Abstract
Drug-resistant pathogens are a growing problem, and novel strategies are needed to combat this threat. Among the most significant of these resistant pathogens is Mycobacterium tuberculosis, which is an unusually difficult microbial target due to its complex membrane. Here, we design peptides for specific activity against M. tuberculosis using a combination of “database filtering” bioinformatics, protein engineering, and de novo design. Several variants of these peptides are structurally characterized to validate the design process. The designed peptides exhibit potent activity (MIC values as low as 4 μM) against M. tuberculosis and also exhibit broad activity against a host of other clinically relevant pathogenic bacteria such as Gram-positive bacteria (Streptococcus) and Gram-negative bacteria (Escherichia coli). They also display excellent selectivity, with low cytotoxicity against cultured macrophages and lung epithelial cells. These first-generation antimicrobial peptides serve as a platform for the design of antibiotics and for investigating structure-activity relationships in the context of the M. tuberculosis membrane. The antimicrobial peptide design strategy is expected to be generalizable for any pathogen for which an activity database can be created.
INTRODUCTION
Antibiotic resistance is increasing at an alarming rate, especially for Mycobacterium tuberculosis, the drug-resistant form of which infected an estimated 400,000 people in 2012 according to the World Health Organization (WHO) (1). Alternatives to traditional antibiotics, which act through diverse mechanisms, are urgently needed to combat these resistant bacteria. Disruption of bacterial, but not mammalian, outer membrane integrity with peptides is one such strategy to destroy pathogenic bacteria in a highly selective manner (2). Design strategies to develop potent, stable antimicrobial peptides (AMPs) stemming from a fundamental understanding of their mechanism of cell disruption are urgently needed.
M. tuberculosis is an interesting target pathogen due to its unique membrane properties, which defies simple classification as a Gram-positive or Gram-negative bacterium. M. tuberculosis presents a complex asymmetric outer membrane containing an inner layer of mainly mycolic acids arranged orthogonal to the cell envelope and an outer layer consisting of a large variety of other lipids (3). The mycolic acid chains are covalently bound to arabinogalactan and form a rigid inner layer, which is thought to become more fluid moving outwards (4). The high lipid content and covalent bonding are thought to contribute to the high degree of impermeability of the mycobacterial membrane, which assists the organism with resistance to lipophilic therapeutics (5). Transport of hydrophilic molecules is mediated by outer membranes proteins, of which few are known and even fewer are characterized (6–8). Despite these daunting membrane properties, AMPs with activity against M. tuberculosis have been reported (9–12).
Many AMPs are short, cationic peptides that adopt an alpha-helical conformation. Other conformations also exist but are usually larger peptides or are stabilized by disulfide bonds (13). Upon discovery of naturally occurring AMPs, many were tested for activity against M. tuberculosis, including human and rabbit defensins and porcine protegrins. The most potent of these displayed >90% killing of M. tuberculosis at 50 μg/ml (15 μM human defensin peptide 1, 13 μM rabbit neutrophil peptide 2, and 23 μM protegrin 1) and acted by a mechanism that produced visible lesions on the mycobacterial outer membrane (11). Subsequently, several of the broadly active natural peptides were modified and tested against M. tuberculosis with MICs as low as 10 μM (10, 14). Large, entirely synthetic libraries were also tested against M. tuberculosis with MICs reported to be as low as 1 μM (9). In addition, peptoids, which are more resistant to degradation than peptides, with MIC values as low as 6 μM were developed (15).
Despite clear evidence of their efficacy, the mechanism of action remains debated. It is generally accepted that the majority of AMPs act through disruption of microbial membranes, although they may have additional host-related immunomodulating activities (16). Recently, many insights into the motifs that govern the effectiveness of short alpha-helical AMPs have been gained. Activity is dependent on a mixture of hydrophobic and cationic residues, arranged to form an amphipathic peptide (17). It has been proposed that the cationic portion targets the peptide to the negatively charged bacterial membrane, while the hydrophobic portion allows for intercalation into the membrane and subsequent disruption of the membrane via a number of proposed mechanisms (18, 19). This amphipathic character lends itself to design due to the periodicity of the alpha-helical arrangement. Peptides can be visualized in two dimensions by using wheel diagrams (20), and sequences bearing separate cationic and hydrophobic faces can easily be constructed (21).
The majority of previous studies have focused on optimizing naturally occurring peptides, screening large random synthetic libraries to develop potential drug candidates against a specific microbial target, or investigating the general mechanism of action. We propose a combined approach, using bioinformatics and rational design informed by certain mechanistic rules, to develop a set of more potent initial peptides than those found in nature. This strategy assimilates data from both natural peptides as well as those identified by screening large randomly constructed libraries. We have combined a de novo design approach called “database filtering” (22) with protein engineering, rational design, and three-dimensional (3D) modeling to design potent AMPs against a selected microbial target. Database filtering uses a library of peptides with reported activity against the target bacterium to determine a characteristic peptide length, overall charge and hydrophobicity, and commonly occurring residues, resulting in a set of amino acids (22). We then employ rational design, including the use of wheel diagrams, to arrange the set of amino acids in a way that maximizes the amphipathic nature of the peptide. 3D modeling is then employed to verify an alpha-helical conformation and the proper distribution of amino acids to generate the amphipathic surface. We demonstrate this technique against M. tuberculosis, resulting in AMPs with appreciable activity toward the mycobacterium.
MATERIALS AND METHODS
Peptide synthesis.
The following reagents for peptide synthesis were purchased from AGTC Bioproducts (Wilmington, MA): N,N-dimethylformamide (DMF) (99.8% purity), piperidine (99.5%), methyl tert-butyl ether (MTBE) (99.5%), dichloromethane (DCM) (99.9%), and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) (coupling reagent). Triisopropylsilane (TIS) (99%), acetonitrile (ACN) (99%), N-methylmorpholine (NMM) (99%), acetic acid (99.7%), and 1-methyl-2-pyrrolidinone (NMP) (99.5%) were purchased from Sigma-Aldrich. Trifluoroacetic acid (TFA) (99.5%) was purchased from Alfa Aesar (Ward Hill, MA). The following amino acids (>99%) were purchased from 21st Century Biochemicals (Marlborough, MA): 9-fluorenylmethoxy carbonyl (Fmoc)-l-Arg(Pbf)-OH, Fmoc-l-Ile-OH, Fmoc-l-Leu-OH, Fmoc-l-Lys(Boc)-OH, Fmoc-l-Ser(OtBu)-OH, and Fmoc-l-Trp(Boc)-OH. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerate (POPG), and extrusion materials were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). AMPs were produced by using an automated Multiprep RS synthesizer (Intavis AG, Germany) in the laboratory of Pankaj Karande. Fmoc solid-phase chemistry was used to synthesize the peptides from their C termini to their N termini on TentaGel rink amide resin (0.25 mmol/g) (Intavis, Inc.). Presynthesis, the resin was swollen in a DMF-DCM (2:1) solution. Postsynthesis, the resin was washed with DCM, and the peptides were cleaved off by using a TFA-TIS-H2O (88/6/6) cocktail. Bulk TFA was removed by precipitating the peptides in ice-cold MTBE, followed by centrifugation and a second MTBE wash. Peptides were air dried and dissolved in ACN-H2O (1:5) for lyophilization. Lyophilized peptides were stored at −20°C.
