Abstract
Aims/hypothesis
Diabetic peripheral neuropathy (DPN) and diabetic nephropathy (DN) are two common microvascular complications of type 1 and type 2 diabetes mellitus that are associated with a high degree of morbidity. In this study, using a variety of systems biology approaches, our aim was to identify common and distinct mechanisms underlying the pathogenesis of these two complications.
Methods
Our previously published transcriptomic datasets of peripheral nerve and kidney tissue, derived from murine models of type 1 diabetes (streptozotocin-injected mice) and type 2 diabetes (BKS-db/db mice) and their respective controls, were collected and processed using a unified analysis pipeline so that comparisons could be made. In addition to looking at genes and pathways dysregulated in individual datasets, pairwise comparisons across diabetes type and tissue type were performed at both gene and transcriptional network levels to complete our proposed objective.
Results
Gene-level analysis identified exceptionally high levels of concordant gene expression in DN (94% of 2,433 genes), but not in DPN (55% of 1,558 genes), between type 1 diabetes and type 2 diabetes. These results suggest that common pathogenic mechanisms exist in DN across diabetes type, while in DPN the mechanisms are more distinct. When these dysregulated genes were examined at the transcriptional network level, we found that the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway was significantly dysregulated in both complications, irrespective of diabetes type.
Conclusions/interpretation
Using a systems biology approach, our findings suggest that common pathogenic mechanisms exist in DN across diabetes type, while in DPN the mechanisms are more distinct. We also found that JAK–STAT signalling is commonly dysregulated among all datasets. Using such approaches, further investigation is warranted to determine whether the same changes are observed in patients with diabetic complications.
Keywords: Diabetic complications, Diabetic nephropathy, Diabetic peripheral neuropathy, Gene expression profiling, JAK–STAT, Janus kinase–signal transducer and activator of transcription, Microarray, Murine models, Systems biology, Transcriptional network analysis
Introduction
Over 8.8% of the global population, approximately 415 million people, have diabetes [1]. Type 2 diabetes mellitus accounts for ~90–95% of cases and is characterised by insulin resistance, while type 1 diabetes mellitus, a state of insulinopenia, accounts for the remainder [2]. A large proportion of both type 1 and type 2 diabetes patients experience diabetes-related complications. Microvascular complications, including diabetic peripheral neuropathy (DPN) and diabetic nephropathy (DN), are major contributors to the morbidity associated with diabetes, affecting ~60% and ~40% of patients, respectively [3, 4]. The prevalence of these complications is confounded by current treatments being largely ineffective in halting disease progression. DPN is characterised as a chronic, symmetrical, progressive disorder with early symptoms of pain, allodynia and paraesthesias. The longest nerve axons are initially affected and the disease continues to progress in a distal-to-proximal fashion [3]. As DPN advances, eventual loss of all sensory modalities in 60–70% of affected individuals leads to an insensate foot at high risk of infection and subsequent ulcer development [5, 6], thus predisposing patients to non-traumatic lower-limb amputation [7]. Similar to DPN, DN is associated with high morbidity and is the leading cause of end-stage renal disease in the USA [8]. DN usually presents with albuminuria between 10–30 years after diabetes onset, with initial renal abnormalities localised to the glomerulus (Glom), including mesangial expansion, glomerular basement membrane thickening and podocyte loss. In progressive DN, increased fibrosis in the glomeruli and tubulointerstitium accompanies a decline in kidney function and many of these patients ultimately reach end-stage renal disease [9].
To better understand the pathogenic mechanism of DPN and DN, murine models of diabetes that emulate the features of the disease have proven to be invaluable. Such models support basic and translational studies for the discovery of novel therapeutic strategies. Recently, using systems biology approaches, analyses of gene expression profiles from the tissue of affected murine models have provided new insight into the cellular processes that drive pathogenesis. Previously, we reported global transcriptomic changes in the sciatic nerve (SCN) in models of type 1 [10] and type 2 diabetes [11, 12]. Furthermore, we have identified similar changes in the kidney Glom from various DN models [13]. In addition to identifying thousands of differentially expressed genes (DEGs), collectively these studies have identified numerous dysregulated pathways in diabetic complications that have been confirmed experimentally. These processes include inflammation [12, 13], oxidative stress [10, 11], lipid and carbohydrate metabolism [11], peroxisome proliferator-activated receptor (PPAR) signalling [10, 11], Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signalling [13] and vascular endothelial growth factor receptor (VEGFR) signalling [13].
While previous studies identified gene expression changes in individual complication-prone tissues [10–13], no comparison of transcriptomic profiles has been made between the diabetes type or between complication-prone tissues. Our aim in this current study was to identify alterations in the transcriptome, both common and distinct, between the peripheral nerve and kidney in the setting of either type 1 or type 2 diabetes. Furthermore, we sought to perform similar comparisons looking at each diabetes-prone tissue across diabetes type. To accomplish this, our previously published DPN and DN transcriptomic datasets from models of type 1 and type 2 diabetes [10, 11, 13] were compared at both the gene and transcriptional network level.
