Abstract
Traditional therapeutic strategies to control chronic colonization in cystic fibrosis (CF) patients are based on the use of a single nebulized antibiotic. In this study, we evaluated the therapeutic efficacy and dynamics of antibiotic resistance in Pseudomonas aeruginosa biofilms under sequential therapy with inhaled aztreonam (ATM) and tobramycin (TOB). Laboratory strains PAO1, PAOMS (hypermutable), PAOMA (mucoid), and PAOMSA (mucoid and hypermutable) and two hypermutable CF strains, 146-HSE (Liverpool epidemic strain [LES-1]) and 1089-HSE (ST1089), were used. Biofilms were developed using the flow cell system. Mature biofilms were challenged with peak and 1/10-peak concentrations of ATM (700 mg/liter and 70 mg/liter), TOB (1,000 mg/liter and 100 mg/liter), and their alternations (ATM/TOB/ATM and TOB/ATM/TOB) for 2 (t = 2), 4 (t = 4), and 6 days (t = 6). The numbers of viable cells (CFU) and resistant mutants were determined. Biofilm structural dynamics were monitored by confocal laser scanning microscopy and processed with COMSTAT and IMARIS software programs. TOB monotherapy produced an intense decrease in CFU that was not always correlated with a reduction in biomass and/or a bactericidal effect on biofilms, particularly for the CF strains. The ATM monotherapy bactericidal effect was lower, but effects on biofilm biomass and/or structure, including intense filamentation, were documented. The alternation of TOB and ATM led to an enhancement of the antibiofilm activity against laboratory and CF strains compared to that with the individual regimens, potentiating the bactericidal effect and/or the reduction in biomass, particularly at peak concentrations. Resistant mutants were not documented in any of the regimens at the peak concentrations and only anecdotally at the 1/10-peak concentrations. These results support the clinical evaluation of sequential regimens with inhaled antibiotics in CF, as opposed to the current maintenance treatments with just one antibiotic in monotherapy.
INTRODUCTION
Pseudomonas aeruginosa is the major cause of chronic respiratory infections (CRI) and the main driver of morbidity and mortality in patients with cystic fibrosis (CF) (1) and other chronic respiratory diseases, such as chronic obstructive pulmonary disease (COPD) or bronchiectasis (2, 3). The establishment of P. aeruginosa CRI requires a complex adaptive process that includes the selection of an important number of mutations required for long-term persistence and the transition from the planktonic to biofilm mode of growth, a hallmark of chronic infections (4–6). The complex structure of biofilms along with the large heterogeneity of bacterial cells in different physiological states may explain the increased tolerance to antibiotics and host defense mechanisms (7). However, more recent works have demonstrated that, in addition to physiological resistance driven by biofilm architecture, mutational mechanisms play a major role in biofilm antibiotic resistance, especially when mutator strains are involved (8). These mutator strains, most often deficient in the DNA mismatch repair (MMR) system, are highly prevalent (30 to 60%) in patients with P. aeruginosa CRI (9, 10). Furthermore, hypermutation has been found to be linked to high antibiotic resistance rates (11), to adaptation to the CRI setting (12), and to adaptability and diversification processes in biofilms, hampering the eradication of CRI (13).
The most relevant P. aeruginosa mutational resistance mechanisms are those leading to overexpression of the chromosomal β-lactamase AmpC, inactivation of the carbapenem porin OprD, or upregulation of several efflux pumps encoded in its genome. The combinations of resistance mechanisms are frequently additive or synergistic and lead to multidrug resistance; however, antagonistic interactions between resistance mechanisms have been described. A recent work confirmed that overexpression of the MexCD-OprJ efflux pump impairs the backbone of related P. aeruginosa intrinsic mechanisms such as the MexAB-OprM and MexXY-OprM efflux pumps and chromosomal β-lactamase AmpC (14). On the other hand, an imbalance between the MexXY-OprM and MexAB-OprM systems has been reported in CF (15). While overexpression of MexXY-OprM, the most important aminoglycoside resistance-conferring system, is frequently found in P. aeruginosa strains in CF (16), MexAB-OprM, which shares the same porin channel, tends to be lost or inactivated in this context, suggesting an antagonism between these two efflux pumps (15). The inactivation of the MexAB-OprM system confers hypersusceptibility to the substrates of this efflux pump, including aztreonam (ATM) and other antibiotics (15).
Traditional therapeutic strategies to control chronic colonization in CF patients are based on the use of a single nebulized formulation, either administered in a 28-day course (on-off) for tobramycin (TOB) or ATM or maintained as a chronic suppressive therapy for colistin (17). Since those strategies seem to be insufficient to eradicate infection and even to prevent or delay the onset of resistance as explained above, antagonistic interactions might be an innovative approach for designing successful therapeutic strategies. Thus, as treatment with aminoglycosides often involves the selection of mutants that overexpress the MexXY-OprM efflux pump, related to the inactivation of MexAB-OprM (15), treatment with MexXY-OprM substrates (such as TOB) theoretically might lead to hypersusceptibility to MexAB-OprM substrates (such as ATM); then, sequential treatment with TOB followed by ATM would entail a clinical benefit by improving the therapeutic efficacy and diminishing the selection of resistant mutants.
Therefore, the present study evaluated the therapeutic efficacy and emergence of resistant populations, comparing the effect of a regimen with a single antibiotic, either TOB or ATM, with that of the sequential alternation of the two antibiotics. We performed the experiments in an in vitro flow cell biofilm model, an open system where there is an attempt to replicate the in vivo conditions through the control of nutrient delivery, flow, and temperature. This model allows the application of pharmacokinetic/pharmacodynamic (PK/PD) parameters, as well as an in situ and nondestructive follow-up of the structural dynamics of biofilms under treatments through the use of confocal laser scanning microscopy (CLSM). P. aeruginosa phenotypes that are highly adapted and prevalent in CRI, such as mucoid and hypermutable strains, were used. Furthermore, we included two hypermutable clinical isolates deriving from two relevant CF epidemic clones, ST274 (18) and the multidrug-resistant (MDR) Liverpool epidemic strain (LES-1), which are likely the clones of most concern in CF worldwide (19).
MATERIALS AND METHODS
P. aeruginosa strains. (i) Laboratory strains.
The wild-type, piliated, reference strain PAO1 was obtained from the Danish collection (Systems Biology-DTU) (20, 21). Its mutS, mucA, and mutS-mucA knockout mutants and the PAOMS hypermutable, PAOMA mucoid, and PAOMSA mucoid and hypermutablestrains were constructed as in previous work by using the Cre-lox system for gene deletion and antibiotic resistance marker recycling (12, 22).
(ii) Clinical strains.
