Abstract
A precise understanding of the role of miR-223 in human hematopoiesis and in the pathogenesis of acute myeloid leukemia (AML) is still lacking. By measuring miR-223 expression in blasts from 115 AML patients, we found significantly higher miR-223 levels in patients with favorable prognosis, whereas patients with low miR-223 expression levels were associated with worse outcome. Furthermore, miR-223 was hierarchically expressed in AML subpopulations, with lower expression in leukemic stem cell–containing fractions. Genetic depletion of miR-223 decreased the leukemia initiating cell (LIC) frequency in a myelomonocytic AML mouse model, but it was not mandatory for rapid-onset AML. To relate these observations to physiologic myeloid differentiation, we knocked down or ectopically expressed miR-223 in cord-blood CD34+ cells using lentiviral vectors. Although miR-223 knockdown delayed myeloerythroid precursor differentiation in vitro, it increased myeloid progenitors in vivo following serial xenotransplantation. Ectopic miR-223 expression increased erythropoiesis, T lymphopoiesis, and early B lymphopoiesis in vivo. These findings broaden the role of miR-223 as a regulator of the expansion/differentiation equilibrium in hematopoietic stem and progenitor cells where its impact is dose- and differentiation-stage-dependent. This also explains the complex yet minor role of miR-223 in AML, a heterogeneous disease with variable degree of myeloid differentiation.
Recent profiling approaches have revealed tissue-specific and/or developmentally regulated expression patterns that point to a role of miRNAs in complex biological processes such as hematopoietic differentiation [1–12]. In particular, granulopoiesis has been linked to miR-223 [13–15], a highly conserved miRNA with a myeloid-specific expression pattern [16,17]. This prompted multiple studies into possible targets and regulatory circuits in normal [17] and malignant hematopoiesis, such as acute myeloid leukemia (AML) [5,14,18,19], revealing a tight control by differentiation factors such as PU.1 and C/EBPs [20]. By using a reporter vector, we measured functional miR-223 activity across multiple murine and human hematopoietic subpopulations with single cell resolution [21,22]. We found miR-223 activity already in hematopoietic stem cells (HSCs) and progenitors (HSPCs), with a progressive increase during myeloid differentiation and a loss of activity in the erythroid and lymphoid lineage. Genetic depletion of miR-223 in mice did not abrogate myeloid differentiation but led to a significant increase in myeloid progenitor cells [17,23]. Besides regulating homeostasis between the HSPCs and the terminally differentiated myeloid compartment, additional roles of miR-223 became evident, such as preventing excessive neutrophil or macrophage activation [17,23,24]. However, much less is known about the role of miR-223 in human normal and malignant hematopoiesis. Lentiviral overexpression of miR-223 led to increased myeloid differentiation of primary AML cells in vitro through activation of the epigenetic machinery as well as bona fide targets such as NFI-A [14,18,25]. In addition, it was shown that E2F1 and miR-223 constitute a negative feedback loop potentially leading to increased cell cycle progression in AML cells [19]. This implies that altered miR-223 expression may contribute to leukemogenesis by targeting differentiation and proliferation. Considering the in vitro nature of these findings, it remains unclear whether miR-223 is actually a relevant factor for the in vivo onset of AML and how it contributes to the leukemic properties of various driver mutations in different AML models. Only very recent studies have begun to address its function in primary hematopoietic cells from human cord blood (CB) using in vitro culture and differentiation assays. In vitro experiments suggested that miR-223 positively regulates granulocytic differentiation [14,21,25–27]. It still remains unclear how these findings relate to the phenotype of miR-223 knockout mice, how modulation of miR-223 levels influence steady-state hematopoiesis in humans, and what role this miRNA has in regulating hematopoietic stem- and progenitor-cell function and homeostasis. Understanding these aspects is fundamental to assess the pathogenetic role of miR-223 in myeloid malignancy.
In this work, we characterized miR-223 expression in primary AML samples from patients stratified into prognostically well defined subgroups and found significantly higher miR-223 levels in patients with favorable prognosis and a more differentiated blast cell phenotype. Absence of miR-223 expression did not significantly alter the disease onset but modulated leukemia initiating cell (LIC) frequency in some experimental AML mouse models. We relate these data to the physiologic function of miR-223 in human hematopoiesis by studying knockdown and over-expression phenotypes in primary CB-derived HSPCs using both in vitro and in vivo xenotransplantation models, showing for the first time, to our knowledge, that miR-223 serves as a rheostat to positively regulate multilineage commitment and to constrain the proliferation of human myeloid progenitors, thus allowing differentiation to proceed.
Material and methods
miR-223 knockdown and overexpression
The miR-223 sponge vector has been previously described [23] (see Supplementary Methods, online only, available at www.exphem.org). High titer lentiviral vectors (LVs; 5–10 × 109 transducing units/mL) were produced as previously described [23]. Successful knockdown of miR-223 in the progeny of CD34+ HSPCs by the miR-223 sponge was verified by quantification of the validated miR-223 target, NFIA, and by Western blot as previously described [23]. Overexpression was verified by quantitative reverse transcription polymerase chain reaction (qPCR) for mature miR-223 as described below.
Transduction and culture of CD34+ cord blood HSPCs
CD34+ human CB cells were purchased from Lonza (Milan, Italy). After thawing, cells were plated at a density of 106 cells/mL and prestimulated for 12–16 hours in Stem Span medium (StemCell Technologies) containing the following cytokines: stem cell factor (100 ng/mL), Fms-related tyrosine kinase 3 ligand (FLT3L; 100 ng/mL), thrombopoietin (50 ng/mL), and interleukin 6 (IL-6; 20 ng/mL). Five-hundredfold concentrated LVs were added at a concentration of 108 transducing units/mL, and the culture was incubated for 24 hours. This protocol yielded 10–20 vector integrations per cell (as measured by quantitative real-time PCR [qPCR] [23]), and 73 ± 6.4% of green fluorescent protein (GFP)-expressing cells at day 7 of serum-free culture.
