Abstract
Environmental and occupational pulmonary diseases impose a substantial burden of morbidity and mortality on the global population. However, it has been long observed that only some of those who are exposed to pulmonary toxicants go on to develop disease; increasingly, it is being recognized that genetic differences may underlie some of this person-to-person variability. Studies performed throughout the globe are demonstrating important gene-by-environment interactions for diseases as diverse as chronic beryllium disease, coal workers’ pneumoconiosis, silicosis, asbestosis, bysinnosis, occupational asthma, and pollution-associated asthma. These findings have, in many instances, elucidated the pathogenesis of these highly complex diseases. At the same time, however, translation of this research into clinical practice has, for good reasons, proceeded slowly. No genetic test has yet emerged with sufficiently robust operating characteristics to be clearly useful or practicable in an occupational or environmental setting. Additionally, occupational genetic testing raises serious ethical and policy concerns. Therefore, the primary objective must remain ensuring that the workplace and the environment are safe for all.
Keywords: Genetic susceptibility, occupational disease, environmental disease, lung disease, gene-by-environment interactions
Situated at the interface of organism and nature, the lung is particularly susceptible to toxins in the environment and the workplace. Above a certain exposure threshold, all individuals will suffer deleterious consequences from such toxins. Below that threshold, however, the response varies from individual to individual. Why do some develop occupational and environmental disease, while others with apparently similar exposures remain healthy? Though additional environmental cofactors no doubt play a role, a widening body of evidence demonstrates that genetic variation is important in modifying individual susceptibility to these diseases.
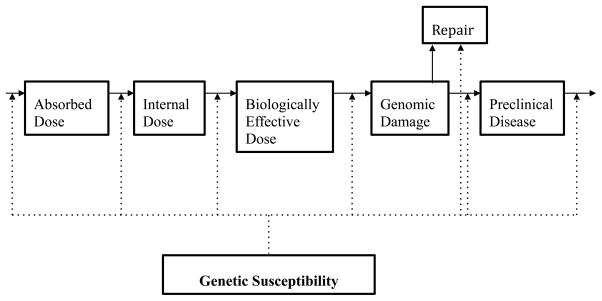
In this review, we will explore how this relationship plays out in the case of multiple environmental and occupational lung diseases, including chronic beryllium disease, other pneumoconioses, asthma, and byssinosis (a summary of notable gene–environment interactions and their putative mechanism can be found in Table 1). Through an exploration of how genes predispose to disease, several points will become clear. First, we will see that there are considerable points of commonality in the pathogenesis of these varied illnesses. The role of oxidative stress, and the individual capacity to counter it, will emerge again and again as a key mediator of disease. The role of the innate immune response— and the manner in which polymorphisms in relevant genes might affect it—is likewise a point of connection. Finally, the complexity of occupational and environmental lung diseases will become evident. No one gene is “high penetrance” (or, in epidemiologic terms, relative risk), and thus disease producing. Instead, these genes confer susceptibility in the presence of exposure; multiple gene–gene and gene–environment interactions may further modify the individual response to a particular exposure. As Fig. 1 demonstrates, genetic susceptibility may play a role in essentially every step in the pathway between exposure and disease development.
Table 1.
Selected Gene-Environment Interactions
| Clinical disease | Exposure | Genes/polymorphisms with possible environment-interaction | Putative mechanism of effect modification |
|---|---|---|---|
| Chronic Beryllium Disease | Beryllium | HLA-DPB1Glu69 | Mediates beryllium presentation by antigen presenting cells (APCs), perhaps by providing a Beryllium binding site. |
| HLA-DRPHe47 | May mediate beryllium presentation. | ||
| TNF2-308A | May increase production of Beryllium-stimulated TNF-α production. | ||
| Asbestosis | Asbestos fibers | GST genes, NAT2 slow-acetylator genotype, SOD2 | Genes with antioxidant activity might modify the response to asbestos-fiber induced oxidative-stress. |
| Silicosis | Silica | IL-1R, TNF-α | Mediation of silica-induced pro-inflammatory cytokine production |
| Byssinosis | Endotoxin | Tyr113His | Decreased microsomal epoxide hydrolase activity could slow detoxication of endotoxin-induced reactive oxygen species (ROS). |
| TNF 308A | Amplification of endotoxin-induced TNF-α upregulation. | ||
| Asthma | Diisocyanates (DA) | HLA-DQB1*0503 | Mediation of diisocyanate presentation by APCs, increasing risk of DA |
| IL4RA | Mediation of the Th2-specific immune response to diisocyanates | ||
| GST genes | Modification of diisocyanate and metabolite conjugation by glutathione | ||
| NAT1 “slow acetylator” genotype | Decreased antioxidant activity | ||
| CTNNA3 | Hypothesized that polymorphisms in this component of the epithelial junctional complex could contribute to abnormal airway epithelial barrier and modified response to DA | ||
| Air pollution | NQO1 Ser 187 | The Ser polymorphism reduces enzyme activity, decreasing NQO1 reduction of quinones, and thereby protecting against oxidative stress. | |
| GSTM1 null | Abolished GST1 activity reduces ability to counter ozone-induced oxidative stress. | ||
| GSTP1 | Modifies response to oxidative stress | ||
| EPHX1 | Modifies the metabolic response to polyaromatic hydrocarbons | ||
| HMOx-1 | Modification of antioxidant response | ||
| TNF-308 GG | Reduces TNF expression, which might attenuate ozone-induced TNF production | ||
| TLR2, TLR4 | Modify air pollution induced innate inflammatory response |
Figure 1.
Genetic susceptibility in occupational disease development (Adapted from Christiani et al.8)
Before proceeding, it is worthwhile commenting on the benefits of studying genetic susceptibility to occupational and environmental lung disease. We can group the potential utility of such research in two categories. First, as we will see in this review, an understanding of genetic susceptibility helps elucidate the pathogenesis of disease. Such an understanding may, in the future, be useful in determining targets for therapy. In the case of pollution-associated asthma, for instance, such work has helped underscore which asthmatics might most benefit from increased antioxidant intake.1,2
Second, an understanding of genetic susceptibility may help identify “at-risk” individuals. This, in turn, could provide information about which individuals should avoid exposure altogether (if that is a possibility), or perhaps about how individual exposure limits might be determined on the basis of genetic susceptibility. Here, however, we are on tenuous ground.3 A genetic test must have sufficient operating characteristics—high sensitivity, specificity, or both—to be clinically useful, and genes which confer only modestly increased susceptibility to disease are unlikely to reach that standard. In addition, significant ethical concerns arise with the genetic testing of workers.3 Such factors must be seriously considered before any translation of genetic testing into the workplace and clinic is advocated.
Gene–Beryllium Interactions
Chronic beryllium disease (CBD) is an occupational lung disease resulting from an exaggerated immunologic response to beryllium exposure. Beryllium is used widely in industry on account of its physical properties, including its high tensile strength and melting point, its ability to resist corrosion, and its low density.4,5 These properties make it useful in various industries worldwide, such as aerospace, defense, atomic energy, electronics, and telecommunications.4,6 Though the introduction of exposure standards has dramatically reduced the incidence of acute beryllium toxicity, beryllium workers remain at risk for immune-mediated CBD.7,8 Clinically and pathologically, CBD cannot be distinguished from sarcoidosis; organ damage results from granulomatous inflammation that most commonly involves the lung.
