Abstract
Cholesterol is detrimental to human health in excess but is also essential for normal embryogenesis. Hence, enzymes involved in its synthesis possess many layers of regulation to achieve balanced cholesterol levels. 7-Dehydrocholesterol reductase (DHCR7) is the terminal enzyme of cholesterol synthesis in the Kandutsch-Russell pathway, converting 7-dehydrocholesterol (7DHC) to cholesterol. In the absence of functional DHCR7, accumulation of 7DHC and a lack of cholesterol production leads to the devastating developmental disorder, Smith-Lemli-Opitz syndrome. This study identifies that statin treatment can ameliorate the low DHCR7 expression seen with common Smith-Lemli-Opitz syndrome mutations. Furthermore, we show that wild-type DHCR7 is also relatively labile. In an example of end-product inhibition, cholesterol accelerates the proteasomal degradation of DHCR7, resulting in decreased protein levels and activity. The loss of enzymatic activity results in the accumulation of the substrate 7DHC, which leads to an increased production of vitamin D. Thus, these findings highlight DHCR7 as an important regulatory switch between cholesterol and vitamin D synthesis.
Keywords: cholesterol, cholesterol regulation, enzyme degradation, genetic disease, vitamin D, 7-dehydrocholesterol, DHCR7
Introduction
Cholesterol is an essential molecule for life in higher organisms, but too much or too little can lead to disease. Its synthesis requires a complex series of reactions that involves >20 steps, which must be tightly controlled to balance cholesterol levels. Here, we focus on the post-translational regulation of 7-dehydrocholesterol reductase (DHCR7),2 the terminal enzyme in the Kandutsch-Russell pathway of cholesterol synthesis (1, 2) (Fig. 1A). The alternative terminal enzyme in the Bloch pathway of cholesterol synthesis is 24-dehydrocholesterol reductase (DHCR24) (3). Despite a certain level of redundancy, both the Kandutsch-Russell and Bloch pathways are utilized in different cellular contexts, and are both essential for cell survival (2).
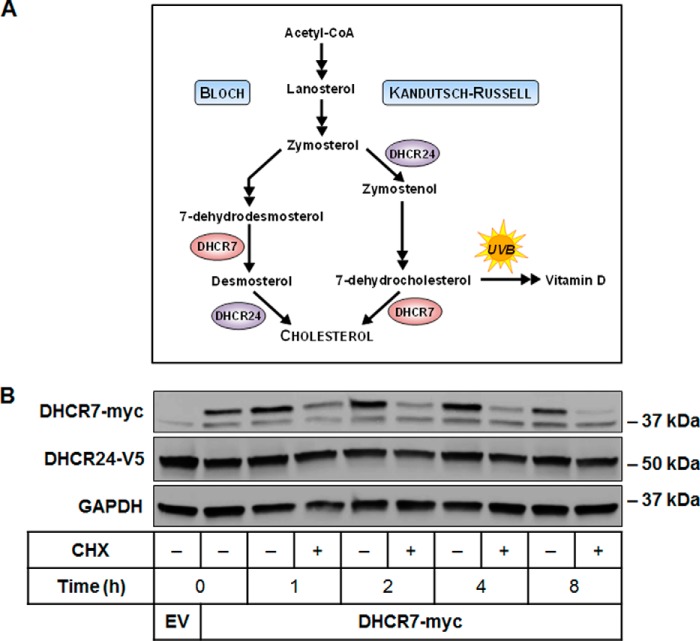
FIGURE 1.
DHCR7 is rapidly turned over compared with DHCR24. A, DHCR7 and DHCR24 catalyze the final steps of cholesterol synthesis via the Kandutsch-Russell and Bloch pathways, respectively. 7-Dehydrocholesterol can be converted to cholesterol by acting as a substrate for DHCR7 or to vitamin D through exposure to ultraviolet B (UVB) light. A simplified version of the modified Kandutsch-Russell pathway is shown as described in detail previously (2). B, CHO-DHCR24-V5 cells were transfected with empty vector (EV) or DHCR7-myc plasmid and treated with or without 10 μg/ml cycloheximide (CHX) for 0–8 h. Protein levels were analyzed using Western blotting with myc, V5, and GAPDH antibodies. Blots are representative of three independent experiments. There is a nonspecific band at 37 kDa in the myc immunoblot.
DHCR7 acts by reducing the C(7–8) double bond of 7-dehydrocholesterol (7DHC) to form cholesterol. A lack of functional DHCR7 results in an accumulation of 7DHC and reduced cholesterol synthesis, resulting in the devastating developmental disorder Smith-Lemli-Opitz syndrome (SLOS) (4, 5). SLOS is characterized by morphogenic and congenital abnormalities, mental retardation, and behavioral problems (6). The incidence of SLOS reportedly ranges between 1 in 10,000 to 70,000 in different populations, with a carrier rate of approximately one percent for pathogenic mutations of SLOS (7). Although the disorder was first identified 50 years ago (4), there is no cure, and clinical and biochemical research into effective therapies is still ongoing (8).
Besides being a precursor for cholesterol, 7DHC is also a precursor in vitamin D synthesis. This reaction is catalyzed by ultraviolet B (UVB) irradiation of the skin (Fig. 1A) and represents the major source of vitamin D in humans. Vitamin D is best known for its role in calcium homeostasis and skeletal health, but its deficiency has also been associated with an increased risk of certain cancers, autoimmune diseases, type II diabetes mellitus, cardiovascular disease, and overall mortality (9–11). With an increasing number of individuals placed on vitamin D supplements despite little evidence of their success in preventing chronic disease (10, 12), there needs to be a greater understanding of how vitamin D synthesis is regulated. Through genome-wide association studies, genetic variants in DHCR7 have been identified as strong determinants of circulating concentrations of 25-hydroxyvitamin D, a marker of vitamin D status (13, 14). Thus, examining DHCR7 regulation will provide insights into both vitamin D and cholesterol synthesis.
The rate-limiting steps of cholesterol synthesis are catalyzed by 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR) and squalene monooxygenase (SM), which are both rapidly turned over in response to sterol triggers (15, 16). In contrast, DHCR24 protein appears relatively stable (17). Because DHCR7 and DHCR24 are regulated similarly at the transcriptional level via sterol-regulatory element-binding protein (SREBP) (18), it is possible that these two alternative terminal enzymes differ at the post-translational level, providing differential regulation of the two pathways. Here, we report for the first time that DHCR7 is rapidly degraded proteasomally in response to cholesterol, which switches flux from cholesterol toward vitamin D synthesis.
Experimental Procedures
Cell Culture
CHO-7 and SRD-1 cell lines (gifts of Drs. Goldstein, Brown, and DeBose-Boyd, University of Texas Southwestern) and HaCaT cells (gift of Dr. Mason, University of Sydney, Australia) were used. HEK-293 cells stably overexpressing SM-N100-GFP-V5 (16) and CHO-7 cells stably overexpressing DHCR24-V5 (CHO-DHCR24) (19) were generated previously. HEK-293 or CHO-7 cells stably overexpressing DHCR7-myc (HEK-DHCR7 or CHO-DHCR7, respectively) were similarly generated for this study. HEK-293 and HaCaT cell lines were maintained in DMEM HG medium supplemented with 10% (v/v) fetal calf serum, and CHO cell lines were maintained in DMEM/F12 media supplemented with 5% (v/v) newborn calf lipoprotein-deficient serum. Cells were seeded at a density of 4.5 × 105 cells per well in a 6-well plate, transfected the following day, and treated the next day. Sterol intermediates were complexed to the carrier methyl-β-cyclodextrin (CD) (20). Treatments were performed in sterol-depleted media that contained the statin compactin (5 μm). All statin treatments were supplemented with mevalonate (50 μm). HEK-SM-N100-GFP-V5 cells were additionally pretreated overnight in the treatment media, as performed previously (16). HEK-293 and HaCaT cell lines were treated in DMEM HG media supplemented with 10% (v/v) fetal calf lipoprotein-deficient serum.
