SUMMARY
Although the structure of lipoic acid and its role in bacterial metabolism were clear over 50 years ago, it is only in the past decade that the pathways of biosynthesis of this universally conserved cofactor have become understood. Unlike most cofactors, lipoic acid must be covalently bound to its cognate enzyme proteins (the 2-oxoacid dehydrogenases and the glycine cleavage system) in order to function in central metabolism. Indeed, the cofactor is assembled on its cognate proteins rather than being assembled and subsequently attached as in the typical pathway, like that of biotin attachment. The first lipoate biosynthetic pathway determined was that of Escherichia coli, which utilizes two enzymes to form the active lipoylated protein from a fatty acid biosynthetic intermediate. Recently, a more complex pathway requiring four proteins was discovered in Bacillus subtilis, which is probably an evolutionary relic. This pathway requires the H protein of the glycine cleavage system of single-carbon metabolism to form active (lipoyl) 2-oxoacid dehydrogenases. The bacterial pathways inform the lipoate pathways of eukaryotic organisms. Plants use the E. coli pathway, whereas mammals and fungi probably use the B. subtilis pathway. The lipoate metabolism enzymes (except those of sulfur insertion) are members of PFAM family PF03099 (the cofactor transferase family). Although these enzymes share some sequence similarity, they catalyze three markedly distinct enzyme reactions, making the usual assignment of function based on alignments prone to frequent mistaken annotations. This state of affairs has possibly clouded the interpretation of one of the disorders of human lipoate metabolism.
INTRODUCTION
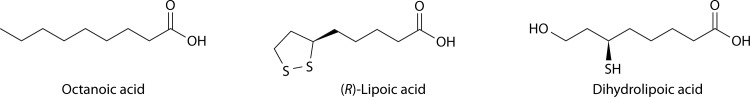
Lipoic acid is an essential cofactor that functions only when covalently bound to specific subunits of the pyruvate dehydrogenase (PDH), 2-oxoglutarate dehydrogenase (OGDH), branched-chain 2-oxoacid dehydrogenase, and acetoin dehydrogenase enzyme complexes. Lipoic acid attachment is likewise required for the glycine cleavage system (called glycine decarboxylase in plants), an enzyme complex that catalyzes reactions very different from those of the 2-oxoacid dehydrogenases. The IUPAC name for lipoic acid is (R)-5-(1,2-dithiolane-3-yl)pentanoic acid, although the cofactor is also called thioctic acid, 6,8-dithiooctanoic acid, and α-lipoic acid. The last name is a relic of the early work in which oxidized and polymerized forms of the molecule were isolated. Lipoic acid acquired its name because it is poorly soluble in water but, like a lipid, is highly soluble in organic solvents (1, 2). Indeed, lipoic acid is a derivative of the short-chain fatty acid octanoic acid (Fig. 1).
FIG 1.
Lipoic acid and related molecules. Lipoic acid is a derivative of octanoic acid in which sulfur atoms are inserted at carbon atoms 6 and 8. The two thiols form the disulfide, which is called lipoic acid (dihydrolipoic acid is the reduced form). The IUPAC names of (R)-lipoic acid and dihydrolipoic acid are (R)-5-(1,2-dithiolan-3-yl)pentanoic acid and 6,8-dimercaptooctanoic acid, respectively.
Lipoic acid was discovered as a cofactor that replaced acetate in supporting the growth of certain lactic acid bacteria (3, 4). Since acetate is volatile in the acidic media favored by these bacteria, much of it was lost upon autoclaving of the growth media, and robust growth required supplementation of the autoclaved medium with acetate. However, acetate-free extracts obtained by acid hydrolysis of liver and yeast also strongly stimulated growth. Extraction into organic solvents concentrated the stimulatory activity, and these preparations were called the acetate replacement factor (1–5). The most straightforward interpretation was that the factor somehow allowed these bacteria to make their own acetate. Another laboratory found that a different lactic acid bacterium, now called Enterococcus faecalis, grew on pyruvate only when the medium was supplemented with a similar acetate-free liver extract, indicating that acetate was synthesized by decarboxylation of pyruvate (6, 7). The two groups then joined forces and, greatly aided by large-scale processing of liver extracts by the Eli Lilly Company, isolated 30 mg of lipoic acid from about 10 tons of liver (8). Lipoic acid was thought to contain a dithiol because pyruvate metabolism in brain tissue was inhibited by trivalent organoarsenic compounds that were known to tightly bind 1,2- and 1,3-dithiols (such the trench warfare agent, Lewisite) and inhibition was reversed by the addition of a 1,2-dithiol (9). The presence of a dithiol was confirmed upon chemical synthesis of lipoate (10).
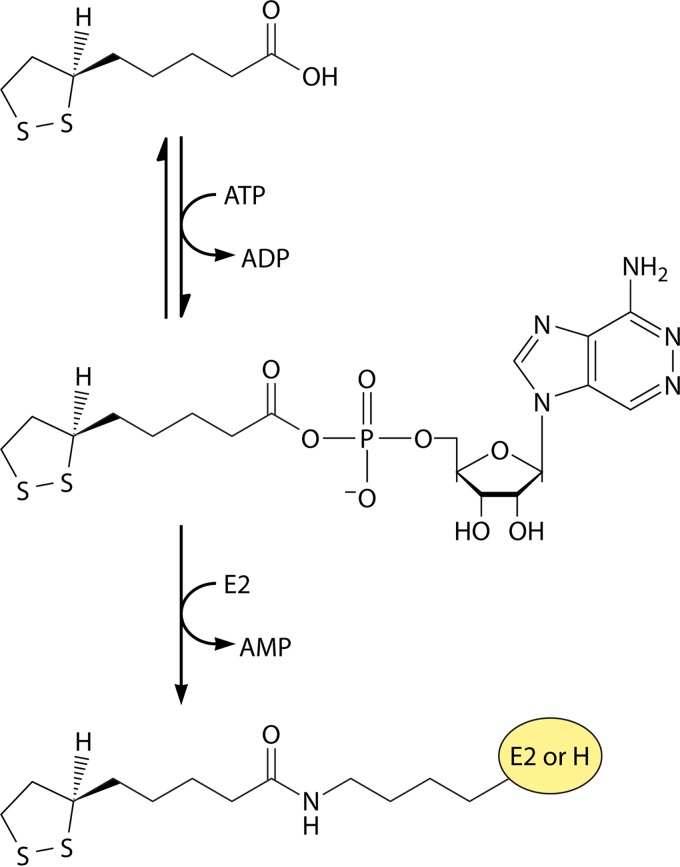
The next question was, how did lipoic acid allow these bacteria to decarboxylate pyruvate to acetate? Lipoic acid added to bacterial cultures at limiting levels could not be recovered. This finding, together with the hydrolytic step required to obtain the compound from natural sources, suggested that lipoic acid was covalently bound to enzyme proteins, and this was demonstrated using 35S-labeled lipoic acid (11). Numerous laboratories had established the other cofactor requirements for the oxidative decarboxylation of 2-oxoacids (see below). Coenzyme A (CoA), NAD, and thiamine diphosphate plus a divalent metal ion and (now) protein-bound lipoic acid were all required. A perplexing result was that the addition of all of these components to cell extracts of Enterococcus faecalis cells grown in the absence of lipoic acid failed to allow pyruvate oxidation (12, 13). However, if the extracts were supplemented with lipoic acid plus ATP and incubated prior to the addition of pyruvate, robust oxidation was observed. Hence, an enzyme activity, now called lipoate protein ligase (Fig. 2), was required to activate the inactive pyruvate oxidation system (13). Lipoate attachment was demonstrated to be through an amide linkage to a specific lysine residue located close to the N terminus of the E2 subunit (2).
FIG 2.
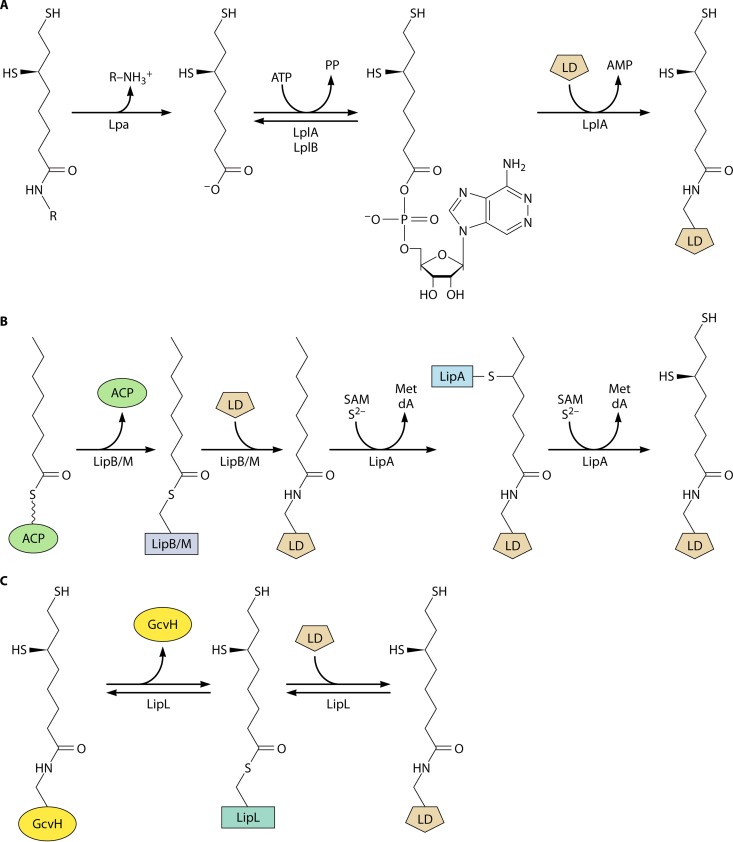
The lipoate ligase reaction. The reaction proceeds in two partial reactions: the formation of lipoyl-adenylate, followed by transfer of the lipoate moiety to the acceptor protein (an E2 or GcvH protein). The lipoyl-adenylate intermediate is tightly bound. The biochemically characterized ligases also use octanoate as a substrate. Attachment is to an LD of a 2-oxoacid dehydrogenase E2 protein or to a glycine cleavage system H protein.
THE ENZYMES THAT REQUIRE ATTACHMENT OF LIPOIC ACID
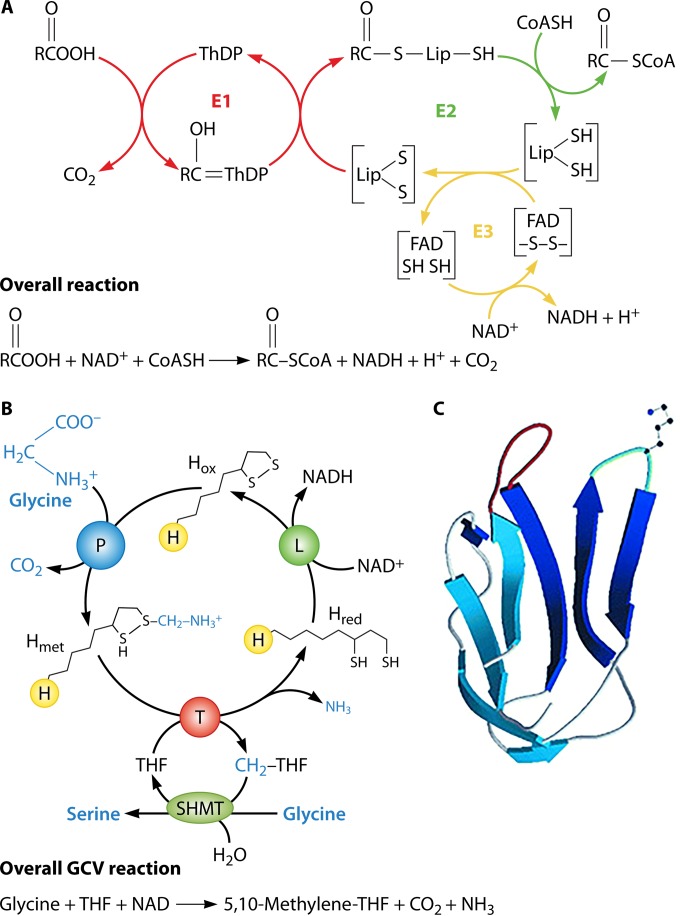
The 2-oxoacid decarboxylases have been the subject of several excellent reviews (1, 2, 14–16), and thus, only an overview will be given here. These enzyme activities reside in very large, stable protein complexes that catalyze the general reaction depicted in Fig. 3A. In the early work, the complexes had the disadvantage that methods to characterize such large complexes and their components were lacking, but this was offset by the advantage that the proteins catalyzing each of the partial reactions of the overall reaction were present in the required stoichiometry (except for the enzymes that modify the proteins with lipoic acid). The pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes are required for entry of carbon into the tricarboxylic acid (TCA) cycle and progression of carbon through the cycle, respectively, whereas the branched-chain 2-oxoacid dehydrogenase complexes are required for degradation of branched-chain amino acids in mammals, for synthesis of branched-chain fatty acids in bacteria, and for synthesis of branched-chain hydrocarbons in plants. In eukaryotes, these enzyme complexes are generally located in the mitochondria or another organelle. Some bacteria also contain a dehydrogenase complex that degrades acetoin (3-hydroxybutanone) to acetyl-CoA and acetaldehyde (acetoin is not an 2-oxoacid, so it cannot be decarboxylated). Acetoin dehydrogenase is closely related to pyruvate dehydrogenase.
FIG 3.
The lipoate-requiring enzymes. (A) The general 2-oxoacid dehydrogenase reaction. ThDP, thiamine diphosphate. (B) The glycine cleavage system (GCV, called glycine decarboxylase in plants). The synthesis of serine from N5,N10-methylene-tetrahydrofolate and a second molecule of glycine by serine hydroxymethyl transferase (SHMT) is not part of the glycine cleavage system but plays an important role in some organisms and may be a key factor in the first role of lipoic acid in metabolism. The E2 subunit of the 2-oxoacid dehydrogenases and the L protein of the glycine cleavage system are the same protein. (C) Structure of the LD of Bacillus stearothermophilus PDH E2 subunit (PDB code 1LAB).
In all 2-oxoacid dehydrogenase complexes, the core of the structure is provided by the E2 subunit, to which the E1 and E3 components are tightly bound. In the pyruvate dehydrogenase (PDH) and 2-oxoglutarate dehydrogenase (OGDH) complexes of Escherichia coli and other Gram-negative bacteria plus the OGDH and branched-chain 2-oxoacid dehydrogenase complexes of mammals (17, 18), the core consists of 24 copies of the E2 chain arranged with octahedral symmetry, whereas in the PDH complexes of mammals and Gram-positive bacteria, the core comprises 60 E2 chains arranged with icosahedral symmetry. In all 2-oxoacid dehydrogenase complexes, the E2 component has a multidomain structure comprising (from the N terminus) lipoyl domain(s), which are ca. 9-kDa domains that are the site(s) of lipoic acid attachment, a small peripheral subunit-binding domain (ca. 4 kDa), and a much larger catalytic domain (ca. 28 kDa) that houses the acyltransferase activity and polymerizes to form the inner core of the complexes. The lipoyl domains (LDs) are separated by long (25 to 30 residue) segments of polypeptide chain, characteristically rich in alanine, proline, and charged amino acids, that form flexible but extended linkers (16). The number of PDH lipoyl domains per E2 subunit varies from one to three. In the PDH complexes of Gram-negative bacteria, the number is usually two or three, whereas all of the OGDH E2o subunits described to date contain a single lipoyl domain, as is the case for the E2b chains of all branched-chain 2-oxoacid dehydrogenase complexes.
Lipoic acid attachment is also required for a reaction very different from those catalyzed by the 2-oxoacid dehydrogenases, the cleavage of glycine to N5,N10-methylene-tetrahydrofolate, CO2, and NH3 (Fig. 3B) (19, 20). This reaction of single-carbon metabolism is catalyzed by a loose complex of four proteins (more properly called a system) and is called the glycine cleavage system in mammals and bacteria and glycine decarboxylase in plants. Like the 2-oxoacid dehydrogenases, these are mitochondrial proteins in eukaryotes. The lipoylated subunit of these complexes is called GcvH or H protein and is a small protein of a size similar to the 2-oxoacid dehydrogenase lipoyl domains. However, rather than carrying an organic acid in thioester linkage as in the 2-oxoacid dehydrogenases, the distal thiol of the H protein lipoate moiety carries aminomethylene. The H protein lipoyl moiety undergoes a cycle of reductive methylamination (catalyzed by the P protein), methylamine transfer (catalyzed by the T protein), and electron transfer (catalyzed by the L protein). Hence, during catalysis, the lipoate moiety is successively methylamined, reduced, and oxidized (Fig. 3B). Glycine cleavage has been most extensively studied in plants and mammals (for excellent reviews, see references 19 and 20), where the N5,N10-methylene-tetrahydrofolate is combined with another molecule of glycine to form serine. In plants, this reaction sequence plays a key role in the photorespiratory cycle. In bacteria, the N5,N10-methylene-tetrahydrofolate can be used in various reactions, including serine synthesis, whereas in mammals, glycine cleavage functions to prevent buildup of toxic levels of glycine. Despite the very different reactions catalyzed, the 2-oxoacid dehydrogenase complexes and the glycine cleavage systems share a subunit (E3 in the dehydrogenases and L in glycine cleavage system) that is responsible for reformation of the lipoate dithiolane ring to allow another catalytic cycle (21, 22).
