Abstract
Background:
PTEN is a tumor suppressor frequently deleted in prostate cancer that may be a useful prognostic biomarker. However, the association of PTEN loss with lethal disease has not been tested in a large, predominantly surgically treated cohort.
Methods:
In the Health Professionals Follow-up Study and Physicians’ Health Study, we followed 1044 incident prostate cancer cases diagnosed between 1986 and 2009 for cancer-specific and all-cause mortality. A genetically validated PTEN immunohistochemistry (IHC) assay was performed on tissue microarrays (TMAs). TMPRSS2:ERG status was previously assessed in a subset of cases by a genetically validated IHC assay for ERG. Cox proportional hazards models adjusting for age and body mass index at diagnosis, Gleason grade, and clinical or pathologic TNM stage were used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for the association with lethal disease. All statistical tests were two-sided.
Results:
On average, men were followed 11.7 years, during which there were 81 lethal events. Sixteen percent of cases had complete PTEN loss in all TMA cores and 9% had heterogeneous PTEN loss across cores. After adjustment for clinical-pathologic variables, complete PTEN loss was associated with lethal progression (HR = 1.8, 95% CI = 1.2 to 2.9). The association of PTEN loss (complete or heterogeneous) with lethal progression was only among men with ERG-negative (HR = 3.1, 95% CI = 1.7 to 5.7) but not ERG-positive (HR = 1.2, 95% CI = 0.7 to 2.2) tumors.
Conclusions:
PTEN loss is independently associated with increased risk of lethal progression, particularly in the ERG fusion–negative subgroup. These validated and inexpensive IHC assays may be useful for risk stratification in prostate cancer.
Phosphatase and tensin homolog (PTEN) is the most commonly inactivated tumor suppressor in prostate cancer (1–5), and its loss is associated with aggressive clinical-pathologic features (6–12) and development of castration resistant disease (13–16). PTEN inactivation may promote castration-resistant tumor growth through upregulation of oncogenic Akt/mTOR signaling (17,18), suppression of androgen receptor (AR) transcription factor activity, and inhibition of AR-regulated negative feedback of Akt (15). Thus, PTEN is a promising potential prognostic biomarker in prostate cancer, and it may identify patients responsive to PI3K/Akt/mTOR inhibitors currently being studied in clinical trials.
PTEN is most commonly lost by deletion, which is frequently a focal and subclonal event in primary prostate tumors (10,19,20), making reliable detection difficult by methods requiring nucleic acid extraction or fluorescence in situ hybridization (FISH). Addressing this issue, our group previously optimized an immunohistochemistry (IHC) assay for in situ PTEN protein detection in prostate cancer (6). Using this IHC assay, PTEN loss has been associated with biopsy upgrading (20), biochemical recurrence (7,9), and metastatic progression in a high-risk cohort (6). However, the association with lethal prostate cancer progression in a population-based, surgically treated cohort has not been tested.
Approximately half of US prostate cancer cases are positive for the TMPRSS2:ERG gene fusion (21), an event that can also be detected using a validated IHC assay (22). Tumor fusion status is not associated with lethal progression in most studies (22), but our group recently presented the first evidence that tumor fusion status may modify the association of prostate cancer risk factors with lethal prostate cancer progression (23). PTEN loss is more common in fusion-positive compared with fusion-negative disease (10,24–27), and PTEN loss almost certainly occurs subsequent to ERG rearrangement (19,28,29). Thus, presence of the TMPRSS2:ERG gene fusion may modify the effects of PTEN loss on disease progression. Indeed, animal models suggest PTEN loss cooperates with TMPRSS2:ERG fusion in tumorigenesis, but results from the few published human studies are varied (11,13,30,31). Clarifying the interaction of TMPRSS2:ERG gene fusion and PTEN loss with respect to disease progression may lead to improved clinical risk stratification, help guide treatment decisions, and improve our understanding of the underlying biological roles of these two somatic events.
We conducted a large patho-epidemiology investigation among prostate cancer patients in the Health Professional Follow-up Study (HPFS) and the Physicians’ Health Study (PHS) addressing: 1) the association of PTEN loss, assessed by a validated IHC protocol, with lethal progression and 2) the potential for TMPRSS2:ERG gene fusion, detected by IHC, to modify the role of PTEN loss in lethal disease progression.
Methods
Study Population
We included 1044 men diagnosed with prostate cancer who were participants in the PHS (n = 245) (32,33) or HPFS (n = 799) (34). The men were diagnosed with cancer between 1983 and 2009 and had available archival prostate tumor materials for evaluation. The PHS was a randomized trial investigating the prevention of cardiovascular disease and cancer among 29 071 male physicians aged followed with annual questionnaires since 1982. The HPFS is an ongoing cohort of 51 529 male health professionals followed with biannual questionnaires since 1986. Incident prostate cancers (ICD-9: 185) were self-reported and confirmed through medical record and pathology report review. Details of the prostate cancer tumor cohorts within these two studies are available elsewhere (23).
