Significance
Amyotrophic lateral sclerosis (ALS; Lou Gehrig’s disease) is a progressive paralyzing condition affecting ∼1:1,000 adults, associated with death of motor neurons, for which there is no effective treatment. ALS is inherited in 10% of cases, whereas the remainder are “sporadic,” yet all behave very similarly. In mice, the condition has been modeled by transgenesis with mutant versions of superoxide dismutase 1 (SOD1) (∼2% of human cases). Mutant SOD1s misfold and form aggregates inside the cytosol of motor neurons in the spinal cord. Recently, a disaggregating machinery has been described consisting of three chaperones, one of which, Hsp110, plays a rate limiting role. Here, we have transgenically overexpressed Hsp110 in motor neurons of a mutant SOD1 strain and observed extended survival.
Keywords: ALS, chaperone, Hsp110, SOD1, survival
Abstract
Recent studies have indicated that mammalian cells contain a cytosolic protein disaggregation machinery comprised of Hsc70, DnaJ homologs, and Hsp110 proteins, the last of which acts to accelerate a rate-limiting step of nucleotide exchange of Hsc70. We tested the ability of transgenic overexpression of a Thy1 promoter-driven human Hsp110 protein, HspA4L (Apg1), in neuronal cells of a transgenic G85R SOD1YFP ALS mouse strain to improve survival. Notably, G85R is a mutant version of Cu/Zn superoxide dismutase 1 (SOD1) that is unable to reach native form and that is prone to aggregation, with prominent YFP-fluorescent aggregates observed in the motor neurons of the transgenic mice as early as 1 mo of age. The several-fold overexpression of Hsp110 in motor neurons of these mice was associated with an increased median survival from ∼5.5 to 7.5 mo and increased maximum survival from 6.5 to 12 mo. Improvement of survival was also observed for a G93A mutant SOD1 ALS strain. We conclude that neurodegeneration associated with cytosolic misfolding and aggregation can be ameliorated by overexpression of Hsp110, likely enhancing the function of a cytosolic disaggregation machinery.
A number of neurodegenerative diseases are associated with misfolding and aggregation of specific proteins in the cytosol of particular neuronal cell types, including α-synuclein in the striatum in Parkinson’s disease (1) and Cu/Zn superoxide dismutase 1 (SOD1) in motor neurons in ∼2% of ALS cases (2). The cytosol is a surprising location in which to find protein misfolding, even of these abundant proteins, because the cytosol is replete with molecular chaperones that, under normal conditions, prevent or reverse misfolding and aggregation of both nascent and preexisting proteins as an essential function. Notably, however, it appears that differentiated neurons fail to exhibit a “heat shock response”—that is, they appear unable to induce expression of chaperones to counteract misfolding under stress. For example, motor neurons facing a misfolded mutant G85R SOD1 in a transgenic G85R SOD1YFP mouse strain fail to exhibit significant induction of chaperone RNAs, with the exception of an approximately twofold induction of one of the cytosolic Hsp110 chaperones (HspH1) (3). Similar lack of induction has been observed for neurons in culture, where undifferentiated neuronal precursors exhibit induction of the major inducible cytosolic chaperone, Hsp70, upon stress exposure, but, once differentiated, fail to do so (4). The lack of chaperone induction in these contexts leaves open the question of whether, for example, forced overexpression of molecular chaperones could prevent or reverse misfolding of such abundant cytosolic proteins as SOD1 in expressing motor neurons.
Recent studies have suggested that mammalian cells contain a cytosolic disaggregase machinery (5) that performs an action homologous to the Hsp104 or ClpB AAA+ hexameric ring assemblies found in the cytoplasm of yeast/plants and bacteria (6), respectively, but which are lacking from the mammalian cytosol. In vitro, this machinery, comprised of a trio of chaperone components including Hsc70, DnaJA or DnaJB class chaperones, and an Hsp110, appears to be able to dissociate amorphous aggregates of luciferase (7) and even amyloid fibrils of α-synuclein (8). Hsc70, notably, is the most abundant molecular chaperone in the motor neuron cytosol and is constitutively expressed, whereas Hsp70 proteins are at least fivefold less abundant. DnaJ proteins are also present at low levels relative to Hsc70, but, as observed recently, both the DnaJA and DnaJB class proteins play a cooperating role in disaggregation (7). Similarly, the three mammalian Hsp110 proteins (HspA4, HspA4L, and HspH1) are present at relatively low levels, but, as noted above, HspH1 was the only chaperone found to be induced in G85R mutant SOD1YFP-expressing motor neurons in vivo (3). Hsp110s have an overall structure that resembles that of an Hsp70 class protein, but their function has been indicated to be that of a nucleotide exchange factor (9, 10), proffering an ATP binding pocket directly to that of Hsc70 (11), releasing ADP from substrate protein-bound Hsc70, and enabling ATP to enter the Hsc70 nucleotide pocket to discharge bound substrate protein. Although nucleotide exchange activity is well-established for Hsp110 proteins, it remains unclear whether they can also directly bind substrate proteins in vivo, as has been observed in vitro in several studies (12, 13). Regardless of whether Hsp110 proteins directly bind substrate proteins in vivo, ADP/ATP exchange appears to be the rate-limiting step in the Hsc70 protein folding cycle, suggesting a critical role for Hsp110 (5).
When mutant misfolded G85R SOD1YFP was immune-captured (through its folded YFP moiety) from the spinal cord of transgenic ALS mice, it brought down Hsc70, DnaJA1, and all three mammalian Hsp110 proteins (14). Thus, this disaggregase complex appears to recognize the mutant misfolded protein, but it is insufficient, even with the twofold induction of endogenous HspH1, to forestall progression to motor deficit and paralysis. Here, we observe that providing additional Hsp110 from a transgene does, in fact, forestall progression of motor disease, indicating that, for this misfolding condition, the supply of additional amounts of a molecular chaperone that affects the presumed rate-limiting step of the disaggregase can have a clinical benefit.
Results
Thy1-Hsp110 Transgene Introduced into G85R SOD1YFP ALS Strain Produces Extended Survival.
