Abstract
The folding of the N-terminal part of the B-domain of staphylococcal protein A (PDB ID: 1BDD, a 46-residue three-α-helix bundle) and the formin-binding protein 28 WW domain (PDB ID: 1E0L, a 37-residue three-stranded anti-parallel β protein) was studied by means of Langevin dynamics with the coarse-grained UNRES force field to assess the influence of hydrodynamic interactions on protein-folding pathways and kinetics. The unfolded, intermediate, and native-like structures were identified by cluster analysis, and multi-exponential functions were fitted to the time dependence of the fractions of native and intermediate structures, respectively, to determine bulk kinetics. It was found that introducing hydrodynamic interactions slows down both the formation of an intermediate state and the transition from the collapsed structures to the final native-like structures by creating multiple kinetic traps. Therefore, introducing hydrodynamic interactions considerably slows the folding, as opposed to the results obtained from earlier studies with the use of Gō-like models.
I. INTRODUCTION
The development of physics-based methods to study protein folding and to predict protein structures continues to be an important field of computational biology.1–9 Coarse-grained models are of great interest in the field because they enable us to extend the time- and size-scale of simulations by orders of magnitude.10–13 However, coarse-grained representation usually makes use of implicit solvent. The potential energy contribution from interactions of the protein molecules with the solvent is usually included in the effective coarse-grained potentials but, apart from this, the solvent influences folding dynamics through the friction and stochastic forces. These are usually accounted for by a Langevin-dynamics treatment, in which each site experiences the non-conservative forces from the solvent individually. However, these forces also lead to a solvent-mediated apparent drag of two objects moving through a liquid, which is known as hydrodynamic interactions (HIs).14–16 These interactions can be accounted for by introducing a non-diagonal matrix of friction coefficients, referred to as, e.g., the Rotne-Prager (RP) tensor17 and its modification, the Rotne-Prager-Yamakawa (RPY) tensor,17,18 whose off-diagonal elements depend on the distances between the objects involved.
The influence of HIs on polymer collapse19–21 was studied by modeling a polymer chain as beads connected by a finitely extendable nonlinear elastic potential, with an effective interaction potential represented by the Lennard-Jones function. This model showed that the introduction of HIs sped up polymer collapse.20,21 This effect was also observed by Chang and Yethiraj19 who found that, without HIs, the compact polymer globule could not be formed, and the system got trapped in a non-ergodic metastable state.
The role of HIs in protein folding has been discussed by Tanaka,14 who suggested that HIs could have a major effect on protein-folding pathways, as well as on the final state. HIs in proteins, modeled by using the RPY tensor, were investigated with Gō-like models and Brownian Dynamics (BD) with the use of the Ermak-McCammon algorithm.22 In that work, HIs were found to speed up folding by a factor of 2 to 3 for proteins composed of 56 to 149 residues. On the other hand, the decrease of the folding rate of the individual secondary-structure elements, the α-helix and the β-hairpin (16-residues-long), was observed.16 Similar results were obtained by Cieplak and Niewieczerzał.15 They suggested that the acceleration is due to faster initial collapse. However, those investigations concerned Gō-like protein models, in which the interaction pattern is highly simplified; the interaction potentials have minima for native long-range interactions and are repulsive-only for non-native interactions while, in reality, the character of long-range residue-residue interactions depends only on the kinds of residues involved.
HIs were also conceptualized as interactions that occur between molecules. With the use of BD simulations and the RPY tensor, Ando and Skolnick found23 that, in the crowded cell interior, the decrease of the diffusion coefficients for macromolecules, compared to infinitely dilute water solution, is most likely caused by HIs and excluded-volume effects. The influence of intermolecular HIs on other biological phenomena such as formation of lipid membranes,24 protein-protein association,25 the sliding motion of protein along DNA,26 and the diffusion processes within an idealized biological cell27 was also studied.
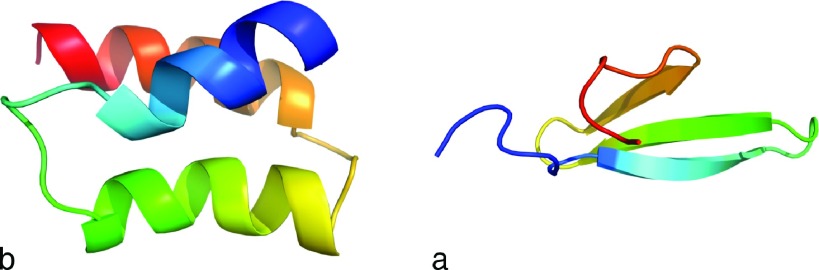
For many years, we have been developing a physics-based coarse-grained force field for the simulation of protein structure and dynamics.28–34 We implemented Langevin dynamics in UNRES35,36 to study protein folding. Despite the medium resolution of the force field resulting from the coarse-grained nature of the UNRES model, it proved successful in protein-structure prediction,37–40 in simulations of the pathways, kinetics, and free-energy landscapes of protein folding,41–44 effect of mutation on protein folding and stability,45,46 as well as in studying biologically important processes.7,47–50 In the study reported here, as an attempt to find a realistic mean-field description of the dynamic effect of the solvent, we introduced HIs in UNRES Langevin dynamics and assessed the effect of these interactions on the pathways and kinetics of folding of two small proteins, the N-terminal domain of staphylococcal protein A, which has the structure of a three-α-helix bundle (PDB ID: 1BDD; 46 residues)51 and a formin-binding protein WW domain, which is a three-stranded anti-parallel β-sheet (PDB ID: 1E0L; 37 residues).52 The experimental structures of these two proteins are shown in Figure 1. These proteins will hereafter be referred to by their PDB IDs.
FIG. 1.
Cartoon representation of model 1 of NMR experimental structures of the (a) 1BDD and (b) 1E0L.
II. METHODS
A. The UNRES force field
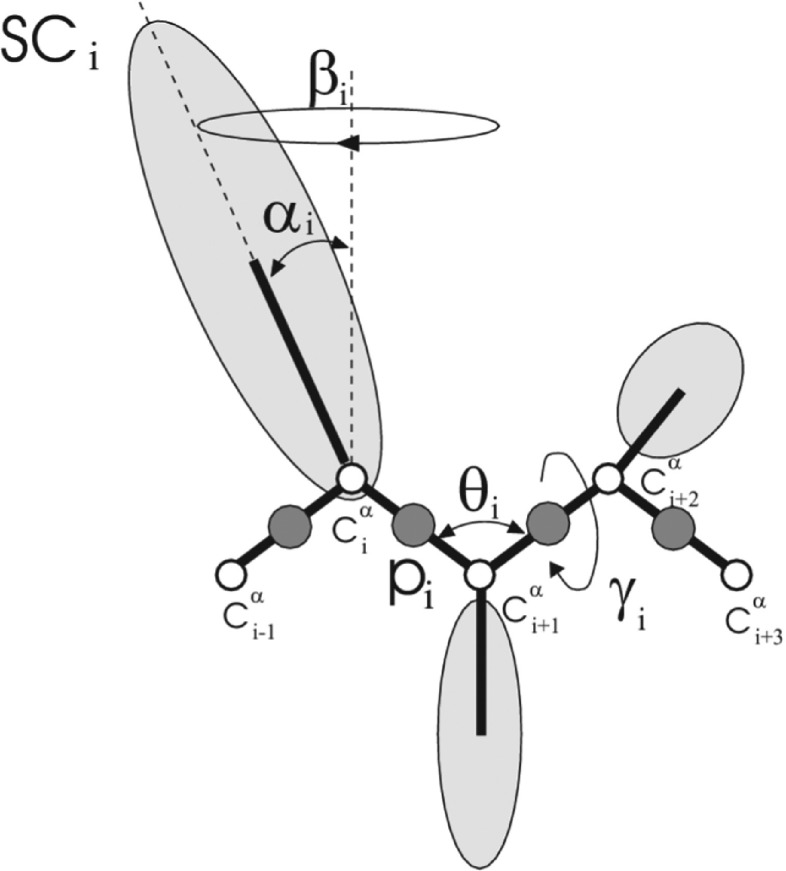
In the UNRES model,28–34 Figure 2, a polypeptide backbone is represented as a sequence of α-carbon (Cα) atoms linked by virtual bonds. A peptide group (p) is placed in the middle of each backbone virtual bond and a side chain (SC) is attached to the respective Cα atom. Only the peptide groups and the side chains are interaction sites and the Cαs serve to define the geometry.
