Abstract
RGS (regulator of G protein signaling) proteins of the R7 subfamily (RGS6, -7, -9, and -11) are highly expressed in neurons where they regulate many physiological processes. R7 RGS proteins contain several distinct domains and form obligatory dimers with the atypical Gβ subunit, Gβ5. They also interact with other proteins such as R7-binding protein, R9-anchoring protein, and the orphan receptors GPR158 and GPR179. These interactions facilitate plasma membrane targeting and stability of R7 proteins and modulate their activity. Here, we investigated RGS7 complexes using in situ chemical cross-linking. We found that in mouse brain and transfected cells cross-linking causes formation of distinct RGS7 complexes. One of the products had the apparent molecular mass of ∼150 kDa on SDS-PAGE and did not contain Gβ5. Mass spectrometry analysis showed no other proteins to be present within the 150-kDa complex in the amount close to stoichiometric with RGS7. This finding suggested that RGS7 could form a homo-oligomer. Indeed, co-immunoprecipitation of differentially tagged RGS7 constructs, with or without chemical cross-linking, demonstrated RGS7 self-association. RGS7-RGS7 interaction required the DEP domain but not the RGS and DHEX domains or the Gβ5 subunit. Using transfected cells and knock-out mice, we demonstrated that R7-binding protein had a strong inhibitory effect on homo-oligomerization of RGS7. In contrast, our data indicated that GPR158 could bind to the RGS7 homo-oligomer without causing its dissociation. Co-expression of constitutively active Gαo prevented the RGS7-RGS7 interaction. These results reveal the existence of RGS protein homo-oligomers and show regulation of their assembly by R7 RGS-binding partners.
Keywords: G protein, G protein-coupled receptor (GPCR), oligomer, protein cross-linking, regulator of G protein signaling (RGS), R7-binding protein (R7BP), immunoprecipitation, subcellular localization
Introduction
G protein-coupled receptors (GPCRs)2 regulate many functions in eukaryotic cells by responding to chemically diverse molecules such as biogenic amines, lipids, and peptides. A large fraction of therapeutic drugs act through GPCRs. Although ligand-bound GPCRs directly interact with several proteins, the canonical signaling mechanism involves heterotrimeric (Gαβγ) G proteins. The receptor promotes the exchange of GDP for GTP on the Gα subunit and the subsequent dissociation of Gα-GTP from Gβγ. Both Gα-GTP and Gβγ modulate the activity of effector enzymes or ion channels, thereby causing alterations in second messenger concentrations, which in turn generate cellular responses (1–3).
GPCR signaling can be terminated by several mechanisms, one of which involves hydrolysis of the Gα-bound GTP and reassembly of the inactive Gαβγ trimer (1). The intrinsic GTPase activity of the Gα subunit is very slow, and for most G proteins it is accelerated by regulator of G protein signaling (RGS) proteins. The GTPase activating (GAP) function of RGS proteins is mediated by the characteristic ∼120-amino acid domain (“RGS box”) present in all members of the RGS family (4–8). The role of RGS proteins in the inactivation of GPCR signaling was demonstrated in numerous studies using mice lacking RGS proteins or harboring RGS-insensitive Gα subunits (9–13). In addition to the RGS box, many RGS proteins contain domains that are not required for the GAP activity and often determine the subcellular localization of RGS proteins (14–18). On the basis of structural homology, the ∼30 members of the RGS family are classified into six subfamilies (8, 19).
RGS proteins that belong to the R7 subfamily include RGS6, RGS7, RGS9, and RGS11, which are highly expressed in neurons (20) and at much lower levels in glands and the heart (21–24). They have a common architecture and are characterized by the presence of four domains: RGS, GGL (Gγ-like), DHEX (DEP helical extension), and DEP (disheveled, EGL10, pleckstrin) (25). The C-terminally located RGS domain can act as a GAP for Gi/o, but not for the Gq family of G proteins (26–30). The R7 RGS proteins regulate sensory transduction and locomotion and reward behavior and other processes via inhibition of dopaminergic, serotonergic, GABAergic, and opioidergic signaling (31–33). However, RGS7 can also regulate the Gq-coupled M3 muscarinic receptor through a mechanism that is independent of the RGS domain (34–37). The centrally located GGL domain is responsible for direct interaction with the divergent Gβ subunit, Gβ5 (27, 38, 39). This obligatory interaction is necessary for stability of both proteins (40). In Gβ5 knock-out mice, expression of the entire R7 protein subfamily is abrogated (41). Likewise, in mice lacking RGS9, Gβ5 is not present in photoreceptors, or its expression is severely diminished in atrial cardiomyocytes of RGS6 knock-out mice (9, 24). The N-terminal DEP and DHEX domains are essential for interaction of R7 proteins with R7-binding protein (R7BP) and R9-anchoring protein (R9AP), which serve as plasma membrane anchors (42–48). The DEP/DHEX regions are also involved in the intramolecular interaction with Gβ5 (25, 47, 49) and GPCRs, including two orphan receptors GPR158 and GPR179. These interactions also enhance the stability and activity of RGS7 (50, 51).
In this study, we used in situ chemical cross-linking as a tool to study protein-protein interactions involving RGS7. Our results show that RGS7 can exist as a homo-oligomer. RGS7 self-association requires the DEP domain and is inhibited by R7BP or active Gαo but not by GPR158. These findings provide a new insight into molecular organization of RGS7 signaling complexes.
Experimental Procedures
Mice and Expression Constructs
Generation and characterization of mice with targeted deletions in R7BP (52), GPR158 (51), and Gβ5 genes have been described (41). C57BL6 mice were used as wild-type controls. Animals of both sexes and at 3–4 months of age were used in these studies. All procedures involving mice were reviewed and approved by the IACUC committee at the University of Miami and Scripps Research Institute.
Full-length untagged RGS7, YFP-RGS7, and Gβ5 were described earlier (49, 53). Triple hemagglutinin (HA)-tagged human RGS7 was purchased from Missouri S&T cDNA Resource Center. Bovine RGS7 and R7BP were subcloned into pmCherry. Triple FLAG-tagged R7BP was a gift from Dr. Kendall Blumer (45). Bovine Gβ5 was PCR-amplified and cloned into pECFP-C1 vector at BglII and HindIII sites. RGS7ΔDEP (lacking amino acids 34–135), RGS7ΔRGS, and RGS7ΔDEP/DHEX (lacking amino acids 1–248) were described earlier (49). YFP-RGS7ΔRGS (amino acids 1–321) and YFP-DEP (amino acids 1–124) (34), and YFP-DEP/DHEX (amino acids 1–248) were previously described (34, 49). GPR158-Myc construct was described previously (50). Wild-type human Gαo, constitutively active Gαo Q205L mutant, and M3 muscarinic receptor were purchased from Missouri S&T cDNA Resource Center.
Cell Culture and Transfection
HEK293T, COS-7, mouse neuroblastoma Neuro-2A (N2A), and rat pheochromocytoma PC-12 Adh cell lines were purchased from ATCC. DRG neurons were isolated and cultured as described previously (53). HEK293T and COS-7 cells were cultured in DMEM with 4.5 g/liter d-glucose, 584 mg/liter l-glutamine and supplemented with 10% FBS and penicillin/streptomycin (100 units/ml penicillin and 100 μg/ml streptomycin). N2A cells were cultured in Iscove's modified Dulbecco's medium + GlutaMAX with 25 mm HEPES, 3.02 g/liter sodium carbonate and supplemented with 10% FBS and penicillin/streptomycin. PC-12 Adh cells were cultured in RPMI 1640 medium with 300 mg/liter glutamine and 25 mm HEPES and supplemented with 10% heat-inactivated horse serum, 5% FBS, and penicillin/streptomycin.
Transfection of all cell lines was carried out using TransIT-LT1 reagent (Mirus) following the manufacturer's protocol. Typically, the cells were seeded either in 10-cm plates or 6-well plates for biochemical experiments and 24- or 12-well plates for microscopy. The ratio of RGS7 to Gβ5 plasmid DNA concentration was 4:1, with a total 15 μg of DNA per 10-cm plate. In experiments involving other proteins, the DNA ratios are mentioned in the figure legends.
Chemical Cross-linking
HEK293T cells were transfected with RGS7 constructs together with Gβ5 and other RGS7-interacting proteins depending on the purpose of the experiment. Two days later, cells were washed two times with phosphate-buffered saline (PBS) and incubated with 1% paraformaldehyde (PFA) in PBS for 30 min. Under these conditions, despite formation of methylene bridges between amino groups, some protein complexes can remain soluble (54). Alternatively, cells were treated with 2 μm disuccinimidyl glutarate (DSG, Sigma) in PBS for 30 min. The reaction was terminated by aspiration of the PFA solution and addition of 0.125 m glycine in PBS. Cells were lysed as described below, and the lysates were subjected to either immunoblotting or immunoprecipitation.
Whole mouse brains were minced with a razor blade to obtain pieces of tissue of ∼1 mm3. They were washed once with PBS and incubated with 1% PFA for 30 or 60 min. After addition of glycine to a final concentration of 0.125 m, the tissue slices were spun down and homogenized in RIPA buffer. Protein concentration was determined using the BCA method (Pierce), and the samples were then subjected to Western blot analysis. The results were identical if freshly dissected or frozen (−80 °C) brains were used.
Immunoprecipitation and Western Blotting
After cross-linking, transfected cells were lysed in RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.8, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate, 1 mm EDTA, 1 mm DTT and protease inhibitors). For immunoprecipitation experiments that did not involve cross-linking, we excluded SDS and deoxycholate from the buffer (IP buffer: 150 mm NaCl, 50 mm Tris-HCl, pH 7.8, 1% Triton X-100, 1 mm EDTA, 1 mm DTT, and protease inhibitors). The cell lysates were centrifuged at 15,000 × g for 15 min at 4 °C. The collected supernatant was incubated with protein A/G beads for 2 h at 4 °C and centrifuged again, and the pre-cleared lysate was then incubated with immobilized antibodies for 2–3 h. Typically, the IP reaction contained 1 ml of lysate (1.5–2.0 mg/ml total protein) and 25 μl of the packed resin. The beads were then washed five times with 1 ml of ice-cold IP buffer and then eluted with 60 μl of SDS-PAGE loading buffer. In the cross-linking experiments, we did not heat up the samples above 25 °C, as heating facilitates hydrolysis of the cross-linked bonds (54). For immunoprecipitation, we used mouse monoclonal anti-FLAG M2 affinity gel (Sigma, A2220), mouse monoclonal anti-HA-agarose (Sigma, A2095), and GFP-trap (Chromotek, gtp-20).
