Abstract
Electrothermal supercharging (ETS) with electrospray ionization produces highly charged protein ions from buffered aqueous solutions in which proteins have native folded structures. ETS increases the charge of ribonuclease A by 34% whereas only a 6% increase in charge occurs for a reduced alkylated form of this protein, which is unfolded and ~66% random coil in this solution. These results indicate that protein denaturation that occurs in the ESI droplets is the primary mechanism for ETS. ETS does not affect the extent of solution-phase hydrogen-deuterium exchange (HDX) that occurs for four proteins that have significantly different structures in solution, consistent with a droplet lifetime that is significantly shorter than observable rates of HDX. Rate constants for HDX of ubiquitin are obtained with a spatial resolution of ~1.3 residues with ETS and electron transfer dissociation of the 10+ charge-state using a single capillary containing a few μL of protein solution in which HDX continuously occurs. HDX protection at individual residues with ETS HDX is similar to that with reagent supercharging HDX and with solution phase NMR, indicating that the high spray potentials required to induce ETS do not lead to HD scrambling.
Introduction
Tandem mass spectrometry (MS) is a powerful technique for identifying proteins and for determining the identities and sites of post-translational modifications. Information about higher-order structure and dynamics of proteins in solution can also be obtained using tandem MS when combined with solution-phase labelling techniques, such as photo-oxidative labelling [1-4], selective noncovalent adduct protein probing mass spectrometry (SNAPP-MS) [5], primary amine acetylation [6-8], and hydrogen-deuterium exchange (HDX) [9-11]. Of these methods, HDX is most widely used and probes primarily protein secondary structure in solution. Rates of exchange of amide hydrogen atoms along the protein backbone are obtained by MS from the 1 Da mass difference where exchange occurs. Backbone hydrogen atoms that undergo slow exchange for deuterium (on the order of several minutes to even days) are “protected” from exchange and are often involved in hydrogen bonding in alpha helix or beta sheet regions of a protein [12]. Conversely, backbone hydrogen atoms that undergo rapid exchange (on the order of milliseconds to seconds) are likely in random coil or other highly flexible regions of a protein [12]. HDX has the advantage that labelling at every amino acid is possible, whereas covalent modifications typically target a more limited subset of residues [13]. However, H/D labeled sites can undergo back exchange with solvent during the time required to analyze the sample [14-16], which is typically not an issue with covalent labelling methods.
HDX is coupled to mass spectrometry in either a bottom-up or top-down approach. In the bottom-up [9, 10] approach, a protein sample is diluted into D2O to initiate exchange. At incremental time points, HDX is “quenched” by adding acid to pH 2-3, where the HDX rate is at a minimum. The protein is subsequently digested with a protease, such as pepsin, which has optimal activity at pH 3, and the HDX of peptide fragments are measured using liquid chromatography (LC)-MS. One of the advantages of bottom-up HDX is that there is no limit to the size of the proteins or protein complexes that can be studied [17-19], but there is a limit to the resolution on the exchange sites that can be obtained using this approach. Information on one exchange site is lost at every proteolytic cleavage site for a given peptide fragment, and the resolution on the exchange sites is typically limited by the size of the peptides that are produced by proteolysis. Tandem MS on the peptides has been used to localize exchange to individual residues [12]. The top-down HDX [11, 20, 21] approach is similar, but does not involve proteolysis in solution prior to electrospray. After HDX is “quenched”, intact protein ions are produced by ESI, and the intact protein is fragmented in the gas phase. With this approach, sites of exchange can be localized to individual amino acids. In tandem MS in either bottom-up or top-down HDX, electron capture or transfer dissociation (ECD or ETD) is typically used to minimize HD scrambling that can occur between residues in a single peptide or protein during fragmentation [20, 22, 23], which can be significant for slow-heating methods, such as collision induced dissociation (CID) [24-26]. For example, Jorgensen and Zubarev [22] showed that for a peptide that exchanges rapidly on one end and slowly on the other, <5% HD scrambling occurs with ECD, but ~92% occurs with CID.
In the top-down HDX approach, it is essential to form high charge-state protein ions in order to maximize sequence coverage and the resolution of HDX information. High charge-state protein ions are more efficiently fragmented in tandem MS than low charge-state ions. Electron capture/transfer efficiencies increase with the extent of charging [27-30], and highly charged ions have more extended conformations with fewer noncovalent interactions, resulting in greater fragmentation and sequence coverage [31, 32]. High charge states for top-down fragmentation are typically produced by denaturing the protein prior to ESI, either by acidification during quenching of HDX, or by the introduction of organic solvents [11, 21]. High charge states can also be formed from aqueous solutions at neutral pH with a variety of supercharging methods. For example, supercharging [33-43] with the reagent m-nitrobenzyl alcohol can be used as a “quench” step [41] that chemically and/or thermally unfolds the protein in the ESI droplet [37-41]. By using supercharging as the HDX quench and to produce high charge state ions, a continuous, real-time measurement of HDX of proteins in buffered aqueous solutions can be obtained from a single nanoESI capillary [41]. This method saves both sample and sample manipulation, and close to single amino acid resolution on exchange rates can be obtained for a small protein.