Expression Plasmids
The protein-coding sequence of human DHCR7 (NM_001360.2) was cloned from cDNA into pGEM T-easy (Promega) and subcloned into pcDNA4 (Life Technologies) with a C-terminal V5-epitope tag and into pcDNA5-FRT (Life Technologies) with a C-terminal myc tag that was used previously (21). The plasmid pcDNA4-DHCR7-V5 was used as the template to generate amino acid mutations via site-directed mutagenesis, whereas pcDNA5-DHCR7-myc-FRT was used to create the stable cell lines for this study. Primer sequences are available upon request.
Plasmid and siRNA Transfections
HEK-293 cells were seeded, and DHCR7 plasmids were transfected the next day for 24 h with 1 μg of DNA to 2 μl of TransIT-2020 reagent (Mirus Bio). Similarly, CHO-DHCR24-V5 cells were transfected with the DHCR7-myc plasmid for 4 h with 1 μg of DNA to 4 μl of Lipofectamine LTX transfection reagent (Life Technologies). Cells were treated the following day as indicated in the figure legends.
For siRNA transfections, HEK-DHCR7 or CHO-DHCR7 cells were transfected for 24 h with 25 nm siRNA using 5 μl of Lipofectamine RNAiMAX transfection reagent (Life Technologies). Then cells were harvested for quantitative real-time-PCR or treated as indicated in the figure legends.
Quantitative Real-Time PCR
After plasmid or siRNA transfection, total RNA was harvested using Tri Reagent and reverse transcribed to cDNA using Superscript III Reverse Transcriptase. Gene expression levels of Insig-1, Insig-2, and DHCR7 were determined and normalized to the housekeeping gene porphobilinogen deaminase (PBGD). Primer sequences for PBGD have been published previously (22, 23). Insig-1 primer sequences were CGCCTGTTGCAGAGGAGCC (forward) and CGAGGTGACTGTCGATACAGGG (reverse), Insig-2 primers were CGCTCTTTCCACCTGATGTG (forward) and CAGTCCAATGGATAGTGCAGCC (reverse), and DHCR7 primers were CACCGGCCGTGCTAGTCTGG (forward) and CAGGCTTGTAGCCCGTTCACCTC (reverse).
Western Blotting
Protein was harvested in modified radioimmunoprecipitation assay buffer, as described previously (24). Protein concentrations were measured and normalized using BCA Protein Assay (Thermo Fisher Scientific), and all samples were equally loaded for 10% (w/v) SDS-PAGE gel separation. Samples were transferred onto nitrocellulose membranes, and equal loading was confirmed with Ponceau S stain. Membranes were blocked with 5% (w/v) skim milk and probed with the following primary antibodies: mouse anti-myc (1:5000 dilution in 0.5% BSA; Santa Cruz, catalog no. SC-40, lot no. A2814), mouse anti-V5 (1:5000 dilution in 5% skim milk; Life Technologies, catalog no. R960-25, lot no. 1658817), rabbit anti-GAPDH (1:1000 dilution in PBST; Cell Signaling Technology, catalog no. 2118, lot no. 8), rabbit anti-calnexin (1:1000 dilution in PBST, Cell Signaling Technology, catalog no. 2433, lot no. 4), and mouse anti-α-tubulin (1:10,000 dilution in 5% BSA, Sigma, catalog no. T5168, lot no. 072M4809). Membranes were then visualized with the enhanced chemiluminescent detection system using the ImageQuant LAS 500 (GE Healthcare). Protein band intensities from Western blots were quantified by densitometry using Image Studio Lite (version 4.0). Locations of molecular weight standards are indicated on the blots.
Gas Chromatography/Mass Spectrometry (GC/MS)
Cells were treated as indicated in the figure legends with 2 μg/ml [2H7]7DHC (Avanti Lipids) complexed to CD for 2 h. If required, cells were treated with UVB, then harvested, and protein was normalized before adding 0.3 μg of 5α-cholestane as an internal standard for GC/MS. Lysates were processed in ethanol, with 10 μm butylated hydroxytoluene and 200 μm EDTA. Lipids were extracted, derivatized, and analyzed in selective ion monitoring mode using a Thermo Trace gas chromatograph coupled with a Thermo DSQII mass spectrometer and Thermo Triplus autosampler (Thermo Fisher Scientific), and data were processed with Thermo Xcalibur Software (Version 2.1.0.1140) as described previously (17). Sterols monitored are detailed in Table 1. Quantification of DHCR7 activity or rate of vitamin D production was based on chromatographic peak areas measured for [2H7]cholesterol relative to [2H7]7DHC or [2H7]vitamin D, respectively.
TABLE 1.
Retention times and ion m/z values monitored in selective ion monitoring mode during gas chromatography/mass spectrometry
| Compound | Retention time | Quantification/confirmation ion (m/z) | Molecular ion (m/z) |
|---|---|---|---|
| min | |||
| 5α-Cholestane | 28.0 | 217 | 372 |
| [2H7]Cholesterol | 33.5 | 336 | 465 |
| [2H7]7DHC | 35.5 | 358 | 463 |
| [2H7]Vitamin D | 30.0 | 358 | 463 |
Data Presentation and Statistical Analysis
Data are presented as the mean ± S.E. Statistical differences were determined by Student's paired t test (two-tailed), where p values of ≤0.05 (*) and ≤0.01 (**) were considered statistically significant.
Results
DHCR7 Is Rapidly Turned Over in Response to Cholesterol
First, we compared the protein stability of DHCR7 and DHCR24 using CHO-7 cells stably overexpressing DHCR24-V5(19). We transiently transfected these cells with DHCR7-myc and treated them with the protein synthesis inhibitor, cycloheximide to study protein turnover. Although DHCR24 protein expression remained stable for 8 h, DHCR7 turnover was rapid (Fig. 1B).
Because cholesterol is produced by DHCR7, we considered whether end-product inhibition occurred. Therefore, we treated cells overexpressing DHCR7 with cholesterol and found that DHCR7 protein expression was indeed reduced by cholesterol treatment in a time-dependent manner (Fig. 2A), consistent with end-product-mediated degradation. Although basal turnover of DHCR7 was very rapid with cycloheximide alone (Fig. 2B), cholesterol treatment further accelerated its degradation (Fig. 2, A and B). A version of DHCR7 tagged at the N terminus was similarly degraded at 8 h (data not shown).
FIGURE 2.
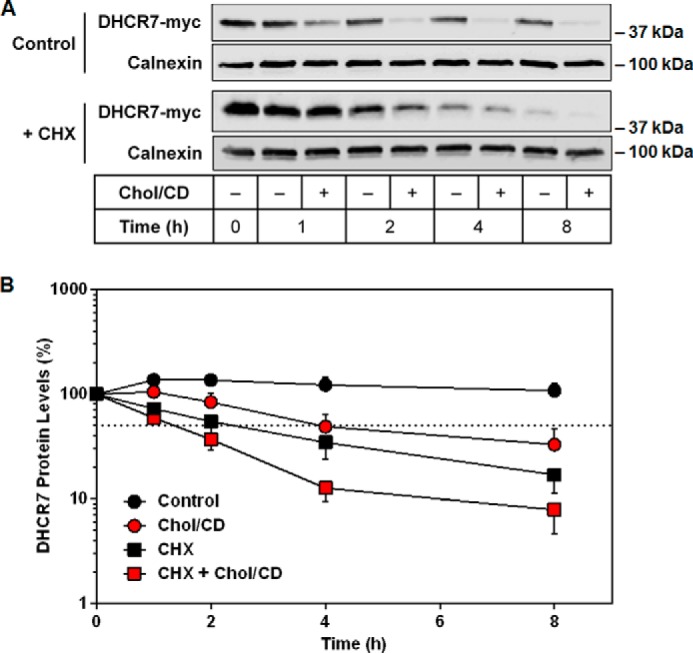
DHCR7 is rapidly turned over in response to cholesterol. A, HEK-DHCR7 cells were treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) and with or without 10 μg/ml cycloheximide (CHX) for 0–8 h. Blots are representative of six independent experiments. Protein levels were analyzed using Western blotting with myc and calnexin antibodies. B, semilog plot of DHCR7 protein levels normalized to the 0-h control conditions from A, which was set to 100%, and is presented as the mean ± S.E. The dotted line indicates that 50% of protein remained.
