Abstract
Regulation of gene expression changes during chondrogenic differentiation by DNA methylation and demethylation is little understood. Methylated cytosines (5mC) are oxidized by the ten-eleven-translocation (TET) proteins to 5-hydroxymethylcytosines (5hmC), 5-formylcytosines (5fC) and 5-carboxylcytosines (5caC) eventually leading to a replacement by unmethylated cytosines (C) i.e. DNA demethylation. Additionally, 5hmC is stable and acts as an epigenetic mark by itself. Here, we report that global changes in 5hmC mark chondrogenic differentiation in vivo and in vitro. Tibia anlagen and growth plate analyses during limb development at mouse embryonic days E 11.5, 13.5 and 17.5 showed dynamic changes in 5hmC levels in the differentiating chondrocytes. A similar increase in 5hmC levels was observed in the ATDC5 chondroprogenitor cell line accompanied by increased expression of the TET proteins during in vitro differentiation. Loss of TET1 in ATDC5 decreased 5hmC levels and impaired differentiation, demonstrating a functional role for TET1-mediated 5hmC dynamics in chondrogenic differentiation. Global analyses of the 5hmC-enriched sequences during early and late chondrogenic differentiation identified 5hmC distribution to be enriched in the regulatory regions of genes preceding the transcription start site (TSS) as well as in the gene bodies. Stable gains in 5hmC were observed in specific subsets of genes including genes associated with cartilage development and in chondrogenic lineage-specific genes. 5hmC gains in regulatory promoter and enhancer regions as well as in gene bodies were strongly associated with activated but not repressed genes, indicating a potential regulatory role for DNA hydroxymethylation in chondrogenic gene expression.
Keywords: 5-hydroxymethylcytosine, Chondrogenesis, Differentiation, DNA demethylation, Epigenetics
Introduction
DNA methylation is a vital epigenetic mark associated with key cellular processes including gene silencing and embryonic development (1). During embryonic development, mesenchymal stem and progenitor cells undergo chondrogenic differentiation to form a cartilaginous template that is later replaced by bone in a process called ‘endochondral ossification’ (2). Although epigenetic processes including DNA methylation and demethylation are increasingly acknowledged for their critical roles in cellular differentiation and development, little is known about their role in chondrogenic differentiation. Such studies can contribute significantly towards an increased understanding of skeletal birth defects as well as help devise better strategies for cartilage tissue engineering.
In the last few years, it has become apparent that DNA methylation is not a static epigenetic mark but is highly dynamic and is governed by a precise molecular network of regulators (3). Methylated cytosines (5mC) can be further oxidized to 5-hydroxymethylcytosines (5hmC), 5-formylcytosines (5fC) and 5-carboxylcyosines (5caC) by Ten-eleven-translocation (TET) proteins (4–6). The TET-oxidized intermediate 5caC is an eventual substrate for the DNA repair enzyme, Thymine DNA Glycosylase (TDG), which can remove the modified base and replace it with an unmodified cytosine, resulting in active DNA demethylation (7, 8). TET proteins have been shown to be critical for embryonic stem cell differentiation, neurogenesis and cognitive functions as well as implicated in multiple cancers. The newly discovered epigenetic modifications, especially 5hmC, are also present stably in DNA with high levels observed in embryonic stem cells (ESC) and neurons (5, 9). Regulatory roles for 5hmC as an epigenetic mark have been suggested by studies of its global distribution in both ESC and neurons and the effects on gene expression patterns (10–12). 5hmC undergoes dynamic changes during neuronal development (12, 13) and appears to regulate neuronal genes during differentiation (14). Although 5hmC is enriched in intragenic regions in both ESC and neurons, there are marked differences in its distribution at the enhancer elements and regulatory regions between ESC and neurons (14, 15).
In view of the recent advances in the understanding of DNA demethylation pathways and increasing evidence for a functional role for stable 5hmC and its derivatives in the epigenome, we sought to investigate the 5hmC dynamics during chondrogenic differentiation. Low levels of DNA methylation were reported in chondrogenic genes including Sox9 and Runx2 in synovial mesenchymal stem cells (MSCs) that remained unchanged upon chondrogenic differentiation (16), while promoter demethylation in the Col10a1 gene was observed upon its activation in hypertrophic chondrocytes (17). Extensive DNA demethylation changes have however been reported in damaged cartilage in Osteoarthritis (OA) (18) and we have recently reported global increase in 5hmC levels in human chondrocytes from OA patients as compared to normal chondrocytes (19). Additionally, a genetic loss of TET1 in mice did not affect viability, however 75% of the newborn mice were smaller than the wild-type mice revealing a skeletal growth defect and a potential regulatory role for TET1-mediated 5hmC dynamics in skeletal development (20). In the present study, we have therefore analyzed the 5hmC dynamics during chondrogenic differentiation utilizing both mice embryos and in vitro chondroprogenitors that mimic the process of chondrogenic differentiation. Mapping the global and locus-specific 5hmC distribution, we provide the first comprehensive resource for DNA hydroxymethylation distribution and its effect on gene expression changes during chondrogenic differentiation.
Materials and Methods
In vivo embryonic growth plate sections
All animal procedures were approved by the Stanford University Administrative Panel on Laboratory Animal Care. B6129SF1/J mice (Jackson Laboratory) were time-mated for these experiments and the females sacrificed at 11.5, 13.5, 17.5 days post-coitus. Embryos were fixed with 4% PFA/PBS at 4°C overnight, limbs were dissected, frozen in O.C.T. (Tissue Tek) and cryosectioned at 10µm.
Global gene expression analyses
For the microarray analysis, two independent replicate samples for day 0 and day 15 were run on Mouse Gene 1.0 ST Arrays (Affymetrix). Data analysis was performed using dChip (http://www.hsph.harvard.edu/cli/complab/dchip/download.htm) as described by the manual, and network analysis of differentially expressed genes was performed using Metacore (Thomson Reuters - http://thomsonreuters.com/metacore/). Further details of microarray analyses and primer sequences can be found in the Supplemental Methods.
Profiling of hydroxymethylated DNA
Total DNA was extracted from ATDC5 cells and was enriched for 5hmC using a biotin-based streptavidin pull down technique (Hydroxymethyl Collector, Active Motif), as per manufacturers guidelines. Libraries were prepared using 300–500ng of 5hmC enriched DNA using the NEBNext DNA Library Prep Master Mix Set for Illumina (NEB) with 1µM of adapter (IDT) per 100ng of input DNA and 12 cycles of PCR and with universal primers (IDT) to amplify the adapter ligated DNA. Libraries were sequenced on an Illumina HiSeq 2000 with single end (1×50 base pair) reads. Details regarding the bioinformatics analyses can be found in the Supplemental Methods.
Validation of the enriched DNA sequencing results was performed using the EpiMark 5hmC and 5mC Analysis Kit (NEB) as per suppliers’ protocol. The EpiMark treated DNA was subjected to quantitative PCR using site-specific primers and the percentage of 5hmC was calculated using the EpiMark comparative Ct method (21, 22).