Bacteria and cell lines.
Slow-growing attenuated tuberculosis (TB) complex mycobacteria, Mycobacterium tuberculosis strains mc26020 and mc26230 (23, 24) and the Mycobacterium bovis BCG Pasteur strain (Trudeau Institute), were used, along with rapidly growing saprophytic Mycobacterium smegmatis, to assess the general range of peptide activity against mycobacteria. The severely attenuated mc26020 strain is a Δlys ΔpanCD mutant that requires lysine and pantothenate to grow, whereas the mc26030 strain is a ΔRD1 ΔpanCD strain that also requires pantothenate and lacks the RD1 virulence genes. All mycobacteria were routinely cultivated at 37°C in liquid Middlebrook 7H9 medium supplemented with 0.5% glycerol, 10% oleic acid-dextrose-albumin-catalase (OADC), and 0.05% Tween 80 (mycomedia), as described previously (25). Additional supplements include 0.2% Casamino Acids and 24 mg/liter pantothenate for mc26020 or mc26230 and 24 mg/liter lysine for mc26020. Other bacterial reference strains include Staphylococcus aureus ATCC 25923, Streptococcus pneumoniae ATCC 49619, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Bacillus subtilis BGSC 1A1. In addition, Streptococcus pyogenes (group A Streptococcus), Streptococcus agalactiae (group B Streptococcus), Streptococcus mutans, and Klebsiella pneumoniae from clinical isolates were obtained from the Bacteriology Laboratory, Wadsworth Center, Department of Health, Albany, NY. All nonmycobacterial strains were grown in Mueller-Hinton (MH) broth (Difco) at 37°C.
J774.16 mouse macrophages were maintained in antibiotic-free Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 20% (vol/vol) fetal bovine serum, 5% (vol/vol) NCTC-109 (Gibco), 1% (vol/vol) nonessential amino acids (NEAA) (Gibco), and 1% (vol/vol) glutamine. A549 human lung epithelial cells were maintained in antibiotic-free Ham's F-12 medium supplemented with 10% fetal bovine serum, as described previously (26).
Bioinformatics and design.
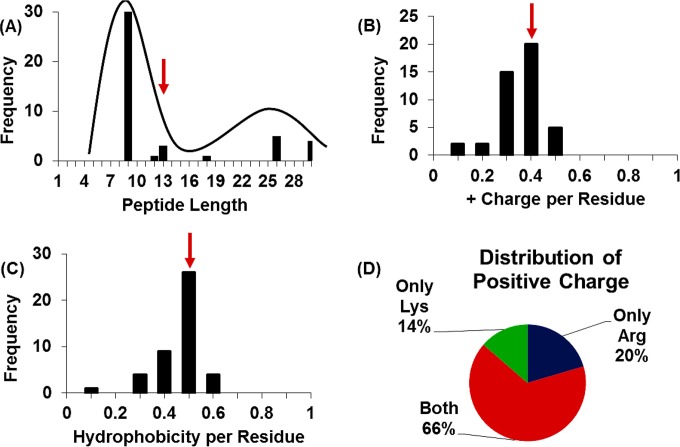
Antimicrobial peptides were designed against M. tuberculosis. The design was based on a previously reported database filtering method (22), a method which outputs the most common peptide length and amino acid composition based on a library of natural and synthetic peptides. It is assumed that if this library is selected to include peptides with high potency against a pathogen of choice, the resulting composition will also have antimicrobial activity. The database filtering method generates a final peptide sequence by ordering the resulting amino acids using additional bioinformatic techniques, but here, we used a structurally informed approach. Briefly, a database was created by using natural peptides from databanks (APD2 [http://aps.unmc.edu/AP/main.php] and CAMP [http://www.camp.bicnirrh.res.in/]) and both natural and synthetic peptides reported in the literature (9–12, 27, 28) that have measured activity against M. tuberculosis (see Table S1 in the supplemental material for a complete list of database peptides). The database was then analyzed to determine the average peptide length, the number of hydrophobic residues, the number of hydrophilic residues per length, and the most represented residues of each type (Fig. 1), resulting in a defined length and amino acid composition.
FIG 1.
Database filtering. (A) Peptide length of the database peptides shows a bimodal distribution centered around 9 and 25 residues. The selected length of the designed AMPs is 13 residues (red arrow), the mean of the entire distribution. (B) Distribution of positive charge per residue length of database peptides. A +0.4 charge per residue, the distribution mean, is selected for the designed AMPs (red arrow). (C) Distribution of the number of hydrophobic residues (Ala, Val, Ile, Leu, Trp, Phe, Met, and Cys) per total peptide length, where 0.5, the mean of the distribution (red arrow), is chosen for the designed AMPs. (D) Distribution of positive charge within database peptides. Histidine was excluded due to the limited number of peptides containing histidine in the database.