High charge-state protein ions can be formed from aqueous buffered solutions with electrothermal supercharging (ETS) [44-46]. In ETS, increasing protein ion charge from aqueous ammonium bicarbonate solutions is as simple as increasing the ESI spray potential and inducing thermal denaturation of the protein in the ESI droplet [44, 45]. However, it is possible that HD scrambling could occur as a result of the energetic source conditions used in ETS in which both high inlet capillary temperature and high ESI potentials are used. Here, we show that ETS is significantly less effective on proteins lacking ordered structure providing additional evidence that ETS unfolds proteins in the ESI droplet. The extent of HDX incorporation in proteins is the same in native MS as ETS MS, indicating that there is no significant back exchange during protein unfolding in the ESI droplet. Top-down fragmentation coupled to ETS HDX MS shows that no measureable HD scrambling occurs during either ETS or ETD, and that ETS HDX MS/MS data provides similar structural information at individual residues that obtained by NMR.
Materials and Methods
Ions were formed by nanoelectrospray ionization (nanoESI) from aqueous solutions containing 100 mM ammonium bicarbonate, pH 7.8, unless otherwise specified. Borosilicate capillaries (1.0 mm o.d./0.78 mm i.d., Sutter Instruments, Novato, CA, USA) pulled to a tip i.d. of ~1 μm using a Flaming/Brown micropipette puller (Model P-87, Sutter Instruments, Novato, CA, USA) were used as nanoESI emitters, and electrospray was initiated by applying a potential of +0.8-0.9 kV for native MS and +1.3 kV for ETS MS to a platinum wire that is in contact with the aqueous solutions. The source capillary temperature was held at 250 °C, resulting in a temperature of ~35 °C at the nanoESI capillary containing the sample solution. A Thermo Linear-Trapping Quadrupole (LTQ) Orbitrap mass spectrometer equipped with electron transfer dissociation (ETD) was used to obtain mass spectral data. ETD was performed by reacting ubiquitin ions with fluoranthene anions for 30 ms. The source chamber was continuously flushed with dry nitrogen gas to reduce back exchange from incorporation of water from air into the ESI droplets. Fragment ions were identified by comparing their m/z values to those of c and z ions calculated using Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm) within an uncertainty of ± 0.005. Rate constants for HDX were obtained from the extent of deuterium incorporation as a function of time for each residue by fitting these data with an exponential function in OriginPro (OriginLab, Northampton, MA, USA). Protection factors (P) are calculated as P = kint/k, where kint is the intrinsic peptide backbone exchange rate constant estimated as that for polyalanine according to the method of Englander and coworkers [47], and k is the measured exchange rate constant at an individual residue of ubiquitin. A value of log(P) = 0.0 is assigned for all residues for which k >1.3 min−1 (or log(P)<3.08), which corresponds to complete exchange within the standard deviation of deuterium incorporation measurements (0.06) prior to the start of data acquisition at 2.2 min.
Lyophilized samples of ribonuclease A (RNase A), holo-myoglobin, apo-myoglobin, and ubiquitin, and all solvents and salts were obtained from Sigma (St. Louis, MO, USA). Amide-capped reduced-alkylated ribonuclease A (raRNase A) was made from RNase A as described previously [37], and ubiquitin was deuterated as described by Sterling et al. [41].
Results and Discussion
ETS charging and protein conformation
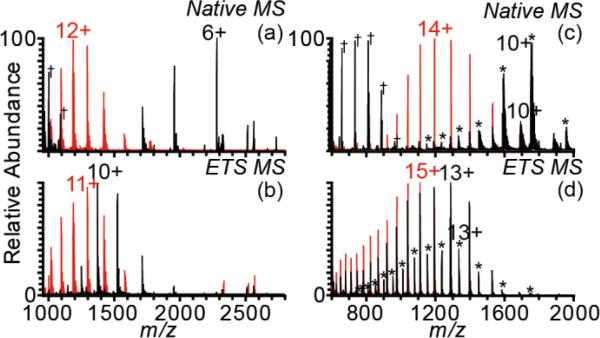
The fidelity of HDX in solution must be maintained throughout measurements aimed at determining where exchange occurs. Heating of ESI droplets and concomitant protein unfolding in the ESI droplet that may occur during ETS [44] could potentially result in some loss of HDX fidelity. The role of protein conformation and unfolding as the primary mechanism of high charging in ETS was investigated by comparing charging obtained for two similar proteins: one that has a native folded structure and one that is disordered and does not have higher order structure. Reduced-alkylated ribonuclease A (raRNase A) is ~66% random coil in solution at pH 6 [48] and is entirely random coil at pH 2-3 [47, 49, 50]. Unmodified ribonuclease A (RNase A) has four disulfide bonds and is folded in solution with ~41% random coil at pH 6 [48]. Ions of both proteins were formed by nanoESI from aqueous 100 mM ammonium bicarbonate, pH 7.8, with native MS (+0.8 kV spray potential) and ETS MS (+1.3 kV spray potential) (Figure 1a,b black and red for RNase A and raRNase A, respectively). With native MS, the width of the charge-state distribution for RNase A is narrow and is centered around ~6+, consistent with a folded conformation in solution. In contrast, the charge-state distribution for raRNase A is broad and centered at ~12+, indicative of its unfolded and largely random coil conformation in solutions around neutral pH. With ETS MS, the charge-state distribution of RNase A shifts to lower m/z, and the average ion charge increases by 34% (Figure 1b; Table 1). In contrast, the charge-state distribution of raRNase A increases by only 6% (Table 1) and shifts from being centered around the 12+ charge state to the 11+ charge state (Figure 1b). Thus, a protein that is extensively disordered in solution does not undergo significant increases in charge in ETS. This indicates that the charge enhancement obtained with ETS is associated with conformational changes of proteins in the ESI droplets.
Figure 1.