Next we used SRD-1 cells (25), a CHO mutant cell line that lacks SREBP-mediated transcriptional regulation, to ensure that the observed effects were post-translational. To measure DHCR7 activity, we metabolically labeled cells with deuterated 7DHC ([2H7]7DHC) and determined conversion to [2H7]cholesterol by GC/MS as we have done previously (21) (Fig. 3, A and B). In the absence of an effective endogenous DHCR7 antibody, this confirmed that the decreased protein levels also resulted in a decrease in DHCR7 activity. Our findings show that cholesterol treatment decreased DHCR7 activity by 50% (Fig. 3C). Furthermore, DHCR7 mRNA levels remained stable after cholesterol treatment in SRD-1 cells (Fig. 3D). Collectively, these results demonstrate that cholesterol stimulates a decrease in DHCR7 protein at the post-translational level, which also caused a decrease in DHCR7 activity.
FIGURE 3.
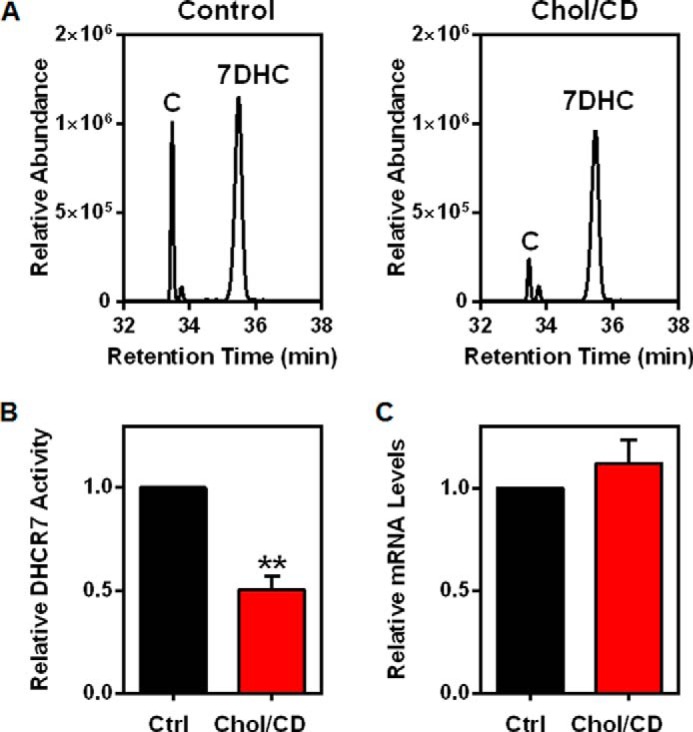
Cholesterol causes a decrease in DHCR7 activity but not DHCR7 mRNA levels. A and B, SRD-1 cells were treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) for 8 h before labeling with [2H7]7DHC/CD for 2 h. Lipid extracts were analyzed for [2H7]cholesterol relative to [2H7]7DHC using GC/MS. A, chromatograms are representative. B, data were normalized to the control condition, which was set to 1, and are presented as mean ± S.E. from five independent experiments. ** indicates statistical significance compared with the control condition, p < 0.01. C, SRD-1 cells were treated with or without 20 μg/ml Chol/CD for 8 h. Gene expression levels of DHCR7 were normalized to the housekeeping gene, PBGD. Data are presented as the mean ± S.E. from four independent experiments performed with triplicate cultures.
Sterol Specificity of DHCR7 Degradation
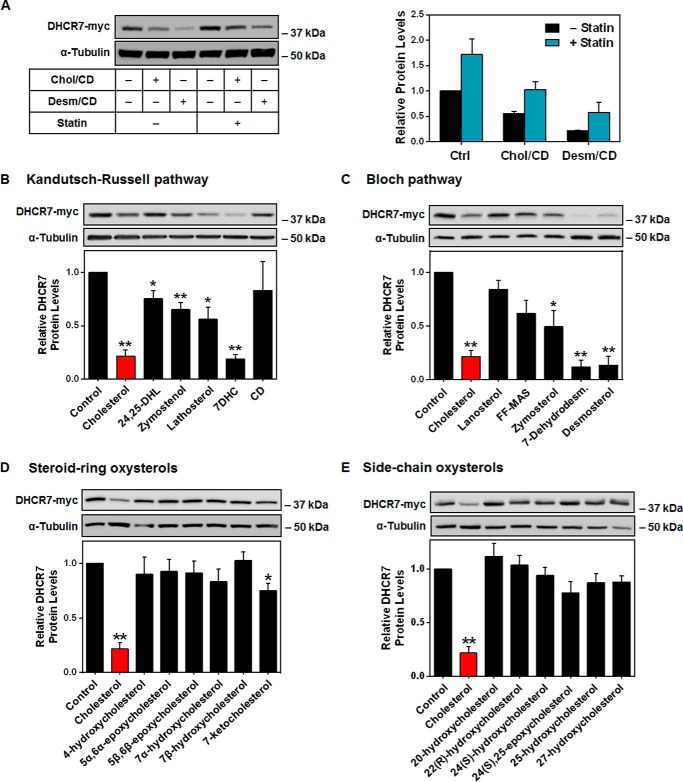
Next, we tested desmosterol, the product of DHCR7 in the Bloch pathway, and found that it also reduced DHCR7 protein levels (Fig. 4A). Conversely, overnight statin pretreatment, which was used to lower sterol status, resulted in higher basal expression of DHCR7 (Fig. 4A).
FIGURE 4.
The effects of various sterols on DHCR7 protein stability. A, HEK-DHCR7 cells were treated with or without 5 μm compactin (statin) overnight and then treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) or desmosterol/cyclodextrin (Desm/CD) for 8 h. Blots are representative, and densitometry is presented as the mean ± S.E. from three independent experiments. HEK-DHCR7 cells were treated with 20 μg/ml concentrations of the indicated sterol from the Bloch pathway, Kandutsch-Russell pathway, or methyl-β-cyclodextrin (CD) (B and C) or treated with 2.5 μm steroid-ring or side-chain oxysterols for 8 h (D and E). Blots are representative, and data are normalized to the control condition, which has been set to 1, and presented as the mean ± S.E. from three (CD, steroid-ring oxysterols), four (Bloch sterols), five (side-chain oxysterols), or six (Kandutsch-Russell sterols) independent experiments. * indicates statistical significance compared with the control condition, p < 0.05; **, p < 0.01. Protein levels were analyzed using Western blotting with myc and α-tubulin antibodies.
HMGCR and SM have distinct sterol response profiles, with SM degradation induced by cholesterol and desmosterol (16), whereas HMGCR responds to earlier intermediates, such as 24,25-dihydrolanosterol, and the oxygenated derivatives of cholesterol, oxysterols (26, 27). We predicted DHCR7 to be degraded similarly to SM in response to sterol products in the pathway. Having shown that both cholesterol and desmosterol accelerate DHCR7 protein degradation, we further investigated sterol specificity by testing a selection of sterol pathway intermediates. There was little sterol specificity with most intermediates causing some decrease in DHCR7 levels (Fig. 4, B and C) and with later products in the pathway tending to display a higher propensity to trigger degradation.
