Highlight
We provide improved biochemical assays for key carboxylases and decarboxylases involved in C3 and C4 photosynthesis. Together with leaf age-related changes in Rubisco activation, these measurements will improve photosynthetic modelling.
Key words: Carbamylation, carbon fixation, CO2-concentrating mechanism, photosynthesis, Rubisco, Rubisco activase.
Abstract
Plants operating C3 and C4 photosynthetic pathways exhibit differences in leaf anatomy and photosynthetic carbon fixation biochemistry. Fully understanding this underpinning biochemical variation is requisite to identifying solutions for improving photosynthetic efficiency and growth. Here we refine assay methods for accurately measuring the carboxylase and decarboxylase activities in C3 and C4 plant soluble protein. We show that differences in plant extract preparation and assay conditions are required to measure NADP-malic enzyme and phosphoenolpyruvate carboxylase (pH 8, Mg2+, 22 °C) and phosphoenolpyruvate carboxykinase (pH 7, >2mM Mn2+, no Mg2+) maximal activities accurately. We validate how the omission of MgCl2 during leaf protein extraction, lengthy (>1min) centrifugation times, and the use of non-pure ribulose-1,5-bisphosphate (RuBP) significantly underestimate Rubisco activation status. We show how Rubisco activation status varies with leaf ontogeny and is generally lower in mature C4 monocot leaves (45–60% activation) relative to C3 monocots (55–90% activation). Consistent with their >3-fold lower Rubisco contents, full Rubisco activation in soluble protein from C4 leaves (<5min) was faster than in C3 plant samples (<10min), with addition of Rubisco activase not required for full activation. We conclude that Rubisco inactivation in illuminated leaves primarily stems from RuBP binding to non-carbamylated enzyme, a state readily reversible by dilution during cellular protein extraction.
Introduction
Plants operating the C3 and C4 pathways contain differing biochemical and anatomical features that facilitate their climatic adaptation to cool-temperate and warm-tropical environments, respectively (Edwards et al., 2010). The rate-limiting CO2 fixation step common to both pathways is catalysed by the photosynthetic enzyme ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (Rubisco, EC 4.1.1.39). Fixation of CO2 to RuBP by Rubisco produces two molecules of 3-phosphoglycerate (PGA) that are cycled through the photosynthetic carbon reduction (PCR) cycle to produce triose-phosphates, the building blocks of carbohydrates needed for plant growth (Raines, 2003).
Rubisco is an imperfect catalyst that is remarkably slow (completing only 2–4 cycles s–1 in leaves) and can fix O2 instead of CO2, with oxygenation of RuBP leading to the production of 2-phosphoglycolate (PGly) whose recycling back to PGA by the photorespiratory pathway spans three cellular compartments and undesirably consumes energy and releases fixed CO2 (Bauwe et al., 2010). Overcoming the catalytic limitations of Rubisco by directed changes to the enzyme or concentrating CO2 around Rubisco to reduce the costs of photorespiration are ongoing bioengineering challenges (Parry et al., 2013; Price et al., 2013; Long et al., 2015). Current efforts to generate or discover plant Rubisco isoforms with joint improvements in specificity for CO2 as opposed to O2 (Sc/o) and carboxylation efficiency (defined as the maximum carboxylation rate (k cat c) divided by the K m for CO2 under ambient O2; K C 21%O2) have yet to yield success (Sharwood and Whitney, 2014), despite such Rubiscos existing in some non-green algae (Andrews and Whitney, 2003).
The evolution of C4 photosynthesis ~35–40 million years ago provided a natural solution to remedy the inefficiency of Rubisco (Sage et al., 2012). The anatomical separation of phosphoenolpyruvate carboxylase (PEPC) in mesophyll cells (MCs) and Rubisco in the bundle sheath cell (BSC) chloroplasts was accompanied by adaptation of biochemical CO2-concentrating mechanisms (CCMs) (Hatch, 1987; Kanai and Edwards, 1999). The C4 pathway involves the hydration by carbonic anhydrase of CO2 to HCO3 − which is combined with phosphoenolpyruvate (PEP) by phosphoenolpyruvate carboxylase (PEPC) into the 4C acid oxaloacetate (OAA) that is converted into malate or aspartate before diffusing into the BSCs where they are decarboxylated, raising the CO2 around Rubisco >4-fold higher than ambient CO2 (von Caemmerer, 2000; Sage, 2004). The 3C decarboxylation product, pyruvate, returns to the MCs for PEP regeneration by pyruvate phosphate dikinase (PPDK) at the cost of two ATP equivalents.
The ratio of the activity between the main carboxylases is a key determinant of the efficiency of the CCM and can be an indication of the CO2 supply to BSCs (von Caemmerer et al., 2014). On the one hand, PEPC activity is much higher than that of Rubisco (2- to 10-fold depending on plant species and environmental conditions) to enable the C4 acid gradient to build and facilitate the diffusion of the C4 acids into the BSCs. On the other hand, the PEPC:Rubisco ratio must be optimized to minimize CO2 leakage from the BSCs, leading to futile cycling involving the CCM (Kromdijk et al., 2008). Futile cycling of the CCM is energetically wasteful for the plant through use of ATP to regenerate PEP.
Based on the main decarboxyating enzymes, C4 plants can be grouped into three biochemical subtypes: NADP-malic enzyme (NADP-ME), NAD-malic enzyme (NAD-ME), and phosphoenolpyruvate carboxykinase (PEPCK) (Hatch, 1987; Kanai and Edwards, 1999). In addition, there is flexibility among some NADP-ME (e.g maize, sorghum, and Panicum antidotale) and NAD-ME (e.g. Cleome angustifolia, Bienertia sinuspersici, and Panicum coloratum) species which also harbour PEPCK (Furbank, 2011; Pinto et al., 2014; Koteyeva et al., 2015). The significance of the dual decarboxylation pathways is not yet fully understood, but evidence suggests that PEPCK allows flexibility to the decarboxylation pathway(s) that may be dependent on environmental cues (Bellasio and Griffiths, 2014; Sharwood et al., 2014). Assaying for PEPCK is often difficult because pure enzyme is required for assaying in the decarboxylase direction due to interference from other C4 pathway enzymes (Ashton et al., 1990). However, assaying for PEPCK activity in the carboxylase direction is also troublesome as PEPC can interfere with this assay, although variations in PEPCK activities can be corroborated by western blot analysis of PEPCK content (Sharwood et al., 2014). Accurately determining the level of maximum PEPCK activity is requisite to determine the level of flexibility in the decarboxylation pathways that may exist.
A conserved feature of Rubisco catalysis is the required priming (activation) of each catalytic site (E) located at the interface of adjoining paired 50kDa large subunits (L2) that arrange as (L2)4 tetrad cores and are capped at either end with tetrads of 15kDa small (S) subunits to form an ~520kDa L8S8 complex (Andersson and Backlund, 2008). Activation proceeds via the slow and reversible binding of non-substrate CO2 to the ε-amino group of a conserved L-subunit Lys201 producing a carbamate (EC) that is rapidly stabilized by Mg2+ to produce a tertiary complex (ECM) capable of RuBP binding and enolization and its subsequent carboxylation or oxygenation (Andersson, 2008). In vivo, the pool of inactive Rubisco comprises decarbamylated catalytic sites (E) and ECM complexes binding inhibitory sugar-phosphate molecules (ECMI) (Parry et al., 2008) (Fig. 1). Examples of these inhibitors include the catalytic misfire product xylulose 1,5-bisphosphate (XuBP) and the ‘nocturnal’ or ‘shade’ inhibitor 2'-carboxy-d-arabinitol 1-phosphate (CA1P) produced under low light and darkness (Gutteridge et al., 1986; Moore and Seemann, 1992; Pearce, 2006; Andralojc et al., 2012). Binding of RuBP to E also leads to the production of catalytically stalled ER complexes (Laing and Christeller, 1976; Jordan and Chollet, 1983). Release of these sugar-phosphate molecules is catalysed by Rubisco activase (RCA) via ATP hydrolysis (Parry et al., 2008; Mueller-Cajar et al., 2014). Following their RCA-facilitated release, the rebinding of XuBP and CA1P is prevented by the enzymes XuBPase (Bracher et al., 2015) and CA1Pase (Salvucci and Holbrook, 1989; Moore and Seemann, 1992). The enzyme CA1Pase is also able to metabolize the inhibitor pentadiulose-1,5-bisphosphate (PDBP; Andralojc et al., 2012), a relatively labile oxygenation byproduct whose inhibitory relevance in vivo remains indeterminate but is a significant contaminant of non-pure RuBP (Kane et al., 1998). Conditions that stimulate Rubisco inactivation include increasing temperature (increased XuBP and PDBP production), low illumination (stimulated CA1P synthesis), and elevated CO2 (possibly increases ER levels) (Crafts-Brandner and Salvucci, 2000; Salvucci and Crafts-Brandner, 2004; Kim and Portis, 2005; Parry et al., 2008).
