Highlight
Complex I dysfunction causes reorganization of cellular respiration and affects metabolic processes in mitochondria, plastids, peroxisomes, and other cellular compartments with drastic consequences for growth and development.
Key words: Arabidopsis thaliana, carbonic anhydrase, complex I, mitochondrial metabolism, photosynthesis, proteomics, respiratory chain.
Abstract
The mitochondrial NADH dehydrogenase complex (complex I) is of particular importance for the respiratory chain in mitochondria. It is the major electron entry site for the mitochondrial electron transport chain (mETC) and therefore of great significance for mitochondrial ATP generation. We recently described an Arabidopsis thaliana double-mutant lacking the genes encoding the carbonic anhydrases CA1 and CA2, which both form part of a plant-specific ‘carbonic anhydrase domain’ of mitochondrial complex I. The mutant lacks complex I completely. Here we report extended analyses for systematically characterizing the proteome of the ca1ca2 mutant. Using various proteomic tools, we show that lack of complex I causes reorganization of the cellular respiration system. Reduced electron entry into the respiratory chain at the first segment of the mETC leads to induction of complexes II and IV as well as alternative oxidase. Increased electron entry at later segments of the mETC requires an increase in oxidation of organic substrates. This is reflected by higher abundance of proteins involved in glycolysis, the tricarboxylic acid cycle and branched-chain amino acid catabolism. Proteins involved in the light reaction of photosynthesis, the Calvin cycle, tetrapyrrole biosynthesis, and photorespiration are clearly reduced, contributing to the significant delay in growth and development of the double-mutant. Finally, enzymes involved in defense against reactive oxygen species and stress symptoms are much induced. These together with previously reported insights into the function of plant complex I, which were obtained by analysing other complex I mutants, are integrated in order to comprehensively describe ‘life without complex I’.
Introduction
Cellular respiration is the fundamental ATP generating process common to most eukaryotes. Mitochondria carry out the final steps of this process and efficiently generate ATP through oxidative phosphorylation (OXPHOS). The mitochondrial OXPHOS system consists of five inner mitochondrial membrane-embedded protein complexes – the four respiratory chain protein complexes (complexes I–IV) and the ATP synthase complex (complex V) – and two mobile electron transporters (ubiquinone and cytochrome c).
The OXPHOS complexes catalyse the electron transfer from NADH or FADH2 to molecular oxygen as the terminal electron acceptor. Electrons are inserted into the mitochondrial electron transport chain (mETC) via NADH generated by glycolysis, the TCA cycle (additionally electrons come from FADH2), and other catabolic processes such as the photorespiration pathway in plants. As first proposed by Peter Mitchell (1961), electron transport at the mETC is coupled to the translocation of protons from the mitochondrial matrix into the intermembrane space. This creates an electrochemical gradient across the inner mitochondrial membrane that results in a proton motive force. Complex V can use this proton gradient to generate ATP, which finally is exported and used for driving energy-demanding processes.
The NADH dehydrogenase complex (complex I) is of special importance for the OXPHOS system because it is the main site for electron insertion into the mETC and can provide up to 40% of the protons for mitochondrial ATP formation (Hunte et al., 2010; Braun et al., 2014). The structure of complex I has been investigated in Escherichia coli, Thermus thermophilus, Bos taurus, and Yarrowia lipolytica (Morgan and Sazanov, 2008; Baradaran et al., 2013; Vinothkumar et al., 2014; Zickermann et al., 2015). Its L-like shape, which originates from two orthogonally arranged ‘arms’, is well conserved in these species. One arm is hydrophobic, embedded in the inner mitochondrial membrane, and termed the ‘membrane arm’. The second arm, which is designated the ‘peripheral arm’, is hydrophilic and is attached to the end of the membrane arm. It protrudes into the mitochondrial matrix. In plants, complex I contains an additional spherical domain, which is attached to the membrane arm at a central position on its matrix-exposed side (Dudkina et al., 2005; Sunderhaus et al., 2006).
Despite overall similarity in shape, complex I from prokaryotes is comparatively small and has a simple subunit composition. The 14 subunits present in the 550kDa complex I of E. coli constitute the ‘minimal set’ of subunits (Weidner et al., 1993; Yagi and Matsuno-Yagi, 2003; Sazanov, 2007). Complex I of eukaryotes is nearly twice as large (1000kDa) (Friedrich and Böttcher, 2004; Brandt, 2006). In addition to the conserved ‘minimal set’ of subunits, eukaryotic complex I includes several accessory subunits (Friedrich, 2001). However, due to the occurrence of some lineage-specific accessory subunits the overall number of complex I subunits varies between different eukaryotes (e.g. Chlamydomonas reinhardtii: 42; Yarrowia lipolytica: 42; Bos taurus: 45; Arabidopsis thaliana: 49) (Cardol et al., 2004; Carroll et al., 2006; Angerer et al., 2011; Peters et al., 2013).
Plant complex I includes nine lineage-specific subunits (Braun et al., 2014), most notably a group of carbonic anhydrases located within the spherical extra domain attached to the membrane arm (Sunderhaus et al., 2006). Within this domain, which is designated the ‘carbonic anhydrase domain’ of plant complex I, three gamma-type carbonic anhydrase (γCA) proteins (γCA1, γCA2, γCA3) and two gamma-type carbonic anhydrase-like (γCAL) proteins (γCAL1 and γCAL2) are located (Perales et al., 2004; Klodmann et al., 2010). Since the γCA domain has a molecular mass of 85kDa it can only include three out of the five γCA/γCAL proteins at the same time (Klodmann et al., 2010). Based on investigations using γCA/γCAL mutants, six possible subunit arrangements have been suggested to occur (Fromm et al., 2016a): each γCA domain contains either the γCAL1 or the γCAL2 protein and additionally two copies of the γCA proteins (γCA1 or γCA2, or both but not γCA3).
The γCA/γCAL subunits have been found to be essential for the early steps of complex I assembly (Meyer et al., 2011; Li et al., 2013). Deletion of the gene encoding Arabidopsis γCA2 causes reduction of complex I (Perales et al., 2005). Deletion or downregulation of more than one gene encoding the γCA/γCAL subunits in Arabidopsis has drastic effects on the amount of complex I (Soto et al., 2015; Fromm et al., 2016b, c). Similarly, deletion of other genes encoding complex I subunits has been reported to cause significant reduction of complex I levels (e.g. deletion of the ndufs4, bir6, and slo3 genes; Kühn et al., 2015; Koprivova et al., 2010; Hsieh et al., 2015). In contrast to vertebrates, plants can withstand complex I reduction and dysfunction because they possess alternative NADH dehydrogenases in the mitochondria, which are important for mitochondrial functioning, especially if plants are growing under unfavorable growth conditions (Rasmusson et al., 2008).
A few Arabidopsis mutants have been described that completely lack mitochondrial complex I. In one mutant the NDUFV1 subunit (also called the 51-kDa subunit of complex I) is missing, causing complete absence of complex I. The mutant displayed increased fluxes through glycolysis and the TCA cycle (Kühn et al., 2015). In a second complex I-deficient Arabidopsis mutant the genes encoding γCA1 and γCA2 are absent. Mutant seeds do not germinate but can be rescued in the presence of high sucrose by an embryo rescue method (Fromm et al., 2016c). Growth of the resulting plants is extremely retarded. In the ca1ca2 mutant the cytochrome c oxidase complex is much induced, probably compensating for the loss of complex I with respect to proton translocation across the inner mitochondrial membrane (Fromm et al., 2016c).
Complex I-deficient mutants offer the unique opportunity to study ‘plant life without complex I’. Here we report a systematic proteomic comparison between ca1ca2 and wildtype Arabidopsis lines that is based on three distinct experimental strategies: 2D IEF/SDS PAGE, 2D BN/SDS PAGE, and a label-free quantitative shotgun proteome approach. MS analyses allowed in-depth insights into the molecular mechanisms compensating for the lack of complex I.
Material and Methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) lines used for this study were of the Columbia ecotype. The SALK_109391 (AT1G19580, ca1/ca1) and SALK_010194 (AT1G47260, ca2/ca2) mutant lines were obtained from The European Arabidopsis Stock Centre (NASC; Loughborough, UK). ca1 and ca2 single mutants were crossed to generate ca1ca2 double-mutants (Fromm et al., 2016c). Plants were grown on 0.5 Murashige and Skoog medium in climate chambers under the following conditions: 8h of light (120 µmol s−1 m−2)/16h of dark, 22 °C, 65% humidity, atmospheric CO2 concentrations. Homozygous ca1ca2 mutants were germinated and rescued by cultivation on 0.5 MS medium containing 3% (w/v) sucrose. After 6 weeks plants were transferred to soil and cultivation was continued under the same conditions without sucrose. Wildtype and double-mutant plants were harvested at the 10-rosette-leaf developmental stage, and leaves were used for proteomic analyses.
Cell cultures of Arabidopsis lines were established as described by May and Leaver (1993). Callus was maintained as suspension culture according to Sunderhaus et al. (2006).
Isolation of mitochondria
Mitochondria from cell culture were purified by differential centrifugation and Percoll density gradient centrifugation as described by Werhahn et al. (2001).
