Abstract
Comparisons of flowering plant genomes reveal multiple rounds of ancient polyploidy characterized by large intragenomic syntenic blocks. Three such whole-genome duplication (WGD) events, designated as rho (ρ), sigma (σ), and tau (τ), have been identified in the genomes of cereal grasses. Precise dating of these WGD events is necessary to investigate how they have influenced diversification rates, evolutionary innovations, and genomic characteristics such as the GC profile of protein-coding sequences. The timing of these events has remained uncertain due to the paucity of monocot genome sequence data outside the grass family (Poaceae). Phylogenomic analysis of protein-coding genes from sequenced genomes and transcriptome assemblies from 35 species, including representatives of all families within the Poales, has resolved the timing of rho and sigma relative to speciation events and placed tau prior to divergence of Asparagales and the commelinids but after divergence with eudicots. Examination of gene family phylogenies indicates that rho occurred just prior to the diversification of Poaceae and sigma occurred before early diversification of Poales lineages but after the Poales-commelinid split. Additional lineage-specific WGD events were identified on the basis of the transcriptome data. Gene families exhibiting high GC content are underrepresented among those with duplicate genes that persisted following these genome duplications. However, genome duplications had little overall influence on lineage-specific changes in the GC content of coding genes. Improved resolution of the timing of WGD events in monocot history provides evidence for the influence of polyploidization on functional evolution and species diversification.
Keywords: whole-genome duplication, grasses, monocots, GC content
Introduction
Paleopolyploidy or ancient whole-genome duplication (WGD) events have occurred across the eukaryotic tree of life and have been hypothesized to have had major impacts on life history innovations and organismal diversification (Levin 1983; Taylor and Raes 2004; Soltis et al. 2009; De Smet and Van de Peer 2012). WGDs have been especially widespread throughout flowering plant history (Cui et al. 2006; Vanneste et al. 2014), and ancient polyploidy has been associated with the origin of seed plants (spermatophyta) and angiosperms (Jiao et al. 2011). In fact, multiple rounds of WGD have been inferred for many plant lineages (Tang et al. 2010; Jiao et al. 2011, 2012; McKain et al. 2012; Yang et al. 2015) including those leading to the model plant species Arabidopsis thaliana (Blanc and Wolfe 2004), the tree species Populus trichocarpa (Tuskan et al. 2006), legumes (Fabaceae; [Cannon et al. 2015]), and grasses (Poaceae; [Paterson et al. 2004; Tang et al. 2010]). Though many recent polyploid lineages have relatively lower net diversification rates than diploid lineages, genome duplications have undoubtedly contributed to molecular, ecological, and phylogenetic diversification during angiosperm evolution (Mayrose et al. 2011; Soltis et al. 2014). However, a mechanistic understanding of how polyploidy has contributed to evolutionary innovations and diversification in angiosperm history requires precise phylogenetic placement of the nature and timing of these ancient WGDs (Soltis et al. 2009).
Polyploid can occur through either somatic genome doubling within meristematic tissue, zygotes, or young embryos, or with the fusion of unreduced gametes (Ramsey and Schemske 1998). Both autopolyploidy (multiple copies of the same genome) and allopolyploidy (multiple copies of different genomes, usually of different species) have prevalence in angiosperm history (Barker et al. 2016). The formation of stable, sexually reproducing polyploid populations is thought to involve two steps: 1) mating between individuals with reduced and unreduced gametes, sometimes producing individuals with odd polyploidy levels, the so-called triploid bridge (Mallet 2007), and 2) subsequent fusion of unreduced gametes producing a stable tetraploid, or higher even-level polyploid, such as hexaploid wheat (Levin 2002). Morphological, physiological, and ecological diversification among polyploid populations can follow as a consequence of differential gene loss or subfunctionalization and neofunctionalization of retained duplicates (i.e., homeologs).
Polyploidy is hypothesized to have played a major role in the evolution of key innovations through the proliferation of novel genes and gene interactions (Ohno 1970; Levin 1983; Van de Peer et al. 2009). Polyploidy was recently shown to be a driver of innovation and novelty in secondary metabolites in the mustard family (Edger et al. 2015), demonstrating a connection between genome duplication, adaptation, and diversification. Additionally, polyploidy has been implicated as spurring epigenetic changes (Yoo et al. 2014), immediate and sustained variation in gene expression (Yoo et al. 2013), and reorganization of gene networks (De Smet and Van de Peer 2012). The process of duplicate gene loss (fractionation) can exhibit biases toward some functional classes (Maere et al. 2005; Doyle et al. 2008; Freeling 2009; Tang et al. 2012), and potentially contribute to phenotypic variation due to differential duplicate retention. In some grass family clades (such as the tribe Andropogoneae), polyploidy has been shown to be a recurring evolutionary process with multiple independent polyploid events occurring over a relatively short period of time (Estep et al. 2014).
Early analyses of the rice genome revealed a WGD event designated as rho (Paterson et al. 2004; Yu et al. 2005) inferred to have predated the divergence of the BOP and PACMAD clades within the grass phylogeny (Grass Phylogeny Working Group 2001; Blanc and Wolfe 2004; Paterson et al. 2004; Schlueter et al. 2004; Yu et al. 2005; Wu et al. 2008; Grass Phylogeny Working Group II 2012; Soreng et al. 2015). The rho event has been implicated as contributing to the success of Poaceae through the role of duplicated MADS-box genes in the development of the spikelet (Preston and Kellogg 2006; Preston et al. 2009) and the role of duplicated genes involved in the development of starch-rich seeds (Wu et al. 2008; Comparot-Moss and Denyer 2009). Despite the rho event’s impact on functional and phylogenetic diversification of the grasses, the absence of genomic-scale analyses for early diverging grass lineages (pre-divergence of BOP and PACMAD clades) and nongrass graminids has left the precise placement of rho uncertain (Paterson et al. 2004; Soltis et al. 2009).
Later comparative genomic analyses performed with refined computational tools elucidated an even older genome duplication in grass genomes designated as sigma that was estimated to have occurred sometime prior to the diversification of Poaceae (Paterson et al. 2009; Tang et al. 2010). Analyses of the Musa genome identified three lineage-specific WGDs within Zingiberales and determined that the rho and sigma events occurred within the Poales lineage but prior to the divergence of the PACMAD and BOP clades (D’Hont et al. 2012).
Based on synteny analysis of the rice and sorghum genomes, Tang et al. (2010) hypothesized a third paleopolyploid event prior to sigma but such an event was not inferred in the Musa genome analyses (D’Hont et al. 2012). Comparative synteny analyses of oil palm (Elaeis guineensis) and eudicot genomes have recently placed this event, designated tau, as early in the evolutionary history of monocots (Jiao et al. 2014), but after the divergence of the Alismatales (Ming et al. 2015). Clearly, polyploidy has contributed to molecular variation and evolution within grass and other monocot lineages, but a full understanding of the impact of the rho, sigma, and tau events requires more precise estimation of the timing of these genome duplications relative to branching points in the monocot phylogeny.
Among the genome-wide molecular changes that may have been influenced by polyploidy, the GC profile of protein-coding genes has been hypothesized to have influenced the synonymous substitution rate of retained duplicates (Wang et al. 2005). A bimodal distribution of GC composition among coding genes, and 5′ to 3′ gradients in GC content of coding genes have long been identified as divergent characteristics of grass genomes relative to eudicot genomes (Carels et al. 1998). There have been conflicting reports on whether these are features of an ancestral monocot genome (Clément et al. 2015) or have been independently derived in the grasses (Carels and Bernardi 2000; Kuhl et al. 2004, 2005) and other plant lineages (Escobar et al. 2011; Serres-Giardi et al. 2012). The functional implications of variation in GC content among monocot lineages have been discussed in light of associations between whole-genome GC content, genome size and architecture, and environmental conditions (Smarda et al. 2014). However, the relative importance of mutational processes, including GC-biased gene conversion (Eyre-Walker and Hurst 2001; Duret and Galtier 2009), and selective processes is controversial (Shi et al. 2006; Tatarinova et al. 2010, 2013; Serres-Giardi et al. 2012; Glémin et al. 2014; Clément et al. 2015). The debate has been fueled by the observed negative correlations between GC-content and gene length, exon number (Carels and Bernardi 2000; Wang et al. 2004; Shi et al. 2006; Tatarinova et al. 2010), and gene body methylation, together with positive correlations with recombination rates and variance in gene expression (Tatarinova et al. 2010; Serres-Giardi et al. 2012). Here, we ask whether ancient polyploidy may have contributed to the evolution of variation in GC composition among coding genes in monocot genomes.