Maintenance/HSPC expansion culture was performed in serum-free stem span medium supplemented with stem cell factor (SCF), FLT3L, thrombopoietin (TPO), and IL-6 (all at 50 ng/mL). Myeloid differentiation culture was performed in Iscove’s Modified Dulbecco’s Medium (IMDM) + 10% fetal calf serum (FCS) + granulocyte colony–stimulating factor (G-CSF) 100 ng/mL (Myelostim, Chugai Sanofi Aventis) + SCF 50 ng/mL for the first week of culture. Clonogenic assays were performed by plating 0.8–1 × 103/mL human HSPCs in a methylcellulose-based medium (MethoCult H4434, StemCell Technologies). After 15 days, colonies were counted as erythroid, myeloid, or mixed erythroid-myeloid by morphologic criteria.
Retroviral infection of murine bone marrow cells
Total bone marrow (BM) from miR-223 knockout (miR-223−/y) and wild-type (miR-223+/y) mice was extracted and prestimulated for 2 days, as previously described [13]. Depletion of miR-223 was confirmed by qPCR. Then, BM cells were either singly transduced with pSF91-MN1–internal ribosome entry site (IRES)-GFP, MSCV-IRES-GFP, MSCV-Hoxa9-IRES-GFP, MSCV-AML1-ETO-IRES-GFP, MSCV-MLL-AF9-IRES-GFP, or simultaneously cotransduced with MSCV-Hoxa9-pgk-neomycin and MSCV-Meis1-IRES-YFP by cocultivation with irradiated (4,000 cG) viral producers, as previously described [28,29]. Except for Hoxa9-Meis1, retrovirally transduced cells were sorted based on expression of GFP by using a FACSAriaIII (Becton Dickinson). Hoxa9-Meis1–transduced cells were sorted for YFP expression and continuously selected with neomycin (1.4 mg/mL).
Bone marrow transplantation assays
Mice were bred and maintained at the British Columbia Cancer Agency Research Center Animal Facility (Vancouver, BC, Canada). All experimental protocols were approved by the University of British Columbia Animal Care Committee. All recipients (C56BL/6 mice) were lethally irradiated (750 cG) and transplanted by tail vein injection along with either sorted Hoxa9-Meis1 or unsorted MN1 (1% GFP+) BM cells, as previously described [29]. Donor-derived engraftment and reconstitution were monitored by flow cytometry for GFP+ and/or YFP+ expression in the peripheral blood of the transplanted animals. Moribund mice were analyzed as previously described [28]. Limiting dilution assays were performed as described above with decreasing numbers of AML cells, as shown below.
Xenotransplantation was performed by intravenous injection of 2 × 105 CD34+ cells into the tail vein of sublethally (200 cG) irradiated, 10-week-old female NOD scid gamma (NSG) mice. Secondary transplantation was performed by purifying human CD34+ cells from the pooled BM recovered from the primary NSG mice using the direct CD34 Microbead Kit (Miltenyi Biotec) and injecting 3 × 105 CD34+ cells per mouse into five sublethally irradiated NSG-3GS recipients (Jackson Laboratories). All experimental procedures were performed in a specific pathogen-free (SPF) environment at the San Raffaele Animal Facility after approval had been obtained by the Institutional Animal Care and Use Committee.
Statistical analysis
Unless otherwise indicated, pairwise comparisons were performed using unpaired Student’s t test, and multiple comparisons were performed using one-way analysis of variance with Bonferroni posttest corrections. The definition of survival endpoints, molecular cytogenetic risk, and statistical analysis were as previously published [30]. The Kaplan-Meier method and log-rank test were used to estimate the distribution of overall survival (OS) and to compare differences between survival curves, respectively. LIC frequencies were calculated with L-calc (Stemcell Technologies). Chimerism (chim) values were transformed into a log-odds scale (lochim = log (% chim/(100% chim))) to allow for proper statistical comparison.
For further details, please refer to Supplementary Methods (online only, available at www.exphem.org).
Results
miR-223 expression differs between prognostic AML subgroups
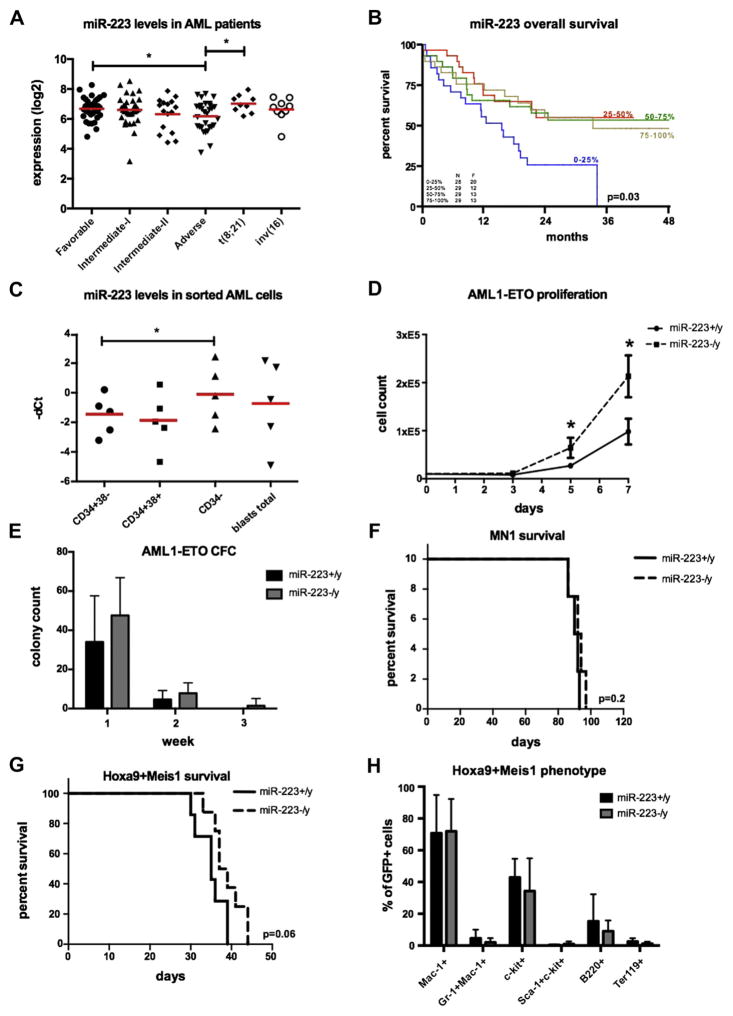
