Abstract
Cardiac fibroblasts support heart function, and aberrant fibroblast signaling can lead to fibrosis and cardiac dysfunction. Yet how signaling molecules drive myofibroblast differentiation and fibrosis in the complex signaling environment of cardiac injury remains unclear. We developed a large-scale computational model of cardiac fibroblast signaling in order to identify regulators of fibrosis under diverse signaling contexts. The model network integrates 10 signaling pathways, including 91 nodes and 134 reactions, and it correctly predicted 80% of independent previous experiments. The model predicted key fibrotic signaling regulators (e.g. reactive oxygen species, tissue growth factor β (TGFβ) receptor), whose function varied depending on the extracellular environment. We characterized how network structure relates to function, identified functional modules, and predicted cross-talk between TGFβ and mechanical signaling, which was validated experimentally in adult cardiac fibroblasts. This study provides a systems framework for predicting key regulators of fibroblast signaling across diverse signaling contexts.
Keywords: fibroblast, signaling networks, systems biology, cardiac fibrosis
Introduction
Cardiac fibroblasts play an important role in cardiac physiology by maintaining the extracellular matrix (ECM), linking with myocytes to participate in electrical propagation, and by actin as a sentinel cell mediating response to cardiac injury[1]. These cells are critical to the heart’s ability to adapt to mechanical, chemical, and electrical changes, and dysregulation of fibroblast activity leads to cardiac pathology. Increased fibrosis in the heart is associated with tissue dysfunction such as arrhythmias, diastolic failure, and systolic failure[2], [3]. Moreover, increased ECM is an independent risk factor for the development of heart failure and is associated with a worse prognosis[4]. In a failing heart, a major source of ECM is the population of myofibroblasts – differentiated fibroblasts characterized by increased contractility (αSMA) and increased expression of collagens, fibronectin, and tissue inhibitors of matrix metalloproteinases (TIMPs), which increase the stiffness of the extracellular matrix. Identifying key drivers of this fibrotic phenotype could be the key to understanding the pathogenesis of heart failure.
Cardiac fibroblasts experience competing cues from growth factors, inflammatory cytokines and mechanical signals, among others, and integrate these diverse signals to produce increases or decreases in matrix turnover. Therefore, appropriate therapeutic strategies to modulate cardiac fibrosis must function within the rich milieu of diverse signaling cues present in the diseased heart, and designing such therapies relies on understanding how cells integrate these signals[5]. Large-scale computational models have been used to describe hypertrophic signaling in cardiac myocytes and have successfully identified signaling mechanisms and key regulatory hubs for cardiac hypertrophy[6]–[8].
In this study we developed a large-scale computational model of the cardiac fibroblast signaling network in order to identify context-dependent drivers of myofibroblast differentiation and extracellular matrix remodeling. The model integrates multiple signaling pathways in order to predict changes in gene expression and protein activity across different signaling contexts. The model identifies a context-dependent functional role for transforming growth factor β receptor (TGFβ-R) and reactive oxygen species (ROS). Additionally, TGFβ-R was found to be important for up-regulation of alpha smooth muscle actin (αSMA) under many signaling contexts. The model predicted that regulation of αSMA by TGFβ-R is dependent on the level of mechanical stimulation, and this novel cross-talk mechanism was experimentally validated in rat cardiac fibroblasts.
Materials and Methods
Model Development
A cardiac fibroblast consensus signaling network was manually reconstructed from previous experimental studies from the literature. This network integrates 10 pathways with 11 mechanical or biochemical stimuli that are altered during cardiac injury or heart failure including: IL1 (interleukin 1), IL6 (interleukin 6), TNFα (tissue necrosis factor α), NE (norepinephrine), NP (natriuretic peptide), β-integrins, TGFβ (tissue growth factor β), angiotensin II, PDGF (platelet derived growth factor), ET1 (endothelin 1), mechanical stimulation, and forskolin.
A review of the literature on cardiac fibroblast signaling was conducted, with a focus on the pathways described above. During literature review, studies were separated for use in validation (see Model Validation section below) if the cell type used was human or rat cardiac fibroblasts and the study investigated input (biochemical or mechanical stimulus) to pathway output (e.g. collagens, αSMA, cell migration, proliferation, and other ECM proteins) responses. Alternatively, studies that focused on direct signaling mechanisms were used to identify interactions to define the structure of the signaling network. Initially, interactions were added based on direct experimental evidence in mammalian cardiac fibroblasts (112 reactions). Then, we performed gap filling of each pathway with intermediate reactions (20 reactions) between those that had support in cardiac fibroblasts if they were well-characterized in other cell types and there was evidence for the interaction in a fibroblast-related cell type. Each reaction in the network is supported by two independent studies, at least one of which was performed in fibroblasts, with a majority of the reactions supported by data in cardiac fibroblasts. Extracellular interactions were included from cell-free measurements. The network includes 91 nodes (mRNA, proteins, and cell processes) connected by 142 reactions. Full documentation of the experimental evidence supporting each reaction is provided in Database S1.
The network reconstruction was converted into a predictive computational model using a previously described logic-based ordinary differential equation modeling approach used previously described[9]. Briefly, the activity of each node is modeled using a normalized Hill ODE with default parameters and logic gating. Default reaction parameters include weight (1), Hill coefficient (1.4), and EC50 (0.6), and species parameters include yinit (0), ymax (1), and τ. The τ parameter (time constant) was scaled according to the type of reaction: 6 minutes for signaling reactions, 1 hour for transcription reactions, and 10 hours for translation reactions. The system of ODEs was auto-generated from Database S1 using the Netflux software available at: https://github.com/saucermanlab/Netflux and implemented in MATLAB.
Model Validation
Literature for validating network input-output relationships (see Table S2) were identified by searching for each network input and output together with the phrase “cardiac fibroblast” in the Pubmed database. Other validation literature was identified while reviewing literature for the development of the network (see above). As a quality and reproducibility control, model validation used only studies that use rat or human cardiac fibroblasts and have at least two agreeing data points for that response (e.g., two methods of measurement, two dosages, two time points, or two independent studies). All supporting studies used in validation were independent of those used to develop the model network. Validation was performed by comparing the qualitative increase, decrease, or no change in output activity of the model simulation to the experimental results. Changes of less than 0.1% were categorized as “no change”.
Sensitivity Analysis
A systematic functional analysis was performed by simulating full knockdown of each node and predicting the change in activity of every node in the network. First the steady-state activity of all nodes was computed under baseline conditions, serving as a control. Then, we knocked down the activity of each node one at a time and subtracted the basal activity levels from the activity in the knocked down case to calculate “Δ Activity”. Influence is measured as the number of nodes with 25% change or greater in activity following knock out of the perturbed node, sensitivity is the number of nodes that will affect the target by a 25% change or greater when knocked out. The collagen sensitivity and αSMA sensitivity are defined as either the change in collagen I activity + change in collagen III activity or the change in αSMA activity respectively when the target node is knocked out. The topology of the fibroblast signaling network was analyzed using the NetworkAnalyzer plugin in Cytoscape [10], [11]. AND relationships were collapsed into their target node using MetaNodes plugin (developed by John Morris, University of California, San Francisco) and network analysis was performed on that topology. The correlation coefficient for matching topological to functional metrics was computed using the fitlm function in MATLAB.