Methods
Study design
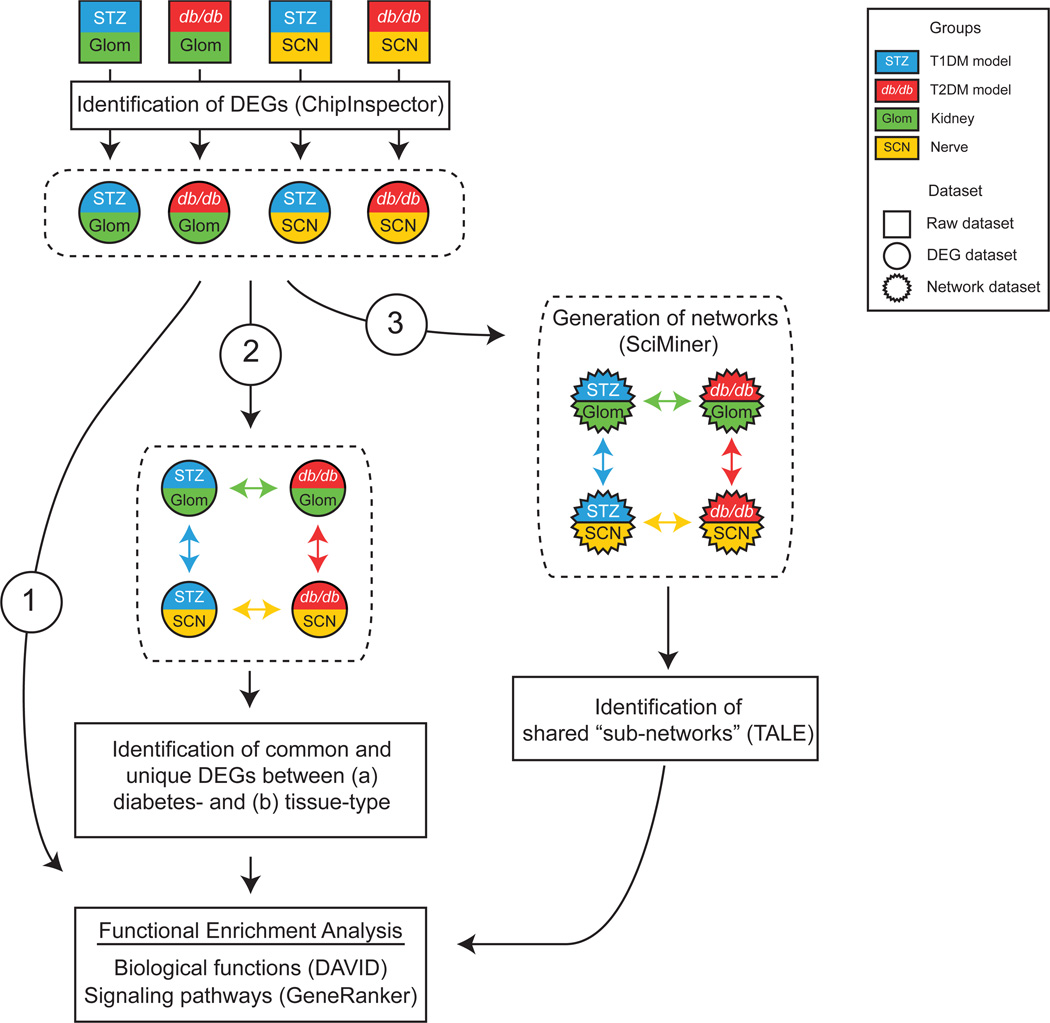
A workflow was designed to optimally analyse our previously published microarray data derived from Glom and SCN from murine models of type 1 and type 2 diabetes and their respective non-diabetic controls [10, 11, 13] (Fig. 1). For comparisons between the gene lists from these individual studies, the original datasets were reanalysed. After acquiring four individual tissue- and disease-specific DEG datasets, a stepwise approach was used to identify over-represented biological functions and signalling pathways among the datasets. Initially, functional enrichment analysis was performed on the individual datasets (Fig. 1 [1]). Subsequently, DEG datasets were compared to identify tissue- and disease-specific changes (Fig. 1 [2]). Finally, to identify gene networks common between tissue or disease, networks were generated from the DEG datasets (Fig. 1 [3]). The resulting networks were compared, resulting in subnetworks that were then subjected to functional enrichment analysis.
Fig. 1.
DEG profiling and network comparison workflow. Previously published microarray datasets were reanalysed using ChipInspector to identify DEG sets for Glom and SCN from murine models of type 1 diabetes (STZ) and type 2 diabetes (db/db) compared with respective non-diabetic controls. Four pairwise comparisons indicated by coloured arrows were made across tissue (green and yellow) and diabetes type (blue and red). Common and unique DEGs and shared transcriptional networks between STZ-Glom, STZ-SCN, db/db-Glom and db/db-SCN were further analysed by DAVID and GeneRanker to identify enriched biological functions and canonical signalling pathways, respectively. Numbered arrows indicate three different analysis approaches from DEG sets. Groups of differentially expressed gene sets and transcriptional networks and their pair-wise comparison schemes are denoted by dashed lines. T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus. The key applies to Figs 1–4
Animals
To model type 1 diabetes, streptozotocin (STZ) was administered to DBA/2J mice at 10 weeks and the study was terminated at either 22 or 34 weeks, for a total of ~12 or ~24 weeks of hyperglycaemia, respectively [10, 13]. As a model of type 2 diabetes, BKS.Cg-Leprdb/db (db/db) mice were euthanised at 24 weeks [11, 13]. Control mice for models of type 1 and type 2 diabetes consisted of healthy DBA/2J and db/+ mice, respectively. As reported previously, each murine model exhibited the typical features of diabetes by study conclusion as well as hallmarks of DN and DPN (electronic supplementary material [ESM] 1 Table 1).
Transcriptomic profiling
Raw transcriptomic data (NCBI Gene Expression Omnibus IDs: GSE33744, GSE11343 and GSE27382) [10, 11, 13] derived from Affymetrix GeneChips were reprocessed using ChipInspector version 2.1 (Genomatix Software, www.genomatix.de) [14] containing up-to-date gene annotation. Microarray image files were analysed using ChipInspector. Microarray gene expression signals were analysed at the single probe level. Significant transcripts were defined using a minimum of five significant probes and a false discovery rate < 1% by the Significance Analysis of Microarrays algorithm using exhaustive comparisons between control and diabetic mice [15]. Four sets of DEGs were obtained based on significant transcripts: STZ-SCN (type 1 diabetes DPN), STZ-Glom (type 1 diabetes DN), db/db-SCN (type 2 diabetes DPN) and db/db-Glom (type 2 diabetes DN).
Diabetes- and tissue-type comparisons of DEG sets
The four DEG sets were compared to examine similarities and differences between tissues (SCN and glomeruli) and diabetes type (type 1 diabetes and type 2 diabetes) at the gene level. Each pair of DEG sets was examined for overlap and concordance of gene expression changes. The percentage of concordant DEGs (i.e. showing the same direction of expression change) and discordant DEGs (i.e. showing the opposite direction of expression change) was also determined. Four pairwise comparisons (STZ-Glom vs STZ-SCN, db/db-Glom vs db/db-SCN, STZ–Glom vs db/db-Glom and db/db-SCN vs db/db-SCN) were performed (Fig. 1 [2]).
Transcriptional network comparisons
Transcriptional networks were generated from each DEG set (Fig. 1 [3]) based on gene–gene associations using a natural language programming strategy to elucidate functional relationships among DEGs (i.e. interactions at the network level). SciMiner, a literature mining system, analysed over 22 million abstracts in PubMed and extracted gene–gene co-citation information for human genes [16]. Transcriptional networks of the four DEG sets were constructed individually after converting mouse genes to human orthologues according to the National Center for Biotechnology Information (NCBI) HomoloGene build 68 and Genomatix annotated ortholog database.
