Abstract
Drosophila telomeres are maintained by transposition to chromosome ends of the HeT-A, TART and TAHRE retrotransposons, collectively designated as HTT. Although all Drosophila telomeres terminate with HTT arrays and are capped by the terminin complex, they differ in the type of subtelomeric chromatin. The HTT sequences of YS, YL, XR, and 4L are juxtaposed to constitutive heterochromatin, while the HTTs of the other telomeres are linked to either the TAS repeat-associated chromatin (XL, 2L, 2R, 3L, 3R) or to the specialized 4R chromatin. We found that mutations in pendolino (peo) cause (telomeric fusions) that preferentially involve the heterochromatin-associated telomeres (Ha-telomeres), a telomeric fusion pattern never observed in the other 10 telomere-capping mutants characterized so far. Peo, is homologous to the E2 variant ubiquitin-conjugating enzymes and is required for DNA replication. Our analyses lead us to hypothesize that DNA replication in Peo-depleted cells results in specific fusigenic lesions concentrated in Ha-telomeres. These data provide the first demonstration that subtelomeres can affect telomere fusion.
Keywords: DNA replication, drosophila, heterochromatin, pendolino (Peo), subtelomeric chromatin, telomere fusion, terminin
Telomeres are nucleoprotein complexes at the ends of eukaryotic chromosomes that perform two essential functions. They counterbalance incomplete replication of terminal DNA, which occurs because conventional polymerases cannot complete the replication of linear DNA. In addition, they form a protective cap at chromosome ends, preventing both DNA damage checkpoint activation and end-to-end fusion events.1-3 This protective function is crucial for maintenance of genome stability, as the dicentric chromosomes generated by telomeric fusions can cause non-disjunction and chromosome breakage during anaphase, ultimately promoting tumor development.4,5
In most organisms, the end replication problem is circumvented by the action of telomerase a reverse transcriptase with an internal RNA template that extends terminal DNA by adding short GC-rich repeats. These repeats specifically bind a discrete number of specialized proteins, which in turn recruit a series of additional factors to form a functional telomere.1,2 In Drosophila, there is no telomerase, and telomeres are elongated by the targeted transposition of three specialized, non-LTR retrotransposons (HeT-A, TART and TAHRE, collectively designated as HTT).6,7 Importantly, in contrast to telomerase-containing organisms, Drosophila telomeres are epigenetically determined structures that can assemble independently of the sequence at the DNA termini.3,8,9
Drosophila telomeres are capped and protected by the terminin complex, which includes HOAP, HipHop, Moi and Ver.3,10-13 All terminin subunits are fast evolving, non-conserved telomere-specific proteins that prevent end-to-end fusion and appear to function only at telomeres. Drosophila telomeres are protected from fusion by a number of other, evolutionarily conserved proteins. These proteins have additional, telomere-unrelated functions and do not localize exclusively to telomeres. They include the HP1a chromatin component,14 the ATM kinase,15-20 each of the subunits of the DNA repair complex Mre11-Rad50- Nbs,15,19,20-22 the E2 ubiquitin conjugating enzyme Eff/UbcD123 and the Woc transcription factor.24 Interestingly, most of the non-terminin proteins (HP1a, ATM, Mre11, Rad50, Nbs) have human counterparts that play roles in human telomere maintenance.3,9
Based on these results, we have previously proposed that concomitant with telomerase loss, Drosophila evolved terminin to bind chromosome ends in a sequence-independent fashion. We have also suggested that terminin is structurally similar and functionally analogous to shelterin,3,11,13,25 the human telomere-capping protein complex consisting of six proteins (TRF1, TRF2, POT1 TIN2, TPP1 and hRAP1) that specifically binds the TTAGGG telomeric repeats in humans.1 We also proposed that Drosophila non-terminin proteins required for telomere capping correspond to ancestral telomere-associated proteins that could not evolve as rapidly as terminin because of the functional constraints imposed by their involvement in other, diverse cellular processes.3,25 This hypothesis implies that non-terminin proteins could have homologues with telomeric functions in other organisms, including humans. This in turn suggests that Drosophila may be an excellent model system for the identification of novel human proteins involved in telomere maintenance.3,25
This hypothesis raises the question of how Drosophila lost telomerase. Most insect orders and species possess telomerase, although they can vary in sequence of the RNA component (TERC) used as template for telomeric DNA (see ref.26 and references therein). Drosophila elongates its telomeres by transposon addition, and mosquitoes and chironomids rely on gene conversion of terminal satellite sequences for telomere maintenance.27,28 Studies on the hymenopteran silkworm Bombyx mori have provided some insights into how telomerase was lost in diptera. The telomeres of B. mori are made of TTAGG repeats, but they also contain two non-LTR transposons that specifically insert into these repeats.29-31 However, in this species, TERT expression is very low and telomerase activity is almost undetectable.32 It is thus conceivable that B. mori is progressively switching from a telomerase-based to a transposon-based mechanism of telomere maintenance, an evolutionary process that has been completed in Drosophila.