FIG. 2.
The UNRES model of the polypeptide chain. Dark circles represent united peptide groups (p), white circles represent the Cα atoms, which serve as geometric points, and ellipsoids represent side chains (SCs). The p’s are located half-way between two consecutive Cα atoms. The virtual-bond angles θ, the virtual-bond dihedral angles γ, and the angles αSC and βSC that define the location of a side chain with respect to the backbone are also indicated.
The UNRES energy function is expressed by
| (1) |
where θi is the ith backbone virtual-bond angle, γi is the ith backbone virtual-bond-dihedral angle, αi and βi are the polar angles defining the location of the united side-chain center of residue i, and di is the length of the ith virtual bond, which is either a Cα⋯Cα virtual bond or a Cα⋯SC virtual bond, Figure 2. Each energy term, U, is multiplied by an appropriate weight, wx, and the terms corresponding to factors of order higher than 1 are additionally multiplied by the respective temperature factors, fn(T),30,53 defined by
| (2) |
where T∘ = 300 K.
In this study, two versions of the UNRES force field that differ in terms of parameterization were used; one was calibrated with the albumin-binding GA module (PDB ID: 1GAB54),30 and the second one was calibrated with the engrailed homeodomain (PDB ID: 1ENH55) and the Fbp28 WW domain (PDB ID: 1E0L52)33 for simulations of 1BDD and 1E0L protein, respectively. Although recently56 we parameterized a version of UNRES with better transferability, these two versions were already used with success in protein-folding studies.42–46
B. Langevin dynamics
The implementation of Langevin dynamics to the UNRES model has been described in earlier work.36 The Langevin equations of motion in virtual bond coordinates are given by Eq. (3). Briefly, the variables are the Cα⋯Cα and Cα⋯SC virtual-bond vectors, denoted by dC∘, dC1, dC2,…, dCn−1 (where denotes the Cartesian coordinates of the ith Cα atom and places the first Cα atom in the coordinate system) and dX2, dX2, … , dXn−1 (where ). These vectors are combined into the vectors of generalized coordinates,
| (3) |
where the inertia-matrix G is defined by
| (4) |
where M is a diagonal matrix of the masses of all the interacting sites, and H is a diagonal matrix (part of the inertia matrix corresponding to the internal stretching motions of the virtual bonds). A is the matrix that transforms the virtual-bond coordinates to Cartesian coordinates. The frictional force in virtual bond coordinates is given by
| (5) |
where, in the absence of HIs, the friction super-matrix is given by
| (6) |
where I is a 3 × 3 identity matrix and the friction coefficient for the ith site and γi is calculated from the Stokes equation as follows:
| (7) |
where Ri and Rw are the radii of the site and water, respectively, and ηw is the water viscosity. Thus, for the purpose of the calculation of the friction and stochastic forces, each site is modeled as a sphere.36
In Langevin stochastic dynamics, the temperature is kept constant (on average) by applying frictional (damping) forces along with the compensating stochastic forces to the force centers. The average stochastic forces are related to the frictional forces by the fluctuation-dissipation theorem.57 This model closely mirrors the physical dynamics of the system, i.e., the stochastic forces model the solvent molecules impinging against the protein, and the frictional forces model the Stokes drag (also due to the solvent) on each interaction site. Therefore, to obtain the information about protein dynamical motions, kinetics, and pathways, this model is an appropriate one to choose.
C. Introduction of hydrodynamic interactions into the UNRES force field
The HIs between unequal virtual-spheres representing the SCs and ps are modeled by the use of the RP tensor, and empirical averages are used for the radii of the spheres.58,59 To compute the hydrodynamic drag, both the SCs and ps will be considered as virtual spheres that experience a Stokes drag from the solvent. This approximation had been used in previous investigations of the effect of HIs on protein folding.16
As each spherical site (SC or p) moves through the solvent, it creates a wake in the solvent that will perturb all surrounding force centers. This perturbing force is referred to as the HI force. It does not alter the form of the UNRES potential since the UNRES potential contains all the conservative forces acting on the coarse-grained model of the protein while the HIs modify the non-conservative damping term in the Langevin equation. A molecular dynamics (MD) study of the motion of a sphere in a viscous Lennard-Jones fluid has shown that the results of continuum hydrodynamics work even if the sphere is comparable in size to that of the fluid molecules.60 This justifies the use of continuum fluid equations to calculate the HIs.
The effective radii for virtual spheres representing the amino-acid side chains have been taken from Levitt.61 The effective radius for the peptide group (p) was set to 5 Å, based on the van der Waals radius of the peptide group. This radius was estimated from calculations of potential of mean force surfaces for interacting peptide groups.62
The force that the solvent exerts on the ith particle is determined, in general, by the instantaneous momentum pi and position ri of all the N particles in the solvent,
| (8) |
The superscript h indicates a hydrodynamic force. In our system, the N particles are virtual spheres representing ps and the SCs. For the fluid dynamics of the protein-solvent HIs, the Navier-Stokes equation, for typical diameters and velocities of the virtual spheres, and typical solvent viscosity, can be linearized allowing one to write the hydrodynamic forces as linear functions of the velocities,63
| (9) |
where Γ is the 3 × 3 microscopic friction tensor. In a very dilute solution, there is approximately no HI between any of the virtual spheres. Therefore, Eq. (5) reduces to
| (10) |
where γ is the friction coefficient for isolated (i.e., hydrodynamically uncoupled) virtual spheres. This is the form for the friction in the unmodified UNRES-MD algorithm.36
To first order HIs, for large separations between the force centers, it can be taken into account by modification of the off diagonal elements as follows:
| (11) |
where the Dijs are the 3 × 3 diffusion tensors, put into a 3N × 3N diffusion super-matrix to relate velocity, vi, of each force center (i.e., virtual sphere), to the hydrodynamic force, , on each virtual sphere, and β = 1/kBT, with Boltzmann constant kB and temperature T.
The friction super-matrix is related to the diffusion super-matrix by a generalization of the Stokes-Einstein relation
| (12) |
From these equations the friction super-matrix is determined.
The random forces are computed by taking the square root of the friction super-matrix36 according to the fluctuation-dissipation theorem57
| (13) |
where δt is the size of the time step, and N(0, 1) is a multi-dimensional normal distribution with zero mean and unit variance.
Two specific forms for the interaction tensor will be considered. The first form assumes that the perturbing force at point j can be considered to be a point force. This assumption, along with the two approximations (Stokes’ equation and incompressibility), enables us to solve for the HI tensor analytically. The result is the Oseen-Burgers tensor64,65
| (14) |
where δ is a unit tensor of dimension 3 × 3, δr is a unit vector pointing from the point where a force makes a disturbance in the solvent to the field point where the resulting disturbance is determined, δr ⊗ δr is a dyad of δr; r is the distance between these two points, and μ is the solvent viscosity. Mathematically, since the perturbing force can be represented as a Dirac-delta function, the Oseen-Burgers tensor is the Green function for the Stokes equation for incompressible flow and fluid dynamics similar to that found in protein dynamics.66
A second form of the interaction tensor extends Eq. (14) to consider the perturbing sphere to have a finite radius, in which case the interaction tensor is the generalized RPY tensor.17 The interaction tensor for spheres of unequal radii ai, aj that are non-overlapping17,18,58 is
| (15) |
In the case of overlapping unequal virtual spheres, one modifies the RP tensor66 to
| (16) |
with an average radius aij proposed on an empirical basis.67 In this paper, a cubic average radius, , is used when the virtual-spheres overlap.
