Abstract
The species Acinetobacter baumannii is one of the most important multidrug-resistant human pathogens. To determine its virulence and antibiotic resistance determinants, the genome of the nosocomial blaNDM-1-positive A. baumannii strain R2090 originating from Egypt was completely sequenced. Genome analysis revealed that strain R2090 is highly related to the community-acquired Australian A. baumannii strain D1279779. The two strains belong to sequence type 267 (ST267). Isolate R2090 harbored the chromosomally integrated transposon Tn125 carrying the carbapenemase gene blaNDM-1 that is not present in the D1279779 genome. To test the transferability of the metallo-β-lactamase (MBL) gene region, the clinical isolate R2090 was mated with the susceptible A. baumannii recipient CIP 70.10, and the carbapenem-resistant derivative R2091 was obtained. Genome sequencing of the R2091 derivative revealed that it had received an approximately 66-kb region comprising the transposon Tn125 embedding the blaNDM-1 gene. This region had integrated into the chromosome of the recipient strain CIP 70.10. From the four known mechanisms for horizontal gene transfer (conjugation, outer membrane vesicle-mediated transfer, transformation, and transduction), conjugation could be ruled out, since strain R2090 lacks any plasmid, and a type IV secretion system is not encoded in its chromosome. However, strain R2090 possesses three putative prophages, two of which were predicted to be complete and therefore functional. Accordingly, it was supposed that the transfer of the resistance gene region from the clinical isolate R2090 to the recipient occurred by general transduction facilitated by one of the prophages present in the R2090 genome. Hence, phage-mediated transduction has to be taken into account for the dissemination of antibiotic resistance genes within the species A. baumannii.
INTRODUCTION
Resistance of microorganisms to antimicrobial compounds poses a public health problem of growing importance. The development and dissemination of antibiotic resistance interfere with treatment regimens and increase costs in health care facilities (1). In particular, infections in immunocompromised patients may turn dramatically due to failure of antimicrobial therapy. Acinetobacter species are strictly aerobic, Gram-negative, coccobacillary rods (2). They were isolated from a variety of habitats, including soil, water, wastewater, and hospital environments (3, 4). Currently, the Acinetobacter genus comprises 52 species (http://www.bacterio.net/acinetobacter.html, http://www.ncbi.nlm.nih.gov/Taxonomy/), some of which are important human pathogens (5). Acinetobacter baumannii is the most prevalent species in clinical environments (4), whereas it occurs more rarely in natural habitats (6). A. baumannii usually is harmless for healthy persons, although some particular A. baumannii strains may cause severe infections in immunocompromised patients, which is a serious problem in intensive care units.
Compared to other Gram-negative pathogens, few virulence determinants have been identified in A. baumannii strains (6). Most A. baumannii virulence determinants belong to the core genome (7). However, a wide spectrum of resistance determinants and its high robustness make A. baumannii a serious nosocomial pathogen (8). It is able to rapidly acquire antibiotic resistance genes (9) by horizontal transfer from other bacteria, thus increasing its own virulence (10).
The multidrug-resistant A. baumannii strain R2090 was isolated from a hospitalized Egyptian patient. To analyze its putative and respective virulence and resistance determinants, the genome of A. baumannii R2090 was completely sequenced. A corresponding genome announcement was published recently (11). Here, pathogenicity and resistance determinants of the isolate were identified. Moreover, the relatedness of strain R2090 to other A. baumannii strains was analyzed to gain epidemiological insights, and the horizontal transmissibility of its blaNDM-1 carbapenemase gene was demonstrated.
MATERIALS AND METHODS
Isolation, cultivation, identification, and total DNA preparation of A. baumannii strains.
A. baumannii strain R2090 was recovered from a rectal swab screening of a patient who was hospitalized in Egypt. A. baumannii strains R2090, CIP 70.10, and R2091 were cultured in Trypticase soy broth medium, and total DNA was extracted and purified using the QIAamp DNA minikit (Qiagen, Hombrechtikon, Switzerland). Identification of strain R2090 was performed by using the API 20NE system (bioMérieux, La Balme-les-Grottes, France). In addition, the sequence of the strain's 16S rRNA gene was established and compared to database entries.
Resistance pattern assessment.
Antibiotic susceptibility testing was evaluated and interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI).
Transfer of the blaNDM-1 resistance gene region to the recipient strain A. baumannii CIP 70.10.
Mating-out assays were performed using A. baumannii R2090 as a donor and A. baumannii CIP 70.10 as the recipient strain. After overnight (o/n) growth in Trypticase soy liquid medium, both strains were diluted (1:10), cultured for 1 h, and then mixed at a ratio of 1 to 4 (donor/recipient) in a total volume of 2 ml. Subsequently, this mixed culture was grown for 3 h at 37°C, and 200 μl of the culture was plated onto selective Mueller-Hinton agar medium supplemented with ticarcillin (100 μg/ml) and rifampin (100 μg/ml). Growing colonies were checked by PCR for the presence of the β-lactamase gene blaNDM-1 and also their resistance phenotypes (resistance to ticarcillin and carbapenems).
Sequencing and annotation of the A. baumannii R2090, CIP 70.10, and R2091 genomes.