Following the classic work of Bowers et al. (2003), we employ a phylogenomic approach to resolve the timing of rho and sigma relative to speciation events within the Poales and tau earlier in monocot history. Transcriptome data for species sampled within each family in Poales were generated and assembled transcripts included in gene trees to expand taxon sampling beyond species with available genome sequences. Species were chosen for our phylogenomic analyses to increase sampling density and uniformity across the Poales phylogeny. Broader taxon sampling is considered to increase the accuracy of phylogeny (Zwickl and Hillis 2002; Leebens-Mack et al. 2005). Gene trees were estimated for all gene families with paralog pairs mapping to syntenic blocks (i.e., syntelogs) within the rice and sorghum genomes, and the timing of duplication relative to speciation events was inferred through interrogation of the gene trees. The combination of synteny-based approach and the phylogenomic approach is the best method to assay the origins of WGD duplicates. At the same time, putative single copy gene families were used to rigorously estimate phylogenetic relationships within the Poales and related monocot lineages, improving precision over previous reconstructions with plastid sequences. Gene trees were reconciled with the inferred species phylogeny to estimate the timing of rho, sigma, and tau and the influence of these events on the evolution of key innovations. Finally, variation in GC composition across the Poales was analyzed and tested to determine if such variation relates to the relative placement of these polyploid events.
Materials and Methods
Taxon Sampling
Relationships within the order Poales have recently been investigated through analyses of gene sequences extracted from plastid genome sequences (Givnish et al. 2010), and we used phylogenies from that study to guide sampling of all major clades and families within the order. RNAs were isolated from mixed vegetative and reproductive tissues for 25 species, and transcriptome assemblies were combined with available genome data sets to carry out the analyses described below. Sample identities and data descriptions can be found in supplementary table S1, Supplementary Material online along with doi numbers for RNA Seq reads, assemblies, multiples sequence alignments, and gene trees deposited in NCBI's Short Read Archive (SRA) (PRJNA313089) database and DRYAD (XXXXX).
RNA Isolation and Sequencing
RNA was isolated from fresh young leaf or apical meristematic tissue using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Samples were kept on liquid nitrogen prior to isolation. RNA was eluted into a final volume of 100 µl of RNase-free water. RNA total mass and quality were estimated using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). Samples were deemed acceptable if RIN (RNA Integrity Number) scores were greater than 8.0.
The TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA) was used to construct paired-end libraries with an average fragment length of 300 base pairs (bp). Sample libraries were sequenced on a GAIIx Genome Analzyer (Illumina), at University of Missouri-Columbia’s DNA Core Facility, and on a HiSeq 2000 (Illumina) at the BGI Americas sequencing facility in Davis, CA.
Transcriptome Assembly
Illumina data generated in this study and downloaded from the NCBI SRA were assembled in the same fashion. FastQC v0.10.1 (Andrews 2010) was used to identify adapter contamination and Cutadapt v1.1 (Martin 2011) was used to clean contaminated sequences. Two mismatches per 10 bp were allowed and a minimum overlap of at least 10 bp was required for contaminant identification and trimming. Cleaned reads were further trimmed using a custom perl script (https://github.com/mrmckain/poales-polyploidy, last accessed March 22, 2016) that trimmed reads from the ends until there were three consecutive bases with a quality score of 20 or more. Finally, reads with a median quality score less than 22, more than three uncalled bases, or trimmed lengths less than 40 bp were removed. We retained the orphaned as well as paired reads for downstream assembly and analyses.
Cleaned and filtered data sets were assembled using Trinity (Release 2012-06-08; Grabherr et al. 2011) with default parameters. Reads were aligned to the Trinity assembly using bowtie v0.12.8 (Langmead et al. 2009) through the alignReads.pl script available in the Trinity distribution. Output from this script was then piped into the run_RSEM.pl script (also packaged with Trinity), which utilizes RSEM v1.2.0 (Li and Dewey 2011) to quantify transcript abundance. Per kilobase of exon per million fragments mapped (FPKM) was estimated for each component (gene), and the percentage of mapped fragments corresponding to each isoform within a component was estimated using the summarize_RSEM_fpkm.pl script packaged within Trinity and an average fragment length setting of 300 bp. Isoforms that had 1% or less of all fragments mapped to a component were removed to create a data set that represented well-supported transcripts. Roche 454 transcriptome reads for E. guineensis (GenBank accession numbers: SRX059258–SRX059263) were assembled with MIRA (Chevreux et al. 2004) using default parameters.
Transcriptome Translation and Gene Family Circumscription
Translation of the transcriptome assemblies was conducted using a series of custom perl scripts (https://github.com/mrmckain/RefSeq, last accessed March 22, 2016). TBLASTX matches to a database of gene models from 22 genomes used in the gene family circumscription developed by the Amborella Genome Project (2013) were identified for each assembled transcript, and matches with an e-value less than 1e-10 were used to guide translations with GeneWise (part of the Wise2 v2.2.0 package; Birney et al. 2004). The longest GeneWise translations for each transcript were used. In rare cases where internal stop codons were identified, they were spliced out of the transcript assemblies. BLASTP searches against the 22-genome amino acid database were then performed with the inferred amino acid sequences in order to assign them into the 22-genome orthogroups (i.e., gene families circumscribed by OrthoMCL (Li et al. 2003) as in the Amborella genome paper (Amborella Genome Project 2013).
Species Tree Estimation
Species trees were estimated using concatenated and coalescence-based approaches. A set of 970 single/low copy gene families were identified in our orthogroups (Duarte et al. 2010) and used as a starting set for identifying strictly defined single copy genes in our taxon set. Peptide sequences within these orthogroups were aligned using MUSCLE v3.8.31 (Edgar 2004), and coding sequences (CDSs) were aligned onto the amino acid alignments using PAL2NAL v13 (Suyama et al. 2006). Alignments were filtered using two criteria. First, columns in alignments were removed if gaps were observed in more than 90% of sequences (rows in the alignment matrix). Second, transcript translations (rows) were removed if they covered less than 30% of the total alignment length for the gene family. Maximum likelihood (ML) gene trees were estimated using RAxML v7.3.0 (Stamatakis 2006) with the general time reversible + Γ substitution model and 500 bootstrap replicates. Gene trees were rooted to outgroup taxa (Amborella trichopoda and Vitis vinifera) found in each gene family. Orthogroup trees were analyzed for gene copy count for each species using a modified version of the clone-reducer script from Estep et al. (2014) (https://github.com/mrmckain/clone_reducer, last accessed March 22, 2016) as used in Cannon et al. (2015).
The primary goal of the species tree estimation was to identify relationships of major lineages (i.e., branches coming off of the backbone of the phylogeny) rather than more shallow relationships. Putative isoforms for a species were collapsed into a consensus sequence if they formed a species-specific clade in the gene tree with a BSV (bootstrap support value) of 50 or greater. Collapsing close isoforms (possible splice variants, alleles, or recent duplicates) allowed us to maximize the number of single copy orthogroups. Further, for the species tree analysis we reduced certain lineages to a single species representative in order to avoid coalescence issues that were potentially lowering the total number of single copy orthogroups. This included reducing the genera with multiple samples (Cyperus, Typha, Juncus, and Mayaca) and the family Centrolepidaceae to either single species or combining them into a single sample. These groups were identified as the hotspots where coalescence of putative isoforms was occurring within the reduced lineage but not always within the species. We repeated the clone reducer methodology after these simplification steps and identified 234 high occupancy orthogroups that maintained genes in single copy across all taxa.
To estimate the concatenation-based species tree, each aligned gene family was concatenated for a species. If the species did not have a representative in the gene family, the taxon was represented by Ns in the alignments for the missing gene. All 234 orthogroups were represented in the concatenated analysis. The topology was reconstructed using RAxML with the GTR + Γ model over 500 bootstrap replicates and rooted relative to A. trichopoda.
The coalescence-based analysis was conducted using ASTRAL v.4.7.6 (Mirarab et al. 2014). Trees that were used in the ASTRAL analysis were generated in RAxML as above. The bootstrapping option of ASTRAL was used for 100 replicates.
A consensus topology was created to represent the shared relationships of both methodologies. Nodes were collapsed where the topology disagreed. This did not affect the estimation of the timing of WGD events.
Ks Frequency Plot Estimation and Estimation of Ks-Derived Putative Paralogs
Ks frequency plots were estimated using the FASTKs pipeline (https://github.com/mrmckain/FASTKs, last accessed March 22, 2016). Translated transcriptome data sets were blasted against themselves using an e-value cutoff of 1e-40. Putative pairs were filtered if they: 1) exhibited 100% identity in their alignment, 2) had less than 300 base pairs of an alignment length, and 3) had less than 40% identity. Amino acid sequences for putative paralog pairs were then aligned using MUSCLE, and back translated to CDS using PAL2NAL. Ks, Ka, and Ka/Ks were estimated for the aligned pairs using codeml in PAML v.4.8 (Yang 2007) using the same parameters used by McKain et al. (2012). Normal mixture models were estimated for Ks values using the mclust v.5.0.2 (Fraley and Raftery 2002; Fraley et al. 2012) as implement in R. Ks plots were used as a secondary confirmation of rho and sigma placement in the species tree (supplementary fig. 3, Supplementary Material online).