Protein gel electrophoresis procedures and staining procedures
One-dimensional Blue Native PAGE (1D BN PAGE) was performed according to Wittig et al. (2006). Mitochondrial membranes were solubilized by digitonin at a concentration of 5g g–1 mitochondrial protein (Eubel et al., 2003). For subsequent SDS PAGE, BN lanes with separated protein complexes were transferred horizontally onto SDS gels. Second-dimension PAGE was carried out as outlined previously (Wittig et al., 2006). Differential gel electrophoresis (DIGE), which is based on labeling of proteins with CyDyes before 2D BN/SDS PAGE, was carried out according to Peters and Braun (2012).
Two-dimensional IEF/SDS PAGE was carried out as described by Mihr and Braun (2003). For the IEF gel dimension, Immobiline DryStrip gels (24cm, non-linear gradient pH 3–11) were used. Focusing took place for 24h at 30 to 8000V using the Ettan IPGphor 3 system (GE Healthcare).
For the second gel dimension, IPG stripes were equilibrated for 15min with DTT (0.4g/40ml) and then 15min with iodoacetamide (1.0g/40ml). SDS PAGE was carried out using the High Performance Electrophoresis (HPE) FlatTop Tower-System (Serva Electrophoresis) using precast Tris-Glycine gels (12.5% polyacrylamide, 24 x 20cm).
Gels were fixed for 2h in 15% (v/v) ethanol, 10% (v/v) acetic acid and stained with Coomassie Brilliant Blue G250 (Neuhoff et al., 1985, 1990).
Comparative proteome analyses were based on gel triplicates and data evaluation using the Delta 2D software 4.3 (Decodon, Greifswald, Germany) according to Berth et al. (2007) and Lorenz et al. (2014).
Protein identification by mass spectrometry
Tryptic digestion of proteins and their identification by mass spectrometry (MS) were performed as described by Klodmann et al. (2010). Peptide separation was carried out by using the EASY-nLC System (Proxeon, Thermo Scientific, Bremen, Germany) and coupled MS analyses by using the MicrOTOF-Q II mass spectrometer (Bruker Bremen, Germany). MS primary data were evaluated using the Proteinscape software package (version 2.1, Bruker, Bremen, Germany), the Mascot Search Engine (Matrix Science, London, UK), the Arabidopsis protein database (www.arabidopsis.org; release TAIR10), and an updated version of a complex I database (Klodmann et al., 2010) that represents a subset of the Arabidopsis TAIR10 database. The threshold Mascot Score was set to 30 or 60 for proteins and 20 for peptides.
Label-free quantitative shotgun mass spectrometry
Sample preparation for ESI-MS/MS
Total proteins of five biological replicates of wt and ca1ca2 leaves were extracted. Then 50 µg of protein were solubilized in 2× sample buffer [4% (w/v) SDS, 125mM Tris-HCl (pH 6.8), 20% (v/v) glycerol, and 0.5% (w/v) bromophenol blue (BPB)] and loaded on a glycine/SDS gel [10% (w/v) acrylamide in stacking gel, 14% (w/v) in separation phase]. To concentrate proteins in a single band the gel run was stopped when the BPB front reached the end of the stacking gel. Gels were then Coommassie-stained and the protein bands were extracted and transferred into low-binding Eppendorf caps (Eppendorf, Wesseling-Berzdorf, Germany). After drying in a vacuum centrifuge (Eppendorf, Wesseling-Berzdorf, Germany), gel pieces were rehydrated in 200 µl reduction solution [20mM DTT, 0.1M ammonium bicarbonate (AmBiC)] for 30min at 56 °C. Afterwards, they were dehydrated again by addition of 200 µl acetonitrile (ACN) for 10min. The supernatant was removed and alkylation of cysteine residues was achieved by incubation in 200 µl alkylating solution (55mM iodoacteamide, 0.1M AmBiC) for 30min in the dark. After ACN-dehydration for 10min the supernatant was removed and 200 µl of 0.1M AmBiC were added. After 15min of incubation the supernatant was removed and gel pieces were dehydrated by addition of ACN. After removal of residual ACN, gel pieces were dried by vacuum centrifugation for 20min. The dried gel pieces were treated with trypsin (Promega, Mannheim, Germany) solution prepared according to the manufacturer’s instruction. Eighty microliters were added to each sample, which were subsequently incubated overnight at 37 °C. Extraction of peptides was initiated by adding 40 µl of 50% (v/v) ACN, 5% (v/v) formic acid (FA) (30min, 37 °C, 800rpm). The tryptic peptide-containing supernatants were collected in new low-binding Eppendorf tubes. The procedure was repeated twice by first adding 40 µl of 50% (v/v) ACN, 1% (v/v) FA, and then 100% (v/v) ACN afterwards. The supernatants for each sample were pooled in the same Eppendorf tubes and subsequently dried using a vacuum centrifuge at 30 °C. For mass spectrometry peptides were absorbed in 20 µl 2% (v/v) ACN, 0.1% (v/v) FA.
ESI-MS/MS
Tandem mass spectrometry (MS/MS) analysis was performed by means of a Q-Exactive (Thermo Fisher Scientific, Dreieich, Germany) mass spectrometer coupled to an Ultimate 3000 (Thermo Fisher Scientific, Dreieich, Germany) UPLC.
Seven microliters of sample solution were drawn from 0.25-ml glass insert vials (Sun-SRI, Rockwood, TN, US) kept at 8 °C in the sample compartment and stored in a 20-µl sample loop before being injected into a 2cm, C18, 5 µm, 100 Å reverse phase trapping column (Acclaim PepMap100, Thermo Fisher Scientific, Dreieich, Germany) at a rate of 4 µl min–1. Peptide separation was achieved on a 50cm, C18, 3 µm, 100 Å reverse phase analytical column (Acclaim PepMap100, Thermo Fisher Scientific, Dreieich, Germany). Peptides were eluted using a non-linear 2% to 30% (v/v) acetonitrile gradient in 0.1% (v/v) formic acid with a flow of 300 nl min–1 over a period of 60 mins and at a set column oven temperature of 35 °C. To clean the column, the ACN concentration was subsequently raised to 95% (v/v) within 10min, where it was kept for another 15min before column equilibration to 2% (v/v) ACN commenced.
Transfer of eluted peptides into the mass spectrometer was achieved by means of a NSI source (Thermo Fisher Scientific, Dreieich, Germany) using stainless steel nano-bore emitters (Thermo Fisher Scientific, Dreieich, Germany) connected to the column outlet by a 50-cm, 0.05mm ID fused silica capillary. During MS analysis spray voltage was set to 2.2kV, capillary temperature to 275 °C, and S-lens RF level to 50%. The MS was run in positive ion mode, MS/MS spectra (top 10) were recorded from 30 mins to 220min. For full MS scans, the number of microscans was set to 1, resolution to 70 000, AGC target to 1e6, maximum injection time to 400ms, number of scan ranges to 1, scan range to 400–1600 m/z, and spectrum data type to ‘profile’. For dd-MS2, the number of microscans was set to 1, resolution to 17 500, AGC target to 1e5, maximum injection time to 250ms, Loop count to 10, MSX count to 1, isolation window to 3.0 m/z, fixed first mass to 100.0 m/z, NCE to 27.0 (stepped NCE deactivated), and spectrum data type to ‘profile’. Data dependent (dd) settings were as follows: underfill ratio, 0.5%; intensity threshold, 2.0e3; apex trigger, 10 to 40s; charge exclusion, unassigned, 1, 5, 5–8, >8; peptide match, preferred; exclude isotopes, on; dynamic exclusion, 45.0s.
MS/MS data were queried against an in-house TAIR10 database, modified to also include common contaminants (keratin, trypsin), MS-standards (BSA, fibrinopeptide) and known modifications of mitochondrial encoded proteins based on RNA-editing (AGIs) using Proteome Discoverer (Thermo Fisher Scientific, Dreieich, Germany). Search runs employed the Mascot (Matrix Science, London, United Kingdom), peptide selector settings employed the following spectrum properties filter: Lower Rt limit, 0; upper RT limit, 0; first scan, 0; last scan, 0; lowest charge state, 1; highest charge state, 5; min. precursor mass, 350Da; max. precursor mass, 5000Da; total intensity threshold, 0; and minimum peak count, 1. The scan event filter was adjusted to the following settings: mass analyser, ftms; ms order, MS2; activation type, HCD; min. collision energy, 0; max. collision energy, 1000; scan type, full; ionization source, nanospray; polarity mode, +. The S/N threshold was set to 1.5. For Mascot, the number of maximum missed cleavage sites was limited to 1, precursor mass tolerance to 10 ppm, and fragment mass tolerance to 0.8Da. Allowed variable modifications were oxidation of methionine residues and N-terminal acetylations. Carbamidomethylation of cysteine residues was selected as fixed modification. For the target decoy PSM validator, strict target FDR was set to 0.01, while 0.05 was selected for relaxed target FDR.
