Abstract
Site-selective functionalization of complex molecules is a grand challenge in chemistry. Protecting groups or catalysts must be used to selectively modify one site among many that are similarly reactive. General strategies are rare such the local chemical environment around the target site is tuned for selective transformation. Here we show a four amino acid sequence (Phe-Cys-Pro-Phe), which we call the “π-clamp”, tunes the reactivity of its cysteine thiol for the site-selective conjugation with perfluoroaromatic reagents. We used the π-clamp to selectively modify one cysteine site in proteins containing multiple endogenous cysteine residues (e.g. antibodies and cysteine-based enzymes), which was impossible with prior cysteine modification methods. The modified π-clamp antibodies retained binding affinity to their targets, enabling the synthesis of site-specific antibody-drug conjugates (ADCs) for selective killing of HER2-positive breast cancer cells. The π-clamp is an unexpected approach for site-selective chemistry and provides opportunities to modify biomolecules for research and therapeutics.
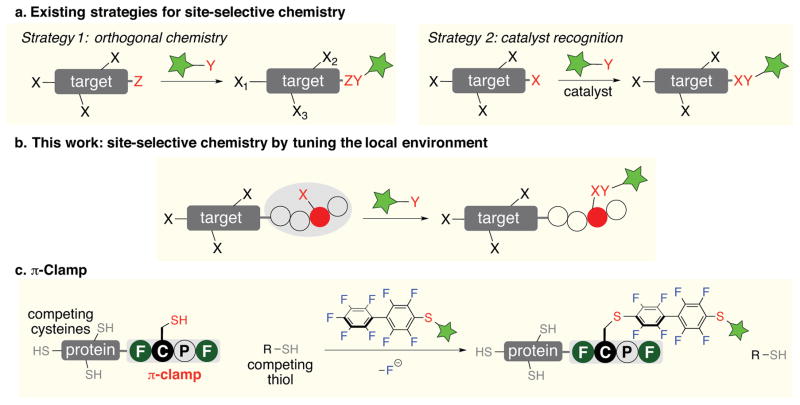
Site-selective chemistry1–5 is essential for creating homogeneously modified biologics6,7, studying protein structure and function8, generating materials with defined composition9, and on-demand modification of complex small molecules10,11. Existing approaches for site-selective chemistry utilize either reaction pairs that are orthogonal to other functional groups on the target of interest (Fig. 1a, strategy 1)12,13 or catalysts that mediate selective reactions at a particular site among many competing ones (Fig. 1a, strategy 2)14–19. These strategies have been widely used in protein modification and have led to the development of multiple bio-orthogonal handles20–25 and enzyme-tag pairs26–31.
Figure 1. π-clamp mediated cysteine conjugation as a new strategy for site-selective chemistry.
a, Existing strategies for site-selective chemistry. Strategy 1: selectivity arises from orthogonal chemistry between site Z and reagent Y. Strategy 2: catalyst mediates the reaction between a particular site X (highlighted in red) and reagent Y. b, This work demonstrates a new strategy for site-selective chemistry by fine-tuning the local chemical environment around the target site. A particular site X (highlighted in red) is tuned to react with reagent Y in the presence of other competing X sites. c, Cysteine residue inside the π-clamp selectively reacts with perfluoroaromatic probes in the presence of other competing cysteine residues and thiol species.
Natural proteins precisely control selective reactions and interactions by building large three-dimensional structures from polypeptides usually much greater than 100 residues.32 For example, enzymes have folded structures where particular amino acids are placed in a specialized active-site environment.33 Inspired by this, we envisioned a new strategy for site-selective chemistry on proteins by fine-tuning the local environment around an amino acid residue in a small peptide sequence (Fig. 1b). This is challenging because peptides are highly dynamic and unstructured thereby presenting a formidable challenge to build defined environments for selective chemical transformations.
Our design efforts leveraged cysteine because Nature has shown its robust catalytic role in enzymes,34,35 and prior efforts indicate the reactivity of a cysteine residue can vary in different protein environments.36 Further, cysteine is the first choice in bioconjugation to modify proteins often via maleimide ligation or alkylation.37,38 However, these traditional cysteine-based bioconjugations are significantly limited because they are not site-specific. When these methods are applied to protein targets with multiple cysteine residues, a mixture of heterogeneous products are generated, as exemplified by recent efforts to conjugate small molecule drugs to antibodies through cysteine-based reactions.39
Small peptide tags that contain multiple cysteine residues have been used for bioconjugation. Tsien and co-workers have developed biarsenic reagents that selectively react with tetra-cysteine motifs in peptides and proteins.40,41 More recently, organic arsenics have been used to modify two cysteine residues generated from reducing a disulfide bond.42 These methods can present challenges with thiol selectivity43 and none report the site-specific modification of one cysteine residue in the presence of many as enzymes or multiple chemical steps must be used to accomplish this feat.44,45 An enzyme-free and one-step method for site-selective cysteine conjugation has yet to be developed.
