Abstract
Human biofluids, especially blood plasma or serum, hold great potential as the sources of candidate biomarkers for various diseases; however, the enormous dynamic range of protein concentrations in biofluids represents a significant analytical challenge for detecting promising low-abundance proteins. Over the last decade, various immunoaffinity chromatographic methods have been developed and routinely applied for separating low-abundance proteins from the high- and moderate-abundance proteins, thus enabling much more effective detection of low-abundance proteins. Herein, we review the advances of immunoaffinity separation methods and their contributions to the proteomic applications in human biofluids. The limitations and future perspectives of immunoaffinity separation methods are also discussed.
Keywords: Biofluid, immunoaffinity chromatography, plasma/serum, immunodepletion, immunoenrichment, biomarker discovery, proteomics, affinity proteomics
1. Introduction
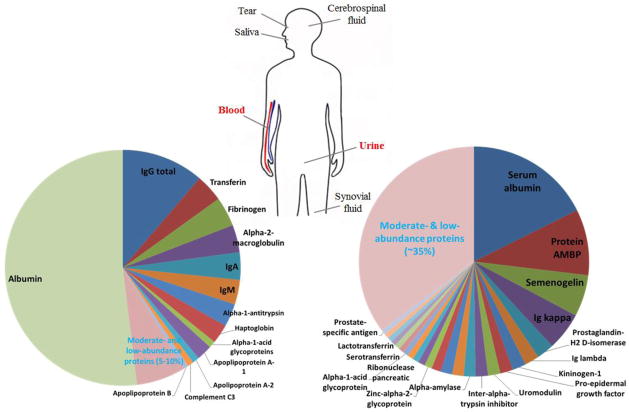
Human biofluids are biological fluids that are excreted or secreted from inside the bodies of living people, including but not limited to blood, urine, cerebrospinal fluid (CSF), saliva, tear, and synovial fluid (See Figure 1). Human biofluids, especially blood plasma/serum and urine, are considered the most promising sources for the discovery of novel biomarkers for disease diagnosis and prognosis based on the notion that these biofluids contain disease-associated proteins secreted or leaked from pathological tissues across the body [1–3]. Comparing to other types of specimens such as tissues, biofluids are often easily obtainable through noninvasive procedures, making it particularly attractive for large-scale clinical and/or longitudinal studies. For these reasons, there has been tremendous interest in profiling the biofluid proteomes for the development of biomarkers for various diseases over the last decade [4–10].
Figure 1.
Biofluids in human. Main sources and origins of human biofluids are depicted; the compositions of protein mass for both plasma/serum (data from in-house protein identification result and protein abundances were based on spectral count) and urine (data from Li et al. [162]) are shown in pie charts on the left and right hand side, respectively.
Blood plasma or serum has been the most popular choice for biomarker development and verification/validation. One of the unique challenges in proteome profiling of plasma/serum lies in its extremely large dynamic range of protein concentrations (up to 10–12 orders of magnitude [4]). The 12 most abundant proteins (e.g., albumin, transferrin, immunoglobulins, etc) account for ~95% of total protein mass in plasma or serum, which leaves thousands of other moderate- and low-abundance proteins (MAPs and LAPs) in only 5% of total protein mass (See Figure 1). The “masking” effect caused by these high-abundance proteins (HAPs) greatly hampers the detection of LAPs such as cytokines and other clinically important proteins that often present at the sub ng/ml level. Besides blood, urine is also an ideal source for biomarker discovery and has been utilized in more and more proteomics studies [9, 11, 12]. Urine is formed in the kidney by ultrafiltration from the blood, which can be easily accessed and collected in large amounts via non-invasive approaches [13]. Comparing to blood, the composition of urine is less dominated by HAPs, which provides a relatively easier access to LAPs (See Figure 1). Other biofluids, such as cerebrospinal fluid, saliva, tear, and synovial fluid, have also been analyzed for potential biomarkers; however in much more limited number of studies comparing to blood and urine [14–16].
Immunoaffinity chromatography (IAC) approaches have become the most commonly utilized strategies for digging deeper into the biofluid proteomes by both global and targeted proteomics [17–19]. IAC represents a specific type of affinity chromatography where the stationary phase is composed of immobilized antibodies or other affinity reagents on solid support matrix. The underlying principle of IAC is based on the selective non-covalent interaction between antibodies (or affinity reagents) and their specific binding targets or antigens. The purpose of IAC separations is to enrich LAPs of interest by either removing HAPs from the complex samples through immunoaffinity depletion (immunodepletion) or directing capturing low-abundance targets of interest through immunoaffinity enrichment (immunoenrichment). Significant advances in IAC methods for both the depletion and enrichment have been made and these methods have been broadly utilized in proteomics applications for many types of biofluids related to various human diseases [18, 20].
2. Overview of Immunoaffinity Chromatography
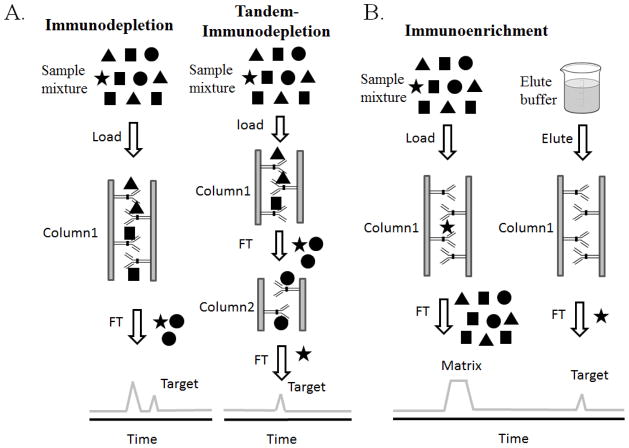
The most common schemes for applying IAC either by immunodepletion or immunoenrichment are illustrated in Figure 2. In these schemes, chromatographic matrices (column) or other resins with immobilized antibodies are used to specifically capture target proteins/peptides, and the resulting flow-through fraction (immunodepletion) or bound fraction (immunoenrichment) is collected for further analyses [21, 22]. The immunodepletion strategy is designed to remove the HAPs and enrich LAPs on a global scale [23]. In immunodepletion, complex samples as plasma/serum are first loaded onto a depletion column, and only specific HAPs are selectively captured by the antibodies immobilized on column, while other LAPs flow through directly and are collected (See Figure 2A). To maximize the detection of LAPs, simultaneous removal of multiple HAPs is desired. Therefore, multiple antibodies are often mixed and immobilized onto column in order to remove multiple HAPs. Furthermore, immunodepletion columns targeting different proteins/peptides can also be used in tandem to enable the removal of a relatively large number of HAPs [21]. On the other hand, the immunoenrichment (or immunoaffinity purification) strategy is more targeted towards specific analytes (e.g., low-abundance proteins, peptides, or PTMs) to enhance the detection of these specific target low abundance analytes of interest, which is ideally suited for coupling with targeted proteomics measurements [24, 25]. In immunoenrichment, only target low-abundance analytes are recognized and bound on column, while all other proteins/peptides are washed away, and the targeted proteins/peptides are further eluted from column for analysis (See Figure 2B).
Figure 2.
The main workflows of immunodepletion (A) and immunoenrichment (B). For immunodepletion, the Star and Circles represents LAPs and MAPs, respectively. For Immunoenrichment, the Star represent the target molecules and other symbols are sample matrix.
The utility of IAC is highly dependent on the quality of affinity reagents, typically antibodies [19]. A “good” antibody for IAC should meet two requirements: (a) a high intrinsic affinity toward target protein, and (b) reversible interactions between antibody and antigen that can be easily de-stabilized. Two main types of antibodies are commonly used in IAC, namely polyclonal and monoclonal antibodies [17, 19]. Polyclonal antibodies are produced as a heterogeneous population of antibodies from multiple clones of B-cells, which can recognize and bind a variety of epitopes on a single antigen with diverse affinity [26]. Sera of immunized animals are the main sources of polyclonal antibodies. Due to their easy accessibility and relative low cost, polyclonal antibodies are widely used for developing IAC methods. However, there are a number of limitations in using polyclonal antibodies. Firstly, sera are usually available in limited supply and could vary from animal to animal. Therefore, it is difficult to obtain consistent quality of antibodies from multiple batches or lots. To obtain high quality antigen-specific antibodies, polyclonal antibodies must be purified against the target antigen via affinity chromatography [19]. However, it is often impractical to acquire a large amount of purified antigen proteins for this purpose. Alternatively, anti-peptide antibodies have been developed to selectively capture the proteolytic peptides of target proteins for their quantification in human biofluids, which is exemplified in the SISCAPA approach (Stable Isotope Standards and Capture by Anti-Peptide Antibodies) [24, 27]. In such case, synthetic peptides conjugated with a carrier protein are used to immunize host animals. Antibodies targeting unique epitopes (or peptide-specific) are produced to specifically recognize the proteolytic peptides. Compared to proteins, synthetic peptides are commercially available in highly purified forms and in large quantities, facilitating the production of a peptide-specific antibody population for IAC analysis. Another limitation of using polyclonal antibodies is that the polyclonal antibodies may consist of a number of antibodies targeted at different epitopes of the same antigen, which makes it difficult to find a favorable eluting condition to simultaneously destabilize multiple types of antigen-antibody interactions that are different in nature [28]. Therefore, potential lack of quantitative elution and low recovery of target protein should be taken into consideration in polyclonal antibody-based IAC applications. For IAC applications in human biofluids for proteomic profiling, polyclonal antibody-based IAC methods have been mainly applied for the selective depletion of HAPs and MAPs where high specificity of capture is preferred over quantitative elution (Table 1).
Table 1.