Estimation of Syntelogs and Gene Tree Estimation for Multicopy Gene Families
Synteny blocks and retained duplicate genes derived from the rho, sigma, and tau WGDs (i.e., syntelogs) were identified in the Oryza sativa and Sorghum bicolor genomes as described by Tang et al. (2010). The rho synteny blocks were identified using a chaining distance of 40 genes. Following this step, we reconstructed the “pre-rho” putative ancestral regions (PARs; Tang et al. 2010) by interleaving the genes between the rho duplicate pairs. A second synteny search was then performed on the pre-rho PARs to identify the sigma + tau synteny blocks using a relaxed chaining distance of 60 genes.
Jiao et al. (2014) inferred tau synteny blocks through comparisons of PARs estimated for oil palm (E. guineensis) and the eudicot, sacred lotus (Nelumbo nucifera). Each of these genomes was known to have included at least one round of paleopolyploidy since divergence and Jiao et al. (2014) estimated PARs for these two genomes using the methods described above. Tau synteny blocks were identified as pairs of oil palm PARs lining up with a single sacred lotus PAR. Syntelog sets identified in the oil palm tau synteny blocks were then compared with rice and sorghum genes to infer a total of 2,248 duplicate gene pairs in the sorghum and rice genomes derived from the tau WGD (Jiao et al. 2014). We performed phylogenomic analyses on orthogroups with theses tau duplicates and compared them with the sigma + tau syntelogs we inferred in analyses of the rice and sorghum pre-rho PARs (see above).
In some instances, syntelog pairs were sorted to different gene families as circumscribed by OrthoMCL (Li et al. 2003; Amborella Genome Project 2013). Because we were interested in identifying the last common ancestral (LCA) node in gene trees for each syntelog set, these orthogroups were combined for alignment and phylogenetic analysis. Gene family alignment and subsequent tree estimation were conducted as described above for species tree estimation. If a gene tree did not contain a nonmonocot outgroup (Amborella or Vitis) homolog, it was dropped from further analysis.
Queries of the resulting gene tree analyses were conducted using PUG (https://github.com/mrmckain/PUG, last accessed March 22, 2016) with the consensus species tree as the guide tree. PUG uses an algorithm that queries putative paralogs in a gene tree identifying species congruence relative to the species tree. Taxa that were removed prior to species tree estimation were inserted back into the tree for estimation of WGD placement. The timing of duplication events relative to the speciation events shown in the consensus species tree (fig. 1) was estimated by querying the rooted gene trees for the species represented in clades defined by an LCA node for each syntelog set. Syntelog sets with poorly supported LCA nodes demonstrating BSVs less than 50 were not used to infer the timing of WGDs. For the remaining syntelog sets, placement of duplication events was based on the species composition within the clade defined by the LCA node in the gene trees and the species composition within its sister lineage. This second criterion for placing duplication events was implemented to minimize the impact of incomplete gene sampling in the transcriptome assemblies. For an LCA node placement to be accepted, the following criteria had to be met: 1) a minimum of two taxa had to be present above the duplication node (i.e., at least one more taxon in addition to rice or sorghum syntelog pairs), 2) no taxa found above the hypothesized WGD position in the species tree could be found in the lineage sister to the putative LCA defined clade in the gene tree, and 3) the lineage sister to the LCA defined clade had to contain at least one taxon from the sister lineage to the hypothesized WGD node in the species tree. Gene tree topologies were also inspected manually to verify the results of the automated analysis. Counts of LCAs (Last Common Ancestor for hypothesized WGD clade) with well supported placement were made for each node in the species tree and split into two categories based on the BSV of the LCA: 1) 50 ≤ BSV < 80 and 2) BSV ≥ 80.
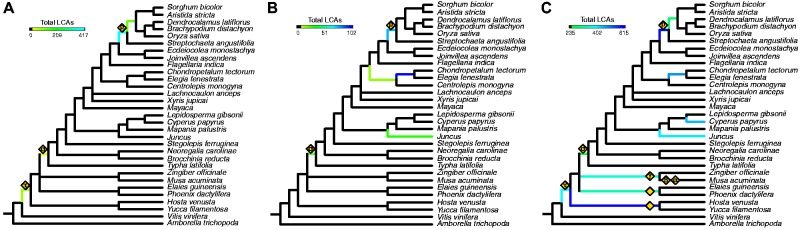
Fig. 1.—
Consensus species tree from concatenated and coalescence-based analysis of 234 single copy orthogroups with results from gene tree querying of putative paralogs. (A) Mapping results of synteny-derived paralogs from the rice and sorghum genomes displayed as total number of unique last common ancestor (LCA) nodes with BSV ≥80. Results show placement of rho, sigma, and tau WGD events. (B) Mapping results of Ks plot-derived paralogs with 22 (number for sigma event) or more total unique LCAs and boostrap values ≥80 for Poales species only. (C) Mapping results of gene tree-derived paralogs with 235 (number for sigma event) or more total unique LCAs and boostrap values ≥80 for monocot species only. Previously published WGD events are identified and placed on the tree, including a shared Zingiberaceae event (gamma), a palm event, and an Agavoideae event, represented as gold diamonds. If previously named, the Greek character representing the event is also displayed. Higher support for rho, sigma, and tau is identified relative to the synteny-derived paralogs and other potential WGD events in Juncus, Cyperus, and Restionaceae are also identified.
For each orthogroup, all possible pair combinations of transcripts/annotated genes for each species, except Amborella and Vitis, were estimated. These pairs were used in a separate analysis with PUG to identify putative WGD events by accumulated signal at all nodes.
GC Content Analysis
GC content was analyzed for all orthogroups with identified putative paralogs pairs and for other orthogroups with at least ten taxa present. A total of 13,798 orthogroups were queried. The PAL2NAL alignments were used for GC estimation. For each orthogroup, sequences were removed if they were not within 1 standard deviation (SD) of the average length of the orthogroup alignment in order to better assess 5′ to 3′ gradients in GC content along complete or nearly complete transcript assemblies.
Total GC percentage was calculated for each transcript along with 5′ to 3′ gradients in GC composition. For each orthogroup, the alignment length was estimated as number of codons and GC composition were estimated for nonoverlapping windows corresponding to 1% of the alignment. For each species, the GC content of each nonoverlapping 1% interval was calculated as the total GC percentage for that window across all orthogroups.
Mean and SD of GC composition (both total and GC3) were estimated for each taxon. A student’s t-test was used to determine if the mean total GC percentage was different from the mean GC3 in each taxon. Hartigan’s dip test (HDT) for unimodality, as implemented in diptest v.0.75-7 in R, was used to test if the distribution of total GC content and GC3 for genes across all taxa exhibited unimodality. For taxa where the unimodality null hypothesis was rejected, an inspection of the distribution of %GC across all orthogroups suggested the data were bimodal. A kmeans clustering analysis of these seven taxa using two clusters identified cluster centers at 46.7% and 63.7% GC for total GC and 45.8% and 82.5% for GC3. Bimodal distributions were particularly evident for GC3, with 18 taxa exhibiting bimodality with kmeans centers at 45.7% and 60.6% GC for total GC and 44.4% and 76.1% for GC3.
For each of the seven taxa found to have a bimodal distribution for total GC composition, kmeans clusters (n = 2) were calculated. Clustered transcripts were used to identify abundance of low GC and high GC transcripts in orthogroups. Orthogroups were then classified into three categories: 1) high GC—75% or more of transcripts for a given taxon in an orthogroup are clustered as high GC, 2) low GC—75% or more of transcripts for a given taxon in an orthogroups are clustered as low GC, and 3) mixed GC—remaining orthogroups that do not fall into other classes. Results from each taxon were compared and sets of true high GC and true low GC were identified across all bimodally distributed taxa. GC composition distributions for all transcripts in both high and low GC classes were compared. We then tested for significant differences in high vs. low GC classification distributions for each taxon and all taxa using t-tests. A contingency test was used to test for significant associations between orthogroup GC content classifications and the retention of tau, sigma, or rho duplicates across orthogroups.
