Rao et al. show that during migration, neurons contain a small population of centrosome-unattached microtubules in the leading process that are capable of sliding. Increasing the proportion of centrosome-unattached microtubules alters neuronal morphology, migration path, and microtubule behavior in the leading process.
Abstract
Contemporary models for neuronal migration are grounded in the view that virtually all functionally relevant microtubules (MTs) in migrating neurons are attached to the centrosome, which occupies a position between the nucleus and a short leading process. It is assumed that MTs do not undergo independent movements but rather transduce forces that enable movements of the centrosome and nucleus. The present results demonstrate that although this is mostly true, a small fraction of the MTs are centrosome-unattached, and this permits limited sliding of MTs. When this sliding is pharmacologically inhibited, the leading process becomes shorter, migration of the neuron deviates from its normal path, and the MTs within the leading process become buckled. Partial depletion of ninein, a protein that attaches MTs to the centrosome, leads to greater numbers of centrosome-unattached MTs as well as greater sliding of MTs. Concomitantly, the soma becomes less mobile and the leading process acquires an elongated morphology akin to an axon.
Introduction
Mammalian brain development involves directed migration of newly born neurons from ventricular zones to faraway destinations where they complete their differentiation (Sidman and Rakic, 1973; Barkovich, 2013). During its journey, which begins after its terminal mitotic division, the migratory neuron consists of a soma, a leading process that remains relatively short, and sometimes also a short trailing process. Microtubules (MTs) that are attached to the centrosome emanate into the leading process and also backward, to engulf the nucleus. Motor-driven forces on the MTs in the leading process yank on the centrosome, pulling it toward and sometimes into the leading process, after which motor-driven forces transport the nucleus along the MTs that engulf it toward the centrosome (Rakic, 1971; Gregory et al., 1988; Solecki et al., 2004; Tanaka et al., 2004). This two-step process occurs repeatedly, resulting in fluid movement of the neuron. Implicit in this model is the idea that all functionally relevant MTs in migrating neurons are centrosome-attached. Another possibility, as yet unexplored, is that some of the MTs are centrosome-unattached and that these MTs are able to slide relative to the centrosome-attached MTs. Such MT movements are known to occur in axons and dendrites (Yu et al., 1996; Slaughter et al., 1997; Wang and Brown, 2002; He et al., 2005; del Castillo et al., 2015a), as well as the mitotic spindle (Sharp et al., 2000), and could theoretically contribute in unstudied ways to the migration of neurons during brain development.
In neurons that have mostly completed their migratory journey, MTs nucleated from the centrosome become detached from the centrosome and are then transported into developing axons and dendrites (Ahmad et al., 1994; Ahmad and Baas, 1995). When few or no MTs remain attached to the centrosome, there is no towing of the centrosome or nucleus, and the soma takes up its permanent residence. Available evidence suggests that this developmental transition occurs in tandem with changes in the distribution of a protein called ninein. In many cell types, including cortical progenitor cells (Shinohara et al., 2013), ninein resides within the pericentriolar material and recaptures MTs after they are released from the centrosome (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Abal et al., 2002). Ninein is concentrated in the pericentriolar material when neurons are migrating, but then redistributes away from the centrosome as the neuron begins to form axons and dendrites (Srivatsa et al., 2015). These observations support a model in which transitions in neuronal phenotype are established in part through regulation of motor-driven sliding of MTs permitted by their detachment from the centrosome (Baas and Falnikar, 2012).
In the present study, we first wished to address the long-standing question of whether migratory neurons have functionally relevant centrosome-unattached MTs and, if so, how their potential sliding contributes to neuronal migration. We then tested the hypothesis that greater MT detachment from the centrosome leads to greater MT sliding, which in turn shifts the neuron toward a phenotype more akin to one that has nearly completed its migratory journey. These issues were addressed using electron tomography (ET) and live-cell imaging of migratory neurons, together with a novel drug treatment for inhibiting MT sliding in neurons. Partial depletion of ninein by RNAi was used to shift the balance of centrosome-attached and -unattached MTs.
Results
Analysis of MT minus-end distribution in migratory neurons
To investigate the distribution of MT minus ends in cultured migratory neurons with sufficient resolution to visualize ends of MTs, ET was used. ET is an advanced version of transmission electron microscopy that allows true ends of MTs to be distinguished from artifactual ends on individual sections through automated serial reconstruction (Höög et al., 2007). Cerebellar granule neurons from p5 rat pups were isolated, plated on laminin-coated coverslips, and allowed to attach to the substratum and undergo migration for 24 h. These neurons, in culture, display the classic morphology and motility of a migrating neuron (Edmondson and Hatten, 1987; Rivas and Hatten, 1995; Falnikar et al., 2013). Some are bipolar, with an elongated leading process and also a retracting trailing process, whereas others are unipolar with only a leading process. In the latter case, the trailing process had presumably fully retracted at the time the cell was observed. The ET technique is limited by practicality to reconstructing regions that are around 8 µm in length and 750 nm in depth. Even so, the volumes that could be reconstructed are sufficient to evaluate whether or not all of the MTs have their minus ends in the region of the centrosome, given the capacity of ET to identify MT ends with certainty. ET was performed on a total of three migrating neurons.
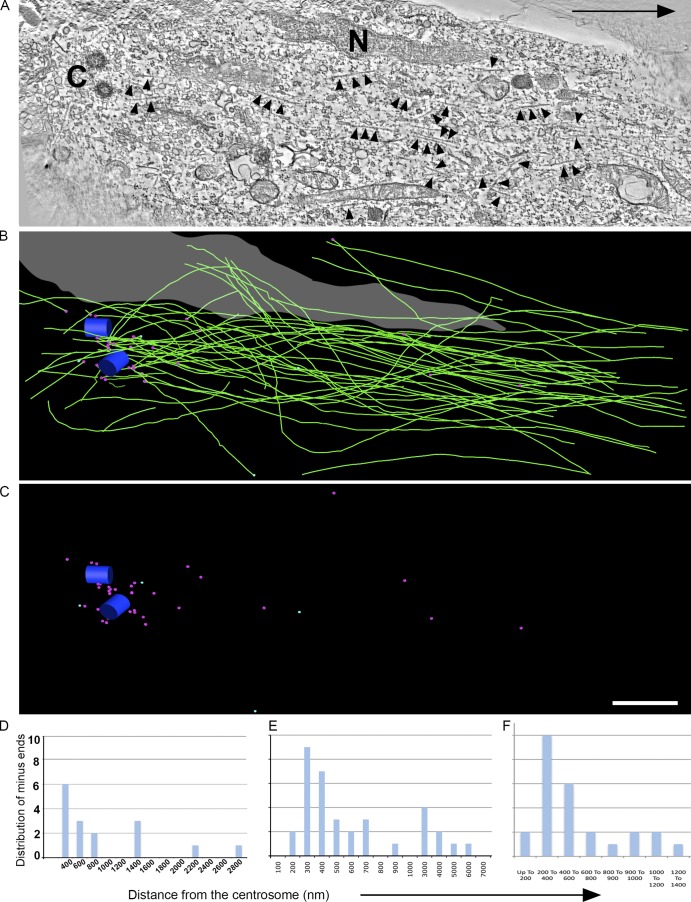
In the first neuron (Fig. 1 A), the centrosome (Fig. 1 A, "C"), a portion of the nucleus (Fig. 1 A, "N"), and MTs (Fig. 1 A, arrowheads) are visible in the selected tomographic section. 75 MTs were found in the analyzed region of this neuron (Fig. 1 B, green lines). Of the 75, minus ends of 35 MTs were present within the analyzed volume (Fig. 1 C, magenta spheres). Of the 35 ends, 27 were within 1 µm from the centrosome (Fig. 1, B and C, blue cylinders represent centrosome) and were considered attached to the centrosome, whereas the remaining eight ends, which were farther away, were considered to be unattached (Fig. 1 E, quantification). Some of the MTs extended behind the centrosome, instead of converging, and are stretched around the nucleus (Fig. 1 B). In this particular reconstruction, four MT plus ends were identified (Fig. 1 C, turquoise spheres).
Figure 1.
ET analysis of MT organization in the centrosomal region of a migrating cerebellar granule neuron. (A) A selected tomographic slice showing MTs (arrowheads), the centrioles (C), and a portion of the nucleus (N). The black arrow indicates the direction of migration. (B) Corresponding 3D model showing the centrioles (blue cylinders), the MTs (green lines), and the nucleus (gray). (C) Model indicating distribution of MT minus ends (magenta spheres) and MT plus ends (turquoise spheres). Bar, 1 µm. (D–F) Quantification of the distribution of MT minus ends with respect to the centrosome as identified by ET of three different migratory neurons. See also Fig. S1.
In the second of these neurons (Fig. S1, A and B), the reconstructed region included a portion of the nucleus as well as the centrosome (Fig. S1, C, D [boxes], and E). MTs are visible in selected tomographic sections (Fig. S1 E, arrowheads). A total of 28 MTs were found in this region (Fig. S1 F, green lines). Of these 28 MTs, 17 MTs had minus ends present within the analyzed volume (Fig. S1 G, turquoise spheres). Some of the MTs (Fig. S1 F) clearly extended beyond the centrosome (Fig. S1, F and G, magenta cylinders). Of the 17 minus ends, 12 ends were within 1 µm of the centrosome and were considered attached to the centrosome, whereas the remaining five were further away and considered unattached (Fig. 1 D, quantification). No plus ends of MTs were identified in the analyzed region.
