Abstract
G protein-coupled receptors (GPCRs) allosterically activate heterotrimeric G proteins and trigger GDP release. Given that there are ~800 human GPCRs and 16 different Gα proteins, does a universal allosteric mechanism govern Gα activation? Here we show that different GPCRs interact and activate Gα proteins through a highly conserved mechanism. Comparison of Gα with the small G protein Ras reveals how the evolution of short segments that can undergo disorder-order transitions decouple regions important for allosteric activation from receptor binding specificity. This might explain how the GPCR-Gα system diversified rapidly, whilst conserving the allosteric activation mechanism.
Introduction
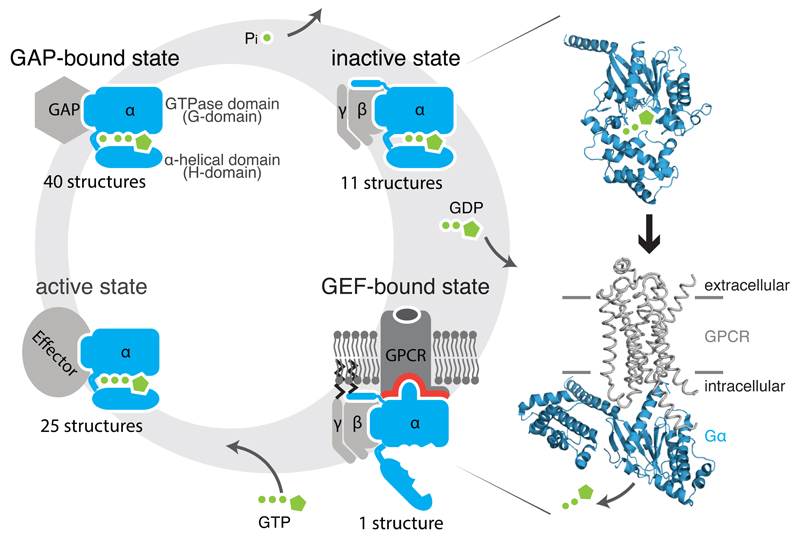
G proteins bind to guanine nucleotides and act as molecular switches in a number of signaling pathways by interconverting between a GDP-bound inactive and a GTP-bound active state1. They are comprised of two major classes2: monomeric small G proteins3 and heterotrimeric G proteins4. While small G proteins and the alpha subunit (Gα) of heterotrimeric G proteins both contain a GTPase domain (G-domain), Gα contains an additional helical domain (H-domain) and also forms a complex with the Gβ and Gγ subunits. Although they undergo a similar signaling cycle (Figure 1), their activation differs in one important aspect. The GDP exchange factors (GEFs) of small G proteins are largely cytosolic proteins, whereas the GEFs of Gα proteins are usually membrane-bound GPCRs. While GEFs of small G proteins interact directly with the GDP binding region1,3, GPCRs bind to Gα at a site almost 30Å away from the GDP binding region5 and allosterically trigger GDP release to activate them.
Figure 1. Gα signaling states and activation.
Heterotrimeric G proteins (1) release GDP upon binding to a GDP Exchange Factor (GEF), which are G protein-coupled receptors, (2) bind GTP and recruit downstream effectors, and (3) hydrolyze GTP promoted by GTPase activating protein (GAP), leading to (4) the inactive, GDP-bound state. Structures of the Gα subunit (blue) bound to GDP (1got; inactive state; top) and bound to the β2-AR (grey) (3sn6; active state; bottom) are shown.
The high-resolution structure of the Gαs-bound β2-adrenergic receptor (β2AR)5 provided crucial insights into the receptor-G protein interface and conformational changes in Gα upon receptor binding6,7. Recent studies described dynamic regions in Gαs6 and Gαi8, the importance of displacement of helix 5 (H5) of Gαs and Gαt by up to 6Å into the receptor5, the extent of helical domain opening during GDP release9,10, and identified residues that contribute to Gαi activation7,10. These studies focused on single, specific Gα proteins, however, in humans there are 16 different Gα genes, with at least 21 isoforms4 that group into four functional subfamilies (Gαs, Gαi, Gαq, Gα12) which each regulate different signaling pathways11. Although they belong to the same protein fold, they have diverged significantly in their sequence such that each Gα protein can be specifically activated by one or several of the ~800 human GPCRs4. Thus a fundamental question is whether there is a universal mechanism of allosteric activation that is conserved across all Gα protein types10. Allosteric communication in proteins is mediated through conformational changes, which is facilitated by the re-organization of non-covalent contacts between residues. Thus studying these contacts can provide detailed insights into the mechanism of allostery12–16. Based on a comprehensive analysis, here we propose that GPCRs interact and activate Gα subunits through a conserved mechanism. We describe molecular details of the key structural transitions and pinpoint residues that constitute the ‘common core’ of Gα activation.
Results
CGN: A numbering scheme for referring equivalent residues in Gα proteins
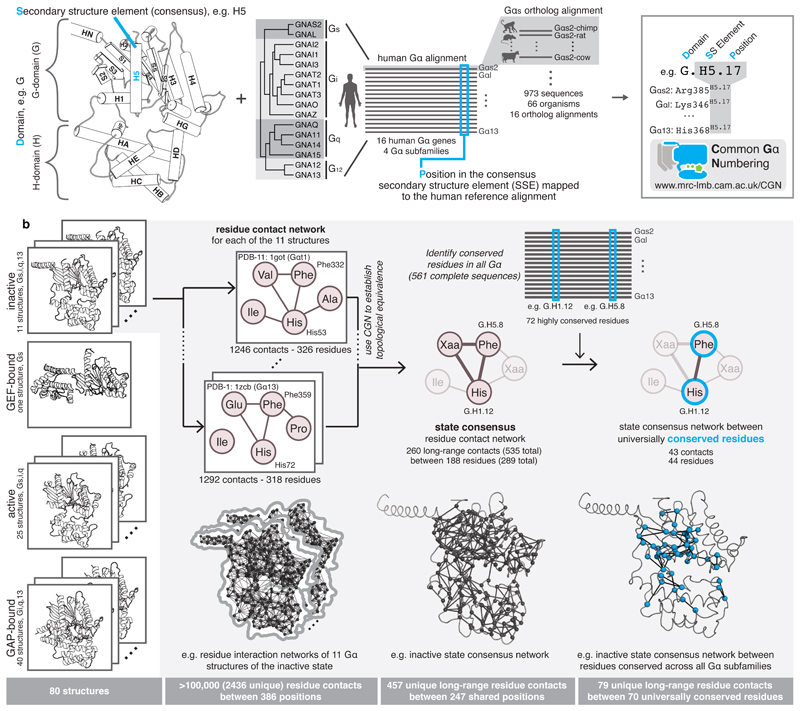
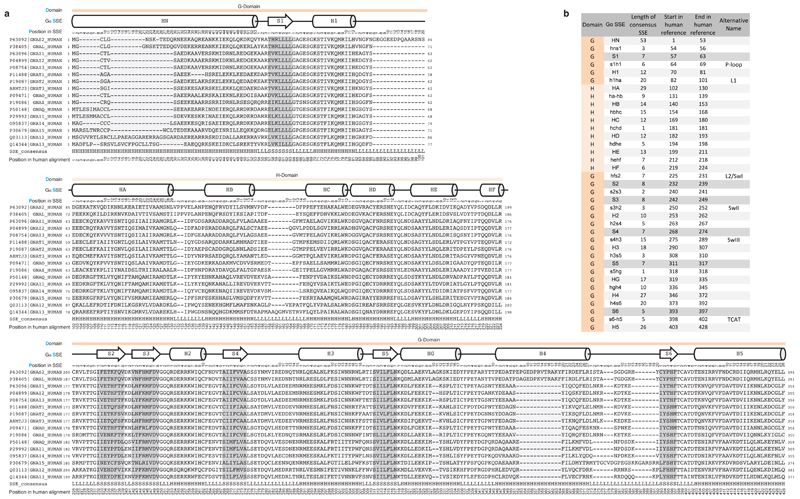
We created a structural and sequence alignment of 80 Gα structures from diverse organisms and 973 sequences from 66 species that have a GPCR-G protein system (auto-activating plant Gα proteins were not considered; Methods). To enable the comparison of any residue/position between different Gα proteins, we devised the Common Gα Numbering (CGN) system (Figure 2a). The CGN provides an ‘address’ for every residue in the D.S.P format, referring to (i) the domain (D), (ii) the consensus secondary structure (S), and (iii) the position (P) within the secondary structure element. For instance, Phenylalanine 336 in Gαi1 will be denoted as Phe336G.H5.8 as it is the eighth residue within the consensus helix H5 of the G-domain. The corresponding position in Gαs2 is Phe376G.H5.8. Loops are labeled in lower case letters of their flanking secondary structure elements (SSE); e.g. s6h5 refers to the loop connecting strand S6 with helix H5 (see Extended Figure 1, Methods and Supplementary Note). A CGN mapping webserver is available at: http://www.mrc-lmb.cam.ac.uk/CGN.
