Abstract
Platelets play a central role in ischemic cardiovascular events. Cardiovascular disease (CVD) is a major cause of death worldwide. Numerous genome-wide association studies (GWASs) have identified loci associated with CVD risk. However, our understanding of how these variants contribute to disease is limited. Using data from the platelet RNA and expression 1 (PRAX1) study, we analyzed cis expression quantitative trait loci (eQTLs) in platelets from 154 normal human subjects. We confirmed these results in silico by performing allele-specific expression (ASE) analysis, which demonstrated that the allelic directionality of eQTLs and ASE patterns correlate significantly. Comparison of platelet eQTLs with data from the Genotype-Tissue Expression (GTEx) project revealed that a number of platelet eQTLs are platelet specific and that platelet eQTL peaks localize to the gene body at a higher rate than eQTLs from other tissues. Upon integration with data from previously published GWASs, we found that the trait-associated variant rs1474868 coincides with the eQTL peak for mitofusin 2 (MFN2). Additional experimental and computational analyses revealed that this eQTL is linked to an unannotated alternate MFN2 start site preferentially expressed in platelets. Integration of phenotype data from the PRAX1 study showed that MFN2 expression levels were significantly associated with platelet count. This study links the variant rs1474868 to a platelet-specific regulatory role for MFN2 and demonstrates the utility of integrating multi-omic data with eQTL analysis in disease-relevant tissues for interpreting GWAS results.
Introduction
Cardiovascular disease (CVD) is a major cause of death worldwide. Thrombotic events, such as myocardial infarction (MI) and stroke, occur when occlusive platelet thrombi form at the site of a ruptured atherosclerotic plaque.1, 2, 3, 4, 5 The critical role of platelets in the pathophysiology of CVD events is further underscored by the standard use of anti-platelet agents in the management of the disease. However, there is little mechanistic understanding to explain why some individuals form occlusive thrombi at the site of ruptured atherosclerotic plaques, whereas other individuals repair the wound without occluding the vessel. Inter-individual variation in platelet reactivity,6 volume, and number7 are likely contributors given that these platelet phenotypes have been prospectively shown to be a risk for recurrent coronary syndromes.8 Abundant evidence suggests that CVD has a genetic component.9, 10, 11, 12, 13 However, the genetic mechanisms underlying CVD risk in the general population are not fully characterized.
Genome-wide associations studies (GWASs) have identified links between CVD risk and genetic variants. However, many disease-associated variants are located in non-coding regions of the genome, making it difficult to identify the mechanism of function.14, 15, 16 Moreover, variations at the DNA level do not pinpoint a tissue of action critical to the development of a higher-level GWAS phenotype such as CVD.17 Integration of expression quantitative trait loci (eQTLs) from relevant tissues with GWAS results has been proposed as an important approach to overcoming these challenges.18 eQTLs can provide mechanistic insights into gene expression in disease-relevant tissues by identifying cell-type-specific regulatory variants. Therefore, eQTL data are considered a key intermediate to connecting expression changes with higher-level phenotypes and clinical outcomes represented by GWAS results.19, 20
Given the role platelets play in the etiology and pathogenesis of CVD, we hypothesized that eQTL analysis in human platelets might connect regulatory variants in platelets with functional expression changes at loci previously linked to CVD by GWASs. Although platelets lack a nucleus, they inherit their transcriptome from their parent bone marrow progenitors, megakaryocytes. Because it is far easier to obtain platelets and because their transcriptome strongly correlates with that of megakaryocytes,21, 22 platelet RNA represents a valuable resource for identifying and characterizing relevant gene expression in thrombotic disorders. We now report a generalizable approach for linking GWAS hits to physiology and disease. This approach integrates cell-type-specific eQTL analysis with primary-tissue multi-omic data from the platelet RNA and expression 1 (PRAX1) study together with other public datasets, such as those of the Genotype-Tissue Expression (GTEx) project23 and ENCODE.24 Our analyses and experiments with rs1474868, previously associated with MI by GWASs,25 revealed that rs1474868 marks the platelet MFN2 (MIM: 608507) eQTL peak and drives expression of a platelet-specific alternative 5′ start site of MFN2. In addition, we identified a significant association between MFN2 expression levels and platelet count. Overall, our findings demonstrate the power of integrating primary-tissue eQTL analysis with multi-omic data to interpret and inform functional genomics of GWASs in CVD and other diseases.
Material and Methods
The PRAX1 Study
The goal of the PRAX1 study was to identify gene-expression correlates responsible for inter-individual variation in platelet function. As described earlier,26, 27 154 healthy individuals (80 of European and 74 of African ancestry) were recruited between 2010 and 2011. For all individuals, demographic information was collected, and platelet functional phenotyping, genome-wide genotyping, and global mRNA profiling were performed. We obtained a complete blood count and mean platelet volume by using an ABX Micros 60 CS (Horiba ABX). Informed consent was obtained from all participants with the approval of the institutional review boards of Baylor College of Medicine in Houston and Thomas Jefferson University in Philadelphia. Research was conducted in accordance with the Declaration of Helsinki.
Gene Expression Data from the PRAX1 Study
We measured gene expression of PRAX1 samples by using the Human Gene 1.0 ST microarray (Affymetrix) as described previously28 (these data are available in the Gene Expression Omnibus under accession number GEO: GSE49921). eQTL mapping analysis was restricted to 5,695 autosomal, uniquely mapping, commonly expressed platelet mRNAs as defined in Table S1 of Simon et al.28
RNA-Seq Data from the PRAX1 Study
A subset of ten subjects from the PRAX1 study were analyzed with RNA-sequencing (RNA-seq) technology in an independent study by Londin et al.29 (The corresponding next-generation sequencing data files are available in the Sequence Read Archive under study identifier SRA: SRP028846.) Next, we converted raw sequencing files from SOLiD to FASTQ format and trimmed adapters with the “cutadapt” tool. To improve mapping accuracy, we performed a two-pass alignment strategy as outlined in Engstrom et al.30 First, we used STAR v.2.3.1 to align reads to the UCSC Genome Browser hg19 reference genome. We then used the resulting file on splice junction loci and the hg19 FASTA sequence to generate sample-specific genomes with the --genomeGenerate and --sjdbFileChrStartEnd functions. Reads were subsequently aligned to each sample-specific hg19 index. As mentioned, these RNA-seq data have been previously published, and no global quantification of gene expression was performed for this current study, given that the RNA-seq data were exclusively used in the allelic-imbalance analysis and detailed follow-up analyses of MFN2 transcription. The RNA-seq data were not used in the eQTL-mapping analysis.
Genotype Data from the PRAX1 Study
DNA from the buffy coats of PRAX1 study subjects was hybridized to the HumanOmni5 array (Illumina) as described earlier26 and genotyped for approximately five million markers. Genotype data were restricted to 1,927,172 markers passing a cohort-specific minor-allele-frequency cutoff of 5%.
eQTL Mapping
We applied a two-pass eQTL-mapping strategy. First, we used cohort genotype- and microarray-based expression data to identify 612 genes containing at least one significant eQTL association (p < 1e−6, multiple linear regression), termed eGenes. Next, using IMPUTE2 and data from the 1000 Genomes Project, we imputed additional genotypes only for those 612 eGenes. Finally, we repeated association testing by using the extended set of both genotyped (368,794) and imputed (1,071,757) variants to fine map the platelet eQTL landscape of these 612 eGenes. We used R statistical software v.3.0.1 in conjunction with the MatrixEQTL package31 to determine significant associations between variant-allele dosage and gene expression levels. We restricted the cis search space to within 500 kb of the transcription start site and used variant-allele dosage (0, 1, and 2) to additively model associations. The following parameters were used: useModel = modelLINEAR, pvOutputThreshold.cis = 1e−6, and cisDist = 5e−5. To account for hidden trans covariates, we used the probabilistic estimation of expression residuals (PEER) framework32 and inferred 15 PEER factors. These PEER factors capture trans variation, such as batch effects or global ethnic differences, present in the gene expression data. Accounting for these trans factors when modeling cis eQTLs improves the power to detect true cis eQTLs by reducing spurious false-positive associations.32 Therefore, we included these 15 PEER factors—in addition to age, gender, and the first two genotype principal components, which completely account for self-identified ethnicity26—as covariates in the model. The same settings were used during the first (genotyped variants) and second (genotyped and imputed variants) passes of the eQTL mapping. Significant eQTLs were defined as variants associated with expression levels of genes at a p value threshold of 1e−6 to account for multiple testing. eGenes were defined as genes associated with at least one significant eQTL.
Imputation
To obtain a more granular view of the eQTL landscape, we imputed additional genotypes across the cis search space of each eGene prior to the second pass of the eQTL mapping. We used IMPUTE2 v.2.3.2 software33 and the 1000 Genomes phase 1 integrated variant-set release as a reference to infer additional genotypes. The imputation software first phases the study genotypes and subsequently imputes additional genotypes by integrating study-subject phasing information with known haplotypes from the 1000 Genomes reference data. We followed the instructions provided on the IMPUTE2 website. In short, original genotypes were extracted for each eGene spanning the 1 Mb cis search space. Genotypes were pre-phased with the --prephase_g function. We downloaded prephased 1000 Genomes phase 1 integrated haplotypes. Using these pre-phased haplotypes as a reference, we imputed samples from the PRAX1 study with the --use_prephased_g function, for which haplotype and legend files for all 1,092 1000 Genomes samples (246 AFR [African], 181 AMR [admixed American], 286 EAS [East Asian], and 379 EUR [European]) and the PRAX1 study genotype were provided with the --h, --l, and --g parameters, respectively. Imputed genotypes called with high confidence (maximum probability > 0.9) were used in subsequent analysis. Accuracy analysis was performed with the IMPUTE2 internal cross-validation system. During each run, IMPUTE2 masks a subset of observed variants and subsequently imputes these genotype data by using flanking markers. The imputation accuracy can be quantified as the percentage of concordance between known and imputed genotypes of these cross-validated variants. In the cross-validation analyses, the percentage of concordance across the 612 eGene regions was very high (minimum = 96.4%, median = 98.7%, maximum = 99.7%), indicating accurate imputation results. Because of the nature of overlapping cis search spaces, some variants were imputed multiple times. In these cases, imputed genotypes with discordant results were excluded.