Little is known about the clinical associations of miR-223 levels in AML patients. Therefore, we assessed miR-223 expression in blasts from 115 AML patients by miRNA hybridization arrays. The patient collective was stratified into genetic risk groups according to guidelines from the European Leukemia Network (ELN) [31]; 36 patients were grouped as favorable, 49 patients as intermediate-I and intermediate-II, and 30 patients as adverse prognosis (Table 1). Comparing miR-223 levels between the ELN genetic risk groups, significantly higher miR-223 levels were detected in the genetically favorable group (n = 36) compared with the adverse risk group (n = 30; p = 0.02) (Fig. 1A; Supplementary Table E1, online only, available at www.exphem.org). Especially, the t(8;21) subgroup (n = 9) exhibited significantly higher miR-223 levels within the genetically favorable group (p = 0.02; Fig. 1A). No statistically significant difference was found for the inv(16) (Fig. 1A) subgroup (n = 9; p = 0.29) or between the intermediate-I (n = 33; p = 0.72) and intermediate-II (n = 16; p = 0.17) groups (Fig. 1A). In addition, a univariate Cox regression analysis demonstrated an inferior survival (p = 0.03) of low miR-223 expressers dichotomized to the 4th quartile (Fig. 1B; Supplementary Figure E1 and Supplementary Table E2, online only, available at www.exphem.org). These findings indicate that miR-223 levels are dynamic between AML subgroups; low miR-223 expression is associated with a worse clinical outcome. In addition, we found a significantly higher expression (p = 0.02) of miR-223 in more committed leukemic bulk cells (CD34−) with respect to CD34+CD38− AML cells in five AML patient samples (Fig. 1C; Supplementary Methods, online only, available at www.exphem.org).
Table 1.
Clinical parameters of screened patients
| Clinical parameters | n |
|---|---|
| Age | |
| Mean years | 45 |
| Range (min–max) | 20–60 |
| WBC count × 109/L | |
| Mean | 43 |
| Range (min–max) | 1.2–215 |
| Sex | |
| Male | 62 |
| Female | 53 |
| ELN genetic risk | |
| Favorable | 36 |
| Intermediate-I (normal karyotype) | 33 |
| Intermediate-II (rest) | 16 |
| Adverse | 30 |
| Cytogenetics | |
| 8 | 2 |
| −5 or −5q or −7 or −7q | 14 |
| −9q | 2 |
| 11q23 | 4 |
| t(6;9) | 0 |
| t(8;21) | 9 |
| t(9;11) | 0 |
| inv(3) or t(3;3) | 3 |
| inv(16) | 9 |
| Normal karyotype | 51 |
| Complex karyotype | 9 |
| Other | 12 |
| Molecular geneticsa | |
| CEBPA single | 0 |
| CEBPA double | 6 |
| FLT3-ITD | 19 |
| FLT3-TKD | 7 |
| NPM1 | 28 |
Determined for patients with normal karyotype.
Figure 1.
(A) miR-223 expression levels are significantly increased in AML patients with a cytogenetically favorable prognosis (ELN classification; p =0.02). (B) Overall survival based on miR-223 expression dichotomized into 4 quartiles. The lowest miR-223 expressers (4th quartile) associated with inferior OS (p =0.03). (C) miR-223 is significantly more highly expressed in CD34− human AML cells (n = 5; p = 0.02). (D) Genetic depletion of miR-223 increased the proliferation rate of BM cells retrovirally transduced with an AML1-ETO construct (n = 6; p = 0.04 and p = 0.02, respectively). (E) Absence of miR-223 allowed colony formation of BM cells retrovirally transduced with an AML1-ETO construct beyond week 2, but had no impact on colony numbers (n = 8; p = 0.09 and p = 0.08, respectively). Survival of BM cells transformed with (F) MN1 (n =4 mice/arm; p =0.2) or (G) Hoxa9-Meis1 (n =7–8 mice/arm; p =0.06) was not significantly changed in the absence of miR-223. (H) Hoxa9-Meis1 leukemias did not exhibit any changes in disease-associated immunophenotypic markers (n =7–8 mice/arm; Mac-1: p = 0.9; Gr-1+Mac-1+: p = 0.2; c-kit+: p = 0.3; sca-1+c-kit+: p = 0.3; b220+: p = 0.29; ter119: p = 0.12). *p < 0.05.
These data support a role for miR-223 in the formation of a leukemic hierarchy by positively regulating myeloid differentiation programs.
miR-223 is dispensable for disease development in murine AML models
Next, we set out to clarify whether the presence of miR-223 contributes to the development of AML in vitro and in vivo. Therefore, we took advantage of a genetically depleted miR-223 mouse (miR-223−/y) model and retrovirally infected miR-223−/y and wild-type miR-223 (miR-223+/y) murine BM cells with oncogenes of increasing leukemia-inducing potency: AML1-ETO [32,33], Hoxa9 [34], MN1 [28], and MLL-AF9 [35] to assess the proliferation rate as well as colony formation in vitro. Except in the case of AML1-ETO, where the absence of miR-223 significantly increased proliferation (Fig. 1D) and showed a consistent trend for higher colony counts during 3 weeks of replating (Fig. 1E), we did not detect significant alterations in proliferation or colony-forming capacity in the miR-223−/y background for the other oncogenes tested (Supplementary Figures E1 and E2, online only, available at www.exphem.org). Notably, AML1-ETO was the only oncogene that did not fully immortalize BM cells, highlighting its weak transforming potential. These results suggest that the tumor-modulating effect of miR-223 is connected to the leukemic potential of the genetic driver lesion. In cells driven by highly leukemogenic oncogenes such as MLL-AF9, multiple pathways regulating proliferation and self-renewal most likely override the effects of miR-223.