Research in recent decades has greatly expanded our understanding of the immune response to CBD, and of the genetic factors that underlie variability in this response. A proportion of beryllium-exposed individuals develop sensitization as the result of a cell-mediated immune response. This response is mediated by the interaction of human leukocyte antigen (HLA) class II on antigen presenting cells (APCs) with CD4+ T cells.5 An individual’s sensitization to beryllium can be assessed by the beryllium lymphocyte proliferation test, in which the patient’s lymphocytes are exposed to beryllium salts.6 A proportion of these sensitized individuals go on to develop CBD, in which a deleterious Th1 immune response results in clinical manifestations of disease.5
However, only a minority of beryllium-exposed individuals develops sensitization, and only some of these individuals go on to develop CBD. Thus, it has been hypothesized that a genetic predisposition might play a role in the pathogenesis of CBD. The role of a specific genetic factor was first demonstrated in an important study by Richeldi et al, who evaluated the association between the HLA-DP gene and CBD.9 In 33 CBD cases and 44 controls, they found a negative association between disease and the HLA-DPB100401 allele, and a positive association with HLA-DPB100201.9 Through sequence analysis, the investigators demonstrated that the primary difference between these two alleles was the presence of a glutamic acid residue at position 69 (Glu69)9: 97% of the CBD cases expressed the HLA-DPB100201-associated glutamic acid at residue 69, as compared with only 30% of controls.9
Subsequent work has shed more light on the complex interaction between genotype, beryllium sensitization and CBD. Though one group found no association between HLA-DPB1Glu69 and sensitization, multiple subsequent studies have found an association between HLA-DPB1Glu69 alleles in patients with both sensitization and disease.10–15 In one study among beryllium workers, having the HLA-DPB1Glu69 marker provided additive predictive effect to a known exposure risk factor, machining, which increases exposure to particulates.16 There is also evidence that it is not only the Glu69 marker but also the presence of specific (and uncommon) Glu69 alleles that puts individuals at the greatest risk.17,18 In addition, genetic dose matters: allele number seems to be an important factor influencing the development of sensitization, the development of disease, and possibly the severity of disease.14,15
Such work clearly demonstrates a role for HLA-DPB1Glu69 in the pathogenesis of beryllium sensitization and disease. Subsequent research has demonstrated that the mechanism for this finding relates to the critical role that Glu69 plays in beryllium presentation. T-cell clones isolated from patients with CBD, for instance, only proliferate in response to beryllium that is presented by cells with the Glu69 marker; changing this one amino acid alone eliminates the response altogether.19 Similarly, using beryllium-specific T-cell lines isolated from the lungs of CBD-affected patients, Fontenot et al found that monoclonal antibodies to the class II MHC molecule HLA-DP selectively inhibited beryllium antigen presentation.20 Ongoing research is exploring the precise mechanism by which glu69 mediates beryllium presentation, but the evidence is clear for a direct interaction between beryllium and the HLA-DPB1Glu69 molecule.21 Several studies have suggested that specific characteristics of the peptide-binding pocket—for instance, a more negative charge—might explain why different alleles result in a differential ability to bind beryllium and mediate disease.18,22,23
Together, this research tells a convincing story about the role that a specific genetic factor—Glu69—plays in the presentation of beryllium and the resultant immune response. However, some 15% of patients with sensitization or disease lack Glu69, suggesting that there must be other pathways by which beryllium can instigate a hypersensitivity response.15 Further light has been shed on this question through investigation of sensitized HLA-DPGlu69-negative patients, with evidence pointing to a role of HLA-DR and HLA-DQ genes in both sensitization and disease progression. Among HLA-DPGlu69-negative patients, for instance, HLA-DRPhe47 has been shown to confer risk for beryllium hypersensitivity; given structural characteristics of the molecule, it is suggested that HLA-DRPhe47 might bind and present beryllium to t cells.24 Other genetic factors could help explain why some patients with HLA-DPB1–69 and beryllium hypersensitivity progress to clinical disease, and others do not.13 Rossman et al, for instance, found associations between particular HLA-DQ and HLA-DR genes in those with CBD compared with those with hypersensitivity without disease, suggesting that these HLA haplotypes might confer risk for progression among the sensitized.13 Other work has supported associations between particular HLA-DR and HLA-DQ phenotypes and beryllium hypersensitivity and disease.10,15
Other candidate susceptibility genes may also play a role. The association between angiotensin-converting enzyme (ACE) activity and CBD disease severity led to the hypothesis that the ACE genotype might predispose to disease; however, no clear association has been found between ACE genotype and disease.25 Because beryllium-induced tumor necrosis factor-α (TNF-α) production might play a role in the pathogenesis of CBD, it has been postulated that polymorphisms in this gene may affect disease development.26 In one study, a polymorphism in the TNF-α promoter gene (TNF2 A allele at -308) was shown to be associated with increased production of beryllium-stimulated TNF-α, while increased TNF-α was associated with markers of disease severity.26 Others have found an association between TNF-α-30802 and beryllium sensitization.10 In addition, it seems that beryllium exposure stimulates mononuclear cells to release TNF-α in patients with and without disease, and even with complete HLA-DP inhibition, suggesting an independent role of TNF-α in CBD development.27
Though this work has profoundly increased our understanding of CBD, the clinical implications of these findings remain unclear. Even the best validated risk factor—HLA-DPB1Glu69—has a low positive predictive value for the development of CBD in the workplace.28 Between imperfect testing characteristics, serious ethical concerns that arise from the genetic screening of workers, the potential for genetic discrimination, and the lack of specific interventions to further protect “at risk” workers, most continue to recommend against the implementation of routine genetic testing in the beryllium industry at present.8,28
Other Pneumoconioses
The variable relationship between dust exposure and the development of the “classic” pneumoconiosis has similarly suggested that genetic predisposition may play in modifying the individual response to these toxicants. Work elucidating the biologic mechanisms of pneumoconiosis has produced several candidate susceptibility genes.29 Nevertheless, many of these associations remain tentative, and no candidate genes approach the stage of clinical utility at present.
Coal dust exposure, for instance, has been associated with elevated levels of inflammatory cytokines (including TNF-α, interleukin [IL]-1, IL-6, and IL-1β) as well as markers of oxidative stress.30 Consistent with this pathophysiology, some groups have found various associations between polymorphisms of the TNF-α gene and coal workers’ pneumoconiosis.31–34 However, a large study of 648 coal workers found no associations between individual polymorphisms in various inflammatory cytokines (including TNF-α) and the development of progressive massive fibrosis.35 Another group, however, found an association between polymorphisms in IL-18 and the progression and prevalence of fibrosis.36 Associations between polymorphisms in mannose-binding lectin (which plays a role in innate immunity) and heat-shock protein 70 (which plays a role in the response to stress and injury) with coal workers’ pneumoconiosis have also been suggested.37,38 Overall, further study is necessary to confirm these associations.
Exposure to asbestos fibers also results in oxidative stress, and this may be an important pathway to the development of fibrosis.39 The GSTM1 null genotype (which reduces antioxidant activity) has been associated with the development of asbestos-induced lung disease in some studies but not in others.40–43 In one study, the combination of a GSTM1 null with NAT2 slow-acetylator genotype had an additive effect in increasing the risk of developing both malignant and nonmalignant asbestos-induced lung disease.43 Possible but variable associations between disease risk and polymorphisms in other genes involved in the response to oxidative stress, such as superoxide dismutase 2 (SOD2), GSTT1, GSTP1, and NLRP3, have also been suggested.41,42,44–46 Gene–gene and other gene– environment interactions (for instance, involving smoking) may also play a role in modifying the risk of asbestosis.47
Gene–environment interactions in the case of silicosis have also been demonstrated. Our understanding of the mechanisms of silica-induced pulmonary toxicity has been greatly elucidated by recent research. Silica results in the production of reactive oxygen species (ROS) both directly on the particle surface and indirectly through downstream effects following phagocytosis.48 Following phagocytosis, silica activates the NALP3 inflammasome complex, which results in the release of proinflammatory cytokines including IL-1β and IL-19 and in the generation of ROS; endotoxin priming and resultant activation of nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) may also play a role.48 The hypothesis that cytokine polymorphisms influence the development and progression of silicosis would be consistent with this understanding of the pathophysiology of the disease.49 Indeed, polymorphisms in genes for the IL-1 receptor (IL-1R) and TNF-α have been associated with the development of silicosis in some, but not all, studies.50–55 A study involving miners in South Africa found an association between rare TNF-α polymorphisms and severe, but not mild, silicosis.52 More recently, a meta-analysis pooling data from nine studies involving 1,267 cases and 1,214 controls from China, the United States, and South Africa found a significant association between the TNF-α-308G/A polymorphism and the risk of silicosis.56 Another study found an association between NF-κB genotype and the risk of silicosis, another biologically plausible susceptibility gene.54
However, in summary, it is clear that further work is needed to better elucidate how genotype modifies the response to silica, coal, and asbestos. At present, adequate protection of all exposed workers is the only appropriate approach.