NanoESI mass spectra of 10 μM RNase A (a,b, black), raRNase A (a,b, red), holomyoglobin (c,d, black, *denotes that the heme is retained), and apo-myoglobin (c,d, red) in 100 mM ammonium bicarbonate acquired under native MS conditions at a potential of +0.9 kV (a,c) and under ETS MS conditions at a potential of +1.3 kV (b,d). Spectra were acquired separately and subsequently overlaid. Some ammonium bicarbonate cluster ions were observed under native MS conditions and are marked by † in the spectra.
Table 1.
Average charge of protein ions produced by nanoESI from 100 mM ammonium bicarbonate solutions. Uncertainties are the standard deviation of three triplicate measurements.
| Native MS (+0.8 kV) | ETS MS (+1.3 kV) | |
|---|---|---|
| RNase A | 7.1 ± 0.1+ | 9.5 ± 0.1+ |
| raRNase A | 11.3 ± 0.3+ | 12.0 ± 0.2+ |
| Myoglobin | *10.5 ± 0.1+ | *15.2 ± 0.2+ |
| 10.5 ± 0.1+ | 15.6 ± 0.3+ | |
| Apo-myoglobin | 13.6 ± 0.4+ | 16.4 ± 0.3+ |
denotes that the heme is retained
The effect of ETS on the charge of apo-myoglobin, a form of myoglobin that does not have a noncovalently attached heme group and that adopts a molten globule-like state in solution [51, 52], was compared to that of the folded form of the protein with a bound heme group, holomyoglobin. A molten globule state has more folded structure compared to that of a random coil state and retains a native-like secondary structure. However, it is a labile and dynamic state with much more conformational flexibility than a tightly folded protein [53]. Ions of holo-myoglobin and apo-myoglobin were formed by nanoESI from ammonium bicarbonate solution under native MS and ETS MS conditions (Figure 1c,d). The charge-state distribution of holo-myoglobin produced by native MS is centered at ~10+ and is predominantly holo-myoglobin (72% of the protein ion population) (Figure 1c, black). The small population of ions without the heme is due to the relatively energetic source conditions (high interface capillary temperature) necessary for optimal ETS [44], resulting in a small amount of ETS even at low spray potentials. The temperature of the solution in the nanoESI capillary is ~35 °C, much lower than the melting temperature of myoglobin (~76.5 °C) [54]. This indicates that loss of heme does not occur prior to droplet formation by ESI. The charge-state distribution of apo-myoglobin is centered around ~14+ (Figure 1c, red). The higher charge for apo-myoglobin compared to holo-myoglobin in native MS is consistent with a more labile conformation associated with a molten globule state. With ETS MS, the average charge of myoglobin formed from the holo-myoglobin solution increases by 48% (Table 1) and shifts from being predominantly holo-myoglobin to mostly apomyoglobin (76% of the protein ion population) (Figure 1d, black). The concomitant loss of heme with increase in charge suggests that ETS causes protein denaturation and subsequent loss of the noncovalently bound heme group in the ESI droplets. For apo-myoglobin, ETS results in a smaller, but still significant increase (21%) (Table 1) in the charge (Figure 1d, red). These data indicate that some unfolding and increase in charge occurs for this molten globule state of the protein, but the increases in charge that are observed are smaller than for the more rigid, folded form of the protein, holo-myoglobin. The observation that both RNase A and holo-myoglobin, which are folded proteins with native or native-like structure in aqueous solution, undergo large increases in charge with ETS, whereas raRNase A and apo-myoglobin, which do not have significant higher order structure in solution, undergo far smaller changes in charge indicates that the mechanism of electrothermal supercharging involves protein denaturation in the ESI droplet. These results are consistent with no increase in charging with ETS for a dendrimer for which unfolding or significant conformational changes are not possible [45].
ETS-HDX and protein conformation
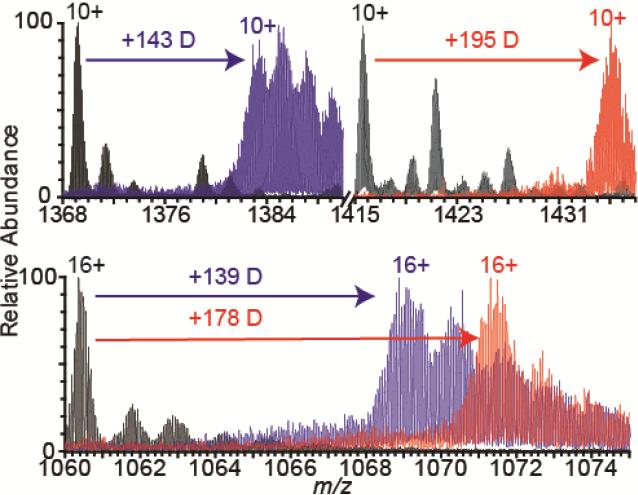
In order to establish whether the fidelity of H/D exchange done under native conditions in solution is adversely affected by the unfolding of proteins in the electrospray droplet that occurs with ETS, HDX of the same four proteins was investigated both with native MS and with ETS. RNase A, raRNase A, holo-myoglobin, and apo-myoglobin were diluted 1:100 to 10 μM in D2O with 100 mM ammonium bicarbonate, pH 7.8, and exchanged at room temperature for 60 minutes prior to electrospray. The deuterium incorporation averaged over all charge states for each protein with both native MS and with ETS is given in Table 2. The average deuterium incorporation for each protein with native MS is the same as that in ETS MS (Table 2). Conformational differences between RNase A and holomyoglobin versus their more disordered counterparts raRNase A and apo-myoglobin, respectively, are indicated by the different number of deuterium incorporated in each (Table 2, Figure 2). For example, the deuterium incorporation of the 10+ charge-state ion of RNase A and raRNase A (Figure 2) produced in ETS MS is 143 and 195 deuterium for RNase A and raRNase A, respectively, corresponding to exchange of 60% and 77%, respectively, of the total number of exchangeable sites on each protein (238 for RNase A and 254 for raRNase A). Similarly, the deuterium incorporation of the 16+ charge-state ion of apo-myoglobin produced by ETS from entirely holo- or apo-myoglobin solutions (Figure 2) is much greater for apo-myoglobin from purely apo-myoglobin solutions (+178 deuterium) than for apo-myoglobin produced from holomyoglobin solutions (+139 deuterium), or 68% and 53%, respectively, of the 262 possible exchangeable sites. Greater deuterium incorporation for raRNase A and apo-myoglobin compared to RNase A and apo-myoglobin produced from holo-myoglobin solutions reflects the greater disorder and structural lability of the former proteins compared to the latter and demonstrates that differences in protein conformation isotopically encoded by HDX are retained during ETS MS.