We have previously described a perfluoroaryl-cysteine SNAr approach for peptide and protein modifications.46–49 The reactions between perfluoroaryl groups and cysteine residues are fast in organic solvent but extremely sluggish in water unless an enzyme is used.47,48 This observation inspired us to develop small peptides to promote the SNAr reaction in an analogous fashion to enzymes.
Results
Here we describe the identification of the π-clamp sequence to mediate site-specific cysteine modification in water without an enzyme, which overcomes the selectivity challenge for cysteine bioconjugation (Fig. 1c). This offers a fundamentally new mode for site-specific chemistry by fine-tuning the microenvironment of a four-residue stretch within a complex protein or peptide. Through a library selection approach (Fig. S26 in the Supplementary Information), we find the sequence Phe-Cys-Pro-Trp within a polypeptide exhibits enhanced reactivity for a perfluoroaryl electrophilic probe (Fig. S1 in the Supplementary Information) via nucleophilic aromatic substitution reaction. This observation is in stark contrast to our prior efforts47 which showed that cysteine residues and perfluoroaryl moieties do not react in water. Thus the Phe-Cys-Pro-Trp sequence appears to radically modify the reactivity of the cysteine thiol. Further mutating the Phe and Trp to Gly eliminated the reaction. Based on these findings and a molecular model of Phe-Cys-Pro-Trp, we hypothesize that the Phe and Trp side chains activate the cysteine thiol and interact with the incoming perfluoroaryl group, while the Pro serves to position the Cys, Phe, and Trp residues into a conformation that promotes the reaction. We refer to this distinctive amino acid sequence Xaa-Cys-Pro-Xaa (Xaa = electron-rich aromatic amino acids including Phe, Trp, or Tyr) as a π-clamp.
To investigate the π-clamp mediated conjugation, we mutated the aromatic residues. Each of 9 peptides (Xaa-Cys-Pro-Xaa-Gly-Leu-Leu-Lys-Asn-Lys, where Xaa was Phe, Trp, or Tyr) were tested for reaction with a perfluoroaryl-probe (2) in 0.2 M phosphate buffer at pH 8.0 and 37 °C with 20 mM TCEP added as the reducing agent. All 9 peptides reacted with probe 2 (rate constants = 0.05 to 0.73 M−1• S−1, see Table S2 in the Supplementary Information). In contrast, the double glycine mutant (1A) formed no product (Fig. 2a, entry 1). The Phe-Phe π-clamp peptide (1E) gave quantitative conversion in 30 minutes (rate constants = 0.73 M−1• S−1, Fig. 2a, entry 5). Single mutations of each Phe to Gly (1B and 1C, Fig. 2a, entries 2 and 3) or converting the L-Pro to D-Pro (1D, Fig. 2a, entry 4) significantly decreased the rate of the arylation reaction. These studies indicate that each amino acid in the π-clamp is essential for product formation.
Figure 2. π-clamp mediated cysteine conjugation on peptides.
a, Mutation studies show Phe-1, Pro-3, and Phe-4 are required for the observed reactivity. TCEP: tris(2-carboxylethyl)phosphine. Yields shown are from LC-MS analysis of the crude reactions at 30 minutes. b, Site-specific conjugation at the π-clamp in the presence of another competing cysteine peptide. Chromatograms shown are total ion currents (TIC) from LC-MS analysis of crude reaction mixtures at 0 minute (black) and 30 minutes (red). The mass spectrum of product 2E is shown as the inset.
π-clamp mediated conjugation is highly selective as indicated by our thiol competition experiments. The π-clamp peptide (1E) was found to undergo quantitative conversion with the perfluoroaryl probe (2) in the presence of a double glycine mutant peptide (1A) that served as the competing thiol species. Only the π-clamp peptide reacted quantitatively to form conjugated product in 30 minutes (Fig. 2b).
To further investigate the π-clamp mediated cysteine conjugation, we carried out additional studies to understand if location mattered and the substrate scope. We found that the π-clamp was efficiently modified irrespective of its position on the polypeptide chain (Fig. 3, Fig. S3 – S6 in the Supplementary Information). π-clamp at the N-terminus (1E), the C-terminus (1N), and the middle (1O) of the polypeptide chain were readily modified with a diverse set of perfluoroaryl-linked probes including peptide, biotin, fluorescein, alkyne, and polyethylene glycol (2 – 6).
