Abstract
The proteins encoded by TELO2, TTI1, and TTI2 interact to form the TTT complex, a co-chaperone for maturation of the phosphatidylinositol 3-kinase-related protein kinases (PIKKs). Here we report six affected individuals from four families with intellectual disability (ID) and neurological and other congenital abnormalities associated with compound heterozygous variants in TELO2. Although their fibroblasts showed reduced steady-state levels of TELO2 and the other components of the TTT complex, PIKK functions were normal in cellular assays. Our results suggest that these TELO2 missense variants result in loss of function, perturb TTT complex stability, and cause an autosomal-recessive syndromic form of ID.
Introduction
Early-onset intellectual disability (ID) describes a common (incidence 1%–3% in the Western world) and highly heterogeneous group of phenotypes.1, 2, 3 It is estimated that variants in >1,000 genes result in the autosomal-recessive forms of ID.3 Customarily, ID is divided into two categories: syndromic forms in which the intellectual problems occur together with a constellation of other phenotypic features and non-syndromic forms in which the only constant manifestation is ID. In practice, this distinction is often difficult to make until a large number of individuals with variants in the same gene are well phenotyped.3
TELO2 (MIM: 611140) is the human ortholog of Tel2, an S. cerevisiae gene identified in a screen for genes involved in maintenance of telomere length.4, 5 Located at 16p13.3, TELO2 has 21 exons and encodes an 837 amino acid protein that interacts physically with TELO2 interacting proteins 1 and 2 (TTI1 and TTI2) to form the TTT complex.6 Homozygosity for a Telo2 knockout allele in mice produces embryonic lethality and S phase cell-cycle arrest in mouse embryonic fibroblasts (MEFs). Mice heterozygous for the Telo2 null allele are viable, fertile, and apparently healthy.7
The TTT complex interacts with Hsp90 and the R2TP complex forming a supercomplex that acts as a co-chaperone for maturation of a set of six phosphatidylinositol 3-kinase-related protein kinases (PIKKs).6, 7, 8, 9 The PIKKs are involved in a variety of key cellular processes, including the double strand DNA breakage response (ATM [MIM: 607585], PRKDC [MIM: 600899]),10, 11, 12 DNA replication stress (ATR [MIM: 601215]),10, 11 growth response to nutrient availability (MTOR [MIM: 601231]),13 nonsense-mediated RNA decay (SMG1 [MIM: 607032]),14, 15 and epigenetic modifications through regulation of histone acetylation (TRRAP [MIM: 603015]).16, 17 Genetically mediated deficiency of various PIKK proteins is associated with specific disease phenotypes: pathogenic biallelic variants in ATM cause ataxia telangiectasia (MIM: 208900),18 those in ATR cause Seckel syndrome 1 (MIM: 210600),19 and those in PRKDC result in immunodeficiency 26 (IMD26 [MIM: 615966]).12, 20 Heterozygosity for missense variants in MTOR have been associated with ID, megalencephaly, and dysmorphic facial features.21, 22 Deregulation in the mTOR pathway is associated with certain cancer syndromes,23 and pathogenic variants in the mTOR interactor TSC1 (MIM: 605284) cause tuberous sclerosis-1 (MIM: 191100), a disorder characterized by abnormally regulated cellular growth.13 Somatic TRRAP variants have been associated with melanoma24 and in mice, homozygosity for a Trrap-null variant results in early embryonic lethality.25, 26
Here we report six individuals from four families with ID and assorted neurological and physical abnormalities. All individuals are compound heterozygotes for rare variants (five missense and one complex allele consisting of a nonsense and splice site variant) in TELO2. Our results indicate that variants in TELO2 are responsible for syndromic ID.
Material and Methods
Subjects
Family 1 was recruited from the Johns Hopkins Hospital Genetics Clinic as part of the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) project. Family 2 was recruited through the Washington University Genetics Clinic based on shared abnormalities in TELO2 detected by clinical WES performed by GeneDx. Family 3 was recruited through the TGEN Center for Rare Childhood Disorders. Family 4 was recruited through the University of Vermont Genetics Clinic after clinical WES performed at GeneDx and subsequent matching of TELO2 as a candidate causative gene through entry into GeneMatcher.27 Our study was approved by the Johns Hopkins Medicine Institutional Review Board and by the IRBs of the other participating institutions. We obtained informed consent from responsible individuals in all four families.
Whole-Exome Sequencing and Analysis
For family 1, we captured the CCDS exonic regions and flanking intronic regions totaling ∼51 Mb by using the Agilent SureSelect XT kit and performed paired end 100 bp reads with the Illumina HiSeq2500 platform. We aligned each read to the 1000 Genomes phase 2 (GRCh37) human genome reference with the Burrows-Wheeler Alignment (BWA) v.0.5.10-tpx.28 Local realignment around indels and base call quality score recalibration were performed with the Genome Analysis Toolkit (GATK)29 v.2.3-9-ge5ebf34. Variant filtering was done via the Variant Quality Score Recalibration (VQSR) method.30 For SNVs, the annotations of MQRankSum, HaplotypeScore, QD, FS, MQ, and ReadPosRankSum were used in the adaptive error model (6 max Gaussians allowed, worst 3% used for training the negative model). HapMap3.331 and Omni2.5 were used as training sites with HapMap3.3 used as the truth set. SNVs were filtered to obtain all variants up to the 99th percentile of truth sites (1% false negative rate). For indels, the annotations of QD, FS, HaplotypeScore, and ReadPosRankSum were used in the adaptive error model (4 max Gaussians allowed, worst 12% used for training the negative model, indels that had annotations more than 10 SD from the mean were excluded from the Gaussian mixture model). A set of curated indels obtained from the GATK resource bundle (Mills_and_1000G_gold_standard.indels.b37.vcf) were used as training and truth sites. Indels were filtered to obtain all variants up to the 95th percentile of truth sites (5% false negative rate). Using the PhenoDB Variant Analysis Tool of PhenoDB,27 we prioritized rare functional variants (missense, nonsense, splice site variants, and indels) that were homozygous or compound heterozygous in all three affected subjects and excluded variants with a minor allele frequency (MAF) > 0.01 in dbSNP 126, 129, and 131, the Exome Variant Server (release ESP6500SI-V2), or 1000 Genomes Project.33, 34, 35 We also excluded all variants found in our in-house controls (CIDRVar 51Mb). We generated lists of homozygous and compound heterozygous variants shared by the affected siblings but heterozygous in the unaffected parents.
