Abstract
The 1918 H1N1 influenza virus was responsible for one of the most deadly pandemics in human history. Yet to date, the structure component responsible for its virulence is still a mystery. In order to search for such a component, the neuraminidase (NA) antigen of the virus was expressed, which led to the discovery of an active form (tetramer) and an inactive form (dimer and monomer) of the protein due to different glycosylation. In this report, the N-glycans from both forms were released and characterized by mass spectrometry. It was found that the glycans from the active form had 26% core-6 fucosylated, while the glycans from the inactive form had 82% core-6 fucosylated. Even more surprisingly, the stalk region of the active form was almost completely devoid of core-6-linked fucose. These findings were further supported by the results obtained from in vitro incorporation of azido fucose and 3H-labeled fucose using core-6 fucosyltransferase, FUT8. In addition, the incorporation of fucose did not change the enzymatic activity of the active form, implying that core-6 fucose is not directly involved in the enzymatic activity. It is postulated that core-6 fucose prohibits the oligomerization and subsequent activation of the enzyme.
Proposed mechanism for how core-fucose prohibits the tetramerization of the 1918 pandemic viral neuraminidase
Only the cross section of the stalk region with two N-linked glycans are depicted for clarity. (A) Carbohydrate-carbohydrate interaction on non-fucosylated monomer allow tetramerization. (B) Core-fucosylation disrupt the interaction and prevent the tetramerization.
Introduction
The 1918 influenza, or “Spanish Flu” virus, caused the most devastating pandemic in recorded human history. Its estimated infection rate lies between 25% to 30% of the 1918 global population with approximately 50-100 million deaths worldwide [1]. Currently, influenza is an ongoing threat to human society [2,3], having caused the recent pandemics of the 2005 Hong Kong avian flu [4] and 2009 swine flu [5]. Research on the 1918 influenza virus may provide key insights for combating the emerging strains of influenza virus. For this reason, the 1918 influenza virus (A/Brevig Mission/1/18) was resurrected from the body of an Alaskan Inuit woman in 2005 [6]. Significant effort has been made to decipher the mechanism of the lethality of this virus since then.
Influenza virus has two surface antigens, hemagglutinin (HA) and neuraminidase (NA) [7]. HA initially recognizes and binds to the terminal sialic acid residue on a host cell surface to mediate viral infection and infusion [5]. NA subsequently hydrolyzes host sialic acid residues to allow release of the progeny virus and spread of the infection [8]. NA is a box-shaped tetrameric glycoprotein with a large and globular head, a stalk region, and a small hydrophobic region that anchors the antigen into the viral lipid membrane [8]. The stalk region of the NA from the 1918 influenza virus contains approximately 50 amino acid residues (aa40 – aa90) and is responsible for the oligomerization of the protein [8].
The active site of NA is highly conserved throughout all influenza viruses [9], and this has allowed the development of two enzyme-inhibitor based anti-flu drugs: Tamiflu and Relenza [10]. Despite this success, our understanding of NA regulation remains limited, particularly regarding the biological functions of NA glycosylation. Previously, the expression of the 1918 influenza viral NA resulted in isolation of both active tetramer and inactive dimer/monomer forms [11]. The inactive form was unable to oligomerize to the active form, suggesting intrinsic molecular differences among the two forms. Subsequently, the N-glycans on the two forms were found to be different. Herein, we further characterize the N-glycans from the two forms by mass spectrometry and in vitro glycosylation study. Mass spectrometry analysis revealed a striking difference between the levels of core-6 fucosylation, where the overall level of core-6-linked fucosylation was significantly lower in the active form accompanied by almost complete absence of core-6-fucose within its stalk region. This discovery was further supported by the data obtained from in vitro incorporation of azido-fucose that was detected via click chemistry [12] and radio-labeled fucose at core-6 position using recombinant fucosyltransferase 8 (FUT8).
Results
Tetramer and Dimer Contain Similar Amount of Total N-Glycan
Previously, the pandemic 1918 H1N1 influenza viral NA was expressed and purified in an active form and an inactive form [11]. The expressed sequence contain seven potential N-glycosylation sites and five of them reside in a single tryptic peptide within its stalk region (Fig. 1A). It was found that the glycosylation was intrinsically different between the two forms. To further investigate this difference, the two forms were first separated on SDS gel after reduction and alkylation (Fig. 1B). It was clear that the two forms had similar mobility around 53 kDa, while the theoretical mass is 48.3 kDa, again suggesting that the difference is structural.