Figure 3. π-clamp functions at distinct positions in polypeptides and is compatible with diverse perfluoroaryl-based probes.
π-clamp at the N-terminus, the C-terminus, and the middle of peptides were readily reacted with perfluoroaryl probes bearing peptide molecule, affinity tag (biotin), fluorescent reporter (fluorescein isothiocyanate, FITC), click chemistry handle (alkyne), and polymer (polyethylene glycol, PEG). Yields shown are from LC-MS analysis of the crude reactions at 60 minutes. *Yields at 120 minutes. See Supplementary Information for LC-MS chromatograms.
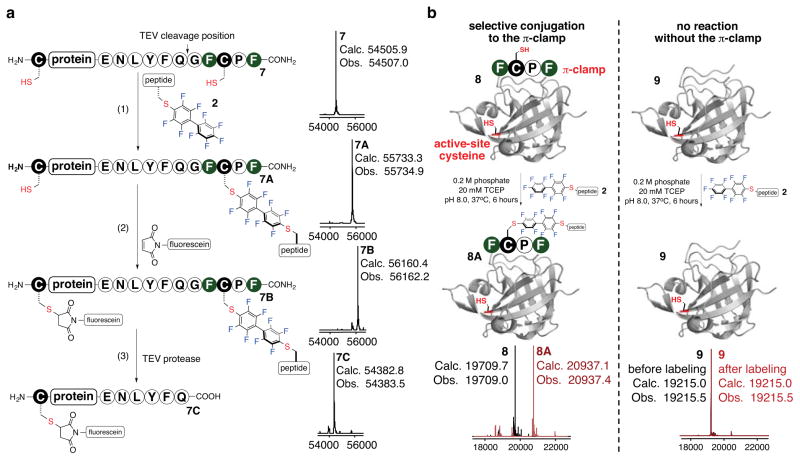
We next investigated the regioselectivity on a 55-kDa protein substrate (Fig. 4a). Model protein 7 was designed to contain an N-terminal cysteine and a C-terminal π-clamp. A protease cleavage site was positioned upstream of the π-clamp thereby allowing for the unequivocal verification of the regioselectivity. Upon reacting the protein (7) with probe 2 for 2 hours, we observed > 95% formation of the mono-labeled product (7A). The N-terminal free cysteine was subsequently labeled with fluorescein-5-maleimide producing the dual-labeled product (7B). Upon protease cleavage, only two products were generated: a protein with maleimide-labeled N-terminal cysteine (7C) and a π-clamp arylated species, confirming the absolute regioselectivity endowed by the π-clamp.
Figure 4. π-clamp mediated site-specific conjugation on proteins with multiple cysteines.
a, Protecting group-free one-pot dual labeling of a 55-kDa protein. The protein used was a fusion protein of the anthrax toxin lethal factor 1–263 (LFN) and diphtheria toxin domain A (DTA). Reaction conditions: (1) 50 μM 7, 1 mM 2, 0.2 M phosphate, 20 mM TCEP, 37 °C, 2 hours. (2) 50 μM 7A, 1 mM fluorescein-5-maleimide, 0.2 M phosphate pH 7.0, room temperature, 10 minutes. (3) 25 μM protein 7B, 0.1 mg/mL TEV protease, 50 mM Tris, 0.1 mM EDTA, 1 mM DTT, pH 8.0, room temperature, 15 hours. TEV: tobacco itch virus; EDTA: ethylenediaminetetraacetic acid; DTT: dithiothreitol; Tris: 2-amino-2-hydroxylmethyl-propane-1, 3-diol. b, Left, quantitative and selective labeling of π-clamp SrtA (PDB entry: 1T2P); right, control shows no labeling of SrtA. Reactions conditions: 38 μM 8 or 9, 1 mM 2, 0.2 M phosphate, 20 mM TCEP, 37 °C, 6 hours.
Next, we site-specifically modified a cysteine-containing transpeptidase Sortase A (SrtA)50 (Fig. 4b). An N-terminal π-clamp SrtA variant (8) reacted with probe 2 to produce > 95% mono-labeled product (8A). The modified variant displayed full catalytic activity (Fig. S10 in the Supplementary Information). No reaction took place with SrtA without the π-clamp (9). In sharp contrast, when the π-clamp-Sortase (8) was reacted with bromoacetamide, a mixture of products was produced with labeling of both cysteine residues (Fig. S9 in the Supplementary Information).
