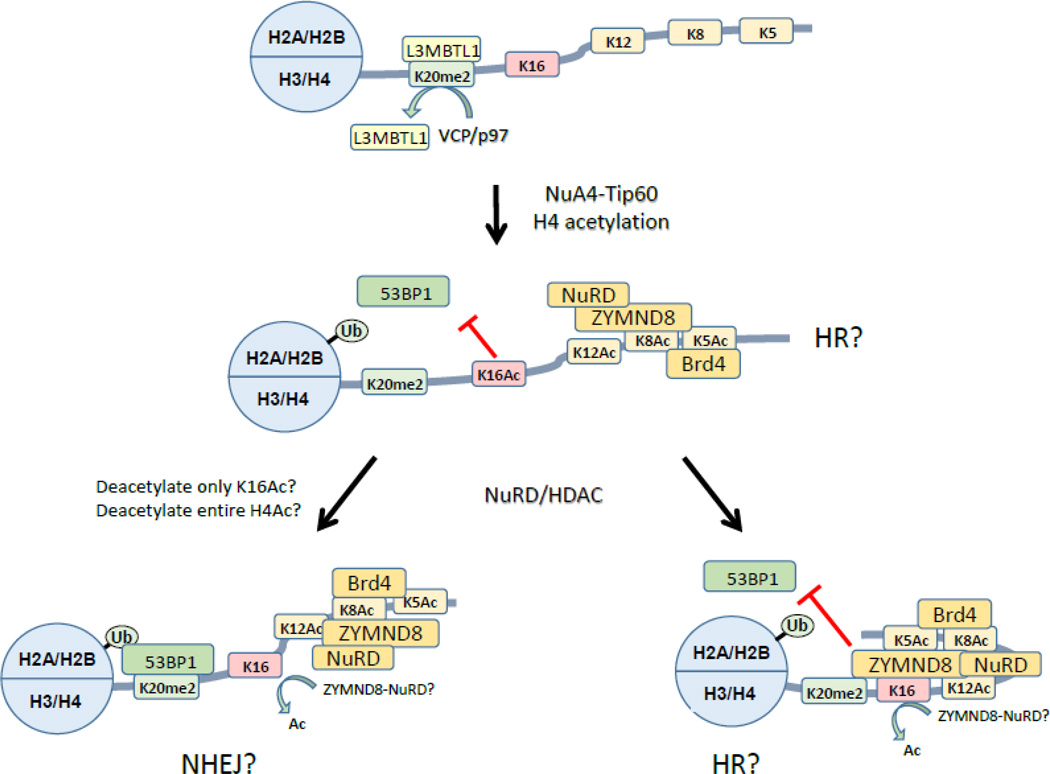
The acetylated H4 tail is emerging as a crowded hub of activity for controlling DSB repair. At least 5 different protein complexes are known to be associated with 3 different histone modifications of H4 during DNA repair (figure 4). 53BP1 loading requires bivalent binding of 53BP1 to both H2AK13/15Ub and H4K20me212. However, the majority of H4K20 is constitutively methylated and associated with proteins including L3MBTL1126 and the JMJD2A demethylase117. DNA damage promotes removal of L3MBTL1 by the VCP/p97 ATPase (figure 4)126 and degradation of JMJD2A117 (reviewed in depth in127) creating unbound H4K20me2 to which 53BP1 can then bind. However, elegant work has shown that 53BP1 binding to H4K20me2 is inhibited when H4K16 is also acetylated23, indicating that NuA4-Tip60 may oppose 53BP1 loading. H4K16Ac blocked 53BP1 loading at DSBs and promoted HR23, indicating that fine tuning of 53BP1 loading via Tip60 regulates end processing of the DNA (figure 4). This is consistent with the idea that NuA4-Tip60 favors HR mediated repair over NHEJ. Further, it suggests that some of the HDACs recruited to DSBs28; 29; 128; 129 may be important for fine tuning H4K16 acetylation during DSB repair.
Figure 4. Potential impact of nucleosome dynamics on ubiquitination and acetylation during DSB repair.
