Summary
VRC01-class antibodies neutralize diverse HIV-1 strains by targeting the conserved CD4-binding site. Despite extensive investigations, crucial events in the early stage of VRC01 development remain elusive. We demonstrated how VRC01-class antibodies emerged within a Chinese donor by antigen-specific single B cell sorting, structural and functional studies, longitudinal antibody and virus repertoire analyses. A monoclonal antibody DRVIA7 with modest neutralizing breadth was isolated that displayed a subset of VRC01 signatures. Structures revealed a VRC01-like angle of approach, but less favorable interactions between DRVIA7 light chain CDR1 and N-terminus with N276 and V5 glycans on gp120. While the DRVIA7 lineage was unable to acquire broad neutralization, longitudinal analysis revealed a repertoire-encoded VRC01 light chain CDR3 signature and VRC01-like neutralizing heavy chain precursors that rapidly matured within two years. Thus, light chain accommodation of the glycan shield should be taken into account in vaccine design targeting this conserved site of vulnerability.
Keywords: antibody maturation, B cell development, broadly neutralizing antibodies, CD4-binding site, HIV infection, glycan shield, gp120, rational vaccine design, virus-host co-evolution, VRC01 class
Graphical abstract
Introduction
The envelope glycoprotein (Env) spike of human immunodeficiency virus type-1 (HIV-1) is a trimer of heterodimers, each containing a receptor-binding protein, gp120, and a transmembrane protein, gp41 (Wyatt and Sodroski, 1998). Dense heterogeneous N-linked glycans and hypervariable loops on the Env surface form barriers to the elicitation of broadly neutralizing antibodies (bNAbs) (Julien et al., 2013; Lyumkis et al., 2013; Pancera et al., 2014). Nonetheless, a small fraction of infected individuals are capable of developing bNAbs after several years of infection (Gray et al., 2011; Li et al., 2007; Mikell et al., 2011; Walker et al., 2010). The identification and characterization of bNAbs and their epitopes are considered paramount to guiding rational vaccine design (Walker and Burton, 2010). Due to the recent advances in antibody isolation, panels of bNAbs with extraordinary breadth and potency have been reported (Burton and Mascola, 2015; Klein et al., 2013; Kwong and Mascola, 2012; Kwong et al., 2013).
Of the conserved HIV-1 epitopes identified thus far, the CD4-binding site (CD4bs) on gp120 appears to be an ideal target for vaccine design (Zhou et al., 2007). However, the CD4bs is recessed between two subunits of the Env trimer, with a narrow range of approach angles by antibodies (Chen et al., 2009; Lyumkis et al., 2013). A class of CD4bs-directed bNAbs, with VRC01 being the first of the class (Wu et al., 2010), has been identified from multiple donors, suggesting that the immune system is capable of recognizing this masked site of vulnerability. Extensive efforts have been made to study the VRC01 class of bNAbs. Co-crystal structures with the gp120 core elucidate a common interaction strategy employed by all VRC01-class bNAbs (Scheid et al., 2011; Wu et al., 2011; Zhou et al., 2010; Zhou et al., 2015; Zhou et al., 2013), while next-generation sequencing (NGS) analysis reveals a converged pattern of maturation towards a set of “VRC01 signatures” (Zhou et al., 2013). These signatures include the preferential use of the IgHV1-2*02 germline allele, a conserved maturation pattern of heavy chains, a stringent 5-aa light chain complementarity determining region (CDR) 3, and specific mutations in the light chain CDR1 (Wu et al., 2011; Zhou et al., 2013). Based on this information, putative VRC01 precursors and intermediates are inferred to guide CD4bs immunogen design (Jardine et al., 2013; Jardine et al., 2015). However, important questions remain unanswered: how much do these VRC01 signatures contribute to the early lineage development? What is the order of molecular events in B cell maturation leading to the VRC01-like neutralizing activity? Do these steps involve overcoming critical barriers that need to be accounted for during immunogen design? The complex patterns of VRC01 lineage development over a 15-year period have been reported (Wu et al., 2015), but bNAbs identified from the earliest time point already possess the mature VRC01 signatures and broad neutralizing activity, indicating that this lineage reached a “plateau” prior to the timeframe studied. Thus, details of early-stage VRC01 development remain unknown.
Here we approached these questions by analyzing samples from an HIV-1-infected Chinese patient (Hu et al., 2012). This donor, DRVI01, was a long-term non-progressor (LTNP) with increasing serum reactivity to CD4bs over a period of five years. We isolated a monoclonal antibody (mAb), DRVIA7, from the fifth year sample, which exhibited a subset of VRC01 signatures with limited neutralizing activity. Structures revealed that DRVIA7 was a VRC01-like antibody but with critical differences in the light chain CDR1 and N-terminus, both of which interacted with gp120 glycans. Functional analysis confirmed that a gp120 glycan patch was a barrier for achieving neutralization breadth. We traced the DRVIA7 antibody lineage temporally within the unbiased B cell repertoires and utilized single genome amplification (SGA) to investigate the viral co-evolution, which confirmed the inhibitory role of the gp120 glycan patch identified. In summary, our study identified a VRC01-class antibody lineage from a Chinese patient, demonstrated that the glycan shield stalled its development of VRC01-like broadly neutralizing activity, and provided functional VRC01 precursors and early intermediates to facilitate B cell lineage-based design of CD4bs immunogens.
Results
Identification and characterization of an HIV-1-infected Chinese donor
The analysis of HIV-1-infected individuals displaying prolonged control of viremia, or LTNPs, presents opportunities to learn how such control is achieved by the immune system (Tomaras et al., 2011; Walker et al., 2010). In this study, we analyzed the HIV-1-neutralizing antibody response in a Chinese LTNP. Historically, two sources have contributed to the HIV epidemic of the last century in China: an early wave of injecting drug users (IDUs) in the southwest Yunnan province that borders on the heroin-producing “Golden Triangle” region in the mid- to late-80s (Ma et al., 1990) and a later outbreak in the former plasma donors (FPDs) through contamination of blood plasma transfusion by the IDU's HIV-1 strain in mid-90s (Li et al., 2012). After ten years of infection without antiviral treatment, some FPD survivors displayed relatively normal CD4+ T cell counts and a certain level of viral control consistent with that of LTNPs. The patient studied here, DRVI01, was selected from the top 5% neutralizers based on serum screening against a panel of more than 20 viruses (Hu et al., 2012). Briefly, DRVI01 entered the study at the age of 40 with informed consent and was procedurally followed up every six months from 2005 to 2010. This donor carried a clade-B′ strain of HIV-1, with a viral load ranging from 74,200 to 310,000 copies/ml and a CD4+ T cell count ranging from 335 to 769 cells/μl during the period studied. Serum neutralization was tested against a global panel containing 12 viruses from diverse clades (Global) (deCamp et al., 2014) and a separate panel containing 25 viruses established at the Division of Research on Virology and Immunology (DRVI) of the China CDC. Over 95% breadth was observed at all the time points studied (Figure 1A; Table S1). The average potency increased from 270 and 246 geometric mean titers (GMTs) in 2005 to 736 and 706 GMTs in 2009 for the two virus panels, respectively. The results indicated rapid development of neutralizing antibodies within this Chinese patient over a 5-year period.