HAPs in plasma targeted by the commonly commercially available immunoaffinity depletion columns.
| Vivapure | Qproteome | ProteoPrep | Mars Hu-6 | Mars Hu-14 | Seppro IgY-14 | ProteoPrep 20 | |
|---|---|---|---|---|---|---|---|
| Albumin | √ | √ | √ | √ | √ | √ | √ |
| IgG | √ | √ | √ | √ | √ | √ | |
| Transferrin | √ | √ | √ | √ | |||
| Fibrinogen | √ | √ | √ | ||||
| IgA | √ | √ | √ | √ | |||
| a-2-Macroglobulin | √ | √ | √ | ||||
| IgM | √ | √ | √ | ||||
| a-1-Antitrypsin | √ | √ | √ | √ | |||
| Complement C3 | √ | √ | √ | ||||
| Haptoglobin | √ | √ | √ | √ | |||
| Apolipoprotein A-I | √ | √ | √ | ||||
| Apolipoprotein A-II | √ | √ | √ | ||||
| Apolipoprotein B | √ | √ | |||||
| a-1-Acid Glycoprotein | √ | √ | √ | ||||
| Transthyretin | √ | ||||||
| Ceruloplasmin | √ | ||||||
| Complement C4 | √ | ||||||
| Complement C1q | √ | ||||||
| IgD | √ | ||||||
| Prealbumin | √ | ||||||
| Plasminogen | √ | ||||||
| Number of depleted HAPs | 1 | 2 | 2 | 6 | 14 | 14 | 20 |
| Vendors | Sartorius | QIAGEN | Sigma Aldrich | Agilent | Agilent | Sigma Aldrich | Sigma Aldrich |
In contrast, monoclonal antibodies are typically produced by fusing myeloma cells with the spleen cells from a mouse that has been immunized with the desired antigen to produce hybridomas. Very high affinity monoclonal antibodies can be identified by screening large numbers of hydridoma supernatants since many therapeutic monoclonal antibodies have reported affinities of ~10−9 M or better [29]. Automated screening of monoclonal antibodies using MS-based platform has also been recently reported [30]. The monoclonal antibodies constitute a homogeneous population that has monovalent affinity since they bind to the same epitope on an antigen [26]. Compared to polyclonal antibodies, monoclonal antibodies have the advantages of consistent specificity and reproducibility. Since all antibodies bind to the same epitope, it is relatively easy to find a gentle elution condition for IAC [19]. Therefore, monoclonal antibodies are preferred for immunoaffinity enrichment applications. Besides antibodies, other affinity reagents such as immobilized metal ions [31], metal oxides [32], lectins [33], and aptamers [34] have also been applied for affinity chromatography-based applications.
In a typical IAC approach, antibodies are immobilized on a solid stationary phase to facilitate the selective and strong binding of antibodies to their target antigens, as well as the subsequent washing (if desired) and elution processes. Conventional matrices for immobilization of antibodies include natural carbohydrate-based materials (e.g., agarose, dextrose and cellulose), synthetic organic polymers and inorganic materials, such as silica and zirconia [17]. Cross-linked agarose is the most popular matrix for IAC, due to its excellent biocompatibility and extraordinary chemical stability in a broad pH range and most solvents. However, agarose beads and other soft gel matrices are not compatible to a highly pressurized system, such as HPLC. In contrast, silica, polystyrene and other highly cross-linked materials have good mechanical strength and stability, thus can be used in a HPLC-based IAC system, namely HPIAC [17]. Various commercial products and kits have been developed over the years for immunoaffinity separation of proteins in human biofluids (Table 1). Immunoaffinity columns are commonly produced with immobilized mammalian IgG or avian immunoglobulin yolk (IgY) antibodies. These two kinds of antibodies can be produced in a relatively high quantity from serum or eggs of immunized animals [19]. IgY antibodies have several advantages over IgG including higher antibody yield and less cross-reactivity toward non-targeted human proteins [18, 35].
3. Immunoaffinity Chromatography Approaches
In the following sections, we will discuss different IAC approaches with some details focusing on immunodepletion and immunoenrichment.
3.1 Immunoaffinity depletion
Immunodepletion of HAPs has become a relatively routine sample preparation strategy for biofluid proteome profiling, in which multiple antibodies are mixed in an optimized ratio and immobilized on solid matrices for removing multiple HAPs simultaneously [18, 36]. While immunodepletion kits in spin column formats, such as Vivapure anti-HAS kit (Sartorius) for removing albumin, Qproteome (Qiagen) for removing albumin and IgG, have been available for several decades now, Pieper et al. were the first to report on the concept of multi-component immunoaffinity subtraction chromatography in an LC column format for reproducible removal of up to 10 plasma HAPs to enhance plasma proteome profiling in 2003 [37]. Two commercial LC column products, Multi-affinity removal system (MARS) Hu-6 by Agilent [36] and ProteomeLab™ IgY12 by Beckman Coulter [35], were shortly made available for removal of 6 and 12 HAPs in blood plasma/serum, respectively; these two immunodepletion columns were further improved into the MARS Hu-14 kit (Agilent) and the Seppro® IgY14 system (Sigma Aldrich), respectively, for removal of top 14 HAPs. Similarly ProteoPrep® 20 (Sigma Aldrich) was developed for removing 20 HAPs in plasma. The detailed list of current commercially available immunoaffinity depletion systems were provided in Table 1. Compared to spin columns, the LC column-based products utilizing automated LC systems provide a number of advantages in effective removal of targeted proteins, such as minimal carryover, good reproducibility, and minimal nonspecific binding [38]. Besides these single-stage depletion systems, an IgY-based SuperMix depletion column has been developed to enable the removal of ~50 MAPs by applying it with IgY12 or IgY14 column in tandem to further enrich LAPs prior to follow-up analysis [21, 39]. In our experience, a typical LC depletion column will offer reproducible depletion for 100–200 biological samples with a shelf life for several years, which provides a great potential for large-scale biomarker discovery and verification studies.
3.1.1 Multi-affinity removal system (MARS)
The MARS column from Agilent Technologies was the first commercially available multi-component immunoaffinity depletion system [36, 40]. Initially, this column consisted of 6 polyclonal IgG antibodies for 6 HAPs including albumin, IgG, IgA, transferrin, α-1-antitrypsin and haptoglobin (so called MARS Hu-6) [41]. Antibodies were immobilized onto column through their Fc regions, which ensured easy protein access to the affinity binding sites with reported depletion efficiency higher than 99% for each target protein [40]. MARS Hu-6 was applied to many proteomics applications in biofluids [42–44]. Later on, MARS Hu-7 column was found to deplete fibrinogen plus the original six HAPs [45]. The most recently product of MARS is the Hu-14 column which allows the depletion of 8 more HAPs including fibrinogen, α-acid glycoprotein, α-macroglobulin, IgM, apolipoproteins A-I & A-II, complement C3 and pre-albumin, approximately 95% of the human plasma proteins [46]. The MARS Hu-14 depletion has also been widely used in recent proteomic applications, including plasma [46–53], urine [54], CSF [55–57], and tissue proximal fluids [58].
3.1.2 IgY-based single-stage and dual-stage depletion systems
The IgY12 depletion system based on avian polyclonal IgY antibodies was developed shortly after the MARS Hu-6, which initially targeted 12 HAPs [20, 35, 59]. The IgY12 system was later improved to IgY14 for removing 14 HAPs in human plasma and the product is now commercialized as Seppro® IgY14 from Sigma Aldrich [39, 60–62]. Both IgY14 and MARS Hu-14 are very popular depletion products for proteomics applications since the performance characteristics of MARS Hu-14 and IgY14 are very comparable with both products offering options of multiple loading capacities (customization options available as well) [63]; however, the IgY antibodies appeared to display the least nonspecific binding [64]. Similar to MARS Hu-14, IgY-14 was broadly applied to proteomics studies, including plasma [65–68] and CSF [69, 70].
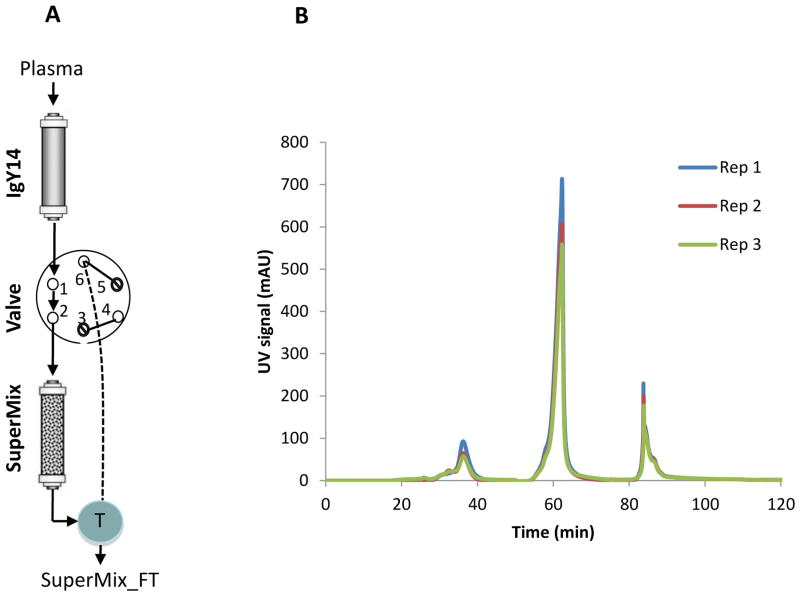
The concept of a SuperMix column was later developed to be applied in tandem with IgY12 or IgY14 so that the detection of LAPs can be further enhanced by depleting an additional number of MAPs [21, 39, 71]. The SuperMix column was developed by immunizing chickens with the protein mixture from IgY14-depleted human blood plasma as mixed antigens, and the purified antibodies were further immobilized and packed into the SuperMix column to target a relatively large number of MAPs [21]. Figure 3 shows an example of the configuration of tandem IgY14 and Supermix depletion and overlays of representative LC chromatograms of three replicates of a reference plasma sample [39]. In our original characterization of the system, we observed that the SuperMix depleted at least 50 MAPs from plasma [21], providing significant enhancement for the detection of LAPs (e.g., a 100-fold enrichment of LAPs comparing to the 20-fold enrichment by the IgY12 alone).