In compact nucleosome arrays, the H4 tail is unacetylated and bound to the acidic patch (omitted for clarity). H4K20me2 is largely bound to the L3MBTL1 repressor, which is removed by the VCP/p97 ATPase during DSB repair. Following H2A.Z exchange and removal, the n-terminal tail of H4 is extensively acetylated on lysines 5, 8, 12 and 16. Several bromodomain proteins, including the ZMYND8-NuRD complex and BRD4 are then recruited to the H4 tail. Note that, because Brd4 and ZMYND8 may bind to multiple acetylated lysines in the H4 tail, each H4 tail is likely to be associated with only one bromodomain protein. Several potential outcomes are proposed. NHEJ: ZMYND8-NuRD may deacetylate H4K16Ac, allowing 53BP1 to access and bind H4K20me2/H2AUb. It is also possible that loading of ZMYND8-NuRD deacetylates the entire H4 tail, leading to loss of ZYMND8 (not shown). HR: Binding of ZMYND8 may shelter H4Ac sites from HDAC activity, so that ZYMND8 may block access of 53BP1 to H4K20me2, even if H4K16Ac is deacetylated.
However, recent work has revealed that the H4 tail is a much more crowded place than previously thought. Several bromodomain proteins, including BRD4130 and ZMYND8131, bind to the acetylated H4 tail after DNA damage (figure 4). BRD4 is proposed to function as a repressor, limiting chromatin expansion at DSBs and favoring a more compact structure adjacent to the DSB130. ZMYND8 recruits the repressive NuRD complex to DSBs131. Further, ZMYND8-NuRD was preferentially recruited to DSBs in transcribed regions and was associated with a general repression of transcription131. This provides an important link to other studies demonstrating that transcriptionally active regions with high levels of H3K36me2 are preferentially repaired by HR44. However, how loading of ZMYD8-NuRD and BRD4 impact 53BP1 loading and the choice between HR and NHEJ remains unclear. Some potential interactions among these proteins are highlighted in figure 4. ZMYND8-NuRD contains HDAC activity, indicating that it may deacetylate the H4 tail131. For example, deacetylation of H4K16Ac by ZMYND8-NuRD may facilitate 53BP1 binding23 (figure 4) and favor NHEJ over HR repair131. In this case, ZMYND8 binding to the H4 tail may protect lysines 5, 8 and 12 from deacetylation. Alternatively, ZMYND8 binding to acetylated H4131 may block access of 53BP1 to H4K20me2 even in the absence of H4K16Ac (figure 4), leading to increased HR. Fine tuning of the acetylation code on H4 may allow cells to regulate binding of e.g. 53BP1 and therefore control choice of DSB repair pathway. Future studies aimed at understanding how 53BP1 and bromodomain proteins, including ZMYND8, compete for binding to the H4 tail are needed to fully understand how H4 tail acetylation contributes to choice of DSB repair pathway. A further complexity is the potential interaction between the acid patch and the H4 tail, which may be heavily influenced by the presence of bromodomain proteins bound to the acetylated tail. Unraveling the complexities of these interactions in different chromatin domains during DSB repair remains an outstanding challenge.
Conclusion
The interaction between chromatin organization and the DNA repair machinery is complex. The initial cellular response to a DSB is the rapid recruitment of repressive complexes onto the chromatin. This repressive chromatin environment is then further processed to create a more open structure associated with increased histone acetylation, loading of bromodomain proteins and specific patterns of histone ubiquitination. DNA damage therefore initiates rapid rewriting of the pre-existing epigenetic signatures on the chromatin at the break site. This rapid change in histone modification signatures likely serves to create a common chromatin structure which can function as a template for the DNA repair machinery. The correct sequential ordering of these changes, including dynamic changes in histone modifications, allows for fine tuning of the repair mechanism. Further, these processes are dynamic, with rapid recruitment and release of various factors occurring during the initial processing of the DSB. Finally, it is not unreasonable to infer that the mechanism of DSB repair will vary greatly depending on the pre-existing chromatin environment, requiring the deployment of distinct remodeling complexes dictated by the underlying chromatin architecture. Of particular importance is the emerging role that the acidic patch on the nucleosome surface plays in DSB repair. The nucleosome surface may function as a central hub during repair, serving to control nucleosome interactions through binding to the H4 tail, providing specificity for the RNF168 ubiquitin ligase as it modifies H2A as well as potentially providing a landing pad for other repair factors. The ability of NuA4-Tip60 and H2A.Z exchange to directly impact the properties and function of the acidic patch may underlie the central remodeling activity of NuA4-Tip60 during DSB repair.
Highlights.
DNA repair requires rapid chromatin remodeling.
Repair of DNA breaks involves transient accumulation of repressive complexes.
Subsequent H2A.Z exchange by NuA4 promotes rapid release of repressors.