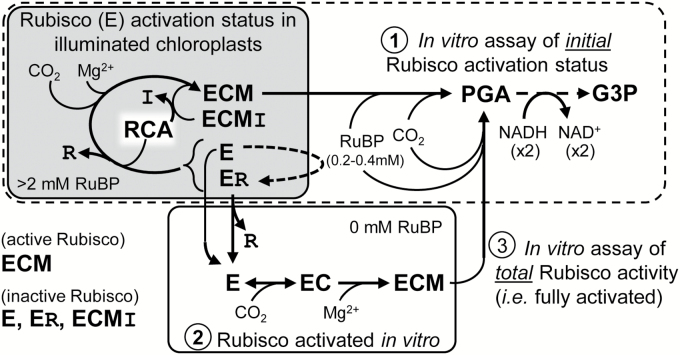
Fig. 1.
Summary of Rubisco activation status in vivo and modulation in vitro. Summary of the NADH-linked assay used to determine Rubisco activation status using rapidly extracted soluble protein from young, non-stressed leaves sampled during illumination [where inactive ECMI complexes containing the Rubisco inhibitors (I) CA1P or XuBP would be negligible]. See text for details of the three assay stages indicated. R, RuBP; RCA, Rubisco activase. Details of the NADH-linked assay are summarized in Supplementary Fig. S1 and Supplementary Table S1.
Extrapolating aspects of cellular biochemistry from leaf gas exchange measurements using available C3 and C4 photosynthesis models is highly reliant on accurately knowing the content and catalytic properties of the carboxylation and decarboxylation enzyme activities (Farquhar et al., 1980; Salvucci and Crafts-Brandner, 2004; Sharkey et al., 2007). Here we appraise and refine the methods for assaying PEPC, PEPCK, and Rubisco activity and activation status using NADH-linked spectrometric assays. We apply these refined assay methods to leaf extracts from C3 and C4 grasses to demonstrate their applicability in accurately measuring variations in the carboxylation/decarboxylation biochemistries of leaves of differing ontogeny.
Materials and methods
Plant seeds and growth conditions
Seeds for Panicum bisulcatum and Megathyrsus maximum were obtained from the Australian Plant Genetic Resources Information System (QLD, Australia) and Queensland Agricultural Seeds Pty. Ltd (Toowoomba, Australia), while the seeds for tobacco (Nicotiana tabacum, cv Petit havana), maize (Zea mays cv Kelvedon Glory), and wheat (Triticum aestivum cv Y70) were sourced locally. The seeds were sown in 2–5 litre pots of commercial self-fertilizing potting mix at 5–7 d intervals to obtain plants of different ages to sample simultaneously. The plants were grown in a glasshouse at set 28/22 oC day/night temperatures under natural illumination during November and December in Canberra, Australia. Plants were watered regularly, with the addition of Hoaglands nutrients to mature plants every 2 d.
Leaf harvesting, protein extraction, and protein assay
Samples of known area (0.3 or 0.5cm2) were harvested using brass cork borers (Met-App Metalware, Melbourne) from different aged leaves on the same day, 5–7h into the light period. The samples were rapidly frozen in liquid nitrogen before storing at –80 °C. For assays of Rubisco activity and content as well as the activity of PEPC and NADP-ME, the soluble leaf protein was extracted using ice-cold 2ml glass homogenizers (Wheaton) into 0.5–1ml of ice cold, N2-sparged extraction buffer [50mM EPPS-NaOH, pH 8.0, 0.5mM EDTA, 2mM DTT, 1% (v/v) plant protease inhibitor cocktail (Sigma-Aldrich), and 1% (w/v) polyvinylpolypyrrolidone (PVPP)] containing 0, 2, 5, or 10mM MgCl2. The lysate was rapidly centrifuged for 0.5, 2, or 5min (16 000 g, 4 °C), and 10 µl of the soluble leaf protein was assayed for initial and total Rubisco activities (see below) and 50 µl to measure Rubisco content by [14C]CABP (2-C-carboxyarabinitol 1,5-bisphosphate) binding as described (Whitney et al., 2001). Protein content was measured against BSA standards using a Coomassie dye binding assay (Pierce). The leaf area extracted in 1ml of buffer was generally 0.3cm2 (T. aestivum), 0.5cm2 (N. tabacum), 0.6cm2 (P. bisulcatum), and 0.9cm2 (P. bisulcatum, M. maximus, and Z. mays)
Phosphoenolpyruvate carboxylase assay
Maximal PEPC activities were measured using an NADH-coupled assay as previously described (Ashton et al., 1990). Extraction was performed using the same extraction buffer as described for Rubisco containing 5mM MgCl2, and samples were incubated at room temperature or on ice for 0, 5, 10, and 20min before adding 10 µl of extract to initiate assays.
NADP-malic enzyme assay
Maximal NADP-ME activity was determined in a coupled NADP assay as previously described (Ashton et al., 1990; Pengelly et al., 2012). Briefly, NADP-ME activity in leaf extracts prepared as described above was assayed in 50mM Tricine-KOH pH 8.3, 5mM malic acid, initiating with 10mM MgCl2.
Phosphoenolpyruvate carboxykinase assay
The maximal activity of PEPCK was measured in the carboxylase direction using a method adapted from Chen et al. (2002) and Walker et al. (2002) as described by Sharwood et al. (2014) in an NADH-coupled assay as depicted in Fig. 2A. To remove interference with PEPC, MgCl2 was excluded from the extraction and assay buffers. PEPC background activity was determined by assaying for PEPC activity as above with and without MgCl2, with no PEPC activity observed when MgCl2 is omitted from the extraction and assay buffer (data not shown). For PEPCK assay, leaf discs were extracted in 50mM HEPES pH 7.0, 5mM DTT, 1% (w/v) PVPP, 2mM EDTA, 2mM MnCl2, and 0.05% Triton. PEPCK activity from leaf extracts was measured in assay buffer [50mM HEPES, pH 7.0, 4% mercaptoethanol (w/v), 100mM KCl, 90mM NaHCO3,1mM ADP, 2mM MnCl2, 0.14mM NADH, and malate dehydrogenase (MDH; 6U; 3.7 μl)] after the addition of 15mM PEP. An optimal PEP concentration was determined in separate assays titrated with 2.5–20mM PEP.
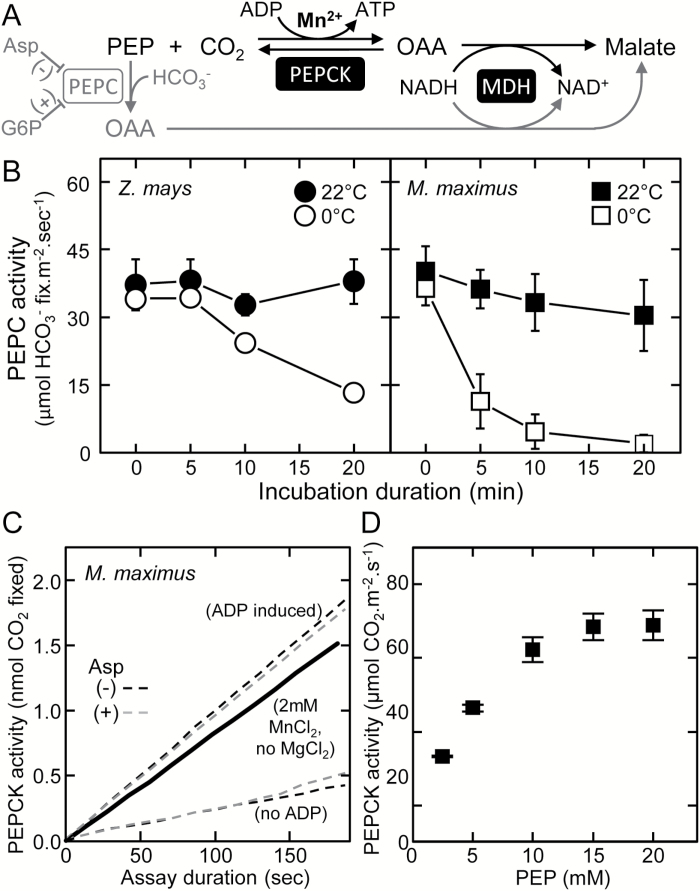
Fig. 2.
Optimizing the measurement of PEPC and PEPCK activities in leaf extract. (A) Summarizing the commonality of the malate dehydrogenase- (MDH) coupled, NADH oxidation-linked assay to quantify the carboxylation of phosphoenolpyruvate (PEP) into oxaloacetate (OAA) by PEP carboxykinase (PEPCK; in black) and PEP carboxylase (PEPC; in gray), an enzyme inhibited (–) by aspartate (Asp; Huber and Edwards, 1975) and activated (+) by glucose-6-phosphate (G6P). (B) Effect of storage temperature (room temperature, 22 °C, or ice, 0 °C) and time on PEPC activity in the soluble protein from young leaves from mature Z. mays and M. maximus plants (n=3 biological replicates ±SD; see leaves m3b and c3b in Fig. 5A for examples). (C) Representative assay of PEP carboxylation by PEPCK measured in leaf soluble protein extract using the no MgCl2 method of Sharwood et al. (2014) ( black line) and the modified ADP method of this study in assays with (gray dashed lines) or without (black dashed lines) 5mM aspartate (a PEPC inhibitor, A). (D) Response of PEPCK activity to [PEP] in M. maximus leaf soluble protein. To prevent PEPC interference ensure: 1) No MgCl2 in assay and extraction buffers, 2) MnCl2 up to 5mM in extraction and assay buffers, 3) pH of extraction and assay buffers <7.0, 4) No glucose-6-phosphate, and 5) include aspartate in assay buffer.
