Abstract
Sirtuins are NAD-dependent lysine deacylases that play critical roles in cellular regulation and are implicated in human diseases. Modulators of sirtuins are needed as tools for investigating their biological functions and possible therapeutic applications. However, the discovery of sirtuin modulators is hampered by the lack of efficient sirtuin assays. Here we report an improved fluorogenic assay for SIRT1, SIRT2, and SIRT3 using a new substrate, a myristoyl peptide with a C-terminal aminocoumarin. The new assay has several advantages, including significantly lower substrate concentration needed, increased signal-to-background ratio, and improved Z′-factor. The novel assay thus will expedite high-throughput screening of SIRT1, SIRT2, and SIRT3 modulators.
Sirtuins, the silent information regulator 2 (Sir2) 1 family of histone deacetylases (HDACs) have seven isoforms (SIRT1–7) in mammals.2 In contrast to the Zn2+-dependent HDACs, sirtuins utilize nicotinamide adenine dinucleotide (NAD) as a co-substrate to perform deacetylation.3 Besides lysine deacetylase activity, sirtuins have also been discovered to exhibit other enzymatic activities. For example, SIRT5 was shown to efficiently catalyze demalonylation, desuccinylation, and deglutarylation,4-7 and SIRT6 is capable of hydrolyzing long-chain fatty acyl group on TNF α.8 By post-translationally modifying various substrate proteins, sirtuins regulate many biological pathways and exert their effects on metabolism,6, 9 genome stability,10 and longevity.11 Therefore, they are considered potential therapeutic targets for a variety of diseases, including diabetes,12 cardiac disease,13 and cancers.14
The Important biological function of sirtuins have sparked interest to develop small molecule modulators that can regulate their activity.15, 16 Certain sirtuin inhibitors showed ability to inhibit cancer cell growth17 and induce cancer cell-specific apoptosis.18 Sirtuin activators could potentially be used to treat diabetes12 and promote longevity,11 although the effects of sirtuin activators are still under debate.19-21 SIRT2-specific inhibitor shows promising effects in Parkinson's disease22 and Huntington's disease.23 Thus, developing sirtuin modulators is of great interest in both basic biological studies and therapeutic applications.
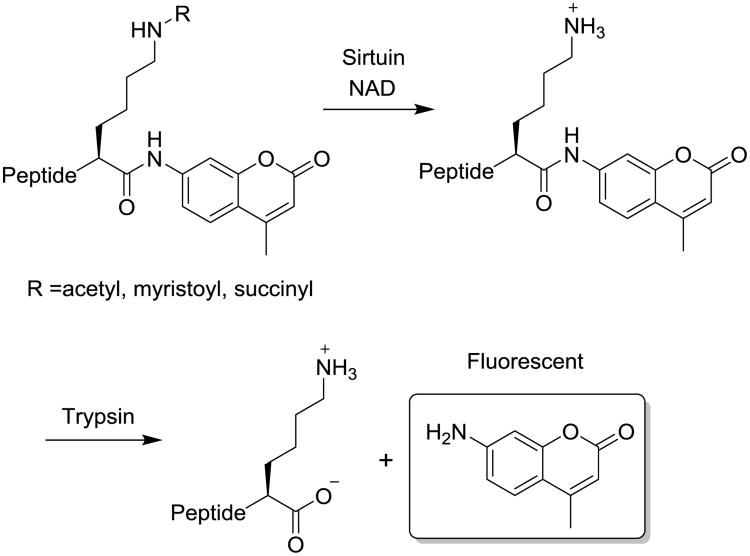
To facilitate the development of potent sirtuin modulators, many assays have been developed for the detection of sirtuin activity,24, 25 including radioisotope-labelled assays,26 high-performance liquid chromatography (HPLC)-based assays,4, 8 mass spectrometry (MS)-based assays,27, 28 and fluorescence-based assays. Fluorescence-based assays which include direct fluorescence assays,29, 30 fluorescence polarization assays,12 and fluorescence resonance energy transfer (FRET)-based assays.31-34 Fluorescence-based assays are most suitable for high-throughput screening of sirtuin modulators.11, 12 The fluorogenic substrates of direct fluorescence assays are based on a peptidic substrate of sirtuin comprising an ε-acylated lysyl moiety and a 7-amino-4-methylcoumarin moiety (AMC) at the C terminus of the peptide chain as a fluorescent tag (Figure 1). 29, 30 The acyl group can be acetyl, myristoyl, or succinyl groups depending on the target sirtuins. In the presence of the co-substrate NAD, sirtuins can remove the acyl modification from the lysine side chain, revealing the free lysine side chain. The resulting lysine peptide can then be cleaved by trypsin to release the fluorophore, allowing facile detection of sirtuin enzymatic activity by reading the fluorescence (Figure 1).