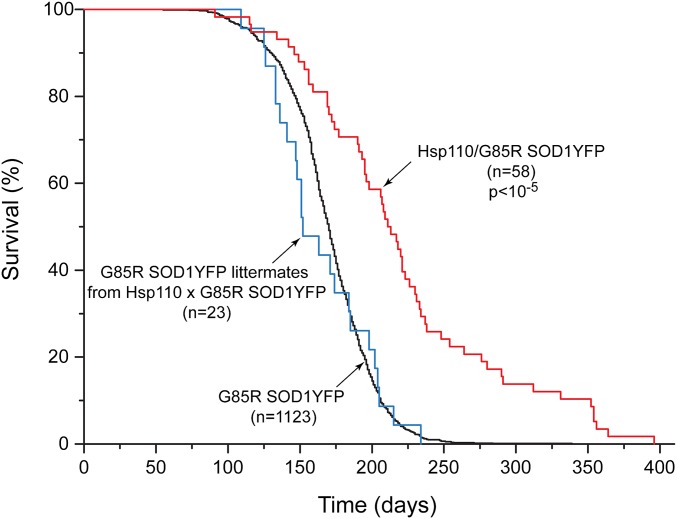
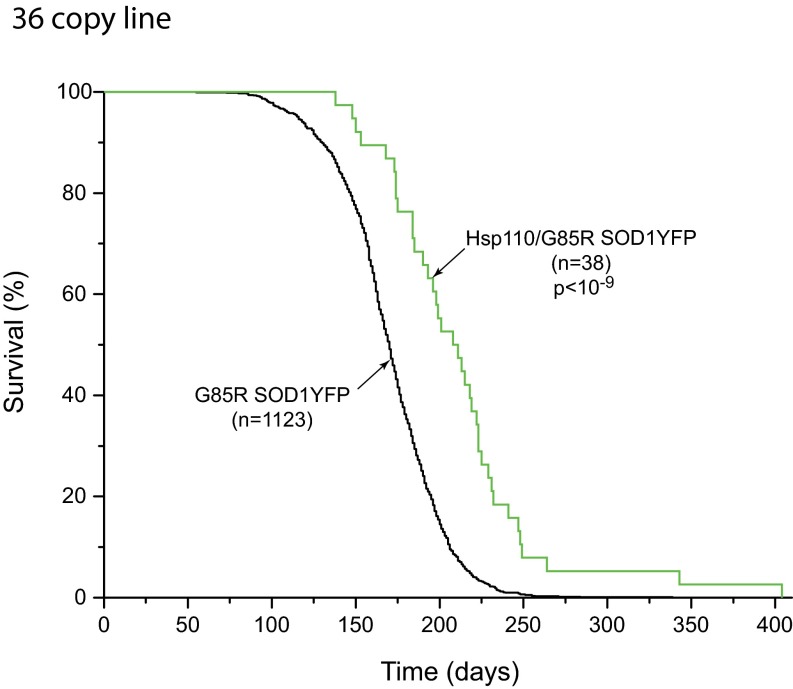
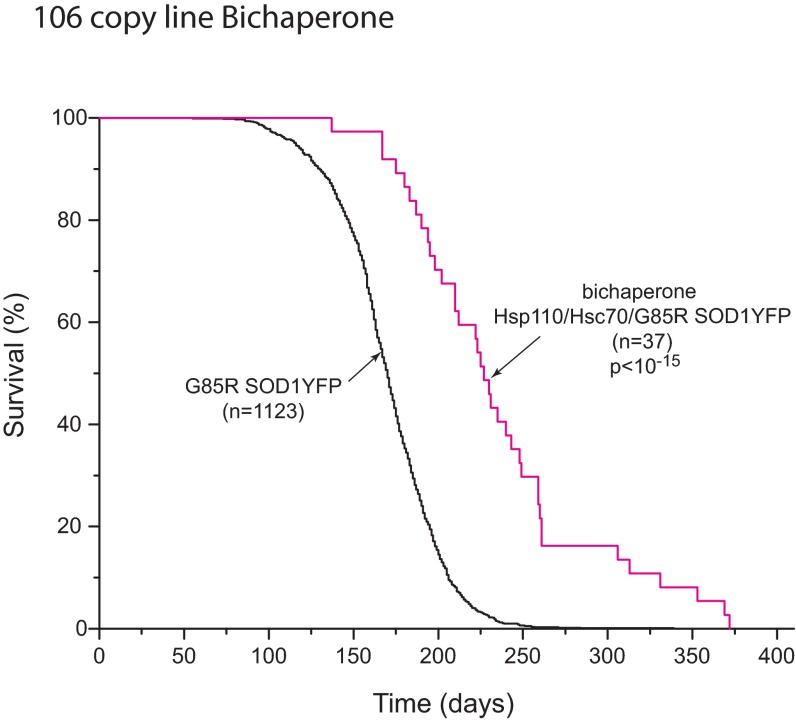
A strain of G85R SOD1YFP homozygous transgenic mice with transgene copy numbers between 210 and 320, on a B6/SJL background, has been followed over several years, observing that all of these mice develop ALS, reaching end-stage lower extremity paralysis by 6.5 mo of age (Fig. 1, black line). We sought to modify the timing of this end-stage phenotype by crossing in various transgenes, initially expressed in B6/SJL, then back-crossing to produce hemizygosity of the modifier and homozygosity of G85R SOD1YFP (210–320 copy number of the latter, ascertained in all mice by real-time PCR). Transgenes including Tet-regulated PGC1α cDNA, Thy1-driven mitochondrial-targeted human catalase cDNA, and Thy1-driven bovine Hsc70 cDNA did not significantly affect the time of survival. In contrast, a transgene composed of the Thy1 promoter driving human Hsp110 cDNA (HspA4L; Apg1) produced a substantial extension of survival (Fig. 1A, red line), with a median survival ∼2 mo longer relative to littermates lacking the Hsp110 transgene (blue line) and ∼1.5 mo longer relative to the cohort of parental G85R SOD1YFP animals (black line). There was a maximum extension of survival of ∼6 mo, approximately doubling the usual life span. A second transgenic line with a lower Thy1-Hsp110 copy number (36 vs. 106 copies) produced almost the same extension of survival (Fig. S1). A third line with both the 106 copy Thy1-Hsp110 transgene and a Thy1-Hsc70 transgene exhibited improvement in survival similar to that of the 106 copy Hsp110 line (Fig. S2).
Fig. 1.
Survival curves of homozygous G85R SOD1YFP mouse strain (black trace) and backcross progeny containing homozygous G85R SOD1YFP and hemizygous 106 copy Thy1-Hsp110 transgene, showing increased survival of transgenic mice with Thy1-Hsp110 and G85R SOD1YFP (red trace) compared with littermates with only G85R SOD1YFP (blue trace). Median survival was extended by 62 d relative to littermates and by 42 d relative to the entire G85R SOD1YFP cohort. n, number of animals of the designated genotype. P value, significance of difference between survival of transgenic Thy1-Hsp110/G85R SOD1YFP (red) and G85R SOD1YFP littermates (blue), determined by log-rank test (see Methods, Survival Curves).
