Abstract
Cephalosporin-resistant Vibrio alginolyticus was first isolated from food products, with β-lactamases encoded by blaPER-1, blaVEB-1, and blaCMY-2 being the major mechanisms mediating their cephalosporin resistance. The complete sequence of a multidrug resistance plasmid, pVAS3-1, harboring the blaCMY-2 and qnrVC4 genes was decoded in this study. Its backbone exhibited genetic homology to known IncA/C plasmids recoverable from members of the family Enterobacteriaceae, suggesting its possible origin in Enterobacteriaceae.
TEXT
Vibrio alginolyticus, like other Vibrionaceae species, such as V. cholerae, V. parahaemolyticus, and V. vulnificus, may cause a range of human infections, such as gastroenteritis, septicemia, and necrotizing fasciitis. Of the pathogenic Vibrio species, V. alginolyticus is one of the top three causes of infections in humans and is genetically homologous to V. parahaemolyticus (1, 2). Resistance to expanded-spectrum cephalosporins and fluoroquinolones has also been reported in V. parahaemolyticus and V. cholerae but not in other Vibrio species such as V. vulnificus and V. alginolyticus (3, 4). Here we report (i) the isolation from food products of V. alginolyticus strains that are resistant to expanded-spectrum cephalosporins and (ii) the resistance mechanisms involved.
V. alginolyticus strains were isolated from food samples, including shrimp, chicken, pork, and beef, in Shenzhen, China, from June 2014 to August 2015 as previously described (5). Suspicious colonies from thiosulfate-citrate-bile salt sucrose and CHROMagar plates were screened by PCR and DNA sequencing and confirmed with API 20E test strips (bioMérieux, Inc.). A total of 23 V. alginolyticus strains were isolated from 515 food samples (2 from 88 chicken samples, 1 from 258 pork samples, 1 from 121 beef samples, and 19 from 48 shrimp samples). These 23 V. alginolyticus strains were subjected to antimicrobial susceptibility testing according to the Clinical and Laboratory Standards Institute (6). These isolates exhibited a high rate of resistance to ampicillin (100%), followed by trimethoprim-sulfamethoxazole (48%); ceftriaxone and cefotaxime (26%); chloramphenicol (13%); cefoxitin and amoxicillin-clavulanic acid (8.7%); and nalidixic acid, ciprofloxacin, ofloxacin, and gentamicin (4.4%). All strains were susceptible to tetracycline, amikacin, and meropenem. Five of six cephalosporin-resistant strains were isolated from shrimp samples in the same market on different dates, whereas one strain, VAS3-1, was isolated from a chicken sample (Table 1). Characterization of these isolates by pulsed-field gel electrophoresis (PFGE) was performed as previously described (7) and revealed that these isolates had distinct PFGE profiles, suggesting a high level of genetic diversity of these isolates even though most of them were recovered from the same market at different times (see Fig. S1 in the supplemental material). The prevalence of β-lactamase genes among the strains was determined by PCR assays as previously described (8) and showed that the blaPER-1 β-lactamase gene is present in strains VA1 and VA6 and that the blaCMY-2 gene is present in VAS3-1. The blaVEB-1 element was detected in strains V2 and V5, but no known β-lactamase gene was found in strain V4 (Table 1). It should be noted that this is the first time these cephalosporinases have been described in V. alginolyticus and that this is the first time the blaVEB-1 element has been recovered from a Vibrio species. Conjugation experiments performed with these strains showed that three strains, namely, VA1, VAS3-1, and VA5, could successfully transfer their cephalosporin resistance phenotypes to a recipient strain, Escherichia coli J53. Plasmid typing results showed that the conjugative plasmid recovered from VAS3-1 belonged to the IncA/C type, whereas conjugative plasmids from VA1 and VA5 were untypeable. Our laboratory has recently recovered an IncA/C-type conjugative plasmid from a V. parahaemolyticus strain (3). We therefore further characterized the IncA/C conjugative plasmid recovered from strain VAS3-1 in this study and investigated whether it is genetically related to the one obtained from V. parahaemolyticus. S1 PFGE and hybridization with a blaCMY-2 probe performed with VAS3-1 showed that the strain contains two blaCMY-2-positive plasmids of ∼180 and 300 kb, respectively, but only the ∼180-kb plasmid was transferrable to the E. coli J53 recipient strain (see Fig. S2 in the supplemental material). This 180-kb conjugative plasmid is larger than the ∼150-kb V. parahaemolyticus plasmid reported in our previous study (3).