For immunoblot analysis, proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and blocked prior to addition of the primary antibodies. To reveal immunoreactive bands, we used Odyssey (LiCOR, Inc.) infrared fluorescence detection system according to the manufacturer's instructions. Primary and secondary antibodies were diluted in the Odyssey blocking buffer supplemented with 0.1% Tween 20. The following primary antibodies were used (the source, catalogue number, and dilution used is shown in parentheses): rabbit affinity purified anti-RGS7 ((41), 1:1,000); rabbit affinity-purified anti-Gβ5 ((55), 1:2,000); rabbit affinity-purified anti-R7BP ((56), 1:1,000); chicken anti-GFP (Abcam_ab13970, 1:5,000); mouse monoclonal anti-FLAG M2 (Sigma_F1804, 1:1,000); affinity-purified rabbit anti-FLAG (Sigma_F7425, 1:1,000); mouse monoclonal anti-HA (Sigma_H3663, 1:10,000); mouse monoclonal anti-mCherry (Abcam_1C51, 1:2,000); mouse monoclonal anti-Myc (Cell Signaling Technologies_2276, 1:1,000); mouse monoclonal anti-actin (EMD Millipore_MAB1501R, 1:2,000); and rabbit anti-Gαo (Santa Cruz Biotechnology _SC-387, 1:1,000). Donkey antibodies against rabbit, mouse, or chicken conjugated to IRDye 680RD or IRDye 800CW (LiCOR) were used at 1:10,000 dilution as the secondary antibody.
Quantitative analysis of scanned blots was done using Odyssey version 1.2 software. Total fluorescence intensity within a protein band was determined by designating a rectangular region incorporating the band. An area of identical size was selected within the same lane as the background, and this value was subtracted from the fluorescence intensity of the band of interest. In the analyses of cross-linked and co-immunoprecipitated proteins, the values obtained for the bands of interest were normalized to the non-cross-linked protein or to the input, respectively. In all these experiments measurements were done in the linear range of the fluorescence signal, and care was taken to avoid saturation and to select appropriate regions representing the background.
Identification of Proteins by Mass Spectrometry (LC-MS/MS)
The immunoprecipitated samples were resolved by SDS-PAGE and stained with silver, and the desired protein bands were excised. To determine the background, we analyzed gel slices from the areas with corresponding molecular weights from the control samples resolved on the same gel. As the control, we used material immunoprecipitated from untransfected HEK293T cells or from beads containing irrelevant antibody (normal IgG). No proteins apart from human keratin background were detected in the control samples.
Gel slices were destained, reduced, alkylated, and digested with trypsin (57). The resulting peptides were extracted, reconstituted in 2% formic acid, and subjected to liquid chromatography and tandem mass spectrometry (LC-MS/MS) analysis (58). LC MS/MS was performed with a Thermo Scientific LTQ Orbitrap Elite hybrid mass spectrometer equipped with an Ultimate nano-LC system and a C-18 column (Acclaim PepMap, 75 μm × 15 cm, 2 μm, 100 Å). Five μl of the tryptic peptide solution were injected and eluted from the column using an acetonitrile, 0.1% formic acid gradient at a flow rate of 0.3 μl/min. The eluates were introduced into the source of the mass spectrometer on line. The microelectrospray ion source was operated at 2.5 kV. The digest was analyzed using the data-dependent multitask capability of the instrument acquiring full scan mass spectra from 300 to 2,000 Da at a resolution of 60,000. These mass spectra were followed by collision-induced dissociation experiments on the eight most abundant ions in the mass spectra. These collision-induced dissociation spectra were performed with collision energy of 35%. The products were analyzed in the Ion Trap mass spectrometer. Protein identification utilized Proteome Discoverer 1.3 (Thermo), the Mascot search engine (Matrix Science, 2.4.1), and the human Uni-Prot/Swiss-Protein database version (SwissProt 2012, 20,309 total human sequences). Database searches were restricted to ≤3 missed tryptic cleavage sites, precursor ion mass tolerance at 50 ppm, fragment ion mass tolerance at 0.8 Da, and a false discovery rate at 1%. Fixed modification was S-carbamidomethyl Cys, and variable modifications included Met oxidation and Asn and Gln deamidation.
To measure the relative protein abundances, we used the following method. First, the numbers of peptide spectra (spectral counting) derived from each protein were extracted from MASCOT search results. The false discovery rate was set at 1%, and the individual peptide MASCOT score had to be above homology or identity score. Then the spectral counting number of each protein was normalized to its total amino acid length. The higher the ratio, the more abundant the protein in the given band.
Immunofluorescence Microscopy
In situ protein immunostaining and microscopy were performed essentially as described previously (53). DRG neurons, N2A, and other cells were cultured on glass coverslips and fixed with 4% paraformaldehyde and 4% sucrose in PBS for 20 min at room temperature. After two washes with PBS, cells were permeabilized and blocked in 0.3% saponin or 0.1% Triton X-100 and 5% normal donkey serum in PBS for 60 min at room temperature. Coverslips were then incubated with primary antibodies for 60 min, and following several washes with PBS, with secondary antibodies. The primary antibody dilutions were as follows: 1:200 for affinity-purified rabbit anti-RGS7 (40), 1:400 for mouse monoclonal anti-FLAG (Sigma_F1408), or 1:400 for mouse monoclonal anti-Myc (Cell Signaling Technologies_2276). Secondary antibodies were Alexa 488- or Texas Red-conjugated donkey anti-rabbit (1:1,000) or anti-mouse (1:500), respectively. Coverslips were then washed with PBS five times and mounted on glass slides. Localization of YFP- and CFP-tagged proteins was analyzed by direct fluorescence as described previously (53).
The images were either acquired on a Leica TC5 SP5 confocal microscope using a ×63 oil immersion objective or a wide-field Nikon Eclipse TE2000 fluorescence microscope using a ×60 oil objective.
Statistical Analysis
All experiments were performed with at least three biological repeats. Data are expressed as means ± S.D. for the indicated number of observations. For comparisons between two determined values, the unpaired two-tailed Student's t test was done. A difference with a p value of less than 0.05 was considered statistically significant.
Results
Subcellular Localization of RGS7 and Its Interaction with R7BP and GPR158
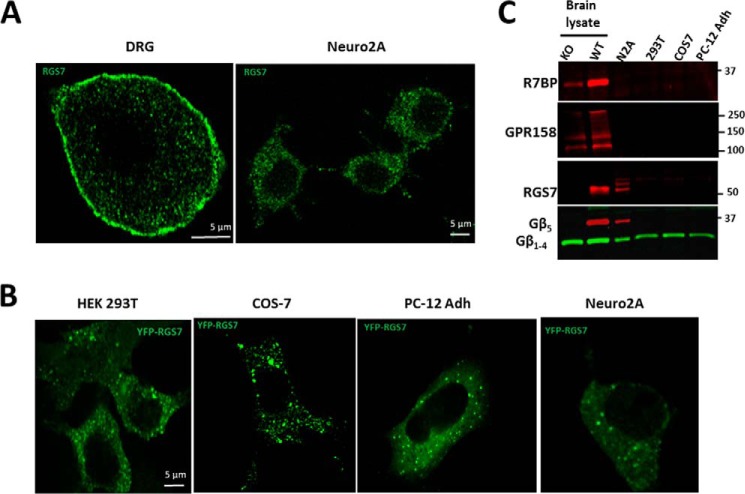
Consistent with our previous report (53), RGS7 immunoreactivity was detected not only at the plasma membrane but also in cytoplasmic granules in primary neurons (Fig. 1A, left panel). However, in the neuroblastoma cell line Neuro2a (N2a), endogenous RGS7 was not detected at the plasma membrane and localized only to the cytoplasm (Fig. 1A, right panel). Likewise, heterologous expression of YFP-RGS7 in CHO K1 (53), HEK293T, COS-7, PC-12 Adh, and N2A cells showed only diffuse cytoplasmic and punctate distribution (Fig. 1B). The most plausible explanation for this localization is that unlike neurons these cell lines do not express the plasma membrane anchors of RGS7, R7BP, or GPR158/GPR179 (Fig. 1C) (44–46, 50, 53, 56). Indeed, consistent with previously published studies (44–46, 50, 56), the Gβ5-RGS7 complex directly binds to R7BP and GPR158 (Fig. 2, A and B) and redistributes from the cytoplasmic puncta to the plasma membrane (Fig. 2, C and D). These results suggest that association of the RGS7 complex with the cytoplasmic granules is weaker than with R7BP or GPR158.
FIGURE 1.
Subcellular localization of endogenous and overexpressed RGS7. A, adult mouse day 6 in vitro DRG neurons (left panel) or N2A cells (right panel) were stained with anti-RGS7 antibodies as described under “Experimental Procedures.” B, indicated cell lines were transfected with plasmids expressing YFP-RGS7 and Gβ5, and YFP was directly imaged by fluorescence microscopy. C, lysates from indicated cell lines were analyzed by Western blot for R7BP, GPR158, RGS7, Gβ5, and G protein β subunit (1–4) expression. Wild-type and Gβ5 knock-out mouse brains were used as a control. Shown are representative immunoblots from two experiments.
FIGURE 2.
R7BP or GPR158 cause redistribution of RGS7 from cytoplasmic granules to plasma membrane. HEK293T cells were co-transfected with taggedRGS7 and Gβ5, with or without FLAG-R7BP (A and B) or GPR158-Myc (A and C) plasmids. A, YFP-RGS7 was immunoprecipitated from the cell lysate, and the eluates were probed for RGS7, Gβ5, and R7BP. B, immunoprecipitated YFP-RGS7 complex was probed for RGS7, Gβ5, and GPR158. C, cells expressing YFP-RGS7 and Gβ5 (top row), or these proteins together with FLAG-R7BP (middle row), or GPR158-Myc (bottom row) were analyzed by confocal microscopy. YFP-RGS7 was detected by epifluorescence, and R7BP or GPR158 were detected using antibodies against FLAG or Myc tag, respectively. D, quantification of the result in C. Regions of interest corresponding to plasma membrane (cell periphery) and cytosol were selected within a cell. Fluorescence intensities within these regions were measured using Leica LAS AF Lite image analysis software. The data show the means ± S.D. of the plasma membrane to cytosol ratio of the YFP-RGS7 fluorescence. ***, p <0.001. For every condition, we analyzed 20–25 cells in each of three independent transfection experiments. The ratio of 1:1 in the absence of R7BP and GPR158 represents the fluorescence distribution for cytosol-localized proteins.