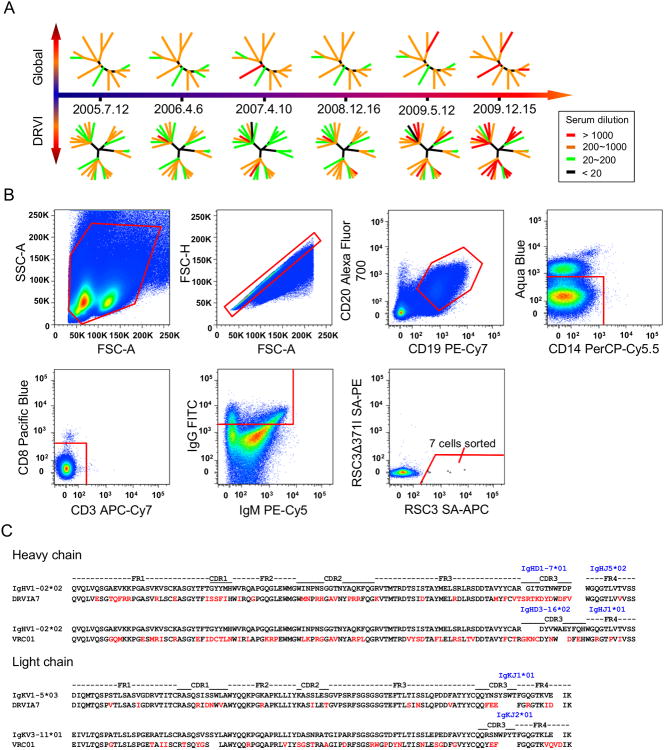
Figure 1. Identification of a VRC01-class antibody DRVIA7 from an HIV-1-infected Chinese donor DRVI01.
(A) The neutralizing ability of DRVI01 sera from six time points from 2005 to 2009 were tested against the global and DRVI panels of pseudoviruses. Dendrograms are shown at each time point representing the pseudovirus panels with nodes representing individual viruses colored according to neutralizing potency. (B) Single B cell sorting of CD4bs-directed monoclonal antibodies by flow cytometry. Around 10 million PBMCs from DRVI01 were incubated with cell markers and sorting probes. Seven B cells that reacted with RSC3, but not RSC3Δ371I, were then sorted into 96 well microtiter plates. FITC, fluorescein isothiocyanate. FSC-H, forward scatter height; FSC-A, forward scatter area; and SSC, side scatter area. (C) Sequence analysis of DRVIA7 and VRC01 heavy and light chains with alignment to respective germline genes. Mature antibody residues that differ from germline are colored in red. See also Table S1 and Figure S1.
Identification of a CD4-binding site-directed antibody, DRVIA7
DRVI01 sera displayed strong CD4bs specificity and some V1V2 (variable regions 1 and 2) reactivity. We isolated CD4bs-directed mAbs from a 2009 sample (Figure 1B). A CD4bs-specific probe pair, RSC3 and RSC3Δ371I, was used to identify antibody-expressing B cells targeting the CD4bs by fluorescence-activated cell sorting (FACS) (Wu et al., 2010). Briefly, peripheral blood mononuclear cells (PBMCs) from DRVI01 were incubated with cell markers and two RSC3 probes. CD4bs-specific memory B cells with the phenotype CD19+CD20+CD14-CD3-CD8-IgG+ RSC3+RSC3Δ371I- were sorted into 96 well microtiter plates. Of seven sorted memory B cells, five were amplified with matching heavy and light chain genes (Figure 1B). After cloning into the IgG1 vector, the full IgG mAbs were expressed transiently in 293F cells. Of the five isolated mAbs, DRVIA1, 3, 4 and 6b showed strong RSC3 binding in ELISA, whereas DRVIA7 bound weakly to RSC3 as characterized by Bio-Layer Interferometry (Figure S1A), and none of the mAbs were reactive to RSC3Δ371I. We further confirmed their CD4bs specificity using a YU2 gp140-foldon (gp140-F) trimer with the D368R mutation (Figure S1B) (Yang et al., 2002). Compared to DRVIA1-6b, DRVIA7 exhibited greater neutralizing ability, but had limited breadth (Figure S1C). Sequence analysis revealed a clonal family consisting of DRVIA1, 3, 4 and 6b, and an outlier DRVIA7 (Figure S1D). The DRVIA1-6b clonal family was characterized by a low level (3.1-6.3%) of somatic hypermutation (SHM), a 19-aa heavy chain CDR3 (HCDR3) loop (Kabat numbering), and a 9-aa κ chain CDR3 loop. By contrast, DRVIA7 displayed VRC01-like sequence features: a SHM of 19.0% for the VH gene and 17% for the Vκ gene, a heavy chain of IgHV1-02*02 allelic origin, a relatively short HCDR3 loop (11aa), and a distinctive 5-aa κ chain CDR3 loop containing a “F91E96” motif (“Y91E96” in VRC01) (Zhou et al., 2013). These findings suggested that DRVIA7 might be a VRC01-class antibody that had acquired a subset of VRC01 signatures but not yet broad neutralizing activity, thus providing an opportunity to study the emergence of VRC01-class antibodies.
Structural differences between DRVIA7 and VRC01-class antibody interactions with gp120 and gp140 trimer primarily at the glycan shield
The limited breadth and potency suggested that some important features present in the mature VRC01 were missing in DRVIA7 (Figure S1C). To elucidate these features, we determined the crystal structure of unliganded DRVIA7 Fab at 2.9 Å resolution (Table S2), which resembled VRC01 and other bNAbs of this class such as VRC03, PGV04, and 12A21 with a Cα root mean square deviation (RMSD) of <1.0 Å over the antibody variable regions (VH and VL) (Figures S2A-B) (Zhou et al., 2013). Using an HIV-1 gp120 from the 93TH057 strain, we next determined the crystal structure of a gp120 core-DRVIA7 Fab complex at 3.4 Å resolution (Figure 2A; Table S2). The gp120 was deglycosylated with endoglycosidase H (EndoH), but only after complex formation to preserve glycan interactions within the antibody-gp120 interface. Overall, DRVIA7 buried 957 Å2 of solvent-accessible gp120 protein surface overlapping the CD4bs. Negative-stain EM and crystal structures confirmed that DRVIA7 bound to the SOSIP gp140 trimer with a similar angle of approach to VRC01 and nearly identical interactions with gp120 (Figures 2B-D; Figures S2C-G) (Derking et al., 2015; Lyumkis et al., 2013; Zhou et al., 2010). For example, 8 of 11 HCDR2 residues and 3 of 4 HCDR3 residues on VRC01 and DRVIA7 that contacted gp120 were conserved between the two antibodies, while their LCDR3 loops adopted identical conformations and contacted a similar set of gp120 residues (Figure S2C). Nevertheless, DRVIA7 and VRC01 heavy chains could still differ in their sensitivity to gp120 mutations. To examine this possibility, we generated a chimeric antibody with DRVIA7 heavy chain and mature VRC01 light chain (DRVIA7H-VRC01L). Indeed, the H54 of DRVIA7 heavy chain made this chimeric antibody less tolerant of the G472A mutation of gp120 than VRC01 (Figures S2H-I). However, the substantial differences in their neutralizing ability (Figure S1C) could not be explained by these primary protein-protein interactions.
