Significance
Efficient cost-effective single-molecule sequencing platforms will facilitate deciphering complete genome sequences, determining haplotypes, and identifying alternatively spliced mRNAs. We demonstrate a single-molecule nanopore-based sequencing by synthesis approach that accurately distinguishes four DNA bases by electronically detecting and differentiating four different polymer tags attached to the terminal phosphate of the nucleotides during their incorporation into a growing DNA strand in the polymerase reaction. With nanopore detection, the distinct polymer tags are much easier to differentiate than natural nucleotides. After tag release, growing DNA chains consist of natural nucleotides allowing long reads. Sequencing is realized on an electronic chip containing an array of independently addressable electrodes, each with a single polymerase–nanopore complex, potentially offering the high throughput required for precision medicine.
Keywords: single-molecule sequencing, nanopore, DNA sequencing by synthesis, polymer-tagged nucleotides, chip array
Abstract
DNA sequencing by synthesis (SBS) offers a robust platform to decipher nucleic acid sequences. Recently, we reported a single-molecule nanopore-based SBS strategy that accurately distinguishes four bases by electronically detecting and differentiating four different polymer tags attached to the 5′-phosphate of the nucleotides during their incorporation into a growing DNA strand catalyzed by DNA polymerase. Further developing this approach, we report here the use of nucleotides tagged at the terminal phosphate with oligonucleotide-based polymers to perform nanopore SBS on an α-hemolysin nanopore array platform. We designed and synthesized several polymer-tagged nucleotides using tags that produce different electrical current blockade levels and verified they are active substrates for DNA polymerase. A highly processive DNA polymerase was conjugated to the nanopore, and the conjugates were complexed with primer/template DNA and inserted into lipid bilayers over individually addressable electrodes of the nanopore chip. When an incoming complementary-tagged nucleotide forms a tight ternary complex with the primer/template and polymerase, the tag enters the pore, and the current blockade level is measured. The levels displayed by the four nucleotides tagged with four different polymers captured in the nanopore in such ternary complexes were clearly distinguishable and sequence-specific, enabling continuous sequence determination during the polymerase reaction. Thus, real-time single-molecule electronic DNA sequencing data with single-base resolution were obtained. The use of these polymer-tagged nucleotides, combined with polymerase tethering to nanopores and multiplexed nanopore sensors, should lead to new high-throughput sequencing methods.
The importance of DNA sequencing has increased dramatically from its inception four decades ago. It is recognized as a crucial technology for most areas of biology and medicine and as the underpinning for the new paradigm of personalized and precision medicine. Information on individuals’ genomes and epigenomes can help reveal their propensity for disease, clinical prognosis, and response to therapeutics, but routine application of genome sequencing in medicine will require comprehensive data delivered in a timely and cost-effective manner (1). Although 35 years of technological advances have improved sequence throughput and have reduced costs exponentially, genome analysis still takes several days and thousands of dollars to complete (1, 2). To realize the potential of personalized medicine fully, the speed and cost of sequencing must be brought down another order of magnitude while increasing sequencing accuracy and read length. Single-molecule approaches are thought to be essential to meet these requirements and offer the additional benefit of eliminating amplification bias (3, 4). Although optical methods for single-molecule sequencing have been achieved and commercialized, the most successful, Pacific Biosciences’ single molecule real-time (SMRT) sequencing by synthesis (SBS) approach, requires expensive instrumentation and the use of fluorescently tagged nucleotides (4, 5).
In the last two decades, there has been great interest in taking advantage of nanopores, naturally occurring or solid-state ion channels, for polymer characterization and distinguishing the bases of DNA in a low-cost, rapid, single-molecule manner (6–9). Three nanopore sequencing approaches have been pursued: strand sequencing in which the bases of DNA are identified as they pass sequentially through a nanopore (6, 7), exonuclease-based nanopore sequencing in which nucleotides are enzymatically cleaved one-by-one from a DNA molecule and monitored as they are captured by and pass through the nanopore (10), and a nanopore SBS approach, described by us, in which identifiable polymer tags are attached to nucleotides and registered in nanopores during enzyme-catalyzed DNA synthesis (11). Common to all these methods is the need for precise control of the reaction rates so that each base is determined in order and without fail. Strand sequencing requires a method for slowing down the passage of the DNA through the nanopore and decoding a plurality of bases within the channel; complex ratcheting approaches, taking advantage of molecular motors, have been developed for this purpose (12, 13) and were used recently to assemble the sequence of phiX174 (14). Exonuclease-based sequencing requires the release of each nucleotide close enough to the pore to guarantee its capture and its transit through the pore at a rate slow enough to obtain a valid ionic current signal (10, 15). In addition, both of these methods rely on distinctions among the four natural bases, two relatively similar purines and two similar pyrimidines. Our nanopore SBS approach has the advantage of using synthetic polymer tags attached to the nucleotides that are designed specifically to produce unique and readily distinguishable ionic current blockade signatures for sequence determination (11). Direct attachment of the polymerase to the nanopore ensures that there is ample time for the polymer tags on the incoming nucleotide to be captured in the nanopore before completion of the DNA polymerase catalytic cycle. The rates of exonucleases are sensitive to sequence context and secondary structure (10, 15–17). In contrast, polymerases have less variable rates than exonucleases and therefore will yield more consistent base identification in nanopore SBS.
We recently demonstrated proof-of-principle for the nanopore SBS sequencing method (11), which combines SBS (18–20) with nanopore-based identification of different-sized polymer tags (8, 9) attached to the nucleotides. One of four different-length PEG tags was attached to the terminal phosphate of each nucleotide. Despite having long tags with 16–36 PEG monomer units, these tagged nucleotides were incorporated efficiently by DNA polymerase. During the phosphoryl transfer step of the DNA polymerase reaction, the tag is released as part of the polyphosphate byproduct, so only the natural nucleotide remains in the growing DNA strand. This tag was detected and identified by monitoring pore current as it passed through a single-protein nanopore (α-hemolysin, hereafter αHL) embedded in a lipid membrane under a voltage gradient. Depending on the length of the PEG tag, the pore current was reduced to different levels, and translocation required different times (11), allowing discrimination of such tags and enabling the identification of each nucleotide incorporated in the SBS process.