TABLE 1.
Antibiotic susceptibility profiles of cephalosporin-resistant V. alginolyticus strains and corresponding recipient transconjugant E. coli strain J53
| Strain | 2015 isolation date | Source | β-Lactamase gene product | MIC (mg/liter)b of: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | CTX | CRO | NAL | CIP | AMK | TET | CHL | GEN | SXT | ||||
| J53 | 1 | 4/2 | 0.03 | 0.015 | 1 | 0.03 | 0.5 | 0.5 | 2 | 0.25 | 0.25/4.75 | |||
| VA1 | 1/14 | Shrimp | PER-1 | >64 | 16/8 | >16 | >16 | 0.5 | 0.06 | 4 | 2 | 0.25 | 32 | >16/304 |
| T-VA1 | PER-1 | >64 | 8/4 | >16 | >16 | 4 | 0.015 | 0.5 | 8 | 2 | 32 | 16/304 | ||
| VA2 | 1/20 | Shrimp | VEB-1 | >64 | 4/2 | >16 | >16 | 32 | >16 | 1 | 8 | 32 | 4 | >16/304 |
| VAS3-1 | 3/14 | Chicken | CMY-2 | >64 | 64/32 | 8 | 16 | 16 | 0.5 | 0.5 | 16 | 32 | 1 | >16/304 |
| T-VAS3-1 | CMY-2 | >64 | 64/32 | 8 | >16 | 8 | 0.5 | 0.5 | 8 | 16 | 1 | 8/152 | ||
| VA4 | 3/4 | Shrimp | NDa | >64 | 4/2 | >16 | >16 | 0.5 | 0.5 | 2 | 4 | >32 | 4 | 8/152 |
| VA-5 | 3/14 | Shrimp | VEB-1 | >64 | 8/4 | >16 | >16 | 2 | 1 | 4 | 0.5 | 1 | 8 | 0.25/4.75 |
| T-VA5 | VEB-1 | >64 | 4/2 | 2 | 2 | 8 | 0.03 | 0.5 | 0.5 | 2 | 8 | 0.25/4.75 | ||
| VA6 | 3/30 | Shrimp | PER-1 | >64 | 4/2 | 16 | 8 | 1 | 0.25 | 4 | 2 | 2 | 4 | 8/152 |
ND, not detected.
AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CRO, ceftriaxone; CTX, cefotaxime; NAL, nalidixic acid; CIP, ciprofloxacin; AMK, amikacin; TET, tetracycline; CHL, chloramphenicol; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole.
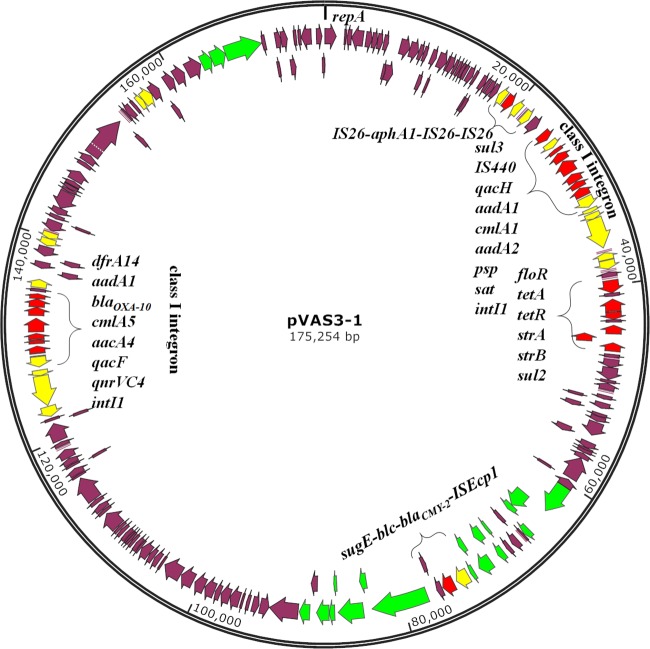
The plasmid from a transconjugant, T-VAS3-1, was sequenced with the Illumina NextSeq 500 platform and the PacBio RSII single-molecule real-time sequencing platform. The completed plasmid sequence was confirmed by PCR and then annotated with the RAST tool and the NCBI Prokaryotic Genome Annotation Pipeline. The plasmid was designated pVAS3-1 and was found to be a circular IncA/C plasmid of 175,254 bp with a G+C content of 52.3%. It contained 216 predicted open reading frames, among which three core regions were disrupted by three resistance-encoding fragments (Fig. 1). The backbone exhibited a high degree of genetic similarity to six other known IncA/C plasmids (99% identity with 82 to 91% coverage) and was found to harbor genes responsible for plasmid replication, horizontal transfer, and maintenance of genetic stability (9, 10) (Fig. 2). All seven plasmids were blaCMY-harboring IncA/C conjugative plasmids, and each contained a mobile mosaic multidrug resistance (MDR) region located between conserved backbone genes ybaA and rhsD. Presumably, with the aid of transposons and other mobile elements, different resistance determinants (rmtE, blaNDM-1, the mer operon, etc.) were found incorporated into the plasmid backbone (9, 11). Resembling other blaCMY-2-positive plasmids, the ISEcp1-blaCMY-2 region was also found to be located between the traA and traC genes in pVAS3-1 (Fig. 1) (12). The blaCMY-2 gene was contained in a mobile element, ISEcp1-blaCMY-2-blc-sugE, that has previously been found mainly in members of the family Enterobacteriaceae, may originate from the Citrobacter freundii chromosome, and is transferrable to other bacterial species through ISEcp1-mediated mobilization and conjugative plasmids (12, 13). It has