Additionally, oxysterols were tested at concentrations shown to degrade HMGCR (28). Side-chain oxysterols, such as 25-hydroxycholesterol, induce HMGCR degradation (29), whereas steroid-ring oxysterols can induce SM degradation (16). Again, we predicted that DHCR7 would follow a similar pattern to SM but found that most oxysterols had no effect. Only 7-ketocholesterol, which can be generated enzymatically from 7DHC (30), significantly decreased DHCR7 levels, albeit to a lesser extent than cholesterol (Fig. 4, D and E). Although DHCR7 is relatively labile and sensitive to degradation via a number of sterol triggers, we focused on the relationship between DHCR7 and cholesterol as it is the most physiologically abundant sterol (31), and therefore, any measurable effects within cells are likely to be largely caused by cholesterol.
Cholesterol-mediated Degradation of DHCR7 Does Not Involve Putative Cholesterol-binding Sites nor the Insig Retention Protein
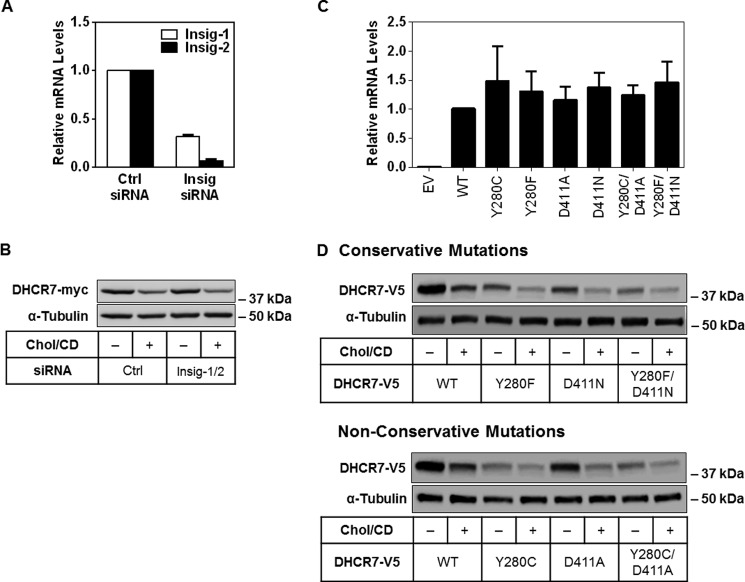
There are at least two ways that DHCR7 may respond to increased cholesterol: first, via a predicted sterol-sensing domain (32, 33), and second, by interacting with putative cholesterol-binding sites (34). In HMGCR, crucial residues were identified in the enzyme's sterol-sensing domain, which must bind to the endoplasmic reticulum retention protein Insig to induce HMGCR degradation (35). DHCR7 has been proposed to contain a sterol-sensing domain (32), although this has not been verified experimentally. To examine if a similar process occurs in the sterol-mediated degradation of DHCR7, we used specific siRNA to knock down both isoforms of Insig (Fig. 5A). Because cholesterol regulation remained after Insig knockdown (Fig. 5A), this indicates that Insig is not required in the cholesterol-mediated degradation of DHCR7.
FIGURE 5.
Degradation of DHCR7 does not involve Insig nor putative cholesterol-binding sites. A, HEK-DHCR7 cells were transfected with 25 nm concentrations of the indicated siRNA for 24 h, then harvested to measure mRNA levels. Gene expression levels of Insig-1 and Insig-2 were normalized to the housekeeping gene, PBGD. Data are normalized to the control siRNA condition, which has been set to 1, and are presented as the mean ± S.E. from three independent experiments performed in triplicate cultures. B, HEK-DHCR7 cells were transfected with 25 nm concentrations of the indicated siRNA for 24 h, then treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) for 8 h. Blots are representative of four independent experiments. C and D, HEK-293 cells were transfected with the indicated DHCR7-V5 plasmid and harvested the next day to measure mRNA levels or treated with or without 20 μg/ml Chol/CD for 8 h. Gene expression levels of DHCR7 were normalized to the housekeeping gene, PBGD. Data are normalized to the control (WT) condition, which has been set to 1 and is presented as mean ± S.E. from three independent experiments performed with triplicate cultures. EV, empty vector. Blots are representative of three independent experiments. Protein levels were analyzed using Western blotting with V5 and α-tubulin antibodies.
Alternatively, mutations of other residues in DHCR7 may confer resistance to cholesterol-mediated degradation. Two cholesterol-binding sites (tyrosine 280 and aspartic acid 411) were recently predicted through solving the crystal structure of a Δ(14)-sterol reductase that is a homologue of human DHCR7 found in the methanotrophic bacterium Methylomicrobium alcaliphilum 20Z (34). Using site-directed mutagenesis, we mutated these residues both singly and together to determine their importance in the degradation of DHCR7. A loss of cholesterol-mediated regulation in these mutants would imply that cholesterol may directly bind to these sites to induce DHCR7 degradation. Despite similar transcript levels indicating equal transfection rates (Fig. 5B), both conservative (Y280F; D411N) and non-conservative mutations (Y280C; D411A) tended to display lower protein expression (Fig. 5C), suggesting that DHCR7 is easily destabilized. The mutant Y280C also represents a mutation identified in SLOS patients (36), which could explain its low protein expression compared with wild type. All mutants maintained regulation by cholesterol (Fig. 5C), suggesting that these sites are not responsible for the cholesterol-mediated degradation of DHCR7.
Low DHCR7 Expression in SLOS Mutations Is Ameliorated by Statin Treatment
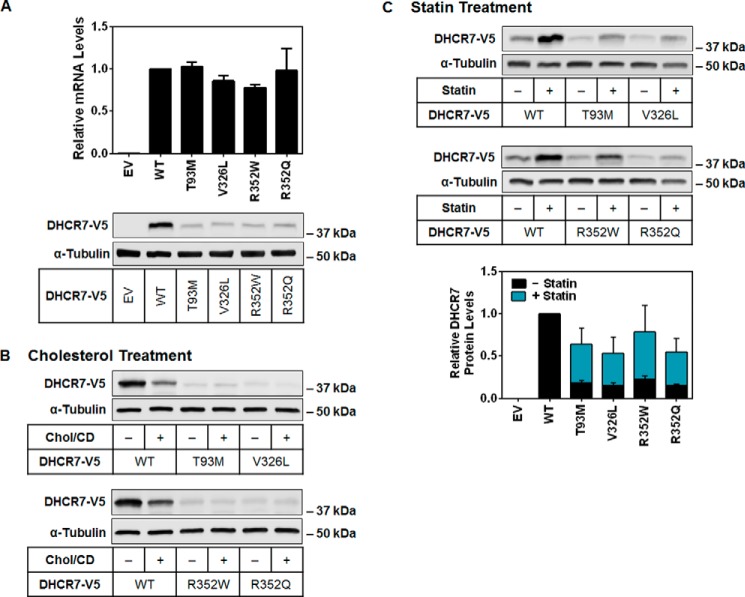
Considering the lowered DHCR7 expression of the Y280C mutant, we sought to specifically examine the effect of common SLOS-related mutations on DHCR7 protein levels. More than 150 known mutations of DHCR7 exist in individuals affected by SLOS (37), which are widely distributed over the gene. Missense mutations compose >85% of the known alleles, although the two most common are a splice acceptor site mutation (IVS8–1G>C) and a nonsense mutation (W151X) (37). As these would abolish DHCR7 expression, we selected the four most common missense mutations to study (T93M, V326L, R352W, and R352Q) (38). Despite equal transcript levels after transient expression in HEK-293 cells, protein expression was significantly reduced with all four mutations (Fig. 6A).