The purpose of using the RP tensor instead of the simpler Oseen-Burgers tensor is to ensure that the friction matrix remains positive-definite. This is important so that we can take the square root of this matrix to find the stochastic forces according to the fluctuation-dissipation theorem as indicated by Eq. (13). It can be proven that the RP tensor is positive-definite.66
The diagonal term of the friction super-matrix (or diffusion super-matrix) is also altered by the inclusion of HIs since the perturbing fluid disturbance propagates away from each virtual-sphere and is reflected off all the other virtual-spheres and propagates back to the original virtual-sphere that caused the disturbance. In effect, there is a self-interaction term. On the other hand, the HIs (solvent interaction) propagate infinitely fast only for incompressible fluids.68 In a real fluid, the HIs will propagate at the speed of sound. For typical distances between the virtual spheres in a folding protein, the HI disturbance will have insufficient time to reflect off of the adjacent virtual spheres and cause a self-interaction; therefore, the diagonal terms in Equation (4) or (9) were left unaltered.
D. Simulation and analysis procedure
To assess the effect of HIs on the folding of 1BDD and 1E0L, multi-trajectory canonical MD simulations with UNRES were carried out for each of them in the classical Langevin mode and with HIs included, at temperatures 300 K for 1BDD and 335 K for 1E0L, respectively, which are below the simulated transition mid-point temperatures equal to 320 K69 and 339 K,45 respectively. In summary, four series of simulations were carried out: 1BDD without HIs, 1BDD with HIs, 1E0L without HIs, and 1E0L with HIs, respectively. Each series of simulation consisted of 200 trajectories run with the 4.89 fs time step. Each trajectory was run for 1 μs UNRES time for 1BDD (both with and without HIs), 1.2 μs UNRES time for 1E0L without HIs, and 5.0 μs UNRES time for 1E0L with HIs, which corresponds to about 1, 1.2, and 5.0 ms of real time, respectively, because of averaging out secondary degrees of freedom in UNRES.35,36 All trajectories were started from an extended structure. To speed up the simulations, the viscosity of water was scaled down by a factor of 100, as in our earlier work.35,36
For each of the four series of simulations, hierarchical cluster analysis using Ward’s minimum variance method70,71 was performed for every 10th trajectory in order to find possible intermediate structures or kinetic traps that might have occurred during simulation. Then the obtained families were classified based on the Cα-RMSD of the mean structure of the cluster from the experimental structure as native (Cα-RMSD below 6 Å), intermediate (Cα-RMSD between 6 and 10 Å) or unfolded (Cα-RMSD higher than 10 Å). For each cluster, the representative conformation was defined as the conformation of the cluster which had the lowest Cα-RMSD from the mean conformation of the cluster. For each of the two proteins, the representatives of the clusters of the intermediate structures corresponding to simulations without (5 clusters for 1E0L and 2 clusters for 1BDD, respectively) and with HIs (8 clusters for 1E0L and 5 clusters for 1BDD, respectively) were then compared to find possible differences between the intermediates obtained with each of the two Langevin-dynamics regimes considered. The cross-cluster Cα-RMSD values are collected in Tables I and II of the supplementary material for 1E0L and 1BDD, respectively, and the structures are superposed in Figure SM9 of the supplementary material. As can be seen from Tables I and II of the supplementary material,72 the RMSD values between the clusters corresponding to one (without HI or with HI) Langevin-dynamics regime do not differ from those corresponding to two different regimes; according to Student’s test,73 the differences between the same- and different-regime RMSDs are insignificant. The structures of the intermediates are also similar (Figure SM9 of the supplementary material).72 It can, therefore, be concluded that the structure of the intermediate does not change remarkably after including HIs. Consequently, for each protein, the representative conformation of the intermediate structure was selected as the conformation closest to the mean conformation over all clusters of intermediate structures; this was the conformation with the lowest sum of cross-cluster Cα-RMSDs (shown in the bottom rows of Tables I and II of the supplementary material).72
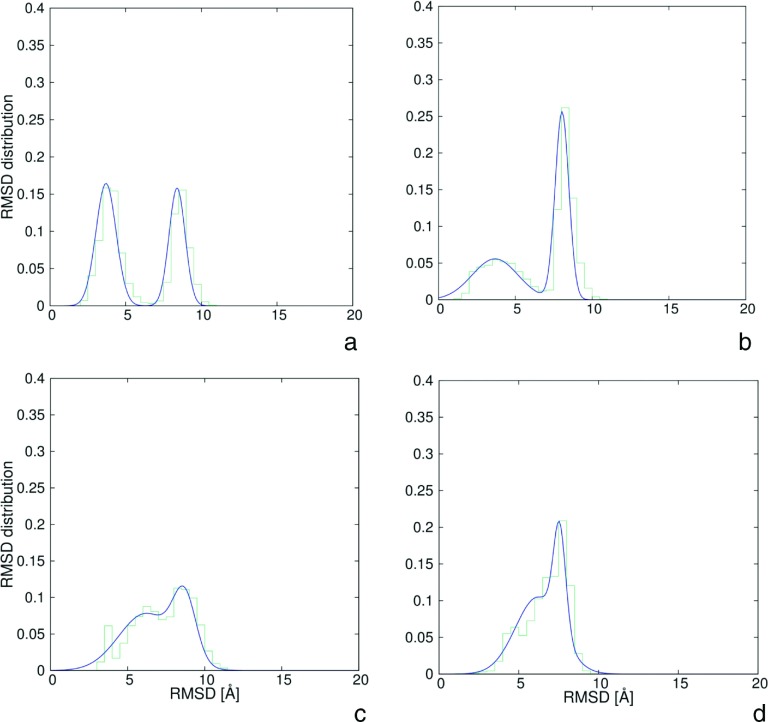
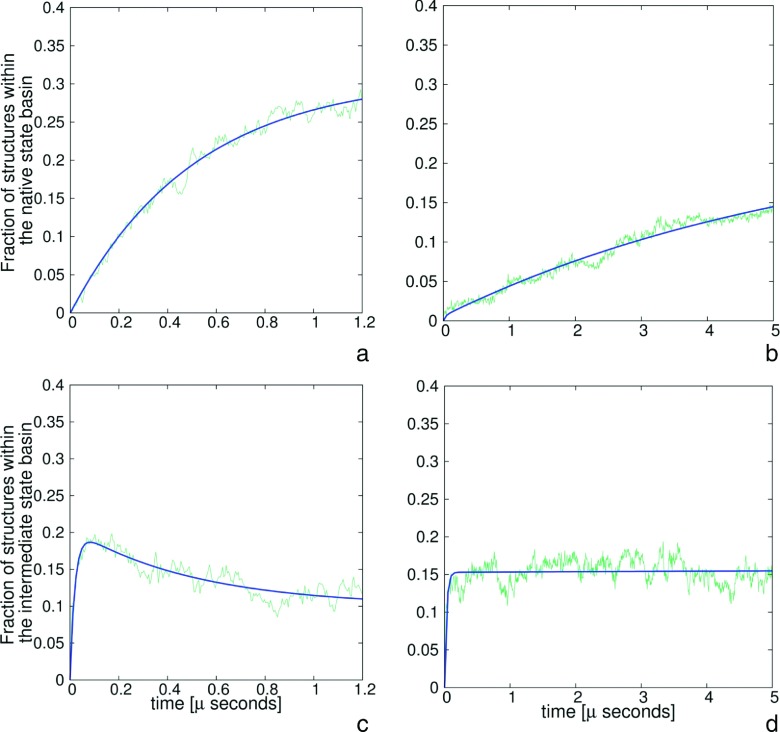
To determine the Cα-RMSD thresholds for the native and intermediate structures, the probability distributions of Cα-RMSD from the respective experimental or intermediate structures were calculated for each quarter of the simulation. For 1BDD (both with and without HIs), the time windows from 0.00 to 0.25 μs, from 0.25 to 0.50 μs, from 0.50 to 0.75 μs, and from 0.75 to 1.00 μs were set. For 1E0L without including the HIs, time windows were set from 0.0 to 0.3 μs, from 0.3 to 0.6 μs, from 0.6 to 0.9 μs, and from 0.9 to 1.2 μs, respectively, while for 1E0L with inclusion of HIs the windows were set from 0.00 to 1.25 μs, from 1.25 to 2.50 μs, from 2.50 to 3.75 μs, and from 3.75 to 5.00 μs, respectively. The Cα-RMSD bin size was set to 0.5 Å. The Cα-RMSD range of values was from 1 Å (the native or intermediate structure) to 42 Å (the extended structure). For 1BDD, the plots of the Cα-RMSD distributions from the native and from the intermediate structure, respectively, corresponding to the last time windows of simulations with inclusion of HIs (for which the intermediates last longer) are shown in Figures 3(a) and 3(b), respectively. For 1E0L, the plots are shown in Figures 3(c) and 3(d), respectively. Detailed plots of the time evolutions of the Cα-RMSD distributions are shown in the supplementary material.72 It can clearly be seen that two peaks are present. The first peak occurs at lower Cα-RMSD and corresponds to structures that are similar to the reference structure (native or intermediate), while the second peak corresponds to structures not similar to the respective reference structure. To facilitate the determination of the cutoffs of the reference-like structures, the distributions were fitted to a sum of two Gaussians (Eq. (17)), with the use of the nonlinear least-squares (NLLS) Marquardt-Levenberg74 algorithm implemented in the GNUPLOT program (http://www.gnuplot.info)
| (17) |
where fg is the fitted distribution at each time window obtained with the optimal parameters, x is the Cα-RMSD, N = 2 is the number of Gaussian terms, Ci is the multiplier for each Gaussian and represents a measure of the population of each Gaussian, and μi and σi are the mean and the standard deviation of each Gaussian, respectively.