Conditional eQTL Analysis
To investigate how many independent eQTL signals contribute to the regulation of a single eGene, we performed conditional eQTL analysis. This analysis followed the procedures outlined in the eQTL Mapping section, but additionally, the genotype of the marker for the corresponding eQTL peak was added as a covariate for each of the 612 eGenes. If no variants remained significantly associated with expression of the corresponding eGene (p < 1e−6) after we accounted for the genotype of the marker at the eQTL peak, we defined the given eGene as regulated by a single genetic signal. If a significant association remained, we interpreted it as evidence of a second independent genetic signal.
Analysis of Allele-Specific Expression
We analyzed allele-specific expression (ASE) with our ten RNA-seq samples from the PRAX1 study. Using the reference genome to map RNA-seq data can be biased by genetic variation, given that reads carrying the non-reference allele are less likely to map correctly.34 This mapping bias is called reference-allele bias and is a non-trivial confounding variable in ASE analysis.35 To overcome this challenge, we aligned RNA-seq reads to inferred personalized genomes on the basis of the genotype data observed in the study. This specialized alignment was used in the ASE analysis only. To generate personalized genomes corresponding to the ten RNA-seq samples from the PRAX1 study, we used IMPUTE2 v.2.3.2 to impute genotypes across 22 autosomal chromosomes (as outlined in the Imputation section); this procedure generates a high-density map of the personal variation present in each sample. Following the recommendations on the IMPUTE2 website regarding the imputation of entire chromosomes, we divided each chromosome into 500 kb intervals and imputed each interval separately. Phasing information obtained from the imputation can be used for inferring parental haplotypes across genetic regions, such as genes. We used the “vcf2diplod” function provided in the AlleleSeq36 software to generate two paternal-genome FASTA files for each sample. Next, we used the --genomeGenerate parameter of STAR v.2.3.1 to generate two separate parental-genome indices for each sample. Because of imputed insertion and deletions, the “liftOver”37 function was needed to convert genomic coordinates between genomes. Next, for each sample, we used STAR v.2.3.1 to align the corresponding FASTQ read file to the two paternal genomes independently. Figure S1 shows that reference-allele bias decreased when RNA-seq reads were aligned to personalized genomes rather than the hg19 reference genome. Next, read pileups were generated with an adapted version of the pipeline outlined in Harvey et al.38 In short, SAMtools39 was used to generate read pileups at 128,163 (genotyped and imputed) sites, which were used in the eQTL-mapping analysis and fell into coding regions according to the UCSC Genome Browser hg19 knownGene table downloaded in January 2015. Next, heterozygous sites within each of the ten PRAX1 study samples were identified and assessed for coverage. Heterozygous coding sites with ten or more reads in a sample were required for the site to be considered in the ASE analysis. To ensure that mapping bias did not affect the calculations, we calculated and recorded the pair of counts for each parental allele from the alignment to the corresponding personalized parental genome. Because a gene can harbor more than one heterozygous coding site in a given sample, we picked the pileup site with the lowest eQTL association p value to examine allelic imbalance. Allelic imbalance was defined as an unequal count of two alleles at a given heterozygous pileup site in a sample. To evaluate the global correspondence between allelic imbalance and the genome-wide global eQTL findings, we used Fisher’s exact test. The null hypothesis states that the directionality of allelic imbalance is independent of the allelic directionality of the eQTL. In other words, the likelihood of observing a higher fraction of the reference allele than of the alternative allele at a given pileup site is independent of the allele implicated with higher expression levels in the eQTL analysis. Directional deviation from this null hypothesis would indicate concordance between allelic imbalances and eQTLs. For the exemplary eGene CXCL5, we developed a separate statistical approach. In order to give equal weight to each sample, we downsampled allelic counts of each heterozygous sample to the minimum sum of allelic counts among all heterozygous samples. Next, we summed these counts into an aggregate pair of counts and performed a binomial test. The null hypothesis for this test states that the likelihoods of observing the reference and alternative alleles are equal, so that reads are generated as coin-flipping trials of a fair coin. Appropriate deviation from this null hypothesis can indicate that allelic imbalance corresponds to the allelic directionality of the eQTL; p values were determined for a one-sided binomial test.
GTEx Data
These data consist of a resource database and associated tissue bank for studying the relationship between genetic variation and gene expression in human tissues. We downloaded the GTEx_Analysis_V6_eQTLs.tar.gz table, containing significant single-tissue variant-gene associations from the GTEx website. These data catalog eQTLs across 46 different tissues. To increase comparability, we restricted analysis to 20 GTEx tissues with a sample size equal to or higher than our PRAX1 study cohort (154) and redefined all GTEx eGenes according to our definition: GTEx eGenes for each tissue were defined as genes containing at least one significant eQTL (p < 1e−6) within 500 kb of the transcription start site. Our definition of an eGene was more stringent than the rules applied by GTEx. Across the 20 tissues analyzed, on average, a subset of 71% of GTEx eGenes remained an eGene according to our definition. An eQTL peak was defined as the genomic location of the most strongly associated eQTL variant(s) within each eGene. Table S3 lists eQTL peaks for GTEx and platelet eGenes. To account for varying sample sizes inherent in the GTEx eQTL dataset, we used Jaccard similarity to compare overlap between platelet and GTEx eGenes. To perform a more granular comparison, we conducted an additional analysis at the variant-gene association level. For each eGene we identified the strongest platelet eQTL association present in each of the GTEx tissues. Next, we compared the sign of the allelic effect of the eQTL association. Concordance between the sign of the allelic effects of eQTL associations in platelets and each GTEx tissue was calculated with Fisher’s exact test. The null hypothesis is that the sign of the allelic effect of the eQTL in platelets is independent of the sign of the allelic effect in the tissue of comparison. Directional deviation from this hypothesis can indicate concordance of allelic effects between the tissues at the variant-gene level.
CVD GWAS Associations
We used the Phenotype-Genotype Integrator25 to download previously published GWAS results. We restricted analyses to variants associated with traits annotated to the Medical Subject Headings (MeSH) category “cardiovascular diseases” and an association p value below 1e−4. These data catalog variant-phenotype associations and were integrated with the variant-eGene associations identified in our eQTL analysis.
Additional Platelet RNA-Seq Data
We downloaded additional platelet RNA-seq data from three previous studies by Rowley et al.,40 Kissopoulou et al.,41 and Eicher et al.42 For the Rowley et al. data, RNA-seq alignments were downloaded according to the instructions outlined in Rowley et al.40 For the Kissopoulou et al. data,41 next-generation sequencing FASTQ files were downloaded from the ArrayExpress website under accession number E-MTAB-1846. Reads were aligned to the hg19 reference genome with STAR v.2.3.1 using default parameters. For the Eicher et al. data,42 next-generation sequencing FASTQ files and corresponding metadata were downloaded from study SRP053296 in the Sequence Read Archive. Sample runs SRR1792698 and SRR1792703 contained errors in the read-pairing annotation and were excluded from further analysis. Reads were aligned to the hg19 reference genome with STAR v.2.3.1 using default parameters. These data comprise RNA-seq data from different library-preparation protocols (total RNA, polyA, and rRNA depleted) and sequencing technologies (Illumina and SOLiD). Using data produced by different groups via different techniques and protocols to validate RNA-seq-based findings limits the chance of false-positive associations due to technical artifacts of the sequencing pipeline. Therefore, we used these independent observations as evidence supporting the expression of exon 2b in platelet MFN2 and an increase in sample size for associative analyses.
In Silico Cross-Tissue Analysis
The Epigenome Roadmap project contains mRNA-seq data from stem cells and primary ex vivo tissues selected to represent the normal counterparts of tissues and organ systems frequently involved in human disease. We used the Genboree Workbench43 to analyze mRNA-seq expression data for the MFN2 region from 69 samples. As input, we provided a customized bed file of 10 bp intervals spanning the MFN2 region. Using this file, we calculated average read density per interval for each sample. Replicates within a sample type were averaged. Read-density averages across these intervals for all samples can be found in Table S4.
PCR Confirmation of MFN2 Exon 2b
Leukocyte-depleted platelet RNA (henceforth referred to as “platelet RNA”) was obtained via density centrifugation and immune depletion of CD45+ cells as described previously.44 500 ng of platelet RNA and SuperScript III (ThermoFisher Scientific) were used for cDNA generation. PCR was performed to amplify and detect the presence of the canonical MFN2 sequence (GenBank: NM_014874.3) spanning exons 1–3 (primers F1 and R1), the platelet MFN2 sequence spanning exons 2b and 3 (primers F2 and R2), and exons 7–10 in ITG2B as a positive control for RNA quality (primers ITG2B-F and ITG2B-R). The primer sequences were as follows: 5′-GGTGACGTAGTGAGTGTGATG-3′ (F1), 5′-CACTTAAGCACTTTGTCACTGC-3′ (R1), 5′-CCCAGCTGACCTGTTTATTTG-3′ (F2), 5′-CTACATCCAGGAGAGCGC-3′ (R2), 5′-AGAGTACTTCGACGGCTACTG-3′ (ITG2B-F), 5′-GCAGCCTCTGGTAGTAGGAA-3′ (ITG2B-R).
5′ Rapid Amplification of cDNA Ends of MFN2
To map the definitive 5′ end of the platelet MFN2 transcript, we used the SMARTer RACE (5′ rapid amplification of cDNA ends) 5′/3′ Kit (Clontech) as described by the manufacturer. In brief, 1 μg platelet RNA was used for synthesis of first-strand cDNA, which added a universal priming sequence to the 5′ end of the first strand via template switching. The 5′ end of the cDNA was then amplified in two rounds of nested PCR using primers that anneal to the universal priming sequence (provided by the manufacturer) and two gene-specific primers: 5′-CACTTAAGCACTTTGTCACTGC-3′ (GSP-1) and 5′-CGGGTAGAGGGCACAGATGGCCATGAGG-3′ (GSP-2). The products of the second round of amplification were run on an agarose gel cloned into the pRACE vector with the provided In-Fusion HD Cloning Kit. The cloned products were then sequenced.
Relative Abundance of MFN2 Isoforms
The relative abundance of MFN2 isoforms was calculated with the splice-loci output generated from the STAR alignments for the RNA-seq data from the PRAX1 study, Kissopoulou et al., and Eicher et al. To estimate the relative abundance of the MFN2 isoform containing exon 2b, we calculated the number of spliced reads that mapped from the 3′ border of exon 2b (chr1: 12,044,353, hg19) to the 5′ border of exon 3 (chr1: 12,049,221, hg19) and divided it by the number of all spliced reads ending at the 5′ border of exon 3. Samples with ten or fewer junction reads ending at the 5′ border of exon 3 were excluded.