Results
Transcriptome Assemblies, Translations, and Gene Family Sorting
In total, over 1.04 million transcripts from 35 species (an average of 36,758 transcripts per taxon) were assembled after adaptor trimming and quality filtering. Transcript assemblies with an average length of 1,005 bp (supplementary table S2, Supplementary Material online) were translated, and sorted into gene families as circumscribed through OrthoMCL clustering of gene models from 22 sequenced and annotated plant genomes including rice and sorghum (Li et al. 2003; Wall et al. 2008; Amborella Genome Project 2013). Resulting orthogroups (i.e., OrthoMCL clusters) including sequences from up to all 35 species included in the study (supplementary table S1, Supplementary Material online) were used for species tree estimation and resolution of timing of ancient WGDs.
Single Copy Gene Family Phylogeny Analysis
Multiple sequence alignments for all 234 orthogroups found to have no more than one gene copy in any of the sampled genome or filtered transcriptomes were concatenated and an ML tree was estimated from the resulting super-matrix (Materials and Methods; supplementary fig. S1, Supplementary Material online). A coalescence-based species tree estimate was generated from the 234 orthogroup trees using ASTRAL (Mirarab et al. 2014; supplementary fig. S2, Supplementary Material online). Minimal discordance was found between the super-matrix and ASTRAL trees, and all relationships with bootstrap values (BSV) greater than 90% were considered reliable and recovered in both analyses (supplementary figs. S1 and S2, Supplementary Material online). These robustly estimated relationships (fig. 1) are largely consistent with previous ML super-matrix analyses of 81 plastid genes (Givnish et al. 2010; Barrett et al. 2016) with a few exceptions. The basal lineage of Poales differs between the nuclear and plastid phylogenies. Although the plastome analyses placed Bromeliaceae sister to the rest of Poales, our nuclear gene analyses recovered strong support for Typhaceae as sister to the rest of Poales in both the concatenated and coalescence-based analysis. Further, Givnish et al. (2010) found strong support for an Eriocaulaceae + Mayacaceae clade and Xyridaceae separate, whereas our analyses suggest Lachnocaulon (Eriocaulaceae) and Xyris (Xyridacae) form in a clade that does not include Mayaca (Mayacaceae). However, the support for a Xyridaceae + Eriocaulaceae clade is very low in the coalescence tree (BSV 44), suggesting rapid diversification at this point within the Poales phylogeny. Finally, we place Joinvillea (Joinvilleaceae) and Ecdeiocolea (Ecdeiocoleaceae) in a clade, sister to the Poaceae (fig. 1) in contrast to the plastome-based resolution of Joinvilleaceae and Ecdeiocoleaceae as successive sister lineages in a grade leading to the Poaceae (Givnish et al. 2010; Barrett et al. 2016). The plastid genome is inherited as a single-locus, so differences between the trees inferred from multiple nuclear genes and plastid genome may be due to incomplete sorting of ancestral variation in the plastid genome haplotypes between speciation events (Maddison 1997). In any event, the discrepancies between our inferred species phylogeny and plastome-based phylogenetic inferences involve single rearrangements across short internodes on the backbone of our species tree estimate. Therefore, discordance among gene histories due to incomplete lineage sorting is expected. We used the well-supported relationships found to be concordant in our nuclear gene super-matrix and coalescence-based analyses (fig. 1) to guide gene tree queries and estimate the timing of the rho, sigma, and tau WGD events, after the reinsertion of removed taxa (Typha angustifolia, Cyperus alternifolius, Aphelia sp.) and the splitting of combined genera (Mayaca—Mayaca fluviatilis and Mayaca sp., Juncus—Juncus effusus and Juncus inflexus).
Delineation of Syntelog Pairs from Sorghum and Oryza Genomes
Following the methodology of Tang et al. (2010), syntenic regions were identified within the genomes of S. bicolor (sorghum) and O. sativa (rice) using a hierarchical approach (see Materials and Methods). A total of 56 syntenic block pairs identified in the rice genome were assigned to the rho event, and 39 rho block pairs were identified in the sorghum genome. These blocks included 4,296 rice syntelog pairs and 3,971 sorghum syntelog pairs. Synteny analysis of the pre-rho blocks resulted in circumscription of 58 sigma + tau blocks in rice and 63 blocks in sorghum. These inferred syntenic blocks associated with sigma or tau include 1,782 and 1,898 syntelog pairs in rice and sorghum genomes, respectively. In addition, a total of 2,248 duplicated rice and sorghum gene pairs were associated with the tau event by Jiao et al. (2014), to add to our exhaustive list of gene pairs for dating purpose. All inferred rho, sigma, and tau syntelog pairs were included in gene tree analyses in order to estimate the timing of duplication relative to speciation events.
Querying of Gene Trees for Placement of WGD Events Using Paralogs from Genomic Data
Orthogroup membership was determined for all 8,267 putative rho and 3,680 sigma + tau syntelog pairs derived from our synteny analysis. In 1,680 instances syntelog pairs were placed in separate orthogroups. In these cases, the homologous orthogroups were combined before sequence alignment and tree estimation. In total, alignments and ML trees were estimated for 2,795 OrthoMCL circumscribed gene families (Amborella Genome Project 2013) including one or more syntelog pairs, 2,089 of which included at least one V. vinifera or A. trichopoda gene that could be used as outgroup sequences to root gene duplications that occurred at any point in monocot history. Collectively, these 2,089 gene trees included a total of 6,484 syntelog pairs mapping to 3,203 unique ancestral nodes representing the LCA homeologous gene pairs.
The 3,203 LCA nodes were filtered based on BSV and grouped into two sets, those with 50≤ BSV<80 and those with BSV≥80 (designated as BSV 50 and BSV 80, respectively). Additionally, trees were queried in relationship to the estimated species tree, and only nodes in the gene tree congruent in topology to the species tree were accepted (see Materials and Methods). After filtering based on support values, 836 unique LCA nodes for 1,348 rho block syntelog pairs and 124 LCA nodes for 318 sigma + tau block pairs for were retained.
The estimated timing of duplication events relative to the speciation events depicted in figure 1A were inferred by determining the collection of species represented in clades defined by the syntelog LCA nodes described above. This analysis strongly suggested that the rho WGD occurred on the branch leading to the last common ancestor of all extant grass species including the basal grass Streptochaeta. Trees with rho syntelog pairs included 411 rho LCA nodes with BSV 80 defining clades with Streptochaeta genes but no genes from taxa outside of the Poaceae (fig. 1A). Another 107 rho syntelog LCA nodes with BSV of 50 or better showed this same pattern. In contrast, just 123 rho LCA nodes (BSV 80; 143 BSV 50) suggested that rho occurred after Streptochaeta diverged from the rest of the grasses but prior to the diversification of the BOP + PACMAD clades (fig. 1A).
As expected, sigma + tau block LCA nodes mapped to two points on the species tree corresponding to the two separate WGD events. Presumed sigma related duplication LCAs were concentrated (26 BSV 80; 11 BSV 50) on the branch leading to the last common ancestor of all extant species within the Poales, whereas the concentration of LCAs mapping to the Asparagales + commelinid clade (50 BSV 80; 14 BSV 50) is interpreted as representing tau (fig. 1A).
This timing of the tau WGD was also inferred by querying orthogroup trees including the homeologs identified by Jiao et al. (2014). Of the 2,248 putative tau pairs from that study, we identified 546 pairs (416 BSV 80; 130 BSV 50) representing 56 unique LCAs. Of these 56 LCAs derived from the Jiao et al. study, 18 overlap with LCAs we identified in the sigma + tau synteny blocks, and all map to the lineage leading to the last common ancestor of Asparagales and the commelinids. This finding is in agreement with results recently reported in the Phalaenopsis equestris (Cai et al. 2014) and pineapple genome (Ming et al. 2015) papers.
In summary, phylogenomic analyses of rho, sigma, and tau syntelog pairs improve precision in the placement of each of these WGD events inferred from analyses of Poaceae genomes and provide a “gold standard” secondary source of WGD estimation.
Querying of Gene Trees for Placement of WGD Events Using Paralogs from Transcriptomic Data
In addition to using the paralogs generated from synteny analyses of rice and sorghum, putative paralogs were estimated from Ks analyses of all of the transcriptomes used in this study (supplementary fig. S3, Supplementary Material online). A total of 20,900 Ks-derived putative paralogs were queried against orthogroups identified as having rho or sigma synteny-derived putative paralogs. Many of the Ks-derived putative paralogs were estimated as isoforms by Trinity, and are more than likely true isoforms or alleles especially for the low Ks pairs. Of all putative paralog pairs, 667 (550 BSV 80; 117 BSV 50) representing 343 unique duplicate LCAs (274 BSV 80; 69 BSV 50) were placed within the species tree after filtering. Concentrations of gene duplications placed at the base of the Restionaceae (unique LCAs: 102 BSV 80; 27 BSV 50), at the base of the restiids (unique LCAs: 15 BSV 80; 2 BSV 50), at the base of Poaceae (unique LCAs: 78 BSV 80; 16 BSV 50), and after the divergence of Juncus from cyperids (unique LCAs: 29 BSV 80; 2 BSV 50; fig. 1B, supplementary figs. S3 and S4, Supplementary Material online). In sum, the Ks plots and phylogenomic analyses provide consistent evidence for three additional WGD events placed on lineages leading to the restiids (including Centrolepidaceae, and Restionaceae), the Restionaceae and Juncus (supplementary fig. S4, Supplementary Material online). As discussed by Cui et al. (2006) and others, Ks plots may not detect ancient genome duplications so these should be interpreted as a minimal set of polyploidy events among the investigated Poales lineages.