FIGURE 6.
Four common SLOS mutations reduce DHCR7 protein stability, which can be rescued by statin treatment. A, HEK-293 cells were transfected with the indicated DHCR7-V5 plasmid and then harvested to measure mRNA or protein levels. Gene expression levels of DHCR7 were normalized to the housekeeping gene, PBGD. Data are normalized to the control (WT) condition, which has been set to 1, and are presented as the mean ± S.E. from three independent experiments performed with triplicate cultures. Blots are representative of three independent experiments. EV, empty vector. B and C, HEK-293 cells were transfected with the indicated plasmid and treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) for 8 h (B) or 5 μm compactin (statin) for 24 h (C). Blots are representative, and densitometry was normalized to the control (WT) condition and is presented as the mean ± S.E. from three independent experiments. Protein levels were analyzed using Western blotting with V5 and α-tubulin antibodies.
This explains why these single point mutations are pathogenic, with ∼90% of protein expression lost, confirming a previous report for V326L and R352W (39). The other mutations tested were R352Q and T93M, which is recognized as a founder mutation for SLOS (40) and observed in ∼10% of patients (37). Because expression levels of the SLOS mutants were so low, cholesterol treatment showed no additional reduction (Fig. 6B). Since we found that statin pretreatment increased the expression of wild-type DHCR7 protein levels (Fig. 4A), we tested whether it could also improve the low expression in these SLOS mutants (Fig. 6C). Indeed, statin treatment ameliorated low DHCR7 expression in the SLOS mutants, causing an ∼3-fold improvement in protein levels.
Cholesterol-mediated Degradation of DHCR7 Occurs via the Proteasome
DHCR7 is a polytopic protein located in the endoplasmic reticulum (41). Proteins, like HMGCR and SM, that are bound to the endoplasmic reticulum (16, 29) are typically targeted for destruction by the ubiquitin-proteasome system (42). We predicted that DHCR7 is similarly degraded and found that inhibition of the proteasome (using MG132 or N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (ALLN)) rescued the cholesterol-induced degradation of DHCR7 (Fig. 7A). Use of ammonium chloride or bafilomycin to inhibit the lysosome, another major degradative compartment in the cell, could not prevent DHCR7 degradation (Fig. 7A). The cholesterol-regulatory domain of SM was similarly preserved by the same proteasomal inhibitors as shown previously (16), albeit to a lesser extent than DHCR7 (Fig. 7A). Considering that the E3 ligase MARCH6 can target both SM and HMGCR for degradation, we employed two independent siRNAs to knock down MARCH6, as we have done previously (28). However, DHCR7 degradation was not rescued (Fig. 7B) suggesting further investigation is needed to identify the E3 ligase(s) involved.
FIGURE 7.
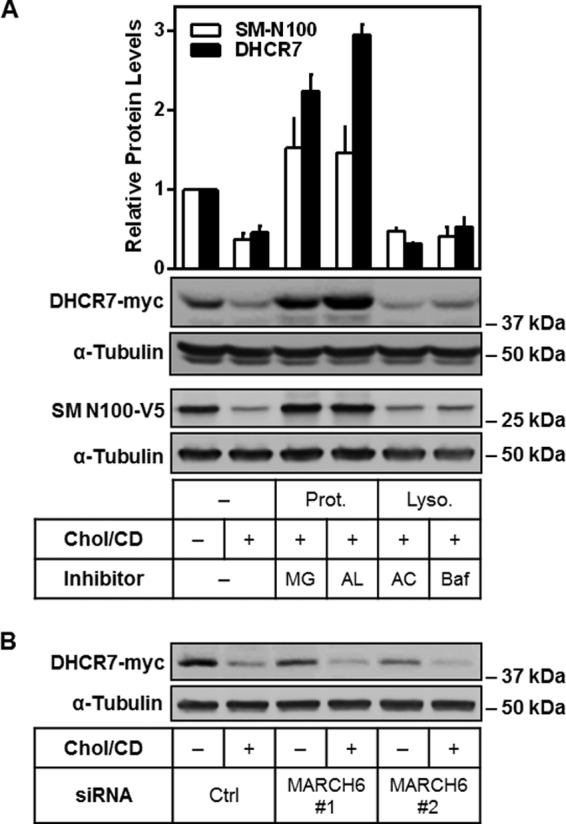
DHCR7 degradation occurs via the proteasome. A, CHO-DHCR7 cells were treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) and proteasomal (Prot., 10 μm MG132 (MG) or 25 mg/ml N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (AL)) or lysosomal (Lyso., 20 mm ammonium chloride (AC) or 10 nm bafilomycin (Baf)) inhibitors for 8 h. HEK-SM-N100-GFP-V5 cells were treated similarly as a positive control (16). Blots are representative, and data were normalized to the control condition, which was set to 1, presented as the mean ± S.E. from three independent experiments. B, CHO-DHCR7 cells were transfected with 25 nm concentrations of the indicated siRNA for 24 h, then treated with or without 20 μg/ml Chol/CD for 8 h. Blots are representative of three independent experiments. Protein levels were analyzed using Western blotting with myc, V5, and α-tubulin antibodies.
Cholesterol-mediated Degradation of DHCR7 Increases Vitamin D Production
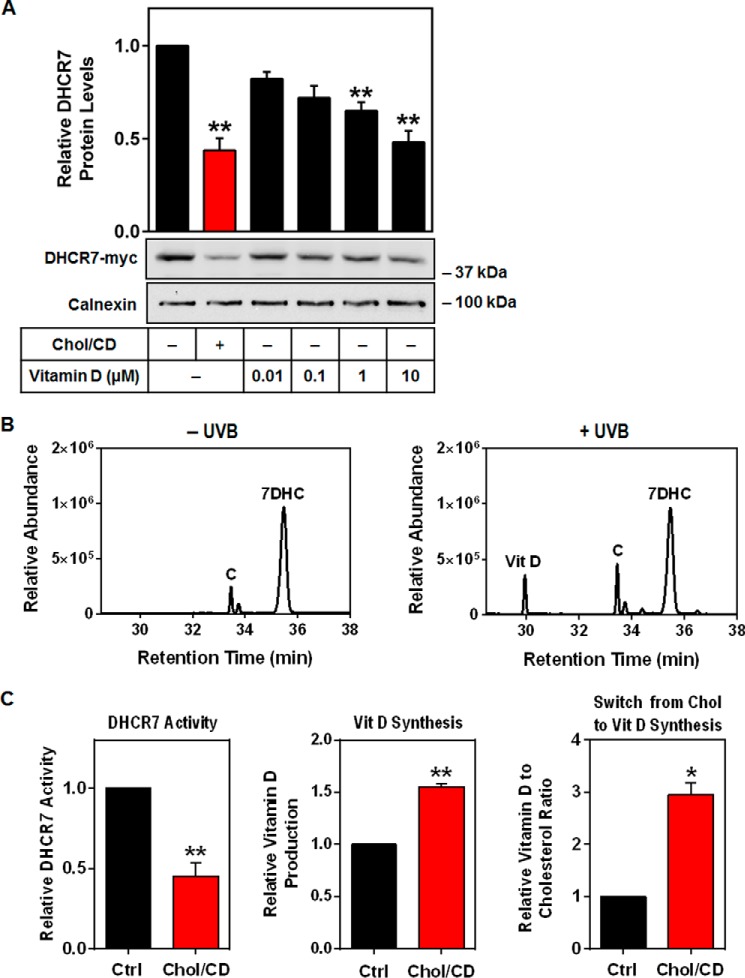
The connection between cholesterol and vitamin D is well known, considering that 7DHC is a precursor in the synthesis of both molecules. The majority of circulating 7DHC exists in the epidermis, where it can be converted to pre-vitamin D via exposure to UVB light, and then to vitamin D (also known as cholecalciferol) (9). This can then move from the skin to the liver and kidneys where it is converted into active forms of vitamin D. Here, we examined the effect of vitamin D (cholecalciferol) on DHCR7 expression and found that it reduces DHCR7 protein levels in a dose-dependent manner (Fig. 8A).