Our recent studies on the Drosophila pendolino (peo) gene33 have provided strong support for our hypothesis on the different evolutionary dynamics of terminin and non-terminin telomere proteins. Strong peo mutants display in average five telomeric fusions/cell and often exhibit multicentric linear chromosomes that resemble little “trains”. The pendolino gene was named after this phenotype, just as caravaggio (cav), modigliani (moi) and verrocchio (ver), which are all names of Italian trains. peo encodes a conserved E2 variant ubiquitin-conjugating enzyme that is enriched in many polytene chromosome bands and interbands, and is therefore a non-terminin protein.33 We have shown that AKTIP and Ft1, the human and mouse orthologues of Peo, are required for telomere maintenance.34 Strikingly, Peo and AKTIP directly bind unrelated terminin and shelterin components, respectively. This finding underscores the importance of Peo and its human orthologues as telomere maintenance factors, as they co-evolved with divergent capping complexes to maintain an interaction with telomeres.
Our studies on peo, in addition to reinforcing the idea that the discovery of additional non-terminin proteins can lead to the identification of new human telomere components, also revealed a new type of “telomere position effect.” The classic telomere position effect (TPE) consists of silencing reporter genes inserted in the proximity of Drosophila telomeres. TPE is reminiscent of the position effect variegation (PEV) induced by Drosophila constitutive heterochromatin, even if most of the genes that modulate PEV have no effect on TPE (see ref.35 and references therein). The new type of “telomere position effect” observed in peo mutants consists of preferential involvement of heterochromatin-associated telomeres in fusion events.
Although all Drosophila telomeres terminate with HTT arrays and are capped by terminin, they differ in the type of subtelomeric chromatin. The HTT arrays of the YL, YS, XR, and 4L telomeres are juxtaposed to constitutive heterochromatin; those of the XL, 2L, 2R, 3L and 3R telomeres are adjacent to clusters of heterogeneous telomere-associated satellite (TAS) DNA sequences,36 while the 4R HTT arrays are placed next to chromatin with both euchromatic and heterochromatic features (Fig. 1). The HTT- and the TAS- associated chromatin types have different silencing properties. The TAS chromatin has the ability to silence white+ (w+) transgenes and is responsible for canonical TPE.36 The HTT chromatin properties are dependent on their chromosomal location. w+ transgenes inserted into the HTTs arrays of the 2L, 3L and 3R arms are not silenced, whereas the expression w+ transgenes within the 4R or YS HTTs is impaired, leading to a variegated eye color phenotype.37,38 In addition, w+ transgenes embedded into the TAS and the YS HTT arrays respond differently to genetic modifiers. The effects on TPE of TAS-associated transgenes are ameliorated by mutation in a specific set of genes, most of which have no effect on PEV (see ref.35 and references therein). In contrast, the expression of transgenes inserted into the YS HTT arrays is enhanced by mutation in typical PEV suppressor genes.38 These results suggest that the chromatin of the HTTs juxtaposed to heterochromatin shares some properties with constitutive heterochromatin and is different from the autosomal HTT and TAS chromatin types (Fig. 1). We therefore designate the YL, YS, XR, and 4 chromosome telomeres as heterochromatin-associated (Ha-telomeres).