Rubisco activity assays
Rubisco activity was measured at 25 °C using an NADH-coupled enzyme assay (Kubien et al., 2011; Supplementary Fig. S1 at JXB online) with the rate of NADH oxidation monitored at 340nm using a diode array spectrophotometer (Agilent model 8453). The RuBP used in the assays was either that synthesized and purified as previously described (Kane et al., 1998) or commercially supplied (Sigma; R0878). Assays were performed in 1ml cuvettes containing 0.48ml of assay buffer [100mM EPPS-NaOH, pH 8.0, 10mM MgCl2, 0.2mM NADH, 20mM NaHCO3, 1mM ATP, pH 7.0, 5mM phosphocreatine, pH 7.0, and 4% (v/v) coupling enzymes] (see Supplementary Table S1). RuBP (0.4mM) was included in the cuvettes used to measure initial Rubisco activity, with 20 µl of soluble leaf protein sample added to start the assays. To measure total Rubisco activities, 20 µl of leaf protein was first activated for 10–15min in RuBP-free assay buffer before initiating Rubisco activity measurements by adding RuBP to 0.4mM. The Rubisco carboxylation rate was determined using the equation:
| (1) |
which uses the extinction coefficient of NADH (6.22×103 M−1 cm−1), the rate of change of absorbance at 340nm per minute (ΔOD340), and accounts for the four NADH molecules oxidized per RuBP carboxylated by Rubisco in the coupled assay (Supplementary Fig. S1). Substrate-saturated Rubisco carboxylase activity in the same leaf soluble protein was measured by 14CO2 fixation assays as described (Sharwood et al., 2008). The carboxylation turnover rate (k cat c) was determined from the Rubisco activity measured by either the NADH-coupled enzyme assay or the 14CO2 fixation assay divided by the Rubisco active site content in the assay as quantified by [14C]CABP binding (Ruuska et al., 1998). Time course measurements of Rubisco activity over 30min at 25 °C were undertaken to confirm the functional integrity of Rubisco in the leaf protein extracts.
Rubisco activase purification and assay
Tobacco RCA was expressed and purified from Escherichia coli as described (Baker et al., 2005). Two-stage assays similar to that described by Barta et al. (2011) were used to assess if sugar-phosphate Rubisco inhibition in tobacco leaf protein extracts influenced measurements of total Rubisco activity. In the first assay stage, 50 μl of leaf extract was incubated in a 0.5ml final volume with 80 µg ml–1 RCA (or BSA in the RCA-free controls) for 2min at 25 °C in ATPase assay buffer [100mM EPPS, pH 8.0, 20mM KCl, 5mM MgCl2, 6% (w/v) polyethylene glycol (PEG) (mol. wt 3350Da)], 2mM PEP, 0.2mM NADH, 2mM ATP, 1% (v/v) of a pyruvate kinase/lactate dehydrogenase mixture (PK, 745U ml−1; LDH, 906U ml−1, Sigma-Aldrich). In the second assay stage, 100 μl of the RCA leaf or BSA leaf protein reactions were added to the NADH-linked Rubisco assays and the total activities compared. Control assays examining tobacco RCA activation of purified tobacco Rubisco ER complexes are described in Supplementary Fig. S2.
Statistical analysis
Statistical analysis was carried out using one- or two-way ANOVA (Statistica, StatSoft Inc. OK, USA). Means were grouped using a post-hoc Tukey test.
Results and Discussion
Optimizing the assay of PEPC and PEPCK activities in total soluble leaf protein
Measuring PEPC and PEPCK activities separately in soluble leaf protein extracts using NADH-linked spectrophotometric assays is complicated by their common requirement for substrate PEP (Fig. 2A) particularly in assays containing Mg2+ and Mn2+ at physiological concentrations (Muhaidat and McKown, 2013). Determining maximal PEPCK activity in soluble leaf protein extracts is typically achieved by assaying the decarboxylation reaction of PEPCK, requiring purified protein free of other C4 enzymes such as PEPC (Ashton et al., 1990; Chen et al., 2002). In contrast, measures of PEPC rates free of PEPCK activity can easily be made by omission of ADP from the assay (Fig. 2A). As in vitro measures of PEPC are sensitive to low temperature storage (Hatch and Oliver, 1978), we examined the PEPC activity in soluble leaf protein from Z. mays and M. maximus stored either at room temperature (22 °C) or on ice (0 °C) (Fig. 2B). When incubated at 22 °C for 20min, there was little or no loss of PEPC activity evident in replica leaf samples from either M. maximus or Z. mays. In contrast, storage of the leaf protein extracts on ice significantly reduced PEPC activities (measured at 25 °C), particularly in M. maximus where >65% of PEPC activity was lost after 5min at 0 °C (Fig. 2B). As a result of its sensitivity to low temperature, all assays of PEPC activity were performed on rapidly extracted (homogenized in <0.5min and centrifuged for 0.5min at 4 oC) leaf soluble protein without storage on ice.
In C4 plants with phosphoenolpyruvate carboxykinase (PCK) physiology (e.g. M. maximus), PEPCK is the dominant decarboxylase enzyme that utilizes ATP hydrolysis during its reversible decarboxylation of OAA (Fig. 2A). In contrast to the alkali pH 8 preference of PEPC (Greenway et al., 1978), the activity of PEPCK is optimal at pH 7 and 80% lower at pH 8.0 (Ray and Black, 1976; Pierre et al., 2004). To minimize, possibly preclude, PEPC activity, the extraction and measurement of PEPCK carboxylase activity was undertaken at pH 7.0 with Mg2+ (required for PEPC activity) omitted and replaced with 2mM Mn2+, a PEPCK cofactor (Fig. 2A). Under these conditions, stable rates of PEPCK were obtained in assays initiated by the addition of PEP (solid line, Fig. 2C) (Sharwood et al., 2014).
Alternative PEPCK analyses were undertaken where the assays were initiated with ADP, not PEP (Fig. 2C, black dashed line). Omission of ADP in control assays produced background rates of apparent PEPC activity (Fig. 2C, lower black dashed line); however, the addition of 5mM aspartate (Fig. 2C, grey dashed line) or 5mM glucose-6-phosphate (and MgCl2) which inhibit and stimulate PEPC activity, respectively (Fig. 2A), had a negligible effect on the measured activities. This suggests that the background ‘no ADP’ PEP carboxylase activities observed arise from PEPCK activity that is utilizing residual ADP in the soluble protein extract. Consequently, we propose that extracting leaf protein in pH 7.0 buffer with no MgCl2 and assaying in an MnCl2-containing buffer at the same pH is sufficient to measure PEPCK activity with little or no contaminating PEPC activity.
Assays containing 2.5–25mM PEP were used to determine that a saturating concentration of 15mM PEP was required for maximal PEPCK activity in soluble leaf protein from 1.3mm2 of M. maximus leaf tissue (Fig. 2D). This saturating PEP concentration is 7-fold higher than the K m for PEP measured for M. maximus PEPCK (Chen et al., 2002) and was the concentration used in all subsequent PEPCK assays.
Measuring Rubisco carbamylation status
The carboxylase-limiting component of the C3 photosynthesis models stemming from those derived by Farquhar et al. (1980) are typically used to derive estimates of V c,max [in units of µmol CO2 fixed m− 2 s−1, that equate to the product of Rubisco sites (µmol CO2 fixed m2) and k cat c (s−1)]. This measure is extrapolated from the response of the CO2 assimilation rate (A) with increasing CO2 measured by leaf gas exchange, and relies heavily on the temperature response measurements of k cat c, K c, the K m for O2 (K o), and CO2/O2 specificity (Sc/o) made for tobacco Rubisco (Sharkey et al., 2007). The universal suitability of these parameters is now in question given the substantial variation observed in the temperature response of these parameters among plant Rubiscos (Walker et al., 2013; Boyd et al., 2015; Perdomo et al., 2015). Moreover, differences in V c,max are primarily attributed to variations in Rubisco content and generally overlook differences in the activation status of Rubisco in vivo, despite its critical influence on estimates of V c,max and in vivo determined k cat c (Bernacchi et al., 2001; Salvucci and Crafts-Brandner, 2004).
As indicated in Fig. 1, within an illuminated leaf chloroplast the catalytic sites of Rubisco (shown as E) are primarily CO2–Mg2+ activated (ECM) and capable of RuBP catalysis. Binding of inhibitory XuBP or, in darkened leaves, CA1P to ECM produces catalytically inactive ECMI complexes whose activation involves RCA-catalysed dissociation of the sugar-phosphate ligands which are then degraded by substrate-specific enzymes (Jordan and Chollet, 1983; Gutteridge et al., 1986; Edmondson et al., 1990; Bracher et al., 2015). Thus the most abundant form of Rubisco inhibition in illuminated chloroplasts is probably the binding of RuBP to non-carbamylated Rubisco (ER) that renders the catalytic site inactive (Jordan and Chollet, 1983).