Functional modules were identified using k-means clustering of the sensitivity analysis in the high TGFβ context. Nodes were clustered based on both influence and sensitivity by concatenating the sensitivity matrix with its transpose. Clustering was performed using MATLAB’s kmeans function using the “correlation” distance measure. The clustering was performed 20 times with different initial centroid positions and nodes were grouped into the module that most frequently appeared. The number of clusters was set at 10 because that gave the highest inter-cluster vs intra-cluster distance without having clusters of single nodes. Functional relationships between modules were derived from the high TGFβ or high mechanical stimulus sensitivity analysis (described above) by summing the influence of all nodes in one module over all nodes in the second module. The line weights indicate the sum of influence of one module over another, with the shape of the target arrow indicating whether the overall relationship is positive or negative.
Cardiac Fibroblast Isolation
Adult rat cardiac fibroblasts were isolated and cultured as previously published[12]. Briefly, Sprague-Dawley rats (6 weeks old, ~ 200g) were sacrificed and the ventricles removed, minced into ~1 mm pieces, and digested using Liberase Blendzyme 3 (Roche, Indianapolis, IN). Successive digestions were centrifuged for 10 min at 400×g and cells were resuspended into culture medium containing Dulbecco’s modified Eagle medium (Sigma-Aldrich, St. Louis, MO) with 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA), 100 U/mL penicillin, 100 g/mL streptomycin, and 2 ng/mL amphotericin B (all Sigma-Aldrich). After incubating in culture flasks for 4 hrs at 37 C and 5% CO2, flasks were rinsed with phosphate-buffered saline (PBS, Sigma-Aldrich) to remove nonadherent cells, and resupplied with culture medium.
After 7 d of culture, fibroblasts were removed from flasks with 0.25% Trypsin-EDTA (Sigma-Aldrich), and seeded into 3D collagen gels as previously published[12]. Briefly, 0.2 M HEPES (Sigma-Aldrich), 10X MEM (Sigma-Aldrich), 3 mg/mL type I bovine collagen (PureCol, Advanced Biomatrix, San Diego, CA) and cells resuspended in low-serum culture medium (1% FBS) at respective ratios of 1:1:8:2 to yield a final collagen concentration of 2 mg/mL and final cell concentrations of 200k cells/mL (for restrained gel conditions) or 133k cells/mL (for floating gel conditions). The cell+collagen gel mixtures were rotated in an incubator for 5 min, then pipetted into 24-well plates (1mL in each well).
In order to apply a high mechanical stimulus cells were seeded into a collagen gel restrained at the boundary, and compared to a free-floating gel (low mechanical stimulus). Restrained gels were poured into non-treated wells and remained adhered to the well bottom and sides; floating gels were poured into wells pre-coated with bovine serum albumin (BSA, Sigma-Aldrich) by incubation in 2% BSA for 1 hr. After 4 hrs of incubation, the floating gels were released from the bottom of the wells with the addition of low-serum culture medium (1% FBS). All gels were then incubated for 2 d in low-serum medium. After 2 d, gels were cultured for an additional 2 d in one of three chemical conditions: low-serum culture medium control, TGFβ-inhibitor treatment (30nM of SD208, Sigma-Aldrich), or TGFβ treatment (100 ng/mL of human TGFβ1, Cell Signaling Technology, Danvers, MA).
Gel Compaction Measurements
Starting immediately after floating the gels, pictures of the floating gels were taken every 24 hrs with a handheld digital camera. Gel outlines were manually traced using ImageJ[13], and relative gel compaction was assessed as the ratio of the area of each gel at a given time point to the initial area of that gel.
Microscopy and Image Analysis
After a total of 4 days of culture, gels were fixed overnight in 4% paraformaldehyde (Sigma-Aldrich), and washed 3× with PBS; cells were then permeabilized in 0.05% TritonX (Sigma-Aldrich) in 1% BSA overnight, stained with monoclonal anti-alpha smooth muscle actin (Sigma-Aldrich) overnight, washed 3× with PBS, stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Life Technologies, Carlsbad, CA), and washed again 3× with PBS. PBS was removed and gels were imaged on an Olympus IX81 inverted microscope with a 10× UPlanSApo 0.40 NA objective (Olympus, Center Valley, PA) and a C9300 cooled CCD digital camera (Hamamatsu, Bridgewater, NJ). An 800 μm × 600 μm area in the central region of every gel was scanned, capturing at least 100 cells per gel.
To quantify αSMA expression, an automated image analysis pipeline was employed in CellProfiler (Broad Institute)[14], [15]. Fibroblast nuclei were identified by DAPI signal, and fibroblast boundaries corresponding to each nuclei were segmented based on the αSMA signal using the “propagate” algorithm. αSMA signal was integrated within each cell’s boundary, and then averaged across all cells in a given gel as a measure of average αSMA expression per cell for that particular gel condition.
Statistics
Fibroblasts were isolated from 7 different rats, each isolation was divided into 18–24 gels, and gels were divided into six experimental groups for a total of 3–4 gels per group per rat (150 gels total, 25 gels per experimental group). αSMA was averaged across the gels within each group and rat, yielding N=7 replicates (one for each rat isolation) across the six experimental conditions. We performed a two-way ANOVA on floating-baseline, floating-SD208, restrained-baseline, and restrained-SD208 groups with post-hoc Bonferroni tests comparing floating-baseline to restrained-baseline, and comparing restrained-baseline to restrained-SD. For the gel compaction assay, we performed a Student’s t-test between floating-control vs. floating-SD208 and between floating-control vs. floating-TGFβ groups with Bonferroni adjustments. Statistical significance was set at p<0.05.
Results
A predictive computational model of cardiac fibroblast signaling
A cardiac fibroblast consensus signaling network was manually reconstructed from previous experimental studies from the literature. Literature papers on cardiac fibroblast signaling were placed into distinct “model development” and “model validation” groups, depending on whether that paper described direct molecular interactions (e.g. smad3 binds to the collagen I promoter) or network input-output relationships (e.g. TGFβ induces collagen I protein expression in cardiac fibroblasts), respectively. The 177 papers in the “model development” literature group were used to define the structure of the cardiac fibroblast signaling network, while the 41 papers in the “model validation” literature group were used to validate model predictions of network function. The detailed procedure for literature review and network reconstruction is provided in Methods.