In the analyzed volume of the third neuron, a total of 55 MTs were identified. 27 of these 55 MTs had their minus ends within the analyzed volume, and several MTs were found to extend beyond the centrosome. 24 of the 27 minus ends were within 1 µm of the centrosome and were considered attached to the centrosome. The remaining three MT ends were considered unattached (Fig. 1 F, quantification).
ET is the gold standard for this kind of analysis and yet is so labor intensive as to preclude large sample numbers. In addition, proximity to the centrosome does not necessarily mean attachment to it. For these reasons, we sought another method to visualize free minus ends of MTs. Calmodulin-regulated spectrin-associated proteins (CAMSAPs) are proteins recently shown to associate with free minus ends of MTs but not with minus ends of MTs that are centrosome-attached (Baines et al., 2009; Goodwin and Vale, 2010; Tanaka et al., 2012; Jiang et al., 2014; Yau et al., 2014). An EGFP fusion for CAMSAP3 was transfected into the cultured neurons, with the expressed protein appearing as speckles. The distribution of CAMSAP speckles was consistent with the distribution of minus ends of MTs considered to be centrosome-unattached from the ET (Fig. S2 A). There was no detectable concentration of EGFP-CAMSAP3 at the centrosome, indicating that the MTs visualized by the ET that were in the proximity of the centrosome were indeed attached to it (Fig. 2 A and Fig. S2 B). EGFP-CAMPSAP3 speckles were observed in the leading process as well, but at extremely low numbers (<10 per leading process; Fig. 2 A). A small number of minus ends of MTs are also consistent with live-cell imaging experiments indicating rapid transport of short MTs down the leading process, with a frequency less than one-tenth of that observed in axons (Falnikar et al., 2011). Thus, CAMSAP localization can be used as an effective means to localize free minus ends of MTs in migratory neurons.
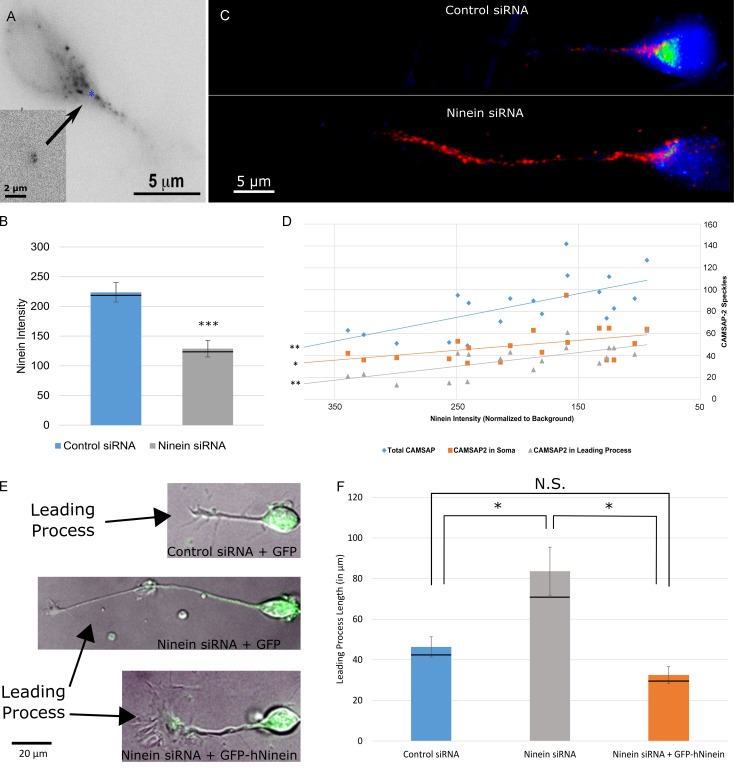
Figure 2.
Ninein depletion increases the number of MT minus ends present in the soma and leading process of migratory neurons. (A) A cultured cerebellar migratory neuron, transfected to express GFP-CAMSAP3 for 2 h. Neurons were cotransfected with RFP-pericentrin to label the centrosome (inset, blue asterisk). (B) Bar graph of ninein intensity under control siRNA and ninein siRNA conditions. n = 30 per group; ***, P < 0.001. Black lines represent median values. (C) 3D z-stack projection of neurons transfected with control siRNA (top) or ninein siRNA (bottom) and coimmunostained for CAMSAP2 (red) and ninein (green). More CAMSAP2 speckles appear in the soma and leading process of ninein-depleted neurons compared with controls. Speckles vary in apparent size because of CAMSAP dynamicity (Jiang et al., 2014). (D) Scatterplot of CAMSAP2 speckles versus ninein intensity. Speckle number in the soma (orange squares), leading process (gray triangles), and the total number of speckles (blue diamonds) were all found to have significant negative correlations with ninein intensity (P < 0.01, 0.028, and 0.01, respectively; n = 20). (E) Merged phase and green channel images of migratory neurons under control siRNA + GFP, ninein siRNA + GFP, and ninein siRNA + GFP-hNinein conditions. Arrows identify the leading process of each neuron. (F) Graph of leading process length quantification under control, ninein siRNA, and ninein siRNA + GFP conditions. Process length is significantly increased after ninein depletion. This change is rescued after ectopic expression of hNinein (n = 10 per group; *, P < 0.05). Black lines represent median values. See also Figs. S2 and S3 and Video 1. Data are represented as mean ± SEM. N.S., not significant.
Ninein depletion affects neuronal morphology and the number of free MT minus ends
Our next goal was to ascertain the effects on neuronal morphology of shifting the balance of centrosome-attached and centrosome-unattached MTs in these migratory neurons. We found that we could accomplish this via partial knockdown of ninein, which we previously documented is present within the pericentriolar material of these neurons (Baird et al., 2004), and which has previously been shown to be essential for anchoring MTs to the centrosome in neuronal progenitor cells (Shinohara et al., 2013). Like most centrosomal proteins, ninein has a long half-life and hence can only be partially knocked down by siRNA (Leo et al., 2016). 3 d after introduction of control or ninein siRNA, cerebellar granule neurons were replated on laminin-coated dishes and coimmunostained for ninein and γ-tubulin. Immunostaining revealed a cloud of ninein staining in a region containing the centrosome, demarcated by γ-tubulin, with no obvious noncentrosomal ninein in the leading process (unpublished data). In the case of neurons transfected with ninein siRNA, fluorescence intensity of ninein immunostaining was reduced by 53 ± 11% compared with mean intensity of neurons transfected with control siRNA (Fig. 2 B).
To directly ascertain whether ninein depletion led to a greater number of MTs with free minus ends, neurons were immunostained for CAMSAP proteins and ninein. For this, an antibody was used for CAMSAP3, which was found to produce a pattern similar to that of ectopically expressed CAMSAP3, and also an antibody for CAMSAP2 that was previously used in work on neurons (Jiang et al., 2014). In this recently published study, CAMSAP2 was shown to be the predominant CAMSAP in vertebrate neurons, with the staining for the antibody confirmed in its specificity by the lack of staining in cells treated with RNAi to reduce the protein. CAMSAP2 staining was used for quantification, but CAMSAP3 staining gave qualitatively the same results. 3D z-stack projections (Fig. 2 C and Video 1) and cross-sectional space plots (Fig. S3, A and B) revealed an increased number of CAMSAP2 speckles both in the soma and in the leading process of cerebellar migratory neurons. This was confirmed by correlation analysis, which showed a significant negative correlation between the intensity of ninein staining and the total number of CAMSAP2 speckles present in the soma and the leading process (Fig. 2 D). Furthermore, a significant negative correlation was found when the speckle number was corrected for the area of the soma and the length of the leading process of the neurons measured (Fig. S3, C and D, respectively).
After partial ninein depletion, the leading processes of migratory neurons were notably longer, taking on an axon-like appearance. The mean leading process length for control neurons was 46.35 ± 4.96 µm, whereas that for ninein-depleted neurons was 83.62 ± 11.85 µm (P < 0.05). Ectopic expression of GFP–human ninein rescued this phenotype, reducing the mean leading process length of neurons treated with ninein siRNA to control values (32.39 ± 4.19 µm), thus indicating that the morphologic phenotype resulted specifically from ninein depletion rather than any off-target effects that the siRNA may have had (Fig. 2, E and F [quantification]).