Fig. S1.
Survival curves of G85R SOD1YFP strain (black trace) and G85R SOD1YFP strain containing a 36 copy Thy1-Hsp110 transgene (green trace), showing increased survival of the Thy1-Hsp110 strain. The prolongation of median survival in relation to the G85R SOD1YFP cohort is slightly reduced relative to the 106 copy Thy1-Hsp110/G85R SOD1YFP mice shown in Fig. 1, 39 d compared with 42 d in Fig. 1.
Fig. S2.
Survival curves of G85R SOD1YFP and bichaperone-containing G85R SOD1YFP strain containing transgenic Thy1-Hsp110 and Thy1-Hsc70 (see SI Methods for derivation). The extension of survival of the bichaperone-containing G85R SOD1YFP mice (red trace) is similar to that of the Thy1-Hsp110/G85R SOD1YFP mice (Fig. 1). Note that with producing founder lines of Thy1-Hsc70, we could not obtain copy numbers above ∼20, thus allowing a copy number of only ∼40 with homozygosis. At the RNA level, analysis of two independent lines, both hemizygous and homozygous, showed an increase of only 50–75% over endogenous mouse Hsc70, and an increase of protein could not be observed by immunostaining or Western blot analysis. Thus, there appears to be a level of regulation that prevents overproduction of Hsc70 and may explain inability here to prolong survival. Alternatively, Hsc70 may be amply present and not comprise a rate-limiting step in the disaggregation cycle.
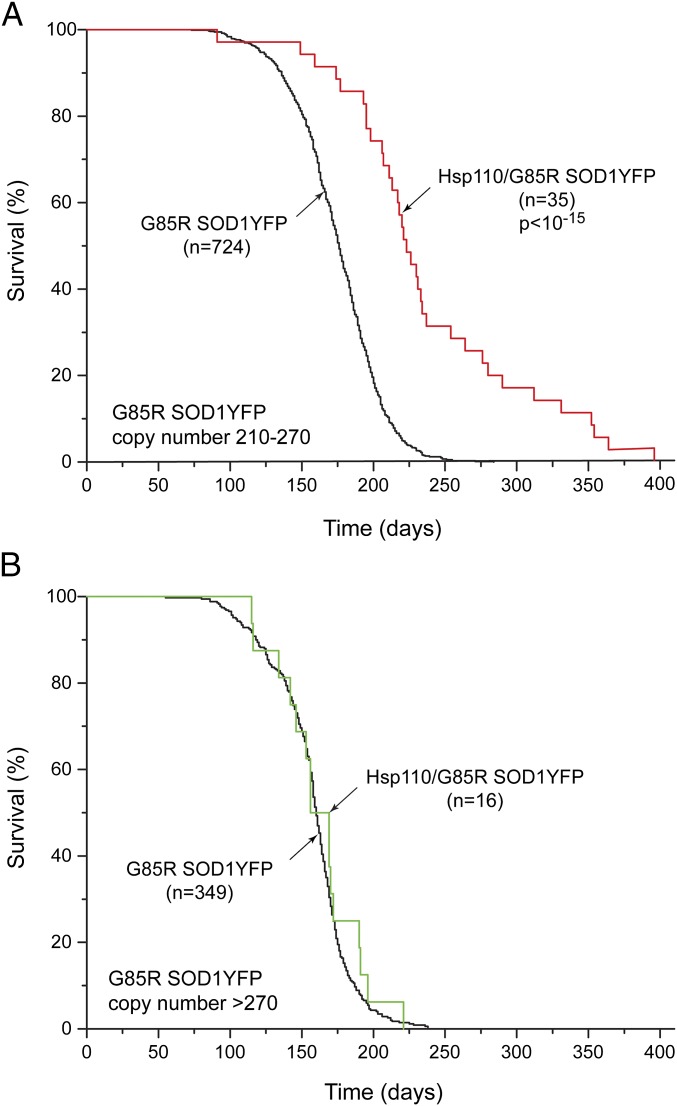
We stratified the survival curve of the 106 copy Thy1-Hsp110/G85R SOD1YFP line in relation to the copy number of G85R SOD1YFP, observing that mice with a copy number of 270 or greater did not achieve improved survival with the Hsp110 transgene compared with the G85R SOD1YFP mice with a copy number of 210–270 (Fig. 2). This difference of effect on survival would suggest that Hsp110 transgene expression (over and above that of endogenous mouse Hsp110s) and participation in the Hsc70/DnaJ/Hsp110 disaggregase system cannot exert a significant effect on mutant G85R SOD1YFP when the mutant protein is expressed beyond the level produced by 270 copies.
Fig. 2.
Stratification of survival of Thy1-Hsp110/G85R SOD1YFP transgenic mice relative to copy number of G85R SOD1YFP. (A) The survival curve of Thy1-Hsp110/G85R SOD1YFP mice with a G85R SOD1YFP copy number between 210 and 270 (red trace) compared with the survival curve of G85R SOD1YFP mice with a copy number between 210 and 270 (black). There is substantial prolongation of survival, similar to that observed in Fig. 1 (e.g., 56 d median survival here vs. 62 d in Fig. 1). (B) The survival curve of Thy1-Hsp110/G85R SOD1YFP transgenic mice with a G85R SOD1YFP copy number greater than 270 (green trace) compared with the survival curve of mice of the G85R SOD1YFP strain with a copy number greater than 270 (black trace). Note that the survival curves are virtually superposable, indicating no beneficial effect of transgenic Thy1-Hsp110 on survival of mice with G85R SOD1YFP copy numbers greater than 270. Note also that the survival curve of the G85R SOD1YFP strain with copy number greater than 270 is slightly shifted to the left relative to the curve of this strain with 210–270 copies as shown in A, indicating that the survival of animals with the larger copy number is decreased (median survival reduced by ∼15 d). n specifies number of mice of the designated genotype, and P values were determined as in Fig. 1.