Results
Different stages of chondrogenic differentiation in vivo are marked by distinct global changes in 5hmC levels
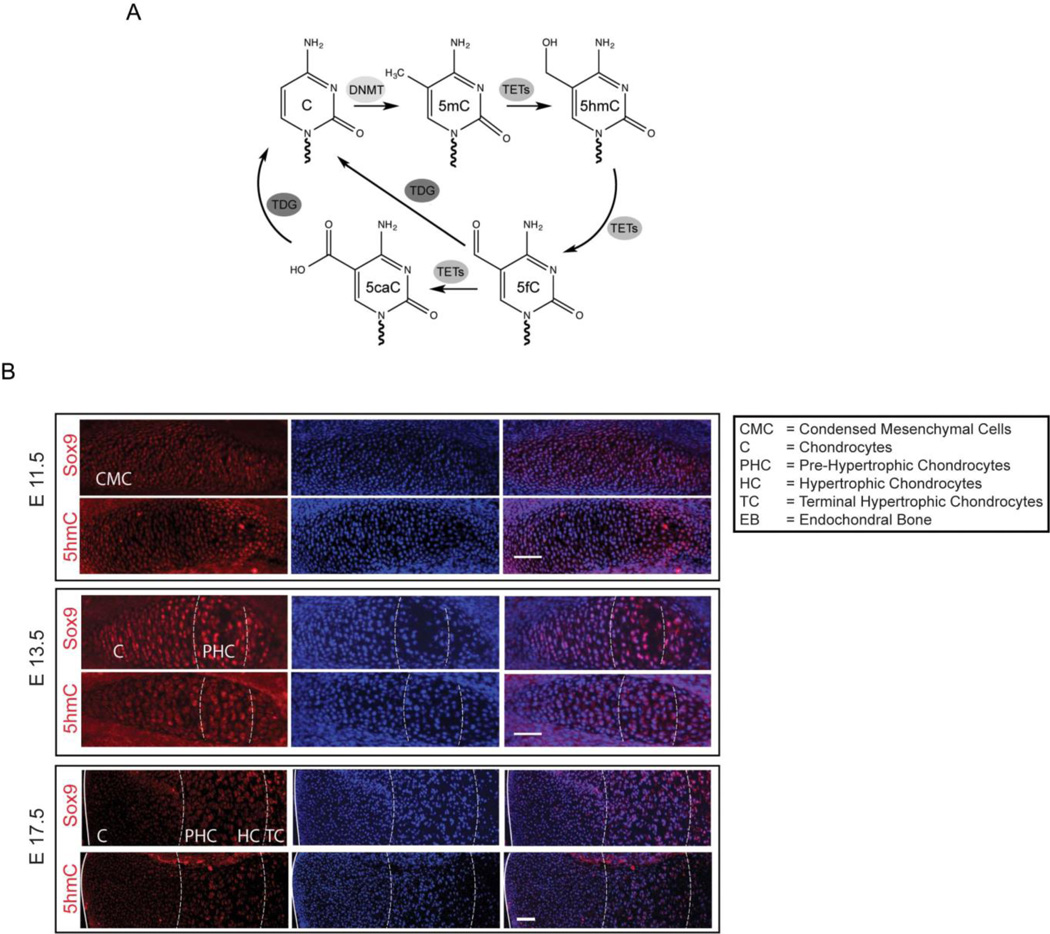
We aimed to understand DNA methylation, hydroxymethylation and demethylation dynamics mediated by the TET proteins (Figure 1A) during chondrogenic differentiation. Towards this aim, we examined global 5hmC levels during embryonic limb development (Figure 1B). In the growth plate, chondrocyte differentiation leads to the formation of a cartilaginous template that is eventually replaced by bone. A cartoon representation of the chondrocyte condensation, maturation and eventual hypertrophy during this process is depicted in Supplemental Figure 1. Mice were time-mated and sacrificed at embryonic days (E) 11.5, 13.5 and 17.5 post-coitus to study the different stages of chondrocyte differentiation. 5hmC levels and expression of the chondrogenic marker, SOX9, were analyzed in the chondrocytes of the tibial embryonic anlagen at E11.5 and growth plate at E13.5 and E17.5 using specific antibodies. Dynamic changes in 5hmC levels were observed in chondrocytes over the entire process of chondrogenic differentiation and hypertrophy (Figure 1B). SOX9 marks the early mesenchymal cells as well as chondroprogenitors and mature chondrocytes. 5hmC levels appeared to increase from E11.5 to E13.5 globally and become enriched in the pre-hypertrophic chondrocytes (PHC). A decrease in global 5hmC levels was then observed in the hypertrophic chondrocytes (HC) in the later stages of growth plate development (E17.5) (Figure 1B). These in vivo results suggested that the global changes in 5hmC likely play a regulatory role in shaping the gene expression patterns in the different stages of chondrogenic differentiation.
Figure 1. Distinct changes in global 5-hydroxymethylcytosine (5hmC) are associated with different stages of chondrogenic differentiation during embryonic limb development.
Also see Supplemental Figure 1.
A. Scheme demonstrating the different pathways and enzyme families responsible for 5hmC generation and turnover. DNA methyltransferases (DNMT) methylate cytosine residues at the C-5 carbon to produce 5-methylcytosine (5mC). DNMT1 is responsible for the maintenance of cytosine methylation marks during cell division, whereas DNMT3A and 3B establish de novo cytosine methylation. The TET family of enzymes (TETs) including TET 1, 2 and 3 convert 5mC to 5-hydroxymethylcytosines (5hmC) and further oxidized products 5-formylcytosines (5fC) and 5-carboxylcyosines (5caC) that can be acted upon by the Base-Excision Repair (BER) glycosylase, TDG.
B. Immunostaining of the developing mouse tibial anlagen at embryonic days E11.5 (days post coitus), and growth plate at E13.5 and E17.5 with antibodies specific to 5hmC and Sox9 (red). Nuclei (blue) are counterstained with DAPI. Scale bar = 50µm.