Greater MT detachment from the centrosome leads to greater mobility within the MT array and reduced migration of the neuron
We would anticipate that greater numbers of centrosome-unattached MTs would lead to greater mobility (i.e., more sliding) of MTs in the leading process of these neurons. The increase in CAMSAP particles in the leading process after partial ninein depletion was our first indication of enhanced MT sliding because more free minus ends of MTs in the leading process would be consistent with greater movement of MTs from the soma into the leading process. To more directly assay for MT sliding, tandem-dimer Eos (tdEos)–tagged tubulin was expressed overnight in neurons into which either control or ninein siRNA had been introduced 2 d before. tdEos is a tag that fluoresces green until it is photoconverted by exposure to 405 nm light, after which it fluoresces red. Two distinct regions of the cell were converted, one in the soma and proximal region of the leading process and another located in the middle one third of the leading process (Fig. 3 A). Once converted, dual-channel live-cell imaging of the neurons was performed. For this set of experiments, the distance was measured between the two converted zones (dz), as well as the total distance (dt) of migration, and the two were compared to determine whether coordination existed between these two parameters as well as to confirm that expression of tdEos-tubulin does not affect migration. After ninein depletion, when red and green images were merged, a greater amount of MT movement was observed in the leading process, as indicated by bright yellow (yellow represents the comingling of red and green signal) regions flanking the proximal and distal borders of the red converted zones in the ninein-depleted neuron. This was not observed in the control neuron (Fig. 3 B and Video 2). Several neurons treated with ninein siRNA showed an elongated leading process that was also notably tortuous along its length, a phenomenon that was never observed in neurons treated with control siRNA (Video 2). We speculate that this tortuosity of the leading process is caused by buckling of centrosome-unattached MTs, with an individual MT buckling because of an abrupt interruption in a bout of sliding.
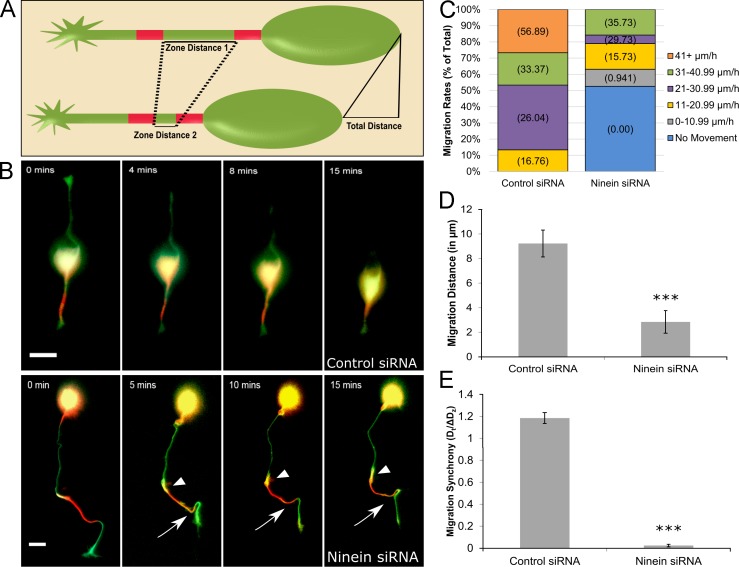
Figure 3.
Ninein depletion compromises neuronal migration. (A) Schematic depicting the experimental paradigm. Under control conditions, the two red converted zones converge by a distance nearly equal to that of the total migration distance. (B, top) Cerebellar migratory neuron cotransfected with tdEos-tubulin and control siRNA. Minimal MT sliding occurs and soma moves forward. (bottom) Cerebellar migratory neuron cotransfected with tdEos-tubulin and ninein siRNA. Abnormal tortuosity of the leading process and increased MT sliding are observed (regions of yellow indicated by arrowheads [proximal] and arrows [distal]). Cells were imaged for 15 min. (C) Stacked bar graph showing migration rate distribution under control siRNA and ninein siRNA conditions. n = 20 per group. Numeral in parentheses represents the mean migration rate per category. (D) Bar graph depicting the total migration distance after treatment with control siRNA or ninein siRNA. ***, P < 0.0001 (n = 20 per group). (E) Bar graph of migration synchrony under control and ninein-depleted conditions. Total migration distance was divided by the change in zone distance. ***, P < 0.0001 (n = 20 per group). See also Video 2. Bars, 15 µm. Data are represented as mean ± SEM.
As for alterations in migration, ninein siRNA treatment caused a large proportion of neurons to become stationary, and those neurons that did migrate after ninein depletion did so at slower rates than neurons treated with control siRNA (Fig. 3 C). Under control conditions, neurons migrated a mean distance of 9.23 ± 1.08 µm over 15 min (Fig. 3 D), and the net change in distance between the two converted zones was nearly equal to the total migration distance, as indicated by a dt/Δdz ratio value of 1.185 ± 0.05. After ninein depletion, mean migration distance was notably hindered (2.84 ± 0.91 µm over 15 min) and what migration did occur generally lacked synchrony, producing a ratio value of 0.024 ± 0.01 (Fig. 3 E). tdEos-tubulin expression did not affect migration rates because the mean rates of migration were not significantly different from neurons that did not express tdEos-tubulin (mean rate of movement for control neurons = 27.1 ± 2.97 µm/h; mean rate of movement for ninein-depleted neurons = 12.378 ± 1.29 µm/h).
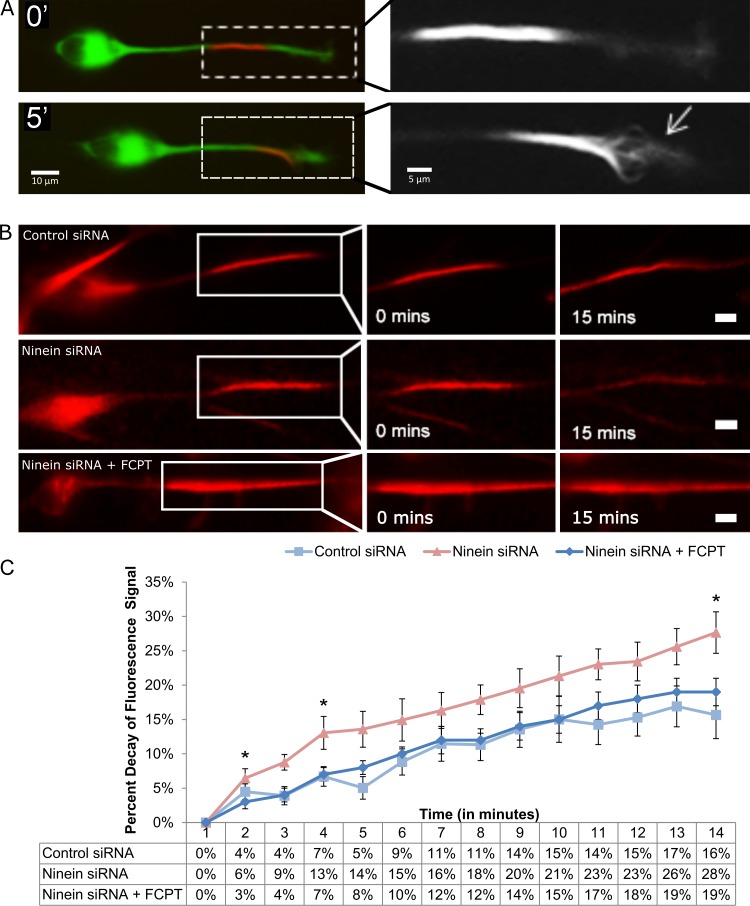
Although MT sliding could be observed in some control migratory neurons, this was rare, with most control neurons not having sufficient sliding of MTs to be directly visualized. Furthermore, MTs in the leading process could occasionally be seen buckling (Fig. 4 A), presumably resulting from sliding that had been interrupted. These observations led us to hypothesize that limited MT sliding occurs in control neurons, but is usually too subtle to directly observe as MT movements with our imaging paradigm. Therefore, to quantitatively compare MT sliding in control and ninein-depleted neurons, tdEos-tubulin was photoconverted in a portion of the leading process of migratory neurons, and the rate of signal decay of the converted tubulin was measured in the red channel, the idea being that more sliding of MTs would result in a greater decay of fluorescence signal. Under control conditions, the tdEos-tubulin signal decreased by 16 ± 3% over 14 min. Loss of tubulin signal jumped to 28 ± 3% after ninein-depletion (Fig. 4 B, top and middle panels, respectively). To ensure that the observed signal decay resulted from MT sliding and not changes in MT stability, we used FCPT, an ATP-competitive inhibitor of kinesin-5 that locks the motor onto the MTs in a rigor-like state that cross-links and immobilizes the two MTs engaged by the motor (Groen et al., 2008). (Kinesin-5 normally acts as a brake on MT movements in neurons [Falnikar et al., 2011; Kahn et al., 2015a], but acts in a regulated fashion that still permits a great deal of MT movement to occur at any given time. By making a rigor complex of most MT-associated kinesin-5 molecules, FCPT is an effective inhibitor of MT sliding.) Fluorescence-decay in ninein-depleted neurons treated with FCPT was reduced from 28 ± 3% to 19 ± 2% (Fig. 4, B [bottom] and C; and Video 3). Similar results were obtained using GSK246053, an ATP-competitive inhibitor of kinesin-5 that is structurally dissimilar to FCPT and yet produces the same effects (Parrish et al., 2007; Groen et al., 2008). These results indicate that the observed increase in fluorescence decay after ninein depletion predominantly results from an increase in MT sliding (Fig. 4 C, quantification).
Figure 4.
Ninein siRNA treatment leads to increased MT sliding in the leading process of migratory neurons. (A) A cerebellar migratory neuron transfected with tdEos-tubulin. (top) A distinguished converted region in the leading process of a neuron treated with control siRNA. Leading process is magnified on the right. (bottom) After 5 min, individual MTs (indicated by the arrow) are observed sliding out of the converted region. (B, top) Migratory neuron treated with control siRNA. Modest signal decay is observed after 15 min. (middle) After treatment with ninein siRNA, significant signal decay can be seen. (bottom) Treating ninein-depleted migratory neurons with FCPT reduces the rate of signal decay to control levels. (C) Quantification of percent decay of fluorescent tubulin signal. Data table shows values under control siRNA (n = 15), ninein siRNA (n = 15), and ninein siRNA + FCPT conditions (n = 15 per group; *, P < 0.05). Bars, 10 µm. Data are represented as mean ± SEM.