STRUCTURES OF LIPOYL DOMAINS
The structures of LDs (Fig. 3C) are highly conserved throughout biology, despite fairly wide variation in amino acid sequences. A demonstration of the structural conservation is that the mammalian (23) and plant (24) domains are readily lipoylated when expressed in Escherichia coli. The LD structures are directly related to the catalytic functions of the domain in substrate channeling and active-site coupling within the complexes. First, although free lipoic acid or its amide are substrates for the E2p and E3 partial reactions in vitro, the lipoylated domain is a better substrate by more than 4 orders of magnitude (25). The attachment of the lipoyl group to the conserved lysine at the tip of the protruding β-turn gives a dramatic reach to the business end. Moreover, the flexible and extended linker regions that connect the lipoyl domain(s) with the catalytic domain contribute increased mobility to the swinging arm. This was shown by deletion of the linker region in a modified single-lipoyl-domain bacterial E2, which caused an almost total loss of overall activity without substantially affecting the individual enzymatic activities (26). Moreover, E1 subunits of different bacterial species can transfer acyl groups only to their cognate E2 protein, thereby providing an accurate substrate channeling mechanism that results in reductive acylation occurring only on the lipoyl group covalently attached to the appropriate E2 subunit (27, 28). Third, although the attached lipoate was once thought to be freely rotating, recent structural data showed that the lipoyl-lysine β-turn of the domain becomes less flexible after lipoylation of the lysine residue (29). The restricted motion of the lipoyl group would facilitate effective E1 and E2 catalysis by presenting the lipoyl group in the preferred orientation to the active sites of E1. This is in agreement with a crystal structure of the E1 component of a BCDH complex (30). In this structure, the active site where thiamine diphosphate binds is located at the bottom of a long funnel-shaped tunnel, suggesting that the lipoyl group attached to the lipoyl domain must be fully extended and accurately positioned to reach the thiamine diphosphate cofactor. The domain amino acid side chains responsible for this specific positioning have been mapped to two residues that flank the lipoyl-lysine (31). Finally, the prominent surface loop connecting β-strands 1 and 2 (which lie close in space to the lipoyl-lysine) is another major determinant of the interactions of the lipoyl domain with E1 (32). Deletion of this loop results in a partially folded domain and almost completely abolishes lipoylation and reductive acylation, indicating that the loop is involved in maintaining the structural integrity of the domain, posttranslational modification, and catalytic function (29). The loop structure may be important for stabilization of the thioester bond of the acyl-lipoyl intermediate (29). The aminomethylene-modified lipoyl moiety of the pea leaf H protein also is protected by the domain structure and does not swing freely (33).
Subgenes encoding the lipoyl domains from a wide range of bacteria have been expressed in E. coli, and sufficient recombinant protein has been obtained for the domain structures to be determined by multidimensional nuclear magnetic resonance (NMR) spectroscopy and X-ray diffraction. The archetypical structure, that of the single apo lipoyl domain of the E2p chain of Bacillus (now Geobacillus) stearothermophilus (Fig. 3C) (34), is composed largely of two four-stranded β-sheets, with the N- and C-terminal residues of the domain close together in space and the lysine residue earmarked for lipoylation in an exposed position in a tight type I β-turn generated by β-strands in the other sheet. There is a well-defined hydrophobic core, the least well-defined regions being the exposed β-turn where the lipoyl-lysine resides and, most notably, the nearby large surface loop that connects the first two β-strands (Fig. 3C). Consistent with the high levels of sequence similarity between lipoyl domains of 2-oxoacid dehydrogenase multienzyme complexes, all other lipoyl domains have the same structural pattern. Given the small differences in the NMR spectra of the lipoylated and unlipoylated forms of E2p domains, the structures of holo and apo domains have been inferred to be substantially the same.
The determination of lipoyl domain structures allowed prediction of the structure of the H protein of the glycine cleavage system (35). H proteins are about 130 residues in length. Although the overall sequence identity was low (<20%), the conservation of key residues indicated that there was likely to be considerable structural similarity between the H protein of glycine cleavage systems and the lipoyl domains of 2-oxoacid dehydrogenase complexes (35). Indeed, the X-ray crystal structure of the lipoylated pea leaf H protein (24, 36) and, subsequently, those of a large number of bacterial lipoyl proteins, agree well with the prediction.
A remarkable finding is that the biotinoyl domains of biotin-dependent enzymes have structures strikingly similar to those of lipoyl domains as predicted by Brocklehurst and Perham (35). As in the case of lipoylation, biotin is ligated to the ε-amino group of a lysine reside located at the tip of a protruding β-turn. The biotinoyl domains of enzymes other than bacterial and plant plastid acetyl-CoA carboxylases most closely resemble lipoyl domains, because they lack the characteristic thumb structure of acetyl-CoA carboxylase domains (37). The structure of the biotin domain of the E. coli acetyl-CoA carboxylase biotinylated subunit (AccB) has been established by X-ray crystallography and NMR spectroscopy (38–40). The structure closely resembles those of the lipoyl domains of the E2 component of 2-oxoacid dehydrogenase complexes and of the H protein of the glycine cleavage system. The high-resolution NMR structure of a biotinoyl domain lacking the thumb structure has also been determined (41), and this structure more closely resembles the lipoyl domain. Depending on the pair of domains chosen for comparison, the root mean square deviation of biotinoyl and lipoyl domain backbone atoms can be as low as 1 Å, and hence, these proteins define a protein family (PF00364). Other work has shown that one of the proline/alanine-rich linker regions that lie between the domains of E. coli PDH can functionally replace the proline/alanine-rich linker region that lies upstream from the biotin domain of E. coli AccB (42), underlining the interrelatedness of the biotin and lipoic acid acceptor proteins. Therefore, nature has used essentially the same structural elements for enzymes that catalyze very different reactions using different covalently attached cofactors.
ATTACHMENT OF LIPOIC ACID
Some bacteria are known to be able to scavenge lipoic acid from the environment, and although this has long been thought to be the case in mammals, recent data argue strongly against this possibility (see below). In bacteria, the source of lipoic acid is the growth medium or the intestinal environment. Lipoic acid attached to dietary proteins can be set free by an intestinal amidase, whereupon bacteria compete for this “free lunch.” Some bacteria also encode an amidase called lipoamidase that is able to release lipoic acid from intact 2-oxoacid dehydrogenase complexes. Indeed, a lipoamidase or equivalent enzyme activity must be responsible for the free lipoic acid available in nature, because lipoic acid is made on its cognate proteins (see below).
Lipoate-protein ligase activity was first described by Reed and coworkers (12) in Enterococcus faecalis and E. coli, and these workers postulated that lipoate was attached to protein by a two-step ATP-dependent reaction with lipoyl-AMP as an activated intermediate (Fig. 2). Although the lipoate-protein ligase was a key reagent in the demonstration of the role of lipoic acid in the 2-oxoacid dehydrogenase reactions, the protein had not been purified to homogeneity, and thus, it was not clear whether or not a single protein catalyzed the activation and transfer steps. This was answered by analysis of the E. coli lplA ligase. The lplA gene was the first gene encoding a lipoate-protein ligase, and LplA was the first such ligase purified to homogeneity (43, 44). Mutants with mutations in lplA were isolated by two different approaches (44, 45). In the first approach, an lipA strain was mutagenized by transposon insertion and the mutagenized cells were supplemented with a mixture of succinate and acetate, which bypasses the lipoate requirement. The supplement was then switched to lipoate and selection for nongrowing cells was performed, followed by plating onto the succinate–acetate-supplemented medium. The resulting colonies were screened for strains able to grow on succinate–acetate-supplemented medium but not on lipoate-supplemented medium (44). Three classes of mutant strains could have resulted from this scheme: strains lacking the ligase (lplA), strains defective in lipoate transport, and mutants lacking the E3 subunit common to all E. coli lipoate-dependent enzymes. Surprisingly, all of the mutants isolated were lplA mutants. The lack of lipoate transport mutants suggests that there may be no lipoate transporter in E. coli (as is believed to be the case for short-chain fatty acids). Given the small size, the hydrophobicity, and the miniscule amount of the cofactor (46) required for growth (lipoate at 0.5 ng/ml gives half-maximal aerobic growth of E. coli lipoate auxotrophs), no transporter may be needed. Indeed, it has been reported that both enantiomers of 35S-lipoate were taken up by E. coli, although only (R)-lipoic acid became attached to the 2-oxoacid dehydrogenases (47). Since a transporter protein would be expected to discriminate between enantiomers, this finding argues strongly against the existence of a lipoate transporter. Mutants with mutations mapping in lplA were also isolated by a direct selection mechanism, resistance to selenolipoic acid. Selenolipoic acid is a growth-inhibitory lipoate analogue in which the sulfur atoms are replaced with selenium (45). These mutants encoded a ligase of somewhat compromised activity that was able to discriminate against the larger selenium atoms.
Purified LplA is a 38-kDa monomeric protein (43). Assays with a fully purified apoprotein acceptor have demonstrated that purified LplA plus lipoate and Mg-ATP are sufficient to reconstitute lipoylation in vitro (43, 44). Thus, it is clear that LplA catalyzes both the ATP-dependent activation of lipoate to lipoyl-AMP and the transfer of this activated lipoyl species to apoprotein, with the concomitant release of AMP (Fig. 2). This conclusion is consistent with the early findings of Reed and coworkers (12), who were unable to fractionate the E. coli activity into lipoate activation and lipoyl transferase components. LplA has been shown to be capable of utilizing octanoate and several lipoate analogs for the posttranslational modification of E2 apoproteins in vivo (44). This rather broad substrate specificity in vivo matches the similarly broad substrate specificity observed in vitro (43). E. coli LplA is a robust enzyme that has become a standard reagent in the study of lipoic acid metabolism and has become a very useful reagent for in vivo protein labeling with diverse fluorophores by substituting other residues for the residue tryptophan-37. Tryptophan-37 is located at the end of the lipoic acid binding pocket and acts as a gatekeeper residue. The mutations enlarge the volume of the active site and allow binding of substrates having different sizes and shapes than lipoic acid (48–52).
Several crystal structures of LplA and of LplA homologues have become available, including structures of E. coli LplA, LplA with bound lipoyl-5′-AMP, and LplA with octanoyl-5′-AMP in a complex with the apo form of GcvH (53). These structures define the LplA structural mechanism. Three large-scale conformational changes occur upon completion of the lipoate adenylation reaction: (i) binding of the adenylate, (ii) movement of the lipoate-binding loops to maintain the lipoyl-5′-AMP reaction intermediate, and (iii) rotation of the C-terminal domain by about 180°. These changes are prerequisites for LplA to accommodate the apoprotein for the lipoate transfer reaction. The invariant Lys133 residue (54) plays essential roles in both the lipoate adenylation and transfer steps. (Note that in a prior E. coli LplA structure, the lipoate molecule was bound in a catalytically inappropriate manner). Two lipoic acid-containing structures of an LplA homologue from the archaeon Thermoplasma acidophilum (55, 56) can be readily superimposed on the E. coli LplA structure, except that the T. acidophilum protein lacks the LplA C-terminal domain. Moreover, the addition of lipoic acid to a complex of the T. acidophilum protein with ATP gave lipoyl-AMP, thereby showing that lipoic acid was bound in a physiologically meaningful manner. Lipoyl-AMP bound in a U-shaped pocket and was well shielded from solvent, explaining the stability of its mixed anhydride linkage. The T. acidophilum LplA was reported to be inactive in catalyzing the overall LplA reaction (55), although lipoyl-AMP synthesis was demonstrated (56). Since T. acidophilum LplA lacks the C-terminal domain of E. coli LplA, this suggested that the missing domain plays a key role in transfer of the lipoyl moiety from lipoyl-AMP to the acceptor domain. Indeed, a second protein had been proposed to interact with T. acidophilum LplA and thereby allow the complete reaction (55), and this proved to be the case (57, 58). Hence, lipoate ligases can be bipartite, and a crystal structure of a bipartite enzyme has recently been reported (59). The lipoate ligase family has recently been shown to include a circularly permuted enzyme from the soil bacterium Streptomyces coelicolor. In this case, the LplA C-terminal domain is found at the N terminus (60). This domain can be moved to the C terminus with some retention of activity or can be expressed as a separate protein, where it is active but only in the presence of the other domain of the protein (60). An interesting proposed mechanism to generate circularly permutated enzymes by partial gene duplication and fold switching has been put forth to explain the structure of the E. coli BirA biotin protein ligase, an enzyme closely related to LplA (see below) (61).
The most curious of the lipoate ligase family proteins are the human (LIPT1) and bovine lipoyltransferases, which relative to LplA are defective enzymes because they cannot perform the first partial reaction, synthesis of lipoyl adenylate (Fig. 2) (62). The large domains of the mammalian lipoyltransferases and E. coli LplA share about 30% identity, whereas the small C-terminal domains show few identical residues. Consistent with the results of the in vitro assays, LIPT1 expression in E. coli failed to replace LplA, and attempts to provide LIPT1 with the small domain from a functional ligase also failed (60). During lipoate activation, the small domain of E. coli LplA moves toward the large domain to form the lipoic acid binding pocket, whereas in the second partial reaction, the small domain rotates away from the large domain by about 180° (63). In contrast, the small domain of the bovine lipoyltransferase is always extended and the β12-α6 region dynamically moves to the ligand side and forms an adenylate binding loop (64). However, in the E. coli LplA structure, the loop equivalent to the adenylate binding loop is disordered due to steric hindrance caused by the altered conformation of the small domain (63). Note that in the synthesis of lipoyl-adenylate, the substrate of LIPT1 has been ascribed to a mitochondrial medium-chain acyl-CoA synthetase known as ACSM1 (65) that contains each of the motifs of this well-studied enzyme family (66, 67). This is an extraordinarily promiscuous enzyme with very broad specifity that includes several xenobiotics (66, 67). However, ACSM1 is reported to convert both the natural (R)- and unnatural (S)-lipoate isomers to their adenylates and also uses GTP to produce lipoyl-guanylate, which is released from the enzyme to serve as a LIPT1 substrate (65). Moreover, ACSM1 uses GTP only to activate lipoate, while acyl-CoA synthesis requires ATP (65). From these considerations in particular, the ability to activate (S)-lipoate makes it very unlikely that ACSM1 acts in lipoate metabolism; stereochemistry is a hallmark of physiological function. Indeed, the nucleotide polymorphisms detected within the ACSM1 gene are associated with schizophrenia and not with central metabolism (68, 69).