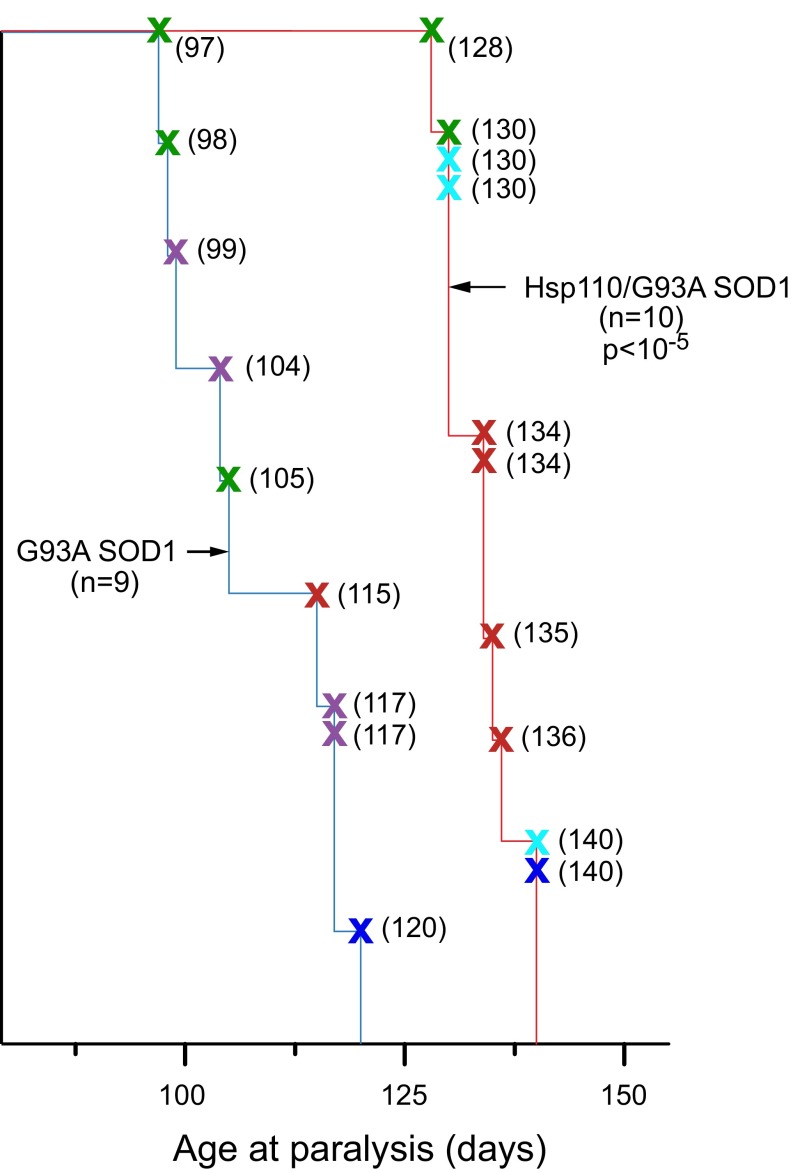
The 106 copy Thy1-Hsp110 transgene was also crossed into G93A SOD1 (hemizygous transgenic) ALS mice, also carried in a B6/SJL background [JAX Tg(SOD1-G93A)1Gur/J]. Here, we examined five litters, comparing survival of G93A progeny with littermates carrying both G93A and Hsp110 transgenes (Fig. S3, blue and red plots, respectively, with survival of the individual mice from each litter represented by specifically colored X’s). As with G85R SOD1YFP, the presence of the Hsp110 transgene was associated with an extension of survival (Fig. S3). The G93A SOD1 progeny (n = 9) developed paralysis by 97–120 d of age, whereas the Thy1-Hsp110/G93A SOD1 mice (n = 10) paralyzed at 128–140 d of age. Median survival of the 106 copy Thy1-Hsp110/G93A SOD1 mice was lengthened by 29 d (105 vs. 134 d).
Fig. S3.
Survival curves of littermates from five different crosses of G93A SOD1 strain with 106 copy Thy1-Hsp110 strain, comparing mice with G93A SOD1 alone (n = 9) (blue trace) to Thy1-Hsp110/G93A SOD1 mice (n = 10) (red trace). The survival of each mouse is designated by a colored X, with the different colors designating the individual crosses and genotyped progeny. Individual times to paralysis are indicated in parentheses. Difference in median survival is 29 d, P < 10−5 by log-rank test. See Results, Thy1-Hsp110 Transgene Introduced into G85R SOD1YFP ALS Strain Produces Extended Survival for additional details.
Transcription and Translation of Hsp110 Transgene in Motor Neurons.
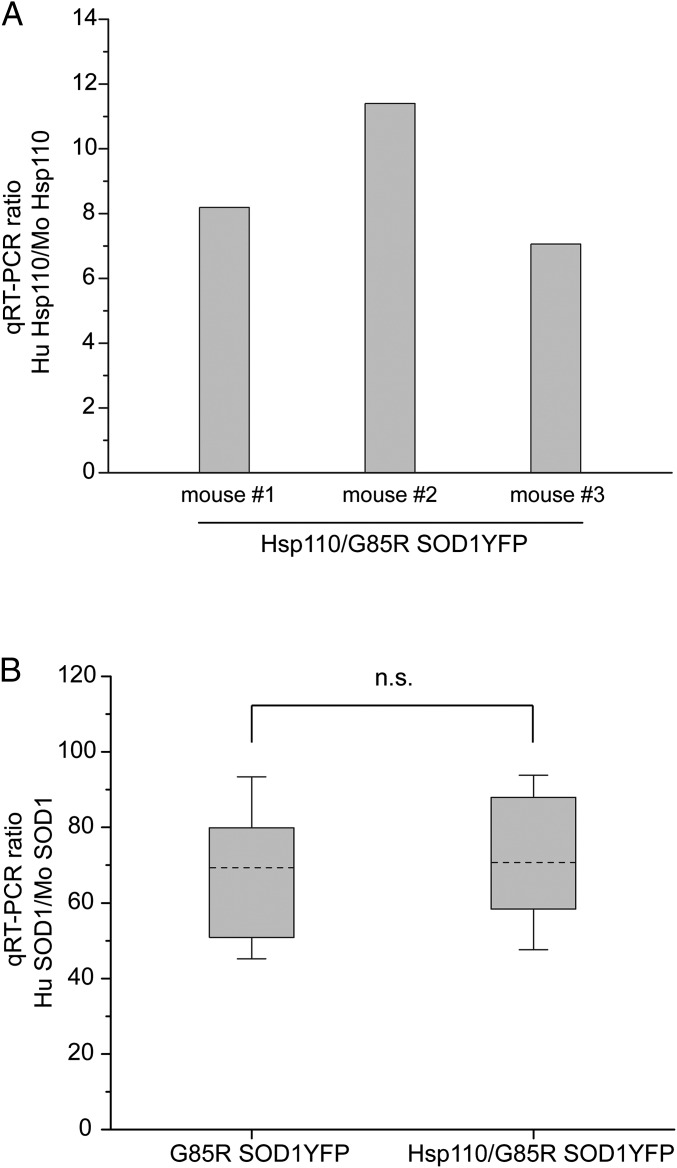
Expression of the 106 copy Thy1-Hsp110 transgene was examined at both the RNA and protein level in motor neurons. At the RNA level, quantitative RT-PCR (qRT-PCR) of RNA isolated from laser-captured motor neurons of the 106 copy Hsp110/G85R SOD1YFP line was carried out, and the amount of human HspA4L RNA was compared with endogenous mouse Hsp110 (HspA4L; Apg1) RNA (Fig. 3A). In three mice examined, the level of transgenic human Hsp110 RNA was 7- to 12-fold greater than that of the endogenous mouse Hsp110 RNA (Fig. 3A).