FIGURE 8.
Cholesterol treatment switches flux from cholesterol to vitamin D production. A, HEK-DHCR7 cells were treated with or without 20 μg/ml cholesterol-cyclodextrin (Chol/CD) or the indicated concentration of cholecalciferol (vitamin D) for 8 h. Blots are representative, and data were normalized to the control condition, which was set to 1, and presented as the mean ± S.E. from four independent experiments. Protein levels were analyzed using Western blotting with myc and calnexin antibodies. B, SRD-1 cells were treated with 20 μg/ml Chol/CD for 8 h before labeling with [2H7]7DHC for 2 h. Lipids were extracted in hexane and treated with or without ultraviolet B light (UVB). Chromatograms are from one in vitro experiment as proof-of-principle for vitamin D production. C, HaCaT cells were treated with or without 20 μg/ml Chol/CD for 8 h before labeling with [2H7]7DHC for 2 h. Cells were harvested for DHCR7 activity or treated with UVB, refreshed and harvested after 16 h to measure vitamin D production and the vitamin D to cholesterol ratio. Lipid extracts were analyzed for [2H7]cholesterol (C) relative to [2H7]vitamin D (Vit D) and [2H7]7DHC (7DHC) levels using GC/MS. Data are normalized to the control condition and are presented as mean ± S.E. from three (vitamin D) or four (DHCR7 activity) independent experiments. * indicates statistical significance compared with the control untreated condition, *, p < 0.05; **, p < 0.01.
In conditions of low DHCR7 levels, the substrate 7DHC accumulates. As 7DHC is a precursor for vitamin D as well as cholesterol, we next tested whether the 7DHC accumulation caused by the cholesterol-dependent degradation of DHCR7 would increase vitamin D synthesis. We used SRD-1 cells as a proof-of-principle to show that vitamin D could be produced and measured with the same GC/MS approach used for sterol analysis (Fig. 3A). To do this we labeled SRD-1 cells with [2H7]7DHC and extracted total lipids, which were subjected to UVB treatment. This showed that UVB efficiently converted [2H7]7DHC to [2H7]vitamin D (Fig. 8B).
Because the skin is where vitamin D is formed as well as a very active site of cholesterol synthesis (43), we employed human keratinocyte (HaCaT) cells (44) that were labeled with [2H7]7DHC and subjected to UVB treatment. Cholesterol treatment resulted in a 55% decrease in DHCR7 activity, shown by the conversion of 7DHC to cholesterol (Fig. 8C), and a 50% increase in vitamin D synthesis (Fig. 8C). To determine the relative fate of [2H7]7DHC, we directly compared cholesterol and vitamin D production. Treatment with cholesterol resulted in a 3-fold increase in vitamin D relative to cholesterol synthesis (Fig. 8C). This demonstrates that cholesterol feeds back on DHCR7, increasing vitamin D production in skin cells.
Discussion
Cholesterol synthesis is controlled at numerous stages (45). DHCR7 inhabits a special place in the pathway, as the gateway to both cholesterol and vitamin D production. As with nearly all cholesterol synthetic enzymes, gene expression of DHCR7 is regulated by SREBP (18, 46). However, the post-translational regulation of DHCR7 has been relatively unexplored to date.
In this study we characterized the effect of common SLOS mutations on DHCR7 stability, providing evidence that statin therapy can at least partially reverse the significant loss of DHCR7 expression in SLOS (Fig. 6C). Second, we show that DHCR7 is rapidly turned over, and its degradation is accelerated by cholesterol (Fig. 2), which occurs via the proteasome (Fig. 7A). We demonstrate that this mechanism acts as a switch to increase vitamin D synthesis as a consequence of 7DHC accumulation (Fig. 8C). Thus, DHCR7 plays a unique role in regulating both cholesterol and vitamin D levels in the cell.
Although we previously found that SM protein is degraded by cholesterol with considerable specificity (16), DHCR7 degradation was induced by numerous sterols (Fig. 4). Besides its products, cholesterol and desmosterol, the substrates of DHCR7, 7DHC, and 7-dehydrodesmosterol, were also particularly effective in stimulating degradation. There are precedents for the substrate-assisted suicide of enzymes (47). However, this likely has physiological relevance only in individuals with high 7DHC levels, such as those affected by SLOS. 7DHC accumulation is considered detrimental in SLOS patients because of the toxic oxidized species it is readily converted into (48), but our data suggest that 7DHC itself may also have a negative effect in further destabilizing any residual DHCR7 (Fig. 4B).
To decrease levels of 7DHC in SLOS patients, statin therapy has been proposed as a means of reducing the cholesterol synthetic pathway entirely (49, 50), and short-term clinical trials using statin as a therapeutic for SLOS have been performed (51). Significantly, we found that statin treatment increased DHCR7 expression in the SLOS mutants tested. Therefore, in addition to reducing 7DHC levels as proposed, statin treatment improves DHCR7 expression transcriptionally (18) and also post-translationally, as shown in this study. Thus, our data identify another benefit of statin use in SLOS patients with missense mutations that destabilize DHCR7.
Based on our findings, we expected that cholesterol would further reduce DHCR7 expression in SLOS mutants, suggesting that cholesterol supplementation may have negative consequences as a treatment for SLOS. Earlier studies have indicated that cholesterol therapy provides initial benefit to SLOS patients in terms of development and behavioral issues (52), but more recent research has shown that this may not improve patient health long term (53, 54). In our system cholesterol had little effect on the SLOS mutants, which could be expected considering that initial DHCR7 expression was already very low (Fig. 6B). This suggests that no additional reduction in DHCR7 activity is likely to occur with cholesterol treatment. Ongoing clinical trials testing cholesterol should be informative about its suitability as a SLOS treatment option (55). However, our data suggesting that statin therapy can help stabilize DHCR7 levels should encourage further investigation into its efficacy in a clinical setting.
We have also identified that DHCR7 is proteasomally degraded, with inhibition of the proteasome causing a 2–3-fold increase of DHCR7 protein expression (Fig. 7A). We previously identified MARCH6 as an E3 ligase involved in the proteasomal degradation of SM and HMGCR (28). However, in this case we excluded its role in targeting DHCR7 for degradation (Fig. 7B). Thus, the identity of the E3 ligase(s) and the specific ubiquitination sites on DHCR7 remain unknown.
Further investigation into the membrane topology of DHCR7 could provide some clues. A recent study by Li et al. (34) made significant advances in predicting the structure of DHCR7. However, we have ruled out the involvement of two putative cholesterol-binding sites that they reported (34) (Fig. 5D). It is possible that the two sites may still act within a larger sterol-sensing domain, which DHCR7 is suspected to possess (33). We showed that this predicted domain does not behave in a similar manner to that in HMGCR (35), in that Insig is not required for DHCR7 degradation (Fig. 5B). Thus, the precise interaction between DHCR7 and cholesterol is yet to be elucidated.