THE OCTANOYL-ACP:PROTEIN N-OCTANOYLTRANSFERASES (LipB AND LipM)
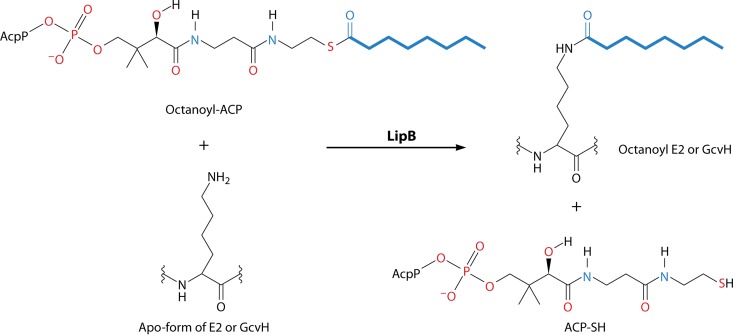
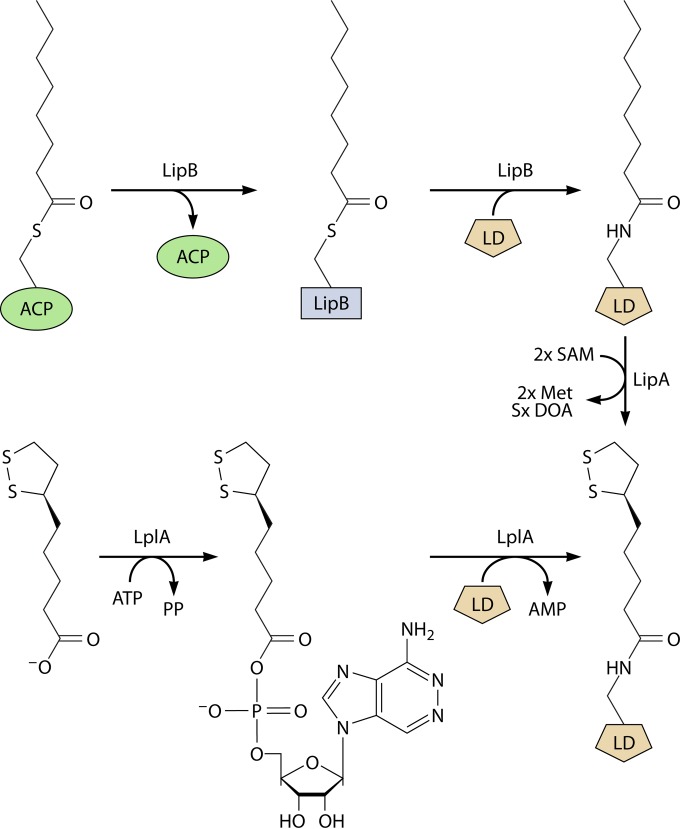
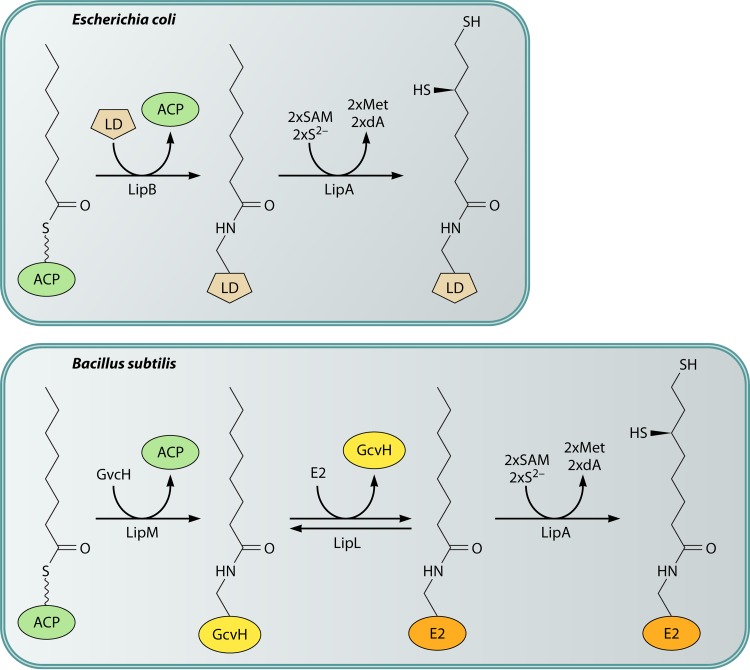
During the characterization of E. coli lplA null mutant strains, compelling evidence was found for a second protein lipoylation pathway that did not require the lplA gene product (44, 45, 70). When independently derived lplA null mutations were introduced into wild-type strains, the resulting mutant strains had active (therefore, lipoylated) 2-oxoacid dehydrogenases. This was directly confirmed by bioassays that showed that lplA null mutants contained normal levels of lipoylated proteins (70). Therefore, E. coli must have an lplA-independent lipoylation pathway. This was first attributed to a second ligase that had been missed in the biochemical analyses. However, no such second ligase could be found (43), and thus, alternative pathways were considered. The most straightforward alternative pathway was one in which the fatty acid synthetic intermediate, octanoyl-acyl carrier protein (octanoyl-acyl carrier protein [ACP]), would be converted either directly or indirectly to lipoylated proteins. That is, lipoate would be synthesized without going through an intermediate that has a free carboxyl group. The carboxyl group would be in the thioester linkage to the prosthetic group of ACP (which is essentially the nonnucleotide portion of CoA attached to serine-36 of AcpP), and this linkage would be attacked by the ε-amino group of the lipoyl domain lysine residue, resulting in the amide-linked coenzyme (as shown for octanoate in Fig. 4). Several lines of evidence demonstrated that this putative protein lipoylation pathway was dependent on the lipB gene product. The lipB gene was originally isolated as a class of E. coli lipoic acid auxotrophs (71). Biochemical evidence demonstrated that lipB was required for an octanoyl-ACP:protein N-octanoyltransferase (72, 73) present in crude extracts that was also active with lipoyl-ACP (74), although this is not its physiological reaction (Fig. 4) (75). This activity was also found in extracts of E. coli lplA null mutants and, unlike LplA, this activity did not require ATP. A temperature-sensitive lipB strain was obtained and found to encode a transferase of decreased thermal stability (73), indicating that lipB encoded the transferase rather than a protein that regulated transferase expression. LipB expression was complicated by the belated finding that a TTG codon, rather than a conserved ATG codon 22 codons downstream, was the initiation codon (73). Finally, hexahistidine-tagged LipB was purified, and the purified protein had high levels of octanoyltransferase activity (73). The untagged protein has also been purified by conventional means (76).
FIG 4.
The LipB reaction. The CoA-derived ACP prosthetic group is shown with the octanoate moiety in blue. AcpP (called AcpA in a few bacteria) is the protein moiety of ACP (ACP is defined as the species carrying the prosthetic group). B. subtilis LipM performs the same reaction via the same mechanism that utilizes a thioester-linked acyl-enzyme intermediate (Fig. 5). This is an unusual pathway in that one protein-bound prosthetic group, 4′-phosphopantheine, allows the synthesis of another, lipoic acid.
Based on these observations, two E. coli pathways for protein lipoylation were proposed (Fig. 5) (70, 74, 77), as follows. When free lipoic acid is available in the medium, E. coli incorporates and attaches extracellular lipoate (78–80) via the LplA-dependent scavenging pathway that proceeds through lipoyl-AMP (Fig. 5). When lipoate is absent from the medium, lipoyl groups must be made by de novo synthesis. An octanoyl group is first transferred from octanoyl-ACP to the apo proteins by LipB. LipA then acts on the protein-bound octanoyl groups to catalyze the sulfur insertion step (Fig. 5). This model explained why lplA null mutants had no growth defect unless the strain also carried a lipA or lipB mutation, as well as the unidirectional redundancy between LipB and LplA functions. Indeed, upon overexpression or mutation, LplA can scavenge the low levels of free cytosolic octanoate, accounting for the leakiness of lipB strains (81). The model also explained the findings of Ali, Guest, and coworkers (82, 83) that high-level production of an E. coli LD domain resulted in the octanoylated protein. It is now clear that these results were due to titration of cellular LipA levels. Strains having null mutations in both lplA and lipB contain no detectable lipoylated proteins, indicating that LplA and LipB are the only E. coli enzymes capable of modifying lipoyl domains (70).
FIG 5.
Comparison of the lipoate assembly and lipoic acid ligation pathways in E. coli. The top scheme shows the transfer of octanoate from ACP to the active site thiol of LipB, followed by the attack on the ε-amino group of the LD lysine residue on the LipB thioester to give octanoyl-LD. LipA then converts this intermediate to lipoate by the consumption of two molecules of S-adenosyl-l-methionine (SAM) and two sulfur atoms derived from the LipA auxiliary [Fe-S] cluster. Two molecules each of methionine (Met) and deoxyadenosine (DOA) are also produced. The bottom scheme shows the lipoate ligase reaction, as described in the legend to Fig. 2. Note that octanoate is also an LplA substrate.
The LipB reaction was shown to proceed via an acyl enzyme intermediate (84). The octanoyl group is transferred from the ACP prosthetic group thiol to the thiol of Cys-169. This protein-bound thioester is then attacked by the ε-amino group of the lipoyl domain lysine to give the lipoyl domain (Fig. 5). The reactivity of the cysteine residue seems to be responsible for the only LipB crystal structures currently available, those of Mycobacterium tuberculosis (85) and Thermus thermophilus (86), in that the proteins crystallized only when the loops containing the cysteine residue were immobilized. The M. tuberculosis LipB (which complemented the growth of E. coli lipB mutant strains) was crystallized in a covalent complex with a decanoic acid. Surprisingly, although the acyl chain was bound to the sulfur atom of a cysteine residue corresponding to Cys-169 of E. coli LipB, the bond was a thioether linkage to C-3 of decanoate rather than a thioester link to the carboxyl group (85). This unexpected finding seems likely to be the result of the addition of the cysteine thiol to the unsaturated bond of trans-2-decenoyl-ACP or cis-3-decenoyl-ACP, a key intermediate in E. coli unsaturated fatty acid biosynthesis. Consistent with this interpretation, no such adduct was seen upon the expression of the protein in Mycobacterium smegmatis, a bacterium that synthesizes unsaturated fatty acids by a pathway that does not involve decenoyl intermediates (85). Based on the crystal structure and mutagenesis studies, LipB is thought to function as a novel cysteine/lysine dyad acyltransferase, in which the dyad residues function as acid/base catalysts (85). Bacillus subtilis LipM catalyzes the same reaction via the same general catalytic mechanism as LipB and will be discussed below. Note that, like LipB (75), LipM isolated from cells is acylated only with octanoate (the octanoyl-enzyme intermediate), indicating that octanoyl-ACP is the sole in vivo substrate (87). The LipM and LipB octanoyltransferases can be added to the list of enzymes that have undergone convergent evolution functionally and mechanistically (88, 89). As previously stated, these enzymes are in the same family and have remarkably similar protein folding patterns (Fig. 6). Despite this, the primary sequences are markedly different. Therefore, it appears that octanoyltransferase activity has evolved from the same protein scaffold through two different paths. This highlights the multitude of possible evolutionary outcomes and explains the rarity of true convergent evolution at the sequence level (88, 89).
FIG 6.
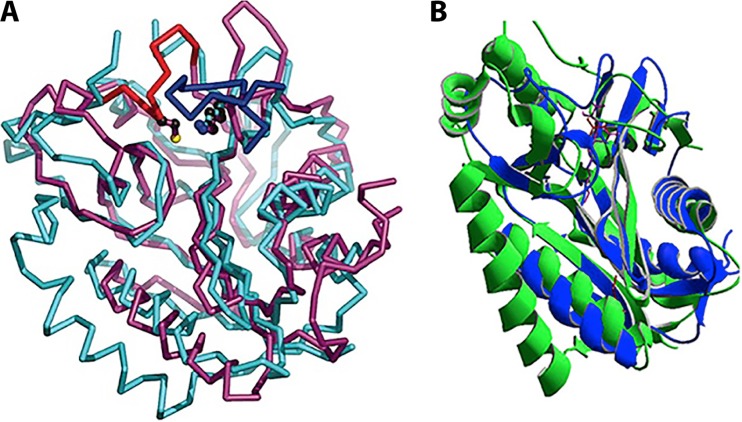
The PFAM 03099 lipoate metabolism enzymes are constructed on a common scaffold. (A) Superimposition of the crystal structures (Cα) of the Mycobacterium tuberculosis LipB octanoyltransferase (PDB code 1W66) (in magenta) with the Thermoplasma acidophilum LplA lipoate ligase (PDB code 2C8M) (in cyan). Note that the structures are circularly permuted. An interesting model to generate circularly permuted proteins has been put forth for a PFAM 03099 biotin ligase (61). (B) Superimposition of the crystal structures (ribbon diagram) of Bacillus halodurans LipL (in green, PDB code 2P5I) with M. tuberculosis LipB (in blue, PDB code 1W66). Note the strong conservation of helices and sheets in both superimpositions.
THE ENZYMES OF LIPOATE ASSEMBLY AND ATTACHMENT ARE MEMBERS OF A MECHANISTICALLY DIVERSE PROTEIN FAMILY
With the exception of the radical S-adenosyl-l-methionine (SAM) enzymes that catalyze sulfur insertion, the enzymes of lipoate assembly and attachment are members of PFAM family PF03099, the cofactor transferase family. Although the proteins of this family share overall-similar folding patterns (Fig. 6), the enzyme reactions catalyzed are mechanistically and functionally diverse (Fig. 7). This circumstance makes assigning functions based on sequence alignments perilous. A prime illustration of the failure of sequence alignments to predict function is the case of LipB, LipM, and LplA. LipBs are octanoyltransferases that transfer octanoate from octanoyl-ACP to H proteins and the E2 subunits of 2-oxoacid dehydrogenases (Fig. 4), whereas LplA is the paradigm lipoate ligase described above (Fig. 2). The E. coli LipB and LplA proteins align with 19% identity over only about 70% of the length of the shorter LipB protein. B. subtilis LipM is also an octanoyltransferase that functionally replaces LipB in E. coli (87, 90). However, LipB and LipM show only 20% identity over less than half their lengths despite the fact that the proteins have the same function and chemical mechanism. In contrast, LipM and LplA show 25% identity over essentially the full length of the shorter LipM. Hence, alignments would argue that LplA and LipM have the same function and that LipB is the functional outlier. However, LplA is the functional outlier. Moreover, the PF03099 family has yet another demonstrated lipoate metabolism activity, the LipL amidotransferase reaction, which transfers lipoate from H proteins to the dehydrogenase E2 subunits (Fig. 7) (90, 91). By sequence, LipL is the closest relative to LipB and retains the conserved structural elements, although it comes from a divergent bacterium and has a different enzyme activity. LipL attacks an amide linkage rather than the much more labile thioester linkage attacked by LipB and LipM (Fig. 7). The PF03099 family encompasses other complications. As mentioned above, a bipartite lipoate ligase is known in which the small domain is encoded by a gene adjacent to the LplA large domain homologue. However, there is no reason that this must be the case. Indeed, there are putative bipartite ligases encoded by genes located far apart in the genome, such as in Pyrococcus furiosus, and there is the circularly permuted ligase mentioned above. Moreover, PF03099 also includes ligases that attach biotin to its cognate proteins. Hence, we are faced with a protein family composed of members constructed on the same structural scaffold but having diverse enzyme activities. These attributes seem an unusually fertile ground for the construction of so-called “moonlighting” proteins having dual enzyme activities (see below).
FIG 7.
The mechanistically diverse PF03099 lipoyl metabolism enzymes. (A) The lipoate ligase reaction catalyzed by B. subtilis LplJ and the LplA proteins of E. coli and S. coelicolor. (B) The octanoyltransferase reaction catalyzed by E. coli LipB and B. subtilis LipM. (C) The amidotransferase reaction catalyzed by the LipL proteins of B. subtilis and L. monocytogenes.
BIOSYNTHESIS OF LIPOIC ACID
Although the functions of lipoic acid in its cognate multienzyme complexes have been thoroughly studied over the past 40 years, the pathways of lipoic acid biosynthesis have only recently been elucidated. Early studies established that octanoic acid (Fig. 1) is the precursor of the lipoic acid carbon chain (92, 93). Analysis of the conversion of specifically labeled forms of octanoic acid to lipoic acid by E. coli cultures showed that the two sulfur atoms are introduced with the loss of only two hydrogen atoms from the chain, one from C-6 and the other from C-8 (92). Additional metabolic feeding studies demonstrated that in E. coli, lipoic acid biosynthesis does not involve either desaturation or hydroxylation of octanoic acid but does result in inversion of stereochemistry at C-6 (92–95). Sulfur is introduced at C-8 with racemization, in agreement with the formation of an intermediate carbon radical at C-8 (92, 94, 95). 8-Thiooctanoic and 6-thiooctanoic acids were both converted to lipoic acid, although 6-thiooctanoic acid was converted only 10 to 20% as efficiently as the other positional isomer (96). (Current knowledge argues that these results were due to LplA-catalyzed ligation of the exogenously fed octanoic acid to the 2-oxoacid dehydrogenases, followed by insertion of the sulfur atoms by the LipA lipoyl synthase, as discussed below.) It should be noted that the octanoyl precursor of lipoate is synthesized by a type II fatty acid synthesis system. Such systems, in which each step of fatty acid synthesis is catalyzed by discrete individual proteins, have been studied most thoroughly in bacteria, where they provide all of the fatty acid moieties (97). In eukaryotes, mitochondria and plant plastids (such as chloroplasts) contain type II fatty acid synthesis systems encoded by nuclear genes with the proteins targeted to the organelle, usually by N-terminal sequences. Organelle fatty acid synthesis seems largely, if not entirely, devoted to providing the octanoyl-ACP for lipoic acid synthesis. The very large polyfunctional proteins that perform the bulk of fatty acid synthesis in fungi, mammals, and some plants play no role in lipoate metabolism.
Genetic studies identified a single E. coli gene responsible for the sulfur insertion steps of lipoic acid biosynthesis, first called lip (80) and now lipA, which encodes the protein called lipoyl synthase (98). E. coli strains having null mutations in lipA cannot synthesize lipoic acid even when provided with the monothiol precursors, suggesting that LipA is responsible for the formation of both C-S bonds (71, 78, 80, 99, 100). Investigations of the mechanism of LipA sulfur insertion were guided by the extensive prior work on biotin synthase (BioB), the enzyme that inserts the sulfur atom of the biotin thiophene ring. BioB makes two C-S bonds to a single sulfur atom by the replacement of two unactivated hydrogen atoms. LipA also makes two C-S bonds but to different sulfur atoms. The similarity in chemistry between the biosynthesis of the lipoate dithiolane ring and the biotin thiophene ring strongly suggested functional parallels in the mechanism of sulfur insertion in the two enzymes. Indeed, the amino acid sequences of the E. coli LipA and BioB proteins show some sequence conservation (40% sequence similarity and 17% sequence identity) (78). BioB is a charter member of what is now a large protein family, the radical SAM (S-adenosyl-l-methionine) family, whose members catalyze a range of reactions that invariably involve difficult chemistry (such as insertion of a sulfur atom into methyl or unactivated methylene groups) accessible only by radical chemistry (101–103). Extensive attempts to demonstrate in vitro sulfur insertion by BioB all failed until Ifuku and coworkers (104) succeeded in showing biotin synthesis from dethiobiotin in a cell extract. The reaction required dethiobiotin, SAM, NADPH, BioB, and an unknown protein or proteins later shown to be flavodoxin and flavodoxin reductase. This breakthrough was soon followed by the demonstration of activity in a defined system containing NADPH, flavodoxin and flavodoxin reductase as the electron transfer system, plus dethiobiotin, SAM, and a BioB preparation in a reducing environment. BioB was found to be a very labile protein that had to be purified and assayed under anaerobic conditions. In this family of proteins, the radical is generated by reductive cleavage of SAM to give a deoxyadenosyl radical plus methionine. The deoxyadenosyl radical then cleaves a C-H bond to generate a carbon radical that allows the chemistry to proceed (see below). The electron donor in the single electron reduction of SAM is a [4Fe-4S] cluster liganded by the cysteine residues of a perfectly conserved CX3CX2C motif.