Table 2.
Average deuterium incorporation of protein ions produced by nanoESI from 100 mM ammonium bicarbonate in D2O after 60 minutes exchange. Uncertainties are the standard deviation of deuterium incorporation for each charge state.
| Native MS (+0.8 kV) | ETS MS (+1.3 kV) | |
|---|---|---|
| RNase A | 148 ± 1 D | 145 ± 5 D |
| raRNase A | 195 ± 4 D | 196 ± 4 D |
| Myoglobin | *137 ± 2 D | *139 ± 2 D |
| 137 ± 2 D | 138 ± 3 D | |
| Apo-myoglobin | 176 ± 3 D | 176 ± 3 D |
denotes that the heme is retained
Figure 2.
Partial nanoESI mass spectra showing the 10+ charge state of RNase A (top left, black) and raRNase A (top right, black) and the 16+ charge state of apo-myoglobin (bottom, black) overlaid with spectra of the same charge states of each protein 60 minutes after dilution into D2O. Blue and red in the bottom spectra correspond to apo-myoglobin produced from holomyoglobin and apo-myoglobin solutions, respectively. All spectra were acquired under ETS MS conditions at +1.3 kV spray potential. The average number of deuterium incorporated for each protein is given above each arrow. An increase in sodium ion adduction is observed from D2O compared to water solutions due to the presence of sodium in the D2O stock.
The same extent of H/D exchange measured in native MS and with ETS indicates that the protein unfolding that occurs in the ESI droplet does not adversely affect the fidelity of the exchange that occurs in bulk solution prior to electrospray. This indicates that protein conformational changes in ETS occur more quickly than H/D exchange occurs. The lifetime of a nanoESI droplet formed under similar conditions was recently measured and is ~27 μs [55]. For exchange to occur on this timescale would require an exchange rate constant on the order of ~6 × 106 min−1. This rate constant is 1000 fold higher than the intrinsic peptide backbone HDX rate constant (~1.6 × 103 min−1) [47] under the conditions used in this experiment. Thus, conformational changes in the ESI droplet during ETS and subsequent ion formation occurs much faster than amide HDX can occur. Although no back exchange can occur in solution during the short lifetime of the electrospray droplet, H/D scrambling may occur as a result of activation and dissociation in the gas phase after ion formation. The extent to which gas-phase scrambling occurs can be investigated by measuring exchange rates at individual amide backbone sites using tandem MS and comparing these rates to those in solution measured by NMR.
ETS-HDX real-time exchange rates for individual residues
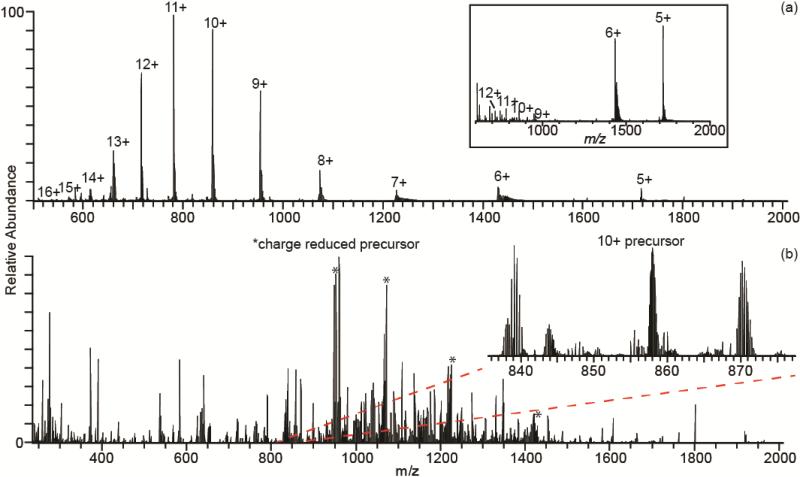
The high charge states that are produced with ETS are efficiently fragmented with ETD [45] resulting in high sequence coverage that is essential to identify individual sites of exchange. ETD of ions of ubiquitin produced with ETS was performed in order to evaluate the efficacy of this method for obtaining real-time H/D exchange rates at individual residues with top-down native MS. Exchange rate constants at individual residues of ubiquitin have been measured with NMR [56-58] and top-down HDX with supercharging reagents [41]. The charge-state distribution of ubiquitin ions produced by ETS from 200 mM ammonium bicarbonate, pH 6, is bimodal, with a low-charge distribution centered around ~5+ and a high-charge distribution centered around ~11+ (Figure 3a). ETD of the 10+ ion, for which top-down HDX kinetics have been obtained previously [41], results in ~90% depletion of the precursor ion (Figure 3b) and cleavage of 70 of the 75 inter-residue bonds (Figure 4). There are 64 c ions and 61 z ions from which 93% sequence coverage is obtained (Figure 4). ETD does not cleave N-terminal to proline residues. Thus, only 72 inter-residue bonds can be cleaved with ETD.