A total of 1,870,214 unique combinations of transcripts or annotated genes were identified across all monocot taxa in sampled orthogroups. This number is inflated to what may be considered paralogs due to existence of isoforms and alleles in transcriptome data. Of these, 36,567 (BSV 80; 51,727 BSV 50) pairs were identified to correspond to nodes in the species tree after filtering. These pairs represent 5,455 unique LCAs (BSV 80; 7,107 BSV 50) spanning almost every node of the species tree. We used the lowest represented known event (sigma, 235 unique LCAs represented in this analysis) to set a minimum threshold for identification of WGD events from the gene tree-derived pairs. This threshold allows for identification of a number of events known from either synteny analysis or other publications including: tau (410 BSV 80), sigma (235 BSV 80), rho (610 BSV 80), Zingerberales gamma event (377 BSV 80)(D’Hont et al. 2012), palm WGD event (345 BSV 80) (D’Hont et al. 2012), and Agavoideae WGD event (615 BSV 80) (McKain et al. 2012; fig. 1C). In addition to these published events, we also have strong evidence for a Juncus event (423 BSV 80), a Cyperus event (463 BSV 80), and a Restionaceae event (499 BSV 80) (fig. 1C). There are two other possible WGD events that fell short of the imposed threshold: a Centrolepidaceae event (200 BSV 80) and a restiid event (184 BSV 80). It is quite possible that our threshold value is simply too conservative and these are true WGD events. Relaxing of the threshold even further would implicate possible events within the cyperids (162 BSV 80), within Bromeliaceae (128 BSV 80), and at the base of the PACMAD clade (121 BSV 80).
Analysis of Changes in GC Content Relative to WGDs
Total GC content (fig. 2A) and GC composition at the third codon position (GC3) calculated for each taxon were highly correlated (table 1, supplementary fig. S5, Supplementary Material online). The highest average %GC found was in Brachypodium distachyon (56.3% GC, SD: 8.9%) and the lowest was in J. effusus (44.8% GC, SD: 6.0%) (table 1). Many of the GC composition distributions of Poales taxa appeared to be bimodal in nature (fig. 2A), but a stringent HDT rejected the unimodal null hypothesis for only seven species (P ≤ 0.05): Lachnocaulon anceps, Aphelia sp., O. sativa, B. distachyon, Dendrocalamus latiflorus, Aristida stricta, and S. bicolor (table 1). Even among these seven species, there is variation in total GC composition and the degree to which the GC-content distributions are bimodal (fig. 2A). When HDTs were conducted on GC3 data, 18 taxa exhibited a bimodal distribution including multiple cyperid taxa, which overall exhibit a much lower GC composition (table 1 and fig. 3B). Interestingly, unimodality in total GC and GC3 could not be rejected for Streptochaeta, suggesting a possible loss of this characteristic in that taxon (fig. 2A) or an independent trend toward bimodality in the BOP + PACMAD clade.
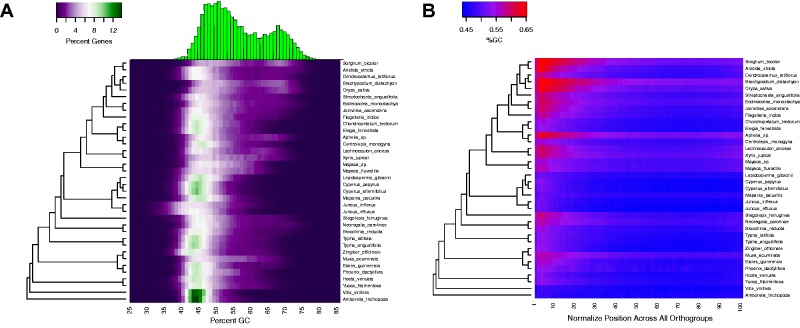
Fig. 2.—
Heatmaps depicting general trends in gene GC composition across all sampled taxa. (A) Distribution of total GC content for all genes across all taxa. The varied GC composition of monocots is highlighted by the differences between low GC species (i.e., Juncus) and high GC species (i.e., Aphelia or grasses). Histogram depicts the GC distribution of Sorghum bicolor demonstrating the heatmap information in a vertical format. (B) Distribution of GC content across genes for all orthogroups and species sampled. A 5′ bias for increased GC percentage is seen.
Table 1.
Statistical Tests for Total GC and GC3 Composition across 13,798 Orthogroups for All Taxa Sampled
| Species | GC.Mean | GC.SD | GC3.Mean | GC3.SD | Hart.GC | Hart.GC3 | T-Test. GCvGC3 |
|---|---|---|---|---|---|---|---|
| Mayaca sp. | 0.505 | 0.063 | 0.551 | 0.132 | 0.712 | 0.163 | 0.000 |
| Amborella trichopoda | 0.463 | 0.040 | 0.461 | 0.084 | 0.997 | 0.822 | 0.171 |
| Aphelia sp. | 0.529 | 0.088 | 0.599 | 0.190 | 0.000 | 0.000 | 0.000 |
| Aristida stricta | 0.508 | 0.095 | 0.547 | 0.201 | 0.003 | 0.000 | 0.000 |
| Brachypodium distachyon | 0.563 | 0.089 | 0.665 | 0.185 | 0.000 | 0.000 | 0.000 |
| Brocchinia reducta | 0.482 | 0.071 | 0.509 | 0.158 | 0.559 | 0.014 | 0.000 |
| Centrolepis monogyna | 0.492 | 0.061 | 0.530 | 0.147 | 0.437 | 0.002 | 0.000 |
| Chondropetalum tectorum | 0.480 | 0.061 | 0.494 | 0.129 | 0.529 | 0.034 | 0.000 |
| Cyperus alternifolius | 0.459 | 0.049 | 0.455 | 0.096 | 0.918 | 0.087 | 0.000 |
| Cyperus papyrus | 0.458 | 0.048 | 0.450 | 0.092 | 0.949 | 0.008 | 0.000 |
| Dendrocalamus latiflorus | 0.500 | 0.085 | 0.534 | 0.189 | 0.009 | 0.000 | 0.000 |
| Ecdeiocolea monostachya | 0.521 | 0.086 | 0.582 | 0.184 | 0.957 | 0.263 | 0.000 |
| Elaies guineensis | 0.487 | 0.071 | 0.503 | 0.155 | 0.762 | 0.118 | 0.000 |
| Elegia fenestrata | 0.476 | 0.060 | 0.489 | 0.130 | 0.412 | 0.008 | 0.000 |
| Flagellaria indica | 0.491 | 0.071 | 0.513 | 0.155 | 0.977 | 0.151 | 0.000 |
| Hosta venusta | 0.472 | 0.060 | 0.481 | 0.142 | 0.760 | 0.012 | 0.000 |
| Joinvillea ascendens | 0.502 | 0.080 | 0.539 | 0.170 | 0.955 | 0.676 | 0.000 |
| Juncus effusus | 0.448 | 0.060 | 0.459 | 0.115 | 0.776 | 0.106 | 0.000 |
| Juncus inflexus | 0.455 | 0.065 | 0.475 | 0.131 | 0.936 | 0.110 | 0.000 |
| Lachnocaulon anceps | 0.518 | 0.083 | 0.572 | 0.179 | 0.045 | 0.000 | 0.000 |
| Lepidosperma gibsonii | 0.471 | 0.050 | 0.484 | 0.096 | 0.667 | 0.011 | 0.000 |
| Mapania palustris | 0.471 | 0.049 | 0.486 | 0.099 | 0.232 | 0.000 | 0.000 |
| Mayaca fluviatilis | 0.498 | 0.069 | 0.538 | 0.146 | 0.300 | 0.018 | 0.000 |
| Musa acuminata | 0.517 | 0.079 | 0.577 | 0.171 | 0.861 | 0.658 | 0.000 |
| Neoregalia carolinae | 0.511 | 0.085 | 0.562 | 0.177 | 0.795 | 0.183 | 0.000 |
| Oryza sativa | 0.542 | 0.096 | 0.618 | 0.200 | 0.000 | 0.000 | 0.000 |
| Phoenix dactylifera | 0.490 | 0.062 | 0.515 | 0.139 | 1.000 | 0.994 | 0.000 |
| Sorghum bicolor | 0.553 | 0.093 | 0.642 | 0.194 | 0.002 | 0.000 | 0.000 |
| Stegolepis ferruginea | 0.497 | 0.081 | 0.533 | 0.164 | 0.193 | 0.004 | 0.000 |
| Streptochaeta angustifolia | 0.506 | 0.083 | 0.552 | 0.183 | 0.830 | 0.113 | 0.000 |
| Typha angustifolia | 0.469 | 0.064 | 0.467 | 0.143 | 0.744 | 0.268 | 0.039 |
| Typha latifolia | 0.479 | 0.068 | 0.494 | 0.147 | 0.908 | 0.179 | 0.000 |
| Vitis vinifera | 0.462 | 0.038 | 0.456 | 0.085 | 0.985 | 0.891 | 0.000 |
| Xyris jupicai | 0.520 | 0.080 | 0.584 | 0.167 | 0.948 | 0.521 | 0.000 |
| Yucca filamentosa | 0.482 | 0.067 | 0.502 | 0.150 | 0.452 | 0.008 | 0.000 |
| Zingiber officinale | 0.468 | 0.072 | 0.470 | 0.156 | 0.426 | 0.056 | 0.229 |
Note.—Bold values represent a significant p-value of < 0.05.