Experimental alterations in MT sliding lead to abnormal MT buckling, process tortuosity, and neuronal migration path
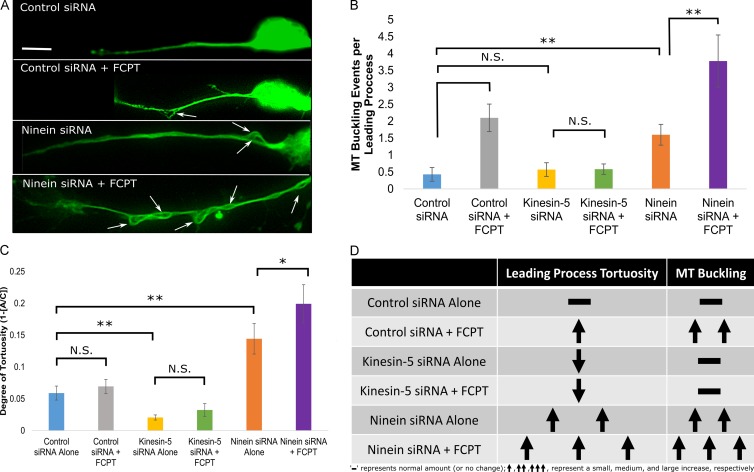
The results presented thus far suggest a scenario wherein only very limited MT sliding normally occurs in the leading process of a migratory neuron, but a great deal more sliding would occur if more of the MTs were centrosome-unattached. In this scenario, greater sliding of MTs leads to a longer leading process, but also less movement of the soma because of reduced numbers of centrosome-attached MTs pulling on the centrosome. Interruptions in MT sliding result in buckling of MTs, which in turn causes tortuosity of the leading process. We next sought to put this scenario to greater experimental scrutiny. To investigate MT buckling, migratory neurons were treated with control siRNA, ninein siRNA, kinesin-5 siRNA, and/or FCPT and were then fixed and stained for tubulin. Control siRNA-treated neurons showed a mean of 0.42 ± 0.22 MT buckling events per leading process. This increased to 1.6 ± 0.31 events per process after ninein depletion, demonstrating an increase in MT buckling when the capacity exists for greater MT sliding (which makes sense if MT buckling is caused by interruptions in MT sliding). Applying FCPT to inhibit MT sliding significantly increased the occurrence of MT buckling after control siRNA (2.1 ± 0.41) and ninein siRNA treatment (3.78 ± 0.78; Fig. 5, A [white arrows indicate buckling] and B [quantification]). This result suggests that MT sliding leads to MT buckling when that sliding is abruptly halted. Confirming the specificity of the drug, pretreatment with kinesin-5 siRNA abolished the FCPT-associated increase in MT buckling (Fig. 5 B).
Figure 5.
Uncontrolled MT sliding alters migratory neuron phenotype and increases MT buckling in the leading process. (A) Cultured rat cerebellar granule neurons subjected to one of the following four conditions: (1) Control siRNA (top); (2) Control siRNA + 50 µM FCPT (top middle); (3) ninein siRNA (bottom middle); and (4) ninein siRNA + 50 µM FCPT (bottom). Neurons were immunostained for tubulin and subsequently analyzed for MT buckling (indicated by white arrows) in the leading process. (B) A bar graph displaying the mean number of MT buckling events present in the leading process of migratory neurons under differing conditions. Treatment with FCPT increases MT buckling per leading process. Kinesin-5 depletion abolishes this effect. (C) A bar graph displaying the mean degree of tortuosity of the leading process of migratory neurons. Neurons treated with control siRNA or control siRNA and FCPT showed equivalent leading process tortuosity. Depleting ninein significantly increases the tortuosity of the leading process, and application of FCPT further exacerbates leading process tortuosity after partial ninein depletion. (D) Table summarizing the observed changes to leading process tortuosity and MT buckling under the tested conditions. (n = 15 per group; brackets indicate statistical analyses between groups). N.S., not significant; *, P < 0.05; **, P < 0.01. Bar, 10 µm. Data are represented as mean ± SEM.
To quantify leading process tortuosity, migratory neurons were treated as previously described, fixed, and stained for tubulin. The arc-chord (AC) ratio of the leading process, which is the distance between the proximal and distal ends of the leading process divided by the total length of the leading process, was calculated, and the resulting value was subtracted from 1 to yield the degree of tortuosity. In this method, the value for a straight line is equal to 0, the value for a circle is infinite (because it has no end), and anything in between would have a value >0. Control siRNA treatment produced migratory neurons with an AC ratio of 0.059 ± 0.011, indicating nearly straight leading processes (Fig. 5 C). After ninein depletion, the leading process exhibited significantly greater tortuosity, yielding an AC value of 0.144 ± 0.022 (Fig. 5 C, P < 0.01). FCPT treatment did not produce a significant effect on leading process tortuosity under control conditions. After ninein depletion, however, tortuosity increased and treatment with FCPT heightened leading process tortuosity and significantly reduced the length of the leading process (Fig. 5 C, P < 0.01; and Fig. S4 A). When kinesin-5 protein levels were reduced, the FCPT effect was lost and leading processes were significantly less tortuous when compared with any other group. These results confirm that tortuosity of the leading process is directly correlated with the cessation of MT sliding because processes with greater MT sliding show greater tortuosity when treated with FCPT. These results are summarized in Fig. 5 D.
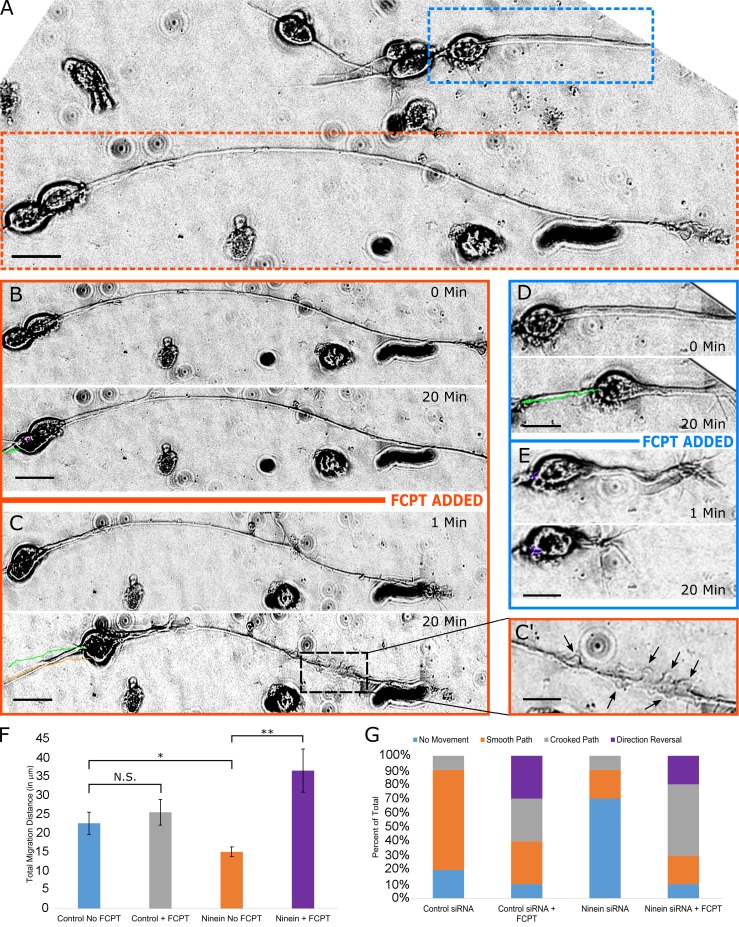
To test our proposed scenario regarding the movement of the soma, we applied FCPT (in live-cell imaging experiments) to either control neurons or neurons that had been partially depleted of ninein. Because we previously found that neurons with longer leading processes after ninein depletion contained more centrosome-unattached MTs, we compared the effect of FCPT treatment on neurons with longer leading processes relative to control or ninein-depleted neurons with shorter leading processes. We predicted that inhibition of MT sliding would result in MT buckling in the leading process as well as tortuosity of the leading process. We also predicted that the soma should resume movement because the centrosome-unattached MTs become cross-linked with the centrosome-attached MTs in the presence of FCPT, thus approximating the situation where all or most of the MTs are directly attached to the centrosome. After addition of FCPT, stationary ninein-depleted neurons (Fig. 6 A, orange box) displayed rapid movement of the soma as well as buckling of MTs in the leading process (Fig. 6, B, C, and C′; and Video 4). In the same dish, a neuron with a short leading process (Fig. 6 A, blue box), presumably caused by less thorough MT detachment from the centrosome, displayed movement of the soma before addition of FCPT (Fig. 6 D), but within minutes of FCPT addition showed an increase in leading process tortuosity as well as compromised path of migration (Fig. 6 E). Under control conditions, the mean total distance of migration was 22.97 ± 2.97 µm over 20 min, with 70% of the neurons exhibiting a smooth path of migration (Fig. 6, F and G). Addition of FCPT did not significantly change the mean migration distance of control neurons (25.632 ± 3.4 µm), but did change migratory behavior, causing an increase in neurons that showed a crooked migratory path or reversed direction of migration (Fig. 6 G). As before, ninein depletion significantly reduced total migration distance (15.11 ± 1.29 µm; Fig. 6 F), with 70% of the cells showing no movement. After the addition of FCPT, 90% of the neurons exhibited movement (mostly crooked paths), and the mean total migration distance increased to 36.7 ± 5.75 µm (Fig. 6, G and F). These results confirmed our prediction that an increase in centrosome-unattached MTs would negatively impact soma movement and prompted us to more thoroughly investigate how MT sliding affects migratory paths.