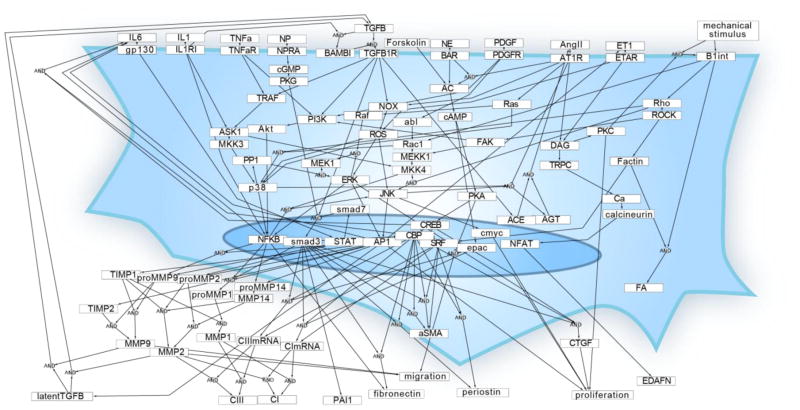
This cardiac fibroblast signaling network (Fig 1) integrates ten signaling pathways previously shown to regulate cardiac fibroblast phenotypes and are up- or down-regulated during cardiac injury or heart failure. The network includes 91 nodes (mRNA, proteins, and cell processes) connected by 142 reactions. Full documentation of the experimental evidence supporting each reaction is provided in Database S1.
Fig. 1. Reconstruction of the cardiac fibroblast signaling network.
Each of the 91 nodes represents a gene product, modification of a gene product, or cell process in the model. Each arrow indicates a reaction based on experimental data of activation or inhibition from cardiac fibroblasts or a fibroblast-related cell line (142 reactions from 177 papers). Where shown, some reactions combine the influence of multiple reactants via AND gate logic. Multiple reactions affecting the same product are combined using OR gate logic.
The network reconstruction was then converted into a predictive computational model using a logic-based ordinary differential equation (ODE) approach that we described previously [9], [16]. Briefly, the normalized activity of each node is modeled using ordinary differential equations, with reactions modeled using saturating Hill functions and continuous OR/AND logic gates. As in previous network models [9], [16], uniform default parameters were used, except that time constants (τ) were scaled to an order of magnitude appropriate for the type of molecule (mRNA, protein, process; see Methods). The baseline condition was defined as 25% signaling activity for all inputs, which represents fibroblasts cultured on a stiff substrate with ligands at basal constitutive levels. Given any combination of the 11 signaling inputs, the model can simulate the dynamic changes in activity for every node in the network.
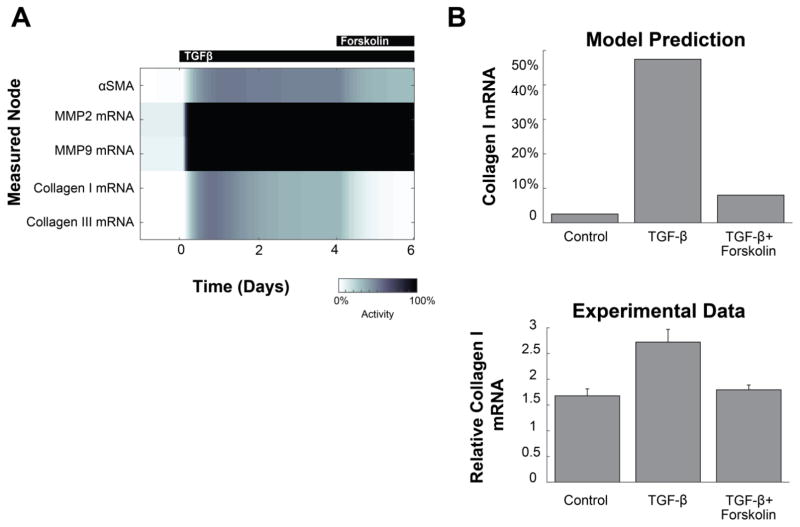
Next, we predicted responses of the fibroblast signaling network to specific stimuli. These predictions were validated against experimental studies performed in rat or human cardiac fibroblasts that were independent from those studies used to reconstruct the signaling network. For example, the effect of a 4-day TGFβ stimulus followed by a 2-day TGFβ + forskolin stimulus was simulated and compared to experimental data from Lu et al [17] (Fig. 2A, with full simulation in Fig. S1). The model predicted that the addition of TGFβ initially increases collagen I mRNA, but forskolin treatment during the last two days partially reverses this increase. This prediction is qualitatively consistent with published data from rat cardiac fibroblasts showing that forskolin attenuates TGFβ-dependent expression of collagen I [17] (Fig. 2B).
Fig. 2. Example of model validation with combined stimuli.
(A) Predicted dynamics of selected outputs in response to TGFβ, followed by a combined TGFβ + forskolin stimulus. Full dynamic prediction shown in Fig. S1 (B) The model prediction is compared to independent experimental data from Lu et al 2013 [17], showing the attenuation of collagen I mRNA by forskolin treatment. The model prediction is expressed as percent of maximal mRNA level. Experimental collagen mRNA is relative to the initial measurement at day 0.
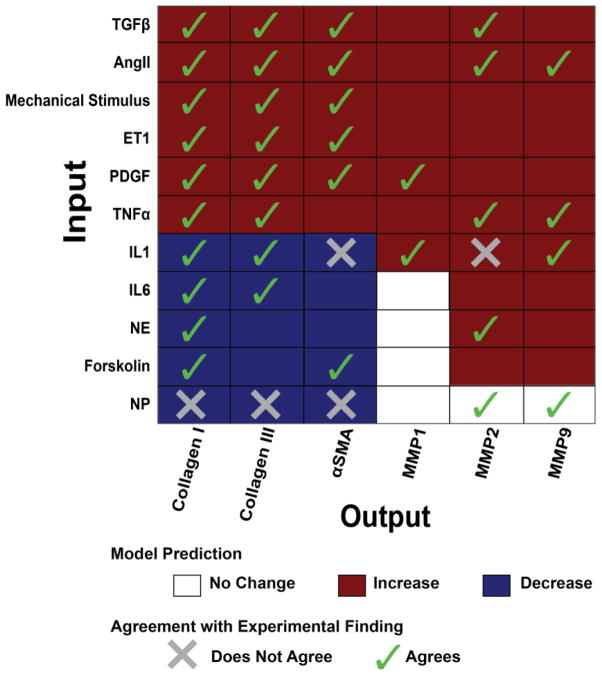
Overall, the model was validated against 82 input-output relationships from 34 papers (see Methods) and accurately predicts 66 of those 82 (80%). Fig. 3 summarizes the predicted relationship of each individual input stimulus to the outputs collagen I, collagen III, αSMA, and the MMPs (matrix metalloproteinases) and the agreement between model predictions and experimental data where available (40 relationships). Validations for the other 42 input-output relationships are shown in Fig. S2, with complete annotation in Database S2. The validation accuracy was robust to a ±50% change in the baseline input levels as shown in Fig. S3.
Fig. 3. Validation of network input-output relationships predicted by the model.
The qualitative response of selected fibrosis-related outputs is shown in response to each of 11 input stimuli. Agreement or disagreement with independent experimental data when available from the literature is indicated as a check or an X, respectively. The model validates 35 of the 40 (88%) predictions shown in this Figure that have experimental data. Overall, the model validates 66 of 82 comparisons (80%), as shown inFigure S2andDatabase S2.