A total of 29 representative P. aeruginosa isolates from a well-characterized collection of cystic fibrosis (CF) strains from Hospital Son Espases (HSE), Palma de Mallorca, Spain (18) were screened for biofilm formation on microtiter plates using the crystal violet (CV) assay (data not shown) described by Christensen et al. (23). Two hypermutable isolates showing robust biofilm formation and belonging to the LES-1 epidemic clone ST146 (146-HSE) and clone ST1089 (1089-HSE) (a hypermutable derivate from the epidemic clone ST274) (18) were selected as relevant representative CF strains for further studies.
All P. aeruginosa strains were fluorescently tagged at the att intergenic neutral chromosomal locus with gfp (green fluorescent protein [GFP]) in mini-Tn7 constructs, as described by Klausen et al. (24).
Antibiotic susceptibility testing.
The MICs of ATM and TOB were determined for each of the six strains in Mueller-Hinton agar (MHA) by Etest, following the standard procedures and using the EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/). Suspensions at a 0.5 or 1 McFarland standard were used for inoculum standardization of regular or mucoid isolates, respectively. The MICs were read after 24 h (or 36 h for slow-growing strains) of incubation at 37°C.
Estimation of spontaneous mutation frequencies.
The frequencies of spontaneous mutation to rifampin (RIF) (300 mg/liter), ATM (4× and 16× MICs), and TOB (4× and 16× MICs) resistance were estimated in all strains as previously described (9). Briefly, independent triplicate 10-ml overnight Mueller-Hinton broth cultures of the P. aeruginosa isolates were collected by centrifugation and resuspended in 1 ml of saline solution. Serial 10-fold dilutions were plated in MHA with and without an antibiotic, and after 36 h of incubation (48 h for slow-growing strains), colonies were counted, and the median fraction of mutants was estimated. The mutS and mutL genes were analyzed through PCR and sequencing following previously described protocols (12).
PK/PD model of biofilm treatment.
Biofilms were grown at 30°C in three-channel flow cells (individual channel dimensions of 1 by 4 by 40 mm) supplied with modified FAB medium (21) supplemented with 0.3 mM glucose. Since the two clinical strains were found to be auxotrophic by culture on glucose minimal medium (M9) plates (25), extra supplementation with 2% Luria-Bertani medium was used (26). The flow system was assembled and prepared as described previously (27). Channels were inoculated with 250 μl of normalized dilutions (1/100 dilution of cultures adjusted to an optical density at 600 nm of 0.1) of saturated bacterial cultures and left without flow for 1 h to allow bacterial adherence. A medium flow was then started at a constant rate of 3 ml h−1 using a Watson-Marlow 205S peristaltic pump.
After 48 h and 96 h of incubation (time 0 [t0]) for the laboratory and clinical strains, respectively, biofilms were challenged with 6 days of ATM or 6 days of TOB as monotherapy or sequential treatment of ATM/TOB/ATM (A/T/A) or TOB/ATM/TOB (T/A/T) for 2 days with each antibiotic for 6 days. The peak concentrations (1,000 mg/liter TOB and 700 mg/liter ATM), which correlate to concentrations measured in the sputum of CF patients after aerosolized drug application (28), were chosen for the experiments with all of the strains (PAO1, PAOMS, PAOMA, PAOMSA, 146-HSE, and 1089-HSE). Additional studies using 1/10-peak concentrations of the antibiotics (100 mg/liter TOB and 70 mg/liter ATM) based on the sputum concentrations approximately 2 h after inhalation (29) were performed with representative strains PAO1, PAOMSA, 146-HSE, and 1089-HSE. The antibiotics were added to fresh medium and renewed daily.
At time points 0 (2- or 4-day-old biofilm), 4 (4 days of treatment or 6-day-old biofilm), and 6 (6 days of treatment or 8-day-old biofilm), biofilms were detached and collected by washing the flow cell channels with 1 ml of glass bead (Sigma) suspension in 0.9% NaCl. Before detachment, the flow cell system was cleaned with antibiotic-free medium for 30 min to minimize the postantibiotic effect. Viable cells and resistant mutants were determined for all antibiotic regimens and control biofilms for each of the six strains. For this purpose, the suspensions and serial 1/10 dilutions were plated in MHA and MHA supplemented with 4× and 16× MICs of TOB or ATM. The detection limit was 5 CFU/ml.
Microscopic analysis.
Biofilm structural dynamics were monitored by CLSM at time points 0, 4, and 6. The dead cells/areas of biofilms were red stained using propidium iodide (PI) to visualize the bactericidal dynamics of the antibiotics in the biofilms. All microscopic observations were performed by using a Zeiss LSM710 CLSM (Carl Zeiss, Jena, Germany) equipped with a multiline argon laser, detector, and filter sets for monitoring GFP expression (excitation, 488 nm; emission, 517 nm), as well as an NeHe laser for simultaneous monitoring of the red fluorescence emitted from the PI (excitation, 543 nm; emission filter, 565 to 615 nm). Images were obtained by using a 63×/1.4 oil PlanApo objective lens.
For assessment of the biomass of biofilms, at least four pictures per channel, flow cell, time point (t0, t4, and t6), and strain were taken and analyzed by using the COMSTAT program (Systems Biology-DTU) (21). Fluorescent signals emitted from the GFP and from PI were considered live and dead biomasses, respectively; total biomass resulted from the addition of both biomasses. The percentage of dead biomass relative to total biomass was calculated. Simulated three-dimensional (3D) images and sections were generated by using the IMARIS software package (Bitplane AG, Zurich, Switzerland). In all cases, the results from at least three independent experiments were considered.
Characterization of resistant mutants.
When appropriate, the resistant mutants obtained from plating detached biofilms in 4× and 16× MICs of ATM and/or TOB were characterized. The MICs of ticarcillin, piperacillin-tazobactam, ceftazidime, cefepime, ATM, ceftolozane-tazobactam, imipenem, meropenem, ciprofloxacin, TOB, amikacin, and colistin were determined by microdilution. The expression levels of ampC, mexB, mexD, mexF, and mexY were determined by real-time reverse transcription (RT)-PCR according to previously described protocols (30, 31). In all cases, the mean values for mRNA expression obtained in two experiments were considered. Overexpression was considered for mutants showing expression values that were ≥3-fold (mexB) or ≥10-fold (ampC, mexY, mexD, and mexF) higher than those for wild-type PAO1 (31).
Statistical analysis.
The structural data for control biofilms versus biofilms treated with each of the four regimens and biofilms under individual treatments versus sequential treatments were compared using Student t tests. A P value of <0.05 was considered statistically significant.
RESULTS
Antimicrobial susceptibility and spontaneous resistance mutation frequencies.