Cotton Textile Workers: Gene–Endotoxin Interactions
“Byssinosis” is an occupational lung disease that has long been described among textile workers. In particular, cotton textile work has been associated with the development of chronic respiratory symptoms and an accelerated decline in forced expiratory volume in 1 second (FEV1).57–59 Endotoxin exposure has been implicated as the key mediator of respiratory disease among these workers. For instance, a dose– response relationship between endotoxin exposure (but not overall dust exposure) and the development of spirometric abnormalities and respiratory symptoms has been described in the setting of acute endotoxin exposure.60,61 In one long-term cohort study of Chinese cotton textile workers, endotoxin exposure emerged as an independent predictor of increased longitudinal loss of FEV1 and forced vital capacity.59
Similar to the other occupational diseases discussed so far, only a minority of those exposed develops disease, suggesting that other factors must play a role in disease causation. There is emerging evidence that genetic factors modify the development of disease, and some of this evidence has helped to elucidate the pathogenesis of byssinosis. In animals and in vivo studies, endotoxin induces the production of ROS.62,63 These ROS may play a role in byssinosis. Because the enzyme microsomal epoxide hydrolase is a key detoxifier of ROS, it has been postulated that polymorphisms that cause differences in enzyme activity might modify disease development. This hypothesis was tested using a 20-year longitudinal cohort of cotton textile workers, in which workers were genotyped for two common genetic polymorphisms, Tyr113His (associated with decreased enzyme activity) and His139Arg (associated with faster enzyme activity).64 A significant interaction between FEV1 decline, endotoxin exposure, and genotype emerged, with a greater decline in lung function from endotoxin exposure among the “slow” activity genotypes.64
Polymorphisms of the TNF gene may also modify the development of byssinosis. TNF is an important cytokine in human health and disease, and has been associated with airway hyperresponsiveness and inflammation.65 Experimental endotoxin inhalation, in turn, may acutely induce increased TNF levels in blood and lung.66,67 Given that polymorphisms of the TNF gene are associated with variable production and expression of TNF, it has been hypothesized that such polymorphisms may modify the relationship between endotoxin exposure and lung disease.68 Using the same 20-year longitudinal cohort of Chinese cotton workers, Zhang et al demonstrated that endotoxin exposure was associated with a greater decline in FEV1 among those with the A allele of the TNF—308G/A polymorphism, which confers greater TNF activity.68
A third approach has been to use genome-wide association to identify single nucleotide polymorphisms (SNPs) that are associated with lung function decline.69 In one exploratory study, two SNPs were found to be significantly associated with FEV1 decline (rs1910047 and rs9469089) and another eight SNPs were identified as suggestive, many of which had biologically plausible functions.69 There was also evidence for multiple gene–gene and gene–environment interactions, with differing effects of endotoxin exposure among subjects with different SNPs.69
To summarize, like many occupational diseases, byssinosis is a complex disease. Although endotoxin has been demonstrated to be the primary environmental toxin responsible for lung disease among cotton workers, emerging evidence points to important gene–endotoxin interactions in disease pathogenesis. These interactions further add to our understanding of the role of oxidative stress and TNF-α in the development of byssinosis. However, currently, none of these genetic factors are sufficiently robust to be used clinically in the identification of at-risk workers.
Occupational Asthma: Gene–Diisocyanate Interactions
Occupational asthma imposes substantial costs and suffering worldwide.70 Diisocyanates are one major cause of occupational asthma. Workers exposed to diisocyanates include producers and handlers of polyurethane foam, urethane adhesives, and urethane molds.71 Only a small proportion of exposed workers develop diisocyanate asthma (DA), which remains a complex and poorly understood disease. Work on gene–environment interactions, however, helps elucidate—at least to some degree—the mechanisms underlying it.
Given that occupational asthma can be thought of as an abnormal immune response to a workplace exposure, polymorphisms in genes involved in antigen presentation might plausibly be thought to modify the response to diisocyanates. Several investigators have explored associations between HLA class II alleles and the development of DA. Bignon et al evaluated differences in HLA class II genetic markers between a group of workers with DA (documented by inhalation challenge) and exposed workers without a history of disease, and identified alleles associated with disease susceptibility (DQB100503 and the combination DQB100201/0301) and protection (DQB100501).72 Balboni et al confirmed the positive association between DA and the DQB100503 allele and the negative association between DA and the DQB100501.73 These two alleles differ by only a single amino acid at position 57 (aspartic acid for DQB100503 and valine for DQB100501); therefore, the investigators calculated the phenotypic frequency of asp57; evidence was found for a gene dose effect, suggesting that it may be of functional significance in disease development.73 Mapp et al also found that DQB100503 conferred risk and DQB100501 protection, and additionally demonstrated that DQA100104 was positively associated, and DQA100101 negatively associated, with DA.74 HLA allele DQB100501 may also confer protection against the development of asthma in sawmill workers exposed to western red cedar.75 In contrast, however, it should be noted that other work has found no significant associations between HLA alleles and disease.76 There is also no evidence for an association between the TNF gene or HLA class I antigens and DA.77
A murine model has provided evidence that T helper 2 (Th2)-specific cytokines might play an important role in the pathogenesis of DA.78 In light of this evidence, Bernstein et al evaluated polymorphisms of Th2 cytokine genes in workers with and without DA.79 They found no association between disease and individual SNPs associated with genes IL4RA, IL-13, and a CD14 promoter.79 However, they did find a significant association between IL4RA polymorphisms and disease in those specifically with hexamethylene diisocyanate exposure.79 They also found evidence of gene–gene interactions, with significant associations emerging when IL4RA polymorphisms were combined with IL-13 and CD14 polymorphisms.79
Diisocyanates and their metabolites are conjugated with glutathione by glutathione S-transferases (GSTs), suggesting that polymorphisms in GST genes that affect enzyme function could modify the relationship between DA and asthma. Consistent with this hypothesis is the finding that an absence of the GSTM1 gene (the null phenotype) is associated with an increased risk of asthma.80 Another study has suggested the possibility that GSTP10Val homozygosity may confer protection against DA and airway hyperresponsiveness in a subset of workers with a prolonged exposure to toluene diiosyanate.81 Like GST, N-acetylcysteine (NAT) plays an important role as an antioxidant in the lung, and there is evidence that the “slow acetylator” NAT1 genotype increases the risk of developing asthma, particularly when combined with a GSTM1 null genotype.82 Together, this evidence underscores the importance of oxidative stress in the development of DA.
A final line of research has used genome-wide association analysis to identify polymorphisms associated with DA. Kim et al conducted a GWA in 84 Korean patients with DA and 263 controls, and found catenin α-3, α-T catenin gene (CTNNA3) to be a potential candidate gene for DA susceptibility.83 This finding was confirmed in a Caucasian population in a larger study by Bernstein et al in 2013.84 The mechanism by which polymorphisms in CTNNA3 might modify disease development, however, remains elusive. Bernstein et al postulate that because CTNNA3 is an important component to the epithelial junctional complex, its reduced expression might theoretically contribute to an abnormal airway epithelial barrier, and therefore to DA, but this remains conjectural.84
To summarize, several gene–environment interactions with biologic plausibility have been identified in the case of DA, including HLA class II genes, genes associated with the Th2 cytokine response, genes associated with antioxidant function, and a gene associated with epithelial junctional integrity. Further work will hopefully elucidate the pathophysiology not just of DA but also of other types of occupational asthma. A better understanding of the pathogenesis of occupational asthma may eventually contribute to our ability to prevent and treat it.