Figure 3.
Immunodepletion using the tandem IgY14-SuperMix configuration. (A) IgY14 and SuperMix columns are connected through a 6-port valve. By switching the valve positions, the SuperMix flow-through (SuperMix_FT), IgY14 bound, and SuperMix bound fractions can be collected consecutively. (B) Representative LC chromatograms of 3 replicates of the tandem IgY14-SuperMix separations during the initial, middle and late stage of the column life of a reference plasma sample.
All the depletion columns (MARS Hu-14, IgY-14, or SuperMix) can be easily implemented and automated using conventional HPLC with the protocol of using the column typically consisting of loading the sample, collecting the flow-through proteins (LAPs), washing, eluting bound proteins, and column regeneration. In our experience, a typical LC depletion column will offer reproducible depletion for 100–200 biological samples with a shelf life for several years, which provides a great potential for large-scale biomarker discovery and verification studies. More recently, a microscale chromatographic depletion system using IgY-14 resin was reported to facilitate more effective depletion of small amounts of human CSF or serum samples, where only ~6 μL serum or 600 μL CSF were used [72].
3.1.3 Combinatorial peptide ligand library (CPLL)
The concept of combinatorial peptide ligand library approach, first commercialized by Bio-Rad Laboratories as ProteoMiner kit, is that the library consists of millions of hexapeptides capable of interacting with most proteins in any given proteome. In principle, each unique hexapeptide binds to a unique protein sequence. Because the bead capacity limits binding capacity, high-abundance proteins quickly saturate their ligands and excess proteins are washed out during the procedure. On the contrary, LAPs are captured on their specific ligands, thereby leading to reduced dynamic range of the biofluid proteome or so called “equalizing of protein concentrations” [73–76]. Several studies have explored the applications of ProteomeMiner in proteome profiling [77–80]. The ProteoMiner kit and depletion columns are conceptually different and complementary. The depletion columns achieve early complete removal of highly abundant proteins, but will suffer non-specific loss of other LAPs. On the other hand, ProteoMiner only offer partial removal of HAPs and reduce the dynamic range given the “equalizing” principle. In a recent study by Gil-Dones et al. [76], it was clearly demonstrated that ProteoMiner was not as effective for enhancing the detection of LAPs as depletion columns. For better depletion of HAPs and enrichment of LAPs, more details such as sample overloading degree, the way of peptide ligand grafted on bead, and chemical modification of peptide ligand library, should be optimized [75].
3.2 Immunoaffinity enrichment
Immunoaffinity enrichment, also called immunoaffinity purification, has been broadly applied as traditional biochemical approaches for enrichment of specific protein targets for decades. In particular, immunoaffinity enrichment approaches have found broad applications in targeted proteomics. For example, the coupling of antibody-based enrichment and targeted mass spectrometry has become a routine approach for developing so called mass spectrometric immunoassays (MSIA) [81, 82]. Many types of affinity reagents have been developed to enable different types of proteomic applications. The affinity reagents can be used to capture specific target proteins by antibodies, or specific classes of proteins such as glycosylated proteins by using lectins [33] or hydrazide resins [83]. The affinity reagents can also be applied to target specific peptides by anti-peptide antibody [24], specific types of PTMs including phosphorylation by immobilized metal ions [31], and metal oxides [32], and other PTMs [84] by antibodies, and specific type of peptides such as cysteine-containing peptides using thiol-affinity resins [85, 86]. Herein, we discuss several major types of immunoaffinity reagents for enrichment other than full antibodies.
3.2.1 Protein ligands
Some natural or recombinant proteins have specific recognition and binding capacity of certain target proteins. For example, bacterial Protein A and Protein G are ideal ligands for immunoglobins by binding to the Fc regions of IgG [87]. A type of recombinant protein ligand, known as Affibody, has also been developed by mimicking or complementing the structures and binding activities of an antibody [88, 89]. Lectins are glycoproteins that have higher affinity for sugar moieties in glycoproteins [90]. The multi-lectin affinity chromatography (M-LAC) has been developed as an effective approach for enriching and analyzing glycoproteins from human biofluids [33, 91]. For instance, M-LAC has been coupled with depletion and other separations to achieve a deep coverage of the plasma proteome by LC-MS/MS [91, 92].
3.2.1 Single chain/domain antibodies
Single chain/domain antibody is another type of protein ligand that consists of a fragment of antibody from a single chain or a single domain [93, 94]. Single chain antibody (Single chain Fv, scFv) was the minimal form of functional antibodies, which comprised of the VH and VL segments in a single polypeptide chain joined by a short flexible linker, and each V-region has three complementarity determining regions (CDRs) in direct contact with antigen; while single domain antibody was an antibody fragment which consisted of a single monomeric variable antibody domain [95]. Single chain/domain antibodies demonstrated several advantages over the traditional whole antibody, including the ease of cloning in bacteria, minimally immunogenic, and better tissue penetration due to the smaller size and better stability [96]. Especially, the advance in phage display technology further facilitates the production of single chain antibodies for proteome research and other applications [97, 98].
Phage display is a technology that enables the detection of interaction between the displayed protein and other test molecules, including protein, peptide and DNA, which could be used for selection of proteins, peptides, or antibodies with affinity and specificity to a molecule or protein of interest from phage libraries [99]. The construction of a phage display library (could be a protein library, peptide library, antibody library) was accomplished by inserting DNA fragments into phage or phagemid genomes which proteins were further expressed on the phage coat. Specifically, the technique was named antibody phage display (APD) when an antibody was displayed, which allowed in vitro selection of antibodies of virtually any specificity, greatly facilitating recombinant production of antibody reagents to create antibody libraries [100]. The use of single-chain antibody library or antibody microarray to screen human serum to discover novel cancer biomarkers has been reported [101, 102].
3.2.3 Anti-peptide or anti-PTM antibodies
The development of immunoaffinity enrichment approaches to targeted specific peptides or specific types of PTMs is another emerging area in proteomics. The first demonstration of using anti-peptide antibodies was reported as the “Stable Isotope Standards and Capture by Anti-Peptide Antibodies” (SISCAPA) approach [24]. SISCAPA has been advanced to become an important technology for enabling the detection of LAPs in biofluids by targeted MS without the need of extensive LC fractionation [25, 103]. SISCAPA also combines the power of peptide level immunoaffinity enrichment with the analytical capability of selected or multiple reaction monitoring (SRM/MRM, which enables sensitive quantitation of corresponding protein biomarkers and targets [103, 104]. Although not extensively applied to biofluids yet, immunoaffinity enrichment using antibodies against specific types of PTMs has also been developed and extensively applied in proteomics studies, including protein phosphorylation [105, 106], ubiquitination [107, 108], acetylation [109, 110], and lately methylation [111].
4. Proteomic applications of IAC in biofluids
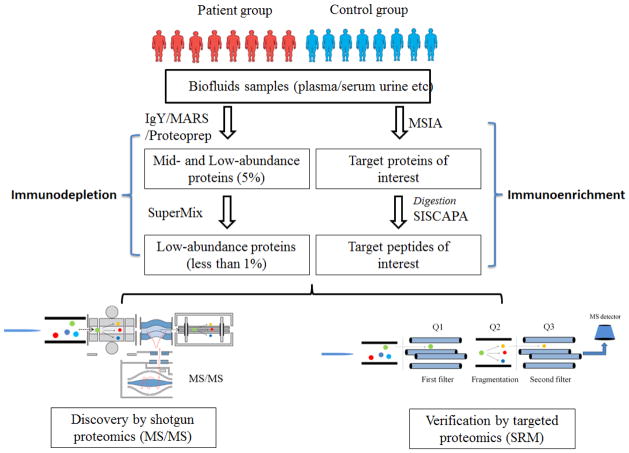
Figure 4 summarizes a general workflow for applying IAC methods in proteomics profiling of human biofluids samples. For any biomarker discovery or verification studies, the experimental design will be the key for its chance of success. As recommended in a recent publication, clearly defined clinical question, selection of subjects, sufficient demographic data, and appropriate statistical approaches including adequate sample size are all important factors in the experimental design [10]. Depletion of HAPs is often required for plasma/serum samples, which can be performed by HPLC using IgY/MARS/ Proteoprep columns. After removal of HAPs, the remaining MAPs and LAPs account for about 5–10% of the original protein mass (~20-fold enrichment of LAPs). If further removal of MAPs is required (typically in plasma/serum), the SuperMix column can be used, which results in a flow-through of ~1% of original protein mass corresponding to ~100-fold enrichment of LAPs. In contrast, urine is less dominated by HAPs. However, pre-concentration methods, such as ultrafiltration, protein precipitation, dialysis, lyophilization, and ultracentrifugation, need to be applied to concentrate urine proteins prior to down streaming processing for proteome analysis [13, 112]. Among these methods, ultrafiltration has been reported to be the best method for concentration and cleanup of protein components from urine [112]. Besides depletion, enrichment strategies such as M-LAC for glycoproteins [91] and SISCAPA [25] can also be applied to enrich LAPs either independently or in combination with the depletion strategies. The enriched samples are then analyzed either by shotgun LC-MS/MS proteomics for discovery studies or by LC-SRM based targeted proteomics for verification studies. In the following sections, we review some examples of applications using different biofluids.
Figure 4.
General workflow of applying immunoenrichment and immunodepletion for a biomarker related study. Biofluid samples (e.g., plasma/serum or urine) are subjected to immunodepletion or immunoenrichment to reduce sample complexity. Immunodepletion is used to remove high abundant species, such as HAPs and MAPs, prior to MS-based proteomic studies. Target proteins or peptides can be selectively enriched through immunoenrichment by using MSIA or SISCAPA approaches, which are especially useful for targeted proteomics.