Chemical Cross-linking and Immunoprecipitation of RGS7 Complexes
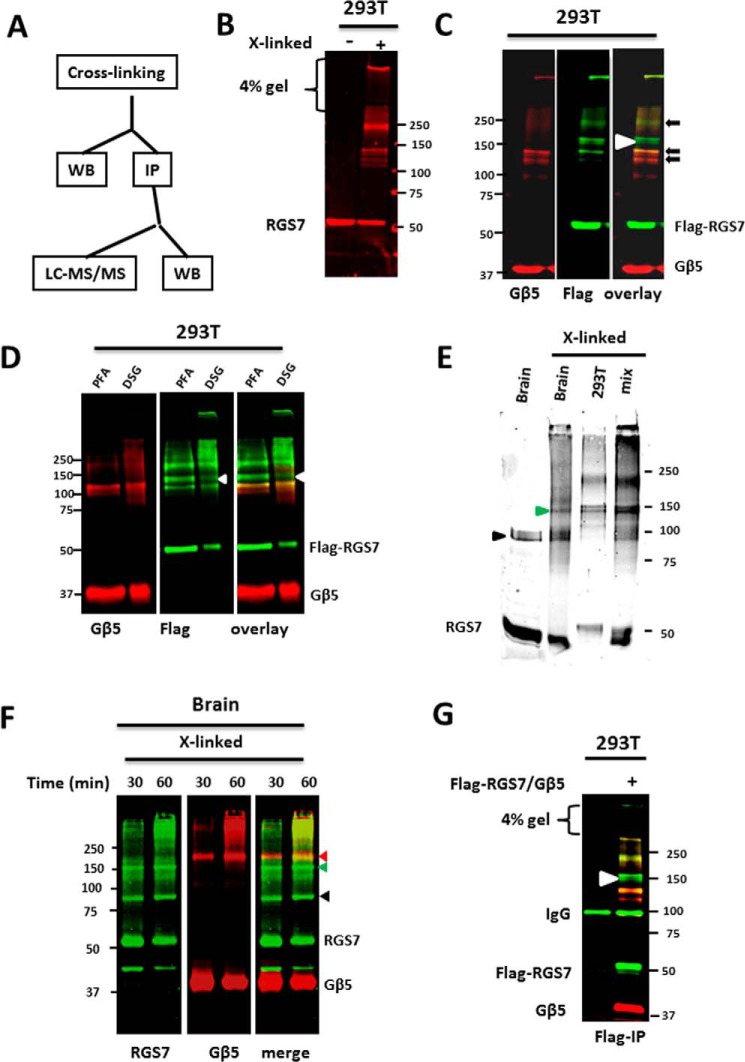
To stabilize the association of Gβ5-RGS7 with potential binding partners, we used covalent cross-linking (Fig. 3). We treated HEK293T cells overexpressing Gβ5-RGS7 or mouse brain slices with a low concentration of formaldehyde (54), and we analyzed the resulting cross-linked RGS7 products by Western blot. At concentrations of PFA over 1% and exposures longer than 1 h most of the immunoreactive proteins did not enter the 4% gel or produced a smear. Under our conditions, cross-linking resulted in formation of distinct bands with molecular masses higher than monomeric RGS7 or Gβ5. This result indicated formation of distinct protein complexes.
FIGURE 3.
Chemical cross-linking of RGS7 complexes in live cells. A, flow chart representing our cross-linking, Western blot (WB), immunoprecipitation (IP), and mass-spectrometry (LC-MS/MS) experiments. B, HEK293T cells expressing RGS7 and Gβ5 were incubated with 1% PFA for 30 min and analyzed by immunoblot with anti-RGS7 antibodies, as described under “Experimental Procedures.” Left lane shows a representative Western blot of the control cells (untreated with PFA) probed with anti-RGS7 antibody. Right lane, lysate from cells treated with PFA. C, immunoblot analysis of lysates from cells expressing FLAG-RGS7 and Gβ5 probed with antibodies against FLAG (green) and Gβ5 (red). Black arrows indicate the cross-linked complexes containing both RGS7 and Gβ5. White arrowhead at ∼150-kDa denotes an RGS7 complex that does not contain Gβ5. D, HEK293T cells were transfected with FLAG-RGS7 and Gβ5 and cross-linked with either PFA or DSG. The resulting products were analyzed by Western blot to compare the patterns of cross-linked proteins using antibodies against FLAG (green) and Gβ5 (red). E, cross-linking of endogenous RGS7 in mouse brain was done using PFA as described under “Experimental Procedures.” To compare the apparent molecular weight of the cross-linked RGS7 products from brain and transfected HEK293T cells, the samples were resolved on 8% gel. The arrowhead points to the band that is identical in electrophoretic mobility. When the two samples were mixed (mix), the intensity of this band increased. F, mouse brain slices were treated with PFA for 30 or 60 min. The proteins were resolved on a 10% gel and analyzed by Western blot using anti-RGS7 and anti-Gβ5 antibodies. Because both primary antibodies were from rabbit, we used the following method. First, the immunoblot was stained using anti-RGS7 antibodies and scanned using the Odyssey instrument (green). The filter was then stripped and probed with anti-Gβ5 antibodies (red). The two images were merged to compare the exact position of RGS7 and Gβ5-positive bands. Red arrowhead denotes the protein cross-linking product that contains both RGS7 and Gβ5. Green arrowhead points to the 150-kDa RGS7 complex. Black arrowhead shows a nonspecific band revealed by the RGS7 antibody in the brain extracts. G, immunoprecipitation of the cross-linked RGS7 complexes from transfected HEK293T cells using anti-FLAG antibody. The eluates from the beads were analyzed by immunoblot and by mass spectrometry (see text and Table 1). Left lane, lysates from untransfected cells were used as a negative control. Shown are representative immunoblots from three (D, E, and F) or more than five (B, C, and G) independent experiments.
In HEK293T cells (Fig. 3B), the bands of ∼120, 130, and 230 kDa were co-stained with the antibodies against Gβ5 and RGS7. Interestingly, an ∼150-kDa doublet positive for RGS7 (white arrowhead in Fig. 3C) was not stained for Gβ5. We used two different antibodies, against the C or N terminus of Gβ5, but neither revealed reactivity in this band (Fig. 3C and data not shown). Thus, the lack of Gβ5 staining is unlikely due to masking the epitope by the chemical modification. Cross-linking with an alternative agent disuccinimidyl glutarate (DSG), which has a four carbon atom spacer “arm” and is also membrane-permeable, resulted in the identical pattern of RGS7 products as with PFA (Fig. 3D). In all subsequent experiments we used PFA.
In the brain, cross-linking also resulted in formation of distinct high molecular mass RGS7 products (Fig. 3, E and F). The overall pattern resembled that in HEK293T cells expressing Gβ5-RGS7 (Fig. 3, B and C). There was more smearing in the high molecular weight region of the gel compared with HEK293T cells, likely because of the higher complexity of the brain tissue. Nevertheless, bands in the 200–300-kDa range containing both Gβ5 and RGS7 were detectable in most experiments. We did not detect the 120- and 130-kDa complexes co-stained with RGS7 and Gβ5 antibodies in the brain samples. Importantly, similarly to the transfected cells, brain samples always contained the Gβ5-negative ∼150-kDa band (Fig. 3F) revealed by the RGS7 antibody. To compare RGS7 products in the brain with those in HEK293T cells, we ran the two samples side-by-side on an 8% SDS-PAGE. As a control for a potential effect of the brain versus cultured cell sample composition on protein electrophoretic mobility, we also analyzed the mixture of the two samples (Fig. 3E). The result clearly showed that the apparent molecular mass of the ∼150-kDa product was indistinguishable from the lower band of the RGS7-positive doublet that occurs in transfected HEK293T cells.
R7 proteins always associate with Gβ5 (40, 41), and so its absence within the 150-kDa band was particularly intriguing. The fact that the molecular mass of the 150-kDa band in the brain and transfected cells was identical indicated that its composition was the same in both systems. We thus sought identification of RGS7-binding partners within this complex. The absence of endogenous R7BP or GPR158 in HEK293T cells allowed us to isolate RGS7 localized in the cytoplasm. In addition, we exploited technical advantages such as affinity tags to facilitate immunoprecipitation, mass spectrometry, and structure-function analysis.
Analysis of the 150-kDa RGS7 Complex by Protein Mass Spectrometry
The material immunoprecipitated from HEK293Tcells co-expressing FLAG-RGS7 and Gβ5 was resolved by SDS-PAGE (Fig. 3G). The 150-kDa band was visualized by silver staining, excised, and digested with trypsin, and the recovered peptides were analyzed by mass spectrometry (Table 1). According to our analysis, about 55% of the detected peptides corresponded to RGS7, and 20% were from Gβ5. In addition, mass spectrometry identified peptides from tubulin, 14-3-3 proteins, SEC23/24C, clathrin heavy chain 1, septin7, septin11, and other proteins. These peptides were absent in the ∼150-kDa gel slice obtained in the control IP experiment using mock-transfected HEK293T cells. However, the abundance of these proteins was at least 10-fold lower than that of RGS7 (Table 1). Furthermore, subsequent Western blot analysis did not detect these proteins in the 150-kDa band (data not shown).
TABLE 1.