Four pairwise comparisons (STZ-Glom vs STZ-SCN, db/db-Glom vs db/db-SCN, STZ–Glom vs db/db-Glom and STZ-SCN vs db/db-SCN) were performed. The large transcriptional networks comprising thousands of gene nodes were compared using a Tool for Approximate Subgraph Matching of Large Queries Efficiently (TALE) [17]. TALE compares network structures and extracts overlapping conserved relations between two networks. We used a mismatch parameter allowing 10% mismatches in generating the neighbourhood of the seed gene nodes as well as in extending the network, and examined the overlap of enriched biological functions and pathways. The validity of the identified enriched biological functions and pathways from the shared networks was examined by generating 1,000 gene sets for each of the four DEG sets containing the same number of genes randomly selected from the 20,016 genes on the microarray. These random gene sets were processed via the same transcriptional network analysis pipeline and the resulting shared subnetworks were examined for overlap of significant canonical pathways. Significant pathways from the real datasets were compared against these simulated datasets and the sizes of the shared networks were evaluated using a Z test.
Functional enrichment analysis
Over-represented biological functions among the four DEG sets were identified using Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) [18, 19]. Biological functions, represented by Gene Ontology (GO; http://geneontology.org/) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://genome.jp/kegg/) pathways, with a Benjamini–Hochberg (BH)-corrected p value < 0.05, were deemed significant. The complete list of 20,016 genes on the Affymetrix Mouse Genome 430 2.0 microarray was used as the background list during the enrichment analysis. Heat maps were generated based on the top ten most over-represented biological functions in each DEG set and clustered based on significance values (log-transformed BH-corrected p values) to visually represent overall similarities and differences between DEGs. Canonical signal transduction pathways were examined using GeneRanker (Genomatix Software, http://genomatix.de), a program that characterises large gene sets based on over 400 canonical signal transduction pathways collected from the NCI-Nature Pathway Interaction Database (http://pid.nci.nih.gov/) [20] and the Cancer Cell Map (http://pathwaycommons.org/) [21].
Results
Identification of changes in gene expression
We previously reported that STZ-treated DBA/2J mice and db/db mice exhibit a significant decrease in nerve conduction velocities and develop severe albuminuria, key features of DPN and DN, respectively (ESM 1 Table 1). Our published nerve and kidney gene expression profiles from murine models of type 1 and type 2 diabetes were examined in order to identify common and distinct gene expression changes across diabetes types and tissues (Fig. 1). As outlined in the methods section, the original transcriptomic datasets (STZ-Glom, db/db-Glom, STZ-SCN and db/db-SCN) were reanalysed using ChipInspector to identify new sets of DEGs containing both upregulated and downregulated genes corresponding to their respective non-diabetic controls. In the STZ-DBA/2J type 1 diabetes model, 4,790 and 4,069 DEGs were identified in glomeruli and SCN, respectively, compared with non-diabetic controls, while in the db/db type 2 diabetes model, 4,897 and 5,068 DEGs were identified in glomeruli and SCN, respectively (Fig. 2). Functional enrichment analyses were used to identify common and unique biological functions (Fig. 1 [1]). DAVID analysis was performed to examine enriched biological functions in terms of GO terms and KEGG pathways, while GeneRanker was used to identify signalling pathways. DAVID analysis revealed that genes related to adenyl nucleotide binding, adenyl ribonucleotide binding and purine ribonucleotide binding were highly enriched across the four DEG sets (ESM 2 Fig. 1). In terms of canonical signalling pathways identified by GeneRanker (ESM 1 Table 2), reelin was the only over-represented signalling pathway across the four DEG datasets. Pathways regulated across three of four DEG datasets included multiple JAK–STAT-related signalling pathways and VEGF signalling pathway.
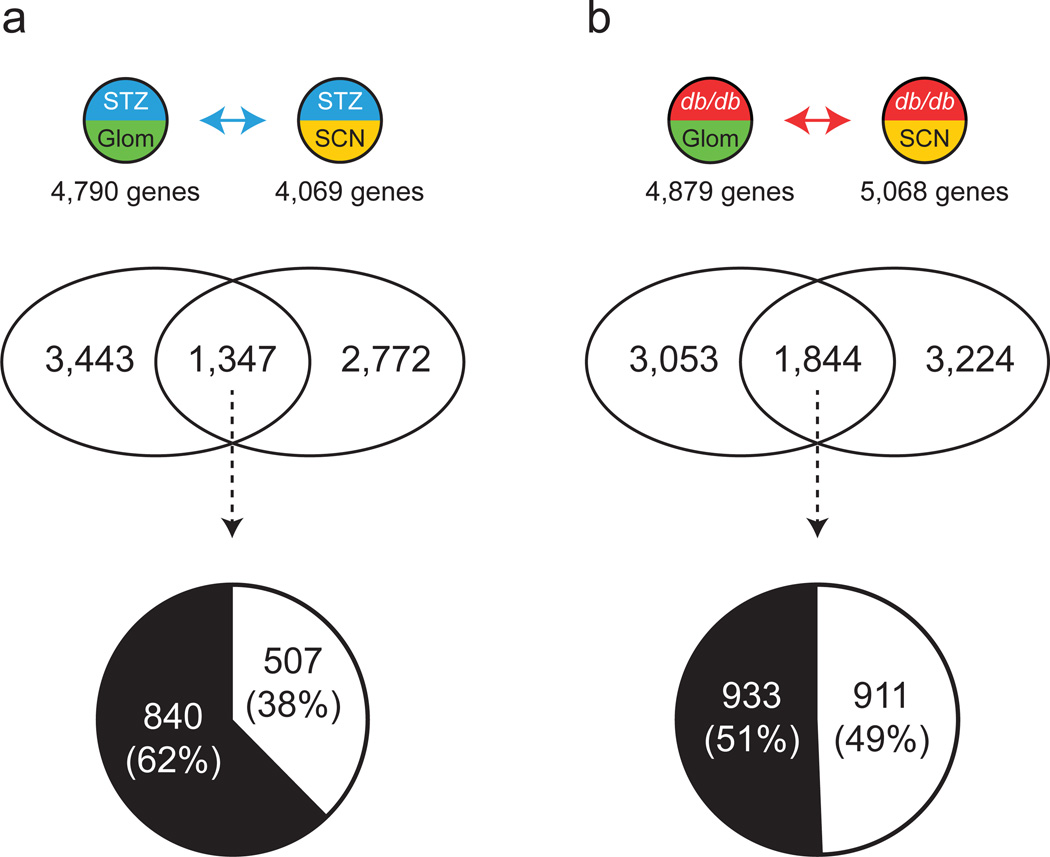
Fig. 2.