Distinct temporal patterns of global 5hmC acquisition during chondrogenic differentiation
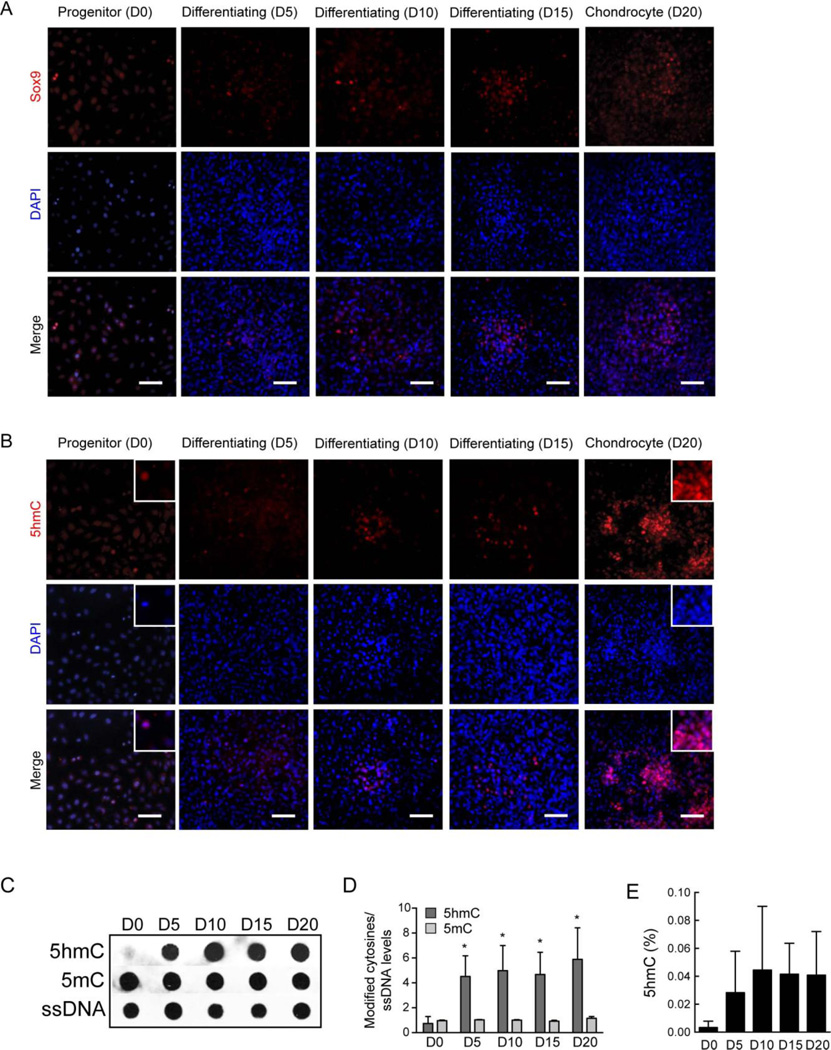
It was challenging to isolate sufficient numbers of cells in homogenous populations of the different stages of chondrocytes in vivo for detailed molecular analyses of 5hmC distribution and function. To overcome this limitation, we utilized a widely used in vitro chondroprogenitor model system that is known to faithfully recapitulate the various stages of in vivo chondrogenic differentiation (23, 24). The ATDC5 chondroprogenitors were derived from a murine teratocarcinoma stem cell line (23) and have the advantages of homogenous and reproducible differentiation towards a chondrocyte fate in sufficient number of cells making detailed molecular analyses possible. Differentiation can be induced synchronously upon addition of Insulin-Transferrin and Selenium (ITS); the chondroprogenitors (day 0) differentiate to fully functional chondrocytes (day 20) that organize themselves in cartilaginous nodules secreting glycosaminoglycans (GAGs) and an extracellular matrix characteristic of chondrocytes (Supplemental Figure 2A). Secreted GAGs were quantified both through the Alcian blue staining and an independent biochemical assay and showed a characteristic increase with chondrogenic differentiation (Supplemental Figure 2B and Supplemental Figure 3A). The differentiated chondrocytes exhibit characteristic gene expression profiles for key chondrogenic markers including Sox5, Sox6, Sox9, type II collagen (Col2a1) and Aggrecan (Acan) (Supplemental Figure 2C, D). Immunostaining of ATDC5 cells over the course of chondrogenic differentiation from chondroprogenitor cells (day 0), through differentiation (days 5, 10 and 15) to differentiated chondrocytes (day 20), showed an increase in SOX9 expression in the mature chondrocytes forming the cartilaginous nodules although a lower level of SOX9 was detected in the chondroprogenitor cells at day 0 as well (Figure 2A). Similar to the embryonic stages E11.5 to E13.5 in mice, we observed a dramatic global increase in 5hmC staining over the course of chondrogenic differentiation in the ATDC5 cells (Figure 2B). 5hmC staining was nuclear and intense staining was observed in the chondrocytes forming the cartilaginous nodules. This in vitro chondrogenic differentiation model therefore mimicked the in vivo scenario and was therefore ideal to further understand the timing and consequences of the global changes in DNA hydroxymethylation and demethylation during differentiation along the chondrogenic lineage.
Figure 2. Chondroprogenitor differentiation in vitro is accompanied by an increase in global 5hmC levels.
Also see Supplemental Figures 2 and 3.
A. Immunostaining of ATDC5 cells over the course of chondrogenesis from progenitor cells (D0 = day 0) to mature chondrocyte (D20 = day 20) with an antibody specific to Sox9 (red). Nuclei (blue) are counterstained with DAPI; merge (violet) is shown in the bottom panel. Scale bar = 30µm.
B. Immunostaining of ATDC5 cells over the course of chondrogenesis from progenitor cells (D0 = day 0) to mature chondrocyte (D20 = day 20) with an antibody specific to 5hmC (red). Nuclei (blue) are counterstained with DAPI; merge (violet) is shown in the bottom panel. Scale bar = 30µm. Insets show higher magnification of selected areas.
C. Representative immunoblot of 5hmC and 5mC levels during chondrogenic differentiation of ATDC5 cells. DNA isolated from cells undergoing differentiation at the indicated time points, was probed with antibodies specific to 5hmC, 5mC and single stranded DNA (ssDNA, as a control for loading).
D. Quantification of the modified cytosine levels as represented in the immunoblot in B, normalized to ssDNA. Data represented as mean ± SD from three independent biological replicates (n = 3). * denotes p < 0.05 compared to the control group at day 0 (D0).
E. Quantification of global 5hmC during percentage chondrogenic differentiation by ELISA. Data represented as mean ±SD (n = 2).
As a next step, we quantitated the 5mC and 5hmC changes during chondrogenic differentiation. Using antibodies specific for 5mC and 5hmC in an immunoblot assay, we assessed the relative levels of these modified cytosines in chondrocyte DNA at days 0, 5, 10, 15 and 20 during chondrogenesis. 5hmC levels showed an increase early in the differentiation process (between day 0 and day 5), which continued to increase over the differentiation process and peaked in the fully differentiated chondrocytes (day 20) (Figure 2C, D and Supplemental Figure 3B, C, D). An ELISA based assay using a different antibody specific to 5hmC quantified and confirmed the increase in 5hmC levels during chondrogenic differentiation (Figure 2E). The percentage of 5hmC increased from 0.003% in chondroprogenitor cells to 0.04% in differentiated chondrocytes.