Context-Dependent Roles of Cardiac Fibroblast Signaling Drivers
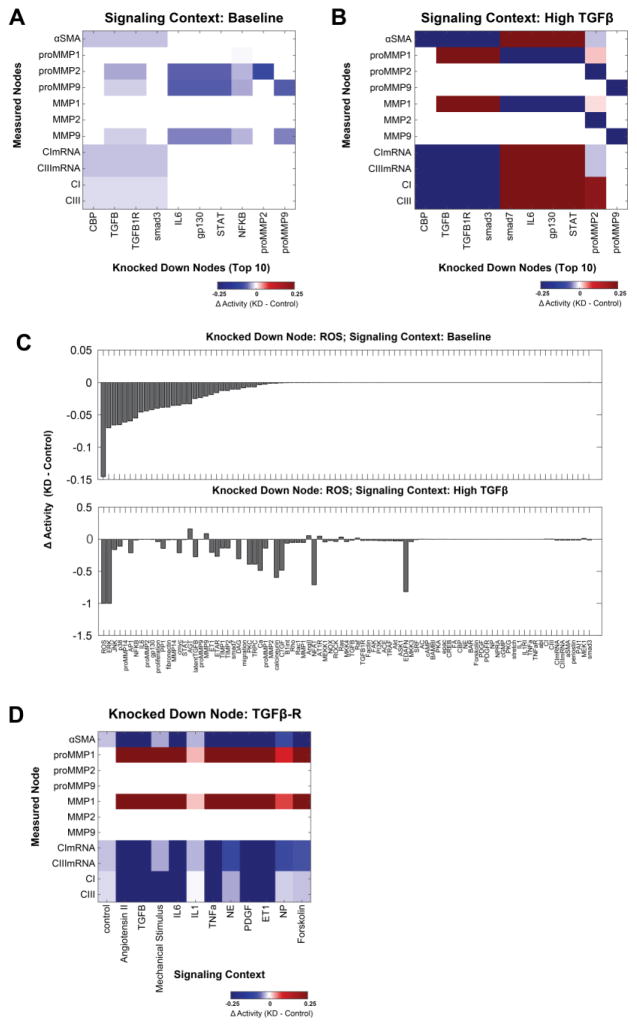
Sensitivity analysis is one way to systematically characterize the functional roles of nodes in a signaling network. We first performed sensitivity analysis under baseline conditions (all inputs at 25%) by simulating complete knockdown of each node in the fibroblast network and quantifying the change in activity of all network nodes in response to each knockdown (Fig. S4). From this analysis, we identified the most influential nodes as those whose knockdown produced the greatest summed magnitude of change in the phenotypic outputs of the network. Fig 4A shows how knockdown of these 10 most influential nodes affected the outputs under baseline conditions. For example, knockdown of interleukin 6 (IL6) was predicted to strongly suppress expression of pro-MMP14 and pro-MMP2, consistent with Dawn et al and Luckett et al [18], [19].
Fig. 4. Sensitivity analysis reveals context-dependent functional roles for regulators of cardiac fibroblast signaling.
Systematic knockdown (KD) simulations (see Fig. S2) revealed that the top 10 most influential differed considerably between (A) baseline and (B) high TGFβ signaling contexts. At baseline all inputs are set to 25%, while for high TGFβ that input is further increased to 90% (see Methods). (C) The response of network nodes to ROS knockdown differs substantially between baseline and high TGFβ contexts. Nodes are rank-ordered by change in activity in the baseline context (note the difference in y-axis scales in upper and lower panels). (D) The effect of TGFβ-R KD on fibrosis-related model outputs varies across baseline and all 11 single-input contexts.
As TGFβ is a well-studied growth factor that is elevated following myocardial infarction [17], [20], we repeated the sensitivity analysis in a high TGFβ context (TGFβ input weight set to 90%, all other inputs at 25%) (Fig. S4B). The role of influential nodes on phenotypic outputs differed substantially between the baseline and high TGFβ contexts (Fig. 4A and B). For example, in the baseline condition proMMP2 and proMMP9 are sensitive to knockdown of IL6 pathway members. However in the high TGFβ context, IL6 pathway members were predicted to regulate proMMP1 but proMMP2 or proMMP9. We also identified key regulators of the overall network. While knockdown of TGFβ receptor (TGFβR) and ROS had broad network effects in both baseline and high TGFβ conditions (Fig. S4), their influence on specific network nodes was highly context-dependent (Fig. 4C). For example, ROS knockdown decreased MMP9 expression under baseline conditions but increased MMP9 activity in the high TGFβ context.
To more fully profile the context-dependent influence of the TGFβ receptor and identify cross-talks between pathways, TGFβR knockdown was simulated in all 12 possible single-stimulus signaling contexts (90% activity of each stimulus, 25% activity of all other inputs). The network response to TGFβR knockdown varied considerably across the 12 signaling contexts (Fig. 4D). In particular, knockdown of TGFβR decreased expression of collagen I, collagen III, and αSMA in all single-stimulus contexts, but to different magnitudes in each context. TGFβR knockdown caused increases in periostin expression in the high NE or high forskolin signaling contexts but decreased expression in the 10 other contexts. Together, these analyses highlight the ability of the model to make predictions about how the influence of regulatory nodes in a signaling network vary as a function of the cell’s environment.
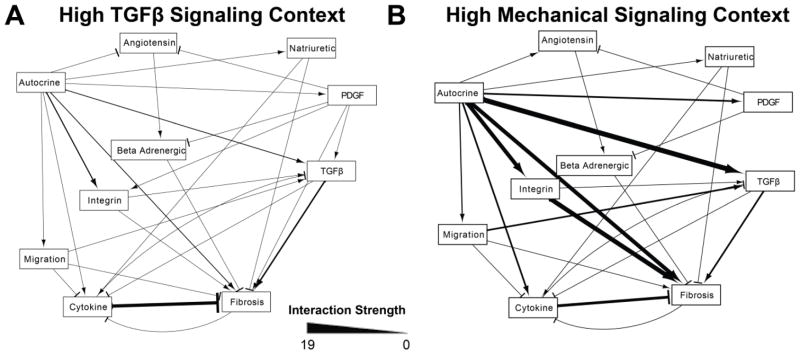
Signaling nodes that have similar function within a network are often thought to form modules which maintain biological robustness and allow for signaling flexibility [21]. Identification of network modules would allow for development of a hierarchical understanding of network function. To predict functional modules in the cardiac fibroblast network, we initially clustered network nodes based on both influence and sensitivity in the baseline signaling context (from Fig. S4A) using k-means clustering. However, we found that clustering using sensitivity analysis from baseline conditions was highly variable due to many signaling nodes having relatively low influence or sensitivity. Therefore, we clustered nodes into functional modules based on both influence and sensitivity in the high TGFβ context (Table 1 and Fig. S5) and computed the strength of functional relationships between modules by summing the influence all nodes in one module had over another, as shown in Fig. 5A.
Table 1. Model-predicted functional modules.