Fig. 3.
qRT-PCR analysis of RNA levels of transgenic Thy1-Hsp110 and G85R SOD1YFP from laser-captured spinal cord motor neurons, measured as ratios of transgene-derived RNA to RNA from corresponding endogenous mouse gene. A shows the ratio of Hsp110 (HspA4L/Apg1) from the human transgene relative to endogenous mouse Hsp110 (HspA4L/Apg1) from motor neuron RNA of three different mice transgenic for Thy1-Hsp110/G85R SOD1YFP. The human transgene-derived Hsp110 RNA was ∼10-fold more abundant than endogenous mouse Hsp110 RNA (see Methods for details). B shows the ratio of transgenic G85R SOD1YFP to endogenous mouse SOD1 for either the G85R SOD1YFP strain or the Thy1-Hsp110/G85R SOD1YFP for three mice of each strain. The data are plotted as a whisker plot, showing no significant difference in the level of G85R SOD1YFP RNA between the G85R SOD1YFP strain and the Hsp110/G85R SOD1YFP strain (P > 0.9 by two-sample t test in Origin). Dashed lines are the means, top and bottom of the boxes are 75th and 25th percentile and whiskers comprise 1.5 SD above and below the mean.
To exclude the possibility that transgenic human Hsp110 expression could be improving survival simply by reducing the level of G85R SOD1YFP RNA expression in motor neurons, the Thy1-Hsp110/G85R SOD1YFP motor neurons were also analyzed for G85R SOD1YFP mRNA levels and compared with motor neurons from G85R SOD1YFP (lacking the Hsp110 transgene), measuring transgene RNA levels in both cases relative to an internal reference of endogenous mouse SOD1 RNA (Fig. 3B). The level of SOD1YFP RNA was unaffected by the presence of the Hsp110 transgene: In both strains, the level of transgenic G85R SOD1YFP RNA was ∼70-fold greater than that of endogenous mouse SOD1 RNA (Fig. 3B).
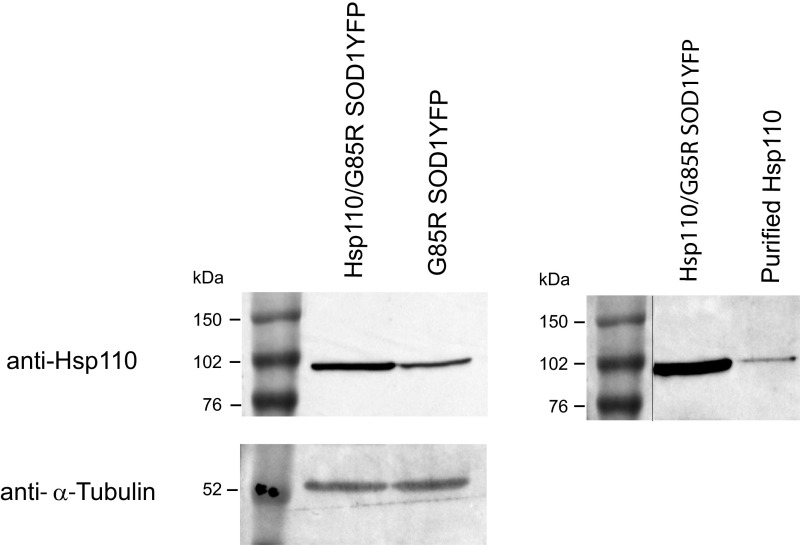
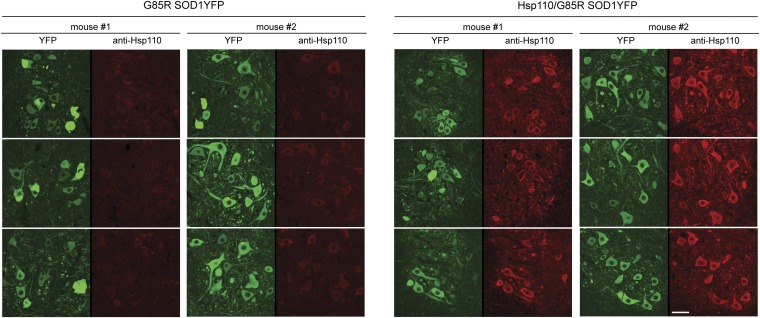
At the protein level, motor neurons were directly inspected for Hsp110 protein by immunostaining with a polyclonal antibody (Santa Cruz Biotechnology) raised against a peptide from the C-terminal region of Hsp110 (also see Fig. S4 for Western blot of total spinal cord). We observed strong anti-Hsp110 immunostaining of motor neurons in Hsp110/G85R SOD1YFP cord cross-sections (Fig. 4, Right, red channel), whereas G85R SOD1YFP animals lacking the Hsp110 transgene showed only a faint signal in their motor neurons (Fig. 4, Left, red channel). Considering that the antibody was produced against a peptide that is identical in human and mouse Hsp110 (HspA4L; Apg1), we conclude that there is considerably more total Hsp110 in the motor neurons of the Thy1-Hsp110/G85R SOD1YFP mice, compared with the endogenous mouse Hsp110 in the G85R SOD1YFP strain. Although Western blot analysis of total spinal cord lysate showed that the Thy1-Hsp110/G85R SOD1YFP mice exhibited ∼2.5-fold more Hsp110 than G85R SOD1YFP mice (Fig. S4), immunostaining suggested that the level of Hsp110 present specifically in spinal cord motor neurons was substantially greater.
Fig. S4.