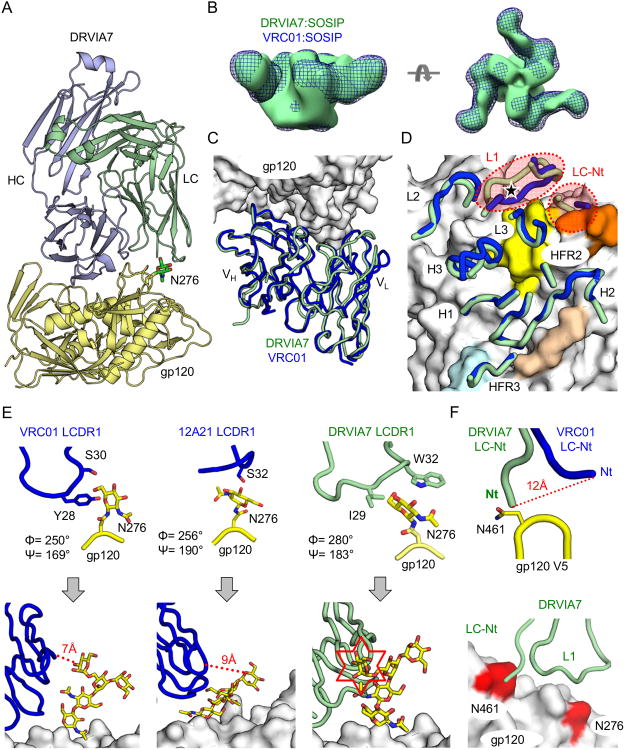
Figure 2. DRVIA7 and VRC01 recognition of gp120 and gp140 differed primarily at light chain interaction with glycans.
(A) Ribbon diagrams of the crystal structure of DRVIA7 Fab (light blue and green) bound to 93TH057 gp120 (yellow) at 3.4 Å resolution are displayed. The N276 glycan is shown in stick-and-ball representation with carbon atoms colored in green, nitrogen atoms colored in blue and oxygen atoms colored in red. (B) Negative-stain EM reconstruction at 21-22 Å of DRVIA7 Fab bound to JRFL SOSIP trimer (green surface) is superposed onto the previously published reconstruction of the BG505 SOSIP trimer bound to VRC01 (blue mesh, EMDB ID: EMDB-6252). (C) Ribbon diagrams of the variable regions of DRVIA7 (green) and VRC01 (blue) are shown after their structures were superposed on gp120 (white molecular surface). (D) The gp120-contacting regions (designated by their CDRs and FR regions) of DRVIA7 and VRC01 are displayed over the molecular surface of gp120 as in (C). The CD4-binding loop on the gp120 surface is colored in beige, the base of the V1/V2 loop in light blue, the V5 loop in orange and the Loop D in yellow. The position of the N276 glycan is indicated by a black star. The light chain CDR1s and N-termini (Nt) display the most structural variance between the two antibodies and are highlighted by red dotted circles. (E) The N276 glycan, from their corresponding gp120 structures, is displayed when bound to VRC01 LCDR1 (blue, left), 12A21 LCDR1 (blue, middle) and DRVIA7 LCDR1 (light green, right). Below, coordinates for Man3GlcNAc2 attached to N276 from a previously published structure (PDBID: 4P9H) are superposed on the protein-proximal GlcNAc visible in the VRC01, 12A21, and DRVIA7 structures. (F) The DRVIA7 and VRC01 light chain N-termini are shown interacting with the gp120 V5 loop. Below, the N276 and V5 loop glycosylation sites are colored in red on the white gp120 surface. Ribbon diagrams of DRVIA7 N-termini and LCDR1 loops are displayed as above. See also Table S2 and Figure S2.
Further analysis revealed that the LCDR1 loops and N-termini of DRVIA7, VRC01, and 12A21, interacted with the glycan shield in subtly different ways (Figure 2E; Figures S3A-C). Specifically, VRC01 had a 2-aa deletion in its LCDR1 and a tyrosine that directed the first GlcNAc of the N276 glycan away from the antibody (Zhou et al., 2010). Similarly, 12A21 LCDR1 appeared to push the GlcNac out of the way. By contrast, two hydrophobic residues in DRVIA7 LCDR1 (I29 and W32) shifted and twisted the GlcNAc in N276 by 5.5 Å and 30° (glycotorsion ϕ), respectively, compared to the VRC01-bound conformation. Superposition of a more intact N276 glycan structure (Scharf et al., 2014) with the gp120-bound DRVIA7, VRC01, and 12A21 structures suggested a potential clash with DRVIA7, but not with VRC01 or 12A21. The DRVIA7 and VRC01 structures also differed at their light chain N-termini, which interacted with an N-linked glycan N461 on the gp120 V5 loop (Figure 2F). Every residue from 460 to 463 was a potential N-linked glycosylation site with 14-44% sequence conservation, and 91% of the 24,602 isolates analyzed here contained at least one glycosylation site in this V5 region. Thus, the N276 glycan and the V5 glycan, although 15 Å apart (Figure 2F), might together play an inhibitory role for DRVIA7 recognition. Indeed, structural modeling showed an unfavorable energy score for the DRVIA7 LCDR1 and N-terminus that interacted with this glycan cluster, which could be substantially reduced by swapping the LCDR1 loop with VRC01 or 12A21 sequences and by truncating the light chain N-terminus (Figure S3D). Of note, the importance of N276 and V5 glycans has already been reported for the in vivo selection and function of CD4bs-directed bNAbs (Freund et al., 2015; Wang et al., 2015). Taken together, these findings suggested that glycan accommodation by the antibody light chain could be a critical determinant of VRC01-class neutralizing ability.
Unfavorable light chain accommodation of the glycan shield limited DRVIA7 neutralizing ability
Following the structural analysis, we examined the effect of glycosylation on the binding of DRVIA7 and DRVIA7H-VRC01L, which showed an identical angle of approach when bound to the SOSIP gp140 trimer, as indicated by negative-stain EM (Figures S2D-G). We first measured their binding towards glycosylated and deglycosylated gp120s of HIV-1 strains YU2, 93TH057, and JRFL. As reflected by the KD (Koff/Kon) values, DRVIA7 exhibited significantly higher affinity towards deglycosylated gp120 while the chimeric antibody displayed an opposite effect, confirming the inhibitory role of glycans for DRVIA7 binding (Figure 3A). Analysis of the binding kinetics revealed that deglycosylation improved the on-rates for both antibodies, while showing a differential effect on the off-rates (Figure 3A; Figure S3E). Specifically, deglycosylation drastically increased the off-rates for DRVIA7H-VRC01L, in comparison to a plateaued dissociation when gp120 was glycosylated. By contrast, deglycosylation showed a lesser effect on the off-rates for DRVIA7. Our results thus highlighted the impact of glycosylation on CD4bs recognition by the VRC01-class bNAbs, consistent with another VRC01-class bNAb, PGV04 (Lyumkis et al., 2013). The specific role of the N276 glycan was further illustrated by antibody binding to YU2 gp120 lacking this glycosylation site (Figure 3B). DRVIA7 showed a more pronounced binding to the mutant gp120, while VRC01 and DRVIA7H-VRC01L recognized both gp120s in a similar manner.
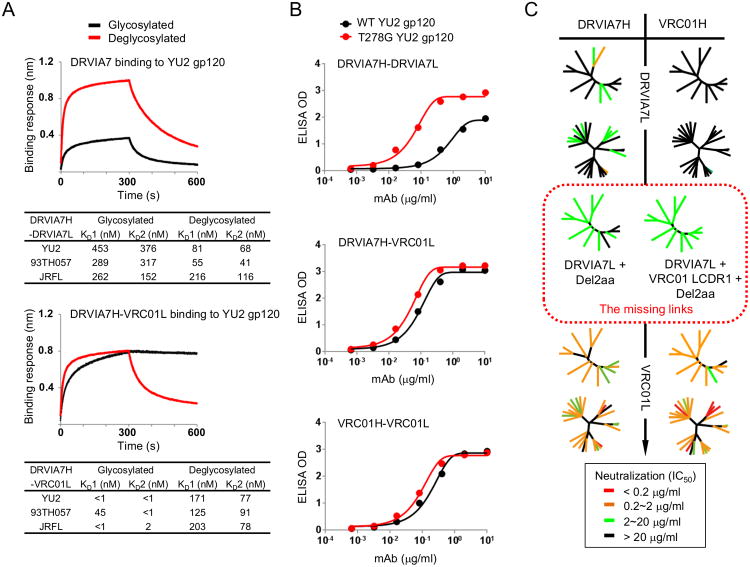
Figure 3. Interaction between DRVIA7 light chain and the N276 glycan determines the neutralizing breadth of the antibody.