Figure 2. The Common Gα Numbering (CGN) system and Gα conserved contact networks.
a, Every position in a Gα is denoted by its domain (D), the consensus secondary structure element where it is present (S), and position (P) within the consensus SSE. Names of SSEs are shown in the cartoon, loops are named with lower case of the flanking SSEs (e.g. h1ha; see Extended Figure 1 for all SSEs). An alignment of all 973 Gα proteins belonging to all the 4 subfamilies from 66 species allowed the identification of equivalent positions, P. An online webserver allows mapping of any Gα sequence or structure to the CGN system. b, Computation of consensus residue contact networks between conserved residues for different Gα signaling states. See Methods, Supplementary Note and Supplementary Data.
Extended Figure 1. Human paralog reference alignment for Common Gα Numbering System.
a, Reference alignment of all canonical human Gα paralogs. The domain (D), consensus secondary structure (S) and position in the SSE of the human reference alignment (P) are shown on top of the alignment. b, Reference table of the definitions of SSEs used in the CGN nomenclature.
Consensus non-covalent contacts for different signaling states
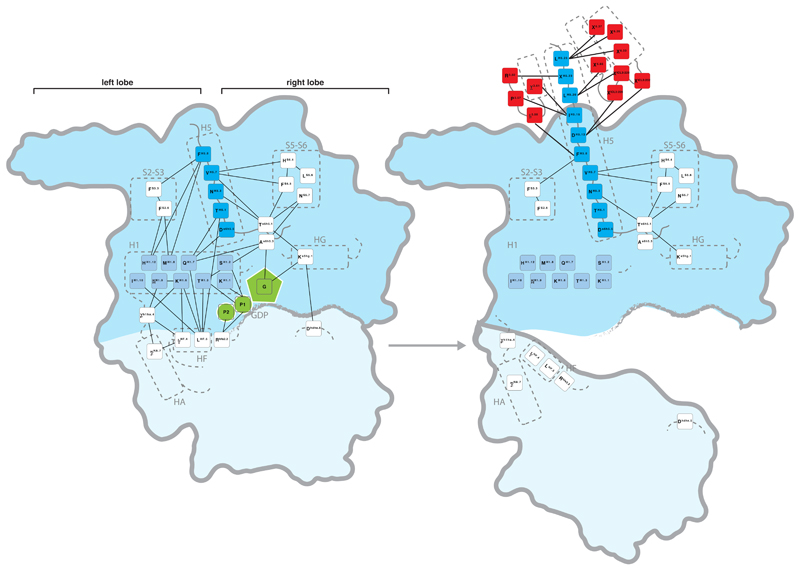
The Gα structures were assigned to the four major signaling states (Figure 1) and the non-covalent contacts between residues were calculated for each structure. We computed the consensus non-covalent residue contacts from all Gα structures of the same signaling state. Using the CGN, we integrated information on evolutionary conservation for every position and derived the consensus contacts mediated by universally conserved residues for each signaling state (Figure 2b). We find that each step of the signaling cycle undergoes contact re-organization to variable extents. Since these conformational changes involve conserved residues, the observed contact re-organization is likely to be universal for all Gα proteins. A description and additional interpretations are provided in the Supplementary Note and Supplementary Material. Below, we describe the major findings pertaining to Gα activation. We first focus on the GPCR-Gα interface and then describe molecular details of how the non-covalent contacts are re-organized and propagated to the GDP binding pocket, leading to GDP release.