IgG molecules modified with small molecule drugs (antibody-drug conjugates, ADCs) are currently used as therapeutic agents.51 However, attaching small molecule agents site-specifically to cysteines in IgGs is as of yet impossible, and thus commercial ADCs are heterogeneous mixtures of conjugates.51 Approaches to engineer cysteine substitutions in antibodies produce mixed disulfides with cysteine or glutathione, thus a fine-tuned reduction-oxidation protocol must be used to afford the free cysteine thiols for selective drug conjugation in the presence of disulfide bonds.52,53
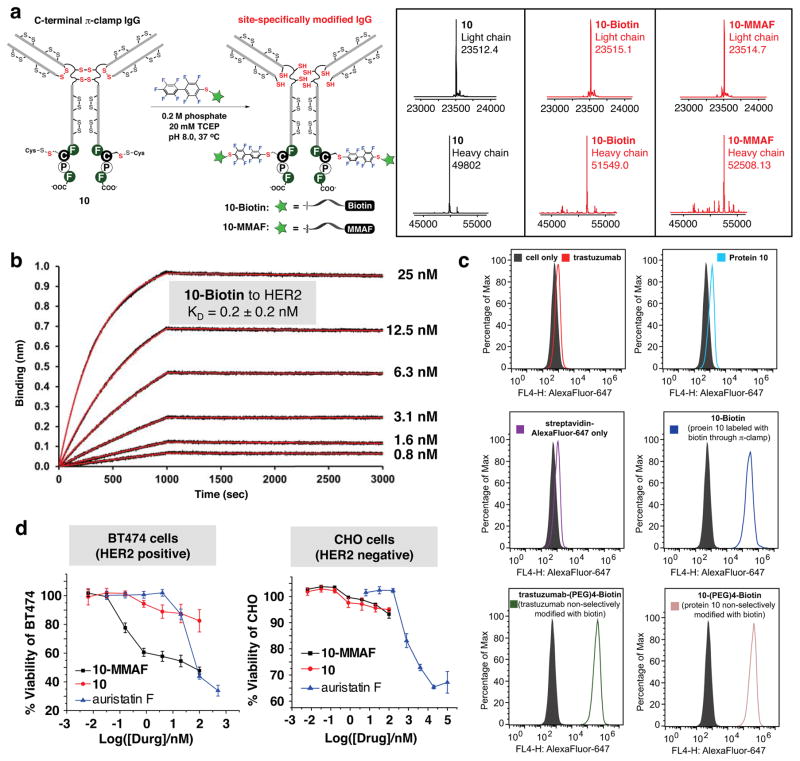
We anticipated that the π-clamp IgG could be used to overcome this specificity problem in ADC synthesis, which is notably challenging because IgGs harbor 32 native cysteine residues. The π-clamp mediated modification on antibodies will be a single-step and site-specific antibody-drug conjugation technology that does not require significant antibody engineering or extra chemical steps52,53. To this end, we inserted the Phe-Cys-Pro-Phe sequence into the C-termini of the heavy chains of trastuzuamab54. Reacting the π-clamp trastuzumab (protein 10) with either a biotin-perfluoroaryl probe (11-Biotin) or a drug-perfluoroaryl probe (11-MMAF) under reducing conditions, we observed facile formation of the heavy chain mono-labeled products (10-Biotin or 10-MMAF) by LC-MS analysis (Fig. 5a). Antibodies without the π-clamp showed no desired modification under the same conditions (Supplementary Fig. S27), highlighting the specificity of the conjugation. Moreover, this selective conjugation reaction works with other antibodies, reacting a π-clamp C225 antibody55,56 with 11-Biotin resulted in only the selective modifications on the π-clamp cysteine residues (Fig. S28), suggesting that the π-clamp could be a general strategy for site-selective antibody modification.
Figure 5. π-clamp mediated site-specific antibody conjugation.