Fig. 1.
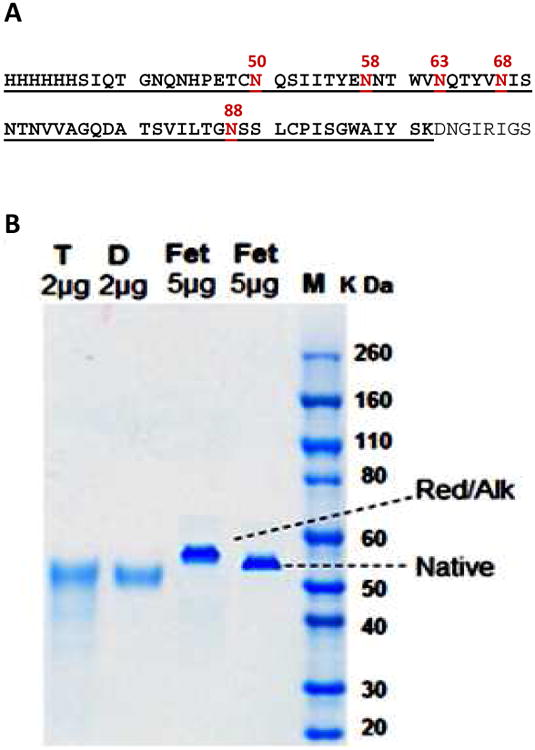
(A) The sequence of the expressed 1918 H1N1 NA contains seven potential N-glycosylation sites. Five sites (N-50 to 88) reside in one large tryptic peptide (6×His tag and S-37 to K-102, underlined) that is a part of the stalk region of the protein. The amino acid sequence numbering is according to GenBank AAF77036. (B) SDS-PAGE of the tetramer NA (T) and dimer NA (D), both reduced and alkylated. Bovine fetuin in native from and reduced/alkylated form was separated in the same gel as a resolution control. The form in reduced/alkylated was up-shifted, due to the carbamidomethylation of six pairs of disulfide bonds.
N-Glycans of the Tetramer and Dimer NA Possess Different Levels of Fucosylation
To further investigate the difference on glycosylation, the N-glycans of the two forms were released and analyzed with MALDI-MS (Fig. 2A and 2B). The mass spectra showed simple pauci- and high-mannose type glycans. In the spectrum of the tetramer, the most dominant peak (m/z 935.36, Hex3HexNAc2) was from a pentasaccharide Man3GlcNAc2 [13], and the second dominant peak at much less abundance (m/z 1081.43, Hex3HexNAc2Fuc1) was from a fucosylated glycan Man3GlcNAc2Fuc1. Surprisingly, in the spectrum of the dimer, the fucosylated glycan (m/z 1081.42, Hex3HexNAc2Fuc1) became the most dominant peak, and the non-fucosylated glycan (m/z 935.33, Hex3HexNAc2) became much less abundant. Since all detected glycans were neutral and similar in size, the relative abundance of individual composition within each sample should be proportional to its peak intensity in each MALDI-MS spectrum [14]. Based on this assumption, the non-fucosylated compositions were calculated to account for 81.89% of the total glycans in the tetramer, and 26.40% of the total glycans in the dimer (Supplemental Table 1).
Fig. 2.
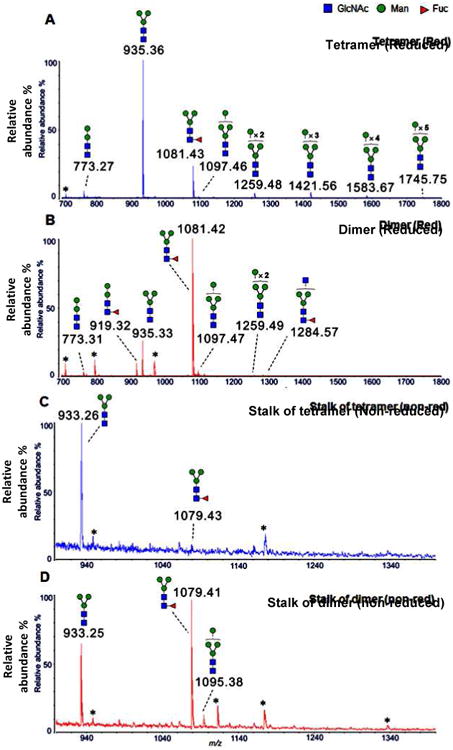
MALDI-MS of N-glycans (reduced to alditols) from tetramer NA (A) and dimer NA (B), and N-glycans (native, non-modified) from the stalk region of tetramer NA (C) and dimer NA (D). The plausible topology of each peak was proposed based on the bio-synthetic knowledge of N-glycosylation and its m/z value. All ions are single sodium adducts. (* Impurities or possible peptides).