Conserved and variable H5 interface residues
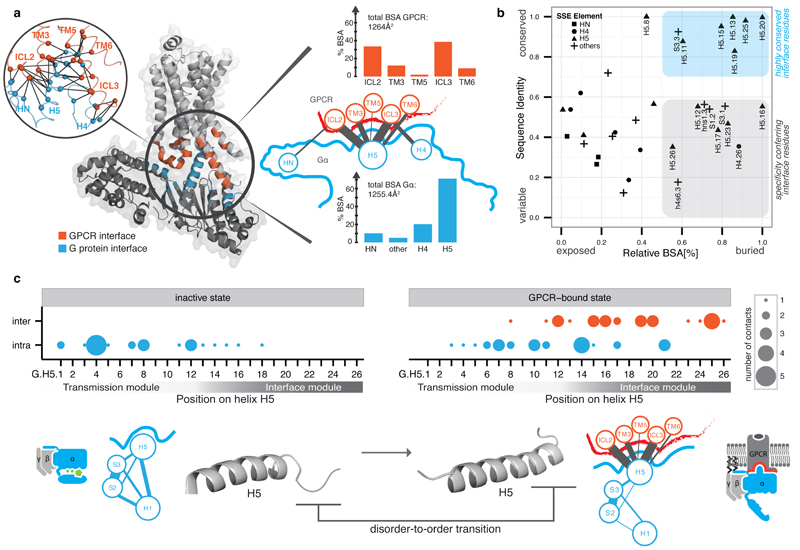
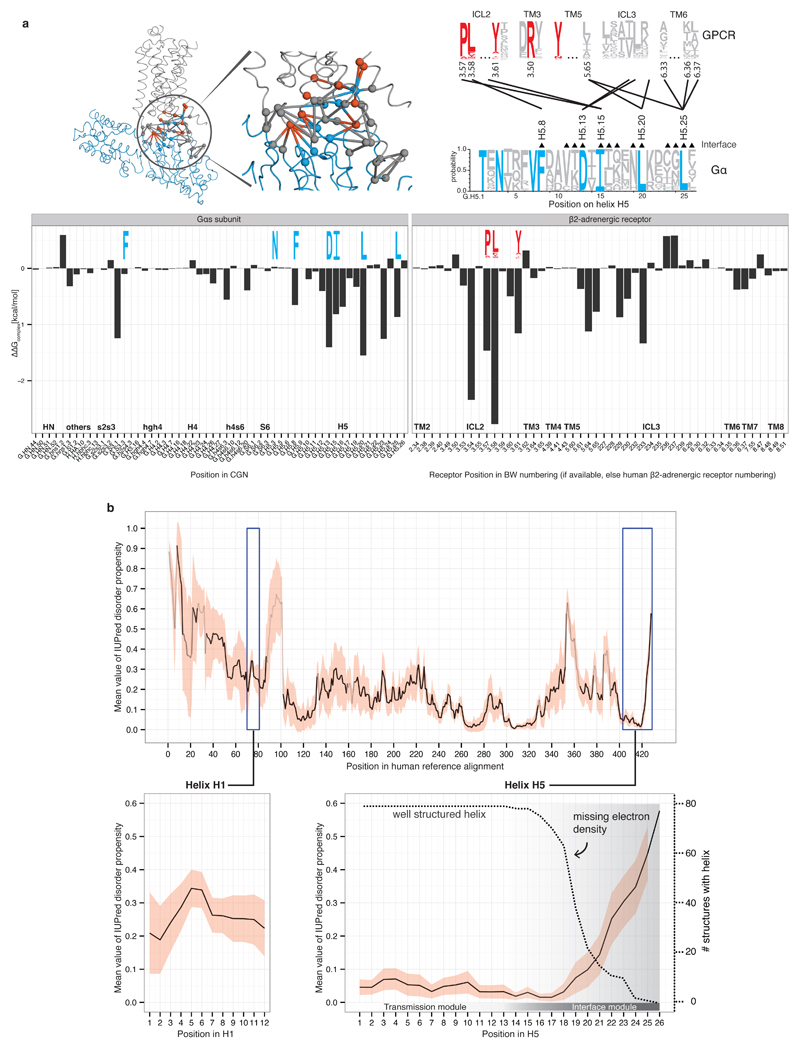
Analysis of the buried surface area (BSA) and residue contacts between the β2AR-Gαs interface5 shows that H5 contributes ~70% (845Å2; 15 residues) of the total BSA. Other SSEs (s2s3/h4hg/H4/h4s6/S6) cover ~20% (289Å2; 14 residues), and the N-terminal membrane-anchored helix HN and its loop with strand S1 contribute ~10% (120Å2; 5 residues) of the total BSA (Figure 3a). H5 is the key interface element that contacts residues in transmembrane helices (TM3, TM5, and TM6) and intracellular loops 2 and 3 (ICL2 and ICL3) of the β2AR5. Contacts from the other Gα regions are mainly restricted to ICL3 of the receptor. An analysis of the contacts of the Gαt C-terminal peptides bound to rhodopsin17–20 shows that conserved residues in H5 tend to interact with the corresponding, topologically equivalent residues in rhodopsin (Supplementary Data).
Figure 3. Helix 5 contains the conserved interface region and is comprised of two modules.
a, Inter Gα–GPCR residue contact network (inset) and buried surface area (BSA) analysis of the heterotrimeric Gαs–β2AR structure (3sn6). The line width between the nodes (SSE) denotes the number of consensus residue contacts. GPCR positions are denoted by extending the BW-numbering system (note parts of ICL3 become extended TM5). b, Scatterplot of Gα sequence conservation and normalized BSA highlights the conserved and variable interface residues. c, Consensus contact rewiring between the inactive and the GPCR-bound state by H5 residues. Positions mediating intra G protein contacts (blue) and receptor-mediated contacts (red) are shown. The circle size represents the number of contacts. H5 can be divided into transmission and interface module. The disorder-to-order transition of the H5 C-terminal region upon receptor binding and SSE contact rewiring are shown.
We mapped evolutionary conservation onto the β2AR-Gαs interface and find that H5 is the only interface region that harbors residues that are highly conserved across species and Gα protein types (~27% of H5; 7 residues; Figure 3b). These residues have significantly higher BSA compared to the non-conserved H5 interface residues. Several highly conserved Gα interface residues interact with conserved interface residues on the GPCR (Extended Figure 2a). Computational energy calculations show that these residues make the highest interface energy contribution, and are likely to be important for complex formation (Extended Figure 2a). Thus, the universally conserved residues on H5 might form the conserved ‘interaction hotspots’ for different Gα proteins to interact with their cognate receptors in a similar binding mode. We also find that two-thirds of the H5 residues are variable but half of these (8 residues) still contact the receptor. Thus H5 harbors distinct sets of interface residues that are either conserved or variable across the different Gα proteins. The variable positions on H5 together with the other interface regions (Figure 3b) could be important for selective coupling to different receptors, as shown for individual Gα proteins21,22. This suggests that the conserved Gα interface positions provide the basis for a common mode of receptor binding while the variable positions might confer selectivity in receptor coupling (Supplementary Note).
Extended Figure 2. Energy estimation of the GPCR-Gα residue contributions and Gα disorder propensity.
a, Energy contribution of single interface residues to the Gαs-β2AR complex calculated with FoldX (T = 298K, pH = 7.0, ion strength = 0.05M). Conserved Gα residues (blue sequence logo) that were identified to form receptor-Gα inter-protein contacts with conserved GPCR residues (red sequence logo) are shown. The contact network between residues of the β2AR and Gαs is shown (red, conserved receptor residue; blue, conserved Gα residue; grey, variable residues; spheres represent Cα positions and links represent non-covalent contact. b, Consensus disorder plot for all Gα proteins. The mean value of the disorder propensity of all full-length Gα sequences (561 sequences) from all 16 Gα types is shown as a black line, the standard deviation at each position is shown as light red ribbon. The color tone of the line indicates the number of gaps at an aligned position (black=no gaps). The left inset shows the disorder propensity of H1. The right inset highlights that H5 is highly structured in its N-terminus, and has increased disorder propensity towards the C-terminus, which is in agreement with the missing electron density in the 79 structures.