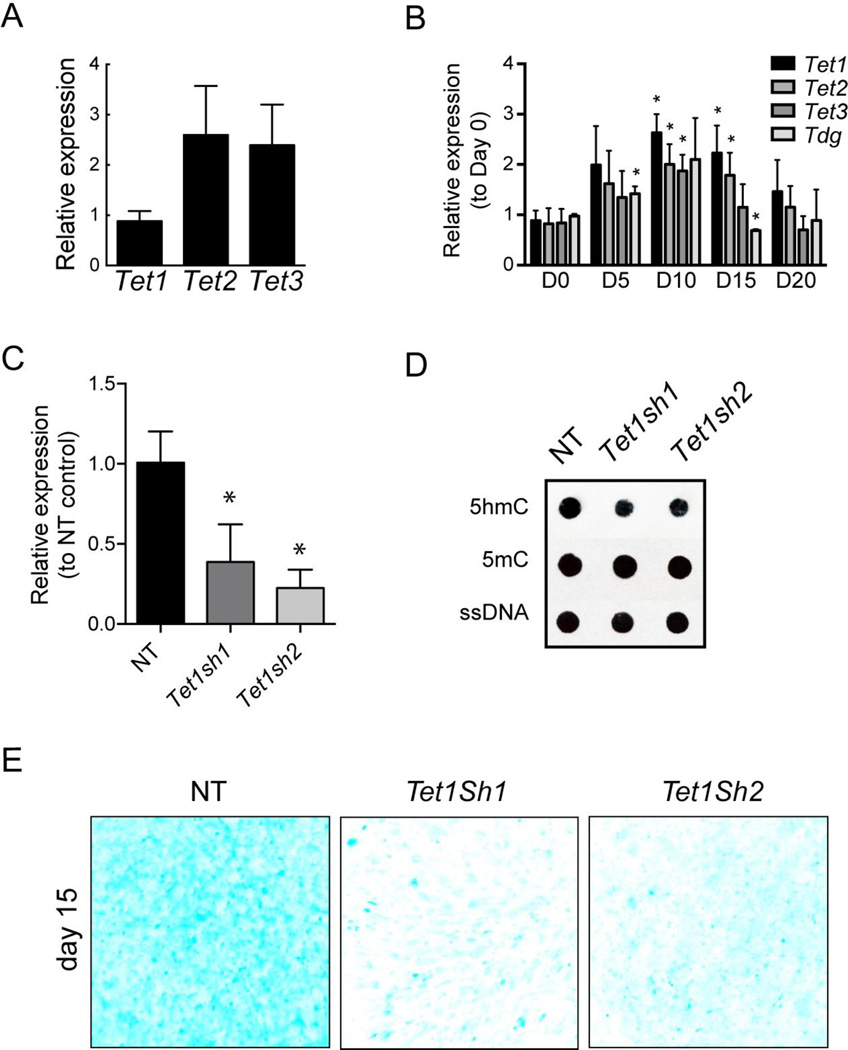
Next, we tested gene expression changes for the DNA demethylation regulators (Figure 1A) during chondrogenic differentiation using quantitative PCR. Of the three TET enzymes, Tet1, 2 and 3, Tet1 had lower mRNA expression than Tet2 and 3 in the chondroprogenitor cells (day 0) (Figure 3A). When compared to gene expression in chondroprogenitors (day 0), all the three Tet family members, as well as the base-excision repair enzyme, Tdg, showed an increase early in differentiation (Figure 3B), consistent with a role for 5hmC and DNA demethylation during the process. The expression of all these regulators peaked around day 10 after which a gradual decline was observed (Figure 3B).
Figure 3. TET1 loss of function leads to the decrease in 5hmC levels and impairment in chondrogenic differentiation.
A. Relative gene expression levels of Tet1, 2 and 3 in the chondroprogenitor cells. Data represented as mean ±SD from three independent biological replicates (n = 3).
B. Gene expression analysis for the Tet family members and Tdg during chondrogenic differentiation of ATDC5 cells. Data represented as mean ±SD from three independent biological replicates (n = 3). * denotes p < 0.05 compared to the control group at day 0 (D0).
C. Relative gene expression levels of Tet1 in ATDC5 cells differentiated after Tet1 knockdown with two different shRNA (Tet1Sh1, Tet1Sh2). Data represented as mean ±SD from three independent biological replicates (n = 3). * denotes p < 0.05 compared to the Non-Target (NT) control group.
D. Representative immunoblot of 5hmC and 5mC in ATDC5 cells differentiated in the presence (NT) or absence of TET1 (Tet1Sh1, Tet1Sh2). DNA isolated from cells at Day 15 of differentiation, was probed with antibodies specific to 5hmC, 5mC and single stranded DNA (ssDNA, as a control for loading).
E. Glycosaminoglycan (GAG) staining of cells with Alcian blue of ATDC5 chondrocytes differentiated in the presence (NT) or absence (Tet1Sh1, Tet1Sh2) of TET1 at day 15. Scale bar = 5mm.
In order to test whether the increase in 5hmC levels had a functional role in chondrogenic differentiation, we performed loss of function studies using an acute Tet1 knockdown. Two distinct shRNAs were used that caused a Tet1 downregulation of 65% (Tet1sh1) and 80% (Tet1sh2) respectively in ATDC5 chondroprogenitors (Figure 3C). The ATDC5 chondroprogenitor cells were transduced with a control non-target shRNA, Tet1 sh1 or Tet1 sh2, selected for the transduced cells by treatment with puromycin and then differentiated as previously described for 15 days. The knockdown was effective as the global 5hmC levels failed to increase to the levels of the control (non-target shRNA) after 15 days of differentiation in the ATDC5 cells transduced with the Tet1 shRNAs (Figure 3D). The 5mC levels were however similar in the ATDC5 cells transduced with non-target, Tet1sh1 and Tet1sh2 after differentiation for 15 days (Figure 3D). Remarkably, loss of TET1 and decreased 5hmC levels led to a clear impairment of chondrogenic differentiation as observed by a reduction in cartilaginous nodule formation and staining of GAG by Alcian blue upon loss of TET1 (Figure 3E). These observations strongly suggest that the increase in 5hmC levels is critical for chondrogenic differentiation.
Stable 5hmC is associated with chondrogenic genes and enriched in gene bodies
In order to understand the functional effect of the 5hmC changes, we mapped the global distribution of 5hmC over the course of chondrogenic differentiation, examining the chondroprogenitors (day 0 of the ATDC5 differentiation time course) and the differentiated chondrocytes (after 20 days of differentiation) to examine the ‘stable’ 5hmC epigenome in chondrocytes. Following chemical conjugation and affinity purification of 5hmC-enriched sequences from chondroprogenitor and chondrocyte DNA as described earlier (25), next generation sequencing (hMe-seq) was used to determine the exact location and distribution of 5hmC in chondroprogenitor cells (day 0), and in fully differentiated chondrocytes (day 20). Upon performing hMe-seq on biological replicates of differentiated chondrocytes from two independent experiments, we observed a high degree of correlation between them with an r-value of ≥ 0.83. Quality checks showed that the hMe-seq sequencing reads were of good quality and of adequate depth (Supplemental Figure 4A and B). A 5hmC gain during chondrogenic differentiation was apparent as a total of 1050 and 56698 5hmC-peaks were identified by merging the reads from the biological replicates in the progenitor (day 0) and the differentiated chondrocytes (day 20) respectively, using the MACS peak calling software (26) (Supplemental Tables 1–2).