(A) Octet association and dissociation curves for DRVIA7 or DRVIA7H-VRC01L towards glycosylated or endoH deglycosylated gp120 YU2 at 1000 nM concentration (above). Binding response (nm shift) is plotted as a function of time (s), with KD values indicated below. (B). ELISA binding for DRVIA7, DRVIA7H-VRC01L and VRC01 against YU2 gp120 (black) and T278G (i.e. no glycan at N276) YU2 gp120 (red), with the optical density (OD) plotted as a function of antibody dilution (μg/ml). (C) The neutralizing activity of DRVIA7, VRC01H-DRVIA7L, DRVIA7H-VRC01L and VRC01 against both the global and DRVI panels of pseudoviruses. In addition, the neutralizing activity of two DRVIA7 variants (circled by red dotted lines), DRVIA7H-DRVIA7L + Del2aa and DRVIA7H-DRVIA7L + VRC01 LCDR1 + Del2aa, against the global panel is shown. Neutralization potencies (Tables S4A-B) are mapped onto dendrogram representations of the virus panels as in Figure 1. See also Figure S3.
We next investigated whether the VRC01-like LCDR1 signature combined with an N-terminus truncation could help DRIVA7 better accommodate the inhibitory glycan patch, and as a result, improve its neutralizing activity (Figure 3C). We have designed three DRVIA7 variants containing a VRC01 LCDR1, a VRC01 LCDR1 and a 2-aa deletion at the light chain N-terminus, and a 12A21 LCDR1 to facilitate the comparison with DRVIA7 and VRC01 (Figure S3D). The N-terminal truncation of DRVIA7 light chain significantly increased the breadth, which was further enhanced by the introduction of VRC01 LCDR1 (Figure 3C, middle). Inclusion of the VRC01 LCDR1, but not the N-terminal deletion, had little effect on the breadth. Lastly, the use of the mature VRC01 light chain substantially improved potency (Figure 3C, bottom left). Our analysis thus demonstrated that proper light chain maturation was critical in the early development of VRC01-class antibodies to accommodate the inhibitory gp120 glycans. Of note, DRVIA7H-VRC01L neutralized 3 out of 16 VRC01-resistant isolates (Figure S3F), suggesting a potential role of heavy chain in strain specificity.
Dynamic antibody evolution revealed by unbiased analysis of DRVI01 B cell repertoires across three time points
We applied the unbiased repertoire analysis (He et al., 2014) to study the DRVI01 memory B cell repertoire across three time points (2006, 2008, and 2009). For each time point, 10-20 million PBMCs were used to prepare antibody H, κ, and λ chain libraries from the 5′-RACE PCR products of the B cell transcripts. In total, NGS on the Ion Torrent Personal Genome Machine (PGM) generated over 11.7 million raw reads. After data processing (He et al., 2014; Wu et al., 2011), 1.52 to 3.89 million antibody chains were obtained for the three time points studied (Table S3).
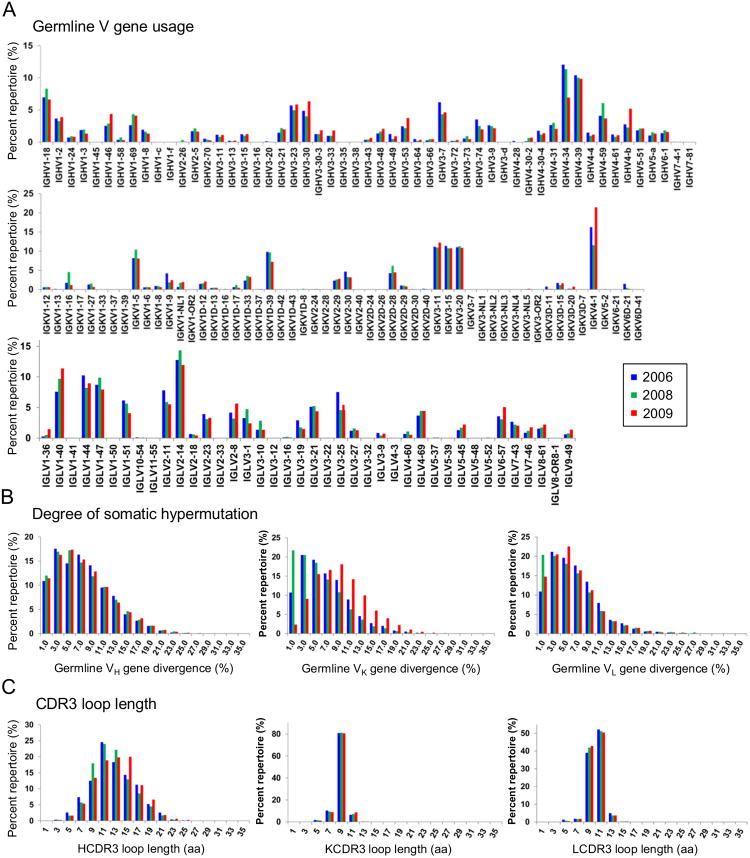
We first analyzed the germline gene usage (Figure 4A). IgHV1-2, the germline family of DRVIA7 heavy chain, accounted for 3.7, 3.3, and 3.9% of the heavy chains at 2006, 2008, and 2009, respectively, while IgHV4-34 and IgHV4-39 were predominantly used across the time points. The low frequency of IgHV1-2 suggested that DRVIA7 did not constitute a major lineage within the repertoire, consistent with the weak binding and neutralization of DRVIA7, which might have limited the B cell expansion. IgKV1-5, the germline family of DRVIA7 light chain, accounted for 8.2, 10.4 and 8.1% of the κ chains at 2006, 2008 and 2009, respectively. We next determined the distribution of germline gene divergence, or SHM (Figure 4B). While the heavy chains displayed a modest 1.5% difference in germline divergence between adjacent time points, the light chains showed a rapid change on a scale of 10-20%. In particular, a large population of germline light chains was found in the 2008 repertoire, constituting nearly 20% of the κ chains. Lastly, in terms of CDR3 loop length, we observed a rather dynamic heavy-chain distribution compared to a steady light-chain distribution (Figure 4C). More heavy chains with 9-aa and 15-aa HCDR3 loops were found in the 2008 and 2009 repertoires, respectively. Thus, the production of a large number of germline light chains that rapidly diverged, coupled with notable shifts of HCDR3 length in the context of a stable level of SHM for heavy chains, demonstrated a dynamic B cell repertoire in this donor.
Figure 4. Unbiased B cell repertoire profiles of donor DRVI01 at three time points.
The distribution is plotted for (A) germline V gene usage for heavy and light (κ and λ) chains, (B) germline gene divergence, or degree of SHM, and (C) CDR3 loop length (H – heavy chain; K – κ chain; L – λ chain). Color coding denotes the time point analyzed, with 2006 shown in blue, 2008 in green, and 2009 in red. See also Table S3.