The members of each functional module identified using k-means clustering of the high TGFβ sensitivity analysis.
| Module | Members |
|---|---|
| PDGF | PDGF, PDGFR, TNFα, TNFαR, p38, PP1, JNK, abl, cmyc |
| Autocrine | ROS, ET1, ETAR, DAG, TRPC, latentTGFβ, Ca, calcineurin, NFAT, ERK, EDAFN, AP1, TIMP1, TIMP2 |
| Migration | Migration, proMMP14, proMMP2, MMP2, MMP14 |
| Natriuretic | NP, NPRA, cGMP, PKG, proliferation |
| Cytokine | smad7, BAMBI, IL6, gp130, STAT, IL1, IL1RI, NFκB, proMMP1, MMP1, fibronectin |
| Mechanical | PKC, mechanical stimulus, β1int, Rho, ROCK, Rac1, MEKK1, FAK, Factin, FA, SRF |
| TGFβ | ACE, NOX, TGFβ, TGFβ-R, PI3K, Akt, TRAF, ASK1, MKK3, MKK4 |
| Angiotensin | AngII, AT1R, AGT, Ras, Raf, MEK1, proMMP9, MMP9 |
| Beta Adrenergic | NE, BAR, forskolin, AC, cAMP, PKA, CREB, epac |
| Fibrosis | CBP, smad3, CTGF, αSMA, PAI1, periostin, CImRNA, CIIImRNA, CI, CIII |
Fig. 5. Relationships between functional modules are context-dependent.
Relationships between functional modules (from Table 2) were quantified by the sum of influence of the members of one module over another in the specified signaling context. A) In the high TGFβ context, the fibrosis module was positively regulated by the TGFβ module and negatively regulated by the cytokine module. B) In the high mechanical signaling context, the autocrine module became a prominent regulator of the mechanical and fibrosis modules.
Because relationships between modules can vary depending on the signaling context [21], we also computed the relationships between functional modules in a high mechanical stimulus context that mimics the mechanical environment during myocardial infarction or volume overload. Fig. 5 compares the relationships between functional modules in the high TGFβ and high mechanical stimulus contexts. This analysis indicated that in conditions of high TGFβ, the TGFβ module promotes and the cytokine module strongly inhibits activation of the fibrosis module, which contains network outputs such as expression of collagen I, collagen III, and αSMA. Intriguingly, the autocrine module became more influential in in the high mechanical stimulus context, predicting an important role for autocrine signals that amplify the fibrotic response to integrin stimulation.
Relationship Between Network Structure and Function
While the above analyses used model simulations to predict function of nodes in the fibroblast signaling network, an alternative approach is to estimate function based on metrics of network topology [22]. Highly connected nodes, as determined by the topology metrics defined in Table 2, are generally expected to be more influential in a network[23], [24]. For example the fibroblast network contains 5 network hubs, defined as nodes with 8+ edges: AP1, smad3, NFκB, CBP, and p38. Topological analysis has been most often applied to large-scale biological networks where a predictive computational model is not available[23], [25]. However, the availability of this large-scale predictive signaling model provides a unique opportunity to examine the relationships between signaling network structure and function.
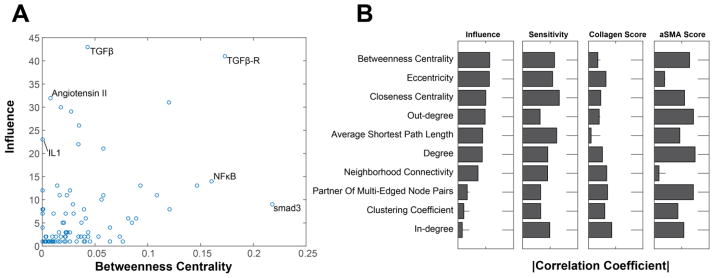
Accordingly, we examined the relationship between metrics of network structure and function as predicted by sensitivity analysis of the model under baseline conditions (from Fig. S2). These functional metrics were: 1) influence, the number of nodes with an activity change of greater than 25% with knockdown of node n; 2) sensitivity, the number of nodes that change the activity of node n by more than 25% when knocked down; 3) collagen sensitivity, the sum of the absolute value of the change in collagen I and collagen III with knockdown of node n; and 4) αSMA sensitivity, the absolute value of the change in αSMA with knockdown of node n. Betweenness centrality, defined as the number of shortest paths from all nodes to all other nodes that pass through node n, is one topological measure of connectivity. As shown in the comparison of betweenness centrality with influence (see Fig. 6A), a few nodes such as the TGFβR had both high topological and functional scores. Yet betweenness centrality was a poor predictor of influence for other nodes such as angiotensin II (underestimating influence) and smad3 (overestimating influence), with only moderate correlation overall (r = 0.64). A similar analysis was performed for all 10 topological metrics compared to influence (Fig. S6), sensitivity, collagen sensitivity, and αSMA sensitivity (Fig. 6B). Overall, functional features were not strongly correlated with topological features, indicating the additional need for predictive signaling models as developed here. Betweenness centrality was the most useful topological metric for predicting overall network influence, while measures of degree (in-degree, out-degree, and edge count, see Table S1) were the most useful for predicting influence over the phenotypic outputs collagen and αSMA.
Fig. 6. Metrics of network topology are insufficient to predict network functions.
(A) Scatter plot showing the relationship of betweenness centrality (a metric of network topology) vs influence (predicted by the model). Influence is calculated by summing the absolute value of the changes in activity with knockdown of the target node. Several nodes of interest have been labeled. (B) The correlation coefficient for each topological feature vs 4 functional features. Sensitivity is the absolute value of the change in activity of the target node for all possible knockdowns. Collagen sensitivity and αSMA sensitivity are the change in collagen or αSMA respectively with knockdown of the target node.