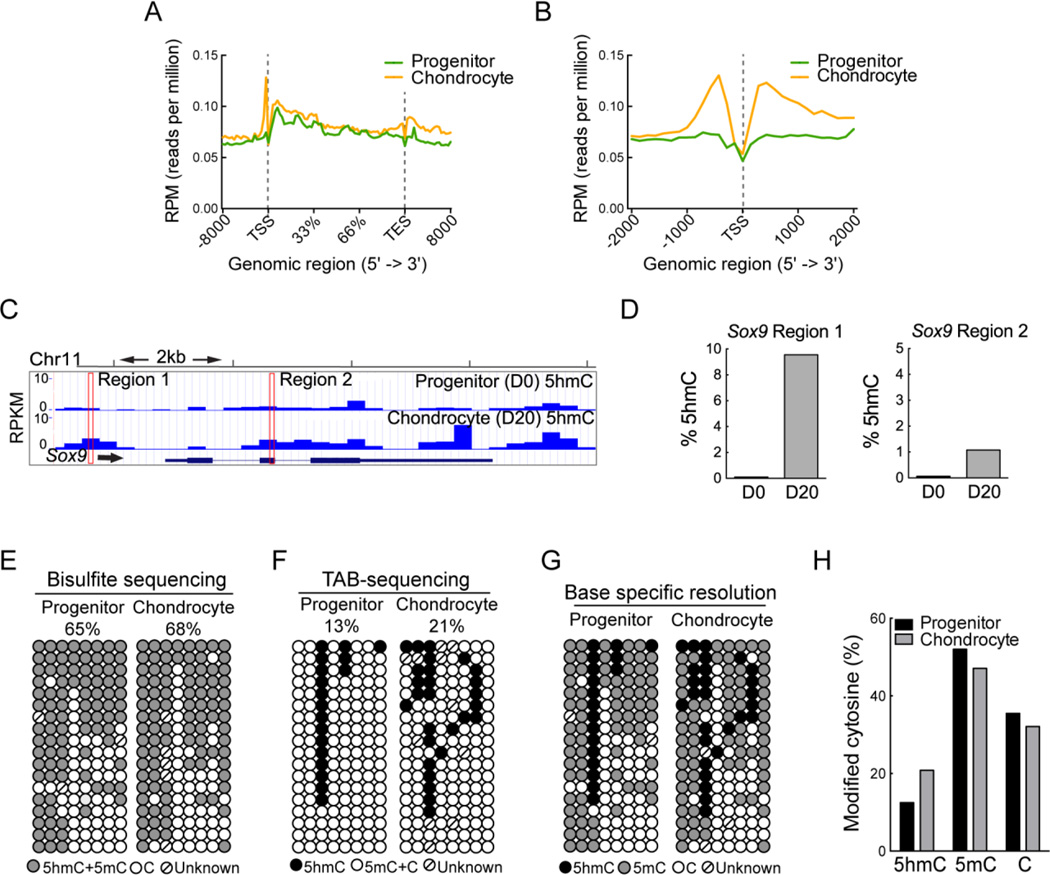
5hmC distribution in both progenitor cells and chondrocytes (Figure 4A, B) was in good agreement with previous reports of 5hmC distribution in ESC, neurons and blood lineages (10, 11, 14, 25, 27, 28). High levels of 5hmC were observed in gene bodies and in the promoters preceding the TSS while low levels of 5hmC were observed around the TSS in progenitors and chondrocytes (Figure 4B). Increased 5hmC over the course of chondrogenic differentiation was associated with many key chondrogenic genes including Sox9, 5 and 6, as well as Col2a1 (Figure 4C and Supplemental Figures 4C, D, E), whereas minimal changes were observed in the housekeeping genes Hprt (Hypoxanthine guanine phosphoribosyl transferase) and Actb (Beta-actin) (data not shown). We validated the differential 5hmC levels in progenitor and differentiated chondrocytes at these key chondrogenic genes identified by hMe-seq data, utilizing an independent locus-specific technique based on glucosylation and restriction enzyme digestion that can accurately distinguish 5mC from 5hmC (19, 21, 22). Increases in 5hmC were observed in both the promoter region and in gene body within the Sox9 gene (Figure 4C). Following this method, we tested a specific CCGG site within two peaks identified by hMe-seq in the Sox9 gene, one in promoter and another intragenic. Increased 5hmC levels were verified at both the 5hmC peaks in the Sox9 gene in differentiated chondrocytes (Figure 4D). In addition, two 5hmC peaks identified in Sox5, one peak in Sox6 and three peaks in Col2a1 were also validated by examining CCGG sites therefore validating the hMe-seq data (Supplemental Figures 4C, D and E).
Figure 4. Mapping global 5hmC distribution during chondrogenic differentiation using hMe-seq.
Also see Supplemental Figure 4, and Supplemental Tables 1–3.
A, B. Composite profiles of the 5hmC distribution during chondrogenesis averaged over the gene body (A) and the transcriptional start site (TSS) (B), respectively.
C. Snapshot of the 5hmC profile over the course of chondrogenesis in a representative genomic region encompassing the chondrogenic gene, Sox9. The outlined regions were used for validation in D.
D. Validation of the 5hmC peaks in the progenitor (day 0) and chondrocyte (day 20) in the Sox9 gene identified by hMe-seq using a restriction digest based approach (See Materials and Methods).
E. Bisulfite sequencing analysis of the region upstream of the Sox9 TSS (outlined in A). Gray circles indicate unconverted cytosines (methylated or hydroxymethylated cytosines); open circles represent unmethylated CpGs and the CpGs with undefined methylation status are marked as unknown.
F. TET-assisted bisulfite (TAB) sequencing for the same region upstream of the Sox9 TSS. Black circles indicate hydroxymethylated CpGs; open circles indicate methylated or unmethylated CpGs and the CpGs with undefined methylation status are marked as unknown.
G. Combined data from the bisulfite and TAB-sequencing for single base resolution of the methylation status of the CpGs in the Sox9 region analyzed. Black circles indicate hydroxymethylated CpGs; gray circles indicate methylated CpGs and open circles indicate unmethylated CpGs. CpGs with undefined methylation status are marked as unknown.
H. Quantification of the percentages of modified cytosines in the progenitor and mature chondrocytes based on D.
In an independent validation, we additionally analyzed the exact 5mC, 5hmC and C distribution at base level resolution in the Sox9 promoter by utilizing TET-Assisted Bisulfite sequencing (TAB-seq) (29). While regular bisulfite sequencing cannot distinguish between 5mC and 5hmC, TAB-seq identifies only 5hmC, hence a combination of both these techniques makes it possible to map 5mC, 5hmC and C in a particular region. Regular bisulfite sequencing showed that the total modified cytosines increased from 65 to 68% from the progenitors to fully differentiated chondrocytes (Figure 4E). 5hmC was detected at 13% of total CpG sites in the promoter of the progenitors and at 21% of sites in the chondrocyte, indicating a stable accumulation of 5hmC over the course of differentiation (Figure 4F). When we combined this with the bisulfite sequencing data, we were able to generate a map of this region with base specific resolution of 5hmC, 5mC and C (Figure 4G). 5mC levels were 52% in the progenitors and 47% in the fully differentiated chondrocytes, showing minimal DNA demethylation in the Sox9 promoter (Figure 4H) in this region.