Inference of rs1474868 Genotype
Because the variant rs1474868 falls into a coding region, we were able to infer the rs1474868 genotype by using the STAR alignments for the Kissopoulou et al.41 and Eicher et al.42 data. Samples with more than one read containing the C or T allele were assigned the corresponding genotype. Samples with a total of fewer than two reads were assigned the unknown status.
Transcription Factor Binding Sites
ENCODE chromatin immunoprecipitation followed by sequencing (ChIP-seq) data on transcription factor binding were downloaded from UCSC Genome Browser. We subsequently restricted all data to experiments carried out in the hematopoietic-derived K562 cells.
MFN2 Luciferase Assays
DNA from two PRAX1 study subjects homozygous for C or T at SNP rs3766744 was used as a template for amplifying two regions surrounding this SNP: (1) a 655 bp region (chr1: 12,043,332–12,043,986, hg19) was amplified with primers 5′-GTTAGCCAGGATGGTCTCGAAC-3′ and 5′-AACTTGCAAGCTGAGATTCCAC-3′, and (2) a 1,928 bp region (chr1: 12,042,341–12,044,268, hg19) was amplified with primers 5′-TTCGCTGGTTGCTTAGAGAG-3′ and 5′-AAGTTCCCTGCCATCAGAAG-3′. The amplified fragments were then cloned upstream of a basal promoter in the pGL4.28-luc vector (Promega). A total of four vectors were produced: MFN2:655(C)-Luc, MFN2:665(T)-Luc, MFN2:1928(C)-Luc, and MFN2:1928(T)-Luc. 2 μg of a MFN2-Luc vector was transfected along with 0.5 μg CMV-β-gal-expressing plasmid into 5 × 105 K562 cells. 24 hr after transfection, lysates were measured for luciferase activity on a FLUOstar OPTIMA plate reader (BMG Labtech) and β-gal activity with a beta-Galactosidase Assay Kit (ThermoFisher). Results are expressed as a ratio of luciferase activity normalized to β-gal activity.
Statistical Analyses
Statistical analyses were conducted with R statistical software. To evaluate the significance of differences in the distribution of eQTL peaks between platelets and GTEx tissues, we performed the Grubbs single-outlier test on the proportion of eQTL peaks mapping into the gene body by using the “grubbs.test” function from the “outliers” R package. We used Spearman’s rank correlation to assess significance between the exon 2b inclusion rate and rs1474868 allele dosage. We used Mann-Whitney tests as implemented in the “wilcox.test” R function to evaluate significance of differential luciferase activity. We used the LDlink online tool to assess linkage disequilibrium. To best represent our study cohort, we restricted linkage-disequilibrium calculations to the 1000 Genomes populations ASW (Americans of African ancestry in southwest USA) and CEU (Utah residents with ancestry from northern and western Europe from the CEPH collection). We used a multiple-linear-regression framework implemented in the R “aov” function to evaluate the significance of the association among platelet count, the rs3766744 genotype, and MFN2 expression levels in the PRAX1 study data. For the rs3766744 variant, the model explained platelet count by using rs3766744 allele dosage after accounting for the marginally significant covariates body mass index, gender, and self-identified ethnicity. For MFN2 expression levels, the model explained platelet count by using MFN2 expression levels after accounting for the same covariates.
Results
Two-Pass cis eQTL Mapping Identifies 612 Platelet eGenes
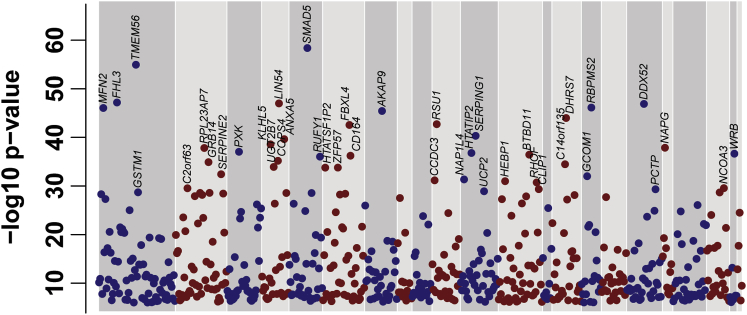
The PRAX1 study profiled mRNA expression levels from platelets and genotypes of 154 healthy human subjects on array platforms.26 To decrease computational burden while increasing genetic resolution, we applied a two-pass strategy for mapping the landscape of human platelet cis eQTLs (see Material and Methods). 5,695 autosomal commonly expressed platelet genes, as defined in Simon et al.,28 were used in the analysis. We identified a total of 44,940 significant eQTLs (p < 1e−6) implicated in expression association with 612 unique genes, termed platelet eGenes. The rates of the number of eGenes per gene tested and significant eQTLs per eGene were comparable to those of other studies with similar sample sizes (Figure S2). Figure 1 presents the distribution of the strongest association p values for each eGene across the 22 autosomal chromosomes tested. To assess how many eQTLs contribute to the association signal of an eGene, we performed conditional eQTL analysis. 560 (92%) eGenes did not contain any significant eQTLs after we accounted for the eQTL peak, indicating that most platelet eGenes are regulated by a single genetic signal. The MatrixEQTL output for all 612 platelet eGenes is contained in Table S1. These results can be interactively queried on our Plateletomics website (see Web Resources).
Figure 1.
Genome-wide View of Platelet eGenes
The y axis shows maximum −log10 p values of eQTLs for 612 platelet eGenes. The x axis corresponds to ordering along 22 autosomal chromosomes. The top 40 eGene symbols have been added to the plot. Color coding of points and backgrounds is used to help separate chromosomes visually.
ASE Analysis Confirms Global eQTL Results In Silico
An eQTL measures the association between genotype and gene expression across samples, whereas ASE measures allelic imbalance at a heterozygous site within a single sample.45 Concordance between the allelic directionality of allelic imbalance and an eQTL can be interpreted as an independent validation of the eQTL.46 Figure S3 contains a detailed schematic describing the relationship between eQTLs and ASE. Therefore, we performed allelic-imbalance analysis (see Material and Methods). In brief, to validate eGenes by using allelic imbalance, we generated read pileups at heterozygous coding sites of all platelet eGenes in all ten RNA-seq samples from the PRAX1 study. A total of 116 (19%) eGenes contained at least one qualifying pileup site with marginal eQTL effect (p < 1e−4) in one or more of the ten samples and provided the opportunity to confirm eQTL effects via allelic-imbalance analysis. We classified all pileup sites of these 116 eGenes according to the allelic directionality of the linked eQTL. Reference and alternatives sites were defined as pileup sites where the linked eQTL showed higher expression of the reference and alternative allele, respectively.
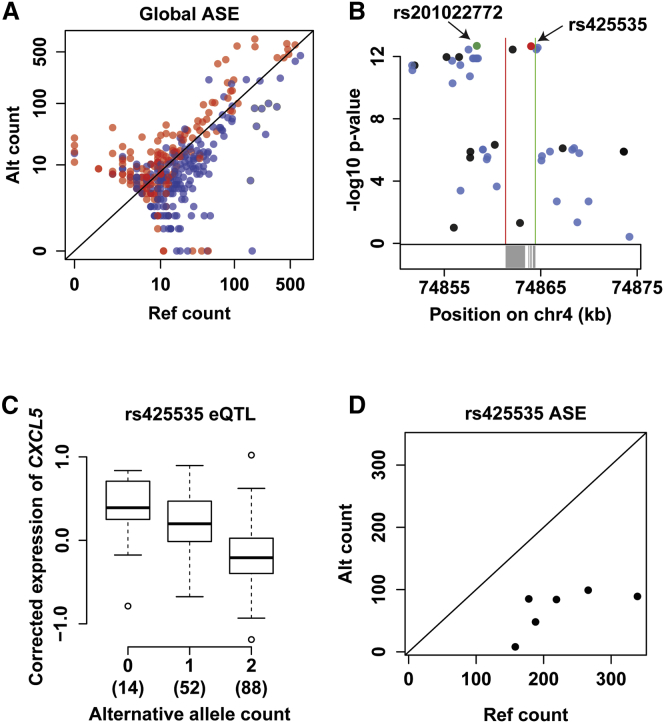
Figure 2A presents the counts of reference and alternative alleles observed at 358 pileup sites corresponding to 116 unique eGenes (Table S2). 266 (75%) alternative and reference sites fell above and below the diagonal line, respectively. The allelic imbalances are in concordance with the allelic directionality of the eQTLs and demonstrate the agreement between eQTLs and ASE (p < 1e−19, Fisher’s exact test).
Figure 2.
Confirmation of eQTLs via ASE
(A) Points represent allele counts at 358 heterozygous coding sites with a marginal eQTL effect (p < 1e−4). Reference and alternatives sites are colored in blue and red, respectively. The black diagonal line indicates the null-hypothesis distribution of equal counts of the reference and alternative alleles.
(B) Manhattan plot of eQTL landscape for eGene CXCL5. The y and x axes correspond to association p values and genomic coordinates, respectively. Blue and black points show imputed and genotyped variants, respectively. The red point shows reference site rs425535, and the green point shows lead eQTL rs201022772. Transcription start and end sites of CXCL5 are indicated by green and red vertical lines, respectively. Gray rectangles below the plot represent CXCL5 exons.
(C) Boxplot shows significant association between CXCL5 expression levels and rs425535 allele dosage (p < 10e−12, multiple linear regression).The x axis shows the alternative allele count. Numbers in parentheses indicate the number of samples within each group. The y axis represents expression levels of CXCL5 corrected for covariates. The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
(D) Reference and alternative allele counts at rs425535 for six heterozygous RNA-seq samples from the PRAX1 study are shown on the x and y axis, respectively.
Figures 2B–2D depict CXCL5 (MIM: 600324), a representative eGene. CXCL5 was chosen because it shows significant allelic imbalance in concordance with the allelic directionality of the eQTL (p < 2.2e−16, binomial test; see Material and Methods for additional details). CXCL5 is a neutrophil-recruiting chemokine that is secreted by platelets. Platelet-released CXCL5 participates in the pathogenesis of coronary artery disease and tumor metastases.47, 48 The Manhattan plot in Figure 2B shows the eQTL landscape of CXCL5. The reference site rs425535 (red point) is in the second exon of the gene and was in linkage disequilibrium with the lead eQTL rs201022772 (green point). High CXCL5 expression levels were associated with the reference allele of variant rs425535 (p < 10e−12, multiple linear regression; Figure 2C). Correspondingly, ASE analysis at rs425535 revealed a higher proportion of the reference allele within all six heterozygous samples (Figure 2D). We interpret these results as in silico ASE validation of the eGene CXCL5.