For families 2 and 4, clinical WES was performed at GeneDx. Candidate variants were validated by Sanger sequencing of PCR amplified products of genomic DNA. Annotated variants are based on RefSeq transcript GenBank: NM_016111.3 and NCBI human genome assembly build 37.
For family 3, WES was performed at TGEN and variants validated by Sanger sequencing.
Telomere Length, Colony Survival, and Mitomycin Sensitivity Assays
Telomere length was measured on peripheral blood lymphocytes by flow cytometry and fluorescence in situ hybridization as previously described.36 Diepoxybutane testing was performed per standard procedure on fresh blood as described.37 EBV-transformed lymphoblastoid cell lines were generated as described.38 Modified colony survival was performed as described,39 and cells were counted on day 8 after irradiation (CK04-01, Dojindo Molecular Technologies). The final surviving fraction was calculated by dividing absorbance at 0.5–2.0 Gy for each sample by absorbance at 0 Gy. Mitomycin C sensitivity was examined as described.40
Immunoblot and Quantitative RT-PCR
We performed immunoblot assays on protein extracted from lymphoblastoid cells and fibroblasts via standard procedures.32 Antibody sources and concentrations were as follows: TTI2 (1:1,000; Bethyl cat# A303-476A; RRID: AB_10948973), TTI1 (1:2,000; Bethyl cat# A303-451A; RRID: AB_10953982), TELO2 (1:2,000; Proteintech cat# 15975-1-AP; RRID: AB_22033337), ATM (1:1,000; Novus Biologicals cat# NB110-55475; RRID: AB_837630), ATR N-19 (1:1,000; Santa Cruz cat# sc-1887; RRID: AB_630893), PRKDC (1:1,000, Thermo Scientific cat# MS-423-P0,), mTOR (1:1,000; Cell Signaling cat# 2983; RRID: AB_10830890), SMG1 (1:1,000; Cell Signaling cat# 9592; RRID: AB_2192936), TRRAP (1:1,000; Cell Signaling cat# 3967; RRID: AB_2209656), and β-actin (1:20,000; Ambion cat# AM4302; RRID: AB_437394). We incubated the membranes with horseradish peroxidase labeled secondary antibodies (goat anti-rabbit IgG-HRP antibody [Santa Cruz cat# sc-2004; RRID: AB_631746] or goat anti-mouse IgG-HRP antibody [Santa Cruz cat# sc-2031; RRID: AB_631737]) diluted 1:20,000, developed the exposed film in a Kodack X-OMAT processor, and analyzed signal intensities with ImageJ software.
To inhibit fibroblast Hsp90, we added 17-allylamino-17-desmethoxygeldanamycin (17-AAG) (Sigma) to the culture medium for the indicated time points and concentrations.41 For quantitative RT-PCR, we isolated total RNA from fibroblasts using Trizol (GIBCO) and subjected 1.25 μg of total cellular RNA to reverse transcription using qScript cDNA SuperMix system (Quanta Biosciences, #95048-100) and PerfeCta SYBR Green FastMix, Rox (95073-012) according to the manufacturer’s protocol. We performed qRT-PCR for TELO2 mRNA in triplicate and normalized to three control genes, GAPDH, HPRT, and YWH4.
Results
Identification of Variants in TELO2
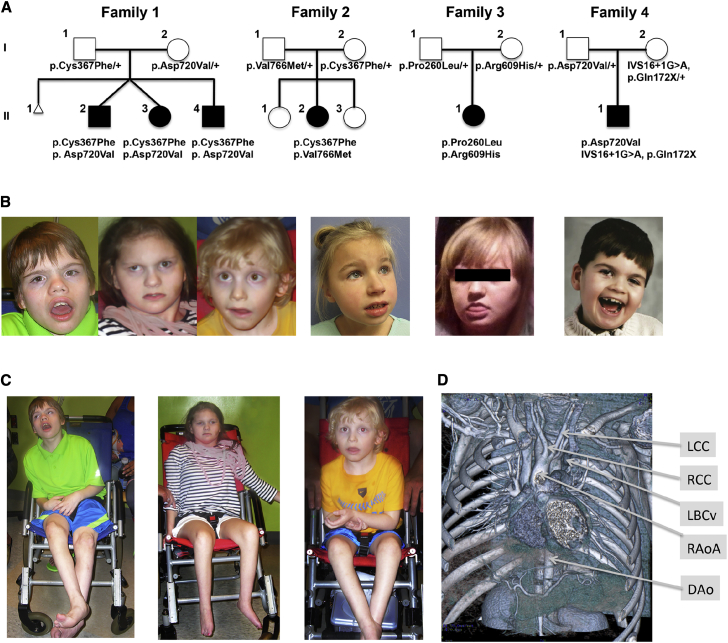
Initially, we identified a single non-consanguineous family (family 1) with three non-ambulatory teenage sibs, all with severe ID, visual and hearing impairments, abnormal movements, and structural abnormalities of the great vessels. Subsequently, we identified three additional unrelated singletons in non-consanguineous families, all with severe ID and clinical features partially overlapping those of the affected individuals in family 1 (Figure 1, Table 1, and the case reports in Supplemental Data). Whole-exome sequencing (WES) of the affected individuals and their parents in family 1 identified rare compound heterozygous missense variants in TELO2: c.1100G>T (p.Cys367Phe) in exon 8 and c.2159A>T (p.Asp720Val) in exon 18 (variant annotation based on RefSeq transcript GenBank: NM_016111.3). These TELO2 variants were the only ones that met