Asthma: Gene–Air Pollution Interactions
Outdoor pollution results in a substantial burden of morbidity and mortality throughout the world, contributing to 3.1% of annual disability-adjusted life years lost globally.85 Air pollution has long been associated with an increase in asthma exacerbations, but some evidence suggests that it might incite new cases as well.86,87 Between urbanization and climate change, it is probable that this problem will worsen in coming decades.88–90
From the mechanistic standpoint, exposure to air pollution can lead to oxidative stress, which in turn has deleterious respiratory consequences.91 In children, for instance, exposure to sulfur dioxide (SO2), nitrogen dioxide (NO2), and particulate matter < 2.5 um (PM2.5) is associated with increased markers of oxidative stress in breath condensate, and also with reduced lung function.92 Exposure to traffic-related air pollutants, including black carbon and NO2, is also associated with increased markers of oxidative stress,93 as is exposure to PM2.5 in trucking industry workers.94
We now have some understanding of how air pollutants cause such oxidative stress, and how this in turn causes respiratory injury. Although low-level exposure to air pollutants might actually induce a protective response through upregulation of genes with antioxidant functions (e.g., NAD (P)H:quinone oxidoreductase 1 [NQO1] and GST), higher level exposures might eventually lead to a deleterious response.95 For instance, NO2, ozone, and PM exposure have all been linked with NF-KB expression; NF-KB, in turn, increases the expression of inflammatory cytokines such as TNF-α, IL-6, and IL-8.95
However, not all individuals respond the same to oxidative stress, and this has led to the hypothesis that genetic susceptibility may play a key role in modifying the pathogenesis of pollution-associated respiratory disease.95 For instance, large person-to-person variability in response to ozone exposure has been seen in the setting of acute ozone exposure.96 Therefore, several investigators have explored how polymorphisms in genes involved in the response to oxidative stress might modify the airway response to air pollution. Candidate antioxidant genes include NQO1, GST (GSTM1 and GSTP1), and EPHX1.
NQO1 and GSTM1 are important phase II detoxifying enzymes that help mediate the response to oxidative stress.97 The combination of a wild-type NQO1 (NQO1 Pro/Pro) genotype and a null GSTM1 genotype has been associated with increased markers of oxidative stress after acute ozone exposure.98,99 Biologically, it is plausible why the null genotype of the antioxidant enzyme GST1—which decreases enzyme activity—would increase susceptibility to oxidative stress. However, why wild-type NQO1—which increases enzyme activity—also increases susceptibility is less clear. One explanation relies on the effects of NQO1 apart from its antioxidant properties: NQO1 catalyzes the reduction of quinones, which themselves generate ROS.97 For this reason, polymorphisms which reduce NQO1 activity might actually be protective against oxidative stress.97
Polymorphisms in these two genes are associated not only with ozone susceptibility but also with disease prevalence. In an early study, David et al evaluated the relationship between the NQO1 Pro187Ser polymorphism and the null GSTM1 polymorphism with asthma in children from Mexico City, an area with high ozone exposure.97 They found that while the NQO1 genotype alone did not affect asthma risk, among those subjects with the GSTM1 null genotype, at least one NQO1 Ser187 allele significantly decreased the risk of asthma.97 In a cohort of asthmatic children who participated in a randomized trial of antioxidants in Mexico (discussed later), a GSTM1 null genotype conferred an increased risk of ozone-associated episodes of difficult breathing (but not reduced lung function).100 Chen et al found an association between the combined GSTM1 null and wild-type NQO1 Pro187Pro genotype and ozone-related decreased lung function in females only.101 In the Children’s Health Study cohort, mean-while, the GSTM1 null genotype was associated with an increased incidence of new-onset asthma during adolescence.102 To summarize, the GSTM1 null genotype—in some studies alone, in some studies in combination with other genotypes—confers an increased risk for ozone susceptibility, asthma, and respiratory symptoms.
Evidence also suggests that nutritional factors may further modify the three-way relationship between genotype, environment, and disease. It has been shown in a randomized controlled trial that supplementation with antioxidant vitamins attenuates the relationship between ozone levels and reduced lung function in asthmatic children in Mexico City.1 But furthermore, the benefit of antioxidant supplementation seems to depend on genotype: GSTM1 null children had both increased susceptibility to the effect of ozone on lung function and increased benefit with antioxidant therapy.2 Consistent with these findings is evidence that deficient antioxidant intake increases the risk of ozone-induced reduced lung function in asthmatic children, and that this effect is potentially modified by the presence of polymorphisms in several antioxidant genes.103 Taken together, these data demonstrate that multiple genetic, environmental, and nutritional factors may interact to modify the respiratory response to oxidative stress in exposed individuals.
Several other candidate genes with antioxidant function have undergone investigation. Many studies, for instance, have pointed to a gene–environment interaction with a common GSTP1 polymorphism that causes a Ile105Val substitution, although there is conflicting evidence as to whether this confers risk or protection.100–102,104–106 Another gene of interest is microsomal epoxide hydrolase (EPHX1), which plays a role in the metabolic response to activated polyaromatic hydrocarbons, which are generated from the burning of fossil fuels.107 In one study which used subjects from the Children’s Health Study population, high EPHX1 activity phenotypes increased the risk for lifetime asthma, an effect that was further modified by the presence of GSTP1 Val 105 alleles and proximity to a roadway.107 Another antioxidant gene of interest is heme oxygenase 1 (HMOX-1). In the Children’s Health Study cohort, functional promoter variants in HMOX-1 were associated with a reduced risk of new-onset asthma, a protective effect which was confined to non-Hispanics and those in low-ozone communities.108
Several other genes outside of the antioxidant system have also been associated with pollution-induced airways disease. For instance, polymorphisms in TNF-α, which is thought to play a role in asthma pathogenesis, have been investigated. In one small study, the TNF-α 308 G/G genotype conferred an increased susceptibility to acute changes in FEV1 after inhaled ozone challenge.109 Yet the effect of this genotype, which is thought to reduce TNF-α expression, is unclear. For instance, the TNF-308 GG genotype was actually found to reduce the risk of lifetime wheezing and asthma in a group of 3,699 children from the Children’s Health Study.110 In addition, the protective effect of this allele was reduced in communities exposed to high ozone levels, suggesting that high levels of oxidative stress might overcome the protective effect of 308 GG.110 Further supporting this explanation is the finding that the protective effect of 308 GG in high-ozone communities was also reduced by the GSTM1 null genotypes, which (as we have seen) confers greater susceptibility to ozone.110 The protective effect of TNF-308GG (assuming it is confirmed) may, in other words, be overwhelmed in the setting of environmental conditions or other genetic factors. Other gene–environment interactions may also play a role here; the effect of TNF-α polymorphisms, for instance, may be modified by parental smoking.111 Finally, polymorphisms that modify the innate inflammatory response to air pollution have also been associated with asthma. Various variant alleles of Toll-like receptor 2 and 4 (TLR2 and TLR4), for instance, have been found to modify the effect of air pollution on asthma prevalence in children in one prospective birth cohort study in the Netherlands.112
To summarize, air pollution–associated respiratory disease is, to a large degree, mediated by oxidative stress. A body of research has demonstrated a biologically plausible link between common genetic variants and pollution-induced lung function decrements and lung disease.
Furthermore, multiple gene–gene and additional gene– environment interactions also play a role. For instance, antioxidant supplementation reduces, and antioxidant deficiency increases, susceptibility to ozone. In addition, the effect of both of these nutritional factors is further modified by antioxidant gene alleles.2 Together, this evidence not only elucidates our understanding of pollution-associated asthma but also raises important policy questions regarding optimal antioxidant intake among high-risk individuals, particularly during periods of high ozone levels.113
Conclusion
Worldwide, occupational and environmental lung disease causes substantial morbidity, disability, and death, and will continue to do so into the foreseeable future.85 Tackling the formidable challenge of this global burden necessitates furthering our understanding of the pathogenesis of these diseases and of their biological mechanisms. The study of gene–environment interactions is one approach to do this. For instance, through a study of gene–environment interactions, we have learned how the development of CBD depends on an appropriate beryllium-binding site being present on APCs. In the case of pneumoconiosis, byssinosis, pollution-associated asthma, and diisocyanate-induced asthma, we have seen how the ability to counter oxidative stress is critical in determining individual susceptibility to toxin exposure. In the case of pollution-induced asthma, this may have direct implications in terms of antioxidant supplementation and deficiency.