4.1 Plasma/Serum
Plasma/serum is one of the most well studied and characterized biofluids for biomarker discovery and validation. Significant effort and progress has been made in extending and characterizing the plasma/serum proteome using various IAC approaches. The initial application of multi-component depletion by Pieper et al. demonstrated significant enhancement of the detection of plasma LAPs by 2-DE following the removal of major proteins [37]. Liu et al. demonstrated that enhanced detection of LAPs in plasma could be achieved with high reproducibility and specificity by employing immunodepletion strategy [38]. The comparison between different immunodepletion columns has also been carried out [113, 114]. Polaskova et al. compared six different immunodepletion columns by using 2-DE, in which Seppro IgY system demonstrated the best results [114]. To further enhance the detection and coverage of LAPs, Qian et al. reported the application of tandem IgY12 and SuperMix depletion to human plasma by successfully removing up to top 60 abundant proteins in plasma, demonstrating nearly doubled plasma proteome coverage compared to single stage depletion and the effective detection of many LAPs with reported normal concentrations of approximately 100 pg/ml to 100 ng/ml [21]. Moreover, the IAC methods are very flexible to be coupled with downstream fractionation, enrichment, or detection techniques such as 2-DE or 1D or 2D-LC-MS/MS for digging deeper into the plasma proteome. For instance, a number of other techniques, including Cysteine-peptide enrichment [115], N-linked glycopeptide enrichment [116], multi-lectin affinity chromatography [91], off-gel electrophoresis [117], isoelectric focusing [91], have been coupled with immunoaffinity depletion for more extensive plasma proteome profiling.
Besides global profiling, the immunodepletion strategy has been extensively applied as a useful tool in targeted quantification studies of low-abundance plasma proteins in plasma/serum [67, 118–124]. For example, Liu et al. developed a prostate specific antigen assay in serum by combining immunoaffinity depletion and SRM detection, which showed very good correlation with conventional immunoassays result from other independent clinical serum samples [118]. Keshishian et al. demonstrated multiplexed quantitative assays for LAPs with limits of quantification in 1–10 ng/mL range in plasma by coupling depletion with limited fractionation and targeted mass spectrometry [59]. Furthermore, Shi et al. developed a so-called “high-pressure, high-resolution separations coupled with intelligent selection and multiplexing” (PRISM) method for ultrasensitive detection of LAPs in plasma, which demonstrated accurate and reproducible quantification of proteins at concentrations at the 50–100 pg/mL levels in plasma by integrating by coupling IgY14 with PRISM and SRM [67]. In a recently large scale targeted quantification study of cancer-associated proteins in human plasma, Hüttenhain et al. demonstrated reproducible quantification of 182 proteins in MARS Hu-14 depleted plasma, spanning five orders of magnitude in abundance and reaching below a concentration of 10 ng/mL [121].
While immunodepletion is highly effective for enhancing proteome analysis of blood plasma/serum, concerns still exist for its applications in biomarker related studies, especially in the aspect of non-specific loss of LAPs due to the non-characteristic protein binding to HAPs or to antibodies. For example, Keshishian et al. demonstrated that troponin T was largely lost during IgY14 immunodepletion [120]. Yadav et al. systematically analyzed the bound HAPs fraction and showed that 101 proteins could be detected with high confidence, and suggested both bound and depleted fractions should be analyzed from immunodepletion for biomarker discovery using plasma/serum [125].
As an important alternative to immunodepletion, immunoaffinity enrichment has also been broadly applied to plasma/serum related biomarker studies. The utility of antibodies for developing high selective mass spectrometric, targeted immunoassays for clinical important proteins in human plasma/serum has been well demonstrated [81, 82, 126]. The development of peptide-level immunoenrichment as exemplified by SISCAPA has also clearly demonstrated the value for quantification of potential low-abundance biomarkers [24]. Following the initial work by Anderson et al., Whiteaker et al. developed an automated and multiplexed approach for quantification of potential protein biomarkers by combining multiplexed SISCAPA and MRM analysis [103]. SISCAPA was also applied to develop multiplexed assays for a known biomarker of cardiac injury (troponin I), and an emerging cardiovascular marker interleukin 33, which could be detected at 1–10 ng/mL in plasma [127]. SISCAPA has been demonstrated for multiplexing up to 50-plex and able to extend the limits of detection to sub-ng/ml range of protein concentration [103, 104]. Ramirez et al. recently also demonstrated the utility of single chain antibody array for detecting low-abundance cancer biomarkers in human serum [101]. By combining a single chain antibody library and protein microarray for screening ovarian cancer serum for novel potential biomarkers, they identified 19 promising scFvs and verified 6 top candidates using full-length antibodies [101].
The IAC approaches have been routinely coupled with both global discovery and targeted verification for disease-biomarker related studies [114, 128], and the applications include pancreatic cancer [124], ovarian cancer [129, 130], breast cancer [131, 132], renal cancer [133], gastric cancer [134], prostate cancer [135, 136], osteoarthritis [137–139], cardiovascular disease [120, 140], etc.
4.2 Urine
Urine perhaps represents one of the most non-invasive, easily accessible biofluids available for diagnostic or prognostic tests. A number of studies have successfully demonstrated that urinary proteins and metabolites are promising biomarkers for diseases, such as kidney disease, prostate cancer, breast cancer, diabetes, atherosclerosis and osteoarthritis [141, 142]. Compared to plasma/serum, the urine proteome has a more limited dynamic range of protein concentrations and is less dominated by HAPs. Nevertheless, a number of studies have applied immunoaffinity-based separation to extend the proteome coverage and search for potential biomarker candidates in urine. Adachi et al. combined the albumin depletion and LC-MS/MS, which successfully identified more than 1500 proteins in normal urine [143]. Furthermore, by utilizing immunodepletion coupled to HPLC-Chip-MS/MS detection, He et al. identified 1641 urinary proteins with high confidence and provided a panel comprising 18 urinary proteins to assess the human health status from urine samples of 100 male and 100 female healthy donors [144]. Martin-Lorenzo et al. evaluated two immunodepletion approaches including ProteoPrep Immunoaffinity Albumin and IgG Depletion Kit (Sigma) and MARS Hu-14, and they demonstrated that using OasisR HLB cartridge purification in combination with albumin depletion by ProteoPrep kit as the best option for urine proteome profiling from patients with proteinuric renal disease [145]. Magagnotti et al. compared three depletion strategies including IgY, MARS and a home-made depletion column for improving coverage of the human urine proteome, and showed IgY depletion followed by ethanol precipitation was the most efficient method for exploring urine proteome [146]. The immunoaffinity based separation has also been applied to various biomarker discovery and verification studies in urine, such as bladder cancer [54], osteoarthritis [147], diabetic nephropathy [148, 149], and renal transplant [150].
4.3 Other Biofluids
4.3.1 Cerebrospinal fluid (CSF)
CSF is a clear, colorless body fluid found in the brain and spine, and plays critical roles in many physiological processes in the central nervous system. CSF contains abundant small metabolites, peptides, and proteins. Changes in their concentrations directly reflect the internal milieu of the brain. Therefore, CSF is considered as a good candidate for the discovery of new biomarkers to improve early diagnosis of neurological diseases, such as Alzheimer’s disease [151, 152]. CSF has low protein content (e.g., ~0.35 mg/mL) but a wide dynamic range spanning at least 9 orders of magnitude. Due to the low total protein mass available from a typical CSF sample, Cunningham et al. applied a modified IgY14-based spin column to analyze CSF obtained from mice, which led to the identification of 289 proteins [153]. Furthermore, Hyung et al. designed a microscale depletion system using IgY14 resin, which could be used to analyze small volume of CSF and plasma with low flow rate [72]. By combining immunoaffinity separation with high sensitivity and resolution LC-MS/MS detection, Schutzer et al. first established a comprehensive proteome map for normal human cerebrospinal fluid, which consisted of 2630 proteins [154]; using the same analytical strategy they further compared the CSF proteomes of Neurologic Post Treatment Lyme disease (nPTLS) patient, Chronic Fatigue Syndrome (CFS) patient and normal healthy control, and reported on a number of CSF proteins that could be useful for distinguishing nPTLS and CFS [155]. Recently, by combining immunodepletion of 14 HAPs, high-pH reverse-phase separation and high resolution MS/MS detection, Zhang et al. constructed a most up-to-date proteome map for CSF, which consisted of 3256 non-redundant proteins [156]. Immunoaffinity separation has also been applied in CSF for biomarker studies of several other neurological diseases, such as Parkinson’s disease [157], Huntington’s disease [158].
4.3.2 Synovial fluid (SF)
Synovial fluid (SF) is a serum filtrate located among the joint, where it also receives protein contributions from the surrounding tissues, articular cartilage, synovial membrane and bone.
In order to identify potential biomarkers for rheumatoid arthritis and osteoarthritis, Mateos et al. applied immunoaffinity depletion followed by LC-MS/MS detection, which lead to the identification of 136 differential proteins [159]. More recently, by using immunoaffinity depletion combined with extensive fractionation and high resolution MS/MS detection, Balakrishnan et al. presented an in-depth analysis of the synovial fluid proteome from patients with osteoarthritis, which successfully detected 677 proteins [160].
5. Conclusion and perspectives
Significant advances have been made over the last decade in the IAC approaches in the aspects of both immunoaffinity depletion and enrichment. Given the challenge of the enormous dynamic range of protein concentrations in human biofluids, IAC has made tremendous contributions to enable more effective detection of LAPs in these biofluids and identify potential biomarkers for various disease conditions by both global proteome profiling and targeted quantification. The development of targeted MS immunoassays based on either anti-protein antibodies or anti-peptide antibodies has become much more automated and higher throughput [81, 103].