our analytic criteria of rare, function-altering alleles in compound heterozygosity or homozygosity present in all three affected siblings. The mother and father are heterozygous for the p.Asp720Val and p.Cys367Phe variants, respectively (Figure 2A). We validated these variants by Sanger sequencing (Figure S1). Both variants are rare: p.Asp720Val is not reported in the 1000 Genomes Project (2,577 samples, build 20130502, accessed November 2015),34 dbSNP build 131,43 Exome Variant Server release ESP6500SI-V2 (6,503 samples, accessed November 2015), or Exome Aggregation Consortium (ExAC) database (60,706 samples, accessed November 2015); and p.Cys367Phe is absent from the 1000 Genomes Project and Exome Variant Server and has an allele frequency of 0.01959% in the ExAC database and is not described in homozygosity. Both variants are likely to be damaging: p.Cys367Phe is in the N-terminal domain of TELO2 and alters a residue conserved among representative vertebrates (Figure 2B) with a PolyPhen-2 score of 0.997 (probably damaging, score range 0 [benign] to 1.0 [probably damaging])44 and SIFT score of 0.01 (scores ≤ 0.05 predicted damaging, damaging, score range 0–1.045, 46); p.Asp720Val falls in the C-terminal domain of TELO2 and is also highly conserved, with PolyPhen-2 and SIFT scores of 1 and 0, respectively.
Figure 1.
Clinical Phenotype of Individuals with TELO2 Variants
(A) The pedigrees of families 1–4. The TELO2 genotype segregating in each family is shown. Individual II-1 in family 4 has an unaffected paternal half-sibling (not shown).
(B) Current portraits of each affected individual.
(C) Full body pictures of affected individuals in family 1.
(D) Coronal view of chest CT angiogram of II-2 in family 1 showing an incomplete vascular ring with a right ascending aortic arch (RAoA), right common carotid artery (RCC), left common carotid artery (LCC), and descending aorta (DAo). There are also venous anomalies with a retroaortic left brachiocephalic vein (LBCv) that joins the azygous vein and enters the superior vena cava.
Table 1.
Phenotypic Features of Individuals with TELO2 Variants
| Family ID | Family 1 | Family 2 | Family 3 | Family 4 | ||
|---|---|---|---|---|---|---|
| Subject | II-2 | II-3 | II-4 | II-2 | II-1 | II-1 |
| Age (years) | 17 | 17 | 10 | 6 | 17 | 9 |
| Intellectual disability | + | + | + | + | + | + |
| Microcephaly | + | + | + | + | + | + |
| Hearing loss | + | + | + | − | − | − |
| Cortical visual impairment | + | + | + | − | − | + |
| Oral frenuli/ankyloglossia | − | + | + | − | − | − |
| Cleft palate | − | + | − | − | − | − |
| Congenital heart disease | +; double aortic arch, vascular ring, cleft mitral valve | +; double aortic arch, atretic L arch, incomplete vascular ring | +; coarctation of aorta | − | − | − |
| Kyphoscoliosis/scoliosis | − | + | − | − | + | + |
| Brachydactyly & clinodactyly | + | + | + | − | − | clinodactyly |
| 4/5 toe syndactyly | + | + | + | − | − | − |
| Abnormal balance | + | + | + | + | + | + |
| Abnormal sleep pattern | − | − | − | + | + | + |
| Laughter outbursts | + | − | − | − | + | + |
| Movement disorder | + | + | + | + | + | + |
| Seizures | − | − | − | − | + | − |
| Rotatory nystagmus | − | + | − | − | + | − |
| TELO2 genotype | c.1100G>T, c.2159A>T | c.1100G>T, c.2159A>T | c.1100G>T, c.2159A>T | c.1100G>T, c.2296G>A | c.779C>T, c.1826G>A | c.2034+1G>A, c.514C>T, c.2159G>A |
| TELO2 alteration | p.Cys367Phe, p.Asp720Val | p.Cys367Phe, p.Asp720Val | p.Cys367Phe, p.Asp720Val | p.Cys367Phe, p.Val766Met | p.Pro260Leu, p.Arg609His | p.Gln172X, p.Asp720Val |
All major abnormalities present in two or more of the individuals plus selected abnormalities present in only one patient. See Supplemental Data for case descriptions and specific data.