Identification and protein quantification
Q-Exactive raw-files were loaded into the MaxQuant software (Cox and Mann, 2008) and processed using the following group specific parameters: variable modifications, acetyl (N-term), oxidation (M); digestion mode, specific; enzyme, Trypsin/P; max. number of missed cleavages, 2; match type, match from and to; number of threads, 3; max. instrument type, Orbitrap; first search peptide tolerance, 20; main search tolerance, 4.5; peptide tolerance unit, ppm; individual peptide mass tolerance, chosen; isotope match tolerance, 2 (ppm); centroid match tolerance, 8 (ppm); centroid half width, 35 (ppm); time valley factor, 1.4; isotope time correlation, 0.6; theoretical isotope correlation, 0.6; recalibration unit, ppm; use MS1 centroids, not chosen; use MS2 centroids, not chosen; intensity dependent calibration, not chosen; min. peak length, 2; max. charge, 5; min. score for recalibration, 70, cut peaks, chosen; gap scans, 1; advanced peak splitting, not chosen; intensity threshold, 500; intensity determination, value at maximum, label-free quantitation (LFQ) min. ratio count, 2; Fast LFQ, chosen; LFQ min. number of neighbors, 3; LFQ average number of neighbors, 6; number of modifications per peptide, 5; min. time, NaN; max. time NaN; additional var mods for special proteins, not chosen; separate variable modifications for first search, not chosen; separate enzyme for first search, not chosen.
Global parameters were chosen as follows: a fasta file containing all Uni-Prot listed Arabidopsis thaliana protein sequences; fixed modifications, carbamidomethyl (C); re-quantify, not chosen; match between runs, chosen; match time window, 0.7min; alignment time window, 20min; match unidentified features, not chosen; decoy mode, revert; special AAs, KR; include contaminants, chosen; I=L, not chosen; max peptide mass, 4600Da; min. peptide length for unspecific search, 8; max. peptide length for unspecific search, 25; PSM FDR, 0.01; protein fdr, 0.01; Site decoy fraction, 0.01; min. peptide length, 7; min. peptides, 1; min. razor + unique peptides, 1; min. unique peptides, 0; min. score for unmodified peptides, 0; min. score for modified peptides, 40; min. delta score for unmodified peptides, 0; min. delta score for modified peptides, 6; base FDR calculation of delta score, not chosen; razor protein FDR, chosen; split protein groups by taxonomy ID, not chosen; filter labelled amino acids, chosen; second peptides, chosen; dependent peptides, not chosen; min ratio count, 1.5; peptides for quantification, unique + razor; use only unmodified peptides and selected modifications, chosen; modifications used in protein quantification, acetyl (N-term), oxidation (M); discard unmodified counterpart peptide, chosen; separate LFQ in parameter groups, not chosen; stabilize large LFQ ratios, chosen; require MS/MS for LFQ comparisons, chosen; iBAQ, chosen; Log fit, chosen; advanced site intensities, chosen.
LFQ intensities from the corresponding MaxQuant ‘proteinGroups.txt’ file were uploaded into the Perseus software (http://www.biochem.mpg.de/5111810/perseus) to build a quantitation matrix. Data were cleaned from the matrix by applying the following parameters: columns identified only by site, reverse, potential contaminant; mode, remove matching rows; filter mode, reduce matrix. Categorical annotation of rows was performed manually (‘create’) and invalid data were removed by filtering rows based on valid values: min. number of values, 3; mode, in at least one group; values should be greater than 0; filter mode, reduce matrix. Two-sample testing was achieved by means of a t-test using the following parameters: S0; side, both; permutation-based FDR, 0.05, number of randomizations, 250; preserve grouping in randomizations, none; –log10, chosen. The –log10 P-value was calculated and the cut-off for the following analysis was P-value >1.31. Localization of proteins was analysed with SUBAcon (Tanz et al., 2013; Hooper et al., 2014) and the functional context with MapMan (Thimm et al., 2004).
Oxygen consumption measurements
Oxygen consumption of isolated mitochondria was measured using a Clark-type oxygen electrode (Hansatech Instruments, Norfolk, UK) according to Meyer et al. (2009). The reaction buffer included 100 µg mitochondrial protein in 3ml respiration buffer (300mM sucrose, 5mM KH2PO4, 10mM TES, 10mM NaCl, 2mM MgSO4, 0.1% [w/v] BSA, pH 7.2) supplied with 5mM succinate and 500 µM ATP. At stable oxygen consumption, 200 µM ADP was added for measuring ADP-dependent respiration. For estimation of AOX capacity, 500 µM of AOX inhibitor n-propyl gallate (nPG) was added and the O2 consumption rate after adding nPG was subtracted from the ADP-dependent O2 consumption rate.
Results
Comparison of the mitochondrial proteomes of wt and ca1ca2 lines using 2D IEF/SDS PAGE
To investigate the consequences of the absence of complex I on the mitochondrial compartment, comparative proteome analyses of wt and ca1ca2 mitochondria isolated from cell culture lines were performed. Proteins were separated by 2D IEF/SDS PAGE and spot volumes were systematically compared using the Delta 2D software package (Decodon, Greifswald).
Volumes of 121 spots were significantly altered, with a fold change of >1.5 (P-value < 0.01) between wt and ca1ca2 lines. Forty-four spots had higher volume in the ca1ca2 mutant, whereas 77 spot volumes were increased in wt (reduced in ca1ca2) (Fig. 1). All 121 spots were analysed by mass spectrometry. After applying a MASCOT threshold score of 60, overall a total of 288 identified proteins were included in further analyses. More than one protein was identified for several spots. Changes in volume were only assigned to a specific protein if a spot only included one main protein. This further reduced the number of unambiguously changed proteins to 106. Sixty-six of these proteins were of decreased abundance in the mutant and 40 of increased abundance. These proteins were annotated according to their functional context (Supplemantary Table S1 at JXB online).
Fig. 1.
Comparative analysis of the mitochondrial proteomes of Arabidopsis wt and ca1ca2 lines. Mitochondria were isolated as described in the Materials and Methods. Total mitochondrial protein was separated by 2D IEF/SDS PAGE and proteins were stained by Coomassie blue. Three replicates were produced per line and used for the calculation of master gels (Delta 2D software package, Decodon, Germany). The molecular masses of standard proteins are given to the left of the 2-D gel (in kDa). Isoelectric focusing range is from pH 3 (left) to pH 11 (right). Proteins indicated in pink are more abundant in the mutant (>1.5-fold increase); proteins indicated in green are less abundant in the mutant (>1.5-fold decrease). Spots indicated by numbers were identified by mass spectrometry (for results see Supplementary Table S1).
As expected, many proteins of reduced abundance were complex I subunits (15 proteins). We did not find all the complex I proteins because 2D IEF/SDS PAGE does not allow separation of very hydrophobic proteins. Reduction of complex I subunits was on average 5-fold. It was shown previously by Fromm et al. (2016c ) that complex I is completely absent in the mutant (Supplemantary Fig. S1); however, non-assembled subunits may be present in mitochondria. Furthermore, reduction of levels of complex I subunits can be expected to be even higher because nearly all spots included not only one main protein but also in addition some proteins of low abundance, which probably were of unchanged or not much changed abundance. As a consequence, fold-changes in general might be slightly higher than determined in the frame of our study. Other proteins of reduced abundance in the mutant are involved in the central mitochondrial metabolism whereas many proteins of increased abundance play roles in transport or stress response processes (Table 1).
Table 1.
Relative spot volumes of altered proteins involved in defined functional processes in ca1ca2 mutant lines. Proteins were separated by 2D IEF/SDS PAGE (Fig. 1), normalized spot volumes of differential OXPHOS subunits were summed up and relative spot volumes were calculated by the Delta 2D software package
| Functional context (number of proteins of changed abundance) | Relative spot volume with respect to wt plants (%) |
|---|---|
| Stress response (12) | 186 |
| Transport (7) | 159 |
| Processing of nucleic acids (5) | 157 |
| Protein folding and processing (6) | 143 |
| Oxidative phosphorylation without complex I (18) | 133 |
| Uncharacterized (4) | 111 |
| Miscellaneous proteins (6) | 102 |
| Amino acid metabolism (7) | 75 |
| Lipid metabolism (9) | 68 |
| TCA cycle (5) | 45 |
| Carbon fixation (4) | 28 |
Detailed evaluation of the dataset revealed the following. Changes in abundance of TCA cycle enzymes were not uniform. We found subunits of citrate synthase (AT2G44350), malate dehydrogenase (AT3G15020), and succinyl-CoA ligase (AT5G08300) of decreased abundance in the ca1ca2 mutant, whereas a subunit of the oxoglutarate dehydrogenase complex (AT5G55070) was increased. Glutamate dehydrogenase (AT5G18170) was also increased in the mutant. Notably, several subunits of the TIM translocase were increased (TIM8, TIM9, TIM23). Several of the most induced proteins in the mutant are involved in plant stress responses.
Some of the proteins of changed abundance were subunits of the remaining OXPHOS complexes II–V. Again, not all of the subunits were identified because many of them are very hydrophobic and not resolvable by 2D IEF/SDS PAGE. It became apparent that several complex IV subunits are clearly induced. To get a more complete impression on how the mutant is altered with respect to the OXPHOS system, we next compared mitochondrial fractions of wt and ca1ca2 lines using 2D BN/SDS PAGE, a gel electrophoresis system known to be particularly suitable for analysing membrane-bound proteins and protein complexes.