Figure 6.
FCPT restores soma migration of ninein-depleted neurons, but with abnormal characteristics. (A) Differential interference contrast image of cultured cerebellar migratory neurons. Different neuronal morphologies are observed. A neuron with a long leading process is seen at the bottom of A (orange box), whereas a neuron with a shorter leading process is observed at the top right (blue box). (B) A representative migratory neuron with a long leading process was imaged for 20 min under normal conditions. (C) FCPT was added to the same dish and imaged for 20 min. Significant movement of the soma is observed after ninein-depleted neurons are treated with FCPT (C, green and orange lines), and MT buckling can be observed along the leading process (indicated by black arrows in C′). (D) A neuron with a short leading process was treated as previously described. Movement is observed before addition of FCPT. (E) Upon addition of FCPT, MTs buckle in the leading process, and neuronal migration is compromised. (F) Bar graph depicting the total migration distance after treatment with control siRNA or ninein siRNA, with or without FCPT. (G) Stacked bar graph showing the distribution of migration subtypes (no movement, smooth path, crooked path, or direction reversal) after treatment with control siRNA or ninein siRNA, with or without FCPT. Brackets indicate statistical analyses between groups: N.S., not significant; *, P< 0.05; **, P < 0.01; n = 10 per group. Bars, 10 µm. Data are represented as mean ± SEM.
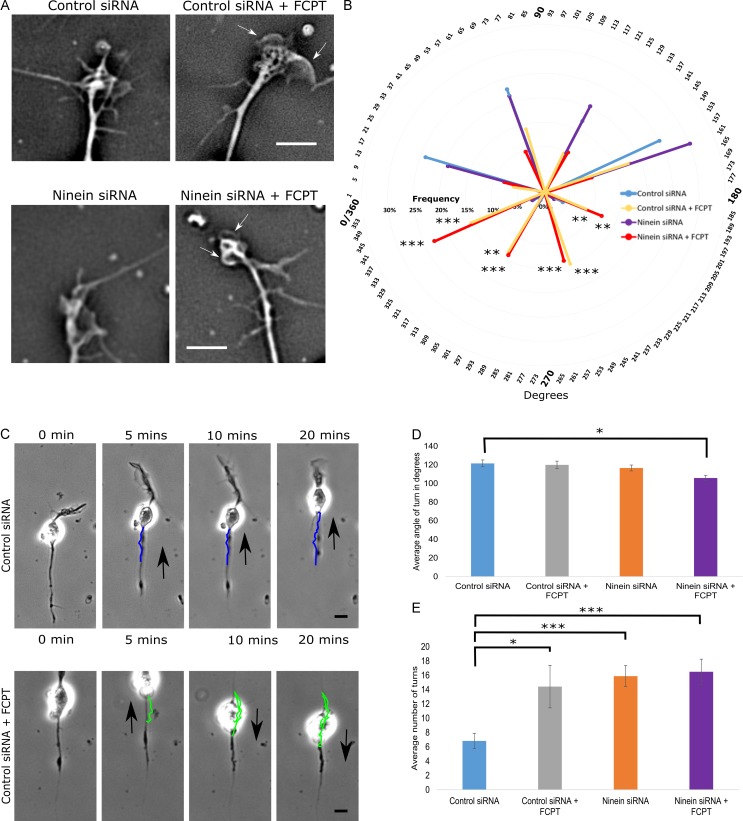
When neurons migrate through the brain, they must maintain an appropriate trajectory so that they do not veer off course. For this reason, we chose to examine MTs at the distal tip of the leading process, which we positioned on a 2D 360° plane over 20 min of imaging. 0–180° (Fig. 7 B, top half of the circle) represents the area directly lateral and in front of the migrating neuron, with 90° serving as the midline. 180–360° represents the area behind the distal tip of the leading process (Fig. 7 B, bottom half of the circle). Under both control and ninein-depleted conditions, MTs were splayed with <10% of the MTs exhibiting positions that were located in the 181–360° region (Fig. 7, A [top and bottom left] and B; and Fig. S4 B, quantification). After FCPT treatment, MTs in the distal tip buckled at extreme angles, with 53% and 63% of MTs positioned over 90° from midline of the distal tip of control and ninein-depleted neurons, respectively (Fig. 7, A [white arrows in top right and bottom right] and B; and Fig. S4 A, quantification). Thus, without their capacity to slide in the distal region of the leading process, even in control neurons, MT buckling results in abnormal MT behaviors that could likely affect the normal path of neuronal migration.
Figure 7.
Altering MT sliding impacts migration path and MT activity at the distal tip of the leading process. (A) Phase images of the distal tips of leading processes after treatment with control siRNA or ninein siRNA with and without FCPT. (B) 360° radar graph showing the mean frequency MTs were positioned in the following bins over 20 min of imaging: 0–45°, 46–90°, 91–135°, 136–180°, 181–225°, 226–270°, 271–315°, or 316–360°. FCPT treatment yielded a significant increase in the number of MTs positioned within the 180–360° region (also shown by white arrows in A). (C) Neuronal migration was tracked over 20 min using time-lapse phase imaging, and the migratory path was traced (colored lines) using Fiji tracking plugin. (top) Control siRNA-treated neuron. (bottom) Control siRNA-treated neuron + FCPT (black arrows indicate direction of migration). (D) Graph showing the mean angle of observed turns during a 20 min bout of migration. Mean turn angle was significantly sharper when ninein-depleted neurons were treated with FCPT. (E) Bar graph displaying the mean number of turns made by migratory neurons during a 20 min bout of migration. Neurons treated with control siRNA showed the fewest number of turns. Treating control neurons with FCPT, depleting ninein, and treating ninein-depleted neurons with FCPT all significantly increased the mean number of turns observed during the 20 min bout of migration. n = 15 per group; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars: (A) 5 µm; (C) 10 µm. Data are represented as mean ± SEM.
To investigate how the observed increase in leading process tortuosity and abnormal MT buckling at the distal tip influences the path of neuronal migration, we used live-cell imaging to capture phase images of migratory neurons over time. Migration was tracked using the manual tracking plugin available in the Fiji program. Neurons were tracked over 20 min, and the number of turns in the migration path and the angles of the turns were measured under control, ninein-depleted, and/or FCPT-treated conditions (Fig. 7 C). Under control conditions, neurons exhibited a mean turn angle of 121.53 ± 3.69° and a mean of 6.83 ± 1.06 turns over 20 min (Fig. 7, D and E, respectively). After treatment of control cells with FCPT or ninein depletion, neurons showed a significant increase in the number of turns, but the mean angle of turns did not significantly change. When ninein-depleted neurons were treated with FCPT, both the angle of turns and the number of turns were altered (Fig. 7, D and E, respectively). These results suggest that MT sliding is important to ensure that neurons remain on their appropriate path of migration.
Discussion
Neurons express a variety of molecular motor proteins that impose forces on MTs that can cause them to slide (i.e., undergo transport). Cytoplasmic dynein has been shown to be an important motor for transporting MTs away from the centrosome and into developing axons and dendrites (Ahmad et al., 1998, 2006; He et al., 2005). Other relevant motor proteins are repurposed from mitosis, where they function to organize MTs into a bipolar spindle and then to separate the half-spindles as cell division progresses (Baas, 1999). In neurons, these motors can generate forces between two MTs, with some motors also able to generate forces between MTs and actin filaments (Hasaka et al., 2004; Myers et al., 2006; Myers and Baas, 2007; Liu et al., 2010). Short MTs can be propelled rapidly over long distances, whereas longer MTs are relatively immobile (Wang and Brown, 2002; Baas et al., 2006). In some cases, particularly during axogenesis and growth cone guidance, the longer MTs can also slide relative to one another or actin-based structures (Slaughter et al., 1997; Schaefer et al., 2008; Liu et al., 2010; del Castillo et al., 2015a,b). The particular role that any motor protein plays is dependent on the length of the MTs with which it interacts, the polarity orientation of the MTs, and whether or not one or both of the MTs are unattached to the centrosome. MT arrays in which all of the MTs are attached to the centrosome would be unable to undergo MT sliding, at least not in the sense that would result in net movements of the MT relative to other structures in the cell. We became interested in migratory neurons because it is generally believed that all MTs in these neurons, at least all functionally relevant MTs, are attached to the centrosome (Rivas and Hatten, 1995; Xie et al., 2003; Solecki et al., 2004; Tanaka et al., 2004; Tsai and Gleeson, 2005). Our first goal was to ascertain whether this is actually true, by documenting for the first time the proportions of centrosome-attached and centrosome-unattached MTs, and then by investigating whether sliding of the centrosome-unattached MTs has any important role to play in neuronal migration.