Platelets Share eGenes with Other Tissues
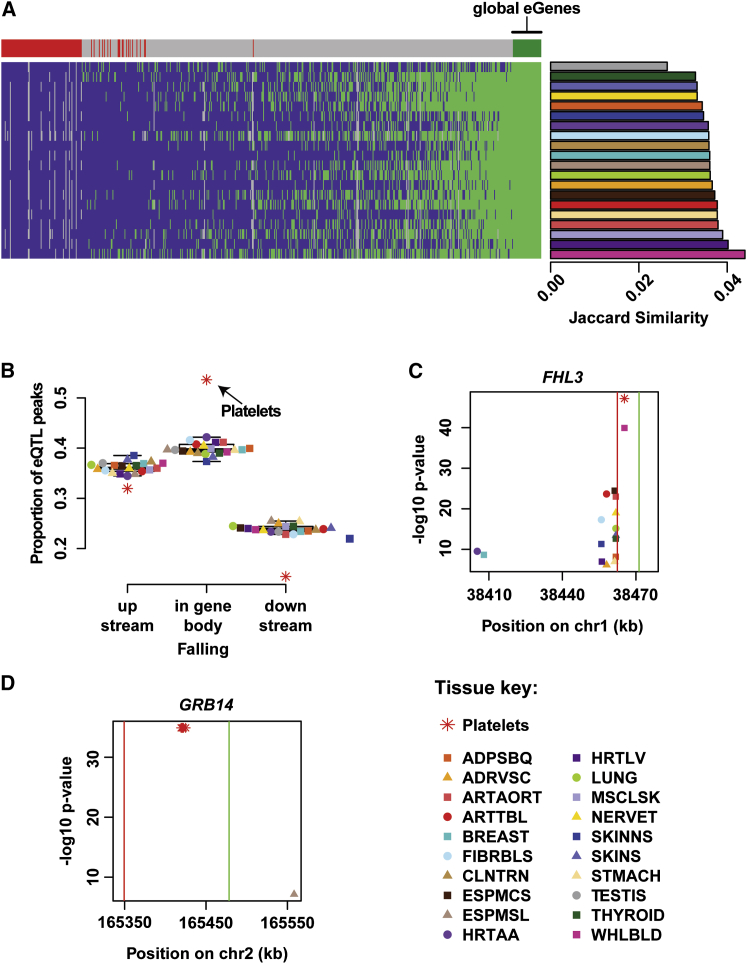
To compare our platelet eQTLs to those identified in other tissues, we reanalyzed GTEx eQTL data23 from 20 different tissues isolated from human cadavers. To improve comparability, we applied our definition of eGenes to the GTEx data (Material and Methods; Table S3). 525 platelet eGenes (86%) were also identified as eGenes in at least one of the GTEx tissues analyzed (p < 2.2e−16, binomial test), additionally validating our eQTL results. To investigate eGenes at a higher resolution, we compared variant-gene associations of our platelet eQTLs to corresponding variant-gene associations tested in the GTEx data. We observed a 75% median concordance rate for the sign of the effect between platelet eQTLs and GTEx tissues, indicating that eGenes commonly share the allelic directionality of their eQTLs (median p value across tissues < 1e−6, Fisher’s exact test; Figure S4). 33 platelet eGenes were classified as eGenes in all 20 GTEx tissues and were therefore considered global eGenes. To evaluate overlap between platelet eGenes and each GTEx tissue separately, we calculated Jaccard similarity. The GTEx tissue “whole blood” shared the highest fraction of eGenes with platelets (258 common eGenes). A total of 110 platelet eGenes were identified as platelet specific because they were observed exclusively in platelets, exempting the GTEx tissue “whole blood,” of which platelets are a component (Figure 3A).
Figure 3.
Comparison of Platelet eQTLs with GTEx
(A) Heatmap columns and rows represent platelet eGenes and GTEx tissues, respectively. The color of the heatmap represents the classification of each platelet eGene as either an eGene (green) or not an eGene (purple) in each of the GTEx tissues. Gray colors indicate genes not tested for eQTLs in the given tissue. In the bar above the heatmap, green and red represent global and platelet-specific eGenes, respectively, and gray indicates platelet eGenes co-occurring in one or more GTEx tissues. The adjacent barplot shows Jaccard similarity comparing the overlap between platelet eGenes and GTEx tissues.
(B) The relative distribution of eQTL peaks across the gene body for all tissues. The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
(C and D) The location of eQTL peaks for eGenes FHL3 and GRB14. The y and x axes correspond to the −log10 p value of eQTL peaks and genomic location, respectively. Green and red vertical lines illustrate the transcription start and stop sites, respectively, for each gene.
Platelet eQTL Peaks Tend to Localize in the Gene Body
To examine eGenes at a more granular level, we defined eQTL peaks as the genomic location of the variant(s) with the strongest association with a given eGene. We identified all eQTL peaks in platelets and GTEx tissues independently. When we compared eQTL peaks to the eGene’s transcription start and stop sites across tissues, platelets had an unusually large proportion of eQTL peaks (54%) mapping to the gene body (p < 1e−8, Grubbs single-outlier test; Figure 3B). This finding suggests that anucleate platelets might be subjected to an expression-regulation architecture that is distinct from that of other tissues with active transcription.
When both GTEx data and platelets have eQTLs for the same eGene, a variety of patterns can emerge. We highlight two exemplary eGenes, such as FHL3 (MIM: 608898), known to play a role in platelet biology. Platelets require Munc13-4 for granule fusion and release,49 and mutations in FHL3 cause defective secretion and familial hemophagocytic lymphohistocytosis.50 Figure 3C depicts the eQTL peaks for the eGene FHL3 across studies. A cluster of eQTL peaks mapped just downstream of the transcription stop site (red vertical line) in multiple tissues, and eQTL peaks fell even further downstream in “heart-atrial appendage” and “breast-mammary tissue.” However, the eQTL peaks for platelets (red asterisk) and the GTEx tissue “whole blood” (purple square) overlapped each other and mapped, distinct from the other GTEx tissues, into the gene body. This observation suggests that the molecular regulation underlying FHL3 expression might be similar in platelets and blood and different from that in other tissues.
Figure 3D depicts the eQTL peaks for an additional exemplary platelet eGene GRB14 (MIM: 601524), which encodes growth factor receptor-bound protein 14. This protein is an adaptor that regulates receptor tyrosine kinase signaling, platelet signaling, and integrin activation.51 The platelet eQTL peak localized to the gene body, whereas in the GTEx tissue “esophagus muscularis,” the eQTL peak fell upstream of the gene.
Overlap between Platelet eQTL Peaks and MI Variants in MFN2
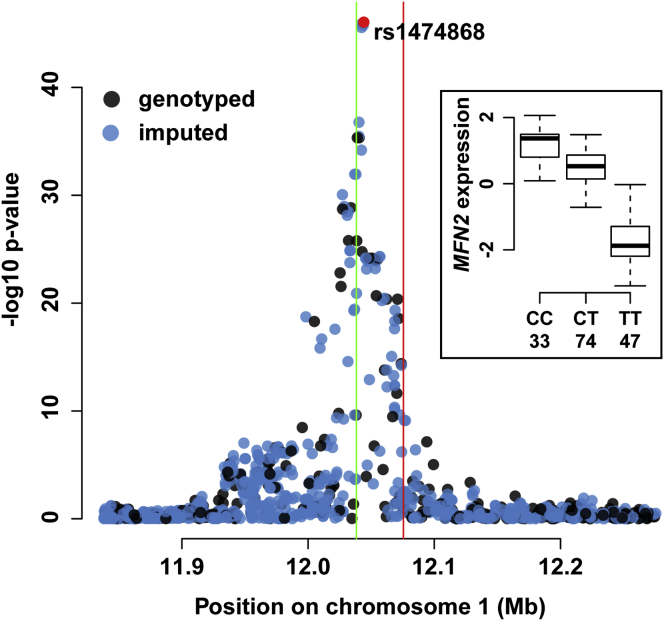
To assess our eQTL findings in the context of CVD GWAS results, we systematically compared our eQTL findings to data from the Genotype and Phenotype Integrator.25 A total of 3,850 unique variants were previously reported to be associated with the risk of CVD in various studies at a p value threshold of 1e−4. After restricting this set of variants to those implicated in our eQTL analysis at a marginal p value cutoff of 1e−4, we identified 40 associations, where platelet eQTLs were previously linked to CVD by GWASs (Table 1). The genetic variant rs1474868 was identified as the variant most strongly associated with platelet MFN2 expression levels (p < 1e−47, multiple linear regression; Figure 4) and was the strongest eQTL of all GWAS CVD-linked variants. The variant rs1474868 was previously associated with MI in the STAMPEED: Cardiovascular Health Study (phs000226) and localized to the second intron of the first transcript isoform (GenBank: NM_014874) of MFN2. Using our ASE approach, we validated the eQTL association between rs1474868 and MFN2 expression levels (Figure S5).
Table 1.