The measurements with PEG tags were made using high salt concentrations, which are not ideal for polymerase reactions. To develop the nanopore SBS approach further and to optimize the tags, we report here the design and synthesis of nucleotides tagged with modified oligonucleotides and their application for nanopore SBS. These tags have structural modifications that create distinct current blockades, measured using an electronic chip-based array of nanopores embedded in lipid bilayer membranes. The tags are attached to the terminal phosphate of 2′-deoxynucleoside-5′-hexaphosphates using Huisgen cycloaddition azide/alkyne coupling chemistry (21). Tagged nucleoside hexaphosphates were chosen because they generally have better activity with DNA polymerases than tagged nucleoside triphosphates (22, 23). With these tagged nucleotides, we demonstrate continuous single-molecule electronic DNA sequencing with single-base resolution by nanopore SBS. The measurement of current is made during the polymerase catalytic cycle when the complementary tagged nucleotide is bound within the complex of DNA polymerase, primer/template, and divalent metal cation and lasts until the completion of the polymerase catalytic step with the release of the tagged polyphosphate product. Once this product is released, the polymer tag is free to leave the pore, ending the blockade signal for that particular DNA synthesis step. To increase the likelihood that each tag will be measured in sequential order, a single polymerase molecule is covalently attached to the nanopore at an appropriate distance to allow fast capture of the tag by the nanopore (Fig. 1A). Each of the four tags has a distinctive structure that interacts with the narrowest constriction in the αHL channel, thereby reducing the ionic current across the channel to different extents (24, 25).
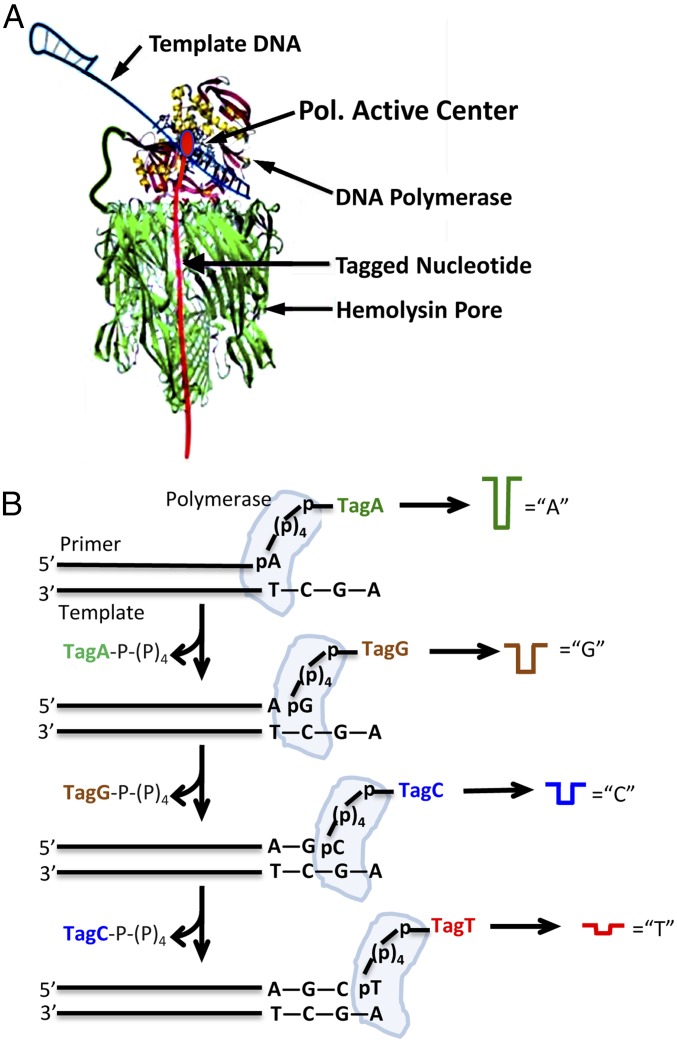
Fig. 1.
Principle of nanopore SBS. (A) Nanopore–polymerase sequencing engine. A single DNA polymerase molecule is covalently attached to an αHL nanopore heptamer. Primer and template DNA (shown as a double-hairpin conformation) along with the tagged nucleotide form a ternary complex with the polymerase. (B) SBS schematic showing the sequential capture and detection of tagged nucleotides by the nanopore as they are incorporated into the growing DNA strand in the polymerase reaction to produce nucleotide-specific current blockades.
Results and Discussion
We previously demonstrated the ability to identify PEG tags of different lengths by their distinct blockade currents in membranes traversed by a single αHL nanopore (8, 11). The instrumentation and testing method used made it possible to record rapid, submillisecond deviations from open current readings (9). Here we used a complementary metal-oxide semiconductor (CMOS) chip containing an independently addressable array of 264 electrodes supporting independent bilayers. In the prototype chip, there was a minimum measurement period of ∼1 ms, which required a revised approach for tag detection.
Fig. 1A shows the nanopore–polymerase sequencing engine with a single Phi29 DNA polymerase molecule (26) covalently attached to αHL (27). Polymerase binds primer and template DNA, along with the complementary tagged nucleotide, allowing placement of the tag within the nanopore. The schematic in Fig. 1B shows the sequential capture and detection of tagged nucleotides by the nanopore as they are being incorporated into the growing primer strand during the polymerase reaction. Preliminary experiments using streptavidin-bound 3′-biotinylated oligonucleotides following published procedures (25) confirmed that it is possible to capture and hold oligonucleotides in chip-based pores and to measure current blockade levels over a period of ∼50–2,000 ms or longer. A survey of oligonucleotide tags, all based on modifications of oligo dT30, was conducted, yielding the four tags shown in Fig. S1 that generate distinguishable current blockade signatures. For sequencing we decided to mimic the experiment that generates the current blockade data shown in Fig. S1 by replacing the streptavidin–tag complex with a DNA polymerase ternary complex that holds the tagged nucleotides (Fig. 2), allowing the monitoring of current while the tags are still attached to the nucleotides. Thus, detection of the tag occurs before nucleotide incorporation by phosphoryl transfer, rather than after the tags are detached from their nucleotides to pass through the nanopore channel. The tag detection can be accomplished if the rate of the nucleotide incorporation is slower than the combined rate of nucleotide tag capture by the nanopore and current blockade measurement. The use of appropriate polymerase mutants and refined reaction conditions can increase the time the tagged nucleotide is present in the ternary complex, and close tethering, via covalent attachment, of the polymerase to the nanopore can result in rapid tag captures on the order of microseconds.
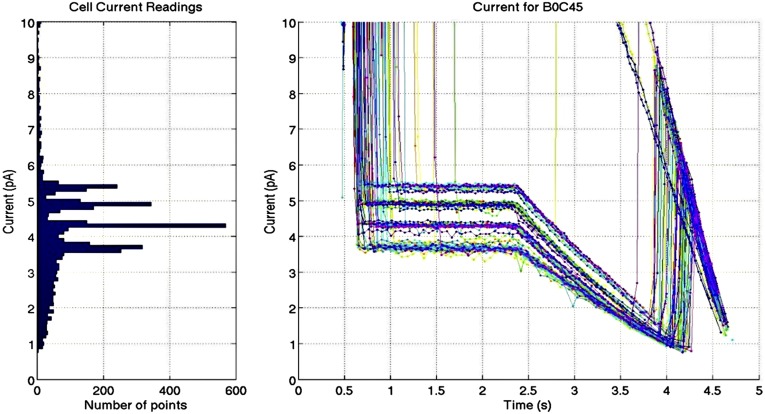
Fig. S1.