Quantitative representation of the peptides detected by LC-MS/MS analysis of the 150-kDa band (Fig. 3, C and D)
Some proteins, mostly tubulins, were excluded from this table. Shown here is the representation of two independent experiments.
| Protein Description | Protein score | Protein mass | Protein matches | Protein sequences | Protein coverage | Protein length | Estimated abundance |
|---|---|---|---|---|---|---|---|
| % | |||||||
| RGS7 | 4638 | 57,917 | 239 | 30 | 58.2 | 495 | 0.549 |
| Gβ5 | 2516 | 44,621 | 80 | 17 | 44.1 | 395 | 0.203 |
| Tubulin β chain | 358 | 50,095 | 20 | 15 | 54.3 | 444 | 0.045 |
| Tubulin β-4B chain | 309 | 50,255 | 18 | 13 | 51 | 445 | 0.040 |
| α-Enolase | 409 | 47,481 | 15 | 11 | 44.7 | 434 | 0.035 |
| Tubulin β-4A chain | 275 | 50,010 | 15 | 11 | 41.2 | 444 | 0.034 |
| Heat shock 70-kDa protein 1A/1B | 396 | 70,294 | 21 | 17 | 42 | 641 | 0.033 |
| Tubulin β-2A chain | 243 | 50,274 | 14 | 11 | 35.1 | 445 | 0.031 |
| Tubulin α-1B chain | 234 | 50,804 | 13 | 9 | 43 | 451 | 0.029 |
| Elongation factor 2 | 483 | 96,246 | 24 | 17 | 32.1 | 858 | 0.028 |
| Heat shock cognate 71-kDa protein | 308 | 71,082 | 17 | 15 | 33.7 | 646 | 0.026 |
| Protein S100-A9 | 107 | 13,291 | 3 | 2 | 24.6 | 114 | 0.026 |
| Heat shock protein HSP 90-β | 278 | 83,554 | 18 | 15 | 26.9 | 724 | 0.025 |
| Peptidyl-prolyl cis-trans isomerase A | 91 | 18,229 | 4 | 4 | 52.1 | 165 | 0.024 |
| Tubulin α-3C/D chain | 165 | 50,612 | 10 | 7 | 30.7 | 450 | 0.022 |
| Eukaryotic initiation factor 4A-I | 151 | 46,353 | 9 | 7 | 25.6 | 406 | 0.022 |
| Elongation factor 1-α1 | 136 | 50,451 | 9 | 6 | 21.4 | 462 | 0.019 |
| d-3-Phosphoglycerate dehydrogenase | 145 | 57,356 | 9 | 8 | 22 | 533 | 0.017 |
| ATP-dependent RNA helicase DDX3X | 321 | 73,597 | 11 | 10 | 22.8 | 662 | 0.017 |
| 14-3-3 protein ζ/δ | 126 | 27,899 | 4 | 4 | 22.4 | 245 | 0.016 |
| 14-3-3 protein ϵ | 138 | 29,326 | 4 | 4 | 24.7 | 255 | 0.016 |
| Septin-7 | 75 | 50,933 | 4 | 3 | 9.4 | 437 | 0.009 |
| Protein transport protein Sec23A | 171 | 87,018 | 7 | 5 | 11 | 765 | 0.009 |
| Rab GDP dissociation inhibitor β | 104 | 51,087 | 4 | 4 | 14.6 | 445 | 0.009 |
| 14-3-3 protein θ | 98 | 28,032 | 2 | 2 | 9.8 | 245 | 0.008 |
| 14-3-3 protein β/α | 51 | 28,179 | 2 | 2 | 8.5 | 246 | 0.008 |
| Septin-11 | 80 | 49,652 | 3 | 2 | 6.5 | 429 | 0.007 |
| Rab GDP dissociation inhibitor α | 71 | 51,177 | 2 | 2 | 8.7 | 447 | 0.004 |
| Protein transport protein Sec24C | 79 | 119,789 | 4 | 4 | 6.1 | 1094 | 0.004 |
| Clathrin heavy chain 2 | 49 | 189,020 | 2 | 2 | 1.8 | 1640 | 0.001 |
| Filaggrin-2 | 66 | 249,296 | 2 | 2 | 2 | 2391 | 0.001 |
The shift in the apparent molecular mass of RGS7 from 55 to ∼150 kDa on SDS-PAGE can only occur upon stoichiometric covalent attachment to another protein(s). Therefore, we hypothesized that RGS7 within the 150-kDa band was bound to itself, i.e. formed a homo-oligomer.
Co-immunoprecipitation Confirms RGS7 Homo-oligomerization
To test this hypothesis, we performed a series of immunoprecipitation experiments with RGS7 fused to different tags (FLAG, HA, Myc, YFP, and mCherry). Unless specified otherwise, Gβ5 was co-expressed with the RGS7 constructs.
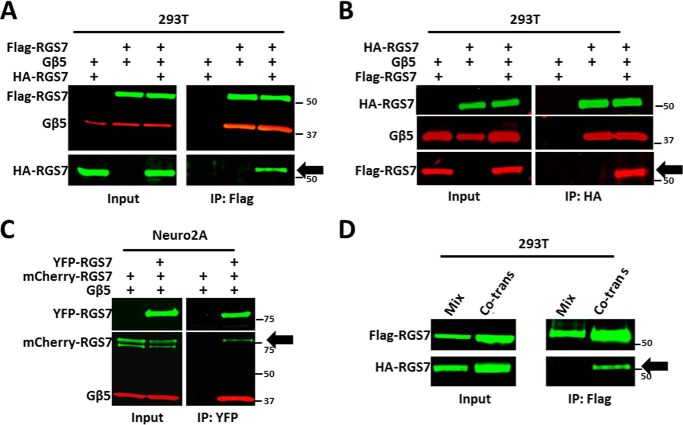
Fig. 4A shows that after co-transfection of HA-tagged RGS7 and FLAG-RGS7, anti-FLAG antibodies can pull down the HA-tagged RGS7 along with FLAG-RGS7 and Gβ5. Quantitative assessment showed that anti-FLAG beads could absorb >90% of FLAG-RGS7, ∼50% of Gβ5, and ∼15% of HA-RGS7. In the reciprocal co-immunoprecipitation using anti-HA antibody, we pulled down FLAG-RGS7 (Fig. 4B). Similar results were obtained with RGS7 tagged with Myc or fluorescent proteins and in different cell lines, including N2A cells (Fig. 4C and data not shown).
FIGURE 4.
Homo-oligomerization of RGS7. A, HEK293T cells were co-transfected with HA-RGS7 and Gβ5, FLAG-RGS7 and Gβ5, or all the three constructs. Cell lysates were subjected to IP using anti-FLAG antibody, as described under “Experimental Procedures.” The eluates from the resin were analyzed using anti-FLAG, HA, and Gβ5 antibodies. B, reciprocal IP using anti-HA beads. Cell transfection and lysis were done as in A; immunoprecipitation was performed using anti-HA antibodies, and the bound material was probed for the presence of FLAG-tagged RGS7. C, mCherry-RGS7 was co-immunoprecipitated with YFP-RGS7 from N2A cell lysates. The experiment was performed essentially as in A or B. D, two separate cell cultures were transfected with either FLAG-RGS7/Gβ5 or HA-RGS7/Gβ5, harvested, and lysed. The two lysates were mixed (mix) prior to co-IP with immobilized anti-FLAG antibody. In parallel, FLAG-RGS7, HA-RGS7, and Gβ5 plasmids were co-transfected (co-trans), and cell lysates were analyzed by co-IP and immunoblot.
Importantly, we found that RGS7-RGS7 co-immunoprecipitation occurred even without chemical cross-linking (Figs. 4–9). This suggested that RGS7 homo-oligomers were rather stable. When we expressed HA-RGS7 and FLAG-RGS7 separately and then mixed cell lysates, no interaction was detected (Fig. 4D). This also indicates that once the oligomeric RGS complex is formed, it remains stable, at least under our experimental conditions.
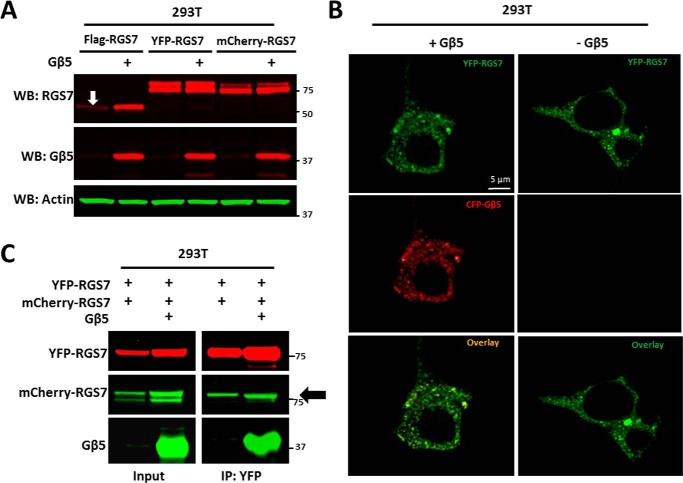
FIGURE 5.
Gβ5 is required for stability but not for homo-oligomerization of RGS7. A, HEK293T cells expressing FLAG-, YFP-, or mCherry-tagged RGS7 with or without Gβ5 were lysed and analyzed by Western blot using antibodies against RGS7, Gβ5, and actin. The white arrow shows FLAG-RGS7 expressed in the absence of Gβ5. B, cells on coverslips were transfected with YFP-RGS7 with or without CFP-Gβ5, then fixed, and YFP and CFP direct fluorescence was detected using a confocal microscope (×63 objective lens). C, lysates from cells expressing YFP-RGS7 and mCherry-RGS7 with or without Gβ5 were subjected to immunoprecipitation using immobilized YFP antibody. The eluates were analyzed by Western blot using YFP, Gβ5, and mCherry antibodies. The black arrow points to the co-immunoprecipitated mCherry-RGS7.
FIGURE 6.
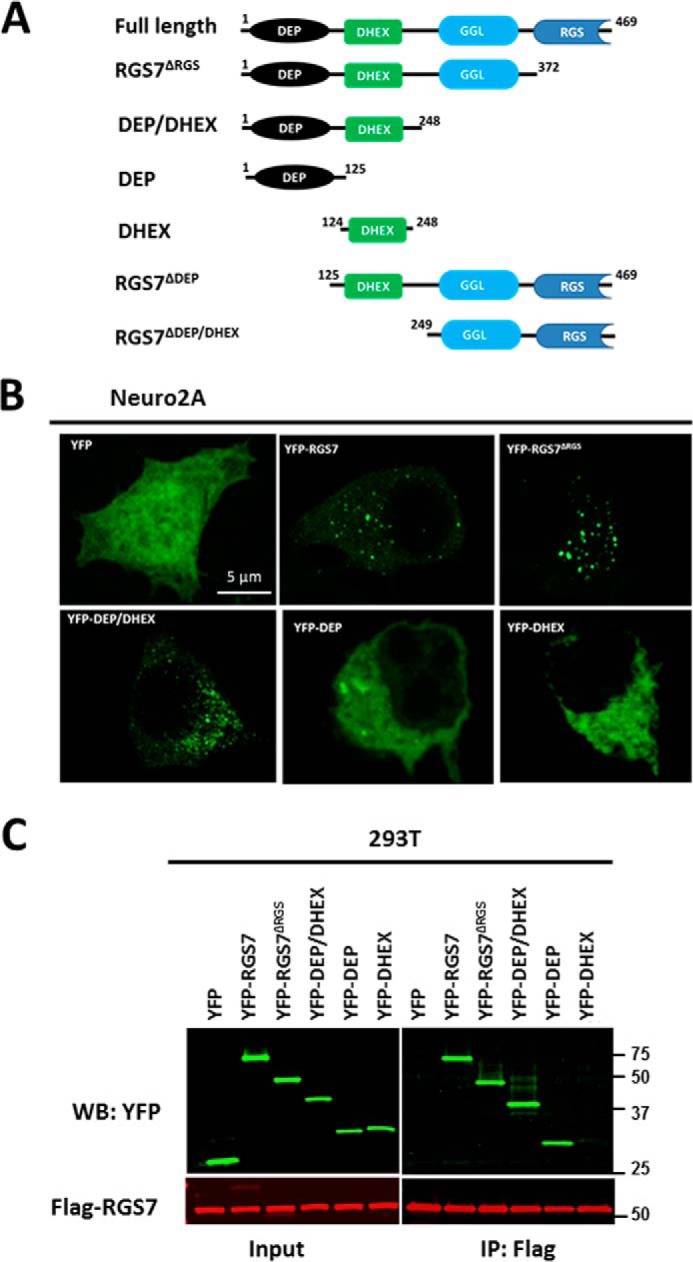
DEP domain is essential for oligomerization of RGS7. A, RGS7 constructs used in the study. B, N2A cells were transfected with plasmids encoding Gβ5 and the indicated YFP-tagged deletion mutants of RGS7. After 24 h, cells were fixed, and YFP epifluorescence was detected by confocal microscopy. C, HEK293T cells expressing FLAG-RGS7, Gβ5 together with YFP, or the indicated YFP-tagged RGS7 mutants were lysed, and the lysates were subjected to immunoprecipitation using anti-FLAG antibody. The eluates from the beads were analyzed by immunoblot using anti-GFP and anti-FLAG antibodies. WB, Western blot.