Figure 1.
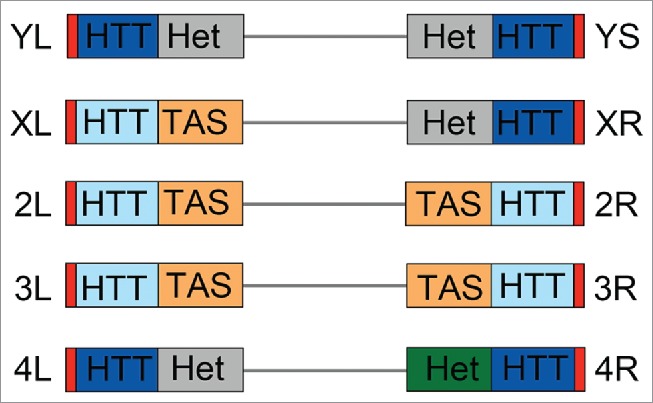
Schematic representation of Drosophila chromosome ends. From a distal to a proximal direction chromosome ends contain the terminin-associated region (red), the HTT array that may either repress (dark blue) or not repress (light blue) the expression of transgenes; the TAS sequences (orange) or different chromatin types: constitutive heterochromatin (Het, gray), or the specialized 4th chromosome chromatin (Het, dark green).
In strong peo mutants that display an average of five telomeric fusions/cell, assessing the relative involvement of individual telomeres in telomeric fusions is quite difficult. However, this can be easily done in weak peo mutants that exhibit approximately one telomeric fusion/cell (Fig. 2A). In these mutants, the vast majority of telomeric fusions involve the XR, the Y and the 4th chromosome telomeres, suggesting that mutations in peo preferentially affect Ha-telomeres and that this effect is partially masked in strong peo mutants where most telomeres are fused.33 A preferential involvement of Ha-telomeres in fusion events is a peculiar feature of peo mutants and has not been observed in the other Drosophila mutants we characterized in the past (eff, Su(var)205, cav, mre11, rad50, nbs, tefu, woc, moi and ver) (Fig. 2B; see also ref.35 and references therein).
Figure 2.
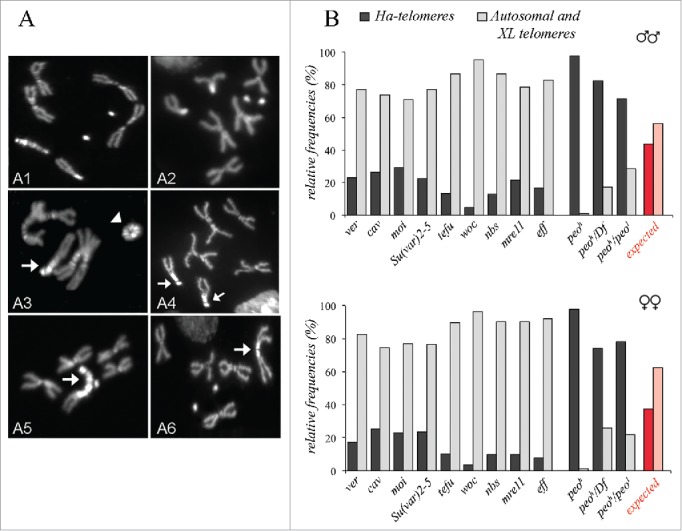
Mutations in peo cause telomeric fusion that preferentially involve Ha-telomeres. (A) Examples of telomeric fusions in peo mutant neuroblasts. (A1-2) Control (Oregon-R) male (A1) and female (A2) metaphases; (A3-A6) metaphases from peo mutant brains showing: (A3) a ring Y chromosome (arrowhead) and a 4 (arrow)-4-XR tricentric chromosome; (A4) two 4-XR telomeric fusions (arrows); (A5) a 4 (arrow)-YS · YL. -XR tricentric chromosome; (A6) an XR-XR telomeric fusion (arrow). (B) Frequencies of telomeric fusions involving Ha-telomeres in different telomere fusion mutants. The red and pink columns represent the frequencies of Ha- (red) and non-HA (autosomal and XL pink) telomeres calculated assuming a random involvement of telomeres in fusion events. The peoh and peo1 are weak and strong peo mutant alleles, respectively.