Yet the limitations of this approach should also be understood. First, it is unlikely that “high penetrance” genes—akin to the BRCA1 mutation—for these complex diseases will be discovered.8 In general, genetic testing for “low penetrance” genes is far less likely to have useful testing characteristics.3 But there are additional challenges with respect to environmental and occupational exposures. For instance, for those environment exposures which impact very large populations (e.g., ozone in Mexico City), genetic testing is unfeasible, even if it had more favorable testing characteristics. Testing of workers is even more problematic. Genetic testing can be considered an invasion of workers’ privacy. Employers are already relying on “workplace wellness initiatives” that incorporate batteries of questionnaires, laboratory tests, and biometric measurements that many find to be intrusive.114 The addition of genetic testing may be perceived as a further violation of privacy and confidentiality. It has the capacity to lead to discrimination against workers perceived to be high risk, who may or may not have other employment opportunities.3 Finally, perhaps the clearest argument relates to the fact that genetic testing could serve as a distraction from the far more important task of ensuring that the right to a safe workplace and environment is realized for us all.3
Contributor Information
Adam Gaffney, Email: agaffney@mgh.harvard.edu.
David C. Christiani, Email: dchris@hsph.harvard.edu.
References
- 1.Romieu I, Sienra-Monge JJ, Ramírez-Aguilar M, et al. Antioxidant supplementation and lung functions among children with asthma exposed to high levels of air pollutants. Am J Respir Crit Care Med. 2002;166(5):703–709. doi: 10.1164/rccm.2112074. [DOI] [PubMed] [Google Scholar]
- 2.Romieu I, Sienra-Monge JJ, Ramírez-Aguilar M, et al. Genetic polymorphism of GSTM1 and antioxidant supplementation influence lung function in relation to ozone exposure in asthmatic children in Mexico City. Thorax. 2004;59(1):8–10. [PMC free article] [PubMed] [Google Scholar]
- 3.Vineis P, Ahsan H, Parker M. Genetic screening and occupational and environmental exposures. Occup Environ Med. 2005;62(9):657–662. 597. doi: 10.1136/oem.2004.019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santo Tomas LH. Beryllium hypersensitivity and chronic beryllium lung disease. Curr Opin Pulm Med. 2009;15(2):165–169. doi: 10.1097/MCP.0b013e3283218341. [DOI] [PubMed] [Google Scholar]
- 5.Samuel G, Maier LA. Immunology of chronic beryllium disease. Curr Opin Allergy Clin Immunol. 2008;8(2):126–134. doi: 10.1097/ACI.0b013e3282f824a4. [DOI] [PubMed] [Google Scholar]
- 6.Seidler A, Euler U, Müller-Quernheim J, et al. Systematic review: Progression of beryllium sensitization to chronic beryllium disease. Occup Med (Lond) 2012;62(7):506–513. doi: 10.1093/occmed/kqs069. [DOI] [PubMed] [Google Scholar]
- 7.Saltini C, Amicosante M, Franchi A, Lombardi G, Richeldi L. Immunogenetic basis of environmental lung disease: lessons from the berylliosis model. Eur Respir J. 1998;12(6):1463–1475. doi: 10.1183/09031936.98.12061463. [DOI] [PubMed] [Google Scholar]
- 8.Christiani DC, Mehta AJ, Yu CL. Genetic susceptibility to occupational exposures. Occup Environ Med. 2008;65(6):430–436. doi: 10.1136/oem.2007.033977. quiz 436, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262(5131):242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 10.Saltini C, Richeldi L, Losi M, et al. Major histocompatibility locus genetic markers of beryllium sensitization and disease. Eur Respir J. 2001;18(4):677–684. doi: 10.1183/09031936.01.00106201. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Farris GM, Newman LS, et al. Beryllium sensitivity is linked to HLA-DP genotype. Toxicology. 2001;165(1):27–38. doi: 10.1016/s0300-483x(01)00410-3. [DOI] [PubMed] [Google Scholar]
- 12.Maier L, Martyny J, Mroz M, et al. Genetic and environmental risk factors in beryllium sensitization and chronic beryllium disease. Chest. 2002;121(3, Suppl):81S. [PubMed] [Google Scholar]
- 13.Rossman MD, Stubbs J, Lee CW, Argyris E, Magira E, Monos D. Human leukocyte antigen Class II amino acid epitopes: susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med. 2002;165(6):788–794. doi: 10.1164/ajrccm.165.6.2104002. [DOI] [PubMed] [Google Scholar]
- 14.McCanlies EC, Ensey JS, Schuler CR, Kreiss K, Weston A. The association between HLA-DPB1Glu69 and chronic beryllium disease and beryllium sensitization. Am J Ind Med. 2004;46(2):95–103. doi: 10.1002/ajim.20045. [DOI] [PubMed] [Google Scholar]
- 15.Maier LA, McGrath DS, Sato H, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171(12):6910–6918. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 16.Richeldi L, Kreiss K, Mroz MM, Zhen B, Tartoni P, Saltini C. Interaction of genetic and exposure factors in the prevalence of berylliosis. Am J Ind Med. 1997;32(4):337–340. doi: 10.1002/(sici)1097-0274(199710)32:4<337::aid-ajim3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, White PS, Petrovic M, et al. Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and -DPA1 alleles. J Immunol. 1999;163(3):1647–1653. [PubMed] [Google Scholar]
- 18.Silveira LJ, McCanlies EC, Fingerlin TE, et al. Chronic beryllium disease, HLA-DPB1, and the DP peptide binding groove. J Immunol. 2012;189(8):4014–4023. doi: 10.4049/jimmunol.1200798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi G, Germain C, Uren J, et al. HLA-DP allele-specific T cell responses to beryllium account for DP-associated susceptibility to chronic beryllium disease. J Immunol. 2001;166(5):3549–3555. doi: 10.4049/jimmunol.166.5.3549. [DOI] [PubMed] [Google Scholar]
- 20.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+ T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci U S A. 2000;97(23):12717–12722. doi: 10.1073/pnas.220430797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amicosante M, Sanarico N, Berretta F, et al. Beryllium binding to HLA-DP molecule carrying the marker of susceptibility to berylliosis glutamate beta 69. Hum Immunol. 2001;62(7):686–693. doi: 10.1016/s0198-8859(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 22.Scott BL, Wang Z, Marrone BL, Sauer NN. Potential binding modes of beryllium with the class II major histocompatibility complex HLA-DP: a combined theoretical and structural database study. J Inorg Biochem. 2003;94(1–2):5–13. doi: 10.1016/s0162-0134(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 23.Snyder JA, Weston A, Tinkle SS, Demchuk E. Electrostatic potential on human leukocyte antigen: implications for putative mechanism of chronic beryllium disease. Environ Health Perspect. 2003;111(15):1827–1834. doi: 10.1289/ehp.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amicosante M, Berretta F, Rossman M, et al. Identification of HLA-DRPhebeta47 as the susceptibility marker of hypersensitivity to beryllium in individuals lacking the berylliosis-associated supratypic marker HLA-DPGlubeta69. Respir Res. 2005;6:94. doi: 10.1186/1465-9921-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier LA, Raynolds MV, Young DA, Barker EA, Newman LS. Angiotensin-1 converting enzyme polymorphisms in chronic beryllium disease. Am J Respir Crit Care Med. 1999;159(4 Pt 1):1342–1350. doi: 10.1164/ajrccm.159.4.9806106. [DOI] [PubMed] [Google Scholar]