Platelet eQTLs Overlap Variants Implicated in CVD GWASs
| SNP | Gene | Gene MIM No. | eQTL p Value | Trait | Study ID |
|---|---|---|---|---|---|
| rs1474868 | MFN2 | 608507 | 8.94e−47 | myocardial infarction | 2873 |
| rs780633 | RSU1 | 179555 | 2.71e−42 | stroke | 2887 |
| rs2701268 | PDK1 | 602524 | 1.66e−21 | stroke | 2886 |
| rs7349405 | CLHC1 (C2orf63) | NA | 7.72e−20 | hypertension | 3041 |
| rs2285515 | FXYD5 | 606669 | 1.96e−19 | Behcet syndrome | 2888 |
| rs6869332 | ELOVL7 | 614451 | 1.06e−14 | stroke | 2887 |
| rs6738196 | PDK1 | 602524 | 1.37e−12 | stroke | 2886 |
| rs3769321 | PDK1 | 602524 | 1.00e−11 | stroke | 2886 |
| rs514659 | ABO | 110300 | 2.85e−10 | cardiovascular diseases | NA |
| rs10863938 | LPGAT1 | 610473 | 2.26e−9 | stroke | 2886 |
| rs10499859 | CD36 | 173510 | 5.12e−9 | hypertension | NA |
| rs712665 | REEP5 | 125265 | 6.46e−9 | stroke | 2886 |
| rs9962325 | NAPG | 603216 | 6.47e−9 | carotid stenosis | 966 |
| rs411356 | REEP5 | 125265 | 8.28e−9 | stroke | 2886 |
| rs505922 | ABO | 110300 | 1.60e−8 | venous thrombosis | NA |
| rs505922 | ABO | 110300 | 1.60e−8 | venous thromboembolism | NA |
| rs4756196 | CD44 | 107269 | 8.27e−8 | stroke | 2887 |
| rs12242391 | TSPAN15 | 613140 | 3.21e−7 | stroke | 2887 |
| rs4937126 | ST3GAL4 | 104240 | 3.30e−7 | coronary artery disease | NA |
| rs2522447 | PAPSS1 | 603262 | 7.90e−7 | coronary disease | 3056 |
| rs102275 | FADS2 | 606149 | 1.69e−6 | hypertrophy, left ventricular | 3052 |
| rs6935954 | HIST1H2BD | 602799 | 2.44e−6 | stroke | 2887 |
| rs102275 | FADS1 | 606148 | 2.77e−6 | hypertrophy, left ventricular | 3052 |
| rs2980996 | PIGN | 606097 | 2.81e−6 | heart failure | 2884 |
| rs174576 | FADS1 | 606148 | 3.54e−6 | hypertrophy, left ventricular | 3052 |
| rs174576 | FADS2 | 606149 | 3.68e−6 | hypertrophy, left ventricular | 3052 |
| rs973126 | PAPSS1 | 603262 | 4.38e−6 | coronary disease | 3056 |
| rs973126 | PAPSS1 | 603262 | 4.38e−6 | coronary disease | 3055 |
| rs9258966 | HLA-H | NA | 4.42e−6 | Behcet syndrome | 2888 |
| rs3094654 | HLA-H | NA | 4.42e−6 | Behcet syndrome | 2888 |
| rs2256919 | ZFP57 | 612192 | 5.73e−6 | Behcet syndrome | 2888 |
| rs6594646 | REEP5 | 125265 | 6.88e−6 | stroke | 2886 |
| rs663354 | PIGN | 606097 | 7.78e−6 | heart failure | 2884 |
| rs2431512 | REEP5 | 125265 | 8.53e−6 | stroke | 2886 |
| rs12719151 | REEP5 | 125265 | 1.15e−5 | stroke | 2886 |
| rs4694656 | CXCL5 | 600324 | 2.18e−5 | heart failure | 2885 |
| rs13325747 | RPN1 | 180470 | 2.65e−5 | heart failure | 2885 |
| rs4601174 | CD164 | 603356 | 2.95e−5 | heart failure | 2885 |
| rs2071653 | ZFP57 | 612192 | 3.91e−5 | Behcet syndrome | 2888 |
| rs1339965 | SMYD3 | 608783 | 4.58e−5 | heart failure | 2885 |
This table contains marginal platelet eQTLs (p < 1e−4) that have been previously associated with CVD. The “Study ID” column contains the study cohort in which the association between SNP and trait was detected. NA stands for “not available.”
Figure 4.
MFN2 eQTL Manhattan Plot
The y and x axes correspond to association p values and genomic coordinates, respectively. Blue and black points show imputed and genotyped variants, respectively. Four variants (points fall on top of each other) were strongly associated with MFN2 expression levels (p < 1e−45, multiple linear regression), and the red point highlights the eQTL peak rs1474868 (genotyped). Transcription start and stop sites are indicated by green and red vertical lines, respectively.
(Inset) The association between rs1474868 allele dosage and MFN2 expression levels (significant at p < 1e−47, multiple linear regression). The y and x axes indicate the corrected MFN2 expression levels and rs1474868 genotype, respectively. Numbers below the genotype labels indicate the number of samples within each group. The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
Platelet RNA-Seq Data Reveal Unannotated Exon 2b in MFN2
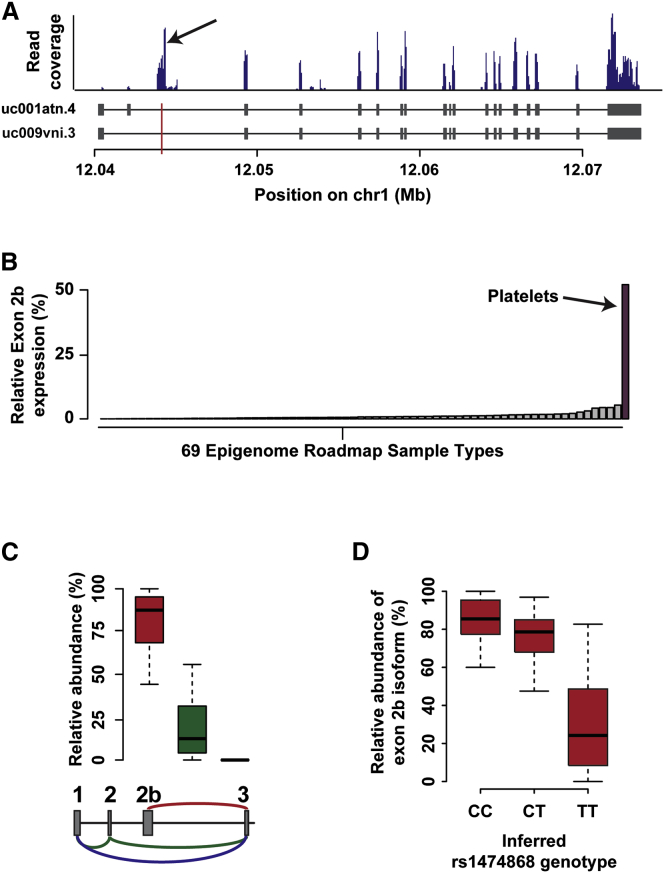
MFN2 encodes mitofusin-2, a critical membrane protein involved in mitochondrial fusion, and normal platelet number and function depend on mitochondrial function.52 Because the presence of a number of transcribed intronic regions was previously noted in human platelets,53 we hypothesized that platelets might contain uncharacterized transcripts that coincide with our platelet eQTL signals. Motivated by this hypothesis, we analyzed human platelet RNA-seq data from multiple groups29, 40, 41, 42 to examine the transcriptional landscape of platelet MFN2. A large number of reads mapped to MFN2 intron 2, overlapping the eQTL peak rs1474868 (Figure 5A and Figure S6). RNA-seq data from mouse platelets40 lacked reads mapping to this region in the MFN2 homolog (Figure S7). We hypothesized that this RNA-seq mapping represents the existence of an unannotated human-platelet-expressed exon, which we call “exon 2b.” We used RT-PCR of RNA from human platelets to confirm the presence of exon 2b in the MFN2 transcript (Figure S8), and sequencing of the RT-PCR products revealed a precise exon 2b location (chr1: 12,043,880–12,044,352, hg19). To test whether exon 2b is present in other tissues, we performed in silico cross-tissue analysis. We were unable to identify comparable expression levels of exon 2b in mRNA-seq data from any of the 69 Epigenome Roadmap sample types queried, suggesting that exon 2b is preferentially expressed in human platelets (Figure 5B).
Figure 5.
Novel Exon in MFN2
(A) Read coverage across MFN2 for a single representative human platelet RNA-seq sample (SRA: SRR957099). Blue bars represent RNA-seq read coverage. Gray rectangles indicate exons of MFN2 transcripts uc001atn.4 and uc009vni.3. The black arrow highlights a pile of reads mapping to an intronic region overlapping rs1474868 (red vertical line).
(B) Average exon 2b expression levels across 69 Epigenome Roadmap mRNA-seq sample types (gray bars) and the Londin et al.29 human platelet RNA-seq data (purple bar). The y axis indicates the exon 2b expression level in relation to that of the most highly expressed MFN2 exon.
(C) Estimated relative abundances of MFN2 isoforms in platelets. The schematic below the plot depicts annotated exons 1–3 and novel platelet exon 2b of MFN2. Red, green, and blue lines indicate mapping of informative RNA-seq junction reads for the isoform containing the novel exon 2b and annotated MFN2 isoforms uc001atn.4 and uc009vnl.3, respectively. The boxplot shows the estimated relative abundance for each of these isoforms across human platelet RNA-seq samples. The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
(D) Estimated relative abundance of the MFN2 isoform containing exon 2b by the inferred rs1474868 genotype. The relative abundance of this isoform was significantly associated with the rs1474868 genotype (p < 1e−6, Spearman’s rank correlation). The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
Exon 2b Is an Alternative Start Site of MFN2
To infer how exon 2b fits into the splicing pattern of MFN2, we examined RNA-seq junction reads from a total of 45 RNA-seq samples of human platelets. We were unable to detect any reads connecting exon 2b to any upstream exon, suggesting that exon 2b is an alternative 5′ start site. Using the 5′ RACE assay, we verified the presence of two separate platelet MFN2 isoforms starting at either exon 2b or exon 1 (Figure S9). Next, we estimated the relative abundance of the MFN2 isoform containing exon 2b. Using junction reads connecting the 5′ border of exon 3 to any upstream exon can unambiguously distinguish between MFN2 isoforms. A median of 86% of junction reads mapped from the 3′ border of exon 2b to the 5′ border of exon 3, suggesting that exon 2b is included in the pre-dominant platelet MFN2 isoform (Figure 5C). Moreover, the relative abundance of this MFN2 isoform was significantly associated with the rs1474868 genotype (p < 1e−6, Spearman’s rank correlation; Figure 5D).
The Variant rs3766744 Alters Transcriptional Activity
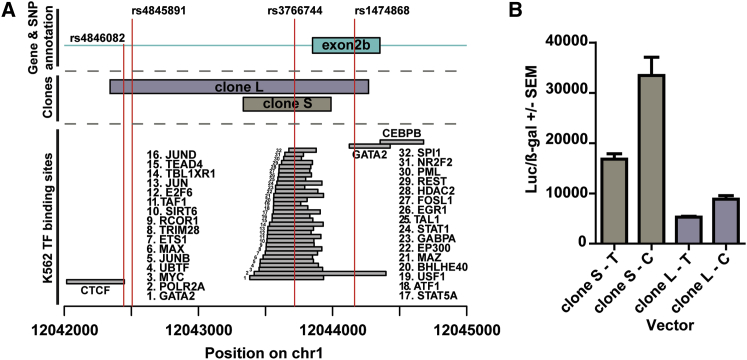
eQTLs have been shown to regulate gene expression by altering transcription factor binding sites;54 therefore, we integrated ENCODE transcription factor ChIP-seq data from K562 myeloid leukemia cells to identify the most likely causal eQTL for MFN2. Three noncoding variants (rs4846082, rs4845891, and rs3766744) were in near perfect linkage disequilibrium with rs1474868 and also strongly associated with MFN2 expression levels (all three p < 1e−45, multiple linear regression). Among this set of variants, we observed an overlap of 32 transcription factor ChIP-seq peaks with rs3766744 (Figure 6A), suggesting a potential mechanism for this variant.