Our ET and CAMSAP studies confirm that most MTs in the migratory neuron are attached to the centrosome. However, these studies also establish that migratory neurons contain a small population of centrosome-unattached MTs that can undergo movements within the neuron as it migrates. Before our present studies, there were some hints of this already emerging in the literature. Our own live-cell imaging experiments have revealed a very small number of short rapidly moving MTs in the leading process of the migratory neuron, about a tenth the number observed in axons (Falnikar et al., 2011). Immunofluorescence analyses suggested that a small number of the long MTs in the leading process did not have their minus ends directly at the site of the centrosome, but rather seemed to reach behind the centrosome in the soma of the neuron (Umeshima et al., 2007; Falnikar et al., 2011). These latter observations are consistent with local regions of antiparallel MTs behind the centrosome that could permit varying degrees of antiparallel sliding. Parallel sliding of the centrosome-attached MTs relative to centrosome-unattached MTs is also possible within the leading process itself.
We previously found that allosteric inhibition of kinesin-5 caused the leading process to grow slightly longer and caused the neuron to migrate faster, presumably as a result of increased MT sliding (Falnikar et al., 2011). Here, we used a different kind of drug that creates a rigor complex of kinesin-5 (Groen et al., 2008) to inhibit the sliding of MTs in migratory neurons. As a result of the MTs being immobilized in this fashion, the leading process became shorter, migration of the neuron deviated from its typical path, and the MTs within the leading process became more buckled. Our studies suggest that buckling of MTs occurs when a MT sliding event is abruptly halted. This can explain why it is that when MT sliding events were experimentally increased (as with partial ninein depletion), a concomitant increase in MT buckling was also observed, and this increase in buckling was further increased in the presence of FCPT, the drug that inhibits MT sliding. Based on the abnormal trajectories of migration that result from inhibition of MT sliding, we posit that the very small degree of MT sliding that normally occurs during neuronal migration is important for providing maneuverability that enables the neuron to stay appropriately on course. Our studies suggest that the MTs in the distal region of the leading process may be most relevant to this, but MT sliding throughout the leading process could also be important for adjusting the path of neuronal migration. The centrosome-attached MTs may also play an important role in preserving the appropriate path of the neuron, by tugging the centrosome and soma along in a motor-regulated manner. Such effects on MTs could be responsive to signaling cascades that enable migration to respond appropriately to environmental cues because motor proteins such as kinesin-5 are activated and deactivated by mechanisms such as phosphorylation that are commonly used in signaling cascades (Nadar et al., 2008; Kahn et al., 2015b).
The other question we wished to address is whether MT sliding enabled by centrosome detachment is a transformational factor in defining neuronal phenotype. In other words, if we start with a migratory neuron and experimentally alter just one thing, namely the proportion of MTs that are unattached to the centrosome, would this be sufficient for the same available forces on the MTs to shift the phenotype of the migratory neuron toward the phenotype of a stationary axon-bearing neuron? We found that we could accomplish this via partial depletion of ninein, a centrosomal protein that recaptures MTs after their release from the centrosome. The level of depletion that we were able to achieve (no more than ∼50% because centrosomal proteins are notoriously long-lived) resulted in a notable increase in centrosome-unattached MTs and a correlated increase in leading process length, tortuosity, migration synchrony, turning, and MT buckling. These results suggest that when the ratio of centrosome-attached to centrosome-unattached MTs is altered, the motor forces transduced onto the MT array in the leading process are still present but are unable to tow the soma, thereby resulting in the observed aberrant phenomena. The leading process of ninein-depleted neurons often became notably longer than the control leading process but did not grow indefinitely as bona fide axons do, suggesting either that more thorough detachment of MTs from the centrosome is necessary for a continuously growing process, or that additional factors are needed for a neuron to transition into a phase of development wherein bona fide axonal development is possible. One such factor might be the up-regulation of MT severing activity by proteins such as katanin and spastin, given that short MTs are generally the ones that move most effectively (Baas et al., 2006).
In summary, the results of our study indicate that only very limited sliding of MTs occurs during neuronal migration and that what little sliding occurs is important for fine-tuning the morphology and migratory behavior of the neuron. In addition, our results indicate that neuronal phenotype is defined in part by the degree of MT sliding permitted by the relative proportions of centrosome-attached and centrosome-unattached MTs. On the basis of our results, summarized in Fig. 8, we would conclude that motor-driven forces exist in the neuron that can drive MT sliding, but these forces are limited in their capacity to do so by attachment of the MTs to the centrosome as well as by regulatable brakes on sliding imposed by proteins such as kinesin-5. The degree of MT sliding in turn influences the migratory behavior of the neuron as well as process length and morphology. We speculate that when all of the MTs are centrosome-detached and especially when they are severed into shorter pieces, MTs can slide to a far greater degree than in any of our experimental scenarios, thus enabling a bona fide axon to grow longer and longer, without tugging along the soma. These ideas are buoyed by the fact that the same motor protein that predominantly transports MTs in the axon, namely cytoplasmic dynein (Ahmad et al., 1998, 2006; He et al., 2005), is also the predominant force-generating motor on MTs during neuronal migration (Xiang et al., 1994; Sheeman et al., 2003; Shu et al., 2004; Tanaka et al., 2004; Tsai and Gleeson, 2005; Willemsen et al., 2012).
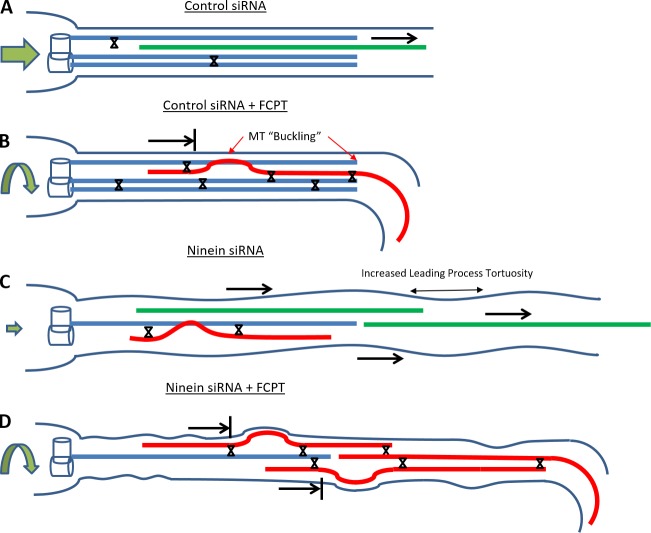
Figure 8.
Schematic summary of MT sliding scenarios in experiments on migratory neurons and the effects on neuronal morphology, migration, and MTs in the leading process. Neurons treated with control siRNA display a minimal amount of sliding (green MT = sliding MT), tortuosity, and MT buckling and migrate in a smooth, consistent path (straight green arrow). When FCPT is used to prevent MT sliding (represented by X), MTs in the leading process of control siRNA-treated neurons show an increase in MT buckling (but do not become tortuous), and their migration path is irregular (curved green arrow). After treatment with ninein siRNA, MT buckling is observed, the leading process becomes tortuous, and migration is compromised (small green arrow). When FCTP is applied to the ninein-depleted neurons, MT buckling and tortuosity are exacerbated, and neuron movement is restored (but with abnormalities in migration path).
Materials and methods
Cell culture and siRNA treatment
Cerebella were harvested from p2–p5 rat pups that were euthanized using hypothermia. The protocol used for neuronal migration was similar to that previously used by other authors (Clark et al., 1995; Bix and Clark, 1998; Hirotsune et al., 1998), except instead of papain solution, the tissue was digested in 0.25% trypsin + 0.05% DNAase made in 1× HBSS, and the trituration media did not contain insulin, human transferrin, or selenium. Cerebellar cells were dissociated, allowed to reaggregate, and plated on a laminin-coated surface. Cells in the aggregate extend processes and cerebellar granule neurons migrate radially away from the aggregates. For siRNA studies, granule neurons were transfected with 10 µM control (AM4636; Ambion) or ninein siRNA (ninein siRNA three-sequence SMARTpool; WD02364621, WD02364621, and WD02364623; Sigma-Aldrich) using the Amaxa Nucleofector II. For the kinesin-5 siRNA groups included in the tortuosity and MT buckling experiments, we followed previously published procedures (Falnikar et al., 2011). In brief, cerebellar cells were transfected with 10 μM of kinesin-5 siRNA and allowed 24 h to reaggregate before being plated on poly-d-lysine/laminin–coated dishes; they were then imaged 12 h after plating.
ET
Dissociated p5 cerebella isolated as described above were cultured overnight on glass coverslips. The next day, cells were fixed using chemical fixation by adding 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 30 min at RT. After rinsing in 0.1 M sodium cacodylate buffer 3× for 10 min and then with 1× PBS, coverslips were stored in 8% sucrose in PBS at 4°C overnight. Cells were then embedded in plastic and serially sectioned into 250-nm-thick sections using an Ultracut UCT microtome (Leica). Poststaining with 2% uranyl acetate in 70% methanol and application of Cationic gold particles (15 nm; Brittish Bio Cell) to be used as fiduciary markers was performed on these sections. For ET, the sample was rotated through 120°, a tilt series was acquired, and the MTs were reconstructed using IMOD software through serial sections (Mastronarde, 1997; Ladinsky et al., 1999).