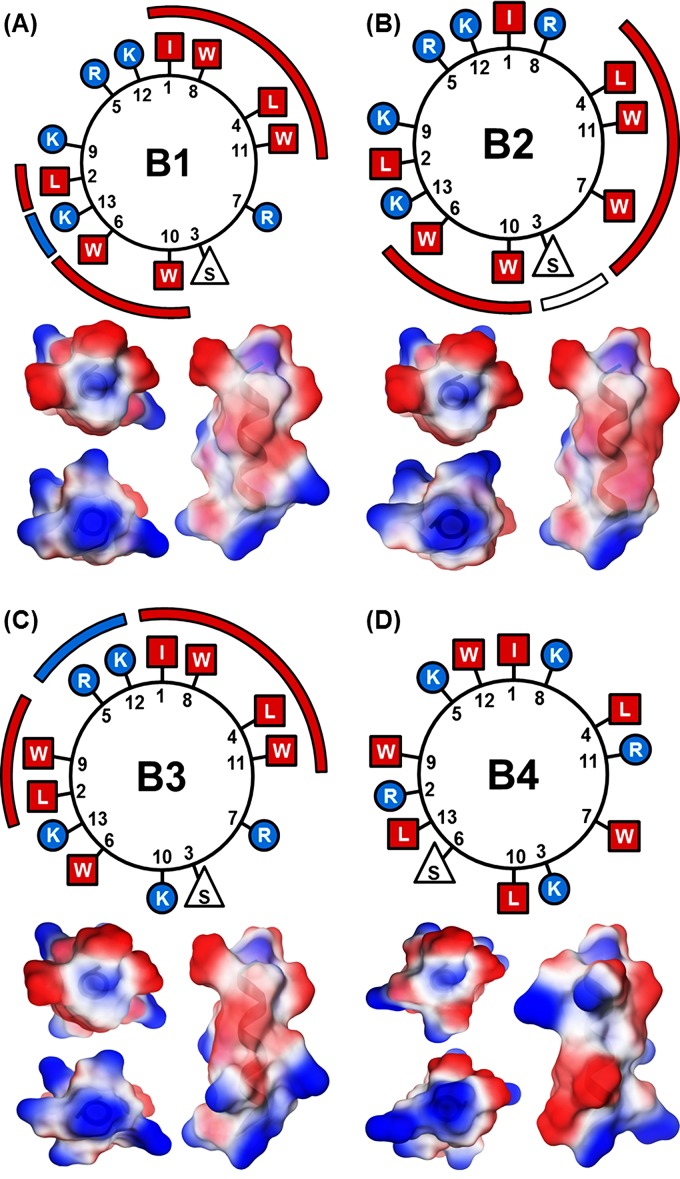
In place of the amino-acid-ordering approach of the database filtering method, which uses bioinformatic-derived sequence motifs, we designed peptides by fitting the resulting set of residues to an alpha-helical template (wheel diagram) (Fig. 2) to produce three amphipathic peptides (the linear sequences for these peptides, along with those of other similar peptides from the literature, are given in Table S2 in the supplemental material). The actual conformation for each designed peptide was predicted by using PepFold (29), and the three-dimensional structures of peptides were modeled by using MOE software (Chemical Computing Group, Montreal, Canada) (Fig. 2).
FIG 2.
AMP visualization. Shown are wheel diagrams for the four designed AMPs located above their respective molecular images. Colored bars are used to emphasize hydrophobic faces (red squares) interrupted by charged (blue circles) or polar (white triangles) residues. Shown are 3D visualizations for the AMPs that correspond with the wheel diagram directly above. For each set of molecular images, the top left image is a top-plan view, the bottom left image is a bottom-up view, and the right image is a side view in the same orientation as the wheel diagrams above. The color scheme (red, hydrophobic; white, polar; blue, charged) is the same throughout the figure.
AMP purification.
AMPs were purified by using a Waters high-performance liquid chromatography (HPLC) instrument (Waters Corporation, Milford, MA) with a C18 column (XBridge BEH130 C18 column, part number 186003568; Waters Corporation). Isocratic elution was performed with 30% acetonitrile at 50°C. Peptide purity increased from <50% to >90% (see Fig. S1 in the supplemental material).
Far-UV circular dichroism.
Lipid films were prepared by dissolving 11.25 μmol POPC and 3.75 μmol POPG in chloroform in a round-bottom flask. Chloroform was evaporated under gentle N2 flow for 10 min and lyophilized overnight to ensure complete evaporation. The lipid film was rehydrated with 3 ml 10 mM phosphate buffer at pH 7.4. Three freeze-thaw cycles were performed, followed by a 31-pass extrusion using a 200-nm membrane (Avanti Polar Lipids, Inc., Alabaster, AL) at 45°C. AMPs were dissolved in 10 mM phosphate buffer at pH 7.4 to a peptide concentration of 500 μM. The peptides and lipids were brought to a final peptide-to-lipid (P:L) molar ratio of 1:10 with a final concentration of 50 μM AMP. Spectra were collected by using a Jasco 815 circular dichroism (CD) spectrometer (Jasco, Easton, MD) with a Spectrosil far-UV quartz cuvette (catalog number 21-Q-1; Starna Cells, Inc., Atascadero, CA). Measurements were taken at 21°C using 10 accumulations. Spectra were read from 260 to 190 nm with a 1.0-nm bandwidth, a sensitivity of 100 mdeg, a response of 1 s, and a scan speed of 100 nm/min. After background subtraction, the mean residual ellipticity (ϴ) was reported as a function of wavelength.
Disk diffusion assay.
Disk diffusion assays were performed with M. tuberculosis mc26020 on Middlebrook 7H10 agar plates supplemented with glycerol, OADC, pantothenate, and lysine as described previously (23). One hundred microliters of a culture at an optical density at 600 nm (OD600) of 0.25 (∼2 × 106 bacteria) was spread onto the surface of the agar plates prior to the addition of 6-mm paper disks (Becton Dickinson) impregnated with 100 μg peptide dissolved in water. Control disks included water (negative) or 10 μg kanamycin (positive). Plates were incubated for 3 weeks at 37°C. All peptides were tested in duplicate. Antibacterial activity was visualized as clear zones around disks (see Fig. S2 in the supplemental material).
Microbroth dilution MIC determinations.