Figure 6.
Differential Regulation of MFN2 Expression by Single-Nucleotide Variant
(A) ENCODE K526 ChIP-seq transcription factor binding peaks clustered near rs1474868. Red vertical lines indicate the location of genetic variants strongly associated with MFN2 expression levels. rs3766744 overlapped a number of transcription factor binding sites. The names of transcription factors are ordered from bottom to top. Exon 2b is displayed as a cyan rectangle. The S and L clones are shown as purple and gray rectangles, respectively.
(B) Differential luciferase activity for the C and T alleles of rs3766744 in the S and L clones. The luciferase levels were higher for the C allele than for the T allele (both p < 0.05, Mann-Whitney test). Error bars correspond to SE.
To evaluate the effect of genotype on transcriptional activity, we generated vectors containing short (S: 655 bp) or long (L: 1,928 bp) DNA fragments flanking rs3766744 upstream of a luciferase reporter gene. Clones of the rs37466744 C and T alleles were generated. Transfection of these reporter constructs into K562 cells resulted in a significant difference of luciferase activity between the C and T alleles for both S and L clones (Figure 6B). These results are in concordance with the allelic directionality of our MFN2 eQTL and ASE findings and support rs3766744 as the causal regulatory variant.
MFN2 Expression Is Associated with Platelet Count
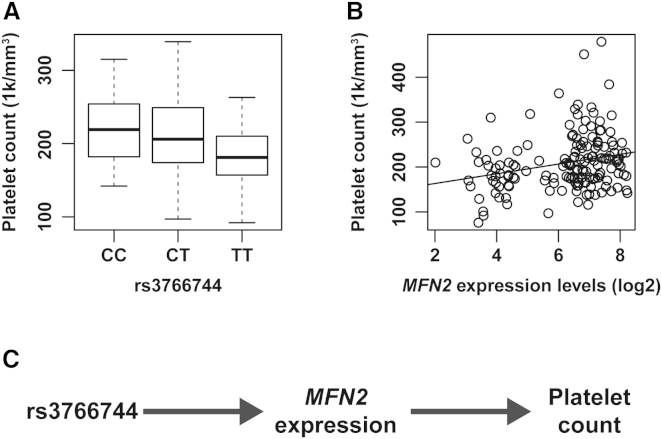
After identifying rs3766744 as the probable causal regulatory variant that controls MFN2 expression levels, we investigated the association among rs3766744 allele dosage, MFN2 expression, and platelet count by using our data from the PRAX1 study. Platelet count was significantly associated with rs3766744 allele dosage (p < 1e−4, multiple linear regression; Figure 7A) and MFN2 expression levels (p < 1e−3, multiple linear regression; Figure 7B). Moreover, after conditioning on MFN2 expression levels, the association between rs3766744 and platelet count disappeared; however, the association between rs3766744 and MFN2 expression levels remained significant after platelet count was accounted for. Together, these conditional association analyses support a causal model where MFN2 expression levels mediate the association between rs3766744 and platelet count (Figure 7C).
Figure 7.
Associations with Platelet Count in the PRAX1 Study Dataset
(A) The association between rs3766744 and platelet count (p < 1e−4, multiple linear regression). The box represents the interquartile range, the horizontal line in the box indicates the median, and the whiskers represent 1.5× the interquartile range.
(B) The association between platelet count and MFN2 expression levels (p < 1e−3, multiple linear regression).
(C) Our proposed model. The variant rs3766744 controls MFN2 expression levels, which in turn affect platelet count.
Discussion
CVD GWASs have been successful at identifying loci associated with disease risk,55, 56 but linking these variants with gene function in relevant tissues has been a challenge.16 Platelets are a critical tissue for this analysis because they are an important contributor to clinical CVD phenotypes and are the target of pharmacologic interventions. In this study, we used platelet transcriptomics and eQTL analyses to uncover platelet-specific gene regulation that corresponds to a CVD GWAS signal in MFN2. This study demonstrates the power of combining tissue-specific eQTL and RNA-seq data with large-scale genomic databases to gain greater insight into the molecular mechanisms underlying GWAS signals.
Validation of Platelet eQTLs
To corroborate the expression associations revealed in our study, we pursued both overlap with external data and ASE analysis of our own genome-wide genotype and transcriptome data. We utilized imputation to provide a more granular view of the eQTL landscape, enhancing our ability to make direct comparisons to markers in other studies.57 Second, the imputation approach also determined a more fine-scale substrate for overlap with ChIP-seq analyses. A third benefit of imputation is the improved ability to perform ASE analysis by extending the set of coding variants linked to eQTLs.
We validated eGenes by analyzing allelic imbalance at heterozygous coding sites that had sufficient read coverage and were also in strong linkage disequilibrium with our eQTL peaks.35 We were able to examine 19% of all platelet eGenes despite the following limitations: (1) only a subset (6.4%) of our cohort was profiled by RNA-seq technology, (2) only a subset (79%) of eGenes contained at least one heterozygous coding site, and (3) only a subset (13%) of pileup sites contained sufficient read coverage for analysis. Globally, the trends indicated a significant concordance between the allelic directionality of eQTLs and RNA-seq allelic-imbalance events in platelet eGenes. Our data suggest that RNA-seq of a subset of subjects for ASE analysis is a useful approach for validating eQTLs developed through microarray studies of gene expression. This approach could be adopted on existing data where microarray studies have determined eQTLs but where subsample RNA-seq could refine candidates for validation and further functional studies.
Characterization of Platelet eQTLs
We integrated and analyzed our primary dataset with GTEx findings to both validate our results and investigate tissue specificity of platelet eQTLs. We acknowledge the difficulties of making comparisons across data produced by different groups on different platforms. Our study has the advantage of highly purified cells obtained from living, healthy subjects. Samples from the PRAX1 study were subjected to a uniform and immediate specimen processing, including RNA isolation and extraction within 1 hr of phlebotomy. Our results indicate that platelets have tissue-specific eQTLs distinct from those identified by GTEx, highlighting the need to extend GTEx studies to additional tissues and biological contexts.
Compared to other cell types, platelet eQTL peaks show an unusually high representation in the gene body (Figure 3D). This finding is intriguing because platelets are anucleate cells free of genomic DNA. RNA messages are inherited from parent megakaryocytes, potentially decreasing the relative impact of transcriptional regulatory regions and increasing the significance of post-transcriptional regulation. This stands in sharp contrast to the other cell types profiled in GTEx, where the transcriptome reflects a balance between transcription and RNA degradation. Further analyses of other anucleate cells, such as erythrocytes, might yield additional insights.
Functional Genomics of Platelet MFN2
Our genome-wide eQTL analysis revealed that rs1474868 marks the eQTL peak of MFN2. The function of MFN2 in platelets and megakaryocytes has not been studied, and our work demonstrates the presence of MFN2 transcripts (Figure 5) and protein (Figure S10) in human platelets. MFN2 is a gene previously reported to function in mitochondrial structure and function.58, 59 Follow-up analyses using RNA-seq data determined that MFN2 has an unannotated alternate first exon, exon 2b, which overlaps rs1474868. Inclusion of exon 2b is predicted to add 34 amino acids to the N terminus of the MFN2 RefSeq coding sequence; we term this extended protein pMFN2 (Figure S11). The anti-MFN2 antibody used to detect MFN2 does not distinguish between MFN2 and pMFN2. We raised a polyclonal antibody against amino acid sequence unique to the predicted pMFN2, but unfortunately it reacted non-specifically with numerous polypeptides and was not useful (data not shown). However, because 86% of MFN2 transcripts contained exon 2b (Figure 5C), we presume the major form observed by western blotting is pMFN2 (Figure S10), but until additional reagents are developed or mass spectrometry of purified pMFN2 is performed, we cannot be certain.
In addition, in silico cross-tissue analysis suggests that this alternate start site is platelet specific. A previous study by Nuernberg et al.60 identified a novel 5′ start site in RNA-seq data from megakaryocytes in the gene DNM3, and we also found this start site in our platelet RNA-seq data (data not shown). Our discovery of an unannotated 5′ alternative start site in platelet MFN2, along with similar observations of platelet-specific exons by others,61 suggests that unannotated lineage-specific alternative 5′ start sites might be common in platelets and platelet progenitor cells. This is an exciting area for further studies.
MFN2 and Platelet Count
We observed a significant association between platelet count and rs3766744, which is in linkage disequilibrium with rs1474868 (Figure 7). Conditional association analysis suggests that this relationship is mediated through differential MFN2 expression driven by allele dosage. Variant rs3766744 is also in linkage disequilibrium with rs2336384 (R2 = 0.49, LDlink), which has previously been linked to platelet count.7 We were able to verify this association in our data from the PRAX1 study (p < 0.05, multiple linear regression). However, in the analysis of these data, the association between rs3766744 and platelet count was stronger than the association between rs1474868 and platelet count, suggesting that the platelet-count association might be driven by rs3766744.
Our working hypothesis is that rs3766744 regulates exon 2b utilization by altering transcriptional enhancer activity, which in turn leads to increased MFN2 expression. Numerous transcription factors were shown to bind to the sequence potentially altered by rs3766744 in K562 cells (Figure 6A). Particularly interesting candidates are GATA2, TAL1, and ETS1, which are known to regulate megakaryocytopoiesis and expression of many platelet-specific genes. We further hypothesize that variation in MFN2 expression alters platelet production given that low Mfn2 expression has been linked to mitochondrial damage in mice,62 and mitochondrial number and function are critical for platelet production by megakaryocytes. Nevertheless, addressing these testable hypotheses will require much more work and the development of new reagents.