Current blockade levels using αHL pores and biotinylated oligonucleotides bound to streptavidin. (Right) Multiple current measurement time courses are overlaid showing the blockade levels obtained once an oligonucleotide tag is held in the pore by the streptavidin with an applied voltage of 80 mV. (Left) A histogram of the current measurements shows four peaks for the four modified oligonucleotides. Open-pore current is ∼20 pA in the presence of 0.3 M KCl. The four oligonucleotides used for the above experiments were dSp8: 5′-TTTTT TTTTT TTSSS SSSSS TTTTT-Biotin-3′ (5.5 pA); dSp3: 5′-TTTTT TTTTT TTTTT SSSTT TTTTT-Biotin-3′ (4.9 pA); dT30: 5′-TTTTT TTTTT TTTTT TTTTT TTTTT-Biotin-3′ (4.4 pA); and FldT2: 5′-TTTTT TTTTT TTTTT FTFTT TTTTT-Biotin-3′ (3.8 pA). Changes in the oligo dT30 sequence are shown in bold, with S referring to the abasic nucleotide, dSp, and F to FldT (Glen Research). The numbers in parentheses refer to the typical blockade currents.
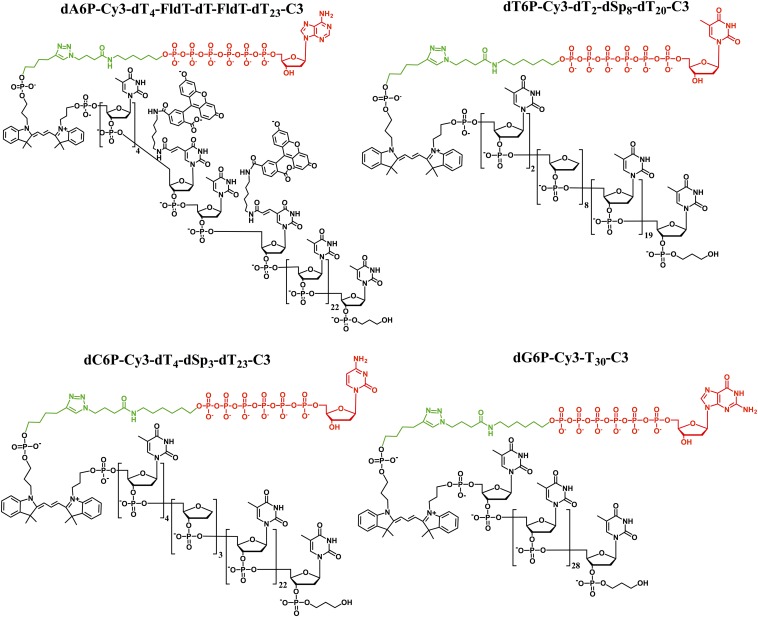
Fig. 2.
Structures of the four polymer-tagged nucleotides. Nucleotides used in this study are 5′-nucleoside hexaphosphates (red) connected to a common linker (green) and an oligonucleotide tag (black) consisting of Cy3 and either an unmodified oligonucleotide chain (dT30) (in dG6P-Cy3-dT30-C3) or oligonucleotides with a variety of modifications including runs of abasic nucleotides (dSp8 and dSp3) (in dT6P-Cy3-dT2-dSp8-dT20-C3 and dC6P-Cy3-dT4-dSp3-dT23-C3) or two thymidines modified with fluorescein (in dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3).
Synthesis and Testing of Tagged Nucleotides for Nanopore SBS Reactions.
We designed and synthesized a wide variety of tagged nucleotides. After characterization of their properties and performance, we selected a subset of four tagged nucleotides that produce distinct and reproducible reduction in nanopore ionic currents with minimum background. The structures of these tagged nucleotides are shown in Fig. 2. The synthetic strategy is to attach an azide group via a chemical linker to the terminal phosphate of the deoxynucleoside-5′-hexaphosphate (dA6P, dC6P, dG6P, and dT6P, the dN6Ps) (Fig. S2). The tags were oligonucleotide-based polymers synthesized with a 5′-alkyne moiety that reacts readily with the desired azido-dN6P by azide-alkyne Huisgen cycloaddition (21, 28, 29) to produce the tagged nucleotides (Fig. 2). The tagged nucleotides were purified by HPLC, and their molecular weights were determined by MALDI-TOF MS (Fig. S3). The tags consisted of oligodeoxynucleotides (e.g., dT30), some of which are interspersed with modified phosphodiester building blocks having specific base or backbone modifications. These modifications include bulky, charged dyes on the nucleotide base and runs of three or eight abasic (dSpacer, hereafter “dSp”) sites (30, 31). Tags were protected at their 3′ end from exonuclease activity with 3′-phospho-propanol. The modifications in the dT30 tag sequence were positioned to be at or near the narrowest constriction in the αHL channel (24), where they can directly and specifically influence ionic current through the pore when the tagged nucleotide is held by polymerase in the ternary complex attached to the pore.
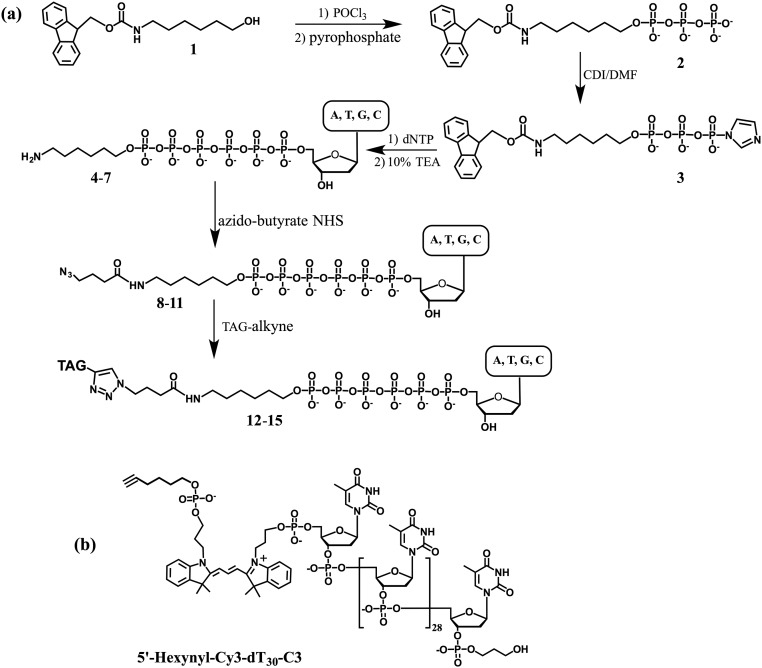
Fig. S2.
(A) Scheme for synthesis of tagged nucleotides (see details in SI Methods). (B) Structure of a TAG-alkyne, using the dT30 tag as an example.
Fig. S3.