Longitudinal tracing of the DRVIA7 lineage over four years of maturation revealed stalled light chain maturation to accommodate glycans
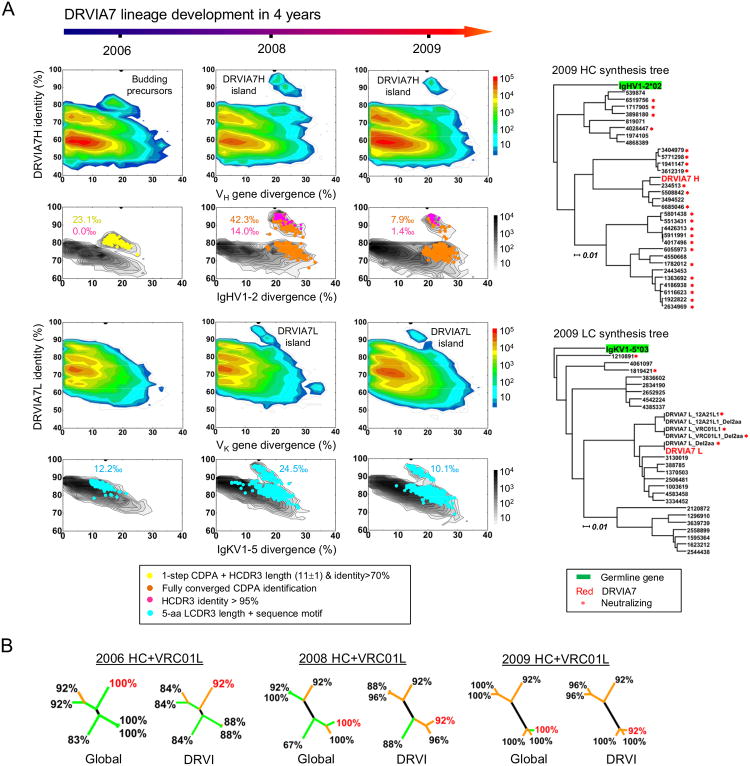
We investigated the DRVIA7 lineage longitudinally by tracing DRVIA7 heavy and light chains in the unbiased repertories obtained from three time points (Figure 5A). We first analyzed the 2009 time point, from which DRVIA7 was identified. The heavy and light chain repertoires with respect to DRVIA7 were visualized using the identity-divergence two-dimensional (2D) plots. As expected, we observed two distinct islands on the 2D plots corresponding to the DRVIA7 somatic population. Specifically, the heavy chain population showed a divergence of 18-27% (on average 23.1%) relative to the IgHV1-2 germline gene with a DRVIA7 identity of 85-95%. In contrast, the light chain population displayed a lower divergence of 13-20% and a DRVIA7 identity of 90-95%. Using the cross-donor analysis (Zhu et al., 2013), we identified 530 VRC01-like heavy chains from the repertoire (Figure S4A), which, when overlaid on the 2D plot, not only covered an island of sequences most similar to DRVIA7, but also extended into the main population (Figure 5A, column 3, panel 2, in orange). These VRC01-like heavy chains accounted for 7.9‰ of the IgHV1-2 gene family, with DRVIA7 somatic variants – defined by an HCDR3 identity of 95% or greater – representing 1.4‰ of the gene family (in magenta). We selected 30 distinct heavy chains (Figure S4B) for phylogenetic analysis, which showed multiple sub-lineages (Figure 5A, top right). When paired with mature VRC01 light chain, most reconstituted antibodies showed broad neutralization, confirming that these heavy chains are indeed VRC01-like (Tables S4-5). We then identified 703 κ chains with the VRC01-class LCDR3 signature (Zhou et al., 2013) (Figure 5A, column 3, panel 4, in cyan), which accounted for 10.1‰ of the IgKV1-5 gene family. Twenty-two distinct light chains (Figure S4B) were selected for phylogenetic analysis. The light chain dendrogram exhibited a branching pattern similar to that of the heavy chains (Figure 5A, bottom right). Taken together, our analysis revealed a large population of VRC01-like antibodies in the 2009 repertoire containing a DRVIA7 lineage that had not yet overcome the glycan barrier.
Figure 5. Longitudinal analysis of the DRVIA7 lineage development.
(A) Identity-divergence analysis of the unbiased heavy (H) and light chain (κ) repertoires of donor DRVI01 at time points 2006, 2008 and 2009, and dendrograms of synthesized heavy and light chains identified from the 2009 repertoire. For the whole-repertoire analysis (rows 1 and 3), sequences are plotted as a function of sequence identity to DRVIA7 and sequence divergence from putative germline genes. Color-coding denotes sequence density. For the analysis of DRVIA7-encoding germline gene families (rows 2 and 4), sequences are plotted as a function of sequence identity to DRVIA7 and sequence divergence from IgHV1-2 and IgKV1-5 for heavy and light chains, respectively. The germline gene family distribution is presented as black contour lines. DRVIA7 and sequences identified based on various criteria are shown on the black contours with per-mille values (‰) labeled accordingly. Dendrograms of functionally tested heavy and light chains are rooted by their respective germline allelic genes, IgHV1-2*2 and IgKV1-5*03, respectively. (B) Neutralization of five selected DRVIA7-like heavy chains paired with mature VRC01 light chain was tested against both global and DRVI panels for each time point, with DRVIA7 heavy chain included as a control (labeled in red). Neutralizing breadth is labeled for each antibody, which is shown as a branch of the dendrogram, and each branch is color-coded by average potency (concentration (μg/ml) <0.1: red; 0.1-1: orange; 1-20: green; and >20: black). See also Tables S4-5 and Figures S4-5.
The 2008 repertoire showed a markedly expanded DRVIA7 lineage with large islands extending from the near-100% identity to the main population on the 2D plots, for both heavy and light chains (Figure 5A, column 2). Compared to 2009, we observed a ∼5-fold expansion of VRC01-like heavy chains defined by cross-donor analysis (42.3‰) and a ∼10-fold expansion of DRVIA7-like heavy chains (14.0‰) defined by an HCDR3 identity of 95% or greater (Figure 5A, column 2, panel 2). These VRC01-like heavy chains showed an average divergence of 22.9%, similar to that observed for 2009 (23.1%). Consistently, we observed a 2.5-fold expansion of IgKV1-5 light chains with the VRC01-class LCDR3 signature (Figure 5A, column 2, panel 4). Of note, a group of near-germline light chains (5-10% divergence) can be seen on the 2D plot, suggesting an active B cell development driven by the light chain selection. Ten distinct VRC01-like heavy chains that were not sequence homologs of the 30 tested 2009 heavy chains, as defined by an identity cutoff of 90%, were selected for functional validation (Figures S4A-B). When reconstituted with VRC01 light chain, they produced neutralization profiles similar to those identified from 2009 (Tables S4-5).
The 2006 repertoire, however, produced a distinct pattern compared to the two later time points. A population of heavy chains characterized by a low SHM level (10-25%, with an average of 17.0%) and a DRVIA7 identity of 85% or less was “budding” from the main population (Figure 5A, column 1, panel 1). These IgHV1-2 sequences appeared not to possess the mature VRC01 signature (Zhu et al., 2013) defined by the converged cross-donor analysis (Figure S4A). However, the sequences identified in the first step of this analysis, accounting for 23.1‰ of the IgHV1-2 gene family, overlapped precisely with the “budding” population on the 2D plot (Figure 5A, column 1, panel 2, in yellow), suggesting that they might be precursors of DRVIA7 heavy chain. Five heavy chains selected from this population (Figure S4B), when reconstituted with VRC01 light chain, exhibited a remarkable neutralization breath of 84% or greater (Figure 5B; Tables S4-5). Light chain analysis revealed that 12.2‰ of the IgKV1-5 gene family possess the VRC01-class LCDR3 signature, suggesting that it is readily encoded by the repertoire. The VJ recombination that gave rise to the 5-aa LCDR3 occurred constantly, as shown by the near-germline light chains in the 2008 repertoire (Figure 5A, column 2, panel 4). Furthermore, 5-aa LCDR3 loops were identified from most light chain germline gene families at each time point.