The NADH-linked spectrophotometric and 14CO2 fixation in vitro assays typically used to measure Rubisco activation status involve three stages (Fig. 1). The first stage involves the rapid measure of Rubisco activity in rapidly extracted soluble leaf protein. This measures the ‘initial’ Rubisco activity. Saturating amounts of RuBP are included to prevent ECM formation from the ER complexes and ensure that the assays are not RuBP limited (Laing and Christeller, 1976). Replica samples of the leaf protein extracts are then allowed to activate fully by incubating in buffer lacking RuBP but containing saturating CO2 and Mg2+. During the second stage, the inconsequential RuBP levels in the extract enable its dissociation from the ER complexes to allow ECM formation. The third stage measures the ‘total’ Rubisco activity rate. In samples from darkened or stressed leaves, the formation of ECMI complexes can cause significant underestimation of the ‘total’ activities (Parry et al., 1997, 2002; Carmo-Silva et al., 2010) a consideration we sought to avoid in this study by sampling healthy, naturally illuminated glasshouse-grown plant material 6–7h into the photoperiod.
Optimizing leaf protein extraction for measuring Rubisco activation status
Analyses undertaken using tobacco leaves highlighted the requirement for speed and inclusion of ~5mM Mg2+ during leaf protein extraction for accurate measurements of initial Rubisco activity (Fig. 3A). As shown in Fig. 1, formation of ECM is initiated by the slow and reversible carbamylation of Lys201 in the catalytic site of E followed by the rapid binding of Mg2+. To avoid Rubisco carbamylation occurring during extraction, the leaf soluble protein is extracted in N2-sparged (i.e. CO2-free) buffer. It was hypothesized that the exclusion of MgCl2 during extraction would undesirably produce EC complexes from which the activating CO2 would dissociate to form inactive E. Indeed, the omission of MgCl2 in the CO2-free extraction buffer led to significantly lower measurements of initial Rubisco activity (70% of maximum activity) after just 0.5min relative to those extracted with 2–10mM MgCl2 (82–85% of maximum activity; Fig. 3A). Extending the extraction (centrifugation) period to 5min reduced all initial Rubisco activity measurements, more so in those extracted without MgCl2 (35% of maximum activity) compared with those extracted with MgCl2 (55–60% of maximum activity). In all treatments, very similar total activity rates were attained, indicating that the varying extraction conditions did not compromise Rubisco integrity (Fig. 3A, circles). These findings caution against the omission of MgCl2 when assaying for initial activities. Furthermore, potential inaccuracies are incurred with extended centrifugation times of >0.5–1min following extraction, emphasizing the need for rapid leaf protein extraction and assay.
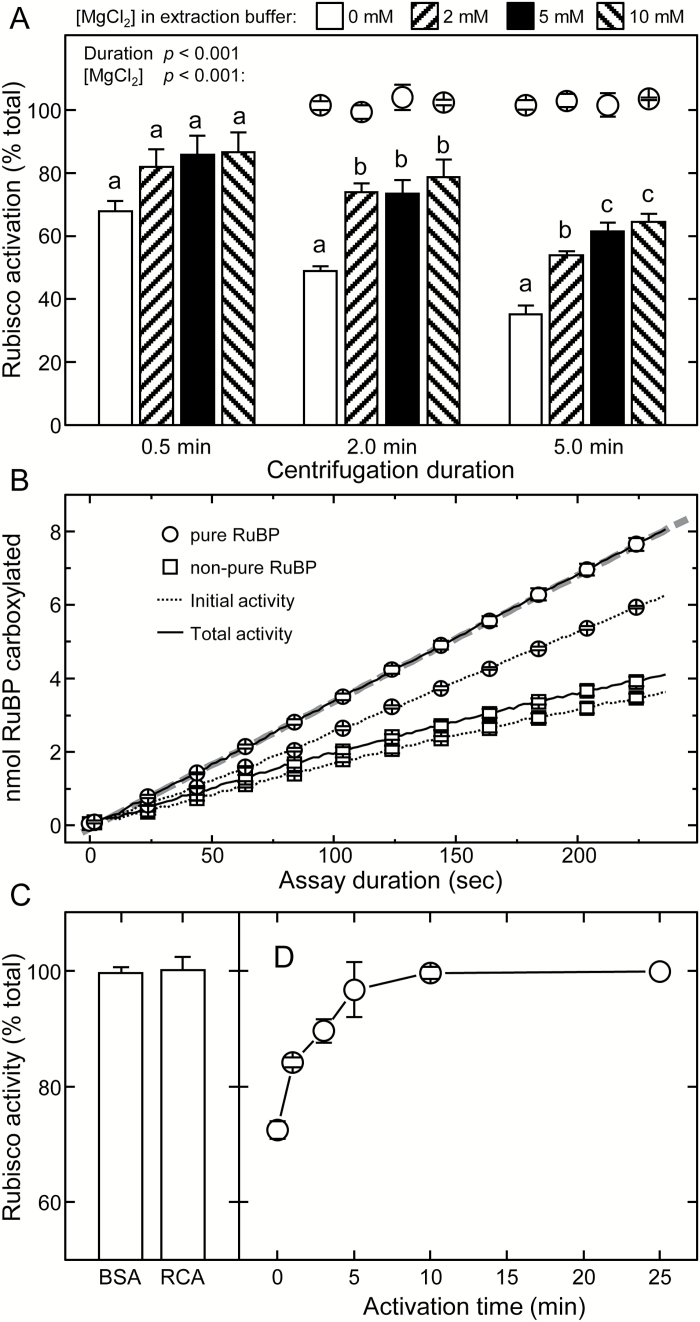
Fig. 3.
Evaluating the experimental methodology for measuring leaf Rubisco activation status. (A) Appraising how MgCl2 inclusion and quickness of soluble leaf protein extraction influences Rubisco activation quantification. NADH-linked assays were performed on N2-frozen replica (n=5) tobacco leaf discs (0.5cm2) taken from a young, nearly fully expanded upper canopy leaf (15cm in diameter) and stored at –80 °C for up to 3 months without effect on recoverable activity. Circles indicate the total activities measured after 10min activation relative to the 0.5min centrifuged sample (B) Representative NADH-linked spectrophotometric measures of initial (dashed lines) and total (solid lines) Rubisco activities made using low purity commercial RuBP (squares) or that purified according to Kane et al. (1998) (circles). Rates correspond to protein from 0.9mm2 of leaf with a Rubisco active site concentration in each assay of ~34.4nM (i.e. a k cat of 2.2s−1). (C) Incubation of leaf protein extract for 2min at 25 °C with 80 μg ml–1 of either BSA or purified tobacco Rubisco activase (RCA) had no effect on the measured rates of fully activated Rubisco, while the same treatment re-activated >80% of inhibited ER complexes formed using a comparable concentration of purified tobacco Rubisco (Supplementary Fig. S2). (D) Change in the activity status of Rubisco in tobacco soluble protein activated at 25 °C for up to 25min. Data in (C) and (D) are the averages (±SE) from analyses with three separate leaf samples expressed as a percentage of the total activities measured after 10min (C) and 25min (D) activation. For (A), the significance level (P) for the [MgCl2] and centrifugation duration factors are shown. Letters indicate the ranking (lowest=a) of means within each centrifugation duration using a post-hoc Tukey test. Values followed by the same letter are not significantly different at the 5% level (P>0.05).
The importance of using purified RuBP
Using pure RuBP devoid of inhibitory impurities such as PDBP is critical for accurately measuring Rubisco catalysis in vitro (Kane et al., 1998; Andralojc et al., 2012). As observed by Scales et al. (2014), by using pure RuBP the measured rates of initial and total activity remain relatively linear over a 4min assay period (Fig. 3B). This (i) indicates that insignificant levels of the catalytic misfire products XuBP or PDBP are produced over the assay period; (ii) indicates that Rubisco activity in the assay is stable; and (iii) confirms the observations of Laing and Christeller (1976) that inclusion of saturating RuBP levels (0.2–0.4mM) in the ‘initial’ assays prevents Rubisco activation. In contrast, Rubisco activities measured in the same extracts using a commercial source of non-pure RuBP (Fig. 3B, squares) declined rapidly, presumably due to PDBP contamination (Andralojc et al., 2012). The use of non-pure RuBP further compromised the quantification of k cat c (2.2s−1 with pure RuBP versus 1.3s−1 with non-pure RuBP) and the calculated activation status of Rubisco (75±2% with pure RuBP versus 83±3% with non-pure RuBP). Use of non-pure RuBP should therefore be avoided in order to avoid underestimating the carbamylation status (Rowland-Bamford et al., 1991; Anwaruzzaman et al., 1996; Ruuska et al., 2004; Sulpice et al., 2007) and activity (Rintamäki et al., 1988; Uemura et al., 2000) of Rubisco.