Comparison of kidney and nerve DEGs. Venn diagrams depict shared and unique DEGs from Glom and SCN in type 1 diabetes (a) and type 2 diabetes (b). The pie charts show percentages of concordant (change in same direction; white) and discordant (change in opposite direction; black) shared DEGs. See Fig. 1 for key
Pairwise comparisons
To ascertain common and distinct processes between complication-prone tissues or between the diabetes-like state of individual tissues, pairwise comparisons between DEG datasets was performed (Fig. 1 [2]) and the overlap of each pairwise comparison was further examined to assess concordance (in terms of direction of change) of common DEGs (Figs 2 and 3). Further breakdown of the concordant and discordant DEGs based on upregulation and downregulation is available in ESM 2 Figs 2 and 3. Subsequently, functional enrichment analysis was performed as outlined above.
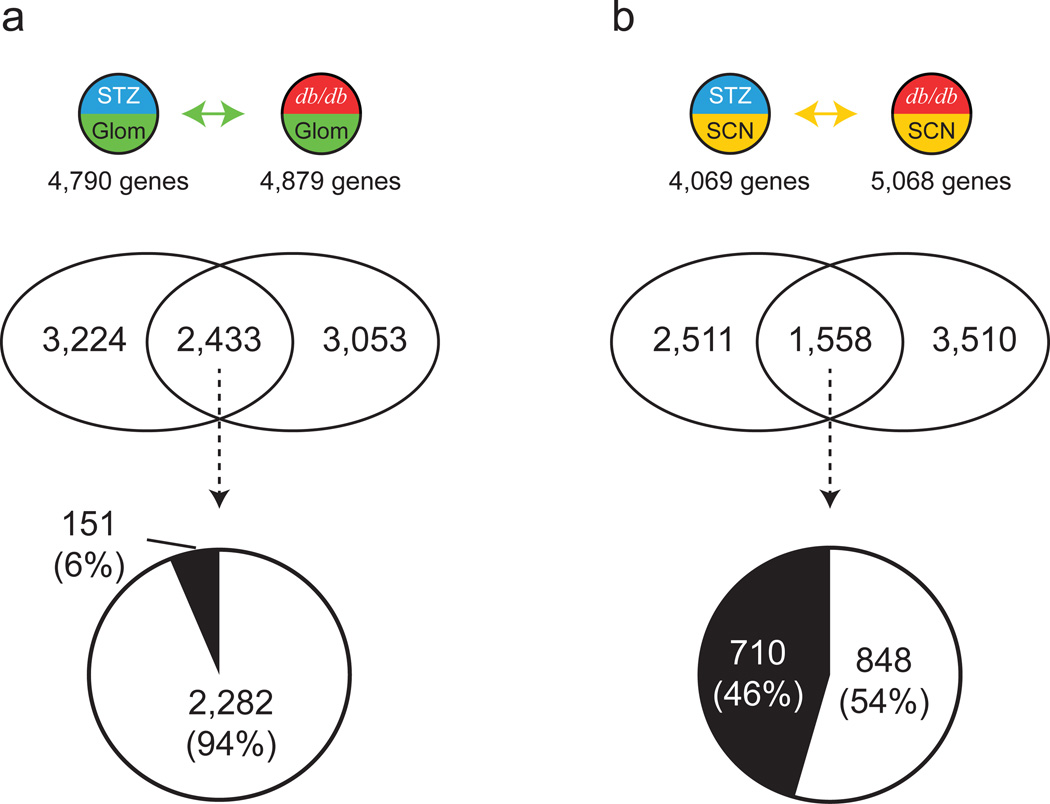
Fig. 3.
Comparison of type 1 diabetes and type 2 diabetes DEGs. Venn diagrams depict shared and unique DEGs from type 1 diabetes and type 2 diabetes in Glom (a) and SCN (b). The pie charts show percentages of concordant (change in same direction; white) and discordant (change in opposite direction; black) shared DEGs. See Fig. 1 for key
Cross-tissue comparison
Comparison of the STZ-Glom and STZ-SCN DEG sets identified 1,347 common DEGs (Fig. 2a and ESM 2 Fig. 2a). Approximately 38% of the common DEGs (n = 507) were concordant, while approximately 62% (n = 840) were discordant. DAVID analysis revealed genes related to mitochondrion to be highly enriched among the concordant DEGs (ESM 2 Fig. 4). In terms of canonical signalling pathways identified by GeneRanker (ESM 1 Table 3), those involved in eicosanoid metabolism, SREBP control of lipid synthesis and IL-3 signalling (JAK1/2, STAT5) were significantly over-represented among the concordant DEGs. Comparison of the db/db-Glom and db/db-SCN DEG sets identified 1,844 common DEGs (Fig. 2b and ESM 2 Fig. 2a), of which approximately 49% (n = 911) were concordant while 51% (n = 933) were discordant. No biological function was significantly over-represented among the concordant DEGs (ESM 2 Fig. 5). In terms of canonical signalling pathways, those involved in cyclin-dependent kinase (CDK) regulation of DNA replication, CD40L, mitogen-activated protein kinase (MAPKinase), NF-κB signalling and regulation of peroxisome proliferator-activated receptor γ coactivator 1-α (PGC1A) were significantly over-represented among the concordant DEGs (ESM 1 Table 4).
Cross-diabetes type comparison
By comparing DEG sets derived from the Glom of type 1 and type 2 diabetes models, STZ-Glom and db/db-Glom, we identified a large number of common DEGs (n = 2,433) (Fig. 3a and ESM 2 Fig. 3a). Approximately 94% (n = 2,282) of these showed concordant changes, while only 6% (n = 151) were discordant. These concordant DEGs had a high correlation in gene expression levels (Pearson correlation coefficient r = 0.955; ESM 2 Fig. 6). ESM 1 Table 5 lists the concordant DN genes with the largest expression changes. Genes related to lipid biosynthetic process and mitochondrion were significantly over-represented among the concordant DEGs (ESM 2 Fig. 7). In terms of canonical signalling pathways (ESM 1 Table 6), the JAK–STAT signalling and integrin pathways were significantly over-represented among the concordant DEGs.