BioB has both a [4Fe-4S] cluster and a [2Fe-2S] cluster. The canonical cluster for iron-sulfur binding, CX3CX2C, is also found in the LipA sequence, leading to early predictions that it was an iron-sulfur protein (78, 99). Spectral studies argued for Fe-S centers, but the data were confusing as to the number of clusters and their coordination. This probably resulted from the LipA species studied,which lacked the first 40 residues due to the selection of an incorrect translational initiation codon. Moreover, LipA was sometimes purified under aerobic conditions. The demonstration that LipA contains two distinct [4Fe-4S] clusters per polypeptide (105) has been confirmed by a recent crystal structure (106).
Direct involvement of LipA in the sulfur insertion reaction of lipoic acid biosynthesis was difficult to demonstrate, due to the lack of an in vitro assay and the assumption that free octanoic acid was the sulfur acceptor. The LipB transferase activity (Fig. 4) and the lipoate auxotrophy of lipB mutants showed that free octanoate was not a substrate and that an ACP-bound intermediate was involved (72). Miller and coworkers (98) were the first to report synthesis of lipoic acid in vitro. This was facilitated by the discovery of LplA and LipB, which allowed the development of a defined in vitro lipoic acid synthesis system plus an assay that was much more sensitive and quantitative than prior assays (98). Lipoic acid synthesis was assayed indirectly by using the apo form of the pyruvate dehydrogenase complex (apo-PDH) as a lipoyl-accepting protein, purified LipA, and either (i) purified LplA, ATP and octanoic acid as a substrate (for lipoic acid synthesis) or (ii) LipB and octanoyl-ACP as the substrate. Activation of apo-PDH by lipoylation was monitored spectrophotometrically via reduction of an NAD+ analogue. The rate of reduced pyridine dinucleotide formation was directly dependent upon the amount of lipoylated PDH. This assay showed that LipA is responsible for both sulfur insertions and that octanoyl-ACP (or a derivative of octanoyl-ACP) but not octanoic acid was a LipA substrate. Moreover, this work showed that, as suspected, the LipA reaction required iron-sulfur clusters and SAM to perform the radical chemistry (98). The advantage of the assay was its sensitivity, which was due to the amplification of each lipoyl attachment by PDH activation. The disadvantage of the assay was its indirect nature and detection of apo-PDH lipoylation rather than of the primary lipoyl protein species per se. The first model to explain these data was a conservative one in which LipA catalyzed sulfur insertion into the C-6 and C-8 positions of the octanoyl moiety of octanoyl-ACP. LipB would then transfer the lipoyl moiety of the lipoyl-ACP to the target protein lysine residues. However, all attempts to isolate the putative lipoyl-ACP intermediate were unsuccessful (98). Thus, the exact identity of the acylated substrate of the LipA reaction was unknown until the proposed conservative pathway was shown to be incorrect.
LIPOIC ACID SYNTHESIS PROCEEDS BY AN EXTRAORDINARY PATHWAY
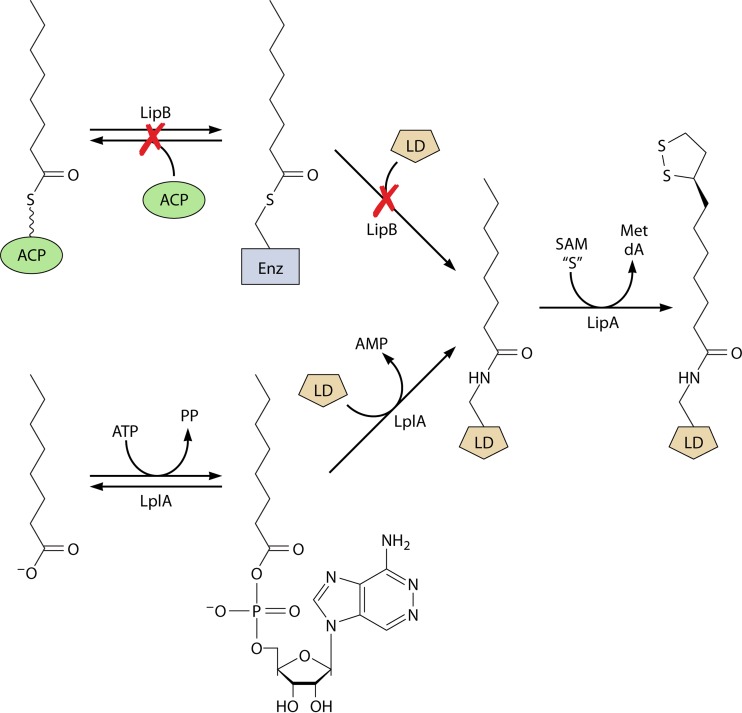
Genetic analysis provided the first evidence that the octanoyl domain rather than octanoyl-ACP was the substrate for sulfur insertion. Mutants with null mutations in lipB were found to grow well when supplemented with octanoic acid in place of lipoic acid (77). Octanoate supplementation of lipB strains required the functions of both lipoyl synthase and lipoyl ligase. Moreover, growth was specific to octanoate—fatty acids of 6, 7, 9, and 10 carbons were inactive (77). These observations argued for the existence of an LplA-dependent pathway that bypassed LipB function in the presence of octanoate. In the postulated bypass pathway (Fig. 8), LplA would attach octanoate obtained from the growth medium to the unmodified E2 domains of the 2-oxoacid dehydrogenase E2 subunits. LipA would then insert two sulfur atoms into the covalently bound octanoyl moiety and thereby convert the octanoyl-E2 domains to lipoyl-E2 domains in situ. Hence, lipoic acid would be assembled on its cognate proteins. The resulting active 2-oxoacid dehydrogenases would account for the growth on octanoate of strains that lacked LipB. This pathway was tested in vivo (77) by accumulating the octanoyl-E2 domain in the absence of the LipA lipoic acid synthase, the octanoate moiety being deuterium labeled. Lipoyl synthase (LipA) was then restored, and the conversion of the octanoyl-E2 domain to the lipoyl domain was monitored by mass spectrometry. In the cultures in which lipoic acid synthase activity was restored, a readily detectable conversion of the E2 domain modified with d15-labeled octanoate to a species of 60 additional mass units was seen (77). This was exactly the increase in mass (gain of two sulfur atoms of mass 32 and loss of two deuterium atoms of mass 2) expected for conversion of the d15-labeled octanoyl-E2 domain to the d13-labeled lipoyl domain. In a variation of the first protocol, the d15-labeled octanoic acid was removed by washing the cells and replaced with normal (nondeuterated) octanoate. This experiment gave essentially the same result: the d15-labeled octanoyl-E2 domain was converted to d13-labeled lipoyl-E2 domain (77). A modification of these experiments also showed that octanoyl-PDH accumulated in a lipA strain was converted to its active form upon restoration of LipA activity (77). The conversion of the octanoyl domain to the lipoyl domain was also observed in vitro (77), although the extent of conversion was much less than stoichiometric with LipA. These results have been confirmed using either octanoyl-H protein or synthetic octanoyl peptides as the substrate (107–109). As mentioned above, lipoyl synthase is a member of the radical SAM enzyme superfamily, which utilizes a reduced iron-sulfur cluster and SAM to generate 5′-deoxyadenosyl radicals for further radical-based chemistry. In the lipoyl synthase reaction, the role of the 5′-deoxyadenosine radicals is to remove one hydrogen atom each from the C-6 and C-8 positions of the octanoyl moiety, thereby allowing for subsequent sulfur insertion (98, 108). Consistent with this prediction, two molecules of SAM are required to synthesize one molecule of lipoyl cofactor (108). This stoichiometry is similar to that obtained in the BioB reaction and suggests that the abortive cleavage of SAM observed in these systems might result from some innate reactivity associated with this subclass of radical SAM enzymes (108).
FIG 8.
Bypass of LipB function by LplA-catalyzed attachment of exogenous octanoate to lipoyl domains (LDs) in E. coli.
As in the case of biotin, the source of the sulfur atoms of lipoic acid was thought to be an [Fe-S] cluster distinct from the SAM radical [4Fe-4S] cluster. In the first successful in vitro lipoyl synthesis assays, lipoylated protein was formed in the absence of exogenous sulfur-containing compounds (98, 108). This suggested that, like BioB, the protein itself has some mobilizable sulfur atoms that originate from an Fe-S cluster, a persulfide, or some other species. One of two LipA [4Fe-4S] clusters is coordinated by the radical SAM CXXXCXXC motif that functions in 5′-dA radical generation. The second cluster is coordinated by a CXXXXCXXXXXC motif plus a serine residue that is found only in lipoyl synthases (106). This second cluster (now generally called the auxiliary cluster) was suggested to be the source of the sulfur atoms (as sulfide). This has been addressed (analogous to BioB) by preparation of LipA from E. coli cells grown on an isotopically labeled (34S) sulfur source (107). As expected, the lipoic acid formed using this enzyme in vitro was isotopically labeled with 34S. Moreover, when the reactions were performed with equimolar amounts of 32S-labeled LipA and 34S-labeled LipA, the lipoic acid molecules formed contained either two 32S atoms or two 34S atoms (107). Hence, both sulfur atoms are donated by the same polypeptide, which precludes the possibility that monothiolated species are released and the second sulfur atom is inserted following rebinding to LipA. If release and rebinding had occurred, half of the lipoic acid formed would contain one atom each of 32S and 34S. Such mechanistic experiments were facilitated by the use of peptides containing an octanoyl lysine residue as the sulfur acceptor. An octanoylated tetrapeptide suffices in the Sulfolobus solfataricus LipA assay (110). This system showed that LipA catalyzes lipoate biosynthesis in a stepwise manner, with sulfur first being inserted at C-6 of the octanoyl chain (109). Moreover, this intermediate remained tightly bound to LipA, consistent with the finding that both sulfur atoms are derived from the same LipA polypeptide. Indeed, biochemical evidence for a covalent cross-link between a sulfur atom of the LipA auxiliary cluster and C-6 of a protein-bound octanoate moiety provides strong support for the model (111). The original model (98), in which reductive cleavage of a SAM molecule is used to abstract a hydrogen atom from C-6 to give a carbon radical which attacks an auxiliary cluster sulfur atom, has stood the test of time (109). Sulfur insertion at C-8 involves cleavage of the second SAM molecule to generate a carbon radical at C-8 that attacks a sulfur atom of the partially degraded auxiliary cluster (111).
Very recently, the first crystal structure of a lipoyl synthase, that of the thermophilic cyanobacterium Thermosynechococcus elongates, was reported by Harmer and coworkers (106). The structure shows the expected two [4Fe-4S] clusters per monomer (Fig. 9). The protein topology varies somewhat from the common partial TIM barrel (β6α6 rather than β8α8) of radical SAM enzymes in that an additional β-sheet strand is present. The two [4Fe-4S] clusters are located at opposite ends inside the barrel. The cysteine residues that bind the SAM reduction cluster (the classical radical SAM cluster) are typical of the enzyme family, whereas the cysteine residues that bind the auxiliary cluster lie on an N-terminal extension. The surprise is that the auxiliary cluster is also liganded by a serine residue located close to the carboxyl terminus. The authors report that the replacement of this serine residue with cysteine or alanine results in complete loss of the ability to synthesize lipoyl peptide. Moreover, the mutant proteins showed significantly less uncoupled SAM turnover than the wild-type protein (106). Since uncoupled SAM turnover is the function of the radical SAM [4Fe-4S] cluster, it seems that the two clusters must somehow communicate. The serine residue is conserved in all LipA proteins of demonstrated activity, consistent with the mutagenesis results. The authors propose that the deprotonated serine side chain might be involved in evicting the partially ligated iron atom produced in the first carbon radical attack on the auxiliary cluster, and this would be consistent with spectroscopic data indicating that an iron ion is lost from the auxiliary cluster during synthesis of the 6-thiooctanoyl intermediate (111). Eviction of the iron atom would allow C-8 of the octanoyl moiety access to a sulfide ion for the second sulfur insertion (106). Recently, the C-6 substrate radical has been trapped and characterized by electron paramagnetic resonance methods, which demonstrated that the acyl chain and auxiliary cluster are sufficiently close for the reaction to proceed (112).
FIG 9.
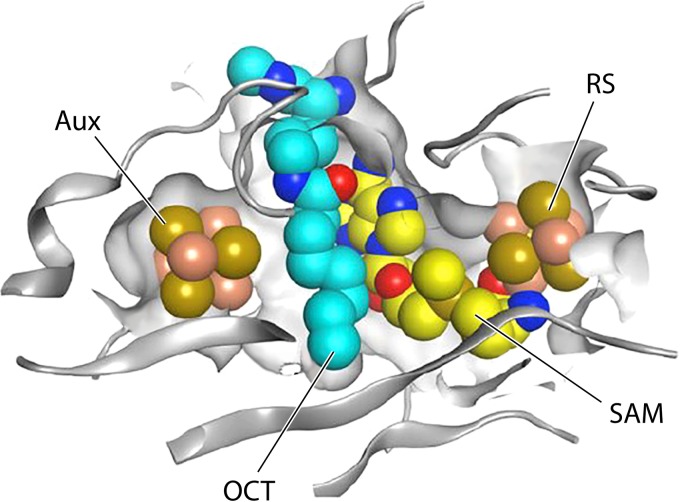
The Thermosynechococcus elongates LipA structure (106). The auxiliary cluster (Aux) is to the left, and the radical SAM (RS) cluster is to the right. The SAM was modeled in based on the binding of the SAM decomposition product, methylthioadenosine, and the complex of another radical SAM enzyme with SAM, whereas the octanoate moiety was docked into a channel between the SAM molecule and the auxiliary cluster (106). The docking is supported by the demonstration that C-6 of octanoate is sufficiently close to the auxiliary cluster for sulfide extraction to occur (112). (Reprinted from reference 106 with permission.)
A remaining question is whether LipA is an enzyme. Enzymes are catalytic by definition, and in vitro, the best LipA preparations synthesize less than one equivalent of lipoate per molecule of LipA protein (106, 111). This raises the possibility (long debated for BioB) that a protein molecule is sacrificed for each molecule of product formed (113). If no repair of the donor auxiliary cluster occurs, LipA would be a reactant or substrate rather than an enzyme. However, BioB has been shown to be catalytic in vivo (114), and the first instance of BioB undergoing multiple turnovers in vitro has been reported (115). New data eliminate the possibility of enzyme self-sacrifice. The recently developed ribosome profiling technique (116) gives the first quantitation of the cellular contents of BioB and LipA in E. coli and their rates of synthesis (these proteins are of such low abundance that neither has been detected by mass spectroscopy-based proteomics). The profiling data indicate that both radical SAM proteins are catalytic in vivo. For E. coli cells grown at a steady state in a minimal medium that requires the function of the lipoate-dependent dehydrogenases, 1,100 LipA molecules must modify 25,000 2-oxoacid dehydrogenase molecules plus 4,000 molecules of GcvH, whereas 900 BioB molecules must modify 5,600 molecules of the biotinylated protein (AccB) required for fatty acid synthesis. Since the rates of synthesis of LipA and BioB are not exceptional (116), it seems clear that the auxiliary clusters of LipA and BioB must be repaired following their catalytic cycles. However, it is unknown how repair is accomplished. E. coli has two pathways for building [Fe-S] clusters. Each pathway permits growth in a chemically defined minimal medium in the absence of the other, and no defects in the synthesis of either lipoate or biotin are seen in strains lacking either function. Mutants lacking both pathways are nonviable, thus precluding in vivo testing of the possibility that an unknown process is responsible for repair of the LipA and BioB auxiliary clusters.