Understanding why Ha-telomeres are prone to fuse in a peo mutant background is not straightforward. All telomeres of peo mutants recruit similar amounts of terminin proteins and HP1a, ruling out the possibility that Ha-telomeres are specifically uncapped, and thus particularly subjected to fusion events. It is possible that the peculiar pattern of telomeric fusions observed in peo mutants reflects problems in DNA replication. Peo mutant brains display fewer nuclei undergoing DNA replication compared with wild type brains. Moreover, peo mutant brains do not differ from control brains in the relative frequency of nuclei in early, mid or late S phase of the cell cycle, suggesting that heterochromatin replicates normally in a peo mutant background.33 These data suggest that the wild type function of peo is required for general DNA synthesis and not for completion of specific sub-phases of DNA replication. Thus, it is unlikely that mutations in peo specifically affect replication of Ha-telomeres, which presumably replicate at the end of the S phase, together with the bulk of heterochromatin.39 It is also unlikely that a general defect in DNA replication results in the characteristic telomere fusion pattern observed in peo mutants, as chemical treatments and mutations that block DNA synthesis in Drosophila brains do not cause telomeric fusions. Thus, our current model suggests that Peo is required for general DNA replication, including telomere replication, and that loss of Peo results in replication-dependent fusigenic lesions concentrated in the Ha-telomeres.33 However, we cannot completely exclude the possibility that peo has a dual function, being independently required for DNA replication and telomere capping.
Conclusions and Perspectives
We have shown that mutations in peo cause telomeric fusions that preferentially involve the telomeres juxtaposed to constitutive heterochromatin. This is a highly specific fusion pattern, as none of the other Drosophila mutations that cause telomeric fusions preferentially affect the Ha-telomeres. To the best of our knowledge, these findings provide the first demonstration that (i) subtelomeres can regulate telomere fusion, and that (ii) individual telomeres are diversely fusigenic in response to mutations that disrupt different components of the telomere capping machinery. We believe that these results will stimulate further studies on the role of subtelomeres in the maintenance of telomere and genome stability, particularly in systems where the telomeres are associated with different chromatin types.
Recent studies have shown that subtelomeric heterochromatin can silence genes embedded into the adjacent HTT array.38 This effect is reminiscent of PEV and suggests that some heterochromatic proteins spread into the HTT array, impairing the expression of the inserted transgenes. These findings open the way to future studies aimed at the identification of the factors that modify the specific sensitivity of Ha-telomeres to the loss of peo function. We are currently investigating whether some of the many genes that modulate PEV are also able to modify the pattern of telomeric fusions elicited by mutations in peo. These studies should help in our understanding of the molecular mechanisms underlying telomeric fusion formation in peo mutants, and will possibly lead to the identification of proteins with specific roles in heterochromatin replication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to thank Laura Ciapponi and Gianni Cenci for thoughtful comments on the manuscript.
Funding
This research was supported by a grant from the Italian Association for Cancer Research to MG (AIRC, IG 16020).