a, site-specific conjugation of biotin or monomethyl auristatin F (MMAF) to π-clamp trastuzumab (protein 10). LC-MS analysis showed site-specific labeling of the π-clamp cysteine residues on the trastuzumab heavy chain. The antibodies were treated with PNGase F to remove the N-linked glycans before LC-MS analysis. Reaction conditions for biotin conjugation: 100 μM 10, 1 mM 11-Biotin, 0.2 M phosphate, 20 mM TCEP, 37 °C, 4 hours. Reaction conditions for MMAF conjugation: 100 μM 10, 1 mM 11-MMAF, 0.2 M phosphate, 20 mM TCEP, 5%DMSO, 37 °C, 16 hours. b, the biotin-conjugated π-clamp trastuzumab (10-Biotin) binds to HER2 in the Octet binding assay (KD = 0.2 ± 0.2 nM). 10-Biotin was immobilized on streptavidin tips and was sampled with serially diluted concentrations of recombinant HER2 (concentrations of HER2 in each experiment are shown next to the curve, and see Supplementary Information for data analysis and fitting). c, 10-Biotin retained binding to BT474 cells (HER2 positive). Cells were treated with 10-Biotin or controls, washed with phosphate buffer saline (PBS) with 0.1% BSA, and then treated with streptavidin-AlexaFluor-647 before analyzed by flow cytometer. 10-(PEG)4-Biotin and trastuzumab-(PEG)4-Biotin were prepared from reacting Biotin-(PEG)4-NHS with protein 10 or trastuzumab, respectively (see Supplementary Information for details). d, 10-MMAF killed BT474 cells (HER2 positive) but was not effective for CHO cells (HER2 negative). EC50 values for BT474 cells were 0.19 nM for 10-MMAF and 41 nM for auristatin F. EC50 value of auristatin F for CHO cells is 1.3 μM. Cells were seeded in 96-well white opaque plate at a density of 5 ×103/well (CHO) or 10 ×103/well (BT474). Cells were allowed to attach for 24 hours at 37 °C and 5% CO2 in humidified atmosphere. Cells were then treated with serial dilutions of auristatin F, 10-MMAF, or 10 for 96 hours (BT474) or 72 hours (CHO, treatment time was shorten to prevent overgrowth). Cell viability was quantified using CellTiter Glo assay and was normalized to cell only. Experiments were done in triplicate for each dose.
Under the developed reaction conditions (0.2 M phosphate, 20 mM TCEP, pH 8.0, at 37 °C), only the inter-chain disulfides and the π-clamp cysteine residues are reduced (Supplementary Fig. S38), and the modified antibodies retained binding affinity to their targets. Biotin modified π-clamp trastuzumab (10-Biotin) showed similar binding affinity to HER2 (KD = 0.2 ± 0.2 nM) compared to native trastuzumab non-selectively modified with a (PEG)4-Biotin (trastuzumab-(PEG)4-Biotin, KD = 0.3 ± 0.1 nM) (Fig. 5b and Supplementary Fig. S31). In addition, both proteins 10 and 10-Biotin readily bound to BT474 cells (HER2-positive) (Fig. 5c and Supplementary Fig. S32 and S33). As another antibody test case, biotin modified C225 antibody (12-Biotin) showed similar binding to A431 cells (EGFR-positive) compared to the native C225 antibody (Supplementary Fig. S34 and S35). Collectively, insertion of the π-clamp into the heavy chains of antibodies and subsequent modification with drugs or probes did not significantly alter the binding properties.
Using the π-clamp mediated cysteine conjugation, we synthesized a site-specific antibody drug conjugate using π-clamp trastuzumab (protein 10) and a monomethyl auristatin F (MMAF) linked to a perfluoroaryl group (11-MMAF, see Supplementary Information for synthesis). LC-MS analysis of the conjugation reaction showed selective labeling of the heavy chain π-clamp cysteine residues (Fig. 5a). The prepared ADC selectively killed BT474 cells (HER2 positive) but was not effective for CHO cells (HER2 negative), indicating that the observed toxicity is receptor-dependent.
To investigate the mechanism of the π-clamp mediated reaction, we first used molecular dynamics (MD) to sample the conformational arrangements of the π-clamp peptide (1E) (Fig. 6a). Simulations indicated that 1E adopts four primary conformations when a cis-Pro is present: a “π-clamp” (S1) with the phenyl rings of Phe-1 and Phe-4 interacting face-on with the Cys-2 thiol; a “half-clamp” (S2) where only the Phe-4 side chain interacts with the Cys-2 thiol; S3 in which the Phe-1 and Phe-4 side chains are stacked together, leaving the Cys-2 thiol exposed; and an open configuration (S4) where all side chains are too far apart to interact. MD simulation for π-clamp peptide (1E) with a trans-Pro indicated two “open” structures with the cysteine thiol not interacting a Phe residue and one structure with Phe-4 side chain interacting with Cys-2 thiol (see Supplementary Fig. S37).
Figure 6. Structure and mechanism of the π-clamp.
a, Four primary structures S1 – S4 were identified from MD simulation of π-clamp peptide 1E. The phenyl rings and cysteine thiol are shown as spheres; the rest of the peptide is drawn as sticks. b, Conjugation to the π-clamp is energetically favored compared to the double glycine mutant. Left, proposed nucleophilic aromatic substitution pathway for arylation at the π-clamp. Right, computed geometries and free energy surface of the nucleophilic aromatic substitution at the π-clamp (red). The free energy surface of the double glycine control is also shown (grey).