Figure 1.
Principle of two-step direct fluorescence assays of sirtuins.
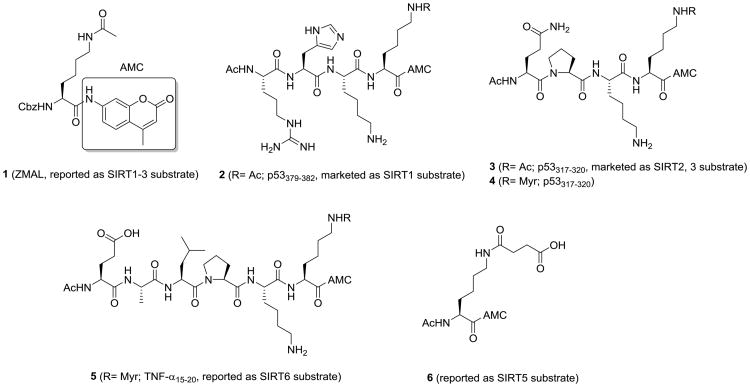
A Cbz-protected lysine with acetyl group and AMC was initially reported as a SIRT1 substrate (1, ZMAL, Figure 2),35 which is then used as the substrate for SIRT2 and SIRT3 assay.36-38 A p53-based peptide with acetyl-lysine and C-terminal AMC was developed for detecting the activity of SIRT1 (2, p53379-382, Figure 2). Later, another p53-based peptide was applied to SIRT2 and SIRT3 assays (3, p53317-320, Figure 2). In addition, a single lysine with succinyl group and AMC was reported as a SIRT5 substrate (6, Figure 2).39 Recently, SIRT1-3 and 6 were found to efficiently remove long chain fatty acyl groups from lysine residues on proteins.8, 40, 41 We further demonstrated that a myristoyl-lysine fluorogenic peptide (5, TNF- α15-20, Figure 2) is a much better substrate for detecting the activity of SIRT6.42
Figure 2.
Fluorogenic substrates for direct fluorescence assays of sirtuins.
One of the criteria for an ideal enzymatic assay is a low Km value for the substrate, which renders the use of low substrate concentrations and decreases the reagent cost.43 The Km value of SIRT2 for a myristoyl peptide (0.24 μM) is much lower than that for an acetyl peptide (19.00 μM).41 Based on this finding, we suspected that a myrsitoyl lysine peptides may be a better substrate and provide a better SIRT1, SIRT2, or SIRT3 assay.
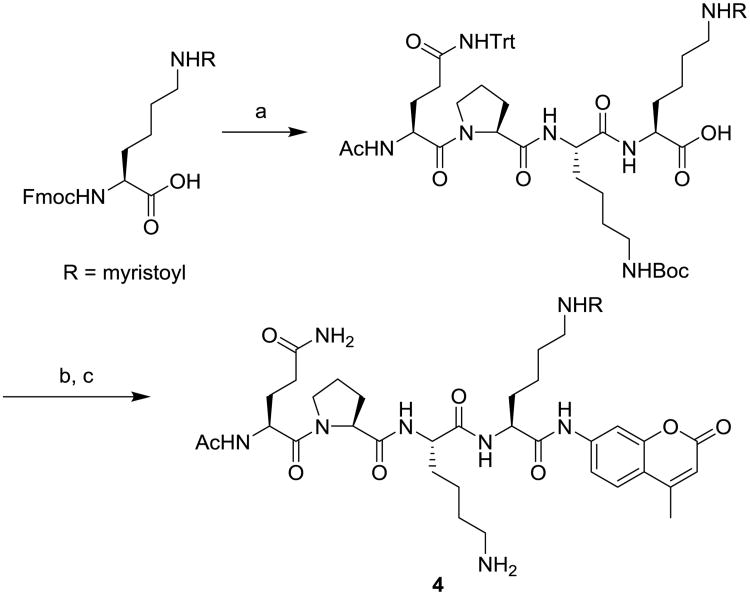
To test this, a p53317-320 peptide with myristol-lysine and C-terminal AMC (substrate 4, Figure 2) was synthesized using standard solid phase peptide synthesis, followed by AMC coupling and deprotection (Scheme 1).42 We first determined the kinetic constants of sirtuins against 4 and our previously reported SIRT6 substrate 5. Using an HPLC-based assay, we measured the initial velocity, V0, as a function of substrate concentration [S] and the data were fitted to the Michaelis–Menten equation (Figure S1) to give the Km and kcat values as shown in Table 1. The Km of myristoyl substrates 4 and 5 for SIRT1, SIRT2, and SIRT3 are much lower than that of the commercial acetyl substrate 3, suggesting that the myristoyl peptides have better affinities toward these sirtuins. The low Km of myristoyl substrates is likely due to the large hydrophobic pockets of SIRT1, 2, and 3, which can accommodate the myristoyl group41.