Table 1 shows the strains used in this work with the MICs of the antibiotics studied and the frequencies of spontaneous mutation to RIF, ATM, and TOB resistance. Despite all strains showing wild-type ATM MICs, they are to be considered intermediate according to EUCAST breakpoints. In other words, EUCAST considers that P. aeruginosa is intrinsically nonsusceptible to aztreonam, mainly due to the basal expression of the MexAB-OprM efflux pump. It should be further noted, however, that the clinical breakpoints available are intended for systemic and not inhaled administration, which provides a much higher antibiotic concentration. On the other hand, all isolates were susceptible to TOB, although the two clinical strains, 146-HSE and 1089-HSE, showed higher MICs due to the overexpression of MexXY-OprM (18). Strain 146-HSE also overexpressed the MexCD-OprJ and MexEF-OprN efflux pumps as previously described resistance (18). The frequencies of spontaneous mutation to RIF resistance on the order of 10−6 confirmed that both clinical strains were hypermutable. Likewise, the mutator strains showed about 2-log higher frequencies of spontaneous mutation to ATM and TOB resistance, although the 16× MIC TOB-resistant mutants were only obtained for the clinical strains (Table 1).
TABLE 1.
Description of the strains used in this work
| Strain | Characteristic(s) | MIC (mg/liter)a |
Spontaneous mutation frequency |
|||||
|---|---|---|---|---|---|---|---|---|
| ATM (S ≤ 1–R >16) | TOB (S ≤4–R >4) | 300 mg/liter RIF | ATM |
TOB |
||||
| 4× MIC | 16× MIC | 4× MIC | 16× MIC | |||||
| PAO1 | Wild-type | 2 | 1 | 3.2 × 10−9 | 1.57 × 10−7 | <10−9 | 4.22 × 10−9 | <10−9 |
| PAOMS | △mutS | 2 | 1 | 1.1 × 10−6 | 9.76 × 10−6 | 8.28 × 10−8 | 8.39 × 10−7 | <10−9 |
| PAOMA | △mucA | 2 | 1 | 6.2 × 10−9 | 1.69 × 10−7 | <10−9 | 2.25 × 10−9 | <10−9 |
| PAOMSA | △mutS-△mucA | 2 | 1 | 1.6 × 10−6 | 3.87 × 10−6 | <10−9 | 1.11−7 | <10−9 |
| 146-HSE | CF strain LES-1, MDR epidemic strain (ST146), hypermutable (△mutS)b | 2 | 4 | 1.1 × 10−6 | 4.2 × 10−5 | 2.4 × 10−6 | 2.4 × 10−5 | 4.7 × 10−6 |
| 1089-HSE | CF strain derived from ST274 epidemic clone (ST1089), hypermutable (△mutS)b | 2 | 4 | 2.2 × 10−6 | 2.8 × 10−6 | 8.8 × 10−8 | 9.2 × 10−7 | 1.2 × 10−8 |
S, susceptible; R, resistant.
Strain 146-HSE shows a nonsynonymous (G778D) mutation in mutS not present in a preceding isogenic nonhypermutable isolate recovered from the same patient. Strain 1089-HSE shows a previously characterized inactivating mutation (4-bp deletion from nucleotide 814) in mutS (18).
Therapeutic efficacy based on the determination of viable cells and antibiotic-resistant mutants.
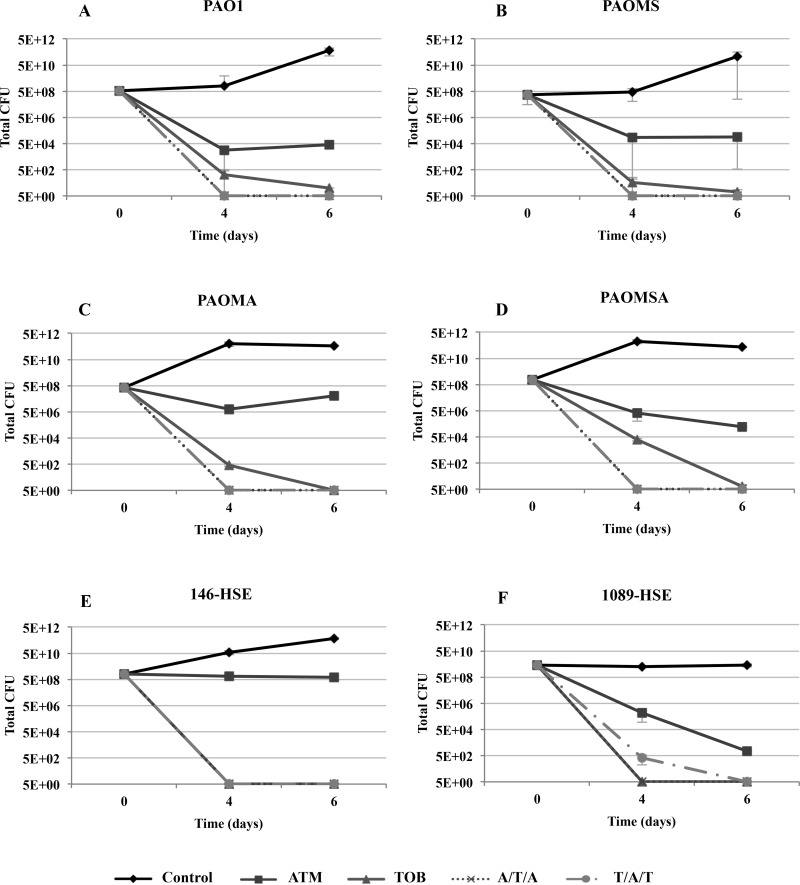
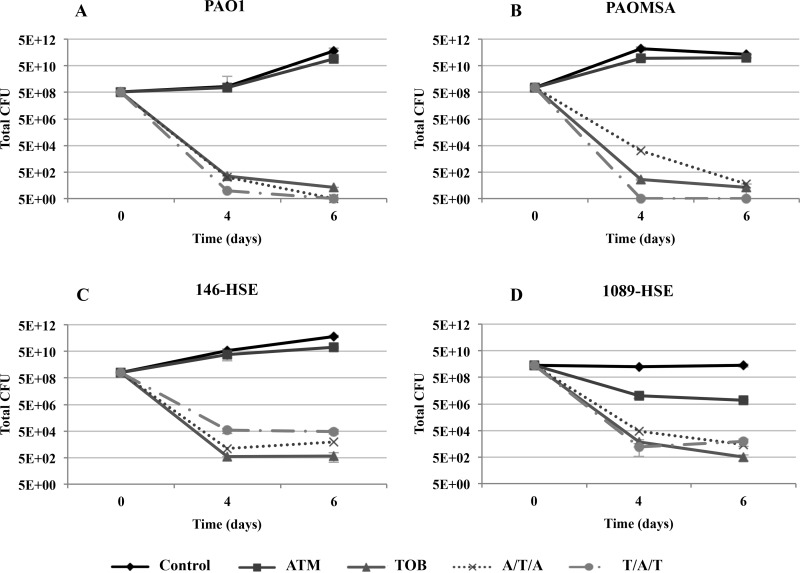
Figures 1 and 2 show the dynamics over time of the bacterial population of biofilms under monotherapy and sequential treatments, with peak and 1/10-peak concentrations, respectively.