Longitudinal lineage tracing thus uncovered the emergence of this VRC01-class antibody. While the DRVIA7-like heavy chains were first detected in 2008, we observed functional precursors in 2006 with a divergence at least 6% lower than that of the 2008 and 2009 sequences. This result placed the “birth date” of the DRVIA7 lineage shortly before 2006. Within four years, the heavy chains of DRVIA7 lineage showed increasing breadth and potency, when paired with mature VRC01 light chain (Figure 5B). In contrast, although the VRC01-class LCDR3 signature was present in the repertoire since 2006, the DRVIA7 light chains were unable to improve upon breadth and potency over time (Table S4B), supporting the notation that the light chain interaction with gp120 glycans is a critical barrier for the VRC01 development. However, the immune system failed to overcome this barrier even though a large pool of germline light chains was generated to accelerate the maturation process (Figure 4B). To better define the range of neutralizing activity of the DRVIA7 lineage, two light chain somatic variants from the 2009 repertoire were reconstituted with the DRVIA7 heavy chain, which displayed neutralization breadths ranging from 17 to 75% against the global panel and 24 to 60% against the DRVI panel (Tables S5C-D). Given the lower level of maturation, antibodies of the DRVIA7 lineage from 2006 should be less broad and potent than those from 2008 and 2009. Nonetheless, DRVIA6b, a clonal lineage utilizing a long HCDR3 loop (19 aa), and therefore a potentially different mode of gp120 binding, began to emerge while DRVIA7 was declining in 2009 (Figure S4C), suggesting an active CD4bs-directed polyclonal B cell response in this Chinese donor.
The identification of DRVIA7 suggested that VRC01-class antibodies might be present in other Chinese LTNPs. This notion was supported by the repertoire analysis of five Chinese LTNPs that showed broad serum neutralization (85-100%) after 7-16 years of infection (Figure S5). Although the heavy chain repertoires obtained from the gene-specific method (He et al., 2014) displayed low frequencies of IgHV1-2 germline gene ranging from 0.3 to 7.2%, cross-donor analysis (Zhu et al., 2013) detected the maturation pattern of VRC01-class heavy chains for two LTNPs. Thus, a systematic, unbiased repertoire profiling will likely identify more VRC01-like antibodies from the HIV-1-infected Chinese cohort.
SGA analysis revealed that rapid HIV-1 envelope evolution outpaced the DRVIA7 antibody lineage
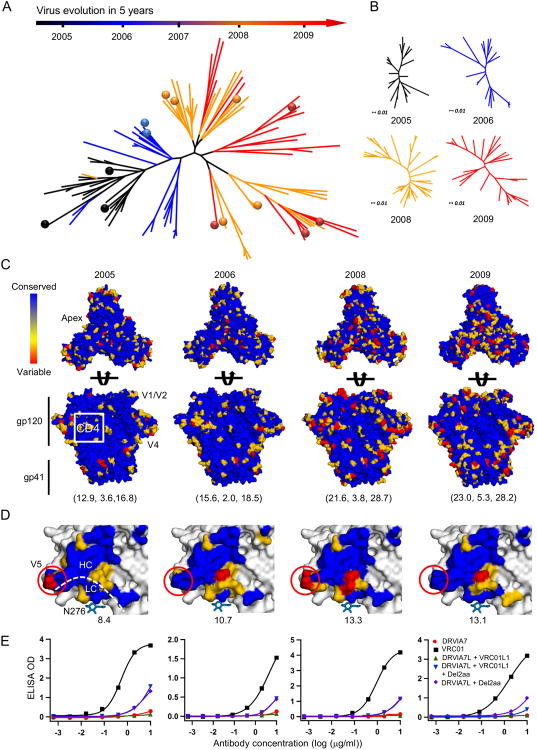
We utilized SGA (Salazar-Gonzalez et al., 2008) to investigate viral diversity and virus-host co-evolution in donor DRVI01. A total of 123 full-length Env genes encoding gp160 were sequenced from plasma viral RNA at 2005, 2006, 2008, and 2009. Of these, 111 non-redundant Env sequences were used to construct a neighbor-joining (NJ) phylogenetic tree (Figure 6A). The phylogenetic branches coincided precisely with the time points from which the Env sequences were derived, showing a clear time axis of viral evolution. Overall, the viruses appeared to diversify rapidly over time, as the genetic distance between phylogenetic branches at 2009 was significantly greater than that at 2005. Consistently, the time point-specific NJ trees also showed a greater intragroup genetic distance for the later time points (Figure 6B).
Figure 6. SGA analysis of virus diversity and co-evolution.
(A) Neighbor-joining (NJ) phylogenetic tree of 111 non-redundant HIV-1 Env sequences isolated from donor DRVI01 by single genome amplification (SGA). These Env sequences, shown as phylogenetic branches, are color-coded by the time point at which the sample was collected. 14 Env sequences selected for experimental validation are labeled as color-coded dots on the tree. (B) NJ tree of non-redundant Env sequences for each individual time point, namely 2005, 2006, 2008, and 2009. The four NJ trees have been rescaled to use the same unit of genetic distance. (C) Trimer models of SGA-derived consensus Env sequences with mutational hotspots labeled on the surface. Top view is shown in panel 1 and side view is shown in panel 2. Molecular surfaces are colored according to sequence conservation. Root-mean-square fluctuation (RSMF) relative to the consensus sequence at each time point is calculated as a diversity score for gp120, gp41, and gp140 and labeled beneath the structural model. (D) The gp120 core models of SGA-derived consensus sequences with CD4bs and mutational hotspots labeled on the molecular surface, which is colored as in (C). The diversity score for gp120 core is labeled beneath each structural model. (E) ELISA binding of VRC01, DRVIA7, and DRVIA7 variants to selected Env trimers at the four time points studied.
We then determined the mutational hotspots by projecting the sequence variation onto a gp140 trimer model of the “consensus” Env sequence derived from each time point (Figure 6C). The root mean square fluctuation (RMSF) relative to the consensus sequence was calculated as a measure of the mutational rate for each Env component. Compared to the 2005 Envs, which showed RMSFs of 12.9, 3.6, and 16.8 for gp120, gp41, and gp140, respectively, the 2009 Envs exhibited significantly more variation, with RMSFs of 23.0, 5.3, and 28.2 for the three Env components. This quantitative analysis was confirmed by the visual inspection of mutational hotspots on the trimer surface. For example, the 2008 and 2009 Envs contained more insertions and substitutions in the V1V2 region near the apex and the V4 loop, whereas the CD4bs remained conserved across time points with only peripheral mutations. By contrast, gp41 showed a moderate increase in mutations at the same time points, suggesting that the viral repertoire was evolving with differential mutational rates for gp120 and gp41.