Based on increasing miR-223 expression levels in differentiated myeloid cells, we hypothesized that genetic depletion of miR-223 might affect the onset of leukemia differently in AML models with or without myeloid differentiation. We chose the widely used Hoxa9-Meis1 model [34,36], which causes AML with myeloid differentiation [36,37] and an MN1-based leukemia model [28], leading to undifferentiated AML in mice with a short disease latency. Therefore, we infected BM cells from miR-223−/y or control miR-223+/y mice with retroviral vectors coding for Hoxa9 and Meis1 (Hoxa9-Meis1) or MN1 and transplanted the transduced cells into irradiated histocompatible C57BL/B6 recipients. All mice that received miR-223−/y (n = 7) or miR-223+/y (n = 8) Hoxa9-Meis1 or that received miR-223−/y (n = 4) or miR-223+/y (n = 4) MN1-transduced cells developed AML, with a mean disease latency of 37 or 34.5 days (p = 0.06) and 89.5 or 91.5 days (p = 0.22), respectively (Figs. 1F and 1G). Encouraged by the trend toward delayed AML onset in the Hoxa9-Meis1 model in the absence of miR-223, we further explored the possibility of miR-223 as modulator of LIC frequency by performing limiting dilution assays (Table 2). Indeed, with this sensitive assay, we found a significantly lower LIC frequency in miR-223−/y Hoxa9-Meis1 AML (frequency: 1 in 48; p = 0.0327) compared with miR-223+/y Hoxa9-Meis1 leukemias (frequency: 1 in 12; Table 2). These minor, but still significant, changes indicate that miR-223 supports but is not essential for the proliferation and self-renewal of LICs in differentiated AML models. Accordingly, no significant differences were seen in Hoxa9-Meis1 leukemias with respect to overall disease characteristics such as white blood cell (WBC) count, red blood cell (RBC), spleen weight, and immuno-phenotypic differentiation markers (Mac-1: p = 0.9; Gr-1+Mac-1+: p = 0.2; c-kit+: p = 0.3; sca-1+c-kit+: p = 0.3; b220+: p = 0.29; ter-119: p = 0.12; Fig. 1H; Supplementary Figure E3, online only, available at www.exphem.org).
Table 2.
Limiting dilution assay of Hoxa9-Meis1 cells
| Cells input | miR-223+/y Hoxa9-Meis1 | miR-223−/y Hoxa9-Meis1 |
|---|---|---|
| Experiment 1 | ||
| 10,000 | 4/4 | 4/4 |
| 1,000 | 4/4 | 4/4 |
| 100 | 4/4 | 4/4 |
| 10 | 3/4 | 1/4 |
| Calculated LIC frequency | 1 in 7 | 1 in 26 p = 0.1474 |
| Experiment 2 | ||
| 10,000 | 4/4 | 4/4 |
| 1,000 | 4/4 | 4/4 |
| 100 | 4/4 | 3/4 |
| 10 | 1/3 | 0/1 |
| Calculated LIC frequency | 1 in 21 | 1 in 76 p = 0.1768 |
| Combined LIC frequency | 1 in 12 | 1 in 48 p = 0.0327 |
Taken together, we found that miR-223 is a weak yet complex modulator of leukemogenesis in murine models of AML and that the effect of miR-223 appears to be dictated by the potency of the oncogenic driver lesion and the differentiation status of the disease.
Inhibition of miR-223 reduces myeloerythroid precursor differentiation
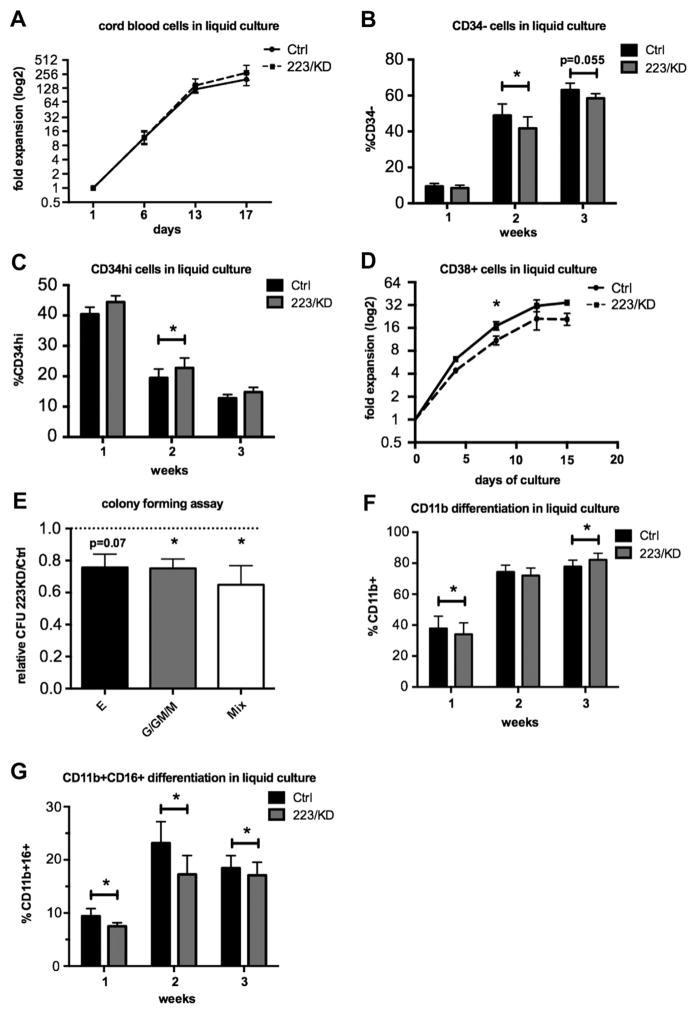
The complex miR-223 phenotypes encountered in the different AML mouse models can hardly be reconciled with the published functions of miR-223 in human hematopoiesis, which mainly relate to its role in promoting myeloid differentiation [25,27]. We therefore set out to further define the role of miR-223 in human primary hematopoietic cells. We transduced CB CD34+ HSPCs with a previously described miR-223 sponge vector [23]. Successful miR-223 knockdown (KD) in huCB was verified by showing upregulation of NFI-A protein by western blot (Supplementary Figure E4A, online only, available at www.exphem.org). We investigated the consequences of miR-223 KD on HSPC proliferation in serum-free liquid culture, which had been optimized for the maintenance and expansion of HSPCs in vitro. During a 3-week culture period, we did not detect significant differences in cell numbers after huCB transduction with the miR-223 sponge, as compared with a scrambled control vector (Fig. 2A). However, there was a minor, yet statistically significant, reduction in the proportion of CD34− cells (Fig. 2B) with a concomitant increase in CD34hi cells, suggesting that dose reductions of miR-223 might reduce spontaneous differentiation in these cultures (Fig. 2C). To distinguish between a possible effect of miR-223 on proliferation associated with self-renewal or differentiation, we sorted CD34hiCD133+ CB cells into a CD38−/low progenitor and a less primitive CD38+ precursor fraction, respectively, and generated growth curves in serum-free liquid culture after transduction with the miR-223 sponge or control vector. Whereas miR-223 KD had no impact on the growth of more primitive CD34hiCD133+CD38−/low cells (Supplementary Figure E4C, online only, available at www.exphem.org), the CD38+ fraction was slowed down in proliferation, confirming an effect of miR-223 on differentiating, intermediate-level precursors rather than on multipotent HSPCs (Fig. 2D).