FIG 1.
Dynamics over time of the bacterial populations of biofilms treated with peak concentrations (700 mg/liter ATM and 1,000 mg/liter TOB). The results represent the medians (symbols) and interquartile ranges (error bars) from at least three independent experiments.
FIG 2.
Dynamics over time of the bacterial populations of biofilms treated with 1/10-peak concentrations (70 mg/liter ATM and 100 mg/liter TOB). The results represent the medians (symbols) and interquartile ranges (error bars) from at least three independent experiments.
Monotherapies.
The ATM peak concentration treatment produced a marked reduction in the biofilm bacterial loads of all laboratory strains (Fig. 1A to D). Regarding the clinical strains, the reduction in viable cells was pronounced for 1089-HSE (Fig. 1F), while for 146-HSE, ATM produced only a slight decrease in the bacterial load over time (Fig. 1E). Treatment with the ATM peak concentration did not select resistant mutants, even in the mutator strains (see Fig. S1 in the supplemental material). Moreover, it was able to eradicate the ATM spontaneous resistant mutants preexisting at t0 for all the strains, except in the case of strain 146-HSE, for which a strong reduction in the number of mutants was documented (see Fig. S1E). However, treatment with the ATM 1/10-peak concentration did not reduce the bacterial load, which remained at approximately the same level as that of the control biofilms (Fig. 2). ATM spontaneous resistant mutants persisted in the mutator strain 1089-HSE (see Fig. S3B and D) and were slightly (1 log compared to control) selected in strain 146-HSE (see Fig. S3C).
On the other hand, the TOB peak concentration produced a major reduction in the bacterial loads of the biofilms from the laboratory strains over time, which fell below the detection limit at t6 (Fig. 1). Surprisingly, the reductions in the bacterial loads were even greater for the clinical strains, reaching the detection limit at t4. Similarly, although smaller, an important reduction was observed with the 1/10-peak concentration (Fig. 2). Both concentrations eradicated the TOB preexisting resistant mutants and did not select resistant mutants over time (see Fig. S2 and S4 in the supplemental material).
Sequential treatments.
Sequential treatments at peak concentrations, in contrast with monotherapies, eradicated biofilm viable cells at t4 in all of the laboratory strains and in the clinical mutator strain 1089-HSE and at t6 for strain 146-HSE (Fig. 1). Furthermore, no resistant mutants were detected in any strain at any time point during the experiments (see Fig. S1 and S2 in the supplemental material). With 1/10-peak concentrations, sequential treatment led to important reductions in the bacterial loads, similar to those with TOB monotherapy (Fig. 2). As for peak concentrations, no resistant mutants were detected in most of the strains (see Fig. S3 and S4); nevertheless, 4× MIC ATM and 4× MIC TOB resistant mutants were selected during sequential treatments in strain 146-HSE (see Fig. S3C and S4C). These resistant mutants showed (Table 2) higher MICs for ceftazidime, cefepime, ATM, carbapenems, and aminoglycosides than the parent strain 146-HSE. In order to explore the underlying resistance mechanisms explaining this increase in MICs, the expression levels of ampC and efflux pumps were studied. However, the resistant mutants did not show modified MexCD-OprJ, MexEF-OprN, or MexXY-OprM expression (overexpression similar to that of the parent strain 146-HSE) and did not overexpress AmpC or MexAB-OprM (data not shown). Thus, the increased MICs were apparently not caused by the classical expected resistance mechanisms.
TABLE 2.
Susceptibility profiles of resistant mutants obtained by plating detached biofilms from sequential treatments
| Strain and treatment | MIC (mg/liter)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | P/T | CAZ | FEP | ATM | C/T | IMI | MER | CIP | TOB | AMI | CST | |
| PAO1 | 8 | 4/4 | 1 | 1 | 2 | 0.5/4 | 4 | 1 | 0.25 | 0.5 | 4 | 2 |
| 146-HSE | >512 | >256/4 | 2 | 32 | 2 | 4/4 | 1 | 1 | 16 | 4 | 32 | 2 |
| T/A/T | ||||||||||||
| MHA | >512 | >256/4 | 8 | >64 | 32 | 4/4 | 2 | 4 | 16 | 8 | 32 | 2 |
| 4× MIC ATM | >512 | >256/4 | 8 | >64 | 128 | 4/4 | 4 | 4 | 16 | 16 | 32 | 2 |
| 16× MIC ATM | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 2 | 4 | 16 | 32 | 64 | 2 |
| 4× MIC TOB | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 2 | 4 | 16 | 32 | 64 | 2 |
| 16× MIC TOB | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 2 | 4 | 16 | >32 | 64 | 8 |
| A/T/A | ||||||||||||
| MHA | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 4 | 4 | >16 | 32 | 32 | 4 |
| 4× MIC ATM | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 2 | 4 | 16 | 32 | 32 | 2 |
| 16× MIC ATM | >512 | >256/4 | 16 | >64 | >128 | 4/4 | 4 | 4 | 16 | 16 | 32 | 4 |
| 4× MIC TOB | >512 | >256/4 | 8 | >64 | 64 | 4/4 | 2 | 2 | 16 | 32 | 64 | 4 |
TIC, ticarcillin; P/T, piperacillin-tazobactam; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; C/T, ceftolozane-tazobactam; IMI, imipenem; MER, meropenem; CIP, ciprofloxacin; TOB, tobramycin; AMI, amikacin; CST, colistin.
Therapeutic efficacy based on biofilm structural dynamics.