High-throughput sequencing on the Illumina MiSeq system and bioinformatic analyses of the obtained genome sequences were accomplished as previously described for A. baumannii R2090 and CIP 70.10 (11, 12). The genome sequence of A. baumannii R2091 was established by applying a strategy similar to that described for strains R2090 and CIP 70.10 (see above). An 8-kb mate-pair sequencing library was constructed for strain R2091 and sequenced on the Illumina MiSeq system. The assembly of obtained sequence reads was performed by using the GS de novo Assembler software (version 2.8; Roche). Genome sequences of all three strains (R2090, CIP 70.10, and R2091) were annotated within the GenDB annotation platform (13) based on the annotation of the reference strain A. baumannii AB307-0294 (GenBank accession no. CP001172) (14). Moreover, a COG annotation was performed by means of WebMGA, with an E value threshold of 1 × 10−20 (15). In addition, the genes discussed in this study were manually annotated, as published recently (16–18), applying pairwise BLASTx comparisons of encoded protein sequences.
Phylogenetic classification and multilocus sequence typing.
Phylogenetic classification based on genome sequence information for the A. baumannii strains sequenced in this study was performed using EDGAR 2.0 (19). For calculation of a core-genome-based phylogenetic tree, the core genes of A. baumannii strains R2090 and CIP 70.10, as well as those of all sequenced A. baumannii reference strains available in the public EDGAR project EDGAR_Acinetobacter, were considered. Furthermore, a Web service (http://enve-omics.ce.gatech.edu/ani/) was used to calculate the average nucleotide identity (ANI) (20).
Moreover, the sequence types (ST) of A. baumannii strains R2090, CIP 70.10, and R2091 were determined using the A. baumannii multilocus sequence typing (MLST) database (http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_abaumannii_pasteur_seqdef&page=sequenceQuery&set_id=2). The MLST is based on the allelic sequences of seven housekeeping genes (Pasteur scheme: cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) and was used for assignment to a clonal complex (CC), as defined previously (10, 21).
Detection of putative pathogenicity determinants and antibiotic resistance genes.
Identification of the putative virulence factors and antibiotic resistance determinants in the genomes of the sequenced isolates was performed using the databases MvirDB (22), ARG-ANNOT (23), CARD (24), and Resfams (25). For the detection of determinants represented in the databases MvirDB and ARG-ANNOT, BLASTp (26) was applied, with an E value threshold of 1 × 10−100. The CARD database was accessed using the Resistance Gene Identifier version 2 (http://arpcard.mcmaster.ca/). Furthermore, a hidden Markov model (HMM) search (27, 28) was performed, with an E value threshold of 1 × 10−100, to identify putative resistance genes of the Resfams Core database. Moreover, the genomes were searched for relaxases and key components of type IV secretion systems using CONJscan-T4SSscan (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::CONJscan-T4SSscan) (29). Additionally, the PHAge Search Tool (PHAST) was used on the nucleic acid level for the prediction of integrated bacteriophages in the chromosomes (30).
Nucleotide sequence accession numbers.
Genome sequences for A. baumannii strains R2090, CIP 70.10, and the CIP 70.10 derivative strain R2091 harboring the β-lactamase gene blaNDM-1 are accessible under the EMBL/GenBank accession no. LN868200 (R2090), LN865143 to LN865144 (CIP 70.10), and LN997846 to LN997847 (R2091).
RESULTS AND DISCUSSION
Origin of the clinical isolate A. baumannii R2090.
Isolate R2090 was recovered during the course of hospital admission from a rectal swab from a colonized Egyptian patient who was transferred to a hospital in France. The obtained isolate showed resistance to all β-lactam antibiotics, including carbapenems at high levels (MICs for imipenem and meropenem, >32 μg/ml). The isolate was considered a colonizing strain, since the patient had not developed any infection symptoms related to A. baumannii.
Sequencing, general features, and phylogenetic analysis of the A. baumannii R2090 genome.
Considering the multidrug resistance pattern of the isolated A. baumannii strain R2090, it was decided to sequence its genome to identify encoded antibiotic resistance and virulence determinants and to analyze its relatedness to known antibiotic-resistant A. baumannii reference strains, thereby addressing epidemiological aspects.
The genome sequence of the isolate A. baumannii R2090 was established by applying the Illumina MiSeq system, the GS de novo assembler (version 2.8; Roche), and the bioinformatics tool Consed (31), as described previously (11), combined with a PCR-based strategy leading to the amplification of a gap-spanning fragment that was sequenced to fill one remaining gap. Annotation of orthologous genes was performed within the annotation tool GenDB 2.4 (13) using the annotated genome sequence of A. baumannii strain AB307-0294 as a reference (14). The features of the R2090 chromosome sequence are depicted in Table 1 and Fig. 1.
TABLE 1.
Genome features of A. baumannii strains R2090, R2091, and CIP 70.10
| Strain/plasmid | Size (bp) | G+C content (%) | Total no. of genes | No. of rRNA operons | No. of tRNAs | No. of CDSs | No. of genes with predicted function |
|---|---|---|---|---|---|---|---|
| R2090 | 3,819,158 | 39.04 | 3,694 | 6 | 73 | 3,603 | 2,615 |
| R2091 | 3,939,746 | 38.97 | 3,705 | 6 | 71 | 3,616 | 2,632 |
| pABR2091 | 7,742 | 37.56 | 13 | 0 | 0 | 13 | 3 |
| CIP 70.10 | 3,928,513 | 38.92 | 3,695 | 6 | 71 | 3,606 | 2,630 |
| pABCIP70.10 | 7,742 | 37.56 | 13 | 0 | 0 | 13 | 3 |
FIG 1.