Live-cell imaging of cerebellar granule neurons
Cerebellar granule neurons were harvested as described above, dissociated, transfected with either control or ninein siRNA, and plated densely on plastic dishes coated with 20 µg/ml poly-l-lysine for 36 h.
tdEos-tubulin experiments
Cells were trypsinized, transfected with tdEos-tubulin (and either control or ninein siRNA), and allowed to reaggregate for 24 h before plating on glass coverslips coated with poly-d-lysine (100 µg/ml) and laminin (25 µg/ml). 12 h after plating, dishes were placed in the live-cell imaging workstation (5% CO2, 37°C), and cerebellar migratory neurons from five randomly selected regions of the dish were chosen for imaging. Photoconversion of the specific regions of the leading process and soma was accomplished using the Mosaic Digital Diaphragm system (Andor). After the 5-s photoconversion light exposure, images were captured every minute in both the red and green channel at 40× magnification for migration synchrony experiments and 63× magnification for fluorescence decay experiments. Both control and ninein siRNA were transfected at a concentration of 10 µM using the Amaxa Nucleofector II. After live-cell imaging was completed, cells were fixed for 10 min in –20°C methanol for morphologic analysis and immunostaining.
Migration turning and distal MT tip imaging experiments
For experiments tracking distal MT activity and turning during migration, cells were cotransfected with control or ninein siRNA and tdEos-tubulin. Cells were plated 24 h after reaggregation and imaged 12 h after plating using a 40× water objective. Phase and green channel images were captured every 30 s for 20 min.
Ectopic expression of human ninein
Neurons were transfected with control siRNA and pmaxGFP (Lonza; 2.5 µg in 10 μl SCN transfection buffer; Lonza). Ninein-depleted neurons were cotransfected with either pmaxGFP (2.5 µg in 10 µl) or GFP-human ninein (2.5 µg in 10 µl; gift from H. Shinohara, Tokyo Medical University, Tokyo, Japan). Cells were plated after 24 h of reaggregation, and leading process length was measured 12 h after plating.
Drug treatments
FCPT and GSK246053 (gifts from T. Mitchison, Harvard Medical School, Boston, MA) were dissolved in DMSO (Sigma-Aldrich) at a concentration of 50 µM, which has been shown to prevent MT sliding (Groen et al., 2008). An equivalent volume of DMSO alone was used in control experiments. Drugs were added with medium 30 min after plating, allowing neurons to settle, and cultures were fixed 6 or 24 h later.
Fluorescence microscopy
In one set of experiments, cerebellar granule neurons were harvested as described above, dissociated, and cotransfected with GFP-CAMSAP3 (gift from M. Takeichi, Institute of Physical and Chemical Research, Kobe, Japan), and RFP-pericentrin (centrosomal marker; gift from S. McConell, Stanford University, Stanford, CA). They were then plated on glass coverslips coated with poly-d-lysine (100 µg/ml) and laminin (25 µg/ml) for live-imaging the next day. For most experiments on CAMPSAPs, endogenous CAMPSAP proteins were visualized by immunofluorescence. For this, cells were transfected with control or ninein siRNA as described earlier. At 72 h after transfection, cells were fixed with –20°C methanol and stained with antibodies against γ-tubulin (Sigma-Aldrich), CAMSAP2 (17880–1-AP; Proteintech), CAMSAP3 (T-14, sc-240596; Santa Cruz Biotechnology, Inc.), and ninein (F-5, sc-376420; Santa Cruz Biotechnology, Inc.). All images were taken using an Axio Observer.Z1 inverted microscope (ZEISS), and image acquisition was controlled using the Zen Blue software (ZEISS), with identical exposure settings. Images used for quantification did not contain pixel saturation, with minimal bleaching during imaging acquisition. Z-stacks images of cultured migratory neurons were acquired using the same system. Optical sections were taken at a 0.19-µm interval size.
Data analyses and statistics
Fluorescence decay measurements were performed using the Zen Blue software package. A region of interest was created in the center of the converted zone of the leading process, and tdEos-tubulin intensity values were measured for each time point. Leading process length measurements, angle measurements, and migration direction analyses were performed using publicly available, open-source plugins in Fiji (National Institutes of Health). Analyses were performed by a blinded rater. All statistical analyses were performed using SPSS 23 (IBM) and graphs were constructed using Excel (Microsoft). Data represent, if not otherwise specified, mean ± SEM. In each experiment, three independent repeats were performed. For statistical analysis, the mean difference was considered to be significant if P < 0.05. Specific statistical analyses are indicated in the figure legends. Correlative analysis was performed by the Spearman correlation test. Multiple group comparison was performed by one-way analysis of variance followed by Bonferroni post hoc analyses. The t test was used for pair analyses.
Online supplemental material
Figs. S1–S4 show data from the ET, ectopic CAMSAP expression, CAMSAP immunostaining, and kinesin-5 and FCPT experiments, respectively. Video 1 shows a 3D z-stack after CAMSAP and ninein staining. Video 2 shows live-cell imaging of neuronal migration. Video 3 shows fluorescent decay under ninein-depleted and ninein-depleted + FCPT conditions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506140/DC1.
Supplementary Material
Acknowledgments
For gifts of DNA constructs and other tools and/or advice, we thank Hiroshi Shinohara, Masatoshi Takeichi of Center for Developmental Biology–Institute of Physical and Chemical Research, Susan McConnell of Stanford University, Maria Diakonova of University of Toledo, Yi-Ren Hong of Kaohsiung Medical University, Erich and Elena Nigg of University of Basel, Michel Bornens of Institut Curie, Mette Mogensen of University of East Anglia, Casper Hoogenraad of Utrecht University, and Timothy Mitchison of Harvard University. We thank Ankita Patil for assistance with data analysis, and Drs. Liang Qiang and Wenqian Yu of Drexel University for critical comments on the manuscript.
The work of the authors is supported by grants to P.W. Baas from the Craig H. Neilsen Foundation (grant 259350), the National Institutes of Health (National Institute of Neurological Disorders and Stroke; grant R01 NS28785), and the U.S. Department of Defense (GW120037 and GW140086). This work is also supported by a Simons Foundation Autism Research Initiative grant to X. Yuan and P.W. Baas from the Simons Foundation (296143). A. Hoenger is supported by a grant from the National Institutes of Health (National Institute of General Medicine Sciences; P41GM103431). A.N. Rao is supported by an F31 Ruth L. Kirschstein National Research Service Award from the National Institutes of Health (1F31NS093748-01A1). A. Falnikar is the recipient of the Doris Willig, MD Award from the Drexel University College of Medicine Institute for Women’s Health and Leadership.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AC
- arc-chord
- CAMSAP
- Calmodulin-regulated spectrin-associated protein
- ET
- electron tomography
- MT
- microtubule
- tdEos
- tandem-dimer Eos
References
- Abal M., Piel M., Bouckson-Castaing V., Mogensen M., Sibarita J.B., and Bornens M.. 2002. Microtubule release from the centrosome in migrating cells. J. Cell Biol. 159:731–737. 10.1083/jcb.200207076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F.J., and Baas P.W.. 1995. Microtubules released from the neuronal centrosome are transported into the axon. J. Cell Sci. 108:2761–2769. [DOI] [PubMed] [Google Scholar]
- Ahmad F.J., Joshi H.C., Centonze V.E., and Baas P.W.. 1994. Inhibition of microtubule nucleation at the neuronal centrosome compromises axon growth. Neuron. 12:271–280. 10.1016/0896-6273(94)90270-4 [DOI] [PubMed] [Google Scholar]
- Ahmad F.J., Echeverri C.J., Vallee R.B., and Baas P.W.. 1998. Cytoplasmic dynein and dynactin are required for the transport of microtubules into the axon. J. Cell Biol. 140:391–401. 10.1083/jcb.140.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F.J., He Y., Myers K.A., Hasaka T.P., Francis F., Black M.M., and Baas P.W.. 2006. Effects of dynactin disruption and dynein depletion on axonal microtubules. Traffic. 7:524–537. 10.1111/j.1600-0854.2006.00403.x [DOI] [PubMed] [Google Scholar]
- Baas P.W. 1999. Microtubules and neuronal polarity: lessons from mitosis. Neuron. 22:23–31. 10.1016/S0896-6273(00)80675-3 [DOI] [PubMed] [Google Scholar]
- Baas P.W., and Falnikar A.. 2012. Re-evaluation of the Neuronal Centrosome as a Generator of Microtubules for Axons and Dendrites. In Centrosome. Schatten H., ed. Humana Press, New York, NY: 309–326. 10.1007/978-1-62703-035-9_18 [DOI] [Google Scholar]
- Baas P.W., Vidya Nadar C., and Myers K.A.. 2006. Axonal transport of microtubules: the long and short of it. Traffic. 7:490–498. 10.1111/j.1600-0854.2006.00392.x [DOI] [PubMed] [Google Scholar]
- Baines A.J., Bignone P.A., King M.D., Maggs A.M., Bennett P.M., Pinder J.C., and Phillips G.W.. 2009. The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol. Biol. Evol. 26:2005–2014. 10.1093/molbev/msp115 [DOI] [PubMed] [Google Scholar]
- Baird D.H., Myers K.A., Mogensen M., Moss D., and Baas P.W.. 2004. Distribution of the microtubule-related protein ninein in developing neurons. Neuropharmacology. 47:677–683. 10.1016/j.neuropharm.2004.07.016 [DOI] [PubMed] [Google Scholar]