Comparison of the mitochondrial proteomes of wt and ca1ca2 lines using 2D BN/SDS PAGE
As previously reported, the ca1ca2 mutant shows changes in the activities of complex II and complex IV (Fromm et al., 2016c ). In order to evaluate changes in the amounts of the OXPHOS complexes II–V in the absence of complex I in more detail, comparative proteome experiments by 2D BN/SDS PAGE were performed with mitochondrial membrane fractions of wt and ca1ca2 cell culture lines. The comparisons were based on two methods: (i) Delta 2D-mediated comparison of 2-D gels, and (ii) fluorophore based comparison (2D DIGE).
Visual inspection of the 2D BN/SDS gels used for Delta 2D comparison clearly revealed the absence of complex I and the I+III2 supercomplex in the mutant (Fig. 2A). On the resulting overlay image (Fig. 2B) the complexes III2 and V are more-or-less unchanged, while the complexes II and IV are clearly increased in the mutant. Average spot volumes were calculated for each complex in the two fractions using the Delta 2D software (Fig. 2B, Table 2). The following amounts of the OXPHOS complexes were found for the mutant (wt=100%): complex II, 133%; complex III, 108%; complex IV, 200%; and complex V, 107%. Proteins within 16 spots were analysed by MS (Supplemantary Table S2) and all revealed the expected identifications (see the 2D BN/SDS GelMap of the Arabidopsis mitochondrial proteome for comparison, https://gelmap.de/1227).
Fig. 2.
Comparative analysis of the mitochondrial membrane proteomes of Arabidopsis wt and ca1ca2 lines. Mitochondria were isolated as described in the Materials and methods. Mitochondrial membrane proteins were separated by 2D BN/SDS PAGE and proteins were stained by colloidal Coomassie (A). Three replicates were produced per fraction and used for the calculation of a master gel (Delta 2D software package, Decodon, Germany) (B). The molecular masses of standard proteins are given to the left of the 2-D gels (in kDa). OXPHOS complexes are boxed in (B); their identities are given above the gels (I, complex I; V, complex V; III2, dimeric complex III; I+III2, supercomplex formed of complex I and dimeric complex III; F1, F1 part of complex V; IV, complex IV; II, complex II). Proteins indicated in pink are less abundant in the mutant (>1.5-fold decrease); proteins indicated in green are more abundant in the mutant (>1.5-fold increase). Spots indicated by numbers were identified by mass spectrometry (for results see Supplementary Table S2).
Table 2.
Relative spot volumes of OXPHOS complexes in ca1ca2 lines. Proteins were separated by 2D BN/SDS PAGE (Fig. 2), normalized spot volumes of differential OXPHOS subunits were summed up and relative spot volumes were calculated by the Delta 2D software package
| OXPHOS complex | relative spot volume with respect to wt plants (%) |
|---|---|
| Complex I | - * |
| Complex II | 133 |
| Complex III | 108 |
| Complex IV | 200 |
| Complex V | 107 |
* not detectable
A more extended comparison of the mitochondrial membrane proteomes of mutant and wt cell lines was carried out based on 2D BN/SDS DIGE (Fig. 3, Supplemantary Table S3). Spots differing in volumes between the two fractions were analysed by MS. After applying a MASCOT threshold score of 60, overall 147 identified proteins were included in further analyses; however, a difference in spot volume only could be assigned to a specific protein if a spot included only one main protein. This further reduced the number of unambiguously changed proteins to 44. These were grouped according to functional context. Changes of individual subunits of OXPHOS complexes were in accordance with the results of the Delta 2D analysis. In addition, several other membrane proteins were found to be of changed abundance in the mutant. Seven subunits of the TIM and TOM transport machineries and one ABC transporter were identified. All were more abundant in the ca1ca2 mutant.
Fig. 3.
Comparative analysis of the mitochondrial membrane proteomes of Arabidopsis wt and ca1ca2 lines by differential gel electrophoresis (DIGE). Mitochondria were isolated as described in the Materials and Methods. Mitochondrial membrane proteins of wt and ca1ca2 were labeled with different CyDyes and separated by 2D BN/SDS PAGE. Proteins were stained by colloidal Coomassie (A). The same gel was used for fluorescence detection of the two CyDyes (B). The molecular masses of standard proteins are given to the left of the 2-D gel (in kDa). The identities of selected mitochondrial protein complexes are given above the gels (I, complex I; V, complex V; III2, dimeric complex III; I+III2, supercomplex formed of complex I and dimeric complex III; F1, F1 part of complex V; IV, complex IV; II, complex II). Proteins indicated in red are less abundant in the ca1ca2 mutant (>1.5-fold decrease) and proteins indicated in green are more abundant in the ca1ca2 mutant (>1.5-fold increase). Proteins given in yellow are not changed in abundance. Spots indicated by numbers were identified by mass spectrometry (for results see Supplementary Table S3). Note: if compared to the comparative experiment shown in Fig. 2, several subunits of OXPHOS complexes appear to be absent in the DIGE approach. This is due to the fact that CyDye labeling takes place at native conditions. As a consequence, only proteins exposed to the surface of protein complexes are labeled. In contrast, image evaluation based on the Delta 2D approach (Fig. 2) allows visualization of all subunits of a protein complex.
Comparison of total protein extracts of wt and ca1ca2 lines by label-free quantitative shotgun proteomics
In addition to effects on the mitochondrial compartment, the consequences of ca1ca2 deletion at the whole-plant level were investigated. Wild type and ca1ca2 mutant plants were harvested at a comparable growth stage and differential protein abundances were analysed by comparative quantitative shotgun MS (note that growth and development of ca1ca2 plants is much delayed; see Fromm et al., 2016c). The experiment was based on five biological replicates. In total, 2233 different proteins were identified. The quantitative analysis of identified proteins was carried out using MaxQuant. After application of a –log10 P-value (P>1.31), 318 proteins of changed amounts were confirmed (Supplemantary Table S4) and were included in further analyses.
The proteins were assigned into categories according to subcellular localization and functional context. Subcellular localization was assigned according to SUBAcon (Fig. 4). Most of the identified proteins are localized in the cytosol (30.4%), followed by plastids (29.4%), mitochondria (15.2%), and other compartments with minor contributions (Fig. 4A). Proteins of increased abundance in ca1ca2 plants are mostly localized in the cytosol (44.1%) and mitochondria (19.8%) (Fig. 4B). In contrast, proteins of decreased abundance in ca1ca2 plants are mostly localized in plastids (62.3%) (Fig. 4C).
Fig. 4.
Subcellular localization of proteins of altered abundances in the ca1ca2 line as obtained by label free quantitative shotgun proteomics. Total protein was extracted from wt and ca1ca2 mutant plants at a similar developmental stage. Proteins were identified and quantified by shotgun MS (for details see the Material and Methods). Predicted localizations of proteins of changed abundances between the two lines were obtained from the SUBA3 database (http://suba3.plantenergy.uwa.edu.au/). (A) Predicted localization of all proteins changed (P-value <0.05). (B) Predicted localization of proteins more abundant in ca1ca2 compared to wt. (C) Predicted localization of proteins less abundant in ca1ca2 mutant compared to wt. Numbers indicate amounts relative to the sum of altered protein species (%).
Assignment of proteins differing in amount between wt and ca1ca2 mutant plants to functional categories was carried out according to TAIR functional annotations (https://www.arabidopsis.org/, TAIR10 genome release) and evaluated by MapMan (Fig. 5, Table 3). Proteins especially induced in mutant plants are involved in glycolysis, fermentation, the TCA cycle, amino acid metabolism, redox regulation, protein folding, as well as stress responses (Supplemantary Table S4). Decreased protein abundances in ca1ca2 plants were mainly found in the functional categories of photosynthesis (photosystem I, photosystem II, Calvin cycle, photorespiration) and tetrapyrrole synthesis (Fig. 5). As expected, complex I subunits were much decreased in the mutant. At the same time, AOX1A (AT3G22370) and alternative NADH dehydrogenase NDB2 (AT4G05020) were clearly induced.
Fig. 5.
Functional annotation of proteins identified by quantitative label-free shotgun MS. The 318 proteins of differential abundances between wt and ca1ca2 mutant plants were grouped into functional BINs using MapMan. BINs are given to the right. Proteins more abundant in ca1ca2 are indicated in green and proteins that are less abundant in red (P-value <0.05). Each square represents one protein. BINs without any identified protein are with a grey dot (for results see Supplementary Table S4).
Table 3.