Figure 2.
(A) CD34+ huCB cells (n = 6 donors) were lentivirally transduced with a miR-223 sponge (223/KD) or control vector, and proliferation was monitored in HSPC maintenance culture conditions (Stem Span serum free expansion medium + SCF, TPO, FLT3L, IL-6). Shown is the mean culture expansion ± SEM, relative to day 1. (B, C) Maintenance culture composition was analyzed by flow cytometry at the indicated time points. Statistical analysis was performed by paired t test after log-odds conversion. (D) CD34+ huCB cells (n = 3 donors) were sorted into a CD133+CD38+ progenitor cell population, transduced with the 223 KD or control LV as described above, and cultured under maintenance conditions. (E) Colony-forming assay was performed on CD34+ CB cells (n = 6 donors) on day 2 after transduction with the 223/KD or control LV. The mean number of erythroid, myeloid, and mixed colonies from three or four technical replicates in the 223/KD group was divided by the mean number in the respective control group to yield a relative colony count, and the mean relative count from six donors ± SEM is shown. Statistical analysis was performed on absolute colony counts using a paired t test. (F, G) 223/KD− or control-LV-transduced CD34+ CB cells (n = 8 donors) were cultured in myeloid differentiation medium (IMDM + 10% FCS + SCF + G-CSF). Culture composition was assessed by flow cytometry at the indicated time points. CD11b+ cells indicate myelocytes and metamyelocytes, whereas CD11b+CD16+ cells correspond to metamyelocytes. Statistical analysis was performed by paired t test after log-odds conversion. Shown is the mean ± SEM. *p < 0.05. Ctrl = control; E = erythroid; G/GM/M = myeloid; Mix = mixed.
We then performed colony-forming assays on CD34+ CB and scored colony type and number after 14 days of culture in complete methylcellulose medium. Whereas colony size was similar between the miR-223 KD and the control group (data not shown), the number of erythroid, myeloid, and mixed colonies was reduced upon miR-223 KD (Fig. 2E), compatible with our hypothesis that less miR-223 reduces differentiation of hematopoietic precursors, resulting in a lower number of colony/burst forming units with erythroid, granulocytic, monocytic, and myeloerythroid bilineage potential (BFU-E, CFU-E, CFU-G, CFU-M, CFU-GM, and CFU-GEMM, respectively). We also investigated the effect of miR-223 on later stages of myeloid differentiation using a unilineage differentiation protocol based on liquid culture in G-CSF–containing medium. Culture growth under myeloid differentiation conditions revealed a small, but statistically significant, reduction in cell numbers in miR-223 sponge treated cells in the first week of culture (Supplementary Figure E4D, online only, available at www.exphem.org). This was accompanied by a reduced accumulation of terminally differentiated cells (CD11b+ myelocytes and CD11b+ CD16+ metamyelocytes; Figs. 2F and 2G). However, the effect decreased with time, and the fraction of CD11b+ cells in the miR-223 sponge group surpassed the control after 3 weeks in culture, suggesting that miR-223 KD delays but does not abrogate myeloid differentiation (Fig. 2F). These results indicate that miR-223 is a rheostat for myeloid differentiation, which is in line with the weak effects that we observed in leukemogenesis.
miR-223 regulates the expansion-differentiation equilibrium in human hematopoietic progenitors
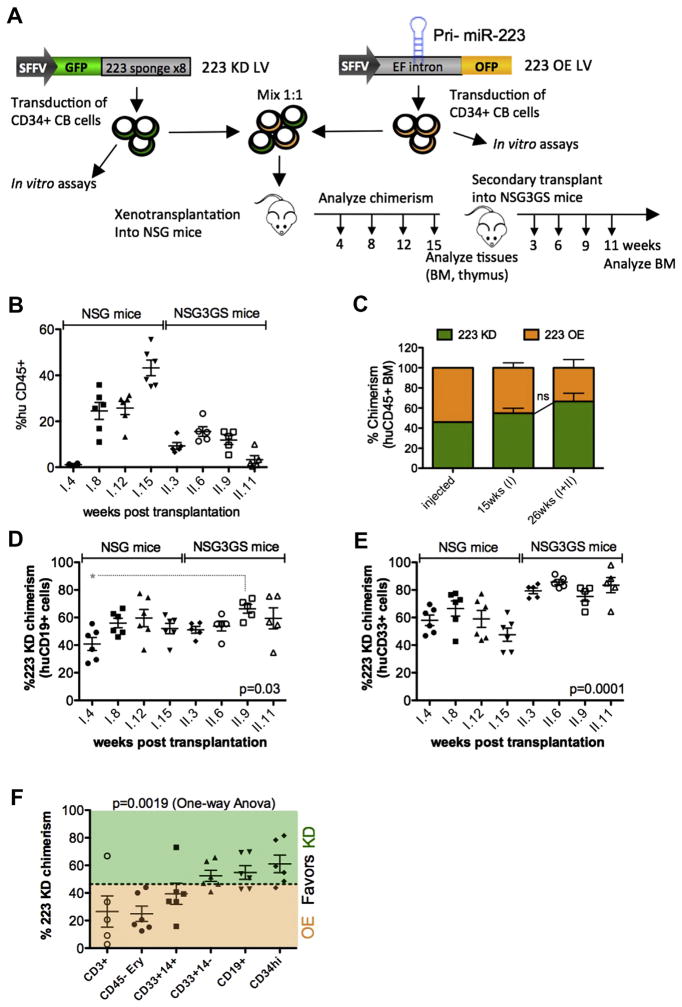
The standard for studying human HSPC functions is the xenotransplantation model using immunodeficient mouse strains such as NSG mice. Therefore, to study the long-term effects of aberrant miR-223 expression on human steady-state hematopoiesis, we transduced huCB CD34+ cells with the miR-223 sponge vector (223 KD) or with a lentiviral miR-223 overexpression (223 OE) vector and performed xenotransplantation (Fig. 3A). Transduced cells could be identified with the help of a coexpressed fluorescent protein: green fluorescent protein (GFP; 223 KD) and orange fluorescent protein (OFP) (223 OE), respectively. 223 OE significantly increased BFU-E and CFU-GEMM numbers but had little effect on myeloid colonies (Supplementary Figure E4E, online only, available at www.exphem.org). This is in line with the 223 KD data because our 223 OE LV induced significant levels of ectopic miR-223 expression in erythroid lineage cells and progenitors, but only modest miR-223 overexpression in the myeloid compartment, where endogenous miR-223 is already highly expressed, as suggested by qPCR (fourfold overexpression on the total colony outgrowth; Supplementary Figure E4F, online only, available at www.exphem.org). Thus, the OE approach will mainly read out the consequences of ectopic miR-223 expression in cell populations where miR-223 is physiologically absent or low, whereas the KD approach will be effective in cell populations physiologically expressing miR-223, making both approaches complementary.