Since the stalk region that is highly enriched with N-glycosylation sites is involved in the oligomerization of the protein [8] and possibly protecting the protein from host cell protease attack [11], different glycosylation on this region is likely to affect these functionalities. For this reasoning, the tryptic peptides of the stalk region of the two forms were further isolated and analyzed with MALDI-MS. Even more surprisingly, the glycans of the stalk region of the active tetramer were found to be almost completely devoid of fucosylation (Fig. 2C), while those of the inactive form were largely fucosylated (Fig. 2D).
Locate the Positioning of Fucosylation via MSn Fragmentation
To locate the position of the fucosylation, the permethylated N-glycans from both forms were subjected to ESI-MSn fragmentation [15,16]. When the fucosylated composition (Hex3HexNAc2Fuc1) from the tetramer was analyzed, the fucose residue was found to be linked to the reducing end GlcNAc of the oligosaccharide Man3GlcNAc2Fuc1 (Supplemental Fig.1). Given that the N-glycans were enzymatically released by PNGase F that lacks activity on core-3-linked fucosylated N-glycans [17], the fucose residue must be at the 6-O position of the reducing end GlcNAc residue.
To this end, the most abundant composition (Hex3HexNAc2) of the tetramer was confirmed to be a typical core pentasaccharide (Man3GlcNAc2) (Supplemental Fig.2). When equivalent compositions from the dimer were analyzed, identical structures were obtained (data not shown). Overall, MS analysis clearly indicated that the level of core-6-linked fucosylation was the major difference between the N-glycans of the two forms of NA.
Probing Core-6 Fucosylation Using Click Chemistry and Tritium-labeled Fucose
Core- 6 fucosylation of the reducing-end GlcNAc on N-glycans is introduced by FUT8 in humans [18]. To confirm the results of mass spectrometry analysis, the levels of core-6 fucosylation were further probed with azido fucose using recombinant human FUT8. The incorporated azido fucose was then conjugated to a biotin residue via a click chemistry reaction [19,20], and detected with streptavidin-conjugated horse radish peroxidase [21,22]. As shown in Fig. 3A, same amount of the tetramer accommodated significantly more azido fucose than the dimer. When the core-6 fucosylation was probed with 3H-fucose using FUT8, again, the active tetramer incorporated significantly more 3H-fucose compared to the dimer (Fig. 3B).
Fig. 3.
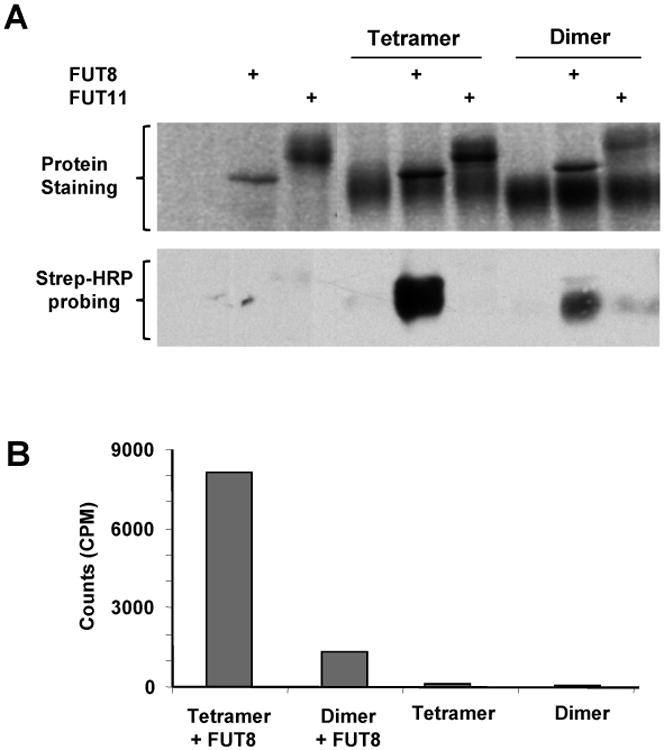
Probing the levels of core fucosylation on the two forms of 1918 H1N1 neuraminidase through in vitro fucosylation. In all experiments, equal amounts of dimer and tetramer were used for comparison. (A) The tetramer and dimer were fucosylated with azido-fucose by FUT8 and FUT11. The substrate specificity of FUT11 was unknown and used as a negative control. The incorporated azido-fucose was probed with streptavidin-HRP via click chemistry reaction. (B) Fucosylation of the dimer and tetramer with 3H-fucose by FUT8. The samples were spotted on glass fiber filters and counted with a liquid scintillation counter.