Clinical and Follow-up Data
We abstracted data on tumor stage, prostate specific antigen (PSA) level at diagnosis, and treatments from medical records and pathology reports. Standardized histopathologic review by study pathologists of H&E slides of each case provided uniform Gleason grading (35). Prostate cancer patients were followed up with written questionnaires to collect detailed information regarding treatments and clinical progression. Biochemical recurrence was reported by participants, the treating physician, or abstracted from medical records, defined as a PSA above 0.2ng/mL postsurgery sustained over two measures. The date of first increase in PSA was considered the date of biochemical recurrence. For prostate cancer cases in the HPFS, treating physicians were contacted to collect clinical course information and to confirm development of metastases. For prostate cancer cases in the PHS, 80% of reports of metastases were confirmed by medical record review; we thus relied on self-report because available records corroborated the self-report of these physician participants in virtually every instance. Cause of death was determined by medical record and death certificate review. Follow-up for mortality in the cohorts is greater than 98%.
Immunohistochemical Assessment of Protein Expression
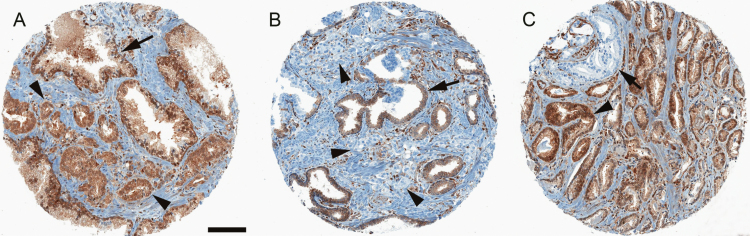
We characterized TMPRSS2:ERG fusion status and PTEN expression on tumor tissue available from a biorepository of archival radical prostatectomy (RP; 95%) and transurethral resection of the prostate (TURP; 5%) tumor specimens. Hematoxylin and eosin slides were reviewed by our study pathologists (MF, SF) to confirm prostate cancer and to identify tumor areas for tissue microarray (TMA) construction. Thirteen TMAs were constructed by sampling at least three 0.6mm cores of tumor per case from the dominant nodule or nodule with the highest Gleason pattern. A detailed description of the immunohistochemical methods is provided in the Supplementary Methods (available online) and previous studies (6,36). Briefly, PTEN IHC was performed using a rabbit antihuman PTEN antibody (Clone D4.3 XP; Cell Signaling Technologies, Danvers, MA), which has been shown to strongly correlate with FISH and high-resolution single-nucleotide polymorphism (SNP) array analysis (6). PTEN scoring was conducted by study pathologists (TLM and CLM) who were both blinded to clinical and outcome data. A tissue core was considered to have PTEN protein loss if the intensity of cytoplasmic and nuclear staining was markedly decreased or entirely lost (0+ intensity) or markedly decreased (1+ intensity) across more than 10% of tumor cells compared with surrounding benign glands and/or stroma, which provide internal positive controls for PTEN protein expression (Figure 1). This simple dichotomous scoring system has been shown to be strongly associated with underlying homozygous genetic deletion of PTEN in prior studies by SNP array and fluorescence in situ hybridization (6) and showed high interobserver reproducibility in the tumor samples from the current study (Supplementary Methods, available online). We previously performed ERG IHC on a subset of tumors (Clone EPR3864, Epitomics, Inc., Burlingame, CA) (22), which is strongly associated with fusion status assessed by FISH (36). We classified tumors as ERG positive (ie, carrying the TMPRSS2:ERG gene fusion) if at least one core stained positive for ERG and ERG negative if all cores stained negative for ERG.
Figure 1.
PTEN immunostaining of prostate cancer tissue microarray (TMA) cores. A) PTEN is intact in small infiltrating cancer glands (arrowheads), with similar cytoplasmic staining intensity (brown pigment) as seen in adjacent larger benign glands (arrow). B) PTEN is homogeneously lost in small infiltrating tumor glands (arrowheads), compared with larger benign glands (arrow) where PTEN is intact. Note intraductal spread of prostate cancer cells with PTEN loss within the larger benign gland (arrow). C) Heterogeneous PTEN loss in tumor sampled on a single TMA core with some cancer glands showing intact PTEN (arrowhead) and others showing PTEN loss (arrow).
Statistical Methods
We compared demographic, clinical characteristics, and prognostic outcomes according to PTEN expression. We used Cox proportional hazards models to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) of the association between PTEN loss with time to development of lethal prostate cancer (defined as prostate cancer death or metastases to bone or other organ) and PSA recurrence. Person-time was calculated from the date of cancer diagnosis to the earliest of the following time points: either development of lethal prostate cancer or PSA recurrence (depending on the endpoint for the analysis), censored at time of death from other causes, or end of follow-up (March 2011 for the PHS and December 2011 for the HPFS). We adjusted for age at diagnosis (years, continuous) and BMI at diagnosis (kg/m2, continuous) in all analyses, and additionally for Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal) and clinical or pathologic stage (T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, and T4 N1 M1; ordinal) in full multivariable analyses. Simple mean imputation was used for individuals missing clinical tumor stage (n = 17). We further evaluated the association of PTEN with lethal progression stratified by tumor stage (T1/T2, N0/Nx, M0/Mx vs T3/T4 or N1 or M1), Gleason score (2 - 7 vs 8 - 10), tissue source (RP vs TURP), and PSA era of cancer diagnosis (diagnosed in or before 1992 vs after 1992). We evaluated PTEN and ERG interaction by cross-classifying tumor PTEN and ERG expression and used the Wald test to evaluate multiplicative interaction terms. The Kaplan-Meier method was used to estimate survival probabilities and the log-rank test to compare groups. For all models, the proportional hazards assumption was evaluated and satisfied by testing the significance of the interaction between PTEN status and follow-up time.