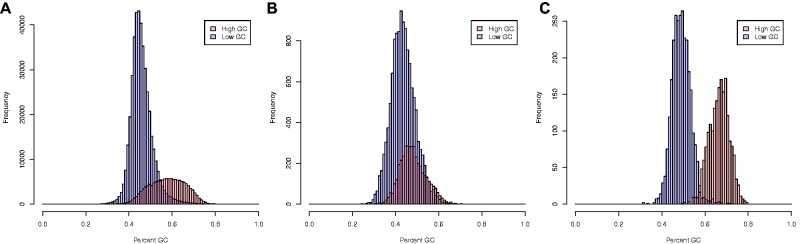
Fig. 3.—
Distributions for percent GC across genes identified to “high GC” (red) or “low GC” (blue) orthogroups for (A) all taxa, (B) Juncus effusus, and (C) Brachypodium distachyon. (A) t-test for distributions of high and low GC orthogroups across all sampled taxa suggests the two sets are distinct (P value = 0.00). (B) The distributions for Juncus effusus demonstrate high overlap of high and low GC orthogroups. t-test of these data suggests that they are distinct sets (P value = 0.00). Juncus effusus transcripts exhibit the lowest overall GC composition across all sampled taxa and transcripts assigned to otherwise high GC composition orthogroups is strikingly lower than the overall distribution of these orthogroups. (C) The distributions for Brachypodium distrachyon exhibit almost nonoverlapping GC values for high and low GC orthogroups. This difference is supported by a t-test (P value = 0.00). Brachypodium distachyon represents the highest GC percentage of all taxa sampled.
Kmeans clusters (n = 2) estimated for the seven taxa identified as having bimodal total GC composition distributions show very similar values for the low and high GC cluster averages (table 2). A total of 13,027 out of 13,798 total orthogroups were found to be either “high GC” or “low GC” across these seven taxa. We identified 6,770 low GC orthogroups and 3,662 high GC orthogroups that were consistent across the seven bimodal taxa. The remaining 2,595 orthogroups varied in GC composition assignments (high GC or low GC) among the seven taxa. Interestingly, contingency tests indicated that orthogroups with high GC content were significantly underrepresented among those retaining syntenic rho, sigma, and tau duplicates in the rice and sorghum genomes (P values 0.027–0.0001; table 3).
Table 2.
Kmeans Clustering of Taxa Exhibiting Bimodal Total GC Composition Distribution
| Species | Low Mean | High Mean |
|---|---|---|
| Aphelia sp. | 0.472 | 0.633 |
| Aristida stricta | 0.460 | 0.651 |
| Brachypodium distachyon | 0.500 | 0.659 |
| Dendrocalamus latiflorus | 0.460 | 0.632 |
| Lachnocaulon anceps | 0.466 | 0.616 |
| Oryza sativa | 0.483 | 0.661 |
| Sorghum bicolor | 0.491 | 0.659 |
Table 3.
Counts of Retained and Lost Paralogs in Gc Classed Orthogroups for Sorghum and Rice with Chi-Squared Test Results
| Paralog Source | Event | High GC | Low GC | Mixed | Chi Sq. | P value |
|---|---|---|---|---|---|---|
| Synteny | Rho retained duplicate | 35 | 279 | 86 | 80.6502 | <0.00001 |
| Synteny | Rho duplicate lost | 3,627 | 6,491 | 2,509 | ||
| Synteny | Sigma retained duplicate | 0 | 24 | 4 | 14.4838 | 0.000716 |
| Synteny | Sigma duplicate Lost | 3,662 | 6,746 | 2,591 | ||
| Synteny | Tau retained duplicate | 5 | 53 | 20 | 18.2902 | 0.000107 |
| Synteny | Tau duplicate lost | 3,657 | 6,717 | 2,575 | ||
| Gene trees | Rho retained duplicate | 51 | 385 | 133 | 109.3626 | <0.00001 |
| Gene trees | Rho duplicate lost | 3,611 | 6,385 | 2,462 | ||
| Gene trees | Sigma retained duplicate | 15 | 163 | 48 | 55.9048 | <0.00001 |
| Gene trees | Sigma duplicate lost | 3,647 | 6,607 | 2,547 | ||
| Gene trees | Tau retained duplicate | 75 | 235 | 95 | 19.256 | 0.000066 |
| Gene trees | Tau duplicate lost | 3,587 | 6,535 | 2,500 |
GC distributions of all transcripts for high and low GC orthogroups were found to exhibit normal distributions (Shapiro test; P value 0.00) that were found to be statistically different from each other (fig. 3A; t-test, P value=0.00). Because orthogroups were classified as high, low, and mixed GC composition based on only the seven taxa with significant bimodal GC composition as indicated by the HDT, we further tested if the variation in GC between the high and low classes was found across all taxa sampled. For each taxon, the high and low GC distributions were found to be significantly distinct (t-test, P value=0.00; fig. 3A). Intra-taxon variation between these classes varied immensely. For example, the difference in GC percentage between the two classes in B. distachyon (highest GC percentage measured) was much greater than difference estimated for J. effusus (lowest GC percentage measured) (fig. 3B and C). The distributions of the same GC classes between these two species were found to be statistically different (t-test, P value=0.00).
As has been described in previous work (Wong et al. 2002; Kuhl et al. 2004, 2005; Tatarinova et al. 2010; Clément et al. 2015), the bimodal GC content distribution observed in grasses and some other monocot genomes is associated with a 5′ to 3′ gradient in the GC-composition of coding sequences. The GC composition gradient is absent or very weak for taxa that were not identified as having multimodal GC3 distributions (fig. 2B).
Discussion
Understanding the effect of polyploidy on the evolution of angiosperms requires both investigation of recent polyploids and characterization of genome evolution following ancient WGDs. Detection and phylogenetic mapping of paleopolyploidy events is a necessary first step in the characterization of genome evolution following ancient WGDs. Methods for detecting WGD events include synteny analysis (Bowers et al. 2003; Tang et al. 2007; Paterson et al. 2012), assessment of frequency distributions for synonymous substitutions (Ks) between duplicate genes (Lynch and Conery 2003; Blanc and Wolfe 2004; Cui et al. 2006; Vanneste et al. 2012), and phylogenomic analyses reconciling duplications in gene trees with known species relationships (Bowers et al. 2003; Jiao et al. 2011; Cannon et al. 2015; Yang et al. 2015). Each of these methods has its strengths and limitations (Bowers et al. 2003; Vanneste et al. 2012). Synteny may decay over time; substitutions at synonymous sites can become saturated, thus reducing the resolution of Ks; and gene tree estimation can be confounded by long-branch attraction artifacts and deep coalescence. These methods can be applied in concert to provide multiple lines of evidence for hypothesized WGD events (Bowers et al. 2003; Paterson et al. 2004, 2010; D’Hont et al. 2012; McKain et al. 2012; Amborella Genome Project 2013; Jiao et al. 2014; Cannon et al. 2015). As first proposed by Bowers et al. (2003), the timing of WGDs identified through synteny analyses can be resolved through phylogenomic analyses including transcript sequences from taxa without reference genome sequences. Such a phylogenomic approach is becoming more reliable as the increasing availability of transcriptome data from taxa well distributed across the tree of life can be used to ameliorate estimation artifacts caused by model misspecification and long-branch attraction (e.g., Leebens-Mack et al. 2005). Further, as we show in this study, increasing the density of taxon sampling within an organismal phylogeny can yield improved precision in estimates of the timing of WGD events.