From the two sets of DEGs derived from the SCN of murine models of diabetes (STZ-SCN and db/db-SCN), we identified 1,558 common DEGs (Fig. 3b and ESM 2 Fig. 3b). Approximately 54% (n = 848) of the common DEGs were concordant, while 46% (n = 710) were discordant. The 20 most upregulated and downregulated genes are listed in ESM 1 Table 7. Functional enrichment analysis revealed that genes related to nucleotide binding were significantly over-represented among the concordant DEGs, while discordant DEGs were highly enriched in categories related to protein transport, vesicle-mediated transport and endoplasmic reticulum (ESM 2 Fig. 8). In terms of canonical signalling pathways, neuroregulin and transcriptional repression by DNA methylation pathways were significantly over-represented among the concordant DEGs, while the insulin pathway, VEGFR3 and VEGFR1 signalling pathways were over-represented among the discordant DEGs (ESM 1 Table 8).
Transcriptional network analysis
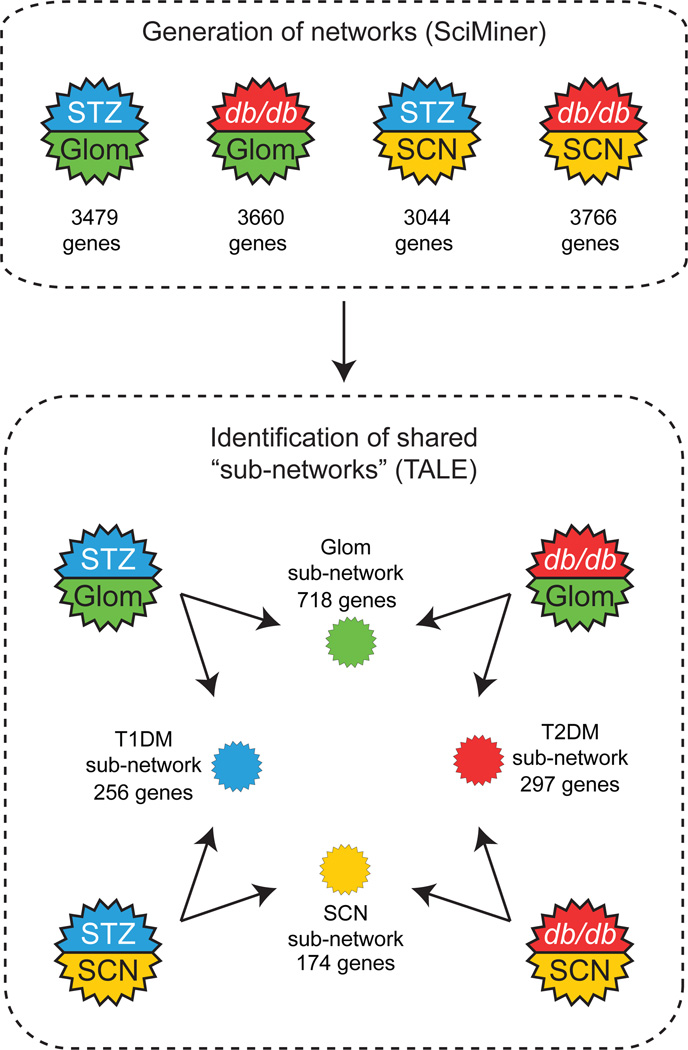
Transcriptional network analysis explores interconnections between DEGs to biologically refine datasets prior to functional enrichment analysis. Thus, the transcriptional networks based on gene–gene associations were generated and subsequently compared across tissue and diabetes type (Figs 1 and 4). Briefly, mouse DEGs were converted to orthologous human genes and networks were generated from each set based on literature-derived gene–gene associations identified by SciMiner [16]. Using a 10% TALE mismatch parameter to limit the shared networks in the four pairwise comparisons, networks ranging from 174–718 genes were identified (Fig. 4), representing approximately 5–15% of the input networks. The network images and Cytoscape network files are available in ESM 2 Fig. 9.
Fig. 4.
Transcriptional network analysis. TALE identified the shared networks between two transcriptional networks generated by SciMiner for the same four pairwise comparisons (STZ-SCN vs STZ-Glom, db/db-SCN vs db/db-Glom, STZ-Glom vs db/db-Glom and STZ-SCN vs db/db-SCN) using a 10% mismatch cut-off. Gene numbers in each shared network are noted. Close-up images of the shared networks are shown in ESM 2 Fig. 9. T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus. See Fig. 1 for key
Following generation of shared subnetworks, functional enrichment analysis was performed using DAVID (ESM 2 Fig. 10) and GeneRanker (Table 1) platforms to identify dysregulated biological functions and signalling pathways, respectively. The over-represented biological functions, in terms of GO and KEGG pathway terms, were highly similar among the shared networks across different comparisons (ESM 2 Fig. 10). GeneRanker identified 11 pathways that were enriched in all four shared subnetworks and 20 additional pathways that were enriched in three of four shared networks (Table 1). These pathways included multiple JAK–STAT- and VEGFR-related signalling pathways, as well as the reelin signalling pathway, suggesting that genes involved in these pathways are commonly dysregulated in both types of tissue and diabetes. To verify whether JAK–STAT signalling was significantly altered in diabetic nerve and kidney, simulated data were randomly generated and processed using the same network-level comparison approach to identify shared transcriptional networks. Between 23% and 38% of the shared networks identified from the simulated data included the JAK–STAT pathway as significant. The number of genes belonging to this pathway in the simulated datasets, however, were significantly fewer than in the real dataset (ESM 2 Fig. 11; Z test p < 0.01), verifying enhanced enrichment of JAK–STAT pathway genes in kidney and nerve in both diabetes types. These network-level data indicate that JAK–STAT pathways are highly dysregulated in both DPN and DN, independent of diabetes type.
Table 1.