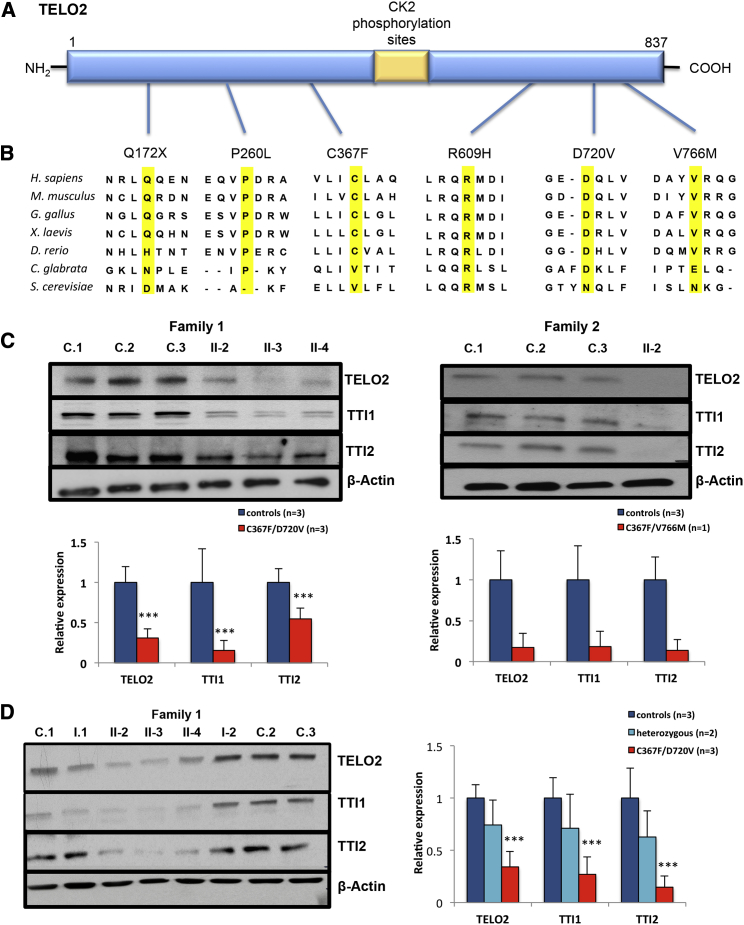
Figure 2.
TELO2 Variants Affect Steady-State Levels of TTT Complex Components
(A) A schematic of the TELO2 protein and the location of the missense variants identified in our study.
(B) Evolutionary conservation of the amino acid residues altered by the missense variants in the indicated affected individuals (highlighted in yellow) and the surrounding TELO2 residues. The species for each sequence is listed on the left. The alignment was determined by ClustalW2 multiple protein alignment.42
(C) Immunoblot analysis of TTT complex components in extracts of primary fibroblasts in family 1 and family 2. The left panel shows decreased protein levels of TELO2, TTI1, and TTI2 in three affected siblings in family 1 (II-2, II-3, and II-4) and three control subjects (C.1, C.2, and C.3). Left lower panel is the quantification of TELO2, TTI1, and TTI2 levels in fibroblast extracts. The error bars show 1 SD around the mean determined in 3 independent experiments as measured by immunoblot analysis. The right panel shows that TELO2, TTI1, and TTI2 levels in extracts of skin fibroblast from the proband (II-2) from family 2 were reduced to about 17%, 18%, and 14% of these in controls in 3 independent experiments. The error bar shows 1 SD in three independent Western blots.
(D) Immunoblot analysis of TTT complex in LCLs in family 1. Three controls LCL (C.1, C.2, and C.3) are shown. I-1 and I-2 are the heterozygous parents of the affected individuals in family 1. The three affected individuals in family 1 (II-2, II-3, and II-4) are shown. TELO2 protein levels were reduced to 31.8% of the mean of control subjects while the levels in the heterozygous parents are reduced to 74% of control subjects (p > 0.1, Student’s t test). Similarly, TTI1 and TTI2 proteins in the heterozygous parents of family 1 showed a decrease as compared to control levels which was not statistically significant (for TTI1, mean = 70%, p > 0.1; for TTI2, mean = 62%, p > 0.1, Student’s t test). In the affected individuals the level of TTI1 is reduced to 17% and TTI2 to 13% of control levels in LCL, respectively (p < 0.05, Student’s t test). The error bar shows 1 SD in 3 independent experiments.
See also Figure S3.
The affected individuals in families 2, 3, and 4 are also compound heterozygotes for rare TELO2 variants. The proband in family 2 is heterozygous for c.1100G>T (p.Cys367Phe), the same variant observed in family 1, and a rare heterozygous missense variant c.2296G>A (p.Val766Met), which has a minor allele frequency of 0.02283% in the ExAC database and is not reported in homozygosity. p.Val766Met variant alters a highly conserved residue in the C-terminal domain of TELO2 and has a PolyPhen-2 score of 0.986 (probably damaging) and a SIFT score of 0.02 (damaging) (Figures 1A, 2A, and 2B). This variant is represented in dbSNP build 138 (rs371675497) but lacks an associated allele frequency. The proband in family 3 is a compound heterozygote for two additional TELO2 variants: c.779C>T (p.Pro260Leu), a substitution with a PolyPhen-2 score of 1 and SIFT score of 0.23, and c.1826G>A (p.Arg609His) with PolyPhen-2 score of 1 and SIFT score of 0. The p.Pro260Leu variant is represented in dbSNP build 138 (rs369656775) without an associated allele frequency, while p.Arg609His is not listed in the dbSNP, 1000 Genomes, EVS, or ExAC databases. The proband in family 4 is a compound heterozygote for p.Asp720Val, the same allele segregating in family 1, and a complex allele, c.514C>T (p.Gln172X) plus c.2034+1G>A (IVS16+1G>A) producing an aberrantly spliced transcript that is likely subject to nonsense-mediated mRNA decay (NMRD) and encodes a severely C-terminal truncated protein. For these reasons, it is almost certainly a null allele. The fact that this individual’s second allele, p.Asp720Val, does not rescue TELO2 function supports our earlier conclusion for family 1 that this missense variant results in loss of function of TELO2.