Scheme 1.
Synthesis of substrates 4. Reagents and conditions: (a) standard solid phase peptide synthesis; (b) ClCOOiBu, N-methylmorpholine, 7-amino-4-methyl-coumarin, DMF; (c) 1% triisopropylsilane in TFA.
Table 1.
Kinetic data for sirtuins against different fluorogenic substrates.
| SIRT1 | SIRT2 | SIRT3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Km (μM) | kcat (10−3s −1) | kcat/Km (s−1M−1) | Km (μM) | kcat (10−3s−1) | kcat/Km (s−1M−1) | Km (μM) | kcat (10−3s−1) | kcat/Km (s−1M−1) | |
| 3 | > 333a | NDa | 1.2 × 102 | > 333a,b | NDa | 18.8 | > 333a,b | NDa | 7.3 |
| 4 | 35.3 ± 4.5 | 17 ± 1.1 | 4.8 × 102 | < 0.50c | 10 ± 0.5 | > 2.0 × 104 | 3.6 ± 1.2 | 2.7 ± 0.3 | 7.5 × 102 |
| 5 | 23.8 ± 3.9 | 19 ± 2.1 | 8.0 × 102 | < 0.50c | 12 ± 0.7 | > 2.4 × 104 | 3.6 ± 1.3 | 3.5 ± 0.4 | 9.9 × 102 |
The Km and kcat could not be determined due to the linear relationship at the concentration range (5.2-333 μM) used, but kcat/Km can be obtained from the slope of the linear plot (Figure S1a).
The value is 51 μM for SIRT2 and 323 μM for SIRT3 provided in the commercial assay kit.
The Km could not be determined due to the detection limit of the HPLC assay. The lowest substrate concentration used was 0.5 μM.
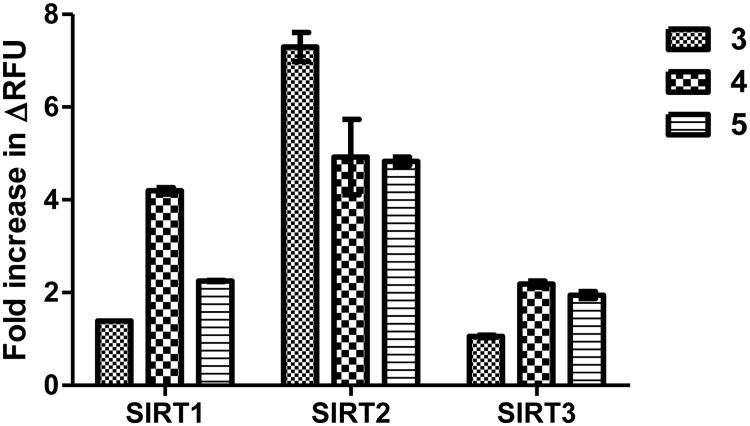
We then compared the fluorescence increase in the two-step sirtuin assay with different fluorogenic substrates (3, 4, and 5). We initially used the conditions specified in the commercial sirtuin assay kit. We incubated 125 μM of the fluorogenic substrates with 1 μM sirtuins (SIRT1, 2, and 3) and 3 mM NAD at 37 °C for 45 min, and then added the stop/developer solution containing trypsin and nicotinamide for 30 min at room temperature to release the fluorophore. For the commercial substrate 3, the increase in fluorescence was less than 10-fold in SIRT1–3 reactions, which was in agreement with previous results.39 For SIRT1 and SIRT3, substrate 4 and 5 gave slightly better fluorescence increase than substrate 3 (Figure 3). However, for SIRT2, substrate 3 gave better increase in fluorescence than substrate 4 and 5 (Figure 3), but substrate 3 also had the highest fluorescence background without addition of sirtuins (Figure S2b).