With these MD structures in hand, we used density functional theory (DFT) to investigate the nucleophilic aromatic substitution energy pathway for structures with a cis-Pro. We found that the half-clamp structure S2 stabilized the arylation product by approximately 5 kcal/mol compared to the double glycine mutant, indicating the important role of Phe-4 in promoting the arylation reaction. This is consistent with our mutation studies showing that Phe-4 alone can partially mediate the arylation reaction (Fig. 2a, Entry 3). The product generated from the open structure (S4) has similar free energy compared to that of the double glycine mutant, further substantiating the hypothesis that the two phenylalanine side chains are important for the arylation reaction with the perfluoroaryl groups.
The most stable product was observed with the “π-clamp” structure (S1) of which the free energy was approximately 7 kcal/mol lower than that of the double glycine mutant. We further found that the activation energy for the formation of the transition state57 (III in Fig. 6b) was decreased by approximately 3 kcal/mol when the π-clamp (S1) was present (see further discussion in the Supplementary Information), presumably because of the phenyl rings recognizing the perfluoroaryl group and activating cysteine sulfur before conjugation. Collectively, these DFT calculations indicated that the π-clamp offers both a kinetic advantage (lower activation energy) and a thermodynamic advantage (lower free energy) over the double glycine mutant for the selective reaction with the perfluoroaryl reagents.
Discussion and Conclusion
Here we describe the discovery of a π-clamp to mediate site-selective cysteine conjugation. The π-clamp is composed of natural amino acids and shares some essential features of large enzymes, yet it mediates a purely abiotic cysteine perfluoroarylation reaction. The π-clamp tunes the reactivity of a cysteine thiol in its “active-site”, recognizes the perfluoroaromatic reaction partner, and decreases the activation energy for the reaction. In addition, the π-clamp has practical applications in protein labeling.4 The reported reaction is site-specific, operational under physiologically relevant conditions, enzyme-free, and as efficient as the commonly used azide-alkyne click chemistry58,59 (π-clamp rate constant: 0.73 M−1• S−1).
Compared to existing bioconjugation techniques38, the advantages of the π-clamp include its (1) small size that offers minimal structural perturbation to the target protein; (2) genetic encodability for straightforward incorporation; (3) ability to perform protecting-group-free dual cysteine modification; (4) and reaction mode that tunes the kinetic parameters to favor the cysteine perfluoroarylation reaction. This mode of reaction is distinct when compared to other advanced cysteine bioconjugations that use entropy to favor conjugation.40–42
The unexpected mode of site-specificity provided by the π-clamp requires further mention. In all existing conjugation methods38, selectivity results from the judicious choice of certain functional groups so that each reaction pair undergoes conjugation in the presence of many other potentially reactive groups. For example, the unnatural handles used for click reactions are orthogonal to other functional groups on the target of interest.12 In contrast, selectivity in the π-clamp mediated conjugation is achieved by fine-tuning the local chemical environment and reactivity as proteins do. This provides a complementary strategy to non-natural amino acid-mediated bioconjugation.60 By fine-tuning the peptide microenvironment to allow for selective modification, the π-clamp significantly expands the chemistry available for selectively tailoring biomolecules.
Acknowledgments
We thank Prof. Steve Buchwald (MIT) and Prof. K. Dane Wittrup (MIT) for comments and suggestions on the manuscript. This work was generously supported by an MIT start-up fund, NIH (R01GM110535) and the Sontag Foundation Distinguished Scientist Award for B.L.P. C.Z. is a recipient of the George Buchi Research Fellowship and the Koch Graduate Fellowship in Cancer Research of MIT. M.W. is an NSF Graduate Research Fellow. We thank the Biological Instrument Facility of MIT for providing the Octet BioLayer Interferometry System (NIF S10 OD016326), the MIT CEHS Instrument Facility for providing flow cytometer (P30-ES002109), and Prof. R. John Collier (Harvard) for contributing select laboratory equipment used in this study.
Footnotes
Author Contributions
C.Z. and B.L.P. conceived the work; C.Z. performed the peptide and protein labeling experiments; M.W., T.Z., and T.V.H. performed the computational studies; N.J.Y. helped with antibody expression and purification; M.S.S. and C.Z. performed the flow cytometer and cell viability assays. C.Z. and B.L.P. wrote the manuscript with input from all other authors; B.L.P. directed the project.
The authors declare no competing financial interests.
A patent application covering this work has been filed by MIT-TLO.
Readers are welcome to comment on the online version of this paper.