5hmC gain is associated with cartilage development and differentiation pathways
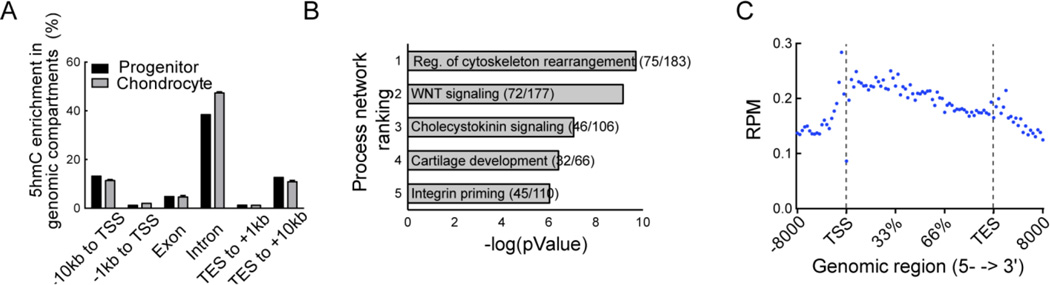
Increases in 5hmC accumulation during differentiation were observed in gene promoters and in gene bodies, with a 1.3-fold increase seen within 1kb of the transcriptional start site (TSS) and in intronic regions (Figure 5A). Next, we analyzed the genes associated with the top 25% of peaks with a 10-fold or greater increase in 5hmC levels in the chondrocyte as compared to the progenitor and identified 4508 genes. Meta-analysis of these genes that acquired 5hmC during differentiation identified pathways essential to cartilage differentiation and development, including Wnt signaling and cytoskeletal rearrangement (Figure 5B). The extremely strong statistical significance (p < 10−8) for these pathways suggests that 5hmC acquisition is regulatory in these pathways critical to chondrogenic differentiation. When we examined the top 25% of genes with a 10-fold gain in 5hmC, the distribution was clearly associated with regions before the TSS and intragenic regions (Figure 5C); a concurrent 10-fold 5hmC loss was minimal, being associated with only 88 genes (Supplemental Figures 5A and B). In these genes, the most enriched pathways consisted of cellular developmental processes, regulation of signaling and cellular differentiation.
Figure 5. Stable 5hmC is enriched in intergenic regions in mature chondrocytes.
Also see Supplemental Figure 5.
A. Proportion of 5hmC in individual genomic compartments in the progenitor and mature chondrocytes respectively. 5hmC peaks observed in the particular compartment are depicted as a percentage of the total 5hmC peaks.
B. The top five pathways identified by network analysis of the top 25% of the subset of genes with a ten-fold or greater 5hmC gain in the mature chondrocytes as compared to progenitor cells. Pathways were ranked by the number of pathway genes with high 5hmC as a ratio of the total number of genes in the pathway (shown in brackets) and by corresponding pValue. Data analysis was performed using Metacore (http://thomsonreuters.com/metacore/).
C. Composite profile of the 5hmC distribution pattern in the top 25% of the subset of genes with a ten-fold or greater 5hmC gain in the mature chondrocytes as compared to progenitor cells.
High 5hmC in gene bodies is associated with gene activation during chondrogenic differentiation
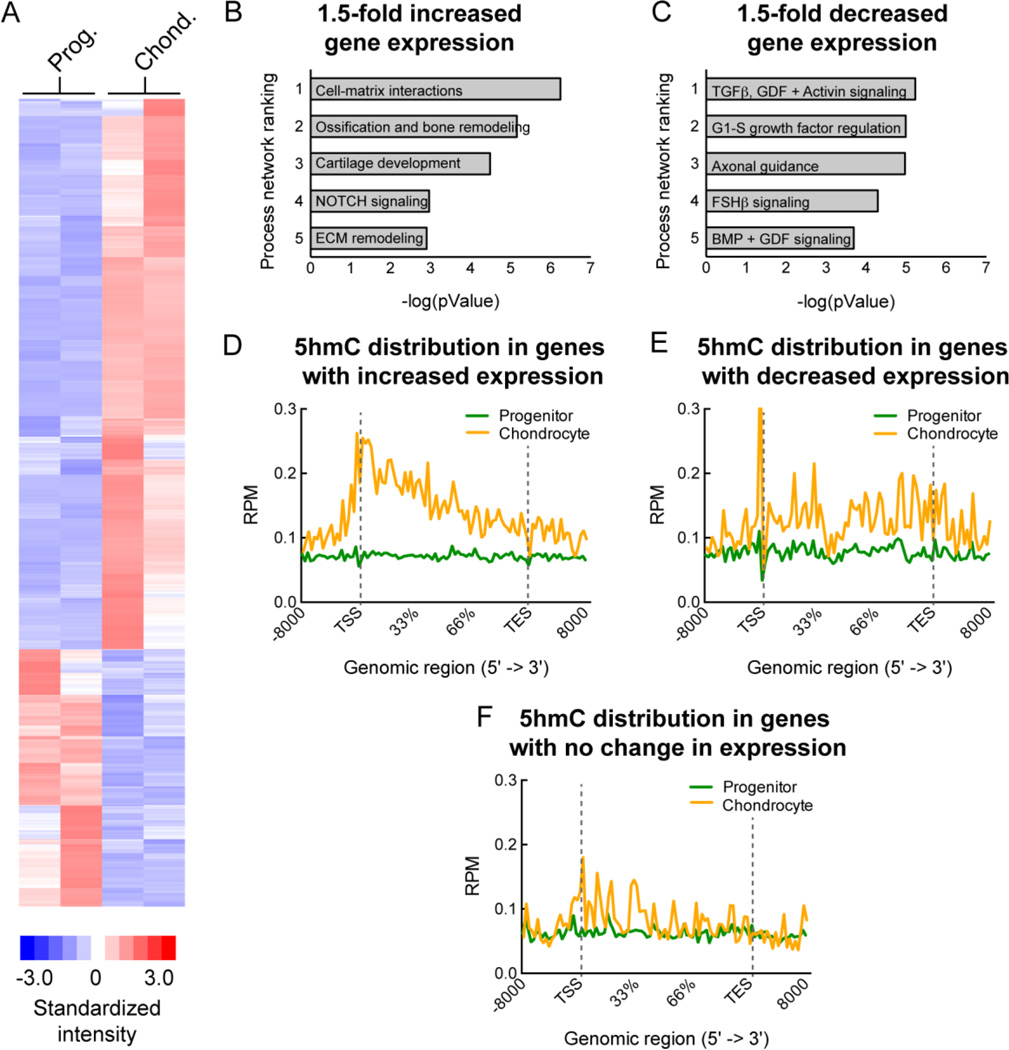
A positive correlation has been observed between 5hmC enrichment in gene bodies and high gene expression in various cell types especially in neurons (14). To understand the relationship between ‘stable’ 5hmC acquisition and gene expression changes during chondrogenic differentiation, we performed global gene expression analyses on progenitor cells and differentiated chondrocytes using exon microarrays. Two biological replicates from independent differentiation experiments showed a good correlation (with r values of ∼ 0.6) (Supplemental Figure 6) and the gene expression changes between the progenitor and differentiated chondrocytes are represented as a heat map (Figure 6A). To assess the distribution of 5hmC in activated and repressed genes, genes were ranked according to fold change in transcript expression in mature chondrocytes compared to the progenitor chondrocytes. Enrichment analysis of the genes with a 1.5-fold or greater increase in expression during chondrogenesis identified similar cartilage development pathways that were identified for genes showing enriched 5hmC (Figure 6B). Other pathways related to limb development were also identified, including ossification and bone remodeling as well as extracellular matrix (ECM) remodeling (Figure 6B). Enrichment analysis of the genes with a 1.5-fold or greater decrease in expression identified signaling pathways known to function in early mesenchymal stem cell differentiation (TGFβ, GDF, Activin A signal transduction pathway) towards adipogenic and osteogenic lineages (30) and other non-chondrogenic lineages (Axonal guidance) (Figure 6C). Composite profiles of the 5hmC patterns in the progenitor and differentiated chondrocytes for the genes with a 1.5-fold or greater increase in expression showed a notable increase in 5hmC levels in the gene bodies, both in introns and exons, in the chondrocytes compared to the progenitors. In addition, an increase in the regulatory regions (including promoters) before the TSS and after the TES was also observed (Figure 6D). The repressed genes on the other hand demonstrated a much smaller difference in 5hmC distribution in the gene body between the chondrocytes and progenitors (Figure 6E), as did the genes that showed no change in expression (Figure 6F). Stable acquisition of 5hmC in gene bodies is therefore associated with activation of gene expression in differentiated chondrocytes. A list of activated genes (1.5X increase) with high 5hmC in the differentiated chondrocytes is provided in Supplemental Table 3.
Figure 6. Stable 5hmC gain is associated with gene activation during chondrogenic differentiation.
Also see Supplemental Figures 6 and 7.
A. A heatmap of global gene expression changes between the progenitor cells (Prog.) and differentiated chondrocytes (Chond.) for two independent biological replicates. Regions of red represent higher than mean expression while regions of blue indicate lower than mean expression.