- Barkovich J. 2013. Complication begets clarification in classification. Brain. 136:368–373. 10.1093/brain/awt001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix G.J., and Clark G.D.. 1998. Platelet-activating factor receptor stimulation disrupts neuronal migration In vitro. J. Neurosci. 18:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V., Moudjou M., Ferguson D.J., Mucklow S., Belkaid Y., Milon G., and Crocker P.R.. 1996. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 109:179–190. [DOI] [PubMed] [Google Scholar]
- Clark G.D., McNeil R.S., Bix G.J., and Swann J.W.. 1995. Platelet-activating factor produces neuronal growth cone collapse. Neuroreport. 6:2569–2575. 10.1097/00001756-199512150-00029 [DOI] [PubMed] [Google Scholar]
- del Castillo U., Lu W., Winding M., Lakonishok M., and Gelfand V.I.. 2015a Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr. Biol. 25:200–205. 10.1016/j.cub.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo U., Winding M., Lu W., and Gelfand V.I.. 2015b Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in neurons. eLife. 4 10.7554/eLife.10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson J.C., and Hatten M.E.. 1987. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J. Neurosci. 7:1928–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A., Tole S., and Baas P.W.. 2011. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol. Biol. Cell. 22:1561–1574. 10.1091/mbc.E10-11-0905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A., Tole S., Liu M., Liu J.S., and Baas P.W.. 2013. Polarity in migrating neurons is related to a mechanism analogous to cytokinesis. Curr. Biol. 23:1215–1220. 10.1016/j.cub.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.S., and Vale R.D.. 2010. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 143:263–274. 10.1016/j.cell.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory W.A., Edmondson J.C., Hatten M.E., and Mason C.A.. 1988. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J. Neurosci. 8:1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A.C., Needleman D., Brangwynne C., Gradinaru C., Fowler B., Mazitschek R., and Mitchison T.J.. 2008. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J. Cell Sci. 121:2293–2300. 10.1242/jcs.024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasaka T.P., Myers K.A., and Baas P.W.. 2004. Role of actin filaments in the axonal transport of microtubules. J. Neurosci. 24:11291–11301. 10.1523/JNEUROSCI.3443-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Francis F., Myers K.A., Yu W., Black M.M., and Baas P.W.. 2005. Role of cytoplasmic dynein in the axonal transport of microtubules and neurofilaments. J. Cell Biol. 168:697–703. 10.1083/jcb.200407191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S., Fleck M.W., Gambello M.J., Bix G.J., Chen A., Clark G.D., Ledbetter D.H., McBain C.J., and Wynshaw-Boris A.. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19:333–339. 10.1038/1221 [DOI] [PubMed] [Google Scholar]
- Höög J.L., Schwartz C., Noon A.T., O’Toole E.T., Mastronarde D.N., McIntosh J.R., and Antony C.. 2007. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell. 12:349–361. 10.1016/j.devcel.2007.01.020 [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F., Heck A.J., Hoogenraad C.C., and Akhmanova A.. 2014. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 28:295–309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Kahn O.I., Ha N., Baird M.A., Davidson M.W., and Baas P.W.. 2015a TPX2 regulates neuronal morphology through kinesin-5 interaction. Cytoskeleton (Hoboken). 72:340–348. 10.1002/cm.21234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn O.I., Sharma V., Gonzalez-Billault C., and Baas P.W.. 2015b Effects of kinesin-5 inhibition on dendritic architecture and microtubule organization. Mol. Biol. Cell. 26:66–77. 10.1091/mbc.E14-08-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky M.S., Mastronarde D.N., McIntosh J.R., Howell K.E., and Staehelin L.A.. 1999. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 144:1135–1149. 10.1083/jcb.144.6.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo L., Yu W., and Baas P.W.. 2016. Using siRNA to study microtubule-related proteins in cultured neurons. Methods Cell Biol. 131:163–174. [DOI] [PubMed] [Google Scholar]
- Liu M., Nadar V.C., Kozielski F., Kozlowska M., Yu W., and Baas P.W.. 2010. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J. Neurosci. 30:14896–14906. 10.1523/JNEUROSCI.3739-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120:343–352. 10.1006/jsbi.1997.3919 [DOI] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., and Bornens M.. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Myers K.A., and Baas P.W.. 2007. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 178:1081–1091. 10.1083/jcb.200702074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K.A., He Y., Hasaka T.P., and Baas P.W.. 2006. Microtubule transport in the axon: Re-thinking a potential role for the actin cytoskeleton. Neuroscientist. 12:107–118. 10.1177/1073858405283428 [DOI] [PubMed] [Google Scholar]
- Nadar V.C., Ketschek A., Myers K.A., Gallo G., and Baas P.W.. 2008. Kinesin-5 is essential for growth-cone turning. Curr. Biol. 18:1972–1977. 10.1016/j.cub.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C.A., Adams N.D., Auger K.R., Burgess J.L., Carson J.D., Chaudhari A.M., Copeland R.A., Diamond M.A., Donatelli C.A., Duffy K.J., et al. . 2007. Novel ATP-competitive kinesin spindle protein inhibitors. J. Med. Chem. 50:4939–4952. 10.1021/jm070435y [DOI] [PubMed] [Google Scholar]
- Rakic P. 1971. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J. Comp. Neurol. 141:283–312. 10.1002/cne.901410303 [DOI] [PubMed] [Google Scholar]
- Rivas R.J., and Hatten M.E.. 1995. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 15:981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.W., Schoonderwoert V.T., Ji L., Mederios N., Danuser G., and Forscher P.. 2008. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell. 15:146–162. 10.1016/j.devcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., and Scholey J.M.. 2000. Microtubule motors in mitosis. Nature. 407:41–47. 10.1038/35024000 [DOI] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., and Pellman D.. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372. 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Shinohara H., Sakayori N., Takahashi M., and Osumi N.. 2013. Ninein is essential for the maintenance of the cortical progenitor character by anchoring the centrosome to microtubules. Biol. Open. 2:739–749. 10.1242/bio.20135231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M.D., Xie Z., Gleeson J.G., and Tsai L.H.. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 44:263–277. 10.1016/j.neuron.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Sidman R.L., and Rakic P.. 1973. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 62:1–35. 10.1016/0006-8993(73)90617-3 [DOI] [PubMed] [Google Scholar]
- Slaughter T., Wang J., and Black M.M.. 1997. Microtubule transport from the cell body into the axons of growing neurons. J. Neurosci. 17:5807–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D.J., Model L., Gaetz J., Kapoor T.M., and Hatten M.E.. 2004. Par6alpha signaling controls glial-guided neuronal migration. Nat. Neurosci. 7:1195–1203. 10.1038/nn1332 [DOI] [PubMed] [Google Scholar]
- Srivatsa S., Parthasarathy S., Molnar Z., and Tarabykin V.. 2015. Sip1 downstream Effector ninein controls neocortical axonal growth, ipsilateral branching, and microtubule growth and stability. Neuron. 85:998–1012. 10.1016/j.neuron.2015.01.018 [DOI] [PubMed] [Google Scholar]
- Tanaka N., Meng W., Nagae S., and Takeichi M.. 2012. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc. Natl. Acad. Sci. USA. 109:20029–20034. 10.1073/pnas.1218017109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Serneo F.F., Higgins C., Gambello M.J., Wynshaw-Boris A., and Gleeson J.G.. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165:709–721. 10.1083/jcb.200309025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L.H., and Gleeson J.G.. 2005. Nucleokinesis in neuronal migration. Neuron. 46:383–388. 10.1016/j.neuron.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Umeshima H., Hirano T., and Kengaku M.. 2007. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci. USA. 104:16182–16187. 10.1073/pnas.0708047104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., and Brown A.. 2002. Rapid movement of microtubules in axons. Curr. Biol. 12:1496–1501. 10.1016/S0960-9822(02)01078-3 [DOI] [PubMed] [Google Scholar]
- Willemsen M.H., Vissers L.E., Willemsen M.A., van Bon B.W., Kroes T., de Ligt J., de Vries B.B., Schoots J., Lugtenberg D., Hamel B.C., et al. . 2012. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet. 49:179–183. 10.1136/jmedgenet-2011-100542 [DOI] [PubMed] [Google Scholar]
- Xiang X., Beckwith S.M., and Morris N.R.. 1994. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 91:2100–2104. 10.1073/pnas.91.6.2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Sanada K., Samuels B.A., Shih H., and Tsai L.H.. 2003. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 114:469–482. 10.1016/S0092-8674(03)00605-6 [DOI] [PubMed] [Google Scholar]
- Yau K.W., van Beuningen S.F., Cunha-Ferreira I., Cloin B.M., van Battum E.Y., Will L., Schatzle P., Tas R.P., van Krugten J., Katrukha E.A., et al. . 2014. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 82:1058–1073. 10.1016/j.neuron.2014.04.019 [DOI] [PubMed] [Google Scholar]
- Yu W., Schwei M.J., and Baas P.W.. 1996. Microtubule transport and assembly during axon growth. J. Cell Biol. 133:151–157. 10.1083/jcb.133.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.