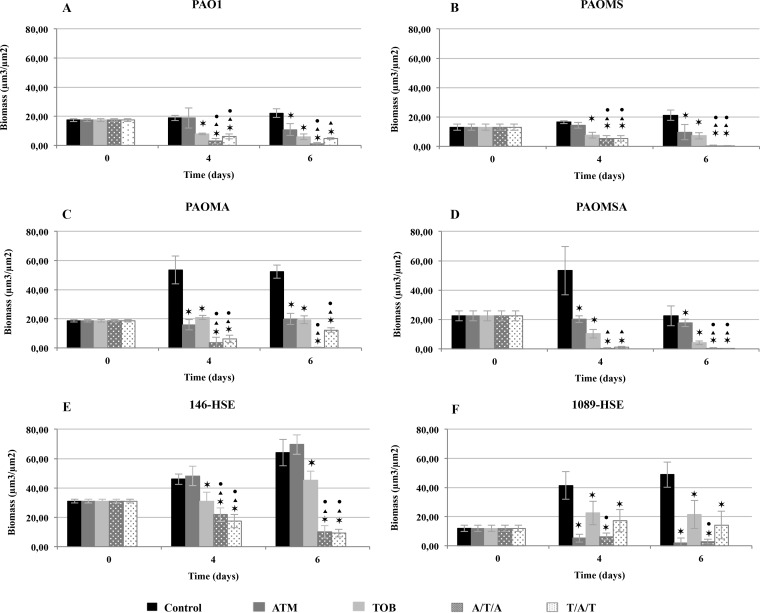
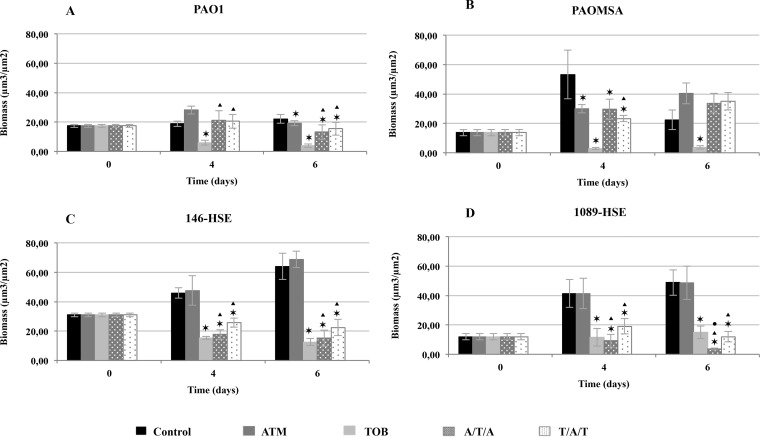
Figures 3 and 4 show biofilm biomass analyses for monotherapy and sequential treatments with peak and 1/10-peak concentrations, respectively. Likewise, Fig. 5 and 6 show 3D image reconstructions of laboratory and clinical strain biofilms, respectively, under monotherapy and sequential treatments with peak and 1/10-peak concentrations.
FIG 3.
Biomass analyses of treated biofilms with peak concentrations (700 mg/liter ATM and 1,000 mg/liter TOB) obtained with the COMSTAT program. The results represent the means (bars) and standard deviations (error bars) from at least three independent experiments. *, statistically significant difference between treatments and the control; ▲, statistically significant difference between ATM and sequential treatments; ●, statistically significant difference between TOB and sequential treatments.
FIG 4.
Biomass analyses of treated biofilms with 1/10-peak concentrations (70 mg/liter ATM and 100 mg/liter TOB) obtained with the COMSTAT program. The results represent the means (bars) and standard deviations (error bars) from at least three independent experiments. *, statistically significant difference between treatments and the control; ▲, statistically significant difference between ATM and sequential treatments; ●, statistically significant difference between TOB and sequential treatments.
FIG 5.
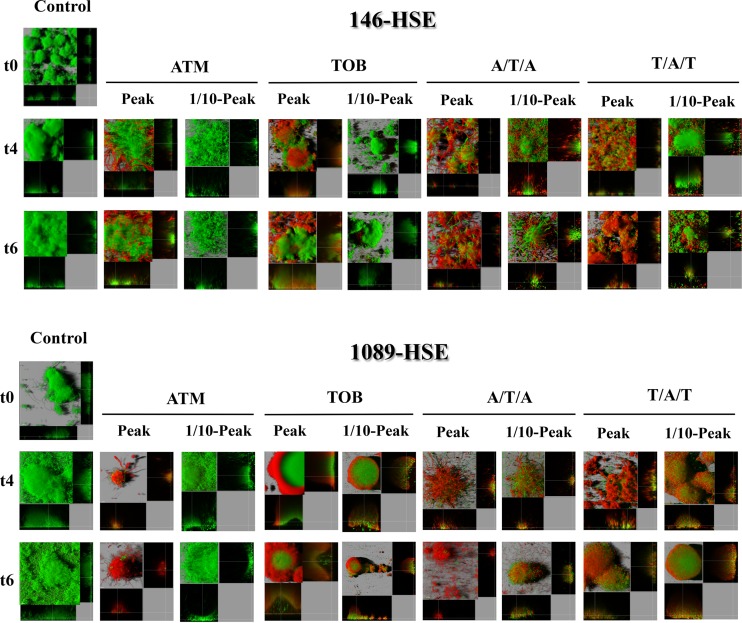
Three-dimensional images and transverse sections of GFP (green)-tagged clinical strain biofilms treated with peak (700 mg/liter ATM and 1,000 mg/liter ATM) and 1/10-peak concentrations (70 mg/liter ATM and 100 mg/liter TOB) and stained with propidium iodide (red). The images obtained at three time points (t0, t4, and t6) are shown.
FIG 6.
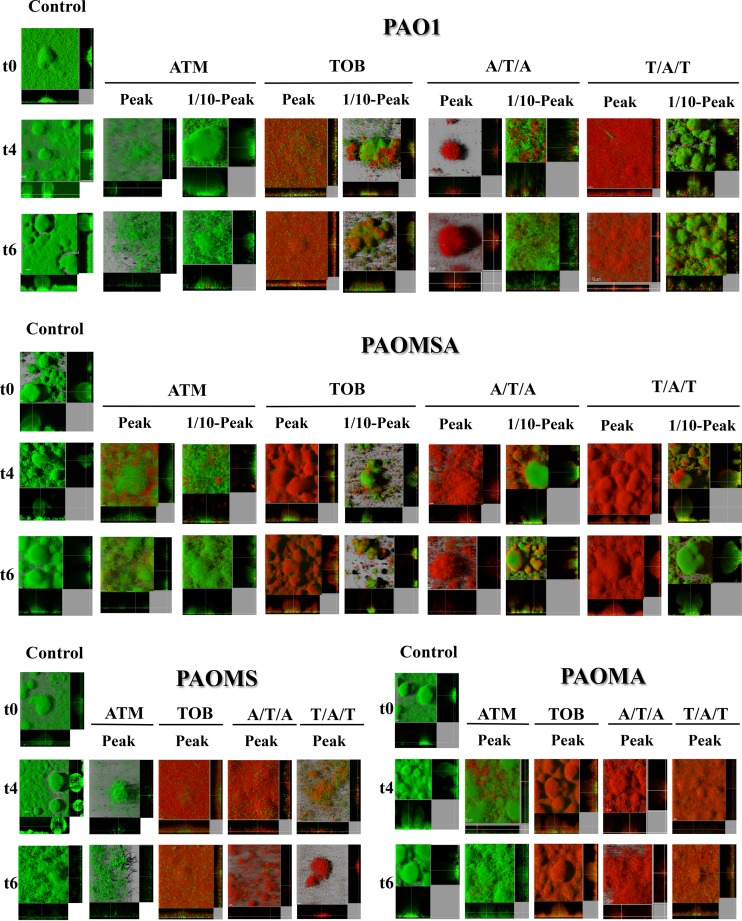
Three-dimensional images and transverse sections of GFP (green)-tagged laboratory strain biofilms treated with peak (700 mg/liter ATM and 1,000 mg/liter ATM) and 1/10-peak concentrations (70 mg/liter ATM and 100 mg/liter TOB) and stained with propidium iodide (red). The images obtained at three time points (t0, t4, and t6) are shown.