Shift to open chromatin at breaks requires H4 acetylation by Tip60.
Transition from repressive to open, acetylated chromatin required for DNA repair.
Dynamic changes in nucleosome organization required to prepare damaged DNA for repair.
Acknowledgements
We thank members of the Price lab for discussion and comments. Supported by NIH grants CA64585, CA93602 and CA177884 to BDP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smerdon MJ. DNA repair and the role of chromatin structure. Current opinion in cell biology. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 3.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Price BD, D'Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 6.Luijsterburg MS, van Attikum H. Chromatin and the DNA damage response: the cancer connection. Molecular oncology. 2011;5:349–367. doi: 10.1016/j.molonc.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol. 2015;117:182–193. doi: 10.1016/j.pbiomolbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 9.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 11.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst) 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Symington LS. Overcoming the chromatin barrier to end resection. Cell Res. 2013;23:317–319. doi: 10.1038/cr.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 17.Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proc Natl Acad Sci U S A. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 20.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 21.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X, Xu Y, Price BD. Acetylation of H2AX on lysine 36 plays a key role in the DNA double-strand break repair pathway. FEBS Lett. 2010;584:2926–2930. doi: 10.1016/j.febslet.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 27.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Molecular and cellular biology. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 2015;11:1486–1500. doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes & development. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 36.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, Oberdoerffer P. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014;8:1049–1062. doi: 10.1016/j.celrep.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. Febs Letters. 2012;586:3920–3925. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nature reviews. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, Lukas C, Bartek J, Andersen JS, Lukas J. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of cell biology. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gursoy-Yuzugullu O, Ayrapetov MK, Price BD. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc Natl Acad Sci U S A. 2015;112:7507–7512. doi: 10.1073/pnas.1504868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci U S A. 2014;111:E728–E737. doi: 10.1073/pnas.1317585111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Liu L, Yang S, Song N, Zhou X, Gao J, Yu N, Shan L, Wang Q, Liang J, Xuan C, Wang Y, Shang Y, Shi L. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci U S A. 2014;111:7096–7101. doi: 10.1073/pnas.1324036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pankotai T, Soutoglou E. Double strand breaks: hurdles for RNA polymerase II transcription? Transcription. 2013;4:34–38. doi: 10.4161/trns.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 47.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nature structural & molecular biology. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 49.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA repair. 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 53.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol. 2014;207:717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 56.Jeggo PA, Downs JA. Roles of chromatin remodellers in DNA double strand break repair. Exp Cell Res. 2014;329:69–77. doi: 10.1016/j.yexcr.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 57.Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- 58.Chambers AL, Brownlee PM, Durley SC, Beacham T, Kent NA, Downs JA. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PLoS One. 2012;7:e32016. doi: 10.1371/journal.pone.0032016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 60.Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, van Attikum H, Llorente B. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robert F, Hardy S, Nagy Z, Baldeyron C, Murr R, Dery U, Masson JY, Papadopoulo D, Herceg Z, Tora L. The Transcriptional Histone Acetyltransferase Cofactor TRRAP Associates with the MRN Repair Complex and Plays a Role in DNA Double-Strand Break Repair. Mol Cell Biol. 2006;26:402–412. doi: 10.1128/MCB.26.2.402-412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. The Journal of cell biology. 2012;199:1067–1081. doi: 10.1083/jcb.201205059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattera L, Escaffit F, Pillaire MJ, Selves J, Tyteca S, Hoffmann JS, Gourraud PA, Chevillard-Briet M, Cazaux C, Trouche D. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene. 2009;28:1506–1517. doi: 10.1038/onc.2008.499. [DOI] [PubMed] [Google Scholar]
- 67.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Penicud K, Behrens A. DMAP1 is an essential regulator of ATM activity and function. Oncogene. 2014;33:525–531. doi: 10.1038/onc.2012.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Negishi M, Chiba T, Saraya A, Miyagi S, Iwama A. Dmap1 plays an essential role in the maintenance of genome integrity through the DNA repair process. Genes Cells. 2009;14:1347–1357. doi: 10.1111/j.1365-2443.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 70.Jha S, Gupta A, Dar A, Dutta A. RVBs are required for assembling a functional TIP60 complex. Mol Cell Biol. 2013;33:1164–1174. doi: 10.1128/MCB.01567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 72.Fischle W. Tip60-ing the balance in DSB repair. Nature Cell Biology. 2009;11:1279–1281. doi: 10.1038/ncb1109-1279. [DOI] [PubMed] [Google Scholar]