Analyses were conducted using SAS version 9.2 (SAS institute Inc., Cary, NC), and all statistical tests were two-sided, with P values below .05 considered statistically significant.
This research project was approved by the institutional review boards at Partners Healthcare and the Harvard School of Public Health. Written, informed consent was obtained from each subject.
Results
The association between PTEN loss and lethal progression did not statistically differ between the two cohorts; we therefore present results for the combined cohorts. Of the 1044 cases with characterized PTEN expression, 16% had complete PTEN loss in all TMA cores, 9% had heterogeneous PTEN loss across cores, and 75% had intact PTEN expression in all cores. Selected clinical and demographic characteristics of the cohort are presented in Table 1. Tumors with complete PTEN loss were statistically significantly more likely to be ERG positive and to have a higher Gleason grade and more advanced clinical and pathologic tumor stage. Moreover, complete PTEN loss tended to be associated with a lower mean BMI at diagnosis, though this was not a statistically significant finding. The frequency of PTEN loss was similar among the RP and TURP specimens. No appreciable association was seen between PTEN expression and age at diagnosis, PSA at diagnosis, or with having a family history of prostate cancer.
Table 1.
Selected clinical characteristics among men with prostate cancer between 1983 and 2009 with measured PTEN in the Health Professional Follow-up Study and the Physicians’ Health Study
| Characteristics | N* | PTEN intact or heterogeneous PTEN loss† (n = 878) |
Complete PTEN loss‡ (n=166) |
P§ |
|---|---|---|---|---|
| Age at diagnosis, mean (SD), y | 1044 | 65.8 (6.0) | 66.3 (5.9) | .26 |
| BMI at diagnosis, mean (SD), kg/m2 | 1043 | 26.0 (3.4) | 25.5 (3.2) | .10 |
| Family history of prostate cancer||, No. (%) | 799 | 143 (21) | 28 (25) | .32 |
| PSA at diagnosis, median (q1, q3), ng/mL | 931 | 6.4 (4.8, 10.0) | 7.0 (4.9, 11.1) | .46 |
| ERG positive, No. (%) | 1019 | 385 (45) | 113 (69) | <.001 |
| Gleason grade, No. (%) | 1044 | <.001 | ||
| <6 | 173 (20) | 11 (7) | ||
| 3+4 | 338 (38) | 35 (21) | ||
| 4+3 | 199 (23) | 55 (33) | ||
| 8+ | 168 (19) | 65 (39) | ||
| Pathologic TNM, No. (%) | 967 | <.001 | ||
| T2 N0/Nx | 621 (76) | 80 (52) | ||
| T3 N0/Nx | 177 (22) | 63 (41) | ||
| T4 N1/M1 | 15 (2) | 11 (7) | ||
| Clinical TNM, No. (%) | 1027 | .01 | ||
| T1,T2 N0/Nx | 830 (96) | 150 (93) | ||
| T3 N0/Nx | 27 (3) | 6 (4) | ||
| T4 N1/M1 | 8 (1) | 6 (4) | ||
| Tissue source, No. (%) | 1044 | |||
| Radical prostatectomy | 831 (95) | 158 (95) | 1.00 | |
| TURP | 47 (5) | 8 (5) | ||
| Cohort, No. (%) | 1044 | |||
| Health Professionals Follow-up Study | 687 (86) | 112 (14) | .003 | |
| Physicians’ Health Study | 191 (78) | 54 (22) |
* Number of men with data available on the respective characteristic. BMI = body mass index; PSA = prostate-specific antigen; TURP = transurethral resections of the prostate.
† At least 1 tissue microarray (TMA) core with PTEN intact.
‡ All TMA cores with PTEN loss.
§ P values are based on the t test for age at diagnosis and body mass index at diagnosis; Fisher’s exact test for ERG expression, family history of prostate cancer, and tissue source; the Wilcoxon-Mann-Whitney test for PSA at diagnosis; the Cochran-Armitage trend test for Gleason grade, clinical tumor stage, and pathologic tumor stage.
|| Family history of prostate cancer in father or brother(s); data are only available in the Health Professionals Follow-up Study.
During a mean follow-up time of 11.7 years, there were 81 (HPFS n = 59 and PHS n = 22) lethal prostate cancer events. Men whose tumors showed complete PTEN loss had a statistically significantly higher risk for lethal progression compared with men whose tumors showed PTEN intact or heterogeneous loss, even after adjusting for clinical-pathologic characteristics (HR = 1.8, 95% CI = 1.2 to 2.9). Similar results were found when comparing tumors with any PTEN loss (heterogeneous or complete) to intact PTEN. However, complete PTEN loss (HR = 1.9, 95% CI = 1.2 to 3.0), but not heterogeneous PTEN loss (HR = 1.2, 95% CI = 0.5 to 2.6), was associated with lethal progression after adjustment for clinical-pathologic characteristics (Table 2). Similar results were found when prostate cancer–specific death was used as the outcome (data not shown).
Table 2.