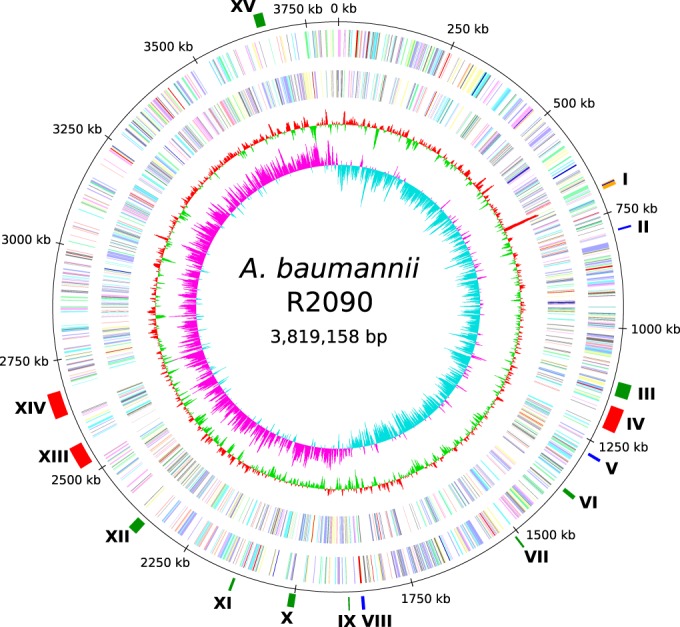
Circular representation of the A. baumannii R2090 chromosome. From the inside to the outside, the circles represent the GC skew (1), the G+C content (2), and the predicted protein-coding sequences (CDSs) on the reverse (3) and forward strand (4), colored according to the assigned Clusters of Orthologous Groups of proteins (COG) classes. Circle 5 displays the chromosome coordinates. In the sixth circle, regions described in the text are marked. These are related to predicted prophages (red), putative antibiotic resistance genes (blue), and virulence determinants (green), as well as the transposon Tn125 (orange). I, transposon Tn125 harboring blaNDM-1 gene; II, resistance-nodulation-division (RND) efflux pump AdeIJK; III, siderophore cluster (acinetobactin); IV, ΦR2090-I; V, RND efflux pump AdeFGH; VI, type I pilus cluster (Csu cluster); VII, type I pilus cluster (truncated); VIII, RND efflux pump AdeABC; IX, siderophore cluster; X, siderophore cluster; XI, type I pilus cluster (truncated); XII, type VI secretion system (truncated); XIII, ΦR2090-II; XIV, ΦR2090-III, XV, N-acyl homoserine lactone (AHL) cluster.
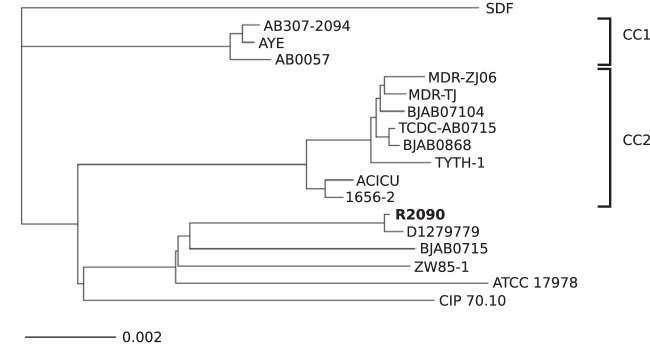
Phylogenetic classification of A. baumannii R2090 was done within the comparative genomics tool EDGAR (19) based on A. baumannii core genes. The calculated phylogenetic tree (Fig. 2) involving 1,885 core genes of 18 A. baumannii strains features three main clusters and two outgroups. Clusters 1 and 2 represent clonal complexes CC1 and CC2 (10, 21), respectively, whereas the strains of cluster 3 and the two outgroup strains (SDF and CIP 70.10) do not belong to clonal complex 1 or 2. The isolate A. baumannii R2090 shows the highest degree of relatedness to strain A. baumannii D1279779 (32). The high similarity of these two strains was also confirmed by average nucleotide identity (ANI) analysis, which resulted in an ANI of 99.92%. Strains R2090 and D1279779 both belong to the ST267, as determined by the Pasteur scheme via the corresponding Web tool PubMLST (21). The reference strain A. baumannii D1279779 originates from an indigenous Australian patient and represents the first fully sequenced community-acquired A. baumannii isolate (32).
FIG 2.
Phylogeny of different A. baumannii strains, as analyzed within the comparative genomics tool EDGAR based on core genes for all isolates. Phylogenetic analyses within EDGAR were essentially done as described previously (16, 19). Strains belonging to a specific clonal complex (CC) are labeled on the right-hand side. Strains SDF, R2090, D1279779, BJAB0715, ZW85-1, ATCC 17978, and CIP 70.10 belong to neither CC1 nor CC2.
Comparative analyses of the two closely related A. baumannii isolates R2090 and D1279779 regarding putative antibiotic resistance and virulence determinants.