Supplementary Information
Materials and Methods
Supplementary Text
Supplementary References and Notes
Tables S1 – S4
Figures S1 – S37
References and Notes
- 1.Carrico IS. Chemoselective modification of proteins: hitting the target. Chem Soc Rev. 2008;37:1423–1431. doi: 10.1039/b703364h. [DOI] [PubMed] [Google Scholar]
- 2.Hackenberger CPR, Schwarzer D. Chemoselective Ligation and Modification Strategies for Peptides and Proteins. Angewandte Chemie International Edition. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 3.Rabuka D. Chemoenzymatic methods for site-specific protein modification. Curr Opin Chem Biol. 2010;14:790–796. doi: 10.1016/j.cbpa.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spicer CD, Davis BG. Selective chemical protein modification. Nat Commun. 2014;5 doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 5.Take aim. Nat Chem. 2012;4:955–955. doi: 10.1038/nchem.1521. [DOI] [PubMed] [Google Scholar]
- 6.Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. mAbs. 2014;6:34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochendoerfer GG. Site-specific polymer modification of therapeutic proteins. Curr Opin Chem Biol. 2005;9:555–560. doi: 10.1016/j.cbpa.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proceedings of the National Academy of Sciences. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishna OD, Kiick KL. Protein- and peptide-modified synthetic polymeric biomaterials. Peptide Science. 2010;94:32–48. doi: 10.1002/bip.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichtor PA, Miller SJ. Combinatorial evolution of site- and enantioselective catalysts for polyene epoxidation. Nat Chem. 2012;4:990–995. doi: 10.1038/nchem.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilcock BC, et al. Electronic tuning of site-selectivity. Nat Chem. 2012;4:996–1003. doi: 10.1038/nchem.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew Chem, Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afagh NA, Yudin AK. Chemoselectivity and the Curious Reactivity Preferences of Functional Groups. Angewandte Chemie International Edition. 2010;49:262–310. doi: 10.1002/anie.200901317. [DOI] [PubMed] [Google Scholar]
- 14.Lewis CA, Miller SJ. Site-Selective Derivatization and Remodeling of Erythromycin A by Using Simple Peptide-Based Chiral Catalysts. Angewandte Chemie International Edition. 2006;45:5616–5619. doi: 10.1002/anie.200601490. [DOI] [PubMed] [Google Scholar]
- 15.Chen MS, White MC. A Predictably Selective Aliphatic C–H Oxidation Reaction for Complex Molecule Synthesis. Science. 2007;318:783–787. doi: 10.1126/science.1148597. [DOI] [PubMed] [Google Scholar]
- 16.Snyder SA, Gollner A, Chiriac MI. Regioselective reactions for programmable resveratrol oligomer synthesis. Nature. 2011;474:461–466. doi: 10.1038/nature10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathak TP, Miller SJ. Site-Selective Bromination of Vancomycin. J Am Chem Soc. 2012;134:6120–6123. doi: 10.1021/ja301566t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wender PA, Hilinski MK, Mayweg AVW. Late-Stage Intermolecular CH Activation for Lead Diversification: A Highly Chemoselective Oxyfunctionalization of the C-9 Position of Potent Bryostatin Analogues. Org Lett. 2004;7:79–82. doi: 10.1021/ol047859w. [DOI] [PubMed] [Google Scholar]
- 19.Peddibhotla S, Dang Y, Liu JO, Romo D. Simultaneous Arming and Structure/Activity Studies of Natural Products Employing O–H Insertions: An Expedient and Versatile Strategy for Natural Products-Based Chemical Genetics. J Am Chem Soc. 2007;129:12222–12231. doi: 10.1021/ja0733686. [DOI] [PubMed] [Google Scholar]
- 20.Saxon E, Bertozzi CR. Cell Surface Engineering by a Modified Staudinger Reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 21.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angewandte Chemie International Edition. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Agard NJ, Prescher JA, Bertozzi CR. A Strain-Promoted [3 + 2] Azide–Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 23.Song W, Wang Y, Qu J, Lin Q. Selective Functionalization of a Genetically Encoded Alkene-Containing Protein via “Photoclick Chemistry” in Bacterial Cells. J Am Chem Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 24.Blackman ML, Royzen M, Fox JM. Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels–Alder Reactivity. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxon E, Armstrong JI, Bertozzi CR. A “Traceless” Staudinger Ligation for the Chemoselective Synthesis of Amide Bonds. Org Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 26.Lin CW, Ting AY. Transglutaminase-Catalyzed Site-Specific Conjugation of Small- Molecule Probes to Proteins in Vitro and on the Surface of Living Cells. J Am Chem Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wollack JW, Silverman JM, Petzold CJ, Mougous JD, Distefano MD. A Minimalist Substrate for Enzymatic Peptide and Protein Conjugation. Chembiochem. 2009;10:2934–2943. doi: 10.1002/cbic.200900566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin J, et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc Natl Acad Sci U S A. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cull MG, Schatz PJ. In: Methods Enzymol. Emr Scott D, Abelson John N, Thorner Jeremy., editors. Vol. 326. Academic Press; 2000. pp. 430–440. [Google Scholar]