Commonly enriched canonical pathways among the representative shared subnetworks
| Pathway description | No. of significant pathways |
Tissue comparison | Diabetes comparison | ||
|---|---|---|---|---|---|
| Glom: STZ vs db/db |
SCN: STZ vs db/db |
STZ: SCN vs Glom |
db/db SCN vs Glom |
||
| AKT(PKB)-Bad signalling (EPO signalling pathway (JAK2 STAT1 STAT3 STAT5)) | 4 | 0.010 | <0.001 | 0.020 | <0.001 |
| AKT(PKB)-Bad signalling (IL-7 signalling (JAK1 JAK3 STAT5)) | 4 | 0.032 | <0.001 | 0.020 | <0.001 |
| Angiopoietin receptor Tie2-mediated signalling | 4 | <0.001 | <0.001 | 0.004 | <0.001 |
| Cytokine receptor degradation signalling (JAK STAT Pathway and Regulation) | 4 | 0.005 | <0.001 | 0.003 | 0.002 |
| Gene expression (VEGF signalling pathway) | 4 | 0.013 | <0.001 | 0.040 | <0.001 |
| IL-6 signalling pathway (JAK1 JAK2 STAT3) | 4 | 0.013 | <0.001 | 0.025 | 0.032 |
| IL6-mediated signalling events | 4 | <0.001 | <0.001 | 0.010 | 0.006 |
| Reelin signalling pathway | 4 | 0.004 | 0.001 | <0.001 | <0.001 |
| SHP2 signalling | 4 | 0.013 | <0.001 | 0.003 | <0.001 |
| TGFBR | 4 | 0.025 | 0.004 | 0.001 | <0.001 |
| VEGFR3 signalling in lymphatic endothelium | 4 | <0.001 | <0.001 | 0.020 | 0.001 |
| Bioactive peptide-induced signalling pathway | 3 | 0.032 | 0.013 | <0.001 | |
| Co-regulation of androgen receptor activity | 3 | 0.032 | 0.050 | 0.005 | |
| Cyclins and cell cycle regulation | 3 | <0.001 | 0.002 | 0.005 | |
| Ephrin B reverse signalling | 3 | 0.032 | 0.006 | 0.010 | |
| HIF-1-α transcription factor network | 3 | 0.001 | 0.013 | 0.002 | |
| IGF-1 pathway | 3 | 0.001 | 0.006 | 0.016 | |
| IGF-1 signalling pathway | 3 | <0.001 | 0.050 | 0.016 | |
| Inhibition of cellular proliferation by Gleevec | 3 | <0.001 | 0.013 | 0.025 | |
| Integrins in angiogenesis | 3 | 0.013 | 0.016 | 0.003 | |
| JAK_STAT_MolecularVariation_1 | 3 | 0.008 | 0.020 | 0.020 | |
| JAK_STAT_MolecularVariation_2 | 3 | <0.001 | <0.001 | 0.016 | |
| KitReceptor | 3 | <0.001 | 0.032 | 0.016 | |
| Regulation of bad phosphorylation | 3 | 0.004 | <0.001 | 0.025 | |
| Signalling events mediated by TCPTP | 3 | 0.003 | 0.016 | 0.025 | |
| Signalling events mediated by VEGFR1 and VEGFR2 | 3 | 0.003 | 0.040 | 0.006 | |
| Syndecan-2-mediated signalling events | 3 | 0.013 | 0.005 | 0.020 | |
| Syndecan-4-mediated signalling events | 3 | <0.001 | 0.002 | 0.005 | |
| IGF-1 receptor and longevity | 3 | <0.001 | 0.025 | 0.001 | |
| Validated targets of C-MYC transcriptional repression | 3 | <0.001 | 0.005 | 0.005 | |
The values reported in this table represent the false discovery rate (FDR) for each comparison. No. of significant pathways: the number of comparisons in which the corresponding signalling pathway was significantly over-represented. C-MYC, c-myc avian myelocytomatosis viral oncogene homolog; EPO, erythropoietin; HIF-1-α, hypoxia-inducible factor 1-alpha; SPH2, SH2 domain-containing tyrosine phosphatase; TCPTP, T-cell protein tyrosine phosphatase; TGFBR, transforming growth factor beta receptor; VEGF, vascular endothelial growth factor
Discussion
Bioinformatics analyses have previously been used to investigate the mechanisms underlying either DN or DPN in the setting of type 1 or type 2 diabetes [10–13, 22]; however, no comparison of transcriptomic profiles has been made between the diabetes type or between complication-prone tissues. As such analyses may identify interesting similarities, we compared our previously published transcriptomic data obtained from murine models of type 1 and type 2 diabetes of DN and DPN at both the gene and transcriptional network levels. We found a large degree of shared DEGs with concordant regulation in DN across both type 1 and type 2 diabetes; in contrast, DEG expression in DPN was more distinct, indicating that DPN pathogenesis may differ fundamentally between type 1 and type 2 diabetes. We also identified dysregulation of the JAK–STAT pathway in both nerve and kidney regardless of diabetes type, suggesting that this pathway may represent a potential common therapeutic target. Overall, these analyses offer critical insight into common and distinct pathogenic mechanisms underlying DN and DPN in type 1 and type 2 diabetes, and further provide a novel foundation for the basis of understanding and designing appropriate therapeutic strategies.
Initial comparisons examined the transcriptomic changes associated with type 1 or type 2 diabetes in either the nerve or kidney at the gene level. For DN, pairwise comparisons of kidney DEG datasets across diabetes type identified a high degree of concordance, with approximately half of all glomerular gene expression changes being shared between diabetes type and 94% of shared DEGs occurring in the same direction (Fig. 3a). This high concordance supports the concept that pathogenic mechanisms between type 1 and type 2 diabetes in DN are conserved [13]. These pathways include: lipid biosynthesis and mitochondria, and JAK–STAT and integrin signalling. Furthermore, these findings agree with observations from our cross-species comparison of gene expression patterns, which was performed between murine models of DN and type 2 diabetes patients with DN [13], that identified dysregulation of JAK–STAT signalling to be common between both species. The current data collectively support the translational relevance of these alterations for both patients with type 1 diabetes and those with type 2 diabetes.
In contrast to our findings in the kidney, DEG dataset comparisons between the peripheral nerve in type 1 and type 2 diabetes revealed a marked difference in transcriptomic profiles between each disease, with fewer total shared genes than DN, and only half of these exhibiting concordant regulation (Fig. 3b). The higher degree of discordance suggests that, unlike DN, DPN pathogenesis differs fundamentally between type 1 and type 2 diabetes, an observation recently described in the literature in the patient population [23]. Of the concordant genes we identified, Map1b, Map4, Nefl and Tubb3 were downregulated, which may reflect SCN structural changes common to type 1 and type 2 diabetes DPN [24].
Functional enrichment analyses identified discordant alterations in peripheral nerve between type 1 and type 2 diabetes for genes involved in insulin signalling, endoplasmic reticulum stress and Golgi apparatus/protein trafficking, suggesting that each may be similarly dysregulated in disease, albeit in a different manner as a consequence of fundamental differences in diabetes type. For example, the canonical insulin pathway (representing insulin resistance) was significantly over-represented in our type 2 diabetes model only (ESM 1 Table 8), consistent with literature implicating insulin resistance in type 2 diabetes DPN pathogenesis, but not in type 1 diabetes, a disease brought about by insulinopenia [25–27]. Thus, the discordant pathways identified in the current analyses offer intriguing insight into differing potential mechanisms in type 1 and type 2 diabetes DPN.