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev of Gent 2008; 42:301-34; http://dx.doi.org/ 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- 2.Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev of Gent 2010; 44:243-69; http://dx.doi.org/ 10.1146/annurev-genet-102108-134841 [DOI] [PubMed] [Google Scholar]
- 3.Raffa GD, Ciapponi L, Cenci G, Gatti M. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2011; 2:383-91; PMID:21989238; http://dx.doi.org/ 10.4161/nucl.2.5.17873 [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Telomere dynamics and genome instability in human cancer. In Telomeres Blackburn EH, Greider CW, eds. (Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press; ) 1995; pp. 265-93 [Google Scholar]
- 5.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science 2002; 297:565-9; PMID:12142527; http://dx.doi.org/ 10.1126/science.297.5581.565 [DOI] [PubMed] [Google Scholar]
- 6.Mason JM, Frydrychova RC, Biessmann H. Drosophila telomeres: an exception providing new insights. Bio Essays 2008; 30:25-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A 2011; 108:20317-24; PMID:21821789; http://dx.doi.org/ 10.1073/pnas.1100278108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cenci G, Ciapponi L, Gatti M. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 2005; 114:135-45; PMID:16012858; http://dx.doi.org/ 10.1007/s00412-005-0005-9 [DOI] [PubMed] [Google Scholar]
- 9.Rong YS. Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma 2008; 117:235-242; PMID:18193446; http://dx.doi.org/ 10.1007/s00412-007-0144-2 [DOI] [PubMed] [Google Scholar]
- 10.Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 2003; 5:82-4; PMID:12510197; http://dx.doi.org/ 10.1038/ncb902 [DOI] [PubMed] [Google Scholar]
- 11.Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci U S A 2009; 106:2271-6; PMID:19181850; http://dx.doi.org/ 10.1073/pnas.0812702106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J 2010; 29:819-29; PMID:20057353; http://dx.doi.org/ 10.1038/emboj.2009.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, Gatti M, Ciapponi L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 2010; 24:1596-601; PMID:20679394; http://dx.doi.org/ 10.1101/gad.574810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 1998; 2:527-38; PMID:9844626; http://dx.doi.org/ 10.1016/S1097-2765(00)80152-5 [DOI] [PubMed] [Google Scholar]
- 15.Bi X, Wei SC, Rong YS. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol 2004; 14:1348-53; PMID:15296751; http://dx.doi.org/ 10.1016/j.cub.2004.06.063 [DOI] [PubMed] [Google Scholar]
- 16.Oikemus SR, McGinnis N, Queiroz-Machado J, Tukachinsky H, Takada S, Sunkel CE, Brodsky MH. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev 2004; 18:1850-61; PMID:15256487; http://dx.doi.org/ 10.1101/gad.1202504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva E, Tiong S, Pedersen M, Homola E, Royou A, Fasulo B, Siriaco G, Campbell SD. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr Biol 2004; 14:1341-7; PMID:15296750; http://dx.doi.org/ 10.1016/j.cub.2004.06.056 [DOI] [PubMed] [Google Scholar]
- 18.Song YH, Mirey G, Betson M, Haber DA, Settleman J. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr Biol 2004; 14:1354-9; PMID:15296752; http://dx.doi.org/ 10.1016/j.cub.2004.06.064 [DOI] [PubMed] [Google Scholar]
- 19.Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A 2005; 102:15167-72; PMID:16203987; http://dx.doi.org/ 10.1073/pnas.0504981102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet 2006; 2:e71; PMID:16710445; http://dx.doi.org/ 10.1371/journal.pgen.0020071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, Engels WR, Gatti M. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol 2004; 14:1360-6; PMID:15296753; http://dx.doi.org/ 10.1016/j.cub.2004.07.019 [DOI] [PubMed] [Google Scholar]
- 22.Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 2006; 173(3):1447-54; PMID:16648644; http://dx.doi.org/ 10.1534/genetics.106.058081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev 1997; 11:863-75; PMID:9106658; http://dx.doi.org/ 10.1101/gad.11.7.863 [DOI] [PubMed] [Google Scholar]