MFN2 and Human Disease
Normal platelet function and lifespan depend on healthy mitochondria as a critical energy source during their 10 day period in circulating blood. There are numerous clinical conditions wherein the platelet count is significantly reduced in association with dysfunctional mitochondria, including inherited and acquired diseases inducing mitochondrial dysfunction.52, 63, 64, 65, 66, 67
Variation in MFN2 has been linked to Charcot-Marie-Tooth disease (CMT) type 2A2 (MIM: 609260) and hereditary motor and sensory neuropathy VIA (MIM: 601152). MFN2 mutations are associated with the disease subtype CMT2A, where diverse coding mutations in MFN2 account for up to 20% of the CMT2A subtype and about 4% of CMT disease overall.68 The CMT2A-linked mutations in MFN2 comprise a diverse collection of variants, including stop-gain and non-synonymous pathogenic changes. Pathogenicity has been connected to more variants in the N terminus of the protein, but variations across the protein have been linked to the disease.69
Not only does MFN2 play a role in Mendelian disease, but there is also evidence that MFN2 might have a function in later-onset complex diseases such as CVD. Common variants in MFN2 have been linked to platelet count7 and the risk of MI25 and hypertension.70 In addition, mutations in MFN2 have also been linked to early-onset stroke.71 Furthermore, it is generally believed that an elevated platelet count is a risk factor for ischemic cardiovascular complications, although this has not been rigorously studied in a prospective fashion. Subjects with very high platelet counts due to acquired, clonal myeloproliferative diseases are at an increased risk of MI and stroke, and standard management most often includes anti-platelet therapies.72
Sharing of Results
We have made our results available by extending our interactive Plateletomics website (see Web Resources). We added a platelet-eQTL browser function, allowing users to quickly and easily query our platelet-eQTL results. This eGene analysis can be particularly useful in cases where wet-bench experiments might give conflicting results if the investigator does not consider the genotype of the donors used.
Conclusion
Our study of platelet eQTLs identified 612 platelet eGenes. Among these, MFN2 harbored the strongest eQTL that corresponded to a previously reported CVD GWAS variant. Integrative analysis demonstrated that the MFN2 eQTL identified an unannotated alternate platelet-specific 5′ start site driving MFN2 expression. Moreover, MFN2 levels were strongly associated with platelet count, a known risk factor for CVD. Our results reinforce using multi-omic data integration of relevant primary tissues to functionalize GWAS signals.
Acknowledgments
This work was funded by National Heart, Lung, and Blood Institute grants HL102482 and R01HL128234, the William M. Keck Foundation, and the Cardeza Foundation for Hematologic Research. L.M.S. is additionally supported by American Heart Association Fellowship 14PRE20480196. Human subject samples for this research were obtained under institutional review board approval at both Baylor College of Medicine in Houston and Thomas Jefferson University in Philadelphia. We thank the Thrombosis Research laboratory at Methodist Hospital in Houston and Angela Bergeron for sample collection and platelet phenotyping. We thank John Belmont for his guidance on platform selection.
Published: April 28, 2016
Footnotes
Supplemental Data include 11 figures and 4 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.007.
Contributor Information
Paul F. Bray, Email: paul.bray@jefferson.edu.
Chad A. Shaw, Email: cashaw@bcm.edu.
Web Resources
1000 Genomes phase I integrated haplotypes, https://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated_SHAPEIT2.html
AlleleSeq, http://alleleseq.gersteinlab.org/
ArrayExpress, https://www.ebi.ac.uk/arrayexpress/
ENCODE, https://www.encodeproject.org/
ENCODE ChIP-seq data on transcription factor binding sites, http://hgdownload.cse.ucsc.edu/goldenpath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wgEncodeRegTfbsClusteredV3.bed.gz
Epigenome Roadmap Project, http://www.roadmapepigenomics.org/
Genboree Workbench, http://genboree.org/site/
Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo/
GTEx Portal, http://www.gtexportal.org/
IMPUTE2, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html
OMIM, http://www.omim.org
Phenotype-Genotype Integrator, http://www.ncbi.nlm.nih.gov/gap/phegeni
Plateletomics, http://www.plateletomics.com/plateletomics/
Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra
Supplemental Data
This table contains 79,544 rows and 12 columns. Each row contains the association between a cis variant and expression levels of a platelet eGene (p < 1e−4). It contains the standard MatrixEQTL output (SNPs, gene, statistic, p value, FDR, and beta) and additional information (rsID, chr, pos, ref, alt, imputed). Description of column headers is contained in the spreadsheet.
This table contains 359 rows and 9 columns. Each row represents a pileup site in one of the 10 Londin et al. RNA-seq samples. Pileup sites were restricted to heterozygous coding sites with a marginal eQTL effect (p < 1e−4) and with 10 or more reads. These data were used in Figure 2 of the manuscript. Description of column headers is contained in the spreadsheet.
This table contains 104,750 rows and 11 columns. Each row represents an eQTL peak in one of the tissues analyzed in this study. The “Platelets” tissue represents data from the PRAX1 study cohort. All other tissues were taken from GTEx V6. These data were used in Figure 3 of the manuscript. Description of column headers is contained in the spreadsheet.
This table contains 3,834 rows and 167 columns. Each row represents mRNA-seq read density across 10 bp intervals spanning the MFN2 region for all mRNA-seq Epigenome samples. These data were used in Figure 5B of the manuscript. Description of column headers is contained in the spreadsheet.
References
- 1.Chandler A.B., Chapman I., Erhardt L.R., Roberts W.C., Schwartz C.J., Sinapius D., Spain D.M., Sherry S., Ness P.M., Simon T.L. Coronary thrombosis in myocardial infarction. Report of a workshop on the role of coronary thrombosis in the pathogenesis of acute myocardial infarction. Am. J. Cardiol. 1974;34:823–833. doi: 10.1016/0002-9149(74)90703-6. [DOI] [PubMed] [Google Scholar]
- 2.Fuster V., Badimon L., Badimon J.J., Chesebro J.H. The pathogenesis of coronary artery disease and the acute coronary syndromes (2) N. Engl. J. Med. 1992;326:310–318. doi: 10.1056/NEJM199201303260506. [DOI] [PubMed] [Google Scholar]
- 3.Lam J.Y., Latour J.G., Lespérance J., Waters D. Platelet aggregation, coronary artery disease progression and future coronary events. Am. J. Cardiol. 1994;73:333–338. doi: 10.1016/0002-9149(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 4.Davies M.J., Thomas A.C., Knapman P.A., Hangartner J.R. Intramyocardial platelet aggregation in patients with unstable angina suffering sudden ischemic cardiac death. Circulation. 1986;73:418–427. doi: 10.1161/01.cir.73.3.418. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald D.J., Roy L., Catella F., FitzGerald G.A. Platelet activation in unstable coronary disease. N. Engl. J. Med. 1986;315:983–989. doi: 10.1056/NEJM198610163151602. [DOI] [PubMed] [Google Scholar]
- 6.Yee D.L., Sun C.W., Bergeron A.L., Dong J.F., Bray P.F. Aggregometry detects platelet hyperreactivity in healthy individuals. Blood. 2005;106:2723–2729. doi: 10.1182/blood-2005-03-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray P.F. Platelet hyperreactivity: predictive and intrinsic properties. Hematol. Oncol. Clin. North Am. 2007;21:633–645, v–vi. doi: 10.1016/j.hoc.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marenberg M.E., Risch N., Berkman L.F., Floderus B., de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 10.Sørensen T.I., Nielsen G.G., Andersen P.K., Teasdale T.W. Genetic and environmental influences on premature death in adult adoptees. N. Engl. J. Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 11.Rissanen A.M. Familial aggregation of coronary heart disease in a high incidence area (North Karelia, Finland) Br. Heart J. 1979;42:294–303. doi: 10.1136/hrt.42.3.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd-Jones D.M., Nam B.H., D’Agostino R.B., Sr., Levy D., Murabito J.M., Wang T.J., Wilson P.W., O’Donnell C.J. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 13.Bray P.F., Mathias R.A., Faraday N., Yanek L.R., Fallin M.D., Herrera-Galeano J.E., Wilson A.F., Becker L.C., Becker D.M. Heritability of platelet function in families with premature coronary artery disease. J. Thromb. Haemost. 2007;5:1617–1623. doi: 10.1111/j.1538-7836.2007.02618.x. [DOI] [PubMed] [Google Scholar]
- 14.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nica A.C., Dermitzakis E.T. Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120362. doi: 10.1098/rstb.2012.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battle A., Montgomery S.B. Determining causality and consequence of expression quantitative trait loci. Hum. Genet. 2014;133:727–735. doi: 10.1007/s00439-014-1446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen R.C., Nap J.P. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 19.Westra H.J., Franke L. From genome to function by studying eQTLs. Biochim. Biophys. Acta. 2014;1842:1896–1902. doi: 10.1016/j.bbadis.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Schadt E.E., Björkegren J.L. NEW: network-enabled wisdom in biology, medicine, and health care. Sci. Transl. Med. 2012;4:115rv1. doi: 10.1126/scitranslmed.3002132. [DOI] [PubMed] [Google Scholar]