Association of PTEN loss with lethal prostate cancer among men diagnosed with prostate cancer between 1983 and 2009 in the Health Professional Follow-up Study and the Physicians’ Health Study
| Measurement | PTEN status | ||
|---|---|---|---|
| PTEN intact or heterogeneous PTEN loss* | Complete PTEN loss† | ||
| No. of lethal events | 53 | 28 | |
| No. total | 878 | 166 | |
| Person-time, y | 10 187 | 1988 | |
| HR (95% CI)‡ | 1.0 | 2.7 (1.7 to 4.3) | |
| HR (95% CI)§ | 1.0 | 1.8 (1.2 to 2.9) | |
| PTEN intact|| | Any PTEN loss¶ | ||
| No. of lethal events | 46 | 35 | |
| No. total | 781 | 263 | |
| Person-time, y | 9124 | 3051 | |
| HR (95% CI)‡ | 1.0 | 2.3 (1.5 to 3.6) | |
| HR (95% CI)§ | 1.0 | 1.7 (1.1 to 2.6) | |
| PTEN intact|| | Heterogeneous PTEN loss# | Complete PTEN loss† | |
| No. of lethal events | 46 | 7 | 28 |
| No. total | 781 | 97 | 166 |
| Person-time, y | 9124 | 1063 | 1988 |
| HR (95% CI)‡ | 1.0 | 1.4 (0.6 to 3.0) | 2.8 (1.8 to 4.5) |
| HR (95% CI)§ | 1.0 | 1.2 (0.5 to 2.6) | 1.9 (1.2 to 3.0) |
* At least 1 tissue microarray (TMA) core with PTEN intact. CI = confidence interval; HR = hazard ratio.
† All TMA cores with PTEN loss.
‡ Adjusted for age at diagnosis and body mass index at diagnosis.
§ Adjusted for age at diagnosis, body mass index at diagnosis, Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal), and clinical tumor stage
(T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, and T4 N1 M1; ordinal).
|| No TMA cores with PTEN loss.
¶ At least 1 TMA core with PTEN loss.
# At least 1 TMA core with PTEN loss and 1 TMA core with PTEN intact.
The association of complete PTEN loss with lethal progression did not statistically differ according to subgroups of clinical characteristics or by tissue specimen. There was a suggestion that PTEN loss may be more associated with lethal progression in advanced pathologic stage tumors (HR = 2.1, 95% CI = 1.1 to 3.9), compared with nonadvanced pathologic stage tumors (HR = 0.7, 95% CI = 0.2 to 2.5), but this was not a statistically significant difference and based on only three lethal events in the nonadvanced group (Table 3). Complete PTEN loss was associated with lethal progression in tumors diagnosed in both the pre-PSA and PSA eras (data not shown). Restricting analyses to cases who had a minimum of three TMA cores scored for PTEN expression did not appreciably alter our findings (data not shown).
Table 3.
Association of PTEN loss with lethal prostate cancer according to clinical characteristics among men diagnosed with prostate cancer between 1983 and 2009 in the Health Professional Follow-up Study and the Physicians’ Health Study
| Clinical characteristics | PTEN intact or heterogeneous PTEN loss* | Complete PTEN loss† |
|---|---|---|
| Nonadvanced clinical tumor stage‡ | ||
| N lethal event | 39 | 22 |
| N total | 830 | 150 |
| HR (95% CI)§ | 1.00 | 1.8 (1.1 to 3.1) |
| Advanced clinical tumor stage|| | ||
| N lethal event | 13 | 6 |
| N total | 35 | 12 |
| HR (95% CI)§ | 1.00 | 1.5 (0.5 to 4.3) |
| Nonadvanced pathologic tumor stage¶ | ||
| N lethal event | 17 | 3 |
| N total | 621 | 80 |
| HR (95% CI)§ | 1.00 | 0.7 (0.2 to 2.5) |
| Advanced pathologic tumor stage# | ||
| N lethal event | 22 | 18 |
| N total | 192 | 74 |
| HR (95% CI)§ | 1.00 | 2.1 (1.1 to 3.9) |
| Gleason grade 2–7 | ||
| N lethal event | 26 | 11 |
| N total | 710 | 101 |
| HR (95% CI)** | 1.00 | 2.5 (1.2 to 5.1) |
| HR (95% CI)†† | 1.00 | 1.6 (0.7 to 3.5) |
| Gleason grade 8–10 | ||
| N lethal event | 27 | 17 |
| N total | 168 | 65 |
| HR (95% CI)** | 1.00 | 1.8 (1.0 to 3.2) |
| HR (95% CI)†† | 1.00 | 1.7 (0.8 to 3.6) |
| Radical prostatectomy | ||
| N lethal event | 41 | 22 |
| N total | 831 | 158 |
| HR (95% CI)‡‡ | 1.00 | 1.5 (0.9 to 2.6) |
| TURP | ||
| N lethal event | 12 | 6 |
| N total | 47 | 8 |
| HR (95% CI)§§ | 1.00 | 2.2 (0.7 to 7.0) |
* At least 1 tissue microarray (TMA) core with PTEN intact. CI = confidence interval; HR = hazard ratio; TURP = transurethral resections of the prostate. † All TMA cores with PTEN loss.
‡ Defined as T1–T2, N0–Nx, M0–Mx.
§ Adjusted for age at diagnosis, body mass index at diagnosis, and Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal).
|| Defined as T3–T4 or N1 or M1.
¶ Defined as T2, N0–Nx, M0–Mx; analyses restricted to radical prostatectomy specimens.
# Defined as T3–T4 or N1 or M1; analyses restricted to radical prostatectomy specimens.
** Adjusted for age at diagnosis, body mass index at diagnosis, and clinical tumor stage (T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, and T4 N1 M1; ordinal).