H5 disorder-order transition breaks H5-H1 contacts
In the 79 structures of Gα not bound to a GPCR, the C-terminal residues of H5 are characterized by missing electron density (Extended Figure 2b). This region undergoes a disorder-to-order transition and extends H5 upon receptor binding as shown for Gαs/t/i5,6,8,20,23. Analysis of H5 from 561 full-length Gα homologs suggests that the higher disorder propensity of the last eight residues compared to the rest of H5 is a universal feature (Extended Figure 2b). Within this disordered region, hydrophobic positions IleH5.15, LeuH5.20 and LeuH5.25 contact the GPCR, fold into a helix and are conserved between human proteins and the yeast homolog Gpa1p (~1200 Mya). This highly conserved peptide motif24 is involved in the disorder-to-order transition upon receptor binding and suggests that this structural transition mediated by the three key conserved interface residues is likely to be a universal feature of all G proteins.
To understand the effect of GPCR binding on contact re-organization within Gα, we analyzed the consensus contacts in the inactive state (11 structures) and the active state (β2AR-Gαs structure). Although we had only one structure of the receptor-G protein complex, identifying changes in the consensus contacts of the inactive state allowed us to focus on residues that are re-organized upon receptor binding (and hence relevant for all Gα types). In all inactive state structures, the N-terminal part of H5 makes extensive contacts with a number of SSEs within the G-domain of Gα. Two universally conserved positions (PheH5.8 and ValH5.7) on H5 contact conserved positions in H1, S2 and S3 (left lobe), and S5 and S6 (right lobe), respectively (Extended Figure 3). In the GPCR-bound state, H5 loses 20% of its intra-Gα contacts (primarily with H1), and gains 27 inter-molecular residue contacts with the GPCR (Figure 3c). Upon receptor binding, the H5 contacts with the right lobe are not lost, but are re-organized to accommodate the structural changes and might be important for the stability of the receptor-bound complex (for discussion, see Supplementary Note and Sun et al25). Thus, H5 is composed of two highly conserved modules with distinct functions, i.e. an interface module important for receptor binding, and a transmission module that harbors intra-G protein contacts which are re-organized upon receptor binding (Figure 3c).
Extended Figure 3. Rewiring of consensus contacts between conserved Gα residues upon receptor binding.
CGN numbers and sequence logo for consensus contacts within Gα in the inactive state (left) and GPCR-bound state (right) are shown. Receptor residues are shown in red, H5 residues in dark blue, H1 residues in light blue and GDP in green. The domains are highlighted with a blue background (G-domain darker blue, H-domain light blue).
In contrast to H5, the non-conserved interface regions from H4/h4s6/h4hg undergo less dramatic reorganization of intra-Gα contacts upon receptor binding (Extended Figure 3). This suggests that the conserved mechanism of allosteric activation is primarily mediated by breaking the contacts between H5 and H1. As the residues that form these contacts are conserved in all 16 Gα types, this contact reorganization upon receptor binding is likely to be universal for Gα proteins.
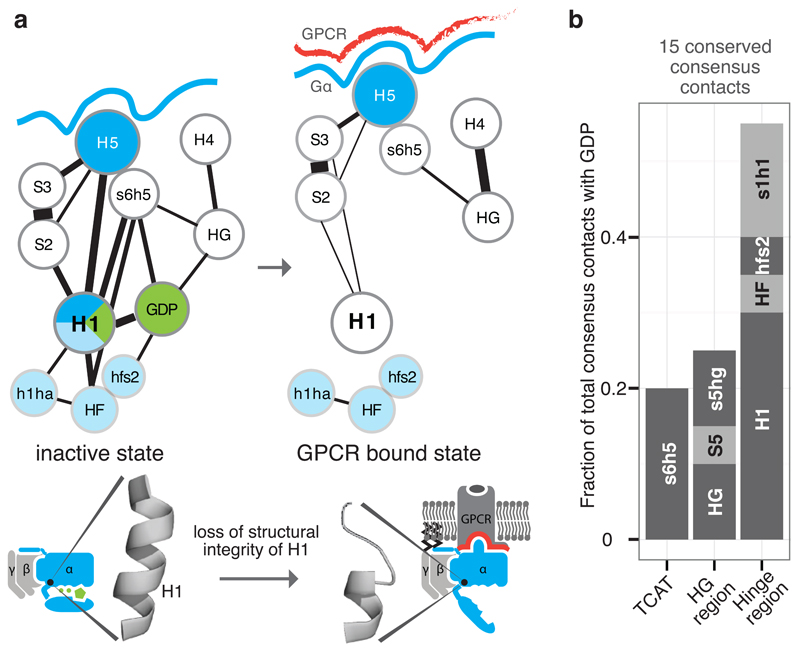
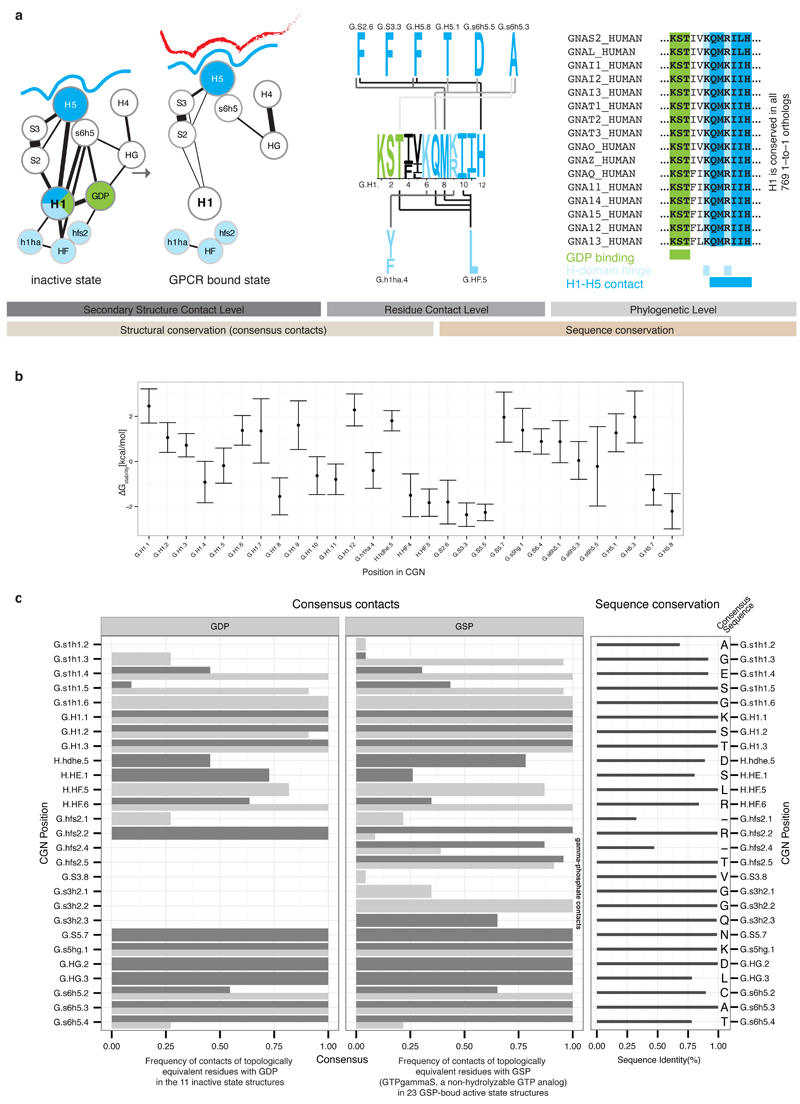
H1 unfolding and GDP release
In the inactive state, H1 acts as a structural ‘hub’ by linking different functional regions of Gα. H1 contacts the N-terminal part of H5 (transmission module), H-domain and GDP through universally conserved residues (Figure 4a). In this manner, H1 links the H-domain and GDP binding site with the conserved residues in the H5 transmission module, which in turn is physically linked to the H5 interface module that binds the receptor. The conserved consensus contacts between H5 and H1 seem essential for the structural integrity of H1. Computational calculations of the per-residue contribution to protein stability for the 79 non-receptor-bound structures are consistent with their role in stabilizing H1 (Extended Figure 4). Upon GPCR binding, the H5-H1 contacts are lost, leading to unfolding of H1. The contact-mediating positions in H1 have missing electron density in the β2AR-Gα structure5 and H/D exchange experiments have shown that this region is dynamic in Gαs upon receptor binding6. Upon becoming flexible, the conserved consensus contacts between H1, GDP and the H-domain hinge region are lost. This results in the loss of a significant fraction of all contacts made with GDP, thereby weakening its binding affinity, (Figure 4b) and H-domain opening. Since the entire sequence of H1, the contacting residues on H5, and the H-domain hinge positions are highly conserved across all Gα proteins, the mechanism of GDP release is likely to be universal for all Gα proteins.