Monotherapies.
The ATM peak concentration led to declines in biomass for all strains, except for 146-HSE (Fig. 3E), which was statistically significant at t6 (Fig. 3). In the clinical strain 1089-HSE, this treatment produced an accentuated reduction in biomass, statistically significant along time and even greater than with TOB monotherapy (Fig. 3F). With ATM treatment, only a slight bactericidal effect was seen for the laboratory strains by PI staining (Fig. 6) and the analysis of dead biomass (see Fig. S5 in the supplemental material). However, for the clinical strains, the bactericidal effect was greater, leading to the death of half of the total biomass in 146-HSE (Fig. 5; see also Fig. S5E) and producing the death of practically the whole biofilm at t6 for 1089-HSE (Fig. 5; see also Fig. S5F). For all strains, the CLSM images showed the characteristic phenotypes of antibiotic damage to biofilms as filamentation, consistent with the high affinity of ATM for penicillin binding protein 3 (PBP3) (32), and flattening or disruption of the mushroom structures (Fig. 5 and 6). Despite the facts that the ATM 1/10-peak concentration did not have a significant effect on biofilm biomass (Fig. 4) and no bactericidal effect was shown (Fig. 5 and 6; see also Fig. S6), filamentation of the biofilm structures may still be observed (Fig. 5 and 6).
The biomass analysis showed that the TOB peak concentration treatment led to a marked statistically significant decline in biomass at t4 that persisted at t6 for all strains (Fig. 3). The CLSM images showed an important bactericidal effect at t4 and t6, which was correlated to the death of approximately 60% of the biofilm (see Fig. S5 in the supplemental material) for all strains. Despite this, the biofilm structure was conserved, and some green (alive) cells in the inner part of the biofilm were still observed, especially in clinical strains (Fig. 5 and 6). Unexpectedly, the biomass decrease was even greater when the biofilms were challenged with a 1/10-peak concentration (Fig. 4), which was correlated not with a greater bactericidal effect on the biofilm (see Fig. S6) but with a disruption of the biofilm structures and a visible loss of biomass (Fig. 5 and 6).
Sequential treatments.
The reduction in biomass compared with that of the untreated controls was statistically significant for both sequential treatments at the peak concentrations at t4 and t6 for all of the strains (Fig. 3). Similarly, comparing each sequential treatment with each monotherapy, the decrease in biomass was statistically significant at t4 and t6 for nearly all strains (Fig. 3), with the exception of 1089-HSE. In the case of this strain, surprisingly, the biomass reduction was greater with the ATM monotherapy, but, on the contrary, both sequential treatments were superior to TOB (Fig. 3F). Regarding the bactericidal effect, the sequential treatments led to the death of practically the complete biofilm in the laboratory strains (see Fig. S5A to D in the supplemental material), and although the effect in the clinical strains was smaller, the percentage of dead biomass was >70% at t6 (see Fig. S5E and F). Likewise, the 3D reconstructed CLSM images revealed a pronounced bactericidal effect and intense filamentation, causing increased destruction of the biofilm structures for most of strains (Fig. 5 and 6).
The biomass analysis of the biofilms exposed to 1/10-peak concentrations of antibiotics during sequential treatments showed a statistically significant decline in biomass compared with those of the untreated controls at t4 for the clinical strains and at t6 for all of the strains (Fig. 4), except PAOMSA (Fig. 4B). Moreover, the reductions in biomass were statistically significant for both alternations compared to those for ATM monotherapy at t4 and t6 for most of the strains (Fig. 4). Regarding the comparison with TOB treatment, both alternations were better only for strain 1089-HSE (Fig. 4D). Similarly to the peak concentration, biofilm filamentation was also observed in CLSM images (Fig. 5 and 6), but a less intense bactericidal effect was demonstrated.
DISCUSSION
Whereas P. aeruginosa eradication cannot be achieved once a CRI is fully established in a CF patient, the aim of antimicrobial therapy is a reduction in bacterial densities in the respiratory tract (17). In the absence of exacerbations, maintenance treatment with inhaled antibiotics is used, due to the high concentrations reached at the site of infection and the minimal systemic effects. Despite the demonstrated efficacy of regimes based on 28-day on/28-day off cycles of nebulized antibiotics (such as TOB or ATM) in the reduction in bacterial loads without favoring the selection of resistant mutants, decreasing antibiotic effects are observed over time (33, 34). This unfortunate outcome may be explained by the extraordinary capacity of P. aeruginosa to develop resistance through chromosomal mutations, the high prevalence of hypermutable strains in CF patients (9), and the innate tolerance of biofilms to antibiotics (including phenotypic and genotypic resistance) (35). Thus, the need for alternative treatment strategies is imperative.
The present study presents an innovative strategy based on sequential treatment cycles with the inhaled antibiotics, ATM and TOB, supported by the antagonistic interactions of resistance mechanisms (14) and the imbalance of intrinsic efflux pumps (MexAB-OprM and MexXY-OprM) (15). Those sequential treatments were compared to monotherapies through evaluation of the dynamics of antibiotic activity and resistance development in biofilms using a flow cell model.