TELO2 Variants Reduce Steady-State Levels of TELO2 Protein
In preliminary studies, we found that there was no alteration in the levels of TELO2 mRNA in total cellular RNA isolated from cultured skin fibroblasts of affected individuals in family 1 (Figure S2). To determine the functional consequences of the TELO2 variants on TELO2 protein, we first evaluated steady-state levels of TELO2 and its interacting partners in cultured lymphoblast cell lines (LCLs) and fibroblasts from the affected individuals in family 1. Using immunoblot assays on protein extracts of both cell types, we found that TELO2 levels were reduced to about 34% of control levels in LCLs and 33% of control levels in fibroblasts harvested in three separate experiments (p < 0.05, Student’s t test, Figures 2C, 2D, and S3). TELO2 levels in extracts of LCL cells from the heterozygous parents of family 1 were nominally reduced (mean of three measurements = 74%), but this reduction was not statistically significant (Figures 2C and 2D). In family 2, TELO2 levels in extracts of cultured skin fibroblasts from the proband were reduced to a mean of 17% of those in control samples (3 independent experiments, range 2%–33%, Figures 2C and 2D). Cultured cells from the affected individuals in families 3 and 4 are not available. We conclude from these results that the TELO2 missense variants in affected individuals from families 1 and 2 destabilize TELO2. Although we were not able to make this measurement in the probands of families 3 and 4, they each share at least one missense allele with the affected individuals in families 1 and 2.
TELO2 Alterations Affect Steady-State Levels of TTI1 and TTI2
We next tested the consequence of the TELO2 variants on steady-state levels of its partner proteins in the TTT complex, TTI1 and TTI2. We found that both proteins were significantly decreased in the affected individuals in family 1 (mean 14.6% and 51.4% of control levels in fibroblast extracts, and mean 27.2% and 14.7% of controls in LCL extracts, respectively, p < 0.05, Student’s t test; Figures 2C, 2D, and S3) and in the proband of family 2 (mean 18% and 14% of control levels in fibroblasts, respectively, 3 independent experiments; Figures 2C and 2D). The heterozygous parents in family 1 exhibited a modest reduction that did not reach statistical significance (70% and 63% of TTI1 and TTI2, respectively, in extracts of LCL, p > 0.1, Student’s t test, 3 independent experiments; Figure 2D). These results indicate that the mutations in TELO2 destabilize the entire TTT complex.
Consequences of TTT Complex Reduction on the Stability and Function of the PIKKs
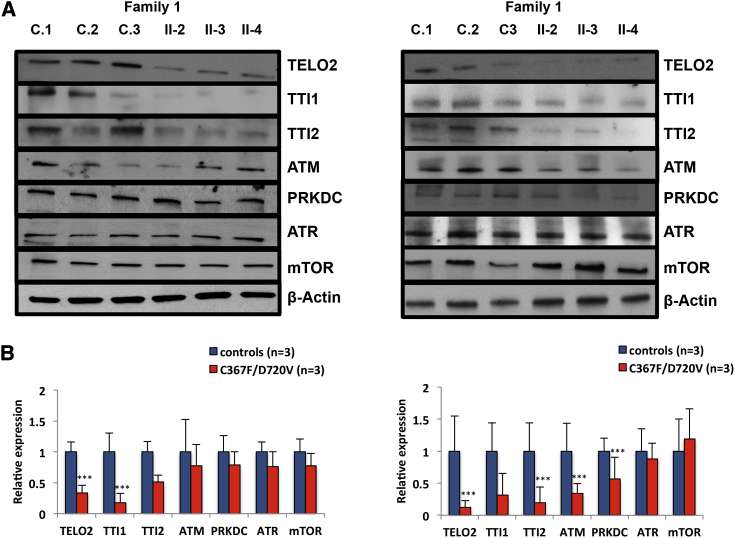
The TTT complex is a component of a large protein super-complex that includes an Hsp90 dimer and the R2TP complex comprising RUVBL1 (MIM: 603449) and RUVB2 (MIM: 604788), RPAP3 (MIM: 611477), and PIH1D1 (MIM: 611480).7, 9, 47 Although the function of the TTT complex is incompletely understood, available evidence suggests that it is necessary for the folding and stability of newly synthesized PIKKs.7, 47, 48, 49 In view of the reduction in TTT complex components in individuals with TELO2 variants, we next measured the steady-state levels of the PIKK proteins ATM, ATR, PRKDC, mTOR, SMG1, and TRRAP in extracts of LCL cells cultured from the affected individuals in family 1. We found no significant change in the levels in affected individuals and their parents compared to controls (Figure S3). We also tested the functional integrity of DNA repair in cells with compromised TTT complex by assessing de novo sensitivity to ionizing radiation, mitomycin C, and diepoxybutane (not shown) but found no abnormalities (Figures S4 and S5). These results suggest that despite reduction of the components of the TTT complex, TELO2 mutant cells retain functional integrity of the PIKKs under these conditions. We also measured telomere length by flow cytometry and fluorescence in situ hybridization in primary lymphocytes and granulocytes from the affected individuals and their parents in family 1 and family 2 and found them to be normal (Figure S6). This result is consistent with what has been thought to be a yeast-specific role of TELO2 in telomere maintenance that is not retained in mammalian cells.7
Hsp90 interacts with the TTT and R2TP complexes to facilitate maturation of the PIKKs.9, 49 Accordingly, we asked whether inhibition of Hsp90 would accentuate the reduction in the TTT complex components. We cultured fibroblasts from three normal control subjects and the three affected individuals in family 1 in medium containing 1 μM of the Hsp90 inhibitor, 17-allylamino-17-desmethoxygeldanamycin (17AAG), for 48 hr.49 The reduction of TELO2 levels we observed in the affected individuals’ cells cultured in standard medium was accentuated by culture in the presence of 17AAG (Figure 3A): levels of TELO2 fell from 33% of control in the absence of 17AAG to 12% of control in the presence of 17AAG (mean values in cultured skin fibroblasts from 3 affected individuals as compared to controls, p < 0.05, Student’s t test). Similarly, the levels of TTI2 fell from 51% of control levels in the absence of 17AAG to 20% of control levels in the presence of 17AAG. TTI1 levels as compared to controls were variable, but the mean actually increased from 14.6% to 31.6% (Figure 3B). This reduction in TELO2 and TTI2 in cells cultured in the presence of 17AAG was associated with significant decreases in protein levels of ATM and PRKDC (means of 34% and 56%, respectively, of levels in control fibroblasts cultured in 17AAG; p < 0.05, Student’s t test; Figure 3B). The levels of ATR and mTOR were not reduced under these conditions. This result suggests that with the additional stress conferred by inhibition of Hsp90, there is further reduction of the TTT complex in the TELO2 mutant cells with corresponding negative effects on the abundance of certain PIKKs.