also recently been recovered in IncA/C plasmids harbored by Aeromonas salmonicida and Vibrio parahaemolyticus (3, 14). In the region upstream of the conserved blaCMY-2 fragment, another conserved MDR region common to the seven plasmids that contained the floR, tet(A/R), strA, strB, and sul2 elements was also detected. In addition, an IS26-aphA1-IS26 fragment and a nonclassic class I integron without the 3′ coding sequence region were also found in the upstream region of pVAS3-1 (Fig. 1). This integron, which harbored six gene cassettes, was interrupted by the IS440 and sul3 elements. Such an integron structure was found in an Enterobacteriaceae species only once before (15). The presence of this unique integron in pVAS3-1 and not in the other six IncA/C plasmids suggests that pVAS3-1 may have originated from one of these six IncA/C plasmids (Fig. 2). Although some researchers have speculated that an aquatic environment may be the source of IncA/C plasmids (16), in view of the above data and the increasing reports of blaCMY-2-positive plasmids in aquatic bacteria such as Aeromonas and Vibrio species (GenBank accession no. KJ909290 and CP007636) in recent years, we believe that dissemination of blaCMY-2 plasmids from Enterobacteriaceae to aquatic pathogens may be the most likely event.
FIG 1.
Gene map of conjugative IncA/C plasmid pVAS3-1. Yellow arrows represent mobile elements. Red arrows represent resistance genes. Blue arrows represent genes related to conjugal transfer functions.
FIG 2.
Whole-plasmid alignment of highly homologous blaCMY-2 IncA/C plasmids recovered from various bacterial species. Plasmid names and sources are on the left; elements located in variable mosaic regions are on the right. GenBank accession numbers: KJ909290, KP056256, JX442976, JN687470, CP007636, and KR091911.
The hypervariable region of pVAS3-1 was found to contain a ca. 20-kb MDR-encoding fragment, including the novel class I integron In1222 (qnrVC4, qacF, aacA4, cmlA5, blaOXA-10, aadA1, and dfrA14), a macrolide resistance determinant (mphA-mrx-mphR), and a mercury resistance operon. The qnrVC4 gene was first identified in Aeromonas punctata in China in a classic class I integron (17). The class I integron found in pVAS3-1 is partially homologous to the one reported in Salmonella enterica serovar Rissen (18) in Thailand and different from the one reported in A. punctata (see Fig. S3 in the supplemental material). qnrVC alleles that encode quinolone resistance determinants that mediate decreased susceptibility to fluoroquinolones have emerged globally (17, 18). To date, seven different qnrVC alleles have been identified, mainly in the Vibrionaceae family; however, these resistance elements have also been recovered in other pathogens, such as Pseudomonas aeruginosa and Acinetobacter baumannii (7, 18, 19), highlighting the possibility that qnrVC alleles can be readily transmitted from aquatic-environment organisms to clinical pathogens.
In conclusion, comparative analysis of the genetic features of pVAS3-1 and plasmids recoverable from other bacterial species indicates that such elements exhibit a high degree of genetic plasticity and that the increasing reports of IncA/C plasmids in aquatic bacteria may be due to the transmission of these plasmids from Enterobacteriaceae.