In agreement with the results of other experimental works (28) and clinical trials (33, 36), our results showed that monotherapy with either ATM or TOB at the peak concentration effectively reduced the bacterial load of biofilms, according to the number of viable cells. Moreover, the treatments not only did not select resistant mutants but also eradicated, even for mutator strains, those spontaneously generated in the biofilms before the initiation of treatment. Accordingly, the microscopic analysis of biofilms under monotherapy showed a general decline in biomass and the filamentation, flattening, or disruption of the structures. Nevertheless, the results for cell viability were not always accurately correlated with a proportional reduction in biomass or death of cells within the biofilm. In the case of ATM, the discrepancies found might be explained by the intense filamentation of bacteria induced in the presence of the antibiotic, which is consistent with the high affinity of ATM for PBP3 (32). Regarding TOB treatment, while it seemed obvious that the marked reduction in the bacterial load (even falling below detection limits) should correspond with a dramatic decrease in live biomass and an important biofilm disruption, the CLSM images showed important areas of live cells in the inner part of apparently eradicated biofilms in terms of viable cells. It has been demonstrated in several studies that oxygen limitation and low metabolic activity (37, 38) in the inner layers of biofilms, along with a slow diffusion of aminoglycosides through the extracellular polymeric matrix (39), determine tolerance to TOB. Therefore, this specific biofilm safeguard against TOB, which cannot reach the slow-growing bacteria in the inter layers, might explain the live biofilm variants that were observed but were nonculturable. Indeed, it has been well described that as a response to some form of stress, bacterial cells can enter into a viable but nonculturable state in which growth on routine bacteriological media fails (40). These findings showed that the analysis of a single parameter does not provide enough information to unravel the complexity of antibiotic effects on biofilms, and cell counts, biomass reduction, and CLSM images should be consider together.
Regarding the CF clinical strains, we found that the ATM peak concentration was extremely effective on strain 1089-HSE but not on the MDR strain 146-HSE. In contrast, the TOB peak concentration had a greater effect on 146-HSE than on 1089-HSE. Although both strains had the same MICs for ATM and TOB, the treatment outcomes were completely different, in agreement with the studies that show a lack of correlation between the susceptibility results and clinical responses (41). Indeed, the success of the antimicrobial therapies depends not only on the MICs but also on the PK/PD indexes, classically calculated for planktonic bacteria (42) without considering the biofilm mode of growth and the nature of the antimicrobial effects on these structured communities. Furthermore, the efficacy differences between the same treatments on the clinical strains might be explained by the imbalance of the efflux pump features of the CF strains (15). It has been reported that treatment with TOB leads to overexpression of MexXY with the consequent competition for the same porin channel, OprM, used by MexAB (15). The low availability of OprM strongly influences the transport activity of these efflux pumps, favoring antibiotic accumulation inside bacteria. The analysis of the strain 1089-HSE efflux pump transcriptional levels revealed overexpression of MexXY, which might explain the decreased activity of TOB compared to that of ATM in this strain (18). In contrast, despite strain 146-HSE also overexpressing MexXY, TOB presented higher activity than ATM, demonstrating the existence of additional, as-yet-uncharacterized, factors modulating the activity of aminoglycosides. The complexity of the antibiotic resistance mechanisms was further shown by the simultaneous overexpression of MexCD-OprJ and MexEF-OprN in strain 146-HSE, although neither of these two efflux pumps seems to play a major role in TOB or ATM resistance (43). These results support previous data (15) and confirm at the same time the complexity of antibiotic resistance mechanisms pointing to the risk of using a single antibiotic as maintenance therapy in CRI caused by P. aeruginosa clinical strains.
Since most of the experiments published in the literature are based on the use of only one concentration, we proposed the evaluation of sputum concentrations after inhalation (peak) and also at a 1/10-peak concentration, which corresponds to the sputum concentration approximately 2 h after inhalation (29). Interestingly, our results revealed that, unexpectedly, the treatment with a 1/10-peak concentration of TOB reduced biofilm biomass more than the high concentration in all cases. Although the decrease in biomass was not correlated with an increase in the bactericidal effect, an important disruption of the biofilm structure was demonstrated. This paradoxical effect has not been previously described, and further studies are needed to elucidate the underlying mechanisms involved. Again, the complex dynamics of biofilms under antibiotic treatment is evident, suggesting a reevaluation of the PK/PD parameters to be adapted to the particular and special biofilm environment (8).
Finally, our results suggest that sequential treatments might be superior to monotherapies. The reduction in viable cells under the detection limits was correlated with a significant decrease of biofilm biomass in contrast to the results with monotherapies. Sequential treatments produced biofilm disruption by the combination of filamentation and the bactericidal effect, resulting in disorganized biofilms with greater areas of dead cells than those challenged with individual treatments. Even in the experiments carried out with 1/10-peak concentrations, treatment alternation produced a decrease in the bacterial load and biomass and generally killed more than 50% of biofilm cells. Our results are consistent with the theory explained above, which proposed that induced selection of antagonistic resistance mechanisms, by the use of a sequential scheme of treatment, results in hypersusceptibility of substrates and in a lower selection of resistant mutants. Moreover, there was no development of resistance during alternation of antibiotics, and spontaneous preexisting resistant mutants were eradicated, with the exception of strain 146-HSE. During sequential treatments with 1/10-peak concentrations, 146-HSE-resistant mutants were selected and nearly all viable cells at t6 were mutants. However, the analysis of resistance mechanisms ruled out classical pathways, including upregulation of AmpC or any of the four major P. aeruginosa efflux pumps. Thus, further study will be required to elucidate the mechanisms involved.
As is the case with most in vitro studies, our work has some limitations. First, despite the fact that the antibiotic concentrations used simulate those achieved during treatment of CF patients, they were fixed and thus did not consider natural clearance kinetics. However, the use of two different concentrations partially compensated for this limitation. Second, the time frame used in this in vitro model is necessarily much shorter than the typical 28-day on/off cycles used in clinical practice. Shorter in vitro than in vivo generation times and faster population turnover might partially compensate for this limitation. Finally, by definition, in vitro models do not accurately mimic the conditions of the CF lung environment. Although the model used needs to be improved to provide a more “real-world” setting, the whole range of antimicrobial activity parameters assessed and the collection of isolates tested, including typical CF phenotypes and relevant CF strains, strengthen the value of this work as a proof-of-concept approach to support the evaluation of sequential therapies.
In summary, the present study represents a step forward for the development of new strategies for combating P. aeruginosa CRI. Our results show that sequential treatment leads, in most of the strains tested, to an enhanced reduction of both viable cells and biofilm biomass along with a patent bactericidal effect and filamentation of biofilm structures. Thus, our results support the potential benefit of sequential regimens with inhaled antibiotics in CF as an alternative to maintenance treatments with single antibiotics and encourage the evaluation of this novel strategy in clinical trials.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Angelic Encarnación, Maria Antònia Llopis Grimalt, and Paloma Llistó Simón, students from the University of the Balearic Islands, for their collaboration in biofilm studies. We also thank Javier Pierola, in charge of the microscopy platform from IdISPa, for his help in optimizing the acquisition of confocal microscopy images.
This work was partially supported by a grant from Gilead Sciences, Inc., and by the Ministerio de Economía y Competitividad of Spain, Instituto de Salud Carlos III cofinanced by the European Regional Development Fund (ERDF), “A Way To Achieve Europe,” through the Spanish Network for the Research in Infectious Diseases (RD06/0008 and RD12/0015).