FIGURE 7.
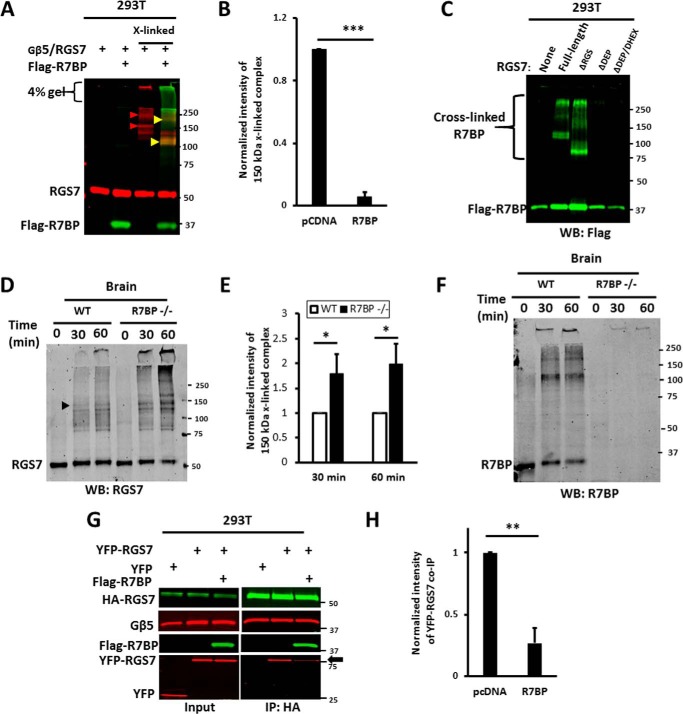
R7BP causes dissociation of RGS7 homo-oligomer. A, HEK293T cells expressing RGS7 and Gβ5, with or without FLAG-R7BP, were subjected to cross-linking as described in the legend to Fig. 3. The lysates were analyzed by immunoblot using a combination of anti-FLAG (green) and RGS7 (red) antibodies. Red arrowheads denote the 150-kDa and 230 RGS7 complexes. Note that in the presence of R7BP these bands disappear. Instead, there are two new bands of ∼110 and ∼200 kDa (yellow arrowheads) revealed with both anti-RGS7 and anti-FLAG antibodies. B, quantification of the 150-kDa band intensity in A. Blots were scanned and analyzed as described under “Experimental Procedures.” The fluorescence intensity determined for the 150-kDa band was normalized to the RGS7 band in the non-cross-linked control sample. The value obtained for 150-kDa band without R7BP (pcDNA) was set to the arbitrary unit of 1.0. Data show means ± S.D. from four independent transfection experiments. ***, p <0.001. C, cells were transfected with FLAG-R7BP and Gβ5 together with or without (none) full-length RGS7 or its deletion mutants. They were subjected to cross-linking with PFA, and the lysates were analyzed by immunoblot using anti-FLAG antibody to detect R7BP. WB, Western blot. D, cross-linking of endogenous RGS7 in the brain tissue from wild-type (WT) and R7BP knock-out (R7BP−/−) mice. Cross-linking with PFA was performed for 30 or 60 min as described under “Experimental Procedures” and the legend to Fig. 3E; shown is a representative immunoblot probed with anti-RGS7 antibody. E, quantification of data represented in D. The intensity of the 150-kDa band (black arrow) was normalized to the non-cross-linked RGS7, and the resulting value for WT mouse was set to 1.0. The data show the means ± S.D. from four independent experiments performed with the tissues from three individual WT (open bars) and three R7BP−/− (black bars) animals. *, p <0.05. The knock-out of R7BP resulted in a 1.8–2.0-fold increase in the 150-kDa band. F, pattern of cross-linked endogenous R7BP revealed by anti-R7BP antibody. G, HEK293T cells were co-transfected with HA-RGS7, YFP-RGS7, and Gβ5, with or without FLAG-R7BP. Cell lysates were immunoprecipitated with anti-HA antibody. The inputs and eluates from IP were probed with the indicated antibodies. The black arrow pointing to YFP-RGS7 highlights the reduction of the interaction between HA-RGS7 and YFP-RGS7 in the presence of R7BP. The RGS7, Gβ5, and R7BP plasmids used for transfection were added in the ratio of 4:1:5, respectively. H, quantification of the data shown in G. YFP-RGS7 band intensity in co-IP eluate was normalized to the intensity of the HA-RGS7 band. The results are expressed as means ± S.D. (n = 4, **, p <0.01).
FIGURE 8.
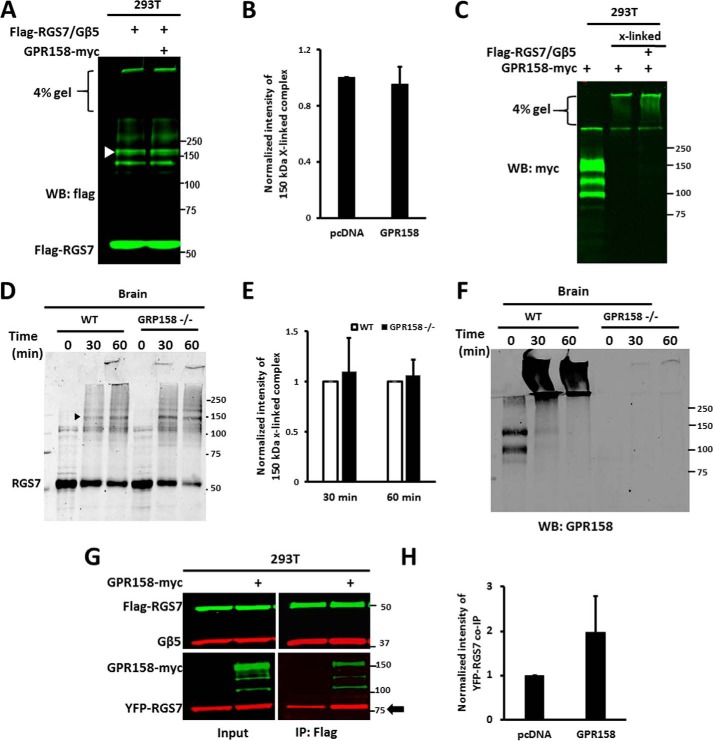
Effect of GPR158 on RGS7 self-association. A, HEK293T cells expressing FLAG-RGS7/Gβ5 with or without GPR158-Myc were treated with PFA. The lysates were analyzed by immunoblot using anti-FLAG antibodies. White arrowhead points to the 150-kDa cross-linked RGS7 product. WB, Western blot. B, 150-kDa band intensity compared between cells with and without GPR158, means ± S.D., n = 3. C, same samples as in A were probed with anti-Myc antibody. D, cross-linking of endogenous RGS7 in the brain tissue from wild-type (WT) and GPR158−/− mice. Cross-linking was performed for 30 or 60 min; shown is a representative immunoblot probed with anti-RGS7 antibody. E, data from D was quantified as described in the legend to Fig. 7E. F, cross-linked brain samples probed with the antibody against GPR158. G, FLAG-RGS7 was immunoprecipitated from HEK293T cells expressing FLAG-RGS7, Gβ5, YFP-RGS7 with or without GPR158Myc. The lysates were analyzed by immunoblot using specific antibodies. The ratio of plasmid DNA in the transfection was 4:1:5 (RGS7, Gβ5, and GPR158, respectively). H, quantification of the results in G. YFP-RGS7 band (co-IP) was normalized to FLAG-RGS7 (IP) as described in the legend to Fig. 7. The observed increase in RGS7 self-association in the presence of GPR158 was not statistically significant (n = 4, p = 0.095).
FIGURE 9.