FIG. 3.
Plots of probability distribution of Cα-RMSD values from the native and intermediate structures (green rugged lines) corresponding to the last time windows of simulations with inclusion of HIs and double-Gaussians fits to these plots (blue smooth lines) (Eq. (17)): (a) for the native structure of 1BDD, (b) for intermediate structure of 1BDD, (c) for the native structure of 1E0L, (d) for intermediate structure of 1E0L.
Based on the Cα-RMSD distributions, the cutoffs on the Cα-RMSDs from the respective experimental structures to assign a conformation to the native-structure family were set at 5.5 Å for 1BDD and 4.5 Å for 1E0L, respectively, and the cutoffs from the respective intermediate structures to assign a conformation to an intermediate-structure family were set at 5.5 Å and 5.0 Å for 1BDD and 1E0L, respectively. The cutoffs used ensured that no structure belongs to both native-like and intermediate-structure families simultaneously.
The fractions of native (fN) or intermediate (fI) structures as a functions of time were calculated for each of the proteins with and without HIs as averages over all 200 trajectories, as given by the following equations, respectively:
| (18) |
| (19) |
where nN(t) is the number of trajectories for which the native conformation appears at time t, nI(t) is the number of trajectories for which the intermediate conformation appears at time t and ntraj = 200 is the total number of trajectories.
Subsequently, biexponential functions (Eqs. (20) and (21)) were fitted, with the use of the nonlinear least-squares (NLLS) Marquardt-Levenberg74 algorithm implemented in the GNUPLOT program (http://www.gnuplot.info), to the simulated fN(t) and fI(t) curves,
| (20) |
| (21) |
where C1 and C2 can be considered as the equilibrium fractions of the native and intermediate states, respectively, and τ1 and τ2 are the respective decay times.
III. RESULTS AND DISCUSSION
A. Effect of HIs on simulated folding
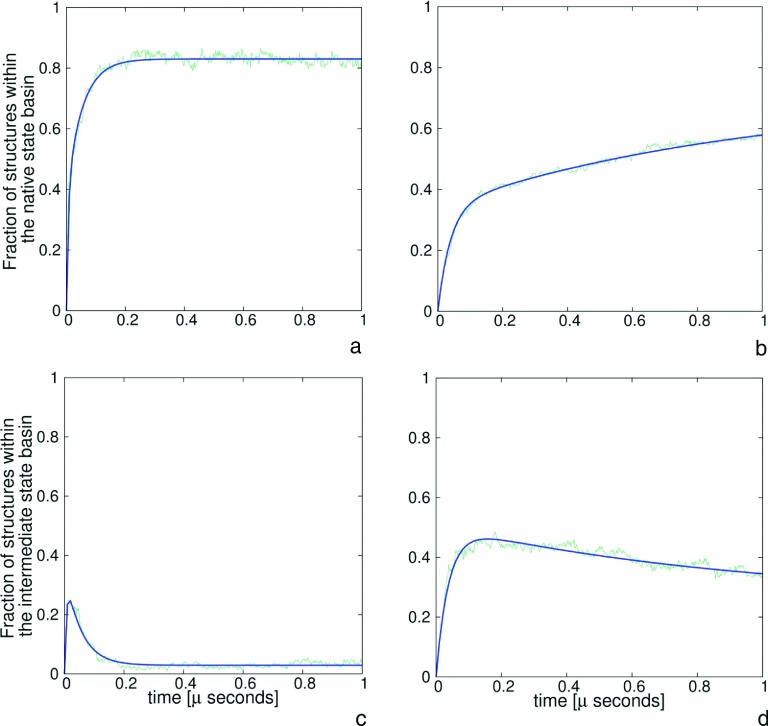
The percentages of trajectories that resulted in folded structures for all four series of simulations are collected in Table I. As can be seen, introducing HIs depletes the fraction of folded trajectories considerably. Thus, introducing HIs slows down the folding. The decrease of folding rate upon introducing HIs is apparent from Figures 4(a), 4(b), 5(a), and 5(b) in which the time evolution of the fraction of the native structure is displayed. Different results were obtained in an earlier study by Frembgen-Kesner and Elcock16 with the use of Gō-like models, in which 11 proteins with sizes comparable to those of 1BDD and 1E0L were investigated, and HIs were accounted for by using the RPY tensor. These researchers found that the overall folding rate increased 2 to 3 times. On the other hand, they also found that the rate of the formation of secondary-structure elements (α-helices and β-hairpins) decreased.
TABLE I.
The percentage of trajectories that folded to the native structure.
| Protein | Percentage of folded trajectories (%) |
|---|---|
| 1BDD without HIs | 85 |
| 1BDD with HIs | 59 |
| 1E0L without HIs | 31 |
| 1E0L with HIs | 15 |
FIG. 4.
Plots of the fractions of native ((a) and (b)) and intermediate ((c) and (d)) structures as a function of time for 1BDD protein without HIs ((a) and (c)) and with HIs ((b) and (d)). Green rugged lines present the simulated data and blue lines correspond to the fits of multi-exponential functions (Eqs. (20) and (21)).
FIG. 5.
Plots of the fractions of native ((a) and (b)) and intermediate ((c) and (d)) structures as a function of time for 1E0L without HIs ((a) and (c)) and with HIs ((b) and (d)). Green rugged lines present the simulated data and blue lines correspond to the fits of multi-exponential functions (Eqs. (20) and (21)).