All antimicrobial susceptibility testing was performed with a final volume of 100 μl in sterile U-shaped 96-well polypropylene microtiter plates. Separate 96-well plates were filled with 100 μl/well of medium broth for the growth of different test organisms. AMPs B1 through B4 and gentamicin (Gent) were prepared at 512 μg/ml in medium broth, and subsequent 2-fold dilutions were performed in 0.1 ml medium broth in the microplates. Nonexperimental wells were filled with sterile distilled water to prevent dehydration in experimental wells. M. tuberculosis (mc26230 or mc26020), M. smegmatis, or BCG bacteria from frozen stocks, initially diluted to an OD600 of 0.1, were each grown for 18 h (M. smegmatis), 7 days (M. tuberculosis mc26230 and BCG) or 9 days (mc26020) at 37°C to late log phase. Following propagation, bacterial cultures were declumped by gentle sonication using a cup horn apparatus maintained at 4°C for a total of 20s with alternating pulses (5 s on/5 s off) at low power (VirSonic 550; Virtis). Declumped bacteria were diluted to an OD600 of 0.01 for inoculation into AMP-containing medium broth at a concentration of 5 μl per well. A well with none of the AMP agents was similarly inoculated as a growth control. The plates were wrapped in aluminum foil and incubated at 37°C for 5 days in a humidified incubator. Following incubation, alamarBlue (resazurin) reagent (10 μl) was added to all experimental wells, and the wells were further incubated for 24 to 48 h. Color changes from blue (inhibition) to pink (bacterial growth) were observed visually, and MICs were defined as the lowest concentration of AMP that prevented a color change (30). For other bacteria, fresh cultures of selected Gram-positive (Staphylococcus aureus ATCC 25923, Streptococcus pneumoniae ATCC 49619, Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus mutans, and Bacillus subtilis BGSC 1A1) and Gram-negative (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Klebsiella pneumoniae) bacteria grown overnight were subcultured to log phase at 37°C for 3.5 to 4 h at 200 rpm in either 5 ml MH broth (Difco) or MH broth supplemented with 5% sheep red blood cells. Bacterial inocula were prepared and then added to 96-well plates containing AMPs as described above for the M. tuberculosis experiments. The OD600 was obtained before and after incubation for each plate by using a spectrophotometer plate reader (Tecan Sunrise). Growth was determined by the change in the OD600 following incubation, and the MIC was defined as the lowest AMP concentration in a well that exhibited no bacterial growth.
Assay for peptide cytotoxicity against mammalian cells.
Cytotoxicity studies were performed by using two types of mammalian cells: cultured J774.16 mouse macrophages and A549 human lung epithelial cells. J774.16 macrophages (maintained in antibiotic-free Dulbecco's modified Eagle's medium containing 20% fetal bovine serum, 5% NCTC-109, 1% nonessential amino acids, and 1% glutamine) or A549 epithelial cells (maintained in antibiotic-free Ham's F-12 medium supplemented with 10% fetal bovine serum) were seeded at a density of ∼1 × 105 cells per ml in Lab-Tek Permanox eight-chamber microscopy slides (Nunc, Inc., Naperville, IL) and incubated for 3 days (J774.16 cells) or 5 days (A549 cells) to a confluence of 70 to 80% before use. Culture medium on macrophages or epithelial cells was then replaced with fresh medium containing the different AMPs at 34 μM or 170 μM, and the cultures were incubated for 24 h at 37°C in 5% CO2. Cells were treated with medium only as a negative control or with 50 μM NaN3 or 1% Triton X-100 as a positive (cytotoxic) control. Cytotoxicity for J774.16 or A549 cells was monitored by using a trypan blue exclusion assay, and the ratio of dead to viable cells was recorded (31). All experiments were done in duplicates with three independent biological repeats.
RESULTS AND DISCUSSION
Peptide design.
A database of 44 peptides was generated from databank and literature sources and analyzed by using the database filtering method (22). The database contained both natural, broadly active AMPs and synthetic AMPs screened specifically against M. tuberculosis. The majority of the synthetic peptides were identified in a single screen performed by Ramon-Garcia et al. (9). The peptides identified by Ramon-Garcia et al. skew the database toward increased hydrophobicity and positive charge; however, these properties were shown by those authors to correlate with increased antimicrobial potency. As a result, these peptides were retained in the database. A full treatment of the influence of the inclusion or exclusion of these peptides on the database can be found in Table S3 in the supplemental material.
The mean length of the database peptides was found to be 13 amino acids, and this length was chosen for our designed peptides. The database showed a distinctly bimodal distribution with a group ∼10 amino acids in length (mostly the synthetic peptides) and another group ∼26 amino acids in length (mostly the natural peptides) (Fig. 1A). Interestingly, a 26-amino-acid alpha-helix is of the approximate length needed to span most of a bacterial membrane. (The translational rise between α-carbons in α-helical peptides is ∼1.5 Å, which is ∼40 Å for a 26-residue peptide. The thickness of a lipid bilayer, a commonly used model for a bacterial membrane, is ∼50 Å.) Concurrent studies on the four de novo-designed peptides suggested that the mechanism of action was barrel stave pore formation in a supported lipid bilayer (B. Murray, unpublished data), which, if translatable to pathogens, would suggest a necessary peptide length based on membrane thickness. Due to the biological relevance of a half-membrane span (13 amino acids), we chose an overall mean length of 13 amino acids rather than the mean length of the first mode (10 amino acids).
The second parameter analyzed was average charge (Fig. 1B). All peptides in the database were cationic. When the overall charge was normalized to the length of the peptide, the mean charge per length was found to be 0.4, a value that was highly consistent across all peptides in the database. Application of this ratio to our peptide of 13 amino acids implies that 5 of the residues should be positively charged.
The average number of hydrophobic residues (Ala, Val, Ile, Leu, Trp, Phe, Met, and Cys) was determined and normalized to the length of each peptide (Fig. 1C). This resulted in a mean value of 0.5 hydrophobic residues per number of total residues. Application of this value to our peptide of 13 amino acids suggested a total of 6.5 hydrophobic residues, which was rounded to 7.
Twelve of the 13 residues were determined to be charged or hydrophobic, leaving 1 uncharged, nonhydrophobic residue. Thus, our peptide should contain 5 positively charged residues (Lys, Arg, or His), 7 hydrophobic residues (Ala, Val, Ile, Leu, Trp, Phe, Met, Gly, or Cys), and 1 uncharged nonhydrophobic residue (Ser, Thr, Tyr, Pro, Asn, or Gln).
To select the identity of the charged residues, the frequency of each residue in the database was analyzed. All peptides in the database contained Lys, Arg, or a combination of the two (Fig. 1D). In contrast, only three peptides in the entire database were found to contain His, so this residue was eliminated. Of peptides containing Arg or Lys, 20% contained only Arg, 14% contained only Lys, and 66% contained both. As a result, a mixture of Lys and Arg was selected, and of our five charged residues, three were selected to be Lys and two were selected to be Arg, because they are asymmetric in their behavior (32).