- 24.Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol Cell 2005; 20:821-31; PMID:16364909; http://dx.doi.org/ 10.1016/j.molcel.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 25.Raffa GD, Cenci G, Ciapponi L, Gatti M. Organization and Evolution of Drosophila Terminin: Similarities and Differences between Drosophila and Human Telomeres. Front Oncol 2013; 3:112; http://dx.doi.org/ 10.3389/fonc.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korandová M, Krůček T, Vrbová K, Čapková Frydrychová R. Distribution of TTAGGG-specific telomerase activity in insects. Chromosome Res 2014; 22:495-503; http://dx.doi.org/ 10.1007/s10577-014-9436-6 [DOI] [PubMed] [Google Scholar]
- 27.Roth CW, Kobeski F, Walter MF, Biessmann H. Chromosome end elongation by recombination in the mosquito Anopheles gambiae. Mol Cell Biol 1997; 17:5176-83; PMID:9271395; http://dx.doi.org/ 10.1128/MCB.17.9.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason JM, Frydrychova RC, Biessmann H. Drosophila telomeres: an exception providing new insights. Bioessays 2008; 30:25-37; PMID:18081009; http://dx.doi.org/ 10.1002/bies.20688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki S, Ishikawa H, Fujiwara H. Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori. Mol Cell Biol 1995; 15:4545-52; PMID:7623845; http://dx.doi.org/ 10.1128/MCB.15.8.4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubo Y, Okazaki S, Anzai T, Fujiwara H. Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in Lepidopteran insects. Mol Biol Evol 2001; 18:848-57; PMID:11319268; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a003866 [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori. Chromosome Res 2005; 13:455-67; PMID:16132811; http://dx.doi.org/ 10.1007/s10577-005-0990-9 [DOI] [PubMed] [Google Scholar]
- 32.Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle). Gene 2006; 376:281-89; PMID:16793225; http://dx.doi.org/ 10.1016/j.gene.2006.04.022 [DOI] [PubMed] [Google Scholar]
- 33.Cenci G, Ciapponi L, Marzullo M, Raffa G, Morciano P, Raimondo D, Burla R, Saggio I, Gatti M. The analysis of pendolino (peo) mutants reveals differences in the fusigenic potential among Drosophila telomeres. PLoS Genetics 2015; 11(6):e1005260; PMID:26110638; http://dx.doi.org/ 10.1371/journal.pgen.1005260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burla R, Carcuro M, Raffa GD, Galati A, Raimondo D, Rizzo A, La Torre M, Micheli E, Ciapponi L, Cenci G, et al.. AKTIP/Ft1, a new shelterin-interacting factor required for telomere maintenance. PLoS Genet 2015; 11(6):e1005167; PMID:26110528; http://dx.doi.org/ 10.1371/journal.pgen.1005167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cipressa F, Romano S, Centonze S, Zur Lage PI, Verni F, Dimitri P, Gatti M, Cenci G. Effete, a Drosophila Chromatin-Associated Ubiquitin-Conjugating Enzyme that Affects Telomeric and Heterochromatic Position Effect Variegation. Genetics 2013; 195(1):147-58; PMID:23821599; http://dx.doi.org/ 10.1534/genetics.113.153320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason J, Villasante A. Subtelomeres in Drosophila and Other Diptera; In: Louis EJ, Becker MM, editors. Subtelomeres: Springer; Berlin Heidelberg; 2014. p. 211-25 [Google Scholar]
- 37.Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, Walter MF, Mason JM. Two distinct domains in Drosophila melanogaster telomeres. Genetics 2005; 171:1767-77; PMID:16143601; http://dx.doi.org/ 10.1534/genetics.105.048827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang SH, Nan R, Accardo MC, Sentmanat M, Dimitri P, Elgin SC. A distinct type of heterochromatin at the telomeric region of the Drosophila melanogaster Y chromosome. PloS One 2014; 9:e86451; PMID:24475122; http://dx.doi.org/ 10.1371/journal.pone.0086451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Ann Rev Genetics 1992; 26:239-75; http://dx.doi.org/ 10.1146/annurev.ge.26.120192.001323 [DOI] [PubMed] [Google Scholar]