While IAC has become an important tool for proteomic analyses of biofluids, there are several caveats that need to be taken into consideration in the experimental design. First, there is a significant concern of non-specific or specific loss of LAPs during immunodepletion. This is especially the case for the SuperMix system since it contains a large number of different antibodies and could deplete or partially deplete many more proteins. Therefore, the SuperMix system was proposed as a fractionation approach rather than a depletion approach. Second, for immunoaffinity enrichment, the quality (specificity, binding affinity) of antibody is still critical. It is still costly to make a good antibody in terms of the time and cost for enriching specific targets. Third, another valid concern for IAC lies in the appropriate loading amount. Determining the optimal loading amount is essential for high efficient binding of target proteins which enables effective depletion as well as high enrichment efficiency. Finally, most IAC approaches are still quite labor intensive and only offer moderate throughput for sample processing. Despite of these concerns, we anticipate the field of immunoaffinity chromatography will continue to advance in the areas of novel affinity reagents, automation for high throughput IAC separations and assay developments, and integration of other advanced devices (e.g., microfluidics-based devices [161]), and mass spectrometry platforms to make even more significant contributions in both global proteomics analyses of human biofluids and large-scale high throughput targeted quantification of biofluid samples for biomarker verification and development.
Highlights.
The enormous dynamic range of protein concentrations in human biofluids represents a significant analytical challenge for discovering low-abundance protein biomarkers.
Immunoaffinity chromatography has become an essential method for enabling the detection of low-abundance proteins in human biofluids.
The advances of immunoaffinity chromatographic methods including immunodepletion and immunoenrichment are reviewed.
Proteomics applications of immunoaffinity approaches to human biofluids are highlighted.
Acknowledgments
Portions of this work were supported by the NIH grant U24-CA-160019 from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), NIGMS Biomedical Technology Research Resource P41GM103493. The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lathrop JT, Anderson NL, Anderson NG, Hammond DJ. Therapeutic potential of the plasma proteome. Curr Opin Mol Ther. 2003;5:250–257. [PubMed] [Google Scholar]
- 3.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 4.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 6.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 7.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 8.Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H. Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. J Proteome Res. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 9.Fliser D, Novak J, Thongboonkerd V, Argiles A, Jankowski V, Girolami MA, Jankowski J, Mischak H. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 10.Mischak H, Allmaier G, Apweiler R, Attwood T, Baumann M, Benigni A, Bennett SE, Bischoff R, Bongcam-Rudloff E, Capasso G, Coon JJ, D’Haese P, Dominiczak AF, Dakna M, Dihazi H, Ehrich JH, Fernandez-Llama P, Fliser D, Frokiaer J, Garin J, Girolami M, Hancock WS, Haubitz M, Hochstrasser D, Holman RR, Ioannidis JP, Jankowski J, Julian BA, Klein JB, Kolch W, Luider T, Massy Z, Mattes WB, Molina F, Monsarrat B, Novak J, Peter K, Rossing P, Sanchez-Carbayo M, Schanstra JP, Semmes OJ, Spasovski G, Theodorescu D, Thongboonkerd V, Vanholder R, Veenstra TD, Weissinger E, Yamamoto T, Vlahou A. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 11.Caubet C, Lacroix C, Decramer S, Drube J, Ehrich JH, Mischak H, Bascands JL, Schanstra JP. Advances in urinary proteome analysis and biomarker discovery in pediatric renal disease. Pediatr Nephrol. 2010;25:27–35. doi: 10.1007/s00467-009-1251-5. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Suarez E, Siwy J, Zurbig P, Mischak H. Urine as a source for clinical proteome analysis: From discovery to clinical application. Biochim Biophys Acta. 2014;1844:884–898. doi: 10.1016/j.bbapap.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Thomas CE, Sexton W, Benson K, Sutphen R, Koomen J. Urine collection and processing for protein biomarker discovery and quantification. Cancer Epidem Biomar. 2010;19:953–959. doi: 10.1158/1055-9965.EPI-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 15.Choi S, Moon SW, Shin JH, Park HK, Jin KH. Label-free biochemical analytic method for the early detection of adenoviral conjunctivitis using human tear biofluids. Anal Chem. 2014;86:11093–11099. doi: 10.1021/ac5025478. [DOI] [PubMed] [Google Scholar]
- 16.Blennow K, Zetterberg H. Cerebrospinal fluid biomarkers for Alzheimer’s disease. J Alzheimers Dis. 2009;18:413–417. doi: 10.3233/JAD-2009-1177. [DOI] [PubMed] [Google Scholar]
- 17.Moser AC, Hage DS. Immunoaffinity chromatography: an introduction to applications and recent developments. Bioanalysis. 2010;2:769–790. doi: 10.4155/bio.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang X, Zhang WW. Affinity separation and enrichment methods in proteomic analysis. J Proteomics. 2008;71:284–303. doi: 10.1016/j.jprot.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Abi-Ghanem DA, Berghman LR. Immunoaffinity Chromatography: A Review. In: Magdeldin DS, editor. Affinity chromatography. InTech; 2012. [Google Scholar]
- 20.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and Challenges in Liquid Chromatography-Mass Spectrometry-based Proteomics Profiling for Clinical Applications. Mol Cell Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7:1963–1973. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rafalko A, Iliopoulos O, Fusaro VA, Hancock W, Hincapie M. Immunoaffinity enrichment and liquid chromatography-selected reaction monitoring mass spectrometry for quantitation of carbonic anhydrase 12 in cultured renal carcinoma cells. Anal Chem. 2010;82:8998–9005. doi: 10.1021/ac101981t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaros JA, Guest PC, Bahn S, Martins-de-Souza D. Affinity depletion of plasma and serum for mass spectrometry-based proteome analysis. Methods Mol Biol. 2013;1002:1–11. doi: 10.1007/978-1-62703-360-2_1. [DOI] [PubMed] [Google Scholar]
- 24.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, Zhao L, Pope ME, Smith D, Rivera KD, Anderson NL, Skates SJ, Pearson TW, Paulovich AG, Carr SA. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012;11:M111 013854. doi: 10.1074/mcp.M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leenaars M, Hendriksen CFM. Critical steps in the production of polyclonal and monoclonal antibodies evaluation and recommendations. Ilar J. 2005;46:269–279. doi: 10.1093/ilar.46.3.269. [DOI] [PubMed] [Google Scholar]
- 27.Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics. 2009;8:995–1005. doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Immunoaffinity Purification. In: Harlow E, Lane D, editors. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York, USA: 1999. pp. 311–343. [Google Scholar]
- 29.Wark KL, Hudson PJ. Latest technologies for the enhancement of antibody affinity. Adv Drug Deliv Rev. 2006;58:657–670. doi: 10.1016/j.addr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Schoenherr RM, Zhao L, Whiteaker JR, Feng LC, Li L, Liu L, Liu X, Paulovich AG. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring-mass spectrometry. J Immunol Methods. 2010;353:49–61. doi: 10.1016/j.jim.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 32.Leitner A. Phosphopeptide enrichment using metal oxide affinity chromatography. Trends Anal Chem. 2010;29:177–185. [Google Scholar]
- 33.Plavina T, Wakshull E, Hancock WS, Hincapie M. Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J Proteome Res. 2007;6:662–671. doi: 10.1021/pr060413k. [DOI] [PubMed] [Google Scholar]
- 34.Romig TS, Bell C, Drolet DW. Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J Chromatogr B Biomed Sci Appl. 1999;731:275–284. [PubMed] [Google Scholar]
- 35.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 36.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5:3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 37.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 38.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi T, Zhou JY, Gritsenko MA, Hossain M, Camp DG, 2nd, Smith RD, Qian WJ. IgY14 and SuperMix immunoaffinity separations coupled with liquid chromatography-mass spectrometry for human plasma proteomics biomarker discovery. Methods. 2012;56:246–253. doi: 10.1016/j.ymeth.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brand J, Haslberger T, Zolg W, Pestlin G, Palme S. Depletion efficiency and recovery of trace markers from a multiparameter immunodepletion column. Proteomics. 2006;6:3236–3242. doi: 10.1002/pmic.200500864. [DOI] [PubMed] [Google Scholar]
- 41.Yocum AK, Yu K, Oe T, Blair IA. Effect of immunoaffinity depletion of human serum during proteomic investigations. J Proteome Res. 2005;4:1722–1731. doi: 10.1021/pr0501721. [DOI] [PubMed] [Google Scholar]