A similar procedure was followed for hydrophobic residues. Ala, Met, Gly, and Cys residues appeared infrequently and were not considered. Of the remaining five possible residues, Trp and Leu appeared most frequently, especially among the subset of peptides that were ∼13 amino acids in length. Ile was also relatively frequent. Thus, four of the seven hydrophobic residues were selected to be Trp, two of the seven were selected to be Leu, and one of the seven was selected to be Ile.
Of the polar residues, Tyr, Asn, and Gln appeared infrequently. Ser appeared more frequently than Thr and Pro and better supports an alpha-helical conformation; thus, the final residue was selected to be Ser.
The overall output of database filtering against M. tuberculosis resulted in a peptide length of 13 amino acids, containing two Arg residues, three Lys residues, four Trp residues, two Leu residues, one Ile residue, and one Ser residue. The peptide sequence was determined by arranging the set of amino acids in an amphipathic manner. This arrangement was additionally constrained to have a charged residue at one terminus, an observation from the database. In total, we developed four candidate peptides, shown in wheel diagram representations in Fig. 2. The helical wheel diagrams demonstrate the amphipathic nature of peptides B1 through B3. Specifically, the helical wheel diagram of peptide B1 shows (i) a hydrophobic face made up of amino acids Ile1, Trp8, Leu4, and Trp11; (ii) a second hydrophobic face made up of amino acids Ile2, Trp6, and Trp10, with a hydrophobic face interruption by amino acid Lys13; and (iii) a hydrophilic face made up of amino acids Lys9, Arg5, and Lys12 (the number after the amino acid letter designates the position of that amino acid). The helical wheel diagram of peptide B2 shows (i) a hydrophobic face made up of amino acids Leu4, Trp11, Trp7, Trp10, and Trp6, with a hydrophobic face interruption by amino acid Ser3, and (ii) a hydrophilic face made up of amino acids Lys13, Lys9, Arg5, Lys12, and Arg8 interrupted by hydrophobic amino acids Leu2 and Ile1. The helical wheel diagram of peptide B3 shows (i) a hydrophobic face made up of amino acids Leu2, Trp9, Ile1, Trp8, Leu4, and Trp11, with a hydrophobic face interruption by amino acids Arg5 and Lys12, and (ii) a hydrophilic face made up of amino acids Arg7, Ser3, Lys10, and Lys13, interrupted by hydrophobic amino acid Trp6. An alpha-helical conformation for each peptide was predicted by using PepFold and visualized by using MOE (29) (Fig. 2).
Peptide structure.
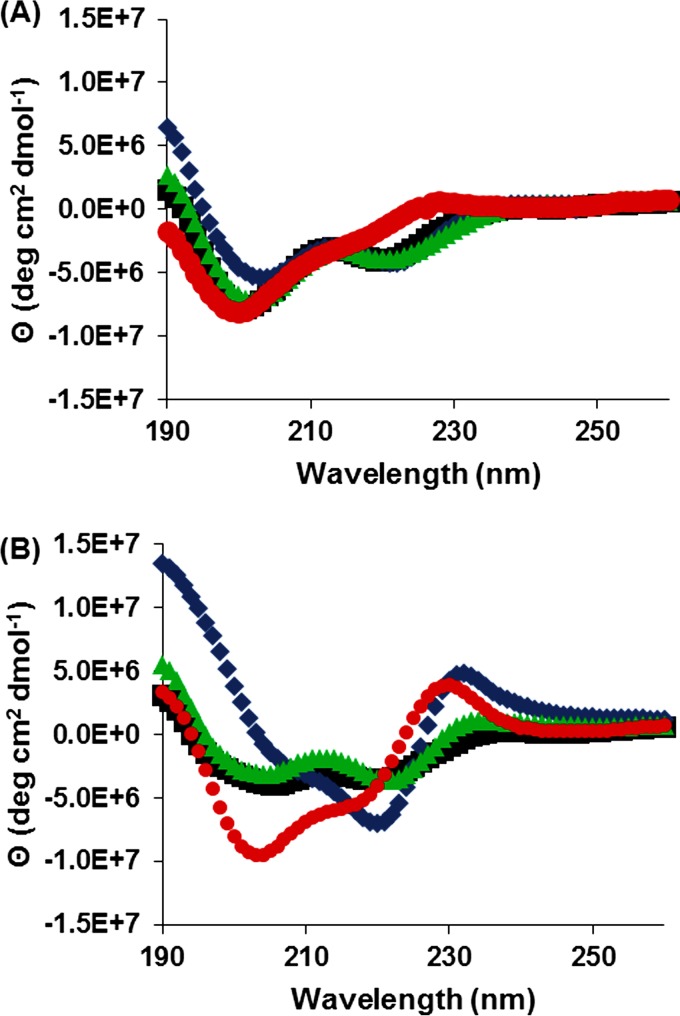
To gain insight into the secondary structure of the designed AMPs, far-UV CD spectroscopy was performed with the AMPs in phosphate buffer (pH 7.0) (Fig. 3A) and in the presence of bacterial mimetic lipid vesicles (POPC-POPG at a 3:1 molar ratio) (Fig. 3B). In phosphate buffer, B1, B2, and B3 produced far-UV CD spectra associated with α-helical peptides, with ϴ minima at 222 nm and 205 nm in addition to an apparent maximum at 190 nm (33, 34). B4, the “scrambled-sequence” control peptide, maintained a spectrum more closely aligned with a random coil/extended structure with a single minimum at 200 nm (Fig. 3A) (35).
FIG 3.
Secondary structure analysis. Shown are far-UV CD spectra for B1 (■), B2 (◆), B3 (▲), and B4 (●) in phosphate buffer (A) and at a 1:10 P:L ratio with POPC-POPG lipids at a 3:1 molar ratio (B). B1 to B3 in phosphate buffer resulted in linear α-helical spectra, while B4's spectrum resembled that of a random coil. In the presence of lipids, B1 and B3 maintained their linear helical spectra, while B2 and B4 produced spectra resembling kinked helices.
In the presence of phosphatidylcholine:phosphatidylglycerol (PC:PG) lipid vesicles, B1 and B3 retained their α-helical structure, with spectra characteristically similar to those in phosphate buffer (Fig. 3A and B). B2 and B4 resulted in completely unique spectra when PC:PG vesicles were present. The B2 spectrum contained an accentuated local maximum at 232 nm, a global minimum at 222 nm, a shoulder at 207 nm, and a global maximum at 190 nm. Conversely, B4 resulted in a slightly blue-shifted spectrum with the same maxima but with the minimum and shoulder locations swapped (Fig. 3B). These spectra contain features associated with kinked proline-rich proteins (232-nm maximum) and helical proteins (222-nm minimum, 207-nm shoulder, and 190-nm maximum), indicating the presence of kinked helices for both B2 and B4 in the presence of bacterial mimetic lipid vesicles (35, 36).