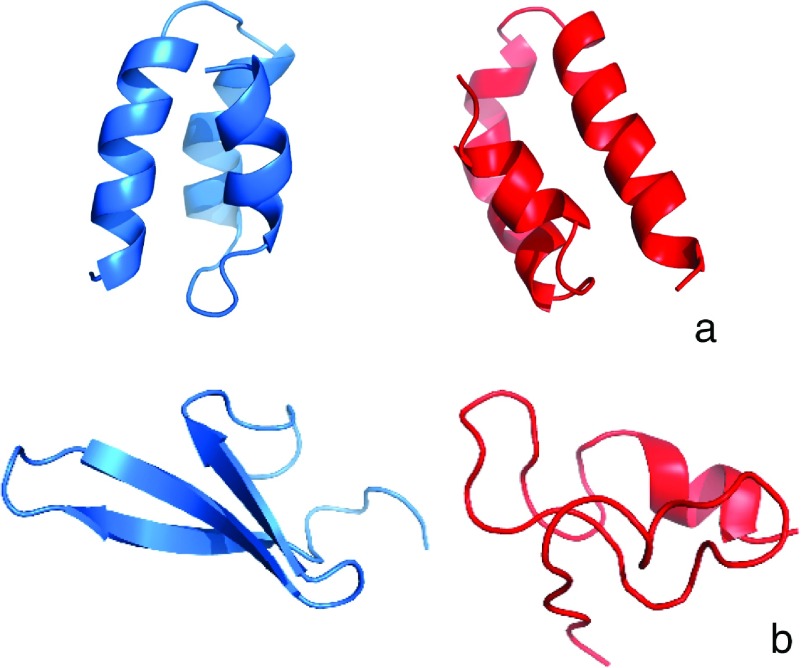
The reason for the slowing-down of the folding when HIs are included is the appearance of intermediates. These structures are shown in Figures 6(a) and 6(b) for 1BDD and 1E0L, respectively. The intermediate in the simulated folding of 1BDD is the mirror image of the native structure (Figure 6(a)); this intermediate was observed in our earlier studies of the folding of protein A with the UNRES force field41–43 and in an all-atom study of this protein with the ECEPP/3 force field.75 For 1E0L, the intermediate is a compact β-sheet-like structure but without regular peptide-group contacts and not well formed secondary structure except for a small α-helical section. This result is consistent with those of the NMR studies of the C-terminal β-hairpin of the IGG protein76–78 and the N-terminal β-hairpin of the formin-binding WW domain79 in which only hairpin-like structures without hydrogen bonds or even short α-helical sections were observed depending on temperature. Also, the C-terminus is bent in the opposite direction compared to the native structure (Figure 6(b)). Such structures were observed in our earlier studies of the free-energy landscapes of the folding of 1E0L and its mutants.44–46
FIG. 6.
Comparisons of the native (blue) and intermediate (red) structures of (a) 1BDD and (b) 1E0L proteins.
Figures 4(c) and 4(d) show the time evolution of the fractions of the intermediate for 1BDD in the plain-Langevin and Langevin-with-HI simulations, while Figures 5(c) and 5(d) show the fractions of the intermediate for 1E0L. It can be seen that, for 1BDD simulations without HIs (Figure 4(c)), the fraction of the intermediate increases sharply during the first 200 ps of simulations and then decreases quickly. Conversely, inclusion of HIs results in very slow decay of the intermediate (Figure 4(d)). For 1E0L, the intermediate decays very slowly even in plain-Langevin simulations (Figure 5(c)) and effectively reaches a steady state when the HIs are included (Figure 5(d)). This result conforms well with the predictions by Tanaka14 that HIs could have a major effect on the selection of the kinetic pathway of protein folding.
The fast formation and slow decay of intermediates after including HIs occur because of the hydrodynamic drag and create kinetic traps. This finding enables us to reconcile our results with the results obtained by Frembgen-Kesner and Elcock16 with the use of Gō-like models. With Gō-like models, attractive interactions occur only between residues that are in contact with the native structure. Therefore, hydrodynamic drag helps to form only the native contacts; non-native interactions are always repulsive and, therefore, the respective contacts will never form. Conversely, in UNRES, the interactions depend only on the kind of residues involved and, therefore, intermediates with non-native interactions are formed quite frequently.
B. Kinetics of the simulated folding
To compare the simulated folding of the two proteins studied without and with the presence of the HIs, first-order kinetic equations (Eqs. (20) and (21)) were fitted to the fractions of native and intermediate structures. All obtained coefficients are summarized in Table II. The faster growing exponential term, characterized by the smaller value of τ, can be identified with the fast, two-state folding pathway while the slow growing exponential term, characterized by the greater value of τ, with the slow, three-state folding pathway, that includes an intermediate state. Thus, the coefficient C1 identifies the fraction of the trajectories that represent the fast rate, while the coefficient C2 identifies the fraction of the slow rate.
TABLE II.
| Protein | τ1 (μs) | τ2 (μs) | C1 | m1 | C2 | m2 |
|---|---|---|---|---|---|---|
| 1BDD without HIs | 6.33 × 10−3 | 5.32 × 10−2 | 0.830 | 0.460 | 0.029 | 12.694 |
| 1BDD with HIs | 3.85 × 10−2 | 9.53 × 10−1 | 0.760 | 0.485 | 0.257 | 1.975 |
| 1E0L without HIs | 2.08 × 10−2 | 5.08 × 10−1 | 0.309 | 0.039 | 0.100 | 2.075 |
| 1E0L with HIs | 2.89 × 10−2 | 5.81 × 100 | 0.246 | 0.024 | 0.156 | 0.980 |
As can be seen from Table II, including HIs results in the increase of the decay times τ corresponding to both the fast and slow rates for both proteins; however the increase of τ2 (corresponding to the slow rates that involve intermediates) is more pronounced than that of τ1. This observation strongly suggests that HIs slow down the simulated folding, especially when intermediates are involved. This conclusion is also consistent with the facts that the C2 coefficients of Eq. (21), which can be regarded as the “equilibrium fractions” of intermediates, are increased upon inclusion of HIs, indicating that the intermediates become persistent when HIs are included (Table II).
IV. CONCLUSIONS
In this work, HIs were implemented in the Langevin-dynamics simulation procedure with the UNRES force field. The influence of HIs on protein folding was investigated with the examples of a small α-helical (1BDD) and a small β-sheet (1E0L) protein. The introduction of HIs was found to slow down the folding compared to plain-Langevin simulations. We found that the decrease of the folding rate resulted from fast formation of persistent intermediates that contain non-native residue-residue contacts, when the HIs were turned on, because of the hydrodynamic drag of interacting sites toward each other. This result seems to contradict the results of an earlier study by Frembgen-Kesner and Elcock16 carried out with the use of Gō-like models, in which it was found that HIs sped up the folding. However, in Gō-like models, the interactions between only the residues that form contacts in the native state are assigned potentials with a minimum, while all other interactions are all-repulsive. Therefore, although HIs facilitate the approach of any objects towards each other, only the native contacts will survive after they are formed. Conversely, in UNRES, the character of interactions depends on the kinds of residues involved and not on the native- or non-native character of the contact between them and, therefore, non-native contacts can survive for a longer time, creating kinetic traps. In reality, the formation of residue-residue contacts is governed by their physicochemical properties and not by the position in the sequence and, therefore, the results obtained in our study seem to be more realistic than those obtained with the use of structure-biased Gō-like models.
On the other hand, HIs are a feature of a macroscopic description of motion through a solvent, which assumes a continuous solvent governed by the Navier-Stokes equation, even though the UNRES sites (peptide groups and side chains) are not much larger in size than a water molecule. For this reason, HIs modeled in a mean-field fashion can result in too fast of an uncorrelated approach of residues, thus generating kinetic traps. The HIs can be more adequate for studies of the behavior of larger systems such as the interior of cells or lipid membrane formation investigated by Ando and Skolnick.23,24
Acknowledgments
We would like to thank Professor Jeffrey Skolnick for suggesting this topic and Dr. Magdalena Mozolewska for many fruitful discussions.