Cross-Talk Between Mechanical and TGFβ Pathways
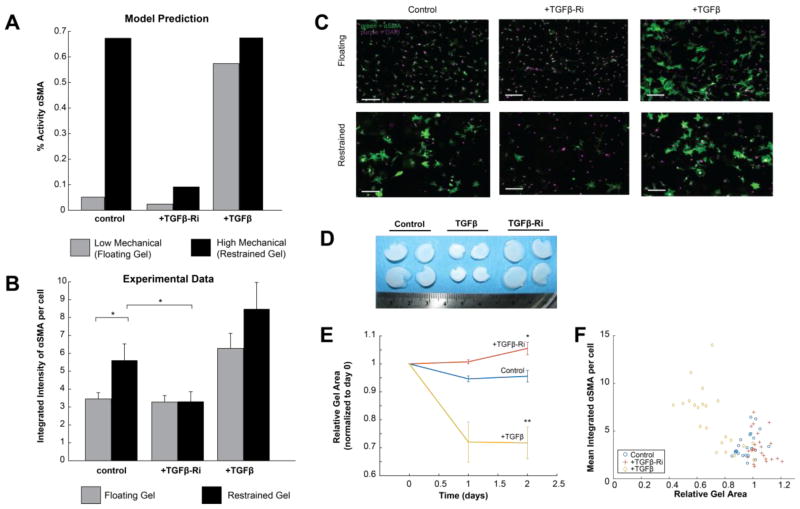
The sensitivity and clustering analyses described above suggested substantial crosstalk between the mechanical stimulus pathway and the TGFβ pathway. Simulated knockdown of the TGFβ receptor lowered expression of αSMA, collagen I, and collagen III in conditions of high mechanical stimulus (Fig. 4D). Furthermore, at a more course-grained level, the TGFβ module was an important regulator of the fibrosis module (which contains important output genes such as collagen I, collagen III, and αSMA) in conditions of both high TGFβ and mechanical stimulus (Fig. 5). This led us to further investigate the potential role for TGFβ in integrin-mediated differentiation of fibroblasts to myofibroblasts. The model predicted that inhibition of the TGFβ receptor would have little effect on αSMA expression in baseline conditions but would attenuate mechanical-induced αSMA expression (Fig. 7A). To experimentally validate this prediction, we cultured rat cardiac fibroblasts in floating and mechanically restrained collagen gels, with and without a TGFβ receptor inhibitor, SD-208 (see Methods). The restraint boundary condition provides mechanical resistance to intrinsic cell contractile forces, enabling cells to produce higher contractile tension and higher corresponding reaction tension in the gel[26]. This restraint has been shown to activate integrin pathways[27]. TGFβ was used as a positive control. As shown in Fig. 7B–C, fibroblasts in the restrained gels had significantly increased αSMA expression, but SD-208 significantly attenuated the expression of αSMA in the restrained gels. Inhibition of the TGFβ receptor in the floating gels did not significantly reduce αSMA expression. Further, the expression of αSMA in floating gels strongly correlated with the degree of gel compaction, a functional measure of cardiac fibroblast contraction (Fig. 7D–E). Together, these experiments semi-quantitatively validate the model prediction that the TGFβ receptor is an important regulator of mechanical-mediated myofibroblast differentiation.
Fig. 7. Experimental validation of predicted role for TGFβ in mechanical-induced expression of αSMA.
(A) The model predicted that high mechanical input would increase αSMA expression, but that this effect would be mitigated by a TGFβ-R inhibitor. TGFβ increased αSMA at both low and high mechanical input. (B) Experimental measurements of αSMA expression as measured by immunofluorescence in adult rat cardiac fibroblasts cultured in floating or restrained gels, in which fibroblasts experienced increased mechanical stimulation. As predicted by the model, restrained gels exhibited increased αSMA expression, which was mitigated by the TGFβ-R inhibitor (TGFβ-Ri) SD208. (C) Example images of cardiac fibroblasts cultured in restrained and floating gels, stained for αSMA (green) and DAPI (purple). Scale bar = 100 μm. (D) Increased compaction of floating gels treated with TGFβ but not with TGFβ-Ri. (E) Floating gels compact over time with TGFβ and become more relaxed over time after treatment with TGFβ-Ri. (F) The final size of floating gels was inversely correlated with αSMA expression. * indicates p<0.05, and ** indicates p<0.01. All error bars indicate standard error of the mean.
Discussion
Here we manually reconstructed a literature-based network of cardiac fibroblast signaling. This network was used to develop a logic-based predictive model of fibroblast signaling, which validated at a rate of 80% in comparison to independent, published studies in cardiac fibroblasts. A comprehensive sensitivity analysis revealed the context-dependent functional roles of nodes in the network, such as ROS and the TGFβ receptor. Betweenness centrality was the topological metric that was most predictive of functional influence, but overall there was a low correlation between topological and functional characteristics. The model predicted substantial crosstalk between TGFβ- and mechanical-induced myofibroblast differentiation, and this prediction was experimentally validated in rat cardiac fibroblasts.
Model validation
While the model validates 80% of input-output relationships for which there is independent data, 16 input-output relationships were incorrectly predicted by the model. Most incorrect predictions were in response to 3 inputs: NP (6), NE (4), and IL1 (4). For example, the model predicted some responses to NP and IL1 where no change was reported experimentally. As NP counteracts fibrotic stimuli, those pathways may have a lower baseline activation than modeled currently. For IL1, validation data exhibited changes mRNA that were either not statistically significant [28] or did not propagate to protein expression as predicted by the model [29]. NE and forskolin both stimulate cAMP but have distinct effects[20], [30], indicating cAMP-independent roles of NE. However these are not yet sufficiently characterized for inclusion in the model. Together, these incorrect predictions highlight areas for future model revision and experiments.
Structure-Function Relationships in a Large Signaling Network
There are several approaches for using biological network reconstructions to identify key regulators of cell signaling. One way to predict the influence of a given node is through network topology analysis. Generally, well-connected nodes (those with high degree or betweenness centrality) are more likely to be essential nodes in the network[23], [24]. We found that, although betweenness centrality was most strongly correlated with influence, topological features were not strongly predictive of functional influence as determined by sensitivity analysis of the logic-based model. This finding is in agreement with other studies which found degree was not able to fully predict essentiality in signaling and metabolic networks[31], [32]. Topological metrics are simplified measures of connectivity, whereas the model utilizes the entire network structure to make functional predictions. This finding argues for the need for large-scale predictive network models as in this study rather than relying on simplified measures of connectivity of individual nodes to identify potential signaling drivers and therapeutic targets.
Context-Dependent Roles of Signaling Molecules
Cardiac fibroblasts play diverse functional roles in sensing and contributing to inflammation, remodeling extracellular matrix, and mediating wound healing. As a result their cellular signaling is highly context-dependent, which has implications for the effect of targeted therapy against fibrosis under these different signaling contexts. The large-scale model provides a unique opportunity to investigate context-dependent signaling roles in the cardiac fibroblast signaling network. TGFβ is known to be up-regulated following cardiac injury and in heart failure, and in vitro it has been established as a strongly pro-fibrotic stimulus on cardiac fibroblasts[17], [20], [33]. Anti-oxidants that suppress ROS have been shown to decrease fibrosis following myocardial infarction and prevent cardiac dilation [34], [35]. In conditions of high TGFβ, the model predicted that suppressing ROS would produce a larger decrease in the TGFβ and ET1 autocrine feedback loops than in the baseline signaling context. Additionally, simulations of ROS suppression predicted decreases in collagen and αSMA activity in the high TGFβ context, consistent with previous studies[36]. Interestingly, ROS suppression in the baseline signaling context decreased MMP-9 activity whereas it increased MMP-9 activity in the high TGFβ context. This is likely due to the effect of ROS knock down on TIMP activity as MMP-9 mRNA levels were predicted to increase with ROS knock down regardless of the signaling context. This has implications for the treatment of heart failure-associated fibrosis with antioxidants as the model predicts antioxidants will be more effective in treating fibrosis under a high TGFβ signaling context (e.g. near a myocardial infarct) than in a baseline context (e.g. in the remote zone).