Assessing the Impact of WGD Events in Monocot History
The Evolution of GC-Profiles in Monocot Genes
The bimodal distribution of GC composition among protein coding genes in cereal grasses (Poaceae) has been studied for nearly two decades (Carels et al. 1998), and Wong et al. (2002) hypothesized that gene and genome duplications may contribute to this pattern. There has been much discussion about the processes responsible for bimodal GC composition distributions (Carels and Bernardi 2000; Wang et al. 2004; Shi et al. 2006; Tatarinova et al. 2010; Serres-Giardi et al. 2012; Tatarinova et al. 2013; Glémin et al. 2014; Clément et al. 2015), but experimental tests of mechanistic hypotheses remain intractable. Inferences have been drawn mainly from correlations between GC-composition and gene length, exon number, gene expression level, synonymous substitution rates, gene body methylation, and local recombination rates (Carels and Bernardi 2000; Wang et al. 2004; Shi et al. 2006; Tatarinova et al. 2010, 2013; Muyle et al. 2011; Serres-Giardi et al. 2012; Glémin et al. 2014; Clément et al. 2015). Our findings support and extend these empirical findings in an evolutionary context. With respect to polyploidy, we find that genes and gene families exhibiting high GC content tend not to retain homeologs following WGD events. Therefore, due to a bias toward low GC in retained genes, whole gene duplications may actually reduce frequency of high GC genes relative to low GC genes. At the same time, some of the grass species exhibit the most bimodal GC content distribution, suggesting that within these lineages any reduction has been offset by some other process. Our data suggest that this bimodality is driven by an increase in the average GC composition of a distinct group of gene families that are consistently classified as high GC across monocot lineages exhibiting bimodality in GC content distribution. These gene families appear to be ancestrally relatively high in GC content, even in species with low total GC composition (fig. 3B).
Greatly improved sampling within Poaceae and Poales reveals multiple reductions and increases in the biomodality of GC composition from what appears to be an ancestrally weak bimodal GC distribution (fig. 2A). Within Poaceae, GC content distributions for Aristida, and Dendrocalamus exhibit a reduction of high GC content genes, and the basal grass lineage, Streptochaeta, has a unimodal GC content distribution (fig. 2A). Sampled species within the cyperid clade (including Cyperaceae and Juncaceae) exhibit weak bimodal or unimodal GC content distribution (fig. 2A and table 1). In agreement with recently published work on strict ortholog sets (Clement et al. 2015), our analyses of homologs within gene families indicate that high GC composition is not randomly distributed across coding genes, but rather it is a presumably an inherited feature of specific gene families. As Clement et al. (2015) inferred for banana, palms, and yam, the declines in GC content in the lineages described above were due to reduction of GC composition in ancestrally high GC gene families. These observations should motivate more focused comparisons of the high GC gene families we have identified in monocot lineages with contrasting GC composition frequencies. Given observed associations between high GC content and higher recombination rates, lower synonymous substitution rates, and absence of gene body methylation in species that do exhibit bimodal GC content distributions, analysis of these features in Aristida, Dendrocalamus (and perhaps all bamboos), Streptochaeta, and the cyperids could be informative and provide insight into processes governing increases and decreases in GC composition of these specific gene families.
The Evolution of Biosynthetic and Developmental Gene Networks
A primary objective of this study was to determine whether rho occurred prior to the diversification of Poaceae or within the family prior to the diversification of the BOP + PACMAD clade (Soltis et al. 2009). An indicator of the impact of rho on the evolution of grasses would be the relationship between duplicated genes, their origin, and their involvement in novel phenotypes. The starch biosynthesis pathway in the endosperm of grasses is unique in that ADP-glucose, a major component of the pathway, is synthesized not only in the plastid like other angiosperms, but in the cytosol as well (Comparot-Moss and Denyer 2009). This alternative pathway dominates over the ancestral plastid pathway for ADP-glucose production (Beckles et al. 2001), suggesting that it is evolutionarily advantageous. The derived, cytosolic pathway is controlled by genes that were duplicated in concert in Brachypodium, Oryza, Setaria, Sorghum, and Zea, placing the origin prior to an ancient speciation event in the BOP + PACMAD clade (Wu et al. 2008; Li et al. 2012). The starch biosynthesis pathway has not been characterized in basal Poaceae species (including Streptochaeta) or the other graminid families—Ecdeiocoleaceae, Joinvilleaceae, and Flagellariaceae—but placement of the rho WGD in the last common ancestor of all extant Poaceae lineages leads to the prediction that basal lineages in the family (e.g., Streptochaeta) potentially harbor the genes necessary for cytosolic production of ADP-glucose (Wu et al. 2008), while members of the other graminid families would not.
Placement of the rho WGD also elucidates the molecular basis for the evolution of morphological characters. The spikelet serves as a key characteristic of the grasses, though the evolution of the structure has long been a subject of research due to the similarity to closely related graminids (Rudall and Stuppy 2005; Sajo and Rudall 2012) and difficulty in determining homology in inflorescence structure between early diverging Poaceae species and the core Poaceae (Sajo et al. 2008; Preston et al. 2009; Sajo and Rudall 2012). The influence of the rho WGD event on the development of the spikelet is suggested through analyses of the MADS-box transcription factor gene family AP1/FUL, which demonstrated that the FUL gene was duplicated prior to the diversification of Poaceae (including Streptochaeta) but after the divergence of Joinvilleaceae (Preston and Kellogg 2006). Interestingly, the paralogs were not maintained in duplicate in either Streptochaeta or Pharus, both of which are early diverging lineages with distinct inflorescence structures compared with core Poaceae (Preston and Kellogg 2006). These findings, in concordance with the placement of rho in the lineage leading to the Poaceae clade, lead us to hypothesize that the retention of duplicate genes led to the development of the spikelet in grasses, though more validation at the functional level looking at paralog retention and expression in the inflorescence across the Poaceae is required to elucidate this complex question.
Synteny analyses (Tang et al. 2010) and a previous phylogenomic analyses including palm and banana genes (D’Hont et al. 2012) suggested that sigma occurred sometime prior to the diversification of Poaceae but after the divergence of Poales from the other commelinid orders. Divergence time analyses suggested that this event occurred approximately 130 Ma (Tang et al. 2010), which would be prior to the diversification of the Poales crown group over 110 Ma (Magallón and Castillo 2009; Magallón et al. 2015) and the divergence of the Poales lineage from other commelinid orders (∼110 Ma; Magallón et al. 2015).
The impact of the sigma event on the evolution of Poales may be seen in the diversity of the group. The order contains approximately 21,000 species, representative of approximately 33% of all monocot species, and is ecological dominant in a number of habitats (Linder and Rudall 2005; Givnish et al. 2010). Of the approximately 21,000 Poales species, three Poales families—Poaceae (∼11,300), Cyperaceae (∼5,700), and Bromeliaceae (∼3,100)—contribute approximately 20,000 species (Givnish et al. 2010). Though the number of species in Poaceae may be partly attributable to the rho event due to diversification in the BOP + PACMAD clade, the other diversifications could be linked to innovations spurred by gene duplications from the sigma event, such as the evolution of the epiphytic habit in Bromeliaceae (Givnish et al. 2011). Investigation into the evolutionary effects of the sigma event will require further identification of duplicated genes and gene networks derived from the event and deep genomic sampling across Poales.
Though this study was primarily aimed at the phylogenomics of Poales and the placement of the rho and sigma WGD events, we were able to detect the tau WGD event characterized by Jiao et al. (2014) and verify its placement prior to the divergence of Asparagales and the commelinid lineage. Analyses within the pineapple genome paper, are consistent with ours in placing the tau WGD before divergence of the commelinid and Asparagales lineages and suggest that it occurred after the divergence of Alismatales from other monocot lineages (Ming et al. 2015). This earlier WGD in monocot history was previously hypothesized by Tang et al. (2010) and supported in our gene tree analyses by 407 unique gene families. A similarly timed duplication of MADS-box genes has been reported to be shared by the Commelinales and Poales, though sampling and support was not high enough to further estimate when exactly the duplication occurred (Litt and Irish 2003). Further sampling of the monocots is needed to more precisely place tau relative to the rapid diversification of monocot lineages early in the history of the group (e.g., Givnish et al. 2010, Magallón et al. 2015) and to understand possible implications of this event on the history of monocots.
Phylogenomic analyses are becoming much more common as high-throughput sequencing becomes the standard. These types of analyses provide insight into the evolution of large portions of the genome of many taxa not previously sampled in genomic studies. Combining phylogenomic analyses with other methods for identifying WGD events, such as synteny analysis, allows for the unambiguous detection of paleopolyploid events and their phylogenetic placement with improved precision and confidence. Understanding when these events occurred in the history of angiosperms will help elucidate the long-term evolutionary patterns associated with polyploidy and further the understanding of how this group of organisms has become so widespread and diverse.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Science Foundation (DEB-0830009 to T.J.G, J.I.D., D.W.S., C.W.D., J.C.P., and J.H.L-M. and DEB-1010905 to M.R.M. and J.H.L.-M.). In addition, we thank the 1kp Initiative lead by Gane Ka-Shu Wong (University of Alberta) for early access to some of their data.