- 42.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 43.Shen Z, Want EJ, Chen W, Keating W, Nussbaumer W, Moore R, Gentle TM, Siuzdak G. Sepsis plasma protein profiling with immunodepletion, three-dimensional liquid chromatography tandem mass spectrometry, and spectrum counting. J Proteome Res. 2006;5:3154–3160. doi: 10.1021/pr060327k. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Clouthier SG, Galchev V, Misek DE, Duffner U, Min CK, Zhao R, Tra J, Omenn GS, Ferrara JL, Hanash SM. Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol Cell Proteomics. 2005;4:618–625. doi: 10.1074/mcp.M400126-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJ, Liebler DC. Depletion of abundant plasma proteins and limitations of plasma proteomics. J Proteome Res. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolotarjova N, Mrozinski P, Chen H, Martosella J. Combination of affinity depletion of abundant proteins and reversed-phase fractionation in proteomic analysis of human plasma/serum. J Chromatogr A. 2008;1189:332–338. doi: 10.1016/j.chroma.2007.11.082. [DOI] [PubMed] [Google Scholar]
- 47.Ernoult E, Bourreau A, Gamelin E, Guette C. A proteomic approach for plasma biomarker discovery with iTRAQ labelling and OFFGEL fractionation. J Biomed Biotechnol. 2010;2010:927917. doi: 10.1155/2010/927917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Z, Yende S, Kellum JA, Robinson RA. Additions to the Human Plasma Proteome via a Tandem MARS Depletion iTRAQ-Based Workflow. Int J Proteomics. 2013;2013:654356. doi: 10.1155/2013/654356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berven FS, Ahmad R, Clauser KR, Carr SA. Optimizing performance of glycopeptide capture for plasma proteomics. J Proteome Res. 2010;9:1706–1715. doi: 10.1021/pr900845m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Majek P, Reicheltova Z, Suttnar J, Cermak J, Dyr JE. Plasma proteome changes associated with refractory cytopenia with multilineage dysplasia. Proteome Sci. 2011;9:64. doi: 10.1186/1477-5956-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zawadzka AM, Schilling B, Held JM, Sahu AK, Cusack MP, Drake PM, Fisher SJ, Gibson BW. Variation and quantification among a target set of phosphopeptides in human plasma by multiple reaction monitoring and SWATH-MS2 data-independent acquisition. Electrophoresis. 2014;35:3487–3497. doi: 10.1002/elps.201400167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Millioni R, Tolin S, Puricelli L, Sbrignadello S, Fadini GP, Tessari P, Arrigoni G. High abundance proteins depletion vs low abundance proteins enrichment: comparison of methods to reduce the plasma proteome complexity. PLoS One. 2011;6:e19603. doi: 10.1371/journal.pone.0019603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CL, Lin TS, Tsai CH, Wu CC, Chung T, Chien KY, Wu M, Chang YS, Yu JS, Chen YT. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J Proteomics. 2013;85:28–43. doi: 10.1016/j.jprot.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 55.Ringman JM, Schulman H, Becker C, Jones T, Bai Y, Immermann F, Cole G, Sokolow S, Gylys K, Geschwind DH, Cummings JL, Wan HI. Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69:96–104. doi: 10.1001/archneurol.2011.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guldbrandsen A, Vethe H, Farag Y, Oveland E, Garberg H, Berle M, Myhr KM, Opsahl JA, Barsnes H, Berven FS. In-depth characterization of the cerebrospinal fluid (CSF) proteome displayed through the CSF proteome resource (CSF-PR) Mol Cell Proteomics. 2014;13:3152–3163. doi: 10.1074/mcp.M114.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroksveen AC, Aasebo E, Vethe H, Van Pesch V, Franciotta D, Teunissen CE, Ulvik RJ, Vedeler C, Myhr KM, Barsnes H, Berven FS. Discovery and initial verification of differentially abundant proteins between multiple sclerosis patients and controls using iTRAQ and SID-SRM. J Proteomics. 2013;78:312–325. doi: 10.1016/j.jprot.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 58.Hoskins ER, Hood BL, Sun M, Krivak TC, Edwards RP, Conrads TP. Proteomic analysis of ovarian cancer proximal fluids: validation of elevated peroxiredoxin 1 in patient peripheral circulation. PLoS One. 2011;6:e25056. doi: 10.1371/journal.pone.0025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandow JE. Comparison of protein enrichment strategies for proteome analysis of plasma. Proteomics. 2010;10:1416–1425. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- 61.Borg J, Campos A, Diema C, Omenaca N, de Oliveira E, Guinovart J, Vilaseca M. Spectral counting assessment of protein dynamic range in cerebrospinal fluid following depletion with plasma-designed immunoaffinity columns. Clin Proteomics. 2011;8:6. doi: 10.1186/1559-0275-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel BB, Barrero CA, Braverman A, Kim PD, Jones KA, Chen DE, Bowler RP, Merali S, Kelsen SG, Yeung AT. Assessment of two immunodepletion methods: off-target effects and variations in immunodepletion efficiency may confound plasma proteomics. J Proteome Res. 2012;11:5947–5958. doi: 10.1021/pr300686k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fratantoni SA, Piersma SR, Jimenez CR. Comparison of the performance of two affinity depletion spin filters for quantitative proteomics of CSF: Evaluation of sensitivity and reproducibility of CSF analysis using GeLC-MS/MS and spectral counting. Proteomics Clin Appl. 2010;4:613–617. doi: 10.1002/prca.200900179. [DOI] [PubMed] [Google Scholar]
- 64.Stempfer R, Kubicek M, Lang IM, Christa N, Gerner C. Quantitative assessment of human serum high-abundance protein depletion. Electrophoresis. 2008;29:4316–4323. doi: 10.1002/elps.200800211. [DOI] [PubMed] [Google Scholar]
- 65.Shield-Artin KL, Bailey MJ, Oliva K, Liovic AK, Barker G, Dellios NL, Reisman S, Ayhan M, Rice GE. Identification of ovarian cancer-associated proteins in symptomatic women: A novel method for semi-quantitative plasma proteomics. Proteomics Clin Appl. 2012;6:170–181. doi: 10.1002/prca.201100008. [DOI] [PubMed] [Google Scholar]
- 66.Rehman I, Evans CA, Glen A, Cross SS, Eaton CL, Down J, Pesce G, Phillips JT, Yen OS, Thalmann GN, Wright PC, Hamdy FC. iTRAQ identification of candidate serum biomarkers associated with metastatic progression of human prostate cancer. PLoS One. 2012;7:e30885. doi: 10.1371/journal.pone.0030885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim JS, Jones N, Moore RJ, Pasa-Tolic L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG, 2nd, Smith RD, Qian WJ. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A. 2012;109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nie S, Lo A, Wu J, Zhu J, Tan Z, Simeone DM, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J Proteome Res. 2014;13:1873–1884. doi: 10.1021/pr400967x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morales DM, Townsend RR, Malone JP, Ewersmann CA, Macy EM, Inder TE, Limbrick DD., Jr Alterations in protein regulators of neurodevelopment in the cerebrospinal fluid of infants with posthemorrhagic hydrocephalus of prematurity. Mol Cell Proteomics. 2012;11:M111 011973. doi: 10.1074/mcp.M111.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuberovic A, Hanrieder J, Hellman U, Bergquist J, Wetterhall M. Proteome profiling of human cerebrospinal fluid: exploring the potential of capillary electrophoresis with surface modified capillaries for analysis of complex biological samples. Eur J Mass Spectrom. 2008;14:249–260. doi: 10.1255/ejms.929. [DOI] [PubMed] [Google Scholar]
- 71.Zhou JY, Petritis BO, Petritis K, Norbeck AD, Weitz KK, Moore RJ, Camp DG, Kulkarni RN, Smith RD, Qian WJ. Mouse-specific tandem IgY7-SuperMix immunoaffinity separations for improved LC-MS/MS coverage of the plasma proteome. J Proteome Res. 2009;8:5387–5395. doi: 10.1021/pr900564f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyung SW, Piehowski PD, Moore RJ, Orton DJ, Schepmoes AA, Clauss TR, Chu RK, Fillmore TL, Brewer H, Liu T, Zhao R, Smith RD. Microscale depletion of high abundance proteins in human biofluids using IgY14 immunoaffinity resin: analysis of human plasma and cerebrospinal fluid. Anal Bioanal Chem. 2014;406:7117–7125. doi: 10.1007/s00216-014-8058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Righetti PG, Boschetti E, Lomas L, Citterio A. Protein Equalizer Technology : the quest for a “democratic proteome”. Proteomics. 2006;6:3980–3992. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- 74.Paulus AFS, Academia K, Thulassiraman V. Accessing Low-Abundance Proteins in Serum and plasma with a Novel, Simple Enrichment and Depletion Method. Bio-Rad Tech Note 5632. 2009 Sample preparation DOI. [Google Scholar]
- 75.Righetti PG, Candiano G, Citterio A, Boschetti E. Combinatorial peptide ligand libraries as a “Trojan Horse” in deep discovery proteomics. Anal Chem. 2015;87:293–305. doi: 10.1021/ac502171b. [DOI] [PubMed] [Google Scholar]