A chromosome-wide alignment of the two A. baumannii isolates R2090 and D1279779 applying progressiveMauve (33), visualized by Mauve (34) and GenomeRing (35), revealed an inversion of about 50 kb around the origin of replication in strain D1279779 in comparison to strain R2090 (Fig. 3). Moreover, structural differences in six regions >10 kb were detected (Fig. 3). Among these, two segments represent two of the three predicted prophage regions (described below). The four other regions comprised predicted mobile genetic elements (insertion sequence [IS] elements, other putative transposable elements, and a phage integrase gene). Regarding possible virulence factors and resistance determinants, two of these regions are of outstanding interest.
FIG 3.
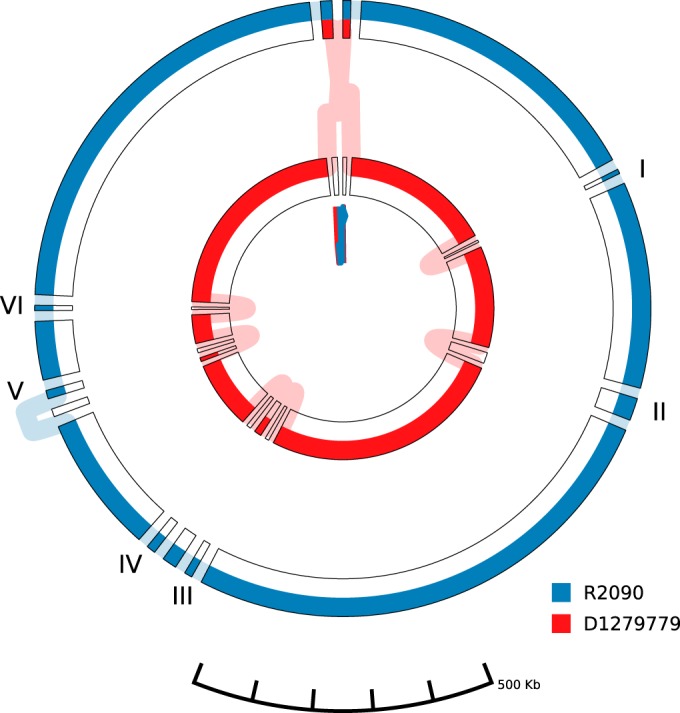
Comparison of the genomes of the A. baumannii isolates R2090 and D1279779. Visualization of the progressiveMauve (35) alignment with default settings of the chromosomes of strains R2090 and D1279799 was done by application of the visualization tool GenomeRing (37). Each chromosome is represented by a colored path (blue, A. baumannii R2090; red, A. baumannii D1279779). The chromosomes' start and end positions are labeled by triangles in the corresponding color. The outer and inner circles represent the forward and reverse orientations, respectively. All subblocks with a minimum block length of 10,000 bp are presented. Insertions in the R2090 chromosome are labeled and denote (I) the transposon Tn125, (II) prophage ΦR2090-I, (III) an ∼17-kb region adjacent to the transposase gene ABR2090_2200, (IV) the truncated type VI secretion system cluster, (V) prophage ΦR2090-III, and (VI) an ∼13-kb region adjacent to an integrase gene (ABR2090_2768). Prophage ΦR2090-II is not displayed in a separate block, as it possesses relatively high similarity to a prophage region in strain D1279779.
An approximately 20-kb region within the genome of strain R2090 is located in the vicinity of a transposase gene (ABR2090_2277). It harbored 13 genes of a putative type VI secretion system (T6SS) (ABR2090_2264 to ABR2090_2276) (36). Although a T6SS cluster was identified in most A. baumannii strains, it is not encoded in strain D1279779 (32, 37). Other conserved putative virulence genes of the species A. baumannii encoding a heme acquisition system (38) and the biofilm-associated protein Bap (39) were not identified in either strain R2090 or D1279779 (32). While the gene encoding the Acinetobacter trimeric autotransporter protein (Ata) (40) is truncated in strain D1279779 (32), the corresponding gene in strain R2090 carries a 627-bp insertion in comparison to the reference gene (A1S_1032) of A. baumannii strain ATCC 17978 (40, 41). Accordingly, Ata seems to be defective in both strains (37). As proposed by Farrugia et al. (32), the reduced biofilm formation activity of strain D1279779 is presumably due to the loss of the genes ata and bap, the T6SS cluster, and the heme acquisition system II. Strains R2090 and D1279779 possess a similar gene layout concerning genes predicted to contribute to biofilm formation. However, no reliable prediction of biofilm formation capabilities can be given by these in silico analyses, since biofilm formation is a multifactor-regulated process. Moreover, R2090 possesses two truncated type I pilus gene clusters (ABR2090_1456 to ABR2090_1457 and ABR2090_2049to ABR2090_2050) (37) also specifying functions in biofilm formation. Further putative virulence factors predicted to be involved in iron acquisition, DNA uptake, and twitching motility do not show significant differences between the two strains (identities of corresponding gene products, ≥99.65%).