Additionally, the TGFβ receptor, which is directly linked to only the TGFβ pathway, was shown to be highly influential in both the baseline context and the high TGFβ context. For this reason, we investigated the role of the TGFβ receptor and, by extension, the involvement of the TGFβ pathway, under the baseline context and all 11 single-input contexts. We found that the TGFβ-R functions to increase collagen I, collagen III, and αSMA under all single-stimulus contexts, but the magnitude of the increase depends on the context. In contrast, the TGFβ-R was predicted to up- or down-regulate periostin in a context-dependent manner. Blocking the TGFβ-R was predicted to decrease periostin under 10 of 12 signaling contexts, but TGFβ-R knockdown was predicted to increase periostin expression in contexts of high β-adrenergic or high forskolin signaling. Together these data demonstrate how a large-scale model that incorporates multiple pathways can be useful for interrogating how fibroblasts respond to different signaling contexts. These results also have implications for how cells in different signaling environments might respond differently to antioxidants (above) or to TGFβ receptor inhibitors. Future studies can use this model to better understand how fibroblasts respond to more complex signaling contexts such as combinatory- or dynamic-stimulus contexts and varied doses of inputs.
Cross-Talk and the Effect on Phenotype
Hypertension is a risk factor for the development of cardiac fibrosis, and understanding how cross-talk between mechanical and chemical stimuli affects the development of a pro-fibrotic phenotype could reveal possible mechanisms of pathogenesis. The model predicted a role for the TGFβ-R in up-regulating collagen and αSMA under a high mechanical stimulus. Therefore we tested this prediction using mechanically restrained or floating gels in order to activate the integrin pathway downstream of mechanical stimulus in the model[27]. Experimentally, we found that the TGFβ-R inhibition abrogates mechanical-induced αSMA up-regulation, validating the model’s prediction. To our knowledge this relationship has not been shown previously, reinforcing the value of large-scale modeling to elucidate novel signaling mechanisms via signal cross-talk. The precise mechanism by which the TGFβ pathway amplifies myofibroblast differentiation in response to integrin stimulation requires further investigation. The model predicts that an autocrine loop involving an increase in TGFβ expression is responsible for sensitizing the fibroblast to differentiation from multiple stimuli including mechanics, angiotensin II and ET1 (data not shown). However, stretch of extracellular matrix has also been shown to increase activation of extracellular stores of latent TGFβ[37]. For example, Sarrazy et al. demonstrated that integrins activate latent TGFβ[38]. Both of these are testable potential mechanisms underlying this cross-talk.
Limitations
As with all modeling approaches, our logic-based ODE approach has inherent limitations. While this model uses default parameters, we have previously shown that this approach still exhibits strong predictive accuracy in comparison to a fully parametrized biochemical model [16]. Further, the model’s validation and predictions are robust to parameter variation (Fig. S3). Availability of more quantitative proteomic data could increase the quantitative and dynamic predictive power of the model. The model structure is not fully comprehensive, focusing instead on the consensus cardiac fibroblast signaling network that meets specified inclusion criteria. However, this provides a framework for future expansion based on new experimental data.
Conclusions
We developed a predictive model of cardiac fibroblast signaling through manual curation of a signaling network, combining 10 pathways that are altered during cardiac injury or heart failure. Sensitivity analysis identified key signaling drivers of fibroblast function, and showed that these drivers vary across diverse signaling contexts. Specifically, TGFβ and ROS were key drivers of fibrosis signaling under both the baseline and high TGFβ context, but their relative effect on different nodes in the network was context-specific. The model also predicted a role for TGFβ in amplifying myofibroblast differentiation and expression of extracellular matrix proteins in response to other signals such as mechanical stimulation. The role for the TGFβ-R in mechanical stimulation-induced αSMA expression was validated experimentally. More generally, we found that functional influence and topological features are not well correlated, revealing the limited ability of topological analysis to predict functionality within a signaling network. The large-scale network modeling approach utilized here enables the prediction of global features of signaling networks that are often non-intuitive from local topological connections alone.
Supplementary Material
Figure S1: Full simulation of TGFβ vs forskolin treatment
Figure S2: Other validated relationships
Figure S3: Robustness of validation accuracy to changes in baseline input levels
Figure S4: Full sensitivity analysis
Figure S5: Clustering analysis
Figure S6: Full comparison of topological features vs influence
Table S1: Definitions of network topology metrics
Database S1: Network model
Database S2: Validation relationships
Highlights.
A computational model of cardiac fibroblast signaling was built and validated.
This model predicted a context-dependent role for regulators of differentiation.
Topological metrics could not fully predict function of fibroblast network nodes.
This model identified novel cross-talk between mechanical and TGFβ signaling.
Acknowledgments
We thank Kellen Chen for assisting in cardiac fibroblast isolation.
Funding: This study was supported by the National Institutes of Health (HL007284, HL094476, HL116449), and an American Heart Association Post-doctoral Fellowship (AHA 14POST20460271).
Footnotes
Author contributions: ACZ developed the computational model and performed all validation and computational analysis. WJR performed all in vitro experiments and analysis. All authors contributed to study design and wrote the manuscript.