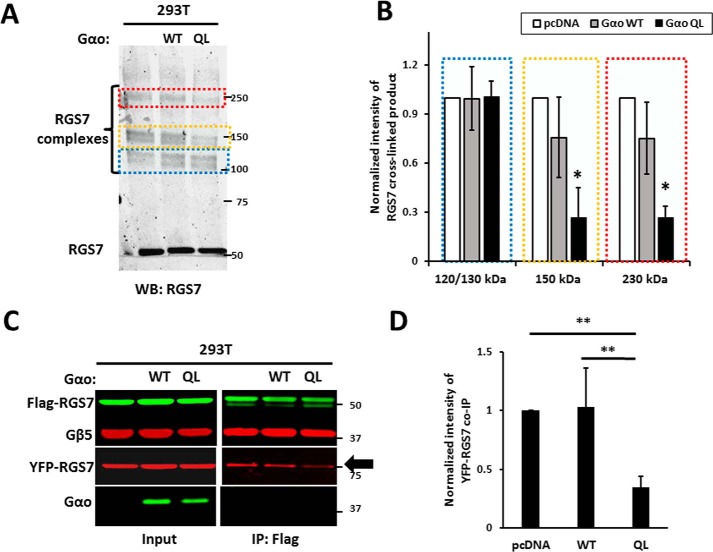
Effect of Gαo subunit on RGS7 homo-oligomerization. A, HEK293T cells transfected with RGS7, Gβ5 with or without wild-type Gαo or its constitutively active Q205L (QL) mutant. The ratio of plasmid DNA in transfection was 4:1:5 (RGS7, Gβ5, Gαo, or pcDNA, respectively). After PFA-induced cross-linking, the cell lysates were analyzed by immunoblot using RGS7 antibody. The areas denoted by the colored broken lines highlight the positions of the 120/130-kDa Gβ5-RGS7 conjugate, the 150-kDa RGS7, and 250-kDa Gβ5-RGS7 oligomers. WB, Western blot. B, quantification of the data in A. The normalized band intensity in the control (pcDNA) was set to 1.0. Data are means ± S.D., n = 3. *, p <0.05. C, lysates from cells expressing FLAG-RGS7, YFP-RGS7, Gβ5, with or without wild-type or mutant Gαo were subjected to immunoprecipitation using FLAG antibody. The eluates were analyzed by immunoblot using FLAG, YFP, Gβ5, and Gαo antibodies. D, quantification of data shown in C. YFP-RGS7 co-IP intensity was normalized to FLAG-RGS7 IP. The results are expressed as means ± S.D.; **, p <0.01.
Role of Gβ5 and Domains of RGS7 in Its Localization and Oligomerization
RGS7 quickly degrades in the absence of Gβ5 (40), but fusion of RGS7 to fluorescent proteins significantly increases its expression level (Fig. 5A) (59). We exploited this phenomenon to test whether the Gβ5 subunit is essential for RGS7 localization and/or oligomerization. As expected, YFP-RGS7 and CFP-Gβ5 co-localized to the same granules (Fig. 5B, left column). However, even in the absence of Gβ5 YFP-RGS7 showed granular localization (Fig. 5B, right column). Furthermore, Gβ5 was not necessary for co-precipitation of mCherry-RGS7 with YFP-RGS7 (Fig. 5C). Thus, although Gβ5 is essential for RGS7 stability, it does not play a direct role in RGS7 oligomerization or granular localization.
The N terminus of RGS7 encompassing both DEP and DHEX domains is required for granular localization in CHO K1 (53), HEK293T (data not shown), and N2A cells (Fig. 6B). Our co-IP assay showed that deletion of the RGS domain or both RGS and GGL domains did not prevent interaction of the RGS7 N terminus with full-length RGS7. Further analysis using YFP-DEP and YFP-DHEX constructs showed that the DEP domain was sufficient for the interaction with full-length RGS7, whereas the DHEX domain alone did not co-precipitate with full-length RGS7 (Fig. 6C).
Effects of R7BP and GPR158 on RGS7 Oligomerization
R7BP anchors R7 proteins to the plasma membrane via the DEP/DHEX domains (Fig. 2, C and D) (44, 45, 47). To test whether R7BP influences RGS7 oligomerization, we studied the effects of R7BP co-expression in HEK293T cells and its deletion in mouse brain on RGS7 cross-linking (Fig. 7). In HEK293T cells, co-transfection of R7BP resulted in the disappearance of the cross-linked 150-kDa band (Fig. 7, A and B). Instead, RGS7 formed a 110-kDa band that also stained positive for R7BP (Fig. 7A, yellow arrowhead), indicating covalent attachment of R7BP to RGS7. Likewise, there was a shift of the 230-kDa band to a smaller complex of ∼200 kDa that also contained R7BP. Analysis of RGS7 deletion mutants showed that the DEP domain was required for the conjugation of R7BP with RGS7 (Fig. 7C). In the mouse brain, the knock-out of Rgs7bp7 gene resulted in a 1.8–2-fold increase in the formation of the 150-kDa RGS7 complex (Fig. 7, D and E). Accordingly, PFA treatment resulted in formation of ∼110 and 200-kDa bands revealed by R7BP antibody. The patterns of these cross-linked products in transfected cells (Fig. 7, A and C) and in mouse brain (Fig. 7F) were similar. In our co-immunoprecipitation assay, R7BP caused a 75 ± 12% decrease in RGS7-RGS7 interaction (Fig. 7, G and H).
GPR158 is another known binding partner of RGS7 (50, 51), which like R7BP causes RGS7 redistribution to the plasma membrane (Fig. 2, C and D) (44, 45, 50, 56). We tested the effect of GPR158 on oligomerization of RGS7 (Fig. 8). GPR158 had no effect on the overall cross-linking pattern and the intensity of the 150- and 230-kDa cross-linked products in transfected cells (Fig. 8, A and B). Likewise, the knock-out of GPR158 had no significant effect on cross-linking of endogenous RGS7 in mouse brain (Fig. 8, D and E). We also found that GPR158 itself formed large cross-linked complexes that could not be resolved by SDS-PAGE (Fig. 8, C and F). We tested the effect of GPR158 on RGS7-RGS7 co-immunoprecipitation (Fig. 8, G and H). In contrast to R7BP, there was an increase in RGS7-RGS7 interaction from cells expressing GPR158, with a high variability between experiments. Thus, although both GPR158 and R7BP can recruit RGS7 to the plasma membranes, only R7BP can prevent RGS7-RGS7 interaction.
We also tested two other reported binding partners of the RGS7 DEP domain, muscarinic M3 receptor (34–37), and snapin (60), but we found that co-transfection of either protein had no effect on subcellular localization or oligomerization of RGS7 (data not shown).
Constitutively Active Gαo Inhibits RGS7 Oligomerization
Gαo is the preferred substrate for RGS7 (28–30), and both wild-type and a GTPase-deficient mutant of Gαo caused redistribution of RGS7 to the plasma membrane (61). To investigate the effect of Gαo on RGS7 oligomerization, we co-expressed the wild-type or constitutively active Gαo together with FLAG- and YFP-tagged RGS7 constructs. Cross-linking experiments showed a reduction of the intensity of the 150- and 230-kDa RGS7 bands in the presence of constitutively active but not wild-type Gαo (Fig. 9, A and B). In contrast, the 120-and 130-kDa RGS7 bands remained unchanged. As determined by co-IP (Fig. 9, C and D), co-expression of constitutively active Gαo caused a 67 ± 9% (n = 3, p value = 0.006) decrease in RGS7-RGS7 interaction. There was no statistical difference between the pcDNA control and wild-type Gαo. These results show that Gαo can prevent RGS7 oligomerization in an activity-dependent manner.
Discussion
The biological function of numerous proteins, including receptors, ion channels, structural proteins, and enzymes, depends on their oligomerization. This process has been demonstrated to affect enzymatic activities, proteolytic stability, subcellular localization, and numerous other functions (62–64). For example, oligomerization was demonstrated for many GPCRs. Although oligomerization may not be essential for the ability of GPCRs to activate G proteins (65–67), its role in receptor trafficking and stability, regulation of ligand affinity, and interaction with intracellular signaling molecules is well documented (68–70). Di- and tetramerization of arrestins was linked with their subcellular localization and functions (71). G proteins themselves and RGS proteins generally have not been reported to homo-oligomerize. We found only one earlier report on dimerization of an RGS protein, where the authors noticed an abnormal electrophoretic mobility of recombinant RGS5 and confirmed it by testing RGS5-RGS5 interaction in a yeast two-hybrid system (72). In the current study, we identified and characterized homo-oligomerization of RGS7.
We discovered oligomerization of RGS7 in the course of our experiments originally aimed at identification of its novel binding partners in cytoplasmic granules. Our approach relied on chemical cross-linking to stabilize the putative complexes. The resulting protein bands were sharp, which was a strong indication of the selectivity of RGS7 conjugation to other proteins. We became particularly interested in the 150-kDa complex because it did not contain Gβ5, the obligatory binding partner of RGS7. The 150-kDa band reproducibly appeared in both native tissue and transfected cells, and its formation was similarly affected by R7BP (Fig. 3). Therefore, we continued our investigation of this complex utilizing the advantages offered by heterologous expression of RGS7 in HEK293T cells. Evidence from these experiments led us to the idea that RGS7 could homo-oligomerize. We confirmed this hypothesis by co-immunoprecipitation of two differentially tagged RGS7 constructs. Within this experimental paradigm, we performed all conceivable controls, including reciprocal IP with alternative tags. We also showed that RGS7-RGS7 interaction occurs within live cells but not after cell lysis (Fig. 4D). The 150-kDa molecular mass of the chemically cross-linked RGS7 product may suggest that this complex contains three 55-kDa RGS7 subunits. However, cross-linking could cause abnormal mobility of the products on SDS-PAGE; therefore, it may be premature to conclude that it is a trimer. It is unlikely that a protein(s) other than RGS7 is present within the 150-kDa band, because the amounts of all additional peptides revealed by proteomic analysis were present in sub-stoichiometric amounts relative to RGS7 (Table 1). At the same time, we certainly do not rule out non-covalent binding of the RGS7 homo-oligomer to other proteins, e.g. Gβ5.
We also observed several cross-linked complexes that contained both RGS7 and Gβ5. At the moment, their origin and composition are unclear. It seems unlikely that each of the detected species represents a distinct complex present in situ. Rather, we propose that cells contain an oligomeric form, for example (Gβ5-RGS7)3. Trapping all the subunits within such a complex would require a high yield of the cross-linking reaction, i.e. increasing time of exposure or concentration of the agent. This inevitably results in non-selective tethering of cellular proteins (smear on the gel). Indeed, upon increasing concentration or time with either PFA or DSG, we generated more non-resolvable products without increasing the yield of the distinct RGS7 products (Fig. 3 and data not shown). Our conditions represent a compromise between non-selective aggregation and incomplete reaction to form pertinent bonds within native RGS7 complexes. Therefore, we theorize that the covalently linked products revealed in our experiments may represent fragments of the larger complex. For example, the 120-kDa band may correspond to Gβ5-RGS72 and the 230-kDa band to Gβ5, 2-RGS73; both products may originate from the larger (Gβ5-RGS7)3 homo-trimer present in situ. Because cross-linked products run abnormally on SDS-PAGE, we cannot determine their subunit composition solely on the basis of their molecular weights. It is also possible that other proteins are attached to the Gβ5-RGS7 adducts, like R7BP present in the 110- and 200-kDa products (Fig. 7A). What seems certain from our results is that RGS7 covalently attaches to another RGS7 and/or to Gβ5 with a much higher probability than to a random protein. Together with our co-IP data, this is a strong argument in favor of RGS7-RGS7 association within cells. Better understanding of the molecular organization and function of these complexes requires additional studies. Here, we started this investigation by asking which structural elements and known binding partners are necessary for oligomerization of RGS7.