Western blot of spinal cord lysate from 106 copy Thy1-Hsp110/G85R SOD1YFP mice and G85R SOD1YFP mice probed with anti-Hsp110 antibody (Left) (SI Methods). Signal is ∼2.5 more intense from the Thy1-Hsp110/G85R SOD1YFP vs. endogenous mouse Hsp110 in G85R SOD1YFP mice, as determined by densitometry. Note that the peptide against which the polyclonal antibody was raised (Santa Cruz Biotechnology) is present in both human Hsp110 (HspA4L/Apg1) and the mouse homolog. (Right) Comigration of Hsp110 from Thy1-Hsp110/G85R SOD1YFP mouse with human Hsp110 (Apg1) overexpressed and purified from Escherichia coli. Note that the original image has been spliced to bring the molecular mass marker lane into apposition to the Hsp110 lanes, indicated by the black line.
Fig. 4.
Expression of Hsp110 protein in motor neurons of Thy1-Hsp110/G85R SOD1YFP strain determined by immunostaining. Twenty-micrometer transverse lumbar sections of fixed spinal cords were prepared from 3-mo-old mice of the G85R SOD1YFP strain or of Thy1-Hsp110/G85R SOD1YFP strain and immunostained with a polyclonal antibody directed against a peptide from the C-terminal region of human Hsp110 (Santa Cruz Biotechnology), as described in SI Methods. (Left) YFP-fluorescent large motor neurons observed in the ventral horn. (Right) Fluorescence of the motor neurons in the red channel from anti-Hsp110 immunostaining, showing weak signals from endogenous mouse Hsp110 in sections from the G85R SOD1YFP strain and strong signals in sections from the Thy1-Hsp110/G85R SOD1YFP strain. This observation is consistent with the presence of additional Hsp110 protein in the motor neurons of the double transgenic mice. See text for details. (Scale bar: 50 µm.)
Discussion
It seems likely that the action of transgenic Hsp110 to improve the survival of both G85R SOD1YFP and G93A SOD1 mice is attributable to an additional supply, beyond the endogenous level, of a molecular chaperone component that functions as part of a disaggregation machinery in the metazoan cytosol, composed of Hsc70, DnaJ, and Hsp110. This machinery has been shown in a number of cases in vitro to dismantle soluble aggregates (7) and recently, also in vitro, to dissociate amyloid fibrils composed of α-synuclein (8). The currently understood action of Hsp110 is to catalyze nucleotide exchange of Hsc70 (removing bound ADP from substrate-bound Hsc70, enabling ATP binding and substrate protein release). Here, the favorable action of providing additional Hsp110 would suggest that this component may be rate-limiting in vivo in the disaggregation of misfolded G85R SOD1YFP inside motor neurons. Notably, G85R mutant forms of SOD1 appear incapable of reaching the native state. In particular, G85R SOD1 subunits fail to be metallated, fail to form the normal disulfide bond, fail to homodimerize, and have no enzymatic activity (15–17). The abundant misfolded states are evidently prone to aggregation in vivo, considering the large fluorescent aggregates observed in motor neurons of G85R SOD1YFP mice as early as weaning age, and, as such, provide a substrate for the Hsc70/DnaJ/Hsp110 disaggregation machinery. In contrast, G93A SOD1 substantially populates the native active state, but it also populates nonnative, aggregation-prone forms (15, 18). This latter mutant form was also affected by the presence of the Hsp110 transgene, judging from the increased survival data, albeit survival was not extended as much as for G85R SOD1YFP.
The potentially more efficient disaggregation system provided by higher levels of Hsp110 from a transgene makes the prediction that formation of visible YFP fluorescent aggregates by G85R SOD1YFP should be reduced in the motor neurons of the double transgenic mice. This prediction was difficult to evaluate because analysis of spinal cord morphology of any given animal precludes measuring its long-term survival. Thus, we were unable to directly correlate the reduction or absence of aggregates at 3 mo of age with the greatest prolongation of survival. [Note that the presence of aggregates in ventral horn motor neurons of G85R SOD1YFP mice is prominent between 1 and 3 mo of age but then is substantially reduced thereafter (19)]. However, our observation from six double transgenic animals killed at 2.5 mo of age for morphology analysis was that two animals of the six exhibited strikingly low levels of aggregate formation. In contrast, we have not observed such absence of aggregation in large numbers of 2- to 3-mo-old G85R SOD1YFP mice. In such animals, we routinely observe prominent aggregate formation affecting many of the ∼20 large motor neurons in each ventral horn of any given 20-μm section of spinal cord. We can only speculate that the double transgenic mice lacking aggregates would have been longest-term survivors.
Concerning the longest-term Hsp110 transgenic survivors, 1 y in the case of the 106 copy transgene, it is possible that the long survival in part relates to polymorphism in the B6/SJL background. That is, whereas transgenic mice were produced by injection into heterozygous zygotes, subsequent breeding might produce population heterogeneity at any given locus for B6 and SJL alleles, which, if particular alleles have influence on disease initiation and progression, could further modify the survival (see, e.g., ref. 20). Arguing against a strong effect of heterogeneity in the Hsp110 transgenic strains, however, is the stratification data, which seem to point toward a strict effect of transgenic Hsp110, such that when the amount of the G85R SOD1YFP mutant substrate exceeds the apparent capacity of Hsp110 to enhance survival (>270 copies), there is complete collapse of the survival curve to that of the G85R SOD1YFP strain (Fig. 2). It remains possible, however, that the longest-surviving Hsp110 transgenic animals might have background polymorphisms that, for example, enhance Hsp110 production.
The loss of beneficial effect of Hsp110 when G85R SOD1YFP exceeds 270 copies also argues that the principal action of the Hsc70/DnaJ/Hsp110 system is upon G85R SOD1YFP as its substrate, rather than other known actions of the chaperone system, e.g., in facilitating clathrin uncoating in the vesicle recycling system. In support of direct action on the mutant protein, in an earlier study (14), we observed physical association of all three mouse Hsp110 proteins, as well as DnaJA1 and Hsc70, with G85R SOD1YFP in coimmunoprecipitation of spinal cord lysates with anti-YFP antisera. This observation thus supports a primary action of Hsp110 on the mutant misfolded protein, most likely mediated via association of Hsp110 with substrate-bound Hsc70.
Recent in vitro studies of Hsp110 in heterologous systems also support a role in detoxifying misfolded protein species. Addition of G85R SOD1YFP to squid axoplasm inhibits anterograde axonal vesicle transport, but purified human Hsp110 (HspA4L) prevents this effect even when added at substoichiometric amounts relative to the misfolded protein, suggesting that it cooperates with endogenous Hsc70 and DnaJ homologs to dissociate toxic oligomeric forms (21). In studies in Drosophila eye, transgenic expression of either Drosophila Hsp110 or human Hsp110 (HspA4L; APG1) was able to rescue toxicity from coexpressed polyQ-expanded proteins (22, 23). As mentioned, in motor neurons of the G85R SOD1YFP line, one of three endogenous mouse Hsp110 RNAs, HspH1, is induced approximately twofold compared with wtSOD1YFP motor neurons, one of the few RNAs observed to be altered (3). This endogenous induction falls well short of the 10-fold Hsp110 (HspA4L) RNA induction observed in the 106 copy Thy1-Hsp110/G85R SOD1YFP line. It remains to be seen whether small molecule inducers of Hsp110 could be identified that might be used therapeutically.