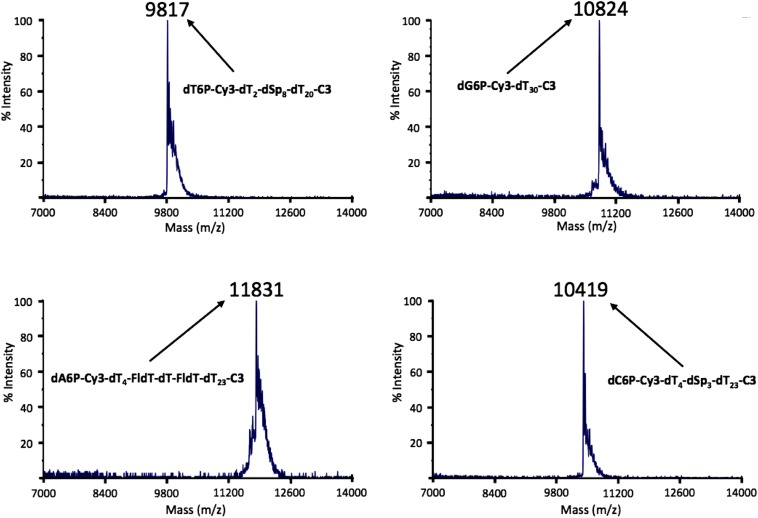
MALDI-TOF MS spectra of nucleotides tagged with oligonucleotide-based polymers. The measured values are indicated in the spectra. The calculated molecular masses of these nucleotides are 9,808 Da for dT6P-Cy3-dT2-dSp8-dT20-C3, 10,826 Da for dG6P-Cy3-dT30-C3, 11,835 Da for dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3, and 10,413 Da for dC6P-Cy3-dT4-dSp3-dT23-C3. All measured values were within 0.1% of the calculated masses.
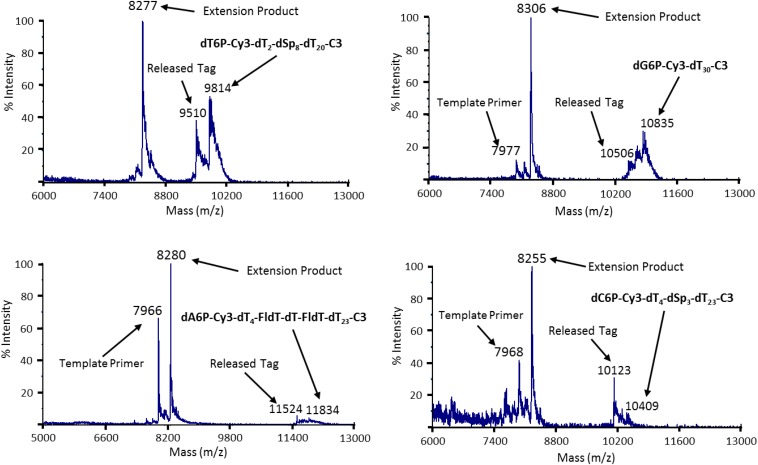
To demonstrate that the nucleotides modified with the polymeric tags were still good substrates for DNA polymerase, single-base extension reactions were performed using Therminator γ DNA polymerase. Primer–loop templates bearing complementary bases opposite the site of incorporation on the primer were used (11). The reaction products were desalted and subjected to MALDI-TOF MS analysis. As shown in Fig. S4, successful incorporations of the tagged nucleotides were revealed by the presence of new peaks with the expected mass of the primer-extension products and peaks indicative of the released pentaphosphate tag, along with peaks representing remaining reactants (primer, tagged nucleotide).
Fig. S4.
Single-base primer extension using four different tagged nucleotides characterized by MALDI-TOF MS. Reactions with the four tagged nucleotides (Fig. 2) along with the appropriate loop–primer template (3′-TCGAGCGCCGCGCCTTGGCGCGGCGC-5′ for T, 3′-GATCGCGCCGCGCCTTGGCGCGGCGC-5′ for G, 3′-CGATGCGCCGCGCCTTGGCGCGGCGC-5′ for A, and 3′-ATCGGCGCCGCGCCTTGGCGCGGCGC-5′ for C) yielded single-base extension products of the expected masses (8,277, 8,306, 8,280, and 8,255 for the T-, G-, A-, and C-tagged nucleotides, respectively). Peaks for the primers, the tagged nucleotides, and the released tagged pentaphosphate products are indicated as well.
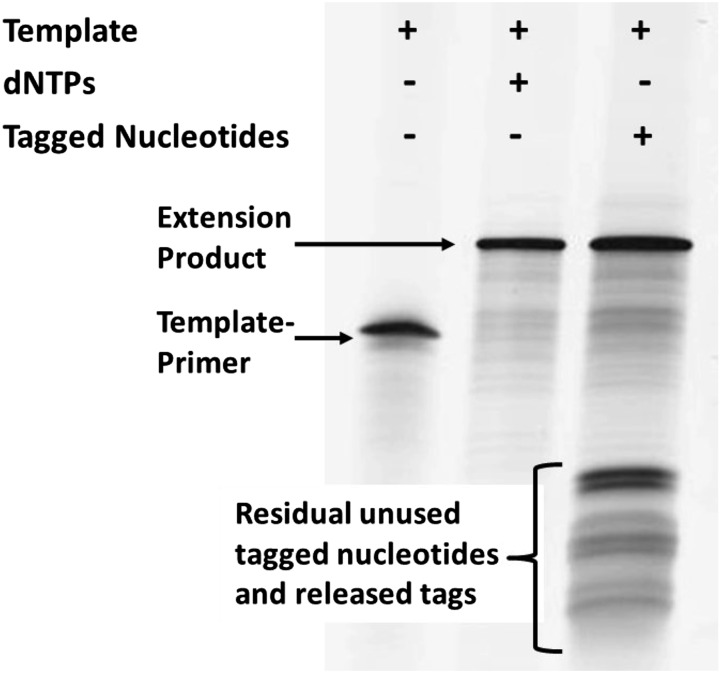
Additional experiments monitoring DNA synthesis on denaturing polyacrylamide gels show that primers can be extended by more than 40 bases using only tagged nucleotides and any of several DNA polymerases. In Fig. 3, we show the result of extension reactions using Bst 2.0 DNA polymerase (an in silico-designed homologue of Bacillus stearothermophilus DNA polymerase I, large fragment from New England Biolabs) at 37 °C, with reactants and products separated on a polyacrylamide-urea gel. The primer/template DNA (83-mer) is self-priming and can be extended by 47 nucleotides. Full-length products were obtained with either all four natural dNTPs or with the four tagged nucleotides depicted in Fig. 2.
Fig. 3.
Full-length polymerase extension products with dNTPs (center lane) or tagged nucleotides (right lane). Reactions were conducted with 3 pmol template (83 bases) and 75 pmol dNTPs or tagged dN6Ps, for 2–10 min at 37 °C, and the products were analyzed on a denaturing polyacrylamide gel.