In vitro activity and toxicity against M. tuberculosis and M. smegmatis.
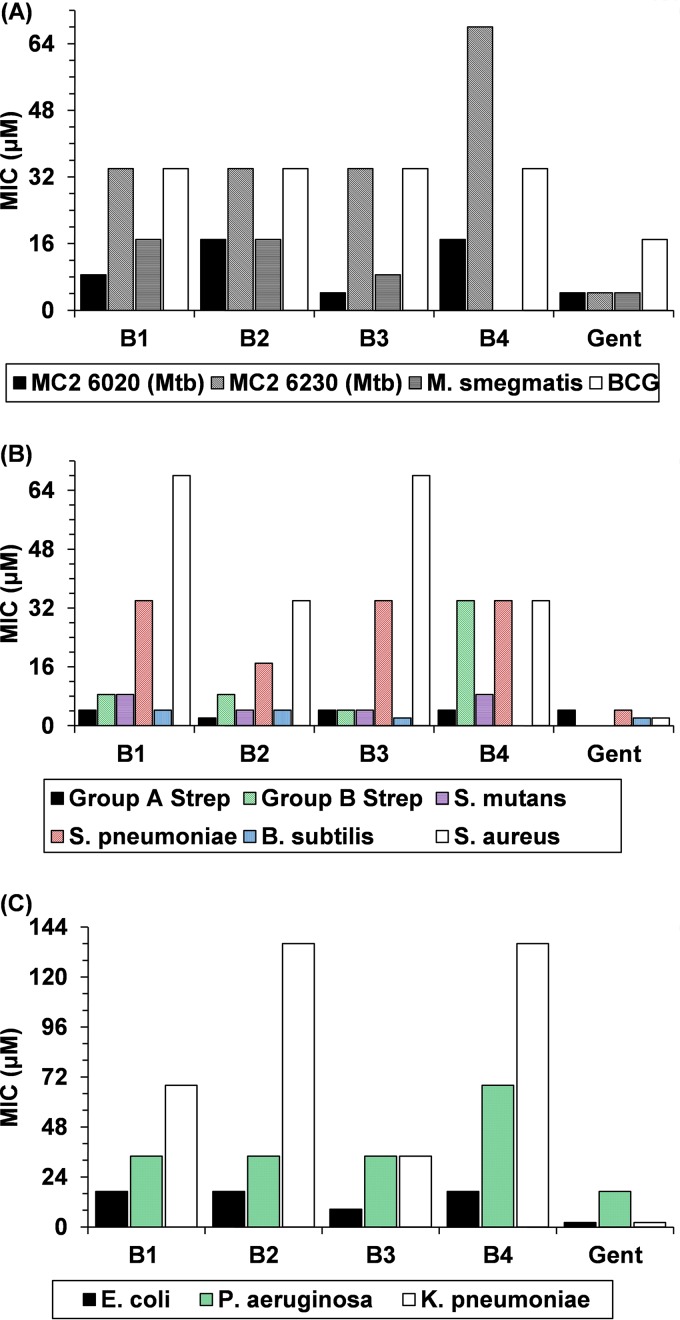
The in vitro activity of the four peptides was measured by using standard methods, including a disk diffusion assay, broth culture, and an alamarBlue assay. All three designed peptides were observed to have antimicrobial properties against mycobacteria in the disk diffusion assay (see Fig. S2 in the supplemental material), and this activity was subsequently quantified with the alamarBlue assay. Figure 4A illustrates MICs of peptides B1 through B3 and the scrambled-sequence control peptide B4 against several mycobacterial species. MICs were determined against two differently attenuated M. tuberculosis strains (mc26020 and mc26230), M. bovis BCG, and M. smegmatis. On a molar basis, B3 was equally as effective as the positive control, gentamicin (Gent), against M. tuberculosis mc26020. This tuberculosis strain is similar to mc26230 but contains the RD1 locus and includes a mutated lysA gene. Peptide B3 was not as effective as gentamicin against M. tuberculosis strain mc26230, M. smegmatis, or BCG but displayed antimicrobial activity, as did peptides B1 and B2. The MIC for B4 against M. smegmatis was not determined. Considering that all four peptides were derived from the same set of amino acids, this result highlights the importance of pairing database filtering with rational design. Although filtering alone resulted in a set of relatively efficacious amino acids, rational ordering of these residues improved the efficacy of peptide B3 against M. tuberculosis by a factor of at least 2 over the control peptide B4. Peptides B1 and B2 also exhibited a lesser but appreciable improvement in activity than that of B4 (Fig. 4A).
FIG 4.
AMP MICs against clinically relevant microbes. Shown are MICs for the attenuated mycobacterial strains (A) and various clinically relevant Gram-positive (B) and Gram-negative (C) bacteria listed in the key. The MICs for B4 against M. smegmatis and B. subtilis were not determined.
In vitro activity and toxicity against other clinically relevant bacteria.
In addition to M. tuberculosis, the designed peptides were tested against several strains of clinically relevant Gram-positive and Gram-negative bacteria (Fig. 4B and C, respectively). In general, the peptides were more potent against Gram-positive species than against Gram-negative species. The MIC values for peptides B1 through B4 against clinically relevant Gram-positive bacteria are reported for six Gram-positive bacteria: group A and B streptococci, Streptococcus mutans, Streptococcus pneumoniae ATCC 49619, Bacillus subtilis BGSC 1A1, and Staphylococcus aureus ATCC 29213. The designed peptides proved very effective against B. subtilis and various strains of Streptococcus with MIC values as low as 2 μM. Although B3 was considerably more effective than B4 against group B Streptococcus, neither peptide was very effective (MIC of 32 μM) against S. pneumoniae, and their activities against group A Streptococcus (MIC of 4 μM for both) and S. mutans (MIC of 4 μM for B3 versus 8 μM for B4) were similar. The MIC for B4 against B. subtilis was not determined. While consistent with previous findings that cationic, amphipathic peptides are broadly active, the range of potencies highlights the need for species-specific database optimization during peptide design.