Figure 3.
NanoESI mass spectrum of ubiquitin with ETS at +1.3 kV spray potential from 200 mM ammonium bicarbonate, pH 6 (a), and ETD mass spectrum of the 10+ ion (b). A nanoESI mass spectrum of the same sample of ubiquitin obtained under the same conditions but minimizing the ETS effect by using a +0.9 kV spray potential is inset in (a).
Figure 4.
Map of sequence coverage of ubiquitin from ETD fragmentation of the 10+ ion.
In order to determine whether the activating source conditions required for ETS (high ESI interface temperature and high spray potentials) lead to HD scrambling within the protein ion, either as a result of ion formation or the ETD process itself, HD exchange was done on ubiquitin. Deuterated ubiquitin (D-ubiquitin, 92% total deuterium incorporated) was diluted 1:70 into 200 mM ammonium bicarbonate in water, pH 6 (pH adjusted with acetic acid), to a final concentration of 3 μM in order to initiate HD exchange. After dilution, the solution was loaded into the nanoESI capillary, and electrothermal supercharging was initiated within 2.2 minutes after mixing. Real time information about the exchange kinetics were obtained from ETD mass spectra of the 10+ charge state. Data were acquired continuously until 21 minutes of exchange, at which point the spray potential was set to 0 kV. All later time points were acquired by increasing the spray potential to reinitiate ion formation by ETS at those times. This was done at the longer exchange times to conserve sample where high-density data is not required to obtain accurate exchange rate constants. In contrast, HDX is quenched at specific time points in more conventional top-down HDX experiments so that multiple experiments are required in order to extract accurate rate constants.
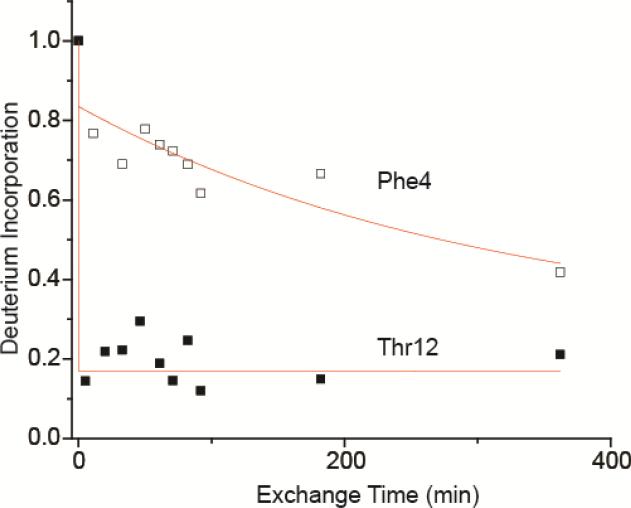
The average deuterium incorporation for an individual residue, n, determined by the difference in mass between fragment ions c(n-1)-c(n-2) or zn-z(n-1), versus reaction time, was used to obtain rate constants of exchange at each site. For example, the difference in the number of deuterium incorporated in c21+ and c31+ corresponds to the average number of deuterium at the backbone peptide bond for residue Phe4, and the difference in the number of deuterium incorporated in c102+ and c112+ corresponds to the average number of deuterium at the backbone peptide bond for residue Thr12 (Figure 5). Phe4 undergoes slow exchange (k = 3.3 × 10−3 min−1, whereas Thr12 undergoes significantly more rapid exchange (k = 4.5 × 10−1 min−1). Due to overlap between isotope distributions for ions at the same m/z, single amino acid resolution of exchange rates were obtained for only 48 out of 75 residues, corresponding to an overall 1.3 residue resolution (weighted average of segment length for which HDX kinetics could be obtained).
Figure 5.
Plot of deuterium incorporation for individual residues Phe4 (open squares) and Thr12 (closed squares) of ubiquitin over three hours’ time.