Disclosures: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Souders CA, Bowers SLK, Baudino TA. Cardiac fibroblast: The renaissance cell. Circulation Research. 2009;105(12):1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreo A, Ambrosio G, De Chiara B, Pu M, Tran T, Mauri F, Raman SV. Influence of myocardial fibrosis on left ventricular diastolic function: noninvasive assessment by cardiac magnetic resonance and echo. Circ Cardiovasc Imaging. 2009;2(6):437–443. doi: 10.1161/CIRCIMAGING.108.838367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong TC, Piehler K, Meier CG, Testa SM, Klock AM, Aneizi AA, Shakesprere J, Kellman P, Shroff SG, Schwartzman DS, Mulukutla SR, Simon MA, Schelbert EB. Association between extracellular matrix expansion quantified by cardiovascular magnetic resonance and short-term mortality. Circulation. 2012;126(10):1206–1216. doi: 10.1161/CIRCULATIONAHA.111.089409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masci PG, Doulaptsis C, Bertella E, Del Torto A, Symons R, Pontone G, Barison A, Droogné W, Andreini D, Lorenzoni V, Gripari P, Mushtaq S, Emdin M, Bogaert J, Lombardi M. Incremental prognostic value of myocardial fibrosis in patients with non-ischemic cardiomyopathy without congestive heart failure. Circ Hear Fail. 2014;7(3):448–456. doi: 10.1161/CIRCHEARTFAILURE.113.000996. [DOI] [PubMed] [Google Scholar]
- 5.Zeigler AC, Richardson WJ, Holmes JW, Saucerman JJ. Computational modeling of cardiac fibroblasts and fibrosis. J Mol Cell Cardiol. 2015 Nov; doi: 10.1016/j.yjmcc.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryall Ka, Holland DO, Delaney Ka, Kraeutler MJ, Parker AJ, Saucerman JJ. Network reconstruction and systems analysis of cardiac myocyte hypertrophy signaling. J Biol Chem. 2012;287(50):42259–68. doi: 10.1074/jbc.M112.382937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryall Ka, Bezzerides VJ, Rosenzweig A, Saucerman JJ. Phenotypic screen quantifying differential regulation of cardiac myocyte hypertrophy identifies CITED4 regulation of myocyte elongation. J Mol Cell Cardiol. 2014 Jul;72:74–84. doi: 10.1016/j.yjmcc.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang JH, Saucerman JJ. Computational models reduce complexity and accelerate insight into cardiac signaling networks. Circulation Research. 2011;108:85–97. doi: 10.1161/CIRCRESAHA.110.223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryall KA, Holland DO, Delaney KA, Kraeutler MJ, Parker AJ, Saucerman JJ. Network reconstruction and systems analysis of cardiac myocyte hypertrophy signaling. J Biol Chem. 2012 Dec;287(50):42259–68. doi: 10.1074/jbc.M112.382937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assenov Y, Ramírez F, Schelhorn SESE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 12.Thomopoulos S, Fomovsky GM, Holmes JW. The development of structural and mechanical anisotropy in fibroblast populated collagen gels. J Biomech Eng. 2005;127(5):742–750. doi: 10.1115/1.1992525. [DOI] [PubMed] [Google Scholar]
- 13.Schneider Ca, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin Da, Chang JH, Lindquist Ra, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for cellprofiler: Modular high-throughput image analysis software. Bioinformatics. 2011;27(8):1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraeutler MJ, Soltis AR, Saucerman JJ. Modeling cardiac β-adrenergic signaling with normalized-Hill differential equations: comparison with a biochemical model. BMC Syst Biol. 2010 Jan;4(1):157. doi: 10.1186/1752-0509-4-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu D, Aroonsakool N, Yokoyama U, Patel HH, Insel Pa. Increase in cellular cyclic AMP concentrations reverses the profibrogenic phenotype of cardiac myofibroblasts: a novel therapeutic approach for cardiac fibrosis. Mol Pharmacol. 2013;84(6):787–93. doi: 10.1124/mol.113.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawn B, Xuan YT, Guo Y, Rezazadeh A, Stein AB, Hunt G, Wu WJ, Tan W, Bolli R. IL-6 plays an obligatory role in late preconditioning via JAK-STAT signaling and upregulation of iNOS and COX-2. Cardiovasc Res. 2004;64(1):61–71. doi: 10.1016/j.cardiores.2004.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luckett LR, Gallucci RM. Interleukin-6 (IL-6) modulates migration and matrix metalloproteinase function in dermal fibroblasts from IL-6KO mice. Br J Dermatol. 2007;156(6):1163–1171. doi: 10.1111/j.1365-2133.2007.07867.x. [DOI] [PubMed] [Google Scholar]
- 20.Swaney JS, Roth DM, Olson ER, Naugle JE, Meszaros JG, Insel PA. Inhibition of cardiac myofibroblast formation and collagen synthesis by activation and overexpression of adenylyl cyclase. Proc Natl Acad Sci U S A. 2005;102(2):437–442. doi: 10.1073/pnas.0408704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 22.Albert R. Network inference, analysis, and modeling in systems biology. Plant Cell. 2007;19(11):3327–3338. doi: 10.1105/tpc.107.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118(Pt 21):4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3(4):713–720. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pržulj N. Protein-protein interactions: Making sense of networks via graph-theoretic modeling. BioEssays. 2011;33(2):115–123. doi: 10.1002/bies.201000044. [DOI] [PubMed] [Google Scholar]
- 26.John J, Throm Quinlan A, Silvestri C, Billiar K. Boundary stiffness regulates fibroblast behavior in collagen gels. Annals of Biomedical Engineering. 2010;38(3):658–673. doi: 10.1007/s10439-009-9856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeldt H. Fibroblast Quiescence and the Disruption of ERK Signaling in Mechanically Unloaded Collagen Matrices. J Biol Chem. 2000 Feb;275(5):3088–3092. doi: 10.1074/jbc.275.5.3088. [DOI] [PubMed] [Google Scholar]
- 28.Turner Na, Warburton P, O’Regan DJ, Ball SG, Porter KE. Modulatory effect of interleukin-1α on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: role of p38 MAP kinase. Matrix Biol. 2010;29(7):613–20. doi: 10.1016/j.matbio.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Nieuwenhoven FA, Hemmings KE, Porter KE, Turner NA. Combined effects of interleukin-1α and transforming growth factor-β1 on modulation of human cardiac fibroblast function. Matrix Biol. Jan;32(7–8):399–406. doi: 10.1016/j.matbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Lai KB, Sanderson JE, Yu CM. Suppression of collagen production in norepinephrine stimulated cardiac fibroblasts culture: differential effect of alpha and beta-adrenoreceptor antagonism. Cardiovasc Drugs Ther. 2009;23(4):271–80. doi: 10.1007/s10557-009-6183-6. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 2006;4(10):1732–1748. doi: 10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahadevan R, Palsson BO. Properties of metabolic networks: structure versus function. Biophys J. 2005;88(1):L07–L09. doi: 10.1529/biophysj.104.055723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bujak M, Frangogiannis NG. The role of TGF-beta signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res. 2007;74(2):184–195. doi: 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, Zhou Y, Zhang Y, Lei J, Wang J. Antioxidant probucol attenuates myocardial oxidative stress and collagen expressions in post-myocardial infarction rats. J Cardiovasc Pharmacol. 2009;54(2):154–162. doi: 10.1097/FJC.0b013e3181af6d7f. [DOI] [PubMed] [Google Scholar]
- 35.Kinugawa S, Tsutsui H, Hayashidani S, Ide T, Suematsu N, Satoh S, Utsumi H, Takeshita A. Treatment with dimethylthiourea prevents left ventricular remodeling and failure after experimental myocardial infarction in mice: role of oxidative stress. Circ Res. 2000;87(5):392–398. doi: 10.1161/01.res.87.5.392. [DOI] [PubMed] [Google Scholar]
- 36.Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, Sorescu D. NAD(P)H oxidase 4 mediates transforming growth factor-??1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ Res. 2005;97(9):900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 37.Klingberg F, Chow ML, Koehler A, Boo S, Buscemi L, Quinn TM, Costell M, Alman BA, Genot E, Hinz B. Prestress in the extracellular matrix sensitizes latent TGF-β1 for activation. J Cell Biol. 2014 Oct;207(2):283–97. doi: 10.1083/jcb.201402006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarrazy V, Koehler A, Chow ML, Zimina E, Li CX, Kato H, Caldarone CA, Hinz B. Integrins αvβ5 and αvβ3 promote latent TGF-β1 activation by human cardiac fibroblast contraction. Cardiovasc Res. 2014;102(3):407–417. doi: 10.1093/cvr/cvu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Full simulation of TGFβ vs forskolin treatment
Figure S2: Other validated relationships
Figure S3: Robustness of validation accuracy to changes in baseline input levels
Figure S4: Full sensitivity analysis
Figure S5: Clustering analysis
Figure S6: Full comparison of topological features vs influence
Table S1: Definitions of network topology metrics
Database S1: Network model
Database S2: Validation relationships