- 26.Maier LA, Sawyer RT, Bauer RA, et al. High beryllium-stimulated TNF-alpha is associated with the -308 TNF-alpha promoter polymorphism and with clinical severity in chronic beryllium disease. Am J Respir Crit Care Med. 2001;164(7):1192–1199. doi: 10.1164/ajrccm.164.7.2012123. [DOI] [PubMed] [Google Scholar]
- 27.Amicosante M, Berretta F, Franchi A, et al. HLA-DP-unrestricted TNF-alpha release in beryllium-stimulated peripheral blood mononuclear cells. Eur Respir J. 2002;20(5):1174–1178. doi: 10.1183/09031936.02.02232001. [DOI] [PubMed] [Google Scholar]
- 28.Silver K, Sharp RR. Ethical considerations in testing workers for the -Glu69 marker of genetic susceptibility to chronic beryllium disease. J Occup Environ Med. 2006;48(4):434–443. doi: 10.1097/01.jom.0000200878.16077.3b. [DOI] [PubMed] [Google Scholar]
- 29.Yucesoy B, Luster MI. Genetic susceptibility in pneumoconiosis. Toxicol Lett. 2007;168(3):249–254. doi: 10.1016/j.toxlet.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Petsonk EL, Rose C, Cohen R. Coal mine dust lung disease. New lessons from old exposure. Am J Respir Crit Care Med. 2013;187(11):1178–1185. doi: 10.1164/rccm.201301-0042CI. [DOI] [PubMed] [Google Scholar]
- 31.Zhai R, Jetten M, Schins RP, Franssen H, Borm PJ. Polymorphisms in the promoter of the tumor necrosis factor-alpha gene in coal miners. Am J Ind Med. 1998;34(4):318–324. doi: 10.1002/(sici)1097-0274(199810)34:4<318::aid-ajim4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Kim KA, Cho YY, Cho JS, et al. Tumor necrosis factor-alpha gene promoter polymorphism in coal workers’ pneumoconiosis. Mol Cell Biochem. 2002;234–235(1–2):205–209. doi: 10.1023/a:1015914409661. [DOI] [PubMed] [Google Scholar]
- 33.Wang XT, Ohtsuka Y, Kimura K, et al. Antithetical effect of tumor necrosis factor-alphagene polymorphism on coal workers’ pneumoconiosis (CWP) Am J Ind Med. 2005;48(1):24–29. doi: 10.1002/ajim.20180. [DOI] [PubMed] [Google Scholar]
- 34.Ates I, Suzen HS, Yucesoy B, Tekin IO, Karakaya A. Association of cytokine gene polymorphisms in CWP and its severity in Turkish coal workers. Am J Ind Med. 2008;51(10):741–747. doi: 10.1002/ajim.20632. [DOI] [PubMed] [Google Scholar]
- 35.Yucesoy B, Johnson VJ, Kissling GE, et al. Genetic susceptibility to progressive massive fibrosis in coal miners. Eur Respir J. 2008;31(6):1177–1182. doi: 10.1183/09031936.00075107. [DOI] [PubMed] [Google Scholar]
- 36.Nadif R, Mintz M, Marzec J, Jedlicka A, Kauffmann F, Kleeberger SR. IL18 and IL18R1 polymorphisms, lung CT and fibrosis: A longitudinal study in coal miners. Eur Respir J. 2006;28(6):1100–1105. doi: 10.1183/09031936.00031506. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Ohtsuka Y, Kimura K, et al. Mannose-binding lectin gene polymorphisms and the development of coal workers’ pneumoconiosis in Japan. Am J Ind Med. 2008;51(7):548–553. doi: 10.1002/ajim.20587. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Jin T, Zhang G, Chen L, Zou W, Li QQ. Polymorphisms in heat-shock protein 70 genes are associated with coal workers’ pneumoconiosis in southwestern China. In Vivo. 2011;25(2):251–257. [PubMed] [Google Scholar]
- 39.Liu G, Cheresh P, Kamp DW. Molecular basis of asbestos-induced lung disease. Annu Rev Pathol. 2013;8:161–187. doi: 10.1146/annurev-pathol-020712-163942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CM, Kelsey KT, Wiencke JK, Leyden K, Levin S, Christiani DC. Inherited glutathione-S-transferase deficiency is a risk factor for pulmonary asbestosis. Cancer Epidemiol Biomarkers Prev. 1994;3(6):471–477. [PubMed] [Google Scholar]
- 41.Franko A, Dodic-Fikfak M, Arneri N, Dolzan V. Glutathione S-transferases GSTM1 and GSTT1 polymorphisms and asbestosis. J Occup Environ Med. 2007;49(6):667–671. doi: 10.1097/JOM.0b013e318065b855. [DOI] [PubMed] [Google Scholar]
- 42.Kukkonen MK, Hämäläinen S, Kaleva S, et al. Genetic susceptibility to asbestos-related fibrotic pleuropulmonary changes. Eur Respir J. 2011;38(3):672–678. doi: 10.1183/09031936.00049810. [DOI] [PubMed] [Google Scholar]
- 43.Hirvonen A, Saarikoski ST, Linnainmaa K, et al. Glutathione S-transferase and N-acetyltransferase genotypes and asbestos-associated pulmonary disorders. J Natl Cancer Inst. 1996;88(24):1853–1856. doi: 10.1093/jnci/88.24.1853. [DOI] [PubMed] [Google Scholar]
- 44.Franko A, Dolzan V, Arnerić N, Dodic-Fikfak M. The influence of genetic polymorphisms of GSTP1 on the development of asbestosis. J Occup Environ Med. 2008;50(1):7–12. doi: 10.1097/JOM.0b013e31815cbab5. [DOI] [PubMed] [Google Scholar]
- 45.Franko A, Dodic-Fikfak M, Arnerić N, Dolzan V. Manganese and extracellular superoxide dismutase polymorphisms and risk for asbestosis. J Biomed Biotechnol. 2009;2009:493083. doi: 10.1155/2009/493083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kukkonen MK, Vehmas T, Piirilä P, Hirvonen A. Genes involved in innate immunity associated with asbestos-related fibrotic changes. Occup Environ Med. 2014;71(1):48–54. doi: 10.1136/oemed-2013-101555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franko A, Dolžan V, Arnerić N, Dodic-Fikfak M. The influence of gene-gene and gene-environment interactions on the risk of asbestosis. Biomed Res Int. 2013;2013:405743. doi: 10.1155/2013/405743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung CC, Yu IT, Chen W. Silicosis. Lancet. 2012;379(9830):2008–2018. doi: 10.1016/S0140-6736(12)60235-9. [DOI] [PubMed] [Google Scholar]
- 49.Yucesoy B, Vallyathan V, Landsittel DP, Simeonova P, Luster MI. Cytokine polymorphisms in silicosis and other pneumoconioses. Mol Cell Biochem. 2002;234–235(1–2):219–224. [PubMed] [Google Scholar]
- 50.Yucesoy B, Vallyathan V, Landsittel DP, et al. Polymorphisms of the IL-1 gene complex in coal miners with silicosis. Am J Ind Med. 2001;39(3):286–291. doi: 10.1002/1097-0274(200103)39:3<286::aid-ajim1016>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 51.Yucesoy B, Vallyathan V, Landsittel DP, et al. Association of tumor necrosis factor-alpha and interleukin-1 gene polymorphisms with silicosis. Toxicol Appl Pharmacol. 2001;172(1):75–82. doi: 10.1006/taap.2001.9124. [DOI] [PubMed] [Google Scholar]
- 52.Corbett EL, Mozzato-Chamay N, Butterworth AE, et al. Polymorphisms in the tumor necrosis factor-alpha gene promoter may predispose to severe silicosis in black South African miners. Am J Respir Crit Care Med. 2002;165(5):690–693. doi: 10.1164/ajrccm.165.5.2010050. [DOI] [PubMed] [Google Scholar]
- 53.Wu F, Qu Y, Tang Y, Cao D, Sun P, Xia Z. Lack of association between cytokine gene polymorphisms and silicosis and pulmonary tuberculosis in Chinese iron miners. J Occup Health. 2008;50(6):445–454. doi: 10.1539/joh.l8006. [DOI] [PubMed] [Google Scholar]
- 54.Wu F, Xia Z, Qu Y, et al. Genetic polymorphisms of IL-1A, IL-1B, IL-1RN, NFKB1, FAS, and FASL, and risk of silicosis in a Chinese occupational population. Am J Ind Med. 2008;51(11):843–851. doi: 10.1002/ajim.20616. [DOI] [PubMed] [Google Scholar]