Insignificant levels of ECMI complexes accumulate in illuminated, non-stressed leaves
The k cat c value of 2.2s−1 was reproducibly quantified for tobacco Rubisco using the NADH-coupled assay. This is ~30% lower than those typically measured by 14CO2 fixation assays (Sharwood et al., 2008; Whitney et al., 2011). Incubation of the leaf protein extracts with purified tobacco RCA or BSA (control) showed no difference in the measured total Rubisco activities (Fig. 3C). In corresponding control assays, the RCA treatment was able to reactivate ER inhibited Rubisco fully over 10min (Supplementary Fig. S2). This indicates that the lower k cat c was not due to residual ECMI complexes in the leaf protein extract. To confirm this, the same tobacco soluble leaf protein was used to quantify k cat c by the 14CO2 assay method of Sharwood et al. (2008). As indicated in Table 1, the expected k cat c of 3.1s−1 for tobacco Rubisco was obtained by the 14CO2 assay. This finding questions the accuracy of the NADH-coupled assay for quantifying Rubisco carboxylase activity, a deficiency also evident in the comparative measurements made by Lilley and Walker (1974). Indeed, published k cat c values determined by the NADH-coupled assay for cyanobacteria (Emlyn-Jones et al., 2006) and plant (Pearce and Andrews, 2003) Rubisco are also 20–25% lower than those measured by 14CO2 fixation (Whitney et al., 1999; Mueller-Cajar and Whitney, 2008). To ensure that the differences were not due to components in the leaf extracts interfering with the coupling enzymes, comparative assays were undertaken in triplicate (technical repeats) using tobacco Rubisco purified by ion exchange chromatography (see the legend to Supplementary Fig. S2). Again, the k cat c values determined by the NADH-coupled assay (1.9±0.2s−1) were 30% lower than that quantified by 14CO2 fixation assays (2.7±0.1s−1). This suggests that substrate limitations for one or more of the enzymes in the NADH-linked assay limit its potential for accurately quantifying k cat c, possibly the rate of 3-PGA reduction (Lilley and Walker, 1974).
Table 1.
Comparative values of Rubisco k cat c at 25 °C quantified by the NADH-linked and 14CO2 fixation assays
| Plant species | Photosynthetic biochemistry | k cat c (±SE s−1) | Significance (P) | |||
|---|---|---|---|---|---|---|
| NADH-linked assay | 14CO2 assay | |||||
| Tobacco | C3 | 2.15±0.02 b | n=24 | 3.07±0.06 b | n=23 | <0.001 |
| P. bisulcatum | C3 | 1.79±0.03 a | n=22 | 2.72±0.11 a | n=7 | <0.001 |
| M. maximus | C4-PCK | 3.85±0.09 e | n=9 | 5.17±0.17 d | n=10 | <0.001 |
| T. aestivum | C3 | 2.70±0.06 c | n=9 | 3.59±0.04 c | n=6 | <0.001 |
| Z. mays | C4-NADP ME | 3.70±0.10 d | n=9 | 5.46±0.10 d | n=6 | <0.001 |
Values (means ±SE) obtained using NADH-linked and 14CO2 assays were compared by one-way ANOVA, and the significance level (P) is shown.
Species’ means obtained by each of the assay types were ranked separately using a post-hoc Tukey test. Values followed by the same letter are not significantly different at the 5% level (P>0.05).
n=number of leaf protein samples (biological replicates) analyzed.
The need for measuring Rubisco activation rate and stability over time
Given that the NADH-coupled assay allows for the continued ‘real-time’ monitoring of both initial and total Rubisco activity (Fig. 3B), it was used to examine the activation rate and stability of Rubisco activity at 25 °C in the soluble protein of leaves from the C3 species P. bisulcatum and T. aestivum (wheat), and the C4 species M. maximus and Z. mays. Linear rates of initial and total Rubisco activities were reproducibly found for each sample (Fig. 4A), with the rates of NADH oxidation significantly lower in the C4 samples due to their low Rubisco contents. As shown in Fig. 4A, the k cat c for both C4 Rubiscos were seen to be higher than those of their C3 counterparts when the activities were corrected for Rubisco content (quantified by [14C]CABP binding).
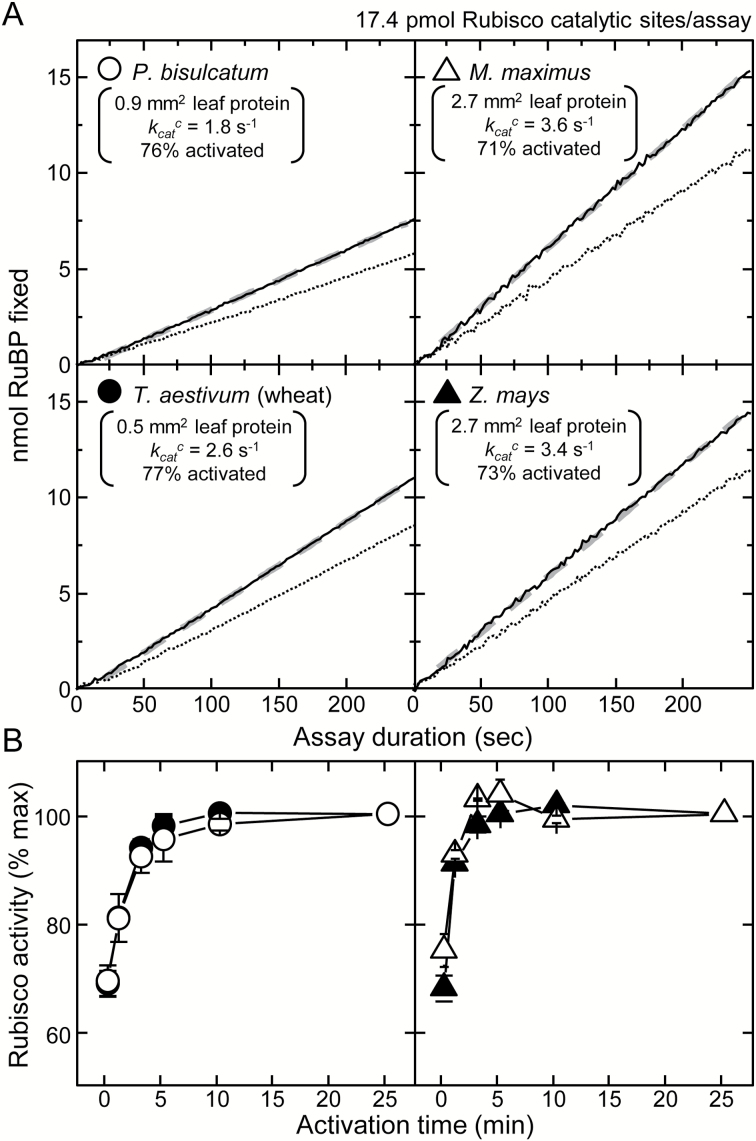
Fig. 4.
Variations in Rubisco content and activation status during leaf ontogeny can account for variations in the in vivo estimates of V c max. (A) Representative NADH-linked assay data showing the linear RuBP carboxylation rates in assays of initial (dashed lines) and total (solid lines, after 10–15min activation) Rubsico activities at 25 °C using soluble leaf protein from both C3 (P. bisulcatum, T. aestivum) and C4 (M. maximus, Z. mays) monocotyledon species. Shown are details of the calculated Rubisco activation status (% of maximum), the derived carboxylation rates (k cat c, quantified from the slope of the fitted linear regression, gray dashed line, divided by Rubisco content quantified by [14C]CABP binding) and the area of leaf protein required to attain the 17.4 pmol Rubsico catalytic sites used to normalize the plotted data to highlight the variations in k cat c between each species (see also Table 1). (B) Response of Rubisco activation and activity in the soluble leaf protein of each species following incubation at 25 °C. Shown is the average (±SE) of analyses from three separate leaf samples for each species expressed as a percentage of the maximum activities measured after 25min activation.
Like tobacco, full activation of Rubisco in the soluble protein extracted from wheat and P. bisulcatum required 10min incubation at 25 ºC (Fig. 4B, circles). In contrast, full activation of Rubisco in the M. maximus and Z. mays leaf protein required only 3–5min (Fig. 4B). Whether the faster rate of Rubisco activation in both C4 species arises from a lower RuBP binding affinity remains a subject for future investigation. Nevertheless, variation in the time needed to activate Rubisco fully in leaf protein extracts questions whether shorter incubation times (e.g. 3min) are sufficient to evaluate Rubisco activation status accurately and extrapolate ECMI levels (Parry et al., 1997, 2002; Carmo-Silva et al., 2010; Galmes et al., 2011; Scales et al., 2014).
The k cat c of C4 plant Rubisco exceeds that of C3 Rubisco
The k cat c values of Rubisco from the C4 species examined in this study were significantly faster relative to that of each C3 plant Rubisco (Table 1). Notably the NADH-coupled assay measures of k cat c were again 30–35% lower than corresponding k cat c measurements made using 14CO2 fixation assays (Table 1). Statistical ranking of the catalytic speed indicated that the C3 Rubisco from P. bisulcatum is slower than that of tobacco, with wheat outperforming both (P<0.001). In contrast to its ancestral P. bisulcatum Rubisco, the k cat c for M. maximus Rubisco is ~2-fold higher but similar to the k cat c of maize Rubisco despite originating from different biochemical subtypes and evolutionary origins (Table 1).