Figure 3.
Effect of Hsp90 Inhibition by 17AAG on Levels of TTT Complex Components and Selected PIKKs
(A) Representative immunoblots of extracts from cultured skin fibroblasts of affected individuals in family 1 (II-2, II-3, and II-4) and three control subjects (C.1, C.2, and C.3) cultured in standard medium (left) or in medium with 1 μM 17AAG for 48 hr (right).
(B) In standard medium (left), the steady-state levels of the TTT complex components and selected PIKKs expressed as percent of the mean of control levels are: TELO2 (33%, p < 0.05), TTI1 (18%, p < 0.05), TTI2 (51%, p = 0.06), ATM (77%, p > 0.1), PRKDC (78%, p > 0.1), ATR (76%, p > 0.1), and mTOR (77%, p > 0.1). In medium with 17AAG, the level of protein expression (right) expressed as percent of the mean of control levels are: TELO2 (12%, p < 0.05), TTI1 (32%, p = 0.056), TTI2 (19.6%, p < 0.05), ATM (34%, p < 0.05), PRKDC (56%, p < 0.05), ATR (87%, p > 0.1), and mTOR (111.8%, p > 0.1). The error bar indicates 1 SD in three independent experiments (Student’s t test).
Discussion
We report six individuals from four unrelated families with overlapping clinical features including global developmental delay, intellectual disability, dysmorphic facial features, microcephaly, abnormal movements, and abnormal auditory and visual function. In addition, the three affected individuals in family 1 have striking developmental abnormalities of the great vessels, a feature not detected in routine evaluations of the affected individuals in families 2–4. Of these, only the affected individual in family 4 was specifically studied for this phenotype, so it is possible that the others might also have some unrecognized abnormality of great vessel anatomy. Focused phenotypic studies of these and additional individuals will be required to determine whether the incidence of these abnormalities is increased by deficiency of TELO2 function.
All affected individuals in our series are compound heterozygous for rare variants (five missense affecting conserved residues and one complex null allele) in TELO2. The missense variants in families 1 and 2 result in a reduction of the steady-state level of TELO2 protein in cultured skin fibroblasts and LBLs (Figure 2C). In the available affected individuals’ cells from families 1 and 2, the levels of the TELO2 interacting proteins, TTI1 and TTI2, are also decreased (Figures 2C and 2D). Interestingly, under standard cell culture conditions, we did not observe alterations in the PIKK proteins (ATM, ATR, PRKDC, mTOR, SMG1, and TRRAP), whose maturation depends on the TTT complex function,6, 7, 8, 9 suggesting that the TELO2 mutations are not sufficient to disrupt their maturation under these conditions. However, with the stress of exposure of the cells to 17AAG, an inhibitor of HSP90, the levels of at least two of the PIKKs (ATM, PRKDC) were reduced in affected individuals’ cells as compared to controls (Figure 3). This result suggests that the further reduction in the TTT complex provoked by 17AAG leads to secondary alterations in the levels of certain PIKKs. We hypothesize that such stresses might occur in certain cells at critical times in development and lead to the phenotypic features in these individuals. Alternatively, disruption of the TTT complex might have heretofore unidentified functions beyond stabilization of PIKK.
Current understanding of TELO2 structure is based on a partial analysis of yeast Tel2 structure49 that shows Tel2 to be a multi-helical protein in which pairs of α helices align with each other to form suprahelical assemblies or α solenoids, characteristic of helical repeat proteins. Yeast Tel2 has 21 NTD α helices and 11 CTD α helices that assemble into NTD and CTD α solenoids, respectively. NTD α helices 15–21 are the most highly conserved structural motifs.49 Pull-down experiments with yeast and mouse TELO2 homologs indicate that the NTD α solenoid interacts with TTI1 and TTI2.49 The 688-residue yeast Tel2 has 81 amino acid identities (11.8%) with the 837-residue human TELO2 (9.9% in the 354-residue NTD and 15.8% in the 261-residue CTD). Although the five missense variants identified in the affected individuals described here all involve highly conserved residues, the consequences of these alterations on the overall structure and function of TELO2 remain to be determined. The p.Cys367Phe variant identified in families 1 and 2 falls in the middle of NTD α helix 18, the region of the NTD α solenoid thought to be involved with binding of TTI1. The p.Pro260Leu variant falls in the middle of the NTD α helix 13. The remaining three missense variants (p.Arg609His, p.Asp720Val, and p.Val766Met) are all located in the CTD α solenoid with only p.Arg609His directly involving an α helix (helix number 29).