Assembly of αHL–Polymerase Complexes and Insertion into Membranes on an Array Chip.
To achieve continuous sequencing, we constructed a sequencing engine having a single DNA polymerase molecule attached directly to the heptameric αHL nanopore (Fig. 1A) using a recombinant technique to make two kinds of αHL monomers, one wild type and the other differing only in carrying a specific peptide domain (32) for attaching polymerase. These two types of monomers were mixed in appropriate ratios, allowing pore heptamers to self-assemble, followed by isolation of the heptamer pore carrying a single-peptide domain. When polymerase engineered to possess the mating protein fragment was added in excess to this heterologous pore, it reacted with the pore monomer tagged with the peptide domain, yielding the desired nanopore–polymerase conjugate (the procedure is described in SI Methods). During nanopore SBS, the polymerase attached to the pore tightly binds the DNA template/primer, completing a single structure that integrates itself into a lipid bilayer configured for monitoring transmembrane conductance and ready for sequencing DNA. When each complementary tagged nucleotide binds the polymerase–primer–template complex, its tag rapidly enters the nanopore channel in response to applied voltage to yield a unique current blockade that is recognized for sequence determination.
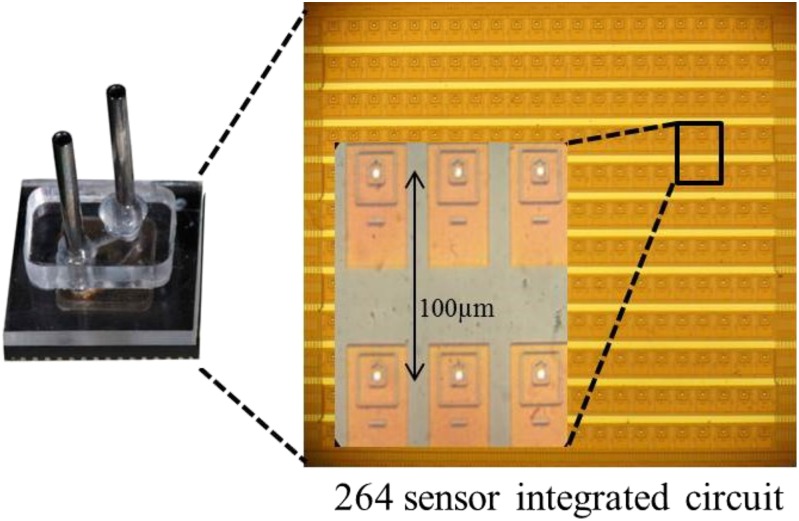
Sequencing experiments were performed using a CMOS nanopore chip (Fig. 4) that has an array of 264 Ag-AgCl electrodes (5-µm diameter) within shallow wells that support lipid bilayer membranes. The electrodes are individually addressable by a computer interface, allowing voltages to be applied only to the membranes in specific wells and thus effectively permitting independent sequence reads at these locations. All reagents are introduced into a simple flow cell above the chip using a computer-controlled syringe pump. Electrical measurements are made asynchronously at least once every millisecond and are recorded on the computer.
Fig. 4.
CMOS chip for nanopore SBS. The CMOS chip (∼1 × 1 mm) consists of an integrated circuit with an array of 264 silver electrodes (5-µm diameter). The chip supports analog-to-digital conversion, reporting electrical measurements from all electrodes independently at a rate of more than 1,000 per second. The flow cell (Left) surrounding the chip (Right) includes sample input and output ports for reagents. The Inset shows details of the architecture of the electrode array and individual sensing cells.
An 83-base synthetic primer/template, which self-anneals at both ends to prevent its entry into the nanopore, was added to the pore–polymerase complex and pumped over the membranes in a sensor chip flow cell in Hepes buffer (pH 7.5) containing 150 mM KCl and 3 mM SrCl2. The applied voltage is adjusted to ensure that, in a majority of cases, one and only one pore is inserted into the membranes of each well. For preliminary experiments using only a single tagged nucleotide and the noncatalytic divalent metal ion (Sr2+), the tagged nucleotide mix was added to the nanopore chip at room temperature, and current recording was started. These experiments established that binding of the tagged nucleotides with the polymerase–nanopore conjugate along with the primer and template to form a ternary complex is sequence-dependent and that the current levels for the four nucleotides are readily distinguished. Representative data for a chemically conjugated polymerase–pore complex are shown in Fig. S5B and Table S1.
Fig. S5.
Capture of single tagged nucleotides by the exo− Bst DNA polymerase–α-HL conjugate. (A) The primer/template DNA used in this experiment. The DNA strand is designed to fold back on itself at both the 5′ and 3′ ends to prevent direct entry into hemolysin pores. The next available base in the template strand is a cytosine, selecting deoxyguanosine monophosphate (dGMP) as the next incorporated nucleotide. (B) Ionic current blockades produced when dG6P-Cy3-dT30-C3 is added to polymerase–hemolysin conjugate pores in the presence of SrCl2 (Sr2+ is a noncatalytic metal ion). Binding of the nucleotide and threading of the tag transiently reduces the current from the open level (∼15 pA) to ∼6 pA. (C) A magnified portion of five blockade events, each lasting about 5–35 ms. Averaging the current levels for ∼500 such events yields an average duration of 29 ms and an average current level of 30–40% of the open-pore current. Similar measurements using suitable primer/template DNAs and the other three tagged nucleotides (Fig. 2), recording both the current blockade level and the duration of all capture events longer than 10 ms, were made also. The results are shown in Table S1. The blockade levels readily distinguish the four tagged nucleotides. (D) Scheme for covalent site-specific attachment of the Bst polymerase to a modified αHL nanopore using a Diels–Alder fast click conjugation reaction. The procedure is described in SI Methods.
Table S1.
Distinct current blockade levels (I/Io) and dwell times (Tdwell) for the tagged nucleotides
| Nucleotide | Current blockade level | Average Tdwell, ms |
| dT6P-Cy3-dT2-dSp8-dT20-C3 | 0.5–0.6 | 17 |
| dC6P-Cy3-dT4-dSp3-dT23-C3 | 0.4–0.5 | 30 |
| dG6P-Cy3-dT30-C3 | 0.3–0.4 | 29 |
| dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3 | 0.2–0.3 | 17 |
The experimental data shown in Fig. S5 and this table were obtained by adding only the complementary (matching) tagged nucleotides in the reaction mixture. For mismatch testing three noncomplementary tagged nucleotides were used instead and produced few blockade events, all of which were shorter than 20 ms, with current levels above 0.6 of the open-pore current level. We estimate that at least 93% of the events observed result from sequence-specific binding within the polymerase/template complex. I, blockade current; Io, open-pore current.
Nanopore SBS.