The MIC values of peptides B1 through B4 against clinically relevant Gram-negative bacteria were measured. MICs were determined against three Gram-negative bacteria: Escherichia coli ATCC 25922, Klebsiella pneumoniae 13-329999g, and Pseudomonas aeruginosa ATCC 27853. For the group of Gram-negative bacteria, the designed peptides proved most effective against E. coli. The designed peptides successfully inhibited the growth of all Gram-negative bacteria tested although generally with MICs higher than those found with the Gram-positive bacteria in our test panel. These results again illustrate the importance of amino acid ordering and indicate that the mechanism of antimicrobial activity may differ between Gram-positive bacteria and M. tuberculosis, which is neither Gram positive nor Gram negative.
Cytotoxicity against mammalian cells.
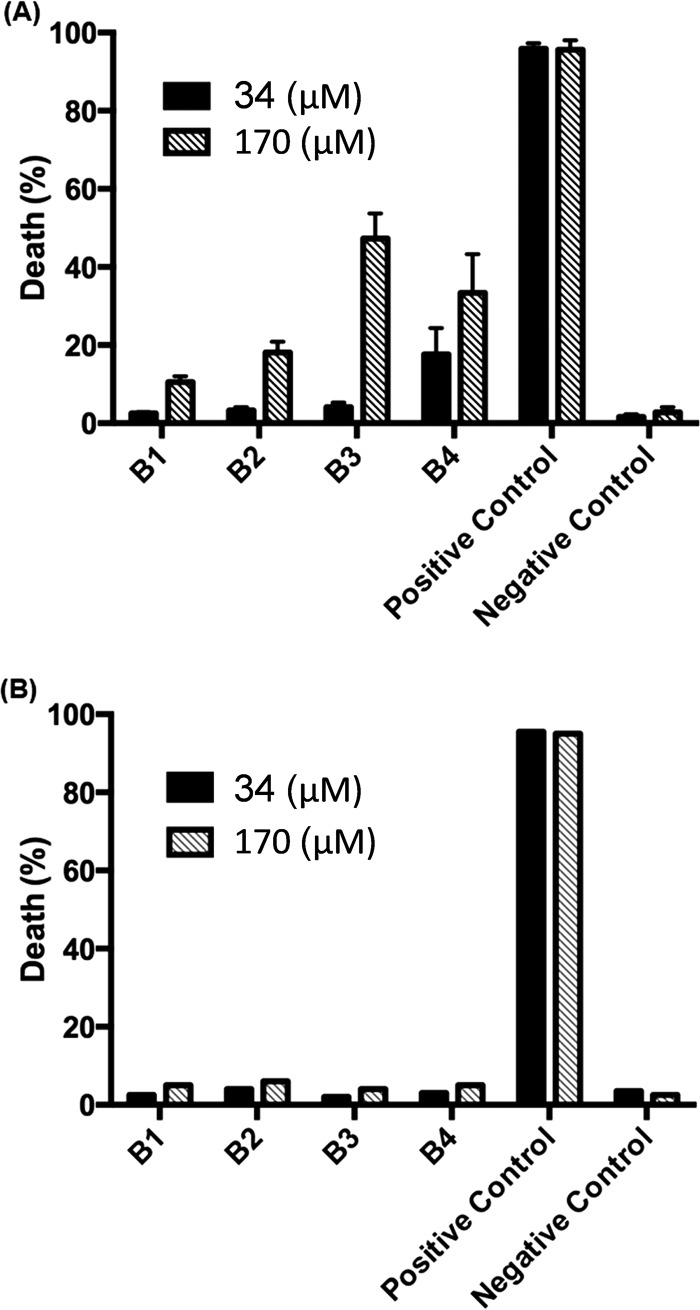
To check for cytotoxicity against mammalian cells, the peptides were tested against J774.16 macrophage cells and lung epithelial cells. All peptides tested were nontoxic to mammalian cells at the MICs required for antimicrobial activity. Figure 5A illustrates the cytotoxicity of peptides B1 through B4 against murine J774.16 macrophages. There was no observable cytotoxicity at a 34 μM peptide concentration (i.e., ≥1× MIC). Twenty percent cell death was observed for peptide B2 at 170 μM, while 47% cell death resulted from incubation with peptide B3 at 170 μM (i.e., ≥5× MIC), both toxic above the value of the negative control (3%). Figure 5B illustrates cytotoxicity of peptides B1 through B4 against human lung epithelial cells. There was no observable cytotoxicity at a 34 μM peptide concentration. Slight cytotoxicity was seen for peptides B2 and B3 at a 170 μM peptide concentration compared with the negative control. The lack of cytotoxicity of the designed peptides is likely due to the designed inclusion of at least one positively charged residue within the hydrophobic face, according to the imperfect-amphipathicity approach described previously by Wimley (37).
FIG 5.
AMP cytotoxicity. Shown is AMP cytotoxicity against J774.16 macrophage cells (A) and A549 lung epithelial cells (B) at 34 μM (black bars) and 170 μM (white striped bars) AMP concentrations. The positive control is 1% Triton X-100 or 50 μM NaN3, while the negative control is cell medium in all cases. Error bars represent 1 standard deviation from three biological replicates.
Conclusions.
We have designed peptides with reasonably effective antimicrobial activity against M. tuberculosis and several other clinically relevant pathogenic bacteria using a combination of database filtering bioinformatics, protein engineering, and de novo design. The designed peptides are short and readily synthesized by using available native amino acids for cheap synthesis, although, if desired, we expect our templates to be compatible with d-form peptides, peptoids, and other peptide mimetic chemistries.
Although guided mainly by the design principle of imperfect amphipathicity in our peptide design, we expect other parameters, such as hydrophobic arrangement along the depth of the peptide, to play a significant role. We are currently using these first-generation peptides as a template to produce peptide variations to probe mechanisms and to develop further design principles. It is expected that our combined method of database filtering bioinformatics, protein engineering, and de novo design has the potential to be applied to any microbe, provided that a library of active AMPs can be generated. The derived AMPs may be used as a strong template for the development of antimicrobials of potential clinical relevance. Further modification of the AMPs using an iterative procedure with our de novo design method and in vivo testing can enable the design of highly selective and potent AMPs against a specific microbial target.
Supplementary Material
ACKNOWLEDGMENTS
B.M. was supported by RPI Georges Belfort Endowed Institute Chair funds (RPI number 140124).