This research was supported by Grant No. BMN 538-8370-B351-14 from the University of Gdask (to A.G.L.), Grant No. DEC-2012/06/A/ST4/00376 (to A.L., A.G.L., and A.K.S.) from the National Science Center of Poland, Grant No. Mistrz 7./2013 from the Foundation for Polish Science (to A.L.), Grant No. GM-14312 from the U.S. National Institutes of Health (to H.A.S.), and by Grant No. MCB10-19767 from the U.S. National Science Foundation (to H.A.S.). This research was supported by an allocation of advanced computing resources provided by the U.S. National Science Foundation (http://www.nics.tennessee.edu/) and by the U.S. National Science Foundation through TeraGrid resources provided by the Pittsburgh Supercomputing Center. Computational resources were also provided by (a) our 588-processor Beowulf cluster at the Baker Laboratory of Chemistry and Chemical Biology, Cornell University, (b) the U.S. National Science Foundation Terascale Computing System at the Pittsburgh Supercomputer Center, (c) the Beowulf cluster at the Department of Computer Science, Cornell University, (d) the Informatics Center of the Metropolitan Academic Network (IC MAN) in Gdask, and (e) the Interdisciplinary Center of Mathematical and Computer Modeling (ICM) at the University of Warsaw.
REFERENCES
- 1.Scheraga H. A., Liwo A., Ołdziej S., Czaplewski C., Pillardy J., Ripoll D. R., Vila J. A., Kaźmierkiewicz R., Saunders J. A., Arnautova Y. A., Jagielska A., Chinchio M., and Nanias M., “The protein folding problem: Global optimization of force fields,” Front. Biosci. 9, 3296–3323 (2004). 10.2741/1482 [DOI] [PubMed] [Google Scholar]
- 2.Kryshtafovych A., Venclovas C., Fidelis K., and Moult J., “Progress over the first decade of CASP experiments,” Proteins: Struct., Funct., Bioinf. 61(S7), 225–236 (2005). 10.1002/prot.20740 [DOI] [PubMed] [Google Scholar]
- 3.Petrey D. and Honig B., “Protein structure prediction: Inroads to biology,” Mol. Cell 20, 811–819 (2005). 10.1016/j.molcel.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 4.Baker D., “Prediction and design of macromolecular structures and interactions,” Philos. Trans. R. Soc., B 361, 459–463 (2006). 10.1098/rstb.2005.1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki J. M., “Protein-structure prediction by recombination of fragments,” ChemBioChem 7, 19–27 (2006). 10.1002/cbic.200500235 [DOI] [PubMed] [Google Scholar]
- 6.Prentiss M. C., Hardin C., Eastwood M. P., Zong C. H., and Wolynes P. G., “Protein structure prediction: The next generation,” J. Chem. Theory Comput. 2, 705–716 (2006). 10.1021/ct0600058 [DOI] [PubMed] [Google Scholar]
- 7.Rojas A., Liwo A., Browne D., and Scheraga H. A., “Mechanism of fiber assembly; treatment of Aβ-peptide peptide aggregation with a coarse-grained united-residue force field,” J. Mol. Biol. 404, 537–552 (2010). 10.1016/j.jmb.2010.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dill K. A., “The protein-folding problem, 50 years on,” Science 338, 1042–1046 (2012). 10.1126/science.1219021 [DOI] [PubMed] [Google Scholar]
- 9.Piana S., Klepeis J. L., and Shaw D. E., “Assessing the accuracy of physical models used in protein-folding simulations: Quantitative evidence from long molecular dynamics simulations,” Curr. Opin. Struct. Biol. 24, 98–105 (2014). 10.1016/j.sbi.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 10.Kolinski A., Gront D., Pokarowski P., and Skolnick J., “A simple lattice model that exhibits a protein-like cooperative all-or-none folding transition,” Biopolymers 69, 399–405 (2003). 10.1002/bip.10385 [DOI] [PubMed] [Google Scholar]
- 11.Czaplewski C., Liwo A., Makowski M., Ołdziej S., and Scheraga H. A., “Coarse-grained models of proteins: Theory and applications,” in Multiscale Approaches to Protein Modeling, edited by Koliński A. (Springer, 2010), Chap. 3. [Google Scholar]
- 12.Wagenmann A. and Geyer T., “Coarse-grained simulations of protein backbone dynamics. I. Local sterics define the dihedral angles,” J. Chem. Theory Comput. 8, 4732–4745 (2012). 10.1021/ct3005529 [DOI] [PubMed] [Google Scholar]
- 13.Jamroz M., Orozco M., Kolinski A., and Kmiecik S., “Consistent view of protein fluctuations from all-atom molecular dynamics and coarse-grained dynamics with knowledge-based force-field,” J. Chem. Theory Comput. 9, 119–125 (2013). 10.1021/ct300854w [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H., “Roles of hydrodynamic interactions in structure formation of soft matter: Protein folding as an example,” J. Phys.: Condens. Matter 17, S2795–S2803 (2005). 10.1088/0953-8984/17/31/004 [DOI] [Google Scholar]
- 15.Cieplak M. and Niewieczerzał S., “Hydrodynamic interactions in protein folding,” J. Chem. Phys. 130, 124906 (2009). 10.1063/1.3050103 [DOI] [PubMed] [Google Scholar]
- 16.Frembgen-Kesner T. and Elcock A. H., “Striking effects of hydrodynamic interactions on the simulated diffusion and folding of proteins,” J. Chem. Theory Comput. 5, 242–256 (2009). 10.1021/ct800499p [DOI] [PubMed] [Google Scholar]
- 17.Rotne J. and Prager S., “Variational treatment of hydrodynamic interaction in polymers,” J. Chem. Phys. 50, 4831–4837 (1969). 10.1063/1.1670977 [DOI] [Google Scholar]
- 18.Yamakawa H., “Transport properties of polymer chains in dilute solution: Hydrodynamic interaction,” J. Chem. Phys. 53, 436–443 (1970). 10.1063/1.1673799 [DOI] [Google Scholar]
- 19.Chang R. and Yethiraj A., “Solvent effects on the collapse dynamics of polymers,” J. Chem. Phys. 114, 7688–7699 (2001). 10.1063/1.1361071 [DOI] [Google Scholar]
- 20.Kikuchi N., Gent A., and Yeomans J. M., “Polymer collapse in the presence of hydrodynamic interactions,” Eur. Phys. J. E 9, 63–66 (2002). 10.1140/epje/i2002-10056-6 [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi N., Ryder J. F., Pooley C. M., and Yeomans J. M., “Kinetics of the polymer collapse transition: The role of hydrodynamics,” Phys. Rev. E 71, 061804 (2005). 10.1103/PhysRevE.71.061804 [DOI] [PubMed] [Google Scholar]
- 22.Ermak D. L. and McCammon J. A., “Brownian dynamics with hydrodynamic interactions,” J. Chem. Phys. 69, 1352–1360 (1978). 10.1063/1.436761 [DOI] [Google Scholar]
- 23.Ando T. and Skolnick J., “Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion,” Proc. Natl. Acad. Sci. U. S. A. 107, 18457–18462 (2010). 10.1073/pnas.1011354107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ando T. and Skolnick J., “On the importance of hydrodynamic interactions in lipid membrane formation,” Biophys. J. 104, 96–105 (2013). 10.1016/j.bpj.2012.11.3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frembgen-Kesner T. and Elcock A. H., “Absolute protein-protein association rate constants from flexible, coarse-grained Brownian dynamics simulations: The role of intermolecular hydrodynamic interactions in barnase-barstar association,” Biophys. J. 99, L75–L77 (2010). 10.1016/j.bpj.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ando T. and Skolnick J., “Sliding of proteins non-specifically bound to DNA: Brownian dynamics studies with coarse-grained protein and dna models,” PLoS Comput. Biol. 10, e1003990 (2014). 10.1371/journal.pcbi.1003990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow E. and Skolnick J., “Effects of confinement on models of intracellular macromolecular dynamics,” Proc. Natl. Acad. Sci. U. S. A. 112, 14846–14851 (2015). 10.1073/pnas.1514757112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liwo A., Ołdziej S., Pincus M. R., Wawak R. J., Rackovsky S., and Scheraga H. A., “A united-residue force field for off-lattice protein-structure simulations. I. Functional forms and parameters of long-range side-chain interaction potentials from protein crystal data,” J. Comput. Chem. 18, 849–873 (1997). [DOI] [Google Scholar]