Literature Cited
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342:9–1. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Barker MS, et al. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210(2):391–398. [DOI] [PubMed] [Google Scholar]
- Barrett CF, et al. 2016. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 209(2):855–870. [DOI] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T. 2001. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 125:818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. GeneWise and genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16:1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. [DOI] [PubMed] [Google Scholar]
- Cai J, et al. 2014. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 47:65–72. [DOI] [PubMed] [Google Scholar]
- Cannon SB, et al. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 32:193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Bernardi G. 2000. Two classes of genes in plants. Genetics 154:1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Hatey P, Jabbari K, Bernardi G. 1998. Compositional properties of homologous coding sequences from plants. J Mol Evol. 46:45–53. [DOI] [PubMed] [Google Scholar]
- Chevreux B, et al. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément Y, Fustier M-A, Nabholz B, Glémin S. 2015. The bimodal distribution of genic GC content is ancestral to monocot species. Genome Biol Evol. 7:336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S, Denyer K. 2009. The evolution of the starch biosynthetic pathway in cereals and other grasses. J Exp Bot. 60:2481–2492. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16:738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. [DOI] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y. 2012. Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol. 15:168–176. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, et al. 2008. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 42:443–461. [DOI] [PubMed] [Google Scholar]
- Duarte JM, et al. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 10:61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 10:285–311. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 112:8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar JS, Glémin S, Galtier N. 2011. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol Biol Evol. 28:2561–2575. [DOI] [PubMed] [Google Scholar]
- Estep MC, et al. 2014. Allopolyploidy, diversification, and the Miocene grassland expansion. Proc Natl Acad Sci U S A. 111:15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. 2001. The evolution of isochores. Nat Rev Genet. 2:549–555. [DOI] [PubMed] [Google Scholar]
- Fraley C, Raftery AE. 2002. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 97:611–631. [Google Scholar]
- Fraley C, Raftery AE, Murphy TB, Scrucca L. 2012. mclust Version 4 for R: normal mixture modeling for model-based clustering, classification, and density estimation.
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60:433–453. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, et al. 2010. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of poales. Ann Missouri Bot Gard. 97:584–616. [Google Scholar]
- Givnish TJ, et al. 2011. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am J Bot. 98:872–895. [DOI] [PubMed] [Google Scholar]
- Glémin S, Clément Y, David J, Ressayre A. 2014. GC content evolution in coding regions of angiosperm genomes: a unifying hypothesis. Trends Genet. 30:263–270. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group. 2001. Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Missouri Bot Gard. 88:373–457. [Google Scholar]
- Grass Phylogeny Working Group II. 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193:304–312. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13:R3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH. 2014. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26:2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl JC, et al. 2004. A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl JC, et al. 2005. Comparative genomic analyses in Asparagus. Genome 48:1052–1060. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J, et al. 2005. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol. 22:1948–1963. [DOI] [PubMed] [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. Am Nat. 122:1–25. [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. New York: Oxford University Press. [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li Q-G, Dunwell JM, Zhang Y-M. 2012. Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol Biol Evol. 29:3227–3236. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder HP, Rudall PJ. 2005. Evolutionary history of Poales. Annu Rev Ecol Evol Syst. 36:107–124. [Google Scholar]
- Litt A, Irish VF. 2003. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2003. The evolutionary demography of duplicate genes. J Struct Funct Genomics. 3:35–44. [PubMed] [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Syst Biol. 46:523–536. [Google Scholar]
- Maere S, et al. 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 102:5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Castillo A. 2009. Angiosperm diversification through time. Am J Bot. 96:349–365. [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207:437–453. [DOI] [PubMed] [Google Scholar]
- Mallet J. 2007. Hybrid speciation. Nature 446:279–283. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Mayrose I, et al. 2011. Recently formed polyploid plants diversify at lower rates. Science 333:1257.. [DOI] [PubMed] [Google Scholar]
- McKain MR, et al. 2012. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am J Bot. 99:397–406. [DOI] [PubMed] [Google Scholar]
- Ming R, et al. 2015. The pineapple genome and the evolution of CAM photosynthesis. Nat Genet. 47:1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, et al. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30:i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Serres-Giardi L, Ressayre A, Escobar J, Glémin S. 2011. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol Biol Evol. 28:2695–2706. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer-Verlag. [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A. 101:9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556. [DOI] [PubMed] [Google Scholar]
- Paterson AH, et al. 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Tang H, Wang X. 2010. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 61:349–372. [DOI] [PubMed] [Google Scholar]
- Preston JC, Christensen A, Malcomber ST, Kellogg EA. 2009. MADS-box gene expression and implications for developmental origins of the grass spikelet. Am J Bot. 96:1419–1429. [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. 2006. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174:421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 1998. Pathyways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 29:467–501. [Google Scholar]
- Rudall P, Stuppy W. 2005. Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am J Bot. 92:1432–1443. [DOI] [PubMed] [Google Scholar]
- Reproductive morphology of the early-divergent grass Streptochaeta and its bearing on the homologies of the grass spikelet. Plant Syst Evol. 275:245–255. [Google Scholar]
- Sajo MG, Rudall PJ. 2012. Morphological evolution in the graminid clade: comparative floral anatomy of the grass relatives Flagellariaceae and Joinvilleaceae. Bot J Linn Soc. 170:393–404. [Google Scholar]
- Sajo M, Longhi-Wagner H, Rudall P. 2008. Reproductive morphology of the early-divergent grass Streptochaeta and its bearing on the homologies of the grass spikelet. Plant Syst Evol. 275:245–255. [Google Scholar]
- Schlueter JA, et al. 2004. Mining EST databases to resolve evolutionary events in major crop species. Genome 47:868–876. [DOI] [PubMed] [Google Scholar]
- Serres-Giardi L, Belkhir K, David J, Glemin S. 2012. Patterns and evolution of nucleotide landscapes in seed plants. Plant Cell 24:1379–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, et al. 2006. Nucleotide substitution pattern in rice paralogues: implication for negative correlation between the synonymous substitution rate and codon usage bias. Gene 376:199–206. [DOI] [PubMed] [Google Scholar]
- Smarda P, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc Natl Acad Sci U S A. 111:E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, et al. 2009. Polyploidy and angiosperm diversification. Am J Bot. 96:336–348. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. 2014. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytol. 202:1105–1117. [DOI] [PubMed] [Google Scholar]
- Soreng RJ, et al. 2015. A worldwide phylogenetic classification of the Poaceae (Gramineae). J Syst Evol. 53:117–137. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Nucleic Acids Res. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH. 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 107:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. 2007. Synteny and collinearity in plant genomes. Science 320:486–488. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. 2012. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA. 2010. GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics 11:308.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinova T, Elhaik E, Pellegrini M. 2013. Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol Evol. 5:1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 38:615–643. [DOI] [PubMed] [Google Scholar]
- Tuskan Ga, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. 2009. The flowering world: a tale of duplications. Trends Plant Sci. 14:680–688. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van De Peer Y. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24:1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Van de Peer Y, Maere S. 2012. Inference of genome duplications from age distributions revisited. Mol Biol Evol. 30:177–190. [DOI] [PubMed] [Google Scholar]
- Wall PK, et al. 2008. PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res. 36:D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Singer GaC, Hickey DA. 2004. Mutational bias affects protein evolution in flowering plants. Mol Biol Evol. 21:90–96. [DOI] [PubMed] [Google Scholar]
- Wang X, Shi X, Hao B, Ge S, Luo J. 2005. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 165:937–946. [DOI] [PubMed] [Google Scholar]
- Wong GK-S, et al. 2002. Compositional gradients in Gramineae genes. Genome Res. 12:851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhu Z, Ma L, Chen M. 2008. The preferential retention of starch synthesis genes reveals the impact of whole-genome duplication on grass evolution. Mol Biol Evol. 25:1003–1006. [DOI] [PubMed] [Google Scholar]
- Yang Y, Moore MJ, Brockington SF, Soltis DE, Wong GK. 2015. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol Biol Evol. 32:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yoo M-J, Liu X, Pires JC, Soltis PS, Soltis DE. 2014. Nonadditive gene expression in polyploids. Annu Rev Genet. 48:485–517. [DOI] [PubMed] [Google Scholar]
- Yoo M-J, Szadkowski E, Wendel JF. 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. 2005. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:1003–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM. 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 51:588–598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.