†† Adjusted for age at diagnosis, body mass index at diagnosis, and pathologic TNM stage (sample size [N lethal/N total] in Gleason grade 2–7 PTEN intact: 22/665, PTEN loss 9/99; Gleason grade 8–10 PTEN intact: 17/148, PTEN loss 12/55).
‡‡ Adjusted for age at diagnosis, body mass index at diagnosis, Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal), and pathologic tumor stage (T2 N0/Nx M0/Mx,
T3 N0/Nx M0/Mx, and T4 N1 M1; ordinal).
§§ Adjusted for age at diagnosis, body mass index at diagnosis, Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal), and clinical tumor stage (T1/T2 N0/Nx M0/Mx,
T3 N0/Nx M0/Mx, and T4 N1 M1; ordinal).
Complete PTEN loss was associated with a higher risk for PSA recurrence in the minimally adjusted model but was attenuated and no longer statistically significant following adjustment for clinical parameters (data not shown).
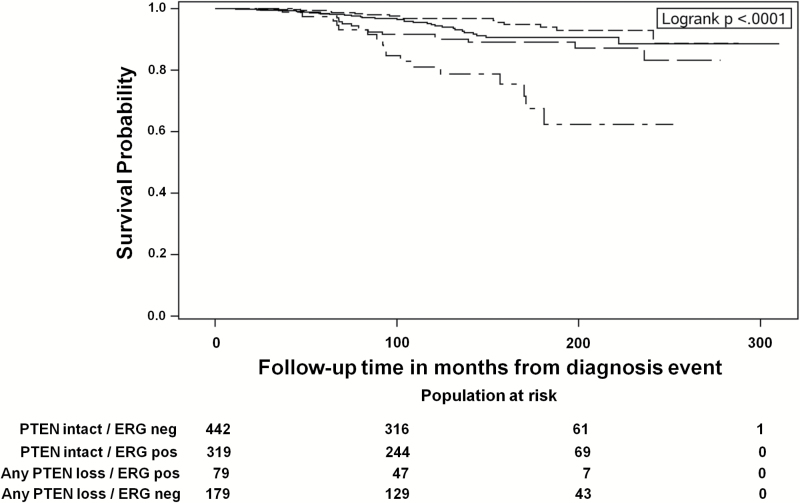
The association of PTEN loss stratified by ERG expression with lethal progression is presented in Figure 2 and Table 4. Compared with ERG-negative cases with intact PTEN, ERG-negative cases with any PTEN loss (complete or heterogeneous) had a statistically higher risk for lethal progression, even after adjustment for Gleason grade and clinical stage (HR = 3.1, 95% CI = 1.7 to 5.7. In contrast, ERG-positive cases with (HR = 1.2, 95% CI = 0.7 to 2.2) or without (HR = 1.0, 95% CI = 0.5 to 1.9) PTEN loss were not at higher risk of lethal progression (P interaction = .05). In a subset of cases with pathologic stage data available, adjusting for pathologic TNM did not appreciably alter these results (P interaction = .02) (Table 4). Results were similar when PTEN loss was classified as PTEN loss in all cores and as PTEN intact for any cores with PTEN expression (Table 5).
Figure 2.
Kaplan-Meier survival plot for association of interaction of PTEN and ERG status with lethal prostate cancer among men diagnosed with prostate cancer between 1983 and 2009 within the Health Professional Follow-up Study and the Physicians’ Health Study.
PTEN intact / ERG negative ———————
PTEN intact / ERG positive – – – – – – – – – –
Any PTEN loss / ERG positive — — — — — —
Any PTEN loss / ERG negative — . — . — . — .
Table 4.
Association of any PTEN Loss stratified by ERG expression with lethal prostate cancer among men diagnosed with prostate cancer between 1983 and 2009 in the Health Professional Follow-up Study and the Physicians’ Health Study
| PTEN/ERG expression | Total observations, No. | Lethal events, No. | Person-years | HR (95% CI)* | P interaction† | HR (95% CI)‡ | P interaction† |
|---|---|---|---|---|---|---|---|
| All cases with PTEN and ERG data | |||||||
| PTEN intact§/ERG negative | 442 | 29 | 4973 | Referent | .23 | Referent | .05 |
| PTEN intact§/ERG positive | 319 | 15 | 3972 | 0.7 (0.4 to 1.2) | 1.0 (0.5 to 1.9) | ||
| Any PTEN loss||/ERG negative | 79 | 17 | 820 | 3.8 (2.1 to 6.9) | 3.1 (1.7 to 5.7) | ||
| Any PTEN loss||/ERG positive | 179 | 18 | 2176 | 1.5 (0.8 to 2.6) | 1.2 (0.7 to 2.2) | ||
| Restricted to cases with pathologic tumor stage data | |||||||
| PTEN intact§/ERG negative | 395 | 21 | 4488 | Referent | .18 | Referent | .02 |
| PTEN intact§/ERG positive | 309 | 12 | 3873 | 0.6 (0.3 to 1.3) | 0.8 (0.4 to 1.7) | ||
| Any PTEN loss||/ERG negative | 76 | 15 | 795 | 4.3 (2.2 to 8.3) | 3.5 (1.8 to 6.9) | ||
| Any PTEN loss||/ERG positive | 171 | 14 | 2136 | 1.4 (0.7 to 2.7) | 0.8 (0.4 to 1.7) | ||
* Adjusted for age at diagnosis and body mass index at diagnosis. CI = confidence interval; HR = hazard ratio.