Considering that the two forms of NA had similar amount of total glycans (Fig. 1B), the results in Fig. 3 again suggest that the active tetramer had significantly more non-fucosylated glycans than the inactive dimer.
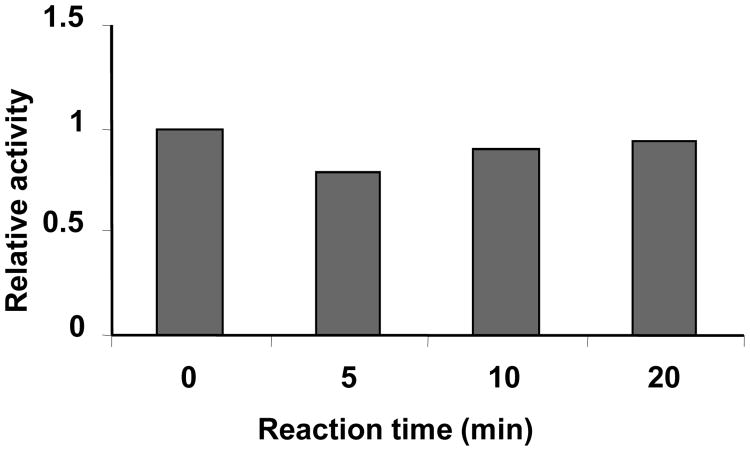
In vitro Core-fucosylation of the Active NA didn't Abolish Its Activity
Given that the active NA had significant lower level of core-fucosylation than that of the inactive form, it is imperative to investigate whether in vitro fucosylation will inactive the active form. To answer this question, the active tetramer was fucosylated using FUT8 for different time lengths before its activity was measured. Surprisingly, the enzymatic activity remained constant along the course (Fig. 4).
Fig. 4.
Effect of in vitro fucosylation on the enzymatic activity of the active form of 1918 H1N1 neuraminidase. The active tetramer was fucosylated in vitro using recombinant human FUT8 for different length of time and then activity was measured. Activity was plotted versus the time of reaction.
Discussion
Previously, 1918 influenza viral NA was expressed as two inconvertible forms, i.e. active and inactive forms, suggesting an intrinsic difference between the two forms. In this report, using mass spectrometry and in vitro fucosylation, we demonstrated that the inactive form is highly core-fucosylated, whereas the active form is mainly devoid of core fucose. However, in vitro core-fucosylation of the active form didn't abolish its activity, suggesting that there is no direct link between core-fucosylation and enzymatic activity.
Core-fucose is known to be involved in several biological events. One well-known example is antibody-dependent cell-mediated cytotoxicity (ADCC), where core-fucose inhibits carbohydrate-carbohydrate interactions between the antibody and its receptor FcγRIIIa [23]. Antibodies lacking core fucosylation show a large increase in affinity for FcγRIIIa, leading to an improved receptor-mediated effector function [24]. Core fucosylation of N-glycans is also required for the binding of EGF to its receptor, suggesting core fucosylation is critical to EGF mediated intracellular signaling [25]. In the case of 1918 influenza viral NA, considering that the stalk region is heavily glycosylated and play roles in the process of oligomerization, it is likely that the core-fucosylation in this region weakened the carbohydrate-carbohydrate interaction so that it prevented the tetramerization but allowed the formation of dimer, and explains the earlier observation that the monomer and dimer were convertible but not the tetramer [11].