Relative protein intensities of altered proteins involved in defined functional processes in ca1ca2 mutant lines analysed by label-free quantitative shotgun MS approach. Proteins were identified and quantified using MaxQuant software
| Functional context (number of proteins of changed abundance) |
Relative protein intensities with respect to wt plants (%) |
|---|---|
| Fermentation (4) | 183 |
| Glycolysis (10) | 173 |
| Protein folding (9) | 172 |
| Oxidative pentose phosphate (2) | 166 |
| Signalling (5) | 162 |
| Redox (8) | 160 |
| TCA cycle (11) | 159 |
| Stress response (14) | 152 |
| Amino acid metabolism (17) | 149 |
| Mitochondrial electron transport without complex I (5) | 144 |
| Cell wall (7) | 140 |
| Cell organisation (14) | 132 |
| Miscellaneous proteins (25) | 121 |
| Lipid metabolism (10) | 120 |
| Secondary metabolism (13) | 114 |
| Protein processing (6) | 110 |
| Protein degradation (18) | 102 |
| Protein synthesis (11) | 95 |
| Co-factor and vitamine metabolism (2) | 89 |
| Development (3) | 89 |
| Metal handling (2) | 89 |
| Protein targeting (7) | 85 |
| Uncharacterized (29) | 81 |
| Carbon metabolism (14) | 79 |
| Transport (15) | 78 |
| Photorespiration (4) | 75 |
| Calvin cycle (9) | 66 |
| Photosystem II (4) | 66 |
| Photosystem I (2) | 66 |
| Tetrapyrrole synthesis (5) | 65 |
| Light reaction others (3) | 64 |
| N-metabolism (4) | 64 |
| Processing of nucleic acids (11) | 63 |
| ATP synthase (plastid) (3) | 61 |
Finally, we analysed the BIN coverage of the identified proteins in order to assess the influence of the absence of complex I on cellular processes (Table 4). We calculated the number of identified proteins in relation to the number of genes that code for proteins of the BINs. The following BINs were most significantly changed in the mutant: fermentation (28.6%), glycolysis (15.4%), nitrogen metabolism (15.4%), TCA cycle (13.8%), photosynthesis (12.6%), tetrapyrrole synthesis (11.1%), mitochondrial ETC (7.5%), and amino acid metabolism (6.8%). The BIN coverage indicates that these cellular process are particularly affected by absence of complex I.
Table 4.
BIN coverage of proteins of altered abundances in ca1ca2 plants as determined by label-free quantitative shotgun MS
| MapMan BIN | BIN name | Sum of genes | Number of differential proteins in wt and ca1ca2 | Number of differential proteins / sum of genes per BIN (%) |
|---|---|---|---|---|
| 5 | Fermentation | 14 | 4 | 28.6 |
| 4 | Glycolysis | 65 | 10 | 15.4 |
| 12 | N-metabolism | 26 | 4 | 15.4 |
| 8 | TCA | 80 | 11 | 13.8 |
| 1 | Photosynthesis | 199 | 25 | 12.6 |
| 19 | Tetrapyrrole synthesis | 45 | 5 | 11.1 |
| 25 | C1 metabolism | 39 | 4 | 10.3 |
| 6 | Gluconeogenese | 10 | 1 | 10.0 |
| 9 | Mitochondrial electron transport | 146 | 11 | 7.5 |
| 13 | Amino acid metabolism | 251 | 17 | 6.8 |
| 7 | Oxidative pentose phosphate pathway | 31 | 2 | 6.5 |
| 2 | Major carbon metabolism | 100 | 5 | 5.0 |
| 21 | Redox | 194 | 8 | 4.1 |
| 3 | Minor carbon metabolism | 124 | 5 | 4.0 |
| 24 | Biodegradation of Xenobiotics | 27 | 1 | 3.7 |
| 15 | Metal handling | 67 | 2 | 3.0 |
| 16 | Secondary metabolism | 438 | 13 | 3.0 |
| 23 | Nucleotide metabolism | 169 | 5 | 3.0 |
| 18 | Co-factor and vitamine metabolism | 79 | 2 | 2.5 |
| 11 | Lipid metabolism | 398 | 10 | 2.5 |
| 31 | Cell organisation | 746 | 14 | 1.9 |
| 26 | Miscellaneous proteins | 1397 | 25 | 1.8 |
| 20 | Stress response | 874 | 14 | 1.6 |
| 34 | Transport | 996 | 15 | 1.5 |
| 29 | Protein folding and processing | 3409 | 51 | 1.5 |
| 10 | Cell wall | 496 | 7 | 1.4 |
| 17 | Hormone metabolism | 495 | 5 | 1.0 |
| 33 | Development | 681 | 3 | 0.4 |
| 30 | Signalling | 1239 | 5 | 0.4 |
| 35 | Not assigned | 7748 | 29 | 0.4 |
| 27 | RNA processing | 2567 | 6 | 0.2 |
| 14 | Sulfur assimilation | 12 | 0 | 0.0 |
| 32 | Micro RNA | 4 | 0 | 0.0 |
| 28 | DNA synthesis | 1352 | 0 | 0.0 |
| 22 | Polyamine metabolism | 18 | 0 | 0.0 |
Changes in protein levels in ca1ca2 plants with respect to wildtype plants as obtained by label-free quantitative shotgun proteomics are summarized in Fig. 6 and in the discussion below.
Fig. 6.
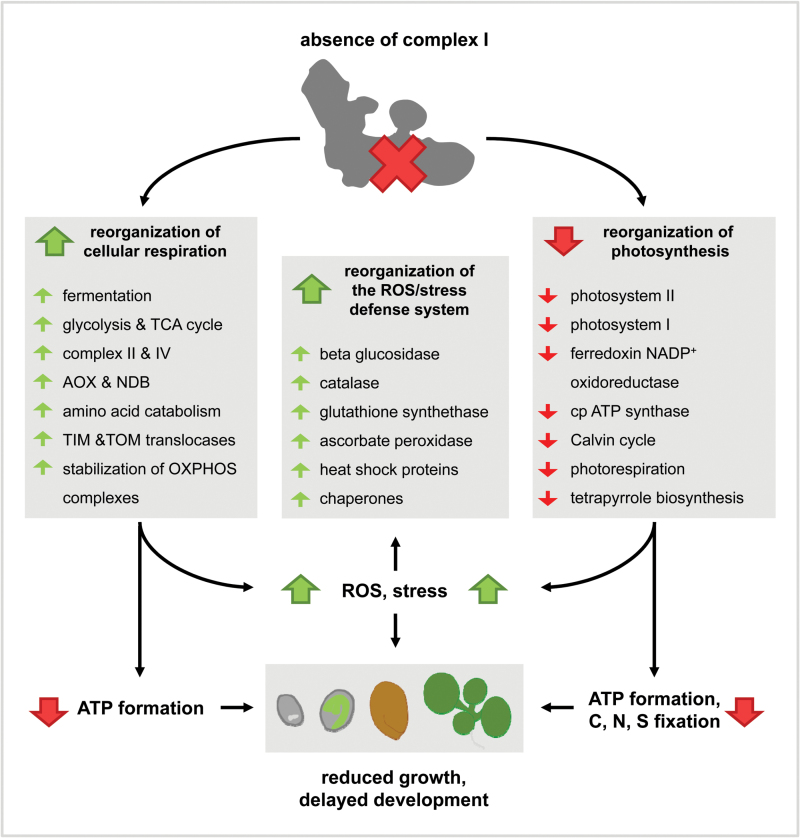
Life without complex I. The figure is based on altered protein levels in ca1ca2 plants relative to wildtype plants as obtained by label-free quantitative shotgun proteomics. Green arrows within the grey boxes indicate increased protein levels in the double-mutant, and red arrows indicate decreased levels. Absence of complex I causes reorganization of the cellular respiration system. Since electron insertion into the first segment of the mETC is not possible, increased electron insertion at later segments takes place (induction of complexes II, IV). This requires increased oxidation of organic substrates (induction of enzymes of glycolysis, the TCA cycle, and amino acid catabolism). Mitochondrial ATP formation most likely is still reduced, which requires increased fermentation. The growth rate of the double-mutant is drastically reduced. This is reflected by reduced amounts of the two photosystems, Calvin cycle enzymes, and enzymes of the tetrapyrrole biosynthesis pathway. Furthermore, altered metabolism and electron transport pathways in the mitochondria and chloroplasts cause increased ROS formation and stress symptoms. Several components of the ROS and stress defense system are induced in the double-mutant, as is the alternative oxidase, a well-known stress indicator in plants. Note: causal events indicated by black arrows do not necessarily indicate primary effects, but may well represent indirect consequences. For further details see the discussion section.
Discussion
In previous studies, the ca1ca2 mutant has been characterized with respect to development, mitochondrial metabolism, and features of the OXPHOS system (Fromm et al., 2016c). In order to systematically monitor the consequences of the double gene deletion on the mitochondrial proteome and the entire leaf proteome, we report here the results of three different experimental approaches, two of which were based on gel electrophoresis and one on gel-free shotgun proteomics. All three experimental systems have advantages and limitations. 2D IEF/SDS PAGE-based analyses excel in investigating prominent hydrophilic proteins. 2D BN/SDS PAGE is strong in analysing membrane proteins and membrane-bound protein complexes, which also are relatively abundant. Shotgun proteome analysis is a less systematic approach that allows the analysis of a very large number of proteins at the same time, and as such it better covers proteins of comparatively low abundance than the two gel-based approaches. Furthermore it should be noted that the 2D IEF/SDS and 2D BN/SDS PAGE approaches were carried out using cell cultures whereas shotgun proteomics was performed on total protein extracts from leaves. Cell suspension cultures are always well supplied with sucrose, whereas plants have to generate sugars by photosynthesis on their own. Therefore, the outcomes of the three experimental approaches need to be compared critically (Tables 1 and 3).
Although the three experimental systems and the sources of the analysed protein fractions differed, all the approaches gave similar results in respect to proteomic alterations taking place in the ca1ca2 double-knock-out mutant. As expected, all the approaches indicated a dramatic reduction of complex I subunits. However, residual levels of non-assembled complex I proteins seem to be present in the mutant’s mitochondria.