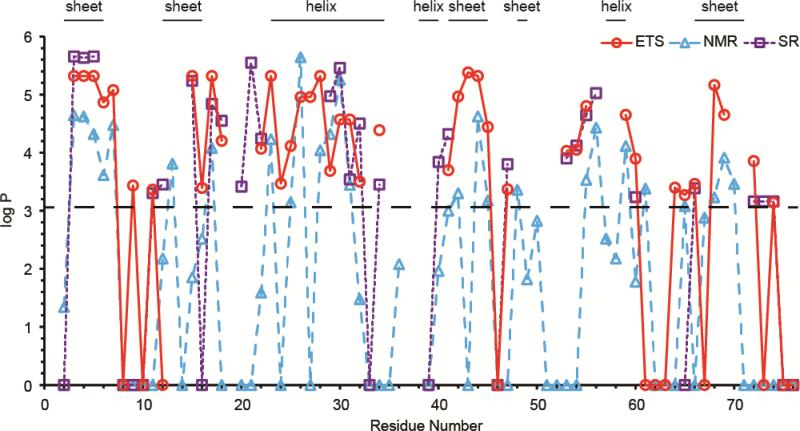
The exchange rates for individual residues were converted to protection factors (P = kint/k) and log(P) at each exchange site, and these data are shown in Figure 6. Overlaid on the top-down ETS HDX data from this work are the log(P) values from the top-down reagent supercharging HDX data measured previously [41], as well as the NMR HDX data from Pan and Briggs [56]. The ETS MS data generally agree with the reagent supercharging MS data and solution phase NMR data, with regions of high or low protection obtained from the ETS MS data corresponding to the same regions of high or low protection measured by the latter two methods. There are some exceptions, however, where MS data shows different protection at an individual residue compared to the NMR data. For example, in the region of residues 71 and 76, the NMR data indicates that there is no protection, whereas the supercharging or ETS MS data indicate that some of these residues have intermediate protection. The dynamic range of the NMR data is greater than that of the MS data. The limit of detection for the MS data is log P ~3.1 so differences in the data sets for smaller values of log P may not be significant. Other discrepancies between data sets occur for individual residues, such as the order of magnitude difference in protection at residues 3-5 between the NMR data and the two MS data sets, and the order of magnitude difference in protection at residue 34 between the ETS and supercharging MS data. These differences may be due to small structural differences in the protein under the different solution conditions (different pH and buffer salts) that were used to obtain the NMR, ETS MS, and supercharging MS data. Regions of high protection in all three data sets correspond to regions of α-helix or β-sheet in the crystal structure of ubiquitin (PDB: 1UBQ). For example, there is a beta sheet between residues 2-7 and a random coil between residues 8-11 in the crystal structure, and these residues have high protection factors (slow exchange) and low protection factors (fast exchange), respectively, in the MS and NMR data sets. There are some residues for which there is high protection for all three data sets in regions that are random coils in the crystal structure. This may be due to the presence of salt bridges or solvent interactions, and discrepancies between NMR or MS HDX data and ordered regions in crystal structure have been reported previously [57, 59, 60]. The similarity of exchange rates measured using ETS MS, reagent supercharging MS and NMR along with the high correlation between exchange rates and ordered regions in the crystal structure demonstrate that there is no significant scrambling of HDX information at individual residues with ETS. These results show that real-time HDX rates of proteins can be obtained using ETS from buffered aqueous solutions in which the protein has a native structure.
Figure 6.
Plot of the logarithm of the protection factor at individual residues of ubiquitin determined with ETS HDX-MS/MS (red circles, solid line), with supercharging HDX-MS/MS (purple squares, dotted line), and with NMR [56] (blue triangles, dashed lines). The horizontal dashed black line indicates the limit of detection for the ETS and supercharging MS data (log P ~ 3.1). At the top are regions of secondary structure in the crystal structure of ubiquitin (PDB: 1UBQ). Single amino acid resolution could not be obtained for residues 61-63 or 75-76, but these segments of the protein underwent immediate exchange for all three or two, respectively, amide hydrogens, indicating no protection at any residue in these segments.
Conclusions
Real-time HDX exchange kinetics that report on the secondary structure of native, folded proteins can be obtained with ETS coupled to top-down MS/MS. ETS causes proteins to unfold in the ESI droplet, the primary mechanism for higher charging with this method. Although the proteins unfold prior to ion formation, this process is sufficiently fast that essentially no back exchange can occur. Thus, information about the higher order structure of the protein is preserved as the ions are transferred into the gas phase. As demonstrated previously, ETD does not produce scrambling of HDX information within a protein in the gas phase [20, 22, 23]. Thus, ETS can be effectively coupled to HDX and tandem MS to obtain residue-specific HDX information on the conformations of proteins from buffered aqueous solutions. An important advantage of this method is that the kinetic data is measured while HDX occurs, which eliminates any quenching steps that are conventionally used. Thus, HDX information on individual residues of a protein can be acquired in real-time using a single nanoESI capillary and only a few hundred nanograms of protein is required.
The charge states that can be produced from proteins using ETS are similar to those produced using the most effective supercharging reagents in native MS [61] and those produced without supercharging reagents from denaturing solutions in which proteins are denatured [45]. ETS has the additional benefits that it is effective on large (~80 kDa) proteins, it eliminates any concerns about potential effects of supercharging reagents on protein conformation in solution prior to electrospray ionization, and high charge states can be produced from aqueous solutions with high buffer concentration where supercharging reagents are less effective [36, 61]. Thus, it should be possible to extend ETS-HDX-MS/MS to larger proteins and even protein complexes than is currently demonstrated.
Acknowledgments
The authors thank the National Institutes of Health (Grant No. R01GM097357) and the National Science Foundation (Graduate Research Fellowship for CAC; Grant No. DGE1106400) for financial support. The authors are grateful to Professor Brian Chait for his pioneering contributions toward understanding effects of protein conformation in electrospray ionization.