We next determined the variation of the DRVIA7 footprint on a gp120 core lacking the V1V2 and V3 loops (Figure 6D). Visual inspection revealed that the mutations mainly occurred at gp120 residues interacting with the DRVIA7 heavy chain- and light chain-contacting interface, which accommodates the V5 loop. Of note, the N276 and V5 glycans appeared to be conserved in all Env sequences analyzed. We selected 14 Envs from the phylogenetic tree (Figure 6A), of which four were synthesized as trimers and used to compare recognition by VRC01 and DRVIA7 variants (Figure 6E). VRC01, but not DRVIA7, bound to all four Envs with high affinity, indicating that the viruses in this donor had actively evolved to escape the DRVIA7 lineage. Such viral escape is commonly observed even for bNAb donors, for example, the VRC01 donor (Wu et al., 2012). Recognition of autologous DRVIA7-resistant Envs was improved after N-terminal truncation of the DRVIA7 light chain, whereas incorporation of the LCDR1 signature alone showed little effect, consistent with the finding that the DRVIA7 lineage began to decline by 2009, likely due to its unfavorable binding and thus poor fitness in B cell selection. Together, SGA and sequence variation analysis revealed a changing face of HIV-1 with a conserved inhibitory glycan patch that raised a significant barrier to the DRVIA7 lineage development during this virus-host co-evolution.
Discussion
VRC01-class antibodies share a common mode of recognition and a set of conserved sequence signatures (Diskin et al., 2011; Wu et al., 2011; Zhou et al., 2010; Zhou et al., 2013). Longitudinal analysis previously uncovered an unexpected complexity of the VRC01 lineage (Wu et al., 2015). However, the early events of VRC01 lineage development prior to the timeframe studied still remain elusive (Wu et al., 2015). In this study, we attempted to fill this knowledge gap by investigating DRVIA7, a VRC01-like antibody from a Chinese donor, which presented a different picture of early VRC01 development than that drawn from previous studies of mature VRC01-class antibodies.
First, the level of heavy chain SHM required for effective HIV-1 neutralization was perhaps lower than previously estimated. It is speculated that a long period of time would be needed to acquire the somatic mutations in the framework and HCDR1/2 regions for full VRC01-like neutralizing activity (Wu et al., 2010; Wu et al., 2015; Wu et al., 2011; Zhou et al., 2013). This leads to a conclusion that heavy chain maturation is a barrier and perhaps the time-limiting step in VRC01 development. In contrast to this paradigm, the DRVIA7 heavy chain precursors identified from 2006 neutralized diverse HIV-1 isolates when paired with mature VRC01 light chain. More importantly, the heavy chain SHM increased from 17.0 to 22.9% within only two years, indicating that the immune system is capable of rapidly maturing the VRC01-like heavy chains.
Second, the 5-aa LCDR3 signature of VRC01-class bNAbs appeared to be rather common in the B cell repertoire, and not as rare as previously perceived. As observed from the VRC01-class bNAbs identified thus far (Scheid et al., 2011; Wu et al., 2011; Zhou et al., 2013; Zhu et al., 2013), the light chain germline gene is less restricted than that of the heavy chain, allowing both κ and λ genes to be used. For DRVIA7, a new germline gene IgKV1-5*01 was used. Our repertoire analysis revealed a stable presence (up to 2% of the repertoire) of light chains with 5-aa CDR3 loops and showed that the immune system could rapidly produce a large number of new germline light chains to facilitate B cell selection. Thus, the LCDR3 signature as a barrier to VRC01 maturation might be readily overcome by an active repertoire.
Third, the interplay of LCDR1 and N-terminus with the N276 and V5 glycans has not been fully appreciated within the context of VRC01-class antibody development. Previous work placed high importance on establishing a favorable angle of approach to avoid clash with neighboring gp120 subunits and the V1V2 loop within the trimeric spike (Chen et al., 2009; Zhou et al., 2015), which appears not to be an issue for DRVIA7. The limited neutralizing activity and the stalled lineage development, however, could be well explained by a less favorable DRVIA7 light-chain interaction with gp120 glycans. A DRVIA7 with reengineered N-terminus and LCDR1 displayed similar breadth to VRC01, indicating that such small changes to accommodate gp120 glycans at N276 and V5 could transform an antibody with limited neutralization breadth to a bNAb.
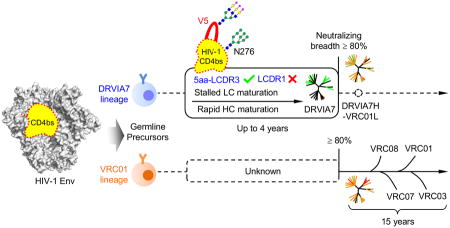
In conclusion, DRVIA7 provided us an opportunity to recapitulate the early events in B cell maturation and to assess the barriers for generation of a VRC01-like antibody. Starting from the naïve B cell with recombined 5-aa LCDR3, the DRVIA7 lineage precursors emerged with neutralization-competent heavy chains in 2006, rapidly peaked by 2008, and began to decline in 2009, all in the face of rapidly diversifying HIV. Despite the attempts by the immune system to overcome the glycan barrier, the maturation of light chain N-terminus and CDR1 did not occur naturally; however, antibody engineering was able to lead to a neutralization breath on par with VRC01. Thus, our case study of a Chinese LTNP provided much sought-after information on the emergence of VRC01-class antibodies that had been sorely lacking. Our findings also supported the notion that redesign of the glycan shield around the CD4bs at N276 and the V5 loop might lead to an effective immunogen to elicit VRC01-like antibodies, providing useful information to facilitate rational vaccine design.
Experimental Procedures
Chinese donor DRVI01 samples
The sera and peripheral blood mononuclear cells (PBMC) described in this study were collected from an HIV-1-infected Chinese patient, DRVI01, from a former plasma donor (FPD) study cohort founded by the Collaboration for AIDS Vaccine Discovery (CAVD) of the Bill and Melinda Gates Foundation, Chinese National Natural Science Foundation, and National Major Project on Infectious Disease. A detailed neutralization analysis has been reported for the samples from this cohort (Hu et al., 2012).
Isolation of monoclonal antibodies from donor DRVI01
Isolation of CD4bs-directed DRVIA mAbs was achieved by single cell FACS using the CD4bs-specific RSC3 and RSC3Δ371I probes (Wu et al., 2010). Memory B cells were sorted on a FACS Aria III cell sorter (BD Biosciences) to obtain single B cells with the phenotype CD19+CD20+CD14-CD3-CD8-IgG+RSC3+RSC3Δ371I-. Single cells were then sorted into 96-well plates followed by RT-PCR, IgG cloning and expression. Details are described in Extended Experimental Procedures.
Expression and purification of DRVIA7 Fab, gp120 and gp140 proteins
All proteins were produced transiently in either FreeStyle™ 293F or GnTI-/- 293S cells as previously described (Kong et al., 2013). Briefly, after 4-7 days, the supernatants of the transfected cells were harvested, and purified with a Galanthus Nivalis lectin column (Vector laboratories, Inc.) for gp120, or a Kappa CaptureSelect column (BAC BV) for DRVIA7 Fab. All proteins were further purified by size exclusion chromatography (SEC).
Crystallographic analysis of DRVIA7 Fab and gp120-bound complex
Crystals were obtained for DRVIA7 Fab and the complex of 93TH057 gp120 core bound to DRVIA7 Fab. The crystals diffracted to 2.9 Å and 3.4 Å resolution, respectively, and their structures were solved by molecular replacement. After refinement, final Rcryst and Rfree values were 23.4% and 28.4% for DRVIA7 Fab, and 25.3% and 30.6% for gp120-bound DRVIA7 Fab complex, respectively (Table S2). Details are described in Extended Experimental Procedures.