- 21.Goodall A.H., Burns P., Salles I., Macaulay I.C., Jones C.I., Ardissino D., de Bono B., Bray S.L., Deckmyn H., Dudbridge F., Bloodomics Consortium Transcription profiling in human platelets reveals LRRFIP1 as a novel protein regulating platelet function. Blood. 2010;116:4646–4656. doi: 10.1182/blood-2010-04-280925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnatenko D.V., Dunn J.J., McCorkle S.R., Weissmann D., Perrotta P.L., Bahou W.F. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101:2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 23.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raney B.J., Cline M.S., Rosenbloom K.R., Dreszer T.R., Learned K., Barber G.P., Meyer L.R., Sloan C.A., Malladi V.S., Roskin K.M. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos E.M., Hoffman D., Junkins H.A., Maglott D., Phan L., Sherry S.T., Feolo M., Hindorff L.A. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet. 2014;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edelstein L.C., Simon L.M., Montoya R.T., Holinstat M., Chen E.S., Bergeron A., Kong X., Nagalla S., Mohandas N., Cohen D.E. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat. Med. 2013;19:1609–1616. doi: 10.1038/nm.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edelstein L.C., Simon L.M., Lindsay C.R., Kong X., Teruel-Montoya R., Tourdot B.E., Chen E.S., Ma L., Coughlin S., Nieman M. Common variants in the human platelet PAR4 thrombin receptor alter platelet function and differ by race. Blood. 2014;124:3450–3458. doi: 10.1182/blood-2014-04-572479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon L.M., Edelstein L.C., Nagalla S., Woodley A.B., Chen E.S., Kong X., Ma L., Fortina P., Kunapuli S., Holinstat M. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood. 2014;123:e37–e45. doi: 10.1182/blood-2013-12-544692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Londin E.R., Hatzimichael E., Loher P., Edelstein L., Shaw C., Delgrosso K., Fortina P., Bray P.F., McKenzie S.E., Rigoutsos I. The human platelet: strong transcriptome correlations among individuals associate weakly with the platelet proteome. Biol. Direct. 2014;9:3. doi: 10.1186/1745-6150-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engström P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Rätsch G., Goldman N., Hubbard T.J., Harrow J., Guigó R., Bertone P., RGASP Consortium Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stegle O., Parts L., Durbin R., Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degner J.F., Marioni J.C., Pai A.A., Pickrell J.K., Nkadori E., Gilad Y., Pritchard J.K. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castel S.E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., Harmanci A., Leng J., Bjornson R., Kong Y., Kitabayashi N. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey C.T., Moyerbrailean G.A., Davis G.O., Wen X., Luca F., Pique-Regi R. QuASAR: quantitative allele-specific analysis of reads. Bioinformatics. 2015;31:1235–1242. doi: 10.1093/bioinformatics/btu802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowley J.W., Oler A.J., Tolley N.D., Hunter B.N., Low E.N., Nix D.A., Yost C.C., Zimmerman G.A., Weyrich A.S. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kissopoulou A., Jonasson J., Lindahl T.L., Osman A. Next generation sequencing analysis of human platelet PolyA+ mRNAs and rRNA-depleted total RNA. PLoS ONE. 2013;8:e81809. doi: 10.1371/journal.pone.0081809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eicher J.D., Wakabayashi Y., Vitseva O., Esa N., Yang Y., Zhu J., Freedman J.E., McManus D.D., Johnson A.D. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27:230–239. doi: 10.3109/09537104.2015.1083543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coarfa C., Pichot C., Jackson A., Tandon A., Amin V., Raghuraman S., Paithankar S., Lee A.V., McGuire S.E., Milosavljevic A. Analysis of interactions between the epigenome and structural mutability of the genome using Genboree Workbench tools. BMC Bioinformatics. 2014;15(Suppl 7):S2. doi: 10.1186/1471-2105-15-S7-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagalla S., Shaw C., Kong X., Kondkar A.A., Edelstein L.C., Ma L., Chen J., McKnight G.S., López J.A., Yang L. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117:5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasin-Brumshtein Y., Hormozdiari F., Martin L., van Nas A., Eskin E., Lusis A.J., Drake T.A. Allele-specific expression and eQTL analysis in mouse adipose tissue. BMC Genomics. 2014;15:471. doi: 10.1186/1471-2164-15-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majewski J., Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Holm T., Damås J.K., Holven K., Nordøy I., Brosstad F.R., Ueland T., Währe T., Kjekshus J., Frøland S.S., Eiken H.G. CXC-chemokines in coronary artery disease: possible pathogenic role of interactions between oxidized low-density lipoprotein, platelets and peripheral blood mononuclear cells. J. Thromb. Haemost. 2003;1:257–262. doi: 10.1046/j.1538-7836.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 48.Labelle M., Begum S., Hynes R.O. Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. USA. 2014;111:E3053–E3061. doi: 10.1073/pnas.1411082111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Q., Wimmer C., Chicka M.C., Ye S., Ren Y., Hughson F.M., Whiteheart S.W. Munc13-4 is a limiting factor in the pathway required for platelet granule release and hemostasis. Blood. 2010;116:869–877. doi: 10.1182/blood-2010-02-270934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldmann J., Callebaut I., Raposo G., Certain S., Bacq D., Dumont C., Lambert N., Ouachée-Chardin M., Chedeville G., Tamary H. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115:461–473. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 51.Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., Boussiotis V.A. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 52.Leaver H.A., Schou A.C., Rizzo M.T., Prowse C.V. Calcium-sensitive mitochondrial membrane potential in human platelets and intrinsic signals of cell death. Platelets. 2006;17:368–377. doi: 10.1080/09537100600757216. [DOI] [PubMed] [Google Scholar]
- 53.Bray P.F., McKenzie S.E., Edelstein L.C., Nagalla S., Delgrosso K., Ertel A., Kupper J., Jing Y., Londin E., Loher P. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castaldi P.J., Cho M.H., Zhou X., Qiu W., Mcgeachie M., Celli B., Bakke P., Gulsvik A., Lomas D.A., Crapo J.D. Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum. Mol. Genet. 2015;24:1200–1210. doi: 10.1093/hmg/ddu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith J.G., Newton-Cheh C. Genome-wide association studies of late-onset cardiovascular disease. J. Mol. Cell. Cardiol. 2015;83:131–141. doi: 10.1016/j.yjmcc.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts R. Genetics of coronary artery disease. Circ. Res. 2014;114:1890–1903. doi: 10.1161/CIRCRESAHA.114.302692. [DOI] [PubMed] [Google Scholar]
- 57.Porcu E., Sanna S., Fuchsberger C., Fritsche L.G. Genotype imputation in genome-wide association studies. Curr. Protoc. Hum. Genet. 2013;Chapter 1:25. doi: 10.1002/0471142905.hg0125s78. [DOI] [PubMed] [Google Scholar]
- 58.Chen H., Chomyn A., Chan D.C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 59.Eura Y., Ishihara N., Yokota S., Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- 60.Nürnberg S.T., Rendon A., Smethurst P.A., Paul D.S., Voss K., Thon J.N., Lloyd-Jones H., Sambrook J.G., Tijssen M.R., Italiano J.E., Jr., HaemGen Consortium A GWAS sequence variant for platelet volume marks an alternative DNM3 promoter in megakaryocytes near a MEIS1 binding site. Blood. 2012;120:4859–4868. doi: 10.1182/blood-2012-01-401893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert S., Weyrich A.S., Rowley J.W. A tour through the transcriptional landscape of platelets. Blood. 2014;124:493–502. doi: 10.1182/blood-2014-04-512756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen W., Xu X., Wang L., Bai G., Xiang W. Low Expression of Mfn2 Is Associated with Mitochondrial Damage and Apoptosis of Ovarian Tissues in the Premature Ovarian Failure Model. PLoS ONE. 2015;10:e0136421. doi: 10.1371/journal.pone.0136421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Illsinger S., Janzen N., Sander S., Schmidt K.H., Bednarczyk J., Mallunat L., Bode J., Hagebölling F., Hoy L., Lücke T. Preeclampsia and HELLP syndrome: impaired mitochondrial function in umbilical endothelial cells. Reprod. Sci. 2010;17:219–226. doi: 10.1177/1933719109351597. [DOI] [PubMed] [Google Scholar]
- 64.Yen H.H., Shih K.L., Lin T.T., Su W.W., Soon M.S., Liu C.S. Decreased mitochondrial deoxyribonucleic acid and increased oxidative damage in chronic hepatitis C. World J. Gastroenterol. 2012;18:5084–5089. doi: 10.3748/wjg.v18.i36.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pereira J., Soto M., Palomo I., Ocqueteau M., Coetzee L.M., Astudillo S., Aranda E., Mezzano D. Platelet aging in vivo is associated with activation of apoptotic pathways: studies in a model of suppressed thrombopoiesis in dogs. Thromb. Haemost. 2002;87:905–909. [PubMed] [Google Scholar]
- 66.Yamada K., Miwa K., Wakita Y., Ikoma Y., Fukui Y., Okuyama M., Kato K., Ichihashi T., Kunishima S., Shamoto M. Familial macrothrombocytopenia with unusually elongated mitochondria. Intern. Med. 1995;34:1140–1143. doi: 10.2169/internalmedicine.34.1140. [DOI] [PubMed] [Google Scholar]
- 67.White J.G., Key N.S., King R.A., Vercellotti G.M. The White platelet syndrome: a new autosomal dominant platelet disorder. Platelets. 2004;15:173–184. doi: 10.1080/09537100410001682805. [DOI] [PubMed] [Google Scholar]
- 68.Fridman V., Bundy B., Reilly M.M., Pareyson D., Bacon C., Burns J., Day J., Feely S., Finkel R.S., Grider T., Inherited Neuropathies Consortium CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J. Neurol. Neurosurg. Psychiatry. 2015;86:873–878. doi: 10.1136/jnnp-2014-308826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiVincenzo C., Elzinga C.D., Medeiros A.C., Karbassi I., Jones J.R., Evans M.C., Braastad C.D., Bishop C.M., Jaremko M., Wang Z. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol. Genet. Genomic Med. 2014;2:522–529. doi: 10.1002/mgg3.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z., Liu Y., Liu J., Liu K., Wen J., Wen S., Wu Z. HSG/Mfn2 gene polymorphism and essential hypertension: a case-control association study in Chinese. J. Atheroscler. Thromb. 2011;18:24–31. doi: 10.5551/jat.5611. [DOI] [PubMed] [Google Scholar]
- 71.Chung K.W., Cho S.Y., Hwang S.J., Kim K.H., Yoo J.H., Kwon O., Kim S.M., Sunwoo I.N., Züchner S., Choi B.O. Early-onset stroke associated with a mutation in mitofusin 2. Neurology. 2008;70:2010–2011. doi: 10.1212/01.wnl.0000312513.96457.7a. [DOI] [PubMed] [Google Scholar]
- 72.Campbell P.J., Green A.R. Management of polycythemia vera and essential thrombocythemia. Hematology (Am Soc Hematol Educ Program) 2005;2005:201–208. doi: 10.1182/asheducation-2005.1.201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table contains 79,544 rows and 12 columns. Each row contains the association between a cis variant and expression levels of a platelet eGene (p < 1e−4). It contains the standard MatrixEQTL output (SNPs, gene, statistic, p value, FDR, and beta) and additional information (rsID, chr, pos, ref, alt, imputed). Description of column headers is contained in the spreadsheet.
This table contains 359 rows and 9 columns. Each row represents a pileup site in one of the 10 Londin et al. RNA-seq samples. Pileup sites were restricted to heterozygous coding sites with a marginal eQTL effect (p < 1e−4) and with 10 or more reads. These data were used in Figure 2 of the manuscript. Description of column headers is contained in the spreadsheet.
This table contains 104,750 rows and 11 columns. Each row represents an eQTL peak in one of the tissues analyzed in this study. The “Platelets” tissue represents data from the PRAX1 study cohort. All other tissues were taken from GTEx V6. These data were used in Figure 3 of the manuscript. Description of column headers is contained in the spreadsheet.
This table contains 3,834 rows and 167 columns. Each row represents mRNA-seq read density across 10 bp intervals spanning the MFN2 region for all mRNA-seq Epigenome samples. These data were used in Figure 5B of the manuscript. Description of column headers is contained in the spreadsheet.