Figure 4. Helix H1 is the key SSE that contacts H5, GDP and the H-domain.
a, Consensus SSE contacts involving H1. The line width between the nodes (SSE) denotes the number of consensus residue contacts. Upon receptor binding, H5 is displaced and crucial contacts between H1 and H5 are lost. This might explain the increased flexibility of H1 in the GPCR-bound state, which results in the loss of GDP contacts and the H-domain hinge region (formed by H1, h1ha, and HF; light blue). b, The extent of GDP consensus contacts mediated by the different SSEs. See Extended Figure 4.
Extended Figure 4. Details of Helix H1 linking H5, GDP and the H-domain.
a, This Figure expands Figure 4 from the main text to provide residue-level details of the role of Helix H1. Residues forming contacts with H5 are shown in blue, with the H-domain in light blue and with GDP in green. Non-covalent consensus contacts between universally conserved residues at the SSE level (left) and per residue-level (center). Lines denote non-covalent contacts between residues. The degree of conservation is shown as sequence logo. Residues are numbered according to the CGN. Helix H1 is almost 100% conserved across all 16 Gα types and forms three structural motifs for interactions with H5, the H-domain and GDP (right). b, Average per residue energy contribution to Gα protein stability as calculated from 79 structures from all four Gα families of the non-receptor bound signaling states using FoldX (T = 298K, pH = 7.0, ion strength = 0.05M). The average energy contribution is shown as dots, the standard deviation as bars. c, Per residue detail of Gα-GDP and Gα-GSP (non-hydrolyzable GTP analog) consensus contacts. The barplot shows the frequency of finding a contact mediated by topologically equivalent positions with GDP/GSP. Side-chain contacts are shown as dark grey bars, main-chain contacts as light grey bars. The degree of conservation of contacting residues (calculated from the 561 complete Gα sequences) is represented in the right panel and the consensus sequence for each position is shown.
Universal model for Gα activation
While variable interface residues in H5 and elsewhere allow specific binding to distinct GPCRs, we find that H5 primarily harbors conserved positions that might allow a common mode of receptor binding and a conserved mechanism of allosteric activation. The contact re-organization between conserved residues links the disorder-to-order transition of H5 upon receptor binding to a change in structural stability of H1, ultimately leading to GDP release. More specifically, H5 is divided into: (a) The N-terminal transmission module, which forms a π- π cluster linking H5, S2, S3 and H1 via universally conserved residues PheH5.8, HisH1.12, PheS2.6 and PheS3.3 in the inactive state and (b) the C-terminal interface module, which undergoes a disorder-to-order transition in the intracellular cavity of the receptor via universally conserved positions. This structural transition results in a displacement of the H5 transmission module, thereby interrupting the π- π cluster. The re-organized residues in the cluster (PheH5.8 and PheS3.3) contact conserved residues within ICL2 of the receptor (extrapolated Ballesteros-Weinstein numbering: 3.58 and 3.57 of β2AR5), as confirmed recently for the CB2 receptor-Gαi complex26. Since the conserved π-π cluster is important for the structural integrity of H1, its disruption leads to an increased flexibility of H1. H1 has a central role in the inactive state by forming contacts both to GDP (Extended Figure 4) via the Walker A motif2,27 and to the H-domain (through a ‘cation-π hinge’ motif; Extended Figure 5). Thus, the increased flexibility due to the partial unfolding of H1 results in GDP release and H-domain opening. The only other conserved inter-domain contact is an ‘ionic latch’ between the C-terminal loop of helix HG of the G-domain and the hChD-loop of the H-domain. This contact is broken upon receptor binding, which might be a result of the reorganization of the right Gα lobe (Extended Figure 3).
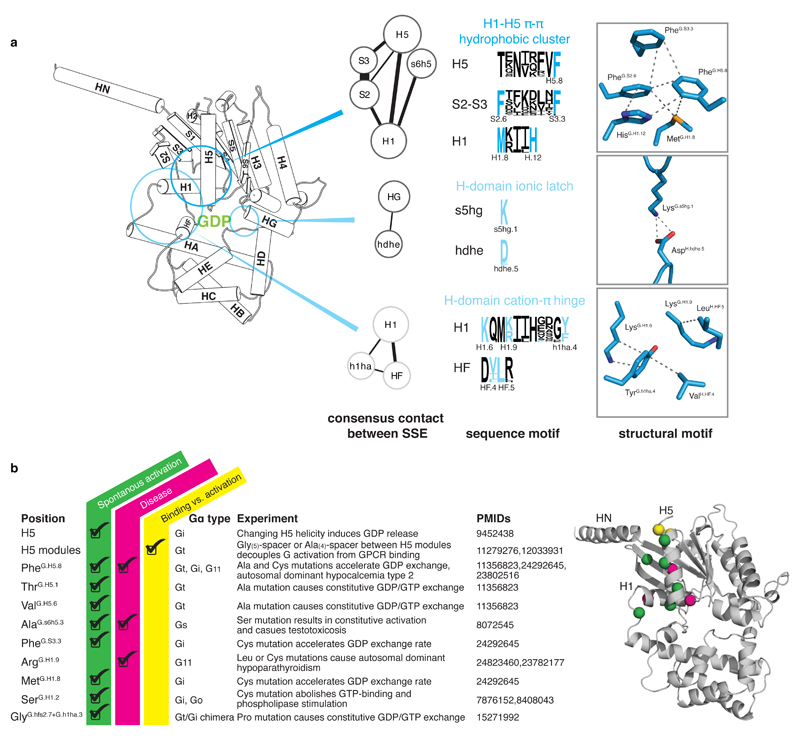
Extended Figure 5. Conserved structural motifs of Gα and known disease and engineered mutations.