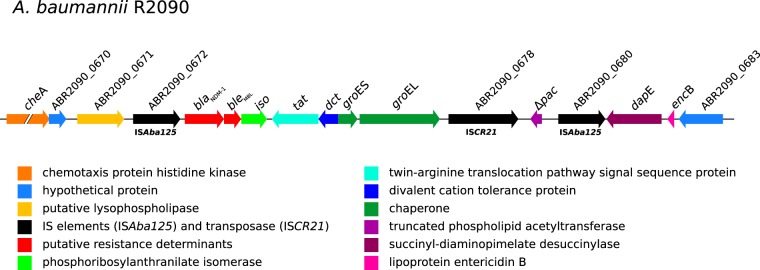
Another 11-kb region (Fig. 3 and 4) lacking in strain D1279779 is flanked by two insertion sequences of ISAba125 (ABR2090_0672 and ABR2090_0680) and carries the metallo-β-lactamase gene blaNDM-1 (ABR2090_0673). This structure corresponds to the previously identified composite transposon Tn125 (42, 43). Due to the presence of the carbapenemase gene blaNDM-1, the isolate was indeed highly resistant to all β-lactams, including carbapenems. Since the blaNDM-1 gene is located on a composite transposon, it was speculated that the whole element is mobile. To test the transmissibility of the predicted mobile element, strain R2090 was mated with another susceptible A. baumannii strain.
FIG 4.
Schematic representation of transposon Tn125 harboring the blaNDM-1 gene. The A. baumannii R2090 chromosomal region (approximately 18.7 kb) harboring the transposon Tn125 is located between coordinates 682069 and 700811. Coding sequences are labeled by either their gene name or locus tag. Lengths of coding sequences are drawn to scale, except for the chemotaxis gene cheA. Annotation of genes was adopted from previous publications (42, 56).
Transfer of the blaNDM-1 resistance region from the clinical A. baumannii strain R2090 to the susceptible A. baumannii recipient strain CIP 70.10.
The most important difference between the clinical A. baumannii strain R2090 and the closely related reference strain D1279779 is the integration of the metallo-β-lactamase resistance region blaNDM-1 in R2090. A corresponding region is missing in strain D1279779. Since the blaNDM-1 gene was located on the mobile genetic element Tn125, the question arose as to whether the element was acquired by horizontal gene transfer (HGT). To test the transmissibility of the blaNDM-1 resistance region, a mating-out experiment between R2090 and the susceptible recipient strain A. baumannii CIP 70.10 was performed. Upon selection on agar medium supplemented with ticarcillin and rifampin, derivative strains arose with a frequency of 1/106 (per recipient titer). PCR analyses and resistance testing confirmed that A. baumannii CIP 70.10 received the blaNDM-1 gene and expressed the corresponding resistance phenotype (resistance to ticarcillin and carbapenems). One representative derivative clone, designated R2091, was chosen for further analysis to determine the extent of the transferred resistance gene region and its integration site in the host chromosome. For this purpose, the complete genome sequences of the derivative R2091 and the corresponding recipient strain CIP 70.10 were established.
Comparative genome sequence analyses of the derivative strain R2091 carrying the blaNDM-1 resistance region and the corresponding susceptible A. baumannii recipient strain CIP 70.10.
The genome sequences of strain R2091 carrying the blaNDM-1 resistance gene and the corresponding susceptible recipient strain CIP 70.10 were established using the Illumina MiSeq system, essentially as described for the clinical A. baumannii isolate R2090 (see above). Sequencing statistics and genome features for the two strains are summarized in Table 1 (11, 12). In comparison to the genome of strain CIP 70.10, that of the derivative strain R2091 was 11,233 bp larger, which almost corresponds to the size of transposon Tn125 carrying the blaNDM-1 gene. The G+C content of the R2091 genome was determined to be 38.97%, and it contained 3,616 coding sequences. The gene contents of strains CIP 70.10 and R2091 are nearly identical, except for 11 additional genes that are present only in the derivative R2091. These accessory genes were located on the transposon Tn125 (see below).
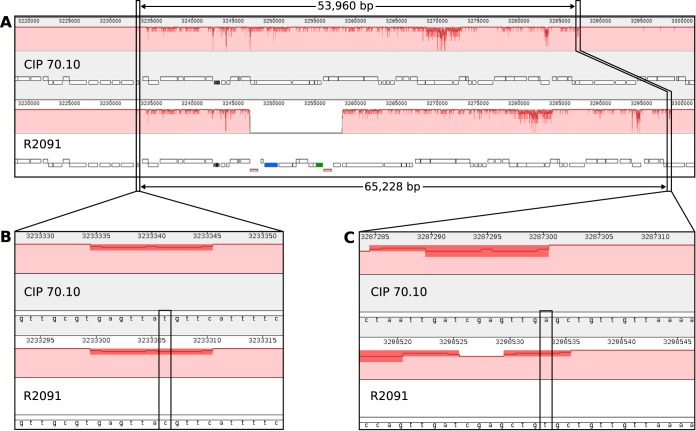
To determine the chromosomal background of the derivative strain R2091 carrying the blaNDM-1 gene, multilocus sequence typing (MLST) and ANI analyses were carried out. The obtained results confirmed that R2091 features the chromosomal background of the recipient strain CIP 70.10. Both strains belong to sequence type 126 (ST126) and showed an ANI of 99.96%, whereas the clinical strain R2090 was assigned to ST267 and displayed an ANI of 97.73% to R2091. However, a chromosome-wide alignment by means of Mauve (34) of strains CIP 70.10 and R2091 (Fig. 5) revealed an approximately 65-kb region in R2091 (coordinates 3233306 to 3298533 bp) featuring a significantly lower ANI of 97.95% (Fig. 3). A large part of the 65-kb segment contains homologous genes, except for an 11-kb region (bp ∼3247000 to 3287300) representing the described transposon Tn125 harboring the blaNDM-1 gene. An alignment of the chromosomes of strains R2090 and R2091 confirmed that the 65 kb originates from the clinical strain R2090 (see Fig. S1 in the supplemental material). Precisely, a 66,182-bp region surrounding the transposon Tn125 was identical between the R2090 and R2091 chromosomes, except for two single nucleotide polymorphisms (SNPs).