Levels of complex II and especially complex IV were much higher in the double-mutant, as were the levels of some of the alternative oxidoreductases of the plant mitochondrial OXPHOS system. This has been previously reported for other complex I mutants (Keren et al., 2012; Hsu et al., 2014). Upregulation of complex IV points to elevated proton translocation at the final segment of the mETC, which could partially compensate for the diminished proton translocation at the first segment of the mETC in the absence of complex I. Indeed, increased in vitro activity of complex IV in the ca1ca2 mutant has been shown previously (Fromm et al., 2016c). An increased oxygen consumption of ca1ca2 mitochondria was observed if succinate was used as substrate (Supplemantary Fig. S2). This might indicate a higher complex II activity, but could also be the consequence of an elevated electron flux through the mETC caused by higher complex IV activity.
In contrast, levels of the complexes III and V were rather similar in the ca1ca2 and wt lines, although relative abundances of some subunits of these complexes were altered (Supplemantary Tables S1 and S2). The overall upregulation of the OXPHOS complexes in mutant cells requires increased import rates of the corresponding nuclear encoded subunits by the TOM and TIM pre-protein import machineries (Murcha et al., 2014). Indeed, levels of TIM and TOM subunits clearly went up in the double-mutant (Fig. 1, Supplemantary Table S3).
The alternative NADH dehydrogenase NDB2 (AT4G05020) and the alternative oxidase AOX1A (AT3G22370) were induced 4- and 8-fold higher in the mutant (Supplemantary Table S4). This corresponds to increased capacity for alternative oxidase in the ca1ca2 mutants (Supplemantary Fig. S2). An increase of AOX has been previously reported for several plant complex I mutants, e.g. the nmat1, opt43, cmsII, ndufv1, and mTerf15 lines (Gutierres et al., 1999; de Longevialle et al., 2007; Keren et al., 2012; Hsu et al., 2014; Kühn et al., 2015; see Table 5 for information on complex I mutants discussed in this section). Indeed, increased oxygen consumption rates have been observed for some complex I mutants, e.g. the ca1ca2 and ndufv1 lines (Kühn et al., 2015; Fromm et al., 2016c).
Table 5.
Summary of complex I mutants in plants
| Name of mutant | Mutation of ... | Complex I depletion (i) or absence of complex I (ii) | Plant species | Reference |
|---|---|---|---|---|
| ca2 | Complex I subunit | i | Arabidopsis thaliana | Perales et al., 2005 & Fromm et al., 2016c |
| ca1ca2 | Complex I subunit | ii | Arabidopsis thaliana | Fromm et al., 2016c |
| cal1cal2i | Complex I subunits | i | Arabidopsis thaliana | Fromm et al., 2016b |
| ca2cal1 or ca2cal2 | Complex I subunits | i | Arabidopsis thaliana | Soto et al., 2015 |
| ndufs4 | Complex I subunit | i | Arabidopsis thaliana | Kühn et al., 2015 |
| ndufv1 | Complex I subunit | ii | Arabidopsis thaliana | Kühn et al., 2015 |
| gldh | Assembly factor | ii | Arabidopsis thaliana | Pineau et al., 2008 |
| indh | Assembly factor | ii | Arabidopsis thaliana | Wydro et al., 2013 |
| opt43 | NAD1 splicing factor | ii | Arabidopsis thaliana | de Longevialle et al., 2007 |
| nMat1 | NAD1 splicing factor | ii | Arabidopsis thaliana | Keren et al., 2012 |
| mTERF15 | NAD2 splicing factor | ii | Arabidopsis thaliana | Hsu et al., 2014 |
| nms1 | NAD4 splicing factor | i | Nicotiana sylvestris | Brangeon et al., 2000 |
| slo3 | NAD7 splicing factor | i | Arabidopsis thaliana | Hsieh et al., 2015 |
| bir6 | NAD7 splicing factor | i | Arabidopsis thaliana | Koprivova et al., 2010 |
| cmsII | Leads to NAD7 deletion | ii | Nicotiana sylvestris | Gutierres et al., 1999 |
| ncs2 | Replacing the 3′-end of NAD4 with sequences from NAD7 |
ii | Zea mays | Karpova et al., 2002 |
Induction of the alternative oxidoreductases of the respiratory chain is known to be an integral part of the general plant stress response (Vanlerberghe, 2013). This, together with the induction of a large number of further stress related proteins [such as beta glucosidases (AT1G66270, AT3G09260, AT3G09260, AT3G16420), catalase 3 (AT1G20620), ascorbate peroxidase 4 (AT4G09010) and glutathione syntethase 2 (AT5G27380)] in the double-mutant strongly indicates that the absence of complex I strongly affects the metabolic balance and the redox homeostasis of the plant cell.
High levels of complex II, complex IV, and alternative oxidoreductases of the mETC require increased provision of electrons to the mETC. Indeed, the enzymes involved in glycolysis were clearly induced in the double-mutants. It has been shown previously for the ndufv1 mutant, which also completely lacks mitochondrial complex I, that complex I deficiency causes a metabolic switch and that flux through glycolysis significantly increases (Kühn et al., 2015). Furthermore, our data point to an extended usage of fermentation to compensate for decreased ATP generation and decreased capacity of NADH oxidation by the complex I-deficient mETC. In contrast, the proteomic changes with respect to the citric acid cycle are more difficult to understand, as some enzymes are reduced whereas others are increased in the ca1ca2 double-mutant. This points to a scenario that the absence of complex I does not induce the entire citric acid cycle, but rather specific segments of the pathway. Indeed, it is known that plant mitochondria have, depending on the physiological state of the respective cell, quite a number of non-cyclic operation modi with respect to the citric acid cycle (Sweetlove et al., 2010).
Electrons for the mETC can also originate from amino acid breakdown (Sweetlove et al., 2010; Schertl and Braun 2014; Hildebrandt et al., 2015). Several proteins involved in mitochondrial amino acid catabolism were identified by our shotgun proteome approach and found to be induced in the ca1ca2 mutant, e.g. alanine, tyrosine, and branched-chain aminotransferases (AT1G17290, AT4G23600, AT3G19710), glutamate dehydrogenase (AT5G18170, AT5G07440), arginase (AT4G08870), and methylmalonate-semialdehyde dehydrogenase (AT2G14170). The latter enzyme is involved in a step of branched-chain amino acid oxidation. The number of electrons provided for the mETC from amino acid breakdown is especially high during oxidation of the branched chain amino acids (Hildebrandt et al., 2015).
Prohibitins and stomatin-like proteins (SLP) were very much increased in the ca1ca2 mutant, as revealed by all three proteome analyses. In animal cells respiratory supercomplexes are stabilized by cardiolipin and SLPs. SLPs can bind cardiolipin and interact with prohibitins (Mitsopoulos et al., 2015). Similar interactions have also been reported for the mitochondria of plants (Gehl et al., 2014; Gehl and Sweetlove, 2014). Knock-out mutants for slp1 have reduced complex I levels and activity, and form lower amounts of supercomplexes, indicating that SLPs and prohibitins can affect the assembly and/or the stability of OXPHOS complexes (Gehl et al., 2014). Complex I subunits that cannot be assembled might be stabilized to a certain degree by prohibitins and SLPs. Furthermore, prohibitins play a role in mitochondrial DNA organization, stress tolerance, and triggering retrograde signals in response to stress and mitochondrial dysfunction (Van Aken et al., 2010).
The phenotype of complex I mutant plants often includes curled leaves and a delayed vegetative and reproductive development (de Longevialle et al., 2007; Meyer et al., 2009; Wang et al., 2012; Kühn et al., 2015; Hsu et al., 2014). The degree of the developmental delay and the curly leaf phenotype are dependent on the amount of residual complex I (Kühn et al., 2015). For example, trace amounts of complex I are sufficient for plants to pass through embryogenesis, whereas mutants lacking complex I, such as cal1cal2, opt43, indh, ndufv1, and ca1ca2, cannot complete this developmental stage and hardly germinate (de Longevialle et al., 2007; Wang et al., 2012; Wydro et al., 2013; Kühn et al., 2015; Fromm et al., 2016c). The growth rate of the ca1ca2 mutant has been reported to be drastically reduced (Fromm et al., 2016c). This is clearly reflected by our shotgun proteome data.
Overall, proteins involved in developmental processes and photosynthesis were very much reduced in the ca1ca2 mutant. More than 60% of the proteins of lower abundance in the ca1ca2 mutant are localized in plastids (Fig. 4C). All detected subunits of the two photosystems (PS) were reduced, as were the ferredoxin-NADP+ oxidoreductase (AT5G66190, AT1G20020) and the subunits of the chloroplast ATP synthase complex. The maize ncsII and ncs6 mutants also have a decrease in PSI while other photosynthetic complexes are unaffected. The chloroplast ultrastructure is abnormal (Roussel et al., 1991; Jiao et al., 2005). Two recently described small twin cystein proteins, which are specifically induced in complex I-deficient plants, also specifically affect chloroplast metabolism (Wang et al., 2016). Furthermore, enzymes involved in the Calvin cycle are of reduced abundance in ca1ca2 plants. This indicates substantial consequences of the absence of complex I on photosynthesis. Additionally, tetrapyrrole synthesis is impaired. Tetrapyrroles are essential for chlorophyll biosynthesis. It has been suggested that reduction of photosynthetic proteins may be caused by impaired chlorophyll synthesis (Brzezowski et al., 2015). Our results are in line with those obtained for the cmsII mutant of Nicotiana sylvestris, which also lacks complex I. In cmsII mutants photosynthetic efficiency is reduced (Sabar et al., 2000; Dutilleul et al., 2003). Defects in the photosystems may result in ROS formation (Schmitt et al., 2014). Higher ROS levels have indeed been observed in the ca1ca2 and other complex I mutants (Keren et al., 2012; Córdoba et al., 2016; Fromm et al., 2016c).