- 29.Liwo A., Czaplewski C., Pillardy J., and Scheraga H. A., “Cumulant-based expressions for the multibody terms for the correlation between local and electrostatic interactions in the united-residue force field,” J. Chem. Phys. 115, 2323–2347 (2001). 10.1063/1.1383989 [DOI] [Google Scholar]
- 30.Liwo A., Khalili M., Czaplewski C., Kalinowski S., Ołdziej S., Wachucik K., and Scheraga H. A., “Modification and optimization of the united-residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins,” J. Phys. Chem. B 111, 260–285 (2007). 10.1021/jp065380a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liwo A., Czaplewski C., Ołdziej S., Rojas A. V., Kaźmierkiewicz R., Makowski M., Murarka R. K., and Scheraga H. A., “Simulation of protein structure and dynamics with the coarse-grained UNRES force field,” in Coarse-Graining of Condensed Phase and Biomolecular Systems, edited by Voth G. (CRC Press, 2008), Chap. 8, pp. 107–122. [Google Scholar]
- 32.He Y., Xiao Y., Liwo A., and Scheraga H. A., “Exploring the parameter space of the coarse-grained unres force field by random search: Selecting a transferable medium-resolution force field,” J. Comput. Chem. 30, 2127–2135 (2009). 10.1002/jcc.21215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozłowska U., Maisuradze G. G., Liwo A., and Scheraga H. A., “Determination of side-chain-rotamer and side-chain and backbone virtual-bond-stretching potentials of mean force from AM1 energy surfaces of terminally blocked amino-acid residues, for coarse-grained simulations of protein structure and folding. II. Results, comparison with statistical potentials, and implementation in the UNRES force field,” J. Comput. Chem. 31, 1154–1167 (2010). 10.1002/jcc.21402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liwo A., Baranowski M., Czaplewski C., Gołaś E., He Y., Jagieła D., Krupa P., Maciejczyk M., Makowski M., Mozolewska M. A., Niadzvedtski A., Ołdziej S., Scheraga H. A., Sieradzan A. K., Ślusarz R., Wirecki T., Yin Y., and Zaborowski B., “A unified coarse-grained model of biological macromolecules based on mean-field multipole-multipole interactions,” J. Mol. Model. 20, 2306 (2014). 10.1007/s00894-014-2306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liwo A., Khalili M., and Scheraga H. A., “Ab initio simulations of protein-folding pathways by molecular dynamics with the united-residue model of polypeptide chains,” Proc. Natl. Acad. Sci. U. S. A. 102, 2362–2367 (2005). 10.1073/pnas.0408885102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalili M., Liwo A., Jagielska A., and Scheraga H. A., “Molecular dynamics with the united-residue model of polypeptide chains. II. Langevin and berendsen-bath dynamics and tests on model α-helical systems,” J. Phys. Chem. B 109, 13798–13810 (2005). 10.1021/jp058007w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J., Liwo A., and Scheraga H. A., “Energy-based de novo protein folding by conformational space annealing and an off-lattice united-residue force field: Application to the 10-55 fragment of staphylococcal protein a and to apo-calbindin D9K,” Proc. Natl. Acad. Sci. U. S. A. 96, 2025–2030 (1999). 10.1073/pnas.96.5.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liwo A., Lee J., Ripoll D. R., Pillardy J., and Scheraga H. A., “Protein structure prediction by global optimization of a potential energy function,” Proc. Natl. Acad. Sci. U. S. A. 96, 5482–5485 (1999). 10.1073/pnas.96.10.5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ołdziej S., Czaplewski C., Liwo A., Chinchio M., Nanias M., Vila J., Khalili M., Arnautova Y. A., Jagielska A., Makowski M., Schafroth H. D., Kaźmierkiewicz R., Ripoll D. R., Pillardy J., Saunders J., Kang Y., Gibson K., and Scheraga H. A., “Physics-based protein-structure prediction using a hierarchical protocol based on the unres force field: Assessment in two blind tests,” Proc. Natl. Acad. Sci. U. S. A. 102, 7547–7552 (2005). 10.1073/pnas.0502655102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Y., Mozolewska M. A., Krupa P., Sieradzan A. K., Wirecki T. K., Liwo A., Kachlishvili K., Rackovsky S., Jagieła D., Ślusarz R., Czaplewski C. R., Ołdziej S., and Scheraga H. A., “Lessons from application of the UNRES force field to predictions of structures of CASP10 targets,” Proc. Natl. Acad. Sci. U. S. A. 110, 14936–14941 (2013). 10.1073/pnas.1313316110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khalili M., Liwo A., and Scheraga H. A., “Kinetic studies of folding of the B-domain of staphylococcal protein a with molecular dynamics and a united-residue (UNRES) model of polypeptide chains,” J. Mol. Biol. 355, 536–547 (2006). 10.1016/j.jmb.2005.10.056 [DOI] [PubMed] [Google Scholar]
- 42.Yin Y., Maisuradze G. G., Liwo A., and Scheraga H. A., “Hidden protein folding pathways in free-energy landscapes uncovered by network analysis,” J. Chem. Theory Comput. 8, 1176–1189 (2012). 10.1021/ct200806n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kachlishvili K., Maisuradze G. G., Martin O. A., Liwo A., Vila J. A., and Scheraga H. A., “Accounting for a mirror-image conformation as a subtle effect in protein folding,” Proc. Natl. Acad. Sci. U. S. A. 111, 8458–8463 (2014). 10.1073/pnas.1407837111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou R., Maisuradze G. G., Sunol D., Todorovski T., Macias M. J., Xiao Y., Scheraga H. A., Czaplewski C., and Liwo A., “Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements,” Proc. Natl. Acad. Sci. U. S. A. 111, 18243–18248 (2014). 10.1073/pnas.1420914111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maisuradze G. G., Zhou R., Liwo A., Xiao Y., and Scheraga H. A., “Effects of mutation, truncation, and temperature on the folding kinetics of a WW domain,” J. Mol. Biol. 420, 350–365 (2012). 10.1016/j.jmb.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maisuradze G. G., Medina J., Kachlishvili K., Krupa P., Mozolewska M. A., Martin-Malpartida P., Maisuradze L., Macias M. J., and Scheraga H. A., “Preventing fibril formation of a protein by selective mutation,” Proc. Natl. Acad. Sci. U. S. A. 12, 13549–13554 (2015). 10.1073/pnas.1518298112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y., Liwo A., Weinstein H., and Scheraga H. A., “PDZ binding to the BAR domain of PICK1 is elucidated by coarse-grained molecular dynamics,” J. Mol. Biol. 405, 298–314 (2011). 10.1016/j.jmb.2010.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gołaś E. I., Maisuradze G. G., Senet P., Ołdziej S., Czaplewski C., Scheraga H. A., and Liwo A., “Simulation of the opening and closing of Hsp70 chaperones by coarse-grained molecular dynamics,” J. Chem. Theory Comput. 8, 1334–1343 (2012). 10.1021/ct200680g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipska A. G., Sieradzan A. K., Krupa P., Mozolewska M. A., D’Auria S., and Liwo A., “Studies of conformational changes of an arginine-binding protein from Thermatoga maritima in the presence and absence of ligand via molecular dynamics simulations with the coarse-grained UNRES force field,” J. Mol. Model. 3, 64 (2015). 10.1007/s00894-015-2609-1 [DOI] [PubMed] [Google Scholar]