B. Enrichment analysis of the genes with a 1.5-fold or greater increase in expression during chondrogenesis (the most activated genes between the progenitor and differentiated chondrocytes). Corresponding pValues are shown. Data analysis was performed using Metacore (http://thomsonreuters.com/metacore/).
C. Enrichment analysis of the genes with a 1.5-fold or greater decrease in expression during chondrogenesis (the most repressed genes between the progenitor and differentiated chondrocytes). Corresponding pValues are shown. Data analysis was performed using Metacore (http://thomsonreuters.com/metacore/).
D. Composite profile of the 5hmC patterns in the progenitor and differentiated chondrocytes for the activated genes during chondrogenic differentiation.
E. Composite profile of the 5hmC patterns in the progenitor and differentiated chondrocytes for the repressed genes during chondrogenic differentiation.
F. Composite profile of the 5hmC patterns in the progenitor and differentiated chondrocytes for the genes with no change in expression during chondrogenic differentiation.
To further investigate the corollary i.e. whether 5hmC enrichment can predict gene activation, we assessed the gene expression levels for the genes associated with the top 25% of peaks with 10-fold or greater increases in 5hmC. Of the 4508 genes associated with high 5hmC we were able to extract gene expression data from the microarray for 3276 genes. Of these 3276 genes, only 8% had 1.5-fold or greater increases in gene expression (see Supplemental Table 3 for the full list of activated genes), 2% had 1.5-fold or greater decreases in gene expression while 90% showed no change in gene expression levels (Supplemental Figure 7A). When we examined the distribution of 5hmC in the subsets of genes with increased, decreased and no change in gene expression over the course of chondrogenesis, a distinct 5hmC accumulation prior to the TSS and throughout the gene body was observed for activated genes as observed beforehand upon examining the 5hmC distribution in activated genes (Supplemental Figure 7B). Conversely, the 5hmC distribution in genes where expression was unchanged or decreased did not show accumulation at these specific genomic locations (Supplementary Figure 7C and D) highlighting that the specific location and context of 5hmC gain is critical for its effect on gene expression.
Discussion
The role of DNA demethylation and in particular, 5hmC, in stem cell differentiation and development has been a subject of intense investigation in the last few years, with critical roles of 5hmC emerging in both ESC and neuronal tissues (12, 14, 27). In this study, we demonstrate that an increase in 5hmC levels accompanies chondrogenic differentiation both in vivo and in vitro. A loss of TET1 that can catalyze oxidation of 5mC to 5hmC leads to both decreased 5hmC levels and an impairment in chondrogenic differentiation. Our data therefore demonstrate a functional role for DNA hydroxymethylation in chondrogenic differentiation highlighting the significance of DNA methylation and its modifications in skeletal development.
DNA methylation has long been associated with gene silencing with the corollary that DNA demethylation can lead to gene activation. Since the TET proteins can catalyze the oxidation of 5mC to 5hmC, leading to either a stable 5hmC gain or further oxidation to 5fC and 5caC and eventual DNA demethylation, it has been questioned whether the stable 5hmC has any direct effects on gene expression. Analyses of stable 5hmC in activated and repressed genes during chondrogenic differentiation clearly showed that activated, but not repressed genes, acquired 5hmC near the TSS and more prominently in the gene bodies. 5hmC acquisition in the intragenic regions was specific to the genes activated during chondrogenic differentiation demonstrating a strong positive effect on transcription. 5hmC accumulation was associated with multiple key chondrogenic lineage factors including the chondrogenic lineage-specific factors, Sox9, 5 and 6, Runx2 and Col2a1. Therefore, 5hmC acquisition appears to play a major regulatory role in cartilage differentiation and development. A similar association of high 5hmC with gene activation has also been observed during neurogenesis (14) and in erythropoiesis (28). 5hmC and TET proteins have been implicated in adult neurogenesis (12, 14) and cognitive functions like spatial learning and memory (31). Similarly, we have recently demonstrated that 5hmC levels and TET1 function are dysregulated in the human cartilage in age-associated Osteoarthritis, suggesting a role for these pathways in adult cartilage function as well (19).
It should be noted that only 8% of the genes that gained substantial 5hmC were activated. These activated genes however showed a broad enrichment of 5hmC near the TSS and in gene bodies immediately following the TSS. In contrast, 90% of the genes that gained 5hmC but did not change in expression showed a random distribution of 5hmC in shorter and localized peaks. These results therefore suggest that a specific distribution of 5hmC is associated with gene activation. In addition, since only high 5hmC does not predict gene activation, other chromatin changes are likely required in conjunction with 5hmC gain for gene activation, as has been noted previously as well (11, 14). It also remains possible that DNA demethylation and 5hmC acquisition act as a ‘dual’ activating mark for gene expression.