Negative stain electron microscopy (EM)
JRFL SOSIP.664 trimers in complex with either DRVIA7 or DRVIA7H-VRC01L Fab were analyzed by negative-stain EM. Data were collected on either an FEI Tecnai T12 electron microscope or an FEI Talos electron microscope. Data processing and 3D reconstruction were performed as previously described (de Taeye et al., 2015). Crystal structures or EM volumes were fit into the 3D reconstructions to generate figures. Details are described in Extended Experimental Procedures.
Ion Torrent PGM sequencing of donor DRVI01 antibody libraries
Antibody library preparation and PGM sequencing of DRVI01 B cell repertoires were performed as previously described (He et al., 2014). Briefly, the 5′-RACE PCR protocol was used to prepare unbiased antibody H, κ, and λ chain libraries for DRVI01, while the gene-specific multiplex PCR protocol was used to prepare antibody libraries for five LTNPs. Details are described in Extended Experimental Procedures.
Bioinformatics analysis of repertoire sequencing data
The Antibodyomics 1.0 pipeline (He et al., 2014; Wu et al., 2011) was used to process the NGS data. Identity-divergence 2D analysis, cross-donor phylogenetic analysis, and CDR3-based lineage tracing were used to dissect the details of DRVIA7 lineage at the three time points studied. Details are described in Extended Experimental Procedures.
HIV-1 neutralization assays
Neutralization assays were performed on TZM-bl reporter cells using various panels of pseudoviruses as previously described (Wu et al., 2010). Neutralization curves were fit by a nonlinear regression analysis to derive IC50 values for DRVI01 serum mapping and functional validation of DRVIA mAbs and variants. Details are described in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are very grateful to R. Stanfield for helpful discussions; M. Elsliger for computer support; H. Tien for crystallization screening; A. Eroshkin for help with calculating N-linked glycosylation frequencies; J. Hou, P. Li and M. Zhu for help with antibody binding assays; D. Li, H. Liang and H. Peng for help with measuring patient CD4+ T-cell counts; Q. Zhao for help with viral load assays; J. Wu and X. Ding for help with patient blood sample collection; and J.P. Verenini for manuscript formatting. X-ray data sets were collected at the Advanced Photon Source (APS) beamline 23ID-B. Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Electron microscopy data were collected at the Scripps Research Institute EM Facility. This work was supported by the National Major projects for Infectious Diseases Control and Prevention (2012ZX10001008) (Y.S.), by the International AIDS Vaccine Initiative Neutralizing Antibody Center and CAVD, by the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID UM1 AI00663) (A.B.W., I.A.W., J.Z.), by the HIV Vaccine Research and Design (HIVRAD) program (P01 AI110657) (A.B.W., I.A.W.), by the Joint Center of Structural Genomics (JCSG) funded by the NIH NIGMS, Protein Structure Initiative (U54 GM094586) (I.A.W.), by the National Major projects for Infectious Diseases Control and Prevention (2012ZX10001008) (Y.S.), by NIH NIAID grants R01 AI102766 (Y.L.) and AI084817 (I.A.W., A.B.W.), by an NIH NIAID development grant from the Center for AIDS Research at the University of California, San Diego (P30AI36214) (Y.L.), and by the SKLID Development grant (2012SKLID103) (Y.S.). A portion of this work was supported by the NIH CIPRA grant (U19AI51915) (Y.S.), and by an American Foundation for AIDS Research Mathilde Krim Fellowship in Basic Biomedical Research (L.K.).
Footnotes
Author Contributions: Project design by L.K., B.J., Y.C., L.H., L.R., J.L., Y.L., I.A.W., J.Z. and Y.S.; neutralizing antibody screening by K.H., L.R. and Y.S.; antibody isolation by Y.C., R.W. and Y.L.; X-ray work and analysis by L.K., Y.H., M.D. and F.G.; EM work by G.O. and A.B.W.; antibody NGS, lineage tracing, and SGA analysis by L.H. and J.Z.; synthesis of antibody genes and construction of expression plasmids by B.J., X.J. and Z.W.; neutralization on the global and DRVI panels by K.H. and L.R.; expression and purification of antibodies and Env proteins by L.K., J.L., X.J., Y.C., R.W. and Y.H.; Env SGA performed by K.H., Y.C. and Y.F.; antibody Fab construct design by Y.C. and R.W.; antibody binding assays by Y.C., L.K., R.W., B.J. and X.J.; epidemiology and cohort maintenance by B.S. and K.H.; VRC01-resistant HIV-1 neutralization by S.O.D, K.M. and J.R.M.; neutralization on the 8-virus panel by Y.C. and R.W.; manuscript written by L.K., Y.L., I.A.W., J.Z. and Y.S. All authors were asked to comment on the manuscript. This is TSRI manuscript number 29101.
Accession Numbers: Coordinates and structure factors for Fab DRVIA7 and its complex with gp120 are deposited with the PDB under accession codes 5CD3 and 5CD5 respectively. The Fab DRVIA7:JRFL SOSIP.664 trimer and Fab DRVIA7H-VRC01L:JRFL SOSIP.664 trimer EM reconstructions are deposited in the EMDB under accession codes EMD-6372 and EMD-6373, respectively.
Competing financial interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, de la Pena AT, Guttman M, Julien JP, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, et al. Global panel of HIV-1 env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2014;88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund NT, Horwitz JA, Nogueira L, Sievers SA, Scharf L, Scheid JF, Gazumyan A, Liu C, Velinzon K, Goldenthal A, et al. A new glycan-dependent CD4-binding site neutralizing antibody exerts pressure on HIV-1 in vivo. PLoS Pathog. 2015;11:e1005238. doi: 10.1371/journal.ppat.1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Sok D, Azadnia P, Hsueh J, Landais E, Simek M, Koff WC, Poignard P, Burton DR, Zhu J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Hong K, Zhao C, Zheng Y, Ma L, Ruan Y, Gao H, Greene K, Sarzotti-Kelsoe M, Montefiori DC, et al. Profiles of neutralizing antibody response in chronically human immunodeficiency virus type 1 clade B′-infected former plasma donors from China naive to antiretroviral therapy. J Gen Virol. 2012;93:2267–2278. doi: 10.1099/vir.0.043802-0. [DOI] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He X, Wang Z, Xing H, Li F, Yang Y, Wang Q, Takebe YM, Shao Y. Tracing the origin and history of HIV-1 subtype B′ epidemic by near full-length genome analyses. AIDS. 2012;26:877–884. doi: 10.1097/QAD.0b013e328351430d. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li Z, Zhao S. HIV infected people were first identified in intravenous drug users in China. Chin J Epidemiol. 1990;11:184–185. [Google Scholar]
- Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, Scheid JF, Lee JH, West AP, Jr, Chen C, Gao H, Gnanapragasam PNP, Mares R, Seaman MS, Ward AB, et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zirkle B, Nie J, Ma J, Gao K, Chen X, Huang W, Kong W, Wang Y. N463 glycosylation site on V5 loop of a mutant gp120 regulates the sensitivity of HIV-1 to neutralizing monoclonal antibodies VRC01/03. J Acquir Immune Defic Syndr. 2015;69:270–277. doi: 10.1097/QAI.0000000000000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang C, O'Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O'Dell S, McKee K, et al. Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yang XZ, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wu X, Zhang B, McKee K, O'Dell S, Soto C, Zhou T, Casazza JP, Mullikin JC, Kwong PD, et al. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc Natl Acad Sci USA. 2013;110:E4088–E4097. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.