How do PEPC, Rubisco content, and activation status vary with leaf age?
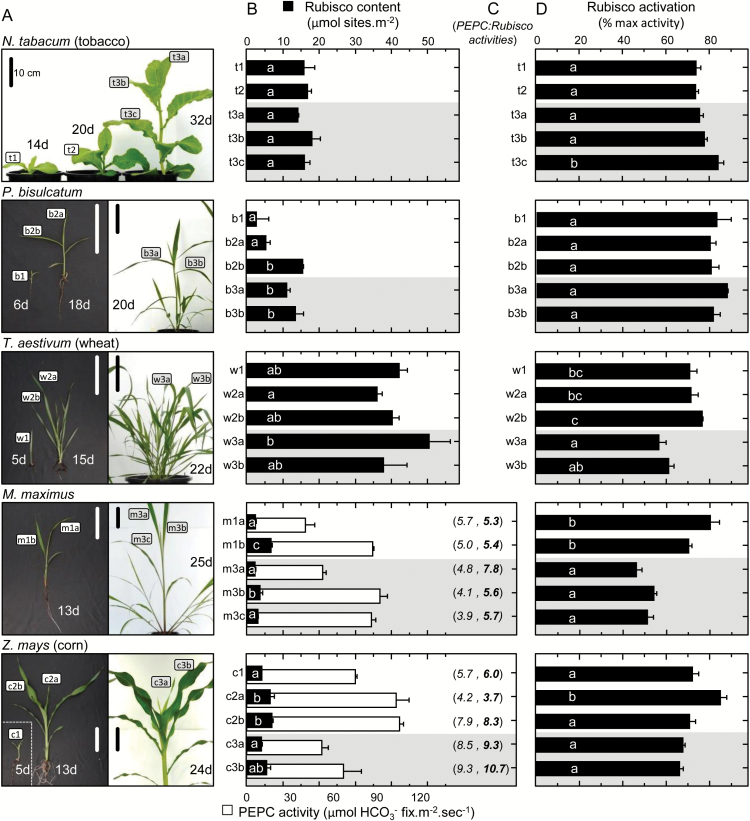
Rubisco comprises a significant but variable N investment in plant leaves. In tobacco, wheat, and rice, Rubisco comprises 20–30% of the leaf N, which is equivalent to 30–60% of the leaf soluble protein (Evans, 1989; Makino et al., 2003; Whitney et al., 2011), while in C4 plants it is 5–10% (Ghannoum et al., 2005). The lower Rubisco requirement of C4 plants stems from their CCM that enables them to operate under near saturating CO2 concentration, which has facilitated the evolution of increased Rubisco k cat c (Ghannoum et al., 2005). Consistent with these findings, the variation in Rubisco content with leaf ontogeny and at different locations in the canopy of M. maximus and maize (Fig. 5A) was 3- to 25-fold lower than that measured in C3 species (tobacco, P. bisulcatum, and wheat, Fig. 5B). Among the C3 plants, significantly higher levels of Rubisco were measured in wheat relative to tobacco and P. bisulcatum, with the latter grass showing the greatest variation in Rubisco content (per leaf area) in the leaves from juvenile and mature plants.
Fig. 5.
Variation in PEPC activity, Rubisco content, and Rubisco activation status with leaf ontogeny and development in C3 and C4 plants. (A) Pictures showing the leaves at differing stages of ontogeny and plant development (as labeled) that were analyzed for (B) Rubisco content (black bars, determined by [14C]CABP binding) and PEPC activity (white bars). (C) For both C4 species, the corresponding PEPC:Rubisco activity ratios are shown in parenthess; the first value is determined from the rates measured using the NADH-linked assays (in italics) and the second value (in bold) takes into account the higher Rubisco k cat c values quantified using 14CO2 assays (Table 1) and (D) the activation status of Rubisco in each leaf analyzed. The age of each plant (days, d) post-cotyledon emergence is indicated with the scale bar=10cm. All data are averages (±SD) of n=3 leaf discs taken from each leaf (or for the juvenile samples b1, c1, and w1, from replica plantlets). Regions shaded gray in (B) and (D) indicates data for leaves sampled from more mature plants. For (B) and (D), letters indicate the ranking (lowest=a) of means within each species using a post-hoc Tukey test. Values followed by the same letter are not significantly different at the 5% level (P>0.05). The levels of Rubisco measured correlate with those previously measured in the leaves of tobacco (Whitney et al., 2011), P. bisulcatum (Pinto et al., 2014), M. maximus (Pinto et al., 2016), and maize (Sharwood et al., 2014).
The variation in Rubisco with leaf ontogeny evident in both C4 species was somewhat mirrored by differences in their PEPC activities (Fig. 5B), resulting in similar PEPC:Rubisco activity ratios of ~3.9–5.7 in M. maximus that were more varied in maize (4.2–7.9 in juvenile plant leaves and 8.5–9.3 in the young leaves of exponentially growing plants; Fig. 5C). This ratio is typically used as an indication of the CO2 supply to the CCM in C4 plants and is normally in balance to minimize leakage of fixed CO2 (von Caemmerer, 2000; von Caemmerer et al., 2014). In both M. maximus and Z. mays, the PEPC:Rubisco ratio tended to increase during ontogeny, in particular when the ratio is adjusted with regard to differences in Rubisco activation status (Fig. 5C). The higher Rubisco:PEPC ratio in mature Z. mays leaves relative to M. maximus may arise from their varying C4 biochemistry and/or evolutionary origin, a consideration beyond the objectives of this study.
Despite being produced in lower abundance, the activation status of C4 Rubisco was similar or lower than that measured in the C3 species. Similarly, low levels of Rubisco activation (~45–55%) have been measured in other C4 species (von Caemmerer et al., 2005; Carmo-Silva et al., 2010) that correlate with those measured in mature M. maximus and Z. mays leaves (Fig. 5D). Higher Rubisco activation levels (~70–80%) were measured in the juvenile C4 plant leaves. These levels matched those measured in tobacco and P. bisulcatum, where little variation in Rubisco activation status was found among the leaves sampled. In contrast, significant variation in Rubisco activation status was observed among the upper wheat panicle leaves, where Rubisco activation was significantly lower than those sampled from juvenile plants (Fig. 5D).
The level of variation in Rubisco content and activation status with leaf ontogeny identified within this explorative study using plants grown under non-stress conditions emphasizes the importance of determining these parameters to compare meaningfully values of V c,max derived by extrapolation from leaf gas exchange (A–Ci curves) for different biological samples. As demonstrated by Whitney and Sharwood (2014), quantifying the leaf Rubisco content is best achieved using the [14C]CABP binding methods as densitometry methods following PAGE separation of Rubisco are highly imprecise, unless appropriately calibrated. Accurate quantification of Rubisco site content in the in vitro assays of Rubisco activity are also critical for quantifying k cat c. This parameter also provides a number of quality checks as reduced measures of k cat c provide a useful indicator of reduced leaf sample viability (as found if ultra-cold temperatures are not maintained during transfer and storage at –80 °C) and incomplete activation (e.g. insufficient activation time and/or presence of significant levels of ECMI complexes in the sample).
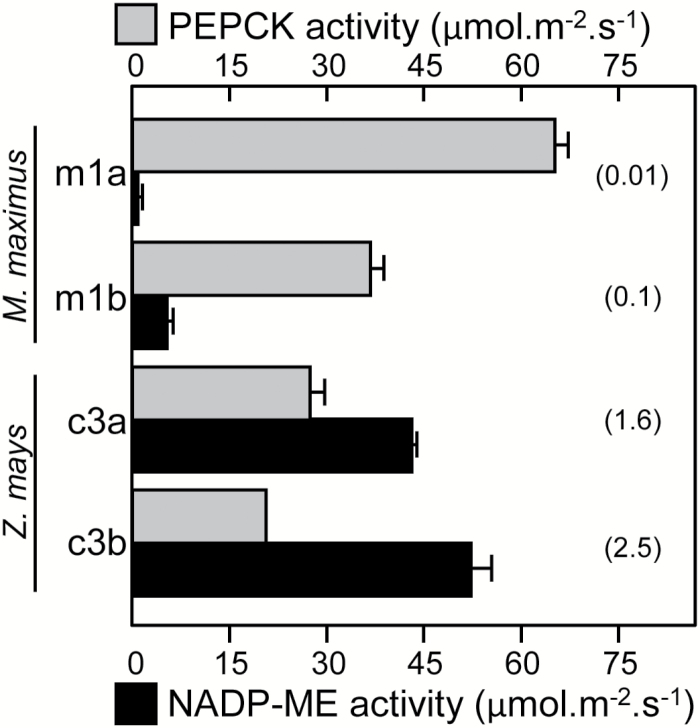
Variation in NADP-ME and PEPCK activities in Z. mays and M. maximus.