Methods
Mouse Strains.
Mouse studies were carried out under a protocol approved by the Yale University Animal Care and Use Committee. The transgenic G85R SOD1YFP strain (containing greater than 210 copies of the transgene via homozygosity, referred to as the 737 line) has been described (14). By 3–4 mo of age, hind-limb clenching and rotarod dysfunction is observed (19), and lower extremity paralysis develops by 6.5 mo of age. The strain transgenic for Thy1-human Hsp110 (HspA4L) was produced by injecting B6/SJL zygotes (Yale Transgenic Mouse Service) with a construct described in SI Methods. Mice hemizygous for the Thy1-Hsp110 transgene were crossed with homozygous G85R SOD1YFP mice, and offspring hemizygous for both Thy1-Hsp110 and G85R SOD1YFP were crossed with G85R SOD1YFP homozygous mice to produce mice hemizygous for Thy1-Hsp110 and homozygous for G85R SOD1YFP. Mice from the second cross that were homozygous for G85R SOD1YFP but lacked the Thy1-Hsp110 transgene were followed as littermate controls for the survival curve of Fig. 1. Two Thy1-Hsp110 strains were examined, one designated “106 copy” (Fig. 1), the other “36 copy” (Fig. S1), based on the copy number of the respective founder mice. Similar copy number was ascertained in progeny mice by real-time PCR. A “bichaperone” strain was produced by crossing (106 copy)Thy1-Hsp110+/−/G85R SOD1YFP+/+ mice with Thy1-Hsc70+/−/G85R SOD1YFP+/+ mice that had been similarly produced and selecting the Thy1-Hsc70+/−/Thy1-Hsp110+/−/G85R SOD1YFP+/+ offspring. The Thy1-Hsp110/G93A mice were produced by crossing hemizygous 106 copy Thy1-Hsp110 female mice with (hemizygous) G93A male mice in the B6/SJL background [the latter the B6SJL-Tg(SOD1-G93A)1Gur/J, 002726, strain from Jackson Laboratory]. Offspring that were double transgenic or contained only the G93A transgene were compared for survival until paralysis. Note that 106 copy Thy1-Hsp110 transgenic mice, either hemizygous or homozygous, are clinically normal and exhibit long-term survival (>18 mo currently).
All mice were genotyped by real-time PCR by using genomic DNA prepared from tail biopsies (DNeasy Blood & Tissue Kit; Qiagen). Mouse ApoB was used as the reference gene. Primer sets for mouse ApoB and human SOD1 were suggested by Jackson Laboratory on their website (https://www.jax.org). Primer sets for the chaperone transgenes consisted of a forward primer in the Thy1.2 sequence and a reverse primer near the beginning of the chaperone coding sequence. Each of these primers was selected manually and confirmed by its efficiency in quantitative PCR, its lack of amplification when template DNA did not contain the respective transgene, and by sequencing the PCR product. To help ensure a consistent ALS phenotype, only G85R SOD1YFP mice with a homozygous copy number >210 were used in the reported experiments.
Survival Curves.
Survival curves (paralysis and euthanasia as end-point) and statistical significance (χ2 and P values) between the survival functions for different strains were generated in OriginPro by using the Kaplan–Meier Estimator function for the log-rank, Breslow, and Tarone–Ware tests. Each gave similar results, and the log-rank test results are reported.
Transgenic constructs, RNA preparation and qRT-PCR, Hsp110 immunostaining, and Western blotting are detailed in SI Methods.
SI Methods
Transgenic Constructs.
Both Hsc70 and HSP110 transgene constructs were produced in the pThy1.2 vector (kindly provided by P. Caroni, Friedrich Miescher Institute, Geneva) (24) for expression in neuronal cells. The Hsp110 construct (human HspA4L) was derived from a plasmid kindly provided by M. Mayer (University of Heidelberg, Heidelberg). The coding sequence was PCR-amplified to place XhoI restriction sites at each end. This fragment was cloned into BlueScript KS, and an internal XhoI site was disrupted by site-directed mutagenesis (QuikChange; Agilent) without changing the encoded amino acid. The resulting construct was digested with XhoI, and the Hsp110 fragment was cloned into XhoI-digested pThy1.2. An endotoxin-free maxiprep (Qiagen) of the resulting Thy1.2-Hsp110 plasmid was digested with NotI and PvuI to release the ∼9,000-bp fragment containing the transgene, which was purified by agarose gel electrophoresis, recovered with a QIAquick Gel Extraction Kit (Qiagen), and used for transgenesis. The Hsc70 construct used the full-length coding sequence of Bov.hsc70.RSET.FL.wt(NarI) (a gift from David McKay, Stanford University, Stanford, CA; Addgene plasmid 12532), PCR-amplified using primers that added an XhoI site to both ends. The XhoI-digested coding sequence was cloned into the XhoI cloning site of the pThy1.2 vector. An endotoxin-free maxiprep (Qiagen) of the resulting Thy1.2-Hsc70 plasmid was digested with NotI and PvuI to release the ∼8,500-bp fragment containing the transgene, which was purified by agarose gel electrophoresis, recovered with a QIAquick Gel Extraction Kit (Qiagen), and used for transgenesis. All constructs were sequenced to confirm their identity.
RNA Preparation and qRT-PCR.
RNA isolation from laser-captured spinal cord motor neurons was carried out as described (3). Total RNA was prepared from ∼2,000 laser-captured motor neurons of three 2- to 4-mo-old G85R SOD1YFP mice and three 2- to 4-mo-old Thy1-Hsp110/G85R SOD1YFP (106 copy line) mice, using the RNeasy Micro kit with on-column DNase digestion (Qiagen). RNA quality was assessed by using an Agilent 2100 BioAnalyzer and a RNA6000 Pico chip; RIN values >7.5 were routinely observed.