Cross-complication comparisons revealed relatively low concordance between DN and DPN DEG datasets, independent of diabetes type. These findings suggest significant dysregulation of gene expression, likely reflecting distinct tissue-specific responses to diabetes (Fig. 2). In contrast to single gene expression comparisons, our transcriptional network-based comparisons identified a number of shared networks between the four DEG datasets. These networks were highly conserved in terms of enriched biological functions (ESM 2 Fig. 10) and signalling pathways (Table 1). Network-based comparisons also identified enriched functions and signalling pathways related to cell survival, proliferation, migration, blood vessel development, VEGFR3 endothelial signalling and angiopoietin signalling in both DN and DPN. While not new to our knowledge base, these categories provide reassurance for the long-standing idea that DPN and DN are ‘microvascular’ complications [28].
Of particular interest, our network-based comparisons identified enriched pathways related to JAK–STAT signalling as being commonly dysregulated across all DEG sets, suggesting a role in DN and DPN pathogenesis, irrespective of diabetes type. JAK–STAT is a stress-sensitive pathway activated by inflammatory mediators [29], oxidised LDL [30] and hyperglycaemia-induced reactive oxygen species [31], all drivers of DN and DPN pathogenesis. With JAK–STAT pathways being highly enriched in shared networks between patients and murine models of DN [13], and with rodent models of both type 1 and type 2 diabetes DN displaying improved kidney function following JAK–STAT inhibitor treatment [32, 33], this signalling pathway appears to be a viable therapeutic target. Indeed, clinical trials targeting the JAK–STAT pathway are currently underway for treatment of DN [34]. There is also evidence demonstrating a role for JAK–STAT signalling in DPN progression. Dysregulated JAK–STAT signalling has been identified in dorsal root ganglia of a rat model of type 1 diabetes and this may contribute to mitochondrial dysfunction resulting in peripheral nerve fibre degeneration [35, 36]. This pathway is also implicated in Wallerian degeneration in rat sciatic nerve after injury [37]. However, with evidence suggesting high discordancy in DPN between type 1 and type 2 diabetes, it is clear that further investigation is needed to fully elucidate the involvement of this pathway in DPN pathogenesis.
With data being drawn from different studies for these ad hoc analyses, there are several experimental limitations for which we could not control. This includes mice originating from different genetic backgrounds and studying mice at different ages. Furthermore, the number of mice is small and experiments were performed at different occasions. Ideally, this study should be repeated to minimise these compounding aspects, something which the results from our current findings warrant. Another limitation of the current study is that generation of the shared networks was based on gene–gene associations that were generated using a natural language programming strategy. As genes that are more comprehensively well-studied are more likely to be included in the resulting transcriptional networks than less well-studied genes, the network does not necessarily accurately reflect the actual gene expression network. The general accuracy of this approach, however, is still reasonable based on previously reported associations [13]. Indeed, our data using this relatively unbiased approach confirm the involvement of pathways that have been identified to be important in both DPN and DN [25, 38, 39].
In summary, our gene/network-level comparison of transcriptomic data suggest that the pathogenic mechanisms in DN are highly conserved across type 1 and type 2 diabetes, while those of DPN are largely distinct. These findings support the contention that diabetes type-specific therapies may be required to treat these common diabetic complications, whereby targeting lipid biosynthesis, cholesterol process and the JAK–STAT pathway may be beneficial for type 1 diabetes complications and, alternatively, targeting MAPKinase, NF-κB, PGC1A and the JAK–STAT pathway may be of greater benefit in type 2 diabetes. Notably, JAK–STAT pathway dysregulation in both DN and DPN, regardless of diabetes type, suggests that therapies targeting this pathway may be beneficial for both complications. Overall, the identified shared and distinct alterations in DN and DPN provide important insight into both common and unique pathogenic mechanisms and potential therapeutic targets for type 1 and type 2 diabetes complications, and future studies examining these pathways and targets are warranted.
Supplementary Material
Acknowledgments
The authors thank S. Sakowski Jacoby at University of Michigan for her expert editorial advice.
Funding
This work was supported by the US National Institutes of Health (NIH 1 R24 DK082841, to FCB, MK and ELF; NIH 1 DP3 DK094292, to FCB and ELF), the American Diabetes Association and the Novo Nordisk Foundation (to ELF), the Program for Neurology Research and Discovery at University of Michigan, the A. Alfred Taubman Medical Research Institute, Juvenile Diabetes Research Foundation Postdoctoral Fellowships (to JH and LMH) and the Applied Systems Biology Core of the George M. O’Brien Michigan Kidney Translational Core Center (NIH P30 DK081943).
Abbreviations
- BH
Benjamini–Hochberg
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEG
Differentially expressed gene
- DN
Diabetic nephropathy
- DPN
Diabetic peripheral neuropathy
- Glom
Glomerulus
- GO
Gene Ontology
- JAK
Janus kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPKinase
Mitogen-activated protein kinase
- PGC1A
Peroxisome proliferator-activated receptor γ coactivator 1-α
- PPAR
Peroxisome proliferator-activated receptor
- SCN
Sciatic nerve
- STAT
Signal transducer and activator of transcription
- STZ
Streptozotocin
- TALE
Tool for Approximate Subgraph Matching of Large Queries Efficiently
- VEGFR
Vascular endothelial growth factor receptor
Footnotes
Some of the data were presented as abstracts at the 2015 Peripheral Nerve Society Biennial Meeting and the American Diabetes Association 75th Scientific Sessions in 2015.
Duality of interest
The authors declare that there is no conflict of interest associated with this manuscript.