Figure 3.
(A) Experimental scheme. CB CD34+ cells (from four donors) were transduced with the indicated lentiviral vectors, mixed in a 1:1 ratio, and competitively transplanted into six NSG mice. GFP/OFP chimerism in the blood was analyzed at the indicated time points, and mice were euthanized at 15 weeks for analysis of hematopoietic organs. CD34+ cells were enriched from the pooled BM of the primary mice, and secondary transplantation was performed into NSG-3GS mice, where GFP/OFP chimerism was followed for another 11 weeks. (B) Total human CD45+ engraftment in the blood of xenotrans-planted mice is shown at the indicated time points (weeks) post primary (I.) or secondary (II.) transplantation. (C) GFP (miR-223 KD) over OFP (miR-223 OE) chimerism was measured by FACS at the time of transplantation (0 weeks) and in the huCD45+ fraction in the BM of primary (I.) NSG transplants and in the BM of secondary (II.) NSG-3GS transplants. Chimerism was calculated as follows: GFPchim = %GFP/(%GFP+%OFP); OFPchim = %OFP/(%GFP+% OFP); Transduction levels were >90% for each individual vector. D–E GFP (223 KD) chimerism in CD19+ B cells (D) or CD33+ myeloid cells (E) in the PB of primary and secondary transplants at the indicated time points (weeks). Shown is the mean GFP chimerism ± SEM (n = 6 primary mice and 5 secondary mice). Statistical analysis was performed by one-way analysis of variance, and p values from Bonferroni posttest are indicated in the plot. (F) Shown is the GFP chimerism in the indicated human-cell subpopulation. Subpopulations were identified in the BM of transplanted NSG mice at 15 weeks, except for CD3+ T cells, which were purified from the thymus. GFP chimerism was calculated as described in (C). *p <0.05. EF intron = First intronic sequence of the human elongation factor 1 α gene; I. = primary; II. = secondary; SFFV = spleen focus-forming virus enhancer/promoter. (Color version available online.)
Twenty-four hours after transduction, equal numbers of 223 OE and 223 KD CD34+ CB cells were pooled and transplanted into sublethally irradiated NSG mice, where GFP(miR-223 KD)/OFP(miR-223 OE) chimerism was followed over time (Fig. 3A). After 15 weeks, mice were euthanized, hematopoietic tissues were analyzed, and huCD34+ cells were purified from the BM to perform secondary transplantation into NSG-3GS recipients, where the xenograft was followed for another 11 weeks. The NSG-3GS model, which expresses human IL-3, G-CSF, and SCF, was chosen because it better supports myeloid differentiation and enables higher human cell engraftment, thus allowing superior readouts, especially in the peripheral blood [38].
Mean human CD45 engraftment in the PB reached up to 43% in the primary recipients and 15% in the secondary mice (Fig. 3B). Within the human CD45+ compartment, GFP/OFP chimerism was well balanced throughout 26 weeks of observation, with a trend toward increasing contribution from 223 KD cells (46.0% at t0, 51.0% at 15 weeks, 61.6% at 26 weeks) that did not reach statistical significance (Fig. 3C). The CD19+ B-cell compartment showed an undulating pattern of chimerism that fluctuated around a mean of 54% KD/46% OE cells (Fig. 3D). Interestingly, 223 OE cells contributed significantly more to the B-cell compartment during the first month after transplantation (OE: 59.2% ± 4.7%; KD: 40.8%) as compared with later time points and the CD33+ myeloid compartment, where chimerism was in slight favor of the 223 KD cells in the primary transplant (Fig. 3E). At sacrifice (15 weeks after primary transplantation), we also noted a miR-223 OE-biased chimerism in CD45− erythroid precursors (OE: 75.1 ± 5.5%; KD: 24.9%), in CD14+ monocytes (OE: 60.6 ± 7.6%; KD: 39.4%), and, surprisingly, in the T-cell compartment (OE: 73.4 ± 11.3%; KD: 26.6%), whereas there was a tendency toward a miR-223 KD bias in BM CD34+ cells (KD: 61.1 ± 6.4%; OE: 39.9%; Fig. 3F). Strikingly, 223 KD cells dominated the CD33+ myeloid graft in the NSG-3GS secondary transplant, constituting on average 81% of this compartment (Fig. 3E). Taken together, these data suggest that miR-223 has a minor impact on multipotent HSPCs but rather acts at the level of committed progenitors in a developmental-stage-specific manner. Reductions in miR-223 levels favor the expansion of early myeloid progenitors, whereas premature miR-223 upregulation pushes myeloid precursors into terminal differentiation. Moreover, our data suggest that miR-223 has a broader role in inducing commitment to differentiation, which is not limited to myeloid precursors but also seen in B-cell, T-cell, and erythroid precursors.