- 76.Gil-Dones F, Darde VM, Alonso-Orgaz S, Lopez-Almodovar LF, Mourino-Alvarez L, Padial LR, Vivanco F, Barderas MG. Inside human aortic stenosis: a proteomic analysis of plasma. J Proteomics. 2012;75:1639–1653. doi: 10.1016/j.jprot.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 77.Mustafa MG, Petersen JR, Ju H, Cicalese L, Snyder N, Haidacher SJ, Denner L, Elferink C. Biomarker discovery for early detection of hepatocellular carcinoma in hepatitis C-infected patients. Mol Cell Proteomics. 2013;12:3640–3652. doi: 10.1074/mcp.M113.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milan E, Lazzari C, Anand S, Floriani I, Torri V, Sorlini C, Gregorc V, Bachi A. SAA1 is over-expressed in plasma of non small cell lung cancer patients with poor outcome after treatment with epidermal growth factor receptor tyrosine-kinase inhibitors. J Proteomics. 2012;76(Spec No):91–101. doi: 10.1016/j.jprot.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 79.Ruhaak LR, Nguyen UT, Stroble C, Taylor SL, Taguchi A, Hanash SM, Lebrilla CB, Kim K, Miyamoto S. Enrichment strategies in glycomics-based lung cancer biomarker development. Proteomics Clin Appl. 2013;7:664–676. doi: 10.1002/prca.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fonslow BR, Carvalho PC, Academia K, Freeby S, Xu T, Nakorchevsky A, Paulus A, Yates JR., 3rd Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J Proteome Res. 2011;10:3690–3700. doi: 10.1021/pr200304u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krastins B, Prakash A, Sarracino DA, Nedelkov D, Niederkofler EE, Kiernan UA, Nelson R, Vogelsang MS, Vadali G, Garces A, Sutton JN, Peterman S, Byram G, Darbouret B, Perusse JR, Seidah NG, Coulombe B, Gobom J, Portelius E, Pannee J, Blennow K, Kulasingam V, Couchman L, Moniz C, Lopez MF. Rapid development of sensitive, high-throughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clin Biochem. 2013;46:399–410. doi: 10.1016/j.clinbiochem.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madian AG, Rochelle NS, Regnier FE. Mass-linked immuno-selective assays in targeted proteomics. Anal Chem. 2013;85:737–748. doi: 10.1021/ac302071k. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 84.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, Gaffrey MJ, Su D, Liu T, Camp DG, II, Smith RD, Qian W-J. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat Protoc. 2014;9:64–75. doi: 10.1038/nprot.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu T, Qian WJ, Strittmatter EF, Camp DG, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 87.Guss B, Eliasson M, Olsson A, Uhlen M, Frej AK, Jornvall H, Flock JI, Lindberg M. Structure of the IgG-binding regions of streptococcal protein G. EMBO J. 1986;5:1567–1575. doi: 10.1002/j.1460-2075.1986.tb04398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gronwall C, Sjoberg A, Ramstrom M, Hoiden-Guthenberg I, Hober S, Jonasson P, Stahl S. Affibody-mediated transferrin depletion for proteomics applications. Biotechnol J. 2007;2:1389–1398. doi: 10.1002/biot.200700053. [DOI] [PubMed] [Google Scholar]
- 89.Lofblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY. Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010;584:2670–2680. doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 90.Nilsson CL. Lectins: proteins that interpret the sugar code. Anal Chem. 2003;75:348A–353A. doi: 10.1021/ac031373w. [DOI] [PubMed] [Google Scholar]
- 91.Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. A proteomics platform combining depletion, multi-lectin affinity chromatography (M-LAC), and isoelectric focusing to study the breast cancer proteome. Anal Chem. 2011;83:4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gbormittah FO, Hincapie M, Hancock WS. Development of an improved fractionation of the human plasma proteome by a combination of abundant proteins depletion and multi-lectin affinity chromatography. Bioanalysis. 2014;6:2537–2548. doi: 10.4155/bio.14.217. [DOI] [PubMed] [Google Scholar]
- 93.Lobato MN, Rabbitts TH. Intracellular antibodies and challenges facing their use as therapeutic agents. Trends Mol Med. 2003;9:390–396. doi: 10.1016/s1471-4914(03)00163-1. [DOI] [PubMed] [Google Scholar]
- 94.Perez-Martinez D, Tanaka T, Rabbitts TH. Intracellular antibodies and cancer: new technologies offer therapeutic opportunities. Bioessays. 2010;32:589–598. doi: 10.1002/bies.201000009. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka T, Rabbitts TH. Selection of functional single domain antibody fragments for interfering with protein-protein interactions inside cells: a “one plasmid” mammalian two-hybrid system. Methods Mol Biol. 2012;911:175–182. doi: 10.1007/978-1-61779-968-6_11. [DOI] [PubMed] [Google Scholar]
- 96.Hagemeyer CE, von Zur Muhlen C, von Elverfeldt D, Peter K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb Haemost. 2009;101:1012–1019. [PubMed] [Google Scholar]
- 97.Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, Hamid M. scFv antibody: principles and clinical application. Clin Dev Immunol. 2012;2012:980250. doi: 10.1155/2012/980250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hust M, Meyer T, Voedisch B, Rulker T, Thie H, El-Ghezal A, Kirsch MI, Schutte M, Helmsing S, Meier D, Schirrmann T, Dubel S. A human scFv antibody generation pipeline for proteome research. J Biotechnol. 2011;152:159–170. doi: 10.1016/j.jbiotec.2010.09.945. [DOI] [PubMed] [Google Scholar]
- 99.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 100.Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2:3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- 101.Ramirez AB, Loch CM, Zhang Y, Liu Y, Wang X, Wayner EA, Sargent JE, Sibani S, Hainsworth E, Mendoza EA, Eugene R, Labaer J, Urban ND, McIntosh MW, Lampe PD. Use of a single-chain antibody library for ovarian cancer biomarker discovery. Mol Cell Proteomics. 2010;9:1449–1460. doi: 10.1074/mcp.M900496-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schroder C, Jacob A, Tonack S, Radon TP, Sill M, Zucknick M, Ruffer S, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Bauer A, Fellenberg K, Hoheisel JD. Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies. Mol Cell Proteomics. 2010;9:1271–1280. doi: 10.1074/mcp.M900419-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9:184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whiteaker JR, Zhao L, Lin C, Yan P, Wang P, Paulovich AG. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol Cell Proteomics. 2012;11:M111 015347. doi: 10.1074/mcp.M111.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karisch R, Fernandez M, Taylor P, Virtanen C, St-Germain JR, Jin LL, Harris IS, Mori J, Mak TW, Senis YA, Ostman A, Moran MF, Neel BG. Global proteomic assessment of the classical protein-tyrosine phosphatome and “redoxome”. Cell. 2011;146:826–840. doi: 10.1016/j.cell.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oyama M, Kozuka-Hata H, Tasaki S, Semba K, Hattori S, Sugano S, Inoue J, Yamamoto T. Temporal perturbation of tyrosine phosphoproteome dynamics reveals the system-wide regulatory networks. Mol Cell Proteomics. 2009;8:226–231. doi: 10.1074/mcp.M800186-MCP200. [DOI] [PubMed] [Google Scholar]
- 107.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 110.Guan KL, Yu W, Lin Y, Xiong Y, Zhao S. Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat Protoc. 2010;5:1583–1595. doi: 10.1038/nprot.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics. 2014;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tantipaiboonwong P, Sinchaikul S, Sriyam S, Phutrakul S, Chen ST. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005;5:1140–1149. doi: 10.1002/pmic.200401143. [DOI] [PubMed] [Google Scholar]
- 113.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J Proteome Res. 2006;5:1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 114.Polaskova V, Kapur A, Khan A, Molloy MP, Baker MS. High-abundance protein depletion: comparison of methods for human plasma biomarker discovery. Electrophoresis. 2010;31:471–482. doi: 10.1002/elps.200900286. [DOI] [PubMed] [Google Scholar]
- 115.Liu T, Qian WJ, Gritsenko MA, Xiao W, Moldawer LL, Kaushal A, Monroe ME, Varnum SM, Moore RJ, Purvine SO, Maier RV, Davis RW, Tompkins RG, Camp DG, 2 , Smith RD Inflammation, P the Host Response to Injury Large Scale Collaborative Research. High dynamic range characterization of the trauma patient plasma proteome. Mol Cell Proteomics. 2006;5:1899–1913. doi: 10.1074/mcp.M600068-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Heller M, Michel PE, Morier P, Crettaz D, Wenz C, Tissot JD, Reymond F, Rossier JS. Two-stage Off-Gel isoelectric focusing: protein followed by peptide fractionation and application to proteome analysis of human plasma. Electrophoresis. 2005;26:1174–1188. doi: 10.1002/elps.200410106. [DOI] [PubMed] [Google Scholar]
- 118.Liu T, Hossain M, Schepmoes AA, Fillmore TL, Sokoll LJ, Kronewitter SR, Izmirlian G, Shi T, Qian WJ, Leach RJ, Thompson IM, Chan DW, Smith RD, Kagan J, Srivastava S, Rodland KD, Camp DG., 2nd Analysis of serum total and free PSA using immunoaffinity depletion coupled to SRM: correlation with clinical immunoassay tests. J Proteomics. 2012;75:4747–4757. doi: 10.1016/j.jprot.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Molecular & Cellular Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, Carapito C, Farrah T, Deutsch EW, Kusebauch U, Moritz RL, Nimeus-Malmstrom E, Rinner O, Aebersold R. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra194. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anderson L, Hunter CL. Quantitative Mass Spectrometric Multiple Reaction Monitoring Assays for Major Plasma Proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 123.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pan S, Chen R, Crispin DA, May D, Stevens T, McIntosh MW, Bronner MP, Ziogas A, Anton-Culver H, Brentnall TA. Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J Proteome Res. 2011;10:2359–2376. doi: 10.1021/pr101148r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yadav AK, Bhardwaj G, Basak T, Kumar D, Ahmad S, Priyadarshini R, Singh AK, Dash D, Sengupta S. A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: implications in biomarker discovery. PLoS One. 2011;6:e24442. doi: 10.1371/journal.pone.0024442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nelson RW, Krone JR, Bieber AL, Williams P. Mass spectrometric immunoassay. Anal Chem. 1995;67:1153–1158. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 127.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55:1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ray S, Reddy PJ, Jain R, Gollapalli K, Moiyadi A, Srivastava S. Proteomic technologies for the identification of disease biomarkers in serum: advances and challenges ahead. Proteomics. 2011;11:2139–2161. doi: 10.1002/pmic.201000460. [DOI] [PubMed] [Google Scholar]