- 55.Wang YW, Lan JY, Yang LY, Wang De J, Kuang J. TNF-α and IL-1RA polymorphisms and silicosis susceptibility in Chinese workers exposed to silica particles: a case-control study. Biomed Environ Sci. 2012;25(5):517–525. doi: 10.3967/0895-3988.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Li Z, Xue J, Yan S, Chen P, Chen L. Association between tumor necrosis factor-α 308G/A gene polymorphism and silicosis susceptibility: a meta-analysis. PLoS ONE. 2013;8(10):e76614. doi: 10.1371/journal.pone.0076614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beck GJ, Schachter EN, Maunder LR, Schilling RS. A prospective study of chronic lung disease in cotton textile workers. Ann Intern Med. 1982;97(5):645–651. doi: 10.7326/0003-4819-97-5-645. [DOI] [PubMed] [Google Scholar]
- 58.Glindmeyer HW, Lefante JJ, Jones RN, Rando RJ, Weill H. Cotton dust and across-shift change in FEV1 as predictors of annual change in FEV1. Am J Respir Crit Care Med. 1994;149(3 Pt 1):584–590. doi: 10.1164/ajrccm.149.3.8118622. [DOI] [PubMed] [Google Scholar]
- 59.Christiani DC, Wang XR, Pan LD, et al. Longitudinal changes in pulmonary function and respiratory symptoms in cotton textile workers. A 15-yr follow-up study. Am J Respir Crit Care Med. 2001;163(4):847–853. doi: 10.1164/ajrccm.163.4.2006063. [DOI] [PubMed] [Google Scholar]
- 60.Castellan RM, Olenchock SA, Kinsley KB, Hankinson JL. Inhaled endotoxin and decreased spirometric values. An exposure-response relation for cotton dust. N Engl J Med. 1987;317(10):605–610. doi: 10.1056/NEJM198709033171005. [DOI] [PubMed] [Google Scholar]
- 61.Rylander R, Bake B, Fischer JJ, Helander IM. Pulmonary function and symptoms after inhalation of endotoxin. Am Rev Respir Dis. 1989;140(4):981–986. doi: 10.1164/ajrccm/140.4.981. [DOI] [PubMed] [Google Scholar]
- 62.Víctor VM, De la Fuente M. Several functions of immune cells in mice changed by oxidative stress caused by endotoxin. Physiol Res. 2003;52(6):789–796. [PubMed] [Google Scholar]
- 63.Ryan KA, Smith MF, Jr, Sanders MK, Ernst PB. Reactive oxygen and nitrogen species differentially regulate Toll-like receptor 4-mediated activation of NF-kappa B and interleukin-8 expression. Infect Immun. 2004;72(4):2123–2130. doi: 10.1128/IAI.72.4.2123-2130.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hang J, Zhou W, Wang X, et al. Microsomal epoxide hydrolase, endotoxin, and lung function decline in cotton textile workers. Am J Respir Crit Care Med. 2005;171(2):165–170. doi: 10.1164/rccm.200407-888OC. [DOI] [PubMed] [Google Scholar]
- 65.Thomas PS, Yates DH, Barnes PJ. Tumor necrosis factor-alpha increases airway responsiveness and sputum neutrophilia in normal human subjects. Am J Respir Crit Care Med. 1995;152(1):76–80. doi: 10.1164/ajrccm.152.1.7599866. [DOI] [PubMed] [Google Scholar]
- 66.Michel O, Ginanni R, Le Bon B, Content J, Duchateau J, Sergysels R. Inflammatory response to acute inhalation of endotoxin in asthmatic patients. Am Rev Respir Dis. 1992;146(2):352–357. doi: 10.1164/ajrccm/146.2.352. [DOI] [PubMed] [Google Scholar]
- 67.Wesselius LJ, Nelson ME, Bailey K, O’Brien-Ladner AR. Rapid lung cytokine accumulation and neutrophil recruitment after lipo-polysaccharide inhalation by cigarette smokers and nonsmokers. J Lab Clin Med. 1997;129(1):106–114. doi: 10.1016/s0022-2143(97)90167-0. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Hang J, Wang X, et al. TNF polymorphisms modify endotoxin exposure-associated longitudinal lung function decline. Occup Environ Med. 2007;64(6):409–413. doi: 10.1136/oem.2006.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang R, Zhao Y, Chu M, et al. A large scale gene-centric association study of lung function in newly-hired female cotton textile workers with endotoxin exposure. PLoS ONE. 2013;8(3):e59035. doi: 10.1371/journal.pone.0059035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vandenplas O, Toren K, Blanc PD. Health and socioeconomic impact of work-related asthma. Eur Respir J. 2003;22(4):689–697. doi: 10.1183/09031936.03.00053203. [DOI] [PubMed] [Google Scholar]
- 71.Tarlo SM, Lemiere C. Occupational asthma. N Engl J Med. 2014;370(7):640–649. doi: 10.1056/NEJMra1301758. [DOI] [PubMed] [Google Scholar]
- 72.Bignon JS, Aron Y, Ju LY, et al. HLA class II alleles in isocyanate-induced asthma. Am J Respir Crit Care Med. 1994;149(1):71–75. doi: 10.1164/ajrccm.149.1.8111601. [DOI] [PubMed] [Google Scholar]
- 73.Balboni A, Baricordi OR, Fabbri LM, Gandini E, Ciaccia A, Mapp CE. Association between toluene diisocyanate-induced asthma and DQB1 markers: a possible role for aspartic acid at position 57. Eur Respir J. 1996;9(2):207–210. doi: 10.1183/09031936.96.09020207. [DOI] [PubMed] [Google Scholar]
- 74.Mapp CE, Beghè B, Balboni A, et al. Association between HLA genes and susceptibility to toluene diisocyanate-induced asthma. Clin Exp Allergy. 2000;30(5):651–656. doi: 10.1046/j.1365-2222.2000.00807.x. [DOI] [PubMed] [Google Scholar]
- 75.Horne C, Quintana PJ, Keown PA, Dimich-Ward H, Chan-Yeung M. Distribution of DRB1 and DQB1 HLA class II alleles in occupational asthma due to western red cedar. Eur Respir J. 2000;15(5):911–914. doi: 10.1034/j.1399-3003.2000.15e17.x. [DOI] [PubMed] [Google Scholar]
- 76.Rihs HP, Barbalho-Krölls T, Huber H, Baur X. No evidence for the influence of HLA class II in alleles in isocyanate-induced asthma. Am J Ind Med. 1997;32(5):522–527. doi: 10.1002/(sici)1097-0274(199711)32:5<522::aid-ajim13>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 77.Beghé B, Padoan M, Moss CT, et al. Lack of association of HLA class I genes and TNF alpha-308 polymorphism in toluene diisocyanate-induced asthma. Allergy. 2004;59(1):61–64. doi: 10.1046/j.1398-9995.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- 78.Matheson JM, Johnson VJ, Luster MI. Immune mediators in a murine model for occupational asthma: studies with toluene diisocyanate. Toxicol Sci. 2005;84(1):99–109. doi: 10.1093/toxsci/kfi051. [DOI] [PubMed] [Google Scholar]
- 79.Bernstein DI, Wang N, Campo P, et al. Diisocyanate asthma and gene-environment interactions with IL4RA, CD-14, and IL-13 genes. Ann Allergy Asthma Immunol. 2006;97(6):800–806. doi: 10.1016/S1081-1206(10)60972-6. [DOI] [PubMed] [Google Scholar]
- 80.Piirilä P, Wikman H, Luukkonen R, et al. Glutathione S-transferase genotypes and allergic responses to diisocyanate exposure. Pharmacogenetics. 2001;11(5):437–445. doi: 10.1097/00008571-200107000-00007. [DOI] [PubMed] [Google Scholar]
- 81.Mapp CE, Fryer AA, De Marzo N, et al. Glutathione S-transferase GSTP1 is a susceptibility gene for occupational asthma induced by isocyanates. J Allergy Clin Immunol. 2002;109(5):867–872. doi: 10.1067/mai.2002.123234. [DOI] [PubMed] [Google Scholar]
- 82.Wikman H, Piirilä P, Rosenberg C, et al. N-Acetyltransferase genotypes as modifiers of diisocyanate exposure-associated asthma risk. Pharmacogenetics. 2002;12(3):227–233. doi: 10.1097/00008571-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 83.Kim SH, Cho BY, Park CS, et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy. 2009;39(2):203–212. doi: 10.1111/j.1365-2222.2008.03117.x. [DOI] [PubMed] [Google Scholar]