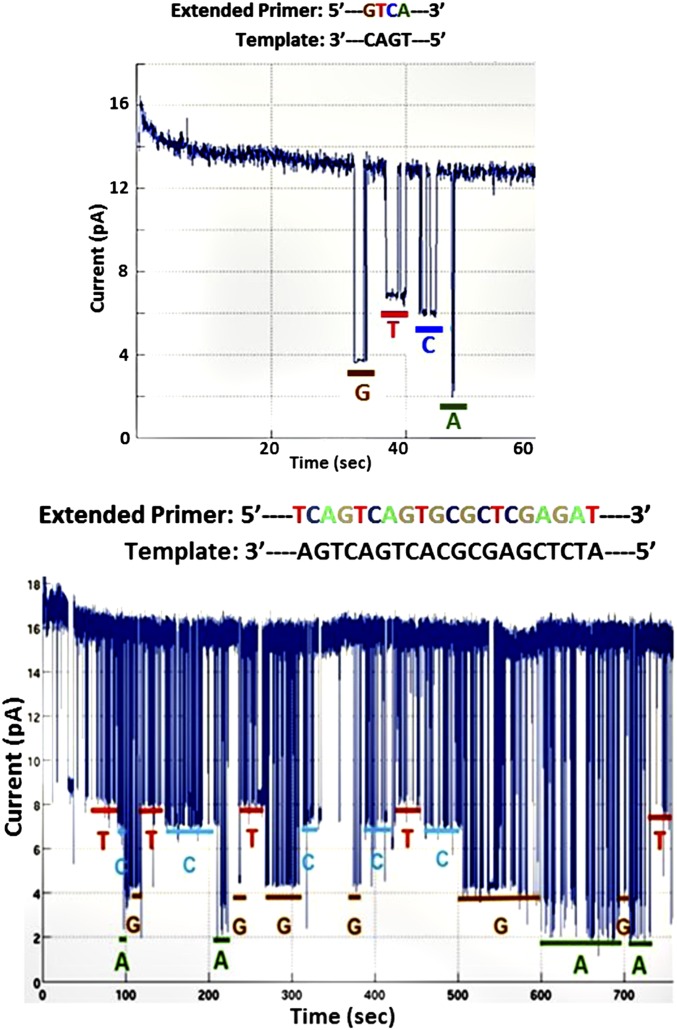
For sequencing experiments, a buffer solution containing 150 mM KCl, 3 mM SrCl2, and either 0.1 mM MnCl2 or 3 mM MgCl2 (catalytic divalent cations) and 3 µM each of the four tagged nucleotides was added to the polymerase–nanopore chip constructed using the peptide-based conjugation method to start the SBS process. Ionic currents were measured by applying 80–120 mV across the membranes for 3–10 min. The sequencing data are shown in Fig. 5. Four distinct transient current levels substantially below the open-pore current level [∼13 pA (Fig. 5, Upper) and ∼16 pA (Fig. 5, Lower)] are observed, indicating pore captures of the tags associated with each nucleotide during each cycle of the polymerase reaction. These data are in accord with the levels observed when only a single tagged nucleotide and only noncatalytic divalent metal (Sr2+) were used (Table S1). In these traces, the correct sequence order for four (Fig. 5, Upper) and 20 bases (Fig. 5, Lower), respectively, was observed.
Fig. 5.
Sequencing results on nanopore array chips. Sequencing reactions were performed with inserted αHL pores conjugated to a single Phi29 DNA polymerase molecule, template, and the four tagged nucleotides. (Upper) Four bases are clearly distinguished (3 mM SrCl2 on the trans side and 0.1 mM MnCl2 on the cis side of the membrane). (Lower) A longer sequence read (3 mM SrCl2 trans, 3 mM MgCl2, 0.7 mM SrCl2 cis).
As expected, using the data in Fig. 5, Lower as an example, the bulkiest tag, in dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3, containing two fluorescein-modified dTs (FldT), gave the lowest current (∼2 pA); the unmodified dT30 tag in dG6P-Cy3-dT30-C3 produced an intermediate current (∼4 pA), whereas higher currents were observed with the less bulky tags containing stretches of an abasic nucleotide, dSp, in dC6P-Cy3-dT4-dSp3-dT23-C3 (∼7 pA) and dT6P-Cy3-dT2-dSp8-dT20-C3 (∼8 pA).
In many cases (Fig. 5, Lower), a nucleotide tag appears to be captured several times in succession (“stuttering” at a given blockade current level) before the tag of the next nucleotide in the sequence is observed. The stuttering is presumably caused by the tagged nucleotide being repeatedly bound to and released from the polymerase before incorporation. Such stuttering can be reduced by changing experimental conditions. As seen in Fig. 6, the use of a 100-mV transmembrane potential and 150 mM KCl, along with 3 mM MgCl2 on the cis side of the membrane and 3 mM SrCl2 on the trans but not on the cis side of the membrane results in less stutter and also decreases the average duration of nucleotide captures. Such adjustment in the relative concentration of catalytic and noncatalytic metal cofactors is one means of making the tagged nucleotide incorporation rate slower than the combined rate of tag capture and detection by the pore. In this example, using a template with alternating homopolymer stretches and carefully considering the duration of each nucleotide capture event, we have obtained nearly perfect sequence reads with minimal stutter. Additional data on homopolymer sequencing are shown in Fig. S6. Optimization of the average duration of nucleotide capture events should allow further increases in sequencing accuracy. Base calling was carried out by manual inspection of the current level of each deflection, ignoring ones with dwell times less than 10 ms.
Fig. 6.
Nanopore SBS results for a 12-base homopolymeric region of DNA. (Upper) A double-hairpin primer template was used to prevent template ends from entering the nanopore. The sequence immediately downstream from the 3′ end encodes 5′-GGGGAAAATTTT-3′. (Lower) Brief reductions in current are indicative of tag capture within the pore during the polymerase reaction, with the depth of the deflection characteristic of the different structures of the four tags. Other deflections in this figure, including the one at center, are brief (<10 ms) and were ignored as noise. The current trace shown is raw data output without noise reduction. Additional representative sequence data are shown in Fig. S6.
Fig. S6.
Additional examples of homopolymer sequence reads. The experiment was carried out as described for Fig. 6 in the main text, using the same tagged nucleotides and polymerase–nanopore complexes. In this case, however, 3 mM SrCl2 and 0.1 mM MnCl2 were present on the trans side and 0.1 mM MnCl2 was present on the cis side of the membrane. Deflections less than 10 ms were discounted. Letters shown in gray (the fourth “T” in the middle trace and the fourth “A” in the lower trace) in the extended primer strand indicate bases that were missed in these reads.
In summary, the sequencing data (Figs. 5 and 6 and Fig. S6) clearly indicate that our nanopore SBS method is capable of generating clear, sequence-dependent electronic signal streams at the single-molecule level in a continuous process that requires no addition or removal of reagents, sequencing an array of multiple DNA templates in parallel in a short time. Although the rate of sequencing and overall accuracy of nanopore SBS remain to be established, the system has many parameters that can be optimized further. These include synthesis of additional tagged nucleotides that offer better-resolved current blockade signatures, improvement of reaction conditions, and the testing of a variety of DNA polymerases that have improved rates, without the use of SrCl2 to moderate the progression of DNA synthesis. With careful attention to these details, along with the construction of integrated circuits with many more electrodes and faster measurement capabilities, we expect to be able to create a high-throughput single-molecule electronic DNA sequencing system.