- 129.Timms JF, Arslan-Low E, Kabir M, Worthington J, Camuzeaux S, Sinclair J, Szaub J, Afrough B, Podust VN, Fourkala EO, Cubizolles M, Kronenberg F, Fung ET, Gentry-Maharaj A, Menon U, Jacobs I. Discovery of serum biomarkers of ovarian cancer using complementary proteomic profiling strategies. Proteomics Clin Appl. 2014;8:982–993. doi: 10.1002/prca.201400063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin B, White JT, Wu J, Lele S, Old LJ, Hood L, Odunsi K. Deep depletion of abundant serum proteins reveals low-abundant proteins as potential biomarkers for human ovarian cancer. Proteomics Clin Appl. 2009;3:853–861. doi: 10.1002/prca.200800141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cohen A, Wang E, Chisholm KA, Kostyleva R, O’Connor-McCourt M, Pinto DM. A mass spectrometry-based plasma protein panel targeting the tumor microenvironment in patients with breast cancer. J Proteomics. 2013;81:135–147. doi: 10.1016/j.jprot.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 132.He W, Tong Y, Wang Y, Liu J, Luo G, Wu J, Zhang J. Serum soluble CD14 is a potential prognostic indicator of recurrence of human breast invasive ductal carcinoma with Her2-enriched subtype. PLoS One. 2013;8:e75366. doi: 10.1371/journal.pone.0075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prieto DA, Johann DJ, Jr, Wei BR, Ye X, Chan KC, Nissley DV, Simpson RM, Citrin DE, Mackall CL, Linehan WM, Blonder J. Mass spectrometry in cancer biomarker research: a case for immunodepletion of abundant blood-derived proteins from clinical tissue specimens. Biomark Med. 2014;8:269–286. doi: 10.2217/bmm.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Humphries JM, Penno MA, Weiland F, Klingler-Hoffmann M, Zuber A, Boussioutas A, Ernst M, Hoffmann P. Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer. Biochim Biophys Acta. 2014;1844:1051–1058. doi: 10.1016/j.bbapap.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 135.Byrne JC, Downes MR, O’Donoghue N, O’Keane C, O’Neill A, Fan Y, Fitzpatrick JM, Dunn M, Watson RW. 2D-DIGE as a strategy to identify serum markers for the progression of prostate cancer. J Proteome Res. 2009;8:942–957. doi: 10.1021/pr800570s. [DOI] [PubMed] [Google Scholar]
- 136.Shi T, Gao Y, Quek SI, Fillmore TL, Nicora CD, Su D, Zhao R, Kagan J, Srivastava S, Rodland KD, Liu T, Smith RD, Chan DW, Camp DG, 2nd, Liu AY, Qian WJ. A highly sensitive targeted mass spectrometric assay for quantification of AGR2 protein in human urine and serum. J Proteome Res. 2014;13:875–882. doi: 10.1021/pr400912c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fernandez-Costa C, Calamia V, Fernandez-Puente P, Capelo-Martinez JL, Ruiz-Romero C, Blanco FJ. Sequential depletion of human serum for the search of osteoarthritis biomarkers. Proteome Sci. 2012;10:55. doi: 10.1186/1477-5956-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fernandez-Puente P, Mateos J, Fernandez-Costa C, Oreiro N, Fernandez-Lopez C, Ruiz-Romero C, Blanco FJ. Identification of a panel of novel serum osteoarthritis biomarkers. J Proteome Res. 2011;10:5095–5101. doi: 10.1021/pr200695p. [DOI] [PubMed] [Google Scholar]
- 139.Takinami Y, Yoshimatsu S, Uchiumi T, Toyosaki-Maeda T, Morita A, Ishihara T, Yamane S, Fukuda I, Okamoto H, Numata Y, Fukui N. Identification of potential prognostic markers for knee osteoarthritis by serum proteomic analysis. Biomark Insigh. 2013;8:85–95. doi: 10.4137/BMI.S11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pedroza-Diaz J, Rothlisberger S. Advances in urinary protein biomarkers for urogenital and non-urogenital pathologies. Biochemia Medica. 2015;25:22–35. doi: 10.11613/BM.2015.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Posada-Ayala M, Zubiri I, Martin-Lorenzo M, Sanz-Maroto A, Molero D, Gonzalez-Calero L, Fernandez-Fernandez B, de la Cuesta F, Laborde CM, Barderas MG, Ortiz A, Vivanco F, Alvarez-Llamas G. Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney Int. 2014;85:103–111. doi: 10.1038/ki.2013.328. [DOI] [PubMed] [Google Scholar]
- 143.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He W, Huang C, Luo G, Dal Pra I, Feng J, Chen W, Ma L, Wang Y, Chen X, Tan J, Zhang X, Armato U, Wu J. A stable panel comprising 18 urinary proteins in the human healthy population. Proteomics. 2012;12:1059–1072. doi: 10.1002/pmic.201100400. [DOI] [PubMed] [Google Scholar]
- 145.Martin-Lorenzo M, Gonzalez-Calero L, Zubiri I, Diaz-Payno PJ, Sanz-Maroto A, Posada-Ayala M, Ortiz A, Vivanco F, Alvarez-Llamas G. Urine 2DE proteome analysis in healthy condition and kidney disease. Electrophoresis. 2014;35:2634–2641. doi: 10.1002/elps.201300601. [DOI] [PubMed] [Google Scholar]
- 146.Magagnotti C, Fermo I, Carletti RM, Ferrari M, Bachi A. Comparison of different depletion strategies for improving resolution of the human urine proteome. Clin Chem Lab Med. 2010;48:531–535. doi: 10.1515/CCLM.2010.109. [DOI] [PubMed] [Google Scholar]
- 147.Li WW, Nemirovskiy O, Fountain S, Rodney Mathews W, Szekely-Klepser G. Clinical validation of an immunoaffinity LC-MS/MS assay for the quantification of a collagen type II neoepitope peptide: A biomarker of matrix metalloproteinase activity and osteoarthritis in human urine. Anal Biochem. 2007;369:41–53. doi: 10.1016/j.ab.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 148.Fisher WG, Lucas JE, Mehdi UF, Qunibi DW, Garner HR, Rosenblatt KP, Toto RD. A method for isolation and identification of urinary biomarkers in patients with diabetic nephropathy. Proteomics Clin Appl. 2011;5:603–612. doi: 10.1002/prca.201000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rao PV, Lu X, Standley M, Pattee P, Neelima G, Girisesh G, Dakshinamurthy KV, Roberts CT, Jr, Nagalla SR. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care. 2007;30:629–637. doi: 10.2337/dc06-2056. [DOI] [PubMed] [Google Scholar]
- 150.Loftheim H, Midtvedt K, Hartmann A, Reisaeter AV, Falck P, Holdaas H, Jenssen T, Reubsaet L, Asberg A. Urinary proteomic shotgun approach for identification of potential acute rejection biomarkers in renal transplant recipients. Transplant Res. 2012;1:9. doi: 10.1186/2047-1440-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Choi YS, Choe LH, Lee KH. Recent cerebrospinal fluid biomarker studies of Alzheimer’s disease. Expert Rev Proteomics. 2010;7:919–926. doi: 10.1586/epr.10.75. [DOI] [PubMed] [Google Scholar]
- 152.Kroksveen AC, Opsahl JA, Aye TT, Ulvik RJ, Berven FS. Proteomics of human cerebrospinal fluid: Discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J Proteomics. 2011;74:371–388. doi: 10.1016/j.jprot.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 153.Cunningham R, Jany P, Messing A, Li L. Protein changes in immunodepleted cerebrospinal fluid from a transgenic mouse model of Alexander disease detected using mass spectrometry. J Proteome Res. 2013;12:719–728. doi: 10.1021/pr300785h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schutzer SE, Liu T, Natelson BH, Angel TE, Schepmoes AA, Purvine SO, Hixson KK, Lipton MS, Camp DG, Coyle PK, Smith RD, Bergquist J. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010;5:e10980. doi: 10.1371/journal.pone.0010980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schutzer SE, Angel TE, Liu T, Schepmoes AA, Clauss TR, Adkins JN, Camp DG, Holland BK, Bergquist J, Coyle PK, Smith RD, Fallon BA, Natelson BH. Distinct cerebrospinal fluid proteomes differentiate post-treatment lyme disease from chronic fatigue syndrome. PLoS One. 2011;6:e17287. doi: 10.1371/journal.pone.0017287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y, Guo Z, Zou L, Yang Y, Zhang L, Ji N, Shao C, Sun W, Wang Y. A comprehensive map and functional annotation of the normal human cerebrospinal fluid proteome. J Proteomics. 2015;119C:90–99. doi: 10.1016/j.jprot.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 157.Lehnert S, Jesse S, Rist W, Steinacker P, Soininen H, Herukka SK, Tumani H, Lenter M, Oeckl P, Ferger B, Hengerer B, Otto M. iTRAQ and multiple reaction monitoring as proteomic tools for biomarker search in cerebrospinal fluid of patients with Parkinson’s disease dementia. Exp Neurol. 2012;234:499–505. doi: 10.1016/j.expneurol.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 158.Fang Q, Strand A, Law W, Faca VM, Fitzgibbon MP, Hamel N, Houle B, Liu X, May DH, Poschmann G, Roy L, Stuhler K, Ying W, Zhang J, Zheng Z, Bergeron JJ, Hanash S, He F, Leavitt BR, Meyer HE, Qian X, McIntosh MW. Brain-specific proteins decline in the cerebrospinal fluid of humans with Huntington disease. Mol Cell Proteomics. 2009;8:451–466. doi: 10.1074/mcp.M800231-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mateos J, Lourido L, Fernandez-Puente P, Calamia V, Fernandez-Lopez C, Oreiro N, Ruiz-Romero C, Blanco FJ. Differential protein profiling of synovial fluid from rheumatoid arthritis and osteoarthritis patients using LC-MALDI TOF/TOF. J Proteomics. 2012;75:2869–2878. doi: 10.1016/j.jprot.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 160.Balakrishnan L, Nirujogi RS, Ahmad S, Bhattacharjee M, Manda SS, Renuse S, Kelkar DS, Subbannayya Y, Raju R, Goel R, Thomas JK, Kaur N, Dhillon M, Tankala SG, Jois R, Vasdev V, Ramachandra Y, Sahasrabuddhe NA, Prasad Ts K, Mohan S, Gowda H, Shankar S, Pandey A. Proteomic analysis of human osteoarthritis synovial fluid. Clin Proteomics. 2014;11:6. doi: 10.1186/1559-0275-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hughes AJ, Lin RK, Peehl DM, Herr AE. Microfluidic integration for automated targeted proteomic assays. Proc Natl Acad Sci U S A. 2012;109:5972–5977. doi: 10.1073/pnas.1108617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li QR, Fan KX, Li RX, Dai J, Wu CC, Zhao SL, Wu JR, Shieh CH, Zeng R. A comprehensive and non-prefractionation on the protein level approach for the human urinary proteome: touching phosphorylation in urine. Rapid Commun Mass Spectrom. 2010;24:823–832. doi: 10.1002/rcm.4441. [DOI] [PubMed] [Google Scholar]