R7 proteins have never been found in the absence of Gβ5 in native tissues because they are extremely unstable without Gβ5 (41). Therefore, it is very unlikely that oligomeric RGS7 can exist without Gβ5 in vivo. However, fusing RGS7 to YFP protects it from degradation in vitro. We found that YFP-RGS7 can localize to cytoplasmic granules (Fig. 5B) and that RGS7-RGS7 co-IP still occurred in the absence of Gβ5 (Fig. 5C), indicating that Gβ5 is not required for RGS7 oligomerization. In other words, it appears that if RGS7 were stable to proteolysis, it could oligomerize in the absence of Gβ5.
Through analysis of deletion mutants, we showed that the DEP domain of RGS7, but not the DHEX, GGL, or RGS domains, was essential for oligomerization (Fig. 6C). In addition, consistent with previous work (53) we found the DEP domain to be essential for localization of Gβ5-RGS7 in the cytoplasmic granules. The involvement of the DEP domain in both localization and oligomerization (Fig. 6) suggests that there might be a cause-and-effect relationship between RGS7 oligomerization and its subcellular localization.
Both R7BP and GPR158 can target RGS7 to the plasma membrane (Fig. 2, C and D) (45, 50, 51). However, we found that they had opposite effects on RGS7-RGS7 co-immunoprecipitation. In contrast to R7BP (Fig. 7), GPR158 did not prevent formation of RGS7 homo-oligomers (Fig. 8). It is unlikely that this was caused by a lower expression level of GPR158 compared R7BP since GPR158 recruited RGS7 to the plasma membrane as effectively as R7BP (Fig. 2, C and D). The ability of GPR158 to bind to oligomeric RGS7 allows us to propose that at the plasma membrane RGS7 can exist as an oligomer bound to GPR158. Binding to R7BP favors the “monomeric” Gβ5-RGS7 heterodimer.
Gαo is the preferred substrate for RGS7 (28–30). The interaction of RGS7 with Gαo presumably occurs at the plasma membrane, and constitutively active Gαo was shown to cause redistribution of RGS7 to the periphery of HEK293T cells (61). In the presence of the constitutively active Gαo Q205L mutant, but not WT Gαo, RGS7-RGS7 co-immunoprecipitation was decreased (Fig. 9, C and D). Although Gβ5 is an obligatory subunit of R7 proteins, interaction of Gα subunits with RGS proteins is dependent on the Gαo-GDP/Gαo-GTP state. It is therefore possible that the oligomeric state of the Gβ5-RGS7 complex could be regulated by upstream signal transduction events.
In summary, our study presents original experimental evidence that an R7 family RGS protein can form a homo-oligomer. Formation of this complex is mediated by its DEP domain and does not require Gβ5. Interaction with R7BP or activated Gαo reduces RGS7-RGS7 association. Future studies will explore the importance of oligomerization in signaling and other functions of this RGS protein.
Author Contributions
J. T. designed, performed, analyzed the experiments, and wrote the paper. Q. W. and A. N. P. contributed to the design of some experiments and preparation of the manuscript. C. O. and K. A. M. participated in the experiments involving mouse knock-out tissues and contributed to writing the paper. G. J. and J. W. C. performed LC-MS/MS analysis. V. Z. S. designed experiments, analyzed the data, and wrote the paper.
Acknowledgments
We thank Drs. Kendall Blumer and William Simonds for providing antibodies and expression constructs used in this study. We also thank Gabriel Gaidosh for advice on microscopy and Drs. Konstantin Levay, Darla Karpinsky-Semper, Lei Zhang, and Evangelos Liapis for their expert technical advice and helpful discussions of the manuscript.
This work was supported by National Institutes of Health Grants R01DK105427 (to V. Z. S.), EY018147 (to J. W. C.), EY14239 (to J. W. C.), DA026405 (to K. A. M.), MH105482 (to K. A. M.), and Center Grant P30-EY014801 (to Bascom Palmer Eye Institute). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GPCR
- G protein-coupled receptor
- RGS
- Regulators of G protein signaling
- R7BP
- R7 binding protein
- Gβ5
- G protein subunit 5
- Gαo
- G protein α subunit oA
- GAP
- GTPase activating protein
- PFA
- paraformaldehyde
- DSG
- disuccinimidyl glutarate
- IP
- immunoprecipitation
- CFP
- cyan fluorescent protein
- GGL
- Gγ-like
- DHEX
- DEP helical extension
- DEP
- disheveled, EGL10, pleckstrin.
References
- 1. Neer E. J. (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80, 249–257 [DOI] [PubMed] [Google Scholar]
- 2. Cabrera-Vera T. M., Vanhauwe J., Thomas T. O., Medkova M., Preininger A., Mazzoni M. R., and Hamm H. E. (2003) Insights into G protein structure, function, and regulation. Endocr. Rev. 24, 765–781 [DOI] [PubMed] [Google Scholar]
- 3. Clapham D. E., and Neer E. J. (1997) G protein βγ subunits. Annu. Rev. Pharmacol. Toxicol. 37, 167–203 [DOI] [PubMed] [Google Scholar]
- 4. Dohlman H. G., Apaniesk D., Chen Y., Song J., and Nusskern D. (1995) Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 3635–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lan K. L., Zhong H., Nanamori M., and Neubig R. R. (2000) Rapid kinetics of regulator of G-protein signaling (RGS)-mediated Gαi and Gαo deactivation. Gα specificity of RGS4 and RGS7. J. Biol. Chem. 275, 33497–33503 [DOI] [PubMed] [Google Scholar]
- 6. Koelle M. R., and Horvitz H. R. (1996) EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84, 115–125 [DOI] [PubMed] [Google Scholar]
- 7. Berman D. M., and Gilman A. G. (1998) Mammalian RGS proteins: barbarians at the gate. J. Biol. Chem. 273, 1269–1272 [DOI] [PubMed] [Google Scholar]
- 8. Ross E. M., and Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 9. Chen C. K., Burns M. E., He W., Wensel T. G., Baylor D. A., and Simon M. I. (2000) Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9–1. Nature 403, 557–560 [DOI] [PubMed] [Google Scholar]
- 10. Krispel C. M., Chen C. K., Simon M. I., and Burns M. E. (2003) Prolonged photoresponses and defective adaptation in rods of Gβ5−/− mice. J. Neurosci. 23, 6965–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heximer S. P., Knutsen R. H., Sun X., Kaltenbronn K. M., Rhee M. H., Peng N., Oliveira-dos-Santos A., Penninger J. M., Muslin A. J., Steinberg T. H., Wyss J. M., Mecham R. P., and Blumer K. J. (2003) Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J. Clin. Invest. 111, 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang X., Fu Y., Charbeneau R. A., Saunders T. L., Taylor D. K., Hankenson K. D., Russell M. W., D'Alecy L. G., and Neubig R. R. (2006) Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol. Cell. Biol. 26, 6870–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neubig R. R. (2015) RGS-insensitive G proteins as in vivo probes of RGS function. Prog. Mol. Biol. Transl. Sci. 133, 13–30 [DOI] [PubMed] [Google Scholar]
- 14. Bernstein L. S., Ramineni S., Hague C., Cladman W., Chidiac P., Levey A. I., and Hepler J. R. (2004) RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11α signaling. J. Biol. Chem. 279, 21248–21256 [DOI] [PubMed] [Google Scholar]
- 15. Shu F. J., Ramineni S., and Hepler J. R. (2010) RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell. Signal. 22, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chidiac P., and Roy A. A. (2003) Activity, regulation, and intracellular localization of RGS proteins. Receptors Channels 9, 135–147 [PubMed] [Google Scholar]
- 17. Saitoh O., and Odagiri M. (2003) RGS8 expression in developing cerebellar Purkinje cells. Biochem. Biophys. Res. Commun. 309, 836–842 [DOI] [PubMed] [Google Scholar]
- 18. Kach J., Sethakorn N., and Dulin N. O. (2012) A finer tuning of G-protein signaling through regulated control of RGS proteins. Am. J. Physiol. Heart Circ. Physiol. 303, H19–H35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng B., De Vries L., and Gist Farquhar M. (1999) Divergence of RGS proteins: evidence for the existence of six mammalian RGS subfamilies. Trends Biochem. Sci. 24, 411–414 [DOI] [PubMed] [Google Scholar]
- 20. Gold S. J., Ni Y. G., Dohlman H. G., and Nestler E. J. (1997) Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J. Neurosci. 17, 8024–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Huang J., Maity B., Gao Z., Lorca R. A., Gudmundsson H., Li J., Stewart A., Swaminathan P. D., Ibeawuchi S. R., Shepherd A., Chen C. K., Kutschke W., Mohler P. J., Mohapatra D. P., Anderson M. E., and Fisher R. A. (2010) RGS6, a modulator of parasympathetic activation in heart. Circ. Res. 107, 1345–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nini L., Zhang J. H., Pandey M., Panicker L. M., and Simonds W. F. (2012) Expression of the Gβ5/R7-RGS protein complex in pituitary and pancreatic islet cells. Endocrine 42, 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Q., Levay K., Chanturiya T., Dvoriantchikova G., Anderson K. L., Bianco S. D., Ueta C. B., Molano R. D., Pileggi A., Gurevich E. V., Gavrilova O., and Slepak V. Z. (2011) Targeted deletion of one or two copies of the G protein β subunit Gβ5 gene has distinct effects on body weight and behavior in mice. FASEB J. 25, 3949–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Posokhova E., Wydeven N., Allen K. L., Wickman K., and Martemyanov K. A. (2010) RGS6/Gβ5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ. Res. 107, 1350–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheever M. L., Snyder J. T., Gershburg S., Siderovski D. P., Harden T. K., and Sondek J. (2008) Crystal structure of the multifunctional Gβ5-RGS9 complex. Nat. Struct. Mol. Biol. 15, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cowan C. W., Fariss R. N., Sokal I., Palczewski K., and Wensel T. G. (1998) High expression levels in cones of RGS9, the predominant GTPase accelerating protein of rods. Proc. Natl. Acad. Sci. U.S.A. 95, 5351–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snow B. E., Krumins A. M., Brothers G. M., Lee S. F., Wall M. A., Chung S., Mangion J., Arya S., Gilman A. G., and Siderovski D. P. (1998) A G protein γ subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gβ5 subunits. Proc. Natl. Acad. Sci. U.S.A. 95, 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Posner B. A., Gilman A. G., and Harris B. A. (1999) Regulators of G protein signaling 6 and 7. Purification of complexes with Gβ5 and assessment of their effects on G protein-mediated signaling pathways. J. Biol. Chem. 274, 31087–31093 [DOI] [PubMed] [Google Scholar]