a, A universally conserved cluster of pi-pi and hydrophobic interactions between S2 (PheG.S2.6) and S3 (PheG.S3.3), H1 (MetG.H1.8 and HisG.H1.12) and H5 (PheG.H5.8) links H5 and H1 in the absence of the receptor. Upon receptor binding, residues within this motif (PheG.H5.8 and PheG.S3.3) interact with the conserved Pro and Leu of ICL2 of the receptor as has been shown for Gαs (3sn6) and Gαi (Mnpotra et al). Interrupting the contacts between H5 and H1 seems to be the trigger for transmitting the signal of GPCR binding to helix H1 (which interacts with GDP and the H-domain.) The only conserved residue contact between the H-domain and the G-domain that is not in the hinge region is formed by a universally conserved salt-bridge (H-domain ionic latch) between the very N-terminal end of HG of the G-domain (LysG.s5hg1) and the loop connecting HD and HE in the H domain (AspH.hdhe.5). The hinge region is formed by H1, the loop between H1 and HA, and HF. H1 interacts via (i) a cation-pi interaction mediated by a universally conserved residue with the loop connecting H1 and HA (LysG.H1.6 and TyrG.h1ha.4) and (ii) a hydrophobic interaction with HF (LysG.H1.9 and LeuH.HF.5). b, Disease and engineered mutations that can be explained by the universal Gα activation model mapped on a Gα protein. Cα position of residues are shown as spheres; mutations at green positions cause spontaneous GDP release by interrupting consensus contacts between conserved residues, thereby ‘mimicking’ the effect of receptor-binding to Gα. Pink positions have also been reported to cause disease by constitutively activating Gα. Insertion of an Ala4 or Gly5 after the yellow position separate the H5 transmission and interface module, thereby allowing GPCR-binding without triggering GDP release.
In addition to H1, residues around HG and within the s6h5 loop (TCAT motif) contact the GDP (Extended Figure 3). The conserved guanine-contacting TCAT motif preserves most of its contacts within Gα upon receptor binding, although TCAT-to-H1 contacts are lost and new contacts are formed between H5, S5, S6, and the TCAT motif (i.e. the re-organized right lobe). Likewise, residues that contact the guanosine moiety, including the interaction between the TCAT motif and HG, s1h1 (P-Loop), S5 and S6, are not significantly re-organized during Gα activation. Whether this arrangement poises Gα for GTP binding (which differs from GDP by a single phosphate and whose physiological concentration exceeds GDP several-fold22), and whether GTP has the capacity to stabilize H1 on its own and trigger Gα release from the receptor, remains to be addressed. An analysis of the GTP-bound Gα reveals that the presence of the third phosphate facilitates additional contacts with the switch regions (Extended Figure 4c).
Conceptually, the GPCR-bound Gα conformation can be considered as a metastable Gα transition state that is stable only due to interactions with the GPCR. Thus, the lost intra-Gα contacts between H1 and the transmission module of H5 are compensated by the helix extension of H5, the gained receptor interface contacts, and some re-organized contacts in the right lobe of Gα (H5 with S5, S6; see role of conserved Y320G.S6.2 in Sun et al.25). In this manner, H5 and H1 act as the primary conduits of information transfer between the receptor interaction interface (input) and the GDP binding site (output). The residues of the structural motifs and functional elements described here are conserved in all the different families of Gα proteins (Extended Figure 5a). Thus the mechanism described here is likely to be universal for activation of all cognate GPCR/Gα protein pairs.
Disease and engineered Gα mutations
We analyzed disease-causing mutations in the human population and found three key positions that are mutated, resulting in constitutive Gα activity: (i) a variant of the transmission module residue PheH5.8 in Gα11 causes autosomal dominant hypocalcemia type 2 by becoming constitutively active28, possibly by destabilizing the contact between H5 and H1, (ii) Gαi variant Alas6h5.3Ser, a position important for H1 stabilization and GDP binding, causes testotoxicosis by constitutively activating adenylate cyclase29, and (iii) Gα11 variant ArgH1.9Cys causes autosomal dominant hypo-parathyroidism30, possibly by affecting H-domain opening and GDP release (see Supplementary Table 3 for a list of mutations). We also analyzed previously performed perturbation experiments on different Gα types using the CGN system and can rationalize how these mutations affect the activation mechanism (Figure 5a and Supplementary Note). Furthermore, comprehensive alanine-scanning mutagenesis performed on Gαi1, coupled with thermostability assays of the mutants in the GDP-bound or nucleotide-free state coupled to rhodopsin25, is also consistent with the analysis performed here (Figure 5b). For example, mutations in the H5 interface module mainly affect Gαi1-rhodopsin complex stability, whereas mutations in the H5 transmission module primarily affect the nucleotide-bound state of Gαi1. Alanine mutations in H1 highly destabilize the GDP-bound state, but not the Gαi1-rhodopsin complex. In addition, mutating residues in the conserved π-π cluster that interact with PheH5.8 significantly affects Gαi1 stability in the GDP-bound form, but not in the receptor bound form (see Sun et al25). Taken together, our analysis, the CGN numbering system and the universal activation mechanism provides a unified framework to relate and interpret a number of independent experimental and disease mutations in different Gα subfamilies.
Figure 5. Mutational studies support the universal Gα activation mechanism.
a, Disease and engineered mutations can be rationalized by the model of Gα activation in different Gα subfamilies. b, Comparison of the stability of Gαβγ-GDP (∆Tm; °C) and the Gαβγ-GPCR complex (∆ relative complex stability; %) by Gαi1 alanine mutagenesis of every position in H1 and H5. *Mutant FH5.8A is not stable in the receptor-free state but can still form the complex with the receptor.