We thank W. R. Jacobs for providing the attenuated M. tuberculosis strains, the Wadsworth Center Bacteriology Laboratory for technical advice and sharing isolates of clinically relevant bacteria, and the Wadsworth Center Media Core for media preparation. We also thank Joseph Grimaldi for assisting with peptide purification and Dmitri Zagorevski for conducting mass spectroscopy on the peptides.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00940-15.
REFERENCES
- 1.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H, Kim SH, Rosenberg EY. 1993. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol Microbiol 8:1025–1030. doi: 10.1111/j.1365-2958.1993.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 4.Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 5.Sarathy JP, Dartois V, Lee EJ. 2012. The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals (Basel) 5:1210–1235. doi: 10.3390/ph5111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. 2008. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 88:526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forrellad MA, McNeil M, de la Paz Santangelo M, Blanco FC, Garcia E, Klepp LI, Huff J, Niederweis M, Jackson M, Bigi F. 2014. Role of the Mce1 transporter in the lipid homeostasis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 94:170–177. doi: 10.1016/j.tube.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danilchanka O, Sun J, Pavlenok M, Maueroder C, Speer A, Siroy A, Marrero J, Trujillo C, Mayhew DL, Doornbos KS, Munoz LE, Herrmann M, Ehrt S, Berens C, Niederweis M. 2014. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci U S A 111:6750–6755. doi: 10.1073/pnas.1400136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramon-Garcia S, Mikut R, Ng C, Ruden S, Volkmer R, Reischl M, Hilpert K, Thompson CJ. 2013. Targeting Mycobacterium tuberculosis and other microbial pathogens using improved synthetic antibacterial peptides. Antimicrob Agents Chemother 57:2295–2303. doi: 10.1128/AAC.00175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linde CM, Hoffner SE, Refai E, Andersson M. 2001. In vitro activity of PR-39, a proline-arginine-rich peptide, against susceptible and multi-drug-resistant Mycobacterium tuberculosis. J Antimicrob Chemother 47:575–580. doi: 10.1093/jac/47.5.575. [DOI] [PubMed] [Google Scholar]
- 11.Miyakawa Y, Ratnakar P, Rao AG, Costello ML, Mathieu-Costello O, Lehrer RI, Catanzaro A. 1996. In vitro activity of the antimicrobial peptides human and rabbit defensins and porcine leukocyte protegrin against Mycobacterium tuberculosis. Infect Immun 64:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Z, Higgins MP, Whitehurst J, Kisich KO, Voskuil MI, Hodges RS. 2011. Anti-tuberculosis activity of alpha-helical antimicrobial peptides: de novo designed L- and D-enantiomers versus L- and D-LL37. Protein Pept Lett 18:241–252. doi: 10.2174/092986611794578288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilcinskas A. 2011. Anti-infective therapeutics from the lepidopteran model host Galleria mellonella. Curr Pharm Des 17:1240–1245. doi: 10.2174/138161211795703799. [DOI] [PubMed] [Google Scholar]
- 14.Sonawane A, Santos JC, Mishra BB, Jena P, Progida C, Sorensen OE, Gallo R, Appelberg R, Griffiths G. 2011. Cathelicidin is involved in the intracellular killing of mycobacteria in macrophages. Cell Microbiol 13:1601–1617. doi: 10.1111/j.1462-5822.2011.01644.x. [DOI] [PubMed] [Google Scholar]
- 15.Kapoor R, Eimerman PR, Hardy JW, Cirillo JD, Contag CH, Barron AE. 2011. Efficacy of antimicrobial peptoids against Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:3058–3062. doi: 10.1128/AAC.01667-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilchie AL, Wuerth K, Hancock RE. 2013. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol 9:761–768. doi: 10.1038/nchembio.1393. [DOI] [PubMed] [Google Scholar]
- 17.Zelezetsky I, Tossi A. 2006. Alpha-helical antimicrobial peptides—using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta 1758:1436–1449. doi: 10.1016/j.bbamem.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 19.Wimley WC. 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tossi A, Sandri L, Giangaspero A. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4–30. doi:. [DOI] [PubMed] [Google Scholar]
- 21.Tossi A, Tarantino C, Romeo D. 1997. Design of synthetic antimicrobial peptides based on sequence analogy and amphipathicity. Eur J Biochem 250:549–558. doi: 10.1111/j.1432-1033.1997.0549a.x. [DOI] [PubMed] [Google Scholar]
- 22.Mishra B, Wang G. 2012. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J Am Chem Soc 134:12426–12429. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambandamurthy VK, Jacobs WR Jr. 2005. Live attenuated mutants of Mycobacterium tuberculosis as candidate vaccines against tuberculosis. Microbes Infect 7:955–961. doi: 10.1016/j.micinf.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR Jr. 2006. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309–6320. doi: 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- 25.McDonough KA, Kress Y, Bloom BR. 1993. Pathogenesis of tuberculosis—interaction of Mycobacterium tuberculosis with macrophages. Infect Immun 61:2763–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonough KA, Kress Y. 1995. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect Immun 63:4802–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S. 2014. CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res 42:D1154–D1158. doi: 10.1093/nar/gkt1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang GS, Li X, Wang Z. 2009. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res 37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thevenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tuffery P. 2012. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol 36:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strober W. 2001. Trypan blue exclusion test of cell viability. Curr Protoc Immunol Appendix 3:Appendix 3B. doi: 10.1002/0471142735.ima03bs21. [DOI] [PubMed] [Google Scholar]
- 32.Ma CD, Wang C, Acevedo-Velez C, Gellman SH, Abbott NL. 2015. Modulation of hydrophobic interactions by proximally immobilized ions. Nature 517:347–350. doi: 10.1038/nature14018. [DOI] [PubMed] [Google Scholar]
- 33.Campagna S, Saint N, Molle G, Aumelas A. 2007. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 46:1771–1778. doi: 10.1021/bi0620297. [DOI] [PubMed] [Google Scholar]
- 34.Blondelle SE, Lohner K, Aguilar M-I. 1999. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: determination and biological specificity. Biochim Biophys Acta 1462:89–108. doi: 10.1016/S0005-2736(99)00202-3. [DOI] [PubMed] [Google Scholar]
- 35.Greenfield NJ. 2006. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc 1:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 37.Wimley WC, Hristova K. 2011. Antimicrobial peptides: successes, challenges and unanswered questions. J Membr Biol 239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.