† P value of PTEN and ERG multiplicative interaction based on the Wald test.
‡ Adjusted for age at diagnosis, body mass index at diagnosis, Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal), and clinical tumor stage (T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, and
T4 N1 M1; ordinal).
§ No tissue microarray (TMA) cores with PTEN loss.
|| At least 1 TMA core with PTEN loss.
Table 5.
Association of complete PTEN loss stratified by ERG expression with lethal prostate cancer among men diagnosed with prostate cancer between 1983 and 2009 in the Health Professional Follow-up Study and the Physicians’ Health Study
| PTEN/ERG expression | Total observations, No. | Lethal events, No. | Person-years | HR (95% CI)* | P interaction † | HR (95% CI)‡ | P interaction † |
|---|---|---|---|---|---|---|---|
| All cases with PTEN and ERG data | |||||||
| Any PTEN intact§/ERG negative | 470 | 32 | 5243 | Referent | .21 | Referent | .12 |
| Any PTEN intact§/ERG positive | 385 | 19 | 4724 | 0.7 (0.4 to 1.2) | 0.9 (0.5 to 1.6) | ||
| Complete PTEN loss||/ERG negative | 51 | 14 | 549 | 4.4 (2.3 to 8.3) | 3.0 (1.6 to 5.7) | ||
| Complete PTEN loss||/ERG positive | 113 | 14 | 1424 | 1.6 (0.9 to 3.1) | 1.3 (0.7 to 2.5) | ||
| Restricted to cases with pathologic tumor stage data | |||||||
| Any PTEN intact§/ERG negative | 423 | 24 | 4759 | Referent | .21 | Referent | .02 |
| Any PTEN intact§/ERG positive | 374 | 15 | 4618 | 0.6 (0.3 to 1.2) | 0.8 (0.4 to 1.7) | ||
| Complete PTEN loss||/ERG negative | 48 | 12 | 524 | 4.7 (2.3 to 9.4) | 3.8 (1.9 to 7.8) | ||
| Complete PTEN loss||/ERG positive | 106 | 11 | 1391 | 1.5 (0.7 to 3.1) | 0.8 (0.4 to 1.7) | ||
* Adjusted for age at diagnosis and body mass index at diagnosis. CI = confidence interval; HR = hazard ratio.
† P value of PTEN and ERG multiplicative interaction based on the Wald test.
‡ Adjusted for age at diagnosis, body mass index at diagnosis, Gleason grade (≤6, 3+4, 4+3, 8–10; ordinal), and clinical tumor stage (T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, and
T4 N1 M1; ordinal).
§ At least 1 tissue microarray (TMA) core with PTEN intact.
|| All TMA cores with PTEN loss.
Discussion
In this large patho-epidemiology investigation, we found that PTEN loss, as assessed by a validated and simple IHC protocol, was associated with a two-fold increase in risk of lethal progression independent of clinical-pathologic parameters. PTEN loss was heterogeneous in about one-third of cases that showed any loss; however, the association with lethal disease was restricted to the men with prostate cancer showing complete loss of PTEN in all sampled tumor glands. PTEN loss (either heterogeneous or complete) was associated with lethal progression primarily among ERG-negative cases and this association was seen after adjustment for pathologic grade and stage. In contrast, PTEN loss among ERG-positive tumors was not associated with lethal progression. To the best of our knowledge, this is the first study to relate combined PTEN and ERG expression to lethal progression in a predominantly surgically treated cohort. Our findings may have important implications for improving our understanding of biologic pathways that drive prostate cancer progression. Moreover, these results suggest that characterizing tumor PTEN and ERG status via simple and relatively inexpensive IHC assays may have clinical utility for distinguishing distinct molecular subtypes of prostate cancer, improving disease risk stratification, and potentially helping to guide treatment decisions.
Most previous studies investigating the prognostic role of PTEN have used biochemical recurrence as the primary outcome, with the majority finding that loss is associated with an increased risk of biochemical recurrence (7,9,11,37–39), but results are not entirely consistent (8,31,40). A limitation of using biochemical recurrence as an outcome is that only a small subset of cases who experience biochemical recurrence will die of prostate cancer (41). Though lethal progression is a more informative endpoint, only a few studies have been able to relate PTEN loss with lethal progression. In the HPFS and PHS, we previously reported that reduced PTEN expression, assessed by a different IHC antibody than that used in the current study (Zymed cat# 18–0256), was associated with an increased risk of lethal progression (HR = 2.4, 95% CI = 1.2 to 4.7). However, following adjustment for clinical characteristics, the association was no longer statistically significant (42). In another study of 675 conservatively managed cases diagnosed by TURP, 18% of cases had PTEN loss by IHC using a similar assay to ours. PTEN loss was associated with a 1.5-fold increased risk of prostate cancer death following adjustment for Gleason grade and PSA (43). Finally, among 217 high-risk surgically treated cases who experienced biochemical recurrence following RP, PTEN loss assessed by our assay was statistically significantly associated with increased risk of metastatic progression (6), though this association did not remain significant after adjusting for grade and stage.