Our study also provides new biochemical evidence connecting DHCR7 and vitamin D. Genetic variants in DHCR7 are reliably associated with vitamin D status (13, 14), indicating that the enzyme plays an important role in vitamin D metabolism. This variation in DHCR7 has been identified as a major adaptation affecting vitamin D metabolism in recent evolutionary history. Kuan et al. (56) determined that the strong selective pressure exerted by polymorphisms associated with DHCR7 assisted early human migration into areas of low sunlight by providing protection from the effects of low vitamin D. However, little was previously known about the biochemical nature of this relationship between DHCR7 and vitamin D. A recent study identified that vitamin D can inhibit DHCR7 activity (57), which suggests that a feed-forward mechanism for vitamin D production could exist. Our data extend this by demonstrating that vitamin D can impede cholesterol synthesis by specifically reducing DHCR7 protein expression in a dose-dependent manner (Fig. 8A). The skin predominantly utilizes a modified Kandutsch-Russell pathway (2), indicating that the pathway can provide a constant source of 7DHC for the production of vitamin D (58). In addition, the highly labile nature of DHCR7, destabilized further by cholesterol, also serves to switch flux toward vitamin D synthesis (Fig. 8C). Therefore, our findings uncover a new control point in cholesterol synthesis that also regulates vitamin D production, with DHCR7 playing a critical role in the homeostasis of two important molecules that are involved in human health and disease.
Author Contributions
A. V. P., W. L., and L. J. S. performed the experiments. A. V. P. and A. J. B. wrote the bulk of the manuscript. All authors contributed to experimental design and analysis of results and approved the final version of the manuscript.
Acknowledgments
We thank Ghazal Sultani and Christine Gana for preliminary work in this study and Martin Bucknall (Bioanalytical Mass Spectrometry Facility, University of New South Wales) and Cecilia Sandoval for technical assistance. We also thank members of the Brown laboratory for critically reviewing this manuscript.
This work was supported by The University of New South Wales Goldstar Award and National Health and Medical Research Council Grant 1060515. The authors declare that they have no conflicts of interest with the contents of this article.
- DHCR7
- 7-dehydrocholesterol (7DHC) reductase
- DHCR24
- 24-dehydrocholesterol reductase
- HMGCR
- 3-hydroxy-3-methylglutaryl CoA reductase
- SLOS
- Smith-Lemli-Opitz syndrome
- SM
- squalene monooxygenase
- SREBP
- sterol-regulatory element-binding protein
- UVB
- ultraviolet B
- CD
- methyl-β-cyclodextrin.
References
- 1. Kandutsch A. A., and Russell A. E. (1960) Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J. Biol. Chem. 235, 2256–2261 [PubMed] [Google Scholar]
- 2. Mitsche M. A., McDonald J. G., Hobbs H. H., and Cohen J. C. (2015) Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife 4, e07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloch K. (1965) The biological synthesis of cholesterol. Science 150, 19–28 [DOI] [PubMed] [Google Scholar]
- 4. Smith D. W., Lemli L., and Opitz J. M. (1964) A newly recognized syndrome of multiple congenital anomalies. J. Pediatr. 64, 210–217 [DOI] [PubMed] [Google Scholar]
- 5. Tint G. S., Irons M., Elias E. R., Batta A. K., Frieden R., Chen T. S., and Salen G. (1994) Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N. Engl. J. Med. 330, 107–113 [DOI] [PubMed] [Google Scholar]
- 6. Waterham H. R., and Wanders R. J. (2000) Biochemical and genetic aspects of 7-dehydrocholesterol reductase and Smith-Lemli-Opitz syndrome. Biochim. Biophys. Acta 1529, 340–356 [DOI] [PubMed] [Google Scholar]
- 7. Cross J. L., Iben J., Simpson C. L., Thurm A., Swedo S., Tierney E., Bailey-Wilson J. E., Biesecker L. G., Porter F. D., and Wassif C. A. (2015) Determination of the allelic frequency in Smith-Lemli-Opitz syndrome by analysis of massively parallel sequencing data sets. Clin. Genet. 87, 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pasta S., Akhile O., Tabron D., Ting F., Shackleton C., and Watson G. (2015) Delivery of the 7-dehydrocholesterol reductase gene to the central nervous system using adeno-associated virus vector in a mouse model of Smith-Lemli-Opitz Syndrome. Mol. Genet. Metab. Rep. 4, 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hossein-nezhad A., and Holick M. F. (2013) Vitamin D for health: a global perspective. Mayo Clin. Proc. 88, 720–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Afzal S., Brøndum-Jacobsen P., Bojesen S. E., and Nordestgaard B. G. (2014) Genetically low vitamin D concentrations and increased mortality: mendelian randomisation analysis in three large cohorts. BMJ 349, g6330–g6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Afzal S., Brøndum-Jacobsen P., Bojesen S. E., and Nordestgaard B. G. (2014) Vitamin D concentration, obesity, and risk of diabetes: a mendelian randomisation study. Lancet. Diabetes Endocrinol. 2, 298–306 [DOI] [PubMed] [Google Scholar]
- 12. Meyer H. E., Holvik K., and Lips P. (2015) Should vitamin D supplements be recommended to prevent chronic diseases? BMJ 350, h321. [DOI] [PubMed] [Google Scholar]
- 13. Wang T. J., Zhang F., Richards J. B., Kestenbaum B., van Meurs J. B., Berry D., Kiel D. P., Streeten E. A., Ohlsson C., Koller D. L., Peltonen L., Cooper J. D., O'Reilly P. F., Houston D. K., Glazer N. L., et al. (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 376, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahn J., Yu K., Stolzenberg-Solomon R., Simon K. C., McCullough M. L., Gallicchio L., Jacobs E. J., Ascherio A., Helzlsouer K., Jacobs K. B., Li Q., Weinstein S. J., Purdue M., Virtamo J., Horst R., et al. (2010) Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 19, 2739–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodwell V. W., Nordstrom J. L., and Mitschelen J. J. (1976) Regulation of HMG-CoA reductase. Adv. Lipid Res. 14, 1–74 [DOI] [PubMed] [Google Scholar]
- 16. Gill S., Stevenson J., Kristiana I., and Brown A. J. (2011) Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13, 260–273 [DOI] [PubMed] [Google Scholar]
- 17. Luu W., Zerenturk E. J., Kristiana I., Bucknall M. P., Sharpe L. J., and Brown A. J. (2014) Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis. J. Lipid Res. 55, 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prabhu A. V., Sharpe L. J., and Brown A. J. (2014) The sterol-based transcriptional control of human 7-dehydrocholesterol reductase (DHCR7): evidence of a cooperative regulatory program in cholesterol synthesis. Biochim. Biophys. Acta 1842, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 19. Zerenturk E. J., Kristiana I., Gill S., and Brown A. J. (2012) The endogenous regulator 24(S),25-epoxycholesterol inhibits cholesterol synthesis at DHCR24 (Seladin-1). Biochim. Biophys. Acta 1821, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 20. Brown A. J., Sun L., Feramisco J. D., Brown M. S., and Goldstein J. L. (2002) Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10, 237–245 [DOI] [PubMed] [Google Scholar]
- 21. Luu W., Hart-Smith G., Sharpe L. J., and Brown A. J. (2015) The terminal enzymes of cholesterol synthesis, DHCR24 and DHCR7, interact physically and functionally. J. Lipid Res. 56, 888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kielar D., Dietmaier W., Langmann T., Aslanidis C., Probst M., Naruszewicz M., and Schmitz G. (2001) Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin. Chem. 47, 2089–2097 [PubMed] [Google Scholar]