Figure 3.
Fold increase in fluorescence with different substrates at 125 μM in the two-step sirtuin assay.
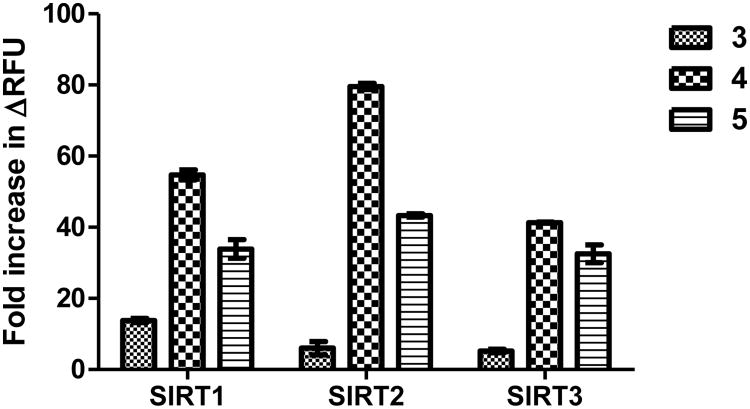
Since the Km values of SIRT2 and SIRT3 against the myristoyl substrates 4 and 5 are less than 10 μM, we then used 10 μM of different fluorogenic substrates to compare the fluorescence increase. The commercial substrate 3 still had the highest fluorescence background (Figure S2c). Substrate 4 and 5 both gave higher fluorescence after SIRT1-3 reactions than substrates 3 (Figure S2c). After dividing by the background signal, substrate 4 gave 55-, 85-, and 43-fold increase in fluorescence in SIRT1, SIRT2, and SIRT3 reactions, respectively (Figure 4). Meanwhile, the commercial substrate 3 only gave 13-, 6-, and 5-fold increase in fluorescence in SIRT1, SIRT2, and SIRT3 reactions, respectively. Substrate 5 also gave higher fluorescence increases in the assays than 3, but compared to substrate 4 the increases were slightly smaller. The new substrate 4 not only works at lower concentration but also gives larger fold increase than the substrate 3 from the commercial assay kit.
Figure 4.
Fold increase in fluorescence with different substrates at 10 μM in the two-step sirtuin assay.
To evaluate the quality of the sirtuin assay for future high-throughput screening use, signal to background (S/B) ratio and Z′-factor were considered.44 S/B ratio represents assay sensitivity, and Z′-factor reflects dynamic range of the assay and the data variation. In the case of fluorogenic assay, fold increase in fluorescence is equivalent to S/B ratio. Although substrate 4 exhibited lower catalytic efficacy than 5 (Table 1), it gave higher S/B ratio and was therefore chosen for further optimization. We varied the concentration of substrate 4 and obtained the S/B ratios and Z′-factors (Table 2). In SIRT1, SIRT2, and SIRT3 assay, 5 μM of substrate 4 gave the best result. The S/B ratio were 75, 165, and 45, and the Z′-factors were 0.98, 0.87, and 0.92 in SIRT1, SIRT2, and SIRT3 assays, respectively. Compared to the use of 125 μM of substrate 3 in the commercial assay kit (S/B and Z′-factor), the significantly improved S/B ratio, the excellent Z′-factor, and the reduction of the working concentration of substrate 4 will better enable high-throughput screening.
Table 2.
Evaluation of the fluorogenic assay by measuring signal-to-background (S/B) ratio and Z′-factor at various substrate concentrations.