We show that the association of PTEN loss with lethal outcomes was strongest among ERG-negative tumors. Few studies have investigated concomitant PTEN and ERG status in relation to disease progression. Similar to earlier reports, we found PTEN loss was substantially enriched for ERG expression (71%) compared with PTEN-positive tumors (46%) (10,24–26). Perhaps explaining this tendency towards co-occurrence, mouse models have suggested that PTEN loss and TMPRSS2:ERG are incapable of driving prostate tumor growth alone, but together these molecular alterations synergize, driving cell migration and invasion (13). Supporting this finding, Yoshimoto et al. (37) and Leinonen et al. (39) found the highest risk for biochemical recurrence was among fusion-positive tumors with PTEN loss. However, a much larger study by Krohn et al. did not find that tumor fusion status modifies the association of PTEN loss with biochemical recurrence (10). Prior to our study, only Reid et al. had related PTEN and ERG status to lethal progression in a conservatively managed TURP cohort. Despite important cohort differences, Reid et al. also found the highest risk for lethal progression among cases with PTEN loss and negative fusion status (HR = 4.9, 95% CI = 2.3 to 10.4) (31), but this group did not replicate this finding in a larger study (43).
Recently, Chen et al. reported on a transgenic mouse model of TMPRSS2:ERG and PTEN loss in which androgen receptor signaling was downregulated in PTEN-negative and fusion-negative tumors (44). These findings present a potential explanation for our findings, in that further repressing androgen signaling in these tumors may be an ineffective treatment strategy. Indeed, Mulholland et al. demonstrated better inhibition of in vitro growth of PTEN-null tumors when AR/androgen blockage was combined with PI3K/Akt/mTOR inhibition (15). However, Chen et al. also found that ERG expression restored androgen signaling among PTEN-null tumors, enabling development of invasive prostate cancer, suggesting PTEN loss and ERG expression cooperate to promote tumor growth (44). Thus, additional research is needed to validate our finding and to clarify how the interplay between PTEN loss and TMPRSS2:ERG fusion affects disease progression.
This study benefits from a large sample size, long follow-up, and prospectively monitored and validated prostate cancer outcomes. Our study pathologists reviewed and provided uniform Gleason grades for all cases reducing measurement error. We used cause-specific death and distant metastasis as the outcome, which are the most clinically relevant endpoints for prostate cancer. The PTEN IHC assay has been validated and is strongly associated with the presence of underlying PTEN gene deletions (6). However, this study does have limitations. We had a limited number of lethal events, especially limiting our statistical power in our subgroup analyses; therefore, these results should be interpreted cautiously. We mainly used prostatectomy tissue in this investigation; therefore our results may not be generalizable to biopsy specimens. However, a recent investigation using the same PTEN assay among active surveillance-eligible case patients found PTEN loss was associated with an increased risk of upgrading at radical prostatectomy (20). Because our study utilized TMAs, tumor heterogeneity is also likely to be an important limitation, as some cases with heterogeneous PTEN loss may have been missed because of tumor sampling, but this would likely attenuate our findings. Importantly, the association of PTEN loss with lethal events among ERG fusion-negative tumors was much stronger than that seen for PTEN loss considered without ERG status, and this association remained strong even when considering tumors with heterogeneous PTEN loss. Finally, white men primarily comprise our cohort (97%), so our results may not be generalizable to other racial/ethnic groups, in which ERG expression and PTEN loss may be less frequent (45–50).
In conclusion, PTEN loss is associated with an increased risk of lethal progression in prostatectomy samples independent of clinical parameters. When considered alone, complete PTEN loss, but not heterogeneous loss, is associated with lethal progression. When considered with ERG, any PTEN loss was associated with lethal progression in ERG-negative cases but not in ERG positive cases. Importantly, both the PTEN and ERG IHC assays are simple to interpret with dichotomous scoring systems, relatively inexpensive compared with DNA-based assays, and currently running on automated platforms in our CLIA-certified pathology laboratory. These findings support the continued development of PTEN and ERG as prognostic biomarkers and, if replicated in large biopsy cohorts, may have important clinical implications for risk stratification and identifying cases likely to benefit from therapies targeting the PI3K/Akt signaling pathway.
Funding
This work was supported by National Institute of Health (grant numbers R01CA136578, R01CA141298, UM1 CA167552, P50 CA090381, R25 CA112355 to REG, and T32 CA09001 to TUA and EME); the Prostate Cancer Foundation Young Investigators Awards to LAM, TLL, JRR, and KMW; the Patrick C. Walsh Prostate Cancer Research Fund; the Swedish Research Council (Reg. No. 2009–7309) to AP and the Royal Physiographic Society in Lund to AP; the American Cancer Society – Ellison Foundation Postdoctoral Fellowship (PF-14-140-01-CCE) to TUA; Office of the Assistant Secretary of Defense for Health Affairs under Award No. W81XWH-14-1-0250 to EME; and the Physicians’ Health Study is supported by grants CA-34944, CA-40360, and CA-097193 from the National Cancer Institute and grants HL-26490 and HL-34595 from the National Heart, Lung, and Blood Institute (Bethesda, MD).
Supplementary Material
We are grateful to the participants and staff of the Physicians’ Health Study and Health Professionals Follow-up Study for their valuable contributions. In addition we would like to thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The Dana-Farber/Harvard Cancer Center Tissue Microarray Core Facility constructed the tissue microarrays in this project, and we would like to thank Chungdak Li for her expert tissue microarray construction. The authors assume full responsibility for analyses and interpretation of these data.