- 30.Fernandez-Suarez M, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotech. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 32.Whitford D. Proteins: Structure and Function. J. Wiley & Sons; 2005. [Google Scholar]
- 33.Walsh C. Enabling the chemistry of life. Nature. 2001;409:226–231. doi: 10.1038/35051697. [DOI] [PubMed] [Google Scholar]
- 34.Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun. 2003;300:1–4. doi: 10.1016/s0006-291x(02)02770-5. [DOI] [PubMed] [Google Scholar]
- 35.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathani RI, et al. A novel approach to the site-selective dual labelling of a protein via chemoselective cysteine modification. Chemical Science. 2013;4:3455–3458. doi: 10.1039/c3sc51333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalker JM, Bernardes GJL, Lin YA, Davis BG. Chemical Modification of Proteins at Cysteine: Opportunities in Chemistry and Biology. Chemistry – An Asian Journal. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- 38.Hermanson GT. In: Bioconjugate Techniques. 3. Hermanson Greg T., editor. Academic Press; 2013. pp. 1–125. [Google Scholar]
- 39.Sun MMC, et al. Reduction–Alkylation Strategies for the Modification of Specific Monoclonal Antibody Disulfides. Bioconjug Chem. 2005;16:1282–1290. doi: 10.1021/bc050201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin BA, Adams SR, Tsien RY. Specific Covalent Labeling of Recombinant Protein Molecules Inside Live Cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 41.Adams SR, et al. New Biarsenical Ligands and Tetracysteine Motifs for Protein Labeling in Vitro and in Vivo: Synthesis and Biological Applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 42.Wilson P, et al. Organic Arsenicals As Efficient and Highly Specific Linkers for Protein/Peptide–Polymer Conjugation. J Am Chem Soc. 2015;137:4215–4222. doi: 10.1021/jacs.5b01140. [DOI] [PubMed] [Google Scholar]
- 43.Stroffekova K, Proenza C, Beam KG. The protein-labeling reagent FLASH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Archiv-European Journal of Physiology. 2001;442:859–866. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- 44.Gautier A, et al. An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Rush JS, Bertozzi CR. New Aldehyde Tag Sequences Identified by Screening Formylglycine Generating Enzymes in Vitro and in Vivo. J Am Chem Soc. 2008;130:12240–12241. doi: 10.1021/ja804530w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spokoyny AM, et al. A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling. J Am Chem Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, Spokoyny AM, Zou Y, Simon MD, Pentelute BL. Enzymatic “Click” Ligation: Selective Cysteine Modification in Polypeptides Enabled by Promiscuous Glutathione S-Transferase. Angew Chem, Int Ed. 2013;52:14001–14005. doi: 10.1002/anie.201306430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C, Dai P, Spokoyny AM, Pentelute BL. Enzyme-Catalyzed Macrocyclization of Long Unprotected Peptides. Org Lett. 2014;16:3652–3655. doi: 10.1021/ol501609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou Y, et al. Convergent diversity-oriented side-chain macrocyclization scan for unprotected polypeptides. Org Biomol Chem. 2014;12:566–573. doi: 10.1039/c3ob42168f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 51.Sievers EL, Senter PD. Antibody-Drug Conjugates in Cancer Therapy. Annu Rev Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 52.Junutula JR, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotech. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 53.Junutula JR, et al. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. J Immunol Methods. 2008;332:41–52. doi: 10.1016/j.jim.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 54.Piccart-Gebhart MJ, et al. Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 55.Spangler JB, Manzari MT, Rosalia EK, Chen TF, Wittrup KD. Triepitopic Antibody Fusions Inhibit Cetuximab-Resistant BRAF and KRAS Mutant Tumors via EGFR Signal Repression. J Mol Biol. 2012;422:532–544. doi: 10.1016/j.jmb.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perrotte P, et al. Anti-epidermal Growth Factor Receptor Antibody C225 Inhibits Angiogenesis in Human Transitional Cell Carcinoma Growing Orthotopically in Nude Mice. Clin Cancer Res. 1999;5:257–264. [PubMed] [Google Scholar]
- 57.Artamkina GA, Egorov MP, Beletskaya IP. Some aspects of anionic. sigma.-complexes. Chem Rev. 1982;82:427–459. [Google Scholar]
- 58.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 59.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lang K, Chin JW. Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]