FIG 5.
Pairwise alignment of the region carrying Tn125 (blaNDM-1) between the A. baumannii strain R2091 and the corresponding recipient strain CIP 70.10 obtained by using Mauve. Chromosome coordinates are plotted on the x axis, and the y axis denotes the percent sequence identity, with 100% representing completely identical sequences. (A) Chromosomal regions of strains CIP 70.10 and R2091 featuring comparatively low sequence similarity. Rectangles below the similarity plot represent coding sequences (CDSs). Colored rectangles denote the insertion sequence (IS) elements ISAba125 (red), a transposase gene (blue), and the gene blaNDM-1 (green). (B and C) Enlarged regions, in which possible homologous recombination events have occurred, are shown. First (B) and last (C) SNPs of the region featuring low sequence similarity are boxed. Beyond these SNPs, the chromosomal sequences of the strains are completely identical, except for very few minor mismatches presumably representing mutation events in one or the other genome or sequencing errors.
These analyses uncovered that a 66-kb region carrying the blaNDM-1 gene had been transferred from the clinical strain R2090 to the recipient strain CIP 70.10, in which it was integrated into the chromosome, presumably through homologous recombination. Four possible mechanisms have to be considered for the observed transfer of the resistance gene region from the donor to the recipient strain: (i) plasmid-mediated conjugative transfer, (ii) outer membrane vesicle (OMV)-mediated transfer, (iii) transformation, or (iv) phage-mediated transduction.
Plasmid-mediated transfer can be excluded, since strain R2090 lacks any plasmid. Moreover, a type IV secretion system (T4SS), required for mating-pair formation and conjugative DNA transfer to a recipient cell (44), is not encoded in the genome of isolate R2090, as determined by application of the bioinformatics tool CONJscan-T4SSscan (29). DNA transfer mediated by outer membrane vesicles (OMV) seems to be unlikely because of the size of the transferred region and its chromosomal location. Some Acinetobacter species are known to be able to transfer DNA via OMVs (45, 46). However, the transferred region in the case of strain R2090 seems to be too large for OMV-mediated transmission, since so far, only 10- to 25-kb fragments were observed to be transferred involving OMVs (45–47). Uptake of free DNA from the medium also is unlikely, although some A. baumannii strains previously were shown to be capable of natural transformation. A. baumannii CIP 70.10 encodes a type IV pilus (T4P) system and a channel formed by the proteins ComA/ComEC implicated in DNA uptake from outside the cell (48). However, transformation efficiency was shown to be very low for strain CIP 70.10 (49). Moreover, DNA uptake of fragments >50 kb by natural transformation currently has to be considered a great challenge for the cell (50), and nucleases may restrict incoming DNA (51).
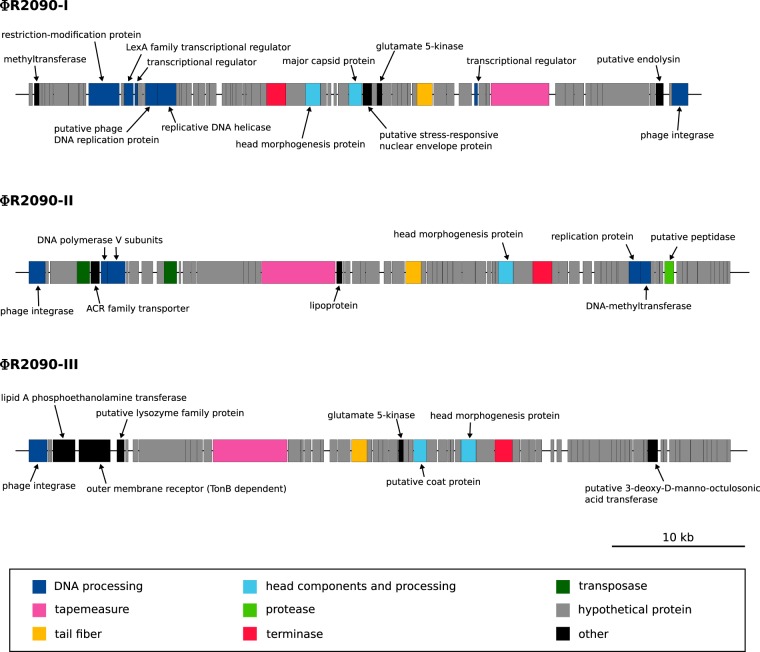
Finally, larger DNA fragments can be transferred by phage-mediated general transduction. Application of the phage search tool PHAST (30) led to the prediction of two intact (from coordinates 1165468 to 1215544 and from 2514187 to 2568847) and one presumptive prophage region (from coordinates 2635367 to 2687628) in the R2090 genome (Fig. 6). The putative prophages show similarity to the reference phage BΦ-B1251 (YMC/09/02/B1251 ABA BP) (52). Overall, 36 of the 62 CDSs of the reference phage were identified at least once in the R2090 chromosome, featuring identities of at least 90% at the amino acid sequence level. Manual analyses of the prophage regions revealed that they are each approximately 49 to 52 kb in size and comprise 69 to 71 CDSs (Fig. 6). The genes in the prophage regions are similar to those in other A. baumannii prophages, which were predicted to be intact (53). Genes known to be essential for phage activity, such as those specifying DNA processing, replication, structural components, morphogenesis, phage integration, and cell lysis, were identified (Fig. 6). Furthermore, the prophage regions ΦR2090-II and ΦR2090-III are bordered by putative attachment sites attL and attR (21 bp and 63 bp in size, respectively). Accordingly, prophage ΦR2090-II was predicted to be intact by means of the bioinformatics tool PHAST (score, 110), whereas ΦR2090-III was classified as questionable (score, 90), despite the predicted attachment sites. Although no valid attachment sites were identified for ΦR2090-I, it was categorized as intact by PHAST (score, 91). However, the activities of these phages remain to be confirmed.