ROS may cause cellular damage and programmed cell death (Li and Xing, 2010), and thus they negatively affect plant development. Our proteome data indicate that seed photosynthesis in ca1ca2 mutant embryos may also be impaired by complex I dysfunction. A higher ROS content has been found in ca1ca2 embryos (Córdoba et al., 2016, Ostersetzer-Brian 2016). Defects in the photosynthetic apparatus should cause decreased synthesis and accumulation of seed storage compounds, which will be further impaired by mitochondrial dysfunction (Schwender et al., 2006). Seed storage compounds such as fatty acids are essential to drive the germination process (Carrie et al., 2013). ca1ca2 mutant embryos depleted in energy-rich components are not able to develop normally during germination, which results in seed abortion (Córdoba et al., 2016, Fromm et al., 2016c).
Reduced photosynthesis affects photorespiration. Indeed, all the identified proteins of the photorespiration pathway were reduced in the ca1ca2 mutant. This also applies for the T and the P subunits of the mitochondrial glycine decarboxylase complex (GDC) (AT1G11860; AT2G26080). Down-regulation of the GDC complex has been reported to be caused either by impaired photosynthesis or by feedback inhibition by an elevated NADH pool in the matrix (Oliver, 1994; Peterhänsel et al., 2010), which may be caused by the absence of complex I.
Besides the absence of the electron transfer function of complex I, which is coupled to proton translocation across the inner mitochondrial membrane, complex I is assumed to include further enzymatic and transport functions (Braun et al., 2014). In particular, the complex I-integrated carbonic anhydrase subunits have been suggested to play a role in recycling mitochondrial CO2 for carbon fixation in the chloroplasts (Zabaleta et al., 2012). Thus it may well be that the ca1ca2 mutant lacks more than just the NADH-ubiquinone-oxidoreductase activity. However, the ca1ca2 mutant very much behaves like other mutants that completely lack complex I, e.g. ndufv1 (Kühn et al., 2015). Furthermore, in mutants lacking complex I due to the absence of other complex I subunits the formation of the carbonic anhydrase domain is also prevented. This makes the ca1ca2 mutant an excellent model for studying the role of mitochondrial complex I in plants, the proteomic level of which has been addressed in this study.
Conclusions
‘Life without complex I’ is not so easy, even in plants that possess some alternative dehydrogenases in the mitochondrial compartment (which, however, do not contribute to the proton gradient across the inner mitochondrial membrane). The metabolic balance of the plant cell is deeply disturbed in the absence of complex I. Complex I dysfunction causes reorganization of cellular respiration and affects metabolic processes in mitochondria, plastids, peroxisomes, and other cellular compartments with drastic consequences for growth and development. Specifically, proteins involved in glycolysis and the TCA cycle are induced, as are subunits of other OXPHOS complexes, especially the complex IV. This requires an upregulation of the TIM and TOM translocases for mitochondrial protein import. Furthermore, alternate electron entry pathways into the mETC are induced, e.g. oxidation of amino acids. Increased flux of electrons through the mETC causes elevated ROS formation in ca1ca2 plants. ATP formation in the chloroplasts is reduced by decreased photosynthesis, e.g. caused by defective chlorophyll biosynthesis in ca1ca2 mutant plants. In summary, plant cells metabolically rearrange in the absence of complex I in order to maintain a minimum level of energy supply and to balance redox homeostasis. At the same time, ca1ca2 mutants suffer from increased ROS production and reduced ATP generation by both, respiration and photosynthesis.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Growth phenotype of ca1ca2 plants, in the absence of complex I.
Figure S2. Respiration through complex II and the AOX capacity of mitochondria derived from Arabidopsis thaliana wildtype (wt) and ca1ca2 double-mutant lines.
Table S1. (a) Proteins of altered abundance in the ca1ca2 mutant as revealed by 2D IEF/SDS PAGE (see Fig. 1). (b) Short version of (a).
Table S2. Identification of proteins from wt and ca1ca2 mutant lines after separation by 2D BN/SDS PAGE and gel evaluation by Delta 2D software (see Fig. 2).
Table S3. Identification of proteins from wt and ca1ca2 mutant lines after analysis by 2D BN/SDS DIGE (see Fig. 3).
Table S4. (a) Proteins of altered abundance in the ca1ca2 mutant as obtained by label-free quantitative shotgun MS analysis. (b) Short version of (a).
Acknowledgements
We gratefully acknowledge the technical assistance of Dagmar Lewejohann and Dr Christin Lorenz. We thank Dr Tatjana Hildebrandt and Dr Ahmed Debez for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (DFG), Forschergruppe 1186 (grant Br1829/10–2).
SF planned and performed experiments, analysed data, and wrote the paper; JS performed experiments, and analysed data; HE analysed data; CP planned experiments; HPB planned experiments and revised the paper.
Glossary
Abbreviations:
- 2D BN/SDS PAGE
two-dimensional blue native/sodium dodecyl sulfate polyacrylamide gel electrophorese
- 2D IEF/SDS PAGE
two-dimensional isoelectric focusing/sodium dodecyl sulfate PAGE
- CA
carbonic anhydrase
- CAL
carbonic anhydrase-like
- complex I
NADH dehydrogenase complex
- DIGE
difference gel electrophorese
- OXPHOS
oxidative phosphorylation
- ROS
reactive oxygen species
- TCA cycle
tricarboxylic acid cycle
- TIM
translocase of inner mitochondrial membrane
- TOM
translocase of outer mitochondrial membrane.
References
- Angerer H, Zwicker K, Wumaier Z, et al. 2011. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. The Biochemical Journal 437, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. 2013. Crystal structure of the entire respiratory complex I. Nature 494, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berth M, Moser F, Kolbe M, Bernhardt J. 2007. The state of the art in the analysis of two-dimensional gel electrophoresis images. Applied Microbiology and Biotechnology 76, 1223–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt U. 2006. Energy converting NADH:quinone oxidoreductase (complex I). Annual Review of Biochemistry 75, 69–92. [DOI] [PubMed] [Google Scholar]
- Brangeon J, Sabar M, Gutierres S, et al. 2000. Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. The Plant Journal 21, 269–280. [DOI] [PubMed] [Google Scholar]
- Braun HP, Binder S, Brennicke A, et al. 2014. The life of plant mitochondrial complex I. Mitochondrion 19, 295–313. [DOI] [PubMed] [Google Scholar]
- Brzezowski P, Richter AS, Grimm B. 2015. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochimica et Biophysica Acta 1847, 968–985. [DOI] [PubMed] [Google Scholar]
- Cardol P, Vanrobaeys F, Devreese B, van Beeumen J, Matagne RF, Remacle C. 2004. Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochimica et Biophysica Acta 1658, 212–224. [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Giraud E, Ng S, Zhang MF, Narsai R, Whelan J. 2013. How do plants make mitochondria? Planta 237, 429–439. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. 2006. Bovine complex I is a complex of 45 different subunits. The Journal of Biological Chemistry 281, 32724–32727. [DOI] [PubMed] [Google Scholar]
- Córdoba JP, Marchetti F, Soto D, Martin MV, Pagnussat GC, Zabaleta E. 2016. The CA domain of the respiratory complex I is required for normal embryogenesis in Arabidopsis thaliana . Journal of Experimental Botany 67, 1589–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana . The Plant Cell 19, 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. 2005. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proceedings of the National Academy of Sciences, USA 102, 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, Paepe R de, Foyer CH, Noctor G. 2003. Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiology 131, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP. 2003. New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiology 133, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T. 2001. Complex I: a chimaera of a redox and conformation-driven proton pump? Journal of Bioenergetics and Biomembranes 33, 169–177. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Böttcher B. 2004. The gross structure of the respiratory complex I: a Lego System. Biochimica et Biophysica Acta 1608, 1–9. [DOI] [PubMed] [Google Scholar]
- Fromm S, Senkler J, Zabaleta E, Peterhänsel C, Braun HP. 2016. a The carbonic anhydrase domain of plant mitochondrial complex I. Physiologia Plantarum : in press, doi: 10.1111/ppl.12424. [DOI] [PubMed] [Google Scholar]
- Fromm S, Göing J, Lorenz C, Peterhänsel C, Braun HP. 2016. b Depletion of the “gamma-type carbonic anhydrase-like” subunits of complex I affects central mitochondrial metabolism in Arabidopsis thaliana . Biochimica et Biophysica Acta 1857, 60–71. [DOI] [PubMed] [Google Scholar]
- Fromm S, Braun HP, Peterhänsel C. 2016. c Mitochondrial gamma carbonic anhydrases are required for complex I assembly and plant reproductive development. New Phytologist : in press, doi: 10.1111/nph.13886. [DOI] [PubMed] [Google Scholar]
- Gehl B, Sweetlove LJ. 2014. Mitochondrial Band-7 family proteins: scaffolds for respiratory chain assembly? Frontiers in Plant Science 5, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl B, Lee CP, Bota P, Blatt MR, Sweetlove LJ. 2014. An Arabidopsis stomatin-like protein affects mitochondrial respiratory supercomplex organization. Plant Physiology 164, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Combettes B, de Paepe R, Mirande M, Lelandais C, Vedel F, Chétrit P. 1999. In the Nicotiana sylvestris CMSII mutant, a recombination-mediated change 5′ to the first exon of the mitochondrial nad1 gene is associated with lack of the NADH:ubiquinone oxidoreductase (complex I) NAD1 subunit. European Journal of Biochemistry 261, 361–370. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araujo WL, Braun HP. 2015. Amino acid catabolism in plants. Molecular Plant 8, 1563–1579. [DOI] [PubMed] [Google Scholar]
- Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH. 2014. SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30, 3356–3364. [DOI] [PubMed] [Google Scholar]
- Hsieh W, Liao J, Chang C, Harrison T, Boucher C, Hsieh M. 2015. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiology 168, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY. 2014. Arabidopsis mTERF15 is required for mitochondrial nad2 Intron 3 splicing and functional complex I activity. PLoS ONE 9, e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Zickermann V, Brandt U. 2010. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451. [DOI] [PubMed] [Google Scholar]
- Jiao S, Thornsberry JM, Elthon TE, Newton KJ. 2005. Biochemical and molecular characterization of photosystem I deficiency in the NCS6 mitochondrial mutant of maize. Plant Molecular Biology 57, 303–313. [DOI] [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ. 2002. Differential expression of alternative oxidase genes in maize mitochondrial mutants. The Plant Cell 14, 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. The Plant Journal 71, 413–426. [DOI] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun HP. 2010. Internal architecture of mitochondrial complex I from Arabidopsis thaliana . The Plant Cell 22, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, des Francs-Small CC, Calder G, Mugford ST, Tanz S, Lee B, Zechmann B, Small I, Kopriva S. 2010. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. The Journal of Biological Chemistry 285, 32192–32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Obata T, Feher K, Bock R, Fernie AR, Meyer EH. 2015. Complete mitochondrial complex I deficiency induces an upregulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiology 168, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Carrie C, Gawryluk RM, Solheim C, Gray MW, Whelan J, Millar AH. 2013. Subcomplexes of ancestral respiratory complex I subunits rapidly turn over in vivo as productive assembly intermediates in Arabidopsis. Journal of Biological Chemistry 288, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xing D. 2010. Mitochondrial pathway leading to programmed cell death induced by aluminum phytotoxicity in Arabidopsis . Plant Signaling and Behavior 5, 1660–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Rolletschek H, Sunderhaus S, Braun HP. 2014. Brassica napus seed endosperm — Metabolism and signaling in a dead end tissue. Journal of Proteomics 108, 382–426. [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ. 1993. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiology 103, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Solheim C, Tanz SK, Bonnard G, Millar AH. 2011. Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. Journal of Biological Chemistry 286, 26081–26092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH. 2009. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiology 151, 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihr C, Braun HP. 2003. Proteomics in plant biology. In: Michael P. ed. Handbook of proteomics methods . Totowa: Humana, 409–416. [Google Scholar]
- Mitchell P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148. [DOI] [PubMed] [Google Scholar]
- Mitsopoulos P, Chang Y, Wai T, Konig T, Dunn SD, Langer T, Madrenas J. 2015. Stomatin-like protein 2 is required for in vivo mitochondrial respiratory chain supercomplex formation and optimal cell function. Molecular and Cellular Biology 35, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DJ, Sazanov LA. 2008. Three-dimensional structure of respiratory complex I from Escherichia coli in ice in the presence of nucleotides. Biochimica et Biophysica Acta 1777, 711–718. [DOI] [PubMed] [Google Scholar]
- Murcha MW, Wang Y, Narsai R, Whelan J. 2014The plant mitochondrial protein import apparatus — The differences make it interesting. Frontiers of Mitochondrial Research 1840, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Stamm R, Eibl H. 1985. Clear background and highly sensitive protein staining with coomassie blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6, 427–448. [Google Scholar]
- Neuhoff V, Stamm R, Pardowitz I, Arold N, Ehrhardt W, Taube D. 1990. Essential problems in quantification of proteins following colloidal staining with coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11, 101–117. [DOI] [PubMed] [Google Scholar]
- Oliver DJ. 1994. The glycine complex from plant-mitochondria. Annual Review of Plant Physiology and Plant Molecular Biology 45, 323–337. [Google Scholar]
- Ostersetzer-Biran O. 2016. Respiratory complex I and embryo development. Journal of Experimental Botany 67, 1205–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP. 2005. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. Journal of Molecular Biology 350, 263–277. [DOI] [PubMed] [Google Scholar]
- Perales M, Parisi G, Fornasari MS, et al. 2004. Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Molecular Biology 56, 947–957. [DOI] [PubMed] [Google Scholar]
- Peterhänsel C, Horst I, Niessen M, Blume C, Kebeish R, Kurkcuoglu S, Kreuzaler F. 2010. Photorespiration. The Arabidopsis book / American Society of Plant Biologists 8, e0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Braun HP. 2012. Comparative analyses of protein complexes by blue native DIGE. Methods in Molecular Biology (Clifton, N.J.) 854, 145–154. [DOI] [PubMed] [Google Scholar]
- Peters K, Belt K, Braun HP. 2013. 3D gel map of Arabidopsis complex I. Frontiers in Plant Science 4, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Layoune O, Danon A, Paepe R de. 2008. L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. The Journal of Biological Chemistry 283, 32500–32505. [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Geisler DA, Møller IM. 2008. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8, 47–60. [DOI] [PubMed] [Google Scholar]
- Roussell DL, Thompson DL, Pallardy SG, Miles D, Newton KJ. 1991. Chloroplast structure and function is altered in the NCS2 maize mitochondrial mutant. Plant Physiology 96, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, De Pape R, de Kouchkovsky Y. 2000. Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris . Plant Physiology 124, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA. 2007. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46, 2275–2288. [DOI] [PubMed] [Google Scholar]
- Schertl P, Braun HP. 2014. Respiratory electron transfer pathways in plant mitochondria. Frontiers in Plant Science 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt FJ, Renger G, Friedrich T, Kreslavski VD, Zharmukhamedov SK, Los DA, Kuznetsov VV, Allakhverdiev SI. 2014. Reactive oxygen species: re-evaluation of generation, monitoring and role in stress-signaling in phototrophic organisms. Biochimica et Biophysica Acta 1837, 835–848. [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB. 2006. Mitochondrial metabolism in developing embryos of Brassica napus . The Journal of Biological Chemistry 281, 34040–34047. [DOI] [PubMed] [Google Scholar]
- Soto D, Córdoba JP, Villarreal F, Bartoli C, Schmitz J, Maurino VG, Braun HP, Pagnussat GC, Zabaleta E. 2015. Functional characterization of mutants affected in the carbonic anhydrase domain of the respiratory complex I in Arabidopsis thaliana . The Plant Journal 85, 831–844. [DOI] [PubMed] [Google Scholar]
- Sunderhaus S, Dudkina NV, Jänsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun HP. 2006. Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. The Journal of Biological Chemistry 281, 6482–6488. [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. 2010. Not just a circle: flux modes in the plant TCA cycle. Trends in Plant Science 15, 462–470. [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. 2013. SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Research 41, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Van Aken O, Whelan J, van Breusegem F. 2010. Prohibitins: mitochondrial partners in development and stress response. Trends in Plant Science 15, 275–282. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC. 2013. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. International Journal of Molecular Sciences 14, 6805–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR, Zhu J, Hirst J. 2014. Architecture of mammalian respiratory complex I. Nature 515, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Fristedt R, Yu X, Chen Z, Liu H, Lee Y, Guo H, Merchant SS, Lin C. 2012. The gamma-carbonic anhydrase subcomplex of mitochondrial complex I is essential for development and important for photomorphogenesis of Arabidopsis. Plant Physiology 160, 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lyu W, Berkowitz O, et al. 2016. Inactivation of mitochondrial complex I induces the expression of a twin-cysteine protein that targets and affects cytosolic, chloroplastidic and mitochondrial function. Molecular Plant : in press, doi: 10.1016/j.molp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. 1993. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. Journal of Molecular Biology 233, 109–122. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun HP. 2001. Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiology 125, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schägger H. 2006. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochimica et Biophysica Acta 1757, 1066–1072. [DOI] [PubMed] [Google Scholar]
- Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J. 2013. The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis. The Plant Cell 25, 4014–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Matsuno-Yagi A. 2003. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry 42, 2266–2274. [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Martin MV, Braun HP. 2012. A basal carbon concentrating mechanism in plants? Plant Science 187, 97–104. [DOI] [PubMed] [Google Scholar]
- Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalbe H, Hunte C, Brandt U. 2015. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.