Different aged leaves sampled from mature M. maximus and Z. mays plants were analysed for maximal NADP-ME and PEPCK activities (Fig. 6). Higher PEPCK activities were measured in the younger leaves from both species, with as much as 2-fold higher activity measured in the Z. mays samples. Conversely, NADP-ME levels were characteristically >10-fold higher in Z. mays, consistent with its C4-NADP-ME photosynthetic biochemistry. While the low NADP-ME activity in M. maximum probably arose from an anaplerotic reaction, the significance of the PEPCK activity in Z. mays is not yet fully understood. Prior analysis of Z. mays exposed to salinity stress and shade treatments showed that there is plasticity in PEPCK contents and activity (Sharwood et al., 2014). This suggests that the PEPCK decarboxylation pathway may serve a role in responding to stressful environmental cues (Bellasio and Griffiths, 2014).
Fig. 6.
Maximal activity of the decarboxylases in C4 grasses. Comparison of PEPCK (gray) and NADP-ME (black) activities in differing aged Z. mays and M. maximus leaves (n=3, ± SE). The leaves analyzed are shown in Fig. 4A. The PEPCK:NADP-ME ratio activities are indicated in parentheses.
Conclusion
In this study, we demonstrate the need for carefully considering the experimental requirements needed to measure accurately, and reproducibly, the activity of key carboxylase and decarboxylase enzymes that are commonly used to evaluate physiological and biochemical parameters between plant samples. Of particular relevance to C3 and C4 photosynthetic modelling studies is how Rubisco content and activation can vary significantly with leaf ontogeny, in particular in C4 plants where Rubisco activation appears characteristically low. Here we show that full Rubisco activity is recoverable in vitro without the need for RCA when extracted with no RuBP. We therefore propose that Rubisco inactivation in the chloroplasts of non-stressed, illuminated leaves is primarily attributable to ER complex formation. Removing Rubisco inhibitors using Na2SO4 and PEG treatments (Parry et al., 1997, 2002; Carmo-Silva et al., 2010; Galmes et al., 2011; Scales et al., 2014) that can potentially harm recoverable activity might therefore be unnecessary using the in vitro assay conditions described in this study.
As summarized in Supplementary Fig. S3, we identified the core requirements for measuring Rubisco activation status (fast extraction, include ~5mM MgCl2, use pure RuBP, activate for 10min), PEPC (pH 8, 22 °C post-extraction), PEPCK (pH 7, >2mM Mn2+ no Mg2+, 15mM PEP), and NADP-ME activities using NADH-linked assays. We highlight how an unresolved limitation in the NADH linked assay underestimates Rubisco k cat c by >20%. We also emphasize the advantage of quantifying Rubisco by [14C]CABP binding to normalize Rubisco activities per active site (i.e. k cat c) as it serves as a quality control indicator of sample integrity and full Rubisco activation. Understandably, the assay and extraction conditions used in this study probably need optimization for other plant samples where additives and conditions (pH, temperature) are required to sustain, or promote, enzyme activities (Supplementary Fig. S3). As shown here by the differing assay requirements of PEPC and PEPCK, this optimization should also assess the compatibility of additives on the activity of each enzyme measured.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Details for the preparation and storage of coupling enzymes used in the NADH-linked spectrophotometric assay of Rubisco activity.
Figure S1. Overview of the NADH-linked enzyme-coupled spectrophotometric assay for measuring Rubisco activity.
Figure S2. Time-dependent activation in vitro of inhibited tobacco Rubisco–RuBP (ER) complexes by RCA.
Figure S3. Core requirements for measuring Rubisco activation status, PEPC, PEPCK, and NADP-ME activities.
Acknowledgements
This research was funded by the Australian Research Council which spans the following grants: DE130101760 (RES), DP120101603 (OG, SMW), and CE140100015 (OG, SMW).
References
- Andersson I. 2008. Catalysis and regulation in Rubisco. Journal of Experimental Botany 59, 1555–1568. [DOI] [PubMed] [Google Scholar]
- Andersson I, Backlund A. 2008. Structure and function of Rubisco. Plant Physiology and Biochemistry 46, 275–291. [DOI] [PubMed] [Google Scholar]
- Andralojc PJ, Madgwick PJ, Tao Y, et al. 2012. 2-Carboxy-d-arabinitol 1-phosphate (CA1P) phosphatase: evidence for a wider role in plant Rubisco regulation. Biochemical Journal 442, 733–742. [DOI] [PubMed] [Google Scholar]
- Andrews TJ, Whitney SM. 2003. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Archives of Biochemistry and Biophysics 414, 159–169. [DOI] [PubMed] [Google Scholar]
- Anwaruzzaman, Nakano Y, Yokota A. 1996. Different location in dark-adapted leaves of Phaseolus vulgaris of ribulose-1,5-bisphosphate carboxylase/oxygenase and 2-carboxyarabinitol 1-phosphate. FEBS Letters 388, 223–227. [DOI] [PubMed] [Google Scholar]
- Ashton AR, Burnell JN, Furbank RT, Jenkins CLD, Hatch MD. 1990. Enzymes of C4 photosynthesis. In: Lea PJ, ed. Methods in Plant Biochemistry , Vol. 3 London: Academic Press, 39–71. [Google Scholar]
- Baker RT, Catanzariti AM, Karunasekara Y, Soboleva TA, Sharwood R, Whitney S, Board PG. 2005. Using deubiquitylating enzymes as research tools. Methods in Enzymology 398, 540–554. [DOI] [PubMed] [Google Scholar]
- Barta C, Carmo-Silva AE, Salvucci ME. 2011. Rubisco activase activity assays. Methods in Molecular Biology 684, 375–382. [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. 2010. Photorespiration: players, partners and origin. Trends in Plant Science 15, 330–336. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Griffiths H. 2014. The operation of two decarboxylases (NADP-ME and PEPCK), transamination and partitioning of C4 metabolic processes between mesophylll and bundle sheath cells allows light capture to be balanced for the maize C4 pathway. Plant Physiology 164, 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi CJ, Singsaas EL, Pimentel C, Portis AR, Long SP. 2001. Improved temperature response functions for models of Rubisco-limited photosynthesis. Plant, Cell and Environment 24, 253–259. [Google Scholar]
- Boyd RA, Gandin A, Cousins AB. 2015. Temperature response of C4 photosynthesis: Biochemical analysis of Rubisco, phosphoenolpyruvate carboxylase and carbonic anhydrase in Setaria viridis . Plant Physiology 169, 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, Sharma A, Starling-Windhof A, Hartl FU, Hayer-Hartl M. 2015. Degradation of potent Rubisco inhibitor by selective sugar phosphatase. Nature Plants 1, 14002. [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Keys AJ, Andralojc PJ, Powers SJ, Arrabaca MC, Parry MA. 2010. Rubisco activities, properties, and regulation in three different C4 grasses under drought. Journal of Experimental Botany 61, 2355–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Walker RP, Acheson RM, Leegood RC. 2002. Phosphoenolpyruvate carboxykinase assayed at physiological concentrations of metal ions has a high affinity for CO2 . Plant Physiology 128, 160–164. [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Salvucci ME. 2000. Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2 . Proceedings of the National Academy of Sciences, USA 97, 13430–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DL, Badger MR, Andrews TJ. 1990. Slow inactivation of ribulosebisphosphate carboxylase during catalysis is caused by accumulation of a slow, tight-binding inhibitor at the catalytic site. Plant Physiology 93, 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Stromberg CA, et al. 2010. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328, 587–591. [DOI] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. 2006. RbcX can function as a Rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant and Cell Physiology 47, 1630–1640. [DOI] [PubMed] [Google Scholar]
- Evans JR. 1989. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78, 9–19. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA. 1980. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90. [DOI] [PubMed] [Google Scholar]
- Furbank RT. 2011. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? Journal of Experimental Botany 62, 3103–3108. [DOI] [PubMed] [Google Scholar]
- Galmes J, Ribas-Carbo M, Medrano H, Flexas J. 2011. Rubisco activity in Mediterranean species is regulated by the chloroplastic CO2 concentration under water stress. Journal of Experimental Botany 62, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Chow WS, Andrews TJ, Conroy JP, von Caemmerer S. 2005. Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiology 137, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway H, Winter K, Luttge U. 1978. Phosphoenolpyruvate carboxylase during development of Crassulacean acid metabolism and during a diurnal cycle in Mesembryanthemum crystallinum . Journal of Experimental Botany 29, 547–559. [Google Scholar]
- Gutteridge S, Parry MAJ, Burton S, Keys AJ, Mudd A, Feeney J, Servaites JC, Pierce J. 1986. A nocturnal inhibitor of carboxylation in leaves. Nature 324, 274–276. [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Hatch MD, Oliver IR. 1978. Activation and inactivation of phosphoenolpyruvate carboxylase in leaf extracts from C4 species. Australian Journal of Plant Physiology 5, 571–580. [Google Scholar]
- Jordan DB, Chollet R. 1983. Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. Journal of BIological Chemistry 258, 13752–13758. [PubMed] [Google Scholar]
- Kanai R, Edwards GE. 1999. The biochemistry of C4 photosynthesis. In: Rowan FS, Russell KM, eds. C4 plant biology . San Diego: Academic Press, 49–87. [Google Scholar]