INVOLVEMENT OF THE GLYCINE CLEAVAGE H PROTEIN IN 2-OXOACID DEHYDROGENASE LIPOYLATION
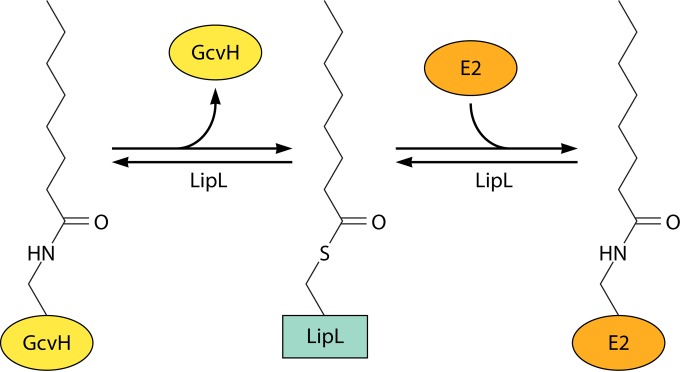
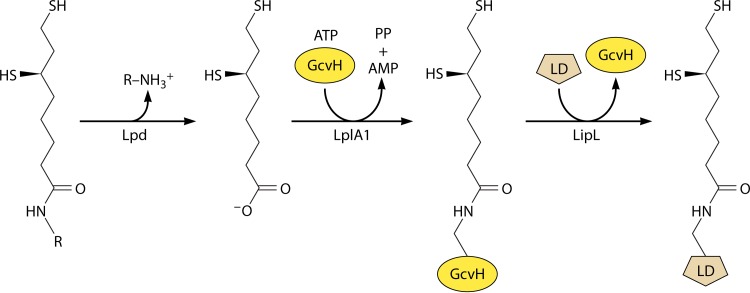
In retrospect, it was fortunate that the lipoate synthetic pathway was first elucidated in E. coli, because the pathway is straightforward and its study provided key insights and reagents. The E. coli lipoyl synthesis pathway requires only two enzymes, the LipB octanoyltransferase and the LipA radical SAM sulfur insertion enzyme (Fig. 5). Additional proteins are required for lipoyl synthesis in other organisms. The first of these more complex lipoyl synthesis pathways was found in Bacillus subtilis, a Gram-positive bacterium that is evolutionarily far removed from the Gram-negative E. coli. B. subtilis encodes a LipA that could functionally replace E. coli LipA, and disruption of the gene resulted in lipoate (or the products of the 2-oxoacid dehydrogenases) being required for growth (117). Although lipoate supplementation should require lipoate ligase activity, it was unclear why this bacterium encoded three putative lipoate ligases and no homologue of the LipB octanoyltransferase (87). This puzzle was approached by screening a plasmid library of B. subtilis genome fragments for the ability to rescue the growth of an E. coli lipB null mutant strain. The screen gave a gene called lipM that was one of the putative lipoate ligase-encoding genes. Biochemical assays showed that the LipM protein had no ligase activity and instead catalyzed octanoyl transfer from octanoyl-ACP (87). Another of the putative ligase genes (now called LplJ) was shown to encode a protein that functionally replaced LplA in E. coli and had only ligase activity (118). However, the protein (called LipL) encoded by the third putative lipoate ligase gene had no octanoyltransferase or ligase activity. LipL was clearly a lipoate synthetic protein, because disruption of the encoding gene resulted in a growth requirement for lipoate. A confounding in vitro finding was that the LipM octanoyltransferase failed to attach octanoate to a domain derived from the B. subtilis pyruvate dehydrogenase E2 subunit. The enzyme was active with an E. coli pyruvate dehydrogenase domain (90). Moreover, the B. subtilis E2 domain was a good acceptor for octanoyl transfer catalyzed by either the E. coli LipB octanoyltransferase or E. coli LplA ligase. Therefore, the B. subtilis proteins were properly folded but failed to interact with one another. Attempts to make a larger functional B. subtilis pyruvate dehydrogenase E2 acceptor domain failed, suggesting that perhaps the E2 subunit had to be assembled into a complex with E1 and/or E3 or that an intermediate protein was required. This led to testing of the B. subtilis glycine cleavage H protein (GcvH) because (as noted above) it is essentially a lipoyl domain and is not part of a stable complex. B. subtilis GcvH turned out to readily accept octanoate either from octanoyl-ACP or from octanoic acid plus ATP in reactions catalyzed by the B. subtilis LipM octanoyltransferase or LplJ ligase, respectively (90). Was GcvH the putative missing intermediate? If so, did another enzyme transfer octanoate (or lipoate) from GcvH to the 2-oxoacid dehydrogenases? A good candidate for this additional enzyme was LipL, the protein encoded by the last of the putative lipoate ligase-encoding genes. LipL was purified and added to a reaction mixture containing the LipM octanoyltransferase, octanoyl-ACP, GcvH, and the B. subtilis pyruvate dehydrogenase domain. The result was octanoyl transfer to the pyruvate dehydrogenase domain, which required both GcvH and LipL (90). When octanoyl-GcvH was incubated with LipL, the octanoate moiety was transferred to a LipL thiol group (Fig. 10). This acyl enzyme intermediate was purified and shown to transfer octanoate to the pyruvate dehydrogenase domain (90). Hence, LipL is an amidotransferase. The LipM thiol attacks the amide bond linking octanoate to GcvH and then relays the octanoate to the 2-oxoacid dehydrogenases.
FIG 10.
The LipL amidotransferase reaction. The active-site thiol of LipL attacks the octanoyl (or lipoyl)-GcvH amide linkage to form an acyl enzyme intermediate, which is then attacked by the ε-amino group of the LD lysine residue. Note that the reactions are reversible, raising the possibility that lipoate could be shuttled among proteins, depending on metabolic conditions.
Much of the B. subtilis lipoate synthesis pathway was based on in vitro biochemistry, and therefore, the pathway required validation in vivo. The predicted properties of mutant strains lacking each of the proteins were tested. First, B. subtilis strains lacking the LplJ ligase should show a growth defect only in strains that also lack LipA. This was the case (118). Second, strains lacking the LipM octanoyltransferase or the LipL amidotransferase should require lipoic acid for growth in the absence of the products of the 2-oxoacid dehydrogenases. These predictions were also confirmed (90, 118). Finally, and most importantly, a strain lacking GcvH, an essential subunit of an enzyme of single-carbon metabolism, should be unable to make lipoylated proteins and thus would require lipoic acid (or the 2-oxoacid dehydrogenase products) for growth (as do the LipM and LipL mutant strains). This was the case (90) and was confirmed by Western blotting of cell extracts from a strain lacking B. subtilis GcvH using an anti-lipoyl protein antibody (Fig. 11). When the strain was grown on medium lacking lipoate but containing the products of 2-oxoacid dehydrogenases, no lipoylated proteins were detected, whereas the addition of lipoic acid to the medium resulted in readily detectable 2-oxoacid dehydrogenase E2 proteins due to lipoate attachment catalyzed by the LplJ lipoate ligase (Fig. 11). Therefore, GcvH has an obligate role in lipoylation of key 2-oxoacid dehydrogenases, as well as its role in glycine cleavage. A comparison of the B. subtilis and E. coli lipoyl assembly pathways is given in Fig. 12. Note that the overall B. subtilis pathway violates the biochemical precept that such transfers proceed by the use of an activated (high-energy) intermediate to synthesize a less activated (low-energy) product. The early part of the pathway obeys the precept, because the thioesters of octanoyl-ACP and the octanoyl-enzyme intermediate formed in the LipB reaction are activated electrophiles. However, the amide bond of octanoyl-GcvH is a low-energy linkage, and thus, attack on that linkage by the LipL thiol to make a thioester is energetically uphill. However, this brand of heresy is also used by the sortase amidotransferases that attach proteins to the cell surfaces of some Gram-positive bacteria (119) and the multisubunit transamidase complexes that attach eukaryotic glycosylphosphatidylinositol anchors to proteins (120).
FIG 11.
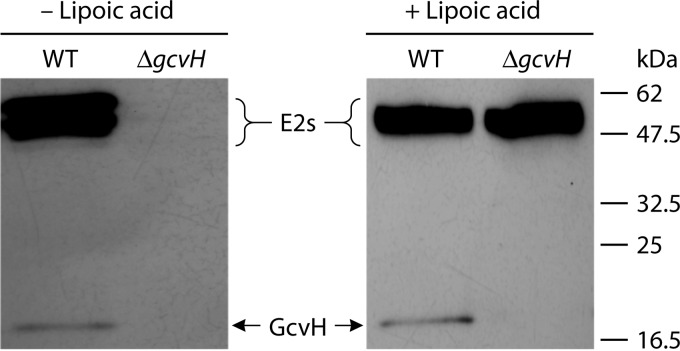
GcvH is required for 2-oxoacid dehydrogenase lipoylation. A strain of B. subtilis lacking the gcvH gene (ΔgcvH) failed to accumulate proteins that react with anti-lipoyl protein antibody when grown without lipoic acid (−LA). In contrast, when cells grown in the presence of lipoate are assayed, the E2 subunits of 2-oxoacid dehydrogenases are modified (all have molecular masses of 43 to 47 kDa).
FIG 12.
Comparison of the lipoate assembly pathways of E. coli and B. subtilis.
Two aspects of this pathway require explanation. First, in bacteria, the glycine cleavage system activity is expressed only in the presence of exogenous glycine. In B. subtilis, the expression of the glycine cleavage system is under the control of a riboswitch. The riboswitch binds glycine, and this complex forms an RNA structure that neutralizes an intrinsic transcription terminator sequence located between the operon promoter and the coding sequence of the first gene (121). Hence, it might be expected that exogenous glycine would be required for protein lipoylation, but this is not the case. The reason is that the riboswitch controls transcription only of the genes encoding the T and P subunits (Fig. 3); the H subunit is encoded at a site far distant from the T and P subunit genes and appears to be expressed as a constitutive (housekeeping) protein, thereby allowing 2-oxoacid dehydrogenase lipoylation to proceed under all growth conditions.
The second aspect to be addressed is the unexpected role of GcvH in lipoylation of the 2-oxoacid dehydrogenases. This seems to be an evolutionary relic. Braakman and Smith (122) have argued persuasively that the glycine cleavage system was the first lipoyl-requiring metabolic pathway to evolve. Their proposal posits that the ancestral function of lipoic acid was to connect glycine/serine metabolism to folate–single-carbon chemistry through the serine/glycine cycle (Fig. 3), which remains (nearly universally) essential to this day. In contrast, the role of lipoic acid in the oxidative TCA cycle or the degradation of branched-chain amino acids likely arose later and, in many organisms, is not essential. This is particularily true in thermophilic chemolithoautotroph bacteria, which derive both biomass and energy from inorganic chemical compounds. These bacteria are thought to represent very early stages in the development of central metabolism and lack 2-oxoacid dehydrogenases. In support of this proposal, archaea and bacteria that lack the glycine cleavage system also lack genes encoding proteins involved in lipoylation or in lipoic acid scavenging. Hence, in this scenario, lipoylation first evolved for glycine cleavage, and upon the advent of 2-oxoacid dehydrogenases, LipL evolved to transfer lipoate to the new enzymes, probably via duplication of LipM and subsequence sequence divergence (see below). The gene encoding the LipM octanoyl transferase neighbors the genes encoding the T and P subunits, arguing that lipM is the original gene from which the genes encoding the LplJ lipoate ligase and LipL amidotransferase arose.
In many bacteria, the two distinct transferase enzymes LipM and LipL were replaced with a single all-purpose LipB transferase, such as those of E. coli and M. tuberculosis, whereas other bacteria, such as B. subtilis, retained the original, more complex pathway. A modification of the B. subtilis pathway is found in the natural lipoate auxotroph Listeria monocytogenes, a pathogenic relative of B. subtilis. L. monocytogenes lacks lipA and lipM and obtains lipoic acid from its mammalian host by use of a lipoamidase that hydrolyzes the amide bond of the host-lipoylated enzymes. This pirated lipoic acid is attached to GcvH using a ligase, and a LipL amidotransferase subsequently transfers the lipoate to the 2-oxoacid dehydrogenases (91). Hence, as seen in the B. subtilis pathway, the attachment of exogenous lipoate to the 2-oxoacid dehydrogenases requires that the lipoate first be attached to GcvH (Fig. 13). The degenerate nature of the L. monocytogenes pathway can probably be explained by the ability to pirate lipoate, which removed selection for retention of the lipoyl assembly genes.
FIG 13.
The Listeria monocytogenes lipoate-scavenging pathway. Lpa denotes a lipoamidase activity encoded by an unknown gene(s) that cleaves lipoate from host proteins or peptides (91). This bacterium encodes two lipoate ligases, but only LplA1 functions during invasion of the host in pathogenesis (164). This is probably due to its more efficient utilization of lipoate (91). LplA1 is largely specific for modification of the H protein of the glycine cleavage system. Following attachment to GcvH, the lipoate can be transferred to the 2-oxoacid dehydrogenases. The transfer reaction has been shown to be reversible (91).
Braakman and Smith (122) also propose that the lipoate ligases that scavenge lipoate from the environment evolved from the octanoyltransferases, thereby explaining the structural relatedness of LipB, LipM, and LplA (and probably LplJ). This proposal seems clear-cut, because lipoate biosynthesis had to evolve before there was lipoate available for scavenging. However, it is possible that the ligases might first have scavenged octanoate.
LIPOYLATION IN EUKARYOTES
Gene and protein nomenclatures differ in bacteria, yeast, and mammals (plant genes and proteins largely follow bacterial nomenclature). The gene names are given in Table 1. The proteins have the same names, although the protein names are not italicized and can contain lowercase letters.
TABLE 1.
Gene names
| Enzyme | Gene name ina: |
|||
|---|---|---|---|---|
| E. coli | B. subtilis | Homo sapiens | S. cerevisiae | |
| Lipoyl synthase | lipA | lipA | LIAS | LIP5 |
| Octanoyltransferase | lipB | lipB | LIPT2 | LIP2 |
| Glycine cleavage H protein | gcvH | gcvH | GCSH | GCV3 |
| Amidotransferase | None | lipL | LIPT1? | LIP3? |
| Lipoate ligase | lplA | lplJ | None? | None? |
The question marks indicate predicted but unproven activities.
Lower Eukaryotes
Lipoylation in yeast has been most thoroughly studied in the laboratory workhorse, Saccharomyces cerevisiae. However, the pathway is almost entirely based on phenotypic and immunoblotting analyses of mutant strains. Very little in vitro biochemistry has been done because the proteins have thus far proven intractable, since they form insoluble inclusion bodies when expressed in E. coli and yeast (123). Several mutants with mutations in lipoate biosynthesis were isolated in the Tzagoloff laboratory (124) as strains having pleiotropic effects on mitochondrial metabolism (all of the lipoyl metabolism proteins are encoded by the nuclear genome but localize to the mitochondria). Mitochondrial metabolism mutants are generally isolated as strains that form very small (petite) colonies when grown on a medium containing glucose. This is due to the inability of the mutant strains to metabolize the ethanol produced in glucose fermentation. Hence, the growth of mutant cells, but not that of wild-type cells, is halted when the supply of glucose in the medium is exhausted. In contrast, the wild-type cells continue growing on ethanol. Petite strains also grow poorly on other carbon sources that require respiration, such as glycerol.
The first of these mutant strains identified as blocked in lipoyl metabolism was one with a mutation in LIP5, which encodes a LipA homologue (125). As expected, the LIP5 strains had very low levels of total protein-bound lipoic acid. Unexpected was that exogenous lipoic acid failed to allow growth of these strains on glycerol (125). A curious sidelight was that these authors were not seeking mutants in lipoyl metabolism but, rather, mutants defective in mitochondrial tRNA processing (see below). Strains with a defect in LIP2, a gene encoding a protein with strong sequence similarity to the E. coli LipB octanoyl transferase, were also isolated while seeking mutant strains defective in other metabolic processes. This occurred in both S. cerevisiae (126) and the related yeast Kluyveromyces lactis (127). These strains were also petite mutants; they could be rescued by ethanol-plus-succinate supplementation but not by lipoate supplementation. Moreover, both strains were unable to use glycine as the sole ammonia source, indicating a glycine cleavage defect. Echoing the LIP5 case, the LIP3 lipoate metabolism gene was isolated while screening for strains defective in mitochondrial RNA processing (128). LIP3 encoded a protein having strong sequence similarity to the E. coli LplA ligase (128). Assay of the lipoylation status of the three S. cerevisiae lipoylated-protein subunits, those of the pyruvate and 2-oxoglutarate dehydrogenases and the H protein of the glycine cleavage system, showed that strains lacking the LIP5 or LIP2 gene had no detectable lipoylated proteins (128). However, strains lacking LIP3 had normal levels of lipoylated H protein but no lipoylated 2-oxoacid dehydrogenases detectable either by Western blotting or by dehydrogenase activity (128). This effect was specific to the H protein in that strains lacking the genes encoding the other glycine cleavage system subunits, P and T, showed normal levels of lipoylated 2-oxoacid dehydrogenases, indicating that cleavage of glycine played no role (128). Moreover, mutant strains in which the H protein lysine targeted for lipoylation was replaced with other residues also blocked all lipoylation. Hence, lipoyl-H protein was the active species and not H protein per se. Note that the mitochondrial RNA processing screen also turned up strains lacking various of the enzymes of the yeast mitochondrial type II fatty acid synthesis pathway and these strains also lacked lipoylated proteins (128). The link between lipoic acid synthesis and mitochondrial RNA processing remains unresolved, although it is thought to be a regulatory checkpoint preventing RNA processing in response to the metabolic state of the cell (129).
In more recent work, the protein encoded by LIP3 (Lip3) was found to allow slow growth of an E. coli strain that lacked both the LplA lipoate ligase and the LipB octanoyl transferase, but only in the presence of octanoate (lipoate did not support growth but was not toxic) (123). Lip3 expression also restored 2-oxoacid dehydrogenase activity and E2 subunit lipoylation. Growth in the presence of octanoate required that the E. coli strain have an active acyl-CoA synthetase, and acyl-CoA synthetase overexpression stimulated growth. Hence, a substrate of Lip3 seemed likely to be octanoyl-CoA, but the growth could also be explained by octanoyl-adenylate released by the synthetase being utilized by Lip3 (123). This was tested in vitro, and Lip3 was found to weakly modify both an E. coli lipoyl domain and yeast H protein (encoded by the GCV3 gene), using octanoyl-CoA as acyl donor. In further investigations, a mitochondrial targeting sequence was added to the N termini of E. coli LplA and LipB. The B. subtilis LipL amidotransferase was also targeted to the mitochondria. The growth of S. cerevisiae strains carrying deletions of the LIP2, LIP3, or GCV3 gene was tested on ethanol medium after transformation with either an empty vector or a plasmid encoding one of the bacterial proteins targeted to mitochondria. The parental wild-type strain grew well on ethanol medium, whereas the LIP2, LIP3, and GCV3 mutant strains transformed with the empty vector failed to grow on the ethanol medium, even when it was supplemented with octanoate. The expression of the E. coli LplA derivative allowed growth of all three yeast mutant strains, indicating that the inactive yeast 2-oxoacid dehydrogenase proteins had been activated by lipoate attachment (123). The growth of the GCV3 mutant strain showed that the H protein is not necessary for respiration but, rather, is required for lipoylation of the 2-oxoacid dehydrogenases. The B. subtilis LipL derivative allowed growth of the LIP3 mutant strain but not of the LIP2 mutant strain, suggesting that Lip2 is the primary source of octanoyl-Gcv3, the substrate of LipL. The E. coli LipB derivative produced octanoyl-Gcv3, as indicated by its ability to allow growth of the LIP2 strain. However, the inability of LipB to allow growth of the GCV3 strain indicates that E. coli LipB cannot efficiently modify S. cerevisiae PDH and OGDH. Finally, the inability of LipB to complement the LIP3 strain indicates that Lip3 acts downstream from the synthesis of octanoyl-Gcv3 catalyzed by either Lip2 or LipB (123).