- 84.Bernstein DI, Kashon M, Lummus ZL, et al. CTNNA3 (α-catenin) gene variants are associated with diisocyanate asthma: a replication study in a Caucasian worker population. Toxicol Sci. 2013;131(1):242–246. doi: 10.1093/toxsci/kfs272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guarnieri M, Balmes JR. Outdoor air pollution and asthma. Lancet. 2014;383(9928):1581–1592. doi: 10.1016/S0140-6736(14)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez L, Declercq C, Iñiguez C, et al. Chronic burden of near-roadway traffic pollution in 10 European cities (APHEKOM network) Eur Respir J. 2013;42(3):594–605. doi: 10.1183/09031936.00031112. [DOI] [PubMed] [Google Scholar]
- 88.Robinson CL, Baumann LM, Romero K, et al. Effect of urbanisation on asthma, allergy and airways inflammation in a developing country setting. Thorax. 2011;66(12):1051–1057. doi: 10.1136/thx.2011.158956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bernstein AS, Rice MB. Lungs in a warming world: climate change and respiratory health. Chest. 2013;143(5):1455–1459. doi: 10.1378/chest.12-2384. [DOI] [PubMed] [Google Scholar]
- 90.Rice MB, Thurston GD, Balmes JR, Pinkerton KE. Climate change. A global threat to cardiopulmonary health. Am J Respir Crit Care Med. 2014;189(5):512–519. doi: 10.1164/rccm.201310-1924PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bowler RP, Crapo JD. Oxidative stress in allergic respiratory diseases. J Allergy Clin Immunol. 2002;110(3):349–356. doi: 10.1067/mai.2002.126780. [DOI] [PubMed] [Google Scholar]
- 92.Liu L, Poon R, Chen L, et al. Acute effects of air pollution on pulmonary function, airway inflammation, and oxidative stress in asthmatic children. Environ Health Perspect. 2009;117(4):668–674. doi: 10.1289/ehp11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patel MM, Chillrud SN, Deepti KC, Ross JM, Kinney PL. Traffic-related air pollutants and exhaled markers of airway inflammation and oxidative stress in New York City adolescents. Environ Res. 2013;121:71–78. doi: 10.1016/j.envres.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neophytou AM, Hart JE, Cavallari JM, et al. Traffic-related exposures and biomarkers of systemic inflammation, endothelial activation and oxidative stress: a panel study in the US trucking industry. Environ Health. 2013;12:105. doi: 10.1186/1476-069X-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Romieu I, Moreno-Macias H, London SJ. Gene by environment interaction and ambient air pollution. Proc Am Thorac Soc. 2010;7(2):116–122. doi: 10.1513/pats.200909-097RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonnell WF. Intersubject variability in human acute ozone responsiveness. Pharmacogenetics. 1991;1(2):110–113. doi: 10.1097/00008571-199111000-00010. [DOI] [PubMed] [Google Scholar]
- 97.David GL, Romieu I, Sienra-Monge JJ, et al. Nicotinamide adenine dinucleotide (phosphate) reduced:quinone oxidoreductase and glutathione S-transferase M1 polymorphisms and childhood asthma. Am J Respir Crit Care Med. 2003;168(10):1199–1204. doi: 10.1164/rccm.200305-684OC. [DOI] [PubMed] [Google Scholar]
- 98.Bergamaschi E, De Palma G, Mozzoni P, et al. Polymorphism of quinone-metabolizing enzymes and susceptibility to ozone-induced acute effects. Am J Respir Crit Care Med. 2001;163(6):1426–1431. doi: 10.1164/ajrccm.163.6.2006056. [DOI] [PubMed] [Google Scholar]
- 99.Corradi M, Alinovi R, Goldoni M, et al. Biomarkers of oxidative stress after controlled human exposure to ozone. Toxicol Lett. 2002;134(1–3):219–225. doi: 10.1016/s0378-4274(02)00169-8. [DOI] [PubMed] [Google Scholar]
- 100.Romieu I, Ramirez-Aguilar M, Sienra-Monge JJ, et al. GSTM1 and GSTP1 and respiratory health in asthmatic children exposed to ozone. Eur Respir J. 2006;28(5):953–959. doi: 10.1183/09031936.06.00114905. [DOI] [PubMed] [Google Scholar]
- 101.Chen C, Arjomandi M, Tager IB, Holland N, Balmes JR. Effects of antioxidant enzyme polymorphisms on ozone-induced lung function changes. Eur Respir J. 2007;30(4):677–683. doi: 10.1183/09031936.00160806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Islam T, Berhane K, McConnell R, et al. Glutathione-S-transferase (GST) P1, GSTM1, exercise, ozone and asthma incidence in school children. Thorax. 2009;64(3):197–202. doi: 10.1136/thx.2008.099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreno-Macías H, Dockery DW, Schwartz J, et al. Ozone exposure, vitamin C intake, and genetic susceptibility of asthmatic children in Mexico City: a cohort study. Respir Res. 2013;14:14. doi: 10.1186/1465-9921-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gilliland FD, Li YF, Saxon A, Diaz-Sanchez D. Effect of glutathione-S-transferase M1 and P1 genotypes on xenobiotic enhancement of allergic responses: randomised, placebo-controlled crossover study. Lancet. 2004;363(9403):119–125. doi: 10.1016/S0140-6736(03)15262-2. [DOI] [PubMed] [Google Scholar]
- 105.Reddy P, Naidoo RN, Robins TG, et al. GSTM1, GSTP1, and NQO1 polymorphisms and susceptibility to atopy and airway hyper-responsiveness among South African schoolchildren. Lung. 2010;188(5):409–414. doi: 10.1007/s00408-010-9246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee YL, Lin YC, Lee YC, Wang JY, Hsiue TR, Guo YL. Glutathione S-transferase P1 gene polymorphism and air pollution as interactive risk factors for childhood asthma. Clin Exp Allergy. 2004;34(11):1707–1713. doi: 10.1111/j.1365-2222.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 107.Salam MT, Lin PC, Avol EL, Gauderman WJ, Gilliland FD. Microsomal epoxide hydrolase, glutathione S-transferase P1, traffic and childhood asthma. Thorax. 2007;62(12):1050–1057. doi: 10.1136/thx.2007.080127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Islam T, McConnell R, Gauderman WJ, Avol E, Peters JM, Gilliland FD. Ozone, oxidant defense genes, and risk of asthma during adolescence. Am J Respir Crit Care Med. 2008;177(4):388–395. doi: 10.1164/rccm.200706-863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang IA, Holz O, Jörres RA, et al. Association of tumor necrosis factor-alpha polymorphisms and ozone-induced change in lung function. Am J Respir Crit Care Med. 2005;171(2):171–176. doi: 10.1164/rccm.200402-194OC. [DOI] [PubMed] [Google Scholar]
- 110.Li YF, Gauderman WJ, Avol E, Dubeau L, Gilliland FD. Associations of tumor necrosis factor G-308A with childhood asthma and wheezing. Am J Respir Crit Care Med. 2006;173(9):970–976. doi: 10.1164/rccm.200508-1256OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu H, Romieu I, Sienra-Monge JJ, et al. Parental smoking modifies the relation between genetic variation in tumor necrosis factor-alpha (TNF) and childhood asthma. Environ Health Perspect. 2007;115(4):616–622. doi: 10.1289/ehp.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kerkhof M, Postma DS, Brunekreef B, et al. Toll-like receptor 2 and 4 genes influence susceptibility to adverse effects of traffic-related air pollution on childhood asthma. Thorax. 2010;65(8):690–697. doi: 10.1136/thx.2009.119636. [DOI] [PubMed] [Google Scholar]
- 113.Yang IA, Fong KM, Zimmerman PV, Holgate ST, Holloway JW. Genetic susceptibility to the respiratory effects of air pollution. Thorax. 2008;63(6):555–563. doi: 10.1136/thx.2007.079426. [DOI] [PubMed] [Google Scholar]
- 114.James J. Health Policy Brief: Workplace Wellness Program. [Accessed in 2014];Health Affairs. 2013 Available at www.healthaffairs.org/healthpolicy-briefs/brief.php?brief_id=93.