Fucosylation is ubiquitous and age related [26]. For example, older people (>60 years old) has higher level of fucosylated haptoglobin compared to younger people (<40 years old) [27]. In a mouse model, it has been reported that the expression of Fut8 is up-regulated with age [28]. If core-fucosylation indeed inhibited the oligomerization and the activation of the neuraminidase of the 1918 pandemic influenza virus, people with higher level of FUT8 activity would have been advantageous for survival during the pandemic. Therefore, together with the age specific expression of core-fucose, the current finding may explain the mystery of the 1918 pandemic influenza, i.e. it caused a relatively lower death rate among the older population, while the younger population experienced a higher death rate [29]. Accordingly, if core-fucosylation is a general mechanism for inhibiting the oligomerization of influenza viral neuraminidases, it will be a good target for influenza drug development.
Materials and Methods
Ammonium bicarbonate, dithiothreitol (DTT), iodoacetamide (IAA), trifluoroacetic acid (TFA), sodium hydroxide (NaOH), dimethyl sulfoxide (DMSO), iodomethane (CH3I), sodium borohydride (NaBH4), 2,5-dihydroxybenzoic acid (DHB), urea, cellulose (medium fibrous), trypsin, HPLC grade ethanol, 1-butanol, and GDP-fucose were from Sigma-Aldrich. Sep-Pak C18 SPE cartridge was from Waters. PNGase F was from New England Biolabs. The NuPAGE series of LDS sample buffer (4×), MOPS SDS running buffer (20×), SimplyBlue SafeStain and 4-12% Bis-Tris Gels were from Life Technologies. 10,000 MWCO centrifugal devices were obtained from Millipore. NA was expressed with N-terminal His tag in sf21 insect cells and purified using nickel-histidine affinity chromatography followed by gel filtration as previously described [11]. Recombinant FUT8, FUT11, GDP-azido-fucose, biotinylated alkyne, and streptavidin-HRP were from R&D Systems.
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
The active tetramer and inactive dimer of the purified 1918 H1N1 NA were first reconstituted in 8M urea/ 0.4M ammonium bicarbonate. Both samples were then reduced by 25 mM DTT (45 min at 50 °C), and alkylated by 40 mM IAA (30 min at 23°C) in a dark environment. An aliquot of 2 μg of each form was mixed with LDS sample buffer and loaded onto a 4-12% Bis-Tris gel. SDS-PAGE was performed using MOPS SDS buffer and protein was stained with SimplyBlue SafeStain according to the vendor's protocol.
Tryptic Digestion and the Collection of Tryptic Peptide Consisting of Stalk Region
The aliquots of both reduced and alkylated forms (tetramer and dimer) of NA were incubated with trypsin overnight at a weight ratio (1/25: trypsin/NA) in a 37 °C oven. The proteolytic digestion was stopped by the addition of TFA into the samples in an ice bath. Large polypeptide-containing stalk regions from both samples were enriched via YM-10 centrifugal filters. Briefly, the mixture of crude tryptic peptides was re-dissolved in 500 μL of HPLC water and transferred into the sample chamber of a centrifugal filter. Small peptides were removed by repeated centrifugation (2×), and the retained fraction in the sample chamber was collected as the stalk region tryptic peptide as described in the Results section. Peptides were further desalted by passage through C18 SPE [30].
Sample Preparation of Released N-glycans
The desalted peptides, either from tryptic digestion of 150 μg of an NA sample or its equivalent amount of purified stalk region, were re-dissolved in 5 μL of 500 mM sodium phosphate (pH 7.5) and 45 μL of HPLC water. Each sample was then digested with 2 μL PNGase F at 37 °C for 1 hour. The released N-glycans were separated from peptides on a second C18 SPE using 5% ACN/0.1 TFA as elution buffer. When required, N-glycans were further reduced to the corresponding alditols by the addition of 200 μL of NaBH4 (10 mg/mL in 0.01M NaOH). The reduced N-glycans were purified using hand-packed cellulose cartridges [15] and permethylated [31] for MSn analysis.