| Substrate 4 concentration (μM) | |||||
|---|---|---|---|---|---|
|
|
|||||
| 2.5 | 5 | 10 | 20 | ||
| SIRT1 | S/B | 21 | 75 | 48 | 30 |
| Z′-factor | -0.45 | 0.98 | 0.83 | 0.68 | |
|
| |||||
| SIRT2 | S/B | 60 | 165 | 80 | 45 |
| Z′-factor | 0.85 | 0.87 | 0.72 | 0.93 | |
|
| |||||
| SIRT3 | S/B | 14 | 45 | 38 | 16 |
| Z′-factor | 0.61 | 0.92 | 0.89 | 0.81 | |
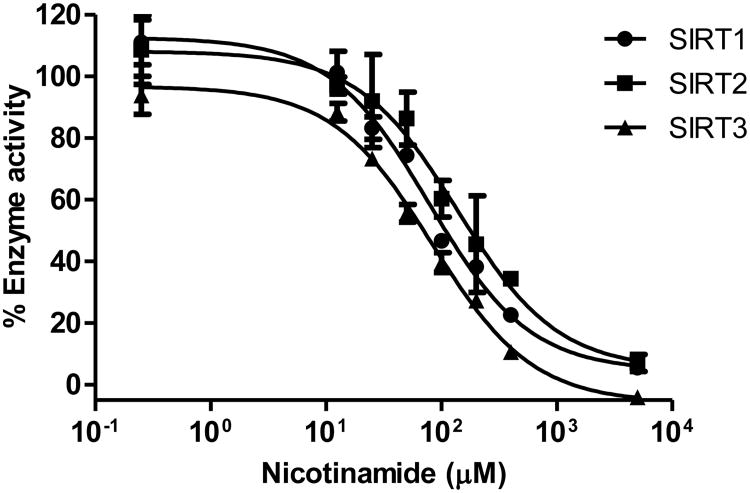
To demonstrate that the new fluorogenic substrate 4 is suitable to detect sirtuin inhibitors, we measured the dose-dependent inhibition of sirtuins by nicotinamide. The dose-response curve was obtained by measuring the fluorescence in various concentration of nicotinamide with 5 μM of substrate 4 and 1 mM of NAD (Figure 5). The IC50 values of nicotinamide for SIRT1, SIRT2, and SIRT3 were determined to be 79, 138, and 84 μM, respectively. The IC50 values were comparable to those previously reported.45, 46 Thus, the new fluorogenic assay provides reliable IC50 values and should be suitable for screening unknown inhibitors of SIRT1/2/3.
Figure 5.
Dose-dependent inhibition of sirtuins by nicotinamide. The IC50 values of nicotinamide for SIRT1, SIRT2, and SIRT3 were determined to be 79, 138, and 84 μM, respectively.
Conclusions
We have developed an improved fluorogenic assay for SIRT1, SIRT2, and SIRT3 using a new myristoyl peptide with C-terminal AMC as the substrate. The new substrate, 4, gave the much excellent S/B ratios and Z′-factors compared to the currently available commercial assay. The new substrate also dramatically decreases the substrate concentration needed, and thus will decrease reagent cost in high-throughput screening. The assay thus will help to discover new sirtuin modulators via high-throughput screening and to facilitate biological and pharmacological studies of sirtuins.
Supplementary Material
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. Proc Natl Acad Sci U S A. 2000;97:6658. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frye RA. Biochem Biophys Res Commun. 2000;273:793. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 3.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 4.Du JT, Zhou YY, Su XY, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin HN. Science. 2011;334:806. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E. Cell Metab. 2013;18:920. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park J, Chen Y, Tishkoff DX, Peng C, Tan MJ, Dai LZ, Xie ZY, Zhang Y, Zwaans BMM, Skinner ME, Lombard DB, Zhao YM. Mol Cell. 2013;50:919. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan MJ, Peng C, Anderson KA, Chhoy P, Xie ZY, Dai LZ, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu GF, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao YM. Cell Metab. 2014;19:605. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du JT, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin HN. Nature. 2013;496:110. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houtkooper RH, Pirinen E, Auwerx J. Nat Rev Mol Cell Biol. 2012;13:225. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing H, Lin H. Chem Rev. 2015;115:2350. doi: 10.1021/cr500457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Nature. 2003;425:191. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 12.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang HY, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushima S, Sadoshima J. Am J Physiol Heart Circ Physiol. 2015;309:H1375. doi: 10.1152/ajpheart.00053.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth M, Chen WY. Oncogene. 2014;33:1609. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villalba JM, Alcain FJ. BioFactors. 2012;38:349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Jing H, Lin HN. Future Med Chem. 2014;6:945–966. doi: 10.4155/fmc.14.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Cancer Res. 2006;66:4368. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 18.Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, Martinez-Chantar ML, Varela-Rey M, Rotili D, Nebbioso A, Ropero S, Montoya G, Oyarzabal J, Velasco S, Serrano M, Witt M, Villar-Garea A, Imhof A, Mato JM, Esteller M, Fraga MF. Oncogene. 2009;28:781. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 19.Borra MT, Smith BC, Denu JM. J Biol Chem. 2005;280:17187. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 20.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. J Biol Chem. 2005;280:17038. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 21.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu XY, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. J Biol Chem. 2010;285:8340. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Science. 2007;317:516. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 23.Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, Kuhn A, Xiang Z, Maxwell MM, Reeves SA, Bates GP, Neri C, Thompson LM, Marsh JL, Kazantsev AG. Proc Natl Acad Sci U S A. 2010;107:7927. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Liu T, Liao S, Li Y, Lan Y, Wang A, Wang Y, He B. Biochem Biophys Res Commun. 2015;467:459. doi: 10.1016/j.bbrc.2015.09.172. [DOI] [PubMed] [Google Scholar]
- 25.Schutkowski M, Fischer F, Roessler C, Steegborn C. Expert Opin Drug Discov. 2014;9:183. doi: 10.1517/17460441.2014.875526. [DOI] [PubMed] [Google Scholar]
- 26.Borra MT, Denu JM. Meth Enzymol. 2004;376:171. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- 27.Rye PT, Frick LE, Ozbal CC, Lamarr WA. J Biomol Screen. 2011;16:1217. doi: 10.1177/1087057111420291. [DOI] [PubMed] [Google Scholar]
- 28.Patel K, Sherrill J, Mrksich M, Scholle MD. J Biomol Screen. 2015;20:842. doi: 10.1177/1087057115588512. [DOI] [PubMed] [Google Scholar]
- 29.Wegener D, Hildmann C, Riester D, Schwienhorst A. Anal Biochem. 2003;321:202. doi: 10.1016/s0003-2697(03)00426-3. [DOI] [PubMed] [Google Scholar]
- 30.Wegener D, Wirsching F, Riester D, Schwienhorst A. Chem Biol. 2003;10:61. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 31.Marcotte PA, Richardson PR, Guo J, Barrett LW, Xu N, Gunasekera A, Glaser KB. Anal Biochem. 2004;332:90. doi: 10.1016/j.ab.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 32.Riester D, Hildmann C, Schwienhorst A, Meyer-Almes FJ. Anal Biochem. 2007;362:136. doi: 10.1016/j.ab.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Huang W, You L, Xie T, He B. Bioorg Med Chem Lett. 2015;25:1671. doi: 10.1016/j.bmcl.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, You L, Huang W, Liu J, Zhu H, He B. Eur J Med Chem. 2015;96:245. doi: 10.1016/j.ejmech.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Heitweg B, Dequiedt F, Verdin E, Jung M. Anal Biochem. 2003;319:42. doi: 10.1016/s0003-2697(03)00276-8. [DOI] [PubMed] [Google Scholar]
- 36.Di Fruscia P, Ho KK, Laohasinnarong S, Khongkow M, Kroll SHB, Islam SA, Sternberg MJE, Schmidtkunz K, Jung M, Lam EWF, Fuchter MJ. Medchemcomm. 2012;3:373. doi: 10.1039/C2MD00290F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rumpf T, Schiedel M, Karaman B, Roessler C, North BJ, Lehotzky A, Olah J, Ladwein KI, Schmidtkunz K, Gajer M, Pannek M, Steegborn C, Sinclair DA, Gerhardt S, Ovadi J, Schutkowski M, Sippl W, Einsle O, Jung M. Nat Commun. 2015;6:6263. doi: 10.1038/ncomms7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer B, Rumpf T, Scharfe M, Stolfa DA, Schmitt ML, He WJ, Verdin E, Sippl W, Jung M. ACS Med Chem Lett. 2012;3:1050. doi: 10.1021/ml3002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madsen AS, Olsen CA. J Med Chem. 2012;55:5582. doi: 10.1021/jm300526r. [DOI] [PubMed] [Google Scholar]
- 40.Feldman JL, Baeza J, Denu JM. J Biol Chem. 2013;288:31350. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Sci Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, He B, Bhargava S, Lin H. Org Biomol Chem. 2013;11:5213. doi: 10.1039/c3ob41138a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.London JW, Shaw LM, Fetterolf D, Garfinkel D. Clin Chem. 1975;21:1939. [PubMed] [Google Scholar]
- 44.Zhang JH, Chung TDY, Oldenburg KR. J Biomol Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 45.Fischer F, Gertz M, Suenkel B, Lakshminarasimhan M, Schutkowski M, Steegborn C. Plos One. 2012;7:e45098. doi: 10.1371/journal.pone.0045098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan X, Lin P, Knoll E, Chakrabarti R. Plos One. 2014;9:e107729. doi: 10.1371/journal.pone.0107729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.