Primer sets spanning an exon-exon junction for each mRNA of interest, except the Thy1-Hsp110 transgene, were either found in the PrimerBank database (https://pga.mgh.harvard.edu/primerbank/) or were designed by using Primer Blast (National Center for Biotechnology Information) against the Mus musculus mRNA RefSeq database. Primers were designed manually for the Thy1-Hsp110 set, one primer across an exon–exon junction in upstream Thy1.2-transcribed noncoding sequence and the other residing in the N-terminal coding sequence of Hsp110 (HspA4L). In preliminary experiments with cDNA prepared from isolated mouse brain RNA, amplified products from each primer pair were subjected to DNA sequencing to confirm their specificity. In the case of the transgenes, their absence from nontransgenic mouse RNA was confirmed by qRT-PCR, and amplified products from transgenic animals were sequenced to confirm their specificity.
First-strand cDNA from each total RNA sample was generated by using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara/Clontech) with a mixture of random hexanucleotide and oligo-dT primers. qRT-PCRs were performed in triplicate on an amount of cDNA corresponding to ∼0.5 ng per well of input motor neuron RNA in a 20-μL reaction volume by using SsoFast EvaGreen Supermix (Bio-Rad) on an MiniOpticon Real-Time PCR System (Bio-Rad). Mouse Gapdh was used as reference for all samples. Because of the small amounts of RNA available, controls without reverse transcriptase in the first-strand synthesis reaction were carried out on only a few samples; no spurious amplification was detected in these samples. Fold change for each transgene and sample was determined by using the ΔCt method. Statistical significance for the human SOD1/mouse SOD1 ratio was calculated in OriginPro by using the two-sample t test.
Hsp110 Immunostaining.
Mice were perfused with 2% (wt/vol) PFA, and spinal cords were dissected and further postfixed in 2% (wt/vol) PFA for 2 d, then rinsed with PBS, and infused with 30% sucrose in PBS (typically 2 d). After cryoprotection, spinal cord pieces were embedded in OCT (Tissue-Tek), frozen on dry ice, and stored at −80 °C. Twenty-micrometer spinal cord cross-sections were prepared on a cryostat (Leica CM3050S) and placed into netwell inserts in a 12-well plate. OCT was washed away with PBS, and free-floating sections were incubated in blocking buffer [1% BSA, 5% (vol/vol) normal donkey serum, 0.3% Triton X-100, 0.3 M glycine in PBS] at 37 °C for 30 min. The sections were transferred to a 24-well plate and incubated with goat anti-HspA4L (sc-249947; Santa Cruz Biotechnologies) at 1:50 in antibody dilution buffer [1% BSA, 5% (vol/vol) normal donkey serum, 0.3% Triton X-100 in PBS] for 30 min at 37 °C and for 2 d at 4 °C on an orbital shaker. At the end of the primary antibody incubation, spinal cord sections were transferred back to netwells, washed in PBS (three 5-min washes), and incubated in the dark with AlexaFluor 568-conjugated donkey anti-goat antibody (A11057, Life Technologies) at 1:500 in antibody dilution buffer for 1 h at room temperature with gentle shaking. After three 5-min washes with PBS, the free-floating spinal cord sections were mounted on Snowcoat X-tra glass slides (Surgipath) by using Vectashield (Vector Laboratories). Images of ventral horn motor neurons were acquired on a Leica SP5 confocal microscope with a 40× 1.25 N.A. oil objective at 1.5 zoom factor, controlled by LAS AF Lite software. The 514-nm laser was used to excite YFP and the 561-nm laser for the AlexaFluor 568.
Western Blotting.
Mice were killed with a lethal dose of ketamine and perfused with ice-cold PBS for 2 min. Spinal cords from 2-mo-old G85R SOD1YFP and 106 copy Thy1-Hsp110/G85R SOD1YFP were quickly dissected and homogenized with a Tissue Ruptor (Qiagen) in 0.5 mL of PBS containing 1 mM EDTA, 1 mM EGTA, 1 mM TCEP, 1 tablet per milliliter Complete Mini protease inhibitor mixture (Roche), and 0.5% anti-foam reagent (Reagent DX, Qiagen). After a 10-min centrifugation at 17,000 × g, the supernatant was collected, and the total protein concentration was determined with the Bio-Rad Protein Assay. Ten micrograms of total soluble protein and 10 ng of purified human HspA4L were resolved on 6% SDS/PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with goat anti-HspA4L (1:200; sc-249947, Santa Cruz Biotechnologies) and rabbit anti–α-tubulin (1:2,000; 2144, Cell Signaling). After incubation with HRP-conjugated secondary antibodies (1:10,000), membranes were developed by using Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific) and imaged by using an ImageQuant LAS400 (General Electric). Gel images were viewed and analyzed by using ImageQuant TL software (General Electric).
Acknowledgments
We thank Howard Hughes Medical Institute for generous support of this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604885113/-/DCSupplemental.
References
- 1.Spillantini MG, et al. α-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay U, et al. RNA-seq profile of spinal cord motor neurons from a presymptomatic SOD1 ALS mouse. PLoS ONE. 2013;8(1):e53575. doi: 10.1371/journal.pone.0053575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oza J, Yang J, Chen KY, Liu AY-C. Changes in the regulation of heat shock gene expression in neuronal cell differentiation. Cell Stress Chaperones. 2008;13(1):73–84. doi: 10.1007/s12192-008-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31(21):4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle SM, Wickner S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci. 2009;34(1):40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Nillegoda NB, et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524(7564):247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X, et al. Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol Cell. 2015;59(5):781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133(6):1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattoo RUH, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288(29):21399–21411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA. 2009;106(5):1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayward LJ, et al. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277(18):15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 16.Jonsson PA, et al. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129(Pt 2):451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 17.Zetterström P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104(35):14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lelie HL, et al. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J Biol Chem. 2011;286(4):2795–2806. doi: 10.1074/jbc.M110.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadzipasic M, et al. Selective degeneration of a physiological subtype of spinal motor neuron in mice with SOD1-linked ALS. Proc Natl Acad Sci USA. 2014;111(47):16883–16888. doi: 10.1073/pnas.1419497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sher RB, et al. A major QTL on mouse chromosome 17 resulting in lifespan variability in SOD1-G93A transgenic mouse models of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(7–8):588–600. doi: 10.3109/21678421.2014.932381. [DOI] [PubMed] [Google Scholar]
- 21.Song Y, et al. Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc Natl Acad Sci USA. 2013;110(14):5428–5433. doi: 10.1073/pnas.1303279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Binari R, Zhou R, Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010;184(4):1165–1179. doi: 10.1534/genetics.109.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo Y, Ren S, Lao U, Edgar BA, Wang T. Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis. 2013;4:e833. doi: 10.1038/cddis.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J Neurosci Methods. 1997;71(1):3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]