Mass Spectrometry
MALDI-MS was carried out on a MALDI-TOF instrument (MALDI-CFR, Shimadzu), with a nitrogen laser at 337 nm. External calibration was performed using the ProteoMass Peptide MALDI-MS calibration kit (Sigma-Aldrich). The mawell fluorescent plate. The trix solution was prepared by dissolving 10 mg of DHB in a volume of 1 mL of 50% acetonitrile containing 2 mM sodium acetate. N-Glycans were directly spotted onto a stainless steel plate and mixed with an equal volume of matrix solution (0.5 - 1 μL). The MALDI-MS spectra were acquired in the positive reflectron mode from 600 to 5000 Da. The laser energy (power) was manually adjusted to obtain best signals (80 - 120). A minimum of 1500 scan were averaged for each spectra using the Spectrum Contents within the application Launchpad (v2.8.4, Shimadzu Biotech). ESI-MS and ESI-MSn of permethylated N-glycans were carried out on a Thermo linear ion trap instrument LTQ (Thermo Fisher Scientific) equipped with direct chip-based infusion (Triversa Nanomate). The ESI source voltage was set at 2.0 - 2.5 kV, and the capillary temperature at 230 °C. MS spectra were acquired from m/z 500 - 1800 with a minimum of 30 scan. MS/MS and MSn spectra were performed using a normalized collision energy at 35%, activation Q at 0.25, and activation time of 30 ms. All scans of one spectrum were accumulated by Xcalibur 2.0.
Neuraminidase Activity Assay
The activity assay of previously described [11] was followed. The NA enzyme was first diluted with an assay buffer (50 mM Tris, 5 mM CaCl2 and 200 mM NaCl at pH 7.5) to a concentration of 1 ng/μL. To start the reaction, 50 μL of the diluted enzyme was mixed with 50 μL of 400 μM substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (Sigma-Aldrich) in a 96-well fluorescent plate. The reaction kinetics was monitored by fluorescence at excitation of 365 nm and emission of 445 nm in a SpectraMax Multi-Mode Microplate Reader (Molecular Device).
In vitro Enzymatic Fucosylation
Enzymatic fucosylation was carried out by combining 1μg of substrate (dimer or tetramer NA) with either 2 μL of GDP-3H-fucose (1.2 μM from Perkin Elmer) or 5 nmol of GDP-fucose, and 0.5 μg of recombinant human FUT8 in 20 μL of 10 mM Tris (pH 7.0) and 10 mM MnCl2. The reaction was then incubated at room temperature for required length of time.
Filter Binding Assay
Following 3H-fucose incorporation, 8 μL from each reaction was spotted on a type GF/C glass fiber (Fisher Scientific). Ethanol (75%) was dropped to these spots to denature the protein. The filters were dried and then thoroughly washed in 200 ml of water in a shaker for 5 minutes. The filters were finally counted with an LS 6500 scintillation counter in Ready Protein™ cocktail (Beckman Coulter).
Probing the Core-6 Fucose on the Two Forms of NA with Azido Fucose
The general procedure of glycoprotein labeling with click chemistry (GLCC) [21] was followed to probe core-6 fucosylation with recombinant human FUT8. The dimer NA at 95 ng/μl or the tetramer NA at 70 ng/μl was incubated with 30 ng/μL of FUT8, or 40 ng/μl of FUT11, plus 0.08 mM GDP-azido-fucose in 25 mM Tris (pH 7.5), 150 mM NaCl, and 4 mM MnCl2 at 37°C for 90 minutes. To initiate the click chemistry reaction, 100 μM biotinylated alkyne, 100 μM CuCl2, and 2 mM ascorbic acid were added to the mixtures, and followed by incubation at room temperature for 1 hour. One μg each of fucosylated NA samples was loaded per well onto a 12% SDS-PAGE gel containing 2,2,2-Trichloroethanol. The gel was run at 50 mA, imaged via the method of Ladner, C.L. et al.[32], and further electro-transferred onto nitrocellulose membrane at 25 volts for 30 minutes. The blot was then blocked with 10% milk, washed thoroughly with TBS buffer (25 mM Tris, pH 7.6, 137 mM NaCl and 0.01% Tween), incubated with 75 ng/mL streptavidin-HRP in 25 mM Tris, 150 mM NaCl, pH 7.5 for 30 minutes, washed three times in TBS for a total of 30 minutes, incubated with ECL chemiluminescent substrate (Thermo Scientific) briefly, and then exposed to an x-ray film for 20 seconds.
Supplementary Material
Expressed 1918 pandemic influenza viral neuraminidase has inactive and active forms.
The inactive form contains high level of core-6 fucose, while the active form lacks such modification.
Core fucose could interfere the oligomerization of the neuraminidase and thus its activation.
This discovery may explain why 1918 pandemic influenza caused higher death rate among young population.