Contribution statement
JH analysed and interpreted data and wrote the manuscript. PDO and LMH interpreted data and wrote the manuscript. VN and BAM analysed data and revised the manuscript. MK, FCB, HVJ and ELF designed the project, oversaw project execution and data analysis and revised the manuscript. All authors approved the final version of the manuscript. JH and ELF are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.International Diabetes Federation. IDF Diabetes Atlas – 7th edition. [accessed 03 October 2015];2015 Available from http://idf.org/diabetesatlas. [PubMed] [Google Scholar]
- 2.Riggs JE. The latency between traumatic axonal injury and the onset of amyotrophic lateral sclerosis in young adult men. Mil Med. 2001;166:731–732. [PubMed] [Google Scholar]
- 3.Edwards JL, Vincent AM, Cheng HT, Feldman EL. Diabetic neuropathy: mechanisms to management. Pharmacol Ther. 2008;120:1–34. doi: 10.1016/j.pharmthera.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molitch ME, DeFronzo RA, Franz MJ, et al. Nephropathy in diabetes. Diabetes Care. 2004;27(Suppl. 1):S79–S83. doi: 10.2337/diacare.27.2007.s79. [DOI] [PubMed] [Google Scholar]
- 5.Boulton AJ, Vinik AI, Arezzo JC, et al. Diabetic neuropathies: a statement by the American Diabetes Association. Diabetes Care. 2005;28:956–962. doi: 10.2337/diacare.28.4.956. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez ER, Oley MA. The management of lower-extremity diabetic ulcers. Managed care interface. 2000;13:80–87. [PubMed] [Google Scholar]
- 7.Gordois A, Scuffham P, Shearer A, Oglesby A, Tobian JA. The health care costs of diabetic peripheral neuropathy in the US. Diabetes Care. 2003;26:1790–1795. doi: 10.2337/diacare.26.6.1790. [DOI] [PubMed] [Google Scholar]
- 8.Geraldes P, King GL. Activation of protein kinase C isoforms and its impact on diabetic complications. Circulation research. 2010;106:1319–1331. doi: 10.1161/CIRCRESAHA.110.217117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palsson R, Patel UD. Cardiovascular complications of diabetic kidney disease. Adv Chronic Kidney Dis. 2014;21:273–280. doi: 10.1053/j.ackd.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiggin TD, Kretzler M, Pennathur S, Sullivan KA, Brosius FC, Feldman EL. Rosiglitazone treatment reduces diabetic neuropathy in streptozotocin-treated DBA/2J mice. Endocrinology. 2008;149:4928–4937. doi: 10.1210/en.2008-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pande M, Hur J, Hong Y, et al. Transcriptional profiling of diabetic neuropathy in the BKS db/db mouse: a model of type 2 diabetes. Diabetes. 2011;60:1981–1989. doi: 10.2337/db10-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien PD, Hur J, Hayes JM, Backus C, Sakowski SA, Feldman EL. BTBR ob/ob mice as a novel diabetic neuropathy model: neurological characterization and gene expression analyses. Neurobiol Dis. 2014;73C:348–355. doi: 10.1016/j.nbd.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgin JB, Nair V, Zhang H, et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013;62:299–308. doi: 10.2337/db11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moll AG, Lindenmeyer MT, Kretzler M, Nelson PJ, Zimmer R, Cohen CD. Transcript-specific expression profiles derived from sequence-based analysis of standard microarrays. PLoS One. 2009;4:e4702. doi: 10.1371/journal.pone.0004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur J, Schuyler AD, States DJ, Feldman EL. SciMiner: web-based literature mining tool for target identification and functional enrichment analysis. Bioinformatics. 2009;25:838–840. doi: 10.1093/bioinformatics/btp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Y, Patel JM. TALE: A Tool for Approximate Large Graph Matching. Proceedings of the 2008 IEEE 24th International Conference on Data Engineering; IEEE Computer Society; 2008. pp. 963–972. [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer CF, Anthony K, Krupa S, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami EG, Gross BE, Demir E, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hur J, Sullivan KA, Pande M, et al. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain : a journal of neurology. 2011;134:3222–3235. doi: 10.1093/brain/awr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callaghan BC, Hur J, Feldman EL. Diabetic neuropathy: one disease or two? Curr Opin Neurol. 2012;25:536–541. doi: 10.1097/WCO.0b013e328357a797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pareyson D, Saveri P, Sagnelli A, Piscosquito G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neuroscience letters. 2015;596:66–77. doi: 10.1016/j.neulet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Vincent AM, Callaghan BC, Smith AL, Feldman EL. Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat Rev Neurol. 2011;7:573–583. doi: 10.1038/nrneurol.2011.137. [DOI] [PubMed] [Google Scholar]
- 26.Kim B, Feldman EL. Insulin resistance in the nervous system. Trends Endocrinol Metab. 2012;23:133–141. doi: 10.1016/j.tem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grote CW, Groover AL, Ryals JM, Geiger PC, Feldman EL, Wright DE. Peripheral nervous system insulin resistance in ob/ob mice. Acta Neuropathol Commun. 2013;1:15. doi: 10.1186/2051-5960-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hur J, Dauch JR, Hinder LM, et al. The metabolic syndrome and microvascular complications in a murine model of type 2 diabetes. Diabetes. 2015;64:3294–3304. doi: 10.2337/db15-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maziere C, Conte MA, Maziere JC. Activation of JAK2 by the oxidative stress generated with oxidized low-density lipoprotein. Free radical biology & medicine. 2001;31:1334–1340. doi: 10.1016/s0891-5849(01)00649-9. [DOI] [PubMed] [Google Scholar]
- 31.Simon AR, Rai U, Fanburg BL, Cochran BH. Activation of the JAK-STAT pathway by reactive oxygen species. The American journal of physiology. 1998;275:C1640–C1652. doi: 10.1152/ajpcell.1998.275.6.C1640. [DOI] [PubMed] [Google Scholar]
- 32.Ortiz-Munoz G, Lopez-Parra V, Lopez-Franco O, et al. Suppressors of cytokine signaling abrogate diabetic nephropathy. Journal of the American Society of Nephrology : JASN. 2010;21:763–772. doi: 10.1681/ASN.2009060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R, Zhong Y, Li X, et al. Role of transcription factor acetylation in diabetic kidney disease. Diabetes. 2014;63:2440–2453. doi: 10.2337/db13-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brosius FC, 3rd, He JC. JAK inhibition and progressive kidney disease. Current opinion in nephrology and hypertension. 2015;24:88–95. doi: 10.1097/MNH.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury SR, Saleh A, Akude E, et al. Ciliary neurotrophic factor reverses aberrant mitochondrial bioenergetics through the JAK/STAT pathway in cultured sensory neurons derived from streptozotocin-induced diabetic rodents. Cell Mol Neurobiol. 2014;34:643–649. doi: 10.1007/s10571-014-0054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleh A, Chowdhury SK, Smith DR, et al. Diabetes impairs an interleukin-1beta-dependent pathway that enhances neurite outgrowth through JAK/STAT3 modulation of mitochondrial bioenergetics in adult sensory neurons. Mol Brain. 2013;6:45. doi: 10.1186/1756-6606-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao D, Li M, Shen D, et al. Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci Bull. 2013;29:321–332. doi: 10.1007/s12264-013-1340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosius FC, Coward RJ. Podocytes, signaling pathways, and vascular factors in diabetic kidney disease. Adv Chronic Kidney Dis. 2014;21:304–310. doi: 10.1053/j.ackd.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro-Gonzalez JF, Mora-Fernandez C, Muros de Fuentes M, Garcia-Perez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol. 2011;7:327–340. doi: 10.1038/nrneph.2011.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.