Discussion
In this work, we investigated the role of miR-223 in acute myeloid leukemia and normal human hematopoiesis in vitro and in vivo by using loss- and gain-of-function approaches in primary cells.
Considering the high abundance of miR-223 in AML cells [39], very little is known about its pathogenetic role in AML. Based on our findings and previous publications miR-223 expression levels vary in AML patients [19,36,37,39,40]. Here, we show for the first time that higher average miR-223 levels are associated with favorable adult AML risk groups. This finding is in line with that of Daschkey et al., who also reported higher miR-223 levels in core-binding factor (CBF) infant AML [37]. Interestingly, Eyholzer et al. highlighted in their analysis miR-223 levels in AML M2 samples comparable to that in monocytes and significantly less than that found in granulocytes [40]. Considering the role of miR-223 as regulator of monocytic and granulocytic differentiation in AML cells, and considering that lentiviral miR-223 overexpression enhances myeloid differentiation in vitro [14,18], it is plausible that AMLs with a favorable prognosis have retained more differentiation capacity than poor-prognosis AMLs. Recent publications have highlighted the adverse prognostic impact of a leukemic stem cell (LSC) signature in AML [41,42], consolidating the link between differentiation status and outcome. Although a formal link between reduced miR-223 expression and LSCs still needs to be confirmed by functional assays, our data support a hierarchical expression of miR-223 in AML, with less miR-223 corresponding to a more LSC-like phenotype and an adverse prognosis. Based on these findings, we speculated that miR-223 is a modulator of leukemic activity in AML. Taking advantage of miR-223−/y bone marrow cells, we found that the absence of miR-223 increased proliferation and enhanced colony formation only in the case of the weakest oncogene, AML1-ETO, adding to the results of Fazi et al. [18]. Unlike Fazi et al, we grouped our AML samples according to a widely used prognostic score based on cytogenetics and molecular genetics, which might explain the observed differences in miR-223 expression levels between our study and that of Fazi et al. Different patient characteristics and technology platforms used for measuring miR-223 might also account for this difference. It will be interesting to measure miR-223 levels in paired diagnosis-relapse samples from AML patients carrying the t(8;21) (AML1-ETO) translocation to better understand the clinical implications of miR-223 downregulation in this specific disease context.
Using two well documented [28,36,43] murine models of AML, we found that disease onset and phenotype were virtually identical in a miR-223−/y or miR-223+/y background, indicating that the presence or absence of miR-223 might not be relevant for the onset of AML, but that it acts as modulator of LIC frequency in myeloid-differentiated AML. Indeed, in HOX-dependent AML models, leukemic stem cells have been characterized as downstream myeloid cells that have acquired self-renewal potential [35], suggesting that these cells might have retained a myeloid molecular program in which the presence of miR-223 enhances proliferative function. Our findings further define a functional role for miR-223 in acute leukemia, but also show that miR-223 deregulation is not a driving event in the onset of AML, instead acting more as a modulator of self-renewal and differentiation. Whether this happens through targeting E2F1 [19] or NFI-A [14] in the presented AML model remains to be determined. AML is a highly heterogeneous disease, and we cannot exclude the possibility that the loss of miR-223 might actively contribute to a differentiation block or increased self-renewal in a subset of AML patients that was not reflected in our experimental mouse models. However, our data also suggest that low miR-223 levels are necessary in some AML subtypes for full leukemic activity. This accords with the fact that only a single AML case has been reported where the miR-223 gene was deleted [44].
To better understand the complex role of miR-223 in AML, we further explored its function in human primary CB HSPCs. In line with previously published data [14,25,27], we confirm that miR-223 promotes myeloid/ granulocytic differentiation. We also found in vitro and in vivo evidence that miR-223 increases erythroid and monocytic differentiation. Different differentiation models and the degree of miR-223 modulation, which was more moderate in our study compared with previous studies [26,27], might account for these discrepancies. Unexpectedly, our in vivo studies using the NSG xenotransplantation model showed increased B and T cells upon miR-223 OE during early lymphoid reconstitution. Interestingly, increased T-cell production upon miR-223 overexpression in mouse HSCs was reported in the first publication describing the in vivo role of miRNAs in hematopoiesis [16], and our data obtained on human CB HSPCs now support this finding. The increased production of differentiated cells from multiple lineages in 223 OE, as opposed to 223 KD, cells points to a biological effect of miR-223 in multi-potent progenitors, where it instructs differentiation. In line with this hypothesis, 223 KD cells were overrepresented in CD34+ progenitor BM cells. Notably, upon secondary transplantation into NSG-3GS mice, the myeloid compartment adjusted to the 223 KD-biased chimerism found in CD34+ cells, whereas chimerism in the lymphoid compartment remained balanced between 223 KD and 223 OE cells. Based on the common model that posttransplant hematopoiesis derives from multipotent HSCs, the stable chimerism in the B-cell compartment seen over 26 weeks argues for a predominant role of miR-223 in instructing commitment and differentiation at the level of hematopoietic progenitors, rather than a direct role of miR-223 in regulating self-renewal and maintenance of hematopoietic stem cells. It is likely that the NSG-3GS model chosen for secondary transplantation highlighted the promoting effect of miR-223 KD on myeloid progenitors. The presence of human cytokines, namely IL3, G-CSF, and SCF, in this model greatly enhances myeloid progenitor development, and huCD33+ cells found in NSG-3GS mice mostly represent granulocyte progenitors, as opposed to monocyte lineage cells seen in the classical NSG model [38]. These data reconcile the finding of increased myeloid progenitor cell numbers in miR-223 knockout/knockdown mice and consolidates the concept that miR-223 is a rheostat which balances self-renewal-type proliferation against commitment and differentiation in hematopoietic progenitors.