- 50.Mozolewska M. A., Krupa P., Scheraga H. A., and Liwo A., “Molecular modeling of the binding modes of the iron-sulfur protein to the Jac1 co-chaperone from Saccharomyces cerevisiae by all-atom and coarse-grained approaches,” Proteins: Struct., Funct., Bioinf. 83, 1414–1426 (2015). 10.1002/prot.24824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouda H., Torigoe H., Saito A., Sato M., Arata Y., and Shimada I., “Three-dimensional solution structure of the B domain of staphylococcal protein A: Comparisons of the solution and crystal structures,” Biochemistry 31, 9665–9672 (1992). 10.1021/bi00155a020 [DOI] [PubMed] [Google Scholar]
- 52.Macias M. J., Gervais V., Civera C., and Oschkinat H., “Structural analysis of WW domains and design of a WW prototype,” Nat. Struct. Biol. 7, 375–379 (2000). 10.1038/75144 [DOI] [PubMed] [Google Scholar]
- 53.Shen H., Liwo A., and Scheraga H. A., “An improved functional form for the temperature scaling factors of the components of the mesoscopic UNRES force field for simulations of protein structure and dynamics,” J. Phys. Chem. B 113, 8738–8744 (2009). 10.1021/jp901788q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansson M. U., de Chateau M., Wikstrom M., Forsen S., Drakenberg T., and Bjorck L., “Solution structure of the albumin-binding GA module: A versatile bacterial protein domain,” J. Mol. Biol. 266, 859–865 (1997). 10.1006/jmbi.1996.0856 [DOI] [PubMed] [Google Scholar]
- 55.Clarke N. D., Kissinger C. R., Desjarlais J., Gilliland G. L., and Pabo C. O., “Structural studies of the engrailed homeodomain,” Protein Sci. 3, 1779–1787 (1994). 10.1002/pro.5560031018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sieradzan A. K., Krupa P., Scheraga H. A., Liwo A., and Czaplewski C., “Physics-based potentials for the coupling between backbone- and side-chain-local conformational states in the united residue (UNRES) force field for protein simulations,” J. Chem. Theory Comput. 11, 817–831 (2015). 10.1021/ct500736a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kubo R., “The fluctuation-dissipation theorem,” Rep. Prog. Phys. 29, 255–284 (1966). 10.1088/0034-4885/29/1/306 [DOI] [Google Scholar]
- 58.de le Torre J. G. and Bloomfield V. A., “Hydrodynamic properties of macromolecular complexes. I. Translation,” Biopolymers 16, 1747–1763 (1977). 10.1002/bip.1977.360160811 [DOI] [PubMed] [Google Scholar]
- 59.Larson R. G., “The rheology of dilute solutions of flexible polymers: Progress and problems,” J. Rheol. 49, 1–70 (2005). 10.1122/1.1835336 [DOI] [Google Scholar]
- 60.Vergeles M., Keblinski P., Koplik J., and Banavar J., “Stokes drag at the molecular-level,” Phys. Rev. Lett. 75, 232–235 (1995). 10.1103/PhysRevLett.75.232 [DOI] [PubMed] [Google Scholar]
- 61.Levitt M., “A simplified representation of protein conformations for rapid simulation of protein folding,” J. Mol. Biol. 104, 59–107 (1976). 10.1016/0022-2836(76)90004-8 [DOI] [PubMed] [Google Scholar]
- 62.Liwo A., Ołdziej S., Czaplewski C., Kozłowska U., and Scheraga H. A., “Parameterization of backbone-electrostatic and multibody contributions to the UNRES force field for protein-structure prediction from ab initio energy surfaces of model systems,” J. Phys. Chem. B 108, 9421–9438 (2004). 10.1021/jp030844f [DOI] [Google Scholar]
- 63.Dhont J. K. G., An Introduction to Dynamics of Colloids (Elsevier, Amsterdam, 1996). [Google Scholar]
- 64.Oseen C., Hydrodynamik (Akademisches Verlag, Leipzig, 1927), Vol. 1. [Google Scholar]
- 65.Burgers J., in Second Report on Viscosity and Plasticity (Academy of Sciences at Amsterdam, Nordemann Publishing Company, 1938), Chap. 3. [Google Scholar]
- 66.Carrasco B. and de le Torre J. G., “Hydrodynamic properties of rigid particles: Comparison of different modeling and computational procedures,” Biophys. J. 76, 3044–3057 (1999). 10.1016/S0006-3495(99)77457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zipper P. and Durchschlag H., “Calculation of hydrodynamic parameters of proteins crystallographic data using multibody approaches,” in Analytical Ultracentrifugation IV, edited byJaenicke R. and Durchschlag H., Progress in Colloid and Polymer Science (Steinkopff Darmstadt, 1997), Vol. 107, pp. 58–71. [Google Scholar]
- 68.Kröger M., Alba-Pérez A., Laso M., and Öttinger H., “Variance reduced Brownian simulation of a bead-spring chain under steady shear flow considering hydrodynamic interaction effects,” J. Chem. Phys. 113, 4767–4773 (2000). 10.1063/1.1288803 [DOI] [Google Scholar]
- 69.Krupa P., Sieradzan A. K., Rackovsky S., Baranowski M., Ołdziej S., Scheraga H. A., Liwo A., and Czaplewski C., “Improvement of the treatment of loop structures in the UNRES force field by inclusion of coupling between backbone- and side-chain-local conformational states,” J. Chem. Theory Comput. 9, 4620–4632 (2013). 10.1021/ct4004977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murtagh F., Multidimensional Clustering Algorithms (Physica-Verlag, Vienna, 1985). [Google Scholar]
- 71.Murtagh F. and Heck A., Multivariate Data Analysis (Kluwer Academic Publishers, 1987). [Google Scholar]
- 72.See supplementary material at http://dx.doi.org/10.1063/1.4948710 E-JCPSA6-144-007619for the structures of intermediates and for the probality distribution of the Cα-RMSD values for the native and intermediate structures of 1BDD and 1E0L proteins with and without the inclusion of HI for each quater of the simulation.
- 73.Nikulin M. S., “Student test,” in Encyclopedia of Mathematics, edited by Rehmann U. (Springer, 2012). [Google Scholar]
- 74.Marquardt D. W., “An algorithm for least-squares estimation of nonlinear parameters,” J. Soc. Ind. Appl. Math. 11, 431–441 (1963). 10.1137/0111030 [DOI] [Google Scholar]
- 75.Vila J. A., Ripoll D. R., and Scheraga H. A., “Atomically detailed folding simulation of the B domain of staphylococcal protein A from random structures,” Proc. Natl. Acad. Sci. U. S. A. 100, 14812–14816 (2003). 10.1073/pnas.2436463100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skwierawska A., Żmudzińska W., Ołdziej S., Liwo A., and Scheraga H. A., “Mechanism of formation of the C-terminal β-hairpin of the B3 domain of the immunoglobulin binding protein G from Streptococcus. Part II. Interplay of local backbone conformational dynamics and long-range hydrophobic interactions in hairpin formation,” Proteins: Struct., Funct., Bioinf. 76, 637–654 (2009). 10.1002/prot.22377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewandowska A., Ołdziej S., Liwo A., and Scheraga H. A., “Mechanism of formation of the C-terminal β-hairpin of the B3 domain of the immunoglobulin binding protein G from Streptococcus. Part IV. Implication for the mechanism of folding of the parent protein,” Biopolymers 93, 469–480 (2010). 10.1002/bip.21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewandowska A., Ołdziej S., Liwo A., and Scheraga H. A., “β-hairpin-forming peptides; models of early stages of protein folding,” Biophys. Chem. 151, 1–9 (2010). 10.1016/j.bpc.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makowska J., Żmudzińska W., Uber D., and Chmurzyński L., “A study of the influence of charged residues on β-hairpin formation by nuclear magnetic resonance and molecular dynamics,” Protein J. 33, 525–535 (2014). 10.1007/s10930-014-9585-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4948710 E-JCPSA6-144-007619for the structures of intermediates and for the probality distribution of the Cα-RMSD values for the native and intermediate structures of 1BDD and 1E0L proteins with and without the inclusion of HI for each quater of the simulation.