SI Methods
Synthesis of the Tagged Nucleotides Shown in Fig. 2 by Coupling dN6P-N3 to 5′-Hexynyl Oligonucleotide Tags.
The overall synthetic scheme is presented in Fig. S2.
Synthesis of Fmoc-6-Aminohexyltriphosphate (Fig. S2, Compound 2).
Fmoc-6-aminohexanol (1 g, 2.94 mmol) (Fig. S2, compound 1) was coevaporated with anhydrous acetonitrile (2 × 20 mL) and then was dissolved in trimethyl phosphate (10 mL). Phosphorous oxychloride (550 µL, 5.88 mmol) was added to this cooled and stirred solution, and the mixture was stirred for 2 h. To the reaction mixture, tributylammonium pyrophosphate [5 eq., 15 mmol, 0.5 M solution in anhydrous N,N-dimethylformamide (DMF)] and tributylamine (15 mmol) were added and stirred for 20 min. The solution was quenched with 0.1 M triethylammonium bicarbonate (TEAB) buffer (200 mL, pH 7.5) and was adjusted to pH ∼7. This solution was loaded on a Sephadex A-25 ion-exchange column and eluted using a 0.1–1.0 M TEAB buffer gradient (pH 7). The appropriate fractions were pooled and further purified by reverse-phase HPLC to provide pure triphosphate (Fig. S2, compound 2). 31P-NMR (D2O): δ, −10.5 [multiplet (m), 2P], −22.8 [triplet (t), 1P].
Synthesis of dN6P-NH2 Nucleotides (Fig. S2, Compounds 4–7).
Fmoc-aminohexyl triphosphate (200 mg, 0.35 mmol) (Fig. S2, compound 2) was coevaporated with anhydrous acetonitrile (2 × 10 mL) and then dissolved in anhydrous DMF (3 mL). Carbonyldiimidazole (4 eq, 1.4 mmol) was added, and the reaction mixture was stirred at room temperature for 4 h to yield compound 3, shown in Fig. S2. Methanol (6 eq, 85 μL) was added, and stirring continued for 30 min. To this reaction mixture a solution of dNTP, tributylammonium salt (0.5 mmol) in 5 mL DMF, and MgCl2 (10 eq, 3.5 mmol) was added. The reaction mixture was stirred for 18 h followed by the addition of 10% triethylamine in water (25 mL) to hydrolyze the Fmoc group. The reaction mixture was stirred further for 16 h; then the precipitated solid was filtered, and the solution was extracted with ether. The aqueous layer was concentrated and purified by reverse-phase HPLC using 0.1 M triethylammonium acetate (TEAC) buffer (pH 7.5) and an acetonitrile gradient to yield the products 4–7 shown in Fig. S2, which were characterized by 31P NMR and MS. 31P-NMR (D2O): δ, −10.6 [broad singlet (bs), 1P], −11.7 [broad singlet (bs), 1P], −23.4 [broad multiplet (bm), 4P]. MALDI TOF MS data: dA6P-NH2 (Fig. S2, compound 4), 832.0 (calculated 830.3); dT6P-NH2 (Fig. S2, compound 5), 826.0 (calculated 821.3); dG6P-NH2 (Fig. S2, compound 6), 848.3 (calculated 846.3); dC6P-NH2 (Fig. S2, compound 7), 807.0 (calculated 806.0).
Synthesis of dN6P-N3 Nucleotides (Fig. S2, Compounds 8–11).
The dN6P-NH2 nucleotides (10 µmol) (Fig. S2, compounds 4–7) were each dissolved in 0.1 M bicarbonate-carbonate buffer (500 µL, pH 8.7), and azidobutyric acid–N-hydroxysuccinimide ester (25 µmol) in 200 µL DMF was added. The reaction mixture was stirred overnight at room temperature and was purified by HPLC using 0.1 M TEAC buffer (pH 7.5) and an acetonitrile gradient to yield the products 8–11 shown in Fig. S2. MALDI TOF MS data: dA6P-N3 (Fig. S2, compound 8), 942.8 (calculated 941.4); dT6P-N3 (Fig. S2, compound 9), 934.6 (calculated 932.3); dG6P-N3 (Fig. S2, compound 10), 960.3 (calculated 957.4); dC6P-N3 (Fig. S2, compound 11), 919.1 (calculated 917.4).
Click Reaction Between dN6P-N3 Nucleotides (Fig. S2, Compounds 8–11) and 5′-Hexynyl Oligonucleotide Tags to Produce Polymer-Tagged dN6P Nucleotides (Fig. S2, Compounds 12–15).
To each 5′-hexynyl-oligonucleotide tag (500 nmol in 200 µL H2O, custom synthesized by TriLink) was added a solution of the corresponding dN6P-N3 nucleotide (750 nmol) followed by the addition of copper bromide (50 µL, 0.1 M solution in 3:1 DMSO/t-BuOH) and Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (100 µL, 0.1 M solution in 3:1 DMSO/t-BuOH). The reaction mixture was stirred at 40 °C for 16 h followed by HPLC purification using 0.1 M TEAC buffer (pH 7.5) and an acetonitrile gradient. The tagged nucleotides 12–15 (Fig. S2) were characterized by MALDI-TOF MS and single-base polymerase extension reaction. MALDI-TOF MS data are shown in Fig. S3: dT6P-Cy3-dT2-dSp8-dT20-C3, 9817 (calculated 9809); dG6P-Cy3-dT30-C3, 10824 (calculated 10827); dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3, 11831 (calculated 11836); dC6P-Cy3-dT4-dSp3-dT23-C3, 10419 (calculated 10414).
Conjugation of Polymerase to the αHL Nanopore.