Taken together, the data argue that Lip3, like B. subtilis LipL, is an amidotransferase that transfers octanoate (and/or lipoate) from the H protein to the 2-oxoacid dehydrogenase E2 subunits. This interpretation fits the data showing that lipoylated Gcv3 is required for lipoylation of the yeast 2-oxoacid dehydrogenases and the finding that the expression of mitochondrion-targeted LipL allowed growth of the yeast LIP3 strain on ethanol. Moreover, it is consistent with analyses of certain human disorders of lipoate metabolism (see below).
Lipoate Metabolism of Pathogenic Protozoa
Lipoic acid synthesis and salvage have been intensively studied in the apicomplexan parasites, Plasmodium and Toxoplasma, which cause malaria and toxoplasmosis, respectively. The apicoplast is a degenerate organelle thought to be the result of endosymbiosis by an algal cell. Apicoplast function is required for the growth of the parasites in that it provides key metabolic pathways, such as fatty acid and lipoic acid synthesis. As is often the case, the original gene complement of the endosymbiont has been transferred to the nucleus. The resulting proteins contain a sequence that targets them to the apicoplast. In both apicomplexan parasites, the LipA homologue is found only in the apicoplast, along with the LipB homologue, the enzymes of fatty acid synthesis, and pyruvate dehydrogenase. The other lipoylated enzyme complexes (OGDH, the branched-chain dehydrogenase, and glycine cleavage system [GCV]) reside in the mitochondrion. This division of labor plays a role in the complex life cycles of these parasites. Synthesis of lipoylated proteins occurs only in the apicoplast. The mitochondrial enzymes acquire lipoic acid by salvage, using two ligases called LplA1 and LplA2. LplA2 is also targeted to the apicoplast and can bypass the loss of LipB in some stages of the complex Plasmodium life cycle. Hence, putting the division of labor and multiple ligases aside, these parasites have lipoate metabolism pathways that are remarkably similar to that of E. coli.
LIPOATE METABOLISM OF HIGHER EUKARYOTES
Plant Lipoate Metabolism
Similarly to the apicomplexan parasites, plants have divided their lipoylated proteins between two organelles, mitochondria and plastids (chloroplasts in green tissue), albeit with some redundancy. In plant cells, PDH, OGDH, branched-chain 2-oxoacid dehydrogenase complexes, and GCV subunits are located in mitochondria, although PDH is also found in plastids. Both organelles carry out fatty acid synthesis and convert octanoyl-ACP to lipoylated proteins (130). LipB activity has been demonstrated in mitochondrial lysates. The expression of the gene called LIP2 in E. coli replaces LipB function (131). A second gene, called LIP2p, encoding a LipB activity specifically targeted to plastids, has also been demonstrated (132). More recent work has shown that plants lacking LIP2p are viable due to a second, nonessential LipB-like activity encoded by LIP2p2, whereas double mutants lacking both plastid isozymes are nonviable (133). LIP1 has the sequence hallmarks of a gene encoding a lipoyl synthase, and its expression was shown to replace LipA in E. coli (134). LIP1 was shown to be targeted to the mitochondria. As in the case of the LipB activities, a distinct LipA activity called LIP1p is targeted to the plastids; this protein has 52% sequence identity with the mitochondrial form (135) and has been shown to be essential (136).
Recent work has shown that the mitochondrial LIP2 is an essential enzyme that is primarily expressed in leaves and other photosynthesizing organs and is not expressed in roots and flowers (136). Note that leaf mitochondria accumulate huge amounts of the GCV proteins to run the photorespiratory cycle, a process essential for oxygenic photosynthesis in which two molecules of glycine are converted to one molecule of serine. In photosynthesizing leaves, these proteins comprise half of the mitochondrial protein content, and thus, very high levels of lipoylation are required for H protein modification. Another essential enzyme targeted to the mitochondria is a ligase that can functionally replace lplA in E. coli (136, 137), although the overall reaction could not be demonstrated in vitro (136). This enzyme is essential in both leaves and roots (136).
Mammalian Lipoate Metabolism
Insights into the importance of lipoylation in mammals primarily come from knockout mouse studies and human genetic defects. All of the mammalian proteins involved in lipoic acid synthesis and utilization are located in the mitochondria, and thus, the complexities of some of the systems discussed above are not present. Studies of knockout mice and human patients give a consistent picture and will be discussed in turn. Although long suspected, it has only recently been demonstrated that mammalian mitochondria contain their own de novo fatty acid synthesis pathway, comprised of proteins encoded by nuclear genes (138–140). A key finding was the demonstration that mitochondrial lysates contain appreciable levels of soluble ACP, a portion of which carried an octanoyl moiety (141). Previously, the mitochondrial ACP was thought to be present only as subunits of complex I of the membrane-bound respiratory chain, where it is called SDAP (named for the amino acid sequence of the prosthetic group attachment side, serine-asparate-alanine-proline, with the serine as the site of attachment) (142). In vitro studies (139, 140) showed that the major product of the mitochondrial pathway is the octanoyl-ACP precursor for lipoyl moieties, which is utilized for the posttranslational modification of the E2 subunits of the 2-oxoacid complexes and the glycine cleavage system H protein. The first lipoyl synthase-deficient mammals were mice in which the LIAS gene had been disrupted (143). LIAS is the mammalian homologue of the LipA sulfur insertion enzyme (Table 1) and functionally replaces LipA in E. coli (144). Homozygous embryos that had disrupted LIAS genes appeared healthy at the blastocyst stage, but their development was globally retarded by 7.5 days of life and all homozygous null embryos died before 9.5 days of life (143). Supplementing the diet of the heterozygous mothers with lipoic acid during pregnancy failed to prevent the death of the homozygous embryos. Therefore, endogenous synthesis of lipoyl groups is essential for mouse embryo development and cannot be bypassed by providing lipoic acid to the mothers. Note that exogenous lipoate is readily taken up into plasma and tissues (lipoic acid has been widely studied as an antioxidant). Several studies have administered radioactive lipoic acid to mammals and followed its fate (145–148). However, lipoate is quickly reduced and degraded by the β-oxidation pathway, and no evidence for attachment of exogenously fed lipoate to proteins has been reported.
The pathway of lipoate modification of mitochondrial 2-oxoacid dehydrogenases and GCSH has also been tested, by using an inducible-knockout mouse model. In this system, the mitochondrial pathway for the de novo synthesis of fatty acyl and lipoyl moieties was controlled by the presence of an inducing compound which caused disruption of the fatty acid synthetic gene, Mcat, the gene encoding the mitochondrial malonyl CoA:ACP transacylase responsible for transfer of malonyl moieties from CoA to the phosphopantetheinyl moiety of ACP (malonyl-ACP is the building block of fatty acid synthesis) (139). Upon the addition of the inducer to disrupt the Mcat gene, the knockout mice fell behind their control littermates in weight gain and progressively lost weight and energy as the experiment proceeded. These symptoms occurred despite the fact that the knockout mice ate more than the control mice. The diet was processed normally and should have provided a rich source of lipoic acid. Enzyme activity and lipoylation assays showed that knockout of the Mcat gene causes a lipoylation defect rather than an altered mitochondrial fatty acid composition (139). The levels of the catalytically active lipoylated forms of all four components of the mitochondrial lipoamide E2 subunits of the three 2-oxoacid dehydrogenases and the H protein component of the glycine cleavage system were reduced in tissues of the knockout mice. Skeletal muscle was particularly affected in that the values were reduced severalfold (139).
Finally, a study in tissue culture cells showed that knockdown of ACP production by the use of small interfering RNAs largely blocked lipoylation and severely decreased 2-oxoacid dehydrogenase activities. Upon knockdown, cell growth progressively slowed and cell death eventually resulted (138). Supplementation of the growth medium with lipoic acid failed to restore protein lipoylation and cell viability, consistent with the results of the in vivo mouse studies.
Human Disorders of Lipoate Synthesis and Attachment
The first patients recognized as having disorders of lipoate metabolism were those with deficiencies of dihydrolipoamide dehydrogenase activity, the E3 subunit of the 2-oxoacid dehydrogenases and the L subunit of the glycine cleavage system (149). Dihydrolipoamide dehydrogenase deficiency is clinically heterogeneous, with effects ranging from early death to chronic fatigue. As expected from a deficiency in 2-oxoacid dehydrogenase function, lactic acid (from reduction of pyruvate) and 2-oxoglutarate accumulate in bodily fluids. Surprisingly, glycine levels are very seldom elevated, an observation which argues that the glycine cleavage system is favored over the 2-oxoacid dehydrogenases when dihydrolipoamide dehydrogenase activity becomes limiting. Knowledge of the lipoate synthetic and attachment pathways in microorganisms has led to recognition of disorders of lipoate attachment. Mutations are found in two genes, that encoding lipoyl synthase (LIAS) and that encoding the defective lipoate ligase activity (LIPT1). As first seen in dihydrolipoamide dehydrogenase deficiency, the four known LIAS patients are clinically heterogeneous (144, 150–152). Two died early in life and two survived, albeit with severe neurological problems. All four patients showed elevated glycine levels in plasma and cerebrospinal fluid plus low or absent glycine cleavage system activity. The administration of lipoic acid failed to reverse the metabolic defects (150). Fibroblasts from the two most severely affected patients had extremely reduced or undetectable levels of lipoylated proteins. The LIAS genes of all four patients contained mutations in highly conserved residues (150), and one of the LIAS alleles was shown to be deficient in restoring the growth of an E. coli lipA mutant strain (144).
In contrast to the LIAS patients, two children with LIPT1 mutations had normal levels of glycine cleavage activity, H protein lipoylation, and glycine in body fluids (153, 154). As seen in the LIAS cases, pyruvate and 2-oxoglutarate dehydrogenase activities were very low. Both patients showed a severe lack of muscle tone and excreted very high levels of lactate (the product of pyruvate reduction) and 2-oxoglutarate (153, 154). The E2 subunits of all three dehydrogenases showed low levels of lipoylation, concomitant with the results of the 2-oxoacid dehydrogenase activity assays, whereas liver tissue glycine cleavage H protein lipoylation appeared to be increased somewhat above control levels in the patient tested (154). The addition of lipoic acid to the growth medium of fibroblasts derived from both patients failed to restore lipoylation (153, 154), whereas the introduction of a wild-type LIPT1 gene restored fibroblast 2-oxoacid dehydrogenase activities (153).
Conclusions and a Unifying Hypothesis
The above-described studies clearly demonstrate that mammals lack the ability to attach exogenous lipoic acid to the essential enzymes that require the lipoyl cofactor for activity. Therefore, endogenous synthesis of lipoylated proteins is essential and the pathway proposed for modification of these enzymes with exogenous lipoic acid in vitro lacks physiological relevance. As discussed above, the ACSM1 enzyme reported to activate the lipoate carboxyl group lacks the properties of a lipoate metabolism enzyme. In contrast, LIPT1 has the hallmarks of an enzyme of lipoate metabolism, including a crystal structure with bound lipoyl-adenylate. I propose that the defective lipoate ligase activity of LIPT1 is a remnant of enzyme evolution that has long misled the field and posit that the relevant activity of the protein is amidotransfer, the reaction catalyzed by the B. subtilis and L. monocytogenes LipL proteins. In recent years, a concept of enzyme evolution called moonlighting has received much support (155–158). A protein can acquire a second (moonlighting) function without concomitantly losing its original function. That is, mutations can enhance the moonlighting function without eliminating the ancestral function. This can arise through mutagenesis followed by gene duplication and further mutation, which could result in two specialist proteins that perform either the original function or the new function. Another probable moonlighting pathway is gene duplication that results in two genes encoding proteins having the original function and one of these genes undergoing mutation to encode a protein having a new function. In the case of LIPT1, its original ancestor would have encoded a fully functional lipoate ligase, such as LplA or LplJ, rather than the defective ligase activity of the present protein. That is, evolution of the gene has resulted in a protein that has lost the ability to activate lipoate while (perhaps temporarily) retaining the ligase lipoyl transfer half-reaction and acquiring amidotransferase activity.
The hypothesis that LIPT1 has amidotransferase activity can fully explain the human LIPT1 deficiency data. These patients (or their derived fibroblasts) accumulate lipoylated H protein and have normal glycine cleavage activity and glycine levels but are unable to synthesize lipoylated 2-oxoacid dehydrogenases (the acceptor proteins are present at normal levels). A parallel argument is that yeast Lip3 protein is also an amidotransferase and the observed octanoyl transfer from octanoyl-CoA is another example of moonlighting. Indeed, the finding that the expression of a version of the B. subtilis LipL amidotransferase targeted to mitochondria allothe wed growth of a yeast LIP3 mutant strain (but not of a LIP2 strain) on ethanol is a strong indication that Lip3 is also an amidotransferase (123). B. subtilis LipL has only amidotransferase activity and lacks ligase and octanoyltransferase activities (90). Biochemical attempts to assay Lip3 for amidotransferase activity were unsuccessful. However, the problematical nature of the yeast proteins engendered technical difficulties that weaken this result. Gcv3 had to be refolded from insoluble inclusions, the acceptor dehydrogenase domain was from E. coli rather than yeast, and only an insensitive gel shift assay could be used (123).
It seems possible that the expression of LplA in mitochondria could bypass the genetic defects of LIAS and LIPT1 patients by restoring the function of their lipoylated enzymes. LplA is known to attach lipoic acid to the human 2-oxoacid dehydrogenase E2 subunits (23, 159) and bovine H protein (160). Indeed, it has been proposed that LIPT1 and yeast Lip3 have the same function (153). If so, an experiment parallel to alleviation of LIPT1 deficiency has been done in yeast. The expression E. coli LplA ligase targeted to mitochondria allowed the growth of the yeast LIP3 mutant strain and, thus, bypassed the loss of Lip3 (123).
An argument against the proposal that LIPT1 and Lip3 (which show 31% identity, found mainly within the large domain) have amidotransferase activity is that when aligned, they lack a conserved residue to replace the key LipL nucleophile, the cysteine thiol that attacks the amide bond. However, a serine residue activated by a catalytic triad as in serine proteases would suffice. Indeed, the alignment of LIPT1 and Lip3 show two highly conserved clusters within the large domain that contain serine residues. It should be noted that serine proteases can act as amidotransferases when water is limiting (161–163).
ACKNOWLEDGMENT
Work from this laboratory was supported by grant AI15056 from the National Institute of Allergy and Infectious Disease.