The TTT Complex and Its Role in PIKK Maintenance
The TTT complex, R2TP and Hsp90 supercomplex is conserved in budding yeast, worms, and mammals.4, 7, 50, 51, 52 Assembly of the TTT complex in yeast involves binding of the Tel2 N-terminal domain (NTD) to Tti1 and Tti2.49 Consistent with our observations, shRNA directed against each component of TTT complex results in reduced levels of the other two components, indicating that the three components depend on each other for stability.6 In mouse, the interaction the TTT complex with Hsp90 and the R2TP complex involves a covalent linkage of phosphorylated residue Ser492 in Tel2 to the Pih1 scaffold component of the R2TP complex and binding of the Tah1 component of R2TP to the homodimeric Hsp90.9, 47 The interaction of this supercomplex with the PIKKs is mediated by non-covalent binding of TTI1 in the TTT complex to the individual PIKKs.6 Once the PIKKs have reached their active conformation, binding to the TTT complex does not appear to be required for their function.49 The consequence of shRNA-mediated reduction in the levels of the individual components of the TTT complex is variable; reduction of TTI1 results in the most dramatic reduction in the PIKKs whereas a similar reduction in TELO2 has the least consequence on the abundance of the PIKKs.6 Moreover, loss of TTT complex function appears to have variable consequences on the abundance of each of the PIKKs, with ATM being most dramatically reduced.7 Whether this property reflects differences in PIKK maturation and/or stability is not known. In general, the cellular consequences of disturbances in PIKK function have the potential to be broad, with abnormalities in DNA damage response, nutritional response, cell cycle progression, and gene expression. For example, in human osteosarcoma cells (U2OS cells), shRNA-induced reduction of the TTT complex to levels <25% of normal is associated with a corresponding reduction in PIKK abundance and checkpoint signaling.6 The importance of these functions is shown by the early embryonic lethality of mouse embryos homozygous for Telo2 knockout alleles and rapid senescence of MEFs made homozygous for Telo2 deficiency.7 This might be related to deficiency of one or more PIKKs alone or combined with other, yet to be determined, functions of TELO2 or the TTT complex.
This model predicts that pathogenic variants in the genes encoding the other members of the TTT complex would have features similar to those with TELO2 deficiency. In this regard, Najmabadi et al.53 described two siblings with non-syndromic ID who were homozygous for a predicted damaging missense variant (p.Arg236His) in TTI2 (MIM: 614426), and Langouet et al.54 reported three affected siblings with ID, short stature, dysmorphic features, and mild lymphocytopenia (MIM: 615541) who were homozygous for a variant in TTI2 c.1307T>A (p.Ile436Asn). The latter variant was associated with decreased steady-state amounts of TTI2 and, secondarily, TELO2 and TTI1 to levels that were about 5% of normal in three affected individuals. The levels of ATM, PRKDC, and mTOR were also decreased in skin fibroblasts from these individuals. Finally, the supplementary material of Najmabadi et al.53 also lists, as a candidate causative variant, a mutation in TTI1 (MIM: 614425) that produces the missense change (p.Pro1020Thr) in a proband with non-syndromic ID.53
In summary, our results in affected individuals in four unrelated families indicate that loss-of-function variants in TELO2 are responsible for a complex human phenotype that includes ID and various other features. The mechanism by which this occurs remains to be determined but could be due to reduced function of one or more of the PIKKs under developmental or to physiological circumstances not replicated in our cultured cell systems or through perturbation of some, as yet undescribed, function of the TTT complex. The former is supported by some overlap in the phenotypic features (ID, growth retardation, abnormal brain growth, movement disorders) of the individuals we describe here with those of individuals with Mendelian phenotypes of the PIKK genes: ataxia telangiectasia, Seckel syndrome 1, and immunodeficiency 26 with or without neurological abnormalities. Moreover, mTOR, another of the PIKKs, has been implicated in regulation of the local translation of dendritic mRNAs, a key aspect of dendrite and spine morphogenesis and synaptic plasticity,55 and mutations in MTOR have recently been associated with ID and macrocephaly.21, 22 We speculate reduction of TTT complex function secondary to TELO2 variants in certain cells and/or developmental stages resulting in a more global but less individually severe dysfunction of the PIKKs as compared to their isolated monogenic deficiency. Alternatively, there might be additional as yet unknown function(s) of TELO2 and the TTT complex that are perturbed in these individuals and account for their phenotype.
Acknowledgments
We are grateful for the families who participated in this study. This work was supported by the NHGRI grant 1U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (D.V.), RO1CA160433, and the Commonwealth Foundation (to M.A.). J.Y., D. Gable, and J.J. are trainees of the Predoctoral Training Program in Human Genetics at Johns Hopkins University and are supported by the NIH training grant T32GM07814.
Published: April 28, 2016
Footnotes
Supplemental Data include seven figures and supplemental case reports and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.014.