- 23. Du X., Kristiana I., Wong J., and Brown A. J. (2006) Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol. Biol. Cell 17, 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faulkner R. A., Nguyen A. D., Jo Y., and DeBose-Boyd R. A. (2013) Lipid-regulated degradation of HMG-CoA reductase and Insig-1 through distinct mechanisms in insect cells. J. Lipid Res. 54, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Sato R., Goldstein J. L., and Brown M. S. (1994) Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 8, 1910–1919 [DOI] [PubMed] [Google Scholar]
- 26. Lange Y., Ory D. S., Ye J., Lanier M. H., Hsu F.-F., and Steck T. L. (2008) Effectors of rapid homeostatic responses of endoplasmic reticulum cholesterol and 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 283, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 27. Song B.-L., Javitt N. B., and DeBose-Boyd R. A. (2005) Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 [DOI] [PubMed] [Google Scholar]
- 28. Zelcer N., Sharpe L. J., Loregger A., Kristiana I., Cook E. C., Phan L., Stevenson J., and Brown A. J. (2014) The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol. Cell Biol. 34, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song B.-L., and DeBose-Boyd R. A. (2004) Ubiquitination of 3-hydroxy-3-methylglutaryl-CoA reductase in permeabilized cells mediated by cytosolic E1 and a putative membrane-bound ubiquitin ligase. J. Biol. Chem. 279, 28798–28806 [DOI] [PubMed] [Google Scholar]
- 30. Shinkyo R., Xu L., Tallman K. A., Cheng Q., Porter N. A., and Guengerich F. P. (2011) Conversion of 7-dehydrocholesterol to 7-ketocholesterol is catalyzed by human cytochrome P450 7A1 and occurs by direct oxidation without an epoxide intermediate. J. Biol. Chem. 286, 33021–33028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McDonald J. G., Smith D. D., Stiles A. R., and Russell D. W. (2012) A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53, 1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bae S. H., Lee J. N., Fitzky B. U., Seong J., and Paik Y. K. (1999) Cholesterol biosynthesis from lanosterol: molecular cloning, tissue distribution, expression, chromosomal localization, and regulation of rat 7-dehydrocholesterol reductase, a Smith-Lemli-Opitz syndrome-related protein. J. Biol. Chem. 274, 14624–14631 [DOI] [PubMed] [Google Scholar]
- 33. Kuwabara P. E., and Labouesse M. (2002) The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18, 193–201 [DOI] [PubMed] [Google Scholar]
- 34. Li X., Roberti R., and Blobel G. (2015) Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature 517, 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 36. Prasad C., Marles S., Prasad A. N., Nikkel S., Longstaffe S., Peabody D., Eng B., Wright S., Waye J. S., and Nowaczyk M. J. (2002) Smith-Lemli-Opitz syndrome: new mutation with a mild phenotype. Am. J. Med. Genet. 108, 64–68 [DOI] [PubMed] [Google Scholar]
- 37. Waterham H. R., and Hennekam R. C. (2012) Mutational spectrum of Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 160, 263–284 [DOI] [PubMed] [Google Scholar]
- 38. Lanthaler B., Wieser S., Deutschmann A., Schossig A., Fauth C., Zschocke J., and Witsch-Baumgartner M. (2014) Genotype-based databases for variants causing rare diseases. Gene 550, 136–140 [DOI] [PubMed] [Google Scholar]
- 39. Fitzky B. U., Witsch-Baumgartner M., Erdel M., Lee J. N., Paik Y. K., Glossmann H., Utermann G., and Moebius F. F. (1998) Mutations in the Δ7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc. Natl. Acad. Sci. U.S.A. 95, 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Witsch-Baumgartner M., Schwentner I., Gruber M., Benlian P., Bertranpetit J., Bieth E., Chevy F., Clusellas N., Estivill X., Gasparini G., Giros M., Kelley R. I., Krajewska-Walasek M., Menzel J., Miettinen, T. et al. (2008) Age and origin of major Smith-Lemli-Opitz syndrome (SLOS) mutations in European populations. J. Med. Genet. 45, 200–209 [DOI] [PubMed] [Google Scholar]
- 41. Holmer L., Pezhman A., and Worman H. J. (1998) The human lamin B receptor/sterol reductase multigene family. Genomics 54, 469–476 [DOI] [PubMed] [Google Scholar]
- 42. Hampton R. Y. (2002) ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 43. Feingold K. R., Wiley M. H., MacRae G., Lear S., Moser A. H., Zsigmond G., and Siperstein M. D. (1983) De novo sterologenesis in the intact rat. Metabolism 32, 75–81 [DOI] [PubMed] [Google Scholar]
- 44. Lehmann B. (1997) HaCaT cell line as a model system for vitamin D3 metabolism in human skin. J. Invest. Dermatol. 108, 78–82 [DOI] [PubMed] [Google Scholar]
- 45. Sharpe L. J., and Brown A. J. (2013) Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288, 18707–18715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., and Goldstein J. L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mbonye U. R., Yuan C., Harris C. E., Sidhu R. S., Song I., Arakawa T., and Smith W. L. (2008) Two distinct pathways for cyclooxygenase-2 protein degradation. J. Biol. Chem. 283, 8611–8623 [DOI] [PubMed] [Google Scholar]
- 48. Xu L., Davis T. A., and Porter N. A. (2009) Rate constants for peroxidation of polyunsaturated fatty acids and sterols in solution and in liposomes. J. Am. Chem. Soc. 131, 13037–13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Correa-Cerro L. S., Wassif C. A., Kratz L., Miller G. F., Munasinghe J. P., Grinberg A., Fliesler S. J., and Porter F. D. (2006) Development and characterization of a hypomorphic Smith-Lemli-Opitz syndrome mouse model and efficacy of simvastatin therapy. Hum. Mol. Genet. 15, 839–851 [DOI] [PubMed] [Google Scholar]
- 50. Haas D., Garbade S. F., Vohwinkel C., Muschol N., Trefz F. K., Penzien J. M., Zschocke J., Hoffmann G. F., and Burgard P. (2007) Effects of cholesterol and simvastatin treatment in patients with Smith-Lemli-Opitz syndrome (SLOS). J. Inherit. Metab. Dis. 30, 375–387 [DOI] [PubMed] [Google Scholar]
- 51. Porter F. D. (2014) Investigation of simvastatin therapy in Smith-Lemli-Opitz syndrome, https://clinicaltrials.gov/ct2/show/NCT00064792, National Library of Medicine, Bethesda, MD [Google Scholar]
- 52. Elias E. R., Irons M. B., Hurley A. D., Tint G. S., and Salen G. (1997) Clinical effects of cholesterol supplementation in six patients with the Smith-Lemli-Opitz syndrome (SLOS). Am. J. Med. Genet. 68, 305–310 [DOI] [PubMed] [Google Scholar]
- 53. Sikora D. M., Ruggiero M., Petit-Kekel K., Merkens L. S., Connor W. E., and Steiner R. D. (2004) Cholesterol supplementation does not improve developmental progress in Smith-Lemli-Opitz syndrome. J. Pediatr. 144, 783–791 [DOI] [PubMed] [Google Scholar]
- 54. Tierney E., Conley S. K., Goodwin H., and Porter F. D. (2010) Analysis of short-term behavioral effects of dietary cholesterol supplementation in Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. A 152A, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oregon Health and Science University (2012) Smith-Lemli-Opitz Syndrome: a Longitudinal Clinical Study of Patients Receiving Cholesterol Supplementation, https://clinicaltrials.gov/ct2/show/NCT01356420 National Library of Medicine, Bethesda, MD [Google Scholar]
- 56. Kuan V., Martineau A. R., Griffiths C. J., Hyppönen E., and Walton R. (2013) DHCR7 mutations linked to higher vitamin D status allowed early human migration to Northern latitudes. BMC Evol. Biol. 13, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zou L., and Porter T. D. (2015) Rapid suppression of 7-dehydrocholesterol reductase activity in keratinocytes by vitamin D. J. Steroid Biochem. Mol. Biol. 148, 64–71 [DOI] [PubMed] [Google Scholar]
- 58. DeLuca H. F. (2008) Evolution of our understanding of vitamin D. Nutr. Rev. 66, S73–S87 [DOI] [PubMed] [Google Scholar]