References
- 1.Pan Y, Piyadasa H, O'Neil JD, Konermann L. Conformational dynamics of a membrane transport protein probed by H/D exchange and covalent labeling: the glycerol facilitator. J. Mol. Biol. 2012;416:400–413. doi: 10.1016/j.jmb.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 2.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 3.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smedley JG, Sharp JS, Kuhn JF, Tomer KB. Probing the pH-dependent prepore to pore transition of Bacillus anthracis protective antigen with differential oxidative protein footprinting. Biochemistry. 2008;47:10694–10704. doi: 10.1021/bi800533t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ly T, Julian RR. Protein–metal Interactions of calmodulin and α-synuclein monitored by selective noncovalent adduct protein probing mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1663–1672. doi: 10.1016/j.jasms.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Glocker MO, Borchers C, Fiedler W, Suckau D, Przybylski M. Molecular characterization of surface topology in protein tertiary structures by amino-acylation and mass spectrometric peptide mapping. Bioconjug. Chem. 1994;5:583–590. doi: 10.1021/bc00030a014. [DOI] [PubMed] [Google Scholar]
- 7.Izumi S, Kaneko H, Yamazaki T, Hirata T, Kominami S. Membrane topology of guinea pig cytochrome P450 17α revealed by a combination of chemical modifications and mass spectrometry. Biochemistry. 2003;42:14663–14669. doi: 10.1021/bi035096z. [DOI] [PubMed] [Google Scholar]
- 8.Bothner B, Schneemann A, Marshall D, Reddy V, Johnson JE, Siuzdak G. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Mol. Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 9.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal. Chem. 2001;73:256 A–265 A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 10.Englander SW. Hydrogen exchange and mass spectrometry: a historical perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: is there a need for a top-down approach? Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wales TE, Eggertson MJ, Engen JR. Considerations in the analysis of hydrogen exchange mass spectrometry data. Methods Mol. Biol. 2013;1007:263–288. doi: 10.1007/978-1-62703-392-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Kaveti S, Engen JR. Extensive deuterium back-exchange in certain immobilized pepsin columns used for H/D exchange mass spectrometry. Anal. Chem. 2006;78:1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- 15.Walters BT, Ricciuti A, Mayne L, Englander SW. Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J. Am. Soc. Mass Spectrom. 2012;23:2132–2139. doi: 10.1007/s13361-012-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng L, Orlando R, Prestegard JH. Amide proton back-exchange in deuterated peptides: applications to MS and NMR analyses. Anal. Chem. 2006;78:6885–6892. doi: 10.1021/ac060948x. [DOI] [PubMed] [Google Scholar]
- 17.Tuma R, Coward LU, Kirk MC, Barnes S, Prevelige PE., Jr Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J. Mol. Biol. 2001;306:389–396. doi: 10.1006/jmbi.2000.4383. [DOI] [PubMed] [Google Scholar]
- 18.Trnka MJ, Burlingame AL. Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene: the power of high resolution electron transfer dissociation for determination of both peptide sequences and their attachment sites. Mol. Cell Proteomics. 2010;9:2306–2317. doi: 10.1074/mcp.M110.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakata E, Stengel F, Fukunaga K, Zhou M, Saeki Y, Förster F, Baumeister W, Tanaka K, Robinson, Carol V. The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol. Cell. 2011;42:637–649. doi: 10.1016/j.molcel.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Abzalimov RR, Kaplan DA, Easterling ML, Kaltashov IA. Protein conformations can be probed in top-down HDX MS experiments utilizing electron transfer dissociation of protein ions without hydrogen scrambling. J. Am. Soc. Mass Spectrom. 2009;20:1514–1517. doi: 10.1016/j.jasms.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan J, Han J, Borchers CH, Konermann L. Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J. Am. Chem. Soc. 2009;131:12801–12808. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 22.Rand KD, Adams CM, Zubarev RA, Jørgensen TJD. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 23.Pan J, Han J, Borchers CH, Konermann L. Electron capture dissociation of electrosprayed protein ions for spatially resolved hydrogen exchange measurements. J. Am. Chem. Soc. 2008;130:11574–11575. doi: 10.1021/ja802871c. [DOI] [PubMed] [Google Scholar]
- 24.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. Is there hydrogen scrambling in the gas phase? Energetic and structural determinants of proton mobility within protein ions. J. Am. Chem. Soc. 2004;126:7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 25.Demmers JAA, Rijkers DTS, Haverkamp J, Killian JA, Heck AJR. Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. J. Am. Chem. Soc. 2002;124:11191–11198. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson PL, Pan J, Wilson DJ, Dempsey B, Lajoie G, Shilton B, Konermann L. Hydrogen/deuterium scrambling during quadrupole time-of-flight MS/MS analysis of a zinc-binding protein domain. Anal. Chem. 2007;79:153–160. doi: 10.1021/ac061261f. [DOI] [PubMed] [Google Scholar]
- 27.Iavarone AT, Paech K, Williams ER. Effects of charge state and cationizing agent on the electron capture dissociation of a peptide. Anal. Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 29.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, McLuckey SA. Electron transfer dissociation: Effects of cation charge state on product partitioning in ion/ion electron transfer to multiply protonated polypeptides. Int. J. Mass Spectrom. 2012;330–332:174–181. doi: 10.1016/j.ijms.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 32.Mikhailov V, Cooper H. Activated ion electron capture dissociation (AI ECD) of proteins: Synchronization of infrared and electron irradiation with ion magnetron motion. J Am Soc Mass Spectrom. 2009;20:763–771. doi: 10.1016/j.jasms.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglass K, Venter A. Investigating the role of adducts in protein supercharging with sulfolane. J. Am. Soc. Mass Spectrom. 2012;23:489–497. doi: 10.1007/s13361-011-0319-1. [DOI] [PubMed] [Google Scholar]