Comparing Gα and monomeric G protein activation
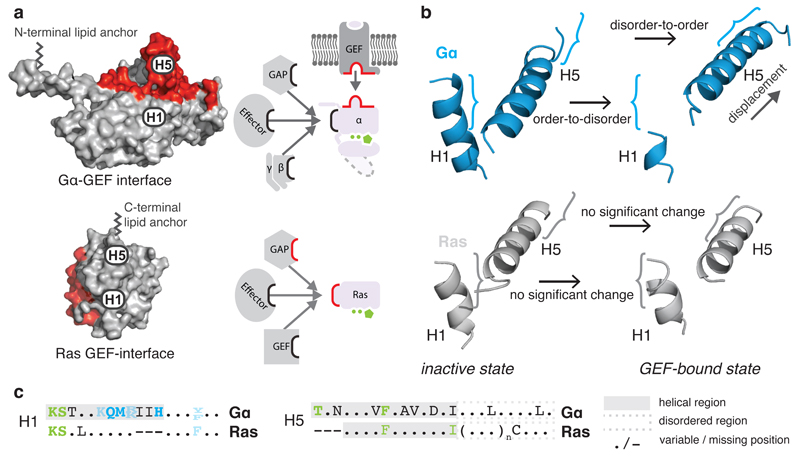
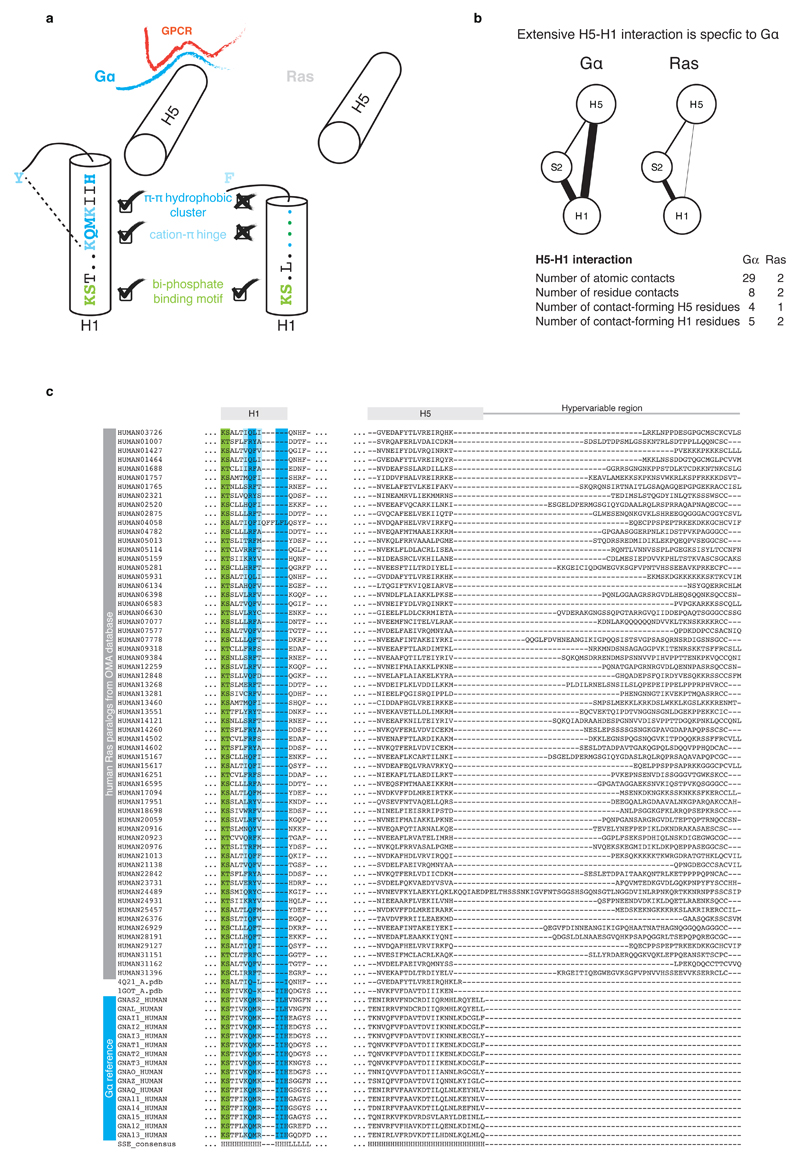
We compared the crystal structures of each equivalent signaling state of Gα and Ras and found that H5 and H1 have significantly changed their functional role. In Ras, H5 has a disordered extension (hypervariable region) that is post-translationally modified for membrane anchoring31. In Gα, the equivalent region forms the GPCR interface module (the N-terminal disordered region of HN is the membrane-anchor32). The nucleotide-binding N-terminal part of H1 is conserved in its sequence and its structural orientation in both Ras and Gα (Figure 6 and Extended Figure 6). In contrast, the central part of H1, which contacts the H-domain hinge, is only conserved in Gα. The C-terminal part of H1 plays a different role in Ras and Gα. While the last turn of H1 in Ras folds back to bind the guanine moiety via a conserved π-π stack, the equivalent region in Gα forms the metastable part of H1 that remains helical in the GDP-bound inactive state due to contacts with the N-terminal transmission module of H5. These contacts are missing in Ras as the C-terminus of H1 and the N-terminus of H5 are each three residues shorter, and H1 in Ras is stable without the interactions with H5. This means that although Ras and Gα are evolutionarily related and share the same architecture, minor but crucial differences in the number and pattern of non-covalent contacts between H5 and H1 have allowed the emergence of an allosteric mechanism for GDP release in Gα. Small extensions in H1 and H5 permit H1 to sense whether H5 is bound to the GPCR. The disordered C-terminal tail of H5 provides both conserved and variable interface residues that allow for a conserved Gα activation mechanism and yet permit the evolution of receptor binding specificity.
Figure 6. H5-H1 interaction permits the allosteric activation mechanism.
a, GEF interaction surfaces (red) for Gα (3sn6) and the small G protein HRas (1bkd) in a superimposed orientation. b, H1 and H5 of the inactive (1got, 4q21) and GEF-bound state (3sn6, 1bkd) for Gα (blue) and HRas (grey). c, Consensus sequences of equivalent residues of H5 and H1 in Gα and Ras. The helical region is highlighted in a grey background and the H5 disordered region is shown in a dashed grey box. See Extended Figure 6.
Extended Figure 6. Helix H5-H1 interaction in Gα provides the allosteric GEF activation mechanism.
a, Schematic representation of structural motifs on H1 that are shared or unique to Gα and Ras. While the part of H1 with the phosphate-binding motif is conserved across both protein families, the C-terminal part is conserved only in Gα. H1 in Gα has three additional residues that allow for extensive residue contacts between H1 and H5. In Ras, these interactions are missing and H5 and H1 are both 3 residues shorter. The consensus sequence and secondary structure of equivalent residues of H1 in Gα and Ras is also depicted. b, Comparison of the residue contact network between topologically equivalent residues in H5 and H1 in the corresponding inactive GDP-bound states of Gα (PDB 1got) and Ras (PDB 4q21). The weight of the link between SSEs denotes the number of atomic contacts. c, Sequence alignments of H1 and H5 of human Gα and Ras paralogs. The sequence alignment was obtained based on cross-referencing the alignments using the structures of Gα and Ras.
Discussion
Our analysis suggests that GPCRs interact and activate Gα subunits through a highly conserved mechanism in which the interruption of the contacts between H1 and H5 is the key step for GDP release. In this sense, while H1 is the molecular switch for GDP release, H5 is the distal trigger that is “pulled” upon receptor binding. Given that Gα proteins belong to the same fold, the existence of evolutionarily conserved residues per se is not surprising. However, the observation that (a) the conserved residues form a network of non-covalent contacts that links the GPCR-binding site with the GDP-binding pocket and that (b) this network of contacts is consistently re-organized upon receptor binding suggest that this mechanism might constitute the common conserved set of structural changes for the allosteric release of GDP (Supplementary Note). While the conserved residue contacts are crucial for Gα activation, non-conserved positions can still be important for allosteric activation in distinct Gα proteins10. Thus, the identified residues are necessary but not sufficient for G protein activation. The variable interface residues, as well as the βγ subunits could play important roles in receptor binding specificity for individual proteins. Thus the conserved universal mechanism likely represents the “skeleton” which can be incorporated into different contexts in different Gα proteins to maintain a conserved mechanism of allosteric activation and yet permit specific binding to the receptor.