Attachment of a polymerase to a single subunit of the αHL heptamer was accomplished using either SpyTag and SpyCatcher peptide domains (32) or chemical conjugation. For SpyTag-based conjugation, an αHL pore coupled to a single polymerase molecule was assembled from three proteins: αHL native subunits, αHL with a C-terminal SpyTag peptide, and ϕ29 polymerase with a C-terminal SpyCatcher peptide. After first generating the 1:6 (SpyTag monomer/native monomers) αHL pore by mixing appropriate ratios of the purified monomers and isolating the 1:6 heptamer by chromatography (33, 34), ϕ29 polymerase-SpyCatcher was added to the assembled αHL to allow formation of the polymerase–hemolysin conjugate. Conjugates with 1:6 stoichiometry and maintaining the required functionalities were purified and used in subsequent experiments (33, 34). For chemical conjugation, we used the Diels–Alder fast click reaction (35) shown in Fig. S5D to attach Bst DNA polymerase to αHL. A Bst polymerase molecule containing a single cysteine was reacted with a 100-fold molar excess of 6-methyl-tetrazine (TA)-PEG4-maleimide (Jena Biosciences) for 60 min at room temperature in 1× PBS, 3 mM EDTA, pH 7.0. In a separate tube, an αHL heptamer containing a single cysteine at position 46 of only one monomer was reacted with a 100-fold molar excess of trans-cyclooctene (TCO)-PEG3-maleimide (Jena Biosciences) under the same conditions. The modified proteins were concentrated on Amicon 10K spin filters and further purified on a G50 column. The TCO-containing pore was reacted overnight at 4 °C with a fivefold molar excess of the TA-modified Bst 2.0 polymerase. The resulting polymerase–nanopore conjugate was concentrated on a 100K spin filter and isolated on a Superdex 200 column. These protein conjugates were characterized by gel electrophoresis before testing on 264-sensor integrated circuit chips.
DNA Polymerase Extension Reactions Using Tagged Nucleotides (Shown in Fig. 2).
Extension reactions were carried out using one of four template–loop primers (sequences are shown in legend of Fig. S4) in which the next available base on the template was A, G, C, or T, allowing extension by a single complementary nucleotide. Each extension reaction was performed at 65 °C for 25 min in a 20-μL volume consisting of 3 μM template–loop primer, 2 units of Therminator γ DNA polymerase (New England Biolabs), and 15 μM of the appropriate tagged dN6P nucleotide shown in Fig. 2. DNA extension products were precipitated with ethanol, purified through C18 ZipTip columns (Millipore), and characterized by MALDI-TOF MS analysis (Fig. S4). Full-length extension reactions were performed as indicated in the legend of Fig. 3.
Methods
Synthesis of the Tagged Nucleotides Shown in Fig. 2 by Coupling 2′-Deoxynucleoside-5′-Hexaphosphate-Azides to 5′-Hexynyl Oligonucleotide Tags.
The overall synthetic scheme for the synthesis of tagged nucleotides by coupling 2′-deoxynucleoside-5′-hexaphosphate-azides (dN6P-N3) to 5′-hexynyl oligonucleotide tags is presented in Fig. S2. The detailed synthetic methods and characterization are presented in SI Methods. DNA polymerase extension reactions using these tagged nucleotides are described in SI Methods.
Nanopore SBS with Tagged Nucleotides.
Sequencing reactions on nanopore chips were performed with inserted αHL pores conjugated to a single Phi29 DNA polymerase molecule, any of a variety of primer/template DNAs similar to that shown in Fig. S5A, the four tagged nucleotides (Fig. 2), 150 mM KCl, and 20 mM Hepes buffer (pH 7.5) at room temperature. Nucleotides consist of dA6P-Cy3-dT4-FldT-dT-FldT-dT23-C3, dG6P-Cy3-dT30-C3, dC6P-Cy3-dT4-dSp3-dT23-C3, and dT6P-Cy3-dT2-dSp8-dT20-C3 (3 µM each), with 3 mM SrCl2 on the trans side and 0.1 mM MnCl2 on the cis side of the membrane (e.g., Fig. 5, Upper), 3 mM SrCl2 trans, 3 mM MgCl2, 0.7 mM SrCl2 cis (Fig. 5, Lower), or 3 mM SrCl2 trans, 3 mM MgCl2 cis (Fig. 6). DC voltage of 100 mV was applied during sequencing.
Acknowledgments
This work was supported by NIH Grant R01 HG007415 and Genia Technologies, Inc.
Footnotes
Conflict of interest statement: The nanopore SBS technology has been exclusively licensed by Genia. In accordance with the policy of Columbia University and the National Institute of Standards and Technology, the coinventors (S. Kumar, M.C., C.T., Z.L., S. Kalachikov, J.J.R., J.J.K., and J.J.) are entitled to royalties through this license. G.M.C. is a member of the Scientific Advisory Board of Genia; other potential conflicts are described at arep.med.harvard.edu/gmc/tech.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601782113/-/DCSupplemental.
References
- 1.Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20(9):1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 4.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 5.Korlach J, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–455. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maitra RD, Kim J, Dunbar WB. Recent advances in nanopore sequencing. Electrophoresis. 2012;33(23):3418–3428. doi: 10.1002/elps.201200272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson JW, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci USA. 2007;104(20):8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JW. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc Natl Acad Sci USA. 2010;107(27):12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, et al. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Sci Rep. 2012;2:684. doi: 10.1038/srep00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32(8):829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiner JE, et al. The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine. J Chem Phys. 2012;137(21):214903. doi: 10.1063/1.4766363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panwar AS, Muthukumar M. Enzyme-modulated DNA translocation through a nanopore. J Am Chem Soc. 2009;131(51):18563–18570. doi: 10.1021/ja904047q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301(5637):1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 18.Ju J, et al. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc Natl Acad Sci USA. 2006;103(52):19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, et al. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc Natl Acad Sci USA. 2008;105(27):9145–9150. doi: 10.1073/pnas.0804023105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lallana E, Riguera R, Fernandez-Megia E. Reliable and efficient procedures for the conjugation of biomolecules through Huisgen azide-alkyne cycloadditions. Angew Chem Int Ed Engl. 2011;50(38):8794–8804. doi: 10.1002/anie.201101019. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, et al. Terminal phosphate labeled nucleotides: Synthesis, applications, and linker effect on incorporation by DNA polymerases. Nucleosides Nucleotides Nucleic Acids. 2005;24(5-7):401–408. doi: 10.1081/ncn-200059823. [DOI] [PubMed] [Google Scholar]
- 23.Sood A, et al. Terminal phosphate-labeled nucleotides with improved substrate properties for homogeneous nucleic acid assays. J Am Chem Soc. 2005;127(8):2394–2395. doi: 10.1021/ja043595x. [DOI] [PubMed] [Google Scholar]
- 24.Movileanu L, Cheley S, Howorka S, Braha O, Bayley H. Location of a constriction in the lumen of a transmembrane pore by targeted covalent attachment of polymer molecules. J Gen Physiol. 2001;117(3):239–252. doi: 10.1085/jgp.117.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007;26(14):3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, et al. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal α-hemolysin into heptameric pore structure. Protein Sci. 2011;20(2):448–456. doi: 10.1002/pro.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14):2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262(21):10171–10179. [PubMed] [Google Scholar]
- 31.Gyarfas B, et al. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 A resolution. ACS Nano. 2009;3(6):1457–1466. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 32.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 2012;109(12):E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis R, Chen R, Bibillo A, Korenblum D, Dorwart M. 2014. Nucleic acid sequencing using tags. Patent Application US20140134616.
- 34.Fuller CW, Kumar S, Ju J, Davis R, Chen R. 2015. Chemical methods for producing tagged nucleotides. US Patent Appl US20150368710.
- 35.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc. 2008;130(41):13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]