Footnotes
‡ For this virtual institution, see http://reipi.org.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00196-16.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infection associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martínez-Solano L, Macià MD, Fajardo A, Oliver A, Martínez JL. 2008. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin Infect Dis 47:1526–1533. doi: 10.1086/593186. [DOI] [PubMed] [Google Scholar]
- 3.Evans SA, Turner SM, Bosch BJ, Hardy CC, Woodhead MA. 1996. Lung function in bronchiectasis: the influence of Pseudomonas aeruginosa. Eur Respir J 9:1601–1604. doi: 10.1183/09031936.96.09081601. [DOI] [PubMed] [Google Scholar]
- 4.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver A, Mena A, Maciá MD. 2008. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections, p 433–444. In Baquero F, Nombela C, Cassell GH, Gutíerrez-Fuentes JA (ed), Evolutionary biology of bacterial and fungal pathogens. ASM Press, Washington, DC. [Google Scholar]
- 6.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton J, Stewart P, Greenberg E. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Macià MD, Perez JL, Molin S, Oliver A. 2011. Dynamics of mutator and antibiotic-resistant populations in a pharmacokinetic/pharmacodynamic model of Pseudomonas aeruginosa biofilm treatment. Antimicrob Agents Chemother 55:5230–5237. doi: 10.1128/AAC.00617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 10.Maciá MD, Blanquer D, Togores B, Sauleda J, Pérez JL, Oliver A. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maciá MD, Borrell N, Segura M, Gómex C, Pérez JL, Oliver A. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:975–983. doi: 10.1128/AAC.50.3.975-983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luján AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM. 2011. Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 6:e27842. doi: 10.1371/journal.pone.0027842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulet X, Moyá B, Juan C, Macià MD, Pérez JL, Blázquez J, Oliver A. 2011. Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob Agents Chemother 55:4560–4568. doi: 10.1128/AAC.00519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vettoretti L, Plésiat P, Muller C, El Garch F, Phan G, Attrée I, Ducruix A, Llanes C. 2009. Unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam S, Oh H, Jalal S, Karpati F, Ciofu O, Høiby N, Wretlind B. 2009. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin Microbiol Infect 15:60–66. doi: 10.1111/j.1469-0691.2008.02097.x. [DOI] [PubMed] [Google Scholar]
- 17.Cantón R, Máiz L, Escribano A, Olveira C, Oliver A, Asensio O, Gartner S, Roma E, Quintana-Gallego E, Salcedo A, Girón R, Barrio MI, Pastor MD, Prados C, Martínez-Martínez MT, Barberán J, Castón JJ, Martínez-Martínez L, Poveda JL, Vázquez C, de Gracia J, Solé A. 2015. Spanish consensus on the prevention and treatment of Pseudomonas aeruginosa bronchial infections in cystic fibrosis patients. Arch Bronconeumol 51:140–150. doi: 10.1016/j.arbres.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 18.López-Causapé C, Rojo-Molinero E, Mulet X, Cabot G, Moyà B, Figuerola J, Togores B, Pérez JL, Oliver A. 2013. Clonal dissemination, emergence of mutator lineages and antibiotic resistance evolution in Pseudomonas aeruginosa cystic fibrosis chronic lung infection. PLoS One 8:e71001. doi: 10.1371/journal.pone.0071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fothergill JL, White J, Foweraker JE, Walshaw MJ, Ledson MJ, Mahenthiralingam E, Winstanley C. 2010. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J Clin Microbiol 48:2053–2059. doi: 10.1128/JCM.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. [DOI] [PubMed] [Google Scholar]
- 21.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 22.Mulet X, Maciá MD, Mena A, Juan C, Pérez JL, Oliver A. 2009. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob Agents Chemother 53:1552–1560. doi: 10.1128/AAC.01264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 22:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Yang L, Liu Y, Markussen T, Høiby N, Tolker-Nielsen T, Molin S. 2011. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol 62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 27.Christensen BB, Sternberg C, Andersen JB, Palmer RJ Jr, Nielsen AT, Givskov M, Molin S. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol 310:20–42. doi: 10.1016/S0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, Griffin EF, Moreau-Marquis S, Schwartzman JD, Stanton BA, O'Toole GA. 2012. In vitro evaluation of tobramycin and aztreonam versus Pseudomonas aeruginosa biofilms on cystic fibrosis-derived human airway epithelial cells. J Antimicrob Chemother 67:2673–2681. doi: 10.1093/jac/dks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson RL, Retsch-Bogart GZ, Oermann C, Milla C, Pilewski J, Daines C, Ahrens R, Leon K, Cohen M, McNamara S, Callahan TL, Markus R, Burns JL. 2006. Microbiology, safety, and pharmacokinetics of aztreonam lysinate for inhalation in patients with cystic fibrosis. Pediatr Pulmonol 41:656–665. doi: 10.1002/ppul.20429. [DOI] [PubMed] [Google Scholar]
- 30.Oh H, Stenhoff J, Jalal S, Wretlind B. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist 9:323–328. doi: 10.1089/107662903322762743. [DOI] [PubMed] [Google Scholar]
- 31.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodríguez C, Moya B, Zamorano L, Suárez C, Peña C, Martínez-Martínez L, Oliver A. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1901. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgopapadakou NH, Smith SA, Sykes RB. 1982. Mode of action of azthreonam. Antimicrob Agents Chemother 21:950–956. doi: 10.1128/AAC.21.6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsey BW, Pepe MS, Quan JM, Otto KL, Montgomery AB, Williams-Warren J, Vasiljev Borowitz -K MD, Bowman CM, Marshall BC, Marshall S, Smith AL. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. Cystic Fibrosis Inhaled Tobramycin Study Group. N Engl J Med 340:23–30. [DOI] [PubMed] [Google Scholar]
- 34.Retsch-Bogart GZ, Quittner AL, Gibson RL, Oermann CM, McCoy KS, Montgomery AB, Cooper P. 2009. Efficacy and safety of inhaled aztreonam lysine for airway pseudomonas in cystic fibrosis. Chest 135:1223–1232. doi: 10.1378/chest.08-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Oermann CM, Retsch-Bogart GZ, Quittner AL, Gibson RL, McCoy KS, Montgomery AB, Cooper PJ. 2010. An 18-month study of the safety and efficacy of repeated courses of inhaled aztreonam lysine in cystic fibrosis. Pediatr Pulmonol 45:1121–1134. doi: 10.1002/ppul.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters MC III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatch RA, Schiller NL. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43:93–100. [PubMed] [Google Scholar]
- 41.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. 2003. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest 123:1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 42.Drusano GL. 2007. Pharmacokinetics and pharmacodynamics of antimicrobials. Clin Infect Dis 45:S89–S95. doi: 10.1086/518137. [DOI] [PubMed] [Google Scholar]
- 43.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.