Nucleotide sequence accession number.
The sequence of plasmid pVAS3-1 has been deposited in GenBank under accession no. KU160531.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Thomas Jové of INTEGRALL for his help in annotating the novel integron In1222 and members of the Sheng lab for their useful discussions.
This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200) and the Health and Medical Research Fund from the Food and Health Bureau, the Government of the Hong Kong SAR (HMRF 13121422 to S.C.).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00300-16.
REFERENCES
- 1.Xie ZY, Hu CQ, Chen C, Zhang LP, Ren CH. 2005. Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Lett Appl Microbiol 41:202–207. doi: 10.1111/j.1472-765X.2005.01688.x. [DOI] [PubMed] [Google Scholar]
- 2.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. 2012. Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin Infect Dis 54(Suppl 5):S391–S395. doi: 10.1093/cid/cis243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Lin D, Chen K, Wong MH, Chen S. 2015. First detection of AmpC β-lactamase bla(CMY-2) on a conjugative IncA/C plasmid in Vibrio parahaemolyticus of food origin. Antimicrob Agents Chemother 59:4106–4111. doi: 10.1128/AAC.05008-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Xie L, Zhang F, Ni Y, Sun J. 2015. Molecular characterization of ISCR1-mediated blaPER-1 in a non-O1, non-O139 Vibrio cholerae strain from China. Antimicrob Agents Chemother 59:4293–4295. doi: 10.1128/AAC.00166-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Pinto A, Terio V, Novello L, Tantillo G. 2011. Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control 22:124–127. doi: 10.1016/j.foodcont.2010.06.013. [DOI] [Google Scholar]
- 6.CLSI. 2005. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; proposed guideline. CLSI document M45-P. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Belotti PT, Thabet L, Laffargue A, Andre C, Coulange-Mayonnove L, Arpin C, Messadi A, M'Zali F, Quentin C, Dubois V. 2015. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J Antimicrob Chemother 70:2237–2340. doi: 10.1093/jac/dkv103. [DOI] [PubMed] [Google Scholar]
- 8.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 9.Lee CS, Li JJ, Doi Y. 2015. Complete sequence of conjugative IncA/C plasmid encoding CMY-2 β-lactamase and RmtE 16S rRNA methyltransferase. Antimicrob Agents Chemother 59:4360–4361. doi: 10.1128/AAC.00852-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother 54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villa L, Guerra B, Schmoger S, Fischer J, Helmuth R, Zong Z, Garcia-Fernandez A, Carattoli A. 2015. IncA/C plasmid carrying blaNDM-1, blaCMY-16, and fosA3 in a Salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany. Antimicrob Agents Chemother 59:6597–6600. doi: 10.1128/AAC.00944-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdet C, Gautier V, Chachaty E, Ronco E, Hidri N, Decre D, Arlet G. 2009. Genetic context of plasmid-carried blaCMY-2-like genes in Enterobacteriaceae. Antimicrob Agents Chemother 53:4002–4006. doi: 10.1128/AAC.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlow M, Hall BG. 2002. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob Agents Chemother 46:1190–1198. doi: 10.1128/AAC.46.5.1190-1198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent AT, Trudel MV, Paquet VE, Boyle B, Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Derome N, Charette SJ. 2014. Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: multidrug resistance, interspecies exchanges, and plasmid reshaping. Antimicrob Agents Chemother 58:7367–7374. doi: 10.1128/AAC.03730-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinué L, Saenz Y, Rojo-Bezares B, Olarte I, Undabeitia E, Somalo S, Zarazaga M, Torres C. 2010. Genetic environment of sul genes and characterisation of integrons in Escherichia coli isolates of blood origin in a Spanish hospital. Int J Antimicrob Agents 35:492–496. doi: 10.1016/j.ijantimicag.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia R, Guo X, Zhang Y, Xu H. 2010. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob Agents Chemother 54:3471–3474. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonseca EL, Vicente AC. 2013. Epidemiology of qnrVC alleles and emergence out of the Vibrionaceae family. J Med Microbiol 62:1628–1630. doi: 10.1099/jmm.0.062661-0. [DOI] [PubMed] [Google Scholar]
- 19.Po KHL, Wong MHY, Chen S. 2015. Identification and characterisation of a novel plasmid-mediated quinolone resistance gene, qnrVC7, in Vibrio cholerae of seafood origin. Int J Antimicrob Agents 45:667–668. doi: 10.1016/j.ijantimicag.2015.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.