Locus-specific analyses of 5hmC and 5mC distribution within regions of the Sox gene trio (Sox 9, 5 and 6) that are the major chondrogenic regulators, demonstrated 5hmC gain at gene bodies for all the three genes. Previous studies of DNA methylation changes in the Sox trio and other chondrogenic regulators during chondrogenesis had utilized techniques like bisulfite sequencing that did not distinguish between 5hmC and 5mC (16) and hence concluded that there were minimal changes in DNA methylation upon chondrogenesis. Our data highlight that although bisulfite sequencing within regions of Sox9, 5 and 6 studied does not show any significant DNA demethylation between the progenitor cells and differentiated chondrocytes, a significant gain of 5hmC is observed in the Sox9 promoter and gene body utilizing both base-resolution TAB-sequencing (that can accurately distinguish between 5mC and 5hmC) and global sequencing data. These findings therefore highlight a potential role for 5hmC as an epigenetic mark associated with various chondrocyte lineage-specific genes including the Sox trio during chondrogenic differentiation.
Gene expression changes in all the Tet family members, Tet 1, 2 and 3, demonstrate a similar pattern during chondrogenic differentiation, with an initial increase followed by a decline. While our data shows a functional role for TET1, it is likely that other TET proteins also contribute to the 5hmC dynamics during chondrogenic differentiation. It is an open question why and how TET-mediated oxidation stalls at particular cytosines to generate ‘stable’ 5hmC while in other cases 5hmC is further oxidized to 5fC or 5caC. It is also unclear whether there are inherent differences in the ability of the TET 1, 2 or 3 proteins in vivo for generation of 5hmC or its oxidized derivatives. Future loss of function studies can be employed to utilize chondrogenic differentiation as a useful model system to answer some of these broader questions.
In conclusion, our study provides a comprehensive resource of genome-wide DNA hydroxymethylation changes in regulating gene expression during chondrogenic differentiation, thereby demonstrating a potential regulatory role for 5hmC. This study highlights the similarities between 5hmC function in progenitor differentiation during chondrogenesis and neurogenesis. Future studies in other somatic tissues and in adult stem cells will help in consolidating and clarifying the precise role of 5hmC and its oxidized derivatives as well as the TET-TDG regulatory network in cell fate determination and differentiation. Further analyses of the role of TET proteins and 5hmC and its oxidized derivatives during cartilage development and in adult cartilage function will lead to a greater fundamental understanding of epigenetic regulation of skeletal tissues.
Supplementary Material
Acknowledgments
We are grateful to Prof. R.L. Smith for the kind gift of ATDC5 chondroprogenitor cells and to Lakshmi Dhulipala for technical assistance. We would like to thank Gary Mantalas of the Stanford University School of Medicine Stem Cell Institute Genome Center for performing the Illumina HiSeq runs and Natalia Kosovilka of the Stanford University School of Medicine Protein and Nucleic Acid Facility for running the microarrays. These studies were supported by NIAMS/NIH grant R03AR066356 to NB; P.S. is supported by the Child Health Research Institute, Lucile Packard Foundation for Children’s Health, and the Stanford CTSA (grant number UL1 TR001085); Y.L. and W.W. are partially supported by NIH grant R01 HG 006018.
Footnotes
Authors’ roles: Study design: NB. Study conduct: ST, PS, MR. Data collection: ST, PS, MR. Data analysis: ST, YL, WW, NB. Data interpretation: ST, YL, PS, WW, NB. Drafting manuscript: ST, NB. Approving final version of manuscript: ST, YL, PS, WW, NB.
References
- 1.Jones PA. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Bhutani N, Burns David M, Blau Helen M. DNA Demethylation Dynamics. Cell. 2011;146(6):866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, NY) 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA Glycosylase Is Essential for Active DNA Demethylation by Linked Deamination-Base Excision Repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriaucionis S, Heintz N. The nuclear DNA base, 5-hydroxymethylcytosine is present in brain and enriched in Purkinje neurons. Science (New York, Ny) 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 11.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szulwach KE, Li X, Li Y, Song C-X, Wu H, Dai Q, et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neuroscience. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Park YK, Kang TW, Lee SH, Rhee YH, Park JL, et al. Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Human Molecular Genetics. 2013;23(3):657–667. doi: 10.1093/hmg/ddt453. [DOI] [PubMed] [Google Scholar]
- 14.Hahn Maria A, Qiu R, Wu X, Li Arthur X, Zhang H, Wang J, et al. Dynamics of 5-Hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Reports. 2013;3(2):291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54–R62. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis & Rheumatism. 2009;60(5):1416–1426. doi: 10.1002/art.24472. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S, Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis & Rheumatism. 2008;58(9):2743–2753. doi: 10.1002/art.23736. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto K, Otero M, Imagawa K, de Andres MC, Coico JM, Roach HI, et al. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1 (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. Journal of Biological Chemistry. 2013;288(14):10061–10072. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor SEB, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A global increase in 5-hydroxymethylcytosine levels marks osteoarthritic chondrocytes. Arthritis & Rheumatology. 2014;66(1):90–100. doi: 10.1002/art.38200. [DOI] [PubMed] [Google Scholar]
- 20.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, et al. Tissue-specific Distribution and Dynamic Changes of 5-Hydroxymethylcytosine in Mammalian Genomes. Journal of Biological Chemistry. 2011;286(28):24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis T, Vaisvila R. High Sensitivity 5-hydroxymethylcytosine Detection in Balb/C Brain Tissue. Journal of Visualized Experiments. 2011;48:e2661. doi: 10.3791/2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atsumi T, Miwa Y, Kimata K, Ikawa Y. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differentiation and Development. 1990;30(2):109–116. doi: 10.1016/0922-3371(90)90079-c. [DOI] [PubMed] [Google Scholar]
- 24.Yao Y, Wang Y. ATDC5: An excellent in vitro model cell line for skeletal development. Journal of Cellular Biochemistry. 2013;114(6):1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 25.Song C-X, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnology. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & Development. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madzo J, Liu H, Rodriguez A, Vasanthakumar A, Sundaravel S, Caces Donne Bennett D, et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Reports. 2014;6(1):231–244. doi: 10.1016/j.celrep.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Hon GC, Szulwach KE, Song C-X, Jin P, Ren B, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nature Protocols. 2012;7(12):2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng F, Boucher S, Koh S, Sastry KSR, Chase L, Lakshmipathy U, et al. PDGF, TGF-, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112(2):295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R-R, Cui Q-Y, Murai K, Lim Yen C, Smith Zachary D, Jin S, et al. Tet1 Regulates Adult Hippocampal Neurogenesis and Cognition. Cell Stem Cell. 2013;13(2):237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.