Taken together, we establish the role of miR-223 in human hematopoiesis as a promoter of lineage commitment and differentiation and highlight its role as modulator, but not as driver, in AML in vivo. To our knowledge, this is the first paper functionally investigating the role of miR-223 in in vivo AML models, as well as in human steady-state hematopoiesis, using a xenotransplantation model.
Supplementary Material
Acknowledgments
We thank Giulia Schira, Nicola Bocchini, Lucia Sergi Sergi, Tiziano Di Tommaso and the TIGET staff for technical and administrative help; the San Raffaele FACS facility for cell sorting; and the Naldini lab for helpful discussions. F Kuchenbauer was supported by grants from Deutsche Krebshilfe (Max-Eder program grant no. 109420), the European Hematology Association (fellowship no. 2010/04), and by the Deutsche Forschungsgemeinschaft (grant no. D.3955 [SFB 1074]). Further financial support was provided by grants to RK Humphries from a Terry Fox Program Project award and the Canadian Institutes of Health Research and to L Naldini from Telethon (TIGET grant), EU (FP7 GA 222878 PERSIST, ERC Advanced Grant no. 249845 TARGETINGGEN ETHERAPY), and the Italian Ministry of Health.
Footnotes
Conflict of interest disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.exphem.2015.05.018.
References
- 1.Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121:4977–4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 2.Petriv OI, Hansen CL, Humphries RK, Kuchenbauer F. Probing the complexity of miRNA expression across hematopoiesis. Cell Cycle. 2011;10:2–3. doi: 10.4161/cc.10.1.14289. [DOI] [PubMed] [Google Scholar]
- 3.Starczynowski DT, Kuchenbauer F, Wegrzyn J, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39:167–178. e164. doi: 10.1016/j.exphem.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Petriv OI, Kuchenbauer F, Delaney AD, et al. Comprehensive micro-RNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A. 2010;107:15443–15448. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuchenbauer F, Morin RD, Argiropoulos B, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695–1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copley MR, Babovic S, Benz C, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Zhang M, Jiang X, et al. miR-223 suppresses differentiation of tumor-induced CD11b(+) Gr1(+) myeloid-derived suppressor cells from bone marrow cells. Int J Cancer. 2011;129:2662–2673. doi: 10.1002/ijc.25921. [DOI] [PubMed] [Google Scholar]
- 10.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 12.Zonari E, Pucci F, Saini M, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122:243–252. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 13.Kuchenbauer F, Mah SM, Heuser M, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–3358. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
- 14.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Leierseder S, Petzold T, Zhang L, Loyer X, Massberg S, Engelhardt S. MiR-223 is dispensable for platelet production and function in mice. Thromb Haemost. 2013;110:1207–1214. doi: 10.1160/TH13-07-0623. [DOI] [PubMed] [Google Scholar]
- 16.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 17.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 18.Fazi F, Racanicchi S, Zardo G, et al. Epigenetic Silencing of the Myelopoiesis Regulator microRNA-223 by the AML1/ETO Oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukao T, Fukuda Y, Kiga K, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 21.Gentner B, Visigalli I, Hiramatsu H, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2:58ra84. doi: 10.1126/scitranslmed.3001522. [DOI] [PubMed] [Google Scholar]
- 22.Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 23.Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang G, Meng C, Guo X, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 25.Zardo G, Ciolfi A, Vian L, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–4046. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 26.Felli N, Pedini F, Romania P, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94:479–486. doi: 10.3324/haematol.2008.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vian L, Di Carlo M, Pelosi E, et al. Transcriptional fine-tuning of microRNA-223 levels directs lineage choice of human hematopoietic progenitors. Cell Death Differ. 2014;21:290–301. doi: 10.1038/cdd.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuser M, Argiropoulos B, Kuchenbauer F, et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in AML patients. Blood. 2007;110:1639–1647. doi: 10.1182/blood-2007-03-080523. [DOI] [PubMed] [Google Scholar]
- 29.Argiropoulos B, Palmqvist L, Yung E, et al. Linkage of Meis1 leukemogenic activity to multiple downstream effectors including Trib2 and Ccl3. Exp Hematol. 2008;36:845–859. doi: 10.1016/j.exphem.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Sun SM, Rockova V, Bullinger L, et al. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. 2013;27:100–106. doi: 10.1038/leu.2012.158. [DOI] [PubMed] [Google Scholar]
- 31.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 32.Kuchenbauer F, Feuring-Buske M, Buske C. AML1-ETO needs a partner: new insights into the pathogenesis of t(8;21) leukemia. Cell Cycle. 2005;4:1716–1718. doi: 10.4161/cc.4.12.2256. [DOI] [PubMed] [Google Scholar]
- 33.Schessl C, Rawat VP, Cusan M, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm BT, Briau M, Austin P, et al. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood. 2011;117:e27–38. doi: 10.1182/blood-2010-07-293332. [DOI] [PubMed] [Google Scholar]
- 37.Daschkey S, Rottgers S, Giri A, et al. MicroRNAs distinguish cytogenetic subgroups in pediatric AML and contribute to complex regulatory networks in AML-relevant pathways. PLoS One. 2013;8:e56334. doi: 10.1371/journal.pone.0056334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller PH, Cheung AM, Beer PA, et al. Enhanced normal short-term human myelopoiesis in mice engineered to express human-specific myeloid growth factors. Blood. 2013;121:e1–e4. doi: 10.1182/blood-2012-09-456566. [DOI] [PubMed] [Google Scholar]
- 39.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 40.Eyholzer M, Schmid S, Schardt JA, Haefliger S, Mueller BU, Pabst T. Complexity of miR-223 regulation by CEBPA in human AML. Leuk Res. 2010;34:672–676. doi: 10.1016/j.leukres.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 42.Metzeler KH, Maharry K, Kohlschmidt J, et al. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27:2023–2031. doi: 10.1038/leu.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heuser M, Yun H, Berg T, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell. 2011;20:39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsingh G, Jacoby MA, Shao J, et al. Acquired copy number alterations of miRNA genes in acute myeloid leukemia are uncommon. Blood. 2013;122:e44–e51. doi: 10.1182/blood-2013-03-488007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.