- 29. Hooks S. B., Waldo G. L., Corbitt J., Bodor E. T., Krumins A. M., and Harden T. K. (2003) RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J. Biol. Chem. 278, 10087–10093 [DOI] [PubMed] [Google Scholar]
- 30. Masuho I., Xie K., and Martemyanov K. A. (2013) Macromolecular composition dictates receptor and G protein selectivity of regulator of G protein signaling (RGS) 7 and 9–2 protein complexes in living cells. J. Biol. Chem. 288, 25129–25142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Slepak V. Z. (2009) Structure, function, and localization of Gβ5-RGS complexes. Prog. Mol. Biol. Transl. Sci. 86, 157–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson G. R., Posokhova E., and Martemyanov K. A. (2009) The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem. Biophys. 54, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stewart A., Maity B., and Fisher R. A. (2015) Two for the price of one: G protein-dependent and -independent functions of RGS6 in vivo. Prog. Mol. Biol. Transl. Sci. 133, 123–151 [DOI] [PubMed] [Google Scholar]
- 34. Sandiford S. L., and Slepak V. Z. (2009) The Gβ5-RGS7 complex selectively inhibits muscarinic M3 receptor signaling via the interaction between the third intracellular loop of the receptor and the DEP domain of RGS7. Biochemistry 48, 2282–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sandiford S. L., Wang Q., Levay K., Buchwald P., and Slepak V. Z. (2010) Molecular organization of the complex between the muscarinic M3 receptor and the regulator of G protein signaling, Gβ (5)-RGS7. Biochemistry 49, 4998–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karpinsky-Semper D., Volmar C. H., Brothers S. P., and Slepak V. Z. (2014) Differential effects of the Gβ5-RGS7 complex on muscarinic M3 receptor-induced Ca2+ influx and release. Mol. Pharmacol. 85, 758–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karpinsky-Semper D., Tayou J., Levay K., Schuchardt B. J., Bhat V., Volmar C. H., Farooq A., and Slepak V. Z. (2015) Helix 8 and the i3 loop of the muscarinic M3 receptor are crucial sites for its regulation by the Gβ5-RGS7 complex. Biochemistry 54, 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cabrera J. L., de Freitas F., Satpaev D. K., and Slepak V. Z. (1998) Identification of the Gβ5-RGS7 protein complex in the retina. Biochem. Biophys. Res. Commun. 249, 898–902 [DOI] [PubMed] [Google Scholar]
- 39. Levay K., Cabrera J. L., Satpaev D. K., and Slepak V. Z. (1999) Gβ5 prevents the RGS7-Gαo interaction through binding to a distinct Gγ-like domain found in RGS7 and other RGS proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 2503–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Witherow D. S., Wang Q., Levay K., Cabrera J. L., Chen J., Willars G. B., and Slepak V. Z. (2000) Complexes of the G protein subunit Gβ5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J. Biol. Chem. 275, 24872–24880 [DOI] [PubMed] [Google Scholar]
- 41. Chen C. K., Eversole-Cire P., Zhang H., Mancino V., Chen Y. J., He W., Wensel T. G., and Simon M. I. (2003) Instability of GGL domain-containing RGS proteins in mice lacking the G protein β-subunit Gβ5. Proc. Natl. Acad. Sci. U.S.A. 100, 6604–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu G., Zhang Z., and Wensel T. G. (2003) Activation of RGS9–1GTPase acceleration by its membrane anchor, R9AP. J. Biol. Chem. 278, 14550–14554 [DOI] [PubMed] [Google Scholar]
- 43. Martemyanov K. A., Lishko P. V., Calero N., Keresztes G., Sokolov M., Strissel K. J., Leskov I. B., Hopp J. A., Kolesnikov A. V., Chen C. K., Lem J., Heller S., Burns M. E., and Arshavsky V. Y. (2003) The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J. Neurosci. 23, 10175–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martemyanov K. A., Yoo P. J., Skiba N. P., and Arshavsky V. Y. (2005) R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 280, 5133–5136 [DOI] [PubMed] [Google Scholar]
- 45. Drenan R. M., Doupnik C. A., Boyle M. P., Muglia L. J., Huettner J. E., Linder M. E., and Blumer K. J. (2005) Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J. Cell Biol. 169, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drenan R. M., Doupnik C. A., Jayaraman M., Buchwalter A. L., Kaltenbronn K. M., Huettner J. E., Linder M. E., and Blumer K. J. (2006) R7BP augments the function of RGS7*Gβ5 complexes by a plasma membrane-targeting mechanism. J. Biol. Chem. 281, 28222–28231 [DOI] [PubMed] [Google Scholar]
- 47. Masuho I., Wakasugi-Masuho H., Posokhova E. N., Patton J. R., and Martemyanov K. A. (2011) Type 5 G protein β subunit (Gβ5) controls the interaction of regulator of G protein signaling 9 (RGS9) with membrane anchors. J. Biol. Chem. 286, 21806–21813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jayaraman M., Zhou H., Jia L., Cain M. D., and Blumer K. J. (2009) R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol. Sci. 30, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Narayanan V., Sandiford S. L., Wang Q., Keren-Raifman T., Levay K., and Slepak V. Z. (2007) Intramolecular interaction between the DEP domain of RGS7 and the Gβ5 subunit. Biochemistry 46, 6859–6870 [DOI] [PubMed] [Google Scholar]
- 50. Orlandi C., Posokhova E., Masuho I., Ray T. A., Hasan N., Gregg R. G., and Martemyanov K. A. (2012) GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol. 197, 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Orlandi C., Xie K., Masuho I., Fajardo-Serrano A., Lujan R., and Martemyanov K. A. (2015) Orphan receptor GPR158 is an allosteric modulator of regulator of G protein signaling 7 (RGS7) catalytic activity with essential role in dictating its expression and localization in the brain. J. Biol. Chem. 290, 13622–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anderson G. R., Lujan R., Semenov A., Pravetoni M., Posokhova E. N., Song J. H., Uversky V., Chen C. K., Wickman K., and Martemyanov K. A. (2007) Expression and localization of RGS9–2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J. Neurosci. 27, 14117–14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liapis E., Sandiford S., Wang Q., Gaidosh G., Motti D., Levay K., and Slepak V. Z. (2012) Subcellular localization of regulator of G protein signaling RGS7 complex in neurons and transfected cells. J. Neurochem. 122, 568–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klockenbusch C., and Kast J. (2010) Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin β1. J. Biomed. Biotechnol. 2010, 927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang J. H., and Simonds W. F. (2000) Copurification of brain G-protein β5 with RGS6 and RGS7. J. Neurosci. 20, RC59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grabowska D., Jayaraman M., Kaltenbronn K. M., Sandiford S. L., Wang Q., Jenkins S., Slepak V. Z., Smith Y., and Blumer K. J. (2008) Postnatal induction and localization of R7BP, a membrane-anchoring protein for regulator of G protein signaling 7 family-Gβ5 complexes in brain. Neuroscience 151, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bollinger K. E., Crabb J. S., Yuan X., Putliwala T., Clark A. F., and Crabb J. W. (2011) Quantitative proteomics: TGFβ(2) signaling in trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci. 52, 8287–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yuan X., Gu X., Crabb J. S., Yue X., Shadrach K., Hollyfield J. G., and Crabb J. W. (2010) Quantitative proteomics: comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol. Cell. Proteomics 9, 1031–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Witherow D. S., Tovey S. C., Wang Q., Willars G. B., and Slepak V. Z. (2003) Gβ5.RGS7 inhibits Gαq-mediated signaling via a direct protein-protein interaction. J. Biol. Chem. 278, 21307–21313 [DOI] [PubMed] [Google Scholar]
- 60. Hunt R. A., Edris W., Chanda P. K., Nieuwenhuijsen B., and Young K. H. (2003) Snapin interacts with the N terminus of regulator of G protein signaling 7. Biochem. Biophys. Res. Commun. 303, 594–599 [DOI] [PubMed] [Google Scholar]
- 61. Takida S., Fischer C. C., and Wedegaertner P. B. (2005) Palmitoylation and plasma membrane targeting of RGS7 are promoted by αo. Mol. Pharmacol. 67, 132–139 [DOI] [PubMed] [Google Scholar]
- 62. Heldin C. H. (1995) Dimerization of cell surface receptors in signal transduction. Cell 80, 213–223 [DOI] [PubMed] [Google Scholar]
- 63. Goodsell D. S., and Olson A. J. (2000) Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 [DOI] [PubMed] [Google Scholar]
- 64. Ali M. H., and Imperiali B. (2005) Protein oligomerization: how and why. Bioorg. Med. Chem. 13, 5013–5020 [DOI] [PubMed] [Google Scholar]
- 65. Whorton M. R., Bokoch M. P., Rasmussen S. G., Huang B., Zare R. N., Kobilka B., and Sunahara R. K. (2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 104, 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Whorton M. R., Jastrzebska B., Park P. S., Fotiadis D., Engel A., Palczewski K., and Sunahara R. K. (2008) Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 283, 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chabre M., and le Maire M. (2005) Monomeric G-protein-coupled receptor as a functional unit. Biochemistry 44, 9395–9403 [DOI] [PubMed] [Google Scholar]
- 68. Park P. S., Filipek S., Wells J. W., and Palczewski K. (2004) Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry 43, 15643–15656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bouvier M. (2001) Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2, 274–286 [DOI] [PubMed] [Google Scholar]
- 70. Ferré S., Casadó V., Devi L. A., Filizola M., Jockers R., Lohse M. J., Milligan G., Pin J. P., and Guitart X. (2014) G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 66, 413–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen Q., Zhuo Y., Kim M., Hanson S. M., Francis D. J., Vishnivetskiy S. A., Altenbach C., Klug C. S., Hubbell W. L., and Gurevich V. V. (2014) Self-association of arrestin family members. Handb. Exp. Pharmacol. 219, 205–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang Z., Gaudio S., Song W., Greenwood M., Jean-Baptiste G., and Greenwood M. T. (2007) Evidence for the dimerization of human regulator of G-protein signalling 5 (RGS5). Cell. Physiol. Biochem. 20, 303–310 [DOI] [PubMed] [Google Scholar]