References
- 1. Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. [DOI] [PubMed] [Google Scholar]
- 4. Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15 (4):356–362. [DOI] [PubMed] [Google Scholar]
- 5. Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18 (1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lotan TL, Gurel B, Sutcliffe S, et al. PTEN Protein Loss by Immunostaining: Analytic Validation and Prognostic Indicator for a High Risk Surgical Cohort of Prostate Cancer Patients. Clin Cancer Res. 2011;17 (20):6563–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antonarakis ES, Keizman D, Zhang Z, et al. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118 (24):6063–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bedolla R, Prihoda TJ, Kreisberg JI, et al. Determining Risk of Biochemical Recurrence in Prostate Cancer by Immunohistochemical Detection of PTEN Expression and Akt Activation. Clin Cancer Res. 2007;13 (13):3860–3867. [DOI] [PubMed] [Google Scholar]
- 9. Chaux A, Peskoe SB, Gonzalez-Roibon N, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25 (11):1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krohn A, Diedler T, Burkhardt L, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181 (2):401–412. [DOI] [PubMed] [Google Scholar]
- 11. Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97 (5):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lotan TL, Gumuskaya B, Rahimi H, et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2013;26 (4):587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41 (5):619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiao J, Wang S, Qiao R, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007;67 (13):6083–6091. [DOI] [PubMed] [Google Scholar]
- 15. Mulholland DJ, Tran LM, Li Y, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19 (6):792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen MM, Abate-Shen C. Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer Res. 2007;67 (14):6535–6538. [DOI] [PubMed] [Google Scholar]
- 17. Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95 (1):29–39. [DOI] [PubMed] [Google Scholar]
- 18. Wu X, Senechal K, Neshat MS, et al. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95 (26):15587–15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gumuskaya B, Gurel B, Fedor H, et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis. 2013;16 (2):209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lotan TL, Carvalho FL, Peskoe SB, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2014;28 (1):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56 (2):275–286. [DOI] [PubMed] [Google Scholar]
- 22. Pettersson A, Graff RE, Bauer SR, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21 (9):1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pettersson A, Lis RT, Meisner A, et al. Modification of the association between obesity and lethal prostate cancer by TMPRSS2:ERG. J Natl Cancer Inst. 2013;105 (24):1881–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44 (6):685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bismar TA, Yoshimoto M, Vollmer RT, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107 (3):477–485. [DOI] [PubMed] [Google Scholar]
- 26. Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41 (5):619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41 (5):524–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krohn A, Freudenthaler F, Harasimowicz S, et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27 (12):1612–1620. [DOI] [PubMed] [Google Scholar]
- 29. Yoshimoto M, Ding K, Sweet JM, et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol. 2013;26 (3):435–447. [DOI] [PubMed] [Google Scholar]
- 30. Chen Y, Chi P, Rockowitz S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19 (8):1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reid AH, Attard G, Ambroisine L, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102 (4):678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gaziano JM, Glynn RJ, Christen WG, et al. Vitamins E and C in the prevention of prostate and total cancer in men: the Physicians’ Health Study II randomized controlled trial. JAMA. 2009;301 (1):52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hennekens CH, Eberlein K. A randomized trial of aspirin and beta-carotene among U.S. physicians. Prev Med. 1985;14 (2):165–168. [DOI] [PubMed] [Google Scholar]
- 34. Giovannucci E, Liu Y, Platz EA, et al. Risk factors for prostate cancer incidence and progression in the health professionals follow-up study. Int J Cancer. 2007;121 (7):1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stark JR, Perner S, Stampfer MJ, et al. Gleason score and lethal prostate cancer: does 3 + 4 = 4 + 3? J Clin Oncol. 2009;27 (21):3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12 (7):590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21 (12):1451–1460. [DOI] [PubMed] [Google Scholar]
- 38. McCall P, Witton CJ, Grimsley S, et al. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99 (8):1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leinonen KA, Saramaki OR, Furusato B, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Caner Epidemiol Biomarkers Prev. 2013;22 (12):2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y, Su J, DingZhang X, et al. PTEN deletion and heme oxygenase-1 overexpression cooperate in prostate cancer progression and are associated with adverse clinical outcome. J Pathol. 2011;224 (1):90–100. [DOI] [PubMed] [Google Scholar]
- 41. Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294 (4):433–439. [DOI] [PubMed] [Google Scholar]
- 42. Zu K, Martin NE, Fiorentino M, et al. Protein expression of PTEN, insulin-like growth factor I receptor (IGF-IR), and lethal prostate cancer: a prospective study. Cancer Epidemiol Biomarkers Prev. 2013;22 (11):1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cuzick J, Yang ZH, Fisher G, et al. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer. 2013;108 (12):2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Y, Chi P, Rockowitz S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19 (8):1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khani F, Mosquera JM, Park K, et al. Evidence for Molecular Differences in Prostate Cancer between African American and Caucasian Men. Clin Cancer Res. 2014;20 (18):4925–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mao X, Yu Y, Boyd LK, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70 (13):5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyagi Y, Sasaki T, Fujinami K, et al. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010;23 (11):1492–1498. [DOI] [PubMed] [Google Scholar]
- 48. Magi-Galluzzi C, Tsusuki T, Elson P, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71 (5):489–497. [DOI] [PubMed] [Google Scholar]
- 49. Rosen P, Pfister D, Young D, et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology. 2012;80 (4):749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rawal S, Young D, Williams M, et al. Low Frequency of the ERG Oncogene Alterations in Prostate Cancer Patients from India. J Cancer. 2013;4 (6):468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.