FIG 6.
Predicted prophage regions within the A. baumannii R2090 chromosome. Putative prophage regions, as identified by application of the bioinformatics tool PHAST (30), each about 50 kb in size, are represented to scale. According to this analysis, ΦR2090-I (ABR2090_1106-1176) and ΦR2090-II (ABR2090_2421-2489) seem to be intact (scores, 91 and 110, respectively), whereas the completeness of ΦR2090-III (ABR2090_2555-2624) is not clear (score, 90). Phage-related genes are colored according to predicted functions of encoded gene products.
It is thus conceivable that after phage-mediated lysis of R2090 donor cells, random DNA fragments were packaged into the capsid of the phage and subsequently transferred to the recipient cell via infection (transduction). Fragments of >100 kb were previously shown to be transferable by phage-mediated transduction (54). DNA introduced into a recipient cell by transduction can be integrated into the chromosome via RecA-dependent recombination (51) and thus may become manifested in the host's genome. Inspection of both ends of the transferred 66-kb fragment comprising the blaNDM-1 gene led to the identification of 180-bp and 774-bp flanking regions that are identical to corresponding regions present in the recipient chromosome of strain CIP 70.10. These sequences presumably are long enough to represent the target sites for homologous recombination (51, 55), leading to the integration of the resistance gene region into the chromosome of the recipient CIP 70.10.
Concluding remarks.
The carbapenem-resistant A. baumannii strain R2090 originated from a colonized Egyptian patient who was admitted to a hospital in France. Interestingly, the isolated strain appeared to be closely related to A. baumannii strain D1279779 belonging to the ST267. The D1279779 reference strain was obtained from an indigenous Australian patient and represents a community-acquired isolate. The close relatedness of an Egyptian and Australian A. baumannii isolate again reflects the dissemination of opportunistic pathogenic bacteria due to long-distance travel opportunities, and it sheds light on A. baumannii epidemiology. The main difference between strains R2090 and D1279779 is that the isolate R2090 harbors a gene encoding the New Delhi metallo-β-lactamase NDM-1, which mediates carbapenem resistance. The presence of this gene severely complicates the treatment of A. baumannii infections by application of β-lactam antibiotics. Basically, it was not possible to deduce how the blaNDM-1 gene was acquired by strain R2090, since it is not located on a mobile plasmid but appeared to be integrated in the host's chromosome. However, the β-lactamase resistance gene is flanked by two different insertion sequence elements (ISAba125 and ISCR21) that potentially enable movement (translocation) of the blaNDM-1 gene. It has been shown experimentally that the resistance gene region could be transferred from the strain R2090 to a susceptible A. baumannii recipient strain. Commonly, mobile plasmids are involved in the horizontal transfer of resistance determinants between bacteria. However, since strain R2090 does not contain any plasmid, another mechanism must account for the resistance gene transfer observed. Natural transformation, outer membrane vesicle-mediated transfer, or transduction facilitated by phages have to be considered for horizontal gene transfer. The first two mechanisms were previously shown to enable the transfer of relatively short DNA fragments. In the case of strain R2090, an approximately 66-kb fragment was transferred to the A. baumannii recipient strain CIP 70.10. Since isolate R2090 possesses three presumably intact prophages integrated in its chromosome, it is very likely that activation of one of these prophages may have facilitated transduction of the blaNDM-1 gene. Although general transduction accomplished by phages is not as efficient as conjugative plasmid-mediated gene transfer, it has to be taken into account for the dissemination of resistance genes among Acinetobacter strains. Hence, phages may contribute to the adaptation of pathogenic Acinetobacter species to selective pressure caused by the presence of antibiotics.
Supplementary Material
ACKNOWLEDGMENTS
The bioinformatics support of the BMBF-funded project Bielefeld-Giessen Center for Microbial Bioinformatics (BiGi) (grant 031A533) within the German Network for Bioinformatics Infrastructure (de.NBI) is gratefully acknowledged. I. Maus and D. Wibberg acknowledge the receipt of a scholarship from the CLIB Graduate Cluster Industrial Biotechnology, cofinanced by the Ministry of Innovation of North Rhine-Westphalia, Germany. This work has also been financed by the University of Fribourg, Switzerland, and by the Swiss National Fund (SNF) under grant 31003A_163432/1.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00124-16.