- Kane HJ, Wilkin JM, Portis AR, Andrews TJ. 1998. Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiology 117, 1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Portis AR. 2005. Temperature dependence of photosynthesis in Arabidopsis plants with modifications in Rubisco activase and membrane fluidity. Plant and Cell Physiology 46, 522–530. [DOI] [PubMed] [Google Scholar]
- Koteyeva NK, Voznesenskaya EV, Edwards GE. 2015. An assessment of the capacity for phosphoenolpyruvate carboxykinase to contribute to C4 photosynthesis. Plant Science 235, 70–80. [DOI] [PubMed] [Google Scholar]
- Kromdijk J, Schepers HE, Albanito F, Fitton N, Carroll F, Jones MB, Finnan J, Lanigan GJ, Griffiths H. 2008. Bundle sheath leakiness and light limitation during C4 leaf and canopy CO2 uptake. Plant Physiology 148, 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, Brown CM, Kane HJ. 2011. Quantifying the amount and activity of Rubisco in leaves. Methods in Molecular Biology 684, 349–362. [DOI] [PubMed] [Google Scholar]
- Laing WA, Christeller JT. 1976. A model for the kinetics of activation and catalysis of ribulose 1,5-bisphosphate carboxylase. Biochemical Journal 159, 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley RM, Walker DA. 1974. An improved spectrophotometric assay for ribulosebisphosphate carboxylase. Biochimica et Biophysica Acta 358, 226–229. [DOI] [PubMed] [Google Scholar]
- Long Stephen P, Marshall-Colon A, Zhu X-G. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. [DOI] [PubMed] [Google Scholar]
- Makino A, Sakuma H, Sudo E, Mae T. 2003. Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant and Cell Physiology 44, 952–956. [DOI] [PubMed] [Google Scholar]
- Moore BD, Seemann JR. 1992. Metabolism of 2'-carboxyarabinitol in leaves. Plant Physiology 99, 1551–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Bracher A. 2014. Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosynthesis Research 119, 191–201. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Whitney SM. 2008. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli . Biochemical Journal 414, 205–214. [DOI] [PubMed] [Google Scholar]
- Muhaidat R, McKown AD. 2013. Significant involvement of PEP-CK in carbon assimilation of C4 eudicots. Annals of Botany 111, 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Khan S, Lea PJ, Keys AJ. 2002. Rubisco activity: effects of drought stress. Annals of Botany 89, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Parmar S, Keys AJ, Habash D, Paul MJ, Alred R, Quick WP, Servaites JC. 1997. Regulation of Rubisco by inhibitors in the light. Plant, Cell and Environment 20, 528–534. [Google Scholar]
- Parry MAJ, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. 2013. Rubisco activity and regulation as targets for crop improvement. Journal of Experiemental Botany 64, 717–730. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Madgwick PJ, Carmo-Silva AE, Andralojc PJ. 2008. Rubisco regulation: a role for inhibitors. Journal of Experimental Botany 59, 1569–1580. [DOI] [PubMed] [Google Scholar]
- Pearce FG. 2006. Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochemical Journal 399, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce FG, Andrews TJ. 2003. The relationship between side reactions and slow inhibition of ribulose-bisphosphate carboxylase revealed by a loop 6 mutant of the tobacco enzyme. Journal of Biological Chemistry 278, 32526–32536. [DOI] [PubMed] [Google Scholar]
- Pengelly JJL, Tan J, Furbank RT, von Caemmerer S. 2012. Antisense reduction of NADP-malic enzyme in Flaveria bidentis reduces flow of CO2 through the C4 cycle. Plant Physiology 160, 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo JA, Cavanagh AP, Kubien DS, Galmés J. 2015. Temperature dependence of in vitro Rubisco kinetics in species of Flaveria with different photosynthetic mechanisms. Photosynthesis Research 124, 67–75. [DOI] [PubMed] [Google Scholar]
- Pierre J-N, Prieto J-L, Gadal P, Vidal J. 2004. In situ C4 phosphoenolpyruvate carboxylase activity and kinetic properties in isolated Digitaria sanguinalis mesophyll cells. Photosynthesis Research 79, 349–355. [DOI] [PubMed] [Google Scholar]
- Pinto H, Powell JR, Sharwood RE, Tissue DT, Ghannoum O. 2016. Variations in nitrogen use efficiency reflect the biochemical subtype while variations in water use efficiency reflect the evolutionary lineage of C4 grasses at inter-glacial CO2 . Plant, Cell and Environment 39, 514–526. [DOI] [PubMed] [Google Scholar]
- Pinto H, Sharwood RE, Tissue DT, Ghannoum O. 2014. Photosynthesis of C3, C3–C4, and C4 grasses at glacial CO2 . Journal of Experimental Botany 65, 3669–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Pengelly JJL, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. 2013. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. Journal of Experimental Botany 64, 753–768. [DOI] [PubMed] [Google Scholar]
- Raines CA. 2003. The Calvin cycle revisited. Photosynthesis Research 75, 1–10. [DOI] [PubMed] [Google Scholar]
- Ray TB, Black CC. 1976. Characterization of phosphoenolpyruvate carboxykinase from Panicum maximum . Plant Physiology 58, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamäki E, Keys AJ, Parry MAJ. 1988. Comparison of the specific activity of ribulose-1,5-bis-phosphate carboxylase-oxygenase from some C3 and C4 plants. Physiologia Plantarum 74, 326–331. [Google Scholar]
- Rowland-Bamford AJ, Baker JT, Allen LH, Bowes G. 1991. Acclimation of rice to changing atmospheric carbon dioxide concentration. Plant, Cell and Environment 14, 577–583. [Google Scholar]
- Ruuska S, Andrews TJ, Badger MR, Hudson GS, Laisk A, Price GD, Caemmerer Sv. 1998. The interplay between limiting processes in C3 photosynthesis studied by rapid-response gas exchange using transgenic tobacco impaired in photosynthesis. Functional Plant Biology 25, 859–870. [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. 2004. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiology 136, 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis New Phytologist 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. 2012. Photorespiration and the evolution of C4 photosynthesis. Annual Reviews of Plant Biology 63, 19–47. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ. 2004. Inhibition of photosynthesis by heat stress: the activation state of Rubisco as a limiting factor in photosynthesis. Physiologia Plantarum 120, 179–186. [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Holbrook GP. 1989. Purification and properties of 2-carboxy-d-arabinitol 1- phosphatase. Plant Physiology 90, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales JC, Parry MAJ, Salvucci ME. 2014. A non-radioactive method for measuring Rubisco activase activity in the presence of variable ATP: ADP ratios, including modifications for measuring the activity and activation state of Rubisco. Photosynthesis Research 119, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL. 2007. Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant, Cell and Environment 30, 1035–1040. [DOI] [PubMed] [Google Scholar]
- Sharwood R, von Caemmerer S, Maliga P, Whitney S. 2008. The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiology 146, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Sonawane BV, Ghannoum O. 2014. Photosynthetic flexibility in maize exposed to salinity and shade. Journal of Experimental Botany 65, 3715–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Whitney SM. 2014. Correlating Rubisco catalytic and sequence diversity within C3 plants with changes in atmospheric CO2 concentrations. Plant, Cell and Environment 37, 1981–1984. [DOI] [PubMed] [Google Scholar]
- Sulpice R, Tschoep H, Von Korf M, Bussis D, Usadel B, Hohne M, Witucka-Wall H, Altmann T, Stitt M, Gibon Y. 2007. Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell & Environment 30, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Uemura K, Shibata N, Anwaruzzaman, Fujiwara M, Higuchi T, Kobayashi H, Kai Y, Yokota A. 2000. The role of structural intersubunit microheterogeneity in the regulation of the activity in hysteresis of ribulose 1, 5-bisphosphate carboxylase/oxygenase. Journal of Biochemistry 128, 591–599. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S. 2000. Biochemical models of leaf photosynthesis. Melbourne: CSIRO Publishing. [Google Scholar]
- von Caemmerer S, Ghannoum O, Pengelly JJL, Cousins AB. 2014. Carbon isotope discrimination as a tool to explore C4 photosynthesis. Journal of Experimental Botany 65, 3459–3470. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Hendrickson L, Quinn V, Vella N, Millgate AG, Furbank RT. 2005. Reductions of Rubisco activase by antisense RNA in the C4 plant Flaveria bidentis reduces Rubisco carbamylation and leaf photosynthesis. Plant Physiology 137, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B, Ariza LS, Kaines S, Badger MR, Cousins AB. 2013. Temperature response of in vivo Rubisco kinetics and mesophyll conductance in Arabidopsis thaliana: comparisons to Nicotiana tabacum . Plant, Cell and Environment 36, 2108–2119. [DOI] [PubMed] [Google Scholar]
- Walker RP, Chen ZH, Acheson RM, Leegood RC. 2002. Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea grass. Plant Physiology 128, 165–172. [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ. 2001. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. The Plant Journal 26, 535–547. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE. 2014. Plastid transformation for Rubisco engineering and protocols for assessing expression. Methods in Molecular Biology 1132, 245–262. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, Galmés J. 2011. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) carboxylation rate in Flaveria . Proceedings of the National Academy of Sciences, USA 108, 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ. 1999. Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiology 121, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.