REFERENCES
- 1.Reed LJ. 1998. From lipoic acid to multi-enzyme complexes. Protein Sci 7:220–224. doi: 10.1002/pro.5560070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed LJ. 2001. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J Biol Chem 276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- 3.Snell EE. 1992. Microorganisms in vitamin and biofactor research. J Nutr Sci Vitaminol (Tokyo) 38:34–39. doi: 10.3177/jnsv.38.Special_34. [DOI] [PubMed] [Google Scholar]
- 4.Snell EE. 1993. From bacterial nutrition to enzyme structure: a personal odyssey. Annu Rev Biochem 62:1–27. doi: 10.1146/annurev.bi.62.070193.000245. [DOI] [PubMed] [Google Scholar]
- 5.Reed LJ. 1966. Chemistry and function of lipoic acid, p 99–126. In Florkin M, Stotz EH (ed), Comprehensive biochemistry, vol 14 Elsevier, New York, NY. [Google Scholar]
- 6.Gunsalus IC. 1953. The chemistry and function of the pyruvate oxidation factor (lipoic acid). J Cell Physiol Suppl 41(Suppl 1):113–136. [DOI] [PubMed] [Google Scholar]
- 7.Gunsalus IC, Struglia L, O'Kane DJ. 1952. Pyruvic acid metabolism. IV. Occurrence, properties, and partial purification of pyruvate oxidation factor. J Biol Chem 194:859–869. [PubMed] [Google Scholar]
- 8.Reed LJ, De BB, Gunsalus IC, Hornberger CS Jr. 1951. Crystalline alpha-lipoic acid; a catalytic agent associated with pyruvate dehydrogenase. Science 114:93–94. doi: 10.1126/science.114.2952.93. [DOI] [PubMed] [Google Scholar]
- 9.Peters RA. 1952. Croonian Lecture: lethal synthesis. Proc R Soc Lond B Biol Sci 139:143–170. doi: 10.1098/rspb.1952.0001. [DOI] [PubMed] [Google Scholar]
- 10.Stokstad ELR. 1954. Discussion: biological activity of analogues and derivatives of thioctic acid. Fed Proc 13:712–714. [PubMed] [Google Scholar]
- 11.Koike M, Reed LJ. 1960. α-Keto acid dehydrogenation complexes. II. The role of protein-bound lipoic acid and flavin adenine dinucleotide. J Biol Chem 235:1931–1938. [PubMed] [Google Scholar]
- 12.Reed LJ, Leach FR, Koike M. 1958. Studies on a lipoic acid-activating system. J Biol Chem 232:123–142. [PubMed] [Google Scholar]
- 13.Reed LJ, Koike M, Levitch ME, Leach FR. 1958. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem 232:143–158. [PubMed] [Google Scholar]
- 14.Perham RN. 1991. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry 30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 15.Perham RN. 1995. Structure and posttranslational modification of lipoyl domain of 2-oxo-acid dehydrogenase multienzyme complexes. Methods Enzymol 251:436–448. doi: 10.1016/0076-6879(95)51147-4. [DOI] [PubMed] [Google Scholar]
- 16.Perham RN. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem 69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 17.Griffin TA, Lau KS, Chuang DT. 1988. Characterization and conservation of the inner E2 core domain structure of branched-chain alpha-keto acid dehydrogenase complex from bovine liver. Construction of a cDNA encoding the entire transacylase (E2b) precursor. J Biol Chem 263:14008–14014. [PubMed] [Google Scholar]
- 18.Hackert ML, Xu WX, Oliver RM, Wall JS, Hainfeld JF, Mullinax TR, Reed LJ. 1989. Branched-chain alpha-keto acid dehydrogenase complex from bovine kidney: radial distribution of mass determined from dark-field electron micrographs. Biochemistry 28:6816–6821. doi: 10.1021/bi00443a006. [DOI] [PubMed] [Google Scholar]
- 19.Douce R, Bourguignon J, Neuburger M, Rebeille F. 2001. The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6:167–176. doi: 10.1016/S1360-1385(01)01892-1. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. 2008. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci 84:246–263. doi: 10.2183/pjab.84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourguignon J, Merand V, Rawsthorne S, Forest E, Douce R. 1996. Glycine decarboxylase and pyruvate dehydrogenase complexes share the same dihydrolipoamide dehydrogenase in pea leaf mitochondria: evidence from mass spectrometry and primary-structure analysis. Biochem J 313:229–234. doi: 10.1042/bj3130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiert PS, Stauffer LT, Stauffer GV. 1990. The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol 172:6142–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn J. 1997. Lipoylation of acyltransferase components of 2-oxo acid dehydrogenase complexes. Methods Enzymol 279:193–202. doi: 10.1016/S0076-6879(97)79023-2. [DOI] [PubMed] [Google Scholar]
- 24.Macherel D, Bourguignon J, Forest E, Faure M, Cohen-Addad C, Douce R. 1996. Expression, lipoylation and structure determination of recombinant pea H-protein in Escherichia coli. Eur J Biochem 236:27–33. doi: 10.1111/j.1432-1033.1996.00027.x. [DOI] [PubMed] [Google Scholar]
- 25.Graham LD, Packman LC, Perham RN. 1989. Kinetics and specificity of reductive acylation of lipoyl domains from 2-oxo acid dehydrogenase multienzyme complexes. Biochemistry 28:1574–1581. doi: 10.1021/bi00430a023. [DOI] [PubMed] [Google Scholar]
- 26.Miles JS, Guest JR, Radford SE, Perham RN. 1988. Investigation of the mechanism of active site coupling in the pyruvate dehydrogenase multienzyme complex of Escherichia coli by protein engineering. J Mol Biol 202:97–106. doi: 10.1016/0022-2836(88)90522-0. [DOI] [PubMed] [Google Scholar]
- 27.Berg A, Vervoort J, de Kok A. 1997. Three-dimensional structure in solution of the N-terminal lipoyl domain of the pyruvate dehydrogenase complex from Azotobacter vinelandii. Eur J Biochem 244:352–360. doi: 10.1111/j.1432-1033.1997.00352.x. [DOI] [PubMed] [Google Scholar]
- 28.Berg A, Westphal AH, Bosma HJ, de Kok A. 1998. Kinetics and specificity of reductive acylation of wild-type and mutated lipoyl domains of 2-oxo-acid dehydrogenase complexes from Azotobacter vinelandii. Eur J Biochem 252:45–50. doi: 10.1046/j.1432-1327.1998.2520045.x. [DOI] [PubMed] [Google Scholar]
- 29.Jones DD, Stott KM, Howard MJ, Perham RN. 2000. Restricted motion of the lipoyl-lysine swinging arm in the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 39:8448–8459. doi: 10.1021/bi992978i. [DOI] [PubMed] [Google Scholar]
- 30.Aevarsson A, Seger K, Turley S, Sokatch JR, Hol WG. 1999. Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes. Nat Struct Biol 6:785–792. doi: 10.1038/11563. [DOI] [PubMed] [Google Scholar]
- 31.Wallis NG, Allen MD, Broadhurst RW, Lessard IA, Perham RN. 1996. Recognition of a surface loop of the lipoyl domain underlies substrate channelling in the pyruvate dehydrogenase multienzyme complex. J Mol Biol 263:463–474. doi: 10.1006/jmbi.1996.0589. [DOI] [PubMed] [Google Scholar]
- 32.Wallis NG, Perham RN. 1994. Structural dependence of post-translational modification and reductive acetylation of the lipoyl domain of the pyruvate dehydrogenase multienzyme complex. J Mol Biol 236:209–216. doi: 10.1006/jmbi.1994.1130. [DOI] [PubMed] [Google Scholar]
- 33.Cohen-Addad C, Pares S, Sieker L, Neuburger M, Douce R. 1995. The lipoamide arm in the glycine decarboxylase complex is not freely swinging. Nat Struct Biol 2:63–68. doi: 10.1038/nsb0195-63. [DOI] [PubMed] [Google Scholar]
- 34.Dardel F, Davis AL, Laue ED, Perham RN. 1993. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. J Mol Biol 229:1037–1048. doi: 10.1006/jmbi.1993.1103. [DOI] [PubMed] [Google Scholar]
- 35.Brocklehurst SM, Perham RN. 1993. Prediction of the three-dimensional structures of the biotinylated domain from yeast pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: a new automated method for the prediction of protein tertiary structure. Protein Sci 2:626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pares S, Cohen-Addad C, Sieker L, Neuburger M, Douce R. 1994. X-ray structure determination at 2.6-A resolution of a lipoate-containing protein: the H-protein of the glycine decarboxylase complex from pea leaves. Proc Natl Acad Sci U S A 91:4850–4853. doi: 10.1073/pnas.91.11.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cronan JE., Jr 2001. The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure is essential and that the domain functions as a dimer. J Biol Chem 276:37355–37364. [DOI] [PubMed] [Google Scholar]
- 38.Athappilly FK, Hendrickson WA. 1995. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure 3:1407–1419. doi: 10.1016/S0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 39.Roberts EL, Shu N, Howard MJ, Broadhurst RW, Chapman-Smith A, Wallace JC, Morris T, Cronan JE Jr, Perham RN. 1999. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry 38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- 40.Yao X, Wei D, Soden C Jr, Summers MF, Beckett D. 1997. Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase. Biochemistry 36:15089–15100. doi: 10.1021/bi971485f. [DOI] [PubMed] [Google Scholar]
- 41.Reddy DV, Shenoy BC, Carey PR, Sonnichsen FD. 2000. High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii. Biochemistry 39:2509–2516. doi: 10.1021/bi9925367. [DOI] [PubMed] [Google Scholar]
- 42.Cronan JE., Jr 2002. Interchangeable enzyme modules. Functional replacement of the essential linker of the biotinylated subunit of acetyl-CoA carboxylase with a linker from the lipoylated subunit of pyruvate dehydrogenase. J Biol Chem 277:22520–22527. [DOI] [PubMed] [Google Scholar]
- 43.Green DE, Morris TW, Green J, Cronan JE Jr, Guest JR. 1995. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem J 309:853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris TW, Reed KE, Cronan JE Jr. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem 269:16091–16100. [PubMed] [Google Scholar]
- 45.Reed KE, Morris TW, Cronan JE Jr. 1994. Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc Natl Acad Sci U S A 91:3720–3724. doi: 10.1073/pnas.91.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbert AA, Guest JR. 1970. Turbidimetric and polarographic assays for lipoic acid using mutants of Escherichia coli. Methods Enzymol 18(Pt A):269–272. doi: 10.1016/0076-6879(71)18314-0. [DOI] [Google Scholar]
- 47.Oehring R, Bisswanger H. 1992. Incorporation of the enantiomers of lipoic acid into the pyruvate dehydrogenase complex from Escherichia coli in vivo. Biol Chem Hoppe Seyler 373:333–335. doi: 10.1515/bchm3.1992.373.1.333. [DOI] [PubMed] [Google Scholar]
- 48.Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. 2008. An engineered aryl azide ligase for site-specific mapping of protein-protein interactions through photo-cross-linking. Angew Chem Int Ed Engl 47:7018–7021. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen JD, Zou P, Ting AY. 2012. Site-specific protein modification using lipoic acid ligase and bis-aryl hydrazone formation. Chembiochem 13:888–894. doi: 10.1002/cbic.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu DS, Nivon LG, Richter F, Goldman PJ, Deerinck TJ, Yao JZ, Richardson D, Phipps WS, Ye AZ, Ellisman MH, Drennan CL, Baker D, Ting AY. 2014. Computational design of a red fluorophore ligase for site-specific protein labeling in living cells. Proc Natl Acad Sci U S A 111:E4551–4559. doi: 10.1073/pnas.1404736111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajasekar A, Anandkumar B, Maruthamuthu S, Ting YP, Rahman PK. 2010. Characterization of corrosive bacterial consortia isolated from petroleum-product-transporting pipelines. Appl Microbiol Biotechnol 85:1175–1188. doi: 10.1007/s00253-009-2289-9. [DOI] [PubMed] [Google Scholar]
- 52.Uttamapinant C, White KA, Baruah H, Thompson S, Fernandez-Suarez M, Puthenveetil S, Ting AY. 2010. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci U S A 107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. 2005. Crystal structure of lipoate-protein ligase A from Escherichia coli. Determination of the lipoic acid-binding site. J Biol Chem 280:33645–33651. [DOI] [PubMed] [Google Scholar]
- 54.Reche PA. 2000. Lipoylating and biotinylating enzymes contain a homologous catalytic module. Protein Sci 9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McManus E, Luisi BF, Perham RN. 2006. Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification. J Mol Biol 356:625–637. doi: 10.1016/j.jmb.2005.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. 2005. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J Biol Chem 280:38081–38089. doi: 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 57.Christensen QH, Cronan JE. 2009. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J Biol Chem 284:21317–21326. doi: 10.1074/jbc.M109.015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posner MG, Upadhyay A, Bagby S, Hough DW, Danson MJ. 2009. A unique lipoylation system in the Archaea. Lipoylation in Thermoplasma acidophilum requires two proteins. FEBS J 276:4012–4022. doi: 10.1111/j.1742-4658.2009.07110.x. [DOI] [PubMed] [Google Scholar]
- 59.Posner MG, Upadhyay A, Crennell SJ, Watson AJ, Dorus S, Danson MJ, Bagby S. 2013. Post-translational modification in the archaea: structural characterization of multi-enzyme complex lipoylation. Biochem J 449:415–425. doi: 10.1042/BJ20121150. [DOI] [PubMed] [Google Scholar]
- 60.Cao X, Cronan JE. 2015. The Streptomyces coelicolor lipoate-protein ligase is a circularly permuted version of the Escherichia coli enzyme composed of discrete interacting domains. J Biol Chem 290:7280–7290. doi: 10.1074/jbc.M114.626879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW. 2006. Co-repressor induced order and biotin repressor dimerization: a case for divergent followed by convergent evolution. J Mol Biol 357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 62.Fujiwara K, Okamura-Ikeda K, Motokawa Y. 1994. Purification and characterization of lipoyl-AMP:N epsilon-lysine lipoyltransferase from bovine liver mitochondria. J Biol Chem 269:16605–16609. [PubMed] [Google Scholar]
- 63.Fujiwara K, Maita N, Hosaka H, Okamura-Ikeda K, Nakagawa A, Taniguchi H. 2010. Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A. J Biol Chem 285:9971–9980. doi: 10.1074/jbc.M109.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujiwara K, Hosaka H, Matsuda M, Okamura-Ikeda K, Motokawa Y, Suzuki M, Nakagawa A, Taniguchi H. 2007. Crystal structure of bovine lipoyltransferase in complex with lipoyl-AMP. J Mol Biol 371:222–234. doi: 10.1016/j.jmb.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 65.Fujiwara K, Takeuchi S, Okamura-Ikeda K, Motokawa Y. 2001. Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J Biol Chem 276:28819–28823. doi: 10.1074/jbc.M101748200. [DOI] [PubMed] [Google Scholar]
- 66.Fujino T, Takei YA, Sone H, Ioka RX, Kamataki A, Magoori K, Takahashi S, Sakai J, Yamamoto TT. 2001. Molecular identification and characterization of two medium-chain acyl-CoA synthetases, MACS1 and the Sa gene product. J Biol Chem 276:35961–35966. doi: 10.1074/jbc.M106651200. [DOI] [PubMed] [Google Scholar]
- 67.Vessey DA, Lau E, Kelley M, Warren RS. 2003. Isolation, sequencing, and expression of a cDNA for the HXM-A form of xenobiotic/medium-chain fatty acid:CoA ligase from human liver mitochondria. J Biochem Mol Toxicol 17:1–6. doi: 10.1002/jbt.10056. [DOI] [PubMed] [Google Scholar]
- 68.Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, Giegling I, Muglia P, Cichon S, Rietschel M, Pietilainen OP, Peltonen L, Bramon E, Collier D, Clair DS, Sigurdsson E, Petursson H, Rujescu D, Melle I, Steen VM, Djurovic S, Andreassen OA. 2010. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res 44:748–753. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Ji W, Li Z, He K, Wang Q, Chen J, Qiang Y, Feng G, Li X, Shen J, Wen Z, Ji J, Shi Y. 2015. Genetic association of ACSM1 variation with schizophrenia and major depressive disorder in the Han Chinese population. Am J Med Genet B Neuropsychiatr Genet 168(Pt B):144–149. doi: 10.1002/ajmg.b.32291. [DOI] [PubMed] [Google Scholar]
- 70.Morris TW, Reed KE, Cronan JE Jr. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol 177:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanden Boom TJ, Reed KE, Cronan JE Jr. 1991. Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol 173:6411–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jordan SW, Cronan JE Jr. 1997. A new metabolic link. The acyl carrier protein of lipid synthesis donates lipoic acid to the pyruvate dehydrogenase complex in Escherichia coli and mitochondria. J Biol Chem 272:17903–17906. [DOI] [PubMed] [Google Scholar]
- 73.Jordan SW, Cronan JE Jr. 2003. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J Bacteriol 185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordan SW, Cronan JE Jr. 1997. Biosynthesis of lipoic acid and posttranslational modification with lipoic acid in Escherichia coli. Methods Enzymol 279:176–183. doi: 10.1016/S0076-6879(97)79021-9. [DOI] [PubMed] [Google Scholar]
- 75.Hassan BH, Cronan JE. 2011. Protein-protein interactions in assembly of lipoic acid on the 2-oxoacid dehydrogenases of aerobic metabolism. J Biol Chem 286:8263–8276. doi: 10.1074/jbc.M110.194191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nesbitt NM, Baleanu-Gogonea C, Cicchillo RM, Goodson K, Iwig DF, Broadwater JA, Haas JA, Fox BG, Booker SJ. 2005. Expression, purification, and physical characterization of Escherichia coli lipoyl(octanoyl)transferase. Protein Expr Purif 39:269–282. doi: 10.1016/j.pep.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 77.Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. 2003. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol 10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 78.Reed KE, Cronan JE Jr. 1993. Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J Bacteriol 175:1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herbert AA, Guest JR. 1975. Lipoic acid content of Escherichia coli and other microorganisms. Arch Microbiol 106:259–266. doi: 10.1007/BF00446532. [DOI] [PubMed] [Google Scholar]
- 80.Herbert AA, Guest JR. 1968. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J Gen Microbiol 53:363–381. doi: 10.1099/00221287-53-3-363. [DOI] [PubMed] [Google Scholar]