- 73.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of Chromatin-Modifying Activities to Phosphorylated Histone H2A at DNA Damage Sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair. Molecular cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerhold CB, Hauer MH, Gasser SM. INO80-C and SWR-C: guardians of the genome. J Mol Biol. 2015;427:637–651. doi: 10.1016/j.jmb.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 76.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. Journal of the Royal Society, Interface / the Royal Society. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian V, Fields PA, Boyer LA. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep. 2015;7:01. doi: 10.12703/P7-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMOH2A. Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 79.Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell. 2014;55:626–639. doi: 10.1016/j.molcel.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 80.Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 81.Renaud-Young M, Lloyd DC, Chatfield-Reed K, George I, Chua G, Cobb J. The NuA4 complex promotes translesion synthesis (TLS)-mediated DNA damage tolerance. Genetics. 2015;199:1065–1076. doi: 10.1534/genetics.115.174490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong S, Han J, Chen H, Liu T, Huen MS, Yang Y, Guo C, Huang J. The human SRCAP chromatin remodeling complex promotes DNA-end resection. Curr Biol. 2014;24:2097–2110. doi: 10.1016/j.cub.2014.07.081. [DOI] [PubMed] [Google Scholar]
- 85.Alatwi HE, Downs JA. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 2015 doi: 10.15252/embr.201540330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishibuchi I, Suzuki H, Kinomura A, Sun J, Liu NA, Horikoshi Y, Shima H, Kusakabe M, Harata M, Fukagawa T, Ikura T, Ishida T, Nagata Y, Tashiro S. Reorganization of damaged chromatin by the exchange of histone variant H2A.Z-2. Int J Radiat Oncol Biol Phys. 2014;89:736–744. doi: 10.1016/j.ijrobp.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 87.Jiang Y, Qian X, Shen J, Wang Y, Li X, Liu R, Xia Y, Chen Q, Peng G, Lin SY, Lu Z. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat Cell Biol. 2015;17:1158–1168. doi: 10.1038/ncb3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PloS one. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 92.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 94.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Potoyan DA, Papoian GA. Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17857–17862. doi: 10.1073/pnas.1201805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Molecular and cellular biology. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, Dimitrov S, Romier C, Hamiche A. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–653. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 98.Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014;24:389–399. doi: 10.1038/cr.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reilly PT, Yu Y, Hamiche A, Wang L. Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. Bioessays. 2014;36:1062–1071. doi: 10.1002/bies.201400058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 101.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. The Journal of biological chemistry. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 102.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Molecular cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 103.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nature structural & molecular biology. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 104.Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S. The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 2013;23:1396–1413. doi: 10.1038/cr.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gospodinov A, Vaissiere T, Krastev DB, Legube G, Anachkova B, Herceg Z. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol Cell Biol. 2011;31:4735–4745. doi: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 108.Bonisch C, Schneider K, Punzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E, Leonhardt H, Mann M, Michaelis J, Schermelleh L, Hake SB. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 2012;40:5951–5964. doi: 10.1093/nar/gks267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vardabasso C, Gaspar-Maia A, Hasson D, Punzeler S, Valle-Garcia D, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, Chung CY, Yao JL, Singh R, Segura MF, Fontanals-Cirera B, Verma A, Mann M, Hernando E, Hake SB, Bernstein E. Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol Cell. 2015;59:75–88. doi: 10.1016/j.molcel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nature structural & molecular biology. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci U S A. 2013;110:16874–16879. doi: 10.1073/pnas.1306160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 115.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Molecular cell biology. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 116.Krukenberg KA, Jiang R, Steen JA, Mitchison TJ. Basal activity of a PARP1-NuA4 complex varies dramatically across cancer cell lines. Cell Rep. 2014;8:1808–1818. doi: 10.1016/j.celrep.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 119.Li ML, Yuan G, Greenberg RA. Chromatin yo-yo: expansion and condensation during DNA repair. Trends Cell Biol. 2014;24:616–618. doi: 10.1016/j.tcb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adkins NL, Niu H, Sung P, Peterson CL. Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol. 2013;20:836–842. doi: 10.1038/nsmb.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 123.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun. 2014;5:3291. doi: 10.1038/ncomms4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D, Miller KM. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet. 2014;10:e1004178. doi: 10.1371/journal.pgen.1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 127.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 128.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]