Acknowledgments
Financial assistance was provided by NIH grants RO1 GM54045 (VNR), and Glycan Connections, LLC. We would like to thank Dr. Gregg Hickey of Bio-techne for proof reading of this manuscript and thank numerous coworkers at Bio-techne who made contributions through product development.
Abbreviations
- HA
hemagglutinin
- NA
neuraminidase
- Hex
hexose
- HexNAc
N-acetyl hexosamine
- Fuc
fucose
- Man
mannose
- GlcNAc
N-acetyl glucosamine
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- ESI
electrospray ionization
- MSn
sequential mass spectrometry fragmentation (n>2)
- FUT8
fucosyltransferase 8
- FUT11
fucosyltransferase 11
- RFU
relative fluorescent unit
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 2.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls H. Pandemic influenza: the inside story. PLoS Biol. 2006;4:e50. doi: 10.1371/journal.pbio.0040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Qi J, Shi Y, Li Q, Gao F, Sun Y, Lu X, Lu Q, Vavricka CJ, Liu D, Yan J, Gao GF. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 2010;1:459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 7.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(4):D49–53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol. 2008;82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 10.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 11.Wu ZL, Ethen C, Hickey GE, Jiang W. Active 1918 pandemic flu viral neuraminidase has distinct N-glycan profile and is resistant to trypsin digestion. Biochem Biophys Res Commun. 2009;379:749–753. doi: 10.1016/j.bbrc.2008.12.139. [DOI] [PubMed] [Google Scholar]
- 12.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Stanley P, Schachter H, Taniguchi N. N-Glycans. Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 14.Zhou H, Warren PG, Froehlich JW, Lee RS. Dual modifications strategy to quantify neutral and sialylated N-glycans simultaneously by MALDI-MS. Anal Chem. 2014;86:6277–6284. doi: 10.1021/ac500298a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Hanneman AJ, Chasteen ND, Reinhold VN. Anomalous N-glycan structures with an internal fucose branched to GlcA and GlcN residues isolated from a mollusk shell-forming fluid. J Proteome Res. 2013;12:4547–4555. doi: 10.1021/pr4006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarentino AL, Plummer TH., Jr Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- 18.Ihara H, Ikeda Y, Toma S, Wang X, Suzuki T, Gu J, Miyoshi E, Tsukihara T, Honke K, Matsumoto A, Nakagawa A, Taniguchi N. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology. 2007;17:455–466. doi: 10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- 19.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Spiteri C, Moses JE. Copper-catalyzed azide-alkyne cycloaddition: regioselective synthesis of 1,4,5-trisubstituted 1,2,3-triazoles. Angew Chem Int Ed Engl. 2010;49:31–33. doi: 10.1002/anie.200905322. [DOI] [PubMed] [Google Scholar]
- 21.Wu ZL, Huang X, Burton AJ, Swift KA. Glycoprotein labeling with click chemistry (GLCC) and carbohydrate detection. Carbohydr Res. 2015;412:1–6. doi: 10.1016/j.carres.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 22.Wu ZL, Huang X, Burton AJ, Swift KA. Probing sialoglycans on fetal bovine fetuin with azido-sugars using glycosyltransferases. Glycobiology. 2016;26:329–334. doi: 10.1093/glycob/cwv109. [DOI] [PubMed] [Google Scholar]
- 23.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281:2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 26.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: detailed analyses of oligosaccharide structures. Proteomics. 2008;8:3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 28.Vanhooren V, Dewaele S, Kuro OM, Taniguchi N, Dolle L, van Grunsven LA, Makrantonaki E, Zouboulis CC, Chen CC, Libert C. Alteration in N-glycomics during mouse aging: a role for FUT8. Aging Cell. 2011;10:1056–1066. doi: 10.1111/j.1474-9726.2011.00749.x. [DOI] [PubMed] [Google Scholar]
- 29.Taubenberger JK. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc. 2006;150:86–112. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Froehlich JW, Briscoe AC, Lee RS. The GlycoFilter: a simple and comprehensive sample preparation platform for proteomics, N-glycomics and glycosylation site assignment. Mol Cell Proteomics. 2013;12:2981–2991. doi: 10.1074/mcp.M113.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 32.Ladner CL, Yang J, Turner RJ, Edwards RA. Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal Biochem. 2004;326:13–20. doi: 10.1016/j.ab.2003.10.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.