Web Resources
1000 Genomes, http://browser.1000genomes.org
Baylor-Hopkins Center for Mendelian Genomics, https://mendeliangenomics.org/
ExAC Browser, http://exac.broadinstitute.org/
GATK resources, https://www.broadinstitute.org/gatk/guide/article?id=1247
GeneMatcher, https://genematcher.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2 v.2.2.2, http://genetics.bwh.harvard.edu/pph2/
SIFT v.1.03, http://sift.bii.a-star.edu.sg/
Supplemental Data
References
- 1.Schalock R.L., Luckasson R.A., Shogren K.A., Borthwick-Duffy S., Bradley V., Buntinx W.H., Coulter D.L., Craig E.M., Gomez S.C., Lachapelle Y. The renaming of mental retardation: understanding the change to the term intellectual disability. Intellect. Dev. Disabil. 2007;45:116–124. doi: 10.1352/1934-9556(2007)45[116:TROMRU]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 3.Musante L., Ropers H.H. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–39. doi: 10.1016/j.tig.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Lustig A.J., Petes T.D. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Runge K.W., Zakian V.A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurov K.E., Cotta-Ramusino C., Elledge S.J. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takai H., Wang R.C., Takai K.K., Yang H., de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 8.Kakihara Y., Houry W.A. The R2TP complex: discovery and functions. Biochim. Biophys. Acta. 2012;1823:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Pal M., Morgan M., Phelps S.E., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., Prodromou C. Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 2014;22:805–818. doi: 10.1016/j.str.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 11.Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 12.Woodbine L., Neal J.A., Sasi N.-K., Shimada M., Deem K., Coleman H., Dobyns W.B., Ogi T., Meek K., Davies E.G., Jeggo P.A. PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. Invest. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Conti E., Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herceg Z., Wang Z.Q. Rendez-vous at mitosis: TRRAPed in the chromatin. Cell Cycle. 2005;4:383–387. doi: 10.4161/cc.4.3.1546. [DOI] [PubMed] [Google Scholar]
- 17.McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D., Cole M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 18.Sandoval N., Platzer M., Rosenthal A., Dörk T., Bendix R., Skawran B., Stuhrmann M., Wegner R.D., Sperling K., Banin S. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum. Mol. Genet. 1999;8:69–79. doi: 10.1093/hmg/8.1.69. [DOI] [PubMed] [Google Scholar]
- 19.Shanske A., Caride D.G., Menasse-Palmer L., Bogdanow A., Marion R.W. Central nervous system anomalies in Seckel syndrome: report of a new family and review of the literature. Am. J. Med. Genet. 1997;70:155–158. doi: 10.1002/(sici)1096-8628(19970516)70:2<155::aid-ajmg10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 20.van der Burg M., Ijspeert H., Verkaik N.S., Turul T., Wiegant W.W., Morotomi-Yano K., Mari P.O., Tezcan I., Chen D.J., Zdzienicka M.Z. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baynam G., Overkov A., Davis M., Mina K., Schofield L., Allcock R., Laing N., Cook M., Dawkins H., Goldblatt J. A germline MTOR mutation in Aboriginal Australian siblings with intellectual disability, dysmorphism, macrocephaly, and small thoraces. Am. J. Med. Genet. A. 2015;167:1659–1667. doi: 10.1002/ajmg.a.37070. [DOI] [PubMed] [Google Scholar]
- 22.Smith L.D., Saunders C.J., Dinwiddie D.L., Altherton A.M., Miller N.A., Soden S.E., Farrow E.G., Abdelmoity A.T., Kingsmore S.F. Exome sequencing reveals de novo germline mutation of the mammalian target of rapamycin (MTOR) in a patient with megalencephaly and intractable seizures. J Genomes Exomes. 2013;2:63–72. [Google Scholar]
- 23.Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Wei X., Walia V., Lin J.C., Teer J.K., Prickett T.D., Gartner J., Davis S., Stemke-Hale K., Davies M.A., Gershenwald J.E., NISC Comparative Sequencing Program Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikiforov M.A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B., Cole M.D. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herceg Z., Hulla W., Gell D., Cuenin C., Lleonart M., Jackson S., Wang Z.Q. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- 27.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L., Jie C., Obie C., Abidi F., Schwartz C.E., Stevenson R.E., Valle D., Wang T. X chromosome cDNA microarray screening identifies a functional PLP2 promoter polymorphism enriched in patients with X-linked mental retardation. Genome Res. 2007;17:641–648. doi: 10.1101/gr.5336307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard T.J., Aken B.L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., McVean G.A.T., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baerlocher G.M., Vulto I., de Jong G., Lansdorp P.M. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat. Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 37.Auerbach A.D. Diagnosis of Fanconi anemia by diepoxybutane analysis. Curr. Protoc. Hum. Genet. 2015;85:1–17. doi: 10.1002/0471142905.hg0807s85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Penno M.B., Pedrotti-Krueger M., Ray T. Cryopreservation of whole blood and isolated lymphocytes for B-cell imortalization. J. Tiss. Cult. Meth. 1993;15:43–48. [Google Scholar]
- 39.Sun X., Becker-Catania S.G., Chun H.H., Hwang M.J., Huo Y., Wang Z., Mitui M., Sanal O., Chessa L., Crandall B., Gatti R.A. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing. J. Pediatr. 2002;140:724–731. doi: 10.1067/mpd.2002.123879. [DOI] [PubMed] [Google Scholar]
- 40.Pinto F.O., Leblanc T., Chamousset D., Le Roux G., Brethon B., Cassinat B., Larghero J., de Villartay J.P., Stoppa-Lyonnet D., Baruchel A. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 2009;94:487–495. doi: 10.3324/haematol.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izumi N., Yamashita A., Hirano H., Ohno S. Heat shock protein 90 regulates phosphatidylinositol 3-kinase-related protein kinase family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci. 2012;103:50–57. doi: 10.1111/j.1349-7006.2011.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 47.Horejsí Z., Takai H., Adelman C.A., Collis S.J., Flynn H., Maslen S., Skehel J.M., de Lange T., Boulton S.J. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 48.Kaizuka T., Hara T., Oshiro N., Kikkawa U., Yonezawa K., Takehana K., Iemura S., Natsume T., Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takai H., Xie Y., de Lange T., Pavletich N.P. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartman P.S., Herman R.K. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics. 1982;102:159–178. doi: 10.1093/genetics/102.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakowski B., Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 52.Shikata M., Ishikawa F., Kanoh J. Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J. Biol. Chem. 2007;282:5346–5355. doi: 10.1074/jbc.M607432200. [DOI] [PubMed] [Google Scholar]
- 53.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 54.Langouët M., Saadi A., Rieunier G., Moutton S., Siquier-Pernet K., Fernet M., Nitschke P., Munnich A., Stern M.H., Chaouch M., Colleaux L. Mutation in TTI2 reveals a role for triple T complex in human brain development. Hum. Mutat. 2013;34:1472–1476. doi: 10.1002/humu.22399. [DOI] [PubMed] [Google Scholar]
- 55.Troca-Marín J.A., Casañas J.J., Benito I., Montesinos M.L. The Akt-mTOR pathway in Down’s syndrome: the potential use of rapamycin/rapalogs for treating cognitive deficits. CNS Neurol. Disord. Drug Targets. 2014;13:34–40. doi: 10.2174/18715273113126660184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.