- 34.Enyenihi AA, Yang HQ, Ytterberg AJ, Lyutvinskiy Y, Zubarev RA. Heme binding in gas-phase holo-myoglobin cations: distal becomes proximal? J. Am. Soc. Mass Spectrom. 2011;22:1763–1770. doi: 10.1007/s13361-011-0182-0. [DOI] [PubMed] [Google Scholar]
- 35.Lomeli SH, Yin S, Loo RRO, Loo JA. Increasing charge while preserving noncovalent protein complexes for ESI-MS. J. Am. Soc. Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterling HJ, Kintzer AF, Feld GK, Cassou CA, Krantz BA, Williams ER. Supercharging protein complexes from aqueous solution disrupts their native conformations. J. Am. Soc. Mass Spectrom. 2012;23:191–200. doi: 10.1007/s13361-011-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterling HJ, Cassou CA, Trnka MJ, Burlingame AL, Krantz BA, Williams ER. The role of conformational flexibility on protein supercharging in native electrospray ionization. Phys. Chem. Chem. Phys. 2011;13:18288–18296. doi: 10.1039/c1cp20277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterling HJ, Daly MP, Feld GK, Thoren KL, Kintzer AF, Krantz BA, Williams ER. Effects of supercharging reagents on noncovalent complex structure in electrospray ionization from aqueous solutions. J. Am. Soc. Mass Spectrom. 2010;21:1762–1774. doi: 10.1016/j.jasms.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sterling HJ, Prell JS, Cassou CA, Williams ER. Protein conformation and supercharging with DMSO from aqueous solution. J. Am. Soc. Mass Spectrom. 2011;22:1178–1186. doi: 10.1007/s13361-011-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterling HJ, Williams ER. Origin of supercharging in electrospray ionization of noncovalent complexes from aqueous solution. J. Am. Soc. Mass Spectrom. 2009;20:1933–1943. doi: 10.1016/j.jasms.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sterling HJ, Williams ER. Real-time hydrogen/deuterium exchange kinetics via supercharged electrospray ionization tandem mass spectrometry. Anal. Chem. 2010;82:9050–9057. doi: 10.1021/ac101957x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teo CA, Donald WA. Solution additives for supercharging proteins beyond the theoretical maximum proton-transfer limit in electrospray ionization mass spectrometry. Anal. Chem. 2014;86:4455–4462. doi: 10.1021/ac500304r. [DOI] [PubMed] [Google Scholar]
- 43.Iavarone AT, Jurchen JC, Williams ER. Supercharged protein and peptide ions formed by electrospray ionization. Anal. Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterling HJ, Cassou CA, Susa AC, Williams ER. Electrothermal Supercharging of Proteins in Native Electrospray Ionization. Anal. Chem. 2012;84:3795–3801. doi: 10.1021/ac300468a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cassou CA, Sterling HJ, Susa AC, Williams ER. Electrothermal supercharging in mass spectrometry and tandem mass spectrometry of native proteins. Anal. Chem. 2013;85:138–146. doi: 10.1021/ac302256d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassou CA, Williams ER. Anions in electrothermal supercharging of proteins with electrospray ionization follow a reverse Hofmeister series. Anal. Chem. 2014;86:1640–1647. doi: 10.1021/ac403398j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins: Struct., Funct., Bioinf. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi S, Kontani T, Yoneda M, Ooi T. A circular dichroic spectral study on disulfide-reduced pancreatic ribonuclease A and its renaturation to the active enzyme. J. Biochem. 1977;82:1127–1133. doi: 10.1093/oxfordjournals.jbchem.a131785. [DOI] [PubMed] [Google Scholar]
- 49.Woodward CK, Rosenberg A. Oxidized RNase as a protein model having no contribution to the hydrogen exchange rate from conformational restrictions. Proc. Natl. Acad. Sci. U.S.A. 1970;66:1067–1074. doi: 10.1073/pnas.66.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pittz EP, Bello J. Interaction of sodium dodecyl sulfate with tyrosyl chromophores in ribonuclease A and model compounds. Arch. Biochem. Biophys. 1971;147:284–298. doi: 10.1016/0003-9861(71)90336-5. [DOI] [PubMed] [Google Scholar]
- 51.Eliezer D, Wright PE. Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 1996;263:531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 52.Lin L, Pinker RJ, Forde K, Rose GD, Kallenbach NR. Molten globular characteristics of the native state of apomyoglobin. Nat. Struct. Biol. 1994;1:447–452. doi: 10.1038/nsb0794-447. [DOI] [PubMed] [Google Scholar]
- 53.Christensen H, Pain RH. Molten globule intermediates and protein folding. Eur. Biophys. J. 1991;19:221–229. doi: 10.1007/BF00183530. [DOI] [PubMed] [Google Scholar]
- 54.Wan LL, Twitchett MB, Eltis LD, Mauk AG, Smith M. In vitro evolution of horse heart myoglobin to increase peroxidase activity. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12825–12831. doi: 10.1073/pnas.95.22.12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mortensen DN, Williams ER. Theta-glass capillaries in electrospray ionization: rapid mixing and short droplet lifetimes. Anal. Chem. 2014;86:9315–9321. doi: 10.1021/ac502545r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan Y, Briggs MS. Hydrogen exchange in native and alcohol forms of ubiquitin. Biochemistry. 1992;31:11405–11412. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 57.Johnson EC, Lazar GA, Desjarlais JR, Handel TM. Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin. Structure. 7:967–976. doi: 10.1016/s0969-2126(99)80123-3. [DOI] [PubMed] [Google Scholar]
- 58.Bougault C, Feng L, Glushka J, Kupče E, Prestegard JH. Quantitation of rapid proton-deuteron amide exchange using Hadamard spectroscopy. J. Biomol. NMR. 2004;28:385–390. doi: 10.1023/B:JNMR.0000015406.66725.30. [DOI] [PubMed] [Google Scholar]
- 59.McAllister RG, Konermann L. Challenges in the interpretation of protein H/D exchange data: a molecular dynamics simulation perspective. Biochemistry. 2015;54:2683–2692. doi: 10.1021/acs.biochem.5b00215. [DOI] [PubMed] [Google Scholar]
- 60.Wang G, Abzalimov RR, Bobst CE, Kaltashov IA. Conformer-specific characterization of nonnative protein states using hydrogen exchange and top-down mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20087–20092. doi: 10.1073/pnas.1315029110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Going CC, Xia Z, Williams ER. New supercharging reagents produce highly charged protein ions in native mass spectrometry. Analyst. 2015;140:7184–7194. doi: 10.1039/c5an01710f. [DOI] [PMC free article] [PubMed] [Google Scholar]