- 81.Hermes FA, Cronan JE. 2009. Scavenging of cytosolic octanoic acid by mutant LplA lipoate ligases allows growth of Escherichia coli strains lacking the LipB octanoyltransferase of lipoic acid synthesis. J Bacteriol 191:6796–6803. doi: 10.1128/JB.00798-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ali ST, Guest JR. 1990. Isolation and characterization of lipoylated and unlipoylated domains of the E2p subunit of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J 271:139–145. doi: 10.1042/bj2710139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ali ST, Moir AJ, Ashton PR, Engel PC, Guest JR. 1990. Octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex in a lipoyl-deficient strain of Escherichia coli. Mol Microbiol 4:943–950. doi: 10.1111/j.1365-2958.1990.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhao X, Miller JR, Cronan JE. 2005. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry 44:16737–16746. doi: 10.1021/bi051865y. [DOI] [PubMed] [Google Scholar]
- 85.Ma Q, Zhao X, Nasser Eddine A, Geerlof A, Li X, Cronan JE, Kaufmann SH, Wilmanns M. 2006. The Mycobacterium tuberculosis LipB enzyme functions as a cysteine/lysine dyad acyltransferase. Proc Natl Acad Sci U S A 103:8662–8667. doi: 10.1073/pnas.0510436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim do J, Lee SJ, Kim HS, Kim KH, Lee HH, Yoon HJ, Suh SW. 2008. Structural basis of octanoic acid recognition by lipoate-protein ligase B. Proteins 70:1620–1625. doi: 10.1002/prot.21843. [DOI] [PubMed] [Google Scholar]
- 87.Christensen QH, Cronan JE. 2010. Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry 49:10024–10036. doi: 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Doolittle RF. 1994. Convergent evolution: the need to be explicit. Trends Biochem Sci 19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 89.Omelchenko M, Galperin M, Wolf Y, Koonin E. 2010. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol Direct 5:31. doi: 10.1186/1745-6150-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. 2011. A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol Microbiol 80:350–363. doi: 10.1111/j.1365-2958.2011.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Christensen QH, Hagar JA, O'Riordan MX, Cronan JE. 2011. A complex lipoate utilization pathway in Listeria monocytogenes. J Biol Chem 286:31447–31456. doi: 10.1074/jbc.M111.273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parry RJ. 1977. Biosynthesis of lipoic acid. 1. Incorporation of specifically tritiated octanoic acid into lipoic acid. J Am Chem Soc 99:6464–6466. doi: 10.1021/ja00461a061. [DOI] [Google Scholar]
- 93.Parry RJ. 1978. Biosynthesis of lipoic acid. 2. Stereochemistry of sulfur introduction as C-6 of octanoic acid. J Am Chem Soc 100:5243–5244. doi: 10.1021/ja00484a073. [DOI] [Google Scholar]
- 94.White RH. 1980. Stoichiometry and stereochemistry of deuterium incorporated into fatty acids by cells of Escherichia coli grown on [methyl-2H3]acetate. Biochemistry 19:9–15. doi: 10.1021/bi00542a002. [DOI] [PubMed] [Google Scholar]
- 95.White RH. 1980. Stable isotope studies on the biosynthesis of lipoic acid in Escherichia coli. Biochemistry 19:15–19. doi: 10.1021/bi00542a003. [DOI] [PubMed] [Google Scholar]
- 96.White RH. 1980. Biosynthesis of lipoic acid: extent of incorporation of deuterated hydroxy- and thiooctanoic acids into lipoic acid. J Am Chem Soc 102:6605–6607. doi: 10.1021/ja00541a059. [DOI] [Google Scholar]
- 97.Beld J, Lee DJ, Burkart MD. 2015. Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Mol Biosyst 11:38–59. doi: 10.1039/C4MB00443D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE Jr, Marletta MA. 2000. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39:15166–15178. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- 99.Hayden MA, Huang I, Bussiere DE, Ashley GW. 1992. The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli. J Biol Chem 267:9512–9515. [PubMed] [Google Scholar]
- 100.Hayden MA, Huang IY, Iliopoulos G, Orozco M, Ashley GW. 1993. Biosynthesis of lipoic acid: characterization of the lipoic acid auxotrophs Escherichia coli W1485-lip2 and JRG33-lip9. Biochemistry 32:3778–3782. doi: 10.1021/bi00065a033. [DOI] [PubMed] [Google Scholar]
- 101.Nicolet Y, Drennan CL. 2004. AdoMet radical proteins—from structure to evolution—alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res 32:4015–4025. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Booker SJ. 2012. Radical SAM enzymes and radical enzymology. Biochim Biophys Acta 1824:1151–1153. doi: 10.1016/j.bbapap.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 103.Booker SJ, Grove TL. 2010. Mechanistic and functional versatility of radical SAM enzymes. F1000 Biol Rep 2:52. doi: 10.3410/B2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ifuku O, Kishimoto J, Haze S, Yanagi M, Fukushima S. 1992. Conversion of dethiobiotin to biotin in cell-free extracts of Escherichia coli. Biosci Biotechnol Biochem 56:1780–1785. doi: 10.1271/bbb.56.1780. [DOI] [PubMed] [Google Scholar]
- 105.Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. 2004. Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry 43:11770–11781. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 106.Harmer JE, Hiscox MJ, Dinis PC, Fox SJ, Iliopoulos A, Hussey JE, Sandy J, Van Beek FT, Essex JW, Roach PL. 2014. Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem J 464:123–133. doi: 10.1042/BJ20140895. [DOI] [PubMed] [Google Scholar]
- 107.Cicchillo RM, Booker SJ. 2005. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J Am Chem Soc 127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- 108.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. 2004. Lipoyl synthase requires two equivalents of S-adenosyl-l-methionine to synthesize one equivalent of lipoic acid. Biochemistry 43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 109.Douglas P, Kriek M, Bryant P, Roach PL. 2006. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl 45:5197–5199. doi: 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- 110.Bryant P, Kriek M, Wood RJ, Roach PL. 2006. The activity of a thermostable lipoyl synthase from Sulfolobus solfataricus with a synthetic octanoyl substrate. Anal Biochem 351:44–49. doi: 10.1016/j.ab.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 111.Lanz ND, Pandelia ME, Kakar ES, Lee KH, Krebs C, Booker SJ. 2014. Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase. Biochemistry 53:4557–4572. doi: 10.1021/bi500432r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lanz ND, Rectenwald JM, Wang B, Kakar ES, Laremore TN, Booker SJ, Silakov A. 2015. Characterization of a radical intermediate in lipoyl cofactor biosynthesis. J Am Chem Soc 137:13216–13219. doi: 10.1021/jacs.5b04387. [DOI] [PubMed] [Google Scholar]
- 113.Booker SJ, Cicchillo RM, Grove TL. 2007. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol 11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi-Rhee E, Cronan JE. 2005. Biotin synthase is catalytic in vivo, but catalysis engenders destruction of the protein. Chem Biol 12:461–468. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 115.Farrar CE, Siu KK, Howell PL, Jarrett JT. 2010. Biotin synthase exhibits burst kinetics and multiple turnovers in the absence of inhibition by products and product-related biomolecules. Biochemistry 49:9985–9996. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martin N, Lombardia E, Altabe SG, de Mendoza D, Mansilla MC. 2009. A lipA (yutB) mutant, encoding lipoic acid synthase, provides insight into the interplay between branched-chain and unsaturated fatty acid biosynthesis in Bacillus subtilis. J Bacteriol 191:7447–7455. doi: 10.1128/JB.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. 2011. A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol Microbiol 80:335–349. doi: 10.1111/j.1365-2958.2011.07597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clancy KW, Melvin JA, McCafferty DG. 2010. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers 94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Orlean P, Menon AK. 2007. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res 48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 121.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 122.Braakman R, Smith E. 2014. Metabolic evolution of a deep-branching hyperthermophilic chemoautotrophic bacterium. PLoS One 9:e87950. doi: 10.1371/journal.pone.0087950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hermes FA, Cronan JE. 2013. The role of the Saccharomyces cerevisiae lipoate protein ligase homologue, Lip3, in lipoic acid synthesis. Yeast 30:415–427. doi: 10.1002/yea.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tzagoloff A, Dieckmann CL. 1990. PET genes of Saccharomyces cerevisiae. Microbiol Rev 54:211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sulo P, Martin NC. 1993. Isolation and characterization of LIP5 A lipoate biosynthetic locus of Saccharomyces cerevisiae. J Biol Chem 268:17634–17639. [PubMed] [Google Scholar]
- 126.Marvin ME, Williams PH, Cashmore AM. 2001. The isolation and characterisation of a Saccharomyces cerevisiae gene (LIP2) involved in the attachment of lipoic acid groups to mitochondrial enzymes. FEMS Microbiol Lett 199:131–136. doi: 10.1111/j.1574-6968.2001.tb10663.x. [DOI] [PubMed] [Google Scholar]
- 127.Chen XJ. 1997. Cloning and characterization of the lipoyl-protein ligase gene LIPB from the yeast Kluyveromyces lactis: synergistic respiratory deficiency due to mutations in LIPB and mitochondrial F1-ATPase subunits. Mol Gen Genet 255:341–349. doi: 10.1007/s004380050505. [DOI] [PubMed] [Google Scholar]
- 128.Schonauer MS, Kastaniotis AJ, Kursu VA, Hiltunen JK, Dieckmann CL. 2009. Lipoic acid synthesis and attachment in yeast mitochondria. J Biol Chem 284:23234–23242. doi: 10.1074/jbc.M109.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schonauer MS, Kastaniotis AJ, Hiltunen JK, Dieckmann CL. 2008. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol Cell Biol 28:6646–6657. doi: 10.1128/MCB.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wada H, Shintani D, Ohlrogge J. 1997. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci U S A 94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wada M, Yasuno R, Jordan SW, Cronan JE Jr, Wada H. 2001. Lipoic acid metabolism in Arabidopsis thaliana: cloning and characterization of a cDNA encoding lipoyltransferase. Plant Cell Physiol 42:650–656. doi: 10.1093/pcp/pce081. [DOI] [PubMed] [Google Scholar]
- 132.Wada M, Yasuno R, Wada H. 2001. Identification of an Arabidopsis cDNA encoding a lipoyltransferase located in plastids. FEBS Lett 506:286–290. doi: 10.1016/S0014-5793(01)02932-5. [DOI] [PubMed] [Google Scholar]
- 133.Ewald R, Hoffmann C, Neuhaus E, Bauwe H. 2014. Two redundant octanoyltransferases and one obligatory lipoyl synthase provide protein-lipoylation autonomy to plastids of Arabidopsis. Plant Biol (Stuttg) 16:35–42. doi: 10.1111/plb.12028. [DOI] [PubMed] [Google Scholar]
- 134.Yasuno R, Wada H. 1998. Biosynthesis of lipoic acid in Arabidopsis: cloning and characterization of the cDNA for lipoic acid synthase. Plant Physiol 118:935–943. doi: 10.1104/pp.118.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yasuno R, Wada H. 2002. The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517:110–114. doi: 10.1016/S0014-5793(02)02589-9. [DOI] [PubMed] [Google Scholar]
- 136.Ewald R, Hoffmann C, Florian A, Neuhaus E, Fernie AR, Bauwe H. 2014. Lipoate-protein ligase and octanoyltransferase are essential for protein lipoylation in mitochondria of Arabidopsis. Plant Physiol 165:978–990. doi: 10.1104/pp.114.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang SG, Jeong HK, Lee E, Natarajan S. 2007. Characterization of a lipoate-protein ligase A gene of rice (Oryza sativa L.). Gene 393:53–61. doi: 10.1016/j.gene.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 138.Feng D, Witkowski A, Smith S. 2009. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem 284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith S, Witkowski A, Moghul A, Yoshinaga Y, Nefedov M, de Jong P, Feng D, Fong L, Tu Y, Hu Y, Young SG, Pham T, Cheung C, Katzman SM, Brand MD, Quinlan CL, Fens M, Kuypers F, Misquitta S, Griffey SM, Tran S, Gharib A, Knudsen J, Hannibal-Bach HK, Wang G, Larkin S, Thweatt J, Pasta S. 2012. Compromised mitochondrial fatty acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS One 7:e47196. doi: 10.1371/journal.pone.0047196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Witkowski A, Joshi AK, Smith S. 2007. Coupling of the de novo fatty acid biosynthesis and lipoylation pathways in mammalian mitochondria. J Biol Chem 282:14178–14185. doi: 10.1074/jbc.M701486200. [DOI] [PubMed] [Google Scholar]
- 141.Cronan JE, Fearnley IM, Walker JE. 2005. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett 579:4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 142.Vinothkumar KR, Zhu J, Hirst J. 2014. Architecture of mammalian respiratory complex I. Nature 515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yi X, Maeda N. 2005. Endogenous production of lipoic acid is essential for mouse development. Mol Cell Biol 25:8387–8392. doi: 10.1128/MCB.25.18.8387-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mayr JA, Zimmermann FA, Fauth C, Bergheim C, Meierhofer D, Radmayr D, Zschocke J, Koch J, Sperl W. 2011. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet 89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shay KP, Moreau RF, Smith EJ, Smith AR, Hagen TM. 2009. Alpha-lipoic acid as a dietary supplement: molecular mechanisms and therapeutic potential. Biochim Biophys Acta 1790:1149–1160. doi: 10.1016/j.bbagen.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Biewenga GP, Haenen GR, Bast A. 1997. The pharmacology of the antioxidant lipoic acid. Gen Pharmacol 29:315–331. doi: 10.1016/S0306-3623(96)00474-0. [DOI] [PubMed] [Google Scholar]
- 147.Bustamante J, Lodge JK, Marcocci L, Tritschler HJ, Packer L, Rihn BH. 1998. Alpha-lipoic acid in liver metabolism and disease. Free Radic Biol Med 24:1023–1039. doi: 10.1016/S0891-5849(97)00371-7. [DOI] [PubMed] [Google Scholar]
- 148.Roy S, Packer L. 1998. Redox regulation of cell functions by alpha-lipoate: biochemical and molecular aspects. Biofactors 7:263–267. doi: 10.1002/biof.5520070324. [DOI] [PubMed] [Google Scholar]
- 149.Brassier A, Ottolenghi C, Boutron A, Bertrand AM, Valmary-Degano S, Cervoni JP, Chretien D, Arnoux JB, Hubert L, Rabier D, Lacaille F, de Keyzer Y, Di Martino V, de Lonlay P. 2013. Dihydrolipoamide dehydrogenase deficiency: a still overlooked cause of recurrent acute liver failure and Reye-like syndrome. Mol Genet Metab 109:28–32. doi: 10.1016/j.ymgme.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 150.Baker PR II, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, Freckmann ML, Shih LY, Wasserstein M, Rasmussen MB, Lund AM, Procopis P, Cameron JM, Robinson BH, Brown GK, Brown RM, Compton AG, Dieckmann CL, Collard R, Coughlin CR II, Spector E, Wempe MF, Van Hove JL. 2014. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain 137:366–379. doi: 10.1093/brain/awt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. 2014. Lipoic acid biosynthesis defects. J Inherit Metab Dis 37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 152.Tsurusaki Y, Tanaka R, Shimada S, Shimojima K, Shiina M, Nakashima M, Saitsu H, Miyake N, Ogata K, Yamamoto T, Matsumoto N. 2015. Novel compound heterozygous LIAS mutations cause glycine encephalopathy. J Hum Genet 60:631–635. doi: 10.1038/jhg.2015.72. [DOI] [PubMed] [Google Scholar]
- 153.Soreze Y, Boutron A, Habarou F, Barnerias C, Nonnenmacher L, Delpech H, Mamoune A, Chretien D, Hubert L, Bole-Feysot C, Nitschke P, Correia I, Sardet C, Boddaert N, Hamel Y, Delahodde A, Ottolenghi C, de Lonlay P. 2013. Mutations in human lipoyltransferase gene LIPT1 cause a Leigh disease with secondary deficiency for pyruvate and alpha-ketoglutarate dehydrogenase. Orphanet J Rare Dis 8:192. doi: 10.1186/1750-1172-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tort F, Ferrer-Cortes X, Thio M, Navarro-Sastre A, Matalonga L, Quintana E, Bujan N, Arias A, Garcia-Villoria J, Acquaviva C, Vianey-Saban C, Artuch R, Garcia-Cazorla A, Briones P, Ribes A. 2014. Mutations in the lipoyltransferase LIPT1 gene cause a fatal disease associated with a specific lipoylation defect of the 2-ketoacid dehydrogenase complexes. Hum Mol Genet 23:1907–1915. doi: 10.1093/hmg/ddt585. [DOI] [PubMed] [Google Scholar]
- 155.Copley SD. 2014. An evolutionary perspective on protein moonlighting. Biochem Soc Trans 42:1684–1691. doi: 10.1042/BST20140245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jeffery CJ. 2015. Why study moonlighting proteins? Front Genet 6:211. doi: 10.3389/fgene.2015.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo M, Schimmel P. 2013. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Espinosa-Cantú A, Ascencio D, Barona-Gómez F, DeLuna A. 2015. Gene duplication and the evolution of moonlighting proteins. Front Genet 6:227 doi: 10.3389/fgene.2015.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Quinn J, Diamond AG, Masters AK, Brookfield DE, Wallis NG, Yeaman SJ. 1993. Expression and lipoylation in Escherichia coli of the inner lipoyl domain of the E2 component of the human pyruvate dehydrogenase complex. Biochem J 289:81–85. doi: 10.1042/bj2890081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fujiwara K, Okamura-Ikeda K, Motokawa Y. 1992. Expression of mature bovine H-protein of the glycine cleavage system in Escherichia coli and in vitro lipoylation of the apoform. J Biol Chem 267:20011–20016. [PubMed] [Google Scholar]
- 161.Bjorup P, Torres JL, Adlercreutz P, Clapes P. 1998. Reaction medium engineering in enzymatic peptide fragment condensation: synthesis of eledoisin and LH-RH. Bioorg Med Chem 6:891–901. doi: 10.1016/S0968-0896(98)00046-7. [DOI] [PubMed] [Google Scholar]
- 162.Reslow M, Adlercreutz P, Mattiasson B. 1988. The influence of water on protease-catalyzed peptide synthesis in acetonitrile/water mixtures. Eur J Biochem 177:313–318. doi: 10.1111/j.1432-1033.1988.tb14378.x. [DOI] [PubMed] [Google Scholar]
- 163.Morihara K. 1987. Using proteases in peptide synthesis. Trends Biotechnol 5:163–170. [Google Scholar]
- 164.Keeney KM, Stuckey JA, O'Riordan MX. 2007. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol 66:758–770. doi: 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]