A comparison to small G proteins revealed how Gα evolved to bind GPCRs at a site that is distal to the GDP binding pocket. Emergence of short regions in H5 and H1 that can undergo structural transitions seem to have been co-opted to make a new GEF (receptor) interface and link it to the GDP binding site. In such a system, displacement of a secondary structure element (H5) upon receptor binding can transmit information by re-organizing key non-covalent contacts between conserved residues that connect different secondary structure elements. Thus a common ancestor of the GTPase fold might have provided the structural framework that can be perturbed by GPCR binding through the interruption of H5-H1 contacts. This ‘new’ allosteric site for GEFs is physically separated from the Gα effector and regulator binding interfaces, and could have provided the basis for the expansion of the GPCR family without affecting the downstream signaling factors.
Our findings suggest that in addition to evolving extensive interfaces and allosteric ‘wires’ as observed in other proteins13,33, another solution for allosteric communication is evolving short segments that undergo disorder-to-order transitions upon a trigger (e.g. receptor binding). This mechanism involves the reorganization of a network of existing contacts to induce conformational changes that affect a distal site without the requirement of directly contacting residues but linked by the same secondary structure (e.g. the interface and transmission module of H5 make distinct sets of contacts but are linked through the protein backbone; like a puppet-on-a-string). Since disordered and loop regions tolerate more sequence changes than structured regions34, an important implication is that such regions allow the separation of binding specificity from a conserved allosteric activation mechanism. Generalizing this principle, we suggest that disordered segments that can undergo structural transitions (regulated folding or unfolding) and thereby re-organize existing networks of contacts within structured regions could play an important role in other protein families and may be exploited in protein engineering applications.
Supplementary Material
Acknowledgements
We thank UF Lang, C Chothia, R Weatheritt, S Balaji, H Harbrecht, A Morgunov, and NS Latysheva for their comments on this work. This work was supported by the Medical Research Council (MC_U105185859; MMB, TF, MK, AJV and CNJR; MC_U105197215; CGT), the Swiss National Science Foundation grants 141898, 133810, 31-135754 (DBV), the MRC Centenary Award (AJV), the AFR scholarship from FNR Luxembourg (CNJR) and the Boehringer Ingelheim Fond (TF). MMB is also a Lister Institute Research Prize Fellow.
Footnotes
Author Contributions
TF collected data, wrote scripts, and performed all the analysis. CNJR and AJV helped TF with analysis. DS and DBV contributed experimental results on alanine scanning. MK prepared the CGN webserver. TF and MMB designed the project, analyzed the results and wrote the manuscript. All authors read and provided their comments on the draft. MMB supervised the project.
References
- 1.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 2.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 3.Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol. 2012;196:189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantharaman V, Abhiman S, de Souza RF, Aravind L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene. 2011;475:63–78. doi: 10.1016/j.gene.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung KY, et al. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preininger AM, Meiler J, Hamm HE. Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: a perspective. J Mol Biol. 2013;425:2288–2298. doi: 10.1016/j.jmb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 9.Westfield GH, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander NS, et al. Energetic analysis of the rhodopsin-G-protein complex links the alpha5 helix to GDP release. Nat Struct Mol Biol. 2014;21:56–63. doi: 10.1038/nsmb.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2012;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 12.Chothia C, Lesk AM. Helix movements and the reconstruction of the haem pocket during the evolution of the cytochrome c family. J Mol Biol. 1985;182:151–158. doi: 10.1016/0022-2836(85)90033-6. [DOI] [PubMed] [Google Scholar]
- 13.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 14.del Sol A, Fujihashi H, Amoros D, Nussinov R. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst Biol. 2006;2:2006 0019. doi: 10.1038/msb4100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Perica T, Teichmann SA. Evolution of protein structures and interactions from the perspective of residue contact networks. Curr Opin Struct Biol. 2013;23:954–963. doi: 10.1016/j.sbi.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Deupi X, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 20.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 21.Slessareva JE, et al. Closely related G-protein-coupled receptors use multiple and distinct domains on G-protein alpha-subunits for selective coupling. J Biol Chem. 2003;278:50530–50536. doi: 10.1074/jbc.M304417200. [DOI] [PubMed] [Google Scholar]
- 22.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 23.Dratz EA, et al. NMR structure of a receptor-bound G-protein peptide. Nature. 1993;363:276–281. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- 24.Tompa P, Davey NE, Gibson TJ, Babu MM. A million peptide motifs for the molecular biologist. Mol Cell. 2014;55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Sun D, et al. Probing Gαi1 Protein Activation at Single Amino Acid Resolution. 2014 doi: 10.1038/nsmb.3070. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mnpotra JS, et al. Structural basis of G protein-coupled receptor-Gi protein interaction: formation of the cannabinoid CB2 receptor-Gi protein complex. J Biol Chem. 2014;289:20259–20272. doi: 10.1074/jbc.M113.539916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 28.Nesbit MA, et al. Mutations affecting G-protein subunit alpha11 in hypercalcemia and hypocalcemia. N Engl J Med. 2013;368:2476–2486. doi: 10.1056/NEJMoa1300253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR. Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 30.Li D, et al. Autosomal dominant hypoparathyroidism caused by germline mutation in GNA11: phenotypic and molecular characterization. J Clin Endocrinol Metab. 2014;99:E1774–1783. doi: 10.1210/jc.2014-1029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Vega M, Burrows JF, Johnston JA. Ubiquitination: Added complexity in Ras and Rho family GTPase function. Small GTPases. 2011;2:192–201. doi: 10.4161/sgtp.2.4.16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldham WM, Hamm HE. Structural basis of function in heterotrimeric G proteins. Q Rev Biophys. 2006;39:117–166. doi: 10.1017/S0033583506004306. [DOI] [PubMed] [Google Scholar]
- 33.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, et al. alpha helix content of G protein or subunit is decreased upon activation by receptor mimetics. Journal of Biological Chemistry. 1998;273:3247–3252. doi: 10.1074/Jbc.273.6.3247. [DOI] [PubMed] [Google Scholar]
- 36.Natochin M, Moussaif M, Artemyev NO. Probing the mechanism of rhodopsin-catalyzed transducin activation. J Neurochem. 2001;77:202–210. doi: 10.1046/j.1471-4159.2001.t01-1-00221.x. [DOI] [PubMed] [Google Scholar]
- 37.Marin EP, Krishna AG, Sakmar TP. Disruption of the alpha5 helix of transducin impairs rhodopsin-catalyzed nucleotide exchange. Biochemistry. 2002;41:6988–6994. doi: 10.1021/bi025514k. [DOI] [PubMed] [Google Scholar]
- 38.Marin EP, Krishna AG, Sakmar TP. Rapid activation of transducin by mutations distant from the nucleotide-binding site: evidence for a mechanistic model of receptor-catalyzed nucleotide exchange by G proteins. J Biol Chem. 2001;276:27400–27405. doi: 10.1074/jbc.C100198200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.