REFERENCES
- 1.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:122–129. doi: 10.1038/nm0204-122. [DOI] [PubMed] [Google Scholar]
- 2.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie J-M, Raoult D, Médigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juni E. 1978. Genetics and physiology of Acinetobacter. Annu Rev Microbiol 32:349–371. doi: 10.1146/annurev.mi.32.100178.002025. [DOI] [PubMed] [Google Scholar]
- 4.Cerqueira GM, Peleg AY. 2011. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060. doi: 10.1002/iub.533. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet PJM, Grimont PAD. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol 36:228–240. doi: 10.1099/00207713-36-2-228. [DOI] [Google Scholar]
- 6.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 7.Touchon M, Cury J, Yoon E-J, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EPC. 2014. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol Evol 6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imperi F, Antunes LCS, Blom J, Villa L, Iacono M, Visca P, Carattoli A. 2011. The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63:1068–1074. doi: 10.1002/iub.531. [DOI] [PubMed] [Google Scholar]
- 11.Krahn T, Wibberg D, Maus I, Winkler A, Nordmann P, Pühler A, Poirel L, Schlüter A. 2015. Complete genome sequence of the clinical strain Acinetobacter baumannii R2090 carrying the chromosomally encoded metallo-β-lactamase gene blaNDM-1. Genome Announc 3(5):e01008-15. doi: 10.1128/genomeA.01008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krahn T, Wibberg D, Maus I, Winkler A, Pühler A, Poirel L, Schlüter A. 2015. Complete genome sequence of Acinetobacter baumannii CIP 70.10, a susceptible reference strain for comparative genome analyses. Genome Announc 3(4):e00850-15. doi: 10.1128/genomeA.00850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. 2003. GenDB: an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, Zhu Z, Fu L, Niu B, Li W. 2011. WebMGA: a customizable Web server for fast metagenomic sequence analysis. BMC Genomics 12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wibberg D, Blom J, Jaenicke S, Kollin F, Rupp O, Scharf B, Schneiker-Bekel S, Sczcepanowski R, Goesmann A, Setubal JC, Schmitt R, Pühler A, Schlüter A. 2011. Complete genome sequencing of Agrobacterium sp. H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J Biotechnol 155:50–62. [DOI] [PubMed] [Google Scholar]
- 17.Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, Linke B, Goesmann A, Grabherr R, Schwab H, Pühler A, Schlüter A. 2012. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol 161:153–166. doi: 10.1016/j.jbiotec.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Eikmeyer F, Hadiati A, Szczepanowski R, Wibberg D, Schneiker-Bekel S, Rogers LM, Brown CJ, Top EM, Pühler A, Schlüter A. 2012. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 68:13–24. doi: 10.1016/j.plasmid.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter F-J, Zakrzewski M, Goesmann A. 2009. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. 2007. MvirDB: a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res 35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain J-M. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson MK, Forsberg KJ, Dantas G. 2015. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 9:207–216. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23:205–211. [PubMed] [Google Scholar]
- 28.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res 8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 32.Farrugia DN, Elbourne LDH, Hassan KA, Eijkelkamp BA, Tetu SG, Brown MH, Shah BS, Peleg AY, Mabbutt BC, Paulsen IT. 2013. The complete genome and phenome of a community-acquired Acinetobacter baumannii. PLoS One 8:e58628. doi: 10.1371/journal.pone.0058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herbig A, Jäger G, Battke F, Nieselt K. 2012. GenomeRing: alignment visualization based on SuperGenome coordinates. Bioinformatics 28:i7–i15. doi: 10.1093/bioinformatics/bts217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr Opin Microbiol 11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilks A, Burkhard KA. 2007. Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep 24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 39.Loehfelm TW, Luke NR, Campagnari AA. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol 190:1036–1044. doi: 10.1128/JB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litrán T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. doi: 10.1128/JB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18:E362–E365. doi: 10.1111/j.1469-0691.2012.03928.x. [DOI] [PubMed] [Google Scholar]
- 43.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Göttig S, Hunfeld K-P, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 44.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 45.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, Ye JY, Weitao T. 2012. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal 2012:128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66:4414–4420. doi: 10.1128/AEM.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mell JC, Redfield RJ. 2014. Natural competence and the evolution of DNA uptake specificity. J Bacteriol 196:1471–1483. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmen R, Hellingwerf KJ. 1997. Uptake and processing of DNA by Acinetobacter calcoaceticus: a review. Gene 192:179–190. doi: 10.1016/S0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 52.Jeon J, Kim J, Yong D, Lee K, Chong Y. 2012. Complete genome sequence of the podoviral bacteriophage YMC/09/02/B1251 ABA BP, which causes the lysis of an OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolate from a septic patient. J Virol 86:12437–12438. doi: 10.1128/JVI.02132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hare JM, Ferrell JC, Witkowski TA, Grice AN. 2014. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS One 9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 55.Nelson RT, Pryor BA, Lodge JK. 2003. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet Biol 38:1–9. doi: 10.1016/S1087-1845(02)00510-8. [DOI] [PubMed] [Google Scholar]
- 56.Bonnin RA, Poirel L, Nordmann P. 2014. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol 9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.